Abstract
The flagellated protozoan parasite Trichomonas vaginalis is the etiologic agent of trichomoniasis, the most common non-viral sexually transmitted infection worldwide. As an obligate extracellular pathogen, adherence to epithelial cells is critical for parasite survival within the human host and a better understanding of this process is a prerequisite for the development of therapies to combat infection. In this sense, recent work has shown S-acylation as a key modification that regulates pathogenesis in different protozoan parasites. However, there are no reports indicating whether this post-translational modification is a mechanism operating in T. vaginalis. In order to study the extent and function of S-acylation in T. vaginalis biology, we undertook a proteomic study to profile the full scope of S-acylated proteins in this parasite and reported the identification of 363 proteins involved in a variety of biological processes such as protein transport, pathogenesis related and signaling, among others. Importantly, treatment of parasites with the palmitoylation inhibitor 2-bromopalmitate causes a significant decrease in parasite: parasite aggregation as well as adherence to host cells suggesting that palmitoylation could be modifying proteins that are key regulators of Trichomonas vaginalis pathogenesis.
Keywords: Parasite, Pathogens, Post-translational modifications, Protein Modification, Microbiology, Infectious disease, Host-Pathogen Interaction, Cell adhesion, Adherence, Palmitoylation, Parasite
The flagellated protozoan parasite Trichomonas vaginalis is the etiologic agent of trichomoniasis, the most common non-viral sexually transmitted infection worldwide with an estimated 276 million new cases annually (1, 2). Although asymptomatic infection is common, multiple symptoms and pathologies can arise in both men and women, including vaginitis, urethritis, prostatitis, low birth weight infants and preterm delivery during pregnancy and infertility (3, 4). T. vaginalis has also emerged as an important cofactor in amplifying the spread of human immunodeficiency virus (HIV) as individuals infected with this parasite have a significantly increased incidence of virus transmission (5, 6). Additionally, chronic infection may increase the risk of cervical and aggressive prostate cancer (7–10). Despite these serious consequences, the underlying biochemical processes that lead to T. vaginalis pathogenesis are poorly understood. In this sense, recent works have unraveled the role of protein S-palmitoylation as a significant post-translational modification (PTM)1 that regulates invasion and motility in different protozoan parasites such as Plasmodium falciparum, Trypanosoma cruzi, Giardia lamblia, and Toxoplasma gondii (11, 12) as well as adherence to host cell in the fungus C. neoformans (13). S-palmitoylation refers to the covalent attachment of a 16-carbon chain palmitic acid to cysteine residues of a protein through a thioester bond. The reaction is conducted by specific enzymes termed Palmitoyl Acyl-Transferases (PATs) (14). PATs were originally identified in yeast (15, 16). Further sequence analysis of PATs of different species revealed that they share a common structure, mainly composed of four predicted TMD, a tetrapeptide of Asp-His-His-Cys (DHHC) inside a Cys rich domain (DHHC-CRD), a short DPG motif upstream the DHHC and another short TTxE motif downstream the DHHC. Mutation of the conserved Cys of the DHHC motif have result in loss of activity for several PATs, and thus, led to the hypothesis that this motif is the active site (17). Both cytosolic and transmembrane proteins can be palmitoylated and this PTM increases the local hydrophobicity of target proteins, having sizable effects on its function and/or localization (18, 19). Specifically, palmitoylation can promote protein association with cellular membranes, affect protein stability, modulate protein-protein interactions as well as alter the conformation of a transmembrane protein affecting its segregation to specific membrane domains (18, 19). Interestingly, it has also been shown to be involved in the regulation of enzymatic activity, gene expression and epigenetic regulatory networks (20–22). Further, because this is the only lipidic reversible PTM identified to date, it can dynamically regulate the function of target proteins (23).
Up to date, there are no reports on the existence and role of protein palmitoylation in T. vaginalis. However, in silico analysis of the genome database (TrichDB) predicts the presence of TvPATs enzymes suggesting that the mechanism is functional in this parasite (11). To analyze the role of protein palmitoylation in T. vaginalis, we undertook a proteomic study to profile the full complement of palmitoylated proteins in the parasite and reported here the identification of 363 proteins involved in a variety of biological processes including protein transport, pathogenesis related and signaling; among others. Additionally, treatment of parasites with the palmitoylation inhibitor 2-bromopalmitate caused a significant decrease in the adherence to host cells. To the best of our knowledge, this is the first study to systematically identify and characterize palmitoylated proteins of Trichomonas parasites and demonstrate a key role regulating the function of proteins that modulate adherence, aggregation and the concomitant pathogenesis of T. vaginalis.
EXPERIMENTAL PROCEDURES
Parasites, Cell Cultures, and Media
Trichomonas vaginalis strain B7RC2 wild type was cultured in TYM medium supplemented with 10% (v/v) horse serum, 10 U/ml penicillin and 10 μg/ml streptomycin (Invitrogen) (24). 100 μg/ml G418 (Invitrogen) was added to culture of the TSP8-HA (TVAG_008950) and TSP9 (TVAG_287570) transfected parasites (25). Parasites were grown at 37 °C and passaged daily. Human HeLa cells (ATCC® CCL2™) were grown in DMEM complemented with 10% (v/v) bovine fetal serum, 10 U/ml penicillin and 10 μg/ml streptomycin (Invitrogen) and cultured at 37 °C/5% CO2.
Isolation of T. vaginalis Palmitoylated Proteins
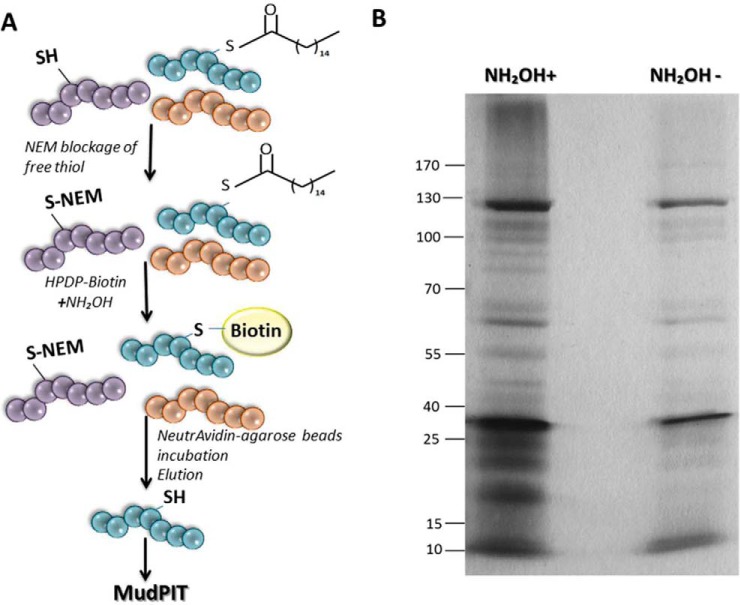
Acyl Biotin Exchange (ABE) of whole parasite lysates was carried out as described by Wan et al. (26). Ten mg of whole parasites lysates were used in each assay. Briefly, 5 × 108 parasites were centrifuged and washed twice in PBS. The pellet was resuspended in lysis buffer (150 mm NaCl, 50 mm Tris, 5 mm EDTA, pH 7.4) and adjusted to 10 mg of whole parasite lysate in 4 ml of lysis buffer containing 10 mm NEM (N-ethylmaleimide) (Pierce) and sonicated 10 seg on/off for 10 periods. Then the concentration of NEM was adjusted to 1 mm for overnight treatment. Then, 25% volume of the final elution was fractionated by SDS-PAGE and nonspecific labeling was checked by silver stain assay. The 75% remaining elution sample was chloroform-methanol precipitated and analyzed by LC-MS/MS technology.
Proteomic Mass Spectrometry Analysis
Precipitated pellet was resuspended in a minimal volume of digestion buffer (100 mm Tris-HCl, pH 8, 8 m urea). Resuspended proteins were reduced and alkylated by the sequential addition of 5 mm tris(2-carboxyethyl)phosphine and 10 mm iodoacetamide as described previously (27). The samples were then digested by Lys-C (Princeton Separations) and trypsin proteases (Promega) (27). First, Lys-C protease (∼1:50 (w/w) ratio of enzyme:substrate) was added to each sample and incubated for 4 h at 37 °C with gentle shaking. The digests were then diluted to 2 m urea by the addition of digestion buffer lacking urea, and trypsin was added to a final enzyme/substrate ratio of 1:20 (w/w) and incubated for 8 h at 37 °C with gentle shaking. Digests were stopped by the addition of formic acid to a final concentration of 5%. Supernatants were carefully removed from the resin and analyzed further by proteomics mass spectrometry.
Digested samples were then analyzed using a shotgun proteomics platform comprised of an on-line reversed phase separation coupled to tandem mass spectrometric analysis of the peptide mixture as described previously (28–30). Briefly, digested samples were loaded onto a fused silica capillary column with a 5-μm electrospray tip and packed in house with 18 cm of Luna C18 3 μm particles (Phenomenex). The column was then placed in line with a Q-Exactive mass spectrometer (Thermo Fisher), and peptides were fractionated using a gradient of increasing acetonitrile. Peptides were eluted directly into the mass spectrometer where MS/MS spectra were collected. The data-dependent spectral acquisition strategy consisted of a repeating cycle of one full MS spectrum (Resolution = 70,000) followed by MS/MS of the twelve most intense precursor ions from the full MS scan (Resolution = 17,500) (31). Raw Data analysis was performed using the IP2 suite of software tools (Integrated Proteomics Applications, San Diego, CA). RAW files were converted to peak lists using RawConverter 1.1.0.19 (http://fields.scripps.edu/downloads.php). Spectra were analyzed using the ProLuCID algorithm v1.4.2 (32) and searching against a fasta protein database consisting of all predicted open reading frames downloaded from TrichDB on January 4, 2015 (33) concatenated to a decoy database in which the amino acid sequence of each entry was reversed (194,950 entries including decoys). The following search parameters were used: (1) precursor ion tolerance was 20 ppm, (2) fragment ion tolerance was 20 ppm, (3) cysteine carbamidomethylation was considered as a static modification, (4) peptides must be fully tryptic, and (5) no consideration was made for missed cleavages. False positive rates for peptide identifications were estimated using a decoy database approach and then filtered using the DTASelect algorithm v2.1.3 (34–36). XCorr and ΔCn cutoffs were identified dynamically using a linear discriminant analysis (34). Proteins identified by at least two fully tryptic unique peptides, each with a false positive rate of less than 5%, were present in the sample. Five different set of samples were independently analyzed. Normalized spectral abundance factor (NSAF) values including shared peptides was calculated for each protein as described and multiplied by 105 to improve readability (27). Proteins that could not be distinguished by uniquely mapping peptides in any given replicate were considered as a group. The letters in the table refer to the different protein groups, corresponding to the minimum number of proteins present. See supplemental Tables S1 and S2 for protein quantification and peptide identification data, respectively.
Experimental Design and Statistical Rationale
Five biological replicates were independently analyzed by mass spectrometry. Each one consisted of a sample treated with NH2OH and a sample without NH2OH as a control. As NH2OH cleave thioester bonds from palmitoylated proteins, the consequent co-incubation with N-[6-(Biotinamido)hexyl]-3′-(2′-pyridyldithio) propionamide (biotin-HPDP; Pierce) allows the enrichment of palmitoylated proteins. As control, samples non-treated with NH2OH allow the detection of nonspecific labeling (Fig. 1A; supplemental Table S2). Mass spectrometry data obtained were analyzed using Microsoft Excel (2010) and Infostat version 2016 (37). Importantly, IDs that were present only in the negative control samples were considered as nonspecific labeling and were eliminated from the analysis. Then, proteins found in at least two of the five biological replicates were included in the further analysis. Statistical analysis of the dataset was carried out as described (38), with specified modifications. Briefly, the natural log of each NSAF value was calculated followed by t test comparison of the ln(NSAF) of the biological replicates treated with NH2OH compared with the ln(NSAF) from the biological replicates of untreated samples (control). To avoid dividing by zero errors in the natural log transformation calculations, 0.5 spectral count was added to spectral counts dataset, and NSAF values were re-calculated. This operation provided the best fit into a Gaussian distribution of the dataset according to Q-Q plot analysis, as the r2 value from the Q-Q plot of the average from ln(NSAF) of the replicates treated and untreated with NH2OH was 0.999 and 0.994 respectively. Proteins were considered as palmitoylated if they met the following criteria: an estimated t test p value less than 0.05 corrected with a FDR q-value threshold of 0.2. A total of 363 proteins out of 1852 total proteins detected followed the criteria, indicating statistically significant enrichment of the protein in the NH2OH treated condition compared with the untreated control (supplemental Table S1).
Fig. 1.
Isolation of palmitoylated proteins. A, Scheme of acyl biotin exchange assay (ABE). B, SDS-PAGE analysis followed by silver staining of proteins isolated with Neutravidin-Agarose by ABE assay. Hydroxylamine treated sample (NH2OH+) and nonspecific labeling control without hydroxylamine (NH2OH-) are shown.
Bioinformatics Analysis
Palmitoylated proteins were identified using online BLAST tool (Basic Local Alignment Search Tool), following by GO term annotation from TrichDB database (33). Prediction of palmitoylated proteins with high threshold was performed using GPS-Lipid 1.0 tool (39). Transmembrane domains were predicted using TMHMM 2.0 tool (40) Phobius (41) and TOPCONS (42). Signal peptides were predicted using both SignalIP 4.1 (43) and Phobius (41) tools. Homologs between T. vaginalis and Plasmodium falciparum, Toxoplasma gondii and Trypanosoma brucei palmitoylated proteins were identified using BLAST 2.2.28 and Venn diagrams were done using RStudio Version 0.99.902 (www.R-project.org). The putative TvPATs sequence alignment was performed using MAFFT V7.123b (44) with l-INS-i algorithm and 1000 iteration. Finally, it was edited using Geneious® 9.0.5 (http://www.geneious.com) (45). 49 sequences were retrieved from TrichDB. Incomplete sequences were eliminated from the analysis.
2-Bromopalmitate (2-BP) Treatment
Viability assays were performed using 1 × 106 B7RC2 parasites grown in 10 ml of TYM media for 16 h and co-incubated with 0, 25, 50, 75, and 100 μm of 2-Bromopalmitate (2-Bromohexadecanoic acid, Sigma). Attachment assays were performed using 1 × 106 B7RC2 parasites grown in 10 ml of TYM media and co-incubated for 16 h with 0, 25, 50, 75, and 100 μm of 2-Bromopalmitate. Aggregation assay were performed with TvTSP8-HA expressing parasites grown to a concentration of 1 × 106 parasites per ml. 0, 50, and 100 μm of 2-BP was added and the reduction of clumps was analyzed after 4 h. As control for all 2-BP treatments, a 0 μm (no-treatment) containing 2% (v/v) DMSO was included.
Analysis of Protein Stability
Parasites pre-incubated with 100 μm 2-BP palmitoylation inhibitor or DMSO during 16 h were incubated with 20 μg/ml cycloheximide (Sigma) at 37 °C. Samples (106 parasites) were taken at times: 0, 3, 6, 9, 12, and 24 h. Parasites were centrifuged and the presence of TvTSP8-HA and MIF proteins were analyzed by SDS-PAGE and Western blotting using anti-HA and anti-MIF antibody. Three independent experiments were performed.
Parasite Viability Assays
Parasites pre-incubated with 0 (DMSO), 25, 50, 75 and 100 μm 2-BP inhibitor were taken at 2, 5, 8, and 24 h postincubation and labeled with the fluorescent exclusion dye propidium iodide (PI, 20 μg/ml) at 4 °C for 10 min. PI fluorescence associated with non-viable cells was measured by flow cytometry (at λ = 544/602 nm) on FACSCalibur (Becton Dickinson), and flow cytometry data were analyzed using Flowing software version 2.4.1 (Perttu Terho, Turku Centre for Biotechnology, Finland; www.flowingsoftware.com). Four independent experiments were performed.
Parasite Aggregation
Parasite aggregation was analyzed under anaerobic conditions when parasites reach a concentration of 1 × 106 parasites/ml using a Nikon E600 epifluorescence microscope with a magnification of 10×. This concentration of parasites was shown in previous assays to be optimal to measure parasite aggregation. A clump was defined as the size corresponding to an aggrupation of at least 5 parasites. Quantification of clumps in thirty 10× magnification fields was performed with Fiji software (46). All experiments were performed with 3 technical replicates per treatment per experiment.
Immunolocalization Experiments
Parasites expressing the hemagglutinin-tag (HA) version of TvTSP8 were incubated at 37 °C on glass coverslips as previously described (47) for 4 h. The parasites were then fixed and permeabilized in cold methanol for 10 min. Cells were then washed and blocked with 5% fetal bovine serum (FBS) in phosphate buffered saline (PBS) for 30 min, incubated with a 1:500 dilution of anti-HA primary anti-body (Covance, Emeryville, CA) diluted in PBS plus 2% FBS, washed with PBS and then incubated with a 1:5000 dilution of Alexa Fluor-conjugated secondary antibody (Molecular Probes). The coverslips were mounted onto microscope slips using ProLong Gold antifade reagent with 4′,6′-diamidino-2- phenylindole (Invitrogen). All observations were performed on a Nikon E600 epifluorescence microscope. Adobe Photoshop (Adobe Systems) and Fiji software (46) were used for image processing.
Subcellular Fractionation Using Optiprep Density Gradient
Parasites transfected with TvTSP8-HA (108 cells) were lysate and subcellular fractionation was performed using Optiprep gradient as described (48). The abundance of TvTSP8 in different fractions was analyzed by Western blotting using an anti-HA antibody (Covance, Emeryville, CA), whereas the presence of soluble TCTP was evaluated using specific anti-TCTP antibody. Two independent experiments were performed.
Attachment Assay
A modified version of an in vitro assay to qualify the binding of T. vaginalis to host cell monolayers (49) was performed. Briefly, HeLa cells were seeded on 12-mm coverslips in 24-well plates at 3 × 105 cells/well in DMEM culture medium (Invitrogen) and grown to confluence at 37 °C in 5% CO2 for 2 days. Cell monolayers were washed before the addition of parasites. T. vaginalis was labeled with 10 mm CellTracker Blue CMAC (7-amino-4-chloromethylcoumarin) (Invitrogen), and 105 labeled parasites in 0.5 ml of DMEM medium were added (1:3 parasite/host cell ratio) for 30 min. Plates were incubated at 37 °C in 5% CO2 for 30 min. Coverslips were subsequently rinsed in PBS to remove unattached parasites, fixed with 4% paraformaldehyde, and mounted on slides with Mowiol (Calbiochem). Thirty 10X magnification fields were analyzed per coverslip. All experiments were performed 3 times with 3 coverslips per treatment per experiment.
Graphics and Statistical Analyses
Specific statistical considerations and the tests used are described separately for each subsection. All analyses used GraphPad Prism for Windows version 7.00. Data are given as mean ± S.E. of the mean (S.E.). Significance was established at p < 0.05.
RESULTS
Isolation of T. vaginalis Palmitoylated Proteins
To profile T. vaginalis palmitoylated proteins, we adapted a method based on an acyl biotin exchange reaction (ABE) on whole-cell lysates (Fig. 1A) (26). Briefly, an incubation step of the whole cell lysate with N-ethylmaleimide (NEM) allows the blockage of all free sulfhydryl groups generating a non-sensitive hydroxylamine (NH2OH) bond that prevents nonspecific labeling. Then, neutral NH2OH was used to specifically cleave thioester bonds from palmitoylated proteins. Consequently, palmitoylated proteins are labeled by coincubation with N-[6-(Biotinamido)hexyl]-3′-(2′-pyridyldithio) propionamide (biotin-HPDP; Pierce) (Fig. 1A). As a control for nonspecific labeling, half of the sample was labeled with biotin-HPDP lacking previous treatment with NH2OH. The resulting samples were then purified using NeutrAvidin-agarose beads (Pierce). After washes to remove contaminating proteins, biotin-labeled proteins retained on the column were eluted by cleavage with DTT, separated by SDS-PAGE and silver stained (Fig. 1B). A range of proteins, varying in molecular mass from >170 to ∼10 kDa were observed, demonstrating the ability of this approach to label and obtain a broad range of proteins (Fig. 1B). Importantly, the control samples where the treatment with NH2OH was omitted contain few proteins that bound non-specifically to the streptavidin column (Fig. 1B). These data suggest that the T. vaginalis biotin-labeled fractions are highly enriched with palmitoylated proteins.
Novel DHHC Domain Containing Proteins Identified In the T. vaginalis Palmitoyl-Proteome May Be Acting As Active Enzymes
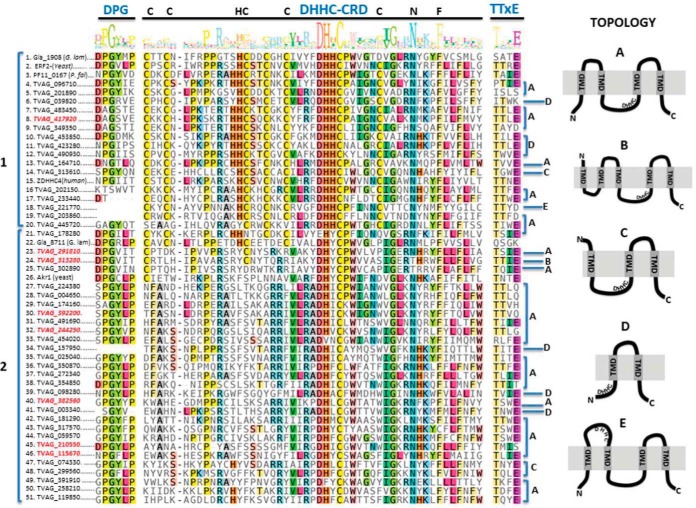
The identity of proteins in the palmitoylated-enriched fractions was determined using protein mass spectrometry. To assess the binding specificity of the neutravidin column, control samples were processed and analyzed identically to the NH2OH treated and biotinylated samples. Proteins with two or more peptides found in at least two of the five biological replicates were included and analyzed using t test. A total of 363 proteins were significantly enriched in the NH2OH treated compared with control samples and were included in the palmitoyl-proteome list (supplemental Table S1). As some proteins use the thioester linkage for chemical reactions other than protein palmitoylation, it is expected to find false positives in ABE assays and indeed, their detection provides some measure of the efficacy of the method. In this regard, we detected two putative ubiquitin-conjugating enzymes (TVAG_038060 and TVAG_191220) which are generally found in palmitoyl-proteomes because they form a thioester linkage during the ubiquitination process (50). Additionally, it is also expected to detect PATs enzymes as these molecules form a transient DHHC-palmitate intermediate during catalysis (17). In this regard, here we identified 8 putative TvPAT-like molecules, indicating that these members of the family could be acting as active enzymes (supplemental Table S1). To gain a better insight about the complete set of T. vaginalis putative PATs we searched for all the DHHC-palmitoyltransferase domain (PF01529) containing proteins in the parasite database (TrichDB). As shown in Fig. 2, the analysis retrieved 45 DHHC-domain containing proteins that display the central DHHC-CRD conserved domain as well as the DPG and TTxE motifs usually found in other organisms. Interestingly, the identified proteins can be distinguished in two major groups (Fig. 2). Group 1 encompasses proteins containing the tetrapeptide motif composed of DHHC as well as other conserved amino acids that form part of the DHHC-CRD (C-x2-C-x9-HC-x2-C-x4-DHHC-x5-C-x4-N-x3-F). Instead, group 2 encloses proteins containing a variant of the classic DHHC where the second histidine of the tetrapeptide is generally replaced by a hydrophobic amino acid (Y, I, L, V, F, or M) and share other conserved amino acids that are absent in the classical PATs. Surprisingly, 7 of the putative PATs proteins obtained in our proteome belong to this novel second group (Fig. 2). The variant DHYC was described before to be functional in the palmitoyl transferases Akr1p from yeast (16) and in gla_8711 from Giardia lamblia (51). The remaining identified PAT, belongs to group 1and contain the classic DHHC tetrapeptide as well as the conserved Cys of the CRD. Almost all the described PATs from yeast and mammals contained four predicted TMD with N-terminal and C-terminal facing the cytosol, and a cytosolic DHHC motif arranged between the TMD 2 and TMD 3 (Topology A in Fig. 2). However, a variety of topologies were displayed in predicted T. vaginalis DHHC proteins (Fig. 2): 33 of them possess the classical topology described before (Topology A in Fig. 2), one of them contain 5 TMD (Topology B in Fig. 2), 3 of them contain 3 TMD (Topology c in Fig. 2), 7 proteins possess only 2 TMD (Topology D in Fig. 2) and one protein contain 4 TMD but with the DHHC motif facing the membrane lumen (Topology E in Fig. 2). All the T. vaginalis PATs identified in the palmitoyl proteome contain the classic 4 TMD except one of them that contain 5 TMD. Further studies are needed to evaluate the functionality of the remaining proteins.
Fig. 2.
Sequence alignment and schematic drawing of T. vaginalis DHHC-containing proteins. Multiple sequence alignment of DHHC proteins shows conserved motifs. The amino acid sequences of the complete set of T. vaginalis DHHC, Giardia lamblia Gla_1908 (XP_001707652) and Gla_8711 (XP_001708375_Gla), yeast ERF2 (KZV09491_ERF2_SacC) and Akr1 (KZV12503_AKR1_SacC), P. falciparum PF11_0167 (001347838_PlaF) and human ZDHHC4 (CAG38542_ZDHHC4_Hum) were aligned using MAFFT (44). The conserved DHHC-CRD domain and the DPG and TTxE motifs are depicted in blue. DHHC proteins can be distinguished in two major groups (1 and 2). The group 1 encompasses proteins containing the tetrapeptide motif composed of DHHC as well as other conserved amino acids that form part of the DHHC-CRD (C-x2-C-x9-HC-x2-C-x4-DHHC-x5-C-x4-N-x3-F). The group 2 encloses proteins containing a variant of the classic DHHC where the second histidine of the tetrapeptide is generally replaced by a hydrophobic amino acid (Y, I, L, V, F, or M) and share other conserved amino acids that are absent in the classical PATs. IDs in red, DHHC proteins identified in the palmitoyl-proteome. A schematic representation of the five different topologies (A, B, C, D and E) found in T. vaginalis DHHC proteins is illustrated.
T. vaginalis Palmitoylated Proteins Are Involved In A Variety Of Biological Functions
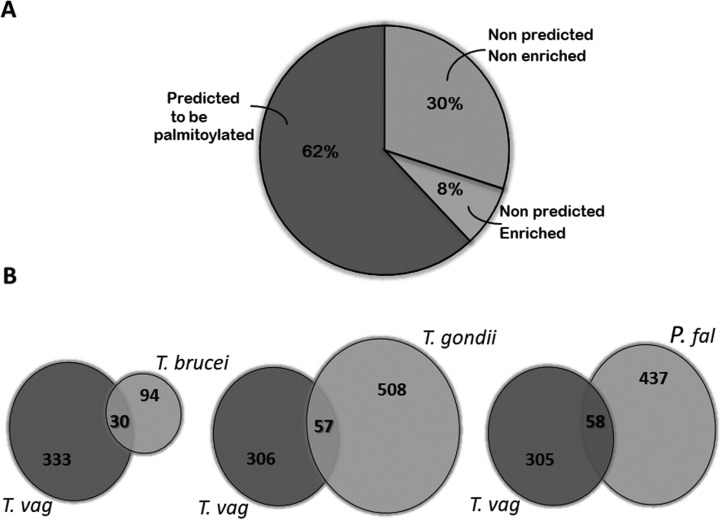
Prediction of palmitoylation with high confidence, using the GPS-Lipid program (www.csspalm.biocuckoo.org), indicates that 226 (∼62%) from the 363 proteins identified by mass spectrometry are predicted to be palmitoylated while the remaining 137 are not (Fig. 3A and supplemental Table S1). GPS-Lipid predicts that ∼25% (15521/60000) of the coding genes in T. vaginalis produce potentially palmitoylated proteins (data not shown). However, given that only about half of the annotated genes are being expressed in the parasite (52) and that palmitoylation is a dynamic PTM, it is hard to stablish a correlation between our proteome and the bioinformatics prediction data. To increase the confidence of our proteomic results, we next search for homologues in palmitoyl proteomes from other protozoan parasites and found that 24% of the molecules identified here are shared with the proteomes analyzed. Specifically, T. vaginalis shared 30 from the 124 palmitoylated proteins found in Trypanosoma brucei, 57 from the 565 of Toxoplasma gondii and 58 from the 495 present in Plasmodium falciparum (12, 53–55) (Fig. 3B and supplemental Table S1). Further, 8% of the identified T. vaginalis proteins that lacks predicted palmitoylated cysteine by GPS-Lipid are present in palmitoyl-proteomes from the other parasites analyzed here (Fig. 3A and 3B). Importantly, some of these homologs form part of the top 20 proteins commonly found in palmitoyl-proteomes of different species (56) such as G-proteins, syntaxin, thioredoxin and vesicle associated membrane proteins (VAMP) and some others represent well-studied palmitoylated proteins such as SNARE, PATs and calnexin among others (supplemental Table S1) (56–58). Taken together, 70% of molecules found in Trichomonas vaginalis palmitoyl-proteome are predicted to be palmitoylated in silico and/or enriched in other palmitoyl-proteomes.
Fig. 3.
Comparison of size and content of different palmitoyl-proteomes with T. vaginalis palmitome. A, Proteins identified by LC-MS/MS analysis. A total of 363 proteins were significantly enriched in five independent experiments analyzed. Of these, 62% of proteins were predicted to be palmitoylated according to GPS-Lipid software (dark gray). Although the remaining proteins were not predicted to be palmitoylated bioinformatically, 8% of these molecules were also detected in palmitoyl-proteomes from other protozoan parasites analyzed in B (white). B, Venn Diagrams depicting overlap between the T. vaginalis and the T. brucei, P. falciparum and T. gondii palmitoyl-proteomes. The numbers in the diagrams indicate the quantity of proteins shared or not by the different palmitoylomes.
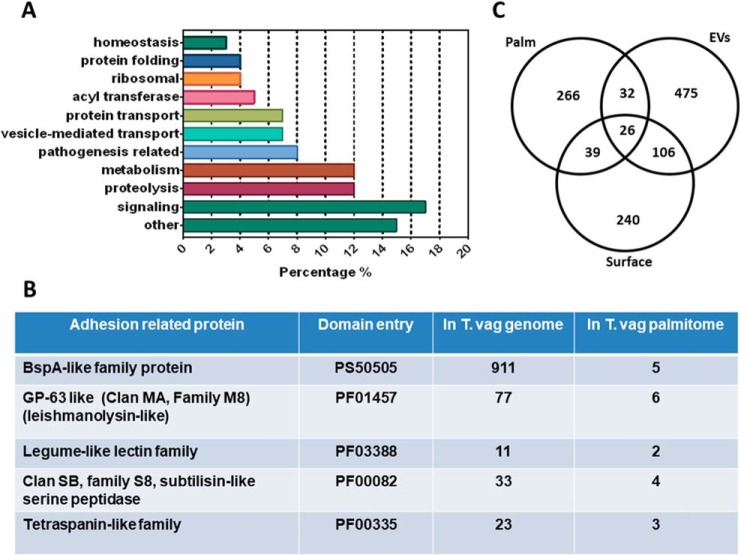
To understand the biological functions associated with protein palmitoylation in T. vaginalis, the identified proteins were sorted into functional groups according to gene ontology term enrichment (GO term enrichment) analysis (Fig. 4A). Of the 363 proteins identified, 47% are proteins with predicted domains whereas the remaining 53% are hypothetical proteins according to BLAST analysis. Among the proteins with identifiable domains that allow the assignment of a predicted function, 17% are molecules related to signaling, 12% are involved in metabolism and proteolysis, 8% are related to pathogenesis of the parasite, and 7% are involved in vesicle transport and protein transport. In less abundant proportions, we found proteins with acyl transferase activity (5%), protein folding and ribosomal (4%), homeostasis (3%) and other cellular activities. Interestingly, we also found 4 entries corresponding to proteins derived from repetitive elements and viral-like encoded proteins (59); supporting the hypothesis that some of the repetitive elements are coding for expressed proteins. Taken together, these data suggest that protein palmitoylation is involved in a wide range of functions in T. vaginalis.
Fig. 4.
Putative palmitoylated proteins sorted into functional groups according to GO Term enrichment analysis. A, Palmitoylated proteins were identified using BLAST analysis and sorted into functional groups using genome annotation. Most representative groups are shown. B, Proteins associated with pathogenesis identified in the T. vaginalis palmitoyl-proteome. The number of genes found in the genome is compared with the number of genes found in the palmitoyl-proteome. C, Venn diagram depicting overlap between T. vaginalis palmitoylated proteins (palm), surface proteins (Surface) and extracellular vesicles (EVs) proteins. The numbers in the diagrams indicate the quantity of proteins shared or not by the different data set.
Proteins that may have a role in T. vaginalis pathogenesis are also present in the palmitoyl-proteome. Interestingly, we identified 5 surface BspA family proteins, 6 GP63-like proteins, 2 Legume-like lectin protein, 4 subtilisin-like serine peptidases, 3 Tetraspanin-like proteins thought to be involved in pathogenesis (60–64) (Fig. 4B). Many of the identified proteins are members of very large gene families (65) however only a few members are detected in our proteome (Fig. 4B), suggesting that only some proteins of the family are palmitoylated and/or that different family members are differentially expressed. For example, 911 putative BspA family proteins predicted to possibly mediate the binding of the parasite to the cell surface have been described, 30% of which have data that support their expression (66) and only 5 were identified in our palmitoyl-proteome. Additionally, we also compared the proteins identified here with the proteomic data obtained from the surface localized proteins and isolated exosomes and microvesicles (EVs) of the parasite (47, 67, 68) (Fig. 4C). Although 211 out of 411 molecules found in the surface proteome are predicted to be palmitoylated with high confidence using GPS-Lipid (data not shown), we only found 65 of them in the palmitoyl-proteome (Fig. 4C and (supplemental Table S1). As palmitoylation is a reversible and dynamic PTM, this is a plausible result. Interestingly, most of the surface proteins found in the palmitoyl-proteome have transmembrane domains (supplemental Table S1). However, the palmitoylated transmembrane proteins from the surface proteome are different from the palmitoylated transmembrane proteins found in EVs (Fig. 4C and supplemental Table S1), supporting the idea of a dynamic composition of the surface membrane of T. vaginalis. Particularly, palmitoylated proteins present in EVs are mostly cytosolic (supplemental Table S1); pointing palmitoylation as a possible signal to direct these molecules to the EVs. Further studies are needed to explore this possibility.
Palmitoylation of TvTSP8 Might Be Contributing to Regulate Aggregation In T. vaginalis
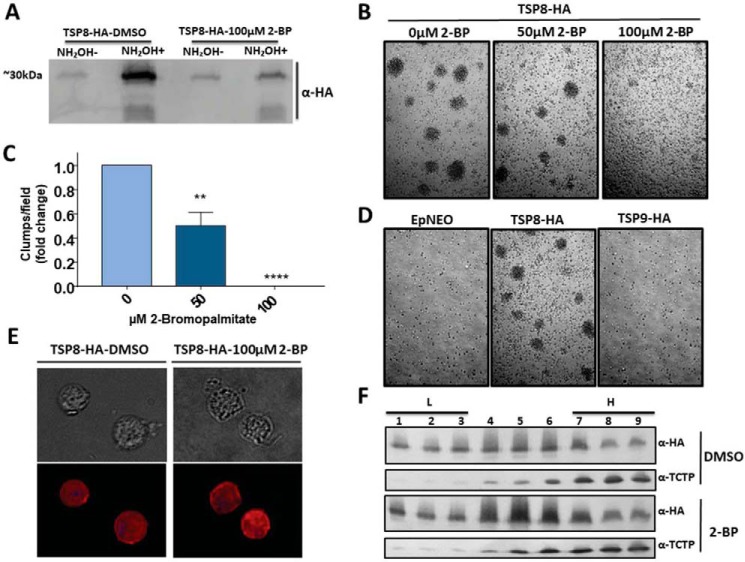
Among the proteins involved in pathogenesis identified in the palmitoyl-proteome, 3 Tetraspanin-like proteins (TvTSP8 (TVAG_008950); TVAG_099160 and TVAG_287570) were found. To validate our palmitoyl-proteome we determined the presence of palmitoylation in TvTSP8 using 2-bromopalmitate (2-BP), a non-metabolizable palmitic acid analogue commonly used to inhibit S-acylation in cells and PAT activity in vitro (69). To this end, using TvTSP8-HA transfected parasites we evaluated whether TvTSP8 palmitoylation is affected on 2-BP treatment by ABE assay followed by Western blotting assay with a monoclonal anti-HA antibody (Fig. 5A). As expected, a clear inhibition of TvTSP8 palmitoylation is observed on treatment with 100 μm of 2-BP (Fig. 5A), demonstrating that 2-BP is functional in T. vaginalis and validating our experimental design as a reliable approach for palmitoylation protein identification.
Fig. 5.
Palmitoylation of TvTSP8 might be contributing to regulate T. vaginalis aggregation. A, ABE assay using TvTSP8-HA overexpressing parasites treated with 100 μm 2-BP (right panel) or negative control treated with vehicle (DMSO) only. 2 mg of protein lysate was labeled with HDPD-biotin and incubated with Neutravidin-Agarose. After washes, 25% of the elution of each sample was loaded on the gel. NH2OH-; without hydroxylamine, NH2OH+; with hydroxylamine. A monoclonal anti-hemmaglutinin (HA) antibody was used to detect HA tagged TvTSP8. B, TvTSP8-HA overexpressing parasites were grown until reach 106cells/ml, followed by treatment with 50 or 100 μm of 2-BP for 4 h. Illustrative pictures were taken at 4 h. C, Quantification of the number of parasite aggregates per fields. 30 fields were analyzed in three independent experiments. Data are expressed as mean fold change related to mock-treated control parasites ± the standard error of the mean (S.E.). ANOVA followed by Tukey's post hoc test were used to determine significant differences. **p < 0,005, ****p < 0,0001. D, Ability to form clumps of TvTSP9 full-length (TVAG_287570) transfected parasites. A representative picture is shown. E, Cells expressing C-terminal HA-tagged versions of TvTSP8 protein treated with 100 μm of 2-BP or DMSO (vehicle) were stained for immunofluorescence microscopy using a mouse anti-HA antibody. The nucleus (blue) was also stained with 4′,6′-diamidino-2-phenylindole. F, TvTSP8-HA transfected parasites were incubated with 100 μm 2BP or DMSO for 16 h, lysed and separated by optiprep density gradient centrifugation. Fractions were collected from the top of the gradient. Equal volume aliquots from each fraction were separated by SDS-PAGE, transferred to PVDF membrane, and immunoblotted using anti-HA or anti-TCTP antibodies. Fractions 1–3 represent the low-density membrane fractions (L), whereas 7–9 the high-density fractions (H).
Interestingly, we have recently demonstrated that the level of expression of TvTSP8 is higher in adherent compared with poorly adherent parasite strains and that exogenous expression of TvTSP8 promoted the ability of a non-clumping strain to aggregate (62). These results suggested a role for surface-localized TvTSP8 in modulating parasite aggregation and cell/cell communication (62); a critical step for establishing infection (60, 70). In other cells, the inhibition of palmitolylation in tetraspanin proteins may lead to an inability to assemble Tetraspanin Enriched Microdomains (TEMs) resulting in impaired functions (71). We examined if the aggregation induced in TvTSP8 transfected parasites is affected on 2-BP treatment. As shown in Fig. 5B and 5C, the treatment with 50 μm of 2-BP reduced clumps formation in 50% whereas the treatment with 100 μm of inhibitor completely abolished the aggregation of these transfected parasites. These results indicate that palmitoylation of TvTSP8, as well as other palmitoylated proteins, contribute to parasite aggregation in transfected parasites. To evaluate if palmitoylation of TSPs is a general key regulator of cell aggregation, another member of the TSP family (TvTSP9) found in the palmitoyl-proteome (TVAG_287570) was cloned fused to an HA-tag and transfected in parasites (72). Nevertheless, the overexpression of TvTSP9 do not promote the formation of clumps (Fig. 5D); suggesting that the role in cell aggregation may be specific for TvTSP8. To evaluate if the effect on parasite aggregation on 2-BP treatment is caused by a change in membrane distribution of TvTSP8, we performed an IFA of TvTSP8-HA transfected parasites using an anti-HA antibody (Fig. 5E). As can be observed in Fig. 5E, the treatment with 2-BP showed no changes in the surface localization of the TvTSP8-HA; an expected result considering that TvTSP8 is a transmembrane protein. Then, we examined if 2-BP treatment of parasites expressing TvTSP8-HA affected protein distribution within the membrane by subcellular fractionation using optiprep gradient (48). To this end, TvTSP8 transfected parasites were treated with 2-BP or DMSO (vehicle) for 16 h, lysated and fractionated using optiprep gradient. Then, different gradient fractions were recovered and analyzed by Western blotting (Fig. 5F). TvTSP8 was recovered in all analyzed fractions and no change in its partition was observed when parasites were treated with 2-BP (Fig. 5F and supplemental Fig. S1). As control, the cytosolic protein TCTP was mostly recovered in the high-density fraction indicating a good separation with relatively little cross-contamination among the different fractions (Fig. 5F).
Protein Palmitoylation Mediates T. vaginalis Aggregation and Adherence to the Host Cell
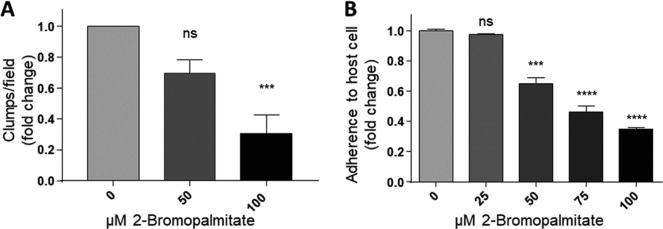
In order to evaluate the role of palmitoylation in a wild type strain, parasites from B7RC2 strain were treated with 2-BP and the effect in parasite/parasite aggregation was evaluated. Similar to data shown in Fig. 4 using transfected parasites, we observed that the B7RC2 strain exhibited a dose-dependent decrease in clump formation in the presence of 2-BP (Fig. 6A).
Fig. 6.
2-Bromopalmitate treatment inhibits T. vaginalis aggregation and attachment to the host cell. A, B7RC2 parasites (106 cells/ml) were treated with 50 or 100 μm of 2-BP for 4 h. Thirty 10× fields were visualized and the number of clumps per field was quantified. Four independent experiments were analyzed. Data are expressed as fold change related to untreated control parasites ± the standard error of the mean (S.E.). ANOVA followed by Tukey's post hoc test were used to determine significant differences. ***p < 0,001, ns; non-significant. B, Parasites were treated 16 h with increasing doses of 2-BP, washed, fluorescently labeled and incubated with HeLa cell monolayers for 30 min at 37 °C. Coverslips were washed to remove unbounded parasites, mounted and attached parasites were quantified by fluorescence microscopy. Data are expressed as fold change related to untreated control parasites ± the standard error of the mean (S.E.). ANOVA followed by Tukey's post hoc test were used to determine significant differences. ***p < 0.001, ****p < 0.0001, ns; non-significant. A representative experiment of three independent experiments is shown.
It has been previously shown that the formation of clumps in cell culture generally correlates with the ability of the strain to adhere and be cytotoxic to host cells (73). Specifically, highly adherent strains tends to aggregate when cultured in the absence of host cells in contrast to poorly adherent strains that generally do not form clumps in vitro (62, 73). Based on this observation and our results demonstrating that treatment with 2-BP led to a decrease in clumps formation (Fig. 6A), we evaluated whether this effect is accompanied with a decrease in parasite adherence to the host cells. Our results demonstrated that treatment with 100 μm of 2-BP resulted in a ∼66% decrease in parasite attachment to host cells when compared with untreated parasites (Fig. 6B). Importantly, although 2-BP might have pleiotropic effects, we have demonstrated that long-term (24 h) incubation of parasites with as high as 100 μm of 2-BP did not affect parasite viability, protein stability (supplemental Fig. S1B and S1C) and cell shape (data not shown). These results demonstrated for first time that protein palmitoylation may be a key factor in regulating the adherence of T. vaginalis to host cells.
DISCUSSION
S-palmitoylation consists in the covalent attachment of a 16-carbon chain palmitic acid to a cysteine residue of a target protein through a thioester bond, regulating thus the localization and/or function of the modified protein because of the consequent change in hydrophobicity (18, 19). Although several Palmitoyl-proteome analyses have been described in the last years, only few have been using protozoan parasites (12, 53–55). Interestingly, in all parasites where the role of palmitoylation has been described, it appears to be involved in vital processes such as invasion and motility (12, 53–55). In this context, here we performed the first large-scale description of palmitoylated proteins of the extracellular human pathogen T. vaginalis, identifying 363 putative palmitoylated proteins. Among the isolated proteins, 8 TvPAT-like molecules were identified. Interestingly, only one of them contain the classical DHHC-CRD domain whereas the remaining seven contain variants of the DHHC-CRD domain. Giardia lambia Gla_8711 contain a DHYC that is expressed in the parasite and participate in the encystation process (51). Particularly, Gla-8711 share some conserved amino acids within the CRD with the classical PATs found in human as well as in T. vaginalis (Fig. 2). Additionally, the DHYC domain has been described in Akr1 from yeast and it has been shown that it is involved in the proper membrane localization of the target kinase Yck2p (74). Unexpectedly, Akr1 share conserved amino acids with the new group found in T. vaginalis. It has been postulated that the conserved cysteines presents in the CRD participate in the formation of a zinc binding domain necessary for the correct function of the enzyme (75). However, because Akr1 do not have a CRD, hence, it is possible to speculate that zinc-binding domain would not be required for general palmitoylation activity.
Using the GPS-Lipid software, only 62% of these proteins are predicted to be palmitoylated with high confidence. However, it should be noted that, with few exceptions, there are many proteins for which the palmitoylated Cys residues are not associated with a defined consensus sequence. It is therefore difficult to predict by sequence alone whether a protein is a substrate for palmitoylation. Although the number of T. vaginalis proteins predicted to be palmitoylated by bioinformatics is lower than expected, 8% of the proteins that were not predicted to be palmitoylated by GPS-Lipid were also present in palmitoyl-proteomes from other protozoan parasites such as Toxoplasma gondii, Trypanosoma brucei and Plasmodium falciparum. Importantly, these shared proteins represent well known palmitoylated proteins (such as G-proteins, SNARE; VAMP, etc.) increasing the confidence of our results.
Interestingly, several proteins identified here were predicted by in silico analyses to be membrane proteins with possible roles in T. vaginalis pathogenesis (47, 60, 61, 65). These include proteins with similarity to BspA proteins of mucosal bacteria known to mediate adherence to host cells which represent the largest gene family encoding potential extracellular proteins in this pathogen (66); metalloproteinases (GP63) implicated in Leishmania virulence (61, 76); subtilisin-like serine peptidases (64), and tetraspanin-like family of proteins (62, 63). Additionally, proteins involved in a wide variety of functions that have yet to be examined and that may play important role in parasite biology were revealed by our analyses. Further validating our approach, molecules commonly found to be palmitoylated such as G-protein α subunits or GTPases family proteins, and SNARE proteins were isolated in our proteome. The work reported here sets the stage for detailed studies to address the roles of the newly identified palmitoylated proteins in T. vaginalis biology.
Significantly, our data also reveal that adherence and aggregation of T. vaginalis is highly dependent on the palmitoylated state of proteins as treatment with 2-BP leads to a decrease in parasite aggregation and adherence to host cell. These observations are consistent with the presence of proteins that might be involved in attachment to host cells in our proteome. T. vaginalis cells are known to form large cell aggregates, which could be important for pathogenesis (77) as highly adherent strains form clumps in cell culture in contrast to poorly adherent strains (62, 73). This observation has also been correlated with the ability of the strain to adhere and be cytotoxic to host cells (73). Results are consistent with our previous observation indicated that transfection of parasites with TvTSP8 promotes the ability of a parasite strain to aggregate (62). Here, we identified TvTSP8 in our proteome and further corroborate its palmitoylated status independently using ABE and immunoblotting assays. Tetraspanins organize laterally in membranes discrete tetraspanin-enriched microdomains (TEMs) (78, 79). TEMs contain primary complexes and tetraspanin homodimers occurring through direct protein-protein interactions (80). These are then brought together into extended secondary complexes, with tetraspanin palmitoylation playing a key role. In this sense, it has been shown that replacement of yuxtamembrane cysteines of CD9 and CD151 by other amino acids in mammalian cells, which abolished the palmitoylation, reduced their association with other tetraspanin proteins affecting the complex formation and the consequent function (80). Alternatively, the palmitoylation of CD81 shown to affect its association with a member of the serine/threonine-binding signaling protein family called 14-3-3s, affecting cell signaling (80). Finally, the palmitoylation of nontetraspanin partner proteins also could affect its incorporation into TEM, affecting its structure (80). In concordance, we observed that treatment with 2-BP of TvTSP8 transfected parasites abolished parasite aggregation and these results agree with reduced palmitoylation of TvTSP8. As no change in the cellular localization, stability and membrane partition of TvTSP8 was observed, it could be speculated that TvTSP8 association with partners and consequent signaling may be affected. Further studies are needed to explore this possibility. However, it is also possible that reduced parasite aggregation on 2-BP treatment is the consequence of reduced palmitoylation of other partner proteins. To evaluate if the observed effect in reduced parasite aggregation is exclusively dependent of palmitoylation of TvTSP8, we engineered a mutant version of the protein that lacks the six juxtamembrane predicted palmitoylated cysteine residues. Unfortunately, after six attempts with varying conditions, we were not able to obtain transfected parasites with this construct. Further investigations of why improper palmitoylation of TvTSP8 leads to a severe growth defect is required.
In summary, our results established for the first time a direct link between palmitoylation and the regulation of parasite aggregation and adherence to host cells, indicating that regulation of palmitoylation can lead to defects in pathogenesis. Thus, the present study encourages further work to pinpoint specific palmitoylated proteins and to further investigate their role in T. vaginalis biology.
DATA AVAILABILITY
The raw mass spectrometric data has been deposited. The location and identifying information are available at ftp://MSV000081060@massive.ucsd.edu.
Supplementary Material
Acknowledgments
We thank our colleagues in the lab for helpful discussions.
Footnotes
* This research was supported with a grant from the ANPCyT grant BID PICT 2013-1184 (NdM) and the NIH (RO1AI103182) (PJJ). NdM and MMC are researchers from the National Council of Research (CONICET) and UNSAM. YRN is a PhD fellow from CONICET. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
 This article contains supplemental Figure and Table.
This article contains supplemental Figure and Table.
1 The abbreviations used are:
- PTM
- post-translational modification
- PATs
- palmitoyl acyl-transferases
- NEM
- N-ethylmaleimide
- NH2OH
- hydroxylamine
- biotin-HPDP
- N-[6-(Biotinamido)hexyl]-3′-(2′-pyridyldithio) propionamide
- ABE
- acyl biotin exchange reaction
- BLAST
- basic local alignment search tool
- GO term
- gene ontology term
- BspA
- basic surface-exposed protein
- GP63
- glycoprotein 63
- 2-BP
- 2-bromopalmitate
- TEMs
- tetraspanin enriched microdomains
- DTT
- dithiothreitol
- SDS-PAGE
- sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SNARE
- SNAP soluble NSF attachment protein receptor
- TvTSP8
- Trichomonas vaginalis tetraspanin 8
- TYM
- trypticase, yeast extract, maltose medium
- DMEM
- Dulbecco's modified eagle medium
- EDTA
- ethylenediaminetetraacetic acid
- PBS
- phosphate-buffered saline
- TrichDB
- Trichomonas genomic resource
- NSAF
- normalized spectral abundance factor
- DMSO
- dimethyl sulfoxide
- PI
- propidium iodide
- CMAC
- CellTracke blue (7-amino-4-clorometilcoumarina)
- SEM
- scanning electron microscope.
REFERENCES
- 1. Gerbase A. C., Rowley J. T., Heymann D. H., Berkley S. F., and Piot P. (1998) Global prevalence and incidence estimates of selected curable STDs. Sex Transm. Infect. 74, S12–S16 [PubMed] [Google Scholar]
- 2. WHO. (2012) Baseline Report on Global Sexually Transmitted Infection Surveillance. World Health Organization, Geneva, Switzerland [Google Scholar]
- 3. Fichorova R. N. (2009) Impact of T. vaginalis infection on innate immune responses and reproductive outcome. J. Reprod. Immunol. 83, 185–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Swygard H., Miller W. C., Kaydos-Daniels S. C., Cohen M. S., Leone P. A., Hobbs M. M., and Sena A. C. (2004) Targeted screening for Trichomonas vaginalis with culture using a two-step method in women presenting for STD evaluation. Sex. Transm. Dis. 31, 659–664 [DOI] [PubMed] [Google Scholar]
- 5. McClelland R. S., Sangare L., Hassan W. M., Lavreys L., Mandaliya K., Kiarie J., Ndinya-Achola J., Jaoko W., and Baeten J. M. (2007) Infection with Trichomonas vaginalis increases the risk of HIV-1 acquisition. J. Infect. Dis. 195, 698–702 [DOI] [PubMed] [Google Scholar]
- 6. Van Der Pol B., Kwok C., Pierre-Louis B., Rinaldi A., Salata R. A., Chen P. L., van de Wijgert J., Mmiro F., Mugerwa R., Chipato T., and Morrison C. S. (2008) Trichomonas vaginalis infection and human immunodeficiency virus acquisition in African women. J. Infect. Dis. 197, 548–554 [DOI] [PubMed] [Google Scholar]
- 7. Gander S., Scholten V., Osswald I., Sutton M., and van Wylick R. (2009) Cervical dysplasia and associated risk factors in a juvenile detainee population. J. Pediatr. Adolesc. Gynecol. 22, 351–355 [DOI] [PubMed] [Google Scholar]
- 8. Stark J. R., Judson G., Alderete J. F., Mundodi V., Kucknoor A. S., Giovannucci E. L., Platz E. A., Sutcliffe S., Fall K., Kurth T., Ma J., Stampfer M. J., and Mucci L. A. (2009) Prospective study of Trichomonas vaginalis infection and prostate cancer incidence and mortality: Physicians' Health Study. J. Natl. Cancer Inst. 101, 1406–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sutcliffe S., Alderete J. F., Till C., Goodman P. J., Hsing A. W., Zenilman J. M., De Marzo A. M., and Platz E. A. (2009) Trichomonosis and subsequent risk of prostate cancer in the Prostate Cancer Prevention Trial. Int. J. Cancer 124, 2082–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Twu O., Dessi D., Vu A., Mercer F., Stevens G. C., de Miguel N., Rappelli P., Cocco A. R., Clubb R. T., Fiori P. L., and Johnson P. J. (2014) Trichomonas vaginalis homolog of macrophage migration inhibitory factor induces prostate cell growth, invasiveness, and inflammatory responses. Proc. Natl. Acad. Sci. U.S.A. 111, 8179–8184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Corvi M. M., Berthiaume L. G., and De Napoli M. G. (2011) Protein palmitoylation in protozoan parasites. Front. Biosci. 3, 1067–1079 [DOI] [PubMed] [Google Scholar]
- 12. Foe I. T., Child M. A., Majmudar J. D., Krishnamurthy S., van der Linden W. A., Ward G. E., Martin B. R., and Bogyo M. (2015) Global Analysis of Palmitoylated Proteins in Toxoplasma gondii. Cell Host Microbe 18, 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Santiago-Tirado F. H., Peng T., Yang M., Hang H. C., and Doering T. L. (2015) A Single Protein S-acyl Transferase Acts through Diverse Substrates to Determine Cryptococcal Morphology, Stress Tolerance, and Pathogenic Outcome. PLoS Pathogens 11, e1004908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Linder M. E., and Deschenes R. J. (2003) New insights into the mechanisms of protein palmitoylation. Biochemistry 42, 4311–4320 [DOI] [PubMed] [Google Scholar]
- 15. Lobo S., Greentree W. K., Linder M. E., and Deschenes R. J. (2002) Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 277, 41268–41273 [DOI] [PubMed] [Google Scholar]
- 16. Roth A. F., Feng Y., Chen L., and Davis N. G. (2002) The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 159, 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mitchell D. A., Mitchell G., Ling Y., Budde C., and Deschenes R. J. (2010) Mutational analysis of Saccharomyces cerevisiae Erf2 reveals a two-step reaction mechanism for protein palmitoylation by DHHC enzymes. J. Biol. Chem. 285, 38104–38114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blaskovic S., Blanc M., and van der Goot F. G. (2013) What does S-palmitoylation do to membrane proteins? FEBS J. 280, 2766–2774 [DOI] [PubMed] [Google Scholar]
- 19. Salaun C., Greaves J., and Chamberlain L. H. (2010) The intracellular dynamic of protein palmitoylation. J. Cell Biol. 191, 1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen X., Du Z., Li X., Wang L., Wang F., Shi W., and Hao A. (2016) Protein palmitoylation regulates neural stem cell differentiation by modulation of EID1 activity. Mol. Neurobiol. 53, 5722–5736 [DOI] [PubMed] [Google Scholar]
- 21. Corvi M. M., Soltys C. L., and Berthiaume L. G. (2001) Regulation of mitochondrial carbamoyl-phosphate synthetase 1 activity by active site fatty acylation. J. Biol. Chem. 276, 45704–45712 [DOI] [PubMed] [Google Scholar]
- 22. Kostiuk M. A., Keller B. O., and Berthiaume L. G. (2010) Palmitoylation of ketogenic enzyme HMGCS2 enhances its interaction with PPARalpha and transcription at the Hmgcs2 PPRE. FASEB J. 24, 1914–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Resh M. D. (2013) Covalent lipid modifications of proteins. Current Biol. 23, R431–R435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clark C. G., and Diamond L. S. (2002) Methods for cultivation of luminal parasitic protists of clinical importance. Clini. Microbiol. Rev. 15, 329–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Delgadillo M. G., Liston D. R., Niazi K., and Johnson P. J. (1997) Transient and selectable transformation of the parasitic protist Trichomonas vaginalis. Proc. Natl. Acad. Sci. U.S.A. 94, 4716–4720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wan J., Roth A. F., Bailey A. O., and Davis N. G. (2007) Palmitoylated proteins: purification and identification. Nat. Protocols 2, 1573–1584 [DOI] [PubMed] [Google Scholar]
- 27. Florens L., Carozza M. J., Swanson S. K., Fournier M., Coleman M. K., Workman J. L., and Washburn M. P. (2006) Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 40, 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Washburn M. P., Wolters D., and Yates J. R. 3rd. (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 29. Wohlschlegel J. A. (2009) Identification of SUMO-conjugated proteins and their SUMO attachment sites using proteomic mass spectrometry. Methods Mol. Biol. 497, 33–49 [DOI] [PubMed] [Google Scholar]
- 30. Melber A., Na U., Vashisht A., Weiler B. D., Lill R., Wohlschlegel J. A., and Winge D. R. (2016) Role of Nfu1 and Bol3 in iron-sulfur cluster transfer to mitochondrial clients. eLife 5, e15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kelstrup C. D., Young C., Lavallee R., Nielsen M. L., and Olsen J. V. (2012) Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole orbitrap mass spectrometer. J. Proteome Res. 11, 3487–3497 [DOI] [PubMed] [Google Scholar]
- 32. Xu T., Park S. K., Venable J. D., Wohlschlegel J. A., Diedrich J. K., Cociorva D., Lu B., Liao L., Hewel J., Han X., Wong C. C., Fonslow B., Delahunty C., Gao Y., Shah H., and Yates J. R. 3rd. (2015) ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J. Proteomics 129, 16–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aurrecoechea C., Brestelli J., Brunk B. P., Carlton J. M., Dommer J., Fischer S., Gajria B., Gao X., Gingle A., Grant G., Harb O. S., Heiges M., Innamorato F., Iodice J., Kissinger J. C., Kraemer E., Li W., Miller J. A., Morrison H. G., Nayak V., Pennington C., Pinney D. F., Roos D. S., Ross C., Stoeckert C. J. Jr, Sullivan S., Treatman C., and Wang H. (2009) GiardiaDB and TrichDB: integrated genomic resources for the eukaryotic protist pathogens Giardia lamblia and Trichomonas vaginalis. Nucleic Acids Res. 37, D526–D530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cociorva D., D L. T, and Yates J. R. (2007) Validation of tandem mass spectrometry database search results using DTASelect. Current protocols in bioinformatics/editoral board, Andreas D. Baxevanis. [et al.] Chapter 13, Unit 13.4 [DOI] [PubMed] [Google Scholar]
- 35. Elias J. E., and Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 36. Tabb D. L., McDonald W. H., and Yates J. R. 3rd. (2002) DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Di Rienzo J. A., Casanoves F., Balzarini M. G., Gonzalez L., Tablada M., and Robledo C. W. (2011) InfoStat. [Google Scholar]
- 38. Zybailov B., Mosley A. L., Sardiu M. E., Coleman M. K., Florens L., and Washburn M. P. (2006) Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5, 2339–2347 [DOI] [PubMed] [Google Scholar]
- 39. Xie Y., Zheng Y., Li H., Luo X., He Z., Cao S., Shi Y., Zhao Q., Xue Y., Zuo Z., and Ren J. (2016) GPS-Lipid: a robust tool for the prediction of multiple lipid modification sites. Sci. Reports 6, 28249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krogh A., Larsson B., von Heijne G., and Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 41. Kall L., Krogh A., and Sonnhammer E. L. (2004) A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036 [DOI] [PubMed] [Google Scholar]
- 42. Tsirigos K. D., Peters C., Shu N., Kall L., and Elofsson A. (2015) The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 43, W401–W407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nielsen H. (2017) Predicting Secretory Proteins with SignalP. Methods Mol. Biol. 1611, 59–73 [DOI] [PubMed] [Google Scholar]
- 44. Katoh K., Misawa K., Kuma K., and Miyata T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., and Drummond A. (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. de Miguel N., Lustig G., Twu O., Chattopadhyay A., Wohlschlegel J. A., and Johnson P. J. (2010) Proteome analysis of the surface of Trichomonas vaginalis reveals novel proteins and strain-dependent differential expression. Mol. Cell. Proteomics 9, 1554–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Macdonald J. L., and Pike L. J. (2005) A simplified method for the preparation of detergent-free lipid rafts. J. Lipid Res. 46, 1061–1067 [DOI] [PubMed] [Google Scholar]
- 49. Bastida-Corcuera F. D., Okumura C. Y., Colocoussi A., and Johnson P. J. (2005) Trichomonas vaginalis lipophosphoglycan mutants have reduced adherence and cytotoxicity to human ectocervical cells. Eukaryotic Cell 4, 1951–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Passmore L. A., and Barford D. (2004) Getting into position: the catalytic mechanisms of protein ubiquitylation. Biochem. J. 379, 513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Merino M. C., Zamponi N., Vranych C. V., Touz M. C., and Ropolo A. S. (2014) Identification of Giardia lamblia DHHC proteins and the role of protein S-palmitoylation in the encystation process. Plos Neglected Tropical Dis. 8, e2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Woehle C., Kusdian G., Radine C., Graur D., Landan G., and Gould S. B. (2014) The parasite Trichomonas vaginalis expresses thousands of pseudogenes and long non-coding RNAs independently from functional neighbouring genes. BMC Genomics 15, 906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Emmer B. T., Nakayasu E. S., Souther C., Choi H., Sobreira T. J., Epting C. L., Nesvizhskii A. I., Almeida I. C., and Engman D. M. (2011) Global analysis of protein palmitoylation in African trypanosomes. Eukaryotic Cell 10, 455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Caballero M. C., Alonso A. M., Deng B., Attias M., de Souza W., and Corvi M. M. (2016) Identification of new palmitoylated proteins in Toxoplasma gondii. Biochim. Biophys. Acta 1864, 400–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jones M. L., Collins M. O., Goulding D., Choudhary J. S., and Rayner J. C. (2012) Analysis of protein palmitoylation reveals a pervasive role in Plasmodium development and pathogenesis. Cell Host Microbe 12, 246–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Blanc M., David F., Abrami L., Migliozzi D., Armand F., Burgi J., and van der Goot F. G. (2015) SwissPalm: Protein Palmitoylation database. F1000Research 4, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hirt R. P., Lal K., Pinxteren J., Warwicker J., Healy B., Coombs G. H., Field M. C., and Embley T. M. (2003) Biochemical and genetic evidence for a family of heterotrimeric G-proteins in Trichomonas vaginalis. Mol. Biochem. Parasitol. 129, 179–189 [DOI] [PubMed] [Google Scholar]
- 58. Lal K., Noel C. J., Field M. C., Goulding D., and Hirt R. P. (2006) Dramatic reorganisation of Trichomonas endomembranes during amoebal transformation: a possible role for G-proteins. Mol. Biochem. Parasitol. 148, 99–102 [DOI] [PubMed] [Google Scholar]
- 59. Pritham E. J., Putliwala T., and Feschotte C. (2007) Mavericks, a novel class of giant transposable elements widespread in eukaryotes and related to DNA viruses. Gene 390, 3–17 [DOI] [PubMed] [Google Scholar]
- 60. Hirt R. P., de Miguel N., Nakjang S., Dessi D., Liu Y. C., Diaz N., Rappelli P., Acosta-Serrano A., Fiori P. L., and Mottram J. C. (2011) Trichomonas vaginalis pathobiology new insights from the genome sequence. Adv. Parasitol. 77, 87–140 [DOI] [PubMed] [Google Scholar]
- 61. Hirt R. P., Noel C. J., Sicheritz-Ponten T., Tachezy J., and Fiori P. L. (2007) Trichomonas vaginalis surface proteins: a view from the genome. Trends Parasitol. 23, 540–547 [DOI] [PubMed] [Google Scholar]
- 62. Coceres V. M., Alonso A. M., Nievas Y. R., Midlej V., Frontera L., Benchimol M., Johnson P. J., and de Miguel N. (2015) The C-terminal tail of tetraspanin proteins regulates their intracellular distribution in the parasite Trichomonas vaginalis. Cell. Microbiol. 17, 1217–1229 [DOI] [PubMed] [Google Scholar]
- 63. de Miguel N., Riestra A., and Johnson P. J. (2012) Reversible association of tetraspanin with Trichomonas vaginalis flagella on adherence to host cells. Cell. Microbiol. 14, 1797–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hernandez-Romano P., Hernandez R., Arroyo R., Alderete J. F., and Lopez-Villasenor I. (2010) Identification and characterization of a surface-associated, subtilisin-like serine protease in Trichomonas vaginalis. Parasitology 137, 1621–1635 [DOI] [PubMed] [Google Scholar]
- 65. Carlton J. M., Hirt R. P., Silva J. C., Delcher A. L., Schatz M., Zhao Q., Wortman J. R., Bidwell S. L., Alsmark U. C., Besteiro S., Sicheritz-Ponten T., Noel C. J., Dacks J. B., Foster P. G., Simillion C., Van de Peer Y., Miranda-Saavedra D., Barton G. J., Westrop G. D., Muller S., Dessi D., Fiori P. L., Ren Q., Paulsen I., Zhang H., Bastida-Corcuera F. D., Simoes-Barbosa A., Brown M. T., Hayes R. D., Mukherjee M., Okumura C. Y., Schneider R., Smith A. J., Vanacova S., Villalvazo M., Haas B. J., Pertea M., Feldblyum T. V., Utterback T. R., Shu C. L., Osoegawa K., de Jong P. J., Hrdy I., Horvathova L., Zubacova Z., Dolezal P., Malik S. B., Logsdon J. M. Jr, Henze K., Gupta A., Wang C. C., Dunne R. L., Upcroft J. A., Upcroft P., White O., Salzberg S. L., Tang P., Chiu C. H., Lee Y. S., Embley T. M., Coombs G. H., Mottram J. C., Tachezy J., Fraser-Liggett C. M., and Johnson P. J. (2007) Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315, 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Noel C. J., Diaz N., Sicheritz-Ponten T., Safarikova L., Tachezy J., Tang P., Fiori P. L., and Hirt R. P. (2010) Trichomonas vaginalis vast BspA-like gene family: evidence for functional diversity from structural organisation and transcriptomics. BMC Genomics 11, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nievas Y. R., Coceres V. M., Midlej V., de Souza W., Benchimol M., Pereira-Neves A., Vashisht A. A., Wohlschlegel J. A., Johnson P. J., and de Miguel N. (2017) Membrane-shed vesicles from the parasite Trichomonas vaginalis: characterization and their association with cell interaction. Cell. Mol. Life Sci. 75, 2211–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Twu O., de Miguel N., Lustig G., Stevens G. C., Vashisht A. A., Wohlschlegel J. A., and Johnson P. J. (2013) Trichomonas vaginalis exosomes deliver cargo to host cells and mediate hostratioparasite interactions. PLoS Pathogens 9, e1003482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Resh M. D. (2006) Use of analogs and inhibitors to study the functional significance of protein palmitoylation. Methods 40, 191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ryan C. M., de Miguel N., and Johnson P. J. (2011) Trichomonas vaginalis: current understanding of host-parasite interactions. Essays Biochem. 51, 161–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hemler M. E. (2008) Targeting of tetraspanin proteins–potential benefits and strategies. Nature Reviews. Drug Discovery 7, 747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Delgadillo M. G., Liston D. R., Niazi K., and Johnson P. J. (1997) Transient and selectable transformation of the parasitic protist Trichomonas vaginalis. Proc. Natl. Acad. Sci. U.S.A., pp. 4716–4720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lustig G., Ryan C. M., Secor W. E., and Johnson P. J. (2013) Trichomonas vaginalis contact-dependent cytolysis of epithelial cells. Infection Immunity 81, 1411–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Feng Y., and Davis N. G. (2000) Akr1p and the type I casein kinases act prior to the ubiquitination step of yeast endocytosis: Akr1p is required for kinase localization to the plasma membrane. Mol. Cell. Biol. 20, 5350–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gonzalez Montoro A., Quiroga R., and Valdez Taubas J. (2013) Zinc co-ordination by the DHHC cysteine-rich domain of the palmitoyltransferase Swf1. Biochem. J. 454, 427–435 [DOI] [PubMed] [Google Scholar]
- 76. Yao C., Donelson J. E., and Wilson M. E. (2003) The major surface protease (MSP or GP63) of Leishmania sp. Biosynthesis, regulation of expression, and function. Mol. Biochem. Parasitol. 132, 1–16 [DOI] [PubMed] [Google Scholar]
- 77. Honigberg B. M. (1990) Host Cell-Trichomonad Interactions and Virulence Assays Using in In Vitro Systems. Trichomonads parasitic in humans, pp. x, 424 p., Springer-Verlag, New York [Google Scholar]
- 78. Berditchevski F., Odintsova E., Sawada S., and Gilbert E. (2002) Expression of the palmitoylation-deficient CD151 weakens the association of alpha 3 beta 1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signaling. J. Biol. Chem. 277, 36991–37000 [DOI] [PubMed] [Google Scholar]
- 79. Hemler M. E. (2003) Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Ann. Rev. Cell Developmental Biol. 19, 397–422 [DOI] [PubMed] [Google Scholar]
- 80. Levy S., and Shoham T. (2005) Protein-protein interactions in the tetraspanin web. Physiology 20, 218–224 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw mass spectrometric data has been deposited. The location and identifying information are available at ftp://MSV000081060@massive.ucsd.edu.