Abstract
In the phytopathogenic fungus Sporisorium scitamineum, sexual mating between two compatible haploid cells and the subsequent formation of dikaryotic hyphae is essential for infection. This process was shown to be commonly regulated by a mitogen-activated protein kinase (MAPK) and a cAMP/PKA signaling pathway in the corn smut fungus Ustilago maydis but remains largely unknown in S. scitamineum. In this study, we identified a conserved putative MAP kinase Kpp2 in S. scitamineum and named it as SsKpp2. The sskpp2Δ mutant displayed significant reduction in mating/filamentation, which could be partially restored by addition of cAMP or tryptophol, a quorum-sensing molecule identified in budding yeast. Transcriptional profiling showed that genes governing S. scitamineum mating or tryptophol biosynthesis were significantly differentially regulated in the sskpp2Δ mutant compared to the WT, under mating condition. Our results demonstrate that the MAP kinase SsKpp2 is required for S. scitamineum mating/filamentation likely through regulating the conserved pheromone signal transduction pathway and tryptophol production.
Keywords: filamentous growth, Kpp2, MAP kinase, mating, Sporisorium scitamineum, tryptophol
Introduction
The basidiomycetous fungus Sporisorium scitamineum causes sugarcane smut that leads to severe economic losses in the major sugarcane growing areas in China, India, and Brazil. There are three distinct stages in the pathogenic life cycle of S. scitamineum, namely the haploid sporidium, the dikaryotic hyphae, and the diploid teliospore stages. The yeast-like and non-pathogenic sporidia are of two opposite mating types, MAT-1 (+) and MAT-2 (−). The compatible sporidia fuse to form dikaryotic hypha, which are capable of infecting the host plant. Diploid teliospores form in planta by nuclear fusion, and go through a round of meiosis to form four haploid sporidia again (Sundar et al., 2012). Sexual mating and the subsequent filamentous growth (filamentation) is a prerequisite for host cane infection. However, our knowledge on molecular basis of S. scitamineum mating/filamentation is very limited.
Mitogen-activated protein kinase (MAPK) signaling pathway is widely conserved in eukaryotic organisms and involved in regulation of cell growth and differentiation, metabolism, stress response including resistance against pathogens in plant or animal hosts (Plotnikov et al., 2011; Meng and Zhang, 2013; Correia et al., 2016). On the other hand, MAPK signaling in fungal pathogens was shown to be closely related to virulence and pathogenicity (Davidson et al., 2003; Roman et al., 2007; Hamel et al., 2012; Jiang et al., 2018). In Ustilago maydis, mating/filamentation was regulated by both cAMP/PKA (protein kinase A) signaling pathway and MAPK signaling cascade Kpp4-Fuz7-Kpp2, with partial overlap, and commonly inducing the a locus that encodes the pheromone precursors and receptors through the transcriptional factor Prf1 (Gold et al., 1994; Muller et al., 1999, 2003; Kaffarnik et al., 2003). The MAP kinase Kpp2 is phosphorylated and activated by the upstream kinase Fuz7, and in turn activates its downstream target protein Prf1, the master transcriptional factor for sexual mating in U. maydis, to initiate sexual mating in response to pheromone signal (Muller et al., 2003). Kpp2 kinase is conserved in its activation loop (A-loop) motif, especially in the TXY motif containing a Threonine (T) and a Tyrosine (Y) residue that are phosphorylated sites and essential for Kpp2 kinase activity (Chen et al., 2001; Pearson et al., 2001).
Quorum-sensing (QS) plays a role in cell-density based coordinating expression of genes, including virulence genes, in bacterial pathogens (Fuqua et al., 1994; Gray, 1997). Investigation on fungal quorum-sensing molecules (QSMs) initiates recently. So far QSMs have been identified in Saccharomyces cerevisiae (Wuster and Babu, 2010), Candida albicans (Hornby et al., 2001; Chen and Fink, 2004), Cryptococcus neoformans (Albuquerque et al., 2013; Tian et al., 2018), Aspergillus nidulans (Williams et al., 2012) and Penicillium sclerotiorum (Raina et al., 2010), regulating fungal morphogenesis, pathogenicity, and/or secondary metabolism (Barriuso et al., 2018). Among these fungal QSMs, aromatic alchohols tyrosol, tryptophol, and phenylethanol regulate filamentous growth or pseudohyphae growth in S. cerevisiae or C. albicans by integrating cell density and nitrogen availability (Chen and Fink, 2006; Wongsuk et al., 2016). These three aromatic alchohols could be produced by Ehrlich pathway (Dickinson et al., 2003; Hazelwood et al., 2008), first step of which is aromatic amino acid deamination catalyzed by aminotransferase Aro8/Aro9, or decarboxylation by DC (amino acid decarboxylase). Following decarboxylation, the aromatic amines could be oxidized by tynA to form the corresponding aldehydes. Particularly, tryptamine is oxidized to indol-3-ylacetaldehyde, which is a direct precursor of both fungal QSM tryptophol and phytohormone auxin/IAA (Indole-3-Acetic Acid; Korasick et al., 2013). Besides as a precursor of tryptophol biosynthesis, tryptophane could also go through kynurenine pathway catalyzed by Bna2/7/4/5/1 and Aro9, to produce an intermediate product quinolinic acid (QA), as a precursor for NAD+ biosynthesis in the budding yeast (Ohashi et al., 2013).
In this study, we identified a conserved KPP2 gene in S. scitamineum, named SsKPP2. The sskpp2Δ mutants were generated in MAT-1 and MAT-2 sporidia background, respectively, and displayed defects in mating/filamentiation and sporidial growth, while no obvious difference in stress response, compared to the wild-type strain. Interestingly, we found that the fungal QSM tryptophol was able to fully restore mating/filamentation between sskpp2Δ and wild-type sporidia while not able to restore that between two sskpp2Δ sporidia. Using qRT-PCR we assessed transcriptional regulation on the conserved mating/filamentation gene loci a and b locus (Kronstad and Leong, 1990; Bolker et al., 1992; Yan et al., 2016; Lu et al., 2017), and genes encoding critical enzymes in tryptophane metabolism. Our results showed that the a locus genes were significantly down-regulated, and two amine oxidase encoding genes TYNA1 and TYNA2 up-regulated, in sskpp2Δ mutant compared to the wild-type strain, under mating condition. We further analyzed the deletion mutants of these two TYNA genes, sstyna1Δ and sstyna2Δ mutants, and found that mating/filamentation was reduced between two sstyna mutant sporidia. Overall, our study indicating an important function of MAPK signaling pathway in S. scitamineum mating/filamentation.
Results
Identification of a Conserved MAP Kinase Gene SsKPP2
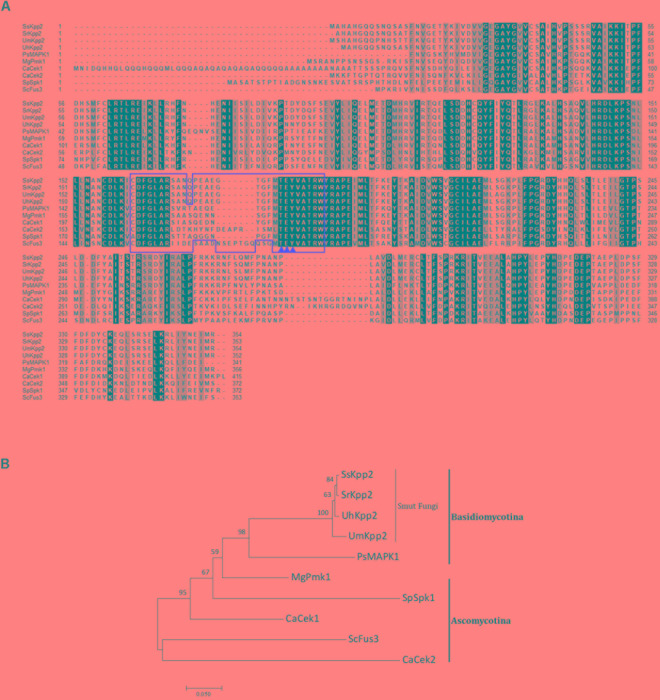
Using the S. cerevisiae Fus3/Kpp2 protein sequence (NP_009537) to search against the S. scitamineum genome available on NCBI website (taxid:49012), via tblastn homology search algorithm1, we retrieved a S. scitamineum MAP kinase protein Kpp2 (SPSC_04357) of 354 amino acids. Therefore, we named this protein as SsKpp2. Alignment of the SsKpp2 protein with other fungal othorlogs showed a high degree of amino acid conservation, including the conserved TXY motif (amino acid 182–184) in the A-loop (amino acid 162–189, Figure 1A boxed region), which is essential for activation of Kpp2 by phosphorylation (Dhanasekaran and Reddy, 1998). Phylogenetic analysis showed that SsKpp2 is highly conserved, especially within smut fungi (Figure 1B).
FIGURE 1.
Amino acid sequences arrangement and phylogenetic analysis with SsKpp2 protein and its orthologs. (A) Amino acid sequences arrangement and phylogenetic analysis with SsKpp2 protein and its fungal orthologs: S. reilianum SrKpp2 (CBQ73711), U. maydis UmKpp2 (AAF15528), U. hordei UhKpp2 (CCF52019), Puccinia striiformis PsMAPK1 (ADL57241), M. oryzae MgPmk1 (AAC49521), C. albicans CaCek1 (XP_715542) and CaCek2 (AAG43110), S. pombe SpSpk1 (NP_594009), and S. cerevisiae ScFus3 (NP_009537). The black and gray shadow denote identical and conserved residues, respectively. The red boxes and three red triangles represent STKc_MAPK domains and predicted dual phosphorylation lip sequences, respectively. (B) Phylogenetic analysis of Kpp2 othorlogs as listed in (A). The tree is calculated with Neighbor-Joining method (Saitou and Nei, 1987) using MEGA 7 (Kumar et al., 2016). Numbers beside each node indicate a percentage of 1000 bootstrap replications, computed using the Poisson correction method (Zuckerkandl and Pauling, 1965).
The sskpp2Δ mutants were generated in wild-type MAT-1 and MAT-2 background, respectively, using a modified PEG-mediated protoplast transformation methodology (Yu et al., 2015). The SsKPP2 open reading frame (ORF) was replaced by the recombinant hygromycin (HPT)-resistant selection marker, derived from two PCR amplified truncated but partially overlapped HPT fragment flanked by 5′- and 3′- untranslated region (UTR) of the SsKPP2 ORF (Supplementary Figure S1A). Transformants K17-8 and K17-10 were confirmed as sskpp2Δ mutants in MAT-1 mating-type background, and K18-5 and K18-10 in MAT-2 background, respectively, by Southern blot (Supplementary Figure S1B). We chose one sskpp2Δ mutant in each mating-type background to assess SsKPP2 gene expression by qPCR, with WT sporidia as control. Our result showed that SsKPP2 was undetectable in the sskpp2Δ mutants (K17-10 and K18-5; Supplementary Figure S1C), further confirming deletion of this gene. In the following, these two mutants were used in various assessment including tolerance to stressful conditions, mating/filamentation, and sporidial (yeast-like) growth.
Assessment of Stress Tolerance
First we examined the tolerance toward various stressful conditions in the wild-type and sskpp2Δ sporidia, including cell wall stress (0.1 mM SDS, or 0.5 mM Congo red), hyperosmotic stress (500 mM NaCl), and oxidative stress (1 mM H2O2). The sskpp2Δ sporidia (K17-10 and K18-5) were slightly more sensitive to oxidative stress, or to the cell wall stress caused by Congo red, compared to the WT, when both were cultured on YePSA medium (MAT-1 and MAT-2, Figure 2). Nitrogen starvation (on minimal medium minus nitrogen source, MM-N) made both WT and the mutant sporidia more sensitive to cell wall stressed raised by Congo red, but not by SDS (Figure 2). In summary, we infer that the SsKpp2-mediated MAPK pathway does not seem to be involved in S. scitamineum hyperosmotic, oxidative, or cell wall integrity (CWI) stress response.
FIGURE 2.
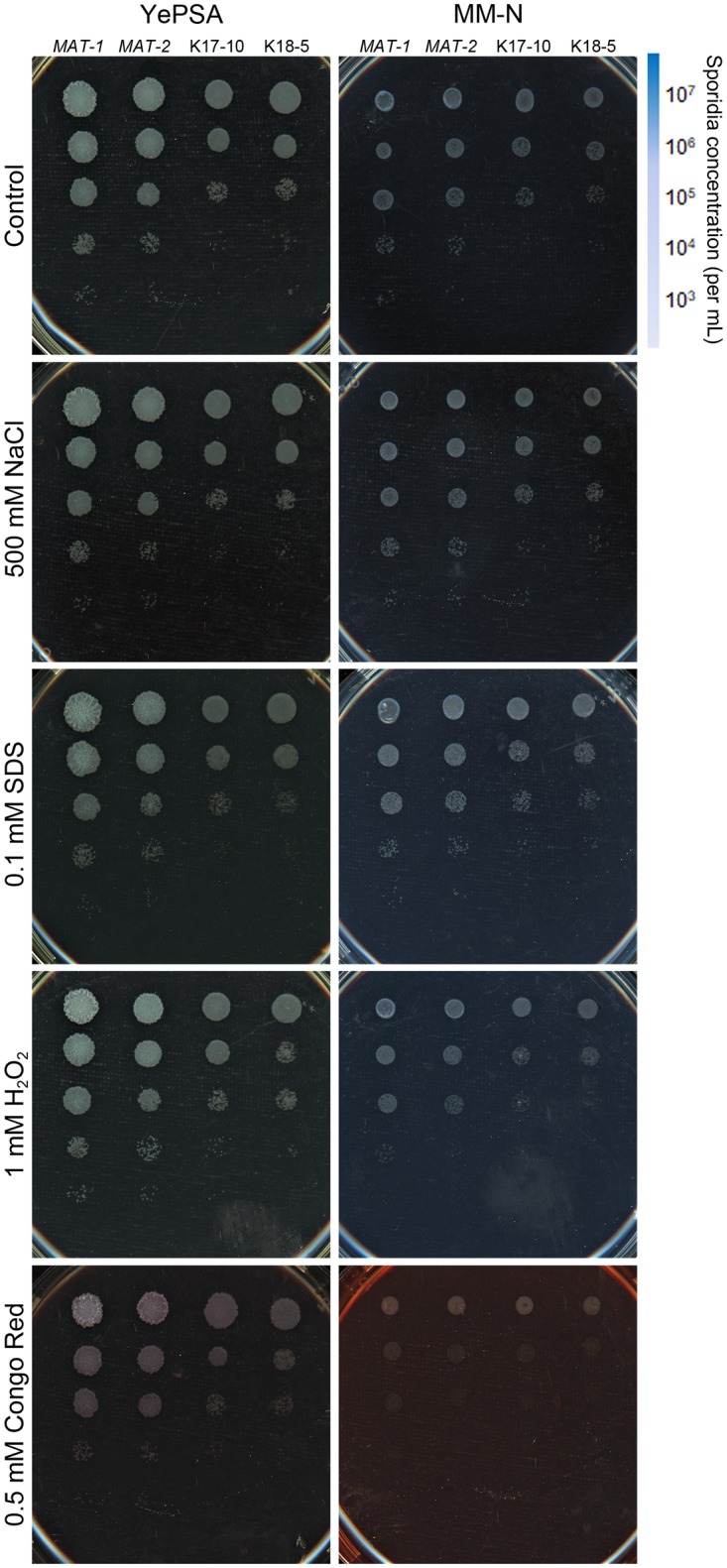
Assessment of tolerance toward stressful conditions. Serially diluted cells of WT or sskpp2Δ mutant (Δ) were spotted onto PDA medium supplemented with H2O2 (1 mM), NaCl (500 mM) or SDS (0.1 mM). Images were taken 72 h post inoculation.
SsKPP2 Is Required for S. scitamineum Mating/Filamentation
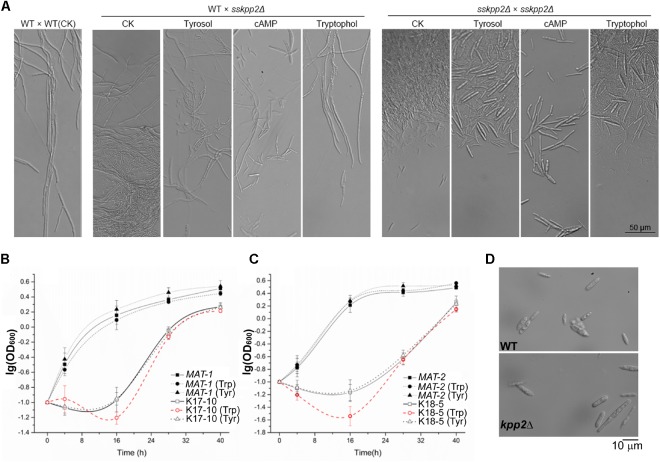
Next we assessed the mating/filamentation of sskpp2Δ mutants mixing with WT sporidia of opposite mating-type, or between two sskpp2Δ mutants. The in vitro culture of mixed WT MAT-1 and MAT-2 sporidia served as positive control, which gave rise to dikaryotic hyphae growth and thus had a fluffy appearance of the colonies (Figure 3A). In contrast, mixed cultured WT MAT-1 with K18-5 (sskpp2Δ in MAT-2 background), or WT MAT-2 with K17-10 (sskpp2Δ in MAT-1 background) both displayed obviously reduced filamentous and radial growth, while their sporidial colony was comparable to that of WT (Figure 3A). Mating/filamenation was completely blocked in the mating culture of sskpp2Δ mutants in two opposite mating-types (Figure 3A). We then tested the effect of cAMP, and two established fungal QSMs tyrosol (Chen et al., 2004) and tryptophol (Wuster and Babu, 2010), respectively, on sskpp2Δ filamenation. Addition of 10 mM cAMP could effectively promote filamentation in the mixed culture between WT and sskpp2Δ mutants and between two sskpp2Δ mutants (Figure 3A), suggesting an overlap between cAMP/PKA and MAPK signaling pathways in regulation of mating/filamentation, similar as reported in U. maydis (Kaffarnik et al., 2003). Tryptophol of concentration ranging from 20 to 200 μM could restore filamentous growth in the mixed culture between WT and sskpp2Δ mutants, but not in the mating culture between two sskpp2Δ mutants (Figure 3B). Tyrosol could not restore sskpp2Δ mating/filamentation (Figure 3C).
FIGURE 3.
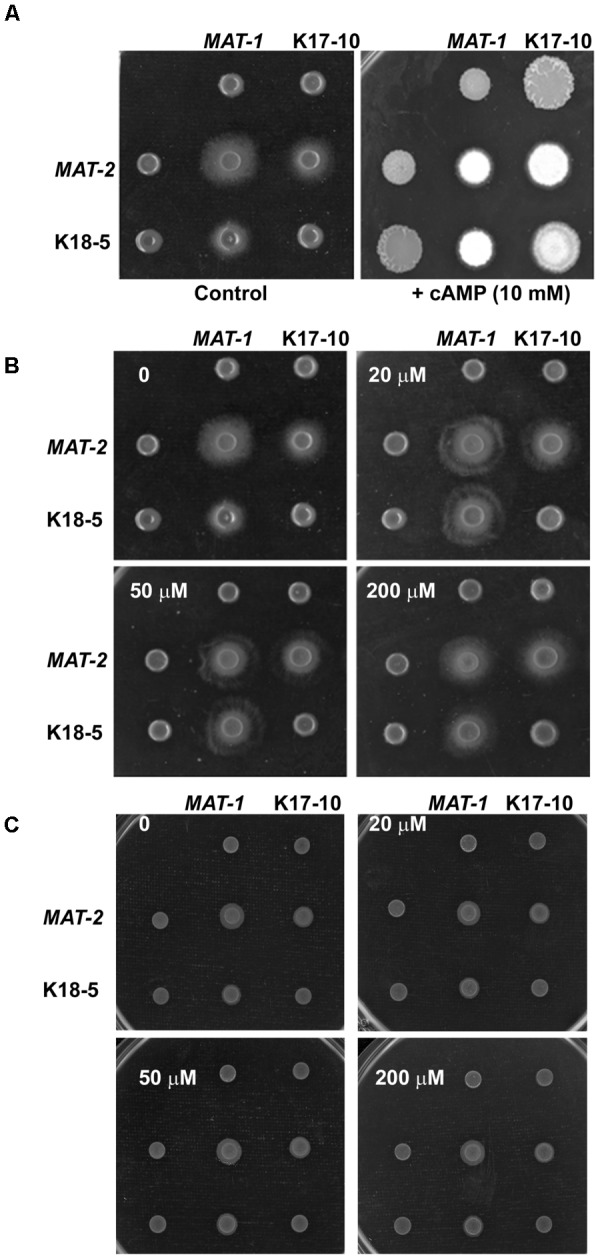
SsKpp2 is essential for S. scitamineum mating/filamentation. Mating/filamentation of WT and sskpp2Δ mutant was assessed on PDA solid medium, with or without addition of 10 μM cAMP (A), 20–200 μM tryptophol (B), or 20–200 μM tyrosol (C). CK denotes untreated control set. Photographs were taken 3 days post inoculation.
Microscopic observation was performed to closely examine the hyphal morphology in the WT and sskpp2Δ mutant. Abundant un-mating sporidia were seen in the WT/sskpp2Δ mating culture, without any treatment (CK; Figure 4A). Meanwhile, hyphae or pseudohyphae were also observed in this WT/sskpp2Δ combination (Figure 4A). In contrast, long and smooth hyphae formed in the WT mating cultured under same condition (Figure 4A). Treatment with tyrosol did not promote hyphae formation in WT/sskpp2Δ culture, while addition of or tryptophol did (Figure 4A). However, in sskpp2Δ/sskpp2Δ culture, even tryptophol could not restore filamentous growth (Figure 4A). Interestingly, addition of cAMP seemed to promote pseudohyphae formation (Figure 4A), which gave rise to the fluffy look of the colonies of WT/sskpp2Δ or sskpp2Δ/sskpp2Δ mating cultures as appeared in Figure 3A. Overall, we found that SsKPP2 is required for S. scitamineum mating/filamentation, which is likely related to tryptophol production.
FIGURE 4.
Effect of aromatic chemicals on S. scitamineum mating/filamentation or sporidial growth. (A) Microscopic imaging of hyphae formed after sexual mating, in WT or sskpp2Δ mutant, supplemented with cAMP (10 mM), tyrosol (20 μM) or tryptophol (20 μM), or without supplemented chemicals (CK) as a control. Images were taken 48 h post inoculation. Scale bar = 50 μm. Sporidial growth was assessed in the WT (solid line) or sskpp2Δ (dashed line) strain of MAT-1 (B) or MAT-2 (C) mating-type background. S. scitamineum sporidia was cultured in liquid YePS medium for 24 h before dilution to reach the concentration of 105 cells per ml. The diluted cells were cultured for a further 40 h in the absence (cube) or presence of tyrosol (20 μM; triangle) or tryptophol (20 μM; circle). The number of cells at each specific time point was acquired by measuring the OD absorption at 600 nm. The growth curve for each culture was prepared by plotting the logarithmic values of OD600 vs. incubation time. Mean ± S.E. are derived from three independent biological repeats, each of which contained three technical repeats. (D) Representative microscopic images of wild-type (WT) and the sskpp2Δ sporidia. n > 40 for each strain examined. Images were taken with Axio Observer Z1 microscope equipped with an sCMOS camera. Scale bar = 10 μm.
We further examined the sporidial growth in WT and the sskpp2Δ mutant, as well as relationship of tyrosol and tryptophol with S. scitamineum cell density. We noticed that the sskpp2Δ mutant was obviously delayed in sporidial growth compared to the WT, in both mating-types (Figures 4B,C). Tyrosol could slightly promote such yeast-like growth in WT but not in the sskpp2Δ mutants (Figures 4B,C). In contrast, tryptophol could slightly suppress sporidial growth in both WT and the sskpp2Δ mutants (Figures 4B,C). This result indicates that the promotion of filametation by tryptophol (Figure 3B) was not due to change in growth rate of haploid sporidia. In summary, we found that SsKpp2 is also involved in regulation on S. scitamineum cell growth (division).
Furthermore, we compared the spordial morphology of the wild-type (MAT-1 and MAT-2) and the sskpp2Δ mutants (K17-10 and K18-5) by microscopy. As shown in Figure 4D, the sskpp2Δ sporidia appeared cigar-shape and identical to those of the wild-type strains. We measured the sporidial size and found that the length of the sskpp2Δ sporidia is significantly (p < 0.001) longer than that of WT, while no difference in the width of WT and sskpp2Δ sporidia (Table 1). These results suggest that SsKpp2 is not required for sporidial morphogenesis.
Table 1.
Measurement of sporidial size of WT (MAT-1 and MAT-2) and sskpp2Δ mutants (K17-10 and K18-5).
| Strain | Size (length × width)∗ |
|---|---|
| MAT-1 | 10.50 ± 1.39A × 3.12 ± 0.46C μm |
| MAT-2 | 13.31 ± 2.22B × 3.35 ± 0.55C μm |
| K17-10 | 11.99 ± 2.29A × 3.31 ± 0.42C μm |
| K18-5 | 14.85 ± 1.87B × 3.31 ± 0.56C μm |
∗numbers represent Average ± S.E., derived from three independent repeats, with >40 sporidia of each strain measured for each repeat. Different capitalized letters represent significant difference statistically.
SsKPP2 Is Involved in Transcriptional Regulation of Mating/Filamentation Genes or Tryptophol Biosynthesis During S. scitamineum Mating/Filamentation
To get a better understanding of the regulatory mechanism on governing S. scitamineum mating/filamentation via SsKpp2-mediated MAPK signaling pathway, we performed a qRT-PCR analysis with genes related to fungal mating/filamentation and tryptophane metabolism (Supplementary Table S1). Our results showed that indeed the a locus genes MFA1 and MFA2 were significantly down-regulated, while the b locus gene bE1, bE2,bW1, and bW1 up-regulated (Figure 5A). The mating/filamentation master regulator, Prf1, was also down-regulated especially in sskpp2Δ/sskpp2Δ culture (Figure 5A). Among the genes regulating tryptophol production, only two TYNA genes (SPSP_0335 and SPSC_0449) were highly up-regulated (Figure 5A). The tynA enzyme catalyzes conversion from tryptamine to indol-3-ylacetaldehyde, the direct precursor for the production of tryptophol in Ehrlich pathway (Hazelwood et al., 2008) or the phytohormone IAA (Korasick et al., 2013). We inferred that up-regulation of TYNA genes in the sskpp2Δ mutant may correlate to its defects in mating/filamentation.
FIGURE 5.
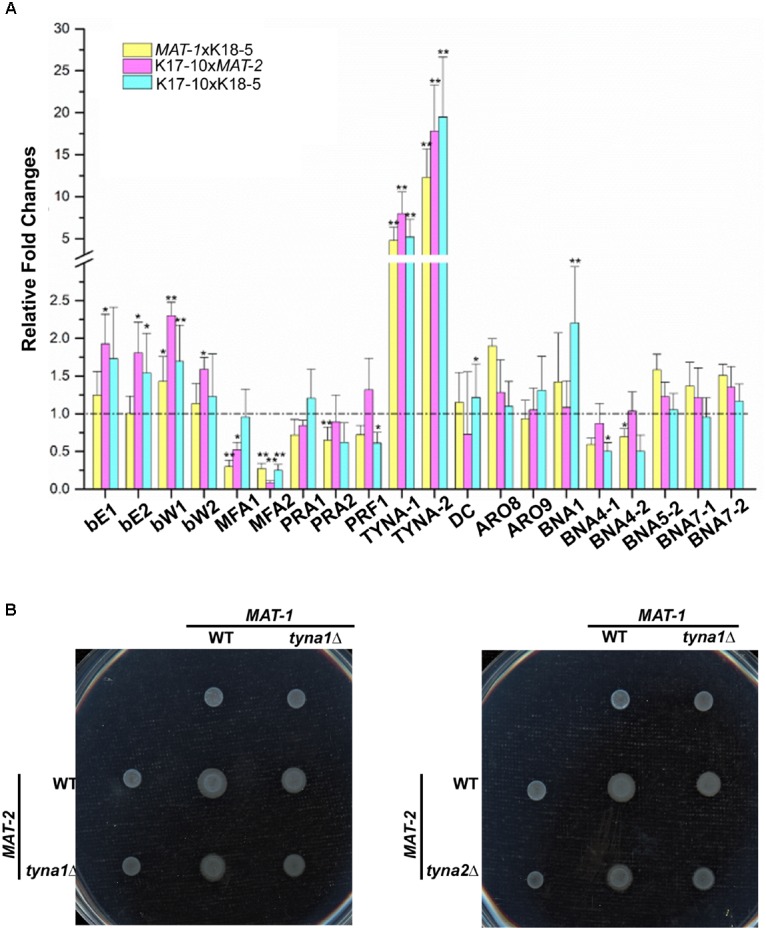
Identification of potential target genes of Sskpp2-mediated MAPK signaling pathway. (A) qRT-PCR analysis with selected genes in the sskpp2Δ mutant in comparison to WT, under mating condition. Relative gene expression fold change was calculated with –ΔΔCt method (Livak and Schmittgen, 2001) with ACTIN as internal control. Mean ± S.E. are derived from two independent biological repeats, each of which contained three replica. ∗ and ∗∗ denote significant difference with p < 0.05 and p < 0.01, respectively. (B) Mating/filamentation of WT, sstyna1Δ and sstyna2Δ mutants was assessed on PDA solid medium. Photographs were taken 3 days post inoculation.
To verify such hypothesis, we generated sstyna1Δ and sstyna2Δ mutant, respectively (Supplementary Figures S2A–C) and tested their mating/filamentation. As both of sstyna1Δ and sstyna2Δ mutant could mating with WT and form filaments only slightly reduced compared to WT mating (MAT-1/MAT-2), mating/filamentation in two mutant sporidia, sstyna1Δ/sstyna1Δ or sstyna1Δ/sstyna2Δ, seemed further reduced (Figure 5B). Overall, we found that SsKpp2 was required for S. scitamineum mating/filamentation, likely via regulation on pheromone signal transduction and tryptohol biosynthesis.
Discussion
Mitogen-activated protein kinase is protein kinase highly conserved in eukaryotic organisms as signal transducers (Kultz and Burg, 1998; Roman et al., 2007). In this study, we identified a S. scitamineum ortholog of KPP2 gene, that encodes a MAP kinase most closely related to other fungal MAP kinases including Pmk1 of M. oryzae, Cek1 and Cek2 of C. albicans, Spk1 of S. pombe, Fus3p of S. cerevisiae, and in several other basidiomycetous fungi especially smut fungi. The phylogenetic analysis showed that SsKpp2 protein is highly conserved among smut fungi, and along with ascomycetes it is conserved in a typical STKc_MAPKs domain (A-loop) with the dual phosphorylation lip sequence TXY is present in all the fungal Kpp2s. Functions in fungal development and pathogenesis was reported with aforementioned Kpp2 othorlogs. For example, M. oryzae Pmk1 for appressoria formation and plant cell-to-cell invation (Zhao et al., 2005; Sakulkoo et al., 2018). C. albicans Cek1 is involved in morphogenesis and hyphal formation (Csank et al., 1998) while Cek2 participates in sexual mating (Chen et al., 2002), and both of them required for dimorphic switch, virulence, and cell wall integrity and partially functionally redundant (Correia et al., 2016). U. maydis Kpp2 regulates sexual mating/filamentation and host infection (Muller et al., 1999). However, Kpp2 kinase has not been identified or functionally characterized in S. scitamineum.
In this current study, we generated and characterized the sskpp2Δ mutants in both mating-types in S. scitamineum and the result showed that deletion of SsKPP2 does not change hyperosmotic, oxidative, or CWI stress response in the sporidial growth stage. However, mating/filamentation was significantly reduced in WT/sskpp2Δ culture and completely blocked in sskpp2Δ/sskpp2Δ culture, indicating that SsKpp2 function is essential for S. scitamineum mating/filamentation. This is consistent with what has been reported in budding yeast, that the MAPK cascade Hog1 is mainly involved in hyperosmotic stress response, and Mpk1/Slt2 cascade in CWI, while Fus3 and Kss1 cascades responsible for pheromone response and/or filamentation (Chen and Thorner, 2007; Waltermann and Klipp, 2010). SsKpp2 function also seems conserved with its U. maydis ortholog (Muller et al., 2003). Transcriptional profiling confirmed that expression of PRF1 orthologous gene, encoding the master regulator of fungal mating/filamentation in response to pheromone signal, was reduced in mating cultures containing sskpp2Δ, so as to the a locus genes, which depend on Prf1 for transcriptional activation (Hartmann et al., 1996). However, at present it remains to be confirmed whether SsKpp2 directly phosphorylates Prf1 for its activation, before we could further investigate the mechanism underlying SsKpp2 regulation on S. scitamineum mating/filamentation. Overall we infer that the MAPK signaling pathway mediated by SsKpp2 may regulate S. scitamineum mating/filamentation through Prf1 activation of pheromone response genes, similar as that in U. maydis.
Two aromatic alcohols, tyrosol and tryptophol, have been reported as fungal QSMs in C. albicans in S. cerevisiae, respectively, in promoting filamentation integrating cell density and/or nitrogen availability (Chen et al., 2004; Wongsuk et al., 2016). Feedback regulation of tryptophol production is mediated by cAMP/PKA signaling pathway in S. cerevisiae while not in C. albicans (Chen and Fink, 2006). It was not reported on any signaling pathway regulating tyrosol production in C. albicans dimorphic switch, but a two-component system and MAPK (Cek1) signaling regulation on production of another fungal QSM farnesol, was reported (Kruppa et al., 2004; Roman et al., 2009). Although the MAPK signal cascade regulation on mating/filamentation in response to pheromone signals were elucidated in U. maydis (Muller et al., 2003), its connection to fungal quorum sensing (if any) has not been reported in this smut fungus, neither was any fungal QSM(s) identified in smut fungi.
Interestingly, in our study we found that addition of tryptophol but not tyrosol could restored mating/filamentation in WT/sskpp2Δ culture, and a transcriptional regulation of TYNA genes involved in tryptophol biosynthesis was indicated by qRT-PCR analysis as well. Mating between two TYNA genes deletion mutants resulted in reduced mating/filamentation, suggesting that tryptophol (or other intermediate product in tryptophane catabolism) may likely play a role in S. scitamineum mating/filamentation and under MAPK signal regulation. We notice that mating/filamentation in sporidia combination of sstyna1Δ/sstyna1Δ or sstyna1Δ/sstyna2Δ seemed further reduced compared to WT/sstyna1Δ or WT/sstyna2Δ combination, indicating that there may be a functional redundancy between these two copies of TYNA genes in S. scitamineum. Such integrated regulation of pheromone signal and potential quorum-sensing signal via MAP kinase function has not been reported in other fungi, although it is pending verification whether tryptophol actually acts as a fungal quorum-sensing molecule in S. scitamineum.
In summary, our study provides evidence that S. scitamineum MAP kinase SsKpp2 is required for proper mating/filamentation, likely via regulation on fungal pheromone response and tryptophol biosynthesis.
Materials and Methods
Growth Conditions and Fungal Strains Used in This Study
Teliospores of sugarcane smut collected from the fields in Guangdong province of China (21°12′ 36″ N; 101°10′ 12″ E) by Yan et al. (2016) was maintained in Z. Jiang’s lab, and the MAT-1 or MAT-2 haploid sporidia isolated from such teliospores were used in this study. The culture medium used in this study include YePSA medium (yeast extract 1%, peptone 2%, sugar 2%, agar 2%), YePS liquid medium (yeast extraction 1%, peptone 2%, sugar 2%, pH7.0), YePS soft medium (yeast extract 1%, peptone 2%, sugar 2%, agar 0.65%), YePSS medium (yeast extract 1%, peptone 2%, sugar 2%, D-sorbitol 18.17%, agar 2%), PDA (Beijing dingguo, HB0233-12) medium (2% agar, PH7.5), and MM-N medium (minimal medium minus nitrogen source, following the established recipe (Sherman, 2002) without addition of nitrogen source). For stress tolerance assessment, WT or sskpp2Δ mutant sporidia of serial diluted concentration from 107 to 103 per mL were inoculated on YePSA or MM-N medium, and allowed to grow in dark at 28°C incubator for 3 days before photographing. For mating/filamentation assay, the equal volume of wild-type or deletion mutant’s haploid sporidia of opposite mating-types were mixed and plated on the solid medium, and kept in dark at 28°C incubator for 2–3 days before photographing. For growth assay, sporidia of S. scitamineum wild-type or sskpp2Δ mutants were cultured in 5 mL of YePS liquid medium at 28°C, with shaking at 200 rpm for 24 h. An aliquot of such cultured sporidia were then diluted to fresh YePS liquid medium, adjusting to cell density of 105 cells per ml, and cultured for another 40 h under the same condition. Measurement of O.D.600 with spectrophotometer (Thermo, NanoDrop 2000C) was performed hourly to monitor the yeast-like (budding) growth of wild-type or mutant strains, with or without addition of chemical reagents as described in the Results.
Chemical Compounds Used in This Study
Tyrosol (Sigma-Aldrich, 188255); tryptophol (Sigema-Aldrich, V900672); cAMP (Sigma-Aldrich, A9501).
Generation of Deletion Constructs
The primers used for generating sskpp2Δ, tyna-1Δ, and tyna-2Δ mutants were listed in Supplementary Table S2. Targeted gene deletion follows the strategy described (Chung et al., 2002; Yang and Chung, 2012), by PCR amplification of two fragments flanking the targeted gene, each of which fused with half partial-overlapping HPT (Hygr gene) sequence. The flanking DNA (1kb 5′- and 3′-) fragments were amplified using wild-type S. scitamineum genomic DNA as template, and the HPT gene with plasmid pEX2 (Yan et al., 2016) as template.
Nucleic Acid Related Manipulation
Fungal genomic DNA was extracted using a HP Fungal DNA Kit (Omega,D3195-01). PCR amplification was performed using Phusion High-Fidelity DNA Polymerase (Thermo Scientific,lot:00528748). DNA fragment elution was performed using Gel Extraction Kit (Omega,D2500-02) and/or Cycle Pure Kit (Omega, D6492-02). In Southern blot assay, restriction enzymes used for digestion of genomic DNA were from NEB [NewEnglandBiolabs (Beijing) Ltd.]. Labeling and Detection Starter Kit I (Roche, 11745832910) was used for labeling of PCR amplified fragments as probe. Amersham Hybond TM-N+ (GE Healthcare, RFN303B) membrane was used for blotting. NBT/BCIP Stock Solution (Roche,11681451001) was used for probed band detection. For total RNA extraction, Qiagen RNeasy Plant Mini kit (74104) was used. Ambion® TURBO DNA-freeTM kit (Invitrogen, AM1907) was used for removing contaminating DNA from RNA preparations. TransScript® First-Strand cDNA Synthesis Super Mix (Transgen, AT301-02) was used for cDNA systhesis. For real-time qPCR we used PowerUpTM SYBR® Green Master Mix (Applied Biosystems, A25742) and the reaction was run on QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific). The primers used for qRT-PCR analysis were listed in Supplementary Table S3.
PEG (Polyethylene Glycol) – Mediated Protoplast Transformation
Polyethylene glycol-mediated protoplast transformation follows the established protocol (Yu et al., 2015) with modification: Wild-type MAT-1 or MAT-2 sporidia was incubated with the lysing enzyme (Sigma L1412) of 4 mg/mL, dissolved in SCS solution (20 mM trisodium citrate and 1 M D-sorbitol, pH 5.8), at 28°C for 30 min for enzyme digestion of fungal cell wall. 40% PEG (Sigema-Aldrich,202444) solution was prepared in 10 mL STC solution (10 mM Tris–HCl, pH 7.5; 1 M D-sorbitol and 100 mM CaCl2). 1∼5 μg of the PCR amplified fragments were mixed with 1 μl heparin solution (15 mg/ml; Beijing dingguo,DH157) and the protoplasts, and incubated with 40% PEG solution on ice for 10 min. The protoplasts were regenerated on the 3-layer regeneration medium composed of top layer of YePS soft medium plus two lays of YePSS medium, with only the bottom YePSS layer containing 400 μg/ml hygromycin B (Calbiochem,CAS:53-84-9) for primary screening of transformants based on antibiotic resistance.
Microscopy
Images were taken with Axio Observer Z1 microscope (Zeiss, Jena, Germany) equipped with an sCMOS camera (PCO Edge, Kelheim, Germany).
Statistic Analysis
Data were expressed as mean ± standard error (SE). Differences among different treatments were analyzed using Student’s t-test formula in Microsoft Excel.
Author Contributions
YD, BC, and ZJ conceived and designed the experiments. BZ, CC, YW, SS, XZ, and SL performed the experiments. YD, CC, BC, and ZJ analyzed the data and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. S. Dixit (South China Agricultural University) for comments on the manuscript and English editing and proofreading.
Funding. This work was supported by the National Basic Research Program of China (973 Program, Grant No. 2015CB150600) and Natural Science Foundation of Guangdong Province, China (Grant No. 2017A030310144). The funder had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02555/full#supplementary-material
References
- Albuquerque P., Nicola A. M., Nieves E., Paes H. C., Williamson P. R., Silva-Pereira I., et al. (2013). Quorum sensing-mediated, cell density-dependent regulation of growth and virulence in Cryptococcus neoformans. mBio 5:e00986-13. 10.1128/mBio.00986-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriuso J., Hogan D. A., Keshavarz T., Martínez M. J. (2018). Role of quorum sensing and chemical communication in fungal biotechnology and pathogenesis. FEMS Microbiol. Rev. 42 627–638. 10.1093/femsre/fuy022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolker M., Urban M., Kahmann R. (1992). The a mating type locus of U. maydis specifies cell signaling components. Cell 68 441–450. 10.1016/0092-8674(92)90182-C [DOI] [PubMed] [Google Scholar]
- Chen H., Fink G. R. (2004). Tyrosol is a quorum-sensing molecule in Candida albicans. Proc. Natl. Acad. Sci. U.S.A. 101 5048–5052. 10.1073/pnas.0401416101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Fink G. R. (2006). Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 20 1150–1161. 10.1101/gad.1411806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Fujita M., Feng Q., Clardy J., Fink G. R. (2004). Tyrosol is a quorum-sensing molecule in Candida albicans. Proc. Natl. Acad. Sci. U.S.A. 101 5048–5052. 10.1073/pnas.0401416101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Chen J., Lane S., Liu H. (2002). A conserved mitogen-activated protein kinase pathway is required for mating in Candida albicans. Mol. Microbiol. 46 1335–1344. 10.1046/j.1365-2958.2002.03249.x [DOI] [PubMed] [Google Scholar]
- Chen R. E., Thorner J. (2007). Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1773 1311–1340. 10.1016/j.bbamcr.2007.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Gibson T. B., Robinson F., Silvestro L., Pearson G., Xu B., et al. (2001). MAP kinases. Chem. Rev. 101 2449–2476. 10.1021/cr000241p [DOI] [PubMed] [Google Scholar]
- Chung K. R., Shilts T., Li W., Timmer L. W. (2002). Engineering a genetic transformation system for Colletotrichum acutatum, the causal fungus of lime anthracnose and postbloom fruit drop of citrus. FEMS Microbiol. Lett. 213 33–39. 10.1111/j.1574-6968.2002.tb11282.x [DOI] [PubMed] [Google Scholar]
- Correia I., Roman E., Prieto D., Eisman B., Pla J. (2016). Complementary roles of the Cek1 and Cek2 MAP kinases in Candida albicans cell-wall biogenesis. Future Microbiol. 11 51–67. 10.2217/fmb.15.127 [DOI] [PubMed] [Google Scholar]
- Csank C., Schroppel K., Leberer E., Harcus D., Mohamed O., Meloche S., et al. (1998). Roles of the Candida albicans mitogen-activated protein kinase homolog. Cek1p, in hyphal development and systemic candidiasis. Infect. Immun. 66 2713–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson R. C., Nichols C. B., Cox G. M., Perfect J. R., Heitman J. (2003). A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 49 469–485. 10.1046/j.1365-2958.2003.03563.x [DOI] [PubMed] [Google Scholar]
- Dhanasekaran N., Reddy E. (1998). Signaling by dual specificity kinases. Oncogene 17 1447–1455. 10.1038/sj.onc.1202251 [DOI] [PubMed] [Google Scholar]
- Dickinson J. R., Salgado L. E., Hewlins M. J. (2003). The catabolism of amino acids to long chain and complex alcohols in Saccharomyces cerevisiae. J. Biol. Chem. 278 8028–8034. 10.1074/jbc.M211914200 [DOI] [PubMed] [Google Scholar]
- Fuqua W. C., Winans S. C., Greenberg E. P. (1994). Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176 269–275. 10.1128/jb.176.2.269-275.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold S., Duncan G., Barrett K., Kronstad J. (1994). cAMP regulates morphogenesis in the fungal pathogen Ustilago maydis. Genes Dev. 8 2805–2816. 10.1101/gad.8.23.2805 [DOI] [PubMed] [Google Scholar]
- Gray K. M. (1997). Intercellular communication and group behavior in bacteria. Trends Microbiol. 5 184–188. 10.1016/S0966-842X(97)01002-0 [DOI] [PubMed] [Google Scholar]
- Hamel L. P., Nicole M. C., Duplessis S., Ellis B. E. (2012). Mitogen-activated protein kinase signaling in plant-interacting fungi: distinct messages from conserved messengers. Plant Cell 24 1327–1351. 10.1105/tpc.112.096156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann H. A., Kahmann R., Bolker M. (1996). The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J. 15 1632–1641. 10.1002/j.1460-2075.1996.tb00508.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelwood L. A., Daran J. M., van Maris A. J., Pronk J. T., Dickinson J. R. (2008). The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 74 2259–2266. 10.1128/aem.02625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornby J. M., Jensen E. C., Lisec A. D., Tasto J. J., Jahnke B., Shoemaker R., et al. (2001). Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 67 2982–2992. 10.1128/AEM.67.7.2982-2992.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Zhang X., Liu H., Xu J. R. (2018). Mitogen-activated protein kinase signaling in plant pathogenic fungi. PLoS Pathog. 14:e1006875. 10.1371/journal.ppat.1006875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffarnik F., Muller P., Leibundgut M., Kahmann R., Feldbrugge M. (2003). PKA and MAPK phosphorylation of Prf1 allows promoter discrimination in Ustilago maydis. EMBO J. 22 5817–5826. 10.1093/emboj/cdg554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick D. A., Enders T. A., Strader L. C. (2013). Auxin biosynthesis and storage forms. J. Exp. Bot. 64 2541–2555. 10.1093/jxb/ert080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad J. W., Leong S. A. (1990). The b mating-type locus of Ustilago maydis contains variable and constant regions. Genes Dev. 4 1384–1395. 10.1101/gad.4.8.1384 [DOI] [PubMed] [Google Scholar]
- Kruppa M., Krom B. P., Chauhan N., Bambach A. V., Cihlar R. L., Calderone R. A. (2004). The two-component signal transduction protein Chk1p regulates quorum sensing in Candida albicans. Eukaryot. Cell 3 1062–1065. 10.1128/ec.3.4.1062-1065.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kultz D., Burg M. (1998). Evolution of osmotic stress signaling via MAP kinase cascades. J. Exp. Biol. 201(Pt 22), 3015–3021. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu S., Shen X., Chen B. (2017). Development of an efficient vector system for gene knock-out and near in-cis gene complementation in the sugarcane smut fungus. Sci. Rep. 7:3113. 10.1038/s41598-017-03233-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Zhang S. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51 245–266. 10.1146/annurev-phyto-082712-102314 [DOI] [PubMed] [Google Scholar]
- Muller P., Aichinger C., Feldbrugge M., Kahmann R. (1999). The MAP kinase kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol. Microbiol. 34 1007–1017. 10.1046/j.1365-2958.1999.01661.x [DOI] [PubMed] [Google Scholar]
- Muller P., Weinzierl G., Brachmann A., Feldbrugge M., Kahmann R. (2003). Mating and pathogenic development of the Smut fungus Ustilago maydis are regulated by one mitogen-activated protein kinase cascade. Eukaryot. Cell 2 1187–1199. 10.1128/EC.2.6.1187-1199.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi K., Kawai S., Murata K. (2013). Secretion of quinolinic acid, an intermediate in the kynurenine pathway, for utilization in NAD + biosynthesis in the yeast Saccharomyces cerevisiae. Eukaryot. Cell 12 648–653. 10.1128/EC.00339-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson G., Robinson F., Beers Gibson T., Xu B. E., Karandikar M., Berman K., et al. (2001). Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 22 153–183. 10.1210/edrv.22.2.0428 [DOI] [PubMed] [Google Scholar]
- Plotnikov A., Zehorai E., Procaccia S., Seger R. (2011). The MAPK cascades: signaling components, nuclear roles and mechanisms of nuclear translocation. Biochim. Biophys. Acta 1813 1619–1633. 10.1016/j.bbamcr.2010.12.012 [DOI] [PubMed] [Google Scholar]
- Raina S., Odell M., Keshavarz T. (2010). Quorum sensing as a method for improving sclerotiorin production in Penicillium sclerotiorum. J. Biotechnol. 148 91–98. 10.1016/j.jbiotec.2010.04.009 [DOI] [PubMed] [Google Scholar]
- Roman E., Alonso-Monge R., Gong Q., Li D., Calderone R., Pla J. (2009). The Cek1 MAPK is a short-lived protein regulated by quorum sensing in the fungal pathogen Candida albicans. FEMS Yeast Res. 9 942–955. 10.1111/j.1567-1364.2009.00545.x [DOI] [PubMed] [Google Scholar]
- Roman E., Arana D. M., Nombela C., Alonso-Monge R., Pla J. (2007). MAP kinase pathways as regulators of fungal virulence. Trends Microbiol. 15 181–190. 10.1016/j.tim.2007.02.001 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- Sakulkoo W., Oses-Ruiz M., Oliveira Garcia E., Soanes D. M., Littlejohn G. R., Hacker C., et al. (2018). A single fungal MAP kinase controls plant cell-to-cell invasion by the rice blast fungus. Science 359 1399–1403. 10.1126/science.aaq0892 [DOI] [PubMed] [Google Scholar]
- Sherman F. (2002). Getting started with yeast. Methods Enzymol. 350 3–41. 10.1016/S0076-6879(02)50954-X [DOI] [PubMed] [Google Scholar]
- Sundar A. R., Barnabas E. L., Malathi P., Viswanathan R. (2012). A Mini-Review on Smut Disease of Sugarcane Caused by Sporisorium scitamineum. London: InTech Open. [Google Scholar]
- Tian X., He G. J., Hu P., Chen L., Tao C., Cui Y. L., et al. (2018). Cryptococcus neoformans sexual reproduction is controlled by a quorum sensing peptide. Nat. Microbiol. 3 698–707. 10.1038/s41564-018-0160-4 [DOI] [PubMed] [Google Scholar]
- Waltermann C., Klipp E. (2010). Signal integration in budding yeast. Biochem. Soc. Trans. 38 1257–1264. 10.1042/BST0381257 [DOI] [PubMed] [Google Scholar]
- Williams H. E., Steele J. C., Clements M. O., Keshavarz T. (2012). γ-Heptalactone is an endogenously produced quorum-sensing molecule regulating growth and secondary metabolite production by Aspergillus nidulans. Appl. Microbiol. Biotechnol. 96 773–781. 10.1007/s00253-012-4065-5 [DOI] [PubMed] [Google Scholar]
- Wongsuk T., Pumeesat P., Luplertlop N. (2016). Fungal quorum sensing molecules: role in fungal morphogenesis and pathogenicity. J. Basic Microbiol. 56 440–447. 10.1002/jobm.201500759 [DOI] [PubMed] [Google Scholar]
- Wuster A., Babu M. M. (2010). Transcriptional control of the quorum sensing response in yeast. Mol. Biosyst. 6 134–141. 10.1039/b913579k [DOI] [PubMed] [Google Scholar]
- Yan M., Zhu G., Lin S., Xian X., Chang C., Xi P., et al. (2016). The mating-type locus b of the sugarcane smut Sporisorium scitamineum is essential for mating, filamentous growth and pathogenicity. Fungal Genet. Biol. 86 1–8. 10.1016/j.fgb.2015.11.005 [DOI] [PubMed] [Google Scholar]
- Yang S. L., Chung K. R. (2012). The NADPH oxidase-mediated production of hydrogen peroxide (H2O2) and resistance to oxidative stress in the necrotrophic pathogen Alternaria alternata of citrus. Mol. Plant Pathol. 13 900–914. 10.1111/j.1364-3703.2012.00799.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Zhang Y., Cui H., Hu P., Yu X., Ye Z. (2015). An efficient genetic manipulation protocol for Ustilago esculenta. FEMS Microbiol. Lett. 362:fnv087. 10.1093/femsle/fnv087 [DOI] [PubMed] [Google Scholar]
- Zhao X., Kim Y., Park G., Xu J. R. (2005). A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell 17 1317–1329. 10.1105/tpc.104.029116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E., Pauling L. (1965). Evolutionary divergence and convergence in proteins. Evol. Genes Proteins 97 97–166. 10.1016/B978-1-4832-2734-4.50017-6 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.