Abstract
Gene expression in mitochondria of Plasmodium falciparum is essential for parasite survival. The molecular mechanisms of Plasmodium organellar gene expression remain poorly understood. This includes the enigmatic assembly of the mitochondrial ribosome from highly fragmented rRNAs. Here, we present the identification of clustered organellar short RNA fragments (cosRNAs) that are possible footprints of RNA-binding proteins (RBPs) in Plasmodium organelles. In plants, RBPs of the pentatricopeptide repeat (PPR) class produce footprints as a consequence of their function in processing organellar RNAs. Intriguingly, many of the Plasmodium cosRNAs overlap with 5′-ends of rRNA fragments. We hypothesize that these are footprints of RBPs involved in assembling the rRNA fragments into a functioning ribosome. A bioinformatics search of the Plasmodium nuclear genome identified a hitherto unrecognized organellar helical-hairpin-repeat protein family that we term heptatricopeptide repeat (HPR) proteins. We demonstrate that selected HPR proteins are targeted to mitochondria in P. berghei and that one of them, PbHPR1, associates with RNA, but not DNA in vitro. A phylogenetic search identified HPR proteins in a wide variety of eukaryotes. We hypothesize that HPR proteins are required for processing and stabilizing RNAs in Apicomplexa and other taxa.
INTRODUCTION
The protozoan parasite Plasmodium spp. is the causative agent of malaria in humans. A 2016 report by the World Health Organization showed 214 million infections with malaria globally, resulting in almost half a million deaths (1). While some antimalarial treatments are available, parasite resistance is a continuing challenge. Plasmodium spp. belong to the family of apicomplexan parasites, most of which contain a non-photosynthetic plastid called the apicoplast (2). As a remnant of a red algae chloroplast, the apicoplast contains its own DNA, as does the Plasmodium mitochondrion. The Plasmodium nuclear genome has a size of 24 MB and contains >5000 genes; by contrast, the apicoplast and mitochondrial genomes are small—35 and 6 kb, respectively.
In keeping with the descendance of the apicoplast from cyanobacteria, the apicoplast genome is organized similarly to bacterial chromosomes. Genes are organized into operons and transcribed as polycistronic precursors, which are heavily processed post-transcriptionally (3,4). Blocking apicoplast gene expression with inhibitors of transcription or translation leads to an immediate or a ‘delayed’ death phenotype, depending on the drug used (5).
The mitochondrial genome organization in Apicomplexa is unique. With a very small genome size of 6 kb and only three protein-coding genes, it is one of the smallest genomes discovered to date (6). In addition to the protein-coding genes, the genome contains highly fragmented rRNA genes instead of full-length rRNA genes. There is strong evidence for translational activity in Plasmodium mitochondria (7,8), suggesting that these rRNA fragments are assembled into functional ribosomes. This inference is based on the essential nature of the electron transport chain in mitochondria (7) and the finding that mutations underlying resistance to atovaquone, a specific inhibitor of the respiratory chain, map to a mitochondrial protein-coding gene (9). Furthermore, tRNAs are imported into mitochondria in Toxoplasma gondii and P. falciparum, providing indirect evidence for mitochondrial translation (10,11).
Transcription of the mitochondrial genome has been shown to lead to polycistronic transcripts, with mRNAs and rRNA fragments transcribed together as one precursor molecule (12,13). Even transcripts representing the entire genome have been detected (12). This expression organization suggests that post-transcriptional regulatory events, such as RNA processing, RNA stabilization, and RNA degradation, play an important role in mitochondria, similar to post-transcriptional gene regulation in the apicoplast (3). Indeed, monocistronic mRNAs have been found for CYTB and COXI (14), which are likely derived from polycistronic precursors via RNA processing. Notably, mitochondrial mRNAs directly abut their neighboring mRNAs or rRNAs, a relationship that has been taken as evidence that polycistronic precursors are precisely cleaved to yield individual mature transcripts (12,13). These processes are expected to require a large number of RNA-binding proteins (RBPs), but most of the components and mechanisms of the RNA-processing machinery remain unknown even in human mitochondria (15,16). All organellar RBPs must originate from the nuclear genome and be transferred to the organelles post-translationally. Few nuclear factors for Plasmodium RNA processing have been characterized to date, and how RNA metabolism is regulated remains largely unknown.
Because Plasmodium, with its apicoplast, has also autotrophic organisms among its ancestry, a comparison with organellar gene expression in plants and algae can shed light on RNA-processing mechanisms in the organelles of this parasite. RNA metabolism in plant organelles is characterized by a massive expansion of nuclear-encoded RBPs. The largest class of organellar RBPs is helical-hairpin-repeat proteins called pentatricopeptide repeat proteins (PPRs) (17). These proteins contain a tandem-repeat motif with up to 35 repetitions. Structurally, each repeat forms two alpha-helical elements that fold back onto each other. Alpha helices from consecutive repeats are stacked to form an extended RNA-interacting surface. Within this surface, each repeat is responsible for binding one base in a single-stranded RNA molecule. Two dedicated amino acids are key for specific RNA base recognition (18). Of the ∼450 predicted PPR proteins in Arabidopsis, approximately two-thirds are localized to the mitochondria, with the remainder are found in the plastid (19). PPR proteins play an important role in RNA processing and transcript stabilization. Binding of a number of PPR proteins to mRNAs acts as a roadblock against exonucleolytic decay. Eventually, only small sequences that are protected by the PPR proteins remain; these can be detected by sequencing small RNAs, providing a method for identifying PPR protein binding sites (20). A protein family with a set of functions and structure similar to PPR proteins, called the octatricopeptide repeat (OPR) family, is more prevalent in single-celled photosynthetic algae, e.g. Chlamydomonas reinhardtii (21). In Plasmodium, there are two annotated PPR proteins (22); one (PF3D7_1406400; PBANKA_1035800) is predicted to localize to the apicoplast and the other (PF3D7_1233300; PBANKA_1448000) to the mitochondrion. To our knowledge, no OPR proteins have been described in Apicomplexa to date.
As a first step toward understanding RNA degradation in P. falciparum, we searched for potential binding sites of helical-hairpin-repeat proteins in Plasmodium organelle RNA metabolism by creating small-RNA sequencing libraries. This effort was complemented by a search for helical-hairpin-repeat proteins. We discovered a novel helical-hairpin-repeat protein family distinct from PPR and OPR proteins, which we termed heptatricopeptide repeat (HPR) proteins, and demonstrated that individual HPRs are targeted to Plasmodium mitochondria. Although these proteins seem particularly abundant in apicomplexan parasites, they were also found in other species within the Alveolata. In fact, while they were not dected in bacteria, HPR proteins are found in most eukaryotic groups analyzed, including humans.
MATERIALS AND METHODS
Ethical statement
This study was carried out in strict accordance with the German ‘Tierschutzgesetz in der Fassung vom 22. Juli 2009’ and the Directive 2010/63/EU of the European Parliament and Council ‘On the protection of animals used for scientific purposes’. The protocol was approved by the ethics committee of the Berlin state authority (Landesamt für Gesundheit und Soziales Berlin, permit numbers G0469/09 and G0294/15). Female NMRI mice were used for all experiments and purchased from Charles River Laboratories (Sulzfeld, Germany).
Cultivation and harvest of Plasmodium falciparum asexual stages
Plasmodium falciparum NF54 was cultivated as described by Trager and Jensen (23). Cultures were split every 2–3 days. Cultures were not synchronized.
Total RNA extraction from Plasmodium falciparum asexual stages
Asexual stages of P. falciparum NF54 in red blood cells were harvested as described above. Cell pellets were resuspended in AIM-Buffer (24) containing 0.1% saponin. After incubation for 5 minutes on ice, cells were centrifuged at 2500 × g for 10 min. Pelleted parasites were washed twice with cold AIM buffer. For RNA extraction, the pellet was resuspended in TRIzol reagent and passed 15–20 times through a syringe (0.55 mm × 25 mm). RNA was extracted with a standard TRIzol-based method according to manufacturer's protocols (Invitrogen).
Preparation of RNA from organelle-enriched fractions by nitrogen cavitation
Organelle-enriched fractions were produced as described (24,25) with minor modifications. The resuspension buffer MSE was supplemented in addition to PMSF and BSA with 1 mg/ml Heparin. After collecting the organelles by centrifugation at 23 000 × g, the pellet was resuspended in MSE buffer. For RNA extraction from organelle fractions, four volumes of TRIzol were added and RNA extraction performed as described above. For detailed information see Supplementary Figure S1.
Sequencing of small RNA fractions
For preparation of sequencing libraries, NEBNext® Multiplex Small RNA Library Prep Set for Illumina® Kit was used according to manufacturer. As input, 1 μg of RNA was used. Size selection was carried out via Gel extraction of RNAs smaller than 100 nt. RNA sequencing on a MySeq platform (Illumina) was outsourced (SMB, Berlin).
Analysis of sequencing libraries
Removal of adapters and quality trimming was performed with cutadapt (26) and the trimmed sequences were mapped to the P. falciparum organellar genomes and the nuclear genome (NCBI apicoplast: DQ642846.1 mitochondria: DQ642845.1 and nuclear genome: ASM276v1) using Bowtie (version 1.1.1) (27) with the following parameters: –a –best –strata –v 2. Using these settings, Bowtie reports all alignments with the highest score and a maximum of two mismatches. Reads mapping to multiple sites of either genomes were excluded so only unique reads were considered in the following analysis. The created read mappings were processed using SAMtools (28), and coverage graphs were extracted using BEDTools (29). Coverage graphs were visualized using the Integrated Genome Browser (30). Additional sequence information for the mitochondrial genome were obtained from PlasmoDB (22) based on previous work (14). To identify sRNA accumulations, only read accumulations with more than 50 reads on the 5′-end were considered.
sRNA gel blot
To verify selected sRNA accumulations, we performed sRNA northern blotting and radiolabeled detection with DNA oligo probes as described before (31). For information on the DNA oligos used see Supplementary Table S1.
Genome wide protein motif search
For the genome wide search of the Helical hairpin repeat proteins in P. falciparum the MEME software suite was used (32). As an input we used OPR proteins described in Chlamydomonas reinhardtii (21). The OPR proteins were downloaded from JGI Genome Portal (33). The proteins were searched for motifs of 37 or 38 amino acid using MEME tools, allowing for any number of repetitions. The discovered motifs were collapsed to a consensus and used as input to search the full P. falciparum proteome with the MAST tool (proteins were downloaded from PlasmoDB). Proteins from the MAST search containing at least two repetitions of the motif and a total e-value <0.0001 were added to the protein list and used together with the first proteins as input in a second MEME search as described above. The discovered motif was used in a second MAST search on the P. falciparum proteome. Additional proteins were added, and refinements of the motifs were repeated until no new proteins were discovered in MAST. To improve the motif further and with the intention to generate a motif representing all apicomplexan HPR proteins, we performed similar searches for Plasmodium berghei and Toxoplasma gondii. The potential proteins from all three organisms were then combined and a final, apicomplexan-specific motif was generated. This Motif was used to search the full genome of additional species.
N-tracts within HPR proteins were searched manually. The average length of N-stretches in the P. falciparum genome is 37 (34). We only considered N-tracts that had at least one stretch of four consecutive Ns and were in total at least ten amino acids long. We allowed single amino acid gaps of non-Ns and double amino acids gaps if they are flanked by two stretches of 2 Ns (e.g. Supplementary Figure S2, PF3D7_1009900). If several stretches of 4 Ns and 3 Ns were separated by up to five other amino acids, they were considered as one coherent N-tract (e.g. PF3D7_092420).
Protein localization
To investigate the localization of selected Plasmodium berghei HPR proteins, we employed an endogenous tagging approach using single homologous recombination (35). To create fluorescently tagged proteins, we amplified the sequence directly upstream of the stop codon (ranging in size from 1326 to 2460 bp) and cloned the amplicon into the pBAT-SIL6 vector system in frame with the mCherry 3xMyc tag sequence (36). Prior to ligation, the vector and the PCR amplicons were digested with EagI and HpaI restriction endonucleases. For primer information see Supplementary Table S1. For mitochondrial visualization, the HSP70 promoter driving high level GFP expression, was excised from PBANKA_051680 and PBANKA_081630 tagging vectors using PvuII in combination with NdeI or PshAI, and exchanged with the promoter and amino-terminus of the mitochondrial HSP70-3 (ranging in size from 1771 to 2008 bp), yielding the mitochondrial marker cassette mito-GFP. All created vectors were verified via Sanger sequencing prior to transfection. Correct insertion into the nuclear genome was investigated via PCR (for scheme, see Supplementary Figure S3). Live cell microscopy was performed on a Zeiss AxioObserver Z1 epifluorescence microscope equipped with a Zeiss AxioCam MRM camera (Zeiss,Oberkochen, Germany). Images were processed using FIJI (37).
Expression of HPR proteins
For the recombinant expression of the PbHPR1:MBP (PBANKA_051680:Maltose binding protein) fusion protein, we used the NEB pMAL™ Protein Fusion and Purification System. The gene was PCR-amplified from P. berhgei genomic DNA, which removed the sequence coding for the predicted N-terminal mitochondrial targeting peptide (N-terminal 126 bp). The prediction was based on an alignment of orthologous proteins from different Plasmodium species. The amplicon was 1524 bp long containing 1368 bp from the CDS and 156 bp of the 3′ UTR. For primer information see Supplementary Table S1. The amplicon was digested with restriction endonucleases BamHI and SalI and ligated into the similarly digested pMAL vector. Sequence integrity was verified by Sanger sequencing. As a control, fusion protein CDC42:MBP (NCBI accession: NP_523414.1) was used in the same vector system. The finished expression vectors were transformed into Escherichia coli BL21-CodonPlus(DE3)-RIPL Expression strain (Agilent). Expression was carried out in LB-Medium supplemented with 2 g/l glucose, 100 μg/ml Carbenicillin and 25 μg/ml Chloramphenicol. Cells were grown at 37°C until the OD600 reached 0.5 absorption. Expression was induced by IPTG (0.3 mM) and cells were grown at 28°C for 16 h. Cells were harvested by centrifugation at 4000 × g for 20 min at 4°C. The cell lysis and purification of the fusion protein was carried out according to instructions of the NEB pMAL™ Protein Fusion and Purification System. The expression was checked by western blotting and immune detection using anti-MBP monoclonal antibody (NEB) as described previously (38). The fusion protein was supplemented with one volume of 50% glycerol and stored at −20°C.
In vitro transcription for RNA pull-down assay
To produce RNA representing the entire P. berghei mitochondrial genome, four fragments of different sizes with small overlaps on either side (fragment positions: 5918–1937; 1874–3577; 6756–4873; 4809–51) were prepared by PCR. To enable strand specific transcription, we added T7-promotors to one of the fragment ends. The fragments were purified and then used as templates for in vitro transcription via T7 RNA polymerase (Thermo Fisher). Transcription was carried out according to the manufacturer's instructions. After incubation at 37°C overnight, the template DNA was digested by adding 4 U of TURBO™ DNase for 30 min at 37°C. The RNA was subsequently extracted with DirectZol-RNA-MiniPrep Kit (Zymo research). Resulting RNA sizes were: 1977 nt, 1704 nt, 1451 nt, 1200 nt. For primer information see Supplementary Table S1.
RNA pull-down
The expressed PbHPR1 and CDC42 fusion proteins were used in an RNA pull-down experiment. All steps were carried out at RT and under rotation. For equilibration, 580 pmol of the each protein solution was incubated in 200 μl Column-Buffer (CB: 20 mM Tris–HCl, 200 mM NaCl, 1 mM EDTA, 1 mM DTT) for 20 min. Strand specific RNA fragments were mixed (6 μg of each) with EDTA (2 mM final concentration), denatured at 95°C for 3 min, and subsequently chilled on ice for 5 min. After taking a sample from the denatured RNA for electrophoresis the RNA products were mixed with the recombinant protein and incubated for 20 min. In parallel, 200 μl of amylose slurry was prepared by washing it five times in CB. After removal of the last wash solution, the pull-down reaction was added. After rotation for additional 20 min, the supernatant was removed from the amylose and the resin was washed twice with 300 μl of CB. For elution, 200 μl of CB supplemented with 10 mM maltose were added to the resin and incubated for 10 min. From each fraction, 10% were used for western blot analysis. RNA was extracted from the supernatant fraction and the pellet via DirectZol-RNA-MiniPrep Kit (Zymo Research). The isolated RNA was separated on a 1% formaldehyde–agarose gel. Northern blotting was carried out as described (38). Radioactive detection with DNA oligoprobes was done as described (31).
Supplemental Movie S1: We predicted the structure of the HPR tract of PbHPR1 (PBANKA_051680; see Supplementary Figure S5 for the exact amino acid sequence of the tract) from P. berghei using i-Tasser (39) and animated it using PyMol (https://pymol.org/2/).
RESULTS
Sequencing of a small RNA fraction from P. falciparum improves mitochondrial rRNA fragment annotation
To investigate the distribution of small RNA fragments in the organelles of P. falciparum, we created a sequencing library from total cellular RNA as well as from RNA purified from enriched P. falciparum organelles (Supplementary Figure S1). The libraries were size-selected for RNA fragments smaller than 100 nucleotides (nt) before sequencing. Massive parallel sequencing created a dataset containing 12 726 675 reads (total RNA) and 10 594 281 reads (enriched RNA), respectively. After removal of linker sequences, the sequencing reads were mapped to the apicoplast and mitochondrial genome (NCBI: apicoplast, DQ642846.1; mitochondria, DQ642845.1) The mitochondrial genome was annotated using coding DNA sequence (CDS) and sequence information provided in the P. falciparum Genome Database (PlasmoDB; ).
Only 0.03% of sequencing reads could be mapped to the apicoplast genome, whereas 8.43% mapped to the mitochondrial genome after sequencing total RNA preparations (0.22% and 19.34% after sequencing enriched RNA). Most apicoplast reads were found to represent known tRNA and rRNA genes. We did not detect a distinct accumulation of small RNAs in either the untranslated regions (UTRs) or CDS of described apicoplast mRNAs, as would be expected for a footprint of a helical-hairpin-repeat protein. However, given the low number of reads obtained, the existence of footprints of RBPs in the apicoplast cannot presently be excluded. Whether the low read number is due to a disadvantage of apicoplast short RNAs during library preparation or is reflecting biological differences, cannot be ascertained at present.
As expected, most of the sequencing reads that mapped to the mitochondrial genome represent already known rRNA fragments (14; Figure 1A). The mapping results were highly similar for the two small RNA sequencing experiments despite the different methods for preparing the RNA (Figure 1A). These mappings confirmed the expression of most rRNAs that had previously been detected only by RNA gel blot hybridization or as rare expressed sequence tags (ESTs) (14; Supplementary Table S2).
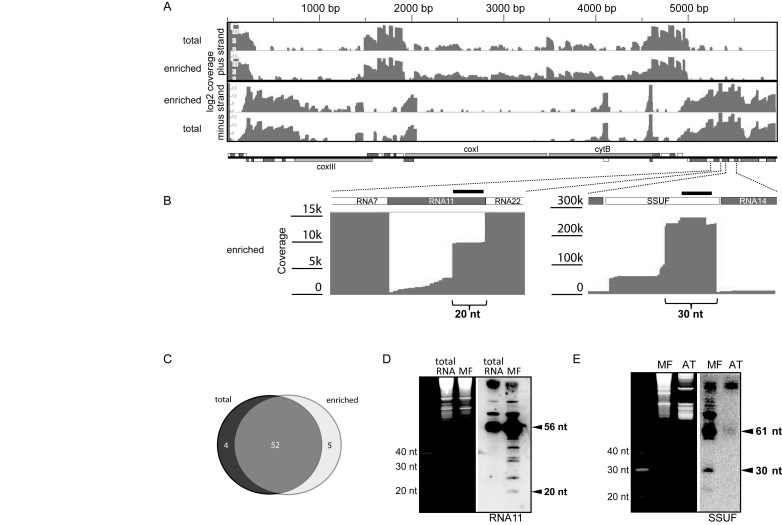
Figure 1.
Mapping of sequenced small RNAs on the P. falciparum mitochondrial genome. (A) Overview of the mapping results for the complete mitochondrial genome of P. falciparum. The two top tracks show the read coverage of the plus strand after small RNA sequencing from total and organelle-enriched parasite RNA preparations, respectively. The bottom tracks shows the mapping results for the minus strand. A schematic representation of the gene content is shown below the coverage graphs with rRNA fragment genes indicated by white and dark grey boxes. (B) Blow-up of two adjacent regions of the mitochondrial genome with coverage graph excerpts. Parenthesis highlight small plateaus of coverage that indicate the presence of short RNAs. Dark grey lines within the coverage graph indicate mismatches at the 3′end of sequencing reads. DNA oligo probes used for the RNA gel blot in D and E are indicated as black bars. (C) Venn diagram summarizing the overlap between cosRNAs found in small sequencing analyses from total RNA (left circle) and RNA from enriched organelles (right circle). The number of cosRANs found by both experiments is indicated by the overlap between the two circles. The numbers of cosRNAs for each set in the Venn-Diagram are shown as well. (D) RNA of an organelle-enriched fraction (MF) and total RNA from P. falciparum were separated on a 15% Urea PAGE gel, stained with Ethidium bromide (left), and blotted onto a membrane. Position of the radio-labeled oligonucleotide to detect the 5′ region of RNA11 is indicated in (B). (E) Same as (D), but hybridization was carried out with an oligo probe against the SSUF rRNA fragment and as a negative control, total leaf RNA from Arabidopsis thaliana (AT) was used.
We also confirmed the expression of five rRNA fragments that had so far only been predicted based on similarities to canonical, unfragmented rRNAs and rare EST clones, but had not been confirmed experimentally (14; Supplementary Table S2). Our mapping also allowed a more precise definition of 5′-ends of rRNA fragments. In seven cases, ends detected by sRNA read mapping lay a few bases up- or downstream of previously annotated ends (Supplementary Table S2). In sum, NGS sequencing of small RNAs helped to specifiy rRNA fragments and thus improved the annotation of the mitochondrial genome of P. falciparum.
Clustered organellar sRNAs are located preferentially at the 5′-end of rRNA fragments in mitochondria
Next, using sRNA miner, a recently developed software for identifying potential footprints of PPR proteins in sRNA sequencing datasets (40), we screened our mapping results for read clusters that could potentially represent footprints of RBPs. Parameters in sRNA miner were set to detect clusters of sRNAs with sharp 5′-ends and less defined 3′-ends, the pattern observed for sequences that mapped to rRNA fragments (see above). This analysis detected a number of read clusters, which are listed in Table 1. Some cosRNAs were identical to rRNA fragments, particularly shorter rRNA fragments, which are expected to be present as full-length transcripts in the size-selected sequencing library and are not further considered here.
Table 1.
cosRNAs mapped to 5′-ends of rRNA fragments
| cosRNA name | rRNA name | Genome position (first and last nt) | Length | Strand | No. of possible 3′ adenine res. added | Read count on 5′-end | comments |
|---|---|---|---|---|---|---|---|
| cosRNA1 | LSUG | 389–355 | 35 | - | - | 6812 | a |
| cosRNA2 | LSUG | 389–333 | 57 | - | - | 6812 | a |
| cosRNA3c | LSUF | 1515–1554 | 40 | + | - | 370 | a |
| cosRNA4 | RNA2 | 1698–1725 | 28 | + | - | 13123 | a |
| cosRNA5c | SSUA | 2030–1972 | 59 | - | - | 1098 | a |
| cosRNA6 | RNA6 | 4849–4868 | 21 | + | 6 | 13 016 | b |
| cosRNA7 | RNA12 | 4893–4944 | 52 | + | - | 2262 | a |
| cosRNA8 | LSUA | 5201–5157 | 45 | - | - | 726 | a |
| cosRNA9 | RNA7 | 5283–5243 | 41 | - | - | 17 260 | a |
| cosRNA10 | RNA7 | 5283–5226 | 58 | - | - | 17 260 | a |
| cosRNA11 | RNA11 | 5340–5320 | 20 | - | 1 | 9989 | a |
| cosRNA12 | SSUF | 5507–5478 | 30 | - | - | 158 228 | a |
| cosRNA13 | LSUE | 5771–5746 | 26 | - | - | 5216 | a |
aMaps to 5′-end of rRNA.
bMaps to 3′-end of rRNA.
cNot found in the NGS datasets for enriched organellar RNA.
Interestingly, most cosRNAs mapped to the 5′-region of rRNA fragments (Figure 1B; Table 1), and their length varied between 18 and 40 nt. In several cases, more than one cosRNA was found to terminate at the same rRNA 5′-end, whereas their 3′-ends differed (Figure 1B). This suggests a step-wise degradation of rRNA fragments. In addition, we found several cosRNAs that mapped to a region immediately upstream of rRNA 5′-ends. Such cosRNAs could have been generated by endonucleolytic cleavage, which yields a mature rRNA fragment and a liberated 5′ leader sequence. When comparing the cosRNA search for the two independent short RNA seq experiments, we found 52 out of 61 cosRNA in both datasets, indicating that cosRNA detection is reproducible. To confirm the presence of cosRNAs, we next performed sRNA gel blot hybridization (Figure 1D,E). Although we could not detect these small RNAs in total RNA preparations, we were able to detect RNAs of the expected size using the RNA preparations from organelle-enriched fractions from P. falciparum cells (Supplementary Figure S1). In the case of the rRNA fragment SSUF, a cosRNA of 30 nt in length was detected by RNA gel blot hybridization, although at much lower levels than the mature SSUF rRNA (61nt). For the rRNA fragment RNA11, we found not only a band of the expected size based on our mapping (20 nt cosRNA), but also several other signals that corresponded to sRNAs not detected as distinct RNAs in the sequencing approach (Figure 1D). The bands between 20 nt and 56 nt could for example be caused by pausings of exonucleases or endonuclease activity. In sum, our analysis demonstrates that mitochondrial cosRNAs preferentially map to the 5′-end of rRNA fragments, whereas 3′-ends are not represented by dedicated cosRNAs.
Three cosRNAs were also found outside of rRNA regions. The first was found at the junction between the COXI and CYTB reading frames and contained a 5′-end that matched the known 5′-end of CYTB mRNA (cosRNA20; Table 2). A second cosRNA matched the 3′-end of COXIII mRNA (cosRNA15; Table 2). The third cosRNA was found in the non-coding region between the rRNA fragment SSUA and the COXIII open reading frame (cosRNA17). It is possible that this cosRNA is the remnant of an SSUA-COXIII co-transcript. Consistent with the low sequencing coverage at this position, we could not detect the cosRNA20 and cosRNA17, respectively, in RNA gel blot hybridizations. Together with the data on cosRNAs derived from rRNA fragments, our analyses demonstrate that sRNAs accumulate in mitochondria. Importantly, they do not exhibit a random distribution, as would be expected for a stochastic RNA degradation process, but were found specifically at selected 5′-ends of rRNAs. In a few instances, cosRNAs were linked to mRNAs as well. Sequences of identified cosRNAs can be found in Supplementary Table S3.
Table 2.
cosRNAs mapped to mRNAs and intergenic regions
| cosRNA name | Genome position (first and last nt) | Length | Strand | possibly 3′ adenine res. added | Read count on 5′-end | comments |
|---|---|---|---|---|---|---|
| cosRNA14 | 166–205 | 40 | + | 5 | 550 | Linked to rRNA |
| cosRNA15 | 787–725 | 63 | - | - | 68 | 3′-end of COXIII mRNA |
| cosRNA16 | 1492–1513 | 22 | + | - | 145 | Linked to rRNA |
| cosRNA17 | 1630–1601 | 30 | - | - | 113 | Intergenic spacer COXIII <> SSUA |
| cosRNA18 | 1638–1725 | 88 | + | - | n.d. | Linked to rRNA |
| cosRNA19 | 2067–2031 | 37 | - | - | 230 | Linked to rRNA |
| cosRNA20 | 3480–3537 | 58 | + | - | 153 | 5′-end of CYTB mRNA |
| cosRNA21a | 4866–4944 | 79 | + | - | ∼120 | Linked to rRNA |
| cosRNA22b | 203–238 | 36 | + | - | 1204 | 3′of RNA23t |
| cosRNA23b | 893–855 | 39 | - | - | 60 | Within the CDS of COXIII 3′ |
| cosRNA24b | 1486–1412 | 75 | - | - | 70 | 5′of COXIII mRNA |
| cosRNA25b | 4697–4719 | 26 | + | 3 | 671 | rRNA4 3′ < > 5′rRNA5 |
aNot found in the NGS datasets for enriched organellar RNA.
bOnly found in the NGS dataset for enriched organellar RNA.
n.d. due to an overlap with reads from the preceding RNA, we could not identify the numbers of reads at the 5′-end of cosRNA18.
Identification of the heptatricopeptide-repeat protein family in Plasmodium
We hypothesized that cosRNAs are caused by the protective action of RBPs, similar to that of plant PPR proteins. In contrast to many other RNA binding proteins like the KH or RRM proteins, PPR proteins, and other related helical-hairpin-repeat proteins usually bind with an extended protein-RNA interaction surface to their target RNAs. This makes them prime candidates for causing cosRNAs in the detected size range and thus the starting point for our search. We therefore initiated a genome-wide bioinformatics screen for proteins with sequence and structural similarity to well-characterized helical-hairpin-repeat proteins. We were unable to identify additional PPR proteins beyond the two candidates already annotated (22). Therefore, we used MEME suite software to generate a protein motif from described OPR-proteins found in C. reinhardtii (21). The initial search motif was almost identical to previously described OPR consensus motifs (Figure 2B). Because helical-hairpin repeats show some length polymorphism, and OPR proteins are longer than PPR proteins (38 versus 35 amino acids), we used the MEME algorithm to generate a search motif that was 37 or 38 amino acids in length. We then screened the complete Plasmodium proteome (from PlasmoDB) for these motifs using the MAST algorithm in the MEME software suite. Potential proteins containing at least two repeats of the input protein motif in close proximity were selected and used to generate an improved protein motif adapted for Plasmodium. This procedure was repeated multiple times, with each iteration adding newly identified proteins to the next motif generation, until no additional protein hits were discovered.
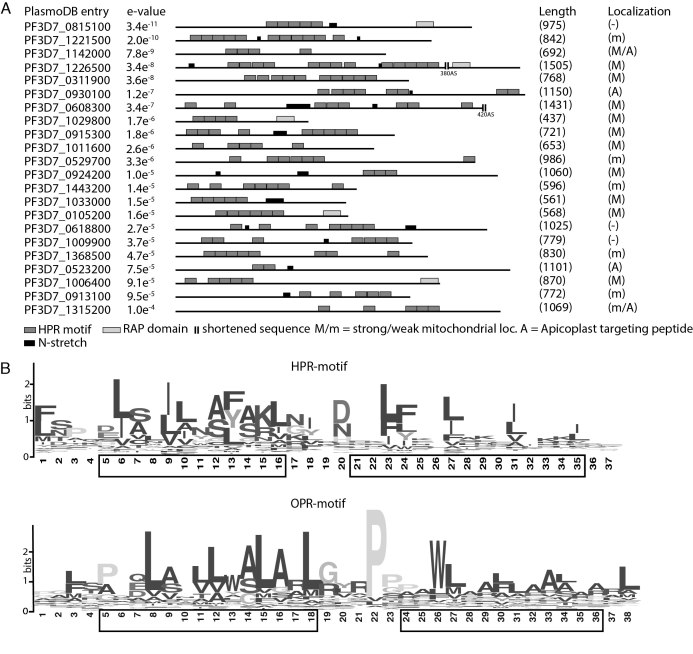
Figure 2.
Identification of apicomplexan HPR proteins. (A) Final results from a MEME-based motif search on the P. falciparum total proteome reveals 22 proteins containing multiple HPR repeats most of which are arranged in tandem repeats. 43 OPR proteins from Chlamydomonas reinhardtii were used for initial motif generation. Each block represents one HPR motif (dark grey), RAP motif (light gray), and N-tracts (black), respectively. Localization was predicted with different algorithms. Probability for mitochondrial localization: strong = >0.5; weak = >0.3 <0.5. (B) The consensus HPR motif (upper panel) generated from 67 proteins from P. falciparum, P. berghei and T. gondii and the OPR motif (lower panel) generated from C. reinhardtii. Predicted helical elements of the motifs are indicated by black boxes.
In our initial searches using an OPR motif with a length of 38 aa, only three hits with few repeats were found; even after four refinement cycles, no additional proteins were discovered. However, after shortening the motif by only one amino acid, we were able to detect multiple repeat-containing proteins, usually with at least some repeats in a tandem organization. By contrast, performing the search with a motif shortened by two amino acids returned no further hits. After four search iterations using the 37-aa motif, we discovered 22 potential candidates containing up to 12 repeats of our search model (Figure 2A). Interestingly four of these candidates contained a C-terminal RAP (RNA-binding domain abundant in apicomplexans) domain. We also searched for non-structured, N-rich tracts that were previously described to be particularly abundant within P. falciparum proteins (34). As expected, several such N-tracts are found in HPR proteins, but none disrupts a HPR motif (Figure 2A, Supplementary Figure S2).
We then refined the model and adapted it to a broader phylogenetic range by including proteins containing the motif identified in P. berghei and T. gondii. This generated a consensus consisting of a total of 299 repeats (Figure 2B). Secondary-structure predictions of this consensus sequence showed two alpha-helical elements, consistent with the structure of PPR and OPR proteins (Figure 2B, lower panel). Because of the 37-aa length of the motif identified, and by analogy to PPR and OPR proteins, we named this motif heptatricopeptide repeat (HPR).
The discovered protein motif showed striking differences compared with the previously described OPR motif (21). Whereas the OPR motif contains 38–40 amino acids, the newly discovered motif is only 37 amino acids long (Figure 2B) and the highly conserved PPPEW domain in OPR proteins was not present in HPR proteins.
A structural model of PbHPR1 and PbHPR2 was generated by I-TASSER (39). The models predicts that the helical repeats continue outside of the predicted HPR motifs. In fact, almost the entire proteins consist of α-helical elements. In case of PbHPR2, this elongated tract of helices shows a slight superhelical twist, whereas PbHPR1 forms a regular half-circle (Supplementary Figure S4, supplementary Movie S1). This structure is reminiscent of the RNA-binding surface of the PPR tract, although the stacking of helical elements along the protein main axis is less regular than in PPR proteins (41–43). There are currently no structural data available for OPR proteins, which at present precludes a structural comparison with this more closely related protein family. We animated the structure of the HPR tract of PbHPR1. For reasons of clarity, we focused on the HPR tract itself, while omitting the remainder of the protein (Supplementary Movie S1 and Supplementary Figure S5). This further highlights the stacking of α-helices and confirms the extended surface formed by HPR proteins. We also calculated surface charge distribution for both proteins. Here, PbHPR1 shows patchy charge distribution of both positive and negative charges. By contrast, PbHPR2 has strong charge differences between the concave and convex surface. While the inner, concave surface is strongly positively charged, the outer, convex surface is almost neutral. Whether the positive charges support RNA interactions of PbHPR2 remains to be tested.
We next used the HPR profile to search plant or algal genomes and thus test, whether the motif picks up PPR and OPR proteins, which are abundant in these taxa. We identified no PPR proteins and very few OPR proteins. We never found other helical hairpin repeat proteins like TPR, HAT, HEAT, Puf or TALE repeat proteins in such searches. Because of these length and composition differences, and since we were unable to reciprocally identify substantial numbers of hits using either the PPR/OPR or HPR motif, we conclude that proteins with the 37-aa motif define a novel class of helical-hairpin-repeat proteins related to OPR and PPR proteins.
HPR proteins are targeted to mitochondria
The cellular localization of all candidate HPR proteins was predicted using TargetP 1.1 (44), MitoProt II (45), PATS (Prediction of Apicoplast-Targeted Sequences) (46), and the PlasmoDB-PlasmoAP algorithm (47) (Figure 2A). For most candidate proteins, the predicted localization was mitochondrial, whereas apicoplast targeting was unlikely - only four candidates show a possible apicoplast targeting peptide (PF3D7_0930100, PF3D7_0523200, PF3D7_1142000, PF3D7_1315200). The latter two show also possible dual targeting with predictions also consistent with a mitochondrial localization. Since our RNA sequencing efforts show no cosRNAs in apicoplasts we focused on proteins predicted to localize to the mitochondria; however future studies with potentially apicoplast targeted HPRs would be interesting.
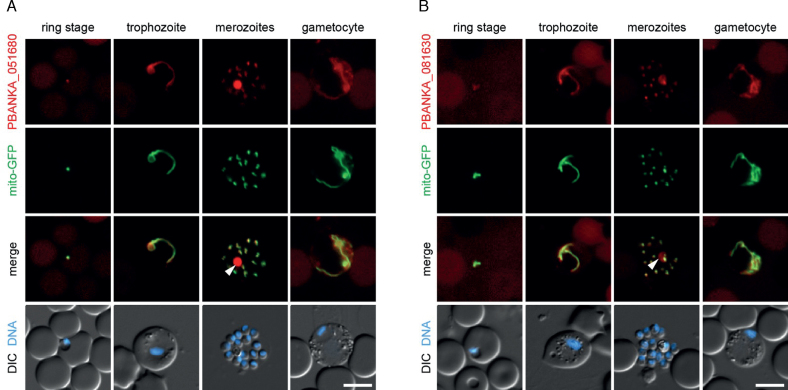
To verify the localization of selected proteins, we created C-terminal mCherry fusion proteins of two selected HPR proteins predicted to be targeted to mitochondria in the murine parasite Plasmodium berghei. The transfection vector also contained a GFP (green fluorescent protein) expression cassette that targets GFP to the mitochondria, thus allowing easy identification of transfected cells using live fluorescence microscopy. We could show that the mCherry signal of both target proteins, PBANKA_051680 (PbHPR1), the ortholog of PF3D7_1033000, and PBANKA_081630, the ortholog of PF3D7_0915300, co-localized with the mitochondrial GFP marker to a distinct organelle (Figure 3). Three further HPR proteins, PBANKA_020810 (ortholog of PF3D7_0105200), PBANKA_051380 (PbHPR2; ortholog of PF3D7_1029800), and PBANKA_143670 (ortholog of PF3D7_1221500), were mCherry-tagged and analyzed together with a cytoplasma-targeted GFP. The mCherry signal is present in worm- or dot-like structures in all three recombinant parasites, which also suggests mitochondrial localization (48; Supplementary Figure S6), although a confirmation with a parallel mitochondrial marker will be eventually required to validate this point. Furthermore, for all five proteins, the mCherry signals are distinct from the blue signals representing nuclei. In sum, this supports the predicted localization, and tentatively suggests that HPR proteins are a family of organellar, primarily mitochondrial, proteins.
Figure 3.
Localization of Plasmodium berghei HPR proteins PBANKA_051680 (PbHPR1) and PBANKA_081630. Life cell microscopy of erythrocytes infected with P.berghei containing HPR:mCherry-3xMyc fusion proteins. Shown are representative microscopy images with Hoechst 33 342 as a marker for the nucleus (DNA) and mitochondrial localized GFP. DIC: differential interference contrast microscopy to visualize cell outlines. Depicted are images of three asexual parasite stages and a female gametocyte. As visible in the overlay of tagged protein (red) and mitochondria (green) both proteins show mitochondrial localization (merge). The white Arrow denotes auto-fluorescence of the hemozoin-containing digestive vacuole, which was also observed with wild type parasites due to prolonged exposure times. Scale bars = 5μm.
The HPR family is widespread in eukaryotes
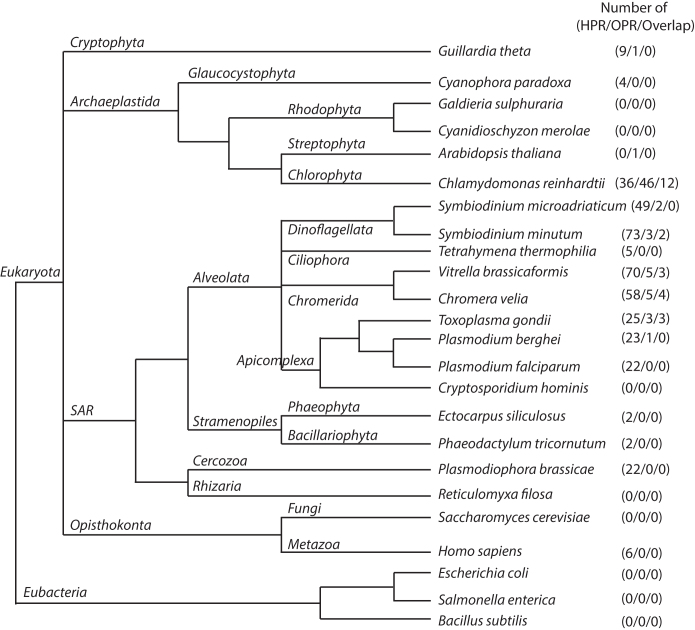
PPR proteins are found throughout the eukaryotic kingdom, but the family has shown particular expansion in land plants. We asked whether HPR proteins show a similar broad distribution, and queried genomes of other apicomplexans using the P. falciparum HPR motif. This uncovered similar numbers of HPR proteins in P. berghei and T. gondii, which were used to generate the HPR consensus representing the three apicomplexan species we investigated (Figure 2B). By contrast, we found no HPR proteins in Cryptosporidium parvum or Cryptosporidium hominis. This remarkable observation is consistent with the fact that Cryptosporidium species have lost their mitochondrial DNA and their unusual mitochondrial compartment is entirely dependent on nuclear-encoded proteins (49). We consider this further evidence that this novel protein family is relevant for the function of mitochondrial gene expression.
Next, we expanded our search to the nearest relatives within the Alveolata using available genomic information. We found similar numbers of HPR proteins, namely 58 and 70, in Chromera velia and Vitrella brassicaformis, respectively (Supplementary Table S4). These phototrophic species within the chromerids form a sister clade of the apicomplexans (50). We found 73 HPR proteins in Symbiodinium minutum, a dinoflagellate (Supplementary Table S4). By contrast, the more distantly related Tetrahymena thermophila, a ciliate, contained fewer than 10 proteins with multiple repeats of the motif (Supplementary Table S4). Notably, ciliates have non-fragmented rRNAs, whereas the apicomplexa, chromerids and dinoflagellates share a fragmented-rRNA organization.
Outside of the Alveolata, we sampled various eukaryotic and prokaryotic genomes. No HPR proteins were found in the three eubacterial genomes sampled. By contrast, we found HPR proteins in all analyzed sister groups of the alveolates with the exception of the single genome sampled from Rhizaria (Figure 4). Cercozoa, Stramenopila, Opisthokonta, and Archaeplastida all include species with HPR proteins. This tentatively suggests that the HPR protein family is an ancient acquisition of the eukaryotic lineage, but a future more extensive sampling of basal eukaryotes will be needed to decide this. Noteworthy, HPR proteins are not found in Rhodophyta and land plants. Given that the green algae Chlamydomonas and the glaucocystophyte Cyanophora paradoxa do have HPR proteins, the loss of HPR proteins in the other Archaeplastida might be secondary.
Figure 4.
Schematic distribution of OPR and HPR proteins in different species. The HPR and OPR motif was used for genome-wide search in different species of Alveolata and related eukaryotic lineages, as well as bacterial outgroups. While the alveolates show little to no OPR proteins, the HPR proteins seems to be the dominant helical-hairpin repeat protein family in this clade. In the single celled algae Chlamydomonas reinhardtii we could detect OPR and HPR proteins simultaneously. Overlap = proteins detected by both, the OPR and HPR motif search. It is important to note that this schematic tree only shows a rough representation of evolutionary branching, but does not depict the evolutionary distance of the species investigated.
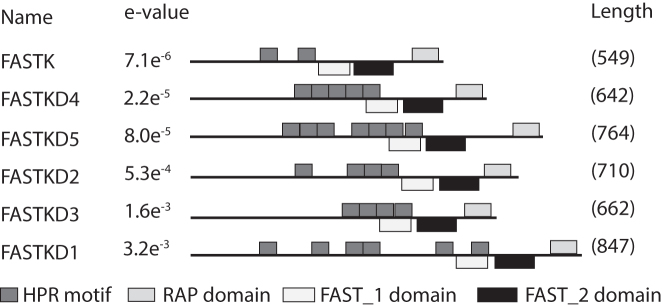
In humans, we found six proteins containing multiple repeats of the motif in humans (Figure 5, Supplementary Figure S7). These proteins, FASTK, FASTKD1, FASTKD2, FASTKD3, FASTKD4/TBRG4 and FASTKD5 have been shown to localize to mitochondria and have important functions in mitochondrial RNA metabolism (51–55). All of them associate with RNA in vivo (56,57). Interestingly, all FASTK proteins contain a C-terminal RAP domain in addition to the HPR tract. A recent study suggested that, whereas the RAP domain is necessary for the catalytic function of FASTKD proteins, RNA binding is facilitated by the region upstream of the RAP domain (53). Noteworthy, structural modelling indicated that the FASTK proteins have a domain dominated by α-helix motifs that resembles the global architecture of the PPR tract (Supplementary Figures S7 and 56). In this area, we found multiple repeats of the HPR motif, suggesting that these motifs could be responsible for specific RNA binding (see Figure 5). In sum, the wide distribution of HPR proteins suggests that they were already present in the ancestor of animals and eukaryotes, similar to PPR proteins, and thus represent an ancient eukaryotic gene family.
Figure 5.
Human proteins showing HPR motif repeats. The human HPR proteins all belong to the FASTKD protein family, namely FASTK (BC000377.2), FASTKD1 (NP_001268405), FASTKD2 (NP_001129666.1), FASTKD3 (NP_076996.2) and FASTKD4/TBRG4 (NP_001248763.1), and FASTKD5 (NP_068598.1). While HPR proteins in apicomplexan show no overlap with known protein domains the FAST_1 domain overlaps with some HPR motifs. However FAST_2 and RAP domain show no overlap. Note that although the HPR search motif is able to detect these repeats e-values are lower than for apicomplexan proteins suggesting stronger degeneration of the motif.
The HPR protein HPR1 binds mitochondrial RNA sequences
We expressed the HPR protein PbHPR 1 (PBANKA_051680) translationally fused to the Maltose binding protein (MBP). The cell cycle protein CDC42 fused to MBP served as negative control. After affinity purification, both proteins were analyzed by coomassie-stained PAGE and immunological analysis (Supplementary Figure S8). This demonstrated that full-length protein was obtained. N-terminal degradation products were found in lesser amounts a well.
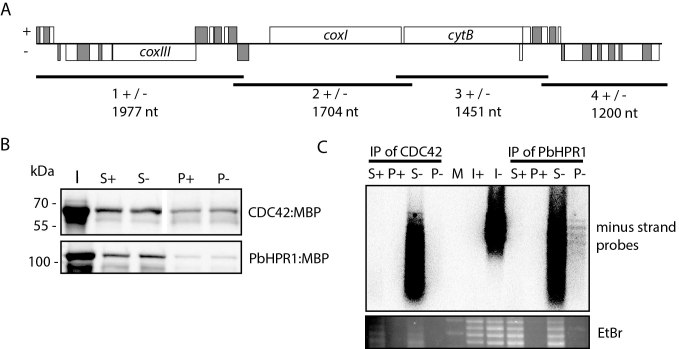
Both proteins were then incubated with in vitro transcripts that represent the entire mitochondrial genome from Plasmodium berghei (Figure 6). Four overlapping in vitro transcripts represent the plus strand and the four reverse complementary RNAs the minus strand of the mitochondrial genome, respectively. The proteins were precipitated using an amylose resin (Figure 6B) and bound RNA was detected by RNA gel blot hybridization using a combo probe specific for the minus strand transcripts. This uncovered signals for all four minus strand RNA species in the pellet of the PbHPR1 immunoprecipitation (Figure 6C). The absence of a signal in the CDC42 pellet demonstrates that the precipitation is not due to contaminating RNA in the pull-down experiment, but is due to the presence of the HPR protein. All four minus strand RNAs are found in the pellet, suggesting that RNA binding of PbHPR1 is unspecific under these conditions. However, under the same conditions, DNA is not bound, demonstrating the preference of PbHPR1 for RNA (Supplementary Figure S9).
Figure 6.
RNA pull-down experiment with PbHPR1. (A) Schematic representation of the eight RNA probes used in the RNA pull-down assay. The mitochondrial genome structure of P. berghei is shown on top with rRNA fragments depicted as unlabelled grey or white rectangles. Genes above the central line are transcribed from right to left, genes below the line in the opposite direction. The black lines below the genome represent the position of the plus strand and minus strand probes. Length of the four probes are given in nucleotides (nt). (B) Immunological analysis of CDC42 and PbHPR1 immunoprecipitation experiments. Fractions of input (I), supernatant (S), and pellet (P) of immunoprecipitation assays were separated by SDS-PAGE, blotted to a nylon membrane, and detected immunologically with an anti-MBP antibody. The minus and plus signs indicate whether the sample is from an immunoprecipitation with plus or minus strand RNAs. (C) RNA gel blot analysis of RNAs co-precipitated with CDC42 and PbHPR1. RNA was extracted from input, supernatant and pellet fractions of immunoprecipitation assays and separated on a denaturing agarose gel. The RNAs were visualized using ethidium bromide staining (EtBr) and afterwards transferred to a nylon membrane and probed with a cocktail of radiolabelled oligonucleotides complementary to the minus strand probes shown in (A). M = molecular weight marker.
DISCUSSION
sRNAs in apicoplasts and mitochondria are indicative of protein-based RNA processing in Plasmodium organelles
Genes in apicoplasts and mitochondria of Plasmodium are transcribed as polycistronic precursors and are processed to shorter, often monocistronic forms (3–4,58–59). In mammalian mitochondria, transcript processing is mediated according to the tRNA punctuation model. tRNAs are situated between coding sequences and serve as targets for RNase P and RNase Z or related enzymes, which cleave at these sites in polycistronic precursors in a process that also releases mature mRNAs (15). This mechanism is maybe also relevant for apicoplast gene expression. However, because mitochondria in the Apicomplexa do not possess tRNAs, processing must occur by another mechanism. We hypothesized that protein-based transcript processing similar to that shown for plant organellar RNAs might operate in Plasmodium organelles. An indicator of protein-mediated protection and processing of transcript ends in plant organelles is the presence of sRNA species—footprints of RBPs—that exactly match the termini of transcripts. We here show that such sRNAs also exist in mitochondria of P. falciparum. Of the three reading frames in mitochondria, one (cosRNA19) was found starting upstream of the CYTB reading frame, and two others (cosRNA14 and cosRNA16) bracketed the COXIII reading frame.
Notably, cosRNA16 and cosRNA19 display identical 5′-ends (TTTATTG), but otherwise show no further sequence similarity. Furthermore, the 5′-end of cosRNA19 coincides with the CYTB transcript end in P. falciparum (13). This end is also conserved in P. yoelii (4). Similarly, the 3′-end of COXIII matches the 3′-end of cosRNA14 (13). This coincidence of cosRNAs and transcript ends is indicative of protein-mediated, post-transcriptional generation of such ends, similar to the process in plant organelles. Given that the 5′-ends of the two cosRNAs are conserved, we speculate that a common mechanism and/or factor is involved in their generation. This inference is further substantiated by the 13 identified cosRNAs that map exactly to the 5′-ends of rRNA fragments.
At 68, 30 and 58 nucleotides, all three cosRNAs adjacent to mRNAs are very long by plant standards. This is also true for several cosRNAs among rRNA fragments (e.g. cosRNA5 and cosRNA10; Table 1). Although some plant cosRNAs are within this range, they are rare, with shorter cosRNAs (∼20 nt) being much more frequent (40). If proteins do contribute to protecting such long mitochondrial RNAs against exonucleolytic decay in Plasmodium (by analogy to PPR proteins in plants), they are expected to act in a different manner. For example, they could act together with other RBPs in a larger complex that is capable of protecting larger transcript regions.
HPR proteins: a novel family of RNA binding proteins that has expanded in the Alveolata
Organellar gene expression depends on a large number of nuclear-encoded RBPs, among which repeat-containing proteins are prevalent (60–62). Structural studies of chloroplast PPR proteins (42–43,63), as well as other related repeat-proteins, such as ARM repeat proteins (64), HEAT repeat proteins (65), Puf-repeat proteins (66), MTERF proteins (67), and TPR proteins (68), have demonstrated that all form a superhelix with an extended surface for macromolecular interactions. Many repeat-containing proteins bind other proteins, but at least PPR and OPR proteins are predominantly involved in RNA interactions. The repeat units of the different classes differ slightly in length: the ARM repeat is 42 aa; the HEAT repeat is 39 aa; the Puf repeat is 36 aa; the TPR is 34 aa; the PPR is 35 aa; and the OPR is 38 aa.
We here identified a 37-aa repeat unit that is predicted to form helical elements. Length, secondary-structure prediction and amino acid composition of the consensus sequences have clear similarities to PPR and OPR proteins. However, there are also striking differences, in particular the absence of the OPR-specific PPPEW motif. In our screens for HPR proteins, using the OPR repeat as a search string retrieved only a few HPR proteins. Conversely, using the HPR consensus to search algal genomes also led to the discovery of only a few OPR proteins. Notably, the phylogenetic distribution of HPR proteins is different from that of OPR and PPR proteins (Figure 4). We found no HPR proteins among embryophytes, where PPR proteins predominate. Similarly, we found few HPRs among green algae, where the OPR family was discovered and is prevalent. A few members are found in animals and humans, as further discussed below. In contrast, HPR proteins are abundant in alveolates like apicomplexans (sporozoa and chromerids) and dinoflagellates, and somewhat less common among ciliates. Apparently, the HPR family has expanded within the Alveolata. Thus, in addition to PPR and OPR families, there is a third family of helical-repeat proteins that shows lineage-specific expansion.
This specific evolutionary distribution, together with consensus sequence idiosyncrasies and the novel repeat length, suggest that the HPR protein family is distinct from the related PPR and OPR protein families. Whether the HPR proteins have evolved functions that separate them from PPR and OPR proteins will be an attractive research direction for future biochemical studies. Intriguingly, we failed to identify any HPR proteins in Cryptosporidium species, which contain a mitochondria-like organelle (mitosome) with a striking absence of organellar DNA, providing a clear phylogenetic link between mitochondrial functions and this novel organellar protein family. It is also possible that functions of PPR, OPR and HPR protein families are similar and it is simply a matter of evolutionary chance whether a helical-repeat protein has 35-, 37- or 38-aa repeats as long as it can mediate specific events in RNA metabolism. Within the PPR protein family, there is a very active set of genes that spawn a novel type (Rf-type) of PPR proteins in a comparatively rapid manner (69,70). An interesting objective of future research would be to search for the origin of HPR proteins and possibly identify evolutionarily active clusters of genes.
Loss of HPR proteins results in profound fitness defects in Plasmodium and Toxoplasma
Recent genome-wide knock-out screens of P. berghei and T. gondii inevitably included many HPR proteins and provide a first estimate of their importance in parasite growth (71,72). In T. gondii, the genome-wide screen included knockouts of each HPR, most of which were strongly selected against (71). Strikingly, six of the 22 HPRs were among the top 10% most strongly counter-selected genes in T. gondii, and on average, HPR proteins were pronouncedly more strongly counter-selected than the average of all genes analyzed (Supplementary Table S4). A similar picture emerges from a P. berghei screen, which covered approximately half of the known genes in this organism. Of the 9 out of 23 P.berghei HPR genes analyzed in this screen, 7 were considered essential (72). In sum, HPR proteins are essential components of apicomplexan parasites. Most HPRs are predicted to be transported into mitochondria, a prediction substantiated by our own targeting analyses as well as by other studies (51,53). Given that organelle functions and the expression of organellar genes are essential for parasite survival (7,73–75), we infer that HPRs play important roles in such essential aspects of organelle biology. Our analysis of the HPR protein family, together with published data, suggests that this role is tightly linked to organellar RNA metabolism (see below).
HPR proteins function in organellar RNA metabolism
As outlined above, HPR proteins are most closely related to OPR proteins. Most, if not all, OPR proteins are involved in RNA metabolism and associate directly with RNA (21,76). OPRs function in translation (77–79), RNA stabilization (79,80), RNA degradation (81), and RNA processing and splicing (82,83). Whereas OPRs in algae have a variety of mRNAs as targets, the transcriptome of the Apicomplexa mitochondrion is truly simple. What then are the target RNAs and functions of the 22 (or more) HPR proteins in P. falciparum, given that there are only three potential mRNAs targets? One way to identify potential RNA target sites of helical-repeat proteins is to determine sRNAs in a transcriptome. This led to the surprising discovery that, aside from a few sRNAs associated with the three mitochondrial mRNAs, there are 12 cosRNAs that match the 5′-ends of rRNA fragments. One potential explanation for these cosRNAs is that RBPs like the HPR proteins associate with the 5′-termini of rRNAs and protect them against exonucleolytic degradation. Eventually, rRNAs are degraded, with the exception of the region protected by the HPR proteins. If this model is correct, cosRNAs would be only degradation products, a situation similar to the manner in which a number of PPR proteins generate short RNAs (40). While HPR proteins as structural relatives of PPR proteins are intriguing candidates for cosRNA generators, other RNA binding proteins, i.e. ribosomal proteins could contribute to cosRNA generation as well.
How the ends of the different rRNA fragments are generated and how they are assembled into the mitochondrial ribosome remains somewhat enigmatic. One possibility is that HPR proteins assist in their biogenesis as well as in their stabilization and assembly. This hypothesis is supported by two major observations. First, HPR numbers are correlated with the organization of mitochondrial rRNAs, such that in taxa with highly fragmented mitochondrial rRNAs, larger number of HPR proteins are found. Apicomplexans, such as T. gondii and P. falciparum, as well as the related algae, chromerids and dinoflagellates, have highly fragmented rRNAs (84); within these taxa, we detected larger numbers of HPR proteins in selected species: 25 in T. gondii, 22 in P. falciparum, 70 in V. brassicaformis, and 72 in S. minutum. By contrast, only five HPR proteins were identified among the ciliates, a sister taxon within the Alveolata. Ciliates have a canonical organization of rRNAs, with only two rRNA species (see Figure 4).
One reason for HPR protein family expansion among apicomplexans, which contain mitochondria, might be the more complex rRNA maturation process. The even higher numbers of HPR proteins in algae might be explained by an increase in additional RNA-processing steps, such as RNA editing and trans-splicing, and addition of further genome idiosyncrasies, including gene scrambling and the loss of stop codons (84). OPR and PPR proteins serve a larger number of RNA functions, including RNA editing, RNA splicing, translation and RNA stabilization (60), and we hypothesize that this is also true for HPR proteins. In fact, recent analyses of human mitochondrial HPR proteins supports this idea, demonstrating functions that include rRNA maturation (53-55).
HPR repeats are also found in six human proteins, FASTK, FASTKD1, FASTKD2, FASTKD3, FASTKD4/TBRG4 and FASTKD5 (Figure 5), which have been shown to be involved in mitochondrial RNA metabolism (51–53). More specifically, FASTKD5 is found in mitochondrial RNA granules, where it binds to the rRNA of the small ribosomal subunit and is required for ribosome biogenesis (51). FASTKD5 is also essential for processing of ATP8/6 and COXIII mRNA, as evidenced by the fact that mutants of FASTKD5 give rise to massive accumulation of precursor RNAs for these two messages and fail to accumulate the processed forms (51). FASTKD4 modulates the half-lives of a subset of mitochondrial mRNAs and, like FASTKD5, associates with them in vivo (52). FASTKD4 might also be involved in both cleavage of polycistronic precursor RNAs as well as protection of mRNAs against exonucleolytic decay (53). While HPR repeats are clearly detectable, the motifs contained in these proteins show a higher degree of degeneration compared to alveolates which precludes the generation of a human-specific consensus motif. These analyses demonstrate that HPR proteins are involved in RNA metabolism, which is supported by our finding that pBHPR1 binds RNA, not DNA in vitro. We hypothesize that HPR proteins function in a manner that is very similar to that of OPR and PPR proteins.
Future genetic studies of Plasmodium species or T. gondii using established tools should pave the way toward identification of RNA targets and functions of this exciting novel protein family. Given the critical importance of HPR proteins for the parasite's fitness, these proteins and the processing steps they mediate are obviously potential future targets for therapeutic intervention.
DATA AVAILABILITY
NGS data on small RNAs from plasmodium have been deposited within the gene expression omnibus repository under accession number GSE107625.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Giel van Dooren (ANU Canberra) and Hannes Ruwe (HU Berlin) for critical discussions. The authors thank D. Eyermann, J. Schmidt, D. Tschierske and Drs G. Costa, Y. Reis and E.A. Levashina (Vector Biology Unit, MPIIB Berlin) for providing P. falciparum infrastructure and materials. We also thank Gongwei Wang and José Muino for help with RNA Seq analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [SCHM1689-5/1 to C.S.L.]; international research training program 2290 ‘Crossing boundaries: molecular interactions in malaria’. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
REFERENCES
- 1. World Health Organization World Malaria Report 2016. World Health Organization. 2016; Geneva. [Google Scholar]
- 2. McFadden G.I., Reith M.E., Munholland J., Lang-Unnasch N.. Plastid in human parasites. Nature. 1996; 381:482–482. [DOI] [PubMed] [Google Scholar]
- 3. Nisbet R.E.R., Kurniawan D.P., Bowers H.D., Howe C.J.. Transcripts in the Plasmodium apicoplast undergo cleavage at tRNAs and editing, and include antisense sequences. Protist. 2016; 167:377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Suplick K., Morrisey J., Vaidya A.B.. Complex transcription from the extrachromosomal DNA encoding mitochondrial functions of Plasmodium yoelii. Mol. Cell. Biol. 1990; 10:6381–6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goodman C.D., Su V., McFadden G.I.. The effects of anti-bacterials on the malaria parasite Plasmodium falciparum. Mol. Biochem. Parasitol. 2007; 152:181–191. [DOI] [PubMed] [Google Scholar]
- 6. Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S. et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002; 419:498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Painter H.J., Morrisey J.M., Mather M.W., Vaidya A.B.. Specific role of mitochondrial electron transport in blood-stage Plasmodium falciparum. Nature. 2007; 446:88–91. [DOI] [PubMed] [Google Scholar]
- 8. Pino P., Aeby E., Foth B.J., Sheiner L., Soldati T., Schneider A., Soldati-Favre D.. Mitochondrial translation in absence of local tRNA aminoacylation and methionyl tRNAMet formylation in Apicomplexa. Mol. Microbiol. 2010; 76:706–718. [DOI] [PubMed] [Google Scholar]
- 9. Siregar J.E., Syafruddin D., Matsuoka H., Kita K., Marzuki S.. Mutation underlying resistance of Plasmodium berghei to atovaquone in the quinone binding domain 2 (Qo(2)) of the cytochrome b gene. Parasitol. Int. 2008; 57:229–232. [DOI] [PubMed] [Google Scholar]
- 10. Esseiva A.C., Naguleswaran A., Hemphill A., Schneider A.. Mitochondrial tRNA import in Toxoplasma gondii. J. Biol. Chem. 2004; 279:42363–42368. [DOI] [PubMed] [Google Scholar]
- 11. Sharma A., Sharma A.. Plasmodium falciparum mitochondria import tRNAs along with an active phenylalanyl-tRNA synthetase. Biochem. J. 2015; 465:459–469. [DOI] [PubMed] [Google Scholar]
- 12. Ji Y.-E., Mericle B.L., Rehkopf D.H., Anderson J.D., Feagin J.E.. The Plasmodium falciparum 6 kb element is polycistronically transcribed. Mol. Biochem. Parasitol. 1996; 81:211–223. [DOI] [PubMed] [Google Scholar]
- 13. Rehkopf D.H., Gillespie D.E., Harrell M.I., Feagin J.E.. Transcriptional mapping and RNA processing of the Plasmodium falciparum mitochondrial mRNAs. Mol. Biochem. Parasitol. 2000; 105:91–103. [DOI] [PubMed] [Google Scholar]
- 14. Feagin J.E., Harrell M.I., Lee J.C., Coe K.J., Sands B.H., Cannone J.J., Tami G., Schnare M.N., Gutell R.R.. The Fragmented Mitochondrial Ribosomal RNAs of Plasmodium falciparum. PLoS One. 2012; 7:e38320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lopez Sanchez M., Mercer T., Davies S., Shearwood A.-M., Nygård K., Richman T., Mattick J., Rackham O., Filipovska A.. RNA processing in human mitochondria. Cell Cycle. 2011; 10:2904–2916. [DOI] [PubMed] [Google Scholar]
- 16. Rackham O., Mercer T.R., Filipovska A.. The human mitochondrial transcriptome and the RNA-binding proteins that regulate its expression. Wiley Interdiscip. Rev. RNA. 2012; 3:675–695. [DOI] [PubMed] [Google Scholar]
- 17. Small I.D., Peeters N.. The PPR motif – a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2000; 25:45–47. [DOI] [PubMed] [Google Scholar]
- 18. Cheng S., Gutmann B., Zhong X., Ye Y., Fisher M.F., Bai F., Castleden I., Song Y., Song B., Huang J. et al. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 2016; 85:532–547. [DOI] [PubMed] [Google Scholar]
- 19. Colcombet J., Lopez-Obando M., Heurtevin L., Bernard C., Martin K., Berthomé R., Lurin C.. Systematic study of subcellular localization of Arabidopsis PPR proteins confirms a massive targeting to organelles. RNA Biol. 2013; 10:1557–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruwe H., Schmitz-Linneweber C.. Short non-coding RNA fragments accumulating in chloroplasts: footprints of RNA binding proteins. Nucleic Acids Res. 2012; 40:3106–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rahire M., Laroche F., Cerutti L., Rochaix J.-D.. Identification of an OPR protein involved in the translation initiation of the PsaB subunit of photosystem I. Plant J. 2012; 72:652–661. [DOI] [PubMed] [Google Scholar]
- 22. Aurrecoechea C., Brestelli J., Brunk B.P., Dommer J., Fischer S., Gajria B., Gao X., Gingle A., Grant G., Harb O.S. et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009; 37:D539–D543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Trager W., Jensen J.B.. Human malaria parasites in continuous culture. Science. 1976; 193:673. [DOI] [PubMed] [Google Scholar]
- 24. Takashima E., Takamiya S., Takeo S., Mi-ichi F., Amino H., Kita K.. Isolation of mitochondria from Plasmodium falciparum showing dihydroorotate dependent respiration. Parasitol. Int. 2001; 50:273–278. [DOI] [PubMed] [Google Scholar]
- 25. Kobayashi T., Sato S., Takamiya S., Komaki-Yasuda K., Yano K., Hirata A., Onitsuka I., Hata M., Mi-ichi F., Tanaka T. et al. Mitochondria and apicoplast of Plasmodium falciparum: Behaviour on subcellular fractionation and the implication. Mitochondrion. 2007; 7:125–132. [DOI] [PubMed] [Google Scholar]
- 26. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. 2011; 17:doi:10.14806/ej.17.1.200. [Google Scholar]
- 27. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing, S. . The sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Freese N.H., Norris D.C., Loraine A.E.. Integrated genome browser: visual analytics platform for genomics. Bioinformatics. 2016; 32:2089–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Loizeau K., Qu Y., Depp S., Fiechter V., Ruwe H., Lefebvre-Legendre L., Schmitz-Linneweber C., Goldschmidt-Clermont M.. Small RNAs reveal two target sites of the RNA-maturation factor Mbb1 in the chloroplast of Chlamydomonas. Nucleic Acids Res. 2014; 42:3286–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grigoriev I.V., Nordberg H., Shabalov I., Aerts A., Cantor M., Goodstein D., Kuo A., Minovitsky S., Nikitin R., Ohm R.A. et al. The genome portal of the department of energy joint genome institute. Nucleic Acids Res. 2012; 40:D26–D32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Muralidharan V., Goldberg D.E.. Asparagine repeats in Plasmodium falciparum proteins: good for nothing. PLoS Pathog. 2013; 9:e1003488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Matz J.M., Matuschewski K., Kooij T.W.A.. Two putative protein export regulators promote Plasmodium blood stage development in vivo. Mol. Biochem. Parasitol. 2013; 191:44–52. [DOI] [PubMed] [Google Scholar]
- 36. Kooij T.W.A., Rauch M.M., Matuschewski K.. Expansion of experimental genetics approaches for Plasmodium berghei with versatile transfection vectors. Mol. Biochem. Parasitol. 2012; 185:19–26. [DOI] [PubMed] [Google Scholar]
- 37. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kupsch C., Ruwe H., Gusewski S., Tillich M., Small I., Schmitz-Linneweber C.. Arabidopsis chloroplast RNA binding proteins CP31A and CP29A associate with large transcript pools and confer cold stress tolerance by influencing multiple chloroplast RNA processing steps. Plant Cell. 2012; 10:4266–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roy A., Kucukural A., Zhang Y.. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010; 5:725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ruwe H., Wang G., Gusewski S., Schmitz-Linneweber C.. Systematic analysis of plant mitochondrial and chloroplast small RNAs suggests organelle-specific mRNA stabilization mechanisms. Nucleic Acids Res. 2016; 44:7406–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gully B.S., Shah K.R., Lee M., Shearston K., Smith N.M., Sadowska A., Blythe A.J., Bernath-Levin K., Stanley W.A., Small I.D. et al. The design and structural characterization of a synthetic pentatricopeptide repeat protein. Acta Crystallogr. D. Biol. Crystallogr. 2015; 71:196–208. [DOI] [PubMed] [Google Scholar]
- 42. Gully B.S., Cowieson N., Stanley W.A., Shearston K., Small I.D., Barkan A., Bond C.S.. The solution structure of the pentatricopeptide repeat protein PPR10 upon binding atpH RNA. Nucleic Acids Res. 2015; 43:1918–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yin P., Li Q., Yan C., Liu Y., Liu J., Yu F., Wang Z., Long J., He J., Wang H.W. et al. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature. 2013; 504:168–171. [DOI] [PubMed] [Google Scholar]
- 44. Emanuelsson O., Nielsen H., Brunak S., von Heijne G.. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000; 300:1005–1016. [DOI] [PubMed] [Google Scholar]
- 45. Claros M.G., Vincens P.. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996; 241:779–786. [DOI] [PubMed] [Google Scholar]
- 46. Zuegge J., Ralph S., Schmuker M., McFadden G.I., Schneider G.. Deciphering apicoplast targeting signals – feature extraction from nuclear-encoded precursors of Plasmodium falciparum apicoplast proteins. Gene. 2001; 280:19–26. [DOI] [PubMed] [Google Scholar]
- 47. Foth B.J., Ralph S.A., Tonkin C.J., Struck N.S., Fraunholz M., Roos D.S., Cowman A.F., McFadden G.I.. Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science. 2003; 299:705. [DOI] [PubMed] [Google Scholar]
- 48. van Dooren G.G., Marti M., Tonkin C.J., Stimmler L.M., Cowman A.F., McFadden G.I.. Development of the endoplasmic reticulum, mitochondrion and apicoplast during the asexual life cycle of Plasmodium falciparum. Mol. Microbiol. 2005; 57:405–419. [DOI] [PubMed] [Google Scholar]
- 49. Henriquez F.L., Richards T.A., Roberts F., McLeod R., Roberts C.W.. The unusual mitochondrial compartment of Cryptosporidium parvum. Trends Parasitol. 2005; 21:68–74. [DOI] [PubMed] [Google Scholar]
- 50. Obornik M., Lukes J.. The organellar genomes of chromera and vitrella, the phototrophic relatives of apicomplexan parasites. Annu. Rev. Microbiol. 2015; 69:129–144. [DOI] [PubMed] [Google Scholar]
- 51. Antonicka H., Shoubridge E.A.. Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Rep. 2015; 10:920–932. [DOI] [PubMed] [Google Scholar]
- 52. Wolf A.R., Mootha V.K.. Functional genomic analysis of human mitochondrial RNA processing. Cell Rep. 2014; 7:918–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Boehm E., Zaganelli S., Maundrell K., Jourdain A.A., Thore S., Martinou J.C.. FASTKD1 and FASTKD4 have opposite effects on expression of specific mitochondrial RNAs, depending upon their endonuclease-like RAP domain. Nucleic Acids Res. 2017; 45:6135–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jourdain A.A., Popow J., de la Fuente M.A., Martinou J.C., Anderson P., Simarro M.. The FASTK family of proteins: emerging regulators of mitochondrial RNA biology. Nucleic Acids Res. 2017; 45:10941–10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Simarro M., Gimenez-Cassina A., Kedersha N., Lazaro J.B., Adelmant G.O., Marto J.A., Rhee K., Tisdale S., Danial N., Benarafa C. et al. Fast kinase domain-containing protein 3 is a mitochondrial protein essential for cellular respiration. Biochem. Biophys. Res. Commun. 2010; 401:440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M. et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012; 149:1393–1406. [DOI] [PubMed] [Google Scholar]
- 57. Baltz A.G., Munschauer M., Schwanhausser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M. et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012; 46:674–690. [DOI] [PubMed] [Google Scholar]
- 58. Gardner M.J., Feagin J.E., Moore D.J., Spencer D.F., Gray M.W., Williamson D.H., Wilson R.J.. Organisation and expression of small subunit ribosomal RNA genes encoded by a 35-kilobase circular DNA in Plasmodium falciparum. Mol. Biochem. Parasitol. 1991; 48:77–88. [DOI] [PubMed] [Google Scholar]
- 59. Gardner M.J., Williamson D.H., Wilson R.J.. A circular DNA in malaria parasites encodes an RNA polymerase like that of prokaryotes and chloroplasts. Mol. Biochem. Parasitol. 1991; 44:115–123. [DOI] [PubMed] [Google Scholar]
- 60. Barkan A., Small I.. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014; 65:415–442. [DOI] [PubMed] [Google Scholar]
- 61. Barkan A. Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 2011; 155:1520–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ferreira N., Rackham O., Filipovska A.. Regulation of a minimal transcriptome by repeat domain proteins. Semin. Cell Dev. Biol. 2018; 76:132–141. [DOI] [PubMed] [Google Scholar]
- 63. Shen C., Zhang D., Guan Z., Liu Y., Yang Z., Yang Y., Wang X., Wang Q., Zhang Q., Fan S. et al. Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat. Commun. 2016; 7:11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Huber A., Nelson W., Weis W.. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell. 1997; 90:871–872. [DOI] [PubMed] [Google Scholar]
- 65. Groves M.R., Hanlon N., Turowski P., Hemmings B.A., Barford D.. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell. 1999; 96:99–110. [DOI] [PubMed] [Google Scholar]
- 66. Edwards T., Pyle S., Wharton R., Aggarwal A.. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001; 105:281–289. [DOI] [PubMed] [Google Scholar]
- 67. Spahr H., Samuelsson T., Hallberg B.M., Gustafsson C.M.. Structure of mitochondrial transcription termination factor 3 reveals a novel nucleic acid-binding domain. Biochem. Biophys. Res. Commun. 2010; 397:386–390. [DOI] [PubMed] [Google Scholar]
- 68. Das A.K., Cohen P.T.W., Barford D.. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 1998; 17:1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Castandet B., Araya A.. The nucleocytoplasmic conflict, a driving force for the emergence of plant organellar RNA editing. IUBMB Life. 2012; 64:120–125. [DOI] [PubMed] [Google Scholar]
- 70. Gaborieau L., Brown G.G., Mireau H.. The propensity of pentatricopeptide repeat genes to evolve into restorers of cytoplasmic male sterility. Front. Plant Sci. 2016; 7:1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sidik S.M., Huet D., Ganesan S.M., Huynh M.H., Wang T., Nasamu A.S., Thiru P., Saeij J.P., Carruthers V.B., Niles J.C. et al. A genome-wide CRISPR screen in toxoplasma identifies essential apicomplexan genes. Cell. 2016; 166:1423–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bushell E., Gomes A.R., Sanderson T., Anar B., Girling G., Herd C., Metcalf T., Modrzynska K., Schwach F., Martin R.E. et al. Functional profiling of a plasmodium genome reveals an abundance of essential genes. Cell. 2017; 170:260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Balabaskaran Nina P., Morrisey J.M., Ganesan S.M., Ke H., Pershing A.M., Mather M.W., Vaidya A.B.. ATP synthase complex of Plasmodium falciparum: dimeric assembly in mitochondrial membranes and resistance to genetic disruption. J. Biol. Chem. 2011; 286:41312–41322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ke H., Morrisey J.M., Ganesan S.M., Mather M.W., Vaidya A.B.. Mitochondrial RNA polymerase is an essential enzyme in erythrocytic stages of Plasmodium falciparum. Mol. Biochem. Parasitol. 2012; 185:48–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sheiner L., Vaidya A.B., McFadden G.I.. The metabolic roles of the endosymbiotic organelles of Toxoplasma and Plasmodium spp. Curr. Opin. Microbiol. 2013; 16:452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kleinknecht L., Wang F., Stube R., Philippar K., Nickelsen J., Bohne A.V.. RAP, the sole octotricopeptide repeat protein in Arabidopsis, is required for chloroplast 16S rRNA maturation. Plant Cell. 2014; 26:777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Auchincloss A., Zerges W., Perron K., Girard-Bascou J., Rochaix J.-D.. Characterization of Tbc2, a nucleus-encoded factor specifically required for translation of the chloroplast psbC mRNA in Chlamydomonas reinhardtii. J. Cell Biol. 2002; 157:953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Eberhard S., Loiselay C., Drapier D., Bujaldon S., Girard-Bascou J., Kuras R., Choquet Y., Wollman F.A.. Dual functions of the nucleus-encoded factor TDA1 in trapping and translation activation of atpA transcripts in Chlamydomonas reinhardtii chloroplasts. Plant J. 2011; 67:1055–1066. [DOI] [PubMed] [Google Scholar]
- 79. Lefebvre-Legendre L., Choquet Y., Kuras R., Loubery S., Douchi D., Goldschmidt-Clermont M.. A nucleus-encoded chloroplast protein regulated by iron availability governs expression of the photosystem I subunit PsaA in Chlamydomonas reinhardtii. Plant Physiol. 2015; 167:1527–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang F., Johnson X., Cavaiuolo M., Bohne A.V., Nickelsen J., Vallon O.. Two Chlamydomonas OPR proteins stabilize chloroplast mRNAs encoding small subunits of photosystem II and cytochrome b6 f. Plant J. 2015; 82:861–873. [DOI] [PubMed] [Google Scholar]
- 81. Boulouis A., Drapier D., Razafimanantsoa H., Wostrikoff K., Tourasse N.J., Pascal K., Girard-Bascou J., Vallon O., Wollman F.A., Choquet Y.. Spontaneous dominant mutations in chlamydomonas highlight ongoing evolution by gene diversification. Plant Cell. 2015; 27:984–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Merendino L., Perron K., Rahire M., Howald I., Rochaix J.D., Goldschmidt-Clermont M.. A novel multifunctional factor involved in trans-splicing of chloroplast introns in Chlamydomonas. Nucleic Acids Res. 2006; 34:262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Marx C., Wunsch C., Kuck U.. The octatricopeptide repeat protein Raa8 is required for chloroplast trans splicing. Eukaryot. Cell. 2015; 14:998–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Waller R.F., Jackson C.J.. Dinoflagellate mitochondrial genomes: stretching the rules of molecular biology. Bioessays. 2009; 31:237–245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NGS data on small RNAs from plasmodium have been deposited within the gene expression omnibus repository under accession number GSE107625.