Abstract
Objectives
To investigate epigenetic mechanisms contributing to regulation of cellular renewal and neurogenesis in adult olfactory epithelium (OE).
Study Design
Prospective basic science study.
Methods
Olfactory basal cell cultures were prepared from adult mice per established protocols. in vivo studies were performed using the mouse methimazole lesion–regeneration paradigm. Nasal tissue sections were prepared from adult mice 7 days following lesion, or from unlesioned controls. Polycomb proteins were assessed by Western blot from culture or nasal tissue lysates, and by gene expression studies from cultures. In addition, in vivo expression patterns of Polycomb proteins were examined using immunohistochemistry. Chromosome immunoprecipitation (ChIP) was performed to investigate epigenetic modifications and specific chromatin interactions for Polycomb proteins in olfactory basal cells.
Results
Subunits of Polycomb Repressive Complex 1 (PRC1) and Polycomb Repressive Complex 2 (PRC2) were identified in basal cell cultures and in vivo. In regenerating OE, basal progenitor cells identified by expression of the c–KIT receptor were found to coexpress the PRC2 protein EZH2. Because multiple variants of PRC1 subunits give rise to diverse PRC1 complexes serving different functions, expression of specific PRC1 variants was further examined. We identified PRC1 components including MEL18 (PCGF2) in immature neurons, and confirm BMI1 (PCGF4) expression in mature neurons. Moreover, we identified CBX8 as a neuron–specific PRC1 subunit. ChIP assays from OE cells demonstrated binding of PRC proteins to regulatory regions of specific transcription factors, consistent with PRC–mediated epigenetic silencing mechanisms active in adult OE.
Conclusions
Multiple Polycomb proteins have cell type–specific expression patterns in the adult OE. Findings presented here, together with evidence from prior studies, suggest that PRC–mediated epigenetic silencing contributes to regulation of cellular renewal and tissue homeostasis in the OE. Efforts to define the mechanisms that regulate repair in the OE are essential for development of new therapeutic strategies for olfactory disorders.
Level of Evidence
N/A
Keywords: Polycomb, olfaction, stem cells, regeneration, epigenetics
INTRODUCTION
Anosmia remains a frustrating problem for clinicians and patients, because there are currently limited effective treatment options. A loss of the sense of olfaction may occur due to active rhinosinusitis, prior head trauma, post–viral olfactory disorder, aging related degenerative changes (presbyosmia), or rare genetic disorders such as ciliopathies.1, 2, 3, 4, 5 Olfactory loss related to rhinosinusitis is one situation that may be responsive to medical or surgical treatments.6 In addition, olfactory training therapy has been somewhat beneficial in other clinical conditions.7 However, no specific pharmacologic treatments have been developed for patients with sensorineural anosmia. One barrier to the development of novel therapies has been a limited understanding of the mechanisms that regulate repair and regeneration, ie, tissue homeostasis, in the olfactory epithelium (OE) of the nose.
Olfactory neurons are situated in the OE lining the olfactory cleft, a relatively limited area of the nasal cavity along the superior septum and corresponding medial vertical lamellae of the superior turbinates. Inspired odors are detected by interacting with odorant receptor proteins on the bipolar olfactory neurons.8 The neurons are considered to be vulnerable to damage, for instance from toxins, pathogens, or inflammatory exposure, and are therefore relatively short–lived.9 However, the pseudostratified OE contains a basal germinal layer housing neural stem and progenitor cells,10 providing a means for replacement of damaged neurons. The basal cells have the capacity to generate new olfactory neurons and supporting cells, and are responsive to epithelial damage, thus normally maintaining tissue homeostasis in the OE11, 12 (Fig. 1). Despite this remarkable reparative capacity, certain conditions appear to lead to persistent damage, with the histologic consequence of OE loss or replacement by respiratory non–neuronal epithelium, and the functional consequence of anosmia.3, 13
Figure 1.
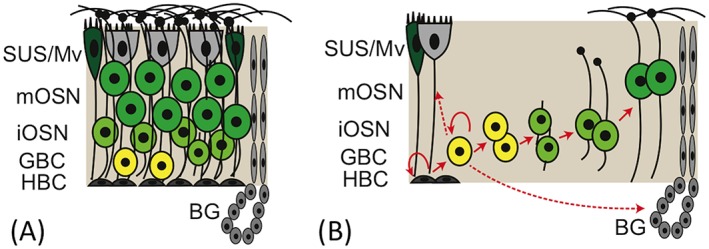
Organization and cellular renewal in the OE. (A) In normal un–damaged OE, cells are pseudostratified with sustentacular (SUS) and microvillar (Mv) cells in the apical layer, fully differentiated mature olfactory sensory neurons (mOSN) in the upper/middle layers, immature neurons (iOSN) deep to the mOSNs, and two progenitor populations in the basal germinal zone: proliferative globose basal cells (GBCs) and reserve horizontal basal cells (HBCs). Also indicated are the acinar and duct structures of Bowman's glands (BG). (B) Following injury, the OE is normally reconstituted from spared basal stem cells, the HBCs and GBCs. Arrows indicate the lineage relationships as OE cells are produced from stem and progenitor cells.
What are the mechanisms that regulate basal stem cells, OE neurogenesis and the response to tissue injury, and might these represent potential therapeutic targets for the prevention or correction of anosmia? Olfactory neurogenesis has been the subject of considerable study, given that the OE is a uniquely robust neural stem cell niche in adult mammals (for review, see Schwob et al.12). While several intrinsic and extrinsic regulatory pathways have been identified,14, 15, 16 many aspects remain incompletely defined. Recently, epigenetic modifiers such as Polycomb group proteins have been identified as master regulators of renewal and differentiation in embryonic and adult stem cells.17 Of interest, we have reported recently that the Polycomb protein BMI1 is present in OE cells, suggesting that Polycomb mechanisms are likely to be important in adult OE neurogenesis.18
BMI1 is known to function as part of a multiprotein complex called Polycomb Repressive Complex 1 (PRC1), which, along with separate PRC2 complexes, classically function to repress the expression of transcription factors and other key regulatory genes in specific cells17, 19, 20 (Fig. 2). For instance, PRCs have been found to be critical for maintaining “stemness” in hematopoietic and intestinal stem cells; repressive epigenetic mechanisms control stem cell functions such as self–renewal or lineage commitment. A key feature of PRCs in embryonic stem cells is to repress developmental regulators, such as Hox or Pax genes, which would lead to lineage differentiation if they were expressed. Here, we further investigate the involvement of Polycomb complex proteins in adult olfactory neurogenesis, in an effort to begin to define possible new therapeutic targets for anosmia.
Figure 2.
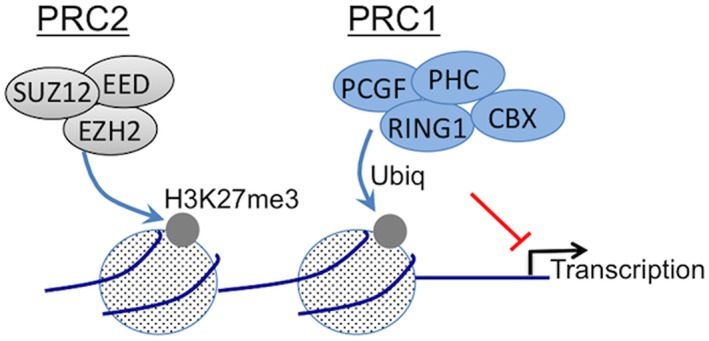
Polycomb complexes silence gene transcription via chromatin structure modulation. As schematized here, Polycomb proteins associate into multiprotein repressive complexes termed PRC1 and PRC2. Polycomb Repressive Complex 2 (PRC2) contains EZH2, which functions as a histone methyl transferase, along with SUZ12 and EED. PRC2 deposits a trimethylation mark at lysine 27 of histone H3 (H3K27me3), which then attracts a canonical PRC1. The composition of PRC1 is variable and complex, suggesting that PRC1 composition affects function. The E3–ligase protein RING catalyzes monoubiquitination of histone H2A at lysine 119 (H2AK119ub) to stabilize transcriptional repression. The experiments here investigate the expression and activity of PRC components in adult OE.
MATERIALS AND METHODS
Animal Use
All experimental procedures were approved by the Institutional Animal Care and Use Committee, and were performed in compliance with NIH Guidelines for the Care and Use of Laboratory Animals. C57BL/6J wild type mice were obtained from Jackson Laboratory (Bar Harbor, ME, USA). Methimazole (Sigma, St. Louis, MO, USA) was dissolved in phosphate buffered saline (PBS) at 5 mg/ml and given at 50 μg/g body weight intraperitoneally (IP).21
Cell Culture
Olfactory tissue was obtained from adult mice, which were euthanized under deep anesthesia, and nasal septal and turbinate mucosa free of bone was harvested and pooled. Basal cell cultures were prepared following our published protocol.18 Briefly, tissue was dissociated in collagenase and dispase solution for 15 min, followed by 3 minutes with 0.125% trypsin. Cells were washed, passed through a 70 μm cell strainer and then GBCs were isolated using APC–conjugated antibody to c–KIT, 1:20 (eBioscience #17–1171, San Diego, CA, RRID:AB_469430) using the EasySep magnetic cell selection kit per instructions (Stem Cell Technologies, Vancouver, BC, Canada). Cells were then plated on vitronectin–coated dishes at approximately 105 cells per well of 6–well plates. Cultures were maintained in NeuroCult NSC Basal Medium, EGF 20ng/mL, FGF2 10ng/mL, heparin 2 µg/mL, (all from Stem Cell Technologies), BMP4 10ng/mL (Peprotech, Rocky Hill, NJ, USA), SB431542 10 uM (Selleck Chemicals, Houston, TX, USA), penicillin–streptomycin (Invitrogen, Carlsbad, CA). Y27632 10 uM (Stem Cell Technologies) was added at the time of splitting cultures, when 80% confluent. Medium was changed every other day. Experiments were performed with cultures below passage 12.
Immunohistochemistry
Cryosections at 10 μm were prepared from adult mouse nasal tissue after perfusion with PBS followed by 4% paraformaldehyde in phosphate buffer. Heat–mediated antigen retrieval was utilized if necessary, using Tris buffer pH 8.0 in a food steamer. For staining, slides were treated in PBS, and blocked using a solution of PBS, 5% normal serum (Jackson ImmunoResearch, West Grove, PA, USA), 4% bovine serum albumin (BSA, Sigma), 5% nonfat dry milk, and 0.3% Triton X–100 (Sigma) for 30 minutes, followed by primary antibody overnight at 4°C (Table I). Slides were rinsed in PBS and incubated with either fluorescent–conjugated secondary antibody or biotinylated secondary (Jackson ImmunoResearch, West Grove, PA, USA) for 30 to 45 minutes in the same blocking solution. For visualization of biotinylated secondary, fluorescein tyramide signal amplification was used (Perkin–Elmer, Waltham, MA, USA). Slides were then rinsed and coverslipped with Vectashield containing 4,6–diamidino–2– phenylindole (DAPI; Vector Labs, Burlingame, CA, USA). Staining was imaged on a Zeiss LSM–710 confocal microscope, and analyzed in ImageJ (http://Imagej.NIH.gov/ij).
Table I.
Primary Antibody Reagents Used in the Present Study.
| Antibody | Supplier | RRID | Concentration |
|---|---|---|---|
| Mouse anti–BMI1 | Abcam #, ab14389, Cambridge, MA, USA | AB_2065390 | 1:750 |
| Rabbit anti–c–KIT | Cell Signaling Technology #3074 | AB_10829442 | 1:30 |
| Rabbit anti EZH2 | Cell Signaling Technology #5246 | AB_10694683 | 1:400 |
| Rabbit anti–SUZ12 | Cell Signaling Technology #3737 | AB_2196850 | 1:400 |
| Rabbit anti–Tri–Methyl_Histone H3 (Lys27) | Cell Signaling Technology #9733 | AB_1147656 | 1:2000 |
| Rabbit anti–CBX8 | Cell Signaling Technology #14696 | AB_2687589 | 1:500 |
| Mouse anti–MEL18 | Santa Cruz Biotechnology #sc–515329 | AB_2687587 | 1:250 |
Western Blotting
Basal cell cultures or mouse nasal septal and turbinate mucosa was collected on ice in RIPA buffer with protease inhibitors (Sigma) and homogenized. Lysate was centrifuged at 14,000 g for 15 minutes and the supernatant stored at −20°C. Protein concentration was measured by Bradford assay. Samples in Laemmli buffer were heated at 100°C for 5 minutes, 25 μg was loaded per lane, separated by 10% bis–Tris PAGE and transferred to PVDF membranes (Bio–Rad). Membranes were blocked in 5% nonfat dried milk in TBS with 0.1% Tween 20 and then incubated overnight with primary antibodies (listed above). Rabbit anti COX IV (Cell Signaling Technology #4850; RRID:AB_2085424) at 1:1000 was used as a normalization control. After incubation with HRP–conjugated secondary antibody diluted in Tris–buffered saline (TBS) with 0.1% Tween 20 and 5% BSA, binding was visualized via chemiluminescence.
Chromosome Immunoprecipitation
ChIP was performed from GBC cultures, using six 10cm dishes per preparation. Crosslinking was performed with fresh paraformaldehyde for 10 min and stopped with glycine. The ChIP–IT Hi–sensitivity kit was used, per protocol (Active Motif, Carlsbad, CA, USA). Briefly, crosslinked chromatin was treated with protease inhibitors and stored at −80°C. Shearing was optimized, using a Bioruptor (Diagenode, Denville, NJ, USA). Approximately 20 x 106 cell equivalents were used per reaction. Immunoprecipitation reactions were performed using validated antibodies: 1 μg anti–H3K27me3 (Cell Signaling Technology #9733) or 2 μg anti–Ring1B (Cell Signaling Technology #5694). Complexes were recovered with Protein G agarose, material was washed, crosslinking was reversed and DNA column purified per protocol. DNA was analyzed using qPCR. Normalization to Input DNA was used as control.22 Input DNA or purified ChIP reaction DNA was used as template. Reactions were prepared using validated commercially developed primers designed to amplify regions known to be bound by Polycomb complexes in embryonic stem cells, for intron 1 of Hoxc10 (#71019, Active Motif) or the 5’ region of Pax2 (#71020, Active Motif), or negative control primers to the promoter region of the beta actin gene, not bound by Polycomb complexes (#71013, Active Motif). PCR reactions were run in triplicate, using SYBRgreen reaction mix on a real time CFX96 cycler (Bio Rad).
RT–qPCR
For quantitative real–time gene expression analysis, total RNA was isolated using column purification (Zymo Research Corp., Irvine, CA, USA). DNase I on–column digestion was performed. First strand cDNA synthesis was performed using Superscript IV (Invitrogen) with oligo–dT primers. Real time qPCR was performed using gene specific primers as indicated (Table II), with SYBRgreen reaction mix (Bio Rad) on a CFX96 cycler (Bio Rad).
Table II.
Primer Sequences Used for RT–qPCR.
| Gene | Primer Direction | mRNA Primer Sequences (5'–3') |
|---|---|---|
| RPO | F | TTCATTGTGGGAGCAGAC |
| R | CAGCAGTTTCTCCAGAGC | |
| EZH2 | F | ATCTGAGAAGGGACCGGTTT |
| R | TGTGCACAGGCTGTATCCTC | |
| RING1A | F | CCTGGACATGCTGAAGAACA |
| R | TCCCGGCTAGGGTAGATTTT | |
| RING1B | F | TTGCGCGGATTGTATTATCA |
| R | GCGCTTCATACTCATCACGA | |
| CBX2 | F | GCAAGCTGGAGTACCTGGTC |
| R | CTGGTTCCTTGAGCTTGGAG | |
| CBX4 | F | GCATCGAAAAGAAGCGGATA |
| R | ACCGGACCTCTCCTTGCTAT | |
| CBX6 | F | GCCGAATCCATCATTAAACG |
| R | TTGGGTTTAGGTCCCCTCTT | |
| CBX7 | F | GAGGCAGAAGCAGACCTGAC |
| R | GCCGCTATTCACAGCTTCTC | |
| CBX8 | F | AGGACGCATGGAATATCTCG |
| R | GGTTTTAGGCTTGGGTCCTC | |
| BMI1 | F | TGTCCAGGTTCACAAAACCA |
| R | TGCAACTTCTCCTCGGTCTT | |
| MEL–18 | F | GAGAATGGAGATGGGGACAA |
| R | ATTCCTTCAGGGGCTCATCT | |
| PHC1 | F | GGGCAACCTATTGCAGGTTA |
| R | GAGCTGAAGCCTGTTGGTTC | |
| PHC2 | F | CCGACTCAGAGATGGAGGAG |
| R | AAAGTCCCACTCGTTTGGTG | |
| PHC3 | F | TACCAGCGGCAGTATTACCC |
| R | TGCAGACTGACAGGAAGGTG |
Statistics
For qPCR analysis, fold–change calculations were performed using the 2–ΔΔCt technique,23 and Ribosomal Protein, large, P0 (RPO) expression was used as a reference. Each cDNA was run in triplicate, and n≥3 independent RNA samples were used for each experimental observation reported. Error bars represent SD, and graphs were prepared using Prism 7.0 (GraphPad Software, Inc.).
RESULTS
PRC2 Components Are Active in Olfactory Basal Cell Cultures and in Vivo
As a first step in determining if PRCs are involved in epigenetic regulation of OE homeostasis, we investigated olfactory PRC2 protein expression. The PRC2 complex is structurally simpler and less heterogeneous than PRC1 (Fig. 2), comprised by EZH2, SUZ12, and EED. Chromatin modification by PRC2 is the initial step in classical gene repression by Polycomb complexes.17, 24, 25, 26 Given the importance of PRCs in pluripotent stem cells, we hypothesized that PRC2 would be active in the globose basal stem cells (GBCs) of the OE. Here, we used an established culture model to purify and expand undifferentiated GBCs for further analysis.18 PRC2 protein expression from GBC–derived cultures and from adult OE tissue extracts was compared by Western blot (Fig. 3A). Western blot indicated that cultures strongly express both SUZ12 and EZH2. Both proteins were also present in adult OE tissue extracts. Furthermore, the repressive histone modification mark H3K27me3, reflective of EZH2 mediated histone methyl–transferase activity, was also identified in both cultures and OE tissue (Fig. 3A). We also confirmed here PRC2 expression in the basal germinal layers of OE tissue sections, as we have reported recently18 (Fig. 3B, C). Protein expression localizes cleanly to the nuclei of GBCs and some immature neurons. Typical basal cell culture morphology is shown (Fig. 3D, E), comprised largely by islands of undifferentiated–appearing cells. We have described in detail the characterization of this culture model previously.18 PRC2 protein expression, along with histone modification marks in OE cultures and in vivo, is consistent with a model in which Polycomb epigenetic mechanisms regulate OE progenitor cells, via transcriptional repression.
Figure 3.
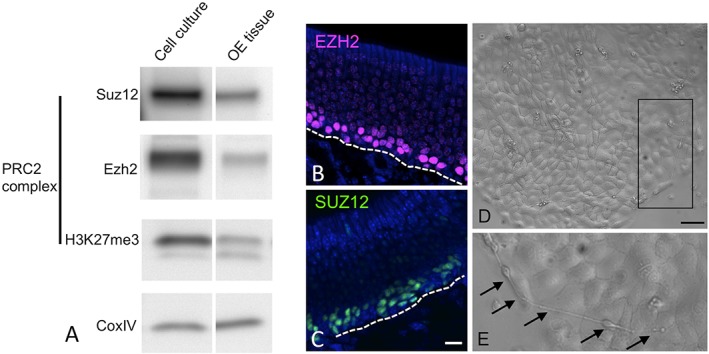
PRC2 components are active in basal cell cultures and normal adult mouse OE. (A) Protein extracts from cultures prepared from purified globose basal stem cells or from adult OE tissue analyzed by Western blot confirm expression of the PRC2 proteins SUZ12 and EZH2, and the H3K27me3 epigenetic mark, indicative of PRC2 enzymatic activity. CoxIV was used as a loading control. By immunohistochemistry, EZH2 (B) and SUZ12 (C) localize selectively to basal progenitor cell layers. Bar=10 μm; dashed line marks the basal lamina; nuclei counterstained with DAPI (blue). (D, E) Light microscopy image of basal cell–derived cultures demonstrates typical undifferentiated–appearing adherent islands; (E) area outlined by Box in D is shown at higher magnification; arrows mark rare cells with bipolar neuronal morphology arising on basal cell islands. Bar=20 μm in D.
PRC2 During Damage–Induced Olfactory Regeneration
To confirm that PRC2 proteins are expressed by stem or progenitor cells in the regenerating OE, tissue sections were prepared from mouse olfactory region one week following experimentally induced damage, using the methimazole model.21 Methimazole injection induces degeneration of the mature cell populations of the OE. Spared basal cells respond by proliferating and reconstituting the OE over the next two weeks.11, 27 One week following lesion, the regenerating OE contains a greatly expanded population of proliferative stem and progenitor cells, many of which express the surface marker c–KIT.27 A mouse model in which the c–KIT+ population is deleted is unable to reconstitute olfactory neurons.27 Immunohistochemical analysis (Fig. 4) confirms that, in the regenerating OE, the key PRC2 protein EZH2 co–localizes extensively with c–KIT, the basal progenitor cell marker, confirming that PRC2–expressing basal stem cells are active during injury–induced adult olfactory regeneration. While there is some c–KIT signal present in the lamina propria of regenerating olfactory mucosa (Fig. 4), likely in axon fascicles, within the epithelium the c–KIT protein localizes cleanly to the cell membrane on the neurogenic GBCs (Fig. 4 inset).27, 28
Figure 4.
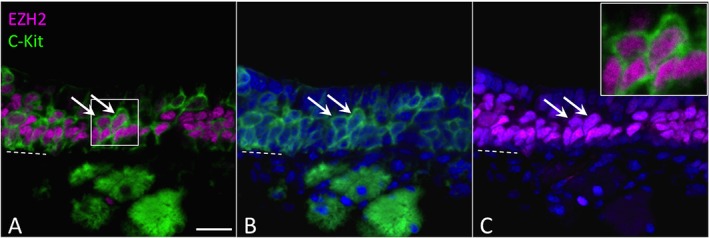
PRC2 proteins are co–expressed with c–KIT, a surface receptor marking globose basal cells, in regenerating OE. Nasal tissue sections prepared from mice 7 days after methimazole–induced lesion were co–stained for c–KIT (green, A, B) and the PRC2 protein EZH2 (magenta, A, C). Most of the c–Kit+ cells are EZH2+ (arrows). Note that c–KIT protein localizes to cell surface while EZH2 localizes to the nucleus; nuclei are co–stained with DAPI (blue in B, C). Previous studies have shown that the c–KIT+ basal cells are required for olfactory neurogenesis27. Dashed line marks basal lamina; bar=20 μm.
PRC1 Subunits Localize to Restricted OE Subpopulations
Epigenetic silencing is crucial to proper cell fate determination in pluripotent or multipotent progenitor cells.26 The specificity of PRC–mediated epigenetic silencing is an active area of research, and evidence suggests that PRC1 heterogeneity is a critical determinant.29, 30, 31 Specifically, canonical PRC1 complexes contain one of five possible CBX subunit variants (Fig. 5A), which are thought to regulate target specificity of a given PRC1, thereby helping to orchestrate the repression of key transcriptional pathways as a multipotent cell determines a lineage choice, ie, neuronal or non–neuronal. In the OE, our previous work has largely focused on the expression of BMI1, a PCGF protein that is a PRC1 component.18 BMI1 was found to be present in a subset of multipotent GBCs, and also in the subset of olfactory neurons that are fully mature, suggesting multiple functions. However, no data have been reported for other PRC1 subunit proteins, such as the CBX variants.
Figure 5.
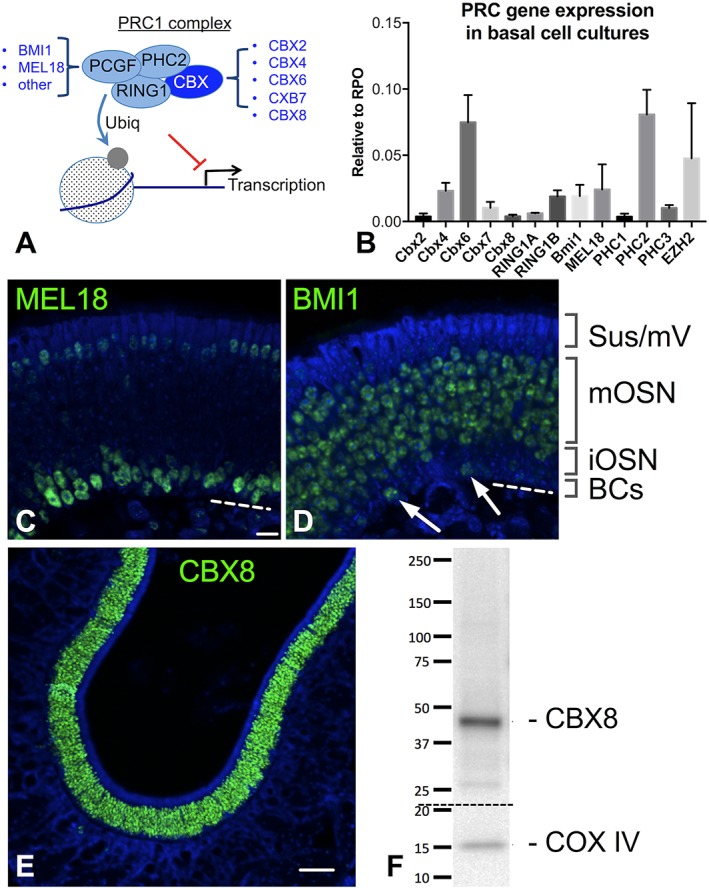
PRC1 subunits localize to restricted OE subpopulations. (A) PRC1 complexes are highly diverse, owing to the multiple variants that exist for each subunit. For instance, a canonical PRC1 contains one of five possible CBX variants and one of six known PCGF variants. (B) Gene expression assays by RT–qPCR from expansion–competent basal cell cultures reveal heterogeneous expression of multiple PRC1 subunits. The PRC2 gene Ezh2, expressed in basal cells, is included for comparison. Gene expression is normalized to RPO, n=3 replicates, bars=SD. (C, D) The PCGF proteins BMI1 (PCGF4) and MEL18 (PCGF2) have complementary expression patterns in adult OE tissue. MEL18 (C) is widely present in globose basal cells (BCs) and immature neurons (iOSN), as well as weakly expressed in the apical sustentacular and microvillar cell layers (Sus/mV); BMI1 (D) is present in mature neurons (mOSN) as well as occasional upstream globose cells (arrows), previously reported to be the SOX2+ subset; note the absence of BMI1 label in the thin iOSN region in D, in contrast to MEL18 label in C. (E, F) CBX8 is a neuron–specific PRC1 subunit in the OE; CBX8 is restricted to the neuronal layers by immunohistochemistry (E); Western blot (F) confirms antigen of correct size is detected by anti–CBX8 from OE extracts. Nuclei are stained with DAPI (blue) C–E, Bar=10μm in C, 50μm in E.
Here, we initially screened cultured GBCs for PRC1 gene expression, by RT–qPCR (Fig. 5B). We detected varying levels of expression of several of the PRC1 subunits, including multiple Cbx orthologs. Although cultures consist largely of undifferentiated basal cell islands, the GBC cultures do contain heterogeneity due to a low level of spontaneous differentiation that may occur18 (see Fig. 3E). Therefore, the detection of multiple PRC1 variants would be expected, assuming that PRC1 composition changes with cell lineage or maturation state. BMI1 (Pcgf4) expression was verified, as reported previously, along with expression of another PCGF variant, Mel18 (Pcgf2). The identification of other PCGF orthologs is of particular interest, given the restricted pattern of BMI1 expression in the OE reported previously, which suggests the possibility that different subpopulations in the OE might utilize specific PRC1 variants.
PCGF subunit expression was further investigated using immunohistochemistry (Fig. 5C, D). The two PCGF variants were found to have essentially complementary expression patterns. MEL18 was found to localize selectively to nuclei of many GBCs and immature neurons, while BMI1 expression was confirmed in the OMP+ mature neuronal layers and occasional basal cells. We previously described in detail the BMI1 expression pattern in the OE, demonstrating that BMI1 is co–expressed in Sox2+ GBCs, is absent in GAP43+ immature neurons, and is strongly expressed in mature OMP+ neurons.18 Additionally, weaker MEL18 signal was identifiable in the sustentacular and microvillar cell layers at the apical region of the OE (Fig. 5C). It is not possible to fully exclude weak expression of BMI1 in some sustentacular cells. Overall, this complementary PCGF expression pattern is consistent with a model in which appropriate epigenetic silencing may be orchestrated by different PRC1 variants during OE lineage differentiation.
CBX8 Is a Neuron–Specific PRC1 Subunit in the OE
Given the importance of CBX orthologs in regulating lineage decisions in pluripotent embryonic stem cells,31 we next sought to investigate CBX activity in adult OE. By RT–qPCR, Cbx8 was only weakly detected in GBC cultures (Fig. 5B). The cultures contain very few neurons, since culture conditions favor renewal rather than differentiation.18 Minimal Cbx8 transcript expression in these cultures suggested that CBX8 might instead be more strongly associated with lineage differentiation, such as neuron–committed cells and their progeny. We identified a validated CBX8 antibody and found that, in OE tissue sections, CBX8 was very cleanly expressed only in the neuronal lineage (Fig. 5E). At low magnification, the restricted expression pattern is evident, with no signal in the apical sustentacular layer, the deepest basal layers, or the extensive lamina propria area beneath the OE. Furthermore, we verified by Western blot that the antibody to CBX8 recognizes a single band of correct size in adult OE tissue extracts (Fig. 5F). A lineage–restricted expression pattern for a specific CBX ortholog is consistent with findings in pluripotent cell studies, which reported that CBX expression can confer lineage specificity by regulating PRC1 chromatin targets. Further mechanistic studies of olfactory CBX8 are warranted, to test the effects of manipulating CBX8 or its chromatin targets on olfactory neurogenesis.
Polycomb Repressive Complexes Are Functional in OE Cells
We next asked whether PRCs are bound to olfactory basal cell chromatin at regulatory regions of transcription factor genes that are known Polycomb targets. Olfactory basal cell cultures were harvested to obtain chromatin, which was analyzed by chromosome immunoprecipitation assay (ChIP–qPCR). Using antibody specific to the PRC2 tri–methylation mark (H3K27me3) or antibody to the PRC1 protein RING1B, ChIP was performed and purified ChIP DNA was analyzed by qPCR (Fig. 6). RING1B was chosen based on our recent report that it is also expressed in OE,18 and because we identified strong Ring1b mRNA expression in GBC cultures (Fig. 5B). Because PRCs bind chromatin in regulatory regions, generally within <1000 nucleotides of transcriptional start sites,17 qPCR using primers specific for 5’ UTR regions of target genes can be used to determine if ChIP DNA contains the target gene of interest. Here, we found that ChIP assays for both PRC2 (H3K27me3) and PRC1 (RING1B) were highly enriched for specific transcription factor DNA. A key feature of PRCs in embryonic stem cells is to repress developmental regulators, such as Hox or Pax genes, which would induce lineage differentiation if they were allowed to be expressed.17 Accordingly, we found that Hoxc10 and Pax2 were enriched 50– to 100–fold compared to a control DNA region in the beta actin gene, which is never PRC–bound (Fig. 6), using antibody to H3K27me3. Also, >10–fold enrichment was observed using the RING1B antibody, which is in accordance with the performance of these reagents. As a control, input DNA samples had no enrichment for Hoxc10 or Pax2. We interpret the results of these binding assays as evidence for the presence of functional PRC complexes, acting to repress specific regulatory genes in olfactory basal cells.
Figure 6.
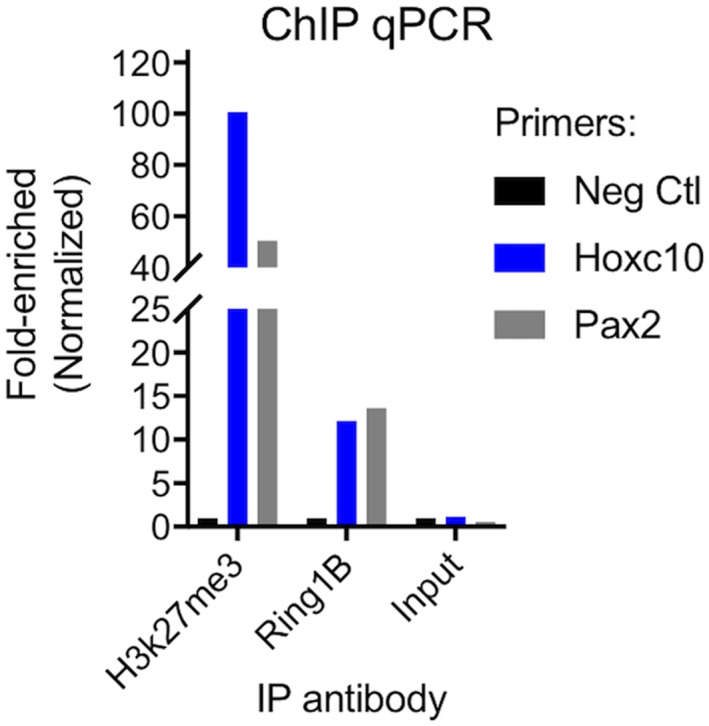
Polycomb repressive complexes are functional in OE cells. ChIP assays were performed on basal cell cultures. Q–PCRs were performed from ChIP DNA with primers to known PRC target genes (Hoxc10 or Pax2) or to a region of the beta actin gene which is not PRC–bound (Neg Ctl). Note that in input DNA, there is no enrichment of target genes compared to negative control; in contrast, PRC target genes are highly enriched in ChIP samples from anti–H3K27me3 or anti–RING1B experiments, consistent with functional Polycomb repressive complexes in OE cells.
DISCUSSION
Epigenetic silencing is crucial to proper cell fate determination in multipotent progenitor cells. Maintenance of the adult OE requires proliferation and appropriate lineage differentiation of multipotent basal progenitor cells.12, 28, 32, 33 In an ongoing effort to understand repair and regeneration processes involved in olfactory maintenance, the present work focuses on examining a family of epigenetic modifiers, Polycomb complex proteins, in adult OE cells. The findings reported here indicate that several key Polycomb proteins are expressed in specific olfactory cell types in normal or regenerating OE. Furthermore, identification of histone modifications (H3K27me3) and results of ChIP assays provide evidence that functional PRC complexes are active in OE cells. Cell type–specific expression of PRC1 subunits such as CBX8 was also identified, suggesting that PRC1–specific epigenetic silencing mechanisms are likely to be involved with lineage differentiation during OE renewal.
Previous studies have indicated that epigenetic mechanisms broadly regulate monoallelic odorant receptor gene expression in maturing olfactory neurons, via lysine–specific demethylase 1 (LSD1) and CoREST (RCOR1).34, 35 It is felt that these mechanisms help to ensure that neurons express only a single odorant receptor gene from about 1000 choices in mice, or 350 choices in humans.8, 36 Another epigenetic modifier, histone deacetylase 2 (HDAC2) also co–localizes in neural precursors and is likely to contribute to this process.37 However, the epigenetic control of multipotential olfactory basal stem and progenitor cells has only recently begun to be appreciated.18
A contribution of Polycomb proteins to regulation of tissue maintenance in the OE is in accordance with other well–studied adult stem cell niches. For instance, BMI1, a PRC1 component, has been extensively studied in adult hematopoietic and intestinal stem cell systems. Fate mapping experiments were used to determine that BMI1 marks a reserve intestinal crypt stem cell population.20 Such studies have also revealed that so–called “facultative” stem cells may respond to tissue injury to be recruited as upstream progenitors. Although such recruitment has been observed in regenerating OE in mice, mechanisms driving this process remain unclear.12 It is intriguing to consider that Polycomb proteins may play a role in redirecting facultative OE stem cells, for instance when GBCs repopulate the reserve stem cell pools following severe injury. In the bone marrow, BMI1 is highly expressed in stem cells and is down regulated upon commitment to differentiation,38 and has been shown to be necessary for self–renewal as well as lineage commitment.19 Given the expression of PRCs in the OE adult stem cell niche, similar regulatory mechanisms are likely to be shared. Indeed, treatment of adult OE basal cell cultures with GSK343, an inhibitor of the PRC2 protein EZH2, led to a dose–dependent reduction in self–renewal.18
The regulation of OE basal cell self–renewal by PRC2, along with the lineage–specific expression of PRC1 subunits in the OE, suggests that PRC–mediated epigenetic silencing is likely to mediate adult OE tissue homeostasis. Experimental approaches such as ChIP–seq, beyond the scope of the present report, will be useful to clarify the genome–wide targets of specific PRCs in the adult OE. Furthermore, because epigenetic modifiers including PRCs are involved in dysregulation of certain cancer cells, novel drugs targeting these proteins are in several stages of development.39 The availability of therapeutic agents with the ability to selectively modulate Polycomb activity may be of interest for investigating Polycomb function and, potentially, regulating tissue renewal in the OE.
CONCLUSION
Taken together, recently published findings regarding Polycomb regulation of OE basal cell culture renewal, along with the expression data and assays reported here, suggest that PRC–mediated epigenetic mechanisms are likely to help regulate renewal and homeostasis in the adult OE.
ACKNOWLEDGMENTS
This work was funded by grants from NIH DC013556 (to B.J.G) and the Triological Society/American College of Surgeons Clinician–Scientist Development Award (B.J.G). The authors have no conflicts of interest to disclose.
Editor's Note: This Manuscript was accepted for publication 25 May 2018.
Triological Society Thesis Paper
BIBLIOGRAPHY
- 1. Doty RL, Shaman P, Applebaum SL, Giberson R, Siksorski L, Rosenberg L. Smell identification ability: changes with age. Science 1984;226:1441–1443. [DOI] [PubMed] [Google Scholar]
- 2. Seiden AM. Postviral olfactory loss. Otolaryngol Clin North Am 2004;37:1159–1166. [DOI] [PubMed] [Google Scholar]
- 3. Holbrook EH, Leopold DA. An updated review of clinical olfaction. Curr Opin Otolaryngol Head Neck Surg 2006;14:23–28. [DOI] [PubMed] [Google Scholar]
- 4. Murphy C, Schubert CR, Cruickshanks KJ, Klein BE, Klein R, Nondahl DM. Prevalence of olfactory impairment in older adults. JAMA 2002;288:2307–2312. [DOI] [PubMed] [Google Scholar]
- 5. Kohli P, Naik AN, Harruff EE, Nguyen SA, Schlosser RJ, Soler ZM. The prevalence of olfactory dysfunction in chronic rhinosinusitis. Laryngoscope 2017;127:309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DeConde AS, Mace JC, Alt JA, Schlosser RJ, Smith TL, Soler ZM. Comparative effectiveness of medical and surgical therapy on olfaction in chronic rhinosinusitis: a prospective, multi–institutional study. Int Forum Allergy Rhinol 2014;4:725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pekala K, Chandra RK, Turner JH. Efficacy of olfactory training in patients with olfactory loss: a systematic review and meta–analysis. Int Forum Allergy Rhinol 2016;6:299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 1991;65:175–187. [DOI] [PubMed] [Google Scholar]
- 9. Carr VM, Farbman AI. The dynamics of cell death in the olfactory epithelium. Exp Neurol 1993;124:308–314. [DOI] [PubMed] [Google Scholar]
- 10. Graziadei GA, Graziadei PP. Neurogenesis and neuron regeneration in the olfactory system of mammals. II. Degeneration and reconstitution of the olfactory sensory neurons after axotomy. J Neurocytol 1979;8:197–213. [DOI] [PubMed] [Google Scholar]
- 11. Leung CT, Coulombe PA, Reed RR. Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat Neurosci 2007;10:720–726. [DOI] [PubMed] [Google Scholar]
- 12. Schwob JE, Jang W, Holbrook EH, et al. Stem and progenitor cells of the mammalian olfactory epithelium: taking poietic license. J Comp Neurol 2017;525:1034–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Loo AT, Youngentob SL, Kent PF, Schwob JE. The aging olfactory epithelium: neurogenesis, response to damage, and odorant–induced activity. Int J Dev Neurosci 1996;14:881–900. [DOI] [PubMed] [Google Scholar]
- 14. Fletcher RB, Das D, Gadye L, et al. Deconstructing olfactory stem cell trajectories at single–cell resolution. Cell Stem Cell 2017;20:817–830 e818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Herrick DB, Lin B, Peterson J, Schnittke N, Schwob JE. Notch1 maintains dormancy of olfactory horizontal basal cells, a reserve neural stem cell. Proc Natl Acad Sci U S A 2017;114:E5589–E5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gokoffski KK, Wu HH, Beites CL, et al. Activin and GDF11 collaborate in feedback control of neuroepithelial stem cell proliferation and fate. Development 2011;138:4131–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006;441:349–353. [DOI] [PubMed] [Google Scholar]
- 18. Goldstein BJ, Goss GM, Choi R, et al. Contribution of Polycomb group proteins to olfactory basal stem cell self–renewal in a novel c–KIT+ culture model and in vivo. Development 2016;143:4394–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oguro H, Yuan J, Ichikawa H, et al. Poised lineage specification in multipotential hematopoietic stem and progenitor cells by the polycomb protein Bmi1. Cell Stem Cell 2010;6:279–286. [DOI] [PubMed] [Google Scholar]
- 20. Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet 2008;40:915–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bergman U, Ostergren A, Gustafson AL, Brittebo B. Differential effects of olfactory toxicants on olfactory regeneration. Arch Toxicol 2002;76:104–112. [DOI] [PubMed] [Google Scholar]
- 22. Kidder BL, Hu G, Zhao K. ChIP–Seq: technical considerations for obtaining high–quality data. Nat Immunol 2011;12:918–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real–time quantitative PCR and the 2(–Delta Delta C(T)) Method. Methods 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- 24. Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 2006;125:301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morey L, Helin K. Polycomb group protein–mediated repression of transcription. Trends Biochem Sci 2010;35:323–332. [DOI] [PubMed] [Google Scholar]
- 26. Vire E, Brenner C, Deplus R, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006;439:871–874. [DOI] [PubMed] [Google Scholar]
- 27. Goldstein BJ, Goss GM, Hatzistergos KE, et al. Adult c–Kit(+) progenitor cells are necessary for maintenance and regeneration of olfactory neurons. J Comp Neurol 2015;523:15–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goss GM, Chaudhari N, Hare JM, et al. Differentiation potential of individual olfactory c–Kit+ progenitors determined via multicolor lineage tracing. Dev Neurobiol 2016;76:241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luis NM, Morey L, Di Croce L, Benitah SA. Polycomb in stem cells: PRC1 branches out. Cell Stem Cell 2012;11:16–21. [DOI] [PubMed] [Google Scholar]
- 30. Morey L, Aloia L, Cozzuto L, Benitah SA, Di Croce L. RYBP and Cbx7 define specific biological functions of polycomb complexes in mouse embryonic stem cells. Cell Rep 2013;3:60–69. [DOI] [PubMed] [Google Scholar]
- 31. Morey L, Pascual G, Cozzuto L, et al. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell 2012;10:47–62. [DOI] [PubMed] [Google Scholar]
- 32. Huard JM, Youngentob SL, Goldstein BJ, Luskin MB, Schwob JE. Adult olfactory epithelium contains multipotent progenitors that give rise to neurons and non–neural cells. J Comp Neurol 1998;400:469–486. [PubMed] [Google Scholar]
- 33. Goldstein BJ, Fang H, Youngentob SL, Schwob JE. Transplantation of multipotent progenitors from the adult olfactory epithelium. Neuroreport 1998;9:1611–1617. [DOI] [PubMed] [Google Scholar]
- 34. Magklara A, Yen A, Colquitt BM, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell 2011;145:555–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lyons DB, Allen WE, Goh T, Tsai L, Barnea G, Lomvardas S. An epigenetic trap stabilizes singular olfactory receptor expression. Cell 2013;154:325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Monahan K, Lomvardas S. Monoallelic expression of olfactory receptors. Annu Rev Cell Dev Biol 2015;31:721–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Coleman JH, Lin B, Schwob JE. Dissecting LSD1–dependent neuronal maturation in the olfactory epithelium. J Comp Neurol 2017;525:3391–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hosen N, Yamane T, Muijtjens M, Pham K, Clarke MF, Weissman IL. Bmi–1–green fluorescent protein–knock–in mice reveal the dynamic regulation of bmi–1 expression in normal and leukemic hematopoietic cells. Stem Cells 2007;25:1635–1644. [DOI] [PubMed] [Google Scholar]
- 39. Comet I, Riising EM, Leblanc B, Helin K. Maintaining cell identity: PRC2–mediated regulation of transcription and cancer. Nat Rev Cancer 2016;16:803–810. [DOI] [PubMed] [Google Scholar]


