Phage-antibiotic synergy (PAS) has been reported for a decade, but the underlying mechanism has never been vigorously investigated. This study shows the presence of PAS from a variety of phage-bacterium-antibiotic pairings. We show that increased phage production resulted directly from a lysis delay caused by the relative shortage of holin in filamented bacterial hosts in the presence of sublethal concentrations of stress-inducing substances, such as antibiotics and reactive oxygen species (ROS).
KEYWORDS: filamentation, holin, lysis delay, phage-antibiotic synergy, recA, stress
ABSTRACT
When phages infect bacteria cultured in the presence of sublethal doses of antibiotics, the sizes of the phage plaques are significantly increased. This phenomenon is known as phage-antibiotic synergy (PAS). In this study, the observation of PAS was extended to a wide variety of bacterium-phage pairs using different classes of antibiotics. PAS was shown in both Gram-positive and Gram-negative bacteria. Cells stressed with β-lactam antibiotics filamented or swelled extensively, resulting in an increase in phage production. PAS was also sometimes observed in the presence of other classes of antibiotics with or without bacterial filamentation. The addition of antibiotics induced recA expression in various bacteria, but a recA deletion mutant strain of Escherichia coli also showed filamentation and PAS in the presence of quinolone antibiotics. The phage adsorption efficiency did not change in the presence of the antibiotics when the cell surfaces were enlarged as they filamented. Increases in the production of phage DNA and mRNAs encoding phage proteins were observed in these cells, with only a limited increase in protein production. The data suggest that PAS is the product of a prolonged period of particle assembly due to delayed lysis. The increase in the cell surface area far exceeded the increase in phage holin production in the filamented host cells, leading to a relatively limited availability of intracellular holins for aggregating and forming holes in the host membrane. Reactive oxygen species (ROS) stress also led to an increased production of phages, while heat stress resulted in only a limited increase in phage production.
IMPORTANCE Phage-antibiotic synergy (PAS) has been reported for a decade, but the underlying mechanism has never been vigorously investigated. This study shows the presence of PAS from a variety of phage-bacterium-antibiotic pairings. We show that increased phage production resulted directly from a lysis delay caused by the relative shortage of holin in filamented bacterial hosts in the presence of sublethal concentrations of stress-inducing substances, such as antibiotics and reactive oxygen species (ROS).
INTRODUCTION
Phage-antibiotic synergy (PAS) refers to the phenomenon where phage production increases in the presence of sublethal concentrations of certain antibiotics (1). For example, increases have been observed in the case of ϕMFP and T4-like phages in Escherichia coli with β-lactam and quinolone antibiotics, prophages of E. coli with ampicillin (2), phiPVP-SE1 (Salmonella enterica serovar Enteritidis), phiPVP-SE2 (Salmonella enterica serovar Enteritidis), phi IBB-PF7A (Pseudomonas fluorescens), and phiIBB-SL58B (Staphylococcus lentus) with ampicillin, cefotaxime, or tetracycline (3), prophages of Pseudomonas aeruginosa with ciprofloxacin (4), phage Sp5 of E. coli with mitomycin C (5), MR-5 and 7 other phages of Staphylococcus aureus with linezolid, tetracycline, or ketolide antibiotics (6), phage T4 of E. coli with cefotaxime (7), phages σ-1 and 001A of P. aeruginosa with ceftriaxone (8), phages KS12 and 14 of Burkholderia cenocepacia with meropenem, ciprofloxacin, or tetracycline (9), and phage EcSw of E. coli with ampicillin, tetracycline, penicillin, or kanamycin (10). It has been suggested that PAS is dependent on bacterial filamentation and sometimes an SOS response (1). Lysogenized phage P1 in E. coli showed PAS in the presence of ciprofloxacin, and a P1 Ref endonuclease amplified the lytic cycle when a bacterial SOS response was induced by DNA damage (11). A synergistic effect was also observed when removing P. aeruginosa and S. aureus biofilms (12, 13). The degree of synergy depended upon the specific antibiotic used (14).
The bacteriophage-mediated lysis of Gram-negative bacteria usually occurs in three steps: phage holins make holes in the inner membrane, phage endolysin degrades the cell wall, and a spanin complex disrupts the outer membrane (15). Importantly, holins accumulate harmlessly in the cytoplasmic membrane until triggered at an allele-specific time to form micron-scale holes, thereby determining the phage lysis time.
Antibiotics prompt an SOS response in bacteria (16, 17, 18), which is usually induced by recA and sometimes accompanies bacterial filamentation by inhibiting ftsZ (19, 20, 21).
Accordingly, this study investigated the PAS effect of various combinations of bacteria, phages, and antibiotics. Stresses other than antibiotics were also tested. We reveal the underlying mechanisms of the PAS effect in relation to stress-induced bacterial filamentation and lysis timing.
RESULTS
We hypothesized that the increase in phage production was related to three features: a change in the size of the production facility, a change in the availability of viral components, and/or a change in particle assembly period. Thus, we tested each possibility.
Change in size of production facility: bacterial morphological changes in the presence of antibiotics.
The test strains were investigated using sublethal doses of 8 different antibiotics (Table 1). In general, all the bacterial strains tested showed some degree of resistance to ampicillin and sulfamethoxazole, while the P. aeruginosa strain showed additional resistance to other antibiotics. Each strain was cultured in the presence of sublethal antibiotic doses and observed by light microscopy for any change in morphology (see Fig. S1 in the supplemental material). Many strains exhibited either bacterial swelling (for cocci) or extensive filamentation (for rods). Under the same conditions, the host bacteria were also infected with various phages, and the plaque sizes measured (Table 2). The bacterial swelling or filamentation was generally accompanied by increased phage production. However, there were some cases where the phage production increased without any bacterial morphological changes (indicated in Table 2).
TABLE 1.
Sublethal doses of antibiotics used with test strains
| Bacterial strain | Concn (μg/ml)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| AP | CTX | KM | TET | CPFX | MitC | TMP | SMX | |
| S. aureus | 64 | 0.5 | 4 | 1 | 0.5 | 0.12 | 0.25 | 128 |
| B. cereus | 65 | 64 | 8 | 0.06 | 0.5 | 0.12 | 32 | 128 |
| E. faecalis | 4 | 0.5 | 16 | 0.5 | 2 | 0.25 | 0.5 | 128 |
| E. coli (K-12) | 8 | 0.01 | 8 | 0.06 | 0.03 | 2 | 0.12 | 128 |
| E. coli (Crooks) | 4 | 0.06 | 8 | 0.25 | 0.01 | 4 | 0.03 | 128 |
| P. aeruginosa | 32 | 32 | 128 | 4 | 0.25 | 4 | 64 | 128 |
AP, ampicillin; CTX, cefotaxime; KM, kanamycin; TET, tetracycline; CPFX, ciprofloxacin; MitC, mitomycin C; TMP, trimethoprim; SMX, sulfamethoxazole.
TABLE 2.
Changes in plaque size and host cell morphology with sublethal antibiotic doses
| Host bacteria | Phage | Phenotypea |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| AP | CTX | KM | TET | CPFX | MitC | SMX | TMP | ||
| Gram positive | |||||||||
| S. aureus | SA11 | + S | ++ S | − | + S | + S | +++ S | − | + S |
| E. faecalis | PBEF7 | +++ S | +++ S | +b | − | + S | ++ S | +b | + S |
| PBEF9 | +++ S | +++ S | +b | − | + S | ++ S | − | + S | |
| B. cereus | PBBC03 | + F | +++ F | + F | + F | +++ F | +++ F | − | − |
| Gram negative | |||||||||
| E. coli K-12 | T4 | ++ F | + F | − | ++ F | ++ F | ++ F | − | ++b |
| E. coli Crooks | PBEC22 | ++ F | ++ F | − | + F | ++ F | ++ F | +b | + Fb |
| PBEC24 | ++ F | ++ F | +b | ++ F | +++ F | ++ F | ++b | + F | |
| PBEC82 | +++ F | ++ F | − | ++ F | +++ F | ++ F | − | + Fb | |
| P. aeruginosa | PA26 | ++ F | ++ F | +b | + F | ++b | ++b | − | ++ F |
| PA22 | ++ F | ++ F | +b | − Fb | ++b | ++b | +b | ++ F | |
| PA25 | + F | +++ F | +b | + F | ++b | +++b | +b | ++ F | |
AP, ampicillin; CTX, cefotaxime; KM, kanamycin; TET, tetracycline; CPFX, ciprofloxacin; MitC, mitomycin C; TMP, trimethoprim; SMX, sulfamethoxazole; S, swelling; F, filamentation; +, >30% increase in plaque diameters; ++, >60% increase in plaque diameters; +++, >90% increase in plaque diameters; −, no change.
Phage production increased in the absence of a bacterial morphological change.
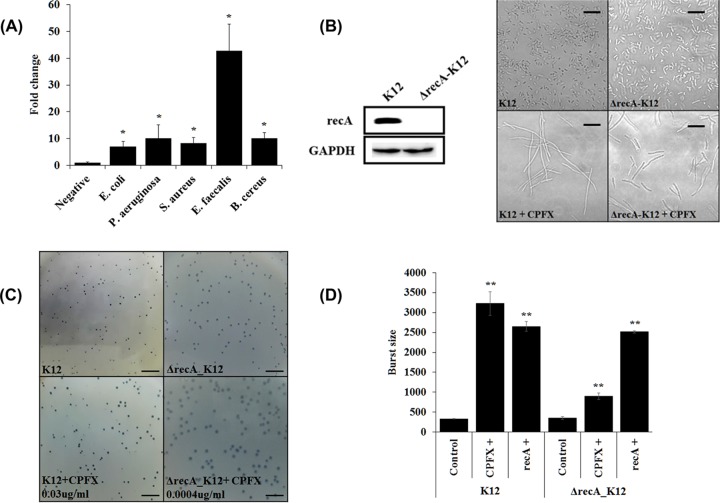
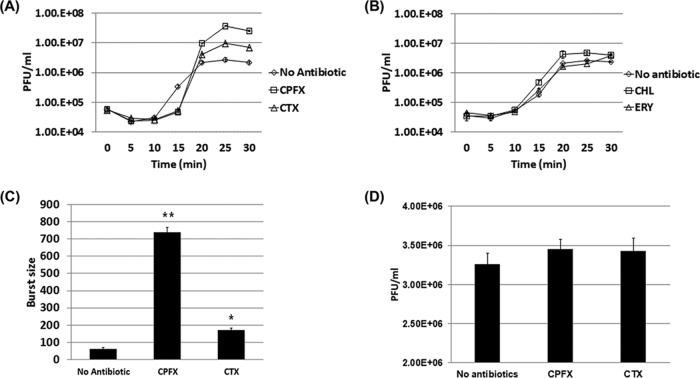
Bacterial filamentation normally results from an SOS response mediated by recA expression (22). In the presence of a sublethal dose of ciprofloxacin, all five bacteria showed an increased expression of recA (Fig. 1A). Under the same conditions, all the bacteria exhibited bacterial filamentation or swelling, except for the P. aeruginosa strain (Table 2), which showed an increased resistance to the various antibiotics tested (Table 1). The genetic and/or physiological alterations in this particular strain may have elicited some unusual responses to filamentation. The P. aeruginosa strain used showed PAS without filamentation when ciprofloxacin was present. Interestingly, the resistance to this antibiotic was not particularly higher. Notwithstanding, the E. coli strain with a recA deletion mutation still showed filamentation, to a lesser degree, in the presence of ciprofloxacin (Fig. 1B). Moreover, the same strain produced larger plaques in the presence of the antibiotic, although at a concentration lower than the sublethal dose (Fig. 1C). The burst size was tested in a recA deletion mutant host (Fig. 1D). When the sublethal dose of ciprofloxacin was added, the burst size of phage T4 from the mutant host increased 3-fold compared to that without the antibiotic. However, the effect was not as great as that seen with the wild-type bacteria. The increase in the burst size recovered when exogenous recA was overexpressed. Taken together, the addition of antibiotics and/or the overexpression of recA led to an increase in burst size. However, it is possible that there may be a different route, other than a recA-mediated SOS response and filamentation, leading to the increased phage production in the presence of antibiotics.
FIG 1.
Relationship between recA expression and phage production. (A) Determination of recA expression level in different bacteria in the presence of a sublethal dose of ciprofloxacin using real-time RT-PCR. Negative, without antibiotic treatment and given a value of 1 for comparison purposes; *, P < 0.05. (B) Bacterial morphological change was observed under a light microscope. E. coli K-12 strain or its recA deletion mutant (ΔrecA K-12, see Materials and Methods) was grown in the presence or absence of the sublethal dose of ciprofloxacin; (left) Western blot analysis showing a lack of RecA protein expression. Scale bar, 10 mm. (C) Observation of plaque sizes when phage T4 infected E. coli K-12 strain or its recA deletion mutant (ΔrecA K-12) in the presence of different doses of ciprofloxacin. Scale bar, 10 mm. (D) Burst sizes measured when phage T4 infected wild-type or recA deletion mutant strain (ΔrecA K-12) of E. coli K-12 in the presence or absence of the sublethal dose of ciprofloxacin. **, P < 0.01.
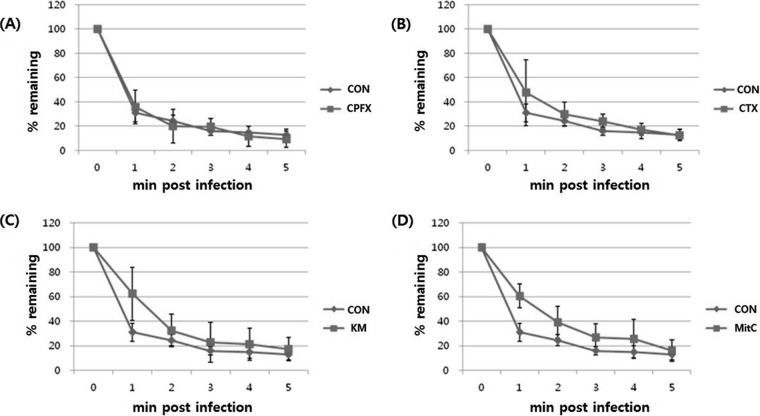
Next, to determine whether the morphological changes to the host bacteria affected phage adsorption, the viral adsorption was checked in the presence of four different antibiotics (Fig. 2). While ciprofloxacin showed no effect on phage adsorption, cefotaxime, kanamycin, and mitomycin C produced an initial retardation of phage adsorption. However, 5 min postinfection, the levels of phage adsorption were the same with or without antibiotics.
FIG 2.
Adsorption of phage T4 to E. coli K-12 strain (MOI = 0.01) in the presence of sublethal concentrations of various antibiotics. After phages were added to a fresh culture of E. coli, the mixture was incubated at 37°C. Aliquots of the culture were taken at the indicated time points and the titer of free phages was measured. Remaining phages were counted every minute for 5 min postinfection. CON, control. (A) CPFX, ciprofloxacin; (B) CTX, cefotaxime; (C) KM, kanamycin; (D) MitC, mitomycin C.
Change in availability of viral components.
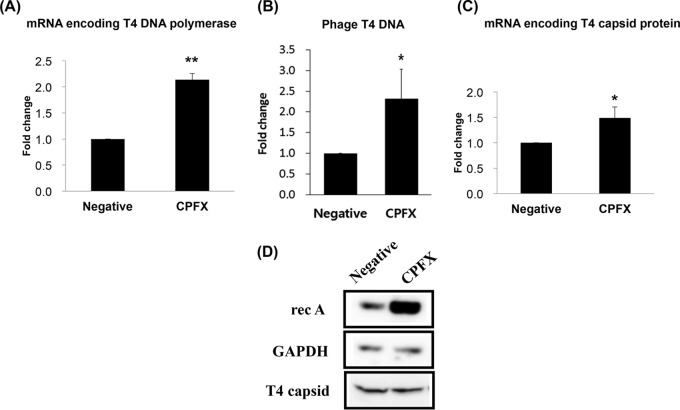
Once inside, an invading phage genome undergoes gene expression and DNA replication, where early gene expression usually occurs first, followed by DNA replication. The virion proteins are then expressed and phage assembly occurs. Therefore, in the present study, we checked for any change in viral component production (Fig. 3). The mRNA encoding the phage DNA polymerase increased 2-fold when ciprofloxacin was added (Fig. 3A), thereby increasing the phage DNA production (Fig. 3B). Polymerase is responsible for the replication of viral genomic nucleic acids, and its availability directly affects the number of new copies of genomic nucleic acid produced (23). Notwithstanding, the mRNA encoding the phage major capsid protein only increased 1.5-fold (Fig. 3C), and the increase in the resulting protein product (T4 capsid protein) in the filamented cells remained unchanged (Fig. 3D). Nonetheless, the production of one phage protein, holin, was increased (see Fig. 5).
FIG 3.
Changes in transcription of phage T4 mRNA and replication of phage DNA in the presence of a sublethal concentration of ciprofloxacin (CPFX) as measured by real-time RT-PCR or real-time PCR. Total RNA or total DNA was isolated from T4-infected E. coli 13 min postinfection and analyzed. (A) mRNA encoding T4 DNA polymerase; (B) phage T4 genomic DNA; (C) mRNA encoding T4 major capsid protein; (D) Western blot analysis of bacterial RecA protein and phage T4 major capsid protein. GAPDH was used as an internal control. *, P < 0.05; **, P < 0.01.
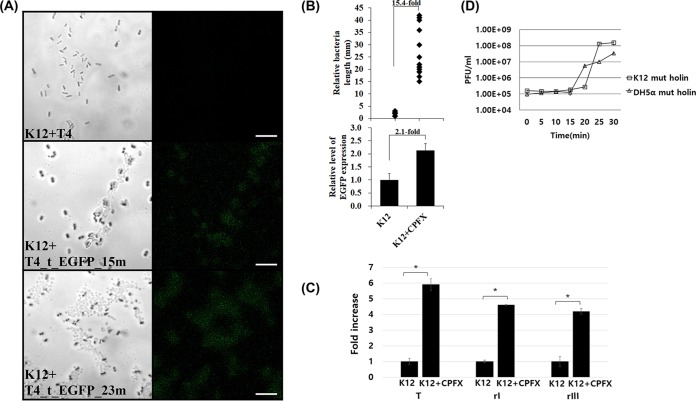
FIG 5.
Underlying mechanism of the delayed lysis in the presence of sublethal doses of CPFX. (A) Observation of T4 holin (t) fused to enhanced green fluorescent protein (EGFP) expressed in bacterial host under a fluorescence microscope at 0, 15, and 23 min postinfection. T4_t_EGFP, engineered phage T4 expressing EGFP-fused holin (see Materials and Methods). Scale bar, 10 mm. (B) Increase in bacterial length due to filamentation and increase in T4 holin production in the presence of the sublethal dose of ciprofloxacin. (C) Real-time RT-PCR measurement of phage T4 holin and antiholins. T, T4 holin; rI and rIII, T4 antiholins. Total RNA was isolated from T4-infected E. coli grown in the presence of ciprofloxacin at 20 min postinfection and subjected to a real-time RT-PCR. *, P < 0.05. (D) One-step multiplication curve of phage T4 in bacterial host expressing the nonsense mutant t (holin) gene (tamA3) from a plasmid. The host is either the suppressor strain (DH5α) or the wild type (K-12). MOI, 0.001.
Change in assembly period: delayed lysis.
The results described above raise a very important question, as a slight increase in the phage component did not explain the much higher increase in the phage burst size, even if we consider an enlarged facility. One hypothesis that could explain this higher increase is that there was a prolonged assembly. To test this hypothesis, a time delay in bacterial lysis was first investigated after phage infection with the sublethal antibiotic concentrations.
At 20 min postinfection without the addition of any antibiotic, the increase in the numbers of phage T4 stopped, indicating that bacterial burst occurred (Fig. 4A). However, in the presence of a sublethal dose of cefotaxime or ciprofloxacin, the increase in the number of phages continued until 25 min postinfection. Thus, the vegetative cycle was prolonged and bacterial lysis was delayed by 5 min. However, in the presence of chloramphenicol or erythromycin, to which the host bacteria showed a higher degree of resistance, no such effect was observed (Fig. 4B). With the lysis delay, the increase in burst size was 2.5-fold (cefotaxime) or 10.5-fold (ciprofloxacin) (Fig. 4C). When the infected cells were lysed at 17 min postinfection by the addition of chloroform, there was little difference between the phage titers from untreated cultures and those from cultures that were treated with antibiotics (Fig. 4D).
FIG 4.
Delayed lysis of T4-infected E. coli K-12 strain in the presence of sublethal doses of different antibiotics. (A) One-step multiplication curve of phage T4 with or without the antibiotics inducing PAS. CPFX, ciprofloxacin; CTX, cefotaxime. An aliquot of the culture was taken, and the titers of free phages were measured at each time point postinfection (MOI = 0.001). (B) One-step multiplication curve of phage T4 with or without the antibiotics not inducing PAS. CHL, chloramphenicol; ERY, erythromycin. MICs were 16 mg/ml and 256 mg/ml for CHL and ERY, respectively. One-half MIC was used as the sublethal dose for this experiment. (C) Burst sizes measured in the presence of antibiotics inducing PAS. *, P < 0.05; **, P < 0.01. (D) Phage titers after a forced lysis at 17 min postinfection with the addition of chloroform (5% [vol/vol]) to the culture.
As a lysis delay was established, the reason for this delay was then investigated further. When the phage components for virion assembly are produced, phage holins and endolysins are translated and create exits for the assembled phages. We hypothesized that phage holins had a certain role in the lysis delay, since these proteins need to be inserted into the bacterial membrane and accumulate to create a hole for the transport of endolysins for degrading the peptidoglycan cell wall (24, 25, 26). A bacterial recombination technique was used to create an engineered phage T4 that harbored a gene encoding the enhanced green fluorescent protein (EGFP) fused to the 3′ end of the holin gene in the T4 genomic DNA. This fusion with EGFP enabled phage holin expression to be quantitated with a fluorometer and its localization traced under a fluorescence microscope. The holin-EGFP was detected in the bacterial cells infected with the engineered T4 (Fig. 5A), and an increased green fluorescence was observed over time. When the bacterial cells filamented in the presence of the sublethal dose of ciprofloxacin, the increase in the bacterial length was 15.4-fold (Fig. 5B). However, the increase in green fluorescence was only 2.1-fold under the same conditions. Although the holin amount was higher in the presence of the antibiotic, the bacterial facility was much longer (and accordingly, had a much larger membrane surface area), leading to a relative shortage of holins to aggregate and form a hole in the bacterial membrane. Furthermore, since antiholins are similarly important for determining the lysis time (27, 28), the transcription levels of the T4 antiholins RI and RIII were also checked under the same conditions. mRNAs encoding T4 holin (T) increased 5.8-fold in the presence of a sublethal dose of ciprofloxacin, while mRNAs encoding antiholins increased 4.8-fold or 4.2-fold for RI or RIII, respectively (Fig. 5C). Although holin expression was a little higher than that of the antiholins, the difference was not big enough to explain the delayed lysis. The differences between the actual amounts of protein would be smaller after translation. To further investigate the importance of delayed lysis due to a shortage of holin molecules, we prepared host bacteria expressing a tamA3 mutant holin (29) from a plasmid. The tamA3 mutation was a C259T point mutation introducing a premature stop codon at the 87th amino acid. The prematurely terminated version of the protein harbors the two transmembrane domains and induces very little lysis but accumulates intracellular phages in a wild-type strain (29). When this mutant holin was expressed from a plasmid in a wild-type host, a delay in lysis and an increased burst size were observed (Fig. 5D). This phenomenon was absent in a suppressor strain.
Influences of stresses other than antibiotics.
An SOS response is induced when bacteria experience stress from antibiotics. However, bacteria are also known to respond to other stresses, including reactive oxygen species (ROS) and heat. Therefore, in this study, we tested whether these two stresses also resulted in increased phage production. In the presence of a sublethal concentration of H2O2, bacterial filamentation was clearly observed (Fig. 6A). Bacterial filamentation also occurred at 45°C, though not as much as in the case of ROS. Accordingly, an increase in phage production was clearly observed in the presence of H2O2, and a slight increase was observed at the elevated temperature (Fig. 6B).
FIG 6.
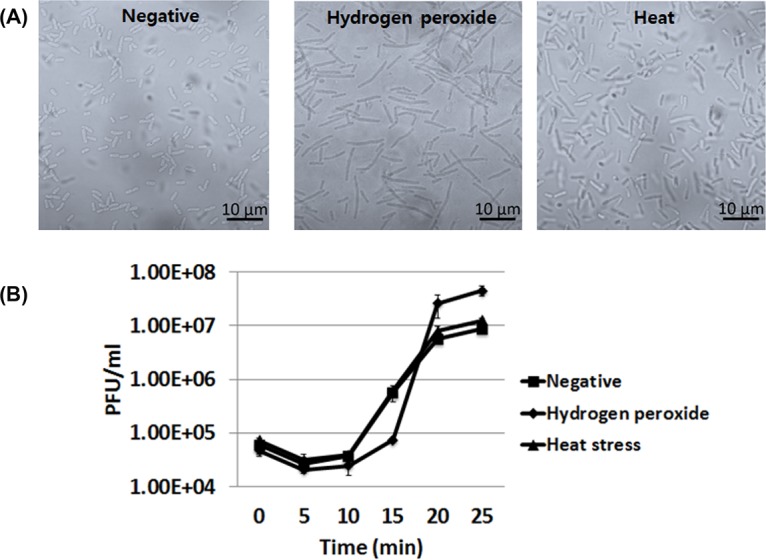
Changes in phage T4 production in the presence of other stresses. (A) Bacterial filamentation in the presence of the sublethal dose of hydrogen peroxide (4.5 mM) or elevated temperature (45°C) was observed under a light microscope. (B) One-step multiplication curve of phage T4 in the presence of stresses other than antibiotics. After phages were added to a fresh culture of host bacteria, an aliquot of the culture was taken and the titers of free phages were measured at each time point postinfection (MOI = 0.001).
DISCUSSION
Among the tested antibiotics, kanamycin and sulfamethoxazole do not lead to notable PAS. Interestingly, most of the bacteria used in this study had a relatively higher resistance to these two antibiotics than to the others tested. Thus, the genetic and/or physiological alterations responsible for this resistance may have led to the limited effect of these antibiotics. The tested bacteria also generally had a higher resistance to ampicillin; yet, PAS was observed in this case. Different resistance mechanisms might lead to different outcomes in PAS. In most cases, PAS was accompanied by bacterial filamentation (rods) or swelling (cocci).
As previously suggested (1), PAS was essentially dependent on an SOS response and bacterial filamentation. However, PAS was also observed when a mutant E. coli with a recA deletion was used as the host. This mutant strain did not induce an SOS response yet still exhibited bacterial filamentation in the presence of the sublethal antibiotic concentration. A filamentation mechanism other than that for an SOS response remains to be identified. Moreover, there were even cases where PAS was observed without bacterial filamentation. Antibiotic resistance may be involved, since the P. aeruginosa strain showing no filamentation exhibited an unusually high resistance to most of the antibiotics tested.
As the production facility increased with bacterial filamentation, more substrates were available for the production of early components (early proteins and genomic DNAs), leading to a notable increase in the production of phage DNA polymerase and phage genomic DNA. Normally, early proteins are not produced to the maximum capacity of the transcription/translation machineries, as they are not in such high demand. The reason for no increase in the phage capsid protein under the same conditions may have been the usual excess production of structural proteins. Essentially, there was little room for an increase in the translation of the capsid protein relative to that of early proteins. Among the late proteins, phage capsid proteins are mostly needed for virion assembly, which requires the highest number of copies. Thus, the higher increase of holins than of the capsid protein was not surprising.
The reason for the increased phage production was not the increased availability of phage components but rather the prolonged assembly period. Generally, phage holins accumulate in the bacterial membrane until a signal triggers oligomerization, followed by pore formation for the transport of endolysins (26). Cell lysis occurs when the holins reach a critical concentration and nucleate to form rafts (30, 31). Antiholins are also produced to counteract holins on the basis of the proton motive force (PMF)-dependent blocking of the holin topological alterations required for triggering lysis (32). In the case of phage T4, the timing of T (holin)-mediated lysis appears to be genetically malleable, and the host density and environmental conditions also affect the lysis timing (33). In the present experiments, more T4 holins accumulated in the presence of sublethal antibiotic concentrations. However, when the membrane surface was significantly increased with cell filamentation, the creation of the membrane raft was slower due to a relative shortage of aggregated holins. As a result, the vegetative cycle continued and lysis was delayed, leading to a prolonged assembly of new phage particles. The increase of transcription of T4 antiholins was slightly lower than that of holins, suggesting that the delayed lysis was not based on the mechanism triggered by antiholins.
Phage-antibiotic synergy (PAS) is a term that describes the enhanced production of phages in the presence of sublethal doses of antibiotics. On the basis of the findings presented here, this phenomenon may also extend to stresses other than antibiotics and thus should be called “stress-induced increase of phage production.” In this study, both ROS and heat stress induced bacterial filamentation and increased phage production. However, the effect of heat was not as strong, since the cellular protein functions of replication, transcription, and translation for supporting phage production are not normal at an elevated temperature.
The PAS effect is an example of a mechanism to which phages have adapted and which enables them to benefit from the variability in their environment. The presence of antibiotics endangers host bacterial survival and ultimately phage propagation. In addition to antibiotics, there are numerous agents that induce stress to bacteria and eventually cause death. Actually, bacteria frequently encounter these situations in nature. To cope with this disadvantageous situation, phages must maximize their production once they find live hosts suitable for infection. The physiology of hosts has already been altered due to the presence of stress, and the resulting PAS enables the phages to take advantage of the situation. It is not uncommon to find viral adaptation to mammalian hosts. For example, viral resistance to interferon responses of the host is well documented for hepatitis C virus (HCV) (34) and myriad other viruses. Viral proteins, including core, E2, NS3/4A, and NS5A of HCV, are responsible for blocking downstream signaling of the JAK-STAT pathway and interferon stimulatory genes. For bacteriophages, the majority of the adaptation is observed at the genomic level. For example, an altered genomic architecture was found in Shigella phage sf6 for faster life cycles (35). The involvement of the clustered regularly interspaced short palindromic repeat (CRISPR) system in bacterium-phage coevolution is another example (36). An expansion of the phage host range via the accumulation of multiple genetic mutations has also been reported (37). The employment of the phage holin concentration relative to host membrane surface area for controlling lysis timing reported in this study is a notable example of beneficial adaptation without the involvement of genetic alterations.
These results suggest that bacterial infections might be treated more efficiently with both a low dose of antibiotic and phages, thanks to the PAS effect. This should also help suppress the emergence of antibiotic-resistant pathogenic bacteria. Additionally, the PAS effect may facilitate the production of therapeutic bacteriophages in less time and at a lower cost.
MATERIALS AND METHODS
Bacterial strains, phages, and growth conditions.
The Gram-positive bacteria included Staphylococcus aureus (ATCC 13301), Enterococcus faecalis (KCTC 2011), and Bacillus cereus (KCTC 1012). The Gram-negative bacteria included Escherichia coli Crooks strain (ATCC 8739), Escherichia coli K-12 strain MG1655 (ATCC 700926), and Pseudomonas aeruginosa (ATCC 13388). All these strains were used as host strains for the bacteriophages. The bacteria were grown in Luria-Bertani (LB) medium at 37°C. All of the phages were obtained from the Bacteriophage Bank of Korea (http://www.phagebank.or.kr/). Phage SA11 was used to infect S. aureus. Phages PBEF7 and PBEF9 were used to infect E. faecalis. Phage T4 was used to infect E. coli MG1655. Another strain (Crooks) of E. coli (ATCC 8739) was infected with phages PBEC22, PBEC24, and PBEC82. P. aeruginosa was infected with phages PA22, PA25, and PA26. E. coli DH5α harbors a supE44 mutation, which confers a weak suppressor activity.
Bacteriophage growth and purification.
Standard phage techniques were used as described previously (38). Briefly, dilutions of phage suspension were mixed with host bacteria in a dilute molten agar matrix (the “top agar” or “overlay”) containing 0.7% (wt/vol) agar in LB broth, which was distributed evenly to solidify on a standard agar plate (the “bottom agar” or “underlay”). Antibiotics were added to both top and bottom agars at the same concentration where needed. After an overnight incubation, plaques were visualized as zones of clearing (or diminished growth) in the bacterial lawn, which grew in the overlay. A burst size was calculated by dividing the number of phages after a single burst by the number of phages initially added to the culture. The phages were purified using centrifugation and a glycerol gradient method, as described elsewhere (39).
Antibiotics.
Ampicillin sodium salt was purchased from Carl Roth, Germany. Cefotaxime sodium salt, tetracycline hydrochloride, and kanamycin monosulfate were purchased from Duchefa Biochemie, Germany. Ciprofloxacin was purchased from Sigma-Aldrich, USA. Each antibiotic was dissolved in distilled water and filter sterilized. Sulfamethoxazole was purchased from Duchefa Biochemie and dissolved in ethanol. Trimethoprim was purchased from Sigma-Aldrich and dissolved in dimethyl sulfoxide (DMSO). Mitomycin C was purchased from Duchefa Biochemie and dissolved in ethanol and NaCl.
Determination of MIC and sublethal dose.
A 1:2 serial dilution of each antibiotic stock solution was added to each bacterial culture in glass tubes. The lowest concentration of antibiotics with no bacterial growth was designated the MIC. A sublethal dose was defined as one-half the MIC.
Phage plaque size measurement.
Phage plaques were allowed to form on a bacterial lawn and the size was measured with a digital caliper.
Construction of engineered T4 expressing EGFP fused to holin.
The gene encoding EGFP was inserted into T4 genomic DNA to create a fusion protein of EGFP and holin based on homologous recombination, as previously reported (40). Briefly, DNA was synthesized to include 50 bp upstream of the T4 holin open reading frame (ORF), T4 holin-EGFP, and 50 bp downstream of the T4 holin ORF (Bioneer, South Korea). Five hundred nanograms of the synthesized DNA was then mixed with competent E. coli cells harboring plasmid pKD46, which was transferred to MicroPulser electroporation cuvettes (Bio-Rad) for electroporation at 2.49 kV for 6.1 ms using a MicroPulser electroporator (Bio-Rad). Thereafter, the bacteria were regenerated at 30°C in a shaking incubator for 3 h. The regenerated cells were then infected with phage T4, and homologous recombination transpired during phage production. The production of phages expressing EGFP was confirmed by a DNA sequence analysis (Solgent, South Korea) of the genomic DNA from the resulting phages.
Construction of ΔrecA strain of E. coli.
The ΔrecA mutant strain of E. coli was constructed by homologous recombination. A kanamycin resistance gene was inserted to replace recA according to the experimental procedure for EGFP insertion to T4. For ΔrecA strain construction, DNA was synthesized to include 55 bp upstream of the recA ORF, the kanamycin resistance gene, and 55 bp downstream of the recA ORF (Bioneer, South Korea), and recA deletion was confirmed by kanamycin selection (50 μg/ml) and sequence analysis (Solgent, South Korea).
Construction of plasmid harboring amA3 mutation in t (holin) gene.
An amA3 mutant (C259T) (29) of the phage T4 t gene (GenBank accession number Y00408) was synthesized (Bioneer, South Korea) and cloned in plasmid vector pBT7C-His (Bioneer, South Korea) at the EcoRI site. The resulting plasmid was used to transform E. coli strain DH5α (supE44) or wild-type MG1655. Expression was induced with the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) at the final concentration of 0.1 mM.
Real-time RT-PCR.
For reverse transcription-PCR (RT-PCR), reverse transcription was performed using a random hexamer and RNA templates isolated from the phage-infected host bacteria. The PCR was performed using SYBR green and a MyiQ single-color real-time PCR detection system (Bio-Rad, USA).
Bacterial imaging.
The bacterial morphologies were observed under a laser scanning microscope using ZEN software (Zeiss LSM 700). For sampling, 5 to 10 μl of the bacteria was dropped on a glass slide and covered with a coverslip. After 20 min of drying in a fume hood, the bacteria were observed using a 100× objective lens. After the bacteria were cultured using a glass-bottom 96-well black plate (Ibidi 89626), the bacteria were fixed with cold acetone and blocked with 1% bovine serum albumin (BSA) in phosphate-buffered saline with Tween 20 (PBST) for 30 min. The enhanced green fluorescent protein (EGFP) was observed using a 488-nm excitation laser line.
EGFP quantification.
The bacteria were cultured in a black 96-well plate (SPL Life Sciences, South Korea). After the cells were infected with EGFP-expressing T4, the fluorescence intensity of the bacteria was measured using a microplate reader (Infinite 200 PRO; Tecan Group Ltd.) with the GFP detection mode.
Western blot analysis.
An immunoblot analysis was performed using anti-RecA (Abcam 63797), anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Thermo MA5-15738), anti-E. coli FtsZ (AgriSera AS07 217), anti-rabbit (Abcam ab6721), or anti-mouse (Abcam ab6728) monoclonal antibody, as previously described (41). Anti-phage T4 antiserum was produced from rabbit (AbClon, South Korea). Briefly, after SDS-polyacrylamide gel electrophoresis, the samples were transferred to a nitrocellulose membrane, followed by blocking, antibody treatment, and chemiluminescence detection.
ROS and heat stress.
The phage growth was conducted as previously described (42). When a fresh culture of the E. coli K-12 strain reached an optical density at 600 nm (OD600) of 0.3, H2O2 was added to a final concentration of 4.5 mM and the culturing was continued to an OD600 of 0.6, at which point phage T4 was added at a multiplicity of infection (MOI) of 0.001. The phage titer was checked every 5 min. For heat stress, a fresh culture of E. coli was grown to an OD600 of 0.3, the incubator temperature was then increased to 45°C, and growth was allowed to an OD600 of 0.6. The temperature was then returned to 37°C, and phage T4 was added at an MOI of 0.001. The phage titer was checked every 5 min.
Statistical analysis.
All the data are presented as the means ± standard deviations (SDs) from triplicate runs of 3 independent experiments. Statistical significance was determined with Student's t tests, and a P value of <0.05 represented significance.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Korean National Research Foundation grants NRF-2015R1A2A2A01003632 and NRF-2017M3A9B8069292, ATC Program grant 10076996 from Ministry of Trade, Industry, and Energy of Korea, and the HUFS Research Fund of 2018.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02085-18.
REFERENCES
- 1.Comeau AM, Tétart F, Trojet SN, Prère MF, Krisch HM. 2007. Phage-antibiotic synergy (PAS): beta-lactam and quinolone antibiotics stimulate virulent phage growth. PLoS One 2:e799. doi: 10.1371/journal.pone.0000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loś JM, Golec P, Wegrzyn G, Wegrzyn A, Loś M. 2008. Simple method for plating Escherichia coli bacteriophages forming very small plaques or no plaques under standard conditions. Appl Environ Microbiol 74:5113–5120. doi: 10.1128/AEM.00306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santos SB, Carvalho CM, Sillankorva S, Nicolau A, Ferreira EC, Azeredo J. 2009. The use of antibiotics to improve phage detection and enumeration by the double-layer agar technique. BMC Microbiol 23:148–157. doi: 10.1186/1471-2180-9-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fothergill JL, Mowat E, Walshaw MJ, Ledson MJ, James CE, Winstanley C. 2011. Effect of antibiotic treatment on bacteriophage production by a cystic fibrosis epidemic strain of Pseudomonas aeruginosa. Antimicrob Agents Chemother 55:426–428. doi: 10.1128/AAC.01257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Islam MR, Ogura Y, Asadulghani M, Ooka T, Murase K, Gotoh Y, Hayashi T. 2012. A sensitive and simple plaque formation method for the Stx2 phage of Escherichia coli O157:H7, which does not form plaques in the standard plating procedure. Plasmid 67:227–235. doi: 10.1016/j.plasmid.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Kaur S, Harjai K, Chhibber S. 2012. Methicillin-resistant Staphylococcus aureus phage plaque size enhancement using sublethal concentrations of antibiotics. Appl Environ Microbiol 78:8227–8233. doi: 10.1128/AEM.02371-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryan EM, Alkawareek MY, Donnelly RF, Gilmore BF. 2012. Synergistic phage-antibiotic combinations for the control of Escherichia coli biofilms in vitro. FEMS Immunol Med Microbiol 65:395–398. doi: 10.1111/j.1574-695X.2012.00977.x. [DOI] [PubMed] [Google Scholar]
- 8.Knezevic P, Curcin S, Aleksic V, Petrusic M, Vlaski L. 2013. Phage-antibiotic synergism: a possible approach to combatting Pseudomonas aeruginosa. Res Microbiol 164:55–60. doi: 10.1016/j.resmic.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Kamal F, Dennis JJ. 2015. Burkholderia cepacia complex phage-antibiotic synergy (PAS): antibiotics stimulate lytic phage activity. Appl Environ Microbiol 81:1132–1138. doi: 10.1128/AEM.02850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Easwaran M, Paudel S, De Zoysa M, Shin HJ. 2015. Functional characterization of a novel lytic phage EcSw isolated from Sus scrofa domesticus and its potential for phage therapy. Mol Cell Probes 29:151–157. doi: 10.1016/j.mcp.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Ronayne EA, Wan YC, Boudreau BA, Landick R, Cox MM. 2016. P1 Ref endonuclease: a molecular mechanism for phage-enhanced antibiotic lethality. PLoS Genet 12:e1005797. doi: 10.1371/journal.pgen.1005797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaudhry WN, Concepción-Acevedo J, Park T, Andleeb S, Bull JJ, Levin BR. 2017. Synergy and order effects of antibiotics and phages in killing Pseudomonas aeruginosa biofilms. PLoS One 12:e0168615. doi: 10.1371/journal.pone.0168615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumaran D, Taha M, Yi Q, Ramirez-Arcos S, Diallo JS, Carli A, Abdelbary H. 2018. Does treatment order matter? Investigating the ability of bacteriophage to augment antibiotic activity against Staphylococcus aureus biofilms. Front Microbiol 9:127. doi: 10.3389/fmicb.2018.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uchiyama J, Shigehisa R, Nasukawa T, Mizukami K, Takemura-Uchiyama I, Ujihara T, Murakami H, Imanishi I, Nishifuji K, Sakaguchi M, Matsuzaki S. 2018. Piperacillin and ceftazidime produce the strongest synergistic phage-antibiotic effect in Pseudomonas aeruginosa. Arch Virol 163:1941–1948. doi: 10.1007/s00705-018-3811-0. [DOI] [PubMed] [Google Scholar]
- 15.Young R. 2014. Phage lysis: three steps, three choices, one outcome. J Microbiol 52:243–258. doi: 10.1007/s12275-014-4087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin TT, Kang HQ, Ma P, Li PP, Huang LY, Gu B. 2015. SOS response and its regulation on the fluoroquinolone resistance. Ann Transl Med 3:358. doi: 10.3978/j.issn.2305-5839.2015.12.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baharoglu Z, Mazel D. 2014. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol Rev 38:1126–1145. doi: 10.1111/1574-6976.12077. [DOI] [PubMed] [Google Scholar]
- 18.Andersson DI, Hughes D. 2014. Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol 12:465–478. doi: 10.1038/nrmicro3270. [DOI] [PubMed] [Google Scholar]
- 19.Vicente M, Gomez MJ, Ayala JA. 1998. Regulation of transcription of cell division genes in the Escherichia coli dcw cluster. Cell Mol Life Sci 54:317–324. doi: 10.1007/s000180050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dajkovic A, Lutkenhaus J. 2006. Z ring as executor of bacterial cell division. J Mol Microbiol Biotechnol 11:140–151. doi: 10.1159/000094050. [DOI] [PubMed] [Google Scholar]
- 21.El-Hajj ZW, Newman EB. 2015. How much territory can a single E. coli cell control? Front Microbiol 6:309. doi: 10.3389/fmicb.2015.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diver JM. 1989. Quinolone uptake by bacteria and bacterial killing. Rev Infect Dis 11(Suppl 5):S941–S946. doi: 10.1093/clinids/11.Supplement_5.S941. [DOI] [PubMed] [Google Scholar]
- 23.Park C, Kee Y, Park J, Myung H. 2002. A nonisotopic assay method for hepatitis C virus NS5B polymerase. J Virol Methods 101:211–214. doi: 10.1016/S0166-0934(01)00443-8. [DOI] [PubMed] [Google Scholar]
- 24.Saier MH, Reddy BL. 2015. Holins in bacteria, eukaryotes, and archaea: multifunctional xenologues with potential biotechnological and biomedical applications. J Bacteriol 197:7–17. doi: 10.1128/JB.02046-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moussa SH, Lawler JL, Young R. 2014. Genetic dissection of T4 lysis. J Bacteriol 196:2201–2209. doi: 10.1128/JB.01548-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savva CG, Dewey JS, Deaton J, White RL, Struck DK, Holzenburg A, Young R. 2008. The holin of bacteriophage lambda forms rings with large diameter. Mol Microbiol 69:784–793. doi: 10.1111/j.1365-2958.2008.06298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Young R. 2016. The last r locus unveiled: T4 RIII is a cytoplasmic antiholin. J Bacteriol 198:2448–2457. doi: 10.1128/JB.00294-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moussa SH, Kuznetsov V, Tran TA, Sacchettini JC, Young R. 2012. Protein determinants of phage T4 lysis inhibition. Protein Sci 21:571–582. doi: 10.1002/pro.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dressman HK, Drake JW. 1999. Lysis and lysis inhibition in bacteriophage T4: rV mutations reside in the holin t gene. J Bacteriol 181:4391–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White R, Chiba S, Pang T, Dewey JS, Savva CG, Holzenburg A, Pogliano K, Young R. 2011. Holin triggering in real time. Proc Natl Acad Sci U S A 108:798–803. doi: 10.1073/pnas.1011921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh A, Dennehy JJ. 2014. Stochastic holin expression can account for lysis time variation in the bacteriophage lambda. J R Soc Interface 11:20140140. doi: 10.1098/rsif.2014.0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pang T, Fleming TC, Pogliano K, Young R. 2013. Visualization of pinholin lesions in vivo. Proc Natl Acad Sci U S A 110:E2054–E2063. doi: 10.1073/pnas.1222283110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramanculov E, Young R. 2001. Genetic analysis of the T4 holin: timing and topology. Gene 265:25–36. doi: 10.1016/S0378-1119(01)00365-1. [DOI] [PubMed] [Google Scholar]
- 34.Qashqari H, Al-Mars A, Chaudhary A, Abuzenadah A, Damanhouri G, Alqahtani M, Mahmoud M, El Sayed Zaki M, Fatima K, Qadri I. 2013. Understanding the molecular mechanism(s) of hepatitis C virus (HCV) induced interferon resistance. Infect Genet Evol 19:113–119. doi: 10.1016/j.meegid.2013.06.025. [DOI] [PubMed] [Google Scholar]
- 35.Dover JA, Burmeister AR, Molineux IJ, Parent KN. 2016. Evolved populations of Shigella flexneri phage Sf6 acquire large deletions, altered genomic architecture, and faster life cycles. Genome Biol Evol 8:2827–2840. doi: 10.1093/gbe/evw177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar MS, Plotkin JB, Hannenhalli S. 2015. Regulated CRISPR modules exploit a dual defense strategy of restriction and abortive infection in a model of prokaryote-phage coevolution. PLoS Comput Biol 11:e1004603. doi: 10.1371/journal.pcbi.1004603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hall JP, Harrison E, Brockhurst MA. 2013. Viral host-adaptation: insights from evolution experiments with phages. Curr Opin Virol 3:572–577. doi: 10.1016/j.coviro.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Kropinski AM, Mazzocco A, Waddell TE, Lingohr E, Johnson RP. 2009. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol Biol 501:69–76. doi: 10.1007/978-1-60327-164-6_7. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Russell DW. 2006. Purification of bacteriophage λ particles by centrifugation through a glycerol step gradient. CSH Protoc 2006:pdb.prot3969. doi: 10.1101/pdb.prot3969. [DOI] [PubMed] [Google Scholar]
- 40.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim MS, Kim S, Myung H. 2014. Degradation of AIMP1/p43 induced by hepatitis C virus E2 leads to upregulation of TGF-β signaling and increase in surface expression of gp96. PLoS One 9:e96302. doi: 10.1371/journal.pone.0096302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim MS, Kim YD, Hong SS, Park K, Ko KS, Myung H. 2015. Phage-encoded colanic acid-degrading enzyme permits lytic phage infection of a capsule-forming resistant mutant Escherichia coli strain. Appl Environ Microbiol 81:900–909. doi: 10.1128/AEM.02606-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.