Abstract
In the mid-20th century, the unicellular and genetically tractable green alga Chlamydomonas reinhardtii was first developed as a model organism to elucidate fundamental cellular processes such as photosynthesis, light perception and the structure, function and biogenesis of cilia. Various studies of C. reinhardtii have profoundly advanced plant and cell biology, and have also impacted algal biotechnology and our understanding of human disease. However, the 'real' life of C. reinhardtii in the natural environment has largely been neglected. To extend our understanding of the biology of C. reinhardtii, it will be rewarding to explore its behavior in its natural habitats, learning more about its abundance and life cycle, its genetic and physiological diversity, and its biotic and abiotic interactions.
Introduction
Chlamydomonas reinhardtii is a single-celled green alga found in temperate soil habitats (Figure 1). It has proven to be such a powerful model for dissecting fundamental processes in biology that investigators have dubbed it the 'green yeast' (Goodenough, 1992; Rochaix, 1995). Ehrenberg described the genus Chlamydomonas in 1833, and Dangeard the species C. reinhardtii in 1888 (Harris et al., 2009). Chlamydomonas was found suitable for genetic studies in the early 20th century (Harris, 2001), while the development of C. reinhardtii as a model organism dates to the 1950s when the first mutants were generated (Harris, 2009).
Figure 1. Structure of a vegetative Chlamydomonas reinhardtii cell.
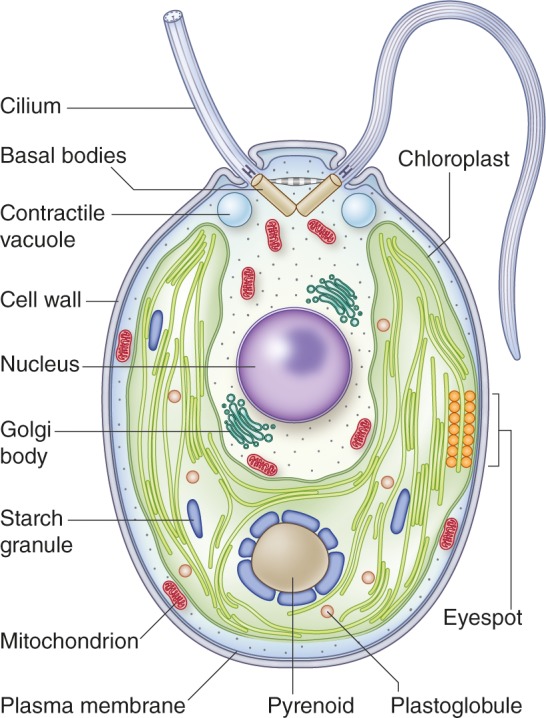
This cell has a 5-10 µm diameter (Gallaher et al., 2015). The two anterior cilia possess a 9+2 microtubule structure characteristic of motile cilia of eukaryotes. The cilia are critical for mating processes and confer motility to the cell (Harris, 2001). A single cup-shaped chloroplast occupies a large proportion of the cell's volume. This organelle houses the machinery for oxygenic photosynthesis and contains the pyrenoid, a structure in which Rubisco is concentrated; the pyrenoid is a component of the carbon concentrating mechanism (CCM) which functions to concentrate inorganic carbon in the cell against a concentration gradient (Mackinder et al., 2016). Close to the cell equator, at the edge of the chloroplast, is the eyespot. This primordial visual system allows the cells to orient their swimming toward or away from the light (phototaxis). Under hypoosmotic conditions, the cytoplasmic water content is maintained by pumping water out of the cell through contractile vacuoles positioned at the cell’s anterior (Komsic-Buchmann et al., 2014). At the base of the cilia are the basal bodies, which are responsible for ciliary assembly (Dutcher and O'Toole, 2016). Other features of the cell include a centrally located nucleus, a proteinaceous cell wall, Golgi bodies within the cup-shaped region formed by the chloroplast, and mitochondria. Image credit: Debbie Maizels.
Various features make C. reinhardtii an excellent laboratory species. It grows vegetatively as a haploid, which allows mutant phenotypes to be expressed immediately. Under optimal conditions, C. reinhardtii grows so rapidly that its numbers can double approximately every 8 hours (Harris, 2001). The fact that it can grow in the dark on acetate-containing medium while retaining a functional photosynthetic apparatus, has allowed even light-sensitive photosynthesis mutants to be isolated (Levine, 1969; Spreitzer and Mets, 1980). The motile cilia of this photosynthetic eukaryote share the same structure and many of the same constituent proteins as those of mammals, and so research into its motility prompted studies that greatly advanced our understanding of cilium dysfunctions in humans (Brown and Witman, 2014). Furthermore, C. reinhardtii can be induced to sexually reproduce in the laboratory, making it easy to introduce multiple traits into a single haploid strain (e.g. to generate double or triple mutants). The power of C. reinhardtii as a model organism was further elevated by the advent of genetic transformation (Boynton et al., 1988; Kindle, 1990; Remacle et al., 2006), the establishment of a full nuclear genome sequence (Merchant et al., 2007), the construction of a genome-wide library of mapped, indexed insertional mutants (Li et al., 2016) and CRISPR-mediated targeted gene disruptions (Ferenczi et al., 2017 and references therein).
Studies of C. reinhardtii have enabled numerous landmark discoveries and advances. One remarkable example is the discovery of intraflagellar transport of granule-like particles (Kozminski et al., 1993) and the roles of motor proteins in the process (Prevo et al., 2017 and references therein). Furthermore, structural analyses of wild type and mutants with defective cilia have massively contributed to our knowledge of the building blocks of these structures, their organization and their function (Goodenough and Heuser, 1985; Silflow and Lefebvre, 2001; Nicastro et al., 2006). These analyses also led to classic studies that demonstrated that abnormal cilia could cause human genetic diseases such as polycystic kidney disease (Pazour et al., 2000; Li et al., 2004). Additionally, acetate-requiring mutants (often unable to perform photosynthesis) have immensely advanced our understanding of photosynthesis, especially the ordering of electron carriers in the photosynthetic electron transport chain (e.g. Gorman and Levine, 1965). Two core proteins of photosystem II (D1 and D2) were first identified in C. reinhardtii (Chua and Gillham, 1977) and later proposed to be key components of this photosystem’s reaction centers (Deisenhofer et al., 1985; Trebst, 1986; Satoh, 2003). More recently, a central role of the STT7 kinase in photosynthetic state transitions (Depège et al., 2003), and a key function of the xanthophyll cycle in nonphotochemical quenching were first established in C. reinhardtii (Niyogi et al., 1997).
The field of optogenetics experienced a recent quantum leap with the discovery of channelrhodopsins in C. reinhardtii. When expressed in other cells, these gated ion channels can be stimulated with light to activate various processes, including neuronal activity (Hegemann and Nagel, 2013). Sophisticated genetic, biochemical and cell biological analyses of C. reinhardtii are currently being performed to understand the cell cycle (Cross and Umen, 2015), basal bodies/centrioles function (Dutcher and O'Toole, 2016), pyrenoid structure (Freeman Rosenzweig et al., 2017), mechanisms associated with photoreceptor function and light acclimation (Minagawa and Tokutsu, 2015; Petroutsos, 2017) and organismal interactions in ecosystems (Thrane et al., 2016). Finally, C. reinhardtii is being exploited to study the evolution of multicellularity, especially with respect to multicellular algal species of the order Volvocales (Hallmann, 2011).
Habitats and biogeography
C. reinhardtii can unambiguously be identified by sequencing internal transcribed spacers (ITS) or various phylogenetically informative genes (Pröschold et al., 2005). Yet many ecological studies have relied on light microscopy to identify Chlamydomonas species (sensu lato – see Box 1). Typically, two anterior cilia and a cup-shaped chloroplast harboring a pyrenoid have been sufficient criteria for a cell to be considered a Chlamydomonas sp. This morphology-based identification may be reliable to the level of genus, but rarely to the species level since many species look very similar. For these reasons, at times we omit species designations and simply note the organism as Chlamydomonas sp. A routine use of genetic taxonomic markers in the future would improve our knowledge of the geographical distribution of C. reinhardtii and related species and allow for more precise classifications.
Box 1.
Taxonomic and laboratory history of C. reinhardtii
Based on traditional taxonomic criteria, the genus Chlamydomonas (sensu lato) contains more than 500 species. In the course of taxonomic revisions, which are still in progress, Chlamydomonas (sensu stricto) is comprised of three species (Pröschold et al., 2018). Accordingly, the taxonomy of some species mentioned in this article, such as C. nivalis or C. euryale, may be revised in the future. Furthermore, our use of the designations 'Chlamydomonas sp.' and 'Chlamydomonas spp.' refers to one or more Chlamydomonas species, respectively, which were typically not classified to the level of species and may not be C. reinhardtii. The majority of the contemporary C. reinhardtii laboratory strains were derived from a single zygote isolated from a potato field in Massachusetts in 1945 (Harris, 2009). The sequencing of 39 common laboratory strains shows that they fall into five genetically distinct lineages from two parents or haplotypes (Gallaher et al., 2015). Under laboratory conditions, mutations accumulate at a rate of ~0.03 division-1 genome-1, corresponding to one mutation every 30 generations (Gallaher et al., 2015). In addition, removal of C. reinhardtii from its natural environment, including cultivation in the laboratory or cryopreservation, may unintentionally select for specific traits. For example, C. reinhardtii is often grown on medium containing ammonium as a nitrogen source, which allowed for the evolution of mutants (nit1, nit2) unable to utilize nitrate (Harris, 2009; Gallaher et al., 2015). For these reasons, the isolates domesticated for decades in the laboratory may only loosely correspond to wild C. reinhardtii strains. Furthermore, we do not know if the laboratory strains are still capable of surviving in the wild. To examine the ecological significance of laboratory findings, it will be important to isolate additional wild C. reinhardtii strains and characterize their behavior both in the field and in culture.
While Chlamydomonas spp. (not identified at the species level) occur widely in temperate, subtropical and tropical soils (Starks et al., 1981), confirmed C. reinhardtii has only been found in temperate soils in Northern America and Japan (Pröschold et al., 2005; Nakada et al., 2010). It occurs in cultivated fields but appears absent from many other habitats, suggesting it prefers nutrient-rich, disturbed soils (Sack et al., 1994). Most contemporary laboratory strains have emanated from a single soil isolate collected in 1945 (Box 1). Light typically penetrates only millimeters into the soil, depending on factors such as the soil structure and moisture content (Tester and Morris, 1987; Ciani et al., 2005). Therefore, photosynthetic microbes are generally most abundant in the upper few millimeters where they can harvest light energy, although in some instances they can be present in soil layers where there is essentially no light (Metting, 1981). Chlamydomonas spp. are even present in biological soil crusts where they help stabilize the surface of drylands, contribute to primary production and potentially act as pioneer species (Büdel et al., 2009).
All unambiguously identified C. reinhardtii isolates were collected from soil habitats (T. Pröschold, personal communication), yet Chlamydomonas spp. are also commonly found in the pelagic zone of lakes, where they sometimes form spring blooms (Similä, 1988; Krivtsov et al., 2000). The term 'pelagic zone' refers to the water column of lakes and oceans not on or near the lake or ocean bottom. Chlamydomonas spp. are usually motile, and although this has an energetic cost, it gives them a competitive advantage in lakes that have stratified into distinct layers as a consequence of seasonal changes in temperature (Striebel et al., 2009). Under conditions of stratification, motile algae often ascend toward the lake surface during the day to optimize their exposure to sunlight. During the night, they tend to descend to access the nutrient-rich environment below the surface. Indeed, this pattern of vertical movement has been observed for the population of Chlamydomonas sp. in a small Finnish lake (Jones, 1988).
Environmental conditions and the availability and distribution of natural resources differ substantially in soils and lakes (Sommer et al., 2012; Coleman et al., 2017). Phosphorous, for example, is likely to be limiting to the growth of organisms in lakes and geologically old soils, while nitrogen limitation is more common in young soils (Schindler, 1977; Vitousek and Howarth, 1991). Light availability and grazing pressure by predators represent additional key environmental differences between soil and lake habitats. Consequently, these two habitats require distinct adaptations and life history strategies to optimize fitness, and it is still an open question as to whether specific pelagic strains of C. reinhardtii exist in lakes (Box 2).
Box 2.
Outstanding questions about the natural history of C. reinhardtii
What are the geographic origins of C. reinhardtii? What are its current geographic and vertical distributions? How do populations of C. reinhardtii quantitatively change over time and what factors impact these changes? What are the major mechanisms of C. reinhardtii dispersal? For example, are aquifers common routes for the transport of C. reinhardtii over long distances?
What is the genetic variability within and between C. reinhardtii populations? What are the relationships among populations of the various Chlamydomonas species?
Do specific pelagic strains of C. reinhardtii exist in lakes? If so, do they have major differences in their life histories, physiologies and genome sequences compared to soil-dwelling strains?
What are the typical division rates of vegetative C. reinhardtii cells in the wild? How frequently does sexual reproduction occur in natural populations? How common are dormant zygospores in the environment, and where do they occur? Are zygospores typical overwintering forms, and do they also have an increased resistance to challenging biotic interactions?
What are the most common biotic interactions of C. reinhardtii in the environment (competing photosynthetic microbes, grazers, bacteria, fungi)? In what ways does C. reinhardtii communicate with its neighbors (e.g. infochemical signals)? What is the metabolic significance of these interactions?
How often and under what situations do cells shed their cilia in nature? Is there a selective advantage of deciliation in response to stress?
Does C. reinhardtii associate with biofilms on soil particles and, if so, how are the algal cells organized within the biofilm community?
Chlamydomonas spp. other than C. reinhardtii are adapted to a wide range of habitats. For example, Chlamydomonas eustigma is an acidophilic species isolated from acid mine drainage (Hirooka et al., 2017), Chlamydomonas euryale is found in temperate marine environments (Burch et al., 2015), Chlamydomonas spp. have been isolated from Antarctic ice (Liu et al., 2006), and some members of the genus Chlamydomonas are carotenoid-rich organisms present on the surface of snow, giving it a red appearance (Remias et al., 2005). A Chlamydomonas sp. has even been identified in the air at 1,100 meters above the ground: this and other algae can be dispersed by wind over extended distances (Brown et al., 1964). Taken together, several reports provide information on the biogeographical distribution of C. reinhardtii and other Chlamydomonas spp. However, there is little knowledge of the abundance and variations of Chlamydomonas spp. in different soil types, the dynamics of these natural populations over daily or seasonal cycles, and their physiological capabilities.
Genomics
The chloroplast and mitochondrial genomes of C. reinhardtii have been sequenced and are 206 and 15.8 kb, respectively (Vahrenholz et al., 1993; Maul et al., 2002; Gallaher et al., 2018). Since the sequence of the nuclear genome was first published (Merchant et al., 2007), the scientific community has focused some effort on elevating the quality of the genome sequence and improving its assembly and annotation (Blaby et al., 2014). The current version 5.5 nuclear genome is 111 Mb, which is similar in size to the genome of the model land plant Arabidopsis thaliana (Blaby et al., 2014). Recently, whole-genome sequences for more than 50 additional laboratory strains and field isolates were generated (Flowers et al., 2015; Gallaher et al., 2015). The sequences of 12 field isolates confirmed earlier reports that with a nucleotide diversity (π) of ~3%, the C. reinhardtii genome is among the most polymorphic of all eukaryotes (Flowers et al., 2015). The field strains, isolated from various locations in the United States and Canada, genetically group into three distinct populations, with gene flow between populations sufficiently low to allow the populations to adapt to their local environments. The low ratio of genome-wide non-synonymous to synonymous substitutions (0.58) further indicates that natural selection efficiently eliminates C. reinhardtii alleles of low fitness (Flowers et al., 2015). Whole-genome and epigenome sequencing has also been used to examine adaptation in the laboratory under changing environmental conditions (Kronholm et al., 2017).
Life cycle and its role in nature
Forming zygotes likely allows C. reinhardtii to survive when conditions become harsh (Harris, 2001; Goodenough et al., 2007). In the laboratory, gametogenesis can be induced by nitrogen starvation (Treier et al., 1989) in conjunction with specific light conditions; both signals may inform the cell of deteriorating environmental conditions (see below). The fusion of haploid gametes results in diploid zygotes that can develop over several days into highly resistant, dormant zygospores (Figure 2). When nitrogen is added back to the medium, the zygotes germinate in the light, undergo meiosis and typically release four haploid cells that resume vegetative growth (Harris, 2001). Dormant zygospores can remain viable in soil for many years (Harris, 2001) and survive freezing (Suzuki and Johnson, 2002), desiccation (Heimerl et al., 2018) and probably other forms of harsh environmental conditions. This extraordinary resistance is associated with the multilayered cell wall of the zygospores, which contains a durable lipid polymer structurally similar to those found in million-year-old microfossils (described for Chlamydomonas monoica; Blokker et al., 1999). Furthermore, sexual reproduction can increase the rate of adaptation of C. reinhardtii to new or changing environmental conditions, particularly if the population and genetic diversity within the population are large (Colegrave, 2002).
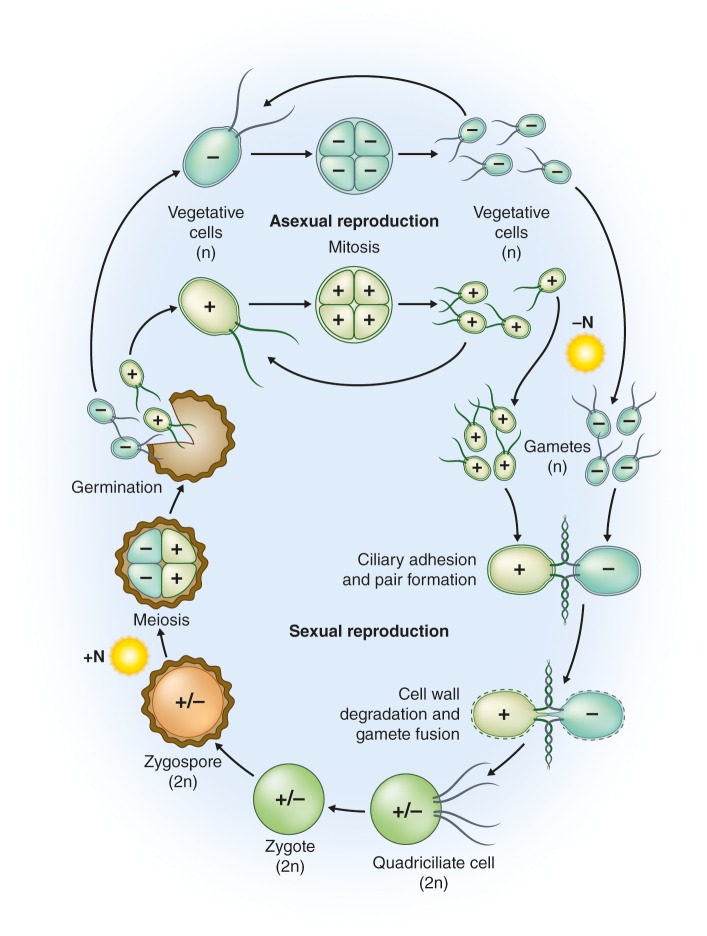
Figure 2. Life cycle of C.reinhardtii.
Haploid (n) vegetative cells occur as two mating types, mt+ and mt-, that divide by mitosis (“Asexual reproduction”; Harris, 2001, Goodenough et al., 2007). Gametogenesis can be induced by nitrogen starvation (-N) in the presence of light, and gametes of opposite mating types can fuse to form diploid (2n) zygotes (“Sexual reproduction”). Within a few hours of fertilization, zygotes resorb their four cilia to become immotile. Over the course of several days these zygotes are remodeled into highly resistant, dormant zygospores. In this process, a strong, multilayered cell wall is formed, and chlorophyll is degraded (Harris, 2001; Goodenough et al., 2007). As a result, mature zygospores appear orange, which reflects their carotenoid content (Lohr, 2009). When environmental conditions improve, the zygote undergoes meiosis to release four haploid cells (sometimes 8 and 16 when mitosis also occurs within the zygote wall; “Germination”). The haploid cells then resume vegetative growth. In the laboratory, zygote germination is induced by the addition of nitrogen (+N) to the medium in the light (Harris, 2001); nitrogen also causes reprogramming of gametes to vegetative cells (Pozuelo et al., 2000). Image credit: Debbie Maizels.
Following gamete fusion, a pair of homeodomain transcription factors initiates the genetic program for zygote development (Kurvari et al., 1998; Lee et al., 2008). The first zygote-specific genes are induced within minutes of gamete fusion, with hundreds of additional genes activated over the next few hours (Lopez et al., 2015; Joo et al., 2017). A gene encoding a polyketide synthase is induced two days after zygote formation and is critical for the zygote-to-zygospore transition, probably because it participates in the biosynthesis of the cell wall lipid polymer (Heimerl et al., 2018). Several stages of the sexual cycle, including gamete formation and maintenance and zygote germination, depend on light and involve regulation by three different photoreceptors (Huang and Beck, 2003; Müller et al., 2017; Zou et al., 2017).
Similar to Saccharomyces cerevisiae (Liti, 2015), we know little about the life cycle of C. reinhardtii in its natural environment. For example, there is no quantitative data on the frequency of sexual reproduction relative to vegetative growth. Yet, nitrogen is thought to become limited more commonly in soils than lakes (Schindler, 1977; Vitousek and Howarth, 1991; Coleman et al., 2017). This notion is congruent with nitrogen limitation being a major cue for zygospore formation in nature, but we are not aware of any data on zygospore induction in natural soil environments. The occurrence of clonal cultures of opposite mating types that are derived from a single zygospore isolated from dry soil provides additional evidence for a critical role of zygospores during desiccation (Harris, 2009). Freezing resistance of zygospores and their more efficient germination under long-day conditions compared to short-day conditions suggests that zygospore formation is an overwintering strategy (Suzuki and Johnson, 2002). If true, the question arises as to whether or not nitrogen limitation and day length are adequate cues to herald the approach of winter, or whether, for example, a decrease in soil moisture content or temperature can also induce zygospore formation in C. reinhardtii.
Physiological and metabolic capabilities
C. reinhardtii not only orients itself with respect to light, but can also swim upward in complete darkness. This negative gravitaxis may facilitate orientation and movement of the cells at night or in the soil environment, potentially helping the alga locate areas with more favorable conditions of illumination following daybreak (Bean, 1977). Furthermore, vegetative cells are attracted to ammonium, nitrite and nitrate (Ermilova and Zalutskaya, 2014 and references therein). Chemotaxis towards ammonium is strongest during the night, whereas phototaxis towards the light is strongest during the day, with both processes regulated by the circadian clock (Bruce, 1970; Byrne et al., 1992). Finally, the hypothesis that the circadian clock depends on gravity or a magnetic field was refuted by experiments performed with C. reinhardtii on a space shuttle under microgravity conditions (Mergenhagen and Mergenhagen, 1987).
Cilia enable C. reinhardtii to swim in an aqueous medium, and also glide on solid surfaces. Gliding motility may be important when C. reinhardtii resides within a thin water film that coats soil particles (Mitchell, 2000). The gliding speed of C. reinhardtii is ~1 µm s-1 (Shih et al., 2013) whereas the average forward swimming speed is 100-200 µm s-1 (Rüffer and Nultsch, 1985). Under various stress conditions, such as acidification of the medium, C. reinhardtii loses or sheds its cilia when a specific break point near the base of the cilium is activated (Quarmby, 2009). When conditions improve, the cilia regenerate. The biological reason for deciliation is still a mystery, but various hypotheses have been put forth (Quarmby, 2009). Deciliation is observed in a wide range of different cell types; for example, inhalation of irritant chemicals can lead to deciliation of respiratory epithelial cells in mammals (Buckley et al., 1984). Therefore, it seems likely that a predetermined break point is an ancient and inherent property of every cilium (Quarmby, 2009). Consequently, deciliation may not confer a selective advantage, but might be a consequence of pathological conditions that cause over-stimulation of the ciliary disassembly process. On the other hand, the ciliary membrane of C. reinhardtii is in direct contact with the environment (not protected by cell wall) and therefore, deciliation may reduce the entrance of noxious compounds into cells. Deciliation may also allow cells to escape when their cilia are stuck to the surface of a predator (Quarmby, 2009). Studying deciliation in the natural environment holds the promise of new insights into selection pressures that led to its evolution.
The ability of C. reinhardtii to grow under heterotrophic and fermentative conditions might be an adaptation to soil environments where there can be both low light and low oxygen. Anoxic/hypoxic conditions are likely to mostly occur at night when there is no photosynthesis to release oxygen and the soil microbes are respiring. Under anoxic conditions, C. reinhardtii can use glycolysis to yield energy, which is sustained by fermentation metabolism and the release of reduced organic compounds (Catalanotti et al., 2013). C. reinhardtii has recently been shown to activate a variety of different pathways that result in the formation of many fermentation products including formate, lactate, acetate, acetyl-CoA, succinate, hydrogen and glycerol (Atteia et al., 2013; Catalanotti et al., 2013; Yang et al., 2015). While some regulatory elements involved in anoxic metabolism are known (Hemschemeier et al., 2013; Huwald et al., 2015; Düner et al., 2018), little is understood about what controls the various pathways associated with fermentation and the ways in which these pathways are integrated.
Biotic interactions
In nature, C. reinhardtii is continuously in contact with other organisms, including competitors, predators, pathogens, parasites, commensals or mutualists. Most molecular details concerning these interactions, which likely involve chemical signaling, nutrient exchange and receptor-mediated processes, have not been examined. In lakes, the various Chlamydomonas spp. successfully compete with many other pelagic algal species for light and nutrients; rapid growth of Chlamydomonas spp. likely compensates for severe grazing losses, such as during periods of rapid proliferation of filter feeders, like water fleas (cladocerans). High rates of algal growth demand high nutrient levels. The concentrations of dissolved nutrients during the growing season are usually highest after spring mixing (Sommer et al., 2012), and therefore, the abundance of Chlamydomonas spp. in temperate lakes often shows a strong peak in spring or summer (Dembowska, 2015). In addition, the absence of filter feeders and the presence of more selective feeders in the soil may result in lower grazing losses and less seasonal differences in abundance patterns.
Predation of C. reinhardtii by zooplankton such as Daphnia, a highly efficient filter feeder (Van Donk et al., 1997), rotifers (Lurling and Beekman, 2006), and protists such as Tetrahymena (Taub and McKenzie, 1973) or Peranema (Figure 3), has been shown to occur in the laboratory. These predators either live exclusively in the pelagic zone of lakes, or at least more commonly in this habitat compared to soils. In the soil, animals such as earthworms or springtails, and protists are typical predators of microscopic algae (Schmidt et al., 2016; Seppey et al., 2017), but there is currently little specific information on predators of C. reinhardtii. The formation of large aggregates of C. reinhardtii cells is a general and probably non-specific defense strategy by which the alga may avoid ingestion. For example, the rotifer Brachionus calyciflorus triggers the formation of so-called palmelloid colonies (Lurling and Beekman, 2006). These colonies are aggregates of C. reinhardtii that may form as a consequence of the failure of the mitotically dividing mother cell to release the daughter cells from its encapsulating cell wall (Khona et al., 2016). This phenomenon may be triggered by stress under conditions in which zygospore formation is not possible (Khona et al., 2016). On the other hand, C. reinhardtii can actively aggregate in the presence of the predatory protist Peranema trichophorum (Sathe and Durand, 2016). A P. trichophorum culture filtrate was able to induce algal aggregation, suggesting that C. reinhardtii senses an unidentified substance (a kairomone) that is released by the predator (Sathe and Durand, 2016).
Figure 3. C. reinhardtii ingested by the predatory protist Peranema trichophorum.
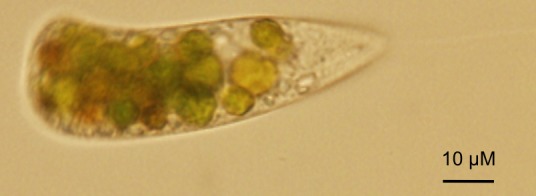
Image credit: Santosh Sathe and Pierre Durand.
C. reinhardtii is also a prey for soil bacteria. The bacterium Pseudomonas protegens can surround and immobilize algal cells (Video 1); it secretes a cyclic lipopeptide that triggers an increase in calcium levels inside C. reinhardtii cells with subsequent deciliation (Aiyar et al., 2017). This antagonistic interaction inhibits algal growth and probably leads to the death of most of the algal cells; the bacteria may acquire trace metals from the dying cells (Aiyar et al., 2017). Furthermore, small molecules from C. reinhardtii activate quorum sensing in Pseudomonas aeruginosa (Rajamani et al., 2008). It will be important to determine if algal cells also produce quorum-sensing mimics that influence P. protegens. Finally, while no viral pathogens of C. reinhardtii have been reported, it seems likely that they exist. The areas of algal-bacterial and algal-viral interactions are fertile for more probing basic research.
Video 1. C. reinhardtii surrounded by the harmful bacteria Pseudomonas protegens (Aiyar et al., 2017).
Video credit: Prasad Aiyar, Severin Sasso and Maria Mittag.
Several beneficial interactions of C. reinhardtii have been described, including interactions with growth-promoting bacteria and even mutualism (e. g. Nikolaev et al., 2008; Lörincz et al., 2010; Kim et al., 2014). These findings provide the basis for future studies that address regulatory mechanisms and identify specific compounds that impact the biology of C. reinhardtii in nature. One compound synthesized by prokaryotes and used by many algae is vitamin B12. Although C. reinhardtii does not depend on vitamin B12 to grow, it can obtain the compound from bacteria and use it as a cofactor in a pathway for methionine biosynthesis that is thermal tolerant (Kazamia et al., 2012; Xie et al., 2013). Indeed, under elevated temperatures, B12-providing bacteria increase the fitness of the alga (Xie et al., 2013). A mutualism was also observed between C. reinhardtii and S. cerevisiae in sealed microtiter plates, with the algae trading reduced nitrogen for CO2 (Hom and Murray, 2014). While the significance of these interactions may be uncertain, they, and many yet to be discovered, likely shape the ways in which C. reinhardtii navigates in a complex biosphere.
Conclusions
Although C. reinhardtii has been studied in the laboratory for many decades, we do not know the extent to which results from the laboratory reflect growth, life cycle and behavior of this alga in nature (Box 2). As a model system, C. reinhardtii is almost exclusively grown as a pure culture, a situation almost never encountered in the 'wild'. Returning a laboratory strain of C. reinhardtii to its native habitat would reveal whether domestication caused it to lose its ability to survive within the dynamic fabric of nature. Molecular analyses of the reintroduced strain could also reveal changes in the cells' physiology that underlie the loss of fitness in natural habitats, as well as other changes potentially associated with its adaptation to laboratory conditions.
Field surveys are often hampered by difficulties in assessing the metabolic state of the cells and in establishing key inter-organismal interactions. However, harnessing the full potential of meta-omics and single-cell technologies could provide a fuller appreciation of the physiological status of cells as they experience environmental fluctuations and the dominant interactions that shape the life of C. reinhardtii. Expanding this understanding will require time-resolved data on the geographical occurrence of C. reinhardtii in different habitats, its genetic potential and population genetics, and dissection of biotic and abiotic interactions. Such studies could then be extended to include analyses performed under controlled laboratory conditions that closely align with conditions encountered in the field, using innovative methods such as microfluidics to mimic conditions of the soil and other complex environments (Stanley et al., 2016).
Acknowledgements
We thank Dr. Thomas Pröschold for helpful comments on this manuscript, Debbie Maizels for preparing Figures 1 and 2, Drs. Santosh Sathe and Pierre Durand for providing Figure 3, and Prasad Aiyar for providing Video 1. Because of space limitations, we were unable to cite many worthy studies relevant to the topic of this article; we apologize to all of our colleagues whose work we did not discuss.
Biographies
Severin Sasso is at the Matthias Schleiden Institute of Genetics, Bioinformatics and Molecular Botany, Friedrich Schiller University, Jena, Germany
Herwig Stibor is in the Department Biology II, Ludwig Maximilian University, Munich, Germany
Maria Mittag is at the Matthias Schleiden Institute of Genetics, Bioinformatics and Molecular Botany, Friedrich Schiller University, Jena, Germany
Arthur R Grossman is at the Carnegie Institution for Science, Stanford, United States
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author contributions
Drafting and revising the article.
Drafting and revising the article.
Drafting and revising the article.
Drafting and revising the article.
Competing interests
No competing interests declared.
Contributor Information
Severin Sasso, Email: severin.sasso@uni-jena.de.
Arthur R Grossman, Email: arthurg@stanford.edu.
Stuart RF King, eLife, United Kingdom.
Peter A Rodgers, eLife, United Kingdom.
Funding Information
This paper was supported by the following grants:
Deutsche Forschungsgemeinschaft SFB 1127 to Severin Sasso, Maria Mittag.
U.S. Department of Energy DE-FG02-07ER64427 to Arthur R Grossman.
U.S. Department of Energy DE-FG02-12ER16338 to Arthur R Grossman.
Deutsche Forschungsgemeinschaft SA 2453/1-1 to Severin Sasso.
National Science Foundation NSF-MCB 0951094 to Arthur R Grossman.
References
- Aiyar P, Schaeme D, García-Altares M, Carrasco Flores D, Dathe H, Hertweck C, Sasso S, Mittag M. Antagonistic bacteria disrupt calcium homeostasis and immobilize algal cells. Nature Communications. 2017;8:1756. doi: 10.1038/s41467-017-01547-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atteia A, van Lis R, Tielens AGM, Martin WF. Anaerobic energy metabolism in unicellular photosynthetic eukaryotes. Biochimica et Biophysica Acta. 2013;1827:210–223. doi: 10.1016/j.bbabio.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Bean B. Geotactic behavior of Chlamydomonas. The Journal of Protozoology. 1977;24:394–401. doi: 10.1111/j.1550-7408.1977.tb04759.x. [DOI] [PubMed] [Google Scholar]
- Blaby IK, Blaby-Haas CE, Tourasse N, Hom EFY, Lopez D, Aksoy M, Grossman A, Umen J, Dutcher S, Porter M, King S, Witman GB, Stanke M, Harris EH, Goodstein D, Grimwood J, Schmutz J, Vallon O, Merchant SS, Prochnik S. The Chlamydomonas genome project: a decade on. Trends in Plant Science. 2014;19:672–680. doi: 10.1016/j.tplants.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokker P, Schouten S, de Leeuw JW, Damsté JSS, van den Ende H. Molecular structure of the resistant biopolymer in zygospore cell walls of Chlamydomonas monoica. Planta. 1999;207:539–543. doi: 10.1007/s004250050515. [DOI] [Google Scholar]
- Boynton JE, Gillham NW, Harris EH, Hosler JP, Johnson AM, Jones AR, Randolph-Anderson BL, Robertson D, Klein TM, Shark KB, Sanford JC. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988;240:1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- Brown Jr RM, Larson DA, Bold HC. Airborne algae: their abundance and heterogeneity. Science. 1964;143:583–585. doi: 10.1126/science.143.3606.583. [DOI] [PubMed] [Google Scholar]
- Brown JM, Witman GB. Cilia and diseases. BioScience. 2014;64:1126–1137. doi: 10.1093/biosci/biu174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce VG. The biological clock in Chlamydomonas reinhardi. The Journal of Protozoology. 1970;17:328–334. doi: 10.1111/j.1550-7408.1970.tb02380.x. [DOI] [Google Scholar]
- Buckley LA, Jiang XZ, James RA, Morgan KT, Barrow CS. Respiratory tract lesions induced by sensory irritants at the RD50 concentration. Toxicology and Applied Pharmacology. 1984;74:417–429. doi: 10.1016/0041-008X(84)90295-3. [DOI] [PubMed] [Google Scholar]
- Büdel B, Darienko T, Deutschewitz K, Dojani S, Friedl T, Mohr KI, Salisch M, Reisser W, Weber B. Southern African biological soil crusts are ubiquitous and highly diverse in drylands, being restricted by rainfall frequency. Microbial Ecology. 2009;57:229–247. doi: 10.1007/s00248-008-9449-9. [DOI] [PubMed] [Google Scholar]
- Burch TA, Adams III WW, Degrenne BLS, Englert CH, Mines BR, Nash PC, Boone EC, Demmig-Adams B. Environmental manipulation of growth and energy carrier release from freshwater and marine Chlamydomonas species. Journal of Applied Phycology. 2015;27:1127–1136. doi: 10.1007/s10811-014-0433-0. [DOI] [Google Scholar]
- Byrne TE, Wells MR, Johnson CH. Circadian rhythms of chemotaxis to ammonium and of methylammonium uptake in Chlamydomonas. Plant Physiology. 1992;98:879–886. doi: 10.1104/pp.98.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotti C, Yang W, Posewitz MC, Grossman AR. Fermentation metabolism and its evolution in algae. Frontiers in Plant Science. 2013;4:150. doi: 10.3389/fpls.2013.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua N-H, Gillham NW. The sites of synthesis of the principal thylakoid membrane polypeptides in Chlamydomonas reinhardtii. The Journal of Cell Biology. 1977;74:441–452. doi: 10.1083/jcb.74.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciani A, Goss K-U, Schwarzenbach RP. Light penetration in soil and particulate minerals. European Journal of Soil Science. 2005;56:561–574. doi: 10.1111/j.1365-2389.2005.00688.x. [DOI] [Google Scholar]
- Colegrave N. Sex releases the speed limit on evolution. Nature. 2002;420:664–666. doi: 10.1038/nature01191. [DOI] [PubMed] [Google Scholar]
- Coleman DC, Callaham Jr MA, Crossley Jr DA. Fundamentals of Soil Ecology. Third edition. Cambridge MA: Academic Press; 2017. [Google Scholar]
- Cross FR, Umen JG. The Chlamydomonas cell cycle. The Plant Journal. 2015;82:370–392. doi: 10.1111/tpj.12795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenhofer J, Epp O, Miki K, Huber R, Michel H. Structure of the protein subunits in the photosynthetic reaction centre of Rhodopseudomonas viridis at 3Å resolution. Nature. 1985;318:618–624. doi: 10.1038/318618a0. [DOI] [PubMed] [Google Scholar]
- Dembowska EA. Seasonal variation in phytoplankton and aquatic plants in floodplain lakes (lower Vistula River, Poland) Wetlands Ecology and Management. 2015;23:535–549. doi: 10.1007/s11273-015-9408-4. [DOI] [Google Scholar]
- Depège N, Bellafiore S, Rochaix J-D. Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science. 2003;299:1572–1575. doi: 10.1126/science.1081397. [DOI] [PubMed] [Google Scholar]
- Düner M, Lambertz J, Mügge C, Hemschemeier A. The soluble guanylate cyclase CYG12 is required for the acclimation to hypoxia and trophic regimes in Chlamydomonas reinhardtii. The Plant Journal. 2018;93:311–337. doi: 10.1111/tpj.13779. [DOI] [PubMed] [Google Scholar]
- Dutcher SK, O'Toole ET. The basal bodies of Chlamydomonas reinhardtii. Cilia. 2016;5:18. doi: 10.1186/s13630-016-0039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermilova E, Zalutskaya Z. Regulation by light of chemotaxis to nitrite during the sexual life cycle in Chlamydomonas reinhardtii. Plants. 2014;3:113–127. doi: 10.3390/plants3010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenczi A, Pyott DE, Xipnitou A, Molnar A. Efficient targeted DNA editing and replacement in Chlamydomonas reinhardtii using Cpf1 ribonucleoproteins and single-stranded DNA. PNAS. 2017;114:13567–13572. doi: 10.1073/pnas.1710597114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers JM, Hazzouri KM, Pham GM, Rosas U, Bahmani T, Khraiwesh B, Nelson DR, Jijakli K, Abdrabu R, Harris EH, Lefebvre PA, Hom EFY, Salehi-Ashtiani K, Purugganan MD. Whole-genome resequencing reveals extensive natural variation in the model green alga Chlamydomonas reinhardtii. The Plant Cell. 2015;27:2353–2369. doi: 10.1105/tpc.15.00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman Rosenzweig ES, Xu B, Kuhn Cuellar L, Martinez-Sanchez A, Schaffer M, Strauss M, Cartwright HN, Ronceray P, Plitzko JM, Förster F, Wingreen NS, Engel BD, Mackinder LCM, Jonikas MC. The eukaryotic CO2-concentrating organelle is liquid-like and exhibits dynamic reorganization. Cell. 2017;171:148–162. doi: 10.1016/j.cell.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaher SD, Fitz-Gibbon ST, Glaesener AG, Pellegrini M, Merchant SS. Chlamydomonas genome resource for laboratory strains reveals a mosaic of sequence variation, identifies true strain histories, and enables strain-specific studies. The Plant Cell. 2015;27:2335–2352. doi: 10.1105/tpc.15.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaher SD, Fitz-Gibbon ST, Strenkert D, Purvine SO, Pellegrini M, Merchant SS. High-throughput sequencing of the chloroplast and mitochondrion of Chlamydomonas reinhardtii to generate improved de novo assemblies, analyze expression patterns and transcript speciation, and evaluate diversity among laboratory strains and wild isolates. The Plant Journal. 2018;93:545–565. doi: 10.1111/tpj.13788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE. Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. The Journal of Cell Biology. 1985;100:2008–2018. doi: 10.1083/jcb.100.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW. Green yeast. Cell. 1992;70:533–538. doi: 10.1016/0092-8674(92)90424-B. [DOI] [PubMed] [Google Scholar]
- Goodenough U, Lin H, Lee J-H. Sex determination in Chlamydomonas. Seminars in Cell and Developmental Biology. 2007;18:350–361. doi: 10.1016/j.semcdb.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Gorman DS, Levine RP. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. PNAS. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmann A. Evolution of reproductive development in the volvocine algae. Sexual Plant Reproduction. 2011;24:97–112. doi: 10.1007/s00497-010-0158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. Chlamydomonas as a model organism. Annual Review of Plant Physiology and Plant Molecular Biology. 2001;52:363–406. doi: 10.1146/annurev.arplant.52.1.363. [DOI] [PubMed] [Google Scholar]
- Harris EH. Introduction to Chlamydomonas and its laboratory use. In: Harris EH, Stern DB, Witman GB, editors. The Chlamydomonas Sourcebook. Oxford: Academic Press; 2009. [Google Scholar]
- Harris EH, Stern DB, Witman GB. The Chlamydomonas Sourcebook. Second edition. Oxford: Academic Press; 2009. [Google Scholar]
- Hegemann P, Nagel G. From channelrhodopsins to optogenetics. EMBO Molecular Medicine. 2013;5:173–176. doi: 10.1002/emmm.201202387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimerl N, Hommel E, Westermann M, Meichsner D, Lohr M, Hertweck C, Grossman AR, Mittag M, Sasso S. A giant type I polyketide synthase participates in zygospore maturation in Chlamydomonas reinhardtii. The Plant Journal. 2018;95:268–281. doi: 10.1111/tpj.13948. [DOI] [PubMed] [Google Scholar]
- Hemschemeier A, Casero D, Liu B, Benning C, Pellegrini M, Happe T, Merchant SS. COPPER RESPONSE REGULATOR1-dependent and -independent responses of the Chlamydomonas reinhardtii transcriptome to dark anoxia. The Plant Cell. 2013;25:3186–3211. doi: 10.1105/tpc.113.115741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirooka S, Hirose Y, Kanesaki Y, Higuchi S, Fujiwara T, Onuma R, Era A, Ohbayashi R, Uzuka A, Nozaki H, Yoshikawa H, Miyagishima S-ya. Acidophilic green algal genome provides insights into adaptation to an acidic environment. PNAS. 2017;114:E8304–E8313. doi: 10.1073/pnas.1707072114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom EFY, Murray AW. Niche engineering demonstrates a latent capacity for fungal-algal mutualism. Science. 2014;345:94–98. doi: 10.1126/science.1253320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Beck CF. Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. PNAS. 2003;100:6269–6274. doi: 10.1073/pnas.0931459100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huwald D, Schrapers P, Kositzki R, Haumann M, Hemschemeier A. Characterization of unusual truncated hemoglobins of Chlamydomonas reinhardtii suggests specialized functions. Planta. 2015;242:167–185. doi: 10.1007/s00425-015-2294-4. [DOI] [PubMed] [Google Scholar]
- Jones RI. Vertical distribution and diel migration of flagellated phytoplankton in a small humic lake. Hydrobiologia. 1988;161:75–87. doi: 10.1007/BF00044102. [DOI] [Google Scholar]
- Joo S, Nishimura Y, Cronmiller E, Hong RH, Kariyawasam T, Wang MH, Shao NC, El Akkad S-E-D, Suzuki T, Higashiyama T, Jin E, Lee J-H. Gene regulatory networks for the haploid-to-diploid transition of Chlamydomonas reinhardtii. Plant Physiology. 2017;175:314–332. doi: 10.1104/pp.17.00731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazamia E, Czesnick H, Nguyen TTV, Croft MT, Sherwood E, Sasso S, Hodson SJ, Warren MJ, Smith AG. Mutualistic interactions between vitamin B12-dependent algae and heterotrophic bacteria exhibit regulation. Environmental Microbiology. 2012;14:1466–1476. doi: 10.1111/j.1462-2920.2012.02733.x. [DOI] [PubMed] [Google Scholar]
- Khona DK, Shirolikar SM, Gawde KK, Hom E, Deodhar MA, D'Souza JS. Characterization of salt stress-induced palmelloids in the green alga, Chlamydomonas reinhardtii. Algal Research. 2016;16:434–448. doi: 10.1016/j.algal.2016.03.035. [DOI] [Google Scholar]
- Kim B-H, Ramanan R, Cho D-H, Oh H-M, Kim H-S. Role of Rhizobium, a plant growth promoting bacterium, in enhancing algal biomass through mutualistic interaction. Biomass and Bioenergy. 2014;69:95–105. doi: 10.1016/j.biombioe.2014.07.015. [DOI] [Google Scholar]
- Kindle KL. High-frequency nuclear transformation of Chlamydomonas reinhardtii. PNAS. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komsic-Buchmann K, Wöstehoff L, Becker B. The contractile vacuole as a key regulator of cellular water flow in Chlamydomonas reinhardtii. Eukaryotic Cell. 2014;13:1421–1430. doi: 10.1128/EC.00163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. PNAS. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov V, Bellinger EG, Sigee DC. Changes in the elemental composition of Asterionella formosa during the diatom spring bloom. Journal of Plankton Research. 2000;22:169–184. doi: 10.1093/plankt/22.1.169. [DOI] [Google Scholar]
- Kronholm I, Bassett A, Baulcombe D, Collins S. Epigenetic and genetic contributions to adaptation in Chlamydomonas. Molecular Biology and Evolution. 2017;34:2285–2306. doi: 10.1093/molbev/msx166. [DOI] [PubMed] [Google Scholar]
- Kurvari V, Grishin NV, Snell WJ. A gamete-specific, sex-limited homeodomain protein in Chlamydomonas. The Journal of Cell Biology. 1998;143:1971–1980. doi: 10.1083/jcb.143.7.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-H, Lin H, Joo S, Goodenough U. Early sexual origins of homeoprotein heterodimerization and evolution of the plant KNOX/BELL family. Cell. 2008;133:829–840. doi: 10.1016/j.cell.2008.04.028. [DOI] [PubMed] [Google Scholar]
- Levine RP. The analysis of photosynthesis using mutant strains of algae and higher plants. Annual Review of Plant Physiology. 1969;20:523–540. doi: 10.1146/annurev.pp.20.060169.002515. [DOI] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, Beales PL, Guay-Woodford LM, Yoder BK, Stormo GD, Katsanis N, Dutcher SK. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/S0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang R, Patena W, Gang SS, Blum SR, Ivanova N, Yue R, Robertson JM, Lefebvre PA, Fitz-Gibbon ST, Grossman AR, Jonikas MC. An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. The Plant Cell. 2016;28:367–387. doi: 10.1105/tpc.15.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G. The fascinating and secret wild life of the budding yeast S. cerevisiae. eLife. 2015;4:e05835. doi: 10.7554/eLife.05835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Huang X, Wang X, Zhang X, Li G. Phylogenetic studies on two strains of Antarctic ice algae based on morphological and molecular characteristics. Phycologia. 2006;45:190–198. doi: 10.2216/03-88.1. [DOI] [Google Scholar]
- Lohr M. Carotenoids. In: Harris EH, Stern DB, Witman GB, editors. The Chlamydomonas Sourcebook. Amsterdam: Academic Press; 2009. pp. 799–817. [Google Scholar]
- Lopez DA, Hamaji T, Kropat J, De Hoff P, Morselli M, Rubbi L, Fitz-Gibbon ST, Gallaher SD, Merchant SS, Umen JG, Pellegrini M. Dynamic changes in the transcriptome and methylome of Chlamydomonas reinhardtii throughout its life cycle. Plant Physiology. 2015;169:2730–2743. doi: 10.1104/pp.15.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lörincz Z, Preininger É, Kósa A, Pónyi T, Nyitrai P, Sarkadi L, Kovács GM, Böddi B, Gyurján I. Artificial tripartite symbiosis involving a green alga (Chlamydomonas), a bacterium (Azotobacter) and a fungus (Alternaria): morphological and physiological characterization. Folia Microbiologica (Praha) 2010;55:393–400. doi: 10.1007/s12223-010-0067-9. [DOI] [PubMed] [Google Scholar]
- Lurling M, Beekman W. Palmelloids formation in Chlamydomonas reinhardtii: defence against rotifer predators? Annales de Limnologie - International Journal of Limnology. 2006;42:65–72. doi: 10.1051/limn/2006010. [DOI] [Google Scholar]
- Mackinder LCM, Meyer MT, Mettler-Altmann T, Chen VK, Mitchell MC, Caspari O, Freeman Rosenzweig ES, Pallesen L, Reeves G, Itakura A, Roth R, Sommer F, Geimer S, Mühlhaus T, Schroda M, Goodenough U, Stitt M, Griffiths H, Jonikas MC. A repeat protein links rubisco to form the eukaryotic carbon-concentrating organelle. PNAS. 2016;113:5958–5963. doi: 10.1073/pnas.1522866113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul JE, Lilly JW, Cui L, dePamphilis CW, Miller W, Harris EH, Stern DB. The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. The Plant Cell. 2002;14:2659–2679. doi: 10.1105/tpc.006155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, Marshall WF, Qu L-H, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H, Lucas SM, Grimwood J, Schmutz J, Cardol P, Cerutti H, Chanfreau G, Chen CL, Cognat V, Croft MT, Dent R, Dutcher S, Fernández E, Fukuzawa H, González-Ballester D, González-Halphen D, Hallmann A, Hanikenne M, Hippler M, Inwood W, Jabbari K, Kalanon M, Kuras R, Lefebvre PA, Lemaire SD, Lobanov AV, Lohr M, Manuell A, Meier I, Mets L, Mittag M, Mittelmeier T, Moroney JV, Moseley J, Napoli C, Nedelcu AM, Niyogi K, Novoselov SV, Paulsen IT, Pazour G, Purton S, Ral JP, Riaño-Pachón DM, Riekhof W, Rymarquis L, Schroda M, Stern D, Umen J, Willows R, Wilson N, Zimmer SL, Allmer J, Balk J, Bisova K, Chen CJ, Elias M, Gendler K, Hauser C, Lamb MR, Ledford H, Long JC, Minagawa J, Page MD, Pan J, Pootakham W, Roje S, Rose A, Stahlberg E, Terauchi AM, Yang P, Ball S, Bowler C, Dieckmann CL, Gladyshev VN, Green P, Jorgensen R, Mayfield S, Mueller-Roeber B, Rajamani S, Sayre RT, Brokstein P, Dubchak I, Goodstein D, Hornick L, Huang YW, Jhaveri J, Luo Y, Martínez D, Ngau WC, Otillar B, Poliakov A, Porter A, Szajkowski L, Werner G, Zhou K, Grigoriev IV, Rokhsar DS, Grossman AR. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–251. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergenhagen D, Mergenhagen E. The biological clock of Chlamydomonas reinhardtii in space. European Journal of Cell Biology. 1987;43:203–207. [PubMed] [Google Scholar]
- Metting B. The systematics and ecology of soil algae. The Botanical Review. 1981;47:195–312. doi: 10.1007/BF02868854. [DOI] [Google Scholar]
- Minagawa J, Tokutsu R. Dynamic regulation of photosynthesis in Chlamydomonas reinhardtii. The Plant Journal. 2015;82:413–428. doi: 10.1111/tpj.12805. [DOI] [PubMed] [Google Scholar]
- Mitchell DR. Chlamydomonas flagella. Journal of Phycology. 2000;36:261–273. doi: 10.1046/j.1529-8817.2000.99218.x. [DOI] [Google Scholar]
- Müller N, Wenzel S, Zou Y, Künzel S, Sasso S, Weiß D, Prager K, Grossman A, Kottke T, Mittag M. A plant cryptochrome controls key features of the Chlamydomonas circadian clock and its life cycle. Plant Physiology. 2017;174:185–201. doi: 10.1104/pp.17.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada T, Shinkawa H, Ito T, Tomita M. Recharacterization of Chlamydomonas reinhardtii and its relatives with new isolates from Japan. Journal of Plant Research. 2010;123:67–78. doi: 10.1007/s10265-009-0266-0. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- Nikolaev YA, Plakunov VK, Voronina NA, Nemtseva NV, Plotnikov AO, Gogoleva OA, Murav’eva ME, Ovechkina GV. Effect of bacterial satellites on Chlamydomonas reinhardtii growth in an algo-bacterial community. Microbiology (Mikrobiologiya) 2008;77:78–83. doi: 10.1134/S0026261708010116. [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Björkman O, Grossman AR. The roles of specific xanthophylls in photoprotection. PNAS. 1997;94:14162–14167. doi: 10.1073/pnas.94.25.14162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, Cole DG. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene Tg737, are required for assembly of cilia and flagella. The Journal of Cell Biology. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroutsos D. Chlamydomonas photoreceptors: Cellular functions and impact on physiology. In: Hippler M, editor. Chlamydomonas: Biotechnology and Biomedicine. Cham (Switzerland): Springer; 2017. pp. 1–19. [Google Scholar]
- Pozuelo M, Merchán F, Macías MI, Beck CF, Galván A, Fernández E. The negative effect of nitrate on gametogenesis is independent of nitrate assimilation in Chlamydomonas reinhardtii. Planta. 2000;211:287–292. doi: 10.1007/s004250000291. [DOI] [PubMed] [Google Scholar]
- Prevo B, Scholey JM, Peterman EJG. Intraflagellar transport: mechanisms of motor action, cooperation, and cargo delivery. The FEBS Journal. 2017;284:2905–2931. doi: 10.1111/febs.14068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pröschold T, Harris EH, Coleman AW. Portrait of a species: Chlamydomonas reinhardtii. Genetics. 2005;170:1601–1610. doi: 10.1534/genetics.105.044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pröschold T, Darienko T, Krienitz L, Coleman AW. Chlamydomonas schloesseri sp. nov. (Chlamydophyceae, Chlorophyta) revealed by morphology, autolysin cross experiments, and multiple gene analyses. Phytotaxa. 2018;362:21–38. doi: 10.11646/phytotaxa.362.1.2. [DOI] [Google Scholar]
- Quarmby LM. Deflagellation. In: Harris EH, Stern DB, Witman GB, editors. The Chlamydomonas Sourcebook. Amsterdam: Academic Press; 2009. pp. 43–69. [Google Scholar]
- Rajamani S, Bauer WD, Robinson JB, Farrow III JM, Pesci EC, Teplitski M, Gao M, Sayre RT, Phillips DA. The vitamin riboflavin and its derivative lumichrome activate the LasR bacterial quorum-sensing receptor. Molecular Plant-Microbe Interactions. 2008;21:1184–1192. doi: 10.1094/MPMI-21-9-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle C, Cardol P, Coosemans N, Gaisne M, Bonnefoy N. High-efficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes. PNAS. 2006;103:4771–4776. doi: 10.1073/pnas.0509501103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remias D, Lütz-Meindl U, Lütz C. Photosynthesis, pigments and ultrastructure of the alpine snow alga Chlamydomonas nivalis. European Journal of Phycology. 2005;40:259–268. doi: 10.1080/09670260500202148. [DOI] [Google Scholar]
- Rochaix J-D. Chlamydomonas reinhardtii as the photosynthetic yeast. Annual Review of Genetics. 1995;29:209–230. doi: 10.1146/annurev.ge.29.120195.001233. [DOI] [PubMed] [Google Scholar]
- Rüffer U, Nultsch W. High-speed cinematographic analysis of the movement of Chlamydomonas. Cell Motility. 1985;5:251–263. doi: 10.1002/cm.970050307. [DOI] [Google Scholar]
- Sack L, Zeyl C, Bell G, Sharbel T, Reboud X, Bernhardt T, Koelewyn H. Isolation of four new strains of Chlamydomonas reinhardtii (Chlorophyta) from soil samples. Journal of Phycology. 1994;30:770–773. doi: 10.1111/j.0022-3646.1994.00770.x. [DOI] [Google Scholar]
- Sathe S, Durand PM. Cellular aggregation in Chlamydomonas (Chlorophyceae) is chimaeric and depends on traits like cell size and motility. European Journal of Phycology. 2016;51:129–138. doi: 10.1080/09670262.2015.1107759. [DOI] [Google Scholar]
- Satoh K. The identification of the photosystem II reaction center: a personal story. Photosynthesis Research. 2003;76:233–240. doi: 10.1023/A:1024933610778. [DOI] [PubMed] [Google Scholar]
- Schindler DW. Evolution of phosphorus limitation in lakes. Science. 1977;195:260–262. doi: 10.1126/science.195.4275.260. [DOI] [PubMed] [Google Scholar]
- Schmidt O, Dyckmans J, Schrader S. Photoautotrophic microorganisms as a carbon source for temperate soil invertebrates. Biology Letters. 2016;12:20150646. doi: 10.1098/rsbl.2015.0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppey CVW, Singer D, Dumack K, Fournier B, Belbahri L, Mitchell EAD, Lara E. Distribution patterns of soil microbial eukaryotes suggests widespread algivory by phagotrophic protists as an alternative pathway for nutrient cycling. Soil Biology and Biochemistry. 2017;112:68–76. doi: 10.1016/j.soilbio.2017.05.002. [DOI] [Google Scholar]
- Shih SM, Engel BD, Kocabas F, Bilyard T, Gennerich A, Marshall WF, Yildiz A. Intraflagellar transport drives flagellar surface motility. eLife. 2013;2:e00744. doi: 10.7554/eLife.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow CD, Lefebvre PA. Assembly and motility of eukaryotic cilia and flagella: lessons from Chlamydomonas reinhardtii. Plant Physiology. 2001;127:1500–1507. doi: 10.1104/pp.010807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Similä A. Spring development of a Chlamydomonas population in Lake Nimetön, a small humic forest lake in southern Finland. Hydrobiologia. 1988;161:149–157. doi: 10.1007/BF00044107. [DOI] [Google Scholar]
- Sommer U, Adrian R, De Senerpont Domis L, Elser JJ, Gaedke U, Ibelings B, Jeppesen E, Lürling M, Molinero JC, Mooij WM, van Donk E, Winder M. Beyond the plankton ecology group (PEG) model: mechanisms driving plankton succession. Annual Review of Ecology, Evolution, and Systematics. 2012;43:429–448. doi: 10.1146/annurev-ecolsys-110411-160251. [DOI] [Google Scholar]
- Spreitzer RJ, Mets LJ. Non-mendelian mutation affecting ribulose-1,5-bisphosphate carboxylase structure and activity. Nature. 1980;285:114–115. doi: 10.1038/285114a0. [DOI] [Google Scholar]
- Stanley CE, Grossmann G, Casadevall i Solvas X, deMello AJ. Soil-on-a-Chip: microfluidic platforms for environmental organismal studies. Lab on a Chip. 2016;16:228–241. doi: 10.1039/C5LC01285F. [DOI] [PubMed] [Google Scholar]
- Starks TL, Shubert LE, Trainor FR. Ecology of soil algae: a review. Phycologia. 1981;20:65–80. doi: 10.2216/i0031-8884-20-1-65.1. [DOI] [Google Scholar]
- Striebel M, Bartholmé S, Zernecke R, Steinlein C, Haupt F, Diehl S, Stibor H. Carbon sequestration and stoichiometry of motile and nonmotile green algae. Limnology and Oceanography. 2009;54:1746–1752. doi: 10.4319/lo.2009.54.5.1746. [DOI] [Google Scholar]
- Suzuki L, Johnson CH. Photoperiodic control of germination in the unicell Chlamydomonas. Naturwissenschaften. 2002;89:214–220. doi: 10.1007/s00114-002-0302-6. [DOI] [PubMed] [Google Scholar]
- Taub FB, McKenzie DH. Continuous cultures of an alga and its grazer. Bulletins from the Ecological Research Committee. 1973;17:371–377. [Google Scholar]
- Tester M, Morris C. The penetration of light through soil. Plant, Cell and Environment. 1987;10:281–286. doi: 10.1111/j.1365-3040.1987.tb01607.x. [DOI] [Google Scholar]
- Thrane JE, Hessen DO, Andersen T. The impact of irradiance on optimal and cellular nitrogen to phosphorus ratios in phytoplankton. Ecology Letters. 2016;19:880–888. doi: 10.1111/ele.12623. [DOI] [PubMed] [Google Scholar]
- Trebst A. The topology of the plastoquinone and herbicide binding peptides of photosystem II in the thylakoid membrane. Zeitschrift für Naturforschung. 1986;41c:240–246. doi: 10.1515/znc-1986-1-235. [DOI] [Google Scholar]
- Treier U, Fuchs S, Weber M, Wakarchuk WW, Beck CF. Gametic differentiation in Chlamydomonas reinhardtii: light dependence and gene expression patterns. Archives of Microbiology. 1989;152:572–577. doi: 10.1007/BF00425489. [DOI] [Google Scholar]
- Vahrenholz C, Riemen G, Pratje E, Dujon B, Michaelis G. Mitochondrial DNA of Chlamydomonas reinhardtii: the structure of the ends of the linear 15.8-kb genome suggests mechanisms for DNA replication. Current Genetics. 1993;24:241–247. doi: 10.1007/BF00351798. [DOI] [PubMed] [Google Scholar]
- Van Donk E, Lürling M, Hessen DO, Lokhorst GM. Altered cell wall morphology in nutrient-deficient phytoplankton and its impact on grazers. Limnology and Oceanography. 1997;42:357–364. doi: 10.4319/lo.1997.42.2.0357. [DOI] [Google Scholar]
- Vitousek P, Howarth R. Nitrogen limitation on land and in the sea: how can it occur? Biogeochemistry. 1991;13:87–115. doi: 10.1007/BF00002772. [DOI] [Google Scholar]
- Xie B, Bishop S, Stessman D, Wright D, Spalding MH, Halverson LJ. Chlamydomonas reinhardtii thermal tolerance enhancement mediated by a mutualistic interaction with vitamin B12-producing bacteria. The ISME Journal. 2013;7:1544–1555. doi: 10.1038/ismej.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Catalanotti C, Wittkopp TM, Posewitz MC, Grossman AR. Algae after dark: mechanisms to cope with anoxic/hypoxic conditions. The Plant Journal. 2015;82:481–503. doi: 10.1111/tpj.12823. [DOI] [PubMed] [Google Scholar]
- Zou Y, Wenzel S, Müller N, Prager K, Jung E-M, Kothe E, Kottke T, Mittag M. An animal-like cryptochrome controls the Chlamydomonas sexual cycle. Plant Physiology. 2017;174:1334–1347. doi: 10.1104/pp.17.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]