SUMMARY
The mitochondrial unfolded protein response (UPRmt) is critical for maintaining mitochondrial protein homeostasis in response to mitochondrial stress, but early steps in UPRmt activation are not well understood. Here, we report a function for SPHK-1 sphingosine kinase in activating the UPRmt in C. elegans. Genetic deficiency of sphk-1 in the intestine inhibits UPRmt activation, whereas selective SPHK-1 intestinal overexpression is sufficient to activate the UPRmt. Acute mitochondrial stress leads to rapid, reversible localization of SPHK-1::GFP fusion proteins with mitochondrial membranes before UPRmt activation. SPHK-1 variants lacking kinase activity or mitochondrial targeting fail to rescue the stress-induced UPRmt activation defects of sphk-1 mutants. Activation of the UPRmt by the nervous system requires sphk-1 and elicits SPHK-1 mitochondrial association in the intestine. We propose that stress-regulated mitochondrial recruitment of SPHK-1 and subsequent S1P production are critical early events for both cell autonomous and cell non-autonomous UPRmt activation.
In Brief
The mitochondrial unfolded protein response (UPRmt) maintains mitochondrial protein homeostasis in response to stress. Kim and Sieburth identify SPHK-1/sphingosine kinase as a positive regulator of the UPRmt that promotes UPRmt activation in response to a variety of mitochondrial stressors. SPHK-1 associates with mitochondria and SPHK-1 mitochondrial association is stress dependent, reversible, and necessary for the UPRmt, indicating that SPHK-1 mitochondrial targeting is an early step in UPRmt activation.
Graphical Abstract

INTRODUCTION
Multicellular organisms have evolved diverse strategies to maintain mitochondrial homeostasis in response to environmental and endogenous stress imposed by changes in their surroundings. The nervous system is emerging as a critical coordinator of organism-wide protection that regulates stress response pathway activation in distal tissues through the release of neuroendocrine signals (Berendzen et al., 2016; Durieux et al., 2011; McCallum et al., 2016; Shao et al., 2016; Taylor and Dillin, 2013). However, little is known about the molecular mechanisms by which signals from the nervous system are perceived by distal tissues to regulate stress responses.
The mitochondrial unfolded protein response (UPRmt) is an evolutionarily conserved adaptive response that functions to maintain mitochondrial protein homeostasis in response to mitochondrial dysfunction caused by mitochondrial DNA damage, impaired mitochondrial protein folding, or perturbations in oxidative phosphorylation. Defects in the UPRmt are implicated in the development of numerous diseases, neurodegeneration, and aging (Durieux et al., 2011; Liu et al., 2014; Martinez et al., 2017; Pellegrino et al., 2014). UPRmt is initiated when the activation of signaling pathways originating in mitochondria leads to epigenetic modifications and transcriptional responses in the nucleus that restore mitochondrial homeostasis. Genetic screens in Caenorhabditis elegans have identified several components of the UPRmt, including the leucine zipper transcription factor ATFS-1, a critical activator of the UPRmt. Under normal conditions, ATFS-1 is imported into mitochondria, where it is degraded by the Lon protease. Upon mitochondrial stress, mitochondrial protein import is compromised, and ATFS-1 translocates into the nucleus, where it regulates the expression of a cascade of genes, including the mitochondrial chaperones HSP-6 and HSP-60 (Nargund et al., 2012, 2015). A second transcription factor, DVE-1, functions with the nuclear protein LIN-65 and the chromatin remolding protein MET-2 to enhance ATFS-1mediated activation of the UPRmt (Tian et al., 2016). Early signaling events in mitochondria that lead to UPRmt activation include the recognition and degradation of misfolded or unassembled mitochondrial matrix proteins by the quality control protease CLPP-1/ClpP into peptide fragments, which are subsequently exported into the cytoplasm through the ABC transporter HAF-1, where they may inhibit mitochondrial protein import (Haynes et al., 2007; Nargund et al., 2012). Besides CLPP-1-mediated proteolysis, protein prenylation by the mevalonate pathway and the generation of the sphingolipid ceramide on mitochondrial membranes define additional critical events in the activation of the UPRmt (Liu et al., 2014).
The UPRmt can be activated either cell autonomously by mitochondrial stress or cell non-autonomously through the release of diffusible factors from neighboring or distant cells (Berendzen et al., 2016; Durieux et al., 2011; Shao et al., 2016). In C. elegans, multiple forms of mitochondrial dysfunction in the nervous system activate the UPRmt in the intestine through the release of specific neuropeptide-like proteins or serotonin from sensory neurons and/or interneurons (Berendzen et al., 2016; Shao et al., 2016). Because increasing or decreasing the strength of signaling by these factors can promote (increasing) or block (decreasing) stress-induced UPRmt activation, the coordination of neuroendocrine signaling by the nervous system is likely to play a critical role in the organism-wide adaptive response to mitochondrial stress. Neuroendocrine-mediated activation of the UPRmt in the intestine requires ATFS-1, but the roles of the other components are less clear, and how neuroendocrine signals lead to activation of the UPRmt in the intestine is not known.
In this study, we identify a function for the sole C. elegans sphingosine kinase ortholog SPHK-1, which catalyzes the conversion of sphingosine (SPH) to sphingosine-1-phosphate (S1P) in the activation of the UPRmt. We find that sphk-1 knockout blocks UPRmt activation induced by diverse mitochondria stressors, whereas SPHK-1 overexpression activates the UPRmt. We find that SPHK-1 is targeted to mitochondria in the intestine in response to stress before the induction of the UPRmt, and mitochondrial SPHK-1 targeting is required for UPRmt activation. SPHK-1 mitochondrial abundance is positively regulated by neuroendocrine signaling, and sphk-1 is required for UPRmt activation elicited by mitochondrial stress originating from the nervous system. Our results indicate that SPHK-1 functions as an early mitochondrial signal to activate the UPRmt in response to both cell autonomous and cell non-autonomous stresses. We propose that SPHK-1::GFP fusion proteins may be useful biomarkers for monitoring early signaling events in mitochondria during UPRmt activation in vivo.
RESULTS
SPHK-1 Functions Cell Autonomously in the Intestine to Selectively Activate the UPRmt
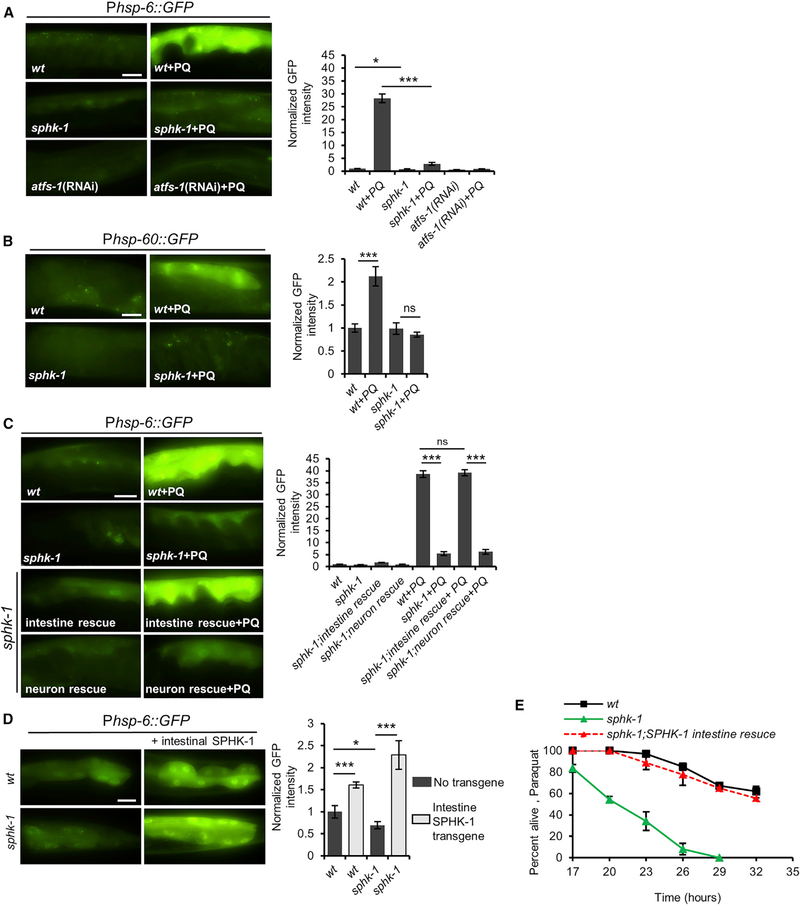
A prior genetic study found that knockdown of sptl-1 serine palmitoyltransferase, which functions in a key step in the biosynthesis of sphingolipids, blocks UPRmt activation. The defect in UPRmt activation was attributed to the depletion of mitochondrial-derived ceramide or its metabolites (Liu et al., 2014). Ceramide is a precursor of SPH, which can subsequently be enzymatically converted into the bioactive lipid second messenger S1P by sphingosine kinase. To investigate the role of sphingosine kinase in the UPRmt, we examined the effects of sphk-1/sphingosine kinase mutants on UPRmt activation in response to paraquat treatment. Paraquat disrupts ETC function at the inner mitochondrial membrane, leading to activation of the UPRmt in mammalian cells, and in C. elegans (Fiorese et al., 2016; Nargund et al., 2012; Runkel et al., 2013). We examined UPRmt activation by quantifying intestinal fluorescence of transgenic animals expressing enhanced GFP (eGFP) under the control of the hsp-6 promoter (Phsp-6::GFP) (Liu et al., 2014; Runkel et al., 2013). The sphk-1(ok1097) mutation deletes nearly the entire coding region of sphk-1 and is predicted to eliminate SPHK-1 activity (Chan et al., 2012). In the absence of stress, Phsp-6::GFP expression was slightly but significantly reduced in sphk-1 mutants compared to wild-type controls (Figure 1A). However, the paraquat-induced increase in Phsp-6::GFP expression was reduced by 88% in sphk-1 mutants, from 28-fold to 3.6-fold (Figure 1A). The paraquat-induced increase in the expression of a second UPRmt reporter, Phsp60::GFP, was completely blocked in sphk-1 mutants (Figure 1B; Nargund et al., 2012; Runkel et al., 2013). The defects in UPRmt activation in sphk-1 mutants were similar to those seen when atfs-1 was knocked down by RNAi (Figure 1A; Haynes et al., 2010). Knockdown of sphk-1 by RNAi or by the independently isolated sphk-1(ju831) missense mutation (McCulloch et al., 2017) attenuated paraquat-induced Phsp-6::GFP expression, albeit not as strongly as sphk-1(ok1097) mutations (Figures S1A and S1B). sphk-1 mutations did not attenuate the activation of the SKN-1/Nrf2-mediated antioxidant response (Pgst-4::GFP reporter induction; Figure S1C) (Inoue et al., 2005) or the endoplasmic reticulum UPR (UPRer, Phsp-4::GFP reporter induction; Figure S1D) (Glover-Cutter et al., 2013). These results indicate that sphk-1 mutants have specific defects in UPRmt activation.
Figure 1. SPHK-1 Positively Regulates UPRmt Activation.
(A) Representative images and fluorescence quantification of intestinal Phsp-6::GFP in young adult wild-type, wild-type in which atfs-1 is knocked down by RNAi, and sphk-1 mutants following treatment with M9 (control) or paraquat (PQ) for 24 hr.
(B) Representative images and quantification of intestinal Phsp-60::GFP in the indicated strains following control or paraquat treatment.
(C) Representative images and quantification of intestinal Phsp-6::GFP in the indicated strains following control or paraquat treatment. Intestinal rescue denotes sphk-1 mutants expressing Pges-1::sphk-1::mCherry transgenes. Neuronal rescue denotes sphk-1 mutants expressing Prab-3::sphk-1::mCherry transgenes.
(D) Representative images and quantification of intestinal Phsp-6::GFP in indicated strains following control or paraquat treatment. Intestinal SPHK-1 denotes animals expressing Pges-1::sphk-1::mCherry transgenes.
(E) Survival curves of indicated strains on plates containing 10 mM paraquat.
Scale bar represents 10 mm. Error bars indicate ± SEMs. The sample sizes (n) and ± SEMs are listed in Table S1. Student’s t test; *p < 0.05, ***p < 0.001, ns, nonsignificant. wt, wild-type.
SPHK-1 is expressed broadly in multiple tissues, including the nervous system, where it functions to promote neurotransmitter release, and the intestine, where its function is unknown (Chan et al., 2012; Chan and Sieburth, 2012). To identify the tissue(s) in which SPHK-1 functions to activate the UPRmt, we introduced transgenes containing sphk-1 cDNA driven by either a neuron-specific promoter, rab-3, or an intestine-specific promoter, ges-1, into sphk-1 mutants expressing Phsp-6::GFP. Intestinal sphk-1 cDNA expression completely restored the paraquat-induced Phsp-6::GFP induction to wild-type levels in sphk-1 mutants, whereas pan-neuronal sphk-1 cDNA expression failed to elicit paraquat-induced Phsp-6::GFP induction in sphk-1 mutants (Figure 1C). Therefore, SPHK-1 functions cell autonomously in the intestine to activate the UPRmt.
To determine whether SPHK-1 activity is sufficient to activate the UPRmt, we selectively overexpressed SPHK-1 in the intestine. We observed a 1.6-fold increase in Phsp-6::GFP expression in animals overexpressing SPHK-1 in the intestine and a 2.3-fold increase in sphk-1 mutants overexpressing SPHK-1 in the intestine but lacking SPHK-1 in all other tissues (Figure 1D). The increase in hsp-6 expression associated with SPHK-1 overexpression was dependent on atfs-1, but not on dve-1, haf-1, or met-2 (Figure S1E). Thus, loss of intestinal SPHK-1 signaling blocks stress-induced UPRmt activation, whereas restricted overexpression of SPHK-1 only in the intestine is sufficient to induce the UPRmt.
The impaired stress-induced UPRmt activation of sphk-1 mutants could be explained by either defective UPRmt activation in response to stress or general defects in mitochondrial stress sensing. To distinguish between these possibilities, we examined survival in the presence of paraquat (Ishii et al., 1990). sphk-1 mutants displayed significantly increased sensitivity to paraquat-induced toxicity compared to wild-type controls, and expression of SPHK-1 in the intestine fully restored normal paraquat sensitivity to sphk-1 mutants (Figure 1E). These results point to a role for SPHK-1 in conferring organism-wide protection to stress by specifically promoting the activation of the UPRmt in the intestine.
SPHK-1 Promotes UPRmt Activation by Diverse Mitochondrial Stressors
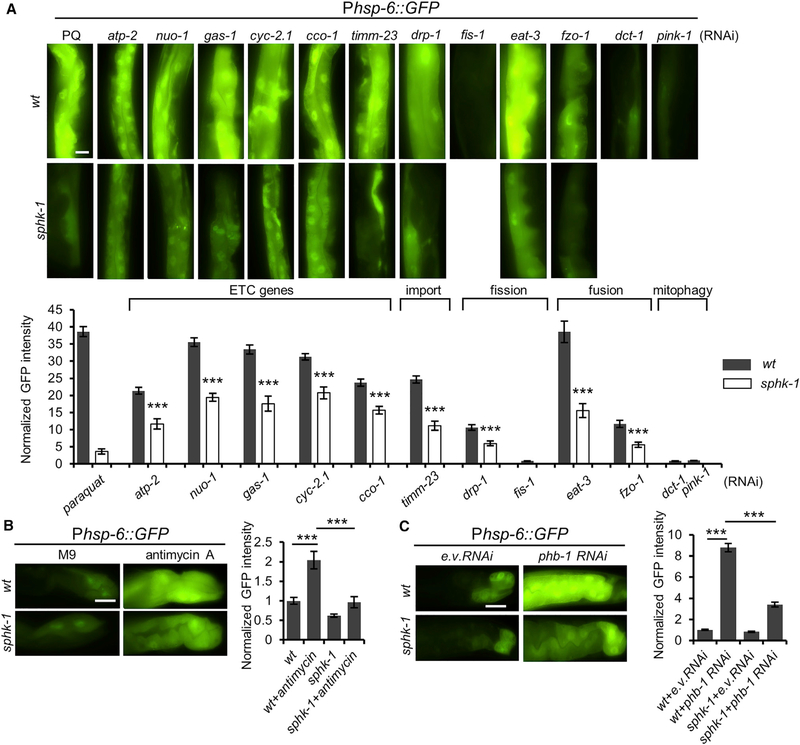
A variety of different pharmacological and genetic manipulations activate the UPRmt, including RNAi-mediated knockdown of components of the electron transport chain (ETC) or treatment with the mitochondrial ETC stressor antimycin A (Liu et al., 2014; Runkel et al., 2013). We found that RNAi knockdown of ETC components (atp-2, nuo-1, gas-1, cyc-2.1, or cco-1) resulted in a robust induction of Phsp-6::GFP expression that was significantly diminished but not eliminated in sphk-1 mutants (Figure 2A) and that acute antimycin A-induced hsp-6 expression was nearly abolished in sphk-1 mutants (Figure 2B). phb-1/prohibitin1 is an evolutionarily conserved mitochondrial protein that regulates mitochondrial biogenesis and morphology and stabilizes the mitochondrial genome, and phb-1 knockdown is a potent activator of the UPRmt (Gatsi et al., 2014). The induction of Phsp-6::GFP expression by phb-1 RNAi knockdown was significantly decreased in sphk-1 mutants compared to wildtype controls (Figure 2C). In addition, we found that the increased Phsp-6::GFP expression associated with RNAi knockdown of genes involved in mitochondrial protein import (timm-23/translocase), mitochondrial fission (drp-1/DRP), or mitochondrial fusion (eat-3/dynamin or fzo-1/mito-fusion) was dependent on SPHK-1. However, disruption of mitophagy genes (dct-1 or pink-1/PINK1) failed to induce Phsp-6::GFP expression (Figure 2A). These results show that SPHK-1 contributes to UPRmt activation in response to diverse mitochondrial stressors as well as alterations in mitochondrial morphology. Moreover, because neurons are largely refractory to gene knockdown by RNAi (Asikainen et al., 2005), the requirement of SPHK-1 in RNAi-induced UPRmt activation suggests that SPHK-1 activates the UPRmt in response to mitochondrial stress arising cell autonomously in the intestine.
Figure 2. SPHK-1 Activates the UPRmt Induced by Diverse Mitochondrial Stressors.
(A) Representative images and quantification of intestinal Phsp-6::GFP in wild-type and sphk-1 mutants treated with paraquat, atp-2, nuo-1, gas-1, cyc-2.1, cco-1 (ETC genes), timm-23 (translocase), drp-1, fis-1 (mito-fission), eat-3, fzo-1 (mito-fusion), or dct-1, pink-1 (mitophagy) RNAi.
(B) Representative images and quantification of Phsp-6::GFP expression in wild-type or sphk-1 mutants in the absence or presence of antimycin A in intestinal cells
(C) Representative images and quantification of Phsp-6::GFP expression in wild-type or sphk-1 mutant intestines treated with empty vector control (e.v.) or phb-1/ prohibitin1 RNAi.
Scale bar represents 10 μm. Error bars indicate ± SEMs. The sample size (n) and ± SEMs are listed in Table S1. Student’s t test; ***p < 0.001.
SPHK-1 Localizes to Intestinal Mitochondria
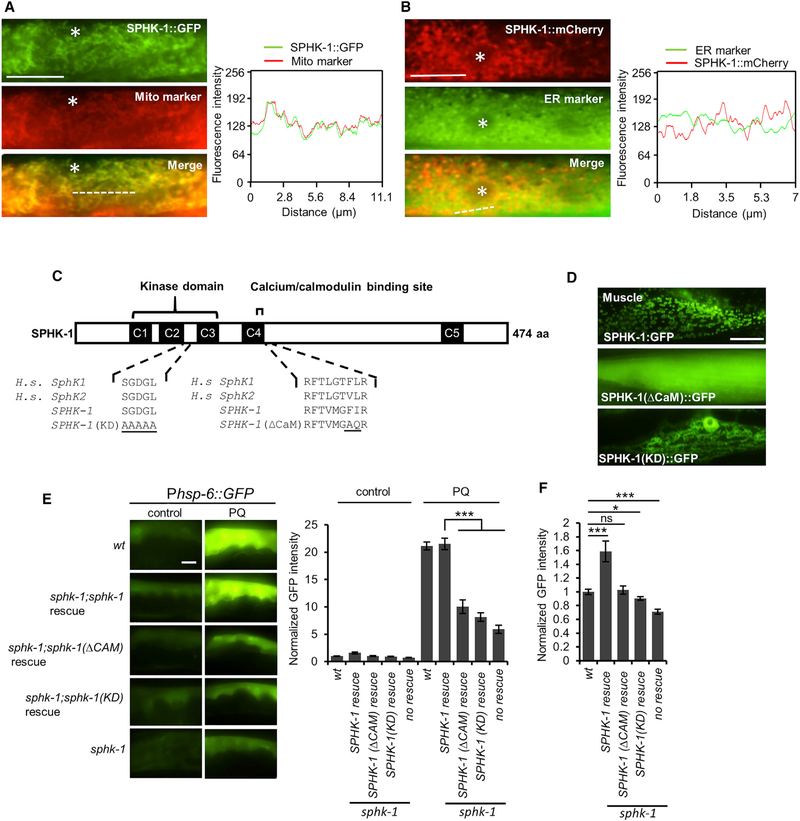
To determine the mechanism by which SPHK-1 activates the UPRmt, we first examined the subcellular localization of SPHK-1 in the intestine. SPHK-1::GFP fusion proteins expressed in the intestine (using the intestine-specific ges-1 promoter) were fully functional in rescuing the UPRmt defects of sphk-1 mutants (Figure S1F), indicating that the localization pattern of SPHK-1::GFP is likely to reflect that of endogenous SPHK-1. SPHK-1::GFP adopted a reticulated fluorescence pattern that extended throughout the cytoplasm and SPHK-1 was excluded from the nucleusand plasma membrane (Figure 3A). Mammalian sphingosine kinases have been reported to associate primarily with two intracellular organelles: mitochondria and the endoplasmic reticulum (ER) (Hatoum et al., 2017; Strub et al., 2011). SPHK-1::GFP co-localized strongly with the outer mitochondrial membrane marker TOMM-20::mCherry (Palikaras et al., 2015) but did not co-localize with the ER marker C34B2.10/signal peptidase (Figures 3A and 3B; Xu et al., 2012). Thus, SPHK-1 selectively associates with mitochondria in intestinal cells.
Figure 3. Mitochondrial Localization and Kinase Activity of SPHK-1 Are Essential for sphk-1-Mediated UPRmt Activation.
(A) Co-localization of SPHK-1::GFP fusion proteins with the outer mitochondrial membrane marker TOMM-20::mCherry in the young adult intestines of sphk-1 mutants. Asterisk indicates an intestinal nucleus (left). Cross-sectional fluorescence intensity graph of GFP and mCherry corresponding to the dotted line at left (right).
(B) Co-localization of intestinal SPHK-1::mCherry fusion proteins with the ER marker GFP::C34B2.10. Asterisk indicates intestinal nucleus (left). Cross-sectionalfluorescence intensity graph of GFP and mCherry corresponding to the dotted line at left (right).
(C) Predicted structure of SPHK-1 showing the kinase domain and calmodulin binding motif (CaM). C1–C5 denotes conserved domains found in human (H.s.) SphK1 and SphK2 proteins. Amino acid substitutions made to generate the kinase dead (KD) and ▵CaM SPHK-1 variants are underlined.
(D) Representative images of sphk-1 mutants expressing SPHK-1::GFP, SPHK-1(▵CaM)::GFP, and SPHK-1(KD)::GFP in muscles.
(E) Representative images and quantification of Phsp-6::GFP fluorescence in the indicated strains in the absence or presence of paraquat (for 24 hr). Rescue denotes sphk-1 mutants expressing the indicated sphk-1 cDNA transgenes under control of the intestinal ges-1 promoter.
(F) Average fluorescence of intestinal Phsp-6::GFP expressed in the indicated strains in the absence of stress is quantified.
The sample sizes (n) and ± SEMs are listed in Table S1. Scale bar represents 10 μm. Error bars indicate ± SEMs. Student’s t test; *p < 0.05, ***p < 0.001.
We examined whether SPHK-1 signaling affects mitochondrial morphology. We found no obvious changes in the distribution or abundance of TOMM-20::mCherry fluorescence in the intestine or in the muscles in sphk-1 mutants compared to wild-type controls (Figures S2A and S2B), suggesting that mitochondrial mass and protein import were largely unaffected in sphk-1 mutants. Overexpression of SPHK-1 led to no obvious defects in intestinal mitochondrial morphology but resulted in a donut-like mitochondrial pattern in muscles in which mitochondria appeared to be fragmented (Figure S2C). Mitochondrial fragmentation is indicative of excessive mitochondrial fission and is associated with UPRmt activation (Labrousse et al., 1999; Pellegrino et al., 2013). Thus, SPHK-1 associates with mitochondrial membranes, and SPHK-1 signaling may increase mitochondrial fragmentation (at least in the muscle), possibly by a mechanism that affects the balance of mitochondrial fusion/fission.
Mitochondrial SPHK-1 Is Required for UPRmt Activation
To determine whether the mitochondrial association of SPHK-1 is important for UPRmt activation, we examined the effects of disrupting mitochondrial SPHK-1::GFP on paraquat-induced Phsp-6::GFP expression. SPHK-1 contains a conserved calcium/calmodulin (CaM)-binding motif that promotes sphingosine kinase association with cellular membranes in response to agonists (Chan et al., 2012; Jarman et al., 2010; Young et al., 2003). Mutation of the CaM-binding site of mammalian SphK1 decreases its association with the plasma membrane without affecting its catalytic activity or folding (Shen et al., 2014). We generated an SPHK-1::GFP fusion protein harboring a mutated CaM motif (SPHK-1(▵CaM)::GFP; Figure 3C) and found that it adopted a uniform localization pattern throughout the cytoplasm that was strikingly different from the mitochondrial localization pattern of wild-type SPHK-1::GFP, both in muscle and intestine (Figures 3D and S2D). Critically, SPHK-1(▵CaM) was unable to fully restore paraquat-induced Phsp-6::GFP expression to sphk-1 mutants (Figure 3E) or to induce Phsp-6::GFP expression above baseline levels in the absence of stress (Figure 3F). These results indicate that mitochondrial localization of SPHK-1 is essential for stress-induced UPRmt activation.
S1P Production by SPHK-1 Activates the UPRmt
The C. elegans genome encodes several conserved enzymes that are predicted to be involved in sphingolipid biogenesis and metabolism (Figures S3A and S3B). We tested the effect of RNAi knockdown of each sphingolipid metabolism enzyme on paraquat-mediated Phsp-6::GFP induction. As expected, knockdown of sptl-1/serine palmitoyltransferase, required early in the S1P biosynthetic pathway, completely abolished paraquat-induced hsp-6 expression, as previously reported (Figure S3B; Liu et al., 2014). F27E5.1/ceramidase catalyzes the biosynthesis of SPH from ceramide. RNAi knockdown of F27E5.1/ceramidase significantly decreased paraquat-induced Phsp-6::GFP expression (Figure S3B). tag-38/S1P lyase is predicted to convert S1P to hexadecenal and phosphoethanolamine. RNAi knockdown of tag-38/S1P lyase enhanced paraquat-induced Phsp-6::GFP expression (Figure S3B). Thus, mutations predicted to decrease S1P levels (sptl-1, sphk-1, or F27E5.1) attenuate UPRmt activation, whereas mutations predicted to increase S1P levels (tag-38/S1P lyase) enhance the UPRmt.
We next generated a kinase-dead (KD) version of SPHK-1 by mutating conserved amino acids in the catalytic domain that are critical for SphK catalytic activity (Figure 3C; Pitson et al., 2002). We found that SPHK-1(KD)::GFP localized to mitochondria (Figure 3D), yet failed to fully restore paraquat-induced Phsp-6::GFP expression to sphk-1 mutants (Figure 3E) or to induce Phsp-6::GFP expression in the absence of stress (Figure 3F). These findings are consistent with the notion that S1P may function as a sphingolipid metabolite that activates the UPRmt.
Mitochondrial SPHK-1 Abundance Is Stress Regulated
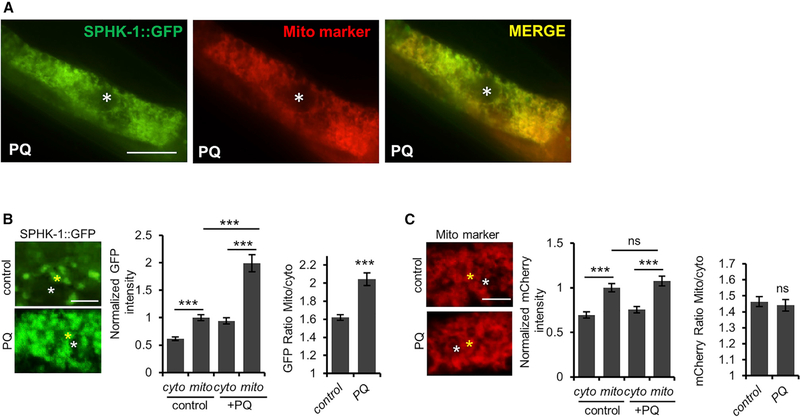
Sphingosine kinase activity is primarily controlled by its regulated targeting to cellular membranes from cytosolic pools (Alemany et al., 2007; Stahelin et al., 2005). We examined whether the mitochondrial abundance of SPHK-1::GFP is regulated by mitochondrial stress. Paraquat treatment for 24 hr produced a significant increase in the ratio of mitochondrial-to-cytosolic SPHK-1::GFP fluorescence compared to untreated controls, as well as an increase in the number of intestinal cells in which the mitochondrial SPHK-1::GFP pattern was detected (referred to as mitochondrial occupancy; Figures 4A, 4B, and S4A). The increase in the mitochondrial-to-cytosolic fluorescence ratio was driven by a large (2-fold) increase in mitochondrial SPHK-1::GFP fluorescence, accompanied by a significantly smaller increase in cytosolic fluorescence. Paraquat does not affect expression levels of the sphk-1::gfp transgene or the mitochondrial mass because paraquat treatment had no effect on the mitochondrial fluorescence intensity of TOMM-20::mCherry, which was expressed using the same intestinal promoter (ges-1) as the sphk-1::gfp transgenes (Figure 4C). Consistent with this observation, prior transcriptome profiling studies found no evidence that sphk-1 expression is stress regulated (Merkwirth et al., 2016; Tian et al., 2016). Chronic activation of the UPRmt by phb-1 or ETC gene knockdown also resulted in significantly increased SPHK-1::GFP mitochondrial abundance, without increasing mitochondrial mass (with the exception of nuo-1 RNAi; Figures S4B–S4F). Arsenite, a mitochondrial toxin that activates the mitochondrial antioxidant response but not the UPRmt, did not increase SPHK-1::GFP mitochondrial fluorescence (Figures S1C, S4G, and S4H), suggesting that mitochondrial SPHK-1 abundance is specifically regulated by stressors that activate the UPRmt. Finally, we found that paraquat treatment had no effect on mitochondrial SPHK-1::GFP abundance in muscles (Figure S5A). We conclude that mitochondrial association of SPHK-1 is positively regulated by multiple stressors that activate the UPRmt and that the regulation of mitochondrial SPHK-1 abundance occurs in a stress- and tissue-specific manner. The increase in the ratio of mitochondrial-to-cytoplasmic SPHK-1::GFP abundance is consistent with the idea that stress regulates the recruitment of SPHK-1 to mitochondria from cytoplasmic pools.
Figure 4. Mitochondrial Stress Promotes Mitochondrial SPHK-1 Accumulation.
(A) Representative intestinal images of animals co-expressing SPHK-1::GFP and mito-marker TOMM-20::mCherry following 24-hr paraquat treatment. Scale barrepresents 10 μm. Asterisk indicates intestinal nucleus.
(B) Representative images of intestinal SPHK-1::GFP in the absence or presence of paraquat. The yellow asterisk marks the reticulated region within the intestinal cells used to quantify mitochondrial SPHK-1::GFP abundance. The white asterisk marks the region in between mitochondria used to quantify cytosolic SPHK1::GFP abundance (left). Average GFP fluorescence intensity of mitochondrial or cytoplasmic SPHK-1::GFP is quantified (center). The ratio of mitochondrial-tocytosolic SPHK-1::GFP fluorescence is quantified (right). Scale bar indicates 2 mm.
(C) Representative images and quantification of TOMM-20::mCherry in intestinal cells in the absence or presence of paraquat. See (B) for details. Scale barindicates 2 μm.
The sample sizes (n) and ±SEMs are listed in Table S1. Error bars indicate ± SEMs. Student’s t test; p < 0.001
Stress-Induced SPHK-1 Mitochondrial Association Precedes UPRmt and Is Reversible
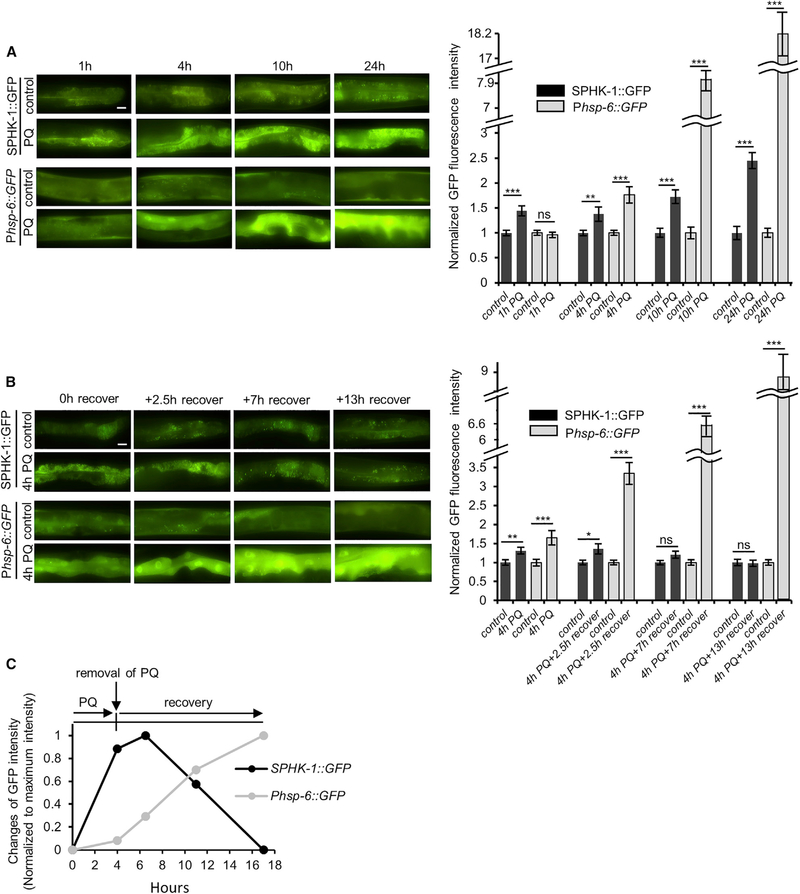
To explore the kinetics of SPHK-1 mitochondrial association during the UPRmt, we first examined mitochondrial SPHK-1::GFP association immediately following paraquat treatment for various periods. We detected a significant increase in SPHK1::GFP mitochondrial fluorescence after just 1 hr of paraquat treatment (Figure 5A). SPHK-1::GFP fluorescence increased further after longer paraquat exposure and reached a maximum of >2-fold after 24 hr of treatment. In contrast, Phsp-6::GFP induction was not detected until 4 hr of treatment and continued to increase until 24 hr of treatment (Figure 5A). The increase in mitochondrial SPHK-1::GFP fluorescence seen after 1 hr was not due to an increase in mitochondrial mass or transgene expression (Figure S5B).
Figure 5. Mitochondrial SPHK-1 Abundance Changes Rapidly in Response to Acute Stress.
(A) Representative images and quantification of intestinal Phsp-6::GFP and SPHK-1::GFP fluorescence in the intestines of animals treated with paraquat for 0, 1, 4, 10, or 24 hr.
(B) Representative images and quantification of intestinal Phsp-6::GFP and SPHK-1::GFP fluorescence in the intestines of animals treated with paraquat for 4 hr following recovery for 0, 2.5, 7, or 13 hr.
(C) Time course of average increases in the intensity of Phsp-6::GFP and SPHK-1::GFP normalized to maximum fluorescence intensity. Acute stress (4-hr paraquat treatment) leads to rapid increases in SPHK-1::GFP mitochondrial association that precede the UPRmt transcriptional response. Removal of stress leads to a more gradual reduction in SPHK-1 mitochondrial association without reducing the UPRmt transcriptional response.
Error bars indicate ± SEMs. The sample size (n) and ± SEMs are listed in Table S1. Scale bar represents 10 mm. Student’s t test; *p < 0.05, **p < 0.01, ***p < 0.001.
To determine whether the paraquat-induced increase in SPHK-1::GFP mitochondria abundance is reversible, we examined how long it would take for mitochondrial SPHK-1::GFP to return to baseline levels following paraquat removal. We found that the mitochondrial SPHK-1::GFP began to decline 7 hr after the removal of paraquat, returning to baseline by 17 hr. In contrast, Phsp-6::GFP induction continued to increase after paraquat removal (Figure 5B). Thus, acute mitochondrial stress leads to the reversible association of SPHK-1 with mitochondria that occurs rapidly (within 1 hr) and begins to decline more slowly (within 4 hr) when stress is removed. In contrast, acute mitochondrial stress leads to a delayed transcriptional response beginning 4 hr after exposure that persists after stress removal for several hours, even after mitochondrial SPHK-1 levels return to baseline (Figure 5C). These results support the idea that SPHK-1 association with mitochondria is stress regulated, reversible, and represents an early event in UPRmt activation that likely occurs via rapid SPHK-1 mitochondrial translocation.
Neuroendocrine Signaling Regulates SPHK-1 Mitochondrial Abundance
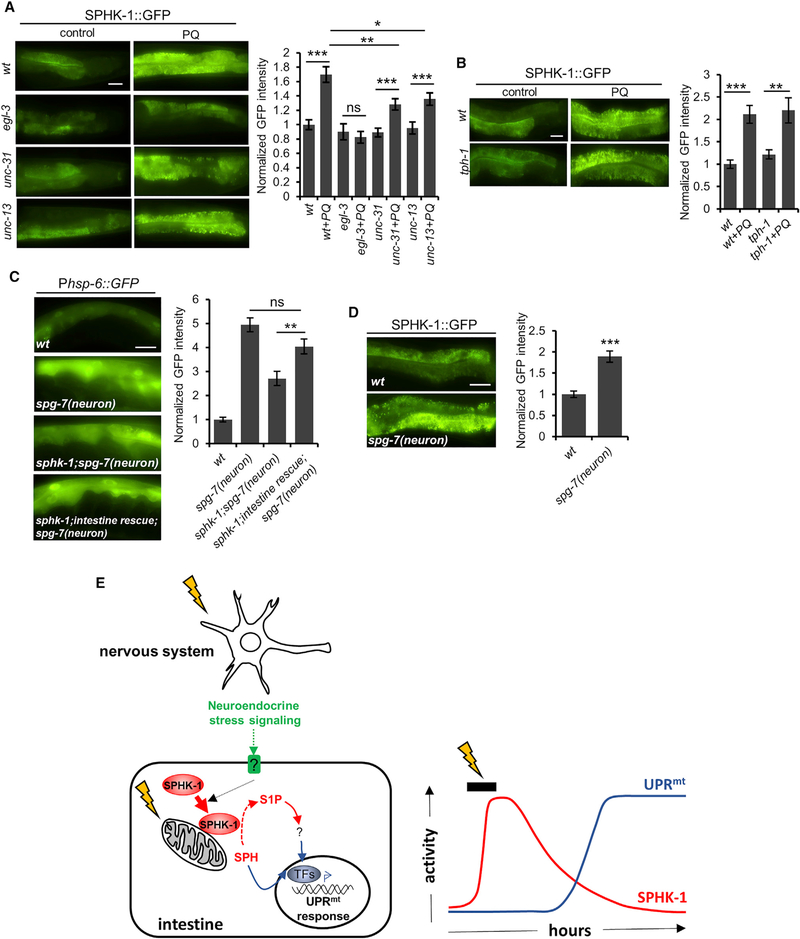
To investigate the role of SPHK-1 in the cell non-autonomous activation of the UPRmt, we examined the effects of neuronal stress signaling on SPHK-1-mediated UPRmt activation in the intestine. Prior studies have shown that EGL-3/pro-protein convertase (PC2), which processes neuropeptide precursors into mature peptides (Li and Kim, 2008), and UNC-31/CAPS, which mediates calcium-dependent exocytosis of neuropeptide- and biogenic amine-containing dense-core vesicles (DCVs) (Speese et al., 2007), promote stress-induced activation of the UPRmt (Berendzen et al., 2016; Shao et al., 2016). In agreement with these studies, we found that egl-3 and unc-31 null mutants reduced but did not eliminate UPRmt activation in response to paraquat (Figure S5C). UNC-13 is a vesicle-priming factor that mediates the exocytosis of synaptic vesicles and DCVs from the nervous system (Richmond and Broadie, 2002; Sieburth et al., 2007). We found that unc-13 mutants also attenuated the paraquat-induced UPRmt activation to an extent similar to that of egl-3 mutants (Figure S5C), reinforcing the notion that neuropeptide and/or neurotransmitter release from neurons is required for paraquat-induced UPRmt activation in the intestine.
To identify the neuronal-derived signals that regulate SPHK-1 signaling in the intestine, we examined SPHK-1::GFP mitochondrial abundance following paraquat treatment in egl-3, unc-31, and unc-13 mutants. SPHK-1::GFP mitochondrial fluorescence was not detectably altered in these mutants in the absence of paraquat (Figure 6A). However, the paraquat-induced increase in mitochondrial SPHK-1::GFP abundance was completely blocked in egl-3 mutants and was significantly attenuated in unc-13 and unc-31 mutants (Figure 6A). In contrast, blocking serotonin biosynthesis using the tph-1/tryptophan hydroxylase mutant, which impairs UPRmt activation (Berendzen et al., 2016), had no effect on stress-induced SPHK-1::GFP mitochondrial targeting (Figure 6B). We conclude that neuropeptides, but not serotonin, activate the UPRmt cell non-autonomously by positively regulating SPHK-1 mitochondrial association in the intestine.
Figure 6. Cell Non-autonomous UPRmt Activation by Neuropeptide Signaling Requires sphk-1 and Recruits SPHK-1 to Mitochondria.
(A) Representative images and quantification of the intestines of wild-type adults, egl-3/PC2, unc-31/CAPS, and unc-13/Munc13 mutants expressing SPHK1::GFP in the absence or presence of paraquat for 24 hr.
(B) Representative images and quantification of the intestines of wild-type and tph-1/tryptophan hydroxylase mutant adults expressing intestinal SPHK-1::GFP in the absence or presence of paraquat for 24 hr.
(C) Representative images and quantification of intestinal Phsp-6::GFP expression in wild-type or sphk-1 mutants expressing rab-3p::Cas9+u6p::spg-7-sg transgenes in which neuronal specific UPRmt is induced by neuronal spg-7 knockdown. Intestinal rescue denotes animals carrying Pges-1::sphk-1::mCherry transgenes
(D) Representative images and quantification of intestinal SPHK-1::GFP fluorescence in wild-type or in animals expressing the neuronal spg-7 knockdown transgene (rab-3p::Cas9+u6p::spg-7-sg).
(E) Working model for UPRmt activation by SPHK-1. Mitochondrial stressors induce SPHK-1 translocation to mitochondrial membranes from the cytoplasm, where SPHK-1 catalyzes the production of S1P from SPH. S1P then activates the UPRmt transcriptional response. Neuroendocrine signaling from the nervous system and cell autonomous mitochondrial stress activates the UPRmt by regulating intestinal SPHK-1 abundance on mitochondria.
Error bars indicate ± SEMs. The sample size (n) and ± SEMs are listed in Table S1. Scale bar represents 10 mm. Student’s t test; *p < 0.05, **p < 0.01, ***p < 0.001.
To directly assess whether SPHK-1 promotes intestinal UPRmt activation in response to neuronal mitochondrial stress, we examined UPRmt activation elicited by neuronal-specific knockdown of spg-7/paraplegin. Pan-neuronal knockdown of spg-7 using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein-9 nuclease (Cas9) elicits robust UPRmt activation in the intestine that is dependent upon egl-3/PC2 (Shao et al., 2016). We found that the increase in Phsp-6::GFP expression by neuronal spg-7 knockdown was significantly reduced in sphk-1 mutants, and that this defect was rescued to wild-type levels by intestine-specific expression of SPHK-1 (Figure 6C). Furthermore, pan-neuronal spg-7 knockdown increased mitochondrial SPHK-1::GFP abundance to an extent similar to that of paraquat treatment (Figure 6D). Collectively, these results indicate that intestinal SPHK-1 is essential for cell non-autonomous UPRmt activation by the nervous system and that neuropeptide release promotes SPHK-1 mitochondrial association in the intestine.
DISCUSSION
This study supports a model whereby SPHK-1 functions as part of an integrated organism-wide response to mitochondrial stress originating either cell autonomously in the intestine or cell nonautonomously in the nervous system that leads to the activation of the UPRmt in the intestine. Cell non-autonomous SPHK-1 regulation occurs through the release of neuropeptides that promote SPHK-1 mitochondrial recruitment in the intestine, presumably through intestinal G protein-coupled receptor (GPCR) signaling. We propose that the rapid association of SPHK-1 with intestinal mitochondria in response to stress leads to the production of S1P from SPH, followed by the subsequent regulation of UPRmt gene expression (Figure 6E).
SPHK-1 Activates the UPRmt
Our results suggest that the strong defects of UPRmt activation in sphk-1 null mutants are not the result of general defects in mitochondrial function or in stress sensing, but rather are due to specific defects in stress-induced initiation of the UPRmt. First, mutants with impaired mitochondrial function (e.g., ETC mutants) are slow growing (Rea et al., 2007), whereas sphk-1 mutants display largely normal rates of development (Chan et al., 2017). Second, mitochondrial abundance, morphology, and protein import do not appear to be grossly altered in sphk-1 mutants (Figures S2A and S2B), and mitochondrial responses to oxidative stress in the intestine are indistinguishable from wild-type responses (Figure S1C). Finally, sphk-1 mutants have normal rates of survival following mitochondrial stress induced by anoxia (Menuz et al., 2009), yet are hypersensitive to paraquat-induced toxicity. Overexpressing SPHK-1 increases hsp-6 expression by 2-fold, but it does not confer detectible resistance on paraquatinduced toxicity (Figure 1E), suggesting that the UPRmt transcriptional response may have to reach a certain threshold to promote organism-wide survival. sphk-1 mutants have shorter lifespans than wild-type controls (Chan et al., 2017), a phenotype that is associated with impaired UPRmt activation (Schulz and Haynes, 2015); but whether the UPRmt defects cause shortened lifespans in sphk-1 mutants remains to be determined. We also found that UPRer activation seems to be significantly enhanced in sphk-1 mutants (Figure S1D), suggesting either an additional inhibitory role of SPHK-1 in UPRer activation or the existence of SPHK-1-mediated crosstalk between the UPRmt and the UPRer.
Our genetic analysis of SPHK-1 reveals that sphk-1(ok1097) null mutants attenuate UPRmt activation to a significantly greater degree than sphk-1(ju831) mutants. The sphk-1(ju831) mutant encodes a P177S substitution located adjacent to the nucleotide binding site in the SPHK-1 kinase domain at a residue that is conserved between C. elegans and mammals but is not conserved in budding yeast (Pitson et al., 2002). Thus, we speculate that residual kinase activity in this strain may account for the weaker UPRmt defects seen in sphk-1(ju831) mutants compared to sphk-1 null mutants (McCulloch et al., 2017). Nonetheless, we observed full rescue of sphk-1(ok1097) with two independent SPHK-1 transgenes (Figures 1C and S1F), providing strong support for the conclusion that SPHK-1 activates the UPRmt. RNAi-mediated knockdown of sphk-1 was only effective in attenuating the UPRmt in the F1 generation (Figure S1A), suggesting that low levels of SPHK-1 or SPHK-1 metabolites (provided maternally or embryonically) are sufficient to activate the UPRmt in adults. The lack of effect on UPRmt revealed by using RNAi in the parental (P0) generation may explain why sphk-1 was not identified in previous large-scale RNAi screens for UPRmt activators (Haynes et al., 2007; Liu et al., 2014; Runkel et al., 2013).
Mitochondrial SPHK-1 Activates the UPRmt
Several results support the idea that SPHK-1 association with mitochondrial membranes is an early event in UPRmt activation. First, SPHK-1 association with intestinal mitochondria is stress dependent, and disruption of SPHK-1 mitochondrial localization (in SPHK-1[DCaM] mutants) strongly attenuates stress-induced UPRmt activation. Second, selective overexpression of SPHK-1 is sufficient to activate the UPRmt in an atfs-1-dependent manner in the absence of stress. Third, SPHK-1 mitochondrial association is dynamic; its abundance increases in response to mitochondrial stress and decreases when stressors are removed or when UPRmt activation is compromised in neuroendocrine signaling mutants. Our kinetic analysis indicates that stressinduced association of SPHK-1 with mitochondria precedes the UPRmt transcriptional response and that the UPRmt transcription remains intact even after SPHK-1 dissociates from mitochondria. We suggest that stress-induced SPHK-1 translocation to mitochondria from cytoplasmic pools and subsequent S1P production is a mechanism by which the UPRmt can be rapidly activated and sustained even if mitochondrial insults are transient.
Our results show that paraquat treatment increases the ratio of mitochondrial-to-cytoplasmic SPHK-1::GFP, which is consistent with the idea that stress promotes the targeting of SPHK-1 to mitochondria from the cytoplasm. We were surprised to find that paraquat treatment results in a small increase in cytoplasmic SPHK-1::GFP fluorescence instead of a decrease, as would be expected if SPHK-1 translocated to the mitochondria. We speculate that light scatter from SPHK-1::GFP associated with neighboring and out-of-plane mitochondria is likely to contribute to the inter-mitochondrial fluorescent signal. Our fluorescence imaging and ultrastructural studies show that mitochondria are very closely spaced throughout the intestine, with the majority being no more than 1 mm apart (Figure 4; Yokota et al., 2002). As a consequence, the increased SPHK-1::GFP abundance on mitochondria following paraquat treatment could lead to increased inter-mitochondrial fluorescence, even if cytosolic SPHK1::GFP abundance decreases.
Regulation of the translocation of sphingosine kinase to cellular membranes is the primary mechanism by which sphingosine kinase is activated. Muscarinic agonist treatment of HEK293 cells results in a rapid, reversible recruitment of approximately half of the SphK1 from the cytosol to the plasma membrane, with a half-time of 3–5 s (ter Braak et al., 2009). Sphingosine kinase membrane recruitment is mediated by both direct interaction of SphK with lipids on membranes (e.g., phosphatidic acid) and interaction of SphK with specific proteins (Siow and Wattenberg, 2011). Notably, calcium and integrinbinding protein-1 (CIB1) has been shown to interact directly with the CaM domain of SphK1 in a calcium-dependent manner, and this interaction is important for SphK1 recruitment to the plasma membrane and S1P production (Jarman et al., 2010). There are four CIB1 homologs in the C. elegans genome, including CALM-1, which regulates SPHK-1 targeting to synapses in motor neurons (Chan and Sieburth, 2012). RNAi knockdown of the intestinal gap-junction protein inx-17 or the inositol phosphate receptor itr-1 attenuates stress-induced UPRmt activation (Liu et al., 2014), suggesting a role for calcium dynamics in regulating the UPRmt. Given the importance of the CaM domain in both SPHK-1 mitochondrial targeting and UPRmt activation by SPHK-1, it will be interesting to determine whether calcium promotes UPRmt activation by regulating SPHK-1 targeting to mitochondria.
S1P Signaling in the UPRmt
The 2014 study by Liu et al. showed that ceramide or its metabolites are involved in UPRmt activation. The observation that knocking out ceramide synthase has minimal effects on UPRmt activation (Figure S3; Liu et al., 2014), and our results showing that knockdown of multiple enzymes directly involved in S1P metabolism are able to alter UPRmt activation lead us to favor the idea that ceramide metabolites (SPH or S1P) may be more important for UPRmt activation than ceramide itself. It is also possible that ceramide and S1P may function cooperatively to activate the UPRmt. C. elegans encodes three ceramide synthase genes (hyl-1, hyl-2, and lagr-1), but RNAi knockdown of these genes alone did not alter paraquat-induced UPRmt activation (Figure S3), suggesting that they may function redundantly to convert ceramide to SPH in the intestine. Consistent with this idea, HYL-1 and HYL-2 are both expressed in the intestine (Hunt-Newbury et al., 2007; Menuz et al., 2009).
How might a small (2-fold) and transient increase in SPHK-1 mitochondrial association lead to a relatively large (>20-fold) and prolonged transcriptional response? We speculate that small increases in SPHK-1 abundance could lead to relatively large increases in S1P through the enzymatic conversion of SPH to S1P on mitochondrial membranes. S1P, in turn, could generate long-lasting effects on the UPRmt by stably associating with cellular targets, leading to enhanced transcriptional response by as-yet-undetermined mechanisms. S1P can act extracellularly as a secreted signaling messenger or as an intracellular signaling molecule. As a secreted signal, S1P signals via a family of five S1PR/EDG GPCRs (Rosen et al., 2013). As an intracellular signal, S1P can directly associate with and modulate the activities of cytoplasmic proteins, including the E3 ubiquitin ligase TRAF2, histone deacetylases (HDAC1 and HDAC2), the mitochondrial mitophagy protein prohibitin (Alvarez et al., 2010; Hait et al., 2009; Wei et al., 2017), and members of the HSP-90 family of heat shock proteins (Park et al., 2016). C. elegans does not encode obvious S1P receptors, so if secreted, S1P is likely to act by a mechanism that is independent of known S1P receptors. We favor the idea that S1P acts as intracellular signaling molecule that binds an as-yet-undefined intestinal target to activate the UPRmt. S1P generated by mitochondrial SphK2 interacts with prohibitin2 in the mitochondria to modulate the assembly and function of cytochrome oxidase, a component of the ETC, and mice lacking SphK2 function display defects in mitochondria respiration (Strub et al., 2011). Our results show that UPRmt activation by phb-1/prohibitin knockdown is dependent on sphk-1 (Figure 2C), suggesting that SPHK-1/S1P may not activate PHB-1 but instead function downstream of PHB-1 to activate the UPRmt. S1P signaling also plays an important role in the innate immune response in epithelial cells in response to ER stress by binding directly to the heat shock proteins GRP94 and HSP90a, resulting in the assembly of a transcriptional complex that activates innate immune response transcriptional targets (Park et al., 2016). Based on these studies, it is tempting to speculate that S1P production may orchestrate the assembly of protein complexes in the mitochondria or cytoplasm that initiates UPRmt activation.
STAR+METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Methyl Viologen Hydrate (paraquat) | Fisher | CAS:1910–42-5 |
| Tunicamycin | Sigma-Aldrich | CAS:11089–65-9 |
| Antimycin A | Sigma-Aldrich | CAS:1397–94-0 |
| Sodium Arsenite | Ricca | CAS:7140–16 |
| Experimental Models: Organisms/Strains | ||
| C. elegans: N2 | Caenorhabditis Genetics Center | WB Cat# N2_(ancestral), RRID:WB-STRAIN:N2_(ancestral) |
| C. elegans: sphk-1(ok1097) | Caenorhabditis Genetics Center | WB Cat# VC916, RRID:WB-STRAIN: VC916 |
| C. elegans: hyl-1(gk203) | Caenorhabditis Genetics Center | WB Cat# VC334, RRID:WB-STRAIN: VC334 |
| C. elegans: egl-3(nr2090) | Caenorhabditis Genetics Center | WormBase: WBVar00091400 (RRID N/A) |
| C. elegans: unc-13(s69) | Caenorhabditis Genetics Center | WB Cat# BC168, RRID:WB-STRAIN: BC168 |
| C. elegans: unc-31(e928) | Caenorhabditis Genetics Center | WB Cat# DA509, RRID:WB-STRAIN: DA509 |
| C. elegans: tph-1(mg280) | Caenorhabditis Genetics Center | WB Cat# MT15434, RRID:WB-STRAIN: MT15434 |
| C. elegans: SJ4100: zcIs13[Phsp-6::GFP] | Caenorhabditis Genetics Center | WB Cat# SJ4100, RRID:WB-STRAIN: SJ4100 |
| C. elegans: SJ4005: zcIs4[Phsp-4::GFP] | Caenorhabditis Genetics Center | WB Cat# SJ4005, RRID:WB-STRAIN: SJ4005 |
| C. elegans: SJ4058: zcIs9 [Phsp-60::GFP + lin-15(+)] | Caenorhabditis Genetics Center | WB Cat# SJ4058, RRID:WB-STRAIN: SJ4058 |
| C. elegans: OJ4113: vjIs138[Pges-1::sphk-1::gfp] | This paper | N/A |
| C. elegans: OJ4143: vjIs148[Pges-1::tomm-20::mCherry] | This paper | N/A |
| C. elegans: OJ2329: vjIs208[Pgst-4::gfp] | This paper | Sean Curran |
| C. elegans: OJ1674: vjIs84[Pmyo-3::invom::rfp] | (Staab et al., 2013) | N/A |
| C. elegans: OJ4647: sphk-1(ok1097);zcIs4 | This paper | N/A |
| C. elegans: OJ4433: sphk-1(ok1097);zcIs9 | This paper | N/A |
|
C. elegans: OJ4127: sphk-1(ok1097);vjEx920[Pges-1:: sphk-1::gfp];zcIs13 |
This paper | N/A |
|
C. elegans: OJ4364: sphk-1(ok1097);vjEx1025[Pges-1:: sphk-1(DCaM)]::gfp;zcIs13 |
This paper | N/A |
|
C. elegans: OJ4367: sphk-1(ok1097);vjEx1459[Prab-3:: sphk-1::mCherry];zcIs13 |
This paper | N/A |
|
C. elegans: OJ4365: sphk-1(ok1097);vjEx1058[Pges-1:: sphk-1(KD)]::gfp zcIs13 |
This paper | N/A |
|
C. elegans: OJ4139: sphk-1(ok1097);vjEx966[Pges-1:: sphk-1::mCherry];zcIs13 |
This paper | N/A |
| C. elegans: OJ4648: sphk-1(ju831);zcIs13 | This paper | N/A |
| C. elegans: OJ4147: vjEx966[Pges-1::sphk-1::mCherry]; zcIs13 | This paper | N/A |
| C. elegans: OJ4398: egl-3(nr2090);zcIs13 | This paper | N/A |
| C. elegans: OJ4397: unc-13(s96);zcIs13 | This paper | N/A |
| C. elegans: OJ4522: unc-31(e928);zcIs13 | This paper | N/A |
| C. elegans: OJ4265: hyl-1(gk203);zcIs13 | This paper | N/A |
| C. elegans: OJ4568: unc-31(e928);vjIs138[Pges-1::sphk-1::gfp] | This paper | N/A |
| C. elegans: OJ4569: unc-13(s69);vjIs138[Pges-1::sphk-1::gfp] | This paper | N/A |
| C. elegans: OJ4570: egl-3(nr2090);vjIs138[Pges-1::sphk-1::gfp] | This paper | N/A |
| C. elegans: OJ5015: tph-1(mg280);vjIs138[Pges-1::sphk-1::gfp] | This paper | N/A |
| C. elegans: OJ2582: vjEx1007[Pges-1::tomm-20::gfp] | This paper | N/A |
| C. elegans: OJ2835: vjEx1072[Pmyo-3::sphk-1(KD)::gfp] | This paper | N/A |
| C. elegans: OJ2577: vjEx1002[Pmyo-3::sphk-1(DCaM)::gfp] | This paper | N/A |
| C. elegans: OJ3331: vjEx1065[Pges-1::tomm-20::mCherry] | This paper | N/A |
|
C. elegans: OJ2806: sphk-1(ok1097);vjEx1065[Pges-1::tomm- 20::mCherry] |
This paper | N/A |
| C. elegans: OJ326: vjEx133[Pmyo-3::sphk-1::gfp] | (Chan and Sieburth, 2012) | N/A |
| C. elegans: LW96: [Prab-3::Cas9+u69::spg-7-sg];zcIs13 | (Shao et al., 2016) | N/A |
| C. elegans: OJ4751: sphk-1;zcIs13;[Prab-3::Cas9+u69::spg-7-sg] | This paper | N/A |
|
C. elegans: OJ5095: sphk-1;vjEx966[Pges-1::sphk-1::mCherry]; [Prab-3::Cas9+u69::spg-7-sg];zcIs13 |
This paper | N/A |
| C. elegans: OJ4924: vjIs138[Pges-1::sphk-1::gfp];[Prab-3::Cas9+ u69::spg-7-sg] | This paper | N/A |
|
C. elegans: OJ1564: hjIs14[Pvha-6::gfp::C34B2.10]; vjEx1564 [Pges-1::sphk-1::mCherry] |
This paper | N/A |
| Oligonucleotides | ||
| For cloning sphk-1 cDNA Forward:ccccccgctagcaaaaatgttcata gtagtggtaac Reverse:ccccccggtaccctaggcagttgatgagaaaacgg | (Chan et al., 2012) | N/A |
| Oligos used for quickchange PCR of sphk-1(KD) cDNA See the reference | (Chan et al., 2012) | N/A |
| Oligos used for quickchange PCR of sphk-1(▵CaM) cDNA See the reference | (Chan et al., 2012) | N/A |
| For cloning tomm-20(n-terminus, 126bp) cDNA Forward: ccccg ctagcaaaaatgtcgga cacaattcttgg Reverse: ccccaccggttccagcctgggcacgtctc | This paper | N/A |
| For phb-1 RNAi clone Forward: ccccccgcatgcaaaagcactttcaag gcgacagta Reverse: ccccccgctagctttgattcgaaggttagtgt |
This paper | N/A |
| Recombinant DNA | ||
| pSK9[Pges-1::sphk-1::gfp] | This paper | N/A |
| pSK21[Pges-1::sphk-1::mCherry] | This paper | N/A |
| pSK70[Prab-3::sphk-1::mCherry] | This paper | N/A |
| pSK24[Pges-1::tomm-20::gfp] | This paper | N/A |
| pSK26[Pges-1::tomm-20::mCherry] | This paper | N/A |
| pSK28[Pges-1::sphk-1(DCaM)::gfp] | This paper | N/A |
| pSK29[Pges-1::sphk 1(KD)::gfp] | This paper | N/A |
| pSK27[Pmyo-3::sphk 1(KD)::gfp] | This paper | N/A |
| pSK25[Pmyo-3::sphk 1(▵CaM)::gfp] | This paper | N/A |
| pSK30[L4440-phb-1] | This paper | N/A |
| Other | ||
| Sequence, gene information | N/A | https://wormbase.org/#012-34-5 |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Derek Sieburth (sieburth@usc.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans strains and maintenance
Strains used in this study were maintained at 20C on NGM plates following standard methods (Brenner, 1974). As a food source, HB101 E.coli bacteria was used because sphk-1 mutants are developmentally healthier than when grown on OP50 E.coli bacteria (Chan et al., 2017). The wild-type reference strain was N2 Bristol. L4 or young adult hermaphrodites were used for all experiments. The following strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources: sphk-1(ok1097), hyl-1(gk203), egl-3(nr2090), unc-13(s69), unc-31(e928), tph-1(mg280). All strains used were outcrossed at least five times with wild-type prior to analysis. The strain information is at https://wormbase.org/#012-34-5.
METHOD DETAILS
Molecular biology
Genes used in this study were cloned from C. elegans wild-type cDNA then inserted into pPD49.26-based vectors using standard molecular biology techniques, unless otherwise indicated. The plasmids and oligos used in this study are listed in the KEY RESCOURCES TABLE. Sequence files of plasmids are available upon request.
Transgenic lines
Transgenic strains were generated by injecting expression constructs (10–25 ng/ul) and the co-injection marker; pJQ70 [Pofm1::mCherry, 40 ng/ul], KP#708[Pttx-3::rfp, 40 ng/ul] or KP#1106 [Pmyo-2::gfp, 10 ng/ul] into N2 or corresponding mutants. Microinjection was performed following standard techniques as previously described (Mello et al., 1991). At least three lines for each transgene were tested and a representative transgene was used for further experiments. The transgenic lines used in this study are listed in the KEY RESCOURCES TABLE.
Microscopy analysis
For all fluorescence microscopy analysis, young adult worms were immobilized by using 2,3-butanedione monoxime (BDM, 30 mg/ mL; Sigma) in M9 buffer then mounted on 2% agarose pads for imaging. Images were captured with the Nikon eclipse 90i microscope equipped with a Nikon PlanApo 40 × or 60× or 100× objective (NA = 1.4) and a PhotometricsCoolsnap ES2 or a Hamamatsu Orca Flash LT+ CMOS camera. To quantify the fluorescence intensity of Phsp-6::GFP, Phsp-4::GFP, and Phsp-60::GFP fluorescent reporters, intestinal cells posterior to the vulva were imaged due to low basal expression in the absence of stress. To analyze Pgst-4::GFP expression, SPHK-1::GFP abundance, and TOMM-20::mCherry or GFP abundance, about 5μm diameter region in posterior intestinal cells were selected for fluorescence intensity quantification. To analyze mitochondrial SPHK-1::GFP and TOMM20::mCherry abundance, average fluoresce intensity from 0.1 mm diameter regions on individual mitochondria from at least 20 animals was measured. For cytosolic abundance, average fluorescence intensity of 0.1 mm diameter regions in intestinal cells between mitochondria was measured. The number of animals and mitochondria analyzed are listed in Table S1. For each experimental sample set, image acquisition was conducted in parallel on the same day. The maximum intensity projection of images was recorded using Metamorph 7.0 software (Universal Imaging/Molecular Devices). For mitochondrial occupancy analysis, only intestinal cells showing reticulated fluorescence pattern were counted. H (high), M (medium) L (low) indicates the percentage of animals showing mitochondrial localized-SPHK-1::GFP more than 2/3 of intestine (H), half of intestine (M), only in a few intestinal cells (L), respectively. For Figure 3E and F, Figure S1F, the fluorescence intensity of SPHK-1::GFP, SPHK-1(▵CaM)::GFP and SPHK-1(KD)::GFP transgenes in intestine was approximately ten fold lower compared to hsp-6::GFP fluorescence, thus they did not significantly impact hsp-6::GFP intensity levels.
RNA Interference
Feeding RNAi knockdown assays were performed following established protocols (Kamath and Ahringer, 2003). Briefly, gravid adult animals were placed on RNAi plates seeded with HT1115(DE3) bacteria transformed with L4440 vector containing the insert of the gene targeted for knockdown or empty L4440 vector as a control, and eggs were collected for 4 hours to obtain age-matched synchronized worm populations. Young adult animals were used for all RNAi assays. RNAi clones were from the Ahringer RNAi library (Kamath and Ahringer, 2003) and dct-1, drp-1, fis-1, pink-1, dve-1, haf-1, met-2, lagr-1, hyl-2, sptl-1, F27E5.1, T27F6.6, tag-38 were confirmed by sequencing.
Stress induction assays
For drug-induced stress, transgenic L4 animals were transferred to fresh NGM plates seeded with HB101 bacteria, then 80 mL of stock solutions of drugs was added to plates for a final concentration of 0.4 mM paraquat, 40 mM arsenite, 80nM Antimycin A. Drugs were dissolved in M9 buffer except Antimycin A, which was dissolved in DMSO. 24 hours later, adults were removed for fluorescence microscopy analysis. For paraquat treatment, 0.4 mM paraquat for 24 hours was used for most assays, unless otherwise indicated. These conditions produced the most significant induction of hsp-6 expression with no toxicity but slightly delayed development. For the analysis of time course-expression of Phsp-6::GFP and SPHK-1::GFP (Figure 5A), 19 to 45 L4 animals were placed on HB101 plates containing 0.4 mM paraquat, and were removed for imaging immediately after 1 hour, 4 hours, 10 hours or 24 hours exposure. For Figure 5B, 22 to 26 L4 animals treated with 0.4 mM paraquat for 4 hours were transferred to new NGM plates without paraquat for 2.5 hours, 7 hours, 13 hours for recovery and then removed for imaging. To synchronize animals for imaging following RNAi treatment, L4 animals were transferred to fresh RNAi plates and imaged 24 hours later. For Phsp-4::GFP induction assays, L4 animals were transferred to seeded NGM plates containing 10mM tunicamycin and imaged 24 hours later. For Pgst-4::GFP imaging, young adult animals are incubated with 5mM arsenite in liquid solution for 1 hour then arsenite was washed out and worms were transferred onto the new NGM plates for 4 hours before imaging. As a control, M9 buffer was used as a control for paraquat and arsenite treatment, and DMSO was used as a control for Antimycin A and tunicamycin treatment. Identifiers of drugs used in this study are listed in the Key Resources Table.
Stress survival assays
For paraquat survival assays (Figure 1E), young adult animals were placed onto NGM plates containing 10mM paraquat for 17 hours. After 17 hours, the percentage of surviving animals was counted every 3 hours over the course of 15 hours. Survival assay were done in experimental duplicate for each biological duplicate. The number of animals tested are indicated in the Table S1.
QUANTIFICATION AND STATISTICAL ANALYSIS
For all assays, the Student’s t test (two-tailed) was used to determine the statistical significance. P values less than 0.05, 0.01 or 0.001 are indicated with asterisks * (p < 0.05), **(p < 0.01), ***(p < 0.001), respectively. Error bars in the figures indicate the standard error of the mean (±SEM). The exact sample sizes (n) and ± SEM values corresponding to all figures are listed in Table S1.
Supplementary Material
Highlights.
SPHK-1/sphingosine kinase functions cell autonomously to activate the UPRmt
SPHK-1 is targeted to mitochondria in response to mitochondrial stress
SPHK-1 mitochondrial targeting is dynamic and occurs before UPRmt activation
SPHK-1 mitochondrial association is positively regulated by neuroendocrine signaling
ACKNOWLEDGMENTS
We thank members of the lab for advice and critical review of the manuscript, and Sean Curran for the Pgst-4::GFP integrant. We thank Yishi Jin and Katherine McCulloch for providing the sphk-1(ju831) strain, Ying Liu for the LW96 strain and Ho Yi Mak for hjIs14. This work was supported by grants from the NIH National Institute of Neurological Disorders and Stroke (NINDS) to D.S. (NS071085 and NS099414). Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures and one table and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.08.037.
REFERENCES
- Alemany R, van Koppen CJ, Danneberg K, Ter Braak M, and Meyer Zu Heringdorf D (2007). Regulation and functional roles of sphingosine kinases. Naunyn Schmiedebergs Arch. Pharmacol. 374, 413–428. [DOI] [PubMed] [Google Scholar]
- Alvarez SE, Harikumar KB, Hait NC, Allegood J, Strub GM, Kim EY, Maceyka M, Jiang H, Luo C, Kordula T, et al. (2010). Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature 465, 1084–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asikainen S, Vartiainen S, Lakso M, Nass R, and Wong G (2005). Selective sensitivity of Caenorhabditis elegans neurons to RNA interference. Neuroreport 16, 1995–1999. [DOI] [PubMed] [Google Scholar]
- Berendzen KM, Durieux J, Shao LW, Tian Y, Kim HE, Wolff S, Liu Y, and Dillin A (2016). Neuroendocrine Coordination of Mitochondrial Stress Signaling and Proteostasis. Cell 166, 1553–1563.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JP, and Sieburth D (2012). Localized sphingolipid signaling at presynaptic terminals is regulated by calcium influx and promotes recruitment of priming factors. J. Neurosci 32, 17909–17920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JP, Hu Z, and Sieburth D (2012). Recruitment of sphingosine kinase to presynaptic terminals by a conserved muscarinic signaling pathway promotes neurotransmitter release. Genes Dev. 26, 1070–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JP, Brown J, Hark B, Nolan A, Servello D, Hrobuchak H, and Staab TA (2017). Loss of Sphingosine Kinase Alters Life History Traits and Locomotor Function in Caenorhabditis elegans. Front. Genet 8, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, and Dillin A (2011). The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorese CJ, Schulz AM, Lin YF, Rosin N, Pellegrino MW, and Haynes CM (2016). The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Curr. Biol 26, 2037–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatsi R, Schulze B, Rodríguez-Palero MJ, Hernando-Rodríguez B, Baumeister R, and Artal-Sanz M (2014). Prohibitin-mediated lifespan and mitochondrial stress implicate SGK-1, insulin/IGF and mTORC2 in C. elegans. PLoS One 9, e107671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter KM, Lin S, and Blackwell TK (2013). Integration of the unfolded protein and oxidative stress responses through SKN-1/Nrf. PLoS Genet. 9, e1003701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, Luo C, Marmorstein R, Kordula T, Milstien S, and Spiegel S (2009). Regulation of histone acetylation in the nucleus by sphingosine-1phosphate. Science 325, 1254–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum D, Haddadi N, Lin Y, Nassif NT, and McGowan EM (2017). Mammalian sphingosine kinase (SphK) isoenzymes and isoform expression: challenges for SphK as an oncotarget. Oncotarget 8, 36898–36929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes CM, Petrova K, Benedetti C, Yang Y, and Ron D (2007). ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev. Cell 13, 467–480. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Yang Y, Blais SP, Neubert TA, and Ron D (2010). The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol. Cell 37, 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A, et al. (2007). High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 5, e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Hisamoto N, An JH, Oliveira RP, Nishida E, Blackwell TK, and Matsumoto K (2005). The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 19, 2278–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Takahashi K, Tomita S, Keino T, Honda S, Yoshino K, and Suzuki K (1990). A methyl viologen-sensitive mutant of the nematode Caenorhabditis elegans. Mutat. Res 237, 165–171. [DOI] [PubMed] [Google Scholar]
- Jarman KE, Moretti PA, Zebol JR, and Pitson SM (2010). Translocation of sphingosine kinase 1 to the plasma membrane is mediated by calcium- and integrin-binding protein 1. J. Biol. Chem 285, 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, and Ahringer J (2003). Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313–321. [DOI] [PubMed] [Google Scholar]
- Labrousse AM, Zappaterra MD, Rube DA, and van der Bliek AM (1999). C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell 4, 815–826. [DOI] [PubMed] [Google Scholar]
- Li C, and Kim K (2008). Neuropeptides In WormBook, Jorgensen EM and Kaplan JM, eds. (The C. elegans Research Community; ). https://doi.org/10.1895/wormbook.1.142.1 . 10.1895/wormbook.1.142.1http://www.wormbook.org. http://www.wormbook.org. [DOI] [Google Scholar]
- Liu Y, Samuel BS, Breen PC, and Ruvkun G (2014). Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature 508, 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez BA, Petersen DA, Gaeta AL, Stanley SP, Caldwell GA, and Caldwell KA (2017). Dysregulation of the Mitochondrial Unfolded Protein Response Induces Non-Apoptotic Dopaminergic Neurodegeneration in C. elegans Models of Parkinson’s Disease. J. Neurosci 37, 11085–11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum KC, Liu B, Fierro-Gonza´ lez JC, Swoboda P, Arur S, Miranda-Vizuete A, and Garsin DA (2016). TRX-1 regulates SKN-1 nuclear localization cell non-autonomously in Caenorhabditis elegans. Genetics 203, 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch KA, Qi YB, Takayanagi-Kiya S, Jin Y, and Cherra SJ 3rd. (2017). Novel Mutations in Synaptic Transmission Genes Suppress Neuronal Hyperexcitation in Caenorhabditis elegans. G3 (Bethesda) 7, 2055–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, and Ambros V (1991). Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuz V, Howell KS, Gentina S, Epstein S, Riezman I, FornallazMulhauser M, Hengartner MO, Gomez M, Riezman H, and Martinou JC (2009). Protection of C. elegans from anoxia by HYL-2 ceramide synthase. Science 324, 381–384. [DOI] [PubMed] [Google Scholar]
- Merkwirth C, Jovaisaite V, Durieux J, Matilainen O, Jordan SD, Quiros PM, Steffen KK, Williams EG, Mouchiroud L, Tronnes SU, et al. (2016). Two Conserved Histone Demethylases Regulate Mitochondrial StressInduced Longevity. Cell 165, 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, and Haynes CM (2012). Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337, 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, and Haynes CM (2015). Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt). Mol. Cell 58, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras K, Lionaki E, and Tavernarakis N (2015). Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 521, 525–528. [DOI] [PubMed] [Google Scholar]
- Park K, Ikushiro H, Seo HS, Shin KO, Kim YI, Kim JY, Lee YM, Yano T, Holleran WM, Elias P, and Uchida Y (2016). ER stress stimulates production of the key antimicrobial peptide, cathelicidin, by forming a previously unidentified intracellular S1P signaling complex. Proc. Natl. Acad. Sci. USA 113, E1334–E1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino MW, Nargund AM, and Haynes CM (2013). Signaling the mitochondrial unfolded protein response. Biochim. Biophys. Acta 1833, 410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino MW, Nargund AM, Kirienko NV, Gillis R, Fiorese CJ, and Haynes CM (2014). Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature 516, 414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitson SM, Moretti PA, Zebol JR, Zareie R, Derian CK, Darrow AL, Qi J, D’Andrea RJ, Bagley CJ, Vadas MA, and Wattenberg BW (2002). The nucleotide-binding site of human sphingosine kinase 1. J. Biol. Chem 277, 49545–49553. [DOI] [PubMed] [Google Scholar]
- Rea SL, Ventura N, and Johnson TE (2007). Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 5, e259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond JE, and Broadie KS (2002). The synaptic vesicle cycle: exocytosis and endocytosis in Drosophila and C. elegans. Curr. Opin. Neurobiol 12, 499–507. [DOI] [PubMed] [Google Scholar]
- Rosen H, Stevens RC, Hanson M, Roberts E, and Oldstone MB (2013). Sphingosine-1-phosphate and its receptors: structure, signaling, and influence. Annu. Rev. Biochem 82, 637–662. [DOI] [PubMed] [Google Scholar]
- Runkel ED, Liu S, Baumeister R, and Schulze E (2013). Surveillance-activated defenses block the ROS-induced mitochondrial unfolded protein response. PLoS Genet. 9, e1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz AM, and Haynes CM (2015). UPR(mt)-mediated cytoprotection and organismal aging. Biochim. Biophys. Acta 1847, 1448–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao LW, Niu R, and Liu Y (2016). Neuropeptide signals cell non-autonomous mitochondrial unfolded protein response. Cell Res. 26, 1182–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Giordano F, Wu Y, Chan J, Zhu C, Milosevic I, Wu X, Yao K, Chen B, Baumgart T, et al. (2014). Coupling between endocytosis and sphingosine kinase 1 recruitment. Nat. Cell Biol 16, 652–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth D, Madison JM, and Kaplan JM (2007). PKC-1 regulates secretion of neuropeptides. Nat. Neurosci 10, 49–57. [DOI] [PubMed] [Google Scholar]
- Siow D, and Wattenberg B (2011). The compartmentalization and translocation of the sphingosine kinases: mechanisms and functions in cell signaling and sphingolipid metabolism. Crit. Rev. Biochem. Mol. Biol 46, 365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speese S, Petrie M, Schuske K, Ailion M, Ann K, Iwasaki K, Jorgensen EM, and Martin TF (2007). UNC-31 (CAPS) is required for dense-core vesicle but not synaptic vesicle exocytosis in Caenorhabditis elegans. J. Neurosci 27, 6150–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staab TA, Griffen TC, Concoran C, Evgrafov O, Knowles JA, and Sieburth D (2013). The conserved SKN-1/Nrf2 stress response pathway regulates synaptic function in Caenorhabditis elegans. PLoS Genetics 9, e1003354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahelin RV, Hwang JH, Kim JH, Park ZY, Johnson KR, Obeid LM, and Cho W (2005). The mechanism of membrane targeting of human sphingosine kinase 1. J. Biol. Chem 280, 43030–43038. [DOI] [PubMed] [Google Scholar]
- Strub GM, Paillard M, Liang J, Gomez L, Allegood JC, Hait NC, Maceyka M, Price MM, Chen Q, Simpson DC, et al. (2011). Sphingosine-1-phosphate produced by sphingosine kinase 2 in mitochondria interacts with prohibitin 2 to regulate complex IV assembly and respiration. FASEB J. 25, 600–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, and Dillin A (2013). XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell 153, 1435–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Braak M, Danneberg K, Lichte K, Liphardt K, Ktistakis NT, Pitson SM, Hla T, Jakobs KH, and Meyer zu Heringdorf D (2009). Galpha(q)mediated plasma membrane translocation of sphingosine kinase-1 and cross-activation of S1P receptors. Biochim. Biophys. Acta 1791, 357–370. [DOI] [PubMed] [Google Scholar]
- Tian Y, Garcia G, Bian Q, Steffen KK, Joe L, Wolff S, Meyer BJ, and Dillin A (2016). Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPR(mt). Cell 165, 1197–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Chiang WC, Sumpter R Jr., Mishra P, and Levine B (2017). Prohibitin 2 Is an Inner Mitochondrial Membrane Mitophagy Receptor. Cell 168, 224–238.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Zhang SO, Cole RA, McKinney SA, Guo F, Haas JT, Bobba S, Farese RV Jr., and Mak HY (2012). The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J. Cell Biol 198, 895–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota S, Togo SH, Maebuchi M, Bun-Ya M, Haraguchi CM, and Kamiryo T (2002). Peroxisomes of the nematode Caenorhabditis elegans: distribution and morphological characteristics. Histochem. Cell Biol 118, 329–336. [DOI] [PubMed] [Google Scholar]
- Young KW, Willets JM, Parkinson MJ, Bartlett P, Spiegel S, Nahorski SR, and Challiss RA (2003). Ca2+/calmodulin-dependent translocation of sphingosine kinase: role in plasma membrane relocation but not activation. Cell Calcium 33, 119–128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.