Abstract
Background
Gene fusions derive from chromosomal rearrangements. The resulting chimeric transcripts are often endowed with oncogenic potential. Furthermore, they serve as diagnostic tools for the clinical classification of cancer subgroups with different prognosis and, in some cases, they can provide specific drug targets. To date, many efforts have been carried out to study gene fusion events occurring in tumor samples. In recent years, the availability of a comprehensive next-generation sequencing dataset for all existing human tumor cell lines has provided the opportunity to further investigate these data in order to identify novel and still uncharacterized gene fusion events.
Results
In our work, we have extensively reanalyzed 935 paired-end RNA-sequencing experiments downloaded from the Cancer Cell Line Encyclopedia repository, aiming at addressing novel putative cell-line specific gene fusion events in human malignancies. The bioinformatics analysis has been performed by the execution of four gene fusion detection algorithms. The results have been further prioritized by running a Bayesian classifier that makes an in silico validation. The collection of fusion events supported by all of the predictive software results in a robust set of ∼1,700 in silico predicted novel candidates suitable for downstream analyses. Given the huge amount of data and information produced, computational results have been systematized in a database named LiGeA. The database can be browsed through a dynamic and interactive web portal, further integrated with validated data from other well-known repositories. Taking advantage of the intuitive query forms, the users can easily access, navigate, filter, and select the putative gene fusions for further validations and studies. They can also find suitable experimental models for a given fusion of interest.
Conclusions
We believe that the LiGeA resource can represent not only the first compendium of both known and putative novel gene fusion events in the catalog of all of the human malignant cell lines but it can also become a handy starting point for wet-lab biologists who wish to investigate novel cancer biomarkers and specific drug targets.
Keywords: database, human gene fusions, malignant cell lines, NGS, gene fusion detection algorithms, chromosomal rearrangements, bioinformatics
Background
Oncogenic gene fusion events result from chromosomal rearrangements that lead to the juxtaposition of two previously separated genes. The accidental joining of DNA of two genes can generate hybrid proteins. It can also result in the misregulation of the transcription of one gene by the cis-regulatory elements (promoters or enhancers) of another, sometimes resulting in the production of oncoproteins that bring the cell to a neoplastic transformation [1]. Not only can gene fusions have a strong oncogenic potential [2] but they can also serve as diagnostic tools for the clinical classification of cancer subgroups with different prognosis and, in some cases, they may provide specific drug targets [3]. For instance, the presence of the PLM-RARA fusion product is a specific hallmark of acute promyelocytic leukemia [4] and represents the first example of gene-fusion targeted therapy [5] that has changed the natural history of this disease. Hence, there are several reasons why studying gene fusions in cancer is very important. In recent years, next-generation sequencing (NGS) technologies have played an essential role in the understanding of the altered genetic pathways involved in human cancers. Today, most studies aiming at fusion discovery use NGS techniques followed by massive bioinformatics analyses. The greatest challenge of these sophisticated algorithms of prediction is the ability to discriminate between artifacts and the actual chromosomal rearrangements that occur [6]. Moreover, each gene fusion predicting software differs in terms of sensitivity and specificity. In the last decade, much effort has been made to catalog gene fusion events, thus resulting in a wide production of databases. At present, a dozen published databases regarding oncogenic fusion genes exists (see Table1 for a summary). Some of them (e.g., FusionCancer, ChiTaRS-3.1) collect in silico predictions of chimeric genes, obtained by analyzing publicly available datasets derived from heterogeneous sources in terms of experimental material (a mix of single-end and paired-end RNA-sequencing [RNA-seq] data, expressed sequence tags) or in terms of the data source (patients and cell lines). Others collect gene fusion events with experimental evidence manually curated from literature collections (e.g., Tumor Cancer Genome Atlas [TCGA], Mitelman, TICdb, COSMIC, ONGene). Here, we focused on the whole catalog of human malignant cell lines, thus obtaining a homogeneous input NGS dataset covering several human malignancies. We performed a massive bioinformatics analysis on 935 paired-end RNA-seq samples derived from 22 different tumor tissues and used a combination of the best-performing gene fusion-detecting algorithms. For ease of understanding, we define the predicted gene fusion event (pGFE) as the entity constituted by the gene fusion couple in a specific cell line and designate the consensus call-set (CCS) as the number of pGFEs supported by all the used algorithms. Starting from this assumption, we obtained 377 ,540 pGFEs, 2,521 of which belong to the CCS. Moreover, since not all of the pGFEs can give rise to oncogenic transformations, the use of a prioritization software is recommended in order to distinguish between real driver mutations and passenger mutations. Therefore, a robust Bayesian classifier has been used to perform an in silico validation of the results. Since one of the main purposes of this big data analysis is to encourage the re-use of our results in order to experimentally validate the in silico predictions, we set up a web portal to collect and systematize these data, LiGeA (cancer cell LInes Gene fusion portAl). It is possible to browse, search, and freely download all the results obtained and described within this article at the LiGeA repository web page available at [7]. To our knowledge, this represents the first compendium of both known and predicted novel gene fusion events in cell lines from 22 different human tumor types.
Table 1:
State-of-the-art of databases reporting gene fusions
| Database name | Short description |
|---|---|
| Tumor Fusion Gene Data Portal [8] | A collection of fusion genes in the TCGA samples |
| TICdb [9] | A collection of 1,374 fusion sequences extracted either from public databases or from published papers (last update: 2013) |
| chimerDB3.0 [10] | A catalog of fusion genes encompassing analysis of TCGA data and manual curations from the literature |
| COSMIC Cell Lines [11] | Gene fusions are manually curated from peer-reviewed publications. Currently, COSMIC includes information on fusions involved in solid tumors but not yet leukemias and lymphomas |
| Mitelman [1] | Reports hundreds of gene fusions associated with clinical reports but does not contain sequence data |
| ChiTaRs-3.1 [12] | A collection of 34,922 chimeric transcripts identified by expressed sequence tags and mRNAs from the GenBank, ChimerDB, dbCRID, TICdb, and Mitelman collections of cancer fusions for several organisms |
| FusionCancer [13] | Includes 591 samples, both single-end and paired-end RNA-seq, published on the sequence read archive database [14] between 2008 and 2014 covering 15 types of human cancers |
| ONGene [15] | Literature-derived database of oncogenes |
Data Description
Methods
We analyzed 935 paired-end RNA-seq experiments available at the Cancer Cell Line Encyclopedia repository [16], for a total of 32 TB of input raw data. The analysis was carried out by using four somatic fusion gene detection algorithms: FusionCatcher [17], EricScript [18], Tophat-Fusion [19], and JAFFA [20]. The choice of the algorithms was driven by the assessment from Kumar et al. [21], who compared 12 methods for the fusion transcripts detection from RNA-seq data and identified these software programs as the ones with the highest positive prediction values. Furthermore, the chosen software programs differ in a variety of aspects and contain several layers of information in their output files, thus giving us the opportunity to collect and interconnect a wide set of complementary data for each pGFE. Following is a short description of each fusion detection tool, accompanied by the used versions and parameters.:
FusionCatcher (FC): FC is a Python-based algorithm. It executes a first mapping run with Bowtie v.1.2.0 [22] and then performs the gene fusion detection based on three aligners: Bowtie2 v.2.2.9 [23], BLAT v.36 [24], and STAR v.2.5.2b [25]. FC takes advantage of National Center for Biotechnology Information viral genomes (v. 2016-01-06) in order to detect exogenous virus material integration into the host genome. Moreover, the FC algorithm compares its own output with a set of published databases, thus providing a detailed list of truly positive and false-positive pGFEs candidates. In our analysis, we downloaded FC v. 0.99.5a and Ensembl genome annotation v.83 and used hg38/GRCh38 as the genome assembly version. The software was executed with default parameters, requiring 111, 620 central processing unit (CPU) core hours, 125 GB of random access memory (RAM), and 20 CPU to complete the execution on our input dataset. Overall, FC detected 25,251 pGFEs involving 8,659 genes.
- Tophat-Fusion (TF): TF uses the TF-post function in order to create a filtered list of gene fusion candidates, starting from the output files obtained by running Tophat with the “–fusion-search” option [26]. The following commands were run subsequently:tophat -o $Sample.output/ -p 20 –fusion-search –keep-fasta-order –bowtie1 –no-coverage-search -r 160 –mate-std-dev 34 –max-intron-length 100000 –fusion-min-dist 100000 –fusion-anchor-length 13 $BOWTIE_INDEX/hg38 $Sample_1.fastq $Sample_2.fastqcd $Sample.output/tophat-fusion-post -p 20 –skip-blast $BOWTIE_INDEX/hg38
Tophat-2.0.12 and samtools 0.1.19 versions were used for this study. This algorithm took about 200,000 CPU core hours, 20 CPU, and 125 GB of RAM in order to complete its runs on the whole input dataset. TF produces several output files, but only the file named “results.txt,” representing the filtered list of predicted gene fusions, was used for subsequent analysis. The results encompassing “Chromosome M” have been manually discarded from the final results in primis because TF and JF were the only two of the four algorithms reporting them and because they represented bona fide false-positive outcomes. Overall, TF highlighted 28,146 pGFEs involving 9,492 genes.
JAFFA (JA): JAFFA (v. 0.9) is a multistep pipeline that takes raw RNA-seq reads and outputs a set of candidate fusion genes along with their cDNA breakpoint sequences. It relies on trimmomatic [27], samtools [28], BLAT [24], bowtie2, bpipe [29], and R software programs [30], as well as on gencode (v. 22), for the annotation and on the Mitelman database for flagging already known gene fusions. For the purpose of this analysis, we used the “Direct” mode pipeline, which is indicated for reads of 100 bp or longer. A total of 1,300,000 CPU core hours, 125 GB of RAM, and 20 CPU were required to successfully complete the analysis. The results encompassing “Chromosome M” have been manually discarded from the final results. Furthermore, only pGFEs supported by at least three spanning reads or flagged as “known” have been retained. Overall, after the filtering process, JA detected 53,400 pGFEs involving 12,256 genes.
- EricScript (ES): ES is developed in R, perl, and bash scripts. It uses the BWA aligner [31] to perform the mapping on the transcriptome reference and samtools v. 0.1.19 to handle SAM/BAM files. Recalibration of the exon-junction reference is performed using BLAT. For the purposes of this project, we used BLAT v.36, R v.3.3.1, bedtools v. 2.24, and ES version 0.5.5. We obtained and built the Ensembl Database v. 84 [32] by using BWA software with the command:bwa index -a bwtsw allseq.fa. A total of 130,900 CPU core hours, 125 GB of RAM, and 20 CPU were required to successfully complete the analysis. We further filtered out ES final results by removing all the predictions for which the software was not able to predict an exact breakpoint position because such pGFEs could not even be experimentally validated. Second, as also applied to FC, TF, and JF results, we retained the pGFEs exhibiting at least three spanning reads over the gene fusion junction. Furthermore, we filtered out all pGFEs with an EricScore value less than 0.85. EricScore is a ranking parameter ranging from 0.5 to 1; greater values correspond to better predictions. Interestingly, by applying these filters, we filtered out almost two-thirds of the initial predictions from EricScript but, at the same time, the CCS did not reduce substantially, thus indicating that the choice of a consensus of predictions is a good strategy to remove false positives and obtain a reliable set of gene fusion candidates to be experimentally validated. Overall, after the filtering process, ES detected 293,220 pGFEs involving 14,740 genes.
Data statistics and validation
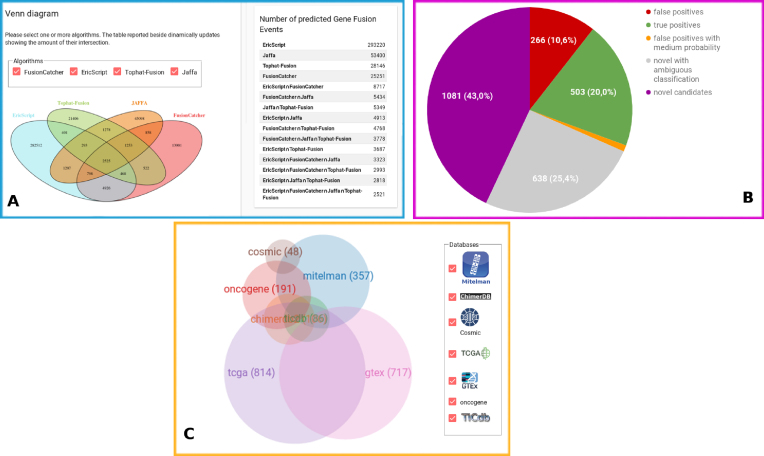
Overall, our extensive analysis results in a CCS of 2,521 pGFEs (Fig.1A) and, respectively, 2,828/9,258 pGFEs supported by exactly three/two methods. As the first validation of our analysis, 661 of the 719 (92%) genes known to be functionally implicated in cancer and collected under the COSMIC gene census are present in our final dataset. As a further validation of our results, about 1/5 of our CCS has already been published or is present in the following databases: chimerdb3, ONGene, COSMIC, TCGA, ticdb, and Mitelman (Fig.1C). Finally, only a small subset of the pGFEs (∼10% of data) present in the CCS has been recognized as false-positive predictions, thus supporting the idea that a combination of algorithms can be of great utility in order to increase the sensitivity and the specificity of the tests. It is worth mentioning that not only did our analysis confirm a large number of known gene fusion events, it also highlighted 1,719 novel putative pGFEs in the CCS that could undergo further downstream analysis (Fig.1B). Therefore, a further step of analysis was run with Oncofuse v.1.1.1 [33] in order to distinguish driver mutations (genomic abnormalities responsible for cancer) from passenger mutations (inert somatic mutations not implicated in carcinogenesis). Oncofuse is considered an in silico validation post-processing step that prioritizes the results obtained from each of the four algorithms. It assigns a functional prediction score to each putative fusion sequence breakpoint identified by the four software programs, thus hinting that pGFEs are worthy of being experimentally validated and studied. Oncofuse supports multiple input formats such as the output from TF and FC. In order to run it on the outputs from ES and JF, a short pre-processing step was executed on these data. As suggested in the Oncofuse manual, the accepted default input format is a tab-delimited file with lines containing 5΄ and 3΄ breakpoint positions. Therefore, these columns were extracted from ES and JF output files and redirected into Oncofuse-accepted input format. Oncofuse was run with default parameters using hg38 as the reference genome.
Figure 1:
(A) Venn diagram showing the intersection of the pGFEs identified by the four algorithms. (B) Distribution of pGFEs in the CCS; 43% (purple) of the CCS has not been previously described in any other database or scientific publication; 10% (red) and 20% (green) of the CCS have been reported in databases from healthy/tumoral samples, thus representing the false-positive/true-positive subset of our analysis; 1% of the CCS (orange) reports tags that classify the pGFE as a false-positive couple with medium probability; 25% (gray) of the results represent novel pGFEs tagged with values that classify them as both false positives and true positives. (C) Venn diagram showing the intersection between the LiGeA CCS and other databases.
Database Description
LiGeA is a database server based on graph-db technology (Neo4j). The portal stores all of the results obtained from each fusion gene predicting algorithm and the prioritization analysis outcome. This database contains not only a mere collection of in silico predictions, it has been integrated with other useful external resources in order to offer a carefully curated web compendium. Here is a short list of the added features:
Whenever the gene fusion couple has already been experimentally validated and published, an extra column with the COSMIC icon is added to the results. By clicking on it, the user will be redirected to an external link containing a manually curated catalog of 212 literature-derived somatic mutations in cancer [34].
Cancer Gene Census is a manually curated catalog of 719 genes for which mutations have been causally implicated in oncogenesis [35]. Whenever one of the two genes involved in the pGFE has already been described to be implicated in cancer, the gene is tagged with an icon. By clicking on it, an external link to the Cancer Gene Census is provided, showing a table of genes included within this category [36].
A legend based on a colorful signature has been added to tag the FC predictions as "validated truly positive couples" (green circle), "validated false-positive couples" (red circle), "false-positive couples with medium probability" (orange circle), and "ambiguous signature" because it is tagged with both positive and negative values (gray circle).
A functional prediction score obtained by extensively running the Oncofuse software is reported as an additional tag to the outputs from each algorithm.
The LiGeA portal is divided into several sections that allow a user-friendly navigation:
Home: On the homepage, the user is provided with a quick overview of the database. A global summary table reports a numeric recapitulation (e.g., the number of genes/transcripts/exons collected into the portal, the number of predicted proteins). Moreover, a histogram shows an abstract of the top 50 involved cell lines. By moving the cursor on the bars, a pop-up opens showing the cell line name and the corresponding number of unique fusion events predicted by all of the algorithms. Information about the algorithm predictions hosted into the portal are supplied with an interactive Venn diagram linked to a dynamic table. Upon user selection of the algorithm/s of interest, both the diagram and the table refresh, thus showing the resulting number of intersections.
Search: This utility allows several searching options to browse and mine genomic fusion events stored in the LiGeA portal (see Table2 for an overview). All the resulting outputs are sorted by the number of algorithms supporting the fusion events, thus showing the most robust set of results on the top of the table. As an additional feature, when specifying the features of interest, it is also possible to choose the minimum number of predicting algorithms. Search results are presented in the form of a paginated table containing those fusion events that satisfy the query parameters; data can also be downloaded in tabular format. Furthermore, by clicking on a given fusion ID, it is possible to access the event-specific page on which relevant information is presented in greater detail (e.g., involved cell line, disease, and genes as well as links to external databases and resources). Two of the nine query forms ("search by fusion information" and "search by virus") are specific annotations-derived FC algorithms. Following is a short description of the provided searching utilities:
"Search by Disease": In this section, all the cell lines derived from the same disease have been grouped together. In this way, it is possible to navigate the gene fusions that putatively cause specific malignancies. The number of cell lines constituting the queried subset is shown next to the pathology name.
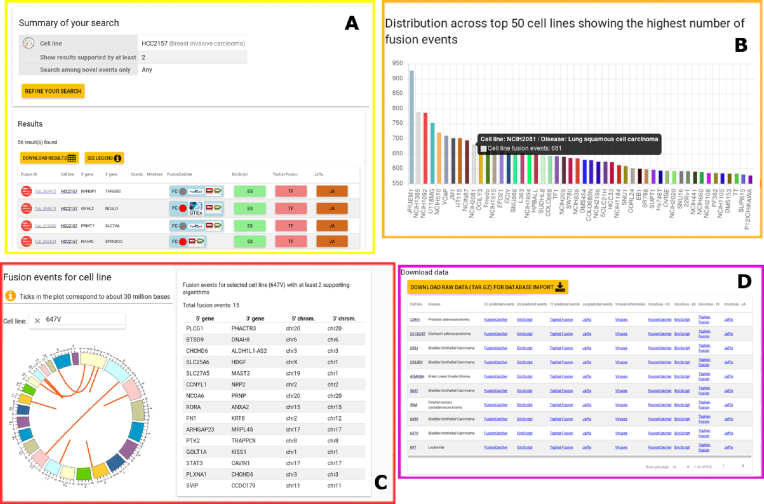
"Search by Cell Line": This module allows you to navigate the database by indicating a specific cell line name. It is possible to tune the results by showing only the novel predictions not yet described in any other database or publication (Fig. 2A).
"Search by Chromosome": This query can be performed by inserting one or two chromosomes involved in the fusion event. The cell line name can be indicated or not.
"Search by Gene": The user can select up to two gene names (Gene Symbol or ENSEMBL ID) and the "cell line" form can be either selected or not. The genes reported in the query form are black if they are involved in pGFE and gray if they are not.
"Search by Transcript": Since the same gene can give rise to different transcripts, it is be reasonable to query which of the transcripts produced by a specific gene are affected by a fusion event. This kind of query can be satisfied by inserting the Ensembl Transcript (ENST) IDs in the specific form.
"Search by Exon": Some of the queries allow you to go into more molecular detail. This search can be done by inserting one or two exon IDs involved in the fusion event. The cell line name can be indicated or not. In this way, it is possible to highlight the specific exons that turn out to be fused in the final result.
"Search by Fusion Information": The pGFEs may have different predicted effects. Indeed, depending on the location of the chromosomal break points, the resulting protein may be in-frame, out-of frame, truncated, and so on. Since the selectable values present in the fusion information form are specific to the FC algorithm, the result of this query returns a table without ES, JA, and TF data. We suggest reviewing the FC manual in order to obtain a full description of all tags.
"Search by Algorithm": This type of query is suitable for users who wish to navigate the outputs from specific software programs, choosing them individually or in combination. Indeed, it is known that some types of fusions, such as those involving immunoglobulins, can be detected by specific software [37].
"Search by Viruses": Additional useful information retrievable from the database relates to virus sequence integration into the host genome. This search utility is virus centered since it is possible to indicate or not the host cell line name. It is possible to select the virus name of interest (whether using GI ID or NC ID). Furthermore, a clickable link redirecting you to the virus genome is also shown on the right side of the table.
Table 2:
Example of possible queries on the LiGeA portal
| Search by | Question | Query |
|---|---|---|
| Disease | What are the gene fusion events present in stomach adenocarcinoma cell lines? | Select "stomach adenocarcinoma" under the "disease" menu. |
| Cell line | What are the novel pGFEs affecting RH30(Sarcoma) cell line? | Select "RH30" under the "cell line" menu and check the box "show only novel results." |
| Chromosome | What are the most suitable fusion partners for chromosome 8? | Select "Chr8" either under the "5’ Chromosome" or the "3’ Chromosome" tab and leave the other forms blank. |
| Gene | How many human cell lines show the PML-RARA fusion event? | Select "PML" under the "5’ gene menu"; select "RARA" from the "3’ gene menu"; leave the "cell line" query form blank. |
| Fusion information | What are all the in-frame pGFEs in the Jurkat cell line? | Select "Jurkat" under the "cell line" menu; select "in-frame" under the "predicted effect" menu. |
| Fusion information | What are the known GFEs predicted to be in-frame in the Jurkat cell line? | Select "Jurkat" under the "cell line" menu; select '‘in-frame" under '‘predicted effect" menu; select "known" under the "fusion description" menu. |
| Algorithm | Show only those GFEs supported by FC and TF in RH30 cell line. | Select "RH30" under the "cell line" query form and check the boxes relative to FC and TF. |
| Viruses | Which cell lines are most affected by hepatitis C virus genome integration? | Select "hepatitis C virus" under the "virus" query form and leave the "cell line" query form blank. |
Figure 2:
An overview of the LiGeA portal. (A) A "Search by Cell Line" example and the corresponding output. (B) An overview of the input dataset. (C) A circos diagram showing the graphical outcome of a "Query by Cell Line" and the corresponding related table. (D) An extract from the Download web page.
Statistics: This section allows you to visually inspect the results. The four submenus are organized as follows:
– "Cell Line Statistics": By choosing the cell line of interest, the resulting circular diagram shows all the chromosome couples involved in GFE predicted by at least two algorithms. The table on the right summarizes the resulting couples of the genes and chromosomes (Fig. 2C).
– "Chromosome Statistics": This page reports a dynamic pie chart showing the number of fusion events per human chromosome. By clicking on each slice of the pie, the related table automatically updates, showing a chromosome summary statistics. Furthermore, information about the number of inter- and intra-chromosomal rearrangements detected by each algorithm is also reported.
– "Disease Statistic"’: The Fusion Statistics pie chart was produced by grouping together the cell lines derived from the same human pathology, thus showing the total number of fusion events normalized by the number of cell lines composing a specific disease. The Virus Statistics panel shows the frequency of exogenous virus integration per human malignancy.
– "Gene Statistics": A word cloud diagram shows the most frequently recurring pGFEs supported by three methods.
– "Database Statistics": This subsection is composed of four panels. The first is related to data in the CCS (Fig.1B), the others relate only to FC and JA results. on this page, it is possible to get information about the number of pGFEs found in known databases (visualized as interactive Venn diagrams and in tabular fashion) and the distribution of predicted effects (histogram view).
Dataset: This page includes a description of the input dataset used for the analysis. Of the more than 1,000 samples available at the Broad Institute portal [16], we downloaded 935 PE RNA-seq samples in fastq format. The SE samples have been discarded since the software programs that were used required it. The histogram in this section shows the number of different cell lines derived from the same diseases (Fig.2B). Furthermore, starting from this section, it is possible to access web pages that resume cell line–specific details (e.g., COSMIC ID, drug resistance, and human disease).
Downloads: From this panel it is possible to download all the processed data described in this article (Fig.2D). Some of the files ("Summary Information" and "Viruses information") are specific products of the FusionCatcher algorithm.
Availability of source code
Project name: LiGeA: a comprehensive database of human gene fusion events
Project home page: http://hpc-bioinformatics.cineca.it/fusion (GitHub project: https://github.com/tflati/fusion)
Operating system(s): Any
Programming language: Python, JavaScript+HTML+CSS
Other requirements: Django 1.10.5, Python 2.7.12, AngularJS 1.5.11
License: GNU GPLv3
Availability of supporting data
The datasets obtained and described within this article are freely downloadable at the LiGeA repository available at http://hpc-bioinformatics.cineca.it/fusion/downloads. Moreover, archival copies of processed files and the source code are available via the GigaScience database, GigaDB [38].
Abbreviations
CCS: consensus call-set; CPU: central processing unit; ES: EricScript; FC: FusionCatcher; JA: JAFFA; LiGeA: cancer cell LInes GEne-fusions portAl; NGS: next-generation sequencing; pGFE: predicted gene fusion event; RAM: random access memory; RNA-seq: RNA sequencing; TCGA: Tumor Cancer Genome Atlas; TF: Tophat-Fusion.
Competing interests
The authors declare that they have no competing interests.
Funding
This work was supported by ELIXIR-IIB, CINECA, and Regione Lombardia. E.G. was supported by the Fondazione Italo Monzino and AIRC (award 17058) funding, awardee M.F..
M.F. was supported by ELIXIR-IIB-CINECA. S.G. was funded by ELIXIR-IIB, program name “Efficient implementation and distribution of HPC bioinformatics resources for Elixir scientific community,” (award 08/AR/2016-IBBE-BA). T.F. was funded by ELIXIR-IIB, program name “Efficient allocation of HPC bioinformatics resources through a federation of Galaxy web-based infrastructures” (award 05/AR/2016-IBBE-BA).
Author contributions
T.C. and M.F. conceived and designed the work. All authors analyzed and interpreted data, wrote the manuscript, and approved the final manuscript.
Supplementary Material
9/29/2017 Reviewed
1/15/2018 Reviewed
10/1/2017 Reviewed
1/16/2018 Reviewed
10/3/2017 Reviewed
1/29/2018 Reviewed
ACKNOWLEDGEMENTS
We acknowledge Andrea Micco for his useful tests on the first prototype of the system. We acknowledge CINECA and the Regione Lombardia Award, under the LISA initiative 2016-2018, for the availability of high-performance computing (HPC) resources and support. In particular, the ELIXIR-ITA HPC@CINECA and ELIXIR-IIB HPC@CINECA initiatives for providing HPC resources to our project.
References
- 1. Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7(4):233–45. [DOI] [PubMed] [Google Scholar]
- 2. Mertens F, Johansson B, Fioretos T, et al. . The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15(6):371–81. [DOI] [PubMed] [Google Scholar]
- 3. Serratì S, De Summa S, Pilato B et al. . Next-generation sequencing: advances and applications in cancer diagnosis. OncoTargets and Therapy. 2016;9:7355–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Borrow J, Goddard A, Sheer D, et al. . Molecular analysis of acute promyelocytic leukemia breakpoint cluster region on chromosome 17. Science. 1990;249(4976):1577–80. [DOI] [PubMed] [Google Scholar]
- 5. Nervi C, Ferrara FF, Fanelli M, et al. . Caspases mediate retinoic acid–induced degradation of the acute promyelocytic leukemia PML/RARα fusion protein. Blood. 1998;92(7):2244–51. [PubMed] [Google Scholar]
- 6. Lou DI, Hussmann JA, McBee RM et al. . High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proc Natl Acad Sci U S A. 2009;110(49):19872–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. LiGeA Portal , http://hpc-bioinformatics.cineca.it/fusion/, Accessed May 2018.
- 8. TCGA, Tumor Fusion Gene Data Portal @ONLINE; http://54.84.12.177/PanCanFusV2//, August 2017.
- 9. Novo F, de Mendíbil I, Vizmanos J. TICdb: a collection of gene-mapped translocation breakpoints in cancer. BMC Genomics. 2007;8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee M, Lee K, Yu N, et al. . ChimerDB 3.0: an enhanced database for fusion genes from cancer transcriptome and literature data mining. Nucleic Acids Research. 2017;45(D1):D784–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. COSMIC, COSMICDatabase-Wellcome Trust Sanger Institute @ONLINE; August 2017. http://cancer.sanger.ac.uk/cell_lines.
- 12. Gorohovski A, Tagore S, Palande V et al. . ChiTaRS-3.1–the enhanced chimeric transcripts and RNA-seq database matched with protein-protein interactions. Nucleic Acids Research. 2017;45(D1):D790–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Y, Wu N, Liu J et al. . FusionCancer: a database of cancer fusion genes derived from RNA-seq data. 2015;12(10):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.SRA, Sequence Read Archive- SRA @ONLINE http://www.ncbi.nlm.nih.gov/sra. Accessed 1st May 2018.
- 15. Liu Y, Sun J, Zhao M. ONGene: a literature-based database for human oncogenes. J Genetics Genomics. 2017;44(2):119–21. [DOI] [PubMed] [Google Scholar]
- 16. CCLE, Broad Institute portal- CCLE Repository https://portals.broadinstitute.org/ccle/home(October 2016).
- 17. Nicorici D, Satalan M, Edgren H, et al. . FusionCatcher - a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv. 2014 http://www.biorxiv.org/content/early/2014/11/19/011650. [Google Scholar]
- 18. Benelli M, Pescucci C, Marseglia G, et al. . Discovering chimeric transcripts in paired-end RNA-seq data by using EricScript. Bioinformatics. 2012;28(24):3232–9. [DOI] [PubMed] [Google Scholar]
- 19. Daehwan K, Salzberg S. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biology. 2011;12(8):R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Davidson NM, Majewski IJ, Oshlack A. JAFFA: high sensitivity transcriptome-focused fusion gene detection. Genome Medicine. 2015;7(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kumar S, Vo AD, Qin F, et al. . Comparative assessment of methods for the fusion transcripts detection from RNA-Seq data. Nature Scientific Reports. 2016;6:21597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Langmead B, Trapnell C, Pop M, et al. . Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10(3):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9(4):357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kent W. BLAT–the BLAST-like alignment tool. Genome Research. 2002;12(4):656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dobin A, Davis CA, Schlesinger F, et al. . STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li H, Handsaker B, Wysoker A et al. . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sadedin SP, Pope B, Oshlack A. Bpipe: a tool for running and managing bioinformatics pipelines. Bioinformatics. 2012;28(11):1525–6. [DOI] [PubMed] [Google Scholar]
- 30. R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria; 2008, http://www.R-project.org, ISBN 3-900051-07-0. [Google Scholar]
- 31. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ensembl DataBase@ONLINE https://docs.google.com/uc?id=0B9s__vuJPvIiUGt1SnFMZFg4TlE&export=download (March 2017).
- 33. Shugay M, Ortiz de Mendíbil I, Vizmanos JL, et al. . Oncofuse: a computational framework for the prediction of the oncogenic potential of gene fusions. Bioinformatics. 2013;29(20):2539–46. [DOI] [PubMed] [Google Scholar]
- 34. COSMIC, COSMIC Database-Wellcome Trust Sanger Institute @ONLINE; March 2017. http://cancer.sanger.ac.uk/cosmic.
- 35. Futreal PA, Coin L, Marshall M et al. . A CENSUS OF HUMAN CANCER GENES. Nature reviews Cancer. 2004;4(3):177–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. COSMIC, COSMIC Gene Census- Wellcome Trust Sanger Institute @ONLINE; February 2018. http://cancer.sanger.ac.uk/census.
- 37. Reshmi SC, Harvey RC, Roberts KG, et al. . Targetable kinase gene fusions in high-risk B-ALL: a study from the Children’s Oncology Group. Blood. 2017;129(25):3352–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gioiosa S, Bolis M, Flati T, et al. . Supporting data for "massive NGS data analysis reveals hundreds of potential novel gene fusions in human cell lines.". GigaScience Database. 2018; 10.5524/100442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
9/29/2017 Reviewed
1/15/2018 Reviewed
10/1/2017 Reviewed
1/16/2018 Reviewed
10/3/2017 Reviewed
1/29/2018 Reviewed