Abstract
Bacterial RNase III plays important roles in the processing and degradation of RNA transcripts. A major goal is to identify the cleavage targets of this endoribonuclease at a transcriptome-wide scale and delineate its in vivo cleavage rules. Here we applied to Escherichia coli grown to either exponential or stationary phase a tailored RNA-seq-based technology, which allows transcriptome-wide mapping of RNase III cleavage sites at a nucleotide resolution. Our analysis of the large-scale in vivo cleavage data substantiated the established cleavage pattern of a double cleavage in an intra-molecular stem structure, leaving 2-nt-long 3′ overhangs, and refined the base-pairing preferences in the cleavage site vicinity. Intriguingly, we observed that the two stem positions between the cleavage sites are highly base-paired, usually involving at least one G-C or C-G base pair. We present a clear distinction between intra-molecular stem structures that are RNase III substrates and intra-molecular stem structures randomly selected across the transcriptome, emphasizing the in vivo specificity of RNase III. Our study provides a comprehensive map of the cleavage sites in both intra-molecular and inter-molecular duplex substrates, providing novel insights into the involvement of RNase III in post-transcriptional regulation in the bacterial cell.
INTRODUCTION
The family of ribonuclease III (RNase III) enzymes is widely spread in both prokaryotes and eukaryotes, with its famous representatives the eukaryotic enzymes Drosha and Dicer (1). A prominent member of the family is RNase III of Escherichia coli, which was discovered ∼50 years ago and extensively studied since (2). While initially the E. coli RNase III was known for its involvement in the processing of RNA precursors transcribed from the early T7 genes and ribosomal RNA (rRNA) genes (3), it is now evident that RNase III can affect the expression level of additional genes by either stabilizing or de-stabilizing the cleaved transcripts [e.g. (4–6) and reviewed in (2)].
RNase III is a divalent-metal-ion-dependent endoribonuclease that cleaves the phosphodiester bond leaving 3′-hydroxyl 5′-phosphomonoester termini (2,7). It works as a symmetric homodimer, where the dimerization generates a valley that can accommodate a double-stranded RNA (dsRNA) substrate of ∼22 base pairs. The two catalytic sites of the enzyme reside within this valley on opposite sides, each facing one of the duplex strands. Each catalytic site can independently cleave the strand it faces, generating a double cleavage. However, single cleavages of only one strand can also take place (2). In the double cleavage, there is usually an offset in the locations of the cleavage positions on the two strands, such that the cleavages leave 2-nt-long 3′ overhangs (2,7). Three regions were symmetrically defined on the double-stranded substrate in the vicinity of each cleavage site, termed by their distance from the cleavage site: proximal (8,9), medial (10) and distal (8,9) boxes. Distinct base-pairing combinations in specific positions within these boxes were found to either favor or disfavor RNase III cleavage and/or binding (7,9). This large body of knowledge of the cleavage site properties was gained mostly by in vitro biochemical studies of specific substrates and by crystallographic studies of RNase III, either RNA-free or complexed with dsRNA substrates. The currently available massive parallel sequencing technologies enable in vivo transcriptome-wide determination of the cleavage sites. Exploitation of these large-scale data can add an in vivo layer to the RNase III cleavage rules and to the scope of its cellular targets, expanding our understanding of the functionality and mechanism of action of this important enzyme.
Tailored RNA-seq-based approaches that rely on the chemical group at the 5′ end of the cleavage products were shown to be very efficient in detecting cleavage sites of distinct endoribonucleases. These approaches distinguish the 5′ end of cleavage products by using a ligase that is capable of ligating an adapter with the appropriate chemical group at its 3′ end. Following library preparation, sequencing and mapping to the respective genome, 5′ ends of cleavage products can be inferred from the mapped read start positions. By comparing the read start counts per position between a wild-type (wt) library and a mutant library of the RNase under study, cleavage positions can be reliably identified at a transcriptome-wide scale. Variations of this approach were successfully applied to the global mapping of cleavage sites of several endoribonucleases. Examples include the toxin MazF (11) and RNase E in E. coli (12), RNase E in Salmonella enterica (13), Rnase J1 and RNase III in Bacillus subtilis (14), and RNase III in Streptococcus pyogenes (15). In the latter study, both 5′ and 3′ ends of cleavage products were mapped. Other approaches for global mapping of RNase III targets that do not involve exact 5′ end mapping were also applied. Examples include co-immunoprecipitation of RNase III in Staphylococcus aureus followed by RNA-seq of the extracted RNA (16), as well as differential transcriptome analysis of wt and RNase III mutant strains in E. coli, previously by microarrays (5) and recently by RNA-seq (17).
Here we carry out a rigorous mapping study of the cleavage positions of RNase III in E. coli at a nucleotide resolution, applying 5′ end mapping to bacteria grown to either exponential or stationary phase, including enrichment of short RNA fragments in some of the library preparations. We present a global map of RNase III targets at both growth phases, substantially expanding the scope of targets across E. coli transcriptome. These exhaustive cleavage data allow an unbiased inference of RNase III cleavage rules in vivo. We substantiate and refine the sequence–structure principles of cleavage in intra-molecular double-strand structures and shed light on the specificity determinants of RNase III cleavage across the transcriptome.
MATERIALS AND METHODS
Strains, growth conditions and RNA extraction
The E. coli K12 MG1655 (genotype: K-12 F– λ–ilvG–rfb-50 rph-1) served as the wt strain and its derivative MG1655 rnc-14::ΔTn10 (18,19) served as the RNase III null mutant. These strains are hereinafter denoted wt and rnc-14, respectively. Over-night cultures were diluted 1/100 in Luria Bertani Broth (LB) and grown at 37°C to an OD600 value of 0.3 (exponential phase) or for 6 h (stationary phase). Bacteria were harvested by centrifugation, the pellet was resuspended in TE buffer, mixed with lysosyme (Sigma Aldrich) in a final concentration of 0.9 mg/ml and immediately frozen in liquid nitrogen. The frozen pellet was subjected to two cycles of thaw and freeze at 37°C and in liquid nitrogen, respectively, and RNA was extracted using Tri-Reagent (Sigma Aldrich).
5′P library construction
Since RNase III cleavage products are mono-phosphorylated (5′P) RNA fragments, we select for fragments that start with 5′P in the library preparation (hereinafter, 5′P library). Total RNA was treated with TURBO DNase (Invitrogen) according to manufacturer instructions. Ten micrograms of the DNase treated RNA was subjected to rRNA depletion using Ribo-Zero magnetic kit (Illumina). To block the 3′ end of the RNA, an adenylated blocking adapter (5rApp/CTGTAGGCACCATCAAT/3ddC; 100 IDT) was ligated using RNA ligase 2 truncated K227Q (New England Biolabs). RNA was then ligated to a 5′ adapter (5InvddT/TCTTTCCCTACACGACGCTCTTCCGATCT; IDT) using RNA ligase 1 (New England Biolabs). This ligation can take place only when the 5′ end of the RNA is mono-phosphorylated. Then, the RNA was randomly trimmed using 5 U of S1 nuclease (Thermo Scientific) at 22°C. Trimming was stopped after 8 min by the addition of ethylenediaminetetraacetic acid (EDTA) to a final concentration of 30 mM. RNA was then ligated to a 3′ RNA adapter (5Phos/GAUCGGAAGAGCACACGUCUGAACUCCAGUCAC/ddC; IDT) using RNA ligase 1. First-strand cDNA was synthesized using the SuperScript III First Strand kit (Invitrogen) and RT primer (GTGACTGGAGTTCAGACGTGTGCTCTTCCGATC). Between each step of library construction, RNA was cleaned-up using 1.8-fold reaction volume (1.8X) RNA Clean XP beads (Beckman Coulter). First-strand complementary DNA (cDNA) was cleaned-up with 1.5X Ampure XP beads (Beckman Coulter). The cDNA was amplified using Illumina compatible indexed primers and cleaned-up with 1X Ampure XP beads. Libraries were quantified by quantitative PCR (qPCR) using KAPA Library Quant Kit (Illumina) prior to single-end 85 cycles sequencing using NextSeq 500 sequencer (Illumina).
To construct the short 5′P libraries, 10 μg of DNase treated RNA was depleted of rRNA as above and then cleaned-up using the RNA Clean and Concentrator 5 kit (Zymo Research) to maintain all RNAs that are >16 nt. The RNA was ligated to a 3′ adenylated adapter (5rApp/GATCGGAAGAGCACACGTCTGAACTCCAGTC/3ddc) using RNA ligase 2 and then to a 5′ adapter (see above) using RNA ligase 1. After each step of ligation, the RNA was cleaned-up using 2.5X RNA Clean XP beads and 7X isopropanol to maintain small RNAs. First strand cDNA synthesis (see above) was followed by cDNA clean-up using 1.8X Ampure XP beads and 2X isopropanol. Polymerase chain reaction amplification with Illumina compatible primers was followed by cDNA clean-up using 1.8X Ampure XP beads. The cDNA libraries were quantified by qPCR using KAPA Library Quant Kit prior to single-end 85 cycles sequencing or paired-end 45 and 40 cycles sequencing using NextSeq 500 sequencer. Schematic representation of the library preparation is presented in Supplementary Figure S1.
Mapping of 5′P library sequencing data to E. coli genome
Raw reads were split into their library of origin using the adapter barcodes and an in-house program. Reads were processed using cutadapt (20) version 1.12 (cutadapt -m 25 -q 15 -a GATCGGAAGAGCA -a CTGTAGGCACCATCAAT -n 5 -e 0.15) and mapped to E. coli K12 MG1655 genome (NC_000913.3) using bwa (21) version 0.7.15-r1140. Note that the short libraries ran with slightly different cutadapt parameters (-m 20 -q 15 -a GATCGGAAGAGCA -n 5 -e 0.15), allowing the retention of shorter reads (-m 20) and excluding the second adapter that was not used in the construction of these libraries. The short stationary phase libraries were originally subjected to paired-end sequencing in order to capture the 3′ ends of the cleaved fragments as well. However, due to lower quality of READ2, we analyzed these sequencing data as single-end, using READ1 only. Run parameters for bwa aln were -n 2 -t 8 -R 200 -l 4 -k 0, and bwa samse was run with the default parameters. Note that the choice of parameters for the processing and mapping software packages was aimed at maximizing the accuracy of the detection of the original 5′ end of the transcripts. Thus, there was no removal of 5′ end adapters (even at the cost of reduced mapping), reads that were mapped with soft-clipping were excluded, and no mismatches were allowed at the first four positions of the mapped read (-l 4 -k 0).
Identification of RNase III cleavage sites
As 5′P fragments are selected for in our library preparation, the genomic position where the first nucleotide of a read is mapped corresponds to the first nucleotide of a RNA fragment that starts with 5′P. Hereinafter, the first nucleotide of a mapped read is denoted ‘read start’. Various cellular processes generate 5′P fragments. To specifically identify those that are produced by RNase III cleavage, we prepared 5′P libraries from wt and from a rnc-14::ΔTn10 mutant in which RNase III is inactive (rnc-14) (usually three biological replicates for each). We then counted the number of read starts that mapped to each position along each of the genome strands. Finally, DESeq2 (22) version 1.14.1 was used to identify positions that differed statistically significant in read start counts between the wt and mutant libraries (Figure 1). At the end of this process each position was assigned two values: (i) Log2FC, which is the log2 of the ratio between the wt and mutant’s DESeq2-normalized read start counts. (ii) padj, the statistical significance of the log2FC value (corrected for multiple hypothesis testing by False Discovery Rate). DESeq2 was applied using the default run parameters with the addition of the pre-filtering parameter set to 1, which only retains positions with at least two reads in at least one library.
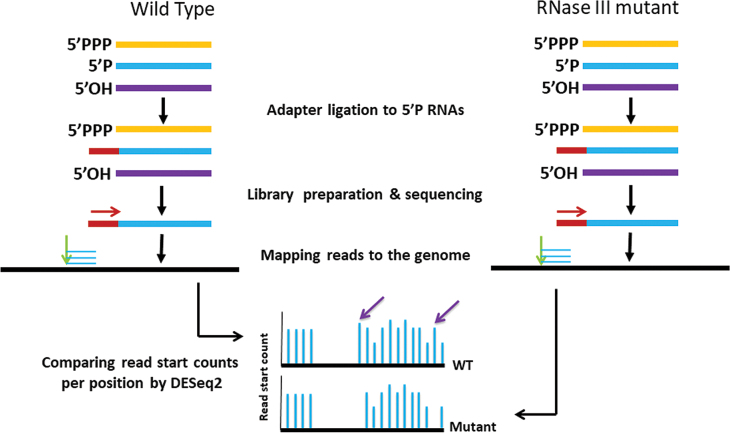
Figure 1.
Identification of RNase III cleavage sites. A schematic illustration of library construction, sequencing, mapping and computational analysis applied for the detection of RNase III cleavage sites. Red and green arrows indicate the sequencing orientation and the genomic position of the mapped read starts, respectively. Purple arrows point at read start positions that appear in the wt but not in the mutant strain, and thus are predicted to be RNase III cleavage sites.
Dataset of previously reported cleavage sites in E. coli
We compiled from the literature a dataset of RNase III cleavage sites in E. coli transcripts, determined by small-scale experiments (Supplementary Data). The data include 36 major and four alternative cleavage sites within intra-molecular RNA double-stranded structures and nine cleavage sites within inter-molecular RNA duplexes.
Collapsing adjacent putative cleavage sites
Cleavage sites were grouped to cleavage regions. A cleavage region included all same strand successive cleavage positions, where each position is at most 15 nt apart from its preceding one. Each such region was finally represented by the position with the highest log2FC value. When multiple sites had equal log2FC values, the most 5′ cleavage site was used as representing the region. Rarely, there were two neighboring cleavage positions at a distance ≤15 nt that were mapped to genomic regions of two different annotations. In those cases, we still selected only one representative cleavage site. However, in the classification of cleavage sites within genomic entities and within transcripts we did take into consideration the alternative annotations.
Computational analysis of dangling ends
This analysis was applied to the set of targets with two major putative cleavage sites.
Generating the intra-molecular double-stranded putative structures
The two cleavage sites were termed C1 and C2, where C1 is the site closest to the 5′ end of the transcript. Assuming that the two cleavage sites reside on the same stem, we expect that the nucleotides flanking the cleavage sites will be capable of base-pairing, and thus we set to predict the potential base pairing of the two regions. Note that C1 and C2 designate the cleavage sites as well as the positions downstream to the cleavage sites. Sequence fragments of 36 nt, flanking each of the cleavage sites, were constructed as follows: for C1: 5′ (20 nt)-C1-(15 nt) 3′; for C2: 5′ (15 nt)-C2-(20 nt) 3′; i.e. 5′ X-20X-19X-18……X-2X-1C1 I1I2……I15 - - - - - - I-15I-14…… I-2I-1 C2 X1 X2……X19X20 3′, where ‘X’ represents the nucleotides upstream to C1 and downstream to C2, and ‘I’ represents the nucleotides between C1 and C2. Fragments with C1-C2 sequential distance <15 were padded symmetrically by ‘N’. RNAduplex -d0 (23) version 2.0.5 was applied to find the potential base pairing of the two regions. The structural distance between C1 and C2 is defined as the number of positions in the duplex (paired or not) between C1 and C2 and determines the length of the dangling ends.
Statistical analysis of the structural distance
We compared the structural distances between C1 and C2 in our double cleavage substrate data to C1-C2 distances in randomly chosen intra-molecular substrates. A total of 100 000 genomic regions (excluding regions defined as antisense to transcribed regions) were randomly chosen. For each region, two sites (C1, C2) with a sequential distance of at least 15 nt residing on the same genomic entity were randomly chosen. Fragments flanking C1 and C2 were defined as described above. RNAduplex -d0 ( 23) was applied to predict stem structure of the random fragments, and the structural distances between C1 and C2 were recorded. The structural distances in each set were divided into 11 categories from a distance of −5 positions to a distance of 5 positions (the −5 and 5 categories included all distances ≤ −5 positions and ≥ 5 positions, respectively). The distributions of structural distances in the two sets were compared by a χ2 test, and structural distances that mostly contributed to the χ2 score were determined.
Score of double cleavage substrates with C1-C2 structural distance of 2 positions
The double-stranded structures of targets with a structural distance of 2 positions were aligned by the cleavage positions C1 and C2. The number of occurrences of all 16 possible nucleotide pairs, as well as of nucleotide-gap or (nucleotide/gap/N)-N, was counted per structural position across all aligned duplexes. All counts were added 0.01 to avoid zero values, and relative frequencies of the nucleotide pairs per position were computed. Assuming position independence, a score of a double cleavage site was defined as the product of the appropriate nucleotide-pair relative frequencies over the 24 positions of the duplex (11 positions at each side of the cleavage sites and the two positions between C1 and C2). For convenience, we represented the score as the log10 of this product.
Distinguishing stem structures that are RNase III substrates from random stem structures
Applying a ‘leave-one-out’ approach, the score for each of the RNase III intra-molecular stem substrates with a C1-C2 structural distance of 2 positions was computed based on the relative frequencies of nucleotide pairs obtained from the alignment of all other intra-molecular stem substrates. To assess the specificity of this sequence–structure signature of the RNase III intra-molecular double-stranded substrates, they were compared to scores of two sets of random substrates: (i) ‘randomly selected stems’—a subset of structures with a C1-C2 structural distance of 2 positions included within the 100 000 randomly selected genomic regions described above (4054 structures). (ii) ‘randomly threaded structures’—a subset of equivalent stem structures with random sequences, generated as follows: We used all intra-molecular stem substrates with a structural distance of 2 positions in our data as structural templates. For each such template, nucleotides were assigned at each structural position, maintaining nucleotide pairings, bulges and internal loops. This procedure was repeated 100 000 times, generating 100 000 random sequences for each structural template. For each structural template, the nucleotides were selected by the following procedure: at base-paired positions, a base pair was selected randomly, keeping a ratio of 0.82:0.18 between Watson–Crick and wobble base pairing. The ratio of 0.82:0.18 was estimated using the number of Watson–Crick and wobble base pairs in the structures of the 100 000 random targets. If a bulge was present in either of the strands at a certain position, the nucleotide facing the gap was chosen at random. If the position had an internal loop, a random nucleotide pair (which cannot base pair) was selected. The score of each of the random substrates was calculated as described above. The distributions of scores of RNase III substrates and random stems were compared by a Wilcoxon test. We used these distributions to generate Receiver Operating Characteristic (ROC) curves by which score thresholds separating the substrate and random stems were determined, allowing a maximal false positive rate of 0.05.
Searching for a potentially missed site of a double cleavage
For each single cleavage target, a window of at most 150 bases upstream and downstream the cleavage site within the transcript (excluding positions at distance <15 nt) was defined as a region that might include a missed cleavage site. For each position in the window, a flanking region was extracted and duplexes were predicted. Stems with a structural distance of 2 positions between the experimentally determined and putative cleavage sites were selected and scored as described for the putative intra-molecular duplexes above. The site that yielded the highest score within the window was recorded along with its DESeq2 padj value. Putative cleavage sites were divided into two reliability categories: (i) High: cleavage sites with DEseq2 padj ≤ 0.1, yielding a score ≥ −25.5. (ii) Moderate/Low: cleavage sites with DEseq2 padj >0.1, yielding a score ≥−25.5 or with DEseq2 padj ≤ 0.1 and −25.5> score ≥−33.8. The −25.5 and -33.8 score thresholds correspond to 0.05 false positive rate for the ‘randomly selected stems’ and ‘randomly threaded structures’ sets, respectively. Putative cleavage sites identified upstream to the experimentally determined cleavage site were considered as C1, while sites downstream to the experimentally determined cleavage site were considered as C2.
Identifying single cleavage sites that could potentially originate from inter-molecular duplexes
For each target with a single cleavage we extracted the flanking regions, as described above for the double cleavage targets. RNAduplex -d0 (23) was applied to all pair-combinations of these fragments to generate inter-molecular putative stems. Note that two structural models were considered for each pair-combination, considering each single cleavage site once as C1 and once as C2. Scores were calculated as described above for duplexes of structural distance of 2 positions, and the best scoring pair-combination for each site was recorded. Pair-combinations with scores above the aforementioned thresholds were considered as putative inter-molecular duplex substrates of RNase III.
Statistical tests
We used χ2, Wilcoxon test (two sided) or Fisher test (two sided) for the various analyses.
RESULTS
5′P libraries were constructed using total RNA extracted from wt or rnc-14 mutant E. coli strains grown to exponential phase (three and four biological replicates for wt and mutant strain, respectively). For details see ‘Materials and Methods’ section, Figure 1 and Supplementary Figure S1. The sequencing reads were mapped to E. coli genome NC_000913.3 (Supplementary Table S1). To identify the 5′P positions generated by RNase III cleavage, we counted the number of read starts mapped to each genomic position and compared these ‘per-position’ counts between the wt and rnc-14 libraries using DESeq2 (22) [see Figure 1 for a schematic overview of the procedure and Figure 2 for an example of an identified cleavage site demonstrated for the previously reported targetarfA (24)]. In total, 4104 genomic positions differed statistically significantly in read start counts between the wt and rnc-14 libraries (DESeq2 adjusted p value for multiple hypothesis testing padj ≤ 0.01). Of these, 1287 positions showed higher counts in the wt compared to the mutant libraries and thus comprised the initial set of putative RNase III cleavage sites.
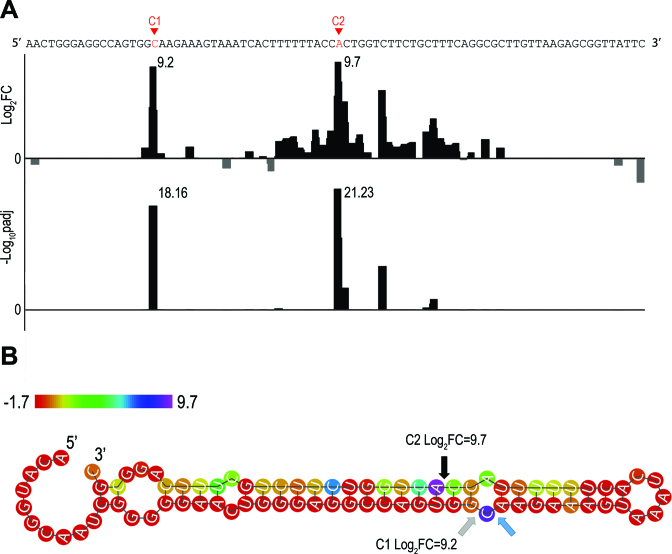
Figure 2.
Example of cleavage site detection within arfA, a known RNase III target (24). (A). Results of the DESeq2 analysis comparing wt and rnc-14 read start counts along the arfA transcript. Shown are the DESeq2 log2 values of the fold change (FC) in read start counts between wt and rnc-14 (Log2FC, upper panel) and the DESeq2 adjusted pvalue for multiple hypotheses testing expressed as -log10, (-Log10padj, lower panel). (B) Secondary structure prediction of arfA transcript. Arrows designate the cleavage sites detected here and by Garza-Sanchez et al. (24): Gray—detected only in our study, blue—detected only by Garza-Sanchez et al. (24) and black—detected in both studies. The two cleavage sites detected in our study determine a structural distance of 2 positions, corresponding to dangling ends of 2 nt on both strands. Nucleotide color indicates the log2FC value. Note that C1 and C2 designate the cleavage sites (Panel B) as well as the positions downstream to the cleavage sites (Panel A).
Rediscovery of known cleavage sites
Previously reported RNase III cleavage sites from small-scale experiments were compiled into two datasets: (i) Known cleavage sites within intra-molecular RNA double-stranded structures (hereinafter, stem structures). (ii) Known cleavage sites within inter-molecular RNA duplexes (Materials and Methods; Supplementary Methods sections).
Detection of known cleavage sites in intra-molecular stem structures
Thirty-six major cleavage sites were previously reported in intra-molecular stem structures (Supplementary Table S2a and b). Seven sites relate to major rRNA and transfer RNA (tRNA) genes encoded by the rRNA operons. As there are multiple copies of the rRNA operons, these seven sites are actually a non-redundant set representing 38 potential cleavage sites in rRNA operons (see Supplementary Table S2a, sites A-G and references therein). The other 29 cleavage sites reside in transcripts of 13 other target genes (Supplementary Table S2b). We detected five of the seven representative rRNA related cleavage sites: The cleavage sites 3′ and 5′ to 16S RNA, 3′ to alaT and 3′ to 5S RNA that were detected in all the respective operon copies and the cleavage sites 3′ to 23S and 3′ to gltT that were detected in 6/7 and 3/4 of the corresponding operon copies, respectively (Supplementary Table S2a). All other 13 RNase III targets were detected by our screen as well. Furthermore, 24 of the 29 cleavage sites in the 13 detected targets overlapped the known sites with a maximal offset of 5 nt (Table 1 and Supplementary Table S2b).
Table 1.
Detection of previously reported cleavage sitesa
| Cleavage sites in intra-molecular duplexes | Cleavage sites/regions in inter-molecular duplexes | Cleavage sites in intra-molecular duplexes reported in Gordon et al. (17) | ||||
|---|---|---|---|---|---|---|
| Sites | Targets | Sites | Targetsb | Sites | targets | |
| Data of known cleavage sites/targets | 29 | 13a | 9 + 2 | Four in ncRNAs and five in ncRNA targets + two in ncRNA regions | 14 | 9 |
| Detected in the analysis of exponential phase datac | 24 | 13 | 3 | Three in ncRNA targets | 12 | 9d |
| Detected in the analysis of exponential phase datac followed by the post-processing procedure | 22 | 13 | 1 | One in a ncRNA target | 10 | 9d |
| Detected in the combined analysis of all datasets | 24 | 13 | 4 + 1 | Four in ncRNA targets + one in ncRNA region | 10 | 9d |
aThe detected known cleavage sites in the rRNA and rnc genes are described in Supplementary Table S2, but are not included here.
bIn total we have five transcript pairs in the data for which at least one cleavage site was found in either the ncRNA or the target of the ncRNA, and an additional pair (gadE/ArrS) in which the cleavage site was previously indirectly deduced. For two ncRNAs, we defined a region that encompasses the complementary region.
cThe data based on the analysis of the 5′P-exp-wt and 5′-exp-mut libraries (Supplementary Table S1).
dIn one of the targets, dctA, we detected a cleavage site but it is 278 nt upstream to the reported one; hence, it is not included in the detected sites and only included in the list of detected targets.
Detection of known cleavage sites in inter-molecular duplexes
This dataset included cleavage sites (or cleavage regions, in case no distinct site was reported in the original paper) in six non-coding RNA (ncRNA)-target duplexes. Four precisely reported cleavage sites reside in the ncRNAs, and five reported cleavage sites (four precisely defined and one deduced) reside in the targets of ncRNAs. We identified three of the five cleavage sites in ncRNA targets, but did not detect any of the reported cleavage sites in the ncRNAs (Table 1 and Supplementary Table S2c). We did, however, identify one cleavage site in the ArrS ncRNA (which was not explicitly determined before), in agreement with the suggestion that ArrS binds in cis to gadE mRNA and regulates its stability in an RNase III-dependent manner (25).
Refining the set of putative RNase III cleavage sites
While all 1287 genomic positions detected by DESeq2 had a statistically significantly higher number of read starts in the wt compared to the mutant libraries, some of these positions had also non-negligible counts of read starts in the rnc-14 mutant libraries, suggesting that they are less likely to be authentic RNase III cleavage sites. Importantly, all but three of the known cleavage sites detected by the DESeq2 analysis had zero or a negligible number of read start counts in the rnc-14 libraries (Supplementary Table S2a–c). Consequently, we excluded from the initial set putative cleavage positions with median read start counts >5 in the rnc-14 libraries. In addition, positions with median read start counts <5 in the wt strain libraries were excluded as well. Lastly, all the cleavage sites that mapped to either the rnc gene or the rRNA operons were excluded from the refined set, as they are highly likely to be affected by the experimental procedure, rRNAs due to rRNA depletion and rnc operon genes due to the effects of the transposon insertion within the rnc gene on the read mapping and on the operon transcription (Supplementary Methods). In total, 485 cleavage sites were excluded by this additional filtering, resulting in a dataset of 802 putative cleavage sites.
Analysis of the 802 sites revealed that 143 (18%) were mapped adjacent to other cleavage sites, and 276 (34%) were at most 15 nt apart from other cleavage sites (Supplementary Figure S2). Notably, multiple adjacent cleavage sites were observed in small-scale experiments as well [e.g. (26)], but often a dominant site was reported. Following the same logic, we represented adjacent cleavage sites by the most dominant cleavage position in the region (‘Materials and Methods’ section), resulting in a total of 526 cleavage sites, out of which 118 positions were representatives of multiple cleavage sites in a region and 408 were isolated sites. While 276 cleavage sites were thus excluded, only two of the known cleavage sites in intra-molecular stem structures were excluded by this process (the two alternative cleavage sites out of the four reported for betT, Table 1; Supplementary Table S2b). The stringent filtering followed by the collapsing of adjacent sites (hereinafter, post-processing procedure) was further assessed using a test set comprising 14 in vitro detected cleavage sites (17) that were not included in our initial compiled set of known targets (see Supplementary Data and Discussion hereinafter). We detected 12 of these sites by our initial analysis, and only 2 out of the 12 were excluded by the post-processing procedure (Table 1 and Supplementary Table S2d). These results support the validity of our data and imply that the post-processing procedure is likely to preserve most of the major RNase III cleavage sites in intra-molecular stem structures.
In the set of known cleavage sites within inter-molecular duplexes, however, our initial detection rate was modest and was further reduced following the post-processing procedure. As some of the inter-molecular duplex cleavage sites reside in genes that are expressed in stationary phase, we reasoned that they might be detected in cells grown to stationary phase and repeated the experiment accordingly. In addition, since all inter-molecular duplexes included small RNAs that upon cleavage are shortened, we reasoned that the inclusion of short RNA fragments in the library preparation might improve their detection. Thus, we constructed an additional set of libraries for both exponential and stationary phase RNA, in which the representation of short RNAs was augmented (‘Materials and Methods’ section). For each of these additional types of libraries (stationary phase regular libraries, and stationary and exponential phase libraries including short RNAs), we generated three biological replicates for both the wt and rnc-14 strains (Supplementary Table S1). For each library type, we applied DESeq2 analysis followed by our post-processing procedure (Supplementary Methods). We then repeated the analysis that selects a representative of multiple cleavage sites in a region across the whole ensemble of putative cleavage sites from all libraries, resulting in a final set of 1003 putative RNase III cleavage sites in 615 targets (Supplementary Table S3 and Supplementary Figure S3). All previously detected known cleavage sites were maintained in the final data set. In addition, eight of the known cleavage sites that were not detected or excluded by the post-processing procedure in the initial analysis were identified in the analysis of the additional libraries and appear in the final set. This included four sites from the set of inter-molecular cleavage sites. As before, most of the latter were detected in the targets of ncRNAs and not in the ncRNAs themselves (Table 1 and Supplementary Table S2c).
Classification of RNase III cleavage sites
Each of the 1003 cleavage sites was assigned its genomic annotation based on EcoCyc (27), (http://ecocyc.org) version 20.0 and classified into one of seven major categories of genomic elements (Figure 3A; Supplementary Methods and Supplementary Table S3). We compared the frequencies of the categories in which the cleavage sites resided to the background distribution of genomic elements by a χ2 test and determined the categories that mostly contributed to the χ2 score. The difference between the distributions was statistically significant (p < 2.2E−16). The cleavage sites in intergenic regions within transcripts, in 5′ untranslated regions (UTRs) and in ncRNAs contributed mostly to the χ2 score, indicating that they are preferentially cleaved by RNase III. Surprisingly, a large number of cleavage sites were identified within coding sequences (CDSs), which were thought to be depleted of RNase III cleavage sites due to topological constrains that the coupling between transcription and translation in bacteria might impose (2). A relatively small number of RNase III cleavage sites in CDSs were also observed in S. pyogenes (15). Gordon et al. also reported cleavage sites falling within coding regions, but did not verify them in their small-scale studies (17). In our study, although high, the fraction of sites that resided in CDSs was found to be similar to the genomic background fraction of CDSs. This might suggest that intermediate RNA degradation products become substrates of RNase III, contributing to the overall degradation machinery of the cell.
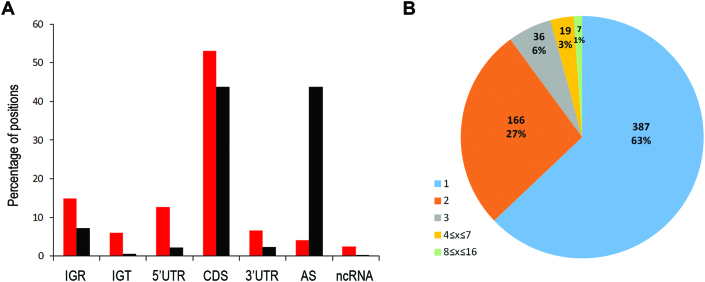
Figure 3.
Classification of RNase III cleavage sites. (A). Distribution of cleavage sites over genomic elements compared to the background distribution. Positions along the genome were classified following EcoCyc (27), (http://ecocyc.org) version 20.0 into seven categories: 5UTR (5′UTR); CDS; 3UTR (3′UTR); AS (unannotated AntiSense); IGR (InterGenic Region); IGT (InterGenic within Transcript) and ncRNA (all annotated non-coding RNAs including small RNA, tRNA and annotated cis antisense). Of note, except for cleavage sites in tRNAs residing in rRNA operons, no cleavage site was annotated as tRNA. Cleavage sites in the vicinity of tRNAs were detected, however, in regions annotated as IGRs. See Supplementary Data for additional annotation details. Red—distribution of genomic elements where RNase III cleavage sites were identified; black—background distribution (counted over all genomic positions). (B) Distribution of number of cleavage sites per target.
We next examined the number of cleavage sites per target (Figure 3B). A total of 387 putative targets (63%) had only a single major cleavage site and 166 putative targets (27%) had exactly two major cleavage sites. These two groups are termed hereinafter SC (Single Cleavage) and DC (Double Cleavage) targets, respectively. A total of 62 targets (10%) had more than two cleavage sites, seven of which seemed to be extensively cleaved, with 8–16 major cleavage sites per target. All the sites but one in this sub-group were detected only in the stationary phase libraries (Supplementary Table S3).
Double cleavage targets
The location of cleavage sites in the putative intra-molecular stem structure supports the classical RNase III cleavage model
The sequential distance between C1 (the cleavage site closest to the target’s 5′ end) and C2 ranged between 16 and 1080 nt with a median of 58 nt. The structural distance was determined by the number of positions (paired or not paired) between C1 and C2 on the predicted stem. The structural distance in the predicted stem–loop structures of the putative targets ranged between −21 and 24 positions, peaking at 2 positions (Figure 4; ‘Materials and Methods’ section). Notably, the structural distance between C1 and C2 determines the length of the dangling ends. It was previously shown that there is heterogeneity of cleavage products leaving 2, 3 and 4 nt dangling ends (2). Consistent with that, we found a variation in the structural distances, with 85, 15 and 3 targets with structural distances of 2, 3 and 4 positions, respectively. Interestingly, while in all the targets with a structural distance of 2 positions the dangling ends of 2 nt were found on both cleavage products, in 80% of the targets with structural distance of 3 positions there was a bulge (mostly upstream to C1), generating asymmetric dangling ends in the two cleavage products, one with 2 nt and the other with 3 nt dangling end.
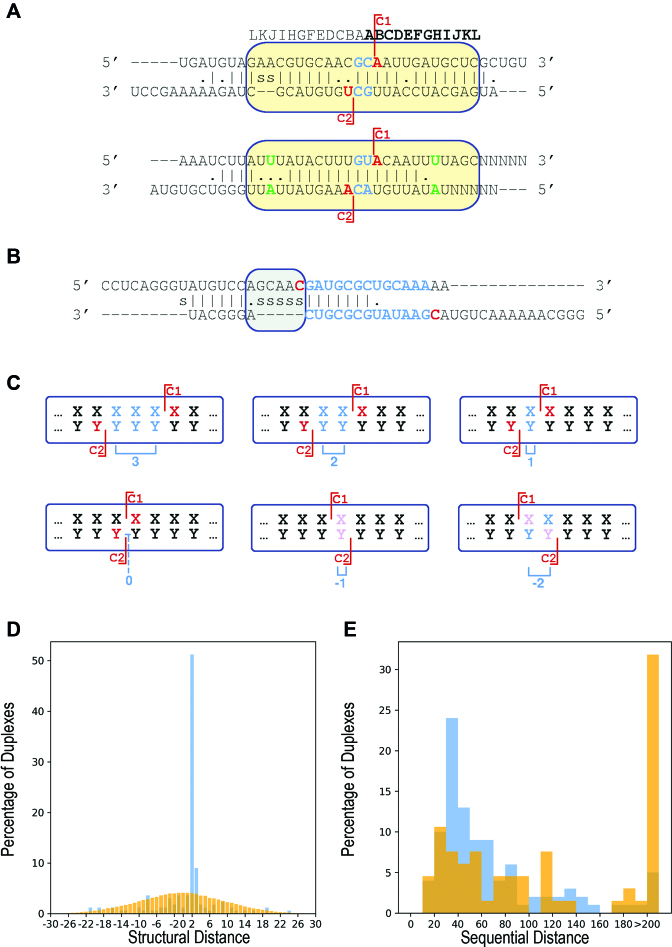
Figure 4.
Structural distance and dangling ends. (A and B). Schematic representations of possible duplexes of double cleavage targets. The letters above the scheme denote the positions relative to the cleavage sites. Each duplex is represented by three lines: upper line and lower line represent the 36-nt sequence fragments flanking C1 5′(20 nt):C1:(15 nt)3′ and C2 5′(15 nt):C2:(20 nt)3′, respectively. Fragments with C1-C2 sequential distance <15 were padded symmetrically by ‘N’. Middle line—the RNAduplex (23) structure prediction for these two regions. ‘|’, ‘.’ and ‘s’ designate base-pair, mismatch and gap (space), respectively. C1 and C2 are colored in red. Positions within the fragments residing out of the RNAduplex predicted structure are heuristically aligned sequentially. Asymmetric loops/bulges are heuristically aligned to the left (e.g. gray rectangle in B). The structural distance (blue) is computed over the duplex positions and is defined as the number of positions in the duplex (paired or not) separating C1 from C2 (see C hereinafter). C1 and C2 can reside outside the major base-paired region in the RNAduplex predicted structure, as in (B). For computing the duplex score, the sequences of all the predicted structures with a structural distance of 2 positions were aligned and the score was computed from the structural region marked with a yellow rectangle in (A). Positions considered to be mismatched by RNAduplex predictions that were counted as paired in our heuristic score computation are colored green. (C) Schematic representation of stem structures obtained by computational folding of the transcript generating structural distances ranging from 3 to −2. Structural distances of 3, 2 and 1 correspond to leaving 3, 2 and 1 nt-long 3′ dangling ends, respectively. Structural distance of 0 yields no dangling end, while the negative structural distances −1 and −2 correspond to leaving 5′ dangling ends of size 1 and 2, respectively. Cleavage sites are designated by red lines and the first nucleotide of each cleaved fragment is colored red. Dangling end nucleotides are colored cyan. Pink colored nucleotides designate positions that are both first in their fragment and are part of a 5′ dangling end corresponding to a negative structural distance. The structural distance is designated in blue below the schemes. Of note, the predicted structural distances yielding non-canonical dangling ends are more likely to reflect separate cleavage events in which only one of the strands is cleaved rather than a dual cleavage event yielding such two products. (D) Structural distance distributions within the stems of 166 double cleavage targets (blue) and in the predicted structures of a set of 100 000 pairs of randomly selected positions in transcripts along the genome (orange). The X-axis ranges between structural distances of −30 to 30 positions, as structural distances beyond these values were negligible. (E) Sequential distance distribution for all double cleavage targets. The double cleavage targets are partitioned to those with canonical structural distances of 2 and 3 positions (blue) and to all those with other structural distances (orange).
We compared the distribution of structural distances to a distribution of structural distances of randomly selected regions and randomly selected C1 and C2 cleavage sites by a χ2 test (‘Materials and Methods’ section). The two distributions differed statistically significantly (p < 2.2E−16), and the main contributors to this difference were the structural distances of 2 and 3 positions (Figure 4D). The predominance of a structural distance of 2 positions is in accord with and in support of the canonical cleavage model. The majority of the remaining double cleavage targets had poor stem predictions with high free energy values, which differed statistically significantly from the free energy values of stems with structural distances of 2 or 3 positions (p ≤ 7.32E−14 by Wilcoxon test). This suggests that although C1 and C2 of these targets reside on the same transcript, they probably do not share a common stem. Interestingly, the sequential distance between C1 and C2 in stems with canonical structural distances of 2 and 3 positions is statistically significantly shorter compared to non-canonical structural distances (Figure 4E, p ≤ 2.17E−04 by Wilcoxon test). Of note, very large sequential distances (up to thousands nt) between two cleavage sites with a structural distance of 2 or 3 positions were observed before (e.g. in 16S RNA and 23S RNA). In our data, however, such large sequential distances between two cleavage sites are infrequent. Taken together, non-canonical structural distances between C1 and C2 along with long C1-C2 sequential distances are highly likely to reflect two cleavage sites that are not formed by the same cleavage event but by two separate cleavage events. These might occur by a different positioning of the stem within RNase III resulting in a single cleavage of only one strand of the duplex. Alternatively, they may result from inclusion of C1 and C2 in other intra- or inter-molecular structures.
The recognition of DC targets by RNase III is highly specific
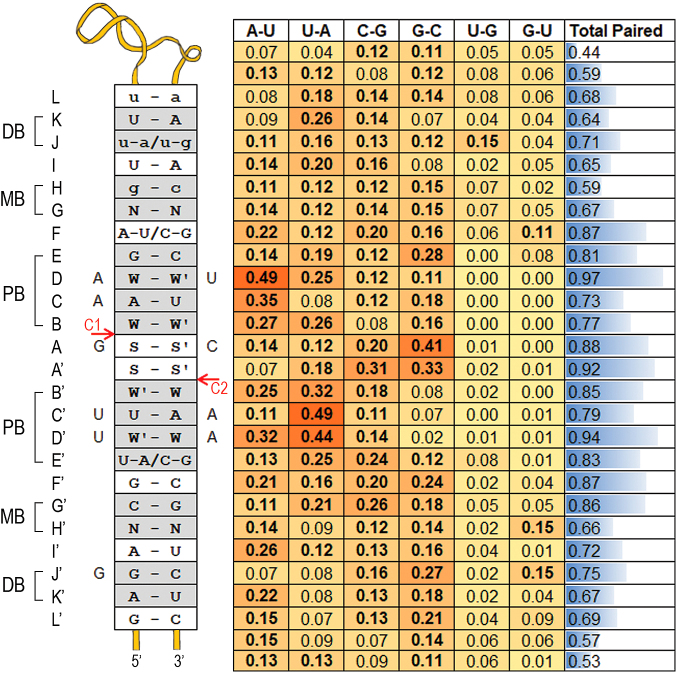
Our results suggest that RNase III cleaves preferentially stem structures in specific positions. How does the enzyme distinguish its targets from other stem structures in the transcriptome? Previous in vitro studies suggested that a specific base-pair pattern in the vicinity of the cleavage sites plays a role in cleavage efficiency (9). Our study allows us to address this question using in vivo cleavage data. To this end, we used the subset of 85 DC targets with a structural distance of 2 positions and aligned their predicted structures using C1 and C2 as anchors (Figure 4). For each structural position along the duplex we counted the number of targets in which this position was paired, and when paired we counted the number of occurrences of the various base pairs (i.e. A-U, U-A, G-C, C-G, G-U and U-G). The observed base-pair preferences (Figure 5) are mostly in accord with the previously reported preferences obtained from in vitro cleavage efficiency measurements of a minimal substrate with various substitutions (9). There is, however, a pronounced difference regarding the two positions between the cleavage sites (positions A and A' in Figure 5). While Pertzev and Nicholson (9) reported them to be mostly unpaired with limited nucleotide preferences, we found that they were predicted to be paired in ∼90% of the targets. We also observed a clear preference for G-C and C-G pairs in these positions.
Figure 5.
Double cleavage structure–sequence motif. All stems of double cleavage targets with a structural distance of 2 positions were aligned using the cleavage positions C1 and C2 as anchors, and base-pair preferences were calculated. For each position, the fraction of targets in which it is paired is presented in a bar format in the rightmost column. The relative frequency of each of the six base pairs A-U; U-A; C-G; G-C; U-G; G-U is presented by a heat map. Note that the frequencies of the six base pairs sum up to the ‘paired’ fraction in the rightmost column. In bold are frequencies of base pairs observed in more than 10% of the targets. The left insert lists the top preferred pairs at each position, provided the pair is found in more than 10% of the targets. Preferred base pairs that are found in <20% of the targets are in lower case. Base pairs were considered of equal probability and are presented separated by a slash if the difference between their relative frequencies was ≤0.02. The C1 and C2 cleavage sites are marked by red arrows. The left and right nucleotides of a pair reside on the ‘C1 strand’ and ‘C2 strand’ of the duplex, respectively. Nucleotides that were present in at least 40% of the duplexes, regardless of their pairing, are marked with a capital letter adjacent to the insert at the matching ‘cleavage strand’ of the duplex. The gray rectangles termed pb, mb and db represent the previously defined proximal (8,9), middle (10) and distal boxes (8,9), respectively. (A-L) and (A'-L') denote the positions relative to the cleavage sites. N: any nucleotide; W: A/U; S: G/C. We use W-W', W'-W and S-S' to designate that the dominant pair was A-U, U-A and G-C, respectively.
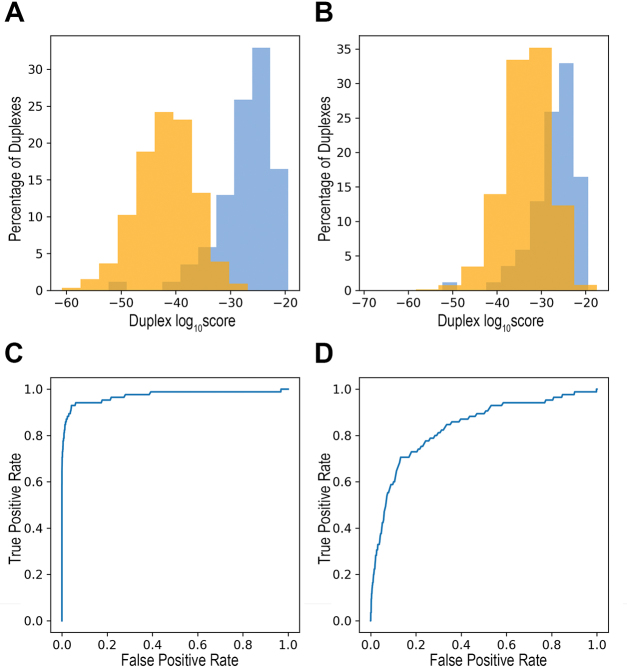
Extending the relative frequencies of base pairs to all 16 possible nucleotide pairs at a stem position (including paired bases, internal loops and bulges), we could define a score for each nucleotide pair at a stem position based on its relative frequency at that position (Figure 5 and Supplementary Table S4). Using these relative frequencies, we computed a score for each duplex with a structural distance of 2 positions, both for the stems in our data of cleavage sites and for random stems (‘Materials and Methods’ section). As shown in Figure 6A, the scores of the double cleavage sites were statistically significantly higher than the scores of the ‘randomly selected stems’ (p < 2.2E−16 by Wilcoxon test). This may be due to either poor stem structures of the random stems and/or incompatibility of their sequences with the nucleotide pair pattern observed in stems of cleavage sites. To specifically evaluate the contribution of the nucleotide pair pattern, we threaded through each of the 85 stems in our data random nucleotides (maintaining nucleotide pairings, bulges and internal loops) and computed their scores (‘Materials and Methods’ section). The distributions of these two sets of targets (Figure 6B) also differed statistically significantly, with the sites of the targets having higher scores (p < 2.2E−16 by Wilcoxon test). These analyses suggest that the base-pairing pattern characterizing RNase III intra-molecular substrates distinguishes them from other RNA stem structures in the E. coli transcriptome (Figure 6C and D), supporting the specificity of RNase III cleavage.
Figure 6.
RNase III cleavage is highly specific. The scores of the 85 double cleavage sites (blue) were statistically significantly higher than: (A) The scores of the 4054 ‘randomly selected stems’ (orange) (p < 2.2E−16 by Wilcoxon test). (B) The scores of 8 500 000 stems generated by maintaining the 85 stem structures and randomly choosing nucleotide pairs, bulges and internal loops (‘randomly threaded stems’, 100 000 times for each stem template, orange) (p < 2.2E−16 by Wilcoxon test). (C and D) ROC curves for the sets in (A) and (B), respectively. Using increasing score thresholds as classifiers, the true positive rate and the false positive rate were recorded. The area under curve is 0.97 and 0.84 for (C) and (D), respectively.
Single cleavage targets
A large fraction of the identified RNase III targets exhibited only a single major cleavage (SC targets). These single cleavages might have originated from intra-molecular stems where the second cleavage did not occur or failed detection, or alternatively they could have taken place in inter-molecular duplexes. Here, we discuss and analyze each of these scenarios.
Putative DC targets mis-annotated as SC targets
Possible failure in detection of one of the cleavage sites of a DC target due to either biological or computational reasons may have resulted in its mis-annotation as SC target. It is possible that the cleavage products were too short to be detected. Alternatively, the mis-identification might have been due to the stringent criteria of our post-processing procedure, or rarely, to mis-annotation of the transcript boundaries. To identify potential DC targets that were misclassified as SC targets, we predicted and scored potential stem structures using the detected single position as anchor (‘Materials and Methods’ section). As a proof of concept, we applied this procedure to a set of 79 out of the 85 putative DC targets, which had structural distance of 2 positions and sequential distance of at most 150 nt. For each DC target the procedure was reciprocally applied, once trying to re-discover C2 as the pair-mate of C1 and once attempting to re-discover C1 as the pair-mate of C2. Indeed, for 71 targets we re-discovered at least one of the cleavage sites, where for 67 targets of those both sites were reciprocally detected. We then applied the same procedure to the SC targets, and identified 46 putative second cleavage positions, six of which were highly indicative of missed DC targets (Supplementary Tables S5 and S6).
Cleavage of inter-molecular duplexes
As RNase III was previously shown to be involved in both cis and trans sense–antisense duplex cleavage [e.g. (16,28)], it is conceivable that at least some of the SC targets originated from sense–antisense duplexes. A total of 41 of the cleavage sites in our data (∼4%) were annotated as ‘AS’ (Figure 3A). A total of 32 of these AS sites (78%) were found in SC targets, and SC targets were statistically significantly enriched with AS annotated sites (p ≤ 2.085E−07 by Fisher Exact test). Moreover, the SC targets were enriched with cleavage sites that overlapped RNase III-regulated sense–antisense regions reported by Lybecker et al. (28) (p ≤ 2.436E−07 by Fisher Exact test). Lybecker et al. developed a high-throughput method for the detection of in vivo functional antisense RNAs by immunoprecipitation of an antibody for double-stranded RNA (28). Conducting their experiment in wt and rnc-14 mutant strains and comparing the results, they identified 316 RNase III-regulated sense–antisense regions. A total of 125 of our putative cleavage sites (spanning 103 different targets) overlapped 87 of the Lybecker et al.’s reported regions. The cleavage sites that overlapped Lybecker et al.’s hybrids were enriched with single cleavage sites (Fisher Exact test p ≤ 7.333E−06).
If both the sense and the antisense transcripts are cleaved by RNase III, we would expect to see cleavage sites residing on opposite strands in close proximity. Thus, we searched in our dataset for two cleavage sites residing on opposite strands that are at most 100 nt apart in their genomic coordinates and detected 91 such sites in 65 targets. Considering all the incidences discussed above, we obtained a total of 118 sites (30%) that could potentially originate from cis sense–antisense duplexes. Our results also suggest that putative RNase III targets made up of sense–antisense duplexes have been usually captured in our experiments by a single cleavage site in only one of the duplex strands.
RNase III cleavage of trans sense–antisense duplexes in E. coli were previously reported (29,30). It is possible that two SC transcripts, encoded at different genomic loci, hybridize to form an inter-molecular duplex that serves as a substrate for RNase III. We attempted to identify such potential duplexes by computationally evaluating the cleavage score of all possible pairs of SC transcript hybrids in our data (‘Materials and Methods’ section). Only seven candidate pairs had a highly reliable score, out of which six overlapped a repetitive extragenic palindrome element (Supplementary Table S7). Although this negligible number could be due to the limitation of our computational analysis, it could also reflect the fact that we truly had a small number of such duplexes in our data, either because they were relatively scarce or because only one of the duplex strands was detected with a cleavage site in our experiment (as observed for the sense–antisense duplexes).
There are ∼100 annotated ncRNAs in EcoCyc (27), (http://ecocyc.org) version 20.0. Putative cleavage sites were detected in 17 of them [a total of 25 cleavage sites, a large fraction of which were identified also by Lybecker et al. (28)]. Interestingly, we detected cleavage sites in genomic elements encoded antisense to 24 ncRNAs. It is of note that 19 of these cleavages were located opposite to the first 15 nucleotides of the ncRNA. Five of the ncRNAs for which we detected a cleavage site in the antisense or in both the ncRNA and the antisense related to type I toxin–antitoxin (TA) systems.
Bacterial TA systems encode a toxin that inhibits cell growth and an antitoxin that prevents the toxin activity [for review, see (31)]. There are various types of antitoxins, where antitoxins of type I are RNA molecules that control the expression levels of the toxin protein by base pairing with the toxin mRNA, affecting its translation and/or stability. RNase III is known to be involved in the degradation of type I TA RNA duplexes [reviewed in (31–34)]. In E. coli MG1655 RNase III cleavage was associated with three of the eight type I TA systems reported in Brantl et al. (32): dinQ/agrB (35), tisB/istR1 (36,37), and ibs/sib (38,39). We detected at least one putative cleavage site in each of the putative duplex regions of each of these three systems, as well as in three additional systems, shoB/OhsC, ldr/Rdl and hok/sok. The latter is a homolog of the R1 plasmid hok/sok TA module, which is known to be cleaved by RNase III (40). The only cleavage site detected in the vicinity of symE/symR resided 165 nt downstream to symR, suggesting it is unrelated to symE/symR, in accord with the observed lack of change in the toxin mRNA level in RNase III mutant (41). No cleavage site was found for ralR/ralA, which is currently the only type I TA system reported to be Hfq dependent (42).
Interestingly, we detected cleavage sites in transcripts related to six other TA systems, where the antitoxin is known to inhibit the toxin by protein–protein interaction: In the transcripts of the antitoxins hipB, ghoS and dinJ of the hipA/hipB, ghoS/ghoT and yafQ/dinJ TA systems, respectively; in an antisense to the antitoxin rnlA of the rnlA/rnlB systems; and in the transcripts of the toxins cptA and yafO of the cptA/sdhE and yafO/yafN systems, respectively. Similar results were reported by Lybecker et al. for type II TA systems (28). It is possible that in addition to the toxin-antitoxin interaction at the protein level, RNase III cleavage may provide an additional layer of regulation at the mRNA level (28).
DISCUSSION
Escherichia coli RNase III was shown to cleave various dsRNA substrates in vitro, including long dsRNA (7) as well as a minimal substrate [e.g. (9)]. In vivo it was shown to cleave both intra-molecular stem structures and cis or trans inter-molecular duplexes (Supplementary Table S2 and references therein). The cleavage can specifically affect the target’s maturation, stability and/or translation (2,16). This entails that RNase III cleavage in vivo should be specific in both the selected targets and the selected cleavage sites within them. Indeed, extensive studies of E. coli RNase III, involving various in vitro biochemical and structural methods, revealed positive and negative determinants that affect substrate binding and/or cleavage [e.g. (9)]. In the current study, we exploited RNA-seq-based technologies to further explore the RNase III in vivo cleavage rules and to provide the global target repertoire of the enzyme in E. coli.
Our methodology specifically identifies RNase III-dependent 5′P transcripts. This is achieved by a strict comparison of the wt and rnc-14 mapped reads, substantiating the confidence of the identified cleavage sites. As any large-scale methodology, it is prone to errors, which may result in both false positive and false negative identifications. Non-technical false positives may occur as there are other cellular enzymes that can generate 5′P RNA fragments (e.g. RppH, RNase E, RNase G and RNase P). Indirect RNase III regulation of the targets or the activity of these other enzymes may result in down-regulation of a gene in the mutant leading to mis-identification of 5′P read starts as direct RNase III cleavage sites. The low number (<5%) of RNase III cleavage sites in our data that overlap (distance ≤5 nt) reported RNase E cleavage sites (12) suggest that at least for RNase E, this is not a major pitfall. False negatives (sites we failed to detect) may occur due to several reasons: (i) The stringent criteria and filters we applied in the computational processing of the data. (ii) Failing of the ligation between the RNA ends and the adapters, resulting from the low efficiency of the ligation reaction, as well as from a ligation bias against structured RNA ends. (iii) Low expression levels of the targets in our experiments. (iv) Cleavage products that are too short to be represented in our libraries. Shorter cleavage products might underlie the slightly worse identification we observed for C1 compared to C2 sites in intra-molecular double cleavage substrates, as well as the difficulty in the detection of cleaved short RNAs in the inter-molecular duplex targets. (v) Cleavage compensation for RNase III in the rnc-14 strain by another enzyme cleaving at the exact same cleavage position as RNase III. This scenario, however, is not likely to happen often as RNase III cleaves double strands and most other enzymes cleave single strands. Notwithstanding the above reservations, the good agreement between previously reported RNase III cleavage sites in the data we compiled and our putative cleavage sites support our data. Furthermore, our data is also supported by recently reported cleavage sites detected in vitro by Gordon et al. (17). Gordon et al. carried out a transcriptome-wide screen for RNase III cleavage sites, comparing expression levels between wt and RNase III mutant strains. They applied in vitro cleavage assays to 18 of their top candidates and found that 11 targets were cleaved and seven were not. They further studied 10 of these 11 putative targets by 5′ RACE and found a total of 15 cleavage sites, detecting at least one cleavage site in each transcript. Since we carried out our cleavage site mapping independently of Gordon et al.’s study, and as all their candidate but one (proP) were not included in our compiled set of known targets, we could use their remainder 17 candidates, (comprising 10 of their 11 targets and 14 of their 15 cleavage sites), as an in vitro test set for our methodology. We detected at least one cleavage site in nine of the 10 cleaved transcripts and identified 10 of the 14 cleavage sites (at a distance of at most 5 nt). Moreover, no cleavage site was detected by our pipeline in the seven transcripts reported by Gordon et al. not to be cleaved. Interestingly, in three out of these seven transcripts we did find a cleavage site that mapped to the corresponding gene, but in a region that was not included in their tested synthetic RNA (Supplementary Methods). Taken together, the support of our data by results of small-scale experiments as well as the consistency of many of the conclusions obtained by our global approach with those from small-scale experiments reinforce the insights gained from our data.
We found that 27% of the identified targets contained two major cleavage sites, ∼50% of which (85 targets) had a structural distance of 2 positions between the two cleavage sites. While it is possible that the two cleavages, identified in the large scale data, originated from two identical RNA molecules each cleaved at only one of its strands, it is much more likely that these targets obey and support the RNase III canonical cleavage mechanism of a double-stranded substrate that is independently cleaved in each strand, with an offset of 2 nt between the two cleavage sites. Alignment of the predicted stems of the 85 targets enabled the inference of in vivo nucleotide pair preferences that contribute to the specificity of RNase III cleavage, as random stems with a structural distance of 2 positions between the randomly determined cleavage sites hardly showed these patterns (Figures 5 and 6; Supplementary Table S4).
In general, there is a good agreement between the in vivo derived base-pair preferences (Figure 5) and the ones obtained by in vitro biochemical experiments on a minimal substrate (9). The most pronounced difference regarded the two stem positions between the two cleavage sites (A and A' in Figure 5). While these positions did not seem to require base pairing or specific nucleotides for the in vitro cleavage of a minimal substrate, they were paired in ∼90% of the putative stems cleaved in vivo, with a strong preference for G-C and C-G pairs (Figure 5). Interestingly, nucleotides at these positions were shown to form hydrogen bonds with RNase III positions D44 in a crystal structure of RNase III from Aquifex aeolicus (43). D44 is one of the most highly conserved residues of RNase III, which interacts with the RNA substrate and is involved in the definition of the scissile bond and in the hydrolysis catalysis. D44 of one subunit, together with the similarly conserved H27 of the second subunit, define the typical cleavage distance of 11 nt on the respective RNA strand. The size determination, however, is attributed mainly to H27. H27 forms a hydrogen bond with the respective strand at position J/J', where we observed a preference for G. Interestingly, the preference for G in the positions involved in hydrogen bonds with H27 and D44 is more pronounced for the nucleotides upstream to C1, where G is observed at the H27 and D44 interacting positions in 40 (50%) and 36 (40%) of the structures, respectively, compared to 29 (34%) of the corresponding positions upstream to C2. Another highly conserved position of RNase III, Q157, forms a hydrogen bond with the nucleotide at the duplex D/D' positions. These positions were shown to be important for binding of RNase III, where G-C and C-G disrupt the binding (9). Not only we identified a very strong preference for A-U/U-A in these positions, but they also were the most base-paired positions, in 97% (D) and 95% (D') of the targets. Thus, our results further refine and expand the determinants of E. coli RNase III specificity in vivo.
The Mini-III RNase is the smallest member of the RNase III superfamily and is unique in lacking the dsRNA-binding domain. The analysis of the cleavage specificity of the Bacilllus subtillis Mini-III RNase (BsminiIII) revealed sequence-specific cleavage of a long dsRNA (44). The detected motif ACC∧U in the vicinity of the cleavage site (marked with ∧) is strikingly similar to the WSSW preferences we found at positions B’A'AB (Figure 5), further supporting our results.
Single cleavage targets comprised 63% of the putative RNase III targets and 39% of all cleavage sites in our data. A single cleavage could originate from an intra-molecular stem for which we either failed to identify the pair-mate cleavage site or the stem had a non-optimal structure resulting in a single-strand nick (2). Alternatively, a single cleavage could originate from either a cis or trans inter-molecular duplex. While for a fraction of the single cleavage sites we could suggest one of these scenarios, we could not fully assess their relative incidence in our data. This limitation is probably due to the variability in the sizes of the RNase III cleavage products, affecting their detection.
The set of putative cis inter-molecular duplex targets included most of the type I TA systems, supporting and expanding the repertoire of type I TA systems cleaved by RNase III (32). In addition, this set of targets included transcripts transcribed antisense to small RNAs (sRNAs). sRNAs are usually considered trans regulators, and our findings, which are in accord with Lybecker et al. (28), suggest that quite a few of them might be involved in functional cis interactions as well. In support of both cis and trans modes of regulation by the same sRNA, we recently reported that ArrS, known to regulate gadE in cis, acts also as a trans regulator through binding to Hfq (45). Furthermore, it was previously reported that sRNAs may down-regulate their targets by base pairing with the mRNA and generating a double strand target for RNase III [e.g. (46)]. The novel sense–antisense inter-molecular duplexes involving sRNAs identified here might shed light on a similar mechanism employed by the sRNAs in cis. The sRNA–RNase III juncture may also have other implications, in the processing of the regulatory RNA to its mature form, as was previously shown for DicF, (47), or in degradation of the regulatory RNA, as suggested for MicA in Salmonella (48).
RNase III protein level and activity have been commonly considered to be reduced in stationary phase (49). We also observed a modest decrease in the steady state levels of the rnc transcript in wt strain between exponential and stationary phases in RNA-seq analyses for the same RNA extractions used for the construction of the 5′P libraries (data not shown). Still, 521 (52%) of the cleavage sites were detected in the stationary phase experiments, 359 of which were detected solely in the stationary phase libraries, including most of the sites involving ncRNAs (Supplementary Figure S3 and Supplementary Table S3). Thus, judging by our data it seems that RNase III does play a role in stationary phase.
RNase III substrate binding does not always result in the cleavage of the substrate [e.g. (50), and reviewed in (2,7)]. It was previously shown that a structural element within the substrate can act as a ‘catalytic’ antideterminant, uncoupling the enzyme binding and catalytic activities (51). Accompanying the method described here by immunoprecipitation-based approaches for the identification of all RNase III bound targets will enable distinguishing between cleaved and non-cleaved targets. Those can be studied, in turn, for sequence and structure properties that enable or prevent cleavage.
Our study provides a rich resource of RNase III target candidates (Supplementary Table S3), which can serve as the basis for further studies and analyses. For example, analyzing all identified RNase III targets overlapping annotated genes in E. coli, we found that usually they are non-operonic, but when operonic, they tend to be the first gene in the operon. Furthermore, for several of the operons preceded by leader peptide genes we observed a cleavage in the leader peptide regions, in addition to cleavage in the first gene of the operon. These included trpL, pheL, pheM and hisL [the latter two were also reported by Gordon et al. (17)]. Interestingly, all these operons are related to translation, as are many other identified RNase III targets in our data, such as ribosomal proteins and elongation factors. Intriguingly, while rRNAs were among the first detected targets of RNase III, our study identified not only the rRNAs themselves as regulated by RNase III, but also some of their modifiers (e.g. 23S methyltransferase and 23S pseudouridine synthase). It is of note that in a global functional analysis of the targets, no functional category was found to be statistically significantly enriched after correction for multiple hypothesis testing. However, when ranking the categories by the number of putative RNase III targets they include, the ‘modifying enzyme’ category ranked among the highest. Out of the 80 genes annotated in EcoCyc as ‘RNA modifiers’ 18 (22%) were detected as RNase III cleavage targets, hinting at putative feed-forward loops involving RNase III, the gene encoding the modifying enzyme and the rRNA. These examples emphasize the value of the resource we provide, setting the ground toward the future challenges in RNase III study: Unraveling the specific regulation mechanism applied to each target; deciphering the functional implications of RNase III binding and cleavage on the targets, ranging from cleavage with little effect to strong effects on target stability and/or translation; integration of the RNase III regulation information into the post-transcriptional regulation network in E. coli. Addressing these goals is expected to expand our understanding of the involvement of RNase III in post-transcriptional regulation in the bacterial cell.
DATA AVAILABILITY
The sequencing data reported in this paper was deposited to ArrayExpress, accession E-MTAB-6704. Link: https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6704.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Peer, N. Grover and N. Friedman for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Israel Science Foundation administered by the Israeli Academy for Sciences and Humanities [876/17]; I-CORE Program of the Planning and Budgeting Committee and The Israel Science Foundation [1796/12]; European Research Council Advanced Grant [322920]. Funding for open access charge: Israel Science Foundation administered by the Israeli Academy for Sciences and Humanities.
Conflict of interest statement. None declared.
REFERENCES
- 1. MacRae I.J., Doudna J.A.. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr. Opin. Struct. Biol. 2007; 17:138–145. [DOI] [PubMed] [Google Scholar]
- 2. Court D.L., Gan J., Liang Y.H., Shaw G.X., Tropea J.E., Costantino N., Waugh D.S., Ji X.. RNase III: genetics and function; structure and mechanism. Annu. Rev. Genet. 2013; 47:405–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dunn J.J., Studier F.W.. T7 early RNAs and Escherichia coli ribosomal RNAs are cut from large precursor RNAs in vivo by ribonuclease 3. Proc. Natl. Acad. Sci. U.S.A. 1973; 70:3296–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gitelman D.R., Apirion D.. The synthesis of some proteins is affected in RNA processing mutants of Escherichia coli. Biochem. Biophys. Res. Commun. 1980; 96:1063–1070. [DOI] [PubMed] [Google Scholar]
- 5. Stead M.B., Marshburn S., Mohanty B.K., Mitra J., Pena Castillo L., Ray D., van Bakel H., Hughes T.R., Kushner S.R.. Analysis of Escherichia coli RNase E and RNase III activity in vivo using tiling microarrays. Nucleic Acids Res. 2011; 39:3188–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sim S.H., Yeom J.H., Shin C., Song W.S., Shin E., Kim H.M., Cha C.J., Han S.H., Ha N.C., Kim S.W. et al. . Escherichia coli ribonuclease III activity is downregulated by osmotic stress: consequences for the degradation of bdm mRNA in biofilm formation. Mol. Microbiol. 2010; 75:413–425. [DOI] [PubMed] [Google Scholar]
- 7. Nicholson A.W. Ribonuclease III mechanisms of double-stranded RNA cleavage. Wiley Interdiscip. Rev. RNA. 2014; 5:31–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang K., Nicholson A.W.. Regulation of ribonuclease III processing by double-helical sequence antideterminants. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:13437–13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pertzev A.V., Nicholson A.W.. Characterization of RNA sequence determinants and antideterminants of processing reactivity for a minimal substrate of Escherichia coli ribonuclease III. Nucleic Acids Res. 2006; 34:3708–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gan J., Tropea J.E., Austin B.P., Court D.L., Waugh D.S., Ji X.. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006; 124:355–366. [DOI] [PubMed] [Google Scholar]
- 11. Schifano J.M., Vvedenskaya I.O., Knoblauch J.G., Ouyang M., Nickels B.E., Woychik N.A.. An RNA-seq method for defining endoribonuclease cleavage specificity identifies dual rRNA substrates for toxin MazF-mt3. Nat. Commun. 2014; 5:3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clarke J.E., Kime L., Romero A.D., McDowall K.J.. Direct entry by RNase E is a major pathway for the degradation and processing of RNA in Escherichia coli. Nucleic Acids Res. 2014; 42:11733–11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chao Y., Li L., Girodat D., Forstner K.U., Said N., Corcoran C., Smiga M., Papenfort K., Reinhardt R., Wieden H.J. et al. . In vivo cleavage map illuminates the central role of RNase E in coding and non-coding RNA pathways. Mol. Cell. 2017; 65:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. DiChiara J.M., Liu B., Figaro S., Condon C., Bechhofer D.H.. Mapping of internal monophosphate 5′ ends of Bacillus subtilis messenger RNAs and ribosomal RNAs in wild-type and ribonuclease-mutant strains. Nucleic Acids Res. 2016; 44:3373–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Le Rhun A., Lecrivain A.L., Reimegard J., Proux-Wera E., Broglia L., Della Beffa C., Charpentier E.. Identification of endoribonuclease specific cleavage positions reveals novel targets of RNase III in Streptococcus pyogenes. Nucleic Acids Res. 2017; 45:2329–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lioliou E., Sharma C.M., Caldelari I., Helfer A.C., Fechter P., Vandenesch F., Vogel J., Romby P.. Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet. 2012; 8:e1002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gordon G.C., Cameron J.C., Pfleger B.F.. RNA sequencing identifies new RNase III cleavage sites in Escherichia coli and reveals increased regulation of mRNA. MBio. 2017; 8:e00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takiff H.E., Chen S.M., Court D.L.. Genetic analysis of the rnc operon of Escherichia coli. J. Bacteriol. 1989; 171:2581–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takiff H.E., Baker T., Copeland T., Chen S.M., Court D.L.. Locating essential Escherichia coli genes by using mini-Tn10 transposons: the pdxJ operon. J. Bacteriol. 1992; 174:1544–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011; 17:10–12. [Google Scholar]
- 21. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lorenz R., Bernhart S.H., Honer Zu, Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garza-Sanchez F., Schaub R.E., Janssen B.D., Hayes C.S.. tmRNA regulates synthesis of the ArfA ribosome rescue factor. Mol. Microbiol. 2011; 80:1204–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aiso T., Kamiya S., Yonezawa H., Gamou S.. Overexpression of an antisense RNA, ArrS, increases the acid resistance of Escherichia coli. Microbiology. 2014; 160:954–961. [DOI] [PubMed] [Google Scholar]
- 26. Lim B., Sim S.H., Sim M., Kim K., Jeon C.O., Lee Y., Ha N.C., Lee K.. RNase III controls the degradation of corA mRNA in Escherichia coli. J. Bacteriol. 2012; 194:2214–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Keseler I.M., Mackie A., Peralta-Gil M., Santos-Zavaleta A., Gama-Castro S., Bonavides-Martinez C., Fulcher C., Huerta A.M., Kothari A., Krummenacker M. et al. . EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 2013; 41:D605–D612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lybecker M., Zimmermann B., Bilusic I., Tukhtubaeva N., Schroeder R.. The double-stranded transcriptome of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:3134–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vecerek B., Beich-Frandsen M., Resch A., Blasi U.. Translational activation of rpoS mRNA by the non-coding RNA DsrA and Hfq does not require ribosome binding. Nucleic Acids Res. 2010; 38:1284–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Afonyushkin T., Vecerek B., Moll I., Blasi U., Kaberdin V.R.. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 2005; 33:1678–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harms A., Brodersen D.E., Mitarai N., Gerdes K.. Toxins, targets, and Triggers: An overview of Toxin-Antitoxin biology. Mol. Cell. 2018; 70:768–784. [DOI] [PubMed] [Google Scholar]
- 32. Brantl S., Jahn N.. sRNAs in bacterial type I and type III toxin-antitoxin systems. FEMS Microbiol. Rev. 2015; 39:413–427. [DOI] [PubMed] [Google Scholar]
- 33. Unterholzner S.J., Poppenberger B., Rozhon W.. Toxin-antitoxin systems: Biology, identification, and application. Mob. Genet. Elem. 2013; 3:e26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fozo E.M., Hemm M.R., Storz G.. Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol. Rev. 2008; 72:579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kristiansen K.I., Weel-Sneve R., Booth J.A., Bjoras M.. Mutually exclusive RNA secondary structures regulate translation initiation of DinQ in Escherichia coli. RNA. 2016; 22:1739–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vogel J., Argaman L., Wagner E.G., Altuvia S.. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 2004; 14:2271–2276. [DOI] [PubMed] [Google Scholar]
- 37. Darfeuille F., Unoson C., Vogel J., Wagner E.G.. An antisense RNA inhibits translation by competing with standby ribosomes. Mol. Cell. 2007; 26:381–392. [DOI] [PubMed] [Google Scholar]
- 38. Fozo E.M., Kawano M., Fontaine F., Kaya Y., Mendieta K.S., Jones K.L., Ocampo A., Rudd K.E., Storz G.. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 2008; 70:1076–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fozo E.M. New type I toxin-antitoxin families from ‘wild’ and laboratory strains of E. coli: Ibs-Sib, ShoB-OhsC and Zor-Orz. RNA Biol. 2012; 9:1504–1512. [DOI] [PubMed] [Google Scholar]
- 40. Franch T., Thisted T., Gerdes K.. Ribonuclease III processing of coaxially stacked RNA helices. J. Biol. Chem. 1999; 274:26572–26578. [DOI] [PubMed] [Google Scholar]
- 41. Kawano M. Divergently overlapping cis-encoded antisense RNA regulating toxin-antitoxin systems from E. coli: hok/sok, ldr/rdl, symE/symR. RNA Biol. 2012; 9:1520–1527. [DOI] [PubMed] [Google Scholar]
- 42. Guo Y., Quiroga C., Chen Q., McAnulty M.J., Benedik M.J., Wood T.K., Wang X.. RalR (a DNase) and RalA (a small RNA) form a type I toxin-antitoxin system in Escherichia coli. Nucleic Acids Res. 2014; 42:6448–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gan J., Shaw G., Tropea J.E., Waugh D.S., Court D.L., Ji X.. A stepwise model for double-stranded RNA processing by ribonuclease III. Mol. Microbiol. 2008; 67:143–154. [DOI] [PubMed] [Google Scholar]
- 44. Glow D., Pianka D., Sulej A.A., Kozlowski L.P., Czarnecka J., Chojnowski G., Skowronek K.J., Bujnicki J.M.. Sequence-specific cleavage of dsRNA by Mini-III RNase. Nucleic Acids Res. 2015; 43:2864–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Melamed S., Peer A., Faigenbaum-Romm R., Gatt Y.E., Reiss N., Bar A., Altuvia Y., Argaman L., Margalit H.. Global Mapping of Small RNA-Target Interactions in Bacteria. Mol. Cell. 2016; 63:884–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Boisset S., Geissmann T., Huntzinger E., Fechter P., Bendridi N., Possedko M., Chevalier C., Helfer A.C., Benito Y., Jacquier A. et al. . Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007; 21:1353–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Faubladier M., Cam K., Bouche J.P.. Escherichia coli cell division inhibitor DicF-RNA of the dicB operon. Evidence for its generation in vivo by transcription termination and by RNase III and RNase E-dependent processing. J. Mol. Biol. 1990; 212:461–471. [DOI] [PubMed] [Google Scholar]
- 48. Viegas S.C., Silva I.J., Saramago M., Domingues S., Arraiano C.M.. Regulation of the small regulatory RNA MicA by ribonuclease III: a target-dependent pathway. Nucleic Acids Res. 2011; 39:2918–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim K.S., Manasherob R., Cohen S.N.. YmdB: a stress-responsive ribonuclease-binding regulator of E. coli RNase III activity. Genes Dev. 2008; 22:3497–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Altuvia S., Locker-Giladi H., Koby S., Ben-Nun O., Oppenheim A.B.. RNase III stimulates the translation of the cIII gene of bacteriophage lambda. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:6511–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Calin-Jageman I., Nicholson A.W.. RNA structure-dependent uncoupling of substrate recognition and cleavage by Escherichia coli ribonuclease III. Nucleic Acids Res. 2003; 31:2381–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data reported in this paper was deposited to ArrayExpress, accession E-MTAB-6704. Link: https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6704.