Abstract
Fungal infections claim an estimated 1.5 million lives every year. Mechanisms that protect from fungal infections are still elusive. Recognition of fungal pathogens relies on C-type lectin receptors (CLR) and their downstream signaling kinase SYK. Here we report that the E3-ubiquitin-ligase CBL-B controls proximal C-type lectin receptor signaling in macrophages and dendritic cells. We show that CBL-B associates with SYK and ubiquitinates SYK, Dectin-1, and Dectin-2 upon fungal recognition. Functionally, CBL-B deficiency results in increased inflammasome activation, enhanced reactive oxygen species production, and increased fungal killing. Genetic deletion of Cblb protects mice from morbidity caused by cutaneous infection and markedly improves survival upon a lethal systemic infection with Candida albicans. Based on these findings, we engineered a cell-permeable CBL-B inhibitory peptide that protects mice from lethal C. albicans infections. We thus describe a key role for Cblb in the regulation of innate anti-fungal immunity and establish a novel paradigm for the treatment of fungal sepsis.
Introduction
Fungal infections constitute a severe health threat, especially to people with an impaired immune status, as observed upon HIV infection1, immune suppression2, or primary immune deficiencies3. Candida species are the second most prevalent agents of fungal infections worldwide, and Candida albicans (C. albicans) is the most frequent pathogen isolated from patients suffering from severe forms of fungal infections in the developed world4. Gaining mechanistic insights into the immune response mounted towards this class of pathogens is thus of considerable interest for uncovering novel treatment options.
The initiation of an immune response towards fungal pathogens relies on the recognition of fungal pathogen associated molecular patterns. Using gene deficient mice, Toll-like receptors (TLR) and C-type lectin receptors (CLRs) have been proven to be essential to fungal recognition5–9. The CLRs Dectin-110, -211, and -312, Mincle13, and mannose receptor14 recognize various constituents of the fungal cell wall including β-glucan10, hyphal mannose 12, and glycolipids15 and initiate protective immunity. While Dectin-1, -2, and -3, and Mincle are essential for immune reactions against disseminated Candidiasis, mannose receptor deficient mice mount a comparably normal immune responses. The mannose receptor appears to be essential for Th17 type immunity, however14,16, and defective Th17 responses have been linked to an impairment in mucosal anti-fungal defense in both mice and men17,18.
Signaling induced by the CLRs Dectin-1, -2, -3, and Mincle critically depends on activation of the spleen tyrosine kinase (SYK)19. SYK is recruited to receptor complexes and activated by phosphorylation via SRC family kinases and auto-phosphorylation20. Activated SYK then triggers the inflammasome21,22, NF-κB signaling23 and reactive oxygen species (ROS) production in phagocytes and neutrophils19. Collectively, the activation of SYK by CLRs causes an immediate activation of fungicidal pathways, and induces cell type specific transcriptional changes to eradicate fungal infections.
The E3-ubiquitin-ligase CBL-B was initially characterized as a negative regulator of the adaptive immune response24,25 by dampening both T-cell- and B-cell-receptor signaling26. Genetic ablation of Cblb24,25 or knock-in of a catalytically inactive CblbC373A/C373A variant27 has thus been reported to cause autoimmunity. Recent observations uncovered additional roles for CBL-B in NK cells28, mast cells29, integrin signaling30, and dendritic cells (DC)31, extending the range of CBL-B functions to cells of the innate immune system. Signaling molecules targeted by CBL-B are often not specific to defined pathways, but can be used by multiple signaling modules. For example, CBL-B mediated negative regulation of SYK activity has been described for several cell types32,33. While the regulation of SYK by CBL-B has been well established in the context of adaptive immune responses, negative regulators of SYK in signal transduction induced by pattern recognition receptors of the CLR superfamily has not been described, yet.
Here we report that Cblb deficiency ameliorates fungal pathology during systemic and cutaneous C. albicans infections. CBL-B acts in phagocytes by regulating the availability of Dectin-1 and -2 and their signaling via SYK. Strikingly, peptide based inhibition of CBL-B increased cellular responses to fungal stimulation in vitro and completely protected mice from morbidity and mortality upon systemic infection with C. albicans.
Results
Loss of CBL-B or CBL-B catalytic activity ameliorates fungal pathology
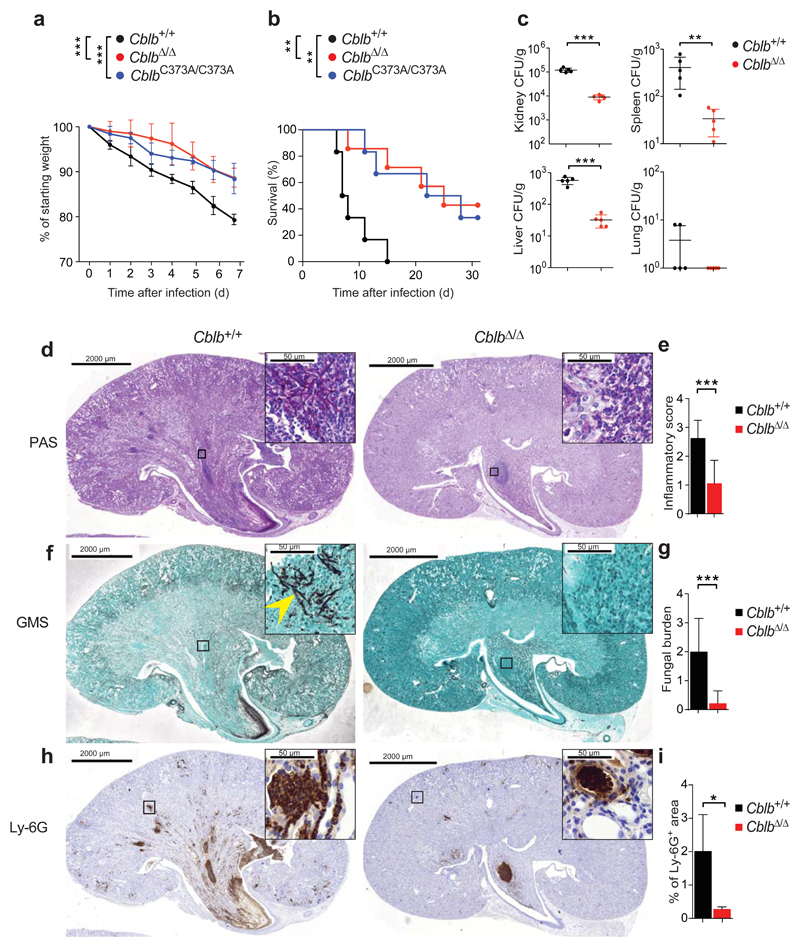
To test for a potential role of CBL-B in anti-fungal immunity, we challenged CblbΔ/Δ, CblbC373A/C373A, carrying catalytically inactive CBL-B27, and Cblb+/+ littermates (Supplementary Fig. 1a) systemically with a lethal dose of C. albicans. Loss of Cblb or CblbC373A/C373A knock-in markedly ameliorated fungal pathology when compared to Cblb+/+ controls (Fig. 1a,b). We also observed significantly reduced fungal burdens in affected organs of CblbΔ/Δ mice, when compared to their Cblb+/+ littermates (Fig. 1c). Histopathologic analysis of kidneys 7 days upon systemic infection confirmed these results; CblbΔ/Δ mice manifested reduced renal inflammation (Fig. 1d,e) and reduced numbers of C. albicans yeasts and hyphae (Fig. 1f,g). Additionally, we detected a significant reduction in neutrophil infiltration into the kidneys of infected CblbΔ/Δ mice when compared to Cblb+/+ littermates (Fig. 1h,i; Supplementary Fig. 1b). Systemic infection with a lethal dose of a clinical isolate of C. dubliniensis yielded similar results with respect to mortality kinetics, when comparing CblbΔ/Δ mice to Cblb+/+ controls (Supplementary Figure 1c). To assess, whether Cblb deficiency caused a broad deregulation of innate immune function not only in fungal infections, we infected Cblb deficient and Cblb+/+ littermates with Listeria monocytogenes (L. monocytogenes). Loss of Cblb did not affect the mortality kinetics of infected mice (Supplementary Fig. 1d). While these results do not exclude a role for Cblb in anti-bacterial immune responses in general, our results show that loss of Cblb results in marked resistance to systemic C. albicans and C. dubliniensis infections.
Figure 1. Cblb regulates anti-fungal immunity.
(a,b) Cblb+/+, CblbΔ/Δ, and CblbC373A/C373A mice were infected intravenously with C. albicans (105 CFU/21.5 g body weight) and monitored over time. Plots depict (a) weight loss as compared to starting weight (P values assessed by two-way ANOVA) or (b) survival after infection (P values assessed with log rank test). n = 6 for Cblb+/+ and CblbC373A/C373A, n = 7 for CblbΔ/Δ mice. (c) C. albicans fungal load in organs 7 days after infection. Dots represents individual mice. (d,e) Periodic-acid-Schiff (PAS) stained kidney sections 7 days after systemic C. albicans infection (d). (e) Combined inflammatory score based on renal immune cell infiltration/inflammation, and tissue destruction. (f,g) GMS stained kidney sections from mice 7 days after C. albicans infection (f). Arrowhead indicates fungal hyphae. (g) Fungal load scored by analysis of GMS stained sections. (h,i) Ly-6G stained kidney sections 7 days after C. albicans infection (h). (i) Plot depicts percentage of kidney area scored as Ly-6G positive. Insets in (d,f,h) show boxed areas at increased magnification. Panels (d-g) n = 4 for Cblb+/+, n = 5 for CblbΔ/Δ mice; panels (h,i) n = 4 for Cblb+/+, n = 4 for CblbΔ/Δ mice; panels (d-i) 4 sections/kidney analyzed. For panels (c,e,g,i) data shown as means ± standard deviation. For panels (a,b) 1 representative of 6, for panel (c-i) 1 representative of 3 independent repeats is shown. *P < 0.05, **P < 0.01, ***P < 0.001 as calculated with Student’s t-test, unless stated otherwise.
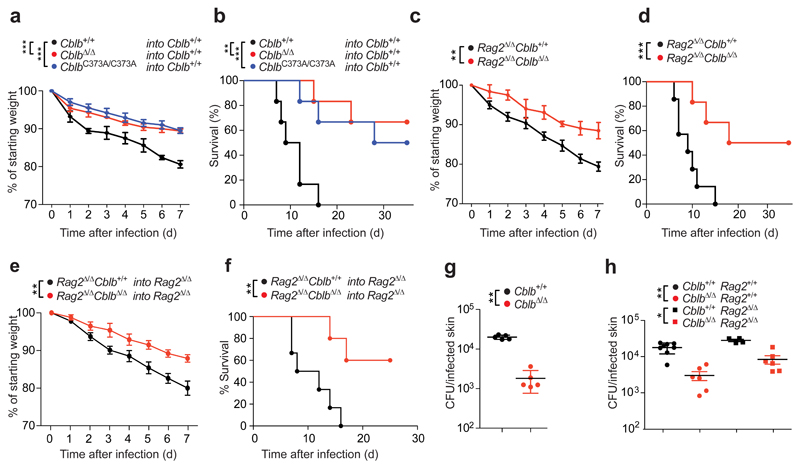
To gain insight into the cellular basis of the observed protective effect, we generated bone marrow chimeric animals by reconstituting lethally irradiated Cblb+/+ mice with syngeneic CblbΔ/Δ, CblbC373A/C373A, or Cblb+/+ bone marrow. Loss of Cblb in radiosensitive hematopoietic cells phenocopied full body Cblb deficiency after systemic C. albicans infection (Fig. 2a,b). To test for a contribution of CBL-B in adaptive immune cells to the observed phenotype, we generated Cblb+/+Rag2Δ/Δ and CblbΔ/ΔRag2Δ/Δ animals. Importantly, Cblb deficiency in the absence of adaptive immune cells closely resembled the full body knock-out phenotype upon lethal C. albicans infection (Fig. 2c,d). Additionally, bone marrow chimeric animals were generated by reconstituting irradiated Rag2Δ/Δ recipients with CblbΔ/ΔRag2Δ/Δ or Cblb+/+Rag2Δ/Δ bone marrow, to restrict Cblb deficiency to innate immune cells. We observed that loss of Cblb confers an innate immune system intrinsic protective effect to systemic infection with C. albicans (Fig. 2e,f).
Figure 2. Cblb in immune cells protects against fungal pathology.
(a,b) Cblb+/+ mice were reconstituted with bone marrow from Cblb+/+, CblbΔ/Δ, or CblbC373A/C373A mice after lethal irradiation. After full reconstitution mice were infected intravenously with C. albicans (105 CFU/21.5 g body weight) and monitored over time. Plots depict (a) weight loss over time after infection, as compared to starting weight (P values assessed by two-way ANOVA) or (b) survival over time after infection (P values assessed with log rank test). n = 6 for all cohorts. (c,d) Cblb+/+Rag2Δ/Δ or CblbΔ/ΔRag2Δ/Δ mice were infected, scored and analyzed as in (a,b) for (c) weight loss and (d) mortality. n = 7 for Cblb+/+Rag2Δ/Δ, n = 6 for CblbΔ/ΔRag2Δ/Δ mice. (e,f) Rag2Δ/Δ mice were reconstituted with bone marrow from Cblb+/+Rag2Δ/Δ or CblbΔ/ΔRag2Δ/Δ mice after lethal irradiation. Eight weeks after reconstitution mice were infected, scored, and analyzed as in (a,b) for (e) weight loss and (f) mortality. n = 6 for Cblb+/+Rag2Δ/Δ into Rag2Δ/Δ, n = 5 for CblbΔ/ΔRag2Δ/Δ into Rag2Δ/Δ mice. (g,h) Cblb+/+, CblbΔ/Δ, Cblb+/+Rag2Δ/Δ, or CblbΔ/ΔRag2Δ/Δ mice were infected on their shaved dorsum with 2 x 108 CFU C. albicans. Plot depicts fungal load in the skin 3 days after infection, plotted as CFU/total infected skin area. Dots represent individual mice. For panels (a,b,e,f,g,h) 1 representative of 3, for panels (c,d) 1 representative of 4 independent experiments is shown. *P < 0.05, **P < 0.01, ***P < 0.001 as calculated with Student’s t-test, unless stated otherwise.
To test for developmental effects of Cblb deficiency on immune cells relevant for anti-fungal defense, we assessed the cellular composition of the bone marrow and spleen of CblbΔ/Δ and Cblb+/+ animals. We observed slightly elevated levels of both bone marrow and splenic neutrophils and splenic NK cells in unchallenged 6 week old mice (Supplementary Fig. 1e,f). To address whether this might affect monocyte or neutrophils organ infiltration, we quantified their recruitment in response to systemic C. albicans challenge 24 h after infection. We observed comparable immune infiltration into kidneys, lungs, and livers of CblbΔ/Δ and Cblb+/+ mice (Supplementary Fig. 2a,b,c). Recognition of cell wall β-glucan by Dectin-1 has been shown to be essential for anti-fungal immune responses10. We detected comparable expression of Dectin-1 on blood monocytes and neutrophils of CblbΔ/Δ and control Cblb+/+ littermates (Supplementary Fig. 2d,e). Moreover, mRNA expression of Dectin-1, Dectin-2, Dectin-3, and Mincle were comparable in splenocytes and bone marrow cells of CblbΔ/Δ and Cblb+/+ animals (Supplementary Fig. 2f,g). Importantly, although CblbΔ/Δ and Cblb+/+ animals exhibited comparable early cellular recruitment, fungal burdens in affected organs were already significantly decreased 24 h after C. albicans infection (Supplementary Fig. 2h), supporting the notion that loss of Cblb results in an increased fungicidal activity of innate immune cells.
To test whether the protective effect of Cblb deficiency could be extended to other routes of infection, we used a model of cutaneous C. albicans infection34. We observed an increased resistance of CblbΔ/Δ mice to skin colonization by C. albicans, when compared to Cblb+/+ controls, as evidenced by a significantly decreased fungal burden (Fig. 2g). Due to the established role of adaptive immune cells in cutaneous anti-fungal immunity34, we performed C. albicans skin infections on a Rag2 deficient background. Importantly, we observed a decreased fungal load in the skin of CblbΔ/ΔRag2Δ/Δ mice, when compared to Cblb+/+Rag2Δ/Δ littermates (Fig. 2h). Thus, loss of Cblb in innate immune cells confers resistance to systemic as well as cutaneous C. albicans infections.
Cblb controls anti-fungal activities of DC and macrophages
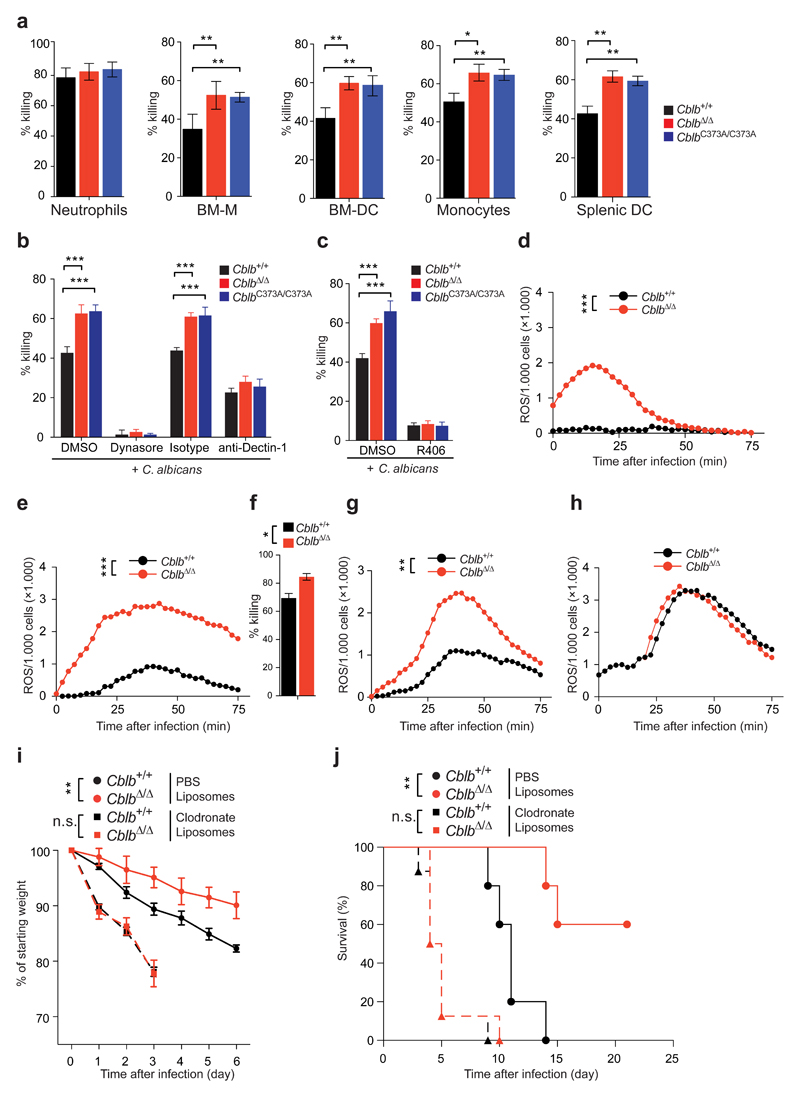
To directly investigate the effect of Cblb deficiency on the main fungicidal immune cell lineages35, we performed in vitro killing assays by co-culturing bone marrow-derived macrophages (BM-M), bone marrow-derived dendritic cells (BM-DC), neutrophils or monocytes isolated from the bone marrow, and splenic DC with C. albicans. We did not observe a significant difference in neutrophil fungicidal activity, when comparing CblbΔ/Δ, CblbC373A/C373A, and Cblb+/+ cells. However, CblbΔ/Δ or CblbC373A/C373A BM-M, BM-DC, bone marrow monocytes and splenic DC showed an increased fungicidal activity, when compared to Cblb+/+ cells (Fig. 3a). Of note, rates of phagocytosis were comparable between BM-M or BM-DC of all three genotypes (Supplementary Fig. 3a,b). Compatible with a more prominent role for Cblb in anti-fungal responses by DC and macrophages, we observed higher levels of CBL-B in BM-DC and BM-M, when compared to neutrophils, both in C. albicans infected and uninfected cells (Supplementary Fig. 3c). Thus, CBL-B controls the fungicidal activity of BM-M, BM-DC, as well as bone marrow monocytes and splenic DC.
Figure 3. Cblb controls anti-fungal activities of dendritic cells and macrophages.
(a) Killing capacity of BM-neutrophils, BM-M, BM-DC, BM-monocytes, or splenic DC as assessed by co-culture with C. albicans. Assays performed in quadruplicates. (b,c) Killing capacity of BM-DC in the presence of (b) the phagocytosis inhibitor Dynasore or an anti-Dectin-1 antibody, or (c) the SYK inhibitor R406. (d,e) ROS production by intraperitoneal immune infiltrates of Cblb+/+ or CblbΔ/Δ mice 24 h after intraperitoneal C. albicans infection (5 * 106 CFU/21.5 g body weight). Plots depict (d) ROS production by immune infiltrates without re-stimulation or (e) with re-stimulation with C. albicans. Experiments performed in triplicates. (f) Killing capacity of immune infiltrates from (d,e). (g,h) ROS production of (g) monocytes/macrophages or (h) neutrophils FACS sorted from infiltrates in (d,e). (i,j) Cblb+/+ or CblbΔ/Δ mice were injected with PBS liposomes or clodronate liposomes 24 h before and 24 h after intravenous infection with C. albicans (105 CFU/21.5 g body weight) and monitored for (i) weight loss, as compared to starting weight (P values assessed by two-way ANOVA), or (j) survival (P values assessed with log rank test). n = 5 for PBS liposome treated, n = 8 for clodronate liposome treated cohorts. For panels (a-c,f,i) data are shown as means ± standard deviation. For panels (a,b) 1 representative of 5, for panels (c-j) 1 representative of 3 independent experiments is shown. *P < 0.05, **P < 0.01, ***P < 0.001 as calculated with Student’s t-test, unless stated otherwise.
Pharmacologic inhibition of phagocytosis using Dynasore abrogated BM-DC mediated killing. Likewise, blocking of Dectin-1 significantly reduced C. albicans killing and abrogated the effects Cblb deficiency (Fig. 3b). In addition, pharmacologic inhibition of SYK with the ATP-competitive SYK inhibitor R406 almost completely abrogated C. albicans killing (Fig. 3c). We next measured whether a lack of CBL-B or CBL-B ligase function affects the ability of the immune lineages above to produce ROS, which is essential for killing of C. albicans36. CblbΔ/Δ or CblbC373A/C373A bone marrow neutrophils did not significantly differ from Cblb+/+ bone marrow neutrophils with respect to ROS production in response to C. albicans (Supplementary Fig. 3d). However, the increased fungicidal activity of CblbΔ/Δ or CblbC373A/C373A BM-M, BM-DC, bone marrow monocytes, and splenic DC correlated with an increased C. albicans-induced ROS production as compared to Cblb sufficient cells (Supplementary Fig. 3e-h). Additionally, we observed, that loss of CBL-B significantly enhanced the production of ROS by BM-DC stimulated with clinical isolates of C. glabrata, C. krusei, and C. dubliniensis (Supplementary Figure 3i,k). Thus, loss of CBL-B enhances the immune response to several clinically relevant fungal pathogens.
C. albicans stimulation of BM-DC activates the Caspase-8 dependent inflammasome21,37. We indeed observed significantly increased Caspase-8 activity (Supplementary Fig. 4a) and increased Dectin-1 dependent IL-1β secretion in C. albicans stimulated CblbΔ/Δ or CblbC373A/C373A BM-DC when compared to Cblb+/+ BM-DC (Supplementary Fig. 4b), which could be blocked by SYK inhibition (Supplementary Fig. 4c). We found similarly increased release of IL-1β by C. albicans-stimulated CblbΔ/Δ or CblbC373A/C373A BM-M, when compared to Cblb+/+ BM-M (Supplementary Fig. 4d).
To monitor the C. albicans induced transcriptional response, we assessed mRNA levels of key pro-inflammatory cytokines. We observed significantly increased transcript levels of IL-1β, IL-6, TNF, IL-23, and IL-12 in CblbΔ/Δ or CblbC373A/C373A BM-DC as compared to Cblb+/+ BM-DC (Supplementary Fig. 4e). Increased transcription correlated with increased secretion of the pro-inflammatory cytokines IL-6 and TNF by CblbΔ/Δ and CblbC373A/C373A BM-DC when compared to Cblb+/+ cells (Supplementary Fig. 4f,g). Blocking Dectin-1 reduced the release of IL-6 and TNF for all three genotypes (Supplementary Fig. 4f,g). Again, pharmacologic inhibition of SYK almost completely abrogated the release of IL-6 and TNF by C. albicans stimulated BM-DC of all three genotypes (Supplementary Fig. 4h,i). C. albicans stimulation also led to a substantially increased release of IL-23 and IL-12 by CblbΔ/Δ and CblbC373A/C373A BM-DC (Supplementary Fig. 4j,k) and IL-6 and TNF by BM-M (Supplementary Fig. 4l,m) when compared to Cblb+/+ cells. Furthermore, we observed increased secretion of IL-1Ra when CblbΔ/Δ and Cblb+/+ C. albicans stimulated BM-DC were compared (Supplementary Fig. 4n).
To further test for the contribution of Dectin-1 to the increased cytokine production by Cblb deficient BM-DC, we stimulated BM-DC with zymosan, a yeast cell wall homogenate, or curdlan, a purified β-glucan. Stimulation with zymosan or the Dectin-1 agonist curdlan resulted in an increased release of IL-1β, IL-6, and TNF by CblbΔ/Δ and CblbC373A/C373A BM-DC, as compared to Cblb+/+ cells (Supplementary Fig. 5a-f). Importantly, we did not observe significant differences in the secretion of IL-6 and TNF in response to stimulation with the TLR4 agonist LPS or the TLR9 agonist CpG, when CblbΔ/Δ and Cblb+/+ BM-DC were compared (Supplementary Fig. 5g-j). Thus, Cblb is a key regulator of Dectin-1/SYK-mediated responses of BM-DC and BM-M to C. albicans infections.
To address whether the observed differences in vitro could be recapitulated in immune cells recruited to inflammatory sites, we challenged CblbΔ/Δ and Cblb+/+ mice by intraperitoneal infection with C. albicans and assessed their anti-fungal activity 24 h later. Loss of Cblb led to an enhanced ROS response by cells infiltrating the peritoneal cavity (Figure 3d,e). Likewise, the fungicidal activity of CblbΔ/Δ infiltrates was increased, when compared to Cblb+/+ infiltrates (Figure 3f). To test for the ability of infiltrating neutrophils or monocytes/macrophages to produce ROS, we FACS purified these cell types from total C. albicans induced infiltrates. We observed increased ROS production by infiltrating CblbΔ/Δ monocytes/macrophages, but not neutrophils, when compared to Cblb+/+ cells (Figure 3g,h). Additionally, we observed an increase in IL-6, TNF, and IL-1Ra in peritoneal lavages from CblbΔ/Δ when compared to Cblb+/+ mice (Supplementary Fig. 5k).
Our data indicated that the protective effect of Cblb deficiency to fungal infection was mediated by phagocytes, but not neutrophils. To test for the role of Cblb in phagocytes, we depleted phagocytes, but not neutrophils, 24 h before and 24 h after systemic infection with C. albicans using clodronate liposomes38,39. While the depletion of phagocytes lead to a significant increase in weight loss when clodronate liposome and PBS liposome treated cohorts were compared38, the difference between Cblb+/+ and CblbΔ/Δ cohorts was abolished due to phagocyte depletion (Figure 3i). Likewise, the survival benefit of CblbΔ/Δ when compared to Cblb+/+ cohorts was abrogated upon phagocyte depletion (Figure 3j). These data show that Cblb deficiency in phagocytic cells is critical for the observed beneficial effect in fungal infections.
Loss of CBL-B enhances CLR-mediated signaling in response to C. albicans
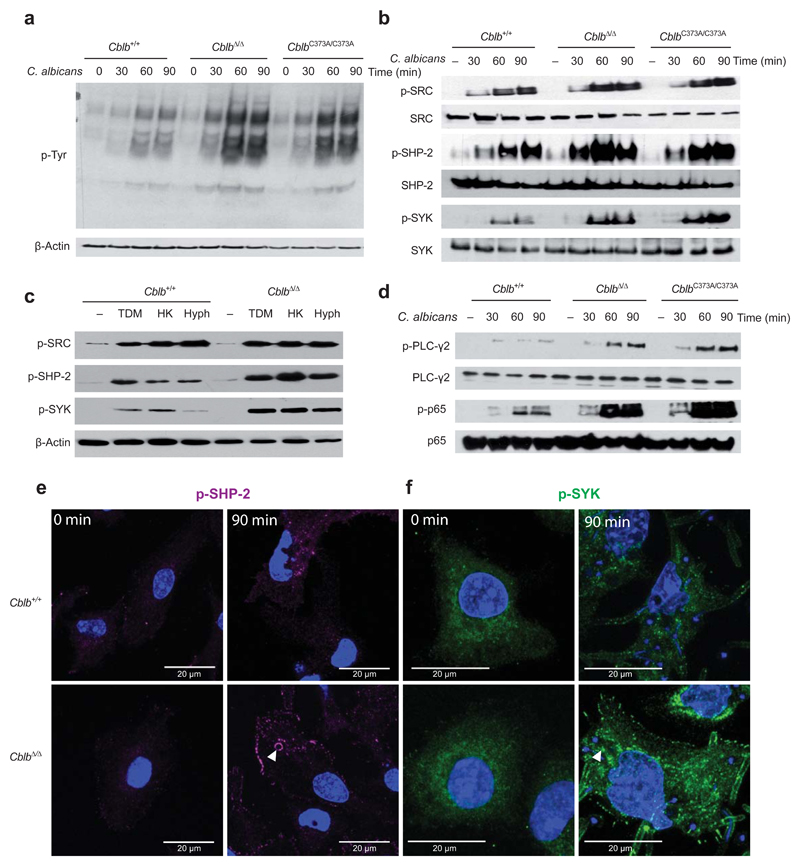
To assess whether loss of CBL-B or CBL-B ligase function altered the signaling response mounted by BM-DC, we analyzed tyrosine phosphorylation in response to C. albicans infection. We observed a strong increase in tyrosine phosphorylation in response to C. albicans stimulation in CblbΔ/Δ and CblbC373A/C373A BM-DC, when compared to control Cblb+/+ BM-DC (Fig. 4a). Canonical signaling by CLRs involves activation of SRC and SHP-2 that subsequently results in the activation of SYK20. We observed comparable SRC tyrosine phosphorylation, but increased levels of phosphorylated SHP-2 and SYK after stimulation of BM-DC with C. albicans (Fig. 4b). The more pronounced effect of SYK inhibition compared to Dectin-1 blockade, prompted us to test for a role of CBL-B in Dectin-1, Dectin-2/3, and Mincle signaling. Although these surface receptors were expressed at comparable levels in CblbΔ/Δ and Cblb+/+ BM-DC (Supplementary Fig. 6a), we observed an increase in SYK and SHP-2 phosphorylation in response to heat killed C. albicans (to activate Dectin-1), C. albicans hyphae (Dectin-2/3), as well as Trehalose-6,6-dimycolate (Mincle) in the absence of Cblb (Fig. 4c). Moreover, we observed an increase in phosphorylation of the distal CLR signaling molecules PLC-γ2 and p65 NF-κB in C. albicans-stimulated CblbΔ/Δ and CblbC373A/C373A BM-DC (Fig. 4d). CblbΔ/Δ BM-DC also showed increased levels of both phosphorylated SHP-2 (Fig. 4e) and phosphorylated SYK (Fig. 4f) following stimulation with C. albicans, as analyzed by confocal microscopy. These data identify a central function for CBL-B in regulating CLR signaling in response to C. albicans infection.
Figure 4. Increased CLR mediated signaling in the absence of Cblb.
(a) Cblb+/+, CblbΔ/Δ or CblbC373A/C373A BM-DC were stimulated with C. albicans for the indicated periods of time and analyzed by Western blotting for levels of phosphorylated tyrosine. (b) BM-DC of the indicated genoytypes were stimulated as in (a) and analyzed for phosphorylated or total amounts of the indicated signaling molecules. (c) Cblb+/+ and CblbΔ/Δ BM-DC were stimulated for 90 min with Trehalose-6,6-dimycolate (TDM), heat killed C. albicans (HK), or C. albicans hyphae (Hyph), and lysates were analyzed for levels of phosphorylated SRC, phosphorylated SHP-2, and phosphorylated SYK by Western blotting. β-Actin is shown as a loading control. (d) BM-DC of the indicated genotypes were stimulated as in (a) and analyzed for phosphorylated or total amounts of the indicated molecules. (e,f) Cblb+/+ or CblbΔ/Δ BM-DC were stimulated with C. albicans for 90 min or left unstimulated, fixed with 4% paraformaldehyde, stained for (e) phosphorylated SHP-2 or (f) phosphorylated SYK, and analyzed by confocal microcopy. Representative images for each genotype and time point are shown. Arrowheads depict phagosome localized (e) phosphorylated SHP-2 or (f) phosphorylated SYK. Cells were counterstained with DAPI (blue). For all panels, 1 representative of 4 independent experiments is shown.
CBL-B targets SYK, Dectin-1, and Dectin-2 for ubiquitination
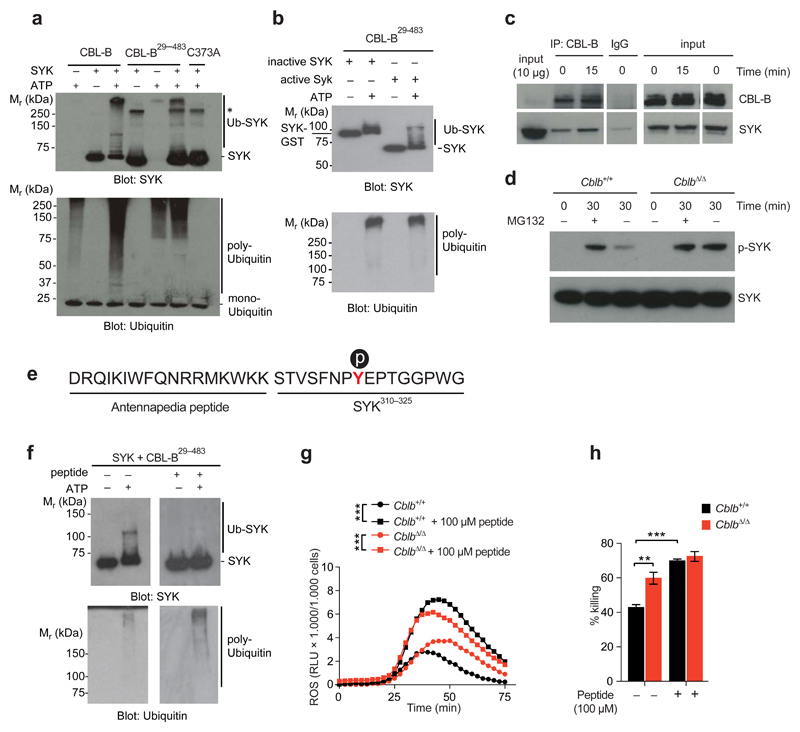
To test whether CBL-B could poly-ubiquitinate CLR signaling molecules, we performed ubiquitination assays. We did not detect poly-ubiquitination of recombinant SHP-2 (not shown), but observed robust poly-ubiquitination of active SYK (Fig. 5a). The chain linkage type formed by CBL-B was analyzed by mass spectrometry and we identified the unique peptide FAGKQLED derived specifically from K48-linked ubiquitin chains (Supplementary Fig. 6b). CBL-B substrate specificity is mainly mediated by its amino-terminal tyrosine kinase binding (TKB) domain. We show that a truncated CBL-B protein (CBL-B29-483), lacking the carboxy-terminal ubiquitin associated (UBA) domain, but containing the TKB and RING finger domain, still exhibited poly-ubiquitination activity towards SYK (Fig. 5a). As expected, the C373A mutant of CBL-B29-483 lacked SYK poly-ubiquitination activity (Fig. 5a). Importantly, non-phosphorylated inactive SYK showed strongly diminished poly-ubiquitination by CBL-B29-483 (Fig. 5b), consistent with CBL-B targeting activated SYK. To test for interactions between SYK and CBL-B, we performed co-immunoprecipitation experiments. We observed increased co-immunoprecipitation of SYK with CBL-B after stimulation of BM-DC with C. albicans (Fig. 5c). Upon treatment with the proteasome blocker MG132, we observed an increase in phosphorylated SYK in lysates prepared from Cblb+/+ BM-DC, but not CblbΔ/Δ BM-DC stimulated with C. albicans (Fig. 5d), indicating that CBL-B mediates its effects via proteasomal targeting. These results show that CBL-B can ubiquitinate active SYK.
Figure 5. CBL-B ubiquitinates SYK.
(a) In vitro ubiquitination of active SYK by recombinant CBL-B, CBL-B29-483, or C373A CBL-B29-483. Assays analyzed by Western blotting for SYK (upper panel) or ubiquitin (lower panel). Note, that CBL-B forms non-substrate attached poly-ubiquitin chains. * denotes unspecific bands. (b) In vitro ubiquitination of inactive SYK (SYK-GST) or active SYK by CBL-B29-483. Assays were analyzed as in (a). (c) Lysates from BM-DC stimulated with C. albicans immunoprecipitated with an anti-CBL-B antibody or IgG control. Precipitates were probed for CBL-B and SYK. Aliquots were blotted for CLB-B and SYK as a loading control. (d) BM-DC were stimulated with C. albicans in the presence or absence of MG132 (10 μM) and analyzed by Western blotting. (e) Sequence of the TKB-binding-peptide. Antennapedia derived, and SYK derived sequences are indicated. Tyrosine317 is shown in red, p symbolizes phosphotyrosine317. (f) In vitro ubiquitination of SYK by CBL-B29-483 in the presence of 100 μM TKB-binding peptide. Assays analyzed by Western blotting as in (a). (g) BM-DC stimulated with C. albicans in the presence of 100 μM TKB-binding peptide and monitored for ROS production. (P values calculated with two-way ANOVA). (h) Killing capacity of BM-DC as assessed by co-culture with C. albicans in the presence of 100 μM TKB-binding peptide. Data shown as means ± standard deviation. P values were calculated with Student’s t-test. For all panels, 1 representative of 3 independent repeats is shown **P < 0.01, ***P < 0.001.
Additionally, we addressed whether Dectin-1, and -2 might also serve as targets for CBL-B mediated degradation. We thus stimulated BM-M with C. albicans or C. albicans hyphae, and tested for Dectin-1, and -2 protein levels, respectively. While Dectin-1, and -2 levels decreased over time in control cells, their expression levels remained constant in the absence of Cblb (Supplementary Fig. 6c,d). To test whether Dectin-1, and -2 are ubiquitinated in response to fungal stimulation, and if so, whether ubiquitination is dependent on CBL-B, we stimulated CblbΔ/Δ and Cblb+/+ cells, immuno-precipitated Dectin-1, or -2 and blotted for ubiquitin. We observed ubiquitination of Dectin-1 and Dectin-2 in response to stimulation with C. albicans or C. albicans hyphae, respectively. This ubiquitination was reduced in the absence of CBL-B (Supplementary Fig. 6e,f). These data show that CBL-B tunes CLR signaling by regulating protein levels of Dectin-1, and -2, as well as by modulating SYK activity.
Inhibition of CBL-B enhances anti-fungal immunity in vitro and in vivo
Based on our data we reasoned that blocking CBL-B-TKB-mediated substrate recognition might de-regulate CLR signaling. We thus designed a fusion peptide consisting of the Drosophila Antennapedia homeodomain39 cell penetrating peptide and a phosphorylated peptide derived from SYK encompassing tyrosine-317 (Y317), reported to be required for CBL-family-ligase mediated targeting of SYK33,40,41 (Fig. 5e). Pre-incubation of CBL-B with this CBL-B-TKB-binding peptide abrogated in vitro ubiquitination of active SYK (Fig. 5f). Additionally, we monitored SYK dependent ROS production in the presence of the CBL-B-TKB-binding peptide. Intriguingly, peptide interference massively increased ROS production by BM-DC in response to C. albicans stimulation (Fig. 5g). The observed effect even surpassed the increase in ROS production in CblbΔ/Δ cells. Since our peptide interference strategy potentially inhibits TKB-mediated substrate recognition by all CBL-family-ligases (CBL, CBL-B, and CBL-C), these data suggest that Cbl-family-ligases may act in a partly redundant manner. Consistent with this hypothesis, we observed an increased sensitivity to CBL-B-TKB-peptide interference in BM-DC lacking CBL-B, when compared to CBL-B sufficient cells (Supplementary Fig. 6g). Notably, the CBL-B-TKB-binding peptide also increased the fungicidal activity of BM-DC upon C. albicans infection (Fig. 5h).
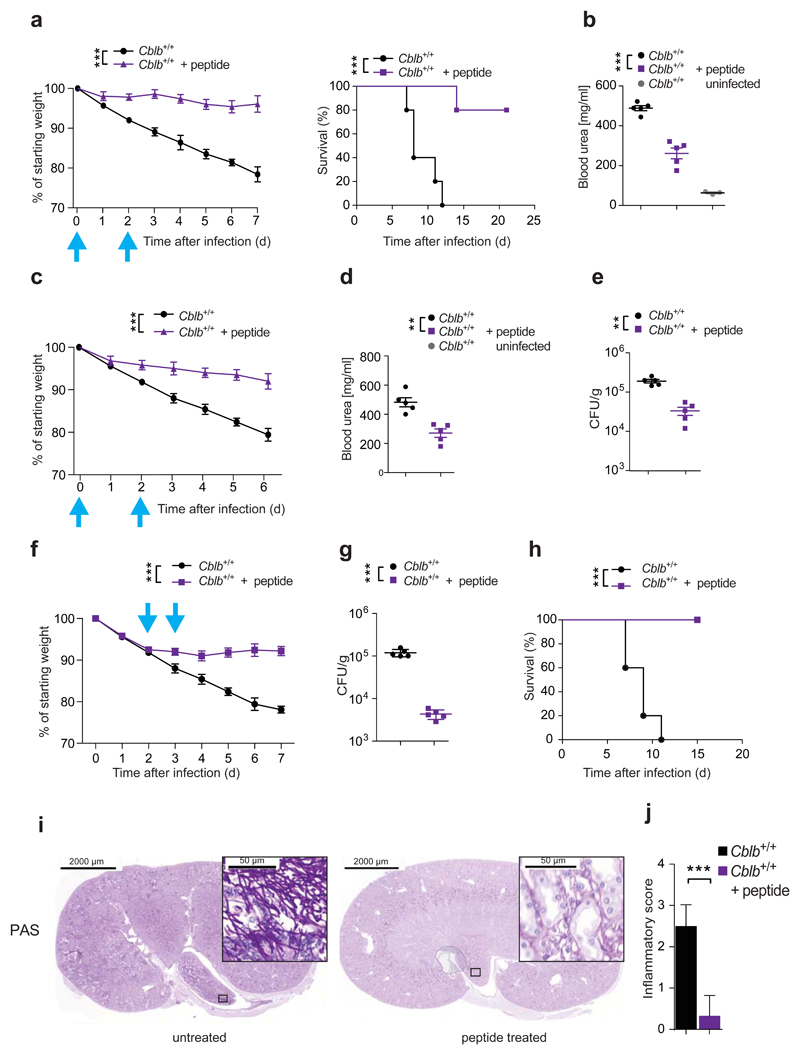
The observed in vitro efficacy of our inhibitory peptide prompted us to investigate its activity in vivo. We thus infected CblbΔ/Δ and Cblb+/+ littermates with C. albicans and treated them twice with the inhibitory CBL-B-TKB-binding peptide (100μg, i.p.; day 0 and day 2), or left them untreated. Strikingly, this treatment had a significant protective effect on Cblb+/+ mice, as evidenced by reduced weight loss and delayed mortality, when compared to the untreated Cblb+/+ controls (Fig. 6a). Moreover, peptide treatment led to a decrease in blood urea nitrogen levels, indicating reduced kidney damage (Fig. 6b). Peptide treatment did not confer any protective effect on CblbΔ/Δ mice (Supplementary Fig. 6h). Reducing the concentration of the TKB-binding-peptide by 5-fold yielded comparable results (Fig. 6c,d; Supplementary Fig. 6i). In line with the decreased morbidity observed upon low dose peptide treatment, we detected a significantly reduced kidney fungal load in peptide treated mice (Fig. 6e). To test for the therapeutic potential of our TKB-binding-peptide in established fungal sepsis, we treated Cblb+/+ mice on day 2 and day 3 (100μg/injection) after infection. Strikingly, this treatment regimen completely blocked further weight loss, significantly reduced kidney fungal load 2 days after the second peptide treatment, and protected mice from mortality (Fig. 6f,g,h). Histopathologic analysis showed significantly reduced renal inflammation (Fig. 6i,j), fungal burden (Supplementary Fig. 6j), and neutrophil infiltration into the kidneys of peptide treated mice when compared to untreated controls (Supplementary Fig. 6k). Thus, the CBL-B-TBK-binding-peptide enhances anti-fungal immunity and can protect from lethal C. albicans sepsis.
Figure 6. CBL-B inhibition enhances anti-fungal immunity.
(a,b) Cblb+/+ mice were infected intravenously with C. albicans (105 CFU/21.5g body weight), and injected intraperitoneally with 100 μg TKB-binding-peptide twice. Plots depict weight loss (left; P values assessed by two-way ANOVA), and survival (right; P values assessed with log rank test). (b) Blood urea concentration 7 days after infection. n = 5 for all cohorts. (c,d,e) Cblb+/+ mice were treated as in (a,b) with 20 μg peptide. Plots depict (c) weight loss (P values assessed by two-way ANOVA; n = 5 for all cohorts), (d) Blood urea concentration and (e) kidney fungal load 7 days after infection. (f,g,h) Cblb+/+ mice infected as in (a,b), were injected intraperitoneally with 100 μg of peptide twice. Plots depict (f) weight loss (P values assessed by two-way ANOVA), (g) kidney fungal load 48 hours after second peptide treatment, and (h) survival (P values assessed with log rank test). (a,c,f) Blue arrows indicate time points of peptide treatment. (i) PAS stained kidney sections from mice treated as in (f) and sacrificed 2 days after second peptide treatment. Boxed areas shown at increased magnification. (j) Inflammatory score of infected kidneys from (i). n = 3 for both cohorts. 4 sections per kidney analyzed. (a,b,c,d,e,f,g,j) Data are shown as means ± SD. (b,d,e,g) Dots represent individual mice. For panels (a,b) 1 representative of 5, for panel (c-j) 1 representative of 3 independent experiments is shown. **P < 0.01, ***P < 0.001 calculated with Student’s t-test.
Discussion
Our study identified a critical role for CBL-B in the regulation of innate anti-fungal immunity. Loss of CBL-B, or CBL-B ligase activity, had a protective effect in systemic or cutaneous C. albicans infections. CBL-B was shown to modulate CLR signaling and dampen the fungicidal activity of phagocytes, but not neutrophils. Mechanistically, CBL-B was found to physically associate with the CLR signalosome via SYK upon fungal activation. Blocking the proteasome mimicked the loss of CBL-B with regard to SYK activation, prompting us to identify CBL-B target molecules. We observed poly-ubiquitination of SYK, as well as Dectin-1, and -2 by CBL-B. CBL-B thus regulates CLR signaling by both affecting the availability of Dectin-1 and -2, as well as dampening proximal signaling via SYK. Based on the SYK-CBL-B interaction we designed a novel CBL-B-TKB-binding peptide to inhibit CBL-B function. We could validate the in vitro and in vivo efficacy of this approach by protecting mice from morbidity and mortality induced by systemic fungal infections.
Genetic networks required for anti-fungal immune responses have been unraveled in elegant studies in mice9,35 and men3,42–47. These studies have failed to provide mechanistic insight into the negative regulation of such immune responses, however. Our study reports CBL-B as the first negative regulator of innate anti-fungal immunity. Current therapeutic strategies exploit the vulnerability of fungi to several classes of anti-fungals48. We propose an alternative approach: directly enhancing immune signaling networks essential for a protective anti-fungal immune response by targeting CBL-B. Our in vitro and in vivo studies show that inhibiting CBL-B using a peptide interference strategy releases the brakes on anti-fungal immunity and completely protects mice from otherwise lethal systemic infections with C. albicans.
While showing beneficial effects in the context of fungal inflammation, inhibiting CBL-B might cause undesirable side effects by deregulating adaptive immunity. It is noteworthy, that a significant association to Type I diabetes (T1D) of a SNP within exon 12 of CBLB49, and T1D patients carrying functionally defective variants of CBL-B have been reported50. Likewise, Cblb deficient mice manifest autoimmunity. It will thus be of relevance to compare deletion of Cblb to the acute and short term blockade reported in our manuscript that might lead to less severe adverse effects due to transient CBL-family inhibition.
Specific inhibition of CBL-family ubiquitin ligase activity has, to the best of our knowledge, not been accomplished, yet. While targeting the TKB domain has been reported in vitro, the in vivo efficacy of this approach was severely limited by the amounts of peptide required51. Based on the molecular mechanism identified in our experiments, we engineered a novel CBL-B-TKB-blocking peptide combined with an enhanced peptide delivery system based on the Antennapedia cell permeable peptide52. Treatment of mice with our inhibitory peptide indeed drastically improved immune function towards C. albicans and protected mice from a lethal infection in a therapeutic setting. Thus, our study not only provides novel insights into the negative regulation of anti-fungal immunity, but also a novel conceptual framework for the design of an efficacious and specific approach to enhance innate anti-fungal immunity.
Online Methods
Mice
CblbΔ/Δ, CblbC373A/C373A and Rag2Δ/Δ deficient mice have been described previously and were bred on a C57BL/6 background. Mouse genotypes were assessed by PCR. Of note, only age- and sex-matched littermates from respective breedings were used for experiments. All mice were bred and maintained in accordance with ethical animal license protocols complying with the current Austrian law.
Generation of bone marrow chimeric mice
To generate bone marrow chimeric mice, 8 week old wild type or Rag2Δ/Δ deficient C57BL/6 mice were split dose irradiated (2 x 6 Gy) and reconstituted 18h after the second irradiation with 3 million donor cells by intravenous injection. Experiments were carried out 8 weeks later to allow for full reconstitution. Reconstitution efficiency under the applied conditions was tested by using congenic CD45.1 recipient mice and staining of splenocytes for CD45.1 (recipient derived) and CD45.2 (donor derived), 8 weeks after reconstitution, and was routinely >98%.
Isolation and differentiation of neutrophils, monocytes, macrophages, and dendritic cells
Neutrophils were isolated from tibias and femurs of mice using a Percoll-gradient (GE Healthcare) as previously described53 or were purified using MACS technology (Miltenyi) after Ly-6G labeling. Bone marrow monocytes were purified by FACS sorting after labeling with antibodies specific for CD11b, Ly-6G, and Ly-6C. Splenic dendritic cells were FACS sorted after labeling with antibodies to CD19, TCRβ, Ly-6G, and CD11c. Purified neutrophils, monocytes, and splenic dendritic cells were cultured in RPMI-1640 medium (PAA). BM-DC or BM-M were obtained by differentiation from bone marrow precursor cells. Peripheral blood monocytes or neutrophils were obtained by sub-mandibular bleeding and collected in potassium EDTA tubes (Sarstedt, Germany). Red blood cells were lysed using an Ammonium-Chloride-Potassium (ACK) buffer.
Flow cytometry
Antibody labeling of leukocytes was carried out in FACS staining buffer (PBS supplemented with 2% FBS and 2mM EDTA) on ice for 30min after blocking of Fc-receptors. Cell viability was assessed with the Fixable Viability Dye eFluor780 (eBioscience). Immune cell recruitment was assessed by Collagenase 4 (Worthington)/DNAse (Roche) digestion of organs, viability staining, Fc-blocking, antibody staining, and subsequent flow cytometric analysis. Samples were acquired with a BD LSRII flow cytometer and data were analyzed using FlowJo software (Treestar). See Supplementary Table 1 for antibodies used in this study.
Measurements of cytokine release, ROS production, Caspase-8 activity
Cytokine secretion by cell culture supernatants or in the peritoneal lavage was assessed with ELISA kits purchased from eBioscience (IL-1β, TNF, IL-6, IL-12, IL-23) or R&D Systems (IL-1Ra) according to the manufacturer’s instructions. Production of reactive oxygen species (ROS) was measured with an assay using luminol as the probe in real-time over 90min as described previously54. Caspase-8 activity was assessed using the CaspGLOW assay (Promega) according to the manufacturer’s instructions.
Phagocytosis assay
For phagocytosis assays C. albicans (strain SC5314) were Alexa 488 labeled in 100mM HEPES buffer (pH 7.5). BM-DC or BM-M were co-cultured with labeled C. albicans at 37°C for 45 minutes. Fluorescence by fungal cells adherent to, but not phagocytosed by, phagocytes, were quenched with trypan blue (Sigma Aldrich), and the rate of phagocytosis was assessed by flow cytometry.
Killing assay
To assess the fungicidal activity, the indicated immune cells were plated in replicates at a density of 1 x 105 cells/well in 96-well plates. Cells were incubated with C. albicans at a multiplicity of infection of 1:500 for 24h. After incubation, phagocytes were fixed by the addition of paraformaldehyde at a final concentration of 4%. Subsequently, C. albicans were stained with Crystal Violet (Sigma) and killing was assessed by a comparison of colony numbers in wells with or without phagocytes.
Mouse infection and clodronate liposome treatment
8-10 week old mice were infected with 105 colony forming units (CFU) of C. albicans (strain SC5314), 106 colony forming units (CFU) of C. dubliniensis, or 105 CFUs of L. monocytogenes (strain EGD) intravenously. C. albicans CFUs were assessed from various organs as described55. For clodronate or PBS liposome treatment experiments, mice were injected intraperitoneally with 100ul/10g mouse weight of liposomes both 24h before and 24h after intravenous C. albicans infections. Clodronate or PBS control liposomes were obtained from clodronateliposomes.org, and handled according to the manufacturer’s instructions. All animal infection experiments were evaluated by the ethics committee of the Medical University of Vienna and approved by the Federal Ministry for Science and Research, Vienna, Austria.
BM-M and BM-DC stimulation experiments
BM-M and BM-DC were stimulated with C. albicans (strain SC5314), Zymosan (Invivogen), Curdlan (Invivogen), TDM (Invivogen), CpG (Invivogen), LPS (Invivogen), or C. albicans hyphae. For inhibition experiments cells were treated with the Syk inhibitor R406 (Selleckchem), anti-Dectin-1 antibody (Invivogen), MG132 (Calbiochem), or Dynasore (Sigma-Aldrich), 30min prior to stimulation (MOI: Immune cell: C. albicans = 1:2). Fungal isolates of Candida krusei, Candida glabrata, and Candida dubliniensis were kindly provided by the Allgemeines Krankenhaus Vienna (AKH). Fungal isolates were tested by PCR to verify the respective species.
Quantitative PCR
mRNA was isolated using the SV-Total RNA Isolation System (Promega) or RNeasy Kit (Qiagen), reverse transcribed using the Reverse Transcription System (Promega) or the iScript cDNA synthesis Kit (Biorad). qPCR was performed with KAPA SYBR Fast Universal (Peqlab). See Supplementary Table 1 for primers used in this study.
Western blotting and Immunoprecipitation
Western blotting was performed according to standard protocols. Blots were blocked for 1 hour with 5% bovine serum albumin in 1 × TBS/0.1% Tween-20, or 4% BSA in 1 × TBS/0.1% Tween-20 for phosphorylated epitopes, followed by overnight incubation at 4°C with primary antibodies. Subsequently, blots were washed three times in 1 × TBS/0.1% Tween-20 for 15 min, followed by incubation with HRP-conjugated secondary antibodies (1:2500; GE Healthcare # NA9340V) for 45 min at room temperature, washed three times in 1 × TBS/0.1% Tween-20 for 15 min and visualized using enhanced chemiluminescence (ECL Plus, Pierce). Anti-β-Actin mAbs were used to control for protein loading. For Cbl-b-immunoprecipitation experiments, cells were stimulated with C. albicans, lysed in RIPA buffer (Cell Signaling Technology) containing phosphatase and proteinase inhibitors (Halt Protease and Phosphatase inhibitor Cocktail; Thermo Scientific), pre-cleared with Protein A/G Plus Agarose beads (Santa Cruz Biotechnology), and incubated overnight with an anti-Cbl-b antibody. Precipitates were analyzed by Western blotting using the indicated antibodies. For Dectin-1 and Dectin-2 immunoprecipitations, BMM of the indicated genotypes were pre-treated with E-64 (10μM) for 30min, infected with C. albicans yeast or hyphae for the indicated periods of time, lysed in RIPA buffer containing 2% SDS on ice for 30min, and diluted with RIPA buffer without SDS for a final SDS concentration of 0.4%. After brief sonication, lysates were immunopreciptitated with an anti-Dectin-1 or anti-Dectin-2 antibody at a concentration of 1μg/ml. Precipitates were analyzed by Western blotting with the indicated antibodies. Please, see Supplementary Table 1 for antibodies used in this study.
In vitro ubiquitination assays
Reaction mixtures for in vitro ubiquitination assays contained 100nM Ube1 (purified in-house), 500nM UbcH5B (Enzo Life Sciences), 150nM full length human Cbl-b (Abnova), or human Cbl-b29-483, or a catalytically inactive human C373A Cbl-b29-483 mutant, and 100mM ubiquitin (Sigma-Aldrich) with 300nM of substrate in reaction buffer (20mM Tris-HCl pH 7.5, 150mM NaCl, 10mM MgCl2 and 1mM DTT). Substrates were active (phosphorylated) human Syk (Merck Millipore) and inactive (non-phosphorylated) human Syk-GST (Life technologies). 1mM ATP was added to initiate the reaction and incubated at 37 °C for 30 min. For TKB binding peptide mediated Cbl-b inhibition, Cbl-b was pre-incubated with the TKB binding peptide at a concentration of 100μM for 20 min at 24 °C, before addition to the reaction mixture. Assays were resolved by SDS-PAGE, transferred on a nitrocellulose membrane (GE Healthcare) and blotted with an anti-ubiquitin antibody, or an anti-Syk antibody. See Supplementary Table 1 for antibodies used in this study.
Production of recombinant CBL-B29-483 and C373A CBL-B29-483
Cblb29-483 and C373A Cblb29-483 coding sequences were cloned into the pGEX-2TK vector to obtain Gluthathione-S-transferase (GST) fusion proteins. Expression in E. coli BL21 was induced with 0.3 mM isopropyl-D-thiogalactopyranoside at OD600 of 1.0 in Terrific Broth medium, and incubated overnight at 20°C. Cells were lysed in lysis buffer (20mM Tris-HCl pH 7.5, 100mM NaCl, 1mM EDTA, 0.5% Triton X-100), sonicated briefly three times, and incubated overnight with Glutathione Sepharose 4B (GE Helathcare) at 4°C. Subsequently, Sepharose beads were washed five times with lysis buffer, and bound proteins were eluted with elution buffer (50mM Tris–HCl, pH 8.0, and 20mM reduced glutathione). Eluates were tested for size and purity with SDS-PAGE and Western blotting.
Histopathology
For histopathology analyses, kidneys were fixed in 10% neutral buffered formalin, processed according to standard procedures, embedded in paraffin, and sectioned. 2 μm thick sections were stained with haematoxylin and eosin (H&E), periodic-acid-Schiff (PAS), or anti-Ly-6G (Biolegend; clone 1A8). Immunohistochemistry was performed with a primary antibody dilution of 1:100 with DAB enhancement after antigen retrieval (BOND Epitope Retrieval kit, Leica). 4 μm sections were stained with Gomori Methenamine Silver (GMS; Merck). Stained slides were scanned using a Pannoramic slide scanner (3D Histech) and evaluated for severity of inflammation and intra-lesional fungal burden. Renal inflammation was scored based upon H&E and PAS stainings (Proportion of renal parenchyma and/or pelvis involved by tubulointerstitial nephritis and/or pyelonephritis) as not significant (score 0), less than 10% (score 1), 10-25% (score 2), 25-50% (score 3), or greater than 50% (score 4). The intralesional fungal burden was based upon PAS and GMS stainings as not significant (score 0), scant presence in less than 10% of inflammatory foci (score 1), mild to moderate presence in 10-25% of inflammatory foci (score 2), moderate to significant presence in 25-50% of inflammatory foci (score 3), or significant presence in more than 50% of inflammatory foci (score 4). Definiens Tissue StudioTM software was applied to evaluate the extent of infiltration by Ly-6G+ cells in affected kidneys. Regions and cells of interest were manually classified in representative areas of slides to train the software algorithm to identify Ly-6G positive target objects and exclude non-target objects. Target objects were identified on the basis of morphology and positive immunohistochemical staining. The optimized algorithm was then applied to analyze all slides automatically. The validity of the analysis was confirmed by histopathologic verification of representative slides from both groups.
Peptide synthesis
Solid phase peptide synthesis (SPPS) was used for TKB binding peptide synthesis on an ABI 433a Peptide Synthesizer (Applied-Biosystems). Peptides were purified with an Ultimate 3000 RP-HPLC system (Thermo Scientific), and analyzed with a 4800 MALDI TOF/TOF analyzer (ABSciex). Lyophilized peptides were resolved in PBS and stored at -80°C.
Immunofluorescence
BM-DC were seeded on µ-slides (IBIDI) 24h before infection with C. albicans. After infection cells were fixed in 4% PFA on ice for 20min, permeabilized (0.2% Triton X-100 in PBS) for 10 min at room temperature, and blocked (3% BSA in PBS) for 1 hour at room temperature. Subsequently, cells were incubated with primary antibodies against SYK (1:100) or SHP-2 (1:100) in 3% BSA at overnight. Cells were then washed in PBS and incubated with an anti-rabbit-Alexa555 antibody (1:500) in 3% BSA at room temperature for 1h. After washing in PBS cells were counterstained with DAPI, and directly imaged with a Zeiss LSM780 Confocal microscope. Please see Supplementary Table 1 for antibodies used in this study.
NanoLC-MS Analysis
The nano HPLC system used for this study was an UltiMate 3000 HPLC RSLC nano system (Thermo Scientific) coupled to a Q Exactive Plus mass spectrometer (Thermo Scientific), equipped with a Proxeon nanospray source (Thermo Scientific). Peptides were loaded onto a trap column (Thermo Fisher Scientific, Amsterdam, Netherlands, PepMap C18, 5 mm × 300 μm ID, 5 μm particles, 100 Å pore size) at a flow rate of 25 μL min-1 using 0.1% TFA as mobile phase. After 10 min the trap column was switched in line with the analytical column (Thermo Fisher Scientific, Amsterdam, Netherlands, PepMap C18, 500 mm × 75 μm ID, 2 μm, 100 Å). Peptides were eluted using a flow rate of 230 nl min-1, and a binary 2h gradient for 165 min. The gradient was started with 98% water/formic acid (99.9/0.1; v/v) and 2% water/acetonitrile/formic acid (19.92/80/0.08; v/v/v), and increased to 35% water/formic acid (99.9/0.1; v/v) and 65% water/acetonitrile/formic acid (19.92/80/0.08; v/v/v) over 120min, followed by a gradient to 10% water/formic acid (99.9/0.1; v/v) and 90% water/acetonitrile/formic acid (19.92/80/0.08; v/v/v) over 5min, kept at this ratio for 5min and subsequently decreased over 2min back to 98% water/formic acid (99.9/0.1; v/v) and 2% water/acetonitrile/formic acid (19.92/80/0.08; v/v/v) for equilibration at 30°C. The Q Exactive mass spectrometer was operated in data-dependent mode, using a full scan (m/z range 370-1650, nominal resolution of 70 000, target value 3E6) followed by MS/MS scans of the 12 most abundant ions. MS/MS spectra were acquired using normalized collision energy 27%, isolation width of 2 and the target value 1E5. Precursor ions selected for fragmentation (charge state 2 and higher) were put on a dynamic exclusion list for 10 sec. The underfill ratio was set to 20% resulting in an intensity threshold of 4E4. The peptide match feature and the exclude isotopes feature were enabled.
Mass spectrometry data analysis
For peptide identification the .RAW-files were loaded into Proteome Discoverer (version 1.4.0.288, Thermo Scientific). All created MS/MS spectra were searched using the search engine node MSAmanda56 against the flybase sequence database (22,256 sequences; 20,222,850 residues) and also against the Swissprot sequence database, using the taxonomy human (20,170 sequences, 11,318,213 residues). The following search parameters were used: Beta-methylthiolation on cysteine was set as a fixed modification, oxidation on methionine, acetylation on lysine, phosphorylation on serine, threonine and tyrosine, deamidation on asparagine and glutamine, and ubiquitinylation on lysine were set as variable modifications. Monoisotopic masses were searched within unrestricted protein masses for tryptic peptides. The peptide mass tolerance was set to ±5 ppm and the fragment mass tolerance to ± 0.03 Da. The maximal number of missed cleavages was set to 2. The result was filtered to 1% FDR using Percolator algorithm integrated in Proteome Discoverer. The localization of the sites of variable modifications within the peptides was performed with the tool ptmRS, integrated in Proteome Discoverer and based on phosphoRS57.
Data Analysis and Statistics
All values in the paper are given as means ± standard deviation unless stated otherwise. All in vitro experiments were reproduced 3-5 independent times. In vivo experiments were reproduced 2-6 times. Figures and statistical analyses were generated using PraphPad Prism software (GraphPad Software). No mice were excluded from the analyses and histopathologic analyses using Definies Tissue studio software were performed blindly. Mice were allocated to experimental groups based upon their genotypes and randomized within their sex- and age matched groups. Since mice were inbred and sex- and age-matched, we assumed similar variance between experimental cohorts. An a priori sample size estimation was not performed. Group sizes were based upon the empirically assessed variability of the model systems or assays used, and groups contained as many mice as possible to minimize type I and type II errors. Data were analyzed using the unpaired two-tailed Student’s t-test, or two way ANOVA, as indicated. For survival analyses, log rank tests were performed. P values < 0.05 were accepted as statistically significant.
Supplementary Material
Acknowledgements
We thank all members of the Penninger laboratory for helpful discussions and technical support. We thank all members of the IMP-IMBA Biooptics service facility, especially G. Schmauss and T. Lendl, for assistance in cell sorting and image quantification. We also like to thank A. Piszczek and M. Zeba from the Vienna Biocenter Core Facilities (VBCF) for excellent histopathology services. This work was supported by a consolidator ERC grant (F.I.), the EC FP7 project “FUNGITECT” (K.K.), the FWF project P-25333 (K.K.), a Marie-Curie “Innovative Training Networks” ImResFun grant (Contract MC-ITN-606786; K.K.), an Advanced ERC grant (J.M.P.), an “Era of Hope/Innovator Award” (J.M.P.), a Helmsley foundation VEO-IBD network grant (J.M.P.), and the Austrian Academy of Sciences (J.M.P).
Footnotes
Accession codes
Data are available via ProteomeXchange with identifier PXD004256.
Author Contributions
G.W., F.Z., K.K., and J.M.P. designed experiments. F.Z., I.K., and R.A.W. performed Western blotting and immunoprecipitation experiments. R.A.W. performed immunohistochemistry. F.Z. performed Candida spp. and L. monocytogenes systemic infections, ELISA, ROS, Caspase-8, and C. albicans killing assays. F.Z. and G.W. performed C. albicans skin infection experiments. G.W. and F.Z. performed qPCR. G.W. and L.T. performed flow cytometry G.W. designed the TKB-binding Antennapedia/SYK fusion peptide. T.A. and F.I. did and analyzed in vitro ubiquitination assays. A.K. performed histopathologic analyses. W.Y.L. provided CblbC373A/C373A mice. G.B. and F.F. provided recombinant CBL-B29-483 and C373A CBL-B29-483. J.M.P and K.K. supervised the study. G.W. and J.M.P. coordinated the study and wrote the manuscript.
Competing financial interests
J.M.P, G.W., F.Z., and K.K. are inventors on a patent application describing the use of a TKB-binding peptide as a therapeutic for the modulation of CBL-B regulated immune responses.
References
- 1.Armstrong-James D, Meintjes G, Brown GD. A neglected epidemic: fungal infections in HIV/AIDS. Trends in microbiology. 2014;22:120–127. doi: 10.1016/j.tim.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Ravikumar S, Win MS, Chai LY. Optimizing Outcomes in Immunocompromised Hosts: Understanding the Role of Immunotherapy in Invasive Fungal Diseases. Frontiers in microbiology. 2015;6:1322. doi: 10.3389/fmicb.2015.01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lanternier F, et al. Primary immunodeficiencies underlying fungal infections. Current opinion in pediatrics. 2013;25:736–747. doi: 10.1097/MOP.0000000000000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown GD, et al. Hidden killers: human fungal infections. Science translational medicine. 2012;4:165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 5.Marr KA, et al. Differential role of MyD88 in macrophage-mediated responses to opportunistic fungal pathogens. Infection and immunity. 2003;71:5280–5286. doi: 10.1128/IAI.71.9.5280-5286.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramirez-Ortiz ZG, et al. Toll-like receptor 9-dependent immune activation by unmethylated CpG motifs in Aspergillus fumigatus DNA. Infection and immunity. 2008;76:2123–2129. doi: 10.1128/IAI.00047-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cambi A, et al. The C-type lectin DC-SIGN (CD209) is an antigen-uptake receptor for Candida albicans on dendritic cells. European journal of immunology. 2003;33:532–538. doi: 10.1002/immu.200310029. [DOI] [PubMed] [Google Scholar]
- 8.Netea MG, et al. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. The Journal of clinical investigation. 2006;116:1642–1650. doi: 10.1172/JCI27114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardison SE, Brown GD. C-type lectin receptors orchestrate antifungal immunity. Nature immunology. 2012;13:817–822. doi: 10.1038/ni.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown GD, Gordon S. Immune recognition. A new receptor for beta-glucans. Nature. 2001;413:36–37. doi: 10.1038/35092620. [DOI] [PubMed] [Google Scholar]
- 11.Saijo S, et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010;32:681–691. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Zhu LL, et al. C-type lectin receptors Dectin-3 and Dectin-2 form a heterodimeric pattern-recognition receptor for host defense against fungal infection. Immunity. 2013;39:324–334. doi: 10.1016/j.immuni.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 13.Wells CA, et al. The macrophage-inducible C-type lectin, mincle, is an essential component of the innate immune response to Candida albicans. Journal of immunology. 2008;180:7404–7413. doi: 10.4049/jimmunol.180.11.7404. [DOI] [PubMed] [Google Scholar]
- 14.van de Veerdonk FL, et al. The macrophage mannose receptor induces IL-17 in response to Candida albicans. Cell host & microbe. 2009;5:329–340. doi: 10.1016/j.chom.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa E, et al. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. The Journal of experimental medicine. 2009;206:2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smeekens SP, et al. The classical CD14(+)(+) CD16(-) monocytes, but not the patrolling CD14(+) CD16(+) monocytes, promote Th17 responses to Candida albicans. European journal of immunology. 2011;41:2915–2924. doi: 10.1002/eji.201141418. [DOI] [PubMed] [Google Scholar]
- 17.Patel DD, Kuchroo VK. Th17 Cell Pathway in Human Immunity: Lessons from Genetics and Therapeutic Interventions. Immunity. 2015;43:1040–1051. doi: 10.1016/j.immuni.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Whibley N, Gaffen SL. Brothers in arms: Th17 and Treg responses in Candida albicans immunity. PLoS pathogens. 2014;10:e1004456. doi: 10.1371/journal.ppat.1004456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mocsai A, Ruland J, Tybulewicz VL. The SYK tyrosine kinase: a crucial player in diverse biological functions. Nature reviews Immunology. 2010;10:387–402. doi: 10.1038/nri2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng Z, et al. Tyrosine phosphatase SHP-2 mediates C-type lectin receptor-induced activation of the kinase Syk and anti-fungal TH17 responses. Nature immunology. 2015;16:642–652. doi: 10.1038/ni.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zwolanek F, et al. The non-receptor tyrosine kinase Tec controls assembly and activity of the noncanonical caspase-8 inflammasome. PLoS pathogens. 2014;10:e1004525. doi: 10.1371/journal.ppat.1004525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross O, et al. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature. 2009;459:433–436. doi: 10.1038/nature07965. [DOI] [PubMed] [Google Scholar]
- 23.Ruland J. CARD9 signaling in the innate immune response. Annals of the New York Academy of Sciences. 2008;1143:35–44. doi: 10.1196/annals.1443.024. [DOI] [PubMed] [Google Scholar]
- 24.Bachmaier K, et al. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature. 2000;403:211–216. doi: 10.1038/35003228. [DOI] [PubMed] [Google Scholar]
- 25.Chiang YJ, et al. Cbl-b regulates the CD28 dependence of T-cell activation. Nature. 2000;403:216–220. doi: 10.1038/35003235. [DOI] [PubMed] [Google Scholar]
- 26.Loeser S, Penninger JM. The ubiquitin E3 ligase Cbl-b in T cells tolerance and tumor immunity. Cell cycle. 2007;6:2478–2485. doi: 10.4161/cc.6.20.4797. [DOI] [PubMed] [Google Scholar]
- 27.Paolino M, et al. Essential role of E3 ubiquitin ligase activity in Cbl-b-regulated T cell functions. Journal of immunology. 2011;186:2138–2147. doi: 10.4049/jimmunol.1003390. [DOI] [PubMed] [Google Scholar]
- 28.Paolino M, et al. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature. 2014;507:508–512. doi: 10.1038/nature12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qu X, et al. Negative regulation of FcepsilonRI-mediated mast cell activation by a ubiquitin-protein ligase Cbl-b. Blood. 2004;103:1779–1786. doi: 10.1182/blood-2003-07-2260. [DOI] [PubMed] [Google Scholar]
- 30.Han C, et al. Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting degradation of MyD88 and TRIF via Cbl-b. Nature immunology. 2010;11:734–742. doi: 10.1038/ni.1908. [DOI] [PubMed] [Google Scholar]
- 31.Wallner S, et al. The role of the e3 ligase cbl-B in murine dendritic cells. PloS one. 2013;8:e65178. doi: 10.1371/journal.pone.0065178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Chiang YJ, Hodes RJ, Siraganian RP. Inactivation of c-Cbl or Cbl-b differentially affects signaling from the high affinity IgE receptor. Journal of immunology. 2004;173:1811–1818. doi: 10.4049/jimmunol.173.3.1811. [DOI] [PubMed] [Google Scholar]
- 33.Sohn HW, Gu H, Pierce SK. Cbl-b negatively regulates B cell antigen receptor signaling in mature B cells through ubiquitination of the tyrosine kinase Syk. The Journal of experimental medicine. 2003;197:1511–1524. doi: 10.1084/jem.20021686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Igyarto BZ, et al. Skin-resident murine dendritic cell subsets promote distinct and opposing antigen-specific T helper cell responses. Immunity. 2011;35:260–272. doi: 10.1016/j.immuni.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drummond RA, Gaffen SL, Hise AG, Brown GD. Innate Defense against Fungal Pathogens. Cold Spring Harbor perspectives in medicine. 2015;5 doi: 10.1101/cshperspect.a019620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aratani Y, et al. Relative contributions of myeloperoxidase and NADPH-oxidase to the early host defense against pulmonary infections with Candida albicans and Aspergillus fumigatus. Medical mycology. 2002;40:557–563. doi: 10.1080/mmy.40.6.557.563. [DOI] [PubMed] [Google Scholar]
- 37.Gringhuis SI, et al. Dectin-1 is an extracellular pathogen sensor for the induction and processing of IL-1beta via a noncanonical caspase-8 inflammasome. Nature immunology. 2012;13:246–254. doi: 10.1038/ni.2222. [DOI] [PubMed] [Google Scholar]
- 38.Qian Q, Jutila MA, Van Rooijen N, Cutler JE. Elimination of mouse splenic macrophages correlates with increased susceptibility to experimental disseminated candidiasis. Journal of immunology. 1994;152:5000–5008. [PubMed] [Google Scholar]
- 39.May MJ, et al. Selective inhibition of NF-kappaB activation by a peptide that blocks the interaction of NEMO with the IkappaB kinase complex. Science. 2000;289:1550–1554. doi: 10.1126/science.289.5484.1550. [DOI] [PubMed] [Google Scholar]
- 40.Zou W, Reeve JL, Zhao H, Ross FP, Teitelbaum SL. Syk tyrosine 317 negatively regulates osteoclast function via the ubiquitin-protein isopeptide ligase activity of Cbl. The Journal of biological chemistry. 2009;284:18833–18839. doi: 10.1074/jbc.M109.012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yankee TM, Keshvara LM, Sawasdikosol S, Harrison ML, Geahlen RL. Inhibition of signaling through the B cell antigen receptor by the protooncogene product, c-Cbl, requires Syk tyrosine 317 and the c-Cbl phosphotyrosine-binding domain. Journal of immunology. 1999;163:5827–5835. [PubMed] [Google Scholar]
- 42.Glocker EO, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. The New England journal of medicine. 2009;361:1727–1735. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng J, et al. Gain-of-function STAT1 mutations impair STAT3 activity in patients with chronic mucocutaneous candidiasis (CMC) European journal of immunology. 2015;45:2834–2846. doi: 10.1002/eji.201445344. [DOI] [PubMed] [Google Scholar]
- 44.van der Graaf CA, et al. Candida-specific interferon-gamma deficiency and toll-like receptor polymorphisms in patients with chronic mucocutaneous candidiasis. The Netherlands journal of medicine. 2003;61:365–369. [PubMed] [Google Scholar]
- 45.Plantinga TS, et al. Human genetic susceptibility to Candida infections. Medical mycology. 2012;50:785–794. doi: 10.3109/13693786.2012.690902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plantinga TS, et al. Early stop polymorphism in human DECTIN-1 is associated with increased candida colonization in hematopoietic stem cell transplant recipients. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2009;49:724–732. doi: 10.1086/604714. [DOI] [PubMed] [Google Scholar]
- 47.Smeekens SP, van de Veerdonk FL, Kullberg BJ, Netea MG. Genetic susceptibility to Candida infections. EMBO molecular medicine. 2013;5:805–813. doi: 10.1002/emmm.201201678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stevens DA. Advances in systemic antifungal therapy. Clinics in dermatology. 2012;30:657–661. doi: 10.1016/j.clindermatol.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 49.Bergholdt R, Taxvig C, Eising S, Nerup J, Pociot F. CBLB variants in type 1 diabetes and their genetic interaction with CTLA4. Journal of leukocyte biology. 2005;77:579–585. doi: 10.1189/jlb.0904524. [DOI] [PubMed] [Google Scholar]
- 50.Kosoy R, Yokoi N, Seino S, Concannon P. Polymorphic variation in the CBLB gene in human type 1 diabetes. Genes and immunity. 2004;5:232–235. doi: 10.1038/sj.gene.6364057. [DOI] [PubMed] [Google Scholar]
- 51.Nakao R, et al. Ubiquitin ligase Cbl-b is a negative regulator for insulin-like growth factor 1 signaling during muscle atrophy caused by unloading. Molecular and cellular biology. 2009;29:4798–4811. doi: 10.1128/MCB.01347-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bechara C, Sagan S. Cell-penetrating peptides: 20 years later, where do we stand? FEBS letters. 2013;587:1693–1702. doi: 10.1016/j.febslet.2013.04.031. [DOI] [PubMed] [Google Scholar]
- 53.Boxio R, Bossenmeyer-Pourie C, Steinckwich N, Dournon C, Nusse O. Mouse bone marrow contains large numbers of functionally competent neutrophils. Journal of leukocyte biology. 2004;75:604–611. doi: 10.1189/jlb.0703340. [DOI] [PubMed] [Google Scholar]
- 54.Bourgeois C, Majer O, Frohner I, Kuchler K. In vitro systems for studying the interaction of fungal pathogens with primary cells from the mammalian innate immune system. Methods in molecular biology. 2009;470:125–139. doi: 10.1007/978-1-59745-204-5_11. [DOI] [PubMed] [Google Scholar]
- 55.Wirnsberger G, et al. Jagunal homolog 1 is a critical regulator of neutrophil function in fungal host defense. Nature genetics. 2014;46:1028–1033. doi: 10.1038/ng.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dorfer V, et al. MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. Journal of proteome research. 2014;13:3679–3684. doi: 10.1021/pr500202e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taus T, et al. Universal and confident phosphorylation site localization using phosphoRS. Journal of proteome research. 2011;10:5354–5362. doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.