The ComRS system can function as a quorum sensing trigger for genetic competence in S. mutans. The signal peptide XIP, which is derived from the precursor ComS, enters the cell and interacts with the Rgg-type cytosolic receptor ComR to activate comX, which encodes the alternative sigma factor for the late competence genes. Previous studies have demonstrated intercellular signaling via ComRS, although release of the ComS or XIP peptide to the extracellular medium appears to require lysis of the producing cells. Here we tested the complementary hypothesis that ComRS can drive comX through a purely intracellular mechanism that does not depend on extracellular accumulation or import of ComS or XIP. By combining single-cell, coculture, and microfluidic approaches, we demonstrated that endogenously produced ComS can enable ComRS to activate comX without requiring processing, export, or import. These data provide insight into intracellular mechanisms that generate noise and heterogeneity in S. mutans competence.
KEYWORDS: Streptococcus mutans, competence, microfluidics, quorum sensing
ABSTRACT
Entry into genetic competence in streptococci is controlled by ComX, an alternative sigma factor for genes that enable the import of exogenous DNA. In Streptococcus mutans, the immediate activator of comX is the ComRS quorum system. ComS is the precursor of XIP, a seven-residue peptide that is imported into the cell and interacts with the cytosolic receptor ComR to form a transcriptional activator for both comX and comS. Although intercellular quorum signaling by ComRS has been demonstrated, observations of bimodal expression of comX suggest that comRS may also function as an intracellular feedback loop, activating comX without export or detection of extracellular XIP. Here we used microfluidic and single-cell methods to test whether ComRS induction of comX requires extracellular XIP or ComS. We found that individual comS-overexpressing cells activate their own comX, independently of the rate at which their growth medium is replaced. However, in the absence of lysis they do not activate comS-deficient mutants growing in coculture. We also found that induction of comR and comS genes introduced into Escherichia coli cells leads to activation of a comX reporter. Therefore, ComRS control of comX does not require either the import or extracellular accumulation of ComS or XIP or specific processing of ComS to XIP. We also found that endogenously and exogenously produced ComS and XIP have inequivalent effects on comX activation. These data are fully consistent with identification of intracellular positive feedback in comS transcription as the origin of bimodal comX expression in S. mutans.
IMPORTANCE The ComRS system can function as a quorum sensing trigger for genetic competence in S. mutans. The signal peptide XIP, which is derived from the precursor ComS, enters the cell and interacts with the Rgg-type cytosolic receptor ComR to activate comX, which encodes the alternative sigma factor for the late competence genes. Previous studies have demonstrated intercellular signaling via ComRS, although release of the ComS or XIP peptide to the extracellular medium appears to require lysis of the producing cells. Here we tested the complementary hypothesis that ComRS can drive comX through a purely intracellular mechanism that does not depend on extracellular accumulation or import of ComS or XIP. By combining single-cell, coculture, and microfluidic approaches, we demonstrated that endogenously produced ComS can enable ComRS to activate comX without requiring processing, export, or import. These data provide insight into intracellular mechanisms that generate noise and heterogeneity in S. mutans competence.
INTRODUCTION
Streptococcus mutans inhabits human oral biofilms and is a primary etiological agent of dental caries (1). Many of the behaviors that facilitate the growth, competition, stress tolerance, and virulence of S. mutans are linked to the regulation of genetic competence, a transient physiological state during which the organism can internalize DNA from its environment (2–7). The competence pathway of S. mutans is complex, as it receives input from extracellular peptide signals and environmental cues as well as regulatory feedback (8–14). Consequently, several elements of the mechanism and dynamics of the pathway are not well understood.
S. mutans initiates entry into the competent state by increasing the transcription of the comX gene (sometimes referred to as sigX), which encodes an alternative sigma factor that is required for the expression of approximately 30 late competence genes (15, 16). Expression of comX can be induced by the peptides CSP (competence-stimulating peptide) and XIP (sig X-inducing peptide), and the efficacy of these peptides is strongly influenced by environmental conditions. CSP is derived by cleavage of a 21-residue peptide from ComC and exported through an ATP-binding cassette transporter. It is further processed to the active 18-residue peptide by the SepM protease (17). Extracellular CSP is detected by the two-component signal transduction system ComDE, with the phosphorylated response regulator ComE activating genes for bacteriocin synthesis and immunity. ComE does not directly activate comX but affects comX indirectly, via a pathway that is not completely understood (18).
The immediate regulator of comX in S. mutans and in streptococci of the salivarius, bovis, and pyogenic groups is the ComRS system. ComR is an Rgg-like cytosolic transcriptional regulator, and the type II ComS of S. mutans is a 17-residue peptide (19). The C terminus of ComS contains XIP, a 7-residue small hydrophobic peptide (SHP). Extracellular XIP is imported by the Opp permease and interacts with ComR to form a transcriptional activator for both comX and comS (19, 20). Notably, the S. mutans competence pathway contains at least two positive-feedback loops, as XIP/ComR activates comS and ComX activates comE expression (19, 20).
An intriguing property of S. mutans competence is that although exogenous CSP and XIP can both activate comX and induce transformability, they do so under different environmental conditions and elicit qualitatively different behaviors in the expression of comX (11). Exogenous CSP elicits a bimodal response in which less than half of the population activates comX, whereas exogenous XIP elicits a unimodal response in which all cells in the population activate comX. Further, CSP activates comX only in complex media containing small peptides; this activation requires that cells carry an intact comS but does not require the Opp permease (11). By contrast, exogenous XIP activates comX only in defined media lacking small peptides (11, 21); this activation requires that cells carry the opp gene but does not require comS (19). Therefore, although exogenous XIP can activate comX in a strain lacking comS, the bimodal comX response to CSP requires an intact comS gene.
The observation that competence in several streptococcal species is directly stimulated by an extracellular ComS-derived peptide suggests that ComRS constitutes a novel type of Gram-positive quorum signaling system, in which the ComS-derived XIP signal is processed and secreted, accumulates in the extracellular medium, and is then reimported. This interpretation in S. mutans is supported by several experimental observations. First, cells that carry opp take up exogenous XIP (in defined medium) and activate comX with high efficiency (19, 21). Second, exogenous synthetic XIP is dramatically more effective in stimulating transformability than is exogenous full-length ComS (19). Third, filtrates of S. mutans cultures grown to an optical density at 550 nm (OD550) of 0.4 in defined medium were able to stimulate a PcomX reporter strain (21). Moreover, liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of supernatants of S. mutans cultures grown to high density (22, 23) detected micromolar quantities of XIP. Fourth, a transposon mutagenesis screen in S. pyogenes identified the widely conserved pptAB ABC transporter as a possible exporter of short hydrophobic peptides of the ComS type (24), raising the possibility that S. mutans may also possess dedicated mechanisms for processing and export of ComS/XIP.
However, such mechanisms have not yet been identified in S. mutans. There is evidence that the Eep membrane protease facilitates the processing of S. thermophilus ComS (25), but Eep did not affect processing of ComS of S. mutans (22). Further, although S. mutans appears to encode a gene product with a fairly high degree of homology to PptAB of S. pyogenes, deletion of the apparent pptAB genes had only a weak effect on competence induction in mid-exponential-phase cultures of S. mutans (24). Consequently, the processing of ComS to XIP remains uncharacterized. The import of XIP presents an additional puzzle for ComRS quorum signaling because the permease Opp is required for XIP to activate comX but is not required for activation by CSP.
We recently demonstrated that XIP can function as a diffusible, intercellular signal (26) in S. mutans. A ComS-producing strain was able to induce comX response in cocultured cells lacking comS, in the absence of physical contact. However, intercellular signaling was greatly impaired by deletion of the atlA gene encoding the primary autolysin. Therefore, lysis and not active transport appears to be the primary route by which S. mutans externalizes the diffusible signal.
Here we used a combination of microfluidic and coculture methods to test the complementary hypothesis that ComRS can also activate comX by an intracellular mechanism, through endogenous ComS production, and that this mechanism does not specifically require processing of ComS/XIP or its extracellular accumulation.
RESULTS
An intact copy of comS alters the comX response to exogenous XIP.
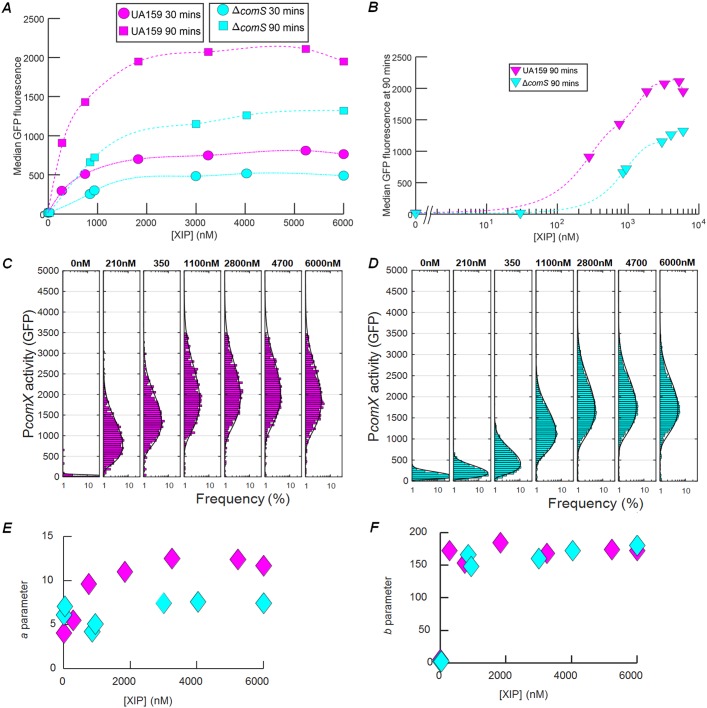
A previous study found that even when a comS deletion strain (mutant ΔcomS) and a wild-type (WT) background strain were supplied with high concentrations of synthetic XIP, the transformation efficiency of the ΔcomS strain was only half that of the wild type (19). Panels A and B of Fig. 1 show results from a similar experiment performed with individual cells carrying a PcomX-gfp plasmid-borne reporter. S. mutans UA159 (wild-type strain) and mutant ΔcomS genetic backgrounds were imaged while adhered within a microfluidic chamber and supplied with a constant flow of defined medium (FMC [see Materials and Methods]) containing synthetic XIP. Although both strains respond to exogenous XIP, the ΔcomS strain consistently showed roughly 1.5-fold-lower PcomX activity than the wild type, even at saturating XIP concentrations. The threshold for comX response occurred at a roughly 2-fold-lower XIP concentration in UA159 than in the ΔcomS strain (Fig. 1B). Therefore, the deletion of comS elevated the threshold for a response to extracellular XIP and reduced the overall response at saturation.
FIG 1.
comS deletion is not fully complemented by synthetic XIP. (A) Comparison of levels of PcomX-gfp activity in S. mutans cells of the UA159 (wild-type) background (magenta) and ΔcomS mutant background (cyan). The median GFP fluorescence is shown for cells that were supplied with a continuous flow of exogenous synthetic XIP in FMC medium in microfluidic chambers. Data are shown at 30 min (circles, dashed-dotted lines) and 90 min (squares, dashed lines) of flow. The smooth curves represent spline fits to the data. (B) Median GFP levels in the two strains at the 90-min time point of the flow experiment. (C and D) Also shown are the histograms of the individual cell PcomX-gfp reporter activity versus exogenous XIP concentration for (C) the UA159 background and (D) the ΔcomS background. Solid black curves in panels C and D show the best-fit gamma probability distribution for each histogram. A cutoff of 40 units of PcomX-gfp fluorescence has been applied to exclude background autofluorescence. (E) Parameter a of the (two-parameter) gamma probability distribution, obtained from the fits in panels C and D, reflecting the ratio of transcription rate to protein degradation rate. (F) Distribution parameter b, reflecting the ratio of the rate of translation to the rate of mRNA degradation. In panels E and F, cyan indicates the ΔcomS mutant and magenta indicates the UA159 background.
The deletion of comS also affected cell-to-cell variability (noise) in comX expression. Panels C and D of Fig. 1 show that the histograms of reporter fluorescence differ in UA159 (wild-type strain) and mutant ΔcomS cells. UA159 showed a generally broader (noisier) comX response than did mutant ΔcomS. We quantified this difference by fitting the histograms to a gamma distribution Γ(n | a,b), a two-parameter probability distribution that can be used to model cell-to-cell variability in n, the copy number for a bacterial protein (27). In a simple physical model, parameter a of the gamma distribution is related to the number of mRNAs produced during the cell division time, while parameter b is related to the number of protein copies produced per mRNA transcript (28). As shown in Fig. 1E and F, the UA159 background had a roughly 2-fold-higher value for parameter a (transcription rate), while the values for parameter b (translation) were similar for the two strains. As this difference persisted even at XIP concentrations exceeding 1 μM, these data show that deletion of comS significantly affected comX expression, even when excess extracellular XIP was provided.
Fluid replacement did not alter induction of comX in a strain overexpressing comS.
To test whether comX activation requires cells to import extracellular XIP from their environment, we examined the effect of fluid flow rate in cells that overexpressed comS from the strong P23 promoter on the plasmid pIB184 (26, 29) while adhered in a microfluidic flow chamber. Cells carrying the 184comS overexpression plasmid produce significantly higher levels of comS mRNA (see Fig. S1 in the supplemental material) and activate comX in defined medium lacking exogenous CSP or XIP (26). We anticipated that if cells were immobilized and supplied with a continuous flow of fresh medium, high flow rates would remove any XIP (or ComS) that was released, leading to diminished comX activity. In order to prevent XIP import via Opp, cells were supplied with a flow of complex medium (brain heart infusion [BHI] medium) lacking XIP or CSP. We loaded 184comS PcomX-rfp cells, which carry both the comS overexpression plasmid and a plasmid-borne PcomX-rfp reporter, into four microfluidic chambers using different flow rates. Chambers were supplied with fresh complex medium flowing at rates between 0.02 ml h−1 and 1 ml h−1. These flow rates were sufficient to completely replace the growth medium within each chamber in time intervals ranging from 6 s to 10 min. We also studied (i) a PcomX-rfp reporter in a UA159 background (negative control) and (ii) a ComS-overproducing strain lacking a start codon (ATG point mutation to AAG) on its chromosomal comS gene (mutant 184comS PcomX-rfp ΔcomS).
Figure 2A shows that PcomX was not activated in the UA159 background (leftmost column). In contrast, the strain overexpressing comS and harboring an intact chromosomal copy of comS showed a highly heterogeneous response, indicating that a subpopulation of these cells strongly activated comX in the flowing complex medium. Further, the rate of fluid flow had no effect on their comX expression. Figure 2B shows the fluorescence of the individual cells for which the signal exceeded the maximum level of PcomX activity (roughly 100 fluorescence units) seen in the UA159 negative control. Rather than declining at high flow rates where the medium was rapidly replaced, the median comX activity in the ComS-overproducing cells actually showed a very slight increase, smaller than the cell-to-cell variability. These data show that overexpression of comS can allow S. mutans to activate comX, even in complex medium which normally does not permit activation of comX transcription by exogenously supplied XIP. The finding that this activation is unaffected by rapid replacement of the medium implies that the comX response in the strain overexpressing comS was not due to accumulation of extracellular XIP (or ComS). Curiously, however, this comX response did require an intact chromosomal comS gene. Figure 2 shows that very few ΔcomS cells activated comX, even though they harbored the comS overexpression plasmid. Together with Fig. 1, these data show that the chromosomal comS gene plays a role in comX activation that is not fully complemented either by saturating concentrations of exogenous XIP or by endogenous overproduction of ComS.
FIG 2.
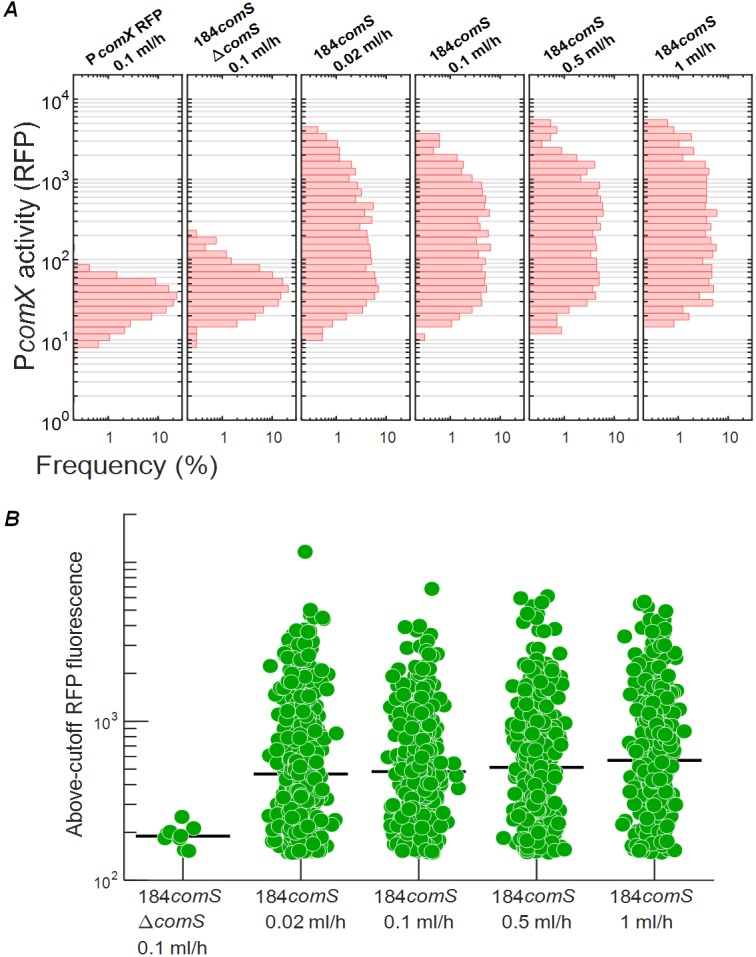
Activation of ComX in a ComS-overexpressing strain is independent of the rate of medium replacement. PcomX-rfp reporter activity is shown in cells growing in microfluidic chambers supplied with continuously flowing fresh complex medium (BHI). (A) Histograms of individual cell PcomX-rfp reporter fluorescence. (First column) wild-type background (negative control) with flow at 0.1 ml h−1. (Second column) ComS-overexpressing 184comS ΔcomS background at 0.1 ml h−1. (Columns 3 to 5) ComS-overexpressing (184comS) background at 0.02 ml h−1, 0.1 ml h−1, 0.5 ml h−1, and 1 ml h−1. (At a flow rate of 1 ml h−1, the medium in each flow chamber is replaced every 6 s.) (B) RFP fluorescence of cells that exceeded the wild type (negative-control) red fluorescence in column 1 of panel A. The black bar indicates the median of data in each channel. (Leftmost column) 184comS ΔcomS background. (Columns 2 to 5) ComS-overexpressing (184comS) background. All RFP measurements were made 4 h after addition of chloramphenicol to the cultures.
Measurement of comS, comX, and comR transcript levels. (A to C) RT-qPCR measurement of (A) comS, (B) comX, and (C) comR transcripts in cultures harvested at an OD600 of 0.5. Each bar indicates the ratio of the median transcript count to the median 16S rRNA count, as measured in multiple biological and technical replicates (see Materials and Methods). WT/BHI, 184comS, and 184comS ΔcomS samples were grown in BHI medium. The remaining samples were grown in FMC. Synthetic XIP was supplied at a 100 nM concentration when used. Error bars indicate the range from the second-lowest to the second-highest value among the replicates for each condition. (D) Comparison of comS mRNA transcripts from S. mutans harboring the ComS overexpression plasmid versus the empty plasmid. Download FIG S1, TIF file, 0.1 MB (79.3KB, tif) .
Copyright © 2018 Underhill et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The presence of comS in the chromosome affects comS mRNA levels.
We used reverse-transcription quantitative PCR (RT-qPCR) to measure comS, comR, and comX transcript copy numbers in mid-exponential-phase cultures of the ComS overexpression, comS deletion, and UA159 strains (Fig. S1). Transcription of comS (Fig. S1A) was significantly increased in cells treated with exogenously added XIP in defined medium compared to controls lacking XIP or growing in complex medium. In addition, cells overexpressing comS in a wild-type genetic background (184comS) and growing in complex medium had comS transcript levels that were significantly higher than those seen with a strain overexpressing comS that lacked the chromosomal copy of comS (strain 184comS ΔcomS). Levels of comR transcripts showed no significant change across strains or conditions (Fig. S1C).
The population density of cells overexpressing comS does not determine the comX response.
To further test whether comS-overexpressing cells release extracellular XIP, we measured the effect of comS-overexpressing (sender) populations on comX activation in comS-deficient (receiver) cells growing in coculture. We mixed sender (184comS PcomX-rfp) and receiver (PcomX-gfp ΔcomS) cultures in different ratios and loaded them into microfluidic chambers containing static medium without exogenous XIP. We anticipated that if the senders released XIP or ComS into the extracellular medium, both senders (red fluorescent protein [RFP] reporter) and receivers (green fluorescent protein [GFP] reporter) would respond by activating comX and that the average activation would increase with the ratio of senders to receivers. We used defined medium (FMC) to facilitate import of ComS/XIP if present.
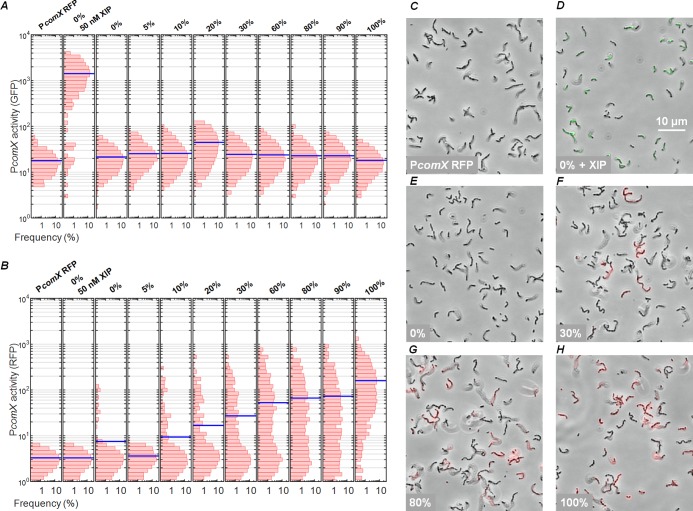
We analyzed the green and red fluorescence of the cocultures to generate histograms of individual levels of cell fluorescence that revealed both the receiver (green) and sender (red) comX responses, shown in Fig. 3A and B, respectively. Representative microscopy images are shown in panels C to H of Fig. 3. In a control chamber containing only receiver cells exposed to 50 nM XIP, the GFP fluorescence was enhanced but RFP fluorescence remained at the baseline level (see Fig. 3D and the second column in Fig. 3A and B). Similarly, a control chamber containing only sender cells showed enhanced RFP fluorescence but only baseline GFP fluorescence (Fig. 3H and rightmost column in Fig. 3A and B). As expected, the median RFP fluorescence of the cocultures (Fig. 3B and F to H) increased at high sender/receiver ratios. The median RFP fluorescence of cells in the coculture increased approximately in proportion to the density of senders, as expected if each sender activated its own comX. RFP fluorescence was constant over a period of 4 h (Fig. S2).
FIG 3.
ComS overexpresser does not induce expression of comX of strain ΔcomS in coculture. (A and B) Histograms of (A) GFP and (B) RFP fluorescence of individual cells in cocultures of sender (184comS PcomX-rfp) and receiver (PcomX-gfp ΔcomS) strains in FMC medium. Strains were grown to equal levels of optical density, mixed in various proportions, and then incubated in microfluidic chambers containing stationary defined medium. Blue lines indicate population medians. Panels A and B show fluorescence of the UA159 background strain containing PcomX rfp reporter, without added XIP (negative control) (first column); of strain PcomX-gfp ΔcomS (receiver) with added 50 nM XIP (positive control) (second column); and of cocultures of sender and receiver, with columns labeled by percentage by volume of 184comS (sender) culture in the initial preparation of the coculture. (C to H) Phase-contrast images of cocultures, overlaid with red and green fluorescence images. (C) PcomX-rfp reporter in UA159 background, with no added XIP (negative control). (D) PcomX-gfp ΔcomS cells with 50 nM added XIP (positive control). (E) PcomX-gfp ΔcomS (receiver) alone, with 0% sender. (F to H) Cocultures containing 30%, 80%, and 100% sender, respectively.
Response of cocultures is time independent. (A) GFP (comX reporter) and (B) RFP (comY) fluorescence of individual cells in cocultures of sender (184comS PcomX-rfp) and receiver (PcomX-gfp ΔcomS) strains of S. mutans. Samples are labeled by percentage by volume of 184comS (sender) culture in the initial preparation of the coculture. Fluorescence was measured immediately (0 h, green) after mixing the coculture or 4 h (magenta) after mixing. The red horizontal bars show the median fluorescence immediately after mixing (0 h); the black vertical bars show the median at 4 h. Data are from the coculture experiment described in the Fig. 3 legend. Download FIG S2, TIF file, 0.2 MB (182.1KB, tif) .
Copyright © 2018 Underhill et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
However, when exogenous XIP was not provided, the GFP fluorescence of cocultures showed no dependence on the sender/receiver ratio. Over the 0% to 100% range of coculture ratios shown in Fig. 3A, the GFP fluorescence histograms did not exceed the baseline level of the negative control (ΔcomS strain alone) (see the third column of Fig. 3A). This GFP response did not change appreciably over a period of 4 h (Fig. S2). Senders did not activate comX in the ΔcomS receivers at any mixing ratio.
These data show that although overexpression of comS stimulated comX within individual cells, over the 4-h time scale of this experiment, this activation did not cause extracellular, diffusible XIP to accumulate to levels sufficient to activate nearby ΔcomS cells.
Growth phase-dependent release of XIP.
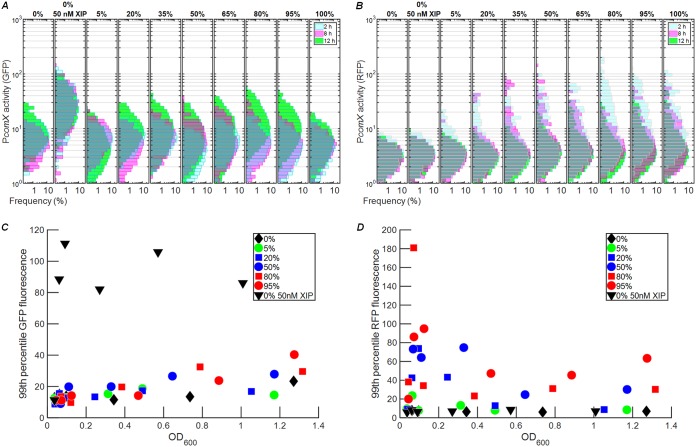
We previously showed that intercellular signaling by S. mutans ComRS can occur late in growth but is impeded by deletion of the atlA gene, which encodes a major autolysin (26, 30). Loss of AtlA inhibits cell lysis, which appears to occur primarily in stationary phase. We therefore tested whether competence signaling from sender (comS-overexpressing) cells to receiver (ΔcomS) cells would be observable in the later phases of growth. We prepared sender/receiver cocultures in different ratios in defined medium (to favor import of extracellular XIP if present). Because low pH suppresses the comX response, we adjusted the pH during the experiment to ensure that cells would remain responsive to comX-activating signals if present (9, 13). Every 2 h, the pH of the cultures was adjusted to 7.0 by addition of 2 N NaOH, the OD600 was recorded, and an aliquot of the culture was collected for fluorescence imaging of comX promoter activity. The GFP fluorescence histograms in Fig. 4A show that comX expression in the ΔcomS strain increased very slightly at 12 h in comparison to expression at 2 h or 8 h. This increase was more pronounced at higher ratios of sender cells to receiver cells, consistent with some release of XIP from lysing senders late in the growth phase. The histograms of Fig. 4B, like those of Fig. 3B, show a strong RFP response (consistent with autoactivation of senders) with a slightly stronger response at earlier times.
FIG 4.
Evidence for release of XIP in cocultures late in growth. (A and B) Histograms of (A) GFP fluorescence and (B) RFP fluorescence of individual cells in cocultures of receiver (PcomX-gfp ΔcomS) and sender (PcomX-rfp 184comS) strains in FMC, following different incubation periods. Labels at the top indicate the volume fraction comprised of the sender strain in the preparation of the coculture. Histogram colors indicate incubation times as follows: cyan, 2 h; magenta, 8 h; green, 12 h. The lower panels show the 99th percentile of the individual cell GFP fluorescence (C) and RFP fluorescence (D) observed in the culture, versus the culture OD600. Exogenous XIP was not added, except in the positive-control (sender-only) sample indicated by the inverted triangles in panels C and D.
The median GFP and RFP signals in the histograms described above do not shift dramatically with either time or coculture ratio. However, the histograms in Fig. 4A suggest moderate, density-dependent increases in receiver (green) fluorescence at 12 h. Panels C and D of Fig. 4 highlight these changes by showing the value of the 99th percentile of red and green fluorescence, respectively, in the cultures versus optical density. As shown in Fig. 4C, the GFP fluorescence of the most active receivers increased slightly at higher OD600 values, a change that was slightly more pronounced at higher sender/receiver ratios. By contrast, Fig. 4D shows no strong trend in the RFP fluorescence of the 99th percentile of senders versus OD600. None of the cocultures exhibited as strong a GFP response as the positive control (receiver plus 50 nM synthetic XIP; Fig. 4C), indicating that even 12 h of growth did not lead to an extracellular XIP accumulation of as large as 50 nM. Overall, these data are consistent with robust self-activation of the comS overexpressing strain, accompanied by a modest level of release of XIP (or ComS) to the extracellular medium during the late stages of the growth curve.
Binding of ComR to the comS and comX promoters in vitro.
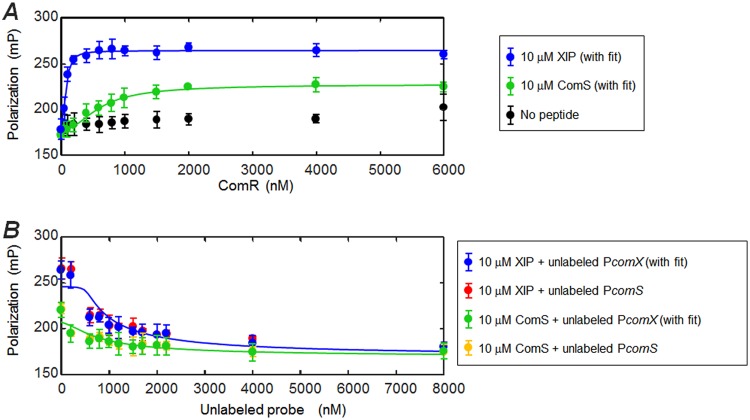
The observation of comX activation in the overexpressing (sender) strain, without significant accumulation of XIP in the medium, implies that activation of comX does not require export and reimport of XIP or ComS if the chromosomal copy of comS is intact. To test whether endogenously produced ComS, acting intracellularly, is sufficient to activate comX, we tested whether unprocessed ComS could enable the binding of ComR to the comS and comX promoter regions in vitro. Figure 5A shows a fluorescence polarization (FP) assay with purified recombinant ComR, synthetic XIP or ComS, and a fluorescently labeled DNA oligomer corresponding to the S. mutans comX promoter region containing the ComR binding site. The assay was performed in the presence and absence of excess (10 µM) ComS or XIP. Because of the excess of peptide (10 μM) relative to fluorescent DNA probe (1 nM), the probe polarization depends primarily on the concentration of ComR added. Figure 5A shows that in the absence of XIP or ComS, ComR caused a weak rise in the fluorescence polarization of the DNA oligomer, indicating relatively weak affinity, as observed for ComR of other streptococci (20). However, in the presence of ComS or XIP the binding isotherm saturated at lower ComR concentrations, indicating formation of a complex with higher affinity for the comX promoter. Histidine tagging of ComR was found to reduce this affinity, as shown in Fig. S3.
FIG 5.
ComS and XIP interact with ComR to bind the comX promoter. A fluorescence polarization assay testing interaction of ComS and XIP peptides with ComR and the PcomX transcriptional activation site was performed. The DNA probe was labeled with a Bodipy FL-X fluorophore. (A) Titration of ComR into a solution containing 10 µM XIP (blue) or full-length ComS (green), labeled DNA probe (1 nM), and 0.05 mg ml−1 salmon DNA. The negative control (black) contained no ComS or XIP peptide. (B) Competition assay in which unlabeled promoter sequence DNA was titrated into a solution containing ComR (1.5 µM), fluorescent DNA probe (1 nM), and peptide (either ComS or XIP, 10 µM). Unlabeled PcomX DNA was used with XIP (blue) and ComS (green). An unlabeled PcomS probe was also tested for its ability to compete with the fluorescent PcomX probe in the presence of XIP (red) and ComS (gold). Solid curves indicate binding and competition behavior predicted by the two-step model described in Materials and Methods, in which peptide (ComS or XIP) first forms a multimeric complex with ComR (k1, n) and a single copy of this complex binds to the (labeled or unlabeled) PcomX DNA (k2). For ComS binding/competition (green), the curves represent the following values: k1 = 3.2 μM, k2 = 2.2 nM, n = 2.5. For XIP binding/competition (blue), the curves represent the following values: k1 = 7.3 μM, k2 = 33 nM, n = 2.4.
Effect of histidine tag on ComR binding of ComS and XIP. A fluorescence polarization study of ComS and XIP interaction with ComR that was N- or C-terminally tagged with 6×-histidine was performed. Polarization is plotted versus [ComR] for (A) C-terminally tagged ComR and (B) N-terminally tagged ComR. In each case, 1 nM fluorescent DNA and 0.05 mg ml−1 salmon DNA were present along with no signal peptides (black), XIP (blue), or ComS (green). The comR gene (S. mutans 61 [SMu.61]) was amplified using gene-specific primers (forward, AAAGAATCCTATGTTAAAAGA; reverse, CACCCTAGGAGACCCATCAAA) and cloned into the pET45b(+) vector bearing an N-terminal 6×His tag. The resulting vector, pET45b(+)his-comRUA159, was transformed into E. coli 10-beta. After sequencing confirmed the correct insertion (using T7 promoter and T7 terminator primers), the vector was transformed into E. coli BL21(DE3) prior to protein purification. Download FIG S3, TIF file, 0.1 MB (103.6KB, tif) .
Copyright © 2018 Underhill et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Figure 5B shows results of a competition assay in which unlabeled (“cold”) PcomX and PcomS DNA oligomers having the same stem-loop structure as the labeled probe were titrated into samples containing 1.5 µM ComR or 10 µM ComS or XIP and the labeled DNA (1 nM). The systematic decrease in polarization is consistent with competition for ComR. The unlabeled PcomS and PcomX probes, which differ by 3 bases, appear to have identical levels of affinity for ComR.
Figure 5A confirms a ComR-dependent interaction between ComS and the DNA probe, although this interaction is weaker than that of XIP. It also shows that, at saturating concentrations of ComR, ComS elicits only half of the total fluorescence polarization that results from an equivalent concentration of XIP. This difference may suggest that ComS and XIP induce qualitatively different interactions between the ComR/peptide complexes and the DNA probe. The solid curves in panels A and B of Fig. 5 were calculated from a two-step, cooperative binding model (see Materials and Methods) in which the dissociation constants for the ComR/peptide complex (k1) and the complex/promoter (k2), as well as the order of multimerization n of the ComR/peptide complex, are variables. Although the data clearly indicate that both ComS and XIP interact with ComR to bind the DNA probe, they do not permit a precise determination of the ComS and XIP interaction parameters. As discussed in Materials and Methods, the data are consistent with a range of parameter values, corresponding roughly to micromolar k1 and nanomolar k2 for both ComS and XIP and to similar cooperativity n values for the two peptides. The curves in Fig. 5 show the model with a roughly 2-fold difference in the ComR dissociation constants for ComS (k1 = 3.2 μM, n = 2.5) and XIP (k1 = 7.3 μM, n = 2.4).
We note that the binding observed with the ComS peptide is likely not attributable to a XIP impurity in the ComS, as ComS and XIP were used at levels of high excess and so a XIP impurity would have caused the blue and green curves in Fig. 5 to be much more qualitatively and quantitatively similar.
ComRS can drive expression from the comX promoter in Escherichia coli.
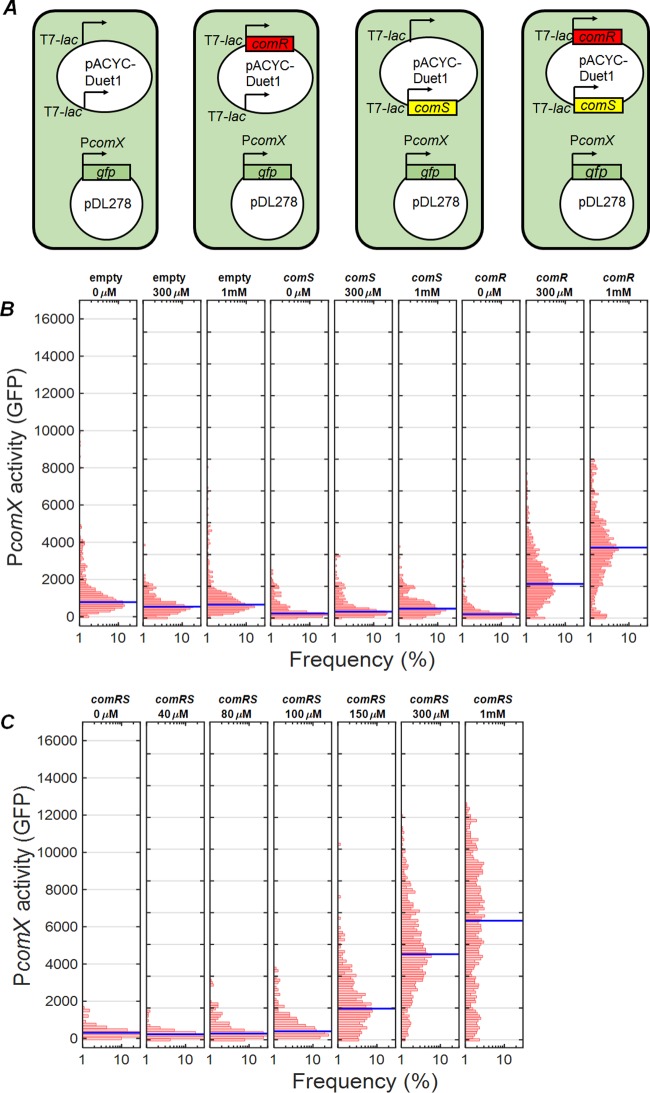
As discussed above, the OppA permease, which imports extracellular XIP, is not required for CSP-driven stimulation of ComRS and comX. In addition, the export of XIP is accomplished by the nonspecific mechanism of lysis. To test whether the ComRS system requires dedicated mechanisms for processing of ComS to XIP, we constructed a dual-plasmid system that placed IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible comS and comR into E. coli cells carrying a PcomX-gfp reporter. Figure 6 shows the GFP activity of E. coli BL21(DE3) cells possessing the pDL278 PcomX-gfp fusion and a pACYC-Duet1 plasmid that either was (i) empty plasmid or else carried inducible (ii) comS, (iii) comR, or (iv) comR plus comS (Fig. 6A). Figure 6B shows GFP activity of the control strains (i to iii) in response to added IPTG. Following induction by IPTG, the comS and empty vector controls showed only baseline GFP expression (Fig. 6B). However, comR alone was able to elicit some expression from the comX reporter, consistent with the weak DNA binding seen at high ComR concentrations (Fig. 5). When both comR and comS were present (Fig. 6C), the median comX expression was roughly 2-fold greater than the level seen with ComR alone at 1 mM IPTG and was roughly 3-fold higher at 300 µM IPTG. These data show that endogenously produced, unprocessed ComS is sufficient to enhance ComR-induced expression from the comX promoter.
FIG 6.
Activation of PcomX by inducible comRS in E. coli. Activity of a PcomX-gfp reporter within individual E. coli cells was measured by fluorescence microscopy. (A) Strains harbored a two-plasmid system that included the PcomX-gfp reporter as well as different combinations of comR and comS under the control of the T7-lac promoter. (B) GFP fluorescence of controls containing the pACYC-Duet1 empty vector, comR alone, or comS alone after 3 h of growth with shaking, following addition of 0, 300 µM, and 1 mM IPTG. (C) GFP fluorescence of the strain possessing both comR and comS after 3 h of growth with shaking in various IPTG concentrations. The blue bar represents the median cell fluorescence.
Data are consistent with an intracellular feedback loop in comS transcription.
The reduced comX response in comS deletion strains in Fig. 1A and 2A shows that an intact comS gene has a stimulatory effect on comX expression that is not entirely matched either by exogenous XIP or by overexpression of ComS from a plasmid. One explanation for this effect is the presence of transcriptional positive feedback, in which endogenously produced ComS retained within the cell interacts with ComR to stimulate transcription from comS and comX (Fig. 7A). Import of extracellular XIP would be expected to stimulate such feedback in cells carrying an intact comS gene, potentially accounting for the stronger comX response to XIP that is observed in the wild-type strain versus the mutant ΔcomS background. This intracellular system is integrated into the upstream CSP pathway through an unknown mechanism known to require bacteriocins, as indicated in Fig. 7B.
FIG 7.
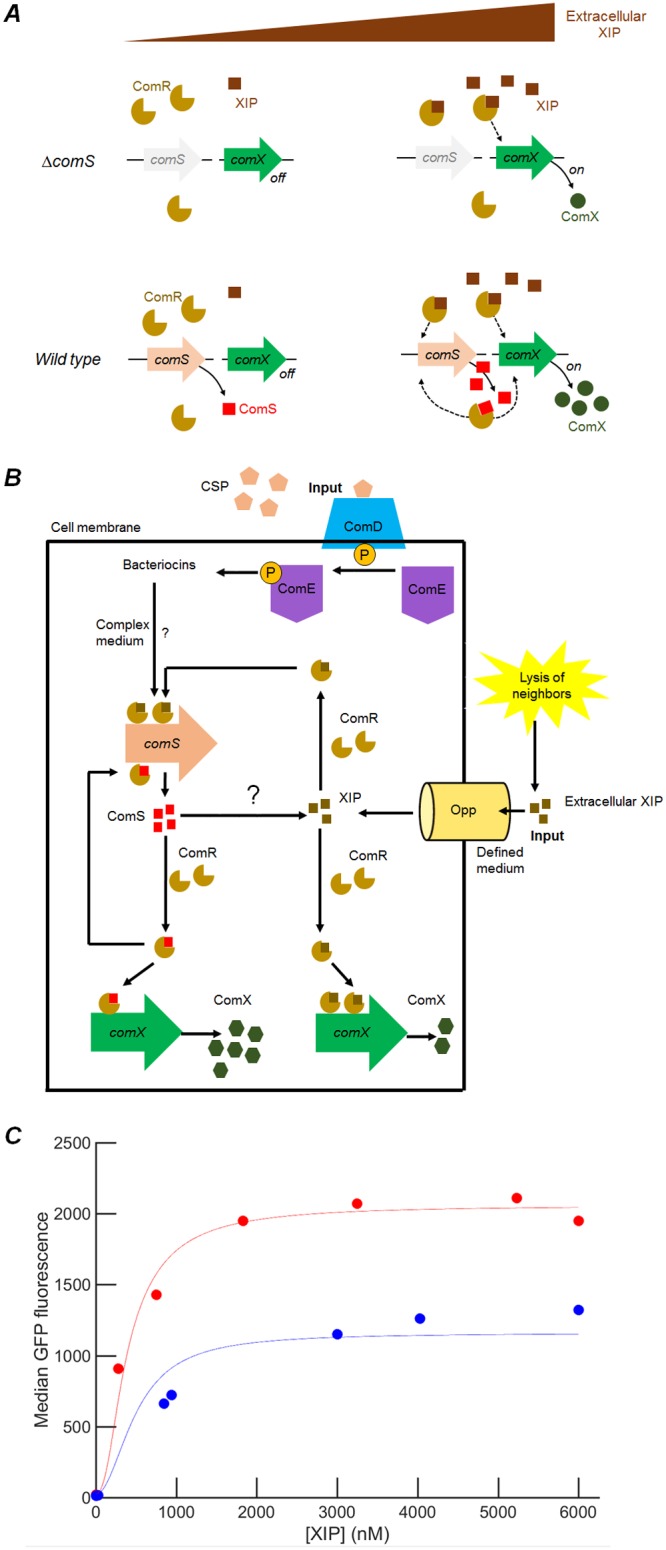
Proposed model for comRS feedback regulation of comX. The model represents comS feedback-enhanced activation of PcomX-gfp reporter by exogenous synthetic XIP. (A) Illustration of the feedback model and the role of comS and extracellular XIP. ComS and XIP both interact with ComR to activate transcription of comS and comX. At low concentrations of extracellular XIP, comS expression is very low and comX is not expressed. At high concentrations of extracellular XIP, XIP is imported efficiently by Opp and interacts with ComR to drive expression of both comS and comR. Endogenously produced ComS is not readily exported in the absence of lysis, and so intracellular accumulation of ComS drives elevated comS and comX expression. Consequently, comX expression at any given XIP concentration is boosted by comS feedback. Cells lacking native comS can respond to synthetic XIP but cannot activate comX to the same level as the wild type. The figure presents the behavior seen in defined medium; in complex medium, extracellular XIP is not imported (11). (B) Integration of this intracellular feedback system into the wider com regulon, involving upstream activator ComE and its response to exogenous CSP through the ComD transmembrane kinase. ComS is allowed to be processed to XIP intracellularly by an unidentified protease. (C) Comparison of model fit with data. Red circles indicate median PcomX-gfp fluorescence of the UA159 background strain supplied with synthetic XIP in microfluidic flow; blue circles indicate the median PcomX-gfp fluorescence of the ΔcomS background. Solid curves represent calculated values from a fit in which 11 parameters were fitted to the microfluidic data, as described in Materials and Methods. The model relates the predicted ComX concentration to the median GFP fluorescence by an offset and scale factor.
We constructed a mathematical model of this mechanism as described in the Materials and Methods and in Text S1 in the supplemental material. The model addresses conditions in a defined medium where extracellular XIP can be imported to the cell and early growth where lysis-driven release of ComS or XIP is not significant. Under these conditions, the model assumes that (i) extracellular XIP is imported to the cell but that (ii) neither ComS nor XIP is exported, though XIP may be intracellularly generated. (iii) Further, both the ComS and XIP complexes of ComR are able to elicit transcription of comS and comX, although not necessarily with equal transcription rates or degrees of multimerization. Assumption iii is motivated by data indicating that ComR binding does not absolutely require processing of ComS to XIP, by differences in the saturating fluorescence polarization achieved with ComS and XIP shown in Fig. 5A, and by the different saturating comX response (to XIP) of the ΔcomS and UA159 backgrounds shown in Fig. 1A. In parametrizing the model, we allowed different orders of multimerization in the ComR/peptide complex that binds DNA: n = 1 (ComS) or n = 2 (XIP) (see Discussion). In addition, the maximal rate of transcription (ComX production) is permitted to depend on whether a ComR/XIP complex or a ComR/ComS complex is bound to the promoter.
Methods used for deterministic modeling of the ComRS activation of comX with intracellular feedback. Download Text S1, DOCX file, 0.01 MB (14.8KB, docx) .
Copyright © 2018 Underhill et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Figure 7C shows the results of solving the steady-state dynamical equations of the model with the parameter values that best reproduce the data (see Materials and Methods). If only XIP (but not full-length ComS) interacts with ComR to activate comX, the model gives identical levels of comX activation in both the ΔcomS and wild-type backgrounds, in contradiction of the data presented in Fig. 1A. However, if both the ComR/XIP complexes and ComR/ComS complexes are permitted to induce transcription, although with different maximal rates, the model reproduces the different comX expression levels in the ΔcomS and wild-type backgrounds. Parameter values (see Table S2 in the supplemental material) were found to preserve the relative orders of magnitude of the dissociation constants of the transcriptional activators found in Fig. 5, with the best fit giving a transcription rate that is between 4-fold and 200-fold greater for the ComS-ComR bound promoter than for XIP-ComR activation of the gene.
DISCUSSION
The ComRS system found in mutans, salivarius, pyogenic, and bovis streptococci has been described alternatively as a quorum sensing mechanism (19) or timing mechanism (25) that directly controls comX, the master regulator of genetic competence. The ComS-derived XIP peptide is readily imported by S. mutans in defined growth medium, where it induces transformability with high efficiency. Key evidence supporting the idea of an intercellular signaling role for XIP includes the detection by LC-MS/MS of XIP in supernates of S. mutans cultures that were grown to high cell densities (22, 23). In addition, filtrates of S. mutans cultures grown to high density induced PcomX activity in reporter strains (21), indicating the presence of an active competence signal in the extracellular medium. A recent coculture study verified that XIP is freely diffusible in aqueous media and that ComS-overexpressing senders are able to activate comX in nearby ΔcomS receiver mutants, with no cell-cell contact being required (26). However, deletion of atlA, which encodes a surface-localized protein associated with envelope biogenesis and autolysis (30), suppressed this intercellular signaling (26). Taken together, these data indicate that the S. mutans ComRS system can provide intercellular competence signaling when autolysis releases sufficient concentrations of ComS or XIP.
We have previously argued that the bimodal expression of comX under conditions of stimulation by CSP is a signature indicating that comX is controlled by an intracellular transcriptional feedback system: endogenously produced ComS (or its product XIP) accumulates within some cells and then interacts with ComR to activate comX. Such feedback mechanisms are often a cause of bimodality in bacterial gene expression, including in other competence pathways (31, 32). The lack of an established mechanism for processing and export of ComS, the sensitivity of comX bimodality to endogenous production (via chromosomal comS) of XIP, and the fact that XIP import via Opp is not required for the bimodal behavior suggest together that purely intracellular signaling in comRS may be possible, at least under some environmental conditions. Such a mechanism would potentially allow individual S. mutans cells within a population to differ in competence behavior, as observed experimentally (14).
The present data support this model by providing several lines of evidence indicating that the chromosomal comS gene plays a role in ComRS activation of comX, regardless of the presence of other XIP or ComS that may be supplied. First, although the response of comX in complex medium supplemented with CSP is different from that seen in defined medium supplemented with XIP, the behavior of comX is affected in both cases by the presence of the intact chromosomal comS gene under the control of its cognate promoter. In complex medium, comS is required in order for CSP to elicit any comX response; in defined media, deletion of comS reduces the level of the comX response (both average and variance) to exogenous XIP and raises the threshold of the XIP concentration required for a response. Further, even if it harbors a comS overexpression plasmid, a comS deletion strain expresses comX much more weakly than does a comS-overexpressing strain that retains its chromosomal comS gene. These data show that the cell’s own native regulation of comS affects its activation of comX, independently of whether it overproduces ComS from a plasmid or imports exogenous XIP via Opp.
Our data also show that even in complex growth medium, which is known to inhibit the uptake of extracellular XIP, ComS-overexpressing (sender) cells activate their own comX gene. This autoactivation is unaffected by very rapid exchange (by flow) of the medium, strongly suggesting that comX activation in these cells does not require accumulation of extracellular XIP. However, this autoactivation requires the presence of a native comS gene in addition to the overexpression plasmid. In addition, even in the ComS overexpression strain, comS mRNA levels were significantly higher when the native comS was present, but comR mRNA levels were unaffected (see Fig. S1 in the supplemental material). These behaviors are consistent with the presence of positive feedback where imported XIP or plasmid-generated ComS enables internal amplification of comS expression.
Finally, the data show that comS-overexpressing cells fail to stimulate ΔcomS cells (receivers) in cocultures under defined medium conditions, which are favorable for import of XIP. As the ΔcomS receivers do respond to exogenously added XIP, these data indicate that the overexpressing cells activate their own comX without releasing significant XIP to the medium. The weak intercellular signaling that is observed in cocultures grown to late growth phases is consistent with eventual lysis of sender cells, possibly linked to autoactivation of the lytic pathway driven by ComX and ComDE.
The finding that diffusive signaling by ComS or XIP between S. mutans cells is inefficient or lacks spatial range is consistent with the conclusion reached by Gardan et al. using S. thermophilus (25). Those authors found that the type I ComS peptide of S. thermophilus was not secreted at detectable levels in a strain that produced it naturally, although an overproducing strain did generate detectable ComS in the medium. They argued that ComS does not diffuse through or accumulate in the medium, although it may be able to signal between cells that are in physical contact. This proximity model for ComS resembles a “self-sensing” quorum system (33) in which the secreted signal is retained at elevated concentrations in the immediate surroundings of the cell, possibly associated with the cell surface, so that the cell responds somewhat more strongly to its own secreted signal than to that of the rest of the population.
Our observations suggest more strongly that intercellular quorum signaling through ComS or XIP was not essential to ComRS control of comX in S. mutans under the conditions examined. A more essential component is the dynamics of the cell’s own comS transcription. A plausible mechanism for the bimodal response of S. mutans comX to CSP stimulation is therefore that CSP stimulates the bistable comRS feedback loop by facilitating, through an indirect mechanism, the positive feedback. It would be sufficient, for example, to inhibit degradation of endogenous ComS, which, depending on the basal transcription levels, would trigger comX activation in at least some cells, leading to the bimodal distribution of comX activity (11). Notably, our data show that overexpression of comS also leads to heterogeneous comX activity, suggesting that it plays a role similar to that played by exogenous CSP by facilitating comS autofeedback.
Since whether S. mutans ComS is processed to XIP inside the cell is unknown, either XIP or ComS could potentially act as the intracellular feedback signal. Although S. mutans competence was shown to be unresponsive to the presence of exogenous full-length ComS (19), this finding may reflect either selectivity by ComR or simply inefficient import of full-length ComS by Opp. ComS is significantly larger (17 residues) than the peptides that are typically transported by ABC transporters. Shanker et al. (34) found that S. mutans ComR is unresponsive to the ComS peptides produced by other streptococcal species, although an eight-residue XIP peptide (ComS10-17) did interact effectively with ComR to bind the comS and comX promoters (20). Our fluorescence polarization data confirm that both ComS and XIP can interact with ComR to bind the comX and comS promoter regions. They also suggest that ComS and XIP may form ComR complexes of different degrees of multimerization, a difference that could have interesting consequences for the nonlinear dynamics of feedback regulation. Our mathematical model for transcriptional autofeedback in the comRS system incorporates the data by assuming that endogenously produced ComS is not released to the environment, although extracellular XIP is imported and supplements the endogenous ComS in interacting with ComR. Unless it is the case that E. coli is equipped to process ComS to XIP or perhaps that ComS cleaves autocatalytically, our finding that E. coli carrying inducible comR and comS can activate a comX reporter plasmid is further evidence that processing of ComS to XIP is not absolutely required.
Other studies show some precedent for such regulation. Structural studies in S. pyogenes have shown that some intracellular Rgg receptor proteins can bind pheromones that differ in length and sequence (35). Crystallographic structures of homologous ComR proteins (34, 36) showed that the SHP binding pocket of the ComR C terminus falls in the tetratricopeptide repeat domain that is responsible for multimerization, while the N terminus helix-turn-helix structure binds DNA after an induced structural rearrangement. The location of the SHP binding pocket could allow the longer ComS to hinder multimerization when bound, resulting in a monomer binding to its target, while XIP does not. Figure S3 shows preliminary evidence that the ComS N terminus affects ComR binding in S. mutans. Neither ComS nor XIP could induce DNA binding by an N-terminally 6 histidine-tagged ComR, whereas XIP (but not ComS) caused DNA binding activity in a C-terminally tagged ComR. These data indicate that steric effects around the SHP binding pocket may influence DNA binding affinities.
Positive feedback occurs in many quorum sensing systems as the accumulation of the chemical signal in the extracellular environment stimulates the cell to produce additional signal or its cognate receptor. For example, in Vibrio fischeri the C8 homoserine lactone autoinducer stimulates expression of ainS, which encodes the autoinducer synthase (37). In Vibrio cholerae, the CAI-1 signal stimulates production of its CqsS receptor (38). In these cases, the extracellular signal concentration, which is the positive-feedback signal, is sensed by large numbers of cells, and so the population responds homogenously. However, if an individual cell responds preferentially to its own signal production, then the feedback signal is specific to the individual cell and the behavior is qualitatively different. Individual feedback can convert a graded (or unimodal) population response to a switched or bimodal response (39). Depending on parameters such as the rate of signal production and the level of noise or the cell density, the response of the cells may then span a range from strongly social or quorum behavior to purely autocrine or self-sensing (33) behavior in which cells respond independently and the population becomes heterogeneous (40). Synthetic biology has exploited this phenomenon in several bacterial quorum sensing systems to amplify the cell’s sensitivity to an exogenous signal. This can lower the quorum circuit’s threshold sensitivity to the signal, and it can also enhance the amplitude of the cell’s full response to that signal. Data in panels A and B of Fig. 1 suggest that the presence of the chromosomal comS gene in S. mutans roughly doubles the amplitude of the comX response and lowers the XIP sensitivity threshold roughly 2-fold. This amplification is comparable to what was accomplished in engineered synthetic systems (41, 42).
As a result, the ComRS system may have two modes of function in S. mutans. During early growth, CSP signaling stimulates the intracellular feedback behavior in ComRS, leading to population bimodality in comX expression levels. Consequently, only cells in a small subpopulation activate the late competence genes. However, in later growth phases or in mature biofilms, stress mechanisms that drive autolysis allow the release of XIP, generating a diffusing signal that is detected by other cells and amplified through the internal feedback mechanism, eliciting a broader competence response in the population. In this sense, XIP may serve to broadcast localized stress conditions, stimulating S. mutans to scavenge DNA resources opportunistically from nearby lysing cells (43, 44).
MATERIALS AND METHODS
Strains and growth conditions.
S. mutans wild-type strain UA159 and mutant reporting/gene deletion strains from glycerol freezer stock were grown in BBL BHI (Becton, Dickinson and Co.) at 37˚C in 5% CO2 overnight. The following antibiotics (concentrations) were used where resistance is indicated in Table 1: erythromycin (10 µg ml−1), kanamycin (1 mg ml−1), and spectinomycin (1 mg ml−1). For experiments performed in defined medium, strains were washed twice by centrifugation followed by removal of supernatant and resuspension in the defined medium FMC (45) (see Text S2 in the supplemental material for the ingredients of FMC). These were then diluted 20-fold into fresh FMC and allowed to grow under the same incubator conditions until an optical density at 600 nm (OD600) of 0.1 was reached. Synthetic XIP (sequence GLDWWSL) was synthesized and purified to 98% purity by NeoBioSci (Cambridge, MA).
TABLE 1.
list of strains and plasmids used
| Strain or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| S. mutans strains | ||
| PcomX-gfp (plasmid) | UA159 harboring PcomX GFP promoter fusion on plasmid pDL278 | 11 |
| PcomX-rfp | UA159 harboring PcomX dsRed RFP promoter fusion on plasmid pDL278 | 26 |
| PcomX-gfp ΔcomS | UA159 comS gene replaced with a nonpolar erythromycin resistance cassette; harboring PcomX GFP promoter fusion on plasmid pDL278 |
11 |
| 184comS PcomX-rfp | UA159 harboring pIB184comS and PcomX dsRed RFP promoter fusion | 26 |
| 184comS PcomX-rfp ΔcomS | UA159 harboring pIB184comS and PcomX dsRed RFP promoter fusion; comS disrupted by point mutation in start codon (ATG to AAG) |
This study |
| E. coli strains | ||
| BL21(DE3) | Used for recombinant protein expression | New England Biolabs, MA |
| 10-beta | Used for propagating plasmids during cloning | New England Biolabs, MA |
| SAMU215 | BL21(DE3) carrying PcomX-gfp on pDL27 | This study |
| SAMU216 | SAMU215 carrying pACYC-Duet1 empty vector | This study |
| SAMU217 | SAMU215 carrying pACYC-Duet1 plasmid with comR on multiple cloning site (MCS) I | This study |
| SAMU218 | SAMU215 carrying pACYC-Duet1 plasmid with comS on MCS II | This study |
| SAMU219 | SAMU215 carrying pACYC-Duet1 plasmid with comR on MCS I and comS on MCS II | This study |
| Plasmids | ||
| pIB184 | Shuttle expression plasmid with the P23 constitutive promoter, Emr | 29 |
| pDL278 | E. coli-Streptococcus shuttle vector, Spr | 46 |
| pET45b(+)his-comRUA159 | pET45b(+) derivative containing the translational fusion PT7lac-6xhis-comRUA159, Apr | This study |
| pACYC-Duet 1 | T7-lac inducible expression vector for coexpression of two proteins, Cmr | Millipore-Sigma, MA |
Emr, erythromycin resistance; Spr, spectinomycin resistance; Apr, ampicillin resistance; Cmr, chloramphenicol resistance.
FMC ingredients. Download Text S2, PDF file, 0.5 MB (527KB, pdf) .
Copyright © 2018 Underhill et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
E. coli strains were grown in LB at 37°C with shaking in an aerobic incubator overnight. The following antibiotics (concentrations) were used where resistance is indicated: ampicillin (10 µg ml−1) and chloramphenicol (34 µg ml−1). For recombinant ComR expression experiments, the next day, the overnight cultures were diluted 100-fold into LB containing ampicillin at the indicated concentration and grown under the same incubator conditions as were used to grow the overnight cultures. For analysis of recombinant comRS expression inducing PcomX-gfp, cells were diluted 100-fold into LB containing chloramphenicol and spectinomycin and grown with shaking at 37°C to an OD600 of 0.1, at which point IPTG was added and the cells were grown with shaking at 30°C until imaging was performed.
Construction of comS point mutant.
The start codon of the comS gene was mutated from ATG to AAG. The mutation was introduced directly into the chromosome by site-directed mutagenesis using a PCR product generated by overlap extension PCR (47). Potential mutants were screened using mismatch amplification mutation analysis (MAMA) PCR (48), as previously described (49, 50). The point mutation was confirmed by PCR and sequencing to ensure that no further mutations were introduced into the comS gene and its flanking regions.
Microfluidic experiments.
Microfluidic experiments were performed using a seven-channel polydimethylsiloxane (PDMS)-cast mixing array device, with fluorescence imaging and single-cell image analysis performed as described previously (11, 51, 52). Cells carrying a PcomX-gfp plasmid-based reporter were grown to ab OD600 of 0.1 from dilution in FMC medium and were sonicated briefly using a Fisher Scientific FB120 sonic dismembrator probe to split large chains. Sonicated cells were then loaded into the device through a syringe capped with a 5-µm-pore-size filter to remove any remaining aggregations. FMC medium containing 1 mg ml−1 spectinomycin and a XIP gradient produced from three inlets containing different concentrations of XIP (0 nM, 600 nM, and 6 µM XIP inlets) passed through a mixing matrix was pumped through the cell chambers at a steady rate of 0.08 ml h−1 to create a constant, different XIP concentration in each cell chamber.
Gamma distributions (two-parameter probability sdistribution describing the amount of protein produced in sequential transcription and translation steps) were fitted to the single-cell fluorescence distributions using Matlab to fit protein production to theoretical descriptions (28). The fit was applied to cells fluorescing at levels above an arbitrary cutoff of 40 units (around the background level) in order to prevent turned-off cells from skewing the distribution. Parameter values were rounded to three significant figures, and data are reported in Table S1 in the supplemental material.
Parameters for gamma distribution fits to single-cell PcomX GFP fluorescence distributions in microfluidic experiments. Download Table S1, DOCX file, 0.01 MB (13.2KB, docx) .
Copyright © 2018 Underhill et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fitted values for the 12 parameters of the feedback activation model and statistical measurement of their robustness from bootstrap process. Download Table S2, DOCX file, 0.01 MB (14KB, docx) .
Copyright © 2018 Underhill et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Flow rate dependence experiment.
In order to measure the flow rate dependence of XIP signaling, we loaded cells into a commercial six-channel microfluidic slide (IBIDI µ-slide VI; IBIDI GmbH). The six channels (channels a to f) contained respectively (channel a) a red fluorescent protein (dsRed) comX reporting strain (PcomX-rfp) control channel flowing fresh BHI at 0.1 ml h−1; (channels b to e) four channels containing comS overexpression strain 184comS PcomX-rfp (comS on plasmid pIB184 under the control of the strong constitutive P23 promoter) with BHI at different flow rates ranging from 0.02 ml h−1 to 1 ml h−1, and (channel f) a 184comS PcomX-rfp strain with a point mutation disrupting the chromosomal comS gene (mutant ΔcomS) under conditions of flow at 0.1 ml h−1. After 2 h, the neat BHI supplied was replaced with BHI supplemented with 50 µg ml−1 chloramphenicol in order to halt further translation and allow any RFP in the cells to fold. This was supplied at a flow rate of 0.1 ml h−1 for all channels. Four hours (the maturation time of our RFP) after chloramphenicol addition, final fluorescence images of the cultures were taken. Due to the bimodal comX activation in BHI, the fluorescence cutoff level was set as the maximum RFP fluorescence observed in the PcomX-rfp negative control. Cells exhibiting RFP fluorescence above this level were collected in an array, and the size of this sample as a percentage of the population and the median of the above-cutoff fluorescence are reported.
Channel coculture experiment.
We loaded cocultures of a PcomX-gfp ΔcomS mutant (responders) with the comS-overexpressing strain 184comS PcomX-rfp (senders) into two commercial microfluidic slides (IBIDI µ-slide VI) using static (not flowing) FMC medium and various ratios of comS overproducers/mutant ΔcomS responders (percentage by volume of cultures at an OD600 of 0.1 subjected to vortex mixing together). Strains PcomX-rfp, PcomX-gfp ΔcomS, PcomX-rfp plus 50 nM XIP, and PcomX-gfp ΔcomS plus 50 nM XIP were used as controls. The end ports of the channels were sealed with mineral oil to prevent drying of the medium in the channels. Images were taken as described for the microfluidic experiments, and analysis was performed similarly. In the case of the controls, XIP was added to planktonic culture and the tube subjected to vortex mixing before pipetting into the slide. Because the population was heterogeneous with respect to both fluorescent reporter type and comX expression, a fluorescence threshold was defined as the maximum RFP fluorescence observed in the PcomX-rfp negative control as described previously. The median of the RFP fluorescence observed above this cutoff level in other samples was used as a measure of how strongly the red cells were activating comX as a function of their number density.
OD dependence of coculture response.
For tests of growth-phase dependence of signaling, cocultures similar to those in microfluidic channel slides were prepared. Overnight cultures were washed and diluted 40× into fresh FMC medium containing erythromycin (10 µg ml−1) and spectinomycin (1 mg ml−1). Once grown to an OD600 of 0.05, these were mixed in various ratios ranging from 0% comS overexpressers to 100% overexpressers, defined by the volume of comS overproducers added divided by the volume of the ΔcomS culture added. Low initial cell densities were used to ensure that the early, middle, and late growth phases were probed for XIP release. Every 2 h, the OD600 of the culture and its pH were measured. The pH was corrected back to 7.0 using 2 N sodium hydroxide in cases in which it had deviated to below 6.5, in order to measure the reactions to any XIP released at late times into the culture. RFP fluorescence and GFP fluorescence were measured by pipetting a small amount of the culture onto a glass coverslip and analyzing single cells. GFP fluorescence at the 99th percentile was then used to determine if XIP was being released to the comS mutants in an OD600-dependent manner.
RT-qPCR measurement of comS, comR, and comX transcripts.
S. mutans cells were diluted 20-fold into BHI medium (strain PcomX-gfp wild type/BHI [WT/BHI], 184comS PcomX-rfp, and 184comS PcomX-rfp ΔcomS samples) or FMC medium (strain PcomX-gfp WT/FMC ± XIP, PcomX-gfp ΔcomS ± XIP, pIB184/WT, and pIB184ComS/WT samples). Where added, XIP was supplied at OD600 = 0.1. Cells were harvested at OD600 = 0.5 by centrifugation and resuspended in RNA protectant buffer for 10 min. Samples were then centrifuged, the supernatant was removed, and the pellets were frozen at −80°C. RNA extraction was performed using a Qiagen RNEasy minikit (Qiagen, USA). RNA sample concentration and purity were measured using a Thermo Scientific NanoDrop One Microvolume UV-Vis spectrophotometer (Thermo Scientific, USA). RNA (1 µg) was then reverse transcribed to cDNA using a Bio-Rad iScript reverse transcription kit with random primers (Bio-Rad, USA). The qPCR was performed on a Bio-Rad CFX96 real-time system using Bio-Rad Sso Advanced Universal SYBR green Supermix with a 50-fold dilution of the cDNA and 500 nM gene-specific primers. Sequences used for the primers are given in Table S3. A standard curve across 8 orders of magnitude of transcript copies (from 108 to 101) was used to determine thetranscript count for each gene. For each sample the comX, comR, and comS transcript counts were then normalized by the 16S rRNA count for the same sample. Figure S1 in the supplemental material shows the median of this ratio, with error bar lengths given by the range from second-lowest to second-highest ratio obtained.
Primer sequences used for RT-qPCR. Download Table S3, DOCX file, 0.02 MB (19.5KB, docx) .
Copyright © 2018 Underhill et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fluorescence polarization.
ComR protein was obtained by cloning the comR gene into the 6×-His tagged site on pET-45b(+) vector in E. coli 10-beta using standard PCR cloning methods. His-ComR was then expressed in E. coli BL21(DE3) by induction with 1 mM IPTG at mid-exponential phase in LB. After 4 h, the cells were lysed using lysozyme in B-PER lysis buffer (Thermo Fisher). Protein was then purified from clarified lysate using nickel-nitrilotriacetic acid (Ni-NTA) agarose affinity chromatography and the histidine tag cleaved using EnterokinaseMax (EKMax) (Invitrogen) at 4°C. The resulting protein solution was dialyzed into phosphate-buffered saline (PBS) (pH 7.4) for experimental use. Native ComR concentrations were measured using the Pierce bicinchoninic acid (BCA) assay (Thermo Scientific), and the purity of the cleaved form was verified by SDS-PAGE run against an uncleaved sample.
Fluorescence polarization assays were performed in a 96-well plate with black bottom and black sides in a Biotek Synergy 2 plate reader (Biotek Instruments Inc.) in the polarization mode. A 5′ Bodipy FL-X-labeled self-annealing stem-loop DNA strand with sequence corresponding to PcomX (sequence 5′-Bodipy FL-X-ATGGGACATTTATGTCCTGTCCCCCACAGGACATAAATGTCCCAT-3′ [synthesized by Thermo Fisher]) was used as the binding aptamer, and a filter set (excitation wavelength, 485 nm; emission wavelength, 528 nm) was used for fluorescence excitation. Labeled DNA probe (1 nM) was added to a previously described reaction buffer (53) supplemented with 1 mM EDTA and 0.05 mg ml−1 salmon DNA. ComR was titrated in concentrations in this buffer alone or in the presence of 10 µM XIP or in the presence of 10 µM comS. The reaction mixtures were incubated at 37°C for 20 min before being read. Synthetic ComS (sequence, MFSILTSILMGLDWWSL) for use in fluorescence polarization was synthesized and purified to 60% purity by Biomatik (Wilmington, DE).
Competing unlabeled probe assays were performed with 1.5 µM ComR in the same buffer containing 1 nM PcomX fluorescent DNA and 10 µM SHP (either ComS or XIP). An unlabeled probe corresponding to either the PcomS probe (sequence, 5′-ACGGGACATAAATGTCCTGTCCCCCACAGGACATTTATGTCCCGT-3′; synthesized by Thermo Fisher) or the PcomX probe described above was titrated into this solution and the decreasing polarization plotted. Reaction mixtures were again incubated at 37°C for 20 min before polarization readings were taken. In all FP experiments, reading was performed three times on the same plate to estimate instrument error. The average polarization was used for plotting and analysis. Details and parameters of the two-step binding model for the FP data are given in Text S3 in the supplemental material.
Methods for modeling of fluorescence polarization binding data of Fig. 5. Download Text S2, DOCX file, 0.01 MB (15.5KB, docx) .
Copyright © 2018 Underhill et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ComRS expression in E. coli.
A recombinant comRS system was produced in E. coli by inserting comR into multiple cloning site I (MCS I) in pACYC-Duet 1 vector and comS into MCS II by standard cloning methods. BL21(DE3) cells containing PcomX-gfp on pDL278 were transformed with the resulting pACYC-Duet 1 comRS plasmid. Controls consisting of empty pACYC-Duet 1 and the vector containing only comR at MCS I and only comS at MCS II were produced in the same way. Cells were diluted 100-fold from overnight cultures into LB containing chloramphenicol and spectinomycin and were grown with shaking at 37°C until they reached an OD600 of 0.1. At that point, IPTG was added in various amounts to induce expression of the T7-lac controlled recombinant system. Following addition of IPTG, the cells were grown with shaking at 30°C and GFP fluorescence was imaged after 3 h.
Mathematical model of comRS control of comX.
Deterministic modeling of comX activation by comRS was performed by least-squares fitting of a chemical equilibrium model to the microfluidic data from the experiments performed for each of the wild-type background PcomX GFP strain and the ΔcomS cells. Details of the model and the robustness analysis are given in the supplemental material, with parameter values given in Table S2.
ACKNOWLEDGMENTS
This work was supported by 1R01 DE023339, R01 DE13239, and T90 DE21990 from the National Institute of Dental and Craniofacial Research.
We thank Minjun Son, Chris Browngardt, Natalie Maricic, Lin Zeng, Hey Min Kim, and Sang-Joon Ahn for helpful discussions, provision of mutant strains, and advice on experimental procedures.
REFERENCES
- 1.Loesche WJ. 1986. Role of Streptococcus mutans in human dental decay. Microbiol Rev 50:353–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Senadheera DB, Cordova M, Ayala EA, Chavez de Paz LE, Singh K, Downey JS, Svensater G, Goodman SD, Cvitkovitch DG. 2012. Regulation of bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans. J Bacteriol 194:1307–1316. doi: 10.1128/JB.06071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn SJ, Lemos JA, Burne RA. 2005. Role of HtrA in growth and competence of Streptococcus mutans UA159. J Bacteriol 187:3028–3038. doi: 10.1128/JB.187.9.3028-3038.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senadheera MD, Lee AW, Hung DC, Spatafora GA, Goodman SD, Cvitkovitch DG. 2007. The Streptococcus mutans vicX gene product modulates gtfB/C expression, biofilm formation, genetic competence, and oxidative stress tolerance. J Bacteriol 189:1451–1458. doi: 10.1128/JB.01161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tremblay YDN, Lo H, Li Y, Halperin SA, Lee SF. 2009. Expression of the Streptococcus mutans essential two-component regulatory system VicRK is pH and growth-phase dependent and controlled by the LiaFSR three-component regulatory system. Microbiology 155:2856–2865. doi: 10.1099/mic.0.028456-0. [DOI] [PubMed] [Google Scholar]
- 6.Ahn SJ, Wen ZT, Burne RA. 2006. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect Immun 74:1631–1642. doi: 10.1128/IAI.74.3.1631-1642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi F, Merritt J, Lux R, Shi W. 2004. Inactivation of the ciaH gene in Streptococcus mutans diminishes mutacin production and competence development, alters sucrose-dependent biofilm formation, and reduces stress tolerance. Infect Immun 72:4895–4899. doi: 10.1128/IAI.72.8.4895-4899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Son M, Ghoreishi D, Ahn SJ, Burne RA, Hagen SJ. 2015. Sharply tuned pH response of genetic competence regulation in Streptococcus mutans: a microfluidic study of the environmental sensitivity of comX. Appl Environ Microbiol 81:5622–5631. doi: 10.1128/AEM.01421-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Q, Ahn SJ, Kaspar J, Zhou X, Burne RA. 2014. Growth phase and pH influence peptide signaling for competence development in Streptococcus mutans. J Bacteriol 196:227–236. doi: 10.1128/JB.00995-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moye ZD, Son M, Rosa-Alberty AE, Zeng L, Ahn SJ, Hagen SJ, Burne RA. 2016. Effects of carbohydrate source on genetic competence in Streptococcus mutans. Appl Environ Microbiol 82:4821–4834. doi: 10.1128/AEM.01205-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Son M, Ahn S, Guo Q, Burne RA, Hagen SJ. 2012. Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX. Mol Microbiol 86:258–272. doi: 10.1111/j.1365-2958.2012.08187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reck M, Tomasch J, Wagner-Döbler I. 2015. The alternative sigma factor SigX controls bacteriocin synthesis and competence, the two quorum sensing regulated traits in Streptococcus mutans. PLoS Genet 11:e1005353. doi: 10.1371/journal.pgen.1005353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Son M, Shields RC, Ahn S, Burne RA, Hagen SJ. 2015. Bidirectional signaling in the competence regulatory pathway of Streptococcus mutans. FEMS Microbiol Lett 362:159. doi: 10.1093/femsle/fnv159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagen SJ, Son M. 2017. Origins of heterogeneity in Streptococcus mutans competence: interpreting an environment-sensitive signaling pathway. Phys Biol 14:015001. doi: 10.1088/1478-3975/aa546c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan R, Rukke HV, Høvik H, Åmdal HA, Chen T, Morrison DA, Petersen FC. 2016. Comprehensive transcriptome profiles of Streptococcus mutans UA159 map core streptococcal competence genes. mSystems 1:e00038-15. doi: 10.1128/mSystems.00038-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fontaine L, Wahl A, Fléchard M, Mignolet J, Hols P. 2015. Regulation of competence for natural transformation in streptococci. Infect Genet Evol 33:343–360. doi: 10.1016/j.meegid.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 17.Hossain MS, Biswas I. 2012. An extracelluar protease, SepM, generates functional competence-stimulating peptide in Streptococcus mutans UA159. J Bacteriol 194:5886–5896. doi: 10.1128/JB.01381-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perry JA, Jones MB, Peterson SN, Cvitkovitch DG, Lévesque CM. 2009. Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol Microbiol 72:905–917. doi: 10.1111/j.1365-2958.2009.06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mashburn-Warren L, Morrison DA, Federle MJ. 2010. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol 78:589–606. doi: 10.1111/j.1365-2958.2010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fontaine L, Goffin P, Dubout H, Delplace B, Baulard A, Lecat-Guillet N, Chambellon E, Gardan R, Hols P. 2013. Mechanism of competence activation by the ComRS signalling system in streptococci. Mol Microbiol 87:1113–1132. doi: 10.1111/mmi.12157. [DOI] [PubMed] [Google Scholar]
- 21.Desai K, Mashburn-Warren L, Federle MJ, Morrison DA. 2012. Development of competence for genetic transformation of Streptococcus mutans in a chemically defined medium. J Bacteriol 194:3774–3780. doi: 10.1128/JB.00337-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan R, Rukke HV, Ricomini Filho AP, Fimland G, Arntzen MO, Thiede B, Petersen FC. 2012. Extracellular identification of a processed type II ComR/ComS pheromone of Streptococcus mutans. J Bacteriol 194:3781–3788. doi: 10.1128/JB.00624-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wenderska IB, Lukenda N, Cordova M, Magarvey N, Cvitkovitch DG, Senadheera DB. 2012. A novel function for the competence inducing peptide, XIP, as a cell death effector of Streptococcus mutans. FEMS Microbiol Lett 336:104–112. doi: 10.1111/j.1574-6968.2012.02660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang JC, Federle MJ. 2016. PptAB exports Rgg quorum-sensing peptides in Streptococcus. PLoS One 11:e0168461. doi: 10.1371/journal.pone.0168461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardan R, Besset C, Gitton C, Guillot A, Fontaine L, Hols P, Monnet V. 2013. Extracellular life cycle of ComS, the competence-stimulating peptide of Streptococcus thermophilus. J Bacteriol 195:1845–1855. doi: 10.1128/JB.02196-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaspar J, Sam U, Shields RC, Reyes A, Rosenzweig S, Hagen SJ, Burne RA. 2017. Intercellular communication via the comX-inducing peptide (XIP) of Streptococcus mutans. J Bacteriol 2017:JB.00404-17. doi: 10.1128/JB.00404-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taniguchi Y, Choi PJ, Li G, Chen H, Babu M, Hearn J, Emili A, Xie XS. 2010. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329:533. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedman N, Cai L, Xie XS. 2006. Linking stochastic dynamics to population distribution: an analytical framework of gene expression. Phys Rev Lett 97:168302. doi: 10.1103/PhysRevLett.97.168302. [DOI] [PubMed] [Google Scholar]
- 29.Biswas I, Jha JK, Fromm N. 2008. Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology 154:2275–2282. doi: 10.1099/mic.0.2008/019265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahn SJ, Burne RA. 2006. The atlA operon of Streptococcus mutans: role in autolysin maturation and cell surface biogenesis. J Bacteriol 188:6877–6888. doi: 10.1128/JB.00536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dubnau D, Losick R. 2006. Bistability in bacteria. Mol Microbiol 61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- 32.Maamar H, Dubnau D. 2005. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol Microbiol 56:615–624. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bareia T, Pollak S, Eldar A. 2018. Self-sensing in Bacillus subtilis quorum-sensing systems. Nat Microbiol 3:83–89. doi: 10.1038/s41564-017-0044-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shanker E, Morrison DA, Talagas A, Nessler S, Federle MJ, Prehna G. 2016. Pheromone recognition and selectivity by ComR proteins among Streptococcus species. PLoS Pathog 12:e1005979. doi: 10.1371/journal.ppat.1005979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aggarwal C, Jimenez JC, Nanavati D, Federle MJ. 2014. Multiple length peptide-pheromone variants produced by Streptococcus pyogenes directly bind Rgg proteins to confer transcriptional regulation. J Biol Chem 289:22427–22436. doi: 10.1074/jbc.M114.583989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talagas A, Fontaine L, Ledesma-Garca L, Mignolet J, Li de la Sierra-Gallay I, Lazar N, Aumont-Nicaise M, Federle MJ, Prehna G, Hols P, Nessler S. 2016. Structural insights into streptococcal competence regulation by the cell-to-cell communication system ComRS. PLoS Pathog 12:e1005980. doi: 10.1371/journal.ppat.1005980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lupp C, Ruby EG. 2004. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J Bacteriol 186:3873–3881. doi: 10.1128/JB.186.12.3873-3881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hurley A, Bassler BL. 2017. Asymmetric regulation of quorum-sensing receptors drives autoinducer-specific gene expression programs in Vibrio cholerae. PLoS Genet 13:e1006826. doi: 10.1371/journal.pgen.1006826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Becskei A, Séraphin B, Serrano L. 2001. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J 20:2528. doi: 10.1093/emboj/20.10.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Youk H, Lim WA. 2014. Secreting and sensing the same molecule allows cells to achieve versatile social behaviors. Science 343:1242782. doi: 10.1126/science.1242782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sayut DJ, Niu Y, Sun L. 2006. Construction and engineering of positive feedback loops. ACS Chem Biol 1:692–696. doi: 10.1021/cb6004245. [DOI] [PubMed] [Google Scholar]
- 42.Nistala GJ, Wu K, Rao CV, Bhalerao KD. 2010. A modular positive feedback-based gene amplifier. J Biol Eng 4:4. doi: 10.1186/1754-1611-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leung V, Dufour D, Lévesque CM. 2015. Death and survival in Streptococcus mutans: differing outcomes of a quorum-sensing signaling peptide. Front Microbiol 6:1176. doi: 10.3389/fmicb.2015.01176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perry JA, Cvitkovitch DG, Lévesque CM. 2009. Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol Lett 299:261–266. doi: 10.1111/j.1574-6968.2009.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terleckyj B, Willett NP, Shockman GD. 1975. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect Immun 11:649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.LeBlanc DJ, Lee LN, Abu-Al-Jaibat A. 1992. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28:130–145. doi: 10.1016/0147-619X(92)90044-B. [DOI] [PubMed] [Google Scholar]
- 47.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 48.Cha RS, Zarbl H, Keohavong P, Thilly WG. 1992. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl 2:14–20. doi: 10.1101/gr.2.1.14. [DOI] [PubMed] [Google Scholar]
- 49.Zeng L, Das S, Burne RA. 2010. Utilization of lactose and galactose by Streptococcus mutans: transport, toxicity, and carbon catabolite repression. J Bacteriol 192:2434–2444. doi: 10.1128/JB.01624-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahn SJ, Kaspar J, Kim JN, Seaton K, Burne RA. 2014. Discovery of novel peptides regulating competence development in Streptococcus mutans. J Bacteriol 196:3735–3745. doi: 10.1128/JB.01942-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeon NL, Dertinger SKW, Chiu DT, Choi IS, Stroock AD, Whitesides GM. 2000. Generation of solution and surface gradients using microfluidic systems. Langmuir 16:8311–8316. doi: 10.1021/la000600b. [DOI] [Google Scholar]
- 52.Kwak IH, Son M, Hagen SJ. 2012. Analysis of gene expression levels in individual bacterial cells without image segmentation. Biochem Biophys Res Commun 421:425–430. doi: 10.1016/j.bbrc.2012.03.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aggarwal C, Jimenez JC, Lee H, Chlipala GE, Ratia K, Federle MJ. 2015. Identification of quorum-sensing inhibitors disrupting signaling between Rgg and short hydrophobic peptides in streptococci. mBio 6:e00393-15. doi: 10.1128/mBio.00393-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Measurement of comS, comX, and comR transcript levels. (A to C) RT-qPCR measurement of (A) comS, (B) comX, and (C) comR transcripts in cultures harvested at an OD600 of 0.5. Each bar indicates the ratio of the median transcript count to the median 16S rRNA count, as measured in multiple biological and technical replicates (see Materials and Methods). WT/BHI, 184comS, and 184comS ΔcomS samples were grown in BHI medium. The remaining samples were grown in FMC. Synthetic XIP was supplied at a 100 nM concentration when used. Error bars indicate the range from the second-lowest to the second-highest value among the replicates for each condition. (D) Comparison of comS mRNA transcripts from S. mutans harboring the ComS overexpression plasmid versus the empty plasmid. Download FIG S1, TIF file, 0.1 MB (79.3KB, tif) .
Copyright © 2018 Underhill et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Response of cocultures is time independent. (A) GFP (comX reporter) and (B) RFP (comY) fluorescence of individual cells in cocultures of sender (184comS PcomX-rfp) and receiver (PcomX-gfp ΔcomS) strains of S. mutans. Samples are labeled by percentage by volume of 184comS (sender) culture in the initial preparation of the coculture. Fluorescence was measured immediately (0 h, green) after mixing the coculture or 4 h (magenta) after mixing. The red horizontal bars show the median fluorescence immediately after mixing (0 h); the black vertical bars show the median at 4 h. Data are from the coculture experiment described in the Fig. 3 legend. Download FIG S2, TIF file, 0.2 MB (182.1KB, tif) .
Copyright © 2018 Underhill et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Effect of histidine tag on ComR binding of ComS and XIP. A fluorescence polarization study of ComS and XIP interaction with ComR that was N- or C-terminally tagged with 6×-histidine was performed. Polarization is plotted versus [ComR] for (A) C-terminally tagged ComR and (B) N-terminally tagged ComR. In each case, 1 nM fluorescent DNA and 0.05 mg ml−1 salmon DNA were present along with no signal peptides (black), XIP (blue), or ComS (green). The comR gene (S. mutans 61 [SMu.61]) was amplified using gene-specific primers (forward, AAAGAATCCTATGTTAAAAGA; reverse, CACCCTAGGAGACCCATCAAA) and cloned into the pET45b(+) vector bearing an N-terminal 6×His tag. The resulting vector, pET45b(+)his-comRUA159, was transformed into E. coli 10-beta. After sequencing confirmed the correct insertion (using T7 promoter and T7 terminator primers), the vector was transformed into E. coli BL21(DE3) prior to protein purification. Download FIG S3, TIF file, 0.1 MB (103.6KB, tif) .
Copyright © 2018 Underhill et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Methods used for deterministic modeling of the ComRS activation of comX with intracellular feedback. Download Text S1, DOCX file, 0.01 MB (14.8KB, docx) .
Copyright © 2018 Underhill et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
FMC ingredients. Download Text S2, PDF file, 0.5 MB (527KB, pdf) .
Copyright © 2018 Underhill et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Parameters for gamma distribution fits to single-cell PcomX GFP fluorescence distributions in microfluidic experiments. Download Table S1, DOCX file, 0.01 MB (13.2KB, docx) .
Copyright © 2018 Underhill et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fitted values for the 12 parameters of the feedback activation model and statistical measurement of their robustness from bootstrap process. Download Table S2, DOCX file, 0.01 MB (14KB, docx) .
Copyright © 2018 Underhill et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primer sequences used for RT-qPCR. Download Table S3, DOCX file, 0.02 MB (19.5KB, docx) .
Copyright © 2018 Underhill et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Methods for modeling of fluorescence polarization binding data of Fig. 5. Download Text S2, DOCX file, 0.01 MB (15.5KB, docx) .
Copyright © 2018 Underhill et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.