Abstract
Fine regulation of the phosphatase and tensin homologue (PTEN) phosphatase dosage is critical for homeostasis and tumour suppression. The 3′-untranslated region (3′-UTR) of Pten mRNA was extensively linked to post-transcriptional regulation by microRNAs (miRNAs). In spite of this critical regulatory role, alternative 3′-UTRs of Pten have not been systematically characterized. Here, we reveal an important diversity of Pten mRNA isoforms generated by alternative polyadenylation sites. Several 3′-UTRs are co-expressed and their relative expression is dynamically regulated. In spite of encoding multiple validated miRNA-binding sites, longer isoforms are largely refractory to miRNA-mediated silencing, are more stable and contribute to the bulk of PTEN protein and signalling functions. Taken together, our results warrant a mechanistic re-interpretation of the post-transcriptional mechanisms involving Pten mRNAs and raise concerns on how miRNA-binding sites are being validated.
INTRODUCTION
Through its dual protein and lipid phosphatase activity, phosphatase and tensin homolog (PTEN) negatively regulates Phosphoinositide 3-kinase (PI3K)/Protein kinase B (Akt) signalling. In doing so, PTEN antagonizes cell proliferation, growth, motility and invasion, senescence and resistance to programmed cell death (1–3). The PTEN gene is finely regulated under physiological conditions by transcriptional and post-transcriptional mechanisms, and is amongst the most frequently mutated genes in cancer (4,5). Even modest changes in PTEN activity have measurable impact on tumorigenesis and tumour progression in mouse models (6–10). In prostate, lung, breast and colon cancer, loss of a single copy of PTEN leads to detectable oncogenic consequences. The remaining copy is typically retained until the development of more aggressive metastatic stages (11,12). Recent evidence suggests a major role for microRNAs (miRNAs) in the post-transcriptional regulation of Pten mRNA (3,13–15). The ∼20-nt-long miRNAs direct the miRNA-induced silencing complex (miRISC) towards mRNA target sites in 3′-untranslated regions (3′-UTRs) of mRNAs, leading to their translational repression, deadenylation and decay (16). Several miRNAs control Pten mRNA (13), and a miRNA–mRNA network involving Pten mRNA at its centre has recently been described (15). Some miRNAs directly target Pten mRNA via several sites in its known 3′-UTR. Targeting of Pten mRNA by miR-26 and miR-29 families, and miR-19a/b, miR-17, miR-20 and miR-92 miRNAs derived from the miR-17∼92 oncogenic polycistron, has been functionally validated in multiple cancer cell types and/or mouse genetic models (17–22). Consistently with an oncogenic role, these miRNAs are often overexpressed and genomically amplified in human tumours (14,23). Moreover, binding of these miRNAs to other mRNA targets was proposed to result in cross-regulation of the Pten mRNA through competition (24).
The output of miRNA-mediated silencing varies widely between mRNA targets and with cell types, but the mechanistic reasons for this are not fully understood. Recent evidence suggests that differences in miRNA-mediated silencing efficacy may in part be due to an under-appreciated diversity in 3′-UTR isoforms (25,26). Landmark papers revealed that a majority of human genes encode more than one 3′-UTR isoform (27–29). This diversity is established mainly via the use of alternative polyadenylation (APA) signals and to a lesser extent by alternative splicing (27). The 3-′UTR isoform expression is dynamic; shorter 3′-UTRs often prevail in proliferating and tumorous cells, whereas longer isoforms are more frequent in differentiated, non-dividing cells (28,30–32). In addition to miRNA-binding sites, 3′-UTR sequences encode other regulatory sequences and structures including binding sites for RNA-binding proteins (RBPs) that affect translation, mRNA stability and localization (33–36). Alternative 3′-UTR isoforms can thus drastically alter the impact of miRNAs, RBPs and structures on gene expression (37–39). The role of alternative 3′-UTR isoforms on the expression and functions of PTEN has not been systematically examined.
Here, we characterize the dynamic diversity of Pten 3′-UTR isoforms and reveal that APAs profoundly regulate PTEN protein dosage. Unexpectedly, long isoforms are mostly refractory to miRNA-mediated silencing. They are more stable and are responsible for the bulk of PTEN protein dosage and signalling functions. We propose that 3′-UTR structures inhibit miRNA-mediated silencing on long Pten 3-′UTRs and that regulatory elements may be selectively activated in specific physiological contexts.
MATERIALS AND METHODS
Cell culture
NIH3T3 (ATCC) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% calf serum (Life). For Figure 1C, Mouse Embryonic Fibroblasts (MEF)s were isolated from mouse mammary glands and placed in culture using methods based on (40). NMuMG cells were cultured in DMEM supplemented with 10% foetal bovine serum (FBS) (Wisent) and 5 μg/ml insulin. T47D cells and HEK 293T cells were cultured in DMEM supplemented with 10% FBS. 22RV1 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% FBS. CaP2 (Pten -/-) and P2 (Pten -/+) mouse prostate cancer epithelial cell lines (41) (kindly provided by Dr Hong Wu, UCLA, USA) were cultured in DMEM supplemented with 10% FBS (Wisent), 25 μg/ml bovine pituitary extract, 6 ng/ml Epidermal Growth Factor (EGF) and 5 μg/ml insulin. All cell lines were incubated in a humidified environment kept at 37°C with 5% CO2. For cell confluency experiments with NIH3T3 cells, cells were plated on 10 cm dishes at different densities and all cells were collected 1 day later for RNA and protein extractions.
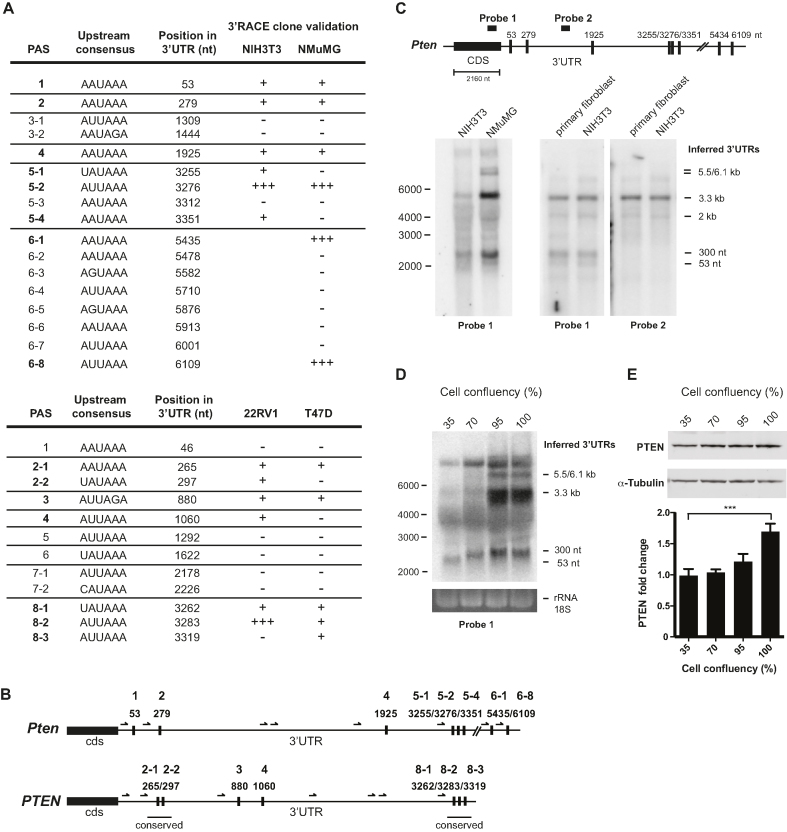
Figure 1.
Alternative polyadenylation signals (PAS) generate an important diversity of Pten mRNA isoforms. (A) Detailed analysis of the 3′RACE clone validation for each putative consensus in mouse (top) and human (bottom) cell lines. + or - indicates if the consensus PAS is validated or not. A majority of clones using a specific consensus for a given position is indicated with +++. (B) Schematic representation of Pten 3′-UTR in mouse (top) and human (bottom) showing the position of validated PAS. Arrows indicate the position of the primers that were used for the 3′RACE assay. (C) Analysis of 3′-UTR isoforms detected by northern blot in murine cell lines. Schematic on top indicates the position of the two probes used for the northern blot. Probe 1 is localized in the CDS, while probe 2 is in the 3′-UTR. Inferred 3′-UTRs matching validated PAS are indicated on the right of the blots, and RNA ladder is indicated on the left. (D and E) The effect of cell confluence on Pten 3′-UTR isoforms, and PTEN protein level was analysed by northern (D) and western (E) blots. RNA size markers are indicated on the left, and inferred 3′-UTR sizes on the right. Western blot quantification was performed from three independent biological replicate experiments using the Odyssey imaging system (LI-COR), and α-Tubulin was used as a loading control. One-way ANOVA analysis was used to calculate P-values (***P ≤ 0.001).
Rapid amplification of 3′ cDNA ends (3′RACE)
The rapid amplification of 3′ cDNA ends (3′RACE) protocol was adapted from (42). The cDNA was prepared using 5 μg of total RNA and an oligo-dT12-anchor primer (Tdo679) using Superscript III reverse transcriptase (Life). The first round of amplification was performed with a gene-specific forward primer and the reverse primer matching the anchor sequence (Tdo680). Conditions for amplification were 98°C/3 min, 64°C/2 min, 72°C/20 min, [94°C/10 s, 64°C/15 s and 72°C/1 min] × 29 cycles, 72°C/5 min. Nested polymerase chain reaction (PCR) was done with a second gene-specific forward primer and the same reverse primer. Conditions for the second amplification were 98°C/3 min, [98°C/10 s, 64°C/15 s and 72°C/5 min] × 30 cycles, 72°C/5 min. Phusion polymerase (NEB) was used for both rounds of amplification. The final PCR products were cloned (StrataClone PCR kit, Agilent Technologies) and sequenced. Sequences with polyadenylation tails longer than 12 nt were considered positives. Sequences of all primers used for this assay are listed in Supplementary Data (Supplementary Tables S3 and S4).
RNA preparation and northern blot
Total RNA from mammalian cells was prepared using the miRNeasy mini kit (Qiagen). One to five micrograms of total RNA or polyA purified RNA (Oligotex, Qiagen) was electrophoresed on a 1% agarose–glyoxal gel, prepared with NorthernMax-Gly gel prep running buffer (Ambion) and transferred to nylon membranes (Hybond XL, GE). Probes were synthesized using a random primer DNA-labelling system (Ambion) with α-32P dATP or T7 MegaScript kit (Ambion) with α-32P UTP. Hybridization was performed at 42 or 68°C overnight in ULTRAHyb hybridization buffer (Ambion) depending on the type of probe used. Following hybridization, membranes were washed, exposed onto an imaging plate and developed using a Typhoon phosphorimager (GE).
3′-UTR isoform specific qRT-PCR
The cDNA for 3′-UTR isoform-specific quantitative reverse transcription polymerase chain reaction (qRT-PCR) (300 or 3.3; see Supplementary Tables and Figures S3 and S4) was prepared using 500 ng of total RNA and an oligo-dT12-anchor primer (Tdo679) using Superscript III reverse transcriptase (Life). Each reverse transcription (RT) reaction was diluted 1:4, and the first round of amplification (PCR1) was performed in the standard PCR machine with the following amplification conditions: 98°C/3 min, [98°C/10 s, 64°C/15 s and 72°C/20 s] × 25 cycles, 72°C/5 min. PCR1 was diluted 1:1000, and a second round of amplification (qPCR2) was performed in the real-time machine using IQ SYBR green supermix (BioRad). qPCR that amplifies specifically the Pten 5.5/6.1 kb isoforms, Pten long, Pten total and the control gene Hprt were performed directly on the RT reactions. Relative differences in RNA levels were calculated with the 2−ΔΔCt method and normalized with Hprt. All primers used for these PCRs are listed in Supplementary Table S6.
miRNA-specific qRT-PCR
For the analysis of miRNAs by qRT-PCR, reverse transcription was performed on 500 ng of totalRNA using miScript II RT kit (Qiagen). RT reactions were diluted 1:40 and qPCR wasperformed using miScript SYBR Green PCR kit (Qiagen) with miScript Primer Assays (Qiagen) specific for miR-92 (MS00005971), miR-19b (MS00005915), miR-20a (MS00001309) and miR-26b (MS00003234). Relative differences in miRNA levels were calculated with the 2−ΔΔCt method and normalized with SNORD25.
Reporter plasmid constructs
The Pten 3.3 kb 3′-UTR was used to generate all Pten isoform-specific 3′-UTR reporters cloned in pmiRGLO vector (Promega). We note that a previous report suggested that different promoter strengths may modulate miRNA output on reporters (43). We chose PGK to drive reporter expression under the mild PGK promoter and to favour an advantageous stoichiometry for miRNA-mediated silencing (44). To mutate each consensus polyadenylation signal (PAS) in the 3′-UTRand produce several reporters individuallyexpressing one of the different Pten3′-UTR isoforms, mini genes were synthesized(GenScript) and subcloned into the Pten3.3 kb 3′-UTR. Each nucleotide at positions2 and 4 of a functional consensus was mutatedto a cytosine (C), except for the consensus at3255 nt, where a small deletion wasintroduced that covers all PAS consensus. Control reporter constructs containing miRNA binding sites (3X-Bulge and the mutated 3X-Bulge) were cloned as described in (44). Mutation of miR-29 binding sites on Pten 3′-UTR (WT) was done by all-around PCR with non-overlapping primers. Nucleotides corresponding to positions 2, 3 and 4 on the miRNA seed were mutated for each binding site on the 3′-UTR of the WT reporter. The following pairs of primers were used for site 1 and 2 mutation. Site 1: (Tdo2572) 5′-AGCATATTGGACGTAGACAAGGCAGCTAGAGTG-3′ and (Tdo2573) 5′-CTTTAAAATGCACAAACAAACAG-3′; Site 2: (Tdo2574) 5′-AACTTATTGGACGTGAAATTGTTCACTAGCTGTGGTC-3′ and (Tdo2575) 5′-ATTGAAAATGGGACTTCACAC-3′. Two sequential all-around PCR were performed to mutate both sites.
Luciferase assay
Cells were plated to 70–80% confluency in 24-well plates and co-transfected with 2.1 fmol of each pmiRGLO construct and 2.5 nM of miRNA mimic miR-26a (MSY0000533), miR-19b (MSY0000513) or control (1027280) (Qiagen), or 33 nM of miRNA inhibitor miR-20a (MIN0000529), miR-29a (MIN0000535), miR-92a (MIN0000539) or control (1027271) (Qiagen) using Lipofectamine 2000 (Invitrogen). Forty-eight hours post-transfection, cells were lysed and processed for luciferase assay using the Dual-Luciferase Reporter Assay System (Promega).
Quantitative northern blot analysis on miRNAs
Standard RNA oligos of miR-19b, miR-20a, miR-26a, miR-92a and miR-29a obtained from Integrated DNA Technology (IDT) were each diluted to concentrations ranging from 1 to 40 pg/μl. Total RNA (2.5/4 μg) from NIH 3T3 cells and the standards were resolved on 15% Tris/Borate/EDTA (TBE)–urea gels (Bio-Rad), transferred onto Hybond-XL membranes (Amersham) and ultraviolet-crosslinked. Hybridization was carried out using 32P-labelled Starfire probes (miRNA StarFire, IDT) at 25°C overnight in ULTRAHyb Oligo Hybridization buffer (Ambion). Following hybridization, membranes were washed, exposed onto an imaging plate (Fujifilm) and developed using a Typhoon phosphorimager (GE). Intensity of the signals was quantified using ImageJ software (NIH).
Western blotting
Protein extracts were prepared using lysis buffer (50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1% IGEPAL CA-630, 1% sodium deoxycholate, 0.1% sodium dodecyl sulphate, 1 mM ethylenediaminetetraacetic acid) supplemented with protease and phosphatase inhibitor cocktails (Sigma and Roche). Proteins were immobilized on Immobilon FL PVDF membranes (Millipore) and probed with the following antibodies diluted in Odyssey blocking buffer (LI-COR): anti-PTEN (D4.3) (1:1000, CST), anti-pAkt S473 (1:250, CST), anti-Akt (1:1000, CST), anti-pS6, S235/236 (D57.2.2E) (1:500, CST), anti-S6 (5G10) (1:1000, CST) and anti-tubulin (DM1A) (1:5000, Abcam). Rabbit and mouse IR dye secondary antibodies (LI-COR Biosciences) were used at 1:10 000. Blots were scanned with the Odyssey imaging system and quantified with the Image Studio Lite software (LI-COR).
mRNA stability assay
At 70–80% confluency, NMuMG cells plated on 10 cm dishes were treated with actinomycin-D (5 μg/ml) (Sigma). Cells were then harvested at various times (0–360 min). RNA was extracted (Qiagen) and 2.5 μg of each were resolved in 1% agarose gel. Northern blotting was performed as described before. Each Pten isoform was quantified by 3′-UTR isoform-specific qRT-PCR. Half-life was calculated with linear regression analysis (GraphPad Prism). All primer sequences are listed in Supplementary Table S6.
Lentivirus preparation and infection
Short hairpin RNA (shRNA) targeting Pten 3′-UTR were designed and cloned according to the instructions provided for the lentiviral pLKO.1-TRC cloning vector. Control shRNA (shc002), shRNAs targeting the coding sequence (CDS; sh931 and sh1159), and shRNA2512 were obtained from Sigma. Sequences of all shRNAs targeting Pten 3′-UTR are listed in Supplementary Table S7. For lentivirus production, HEK 293T cells were co-transfected with 4 μg of shRNA construct and 2 μg of each lentiviral packaging vectors pMD2.G and psPax2 using Lipofectamine 2000 (Invitrogen). Lentiviral particles were collected 36 and 60 h post-transfection, mixed together and filter sterilized. For cell infection, NMuMG and NIH3T3 were plated at 50–70% confluency in 6-well plates and infected with 1 ml of lentiviral preparations supplemented with polybrene at 4 μg/ml. Cells were selected with 1.5 μg/ml (NMuMG) or 2 μg/ml (NIH3T3) of puromycin for 7 days or longer and collected for RNA and protein analyses.
Preparation of stable cells overexpressing miR-19b and miR-26b
MiR-19b and miR-26b miRNA precursors were amplified and cloned in Lenti-miR vector (SBI). Primers are listed in Supplementary Table S8. Lentivirus preparation, infection and selection were done as described for the pLKO.1-TRC cloning vector.
Statistical analyses
All data are presented as mean ± standard deviation. Two-tailed Student t-test and one-way Analysis Of Variance (ANOVA) analysis were used for comparisons between samples and to determine statistical significance. Pearson's correlation coefficient was used to measure the correlation between two sets of samples. Statistical tests were performed with GraphPad Prism.
RESULTS
APAs lead to an important diversity of Pten 3′-UTR isoforms
To capture the diversity of Pten 3′-UTR isoforms, we performed 3′RACE analyses on a panel of murine (NIH3T3, NMuMG) and human (22RV1, T47D) cell lines. Forward primers were positioned to capture APAs ∼15 nt downstream of candidate PAS (Supplementary Tables S1 and S2; Figure 1A). APAs were detected downstream of eight and seven candidate PAS, in mouse and human cell lines, respectively. Amongst them, two clusters of PAS positioned in regions located 300 nt and 3.3 kb (numbered relative to the stop codon) were functional in both mouse and human cell panels (Figure 1A and B). Additional APAs were validated further downstream in mouse Pten 3′-UTR sequences, at 5.5 and 6.1 kb in the NMuMG cell line. Since 3′RACE is semi-quantitative at best, we turned to northern blot analysis to validate the detected 3′-UTR mRNA isoforms. Consistent with 3′RACE results, a probe complementary to the CDS revealed several specific bands, with two major bands consistent with 300 nt and 3.3 kb APA activity, which were also active in human PTEN 3′-UTR (Figure 1B and Figure 1C, left panel). Northern blot analysis with a probe complementary to mid-3′-UTR sequence confirmed the identity of the bands observed (Figure 1C, probe 2). Our results reveal that a diversity of 3′-UTR isoforms are generated through APA usage in mouse and human cells, with conserved isoforms of 300 nt and 3.3 kb in length being dominant in the examined cell lines. These data further indicate that PTEN APA usage is differentially regulated amongst cell types and/or states. Our results are further corroborated by human and mouse GenXpro APA database (Supplementary Table S5) (45).
Cell proliferation can alter APA usage (28). To examine the impact of cell density on PTEN APA usage, we performed northern blot on RNA isolated from cells cultured at different densities. When NIH3T3 cells were recovered at high cell density (>95% confluency), all Pten 3′-UTR isoforms were increased, except the shortest (APA at 53 nt). Northern blots revealed a switch towards more distal Pten mRNA APAs (3.3, 5.5 and 6.1 kb) over the proximal ones (53 and 300 nt) (Figure 1D). Interestingly, the two longest 3′-UTR isoforms (at 5.5/6.1 kb) could only be detected at higher density in these cells (Figure 1D). Strikingly, the switch to distal APAs correlated with a 1.6-fold increase in PTEN protein expression (Figure 1E). Taken together, these results show that Pten 3′-UTR isoforms are dynamically regulated by cell density and suggest that longer 3′-UTRs contribute more to PTEN protein dosage.
Pten mRNA APAs modulate regulation by miR-29
Pten 3′-UTRs encode validated binding sites for a diversity of miRNAs, including members of the miR-26 and miR-29 miRNA families and miRNAs derived from the oncogenic miR-17–92 polycistron (miR-17/20, miR-19a/b and miR-92a) (Figure 2A) (21,22,46–49). It is generally assumed that longer 3′-UTRs should lead to more potent regulation by miRNAs as they encode more miRNA-binding sites, whereas shorter isoforms would escape miRNAs regulation (50,51). We thus set to determine the impact of miRNAs on alternative Pten 3′-UTRs. To this end, we designed luciferase reporters driving individual Pten 3′-UTR isoforms by mutating and/or deleting the sequence of the validated PAS (Figure 2A and ‘Materials and Methods’ section). Expression of 3′-UTR isoform reporters in NIH3T3 was confirmed by northern blot with a probe mapping to firefly luciferase (FL) CDS (Figure 2B). Multiple bands matching the captured 3′-UTR were detected upon transfection of the Pten wild-type (WT) 3′-UTR luciferase reporter construct, consistent with the concurrent use of multiple APAs. Constructs engineered to drive expression of individual Pten 3′-UTR reporters led to a major band matching the expected size (Figure 2B, lower panel). Interestingly, mutation of functional PAS led to usage of other sites, including cryptic sites (although to a minor extent), suggesting that the choice of PAS usage in 3′-UTR biogenesis may be a competitive process (Figure 2B, lower panel). We note that this series of experiments raises important questions on the current standards for miRNA-binding site validation using cloned 3′-UTR reporters (see ‘Discussion’ section).
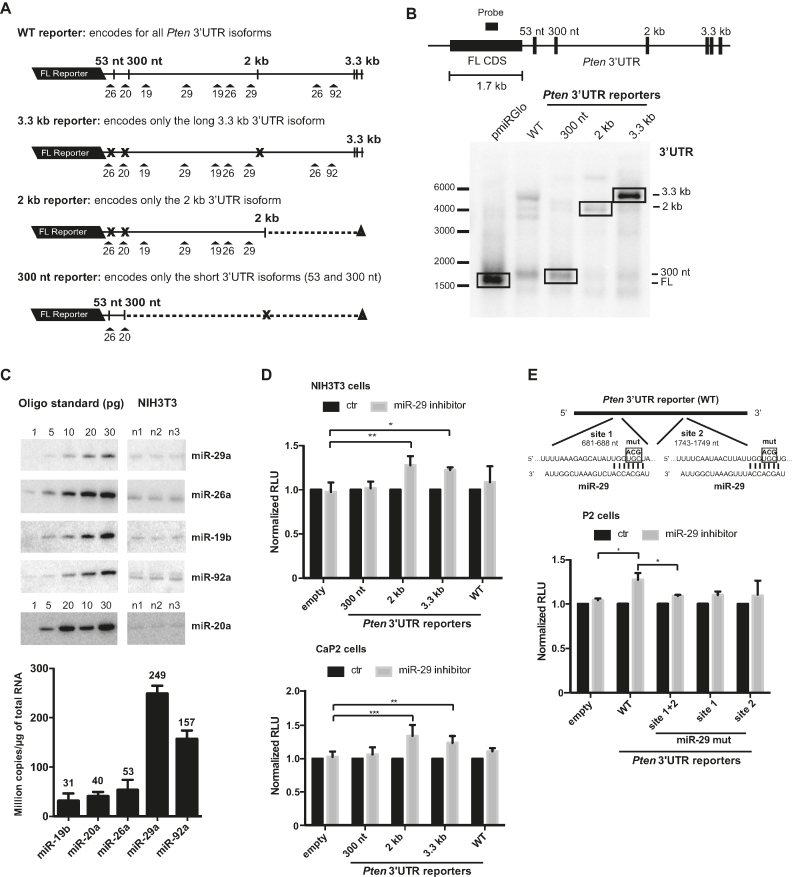
Figure 2.
Pten mRNA APAs modulate regulation by miR-29. (A) Schematic of the reporter constructs with varying Pten 3′-UTR isoforms used for the luciferase assay. Numbers on top indicate the validated PAS and arrows with numbers underneath indicate predicted miRNA sites. The cross (X) shows mutation of the consensus sequence for the indicated PAS to create reporter constructs, which express specific 3′-UTR isoform. (B) Northern blot analysis of the reporter constructs transfected in NIH3T3 cells with a schematic showing the localization of the probe in the FL CDS. (C) Quantitative northern blot analysis with RNA oligo standard in picogram (pg). The bar graph underneath the blot indicates the copy number per microgram of isolated RNA in NIH3T3 cells for each miRNA. Note that samples and corresponding standards were imaged from the same blots. Empty lanes were cropped out for clarity. (D) Luciferase assay in NIH3T3 and prostate CaP2 cells co-transfected with miR-29 inhibitor and isoform-specific Pten 3′-UTR reporter constructs, 300-nt, 2, 3.3 kb and WT or pmiRGlo vector alone (empty). Values are represented as fold change over control treated samples in normalized relative light units (RLU). (E) Schematic of the two miR-29 site positions in Pten 3′-UTR showing the mutations in the miRNA seed sequence (UGC-ACG) (top panel). Luciferase assay in prostate P2 cells co-transfected with miR-29 inhibitor and Pten 3′-UTR reporter (WT) or the same reporter with miR-29 sites mutated, miR-29 mut site 1+2, site 1 or site 2 (bottom panel). All data are presented as mean ± standard deviation from technical triplicates of at least three independent biological experiments. One-way ANOVA analysis was used to calculate P-values (*P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001).
To identify miRNA-binding sites that could be active in NIH3T3, we quantified the expression of previously investigated miRNAs by quantitative northern blot. Amongst miRNAs examined, miR-29a was the most expressed, followed by miR-92a, while miR-19b, miR-20a and miR-26b were 3–8-fold less abundant (Figure 2C). Considering its stronger expression, we further investigated the differential regulation of the Pten 3′-UTR isoforms by miR-29a. Pten 3′-UTRs encode two predicted miR-29 binding sites, which are missing from the shortest (53- and 300-nt 3′-UTRs) but present in all longer, validated 3′-UTR isoforms (Figure 2A). Reporter constructs were co-transfected in NIH3T3 and CaP2 cells with a miR-29 inhibitor or a non-specific control, and reporter expression was quantified by luciferase assays (see ‘Materials and Methods’ section). In both cell lines, we observed a mild but significant 10–15% de-repression of 2 and 3.3 kb 3′-UTR reporters (Figure 2D). As expected, the 300 nt reporter was not affected by miR-29 inhibitors. Interestingly, expression of multiple 3′-UTR isoforms from the WT construct seemed to impair silencing of reporters overall in both NIH3T3 and in CaP2 cells, in comparison with constructs encoding only the 2 and 3.3 kb 3′-UTRs. De-repression of the WT reporter mRNAs was not significant in NIH3T3 or CaP2 cells (Figure 2D). To confirm that the observed repression was due to the direct binding of miR-29 to the Pten 3′-UTRs, the two miR-29-binding sites were mutated individually or together and the reporters were tested in P2 cells, wherein miR-29 silencing of the WT 3′-UTR reporter is significant (Figure 2E, top panel). Mutation of both miR-29 sites completely abolished repression of the WT reporter mRNA in P2 cells (Figure 2E, lower panel).
Together with the dynamic nature of Pten 3′-UTR expression observed above, these results indicate that Pten 3′-UTR APA choice, isoform representation and co-expression can either potentiate or impair miR-29-mediated silencing.
Pten 3′-UTR isoforms are largely refractory to miRNA-mediated silencing
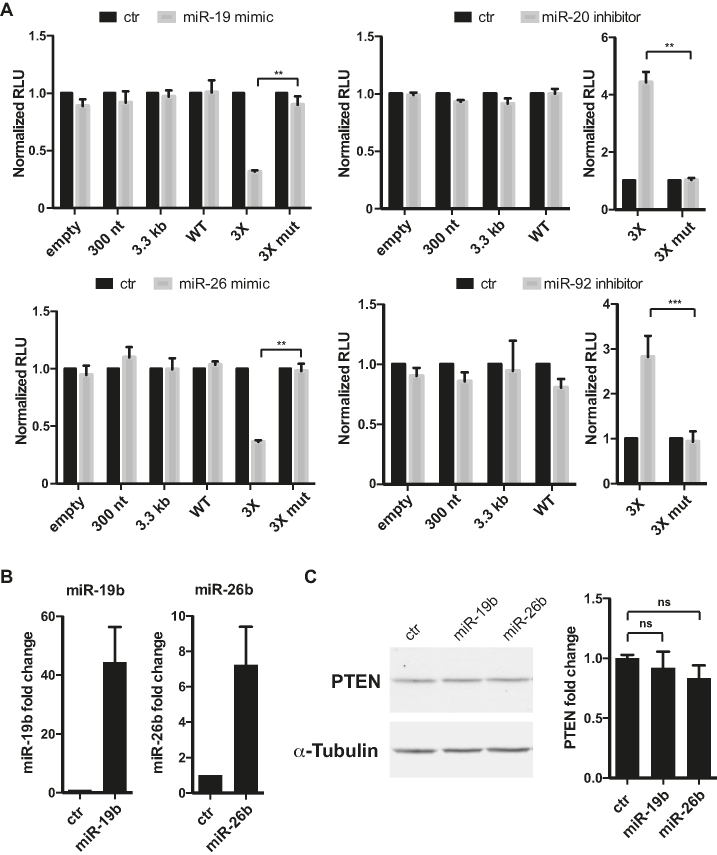
We next examined the response of Pten 3′-UTRs to predicted binding sites for other co-expressed miR-26, miR-19, miR-17/20 and miR-92, which have been validated in human cancer and mouse models (Figure 2A). Constructs encoding each 3′-UTR isoform reporter were co-transfected with miRNA mimics or inhibitors, as previously (44). Surprisingly, no silencing was observed with any of the Pten 3′-UTR reporter constructs for any of the four miRNAs or their families (Figure 3A). Luciferase assays were carried out side by side with positive (3X-Bulge-miRNA) and negative (3X-Mut) control reporters and co-transfected with the corresponding miRNA inhibitor or mimic (Figure 3A). Constructs expressing 3x-miR-19, 17/20, 92 and 26 reporters under the same promoter were efficiently silenced (60–80%), whereas 3XMut reporters were de-repressed to levels comparable to inhibitor co-transfections. The same experiments were repeated for miR-19 and miR-26 in the CaP2 cell line with similar results (Supplementary Figure S1). We further challenged endogenous Pten 3′-UTRs with enforced levels of miR-19b and miR-26b. For this, we transduced NIH3T3 cells with lentiviral particles driving expression of either miRNAs, and isolated stable overexpressing populations (Figure 3B). Despite the overexpression of miR-19b by 30–60-fold and miR-26b by 5–10-fold, no effect on endogenous PTEN protein was observed (Figure 3C). Similar results were observed in NMuMG cells (Supplementary Figure S2). Collectively, these results show that Pten 3′-UTR isoforms are largely refractory to silencing by many, but not all, miRNAs, whether expressed from a reporter construct or endogenously.
Figure 3.
Pten 3′-UTR isoforms are largely refractory to miRNA-mediated silencing. (A) Luciferase assays in NIH3T3 cells co-transfected with isoform-specific Pten 3-′UTR reporter constructs and miRNA mimic or inhibitor. Control 3X-Bulge (3X) and 3X-mutated (3X mut) miR-19, mir-20, miR-26 and miR-92 binding site reporters were used as a positive control with their corresponding miRNA mimic or inhibitor (as indicated). (B) Quantification of mature miRNAs by qRT-PCR following infection of NIH3T3 cells with lentivirus expressing control, miR-26b or miR-19b. (C) Quantification of PTEN by western blot in stable infected cells from three independent biological replicate experiments using the Odyssey imaging system (LI-COR). α-Tubulin was used as a loading control. Two-tailed Student’s t-test or one-way ANOVA were used to calculate P-values (**P ≤ 0.01 and ***P ≤ 0.001). Data is presented as as means +- standard deviation. ns: not significant.
Longer Pten 3′-UTR isoforms are more stable
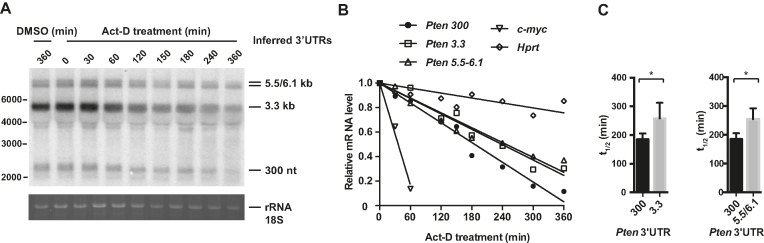
The previously characterized Pten 3′-UTR is thought to be the focus of several post-transcriptional regulation mechanisms. Like many other such mechanisms, miRNA-mediated silencing is in good part effected through mRNA destabilization and decay. We thus sought to determine the half-lives of individual Pten mRNA 3′-UTR isoforms by performing an mRNA decay time course. Replicates of NMuMG cells were treated with actinomycin-D and mRNAs were allowed to decay in vivo without transcription over a course of 360 min. Total RNA was isolated, resolved and Pten mRNAs were detected in high-resolution northern blot with a probe complementary to its CDS (Figure 4A). Approximately 300 nt, 3.3, 5.5 and 6.1 kb Pten 3′-UTR isoforms were readily detectable and their decay traceable within the 6 h time frame, but the group of isoforms derived from the ∼3.3 and 5.5/6.1 kb APAs could not be fully resolved. We thus developed and validated a 3′-UTR isoform-specific qRT-PCR strategy to precisely quantify the 300 nt and 3.3 kb isoforms (Supplementary Figures S3 and S4; ‘Materials and Methods’ section). Half-lives of the longer 5.5/6.1 kb isoforms were quantified by qRT-PCR using primers located downstream of the 3.3 kb APA isoforms. An mRNA with a long half-life, Hprt, was used as normalizing control and the unstable c-Myc mRNA was used to validate actinomycin-D treatment for each experiment (Supplementary Figure S5). The relative expression of Pten 3′-UTR isoforms at each time point was plotted, linear regression of decay courses yielded curves with R2 values >0.8, and each mRNA isoform half-life was calculated (Figure 4B). Statistically, the long 3.3 kb (half-life: 258 min) and 5.5/6.1 kb (half-life: 254 min) 3′-UTR mRNA isoforms had the same half-lives, and both were significantly more stable than the short 300 nt 3′-UTR isoform (186 min) (Figure 4B and C; Supplementary Figure S5). Polysome gradient distribution of the 300 nt, 3.3 and 5.5/6.1 kb isoforms suggest that longer isoforms are translated as efficiently, if not more efficiently than the 300 nt 3′-UTR isoform (performed as in (52); Supplementary Figure S6). These data are consistent with longer Pten 3′-UTRs being refractory to the de-stabilization and translational repression activities mediated by miRNAs, and thus stand at odds with the large number and diversity of binding sites they encode. These results also suggest that Pten 3′-UTR isoforms may distinctly contribute to PTEN protein dosage.
Figure 4.
Longer Pten 3′-UTR isoforms are more stable. (A) Representative northern blot with NMuMG cells showing the decay of each Pten 3′-UTR isoform upon actinomycin-D (Act-D) treatment. Cells were harvested at 0, 30, 60, 120, 150, 180, 240, 300 and 360 min post-treatment. Dimethyl sulfoxide (DMSO) was used to solubilize Act-D and served as a mock control. (B) Each Pten 3′-UTR isoform was quantified with the specific isoform qRT-PCR assay, Pten 300, Pten 3.3 and Pten 5.5/6.1. c-Myc mRNA was used as a positive control for the Actinomycin-D treatment and was also quantified by qRT-PCR. Total mRNA was normalized with Hprt, and mRNA half-life was determined by linear regression analysis. (C) The bar graph shows the half-life in minutes calculated for each Pten 3′-UTR isoform, 300-nt, 3.3, 5.5/6.1 kb and positive control c-Myc from the qRT-PCR analysis. Data shown are means ± standard deviation from four independent biological experiments. A two-tailed Student’s t-test was used (*P ≤ 0.05).
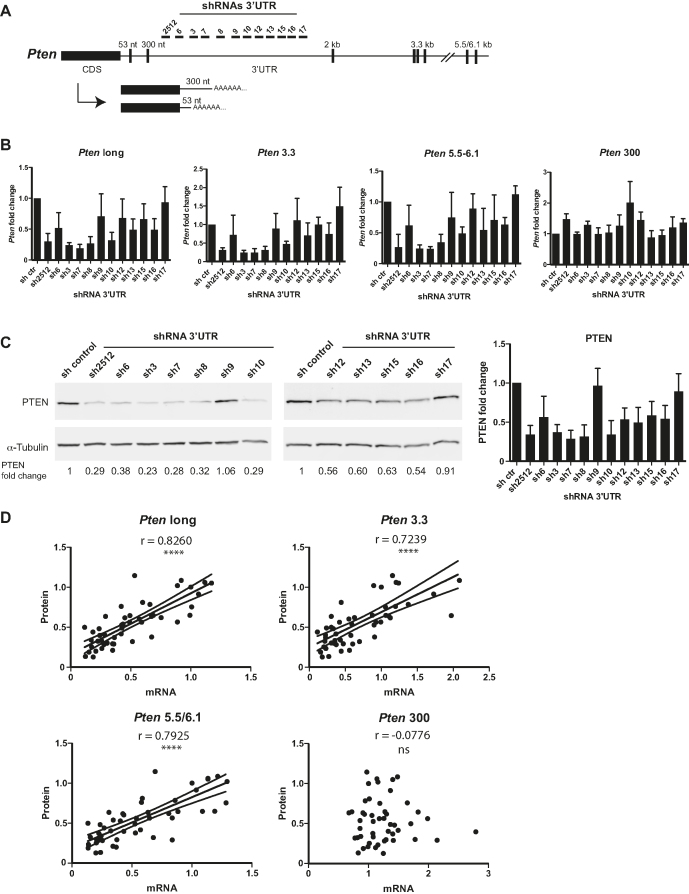
Longer 3′-UTR isoforms dictate PTEN protein dosage
We sought to determine the contributions of Pten 3′-UTR mRNA isoforms to PTEN protein dosage. To this end, we aimed to modulate the longer Pten 3′-UTR mRNA isoforms independently of the shorter ones (53 and 300 nt). We took advantage of a battery of 12 shRNAs targeting sequences designed along the long 3′-UTRs in a region between the PAS located at 300 nt and 2 kb (Figure 5A). Following transduction and selection, we quantified the effect of each shRNA on Pten 3′-UTR isoforms using isoform-specific qRT-PCR assays, as described above. Individual shRNAs impacted differentially on the long isoforms, ranging from 80% knockdown to no significant effect. In contrast and as expected, none of the shRNAs tested significantly reduced the expression of the short 300 nt isoform. Curiously, this isoform increased slightly with some of the shRNAs targeting the long isoforms, possibly through APA competitive effects (Figure 5B, Pten 300). Concurrently, PTEN protein levels were quantified using quantitative western blot (see ‘Materials and Methods’ section). Knockdown of the long isoforms decreased PTEN protein expression to different extents and by up to ∼75% (Figure 5C). By comparing changes in the mRNA isoform and protein expression in the different knockdown lines, we noticed that expression of long 3′-UTR isoforms seemed to correlate best with PTEN protein expression (Figure 5D). Correlation was strong for 3.3 kb (r = 0.7239) and 5.5/6.1 kb isoforms (r = 0.7925), and with the pooled expression of all long isoforms (r = 8260; P < 0.0001). In stark contrast, expression of the short 300 nt isoform did not correlate with PTEN protein levels. These results suggest that the long Pten 3′-UTR isoforms contribute to the very bulk of the PTEN protein dosage, whereas shorter isoforms (53 nt/300 nt) do not contribute significantly. This further lends support to the view that longer Pten 3′-UTR isoform mRNAs are refractory to miRNA-mediated repression in the cells/conditions tested. Because miRNA expression, and 3′-UTR sequences, structures and interactions are intricately linked to cell type, genotype and environmental contexts, we do not claim that this is always the case (see ‘Discussion’ section).
Figure 5.
Longer 3′-UTR isoforms dictate PTEN protein dosage. (A) Schematic showing all 12 shRNAs (shRNA 3′-UTR) used to specifically knockdown the expression of all 3′-UTR isoforms longer than 300 nt without affecting the expression of the short ones, 53 and 300 nt. (B) mRNA expression for Pten long (all isoforms longer than 300 nt) and each specific 3′-UTR isoform, Pten 3.3, Pten 5.5/6.1 and Pten 300, was quantified by qRT-PCR in NMuMG stable cell lines. Data are presented as Pten mRNA fold change for each 3′-UTR shRNA compared to the control shRNA (shc002). (C) The effect of each shRNA 3′-UTR on PTEN protein level was analysed by western blot with α-Tubulin as a loading control. The Odyssey imaging system (LiCOR) was used to quantify PTEN fold change level relative to control shRNA. (D) Pearson’s correlation analysis was performed between the mRNA expression of each Pten 3′-UTR isoform (B) and the corresponding PTEN protein level (C). Pearson’s coefficient (r) values are indicated on the graph with their corresponding P-value (****P ≤ 0.0001). Data shown in (A–C) are means ± standard deviation of four independent biological experiments. In (D), linear regression and 95% confidence interval are presented.
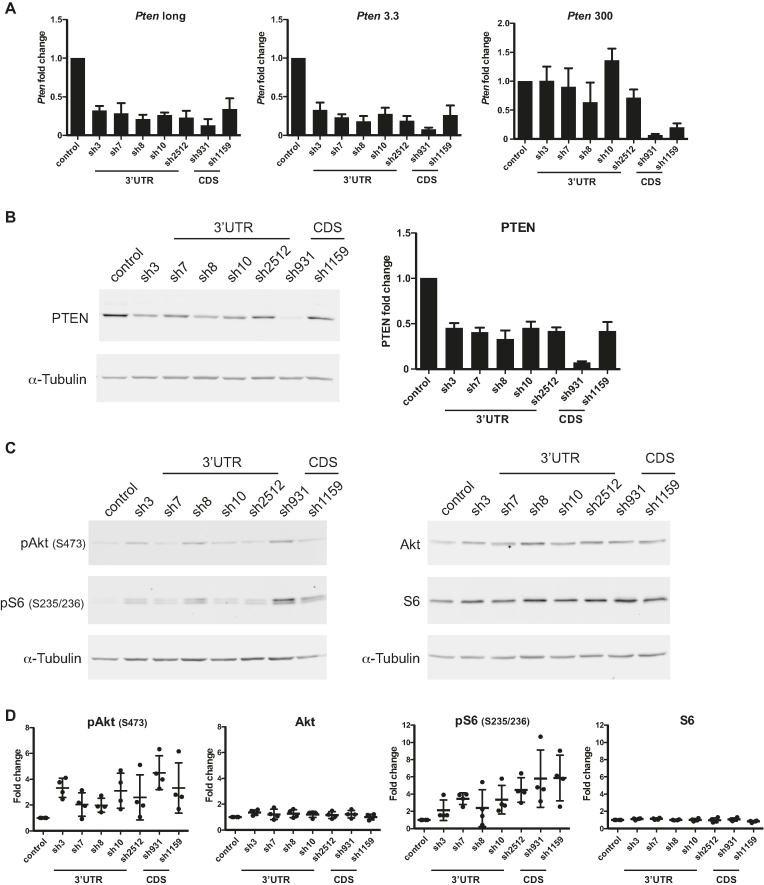
Long Pten 3′-UTR isoforms control PI3K/Akt/mTOR signalling
PTEN is a prominent negative regulator of the PI3K/Akt/mTOR signalling pathway (3,4). Its phosphatase activity reverts the generation of membrane-associated PIP3 by PI3K, which prevents Akt phosphorylation on residues Thr308 and Ser473 and its activation. Downstream events further lead to phosphorylation of ribosomal protein S6 by mammalian Target Of Rapamycin (mTOR) on serine residues 235 and 236. To determine the functional impact of long Pten 3′-UTR isoforms on Akt signalling, we used the most potent 3′-UTR shRNAs to generate NIH3T3 stable cell lines. NIH3T3 were chosen for their very low Akt signalling activity in our culture conditions. Lines were also derived for two shRNAs targeting the CDS of Pten mRNAs (sh931, sh1159) as positive controls. Knockdown efficiency and specificity were verified using isoform-specific qRT-PCR assays as above (Figure 6A). CDS shRNAs decreased the PTEN protein consistently with the extent of knockdown of all mRNAs (Figure 6A and B). 3′-UTR shRNAs led to a 60–75% reduction of long isoforms and left shorter (53 and 300 nt) 3′-UTR isoforms un-diminished. As with NMuMG, knockdown of long 3′-UTR mRNA isoforms was mirrored by a 55–75% decrease in PTEN protein expression. Activation of Akt was verified by comparing western blots on total Akt protein with the signal from phospho-serine 473 antibody. All 3′-UTR shRNA cell lines tested increased Akt phosphorylation (pAkt S473, median increase 2–4-fold) while the level of total Akt protein remained the same (Figure 6C and D). We further examined downstream effects on the cascade by assessing the phosphorylation of S6 ribosomal protein. As with phospho-Akt, phospho-S6 (Ser235/236) was increased (median 2–6-fold) while total S6 protein levels were unchanged (Figure 6C and D).
Figure 6.
Long Pten 3′-UTR isoforms control PI3K/Akt/mTOR signalling. (A) NIH3T3 cells infected with shRNAs targeting either the 3′-UTR or the coding sequencing (3′-UTR or CDS). mRNA expression analysis of specific Pten 3′-UTR isoforms (Pten long, Pten 3.3 and Pten 300) by qRT-PCR. All qRT-PCR data are presented as Pten mRNA fold change relative to shRNA control. (B) PTEN protein quantification analysis and a representative western blot. All bar graph data shown in (A and B) are means ± standard deviation of four independent biological experiments. (C) Representative western blots for the analysis of Akt and S6 phosphorylation status after shRNA infection. (D) Quantification of Akt and S6 phosphorylation status from four independent biological experiments. The Odyssey imaging system (LiCOR) was used to quantify PTEN, pAkt S473, Akt, pS6 (S235/236) and S6 fold change levels relative to shRNA control, with α-Tubulin as a loading control.
These significant changes in Akt and S6 activation without reduction in shorter 3′-UTR isoforms indicate that longer Pten 3′-UTR mRNAs contribute to most of the protein dosage and can account for the bulk of its activity in the PI3K/Akt/mTOR cascade. These results further support the conclusion that longer 3′-UTRs isoforms of Pten mRNA are more stably expressed due to their refractory nature to miRNA repression in the cell types tested.
DISCUSSION
Dosage of PTEN protein is critical for its tumour suppressive functions (10). Whereas recent publications have highlighted the importance of post-transcriptional mechanisms for the control of PTEN expression, no systematic assessment of alternative Pten mRNA 3′-UTRs, or their interplay with miRNAs, had been reported. Here, we show that an important diversity of co-expressed Pten 3′-UTR mRNA isoforms are generated by APA usage and that their relative expression is dynamically controlled by cell density. Most importantly, we found that in spite of the diversity of validated miRNA-binding sites they encode, longer 3′-UTRs are largely refractory to miRNA-mediated silencing and account for the bulk of PTEN protein dosage. This is supported herein through four distinct direct lines of evidence. Firstly, long Pten 3′-UTR isoforms did not respond to inhibition or overexpression of miRNAs whether endogenously or when expressed from reporter constructs. In parallel, reporters encoding 3X miRNA-binding sites responded potently. Secondly, the long 3′-UTR isoforms were significantly more stable than the shorter 300 nt isoform, which encodes much fewer miRNA-binding sites. Thirdly, the extent of knockdown of longer Pten 3′-UTR mRNA isoforms closely correlated with changes in PTEN protein, in spite of co-expression of the shorter 3′-UTR isoforms. Lastly, activation of PI3K/Akt/mTOR signalling was observed upon specific knockdown of longer Pten 3′-UTR isoforms, while shorter ones remained expressed. Shorter Pten 3′-UTR isoforms thus fail to compensate for loss of longer 3′-UTR isoform mRNAs, either functionally or for the overall PTEN protein load.
Several factors could explain why longer Pten 3′-UTR isoforms are refractory to miRNA-mediated regulation. Folding structures of longer 3′-UTRs can impinge on miRNA-binding site accessibility and output (38). A miRNA-binding site encoded near either ends of long 3′-UTRs exerts a more pronounced effect than when positioned in the middle of the 3′-UTR, and this effect is more pronounced for 3′-UTR longer than 1.3 kb (53). 3′-UTR folding structures could affect accessibility to miRISC or the consequent assembly and activation of the effector complexes such as the CCR4–NOT deadenylase. In support for this hypothesis, modelling the folding of Pten 3′-UTR sequences using the MC-Fold algorithm (54) suggests extensive base-pairing around refractory miRNA-binding sites (Supplementary Figure S7). Further along these lines, we previously noticed that an increased distance between miRNA-binding sites and the poly(A) tail could significantly slow the deadenylation of target mRNAs (55). Interestingly, we and others have also shown that re-positioning of miRNA-binding sites closer to poly(A) tails by APAs could potentiate miRNA-mediated silencing (32,55). miRNAs and miRISC are not the only trans-acting factors interacting with 3′-UTRs. Indeed, association of RBPs with 3′-UTRs can also impinge on the efficacy of miRNA sites by either competing with miRNA binding sites or changing the RNA structure (25), and thus contribute to stabilize longer Pten 3′-UTR mRNA isoforms (35).
Several publications, some of which based on robust mouse genetic models, have supported the targeting of Pten mRNAs by miRNAs such as members of the miR-17∼92 polycistron. The reasons why those sites are enabled in vivo are intriguing. We reason that alternative 3′-UTR expression, RBPs and/or miRNA concentrations, have co-evolved to specifically potentiate miRNA-mediated silencing of the Pten 3′-UTRs in those contexts. However, for several other miRNAs reported to target Pten 3′-UTRs, the experimental validation was not as robust. Our results warrant a critical reconsideration of how putative miRNA-binding sites in Pten 3′-UTRs were validated. In most cases, authors would report the identification of putative sites based on prediction algorithms. They would then clone a predicted or previously published 3′-UTR in a luciferase reporter and test their response to overexpressed candidate miRNAs. In some cases, a minimal fragment of the 3′-UTR would be sub-cloned in reporters and tested. In effect, this experimental design ignores the diversity of 3′-UTRs that could prevail in the physiological context (25) and is blind to critical constraints (RBPs, RNA folding) that dictate miRNA-binding site regulation. We reason that any miRNA-binding site validation effort, for Pten or any other gene, should be accompanied by an assessment of 3′-UTR expression in physiological contexts or closely related surrogates. Use of miRNA inhibitors and detection of increased protein expression would demonstrate a function for the endogenous pool of a specific miRNA family. Mutation of candidate target sites should be performed in the context of the full-length 3′-UTR. While variants of Argonaute/miRISC cross-linking/immunoprecipitation strategies (56) certainly can prove that a miRNA-binding site is occupied by miRISC in a fraction of mRNAs, such techniques could not determine which fraction is bound, nor could it prove the sites to drive silencing.
Ultimately, the direct nature of the miRNA–mRNA interaction and its functional impact should be assessed by mutagenesis of the target site itself, ideally in its physiological context, in vivo. This design has recently become more accessible through CRISPR genome edition. Traditional rescue strategies introducing a compensatory mutation in the sequence of exogenously provided miRNA would further enhance the demonstration. The first papers where 3′-UTR sites were edited within the genome have begun to emerge. A systematic, critical revisit of all previously ‘validated’ Pten 3′-UTR miRNA-binding sites would thus be timely.
Supplementary Material
ACKNOWLEDGEMENTS
We apologize to authors whose relevant work may not have been cited in this manuscript. We thank Dr Hong Wu for providing the P2 and CaP2 cells, and Dr Morag Park for support in the isolation of MEFs. We thank Dr Ivan Topisirovic and his lab for assistance with polysome gradients, and Dr Mathieu Flamand for his assistance with the 3′RACE methods.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cancer Research Society (CRS); Canadian Institute of Health Research (CIHR) [MOP-123352 to T.F.D.]; Fonds de la Rercherche en Santé du Québec; Chercheur-Boursier Senior salary award (to T.F.D.); Susan G. Komens fellowship (to C.L.). Funding for open access charge: CIHR [MOP-123352].
Conflict of interest statement. None declared.
REFERENCES
- 1. Vivanco I., Sawyers C.L.. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat. Rev. Cancer. 2002; 2:489–501. [DOI] [PubMed] [Google Scholar]
- 2. Hay N., Sonenberg N.. Upstream and downstream of mTOR. Genes Dev. 2004; 18:1926–1945. [DOI] [PubMed] [Google Scholar]
- 3. Song M.S., Salmena L., Pandolfi P.P.. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 2012; 13:283–296. [DOI] [PubMed] [Google Scholar]
- 4. Bermudez Brito M., Goulielmaki E., Papakonstanti E.A.. Focus on PTEN regulation. Front. Oncol. 2015; 5:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li J., Yen C., Liaw D., Podsypanina K., Bose S., Wang S.I., Puc J., Miliaresis C., Rodgers L., McCombie R. et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997; 275:1943–1947. [DOI] [PubMed] [Google Scholar]
- 6. Alimonti A., Carracedo A., Clohessy J.G., Trotman L.C., Nardella C., Egia A., Salmena L., Sampieri K., Haveman W.J., Brogi E. et al. Subtle variations in Pten dose determine cancer susceptibility. Nat. Genet. 2010; 42:454–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schade B., Rao T., Dourdin N., Lesurf R., Hallett M., Cardiff R.D., Muller W.J.. PTEN deficiency in a luminal ErbB-2 mouse model results in dramatic acceleration of mammary tumorigenesis and metastasis. J. Biol. Chem. 2009; 284:19018–19026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shen-Li H., Koujak S., Szablocs M., Parsons R.. Reduction of Pten dose leads to neoplastic development in multiple organs of Pten (shRNA) mice. Cancer Biol. Ther. 2010; 10:1194–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Berger A.H., Pandolfi P.P.. Haplo-insufficiency: a driving force in cancer. J. Pathol. 2011; 223:137–146. [DOI] [PubMed] [Google Scholar]
- 10. Carracedo A., Alimonti A., Pandolfi P.P.. PTEN level in tumor suppression: how much is too little. Cancer Res. 2011; 71:629–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Trotman L.C., Niki M., Dotan Z.A., Koutcher J.A., Di Cristofano A., Xiao A., Khoo A.S., Roy-Burman P., Greenberg N.M., Van Dyke T. et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003; 1:e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kwabi-Addo B., Giri D., Schmidt K., Podsypanina K., Parsons R., Greenberg N., Ittmann M.. Haploinsufficiency of the Pten tumor suppressor gene promotes prostate cancer progression. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:11563–11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He L. Posttranscriptional regulation of PTEN dosage by noncoding RNAs. Sci. Signal. 2010; 3:pe39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xiao C., Srinivasan L., Calado D.P., Patterson H.C., Zhang B., Wang J., Henderson J.M., Kutok J.L., Rajewsky K.. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 2008; 9:405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sumazin P., Yang X., Chiu H.S., Chung W.J., Iyer A., Llobet-Navas D., Rajbhandari P., Bansal M., Guarnieri P., Silva J. et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011; 147:370–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fabian M.R., Sonenberg N.. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012; 19:586–593. [DOI] [PubMed] [Google Scholar]
- 17. Kim H., Huang W., Jiang X., Pennicooke B., Park P.J., Johnson M.D.. Integrative genome analysis reveals an oncomir/oncogene cluster regulating glioblastoma survivorship. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:2183–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tian L., Fang Y.X., Xue J.L., Chen J.Z.. Four microRNAs promote prostate cell proliferation with regulation of PTEN and its downstream signals in vitro. PLoS One. 2013; 8:e75885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yan B., Guo Q., Fu F.J., Wang Z., Yin Z., Wei Y.B., Yang J.R.. The role of miR-29b in cancer: regulation, function, and signaling. OncoTargets Ther. 2015; 8:539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zaman M.S., Thamminana S., Shahryari V., Chiyomaru T., Deng G., Saini S., Majid S., Fukuhara S., Chang I., Arora S. et al. Inhibition of PTEN gene expression by oncogenic miR-23b-3p in renal cancer. PLoS One. 2012; 7:e50203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Olive V., Jiang I., He L.. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int. J. Biochem. Cell Biol. 2010; 42:1348–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zou H., Ding Y., Wang K., Xiong E., Peng W., Du F., Zhang Z., Liu J., Gong A.. MicroRNA-29A/PTEN pathway modulates neurite outgrowth in PC12 cells. Neuroscience. 2015; 291:289–300. [DOI] [PubMed] [Google Scholar]
- 23. Mi S., Li Z., Chen P., He C., Cao D., Elkahloun A., Lu J., Pelloso L.A., Wunderlich M., Huang H. et al. Aberrant overexpression and function of the miR-17-92 cluster in MLL-rearranged acute leukemia. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tay Y., Kats L., Salmena L., Weiss D., Tan S.M., Ala U., Karreth F., Poliseno L., Provero P., Di Cunto F. et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011; 147:344–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nam J.W., Rissland O.S., Koppstein D., Abreu-Goodger C., Jan C.H., Agarwal V., Yildirim M.A., Rodriguez A., Bartel D.P.. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol. Cell. 2014; 53:1031–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Clancy J.L., Wei G.H., Echner N., Humphreys D.T., Beilharz T.H., Preiss T.. mRNA isoform diversity can obscure detection of miRNA-mediated control of translation. RNA. 2011; 17:1025–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mayr C., Bartel D.P.. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009; 138:673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sandberg R., Neilson J.R., Sarma A., Sharp P.A., Burge C.B.. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008; 320:1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Derti A., Garrett-Engele P., Macisaac K.D., Stevens R.C., Sriram S., Chen R., Rohl C.A., Johnson J.M., Babak T.. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012; 22:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ji Z., Lee J.Y., Pan Z., Jiang B., Tian B.. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:7028–7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Elkon R., Drost J., van Haaften G., Jenal M., Schrier M., Oude Vrielink J.A., Agami R.. E2F mediates enhanced alternative polyadenylation in proliferation. Genome Biol. 2012; 13:R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoffman Y., Bublik D.R., Ugalde A.P., Elkon R., Biniashvili T., Agami R., Oren M., Pilpel Y.. 3′UTR shortening potentiates MicroRNA-Based repression of Pro-differentiation genes in proliferating human cells. PLos Genet. 2016; 12:e1005879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mayr C. Regulation by 3′-Untranslated Regions. Annu. Rev. Genet. 2017; 51:171–194. [DOI] [PubMed] [Google Scholar]
- 34. Mortimer S.A., Kidwell M.A., Doudna J.A.. Insights into RNA structure and function from genome-wide studies. Nat. Rev. Genet. 2014; 15:469–479. [DOI] [PubMed] [Google Scholar]
- 35. Iadevaia V., Gerber A.P.. Combinatorial control of mRNA fates by RNA-Binding proteins and Non-Coding RNAs. Biomolecules. 2015; 5:2207–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wan Y., Qu K., Zhang Q.C., Flynn R.A., Manor O., Ouyang Z., Zhang J., Spitale R.C., Snyder M.P., Segal E. et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014; 505:706–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mayr C. Evolution and biological roles of alternative 3′UTRs. Trends Cell Biol. 2016; 26:227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E.. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007; 39:1278–1284. [DOI] [PubMed] [Google Scholar]
- 39. Tan X., Lu Z.J., Gao G., Xu Q., Hu L., Fellmann C., Li M.Z., Qu H., Lowe S.W., Hannon G.J. et al. Tiling genomes of pathogenic viruses identifies potent antiviral shRNAs and reveals a role for secondary structure in shRNA efficacy. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Soule H.D., McGrath C.M.. A simplified method for passage and long-term growth of human mammary epithelial cells. In Vitro Cell. Dev. Biol. 1986; 22:6–12. [DOI] [PubMed] [Google Scholar]
- 41. Jiao J., Wang S., Qiao R., Vivanco I., Watson P.A., Sawyers C.L., Wu H.. Murine cell lines derived from Pten null prostate cancer show the critical role of PTEN in hormone refractory prostate cancer development. Cancer Res. 2007; 67:6083–6091. [DOI] [PubMed] [Google Scholar]
- 42. Scotto-Lavino E., Du G., Frohman M.A.. 3′ end cDNA amplification using classic RACE. Nat. Protoc. 2006; 1:2742–2745. [DOI] [PubMed] [Google Scholar]
- 43. Wolter J.M., Kotagama K., Pierre-Bez A.C., Firago M., Mangone M.. 3′LIFE: a functional assay to detect miRNA targets in high-throughput. Nucleic Acids Res. 2014; 42:e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mayya V.K., Duchaine T.F.. On the availability of microRNA-induced silencing complexes, saturation of microRNA-binding sites and stoichiometry. Nucleic Acids Res. 2015; 43:7556–7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Muller S., Rycak L., Afonso-Grunz F., Winter P., Zawada A.M., Damrath E., Scheider J., Schmah J., Koch I., Kahl G. et al. APADB: a database for alternative polyadenylation and microRNA regulation events. Database. 2014; 2014:bau076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huse J.T., Brennan C., Hambardzumyan D., Wee B., Pena J., Rouhanifard S.H., Sohn-Lee C., le Sage C., Agami R., Tuschl T. et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009; 23:1327–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Olive V., Bennett M.J., Walker J.C., Ma C., Jiang I., Cordon-Cardo C., Li Q.J., Lowe S.W., Hannon G.J., He L.. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009; 23:2839–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang H., Cao H., Xu D., Zhu K.. MicroRNA-92a promotes metastasis of nasopharyngeal carcinoma by targeting the PTEN/AKT pathway. OncoTargets Ther. 2016; 9:3579–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou W., Shi G., Zhang Q., Wu Q., Li B., Zhang Z.. MicroRNA-20b promotes cell growth of breast cancer cells partly via targeting phosphatase and tensin homologue (PTEN). Cell Biosci. 2014; 4:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Blazie S.M., Geissel H.C., Wilky H., Joshi R., Newbern J., Mangone M.. Alternative polyadenylation directs Tissue-Specific miRNA targeting in caenorhabditis elegans somatic tissues. Genetics. 2017; 206:757–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lianoglou S., Garg V., Yang J.L., Leslie C.S., Mayr C.. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 2013; 27:2380–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gandin V., Sikstrom K., Alain T., Morita M., McLaughlan S., Larsson O., Topisirovic I.. Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. J. Vis. Exp. 2014; doi:10.3791/51455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P.. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007; 27:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Parisien M., Major F.. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008; 452:51–55. [DOI] [PubMed] [Google Scholar]
- 55. Wu E., Thivierge C., Flamand M., Mathonnet G., Vashisht A.A., Wohlschlegel J., Fabian M.R., Sonenberg N., Duchaine T.F.. Pervasive and cooperative deadenylation of 3′UTRs by embryonic microRNA families. Mol. Cell. 2010; 40:558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hafner M., Lianoglou S., Tuschl T., Betel D.. Genome-wide identification of miRNA targets by PAR-CLIP. Methods. 2012; 58:94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.