Abstract
Mutations of the Glu76 residue of canonical histone H2B are frequently found in cancer cells. However, it is quite mysterious how a single amino acid substitution in one of the multiple H2B genes affects cell fate. Here we found that the H2B E76K mutation, in which Glu76 is replaced by Lys (E76K), distorted the interface between H2B and H4 in the nucleosome, as revealed by the crystal structure and induced nucleosome instability in vivo and in vitro. Exogenous production of the H2B E76K mutant robustly enhanced the colony formation ability of the expressing cells, indicating that the H2B E76K mutant has the potential to promote oncogenic transformation in the presence of wild-type H2B. We found that other cancer-associated mutations of histones, H3.1 E97K and H2A.Z.1 R80C, also induced nucleosome instability. Interestingly, like the H2B E76K mutant, the H3.1 E97K mutant was minimally incorporated into chromatin in cells, but it enhanced the colony formation ability. In contrast, the H2A.Z.1 R80C mutant was incorporated into chromatin in cells, and had minor effects on the colony formation ability of the cells. These characteristics of histones with cancer-associated mutations may provide important information toward understanding how the mutations promote cancer progression.
INTRODUCTION
The nucleosome is the basic repeating unit of chromatin, and is composed of two molecules of histones (H2A, H2B, H3 and H4) and about 150 bp of DNA (1,2). Canonical types of histones are incorporated into chromatin during the S-phase of the cell cycle, in a DNA replication-dependent manner (3). To ensure the robust production of the canonical histones during S-phase, multiple copies of the canonical histone genes exist in the genomes of higher eukaryotes (4). In addition, non-allelic isoforms of histones (histone variants) have been identified in many species (5). In humans, each of the histone variants is encoded by either a single gene or a few copies of genes (6).
Many mutations in the histone variant genes have been identified in cancer cells, suggesting that the mutations directly perturb the function of the corresponding histone variants (7–9). Mutations of the histone variant H3.3 Lys27 and Gly34 residues, which are located in the disordered N-terminal tail, have been reported in pediatric gliomas (7,8). The histone tails function as target sites for the modifier and reader proteins for the histone post-translational modifications (10). Interestingly, the production of the H3.3 K27M mutant, in which the H3.3 Lys27 residue is replaced by Met, abrogates the activity of polycomb repressive complex 2 (PRC2), which methylates the H3 Lys27 residue (11). Similar inhibition of methyltransferases by the H3 K9M and K36M mutations has also been reported (12). The effects of these mutations within the histone tails suggest that the Lys-to-Met substitutions on the target sites for histone methylation may disturb the proper histone modification landscape in the epigenome, and thus promote a variety of genomic disorders.
Curiously, mutations in some of the multiple canonical histone genes have been identified in human cancer cells (8,13–16). Many of these mutations occur in the globular ‘histone-fold’ domain. This suggests that these amino acid substitutions may have a direct impact on the nucleosome structure, and could abrogate proper gene regulation by the chromatin architecture (17,18). Therefore, the production of these canonical histone mutants, which may be partly incorporated into chromatin, could dominantly enhance cell growth and the incidence of cancer-associated mutations, probably by changing the physical and structural properties of the nucleosome. However, the impact of the cancer-associated mutations of canonical histones has not been studied yet.
Mutations in one of the seventeen H2B genes, especially HIST1H2BC, HIST1H2BD and HIST1H2BE, have been dominantly found in cancer cells (13–15). Among them, mutations of the H2B Glu76 residue are found in cancer cells with high frequency. Therefore, in the present study, we focused on the H2B E76K mutation, in which the Glu76 residue is replaced by Lys. Our structural and biochemical analyses revealed that, in the nucleosome, the H2B E76K mutation perturbs the interaction between H2B and H4, and profoundly weakens the association of the H2A-H2B dimer with the H3-H4 tetramer. We found that the exogenous production of the H2B E76K mutant robustly enhances the colony formation ability, mimicking the dominant phenotype observed in cancer cells. Interestingly, the other cancer-associated mutations found in histones H3.1 and H2A.Z.1 also induce nucleosome instability. Remarkably, the H3.1 E97K mutant significantly enhanced the colony formation ability, although only small amounts were incorporated into chromatin in cells. In contrast, the H2A.Z.1 R80C mutant minimally affected the colony formation ability of the cells. Therefore, the cancer-associated mutations in histones H2B, H3.1 and H2A.Z.1 seem to affect the nucleosome structure and stability in different manners, and may facilitate the oncogenic transformation of cells.
MATERIALS AND METHODS
Purification of recombinant human histones
Human histones H2A (type 1-B/E), H2B (type 1-J), H3.1, H4, H2A.Z.1, H2B E76K, H2A.Z.1 R80C and H3.1 E97K were expressed and purified as described previously (19,20).
In vitro nucleosome reconstitution and purification
The H2A-H2B dimer, H2A-H2B E76K dimer, H2A.Z.1-H2B-H3.1-H4 octamer, H2A.Z.1 R80C-H2B-H3.1-H4 octamer, H3.1-H4 tetramer and H3.1 E97K-H4 tetramer were reconstituted with purified lyophilized histones, and the reconstituted histone complexes were isolated by Superdex 200 gel filtration chromatography, as described previously (21). For the nucleosome reconstitution, the purified H2A-H2B dimer and the H3-H4 tetramer or the H2A.Z-H2B-H3.1-H4 octamer were mixed with a 146 bp palindromic α-satellite DNA (1,21) in buffer containing 2 M KCl, and the KCl concentration was gradually reduced to 0.25 M, as previously described (21). The reconstituted nucleosomes were further purified by preparative native polyacrylamide gel electrophoresis (PAGE).
Crystallization and structure determination
The purified H2B E76K nucleosome, H2A.Z.1 R80C nucleosome, and H2B wild-type nucleosome were dialyzed against 20 mM potassium cacodylate (pH 6.0) buffer, containing 1 mM ethylenediaminetetraacetic acid (EDTA). For crystallization, 1 μl of the nucleosome samples (equivalent to 3.0 μg DNA/μl) was mixed with 1 μl of 20 mM potassium cacodylate (pH 6.0) buffer, containing 50 mM KCl and 110 mM MnCl2, and equilibrated against a reservoir solution of 20 mM potassium cacodylate (pH 6.0), 40 mM KCl and 70 mM MnCl2. The crystals of the H2B E76K nucleosome and the H2B wild-type nucleosome were cryoprotected with a 30% polyethylene glycol 400 solution, containing 20 mM potassium cacodylate (pH 6.0), 36 mM KCl, 63 mM MnCl2 and 5% trehalose, and were flash-cooled in liquid nitrogen. The X-ray diffraction data of the H2B wild-type nucleosome were collected at the beamline BL1A (wavelength: 1.10000 Å) at the Photon Factory (Tsukuba, Japan). The data of the H2B E76K nucleosome and the H2A.Z.1 R80C nucleosome were collected at the beamline BL41XU (wavelength: 1.00000 Å) at SPring-8 (Harima, Japan). The diffraction data were scaled and processed using the HKL2000 and CCP4 programs (22,23). The structures of the nucleosomes were determined by the molecular replacement method, using the PHASER program (24). For the H2B E76K nucleosome and the H2B wild-type nucleosome, the human nucleosome structure (PDB ID: 2CV5) was used as the search model for molecular replacement (25). For the H2A.Z.1 R80C nucleosome, the human H2A.Z.1 nucleosome structure (PDB ID: 3WA9) was used as the search model (26). The atomic coordinates were refined using the PHENIX and Coot programs (27,28). Structural graphics rendering and root mean square deviation (rmsd) value calculations were performed using the PyMOL program (http://pymol.org). The atomic coordinates of the H2B E76K nucleosome, the H2A.Z.1 R80C nucleosome and the H2B wild-type nucleosome have been deposited in the Protein Data Bank, with the PDB IDs: 5Y0D, 5Z30 and 5Y0C, respectively.
Thermal stability assay of nucleosomes
The stabilities of the purified nucleosomes were evaluated by a thermal stability assay, as previously described (29,30). This method monitors the fluorescence signal from SYPRO Orange, which binds hydrophobically to the histones released from the nucleosome by thermal denaturation. The thermal stability assay was performed in 19.6 mM Tris–HCl (pH 7.5) buffer, containing 0.9 mM dithiothreitol (DTT), 100 mM NaCl and SYPRO Orange (x5). The nucleosome concentrations were equivalent to 0.225 μg DNA/μl in the experiments shown in Figures 2 and 6, and to 0.135 μg DNA/μl in the experiments shown in Figure 7. The fluorescence signals of the SYPRO Orange were detected with a StepOnePlus Real-Time PCR unit (Applied Biosystems), using a temperature gradient from 26 to 95°C, in steps of 1°C/min. Raw fluorescence data were adjusted to normalized % values as (F(T) - F26°C)/(F95°C - F26°C), where F(T), F26°C and F95°C indicate each fluorescence at a particular temperature, the fluorescence at 26°C and the fluorescence at 95°C, respectively.
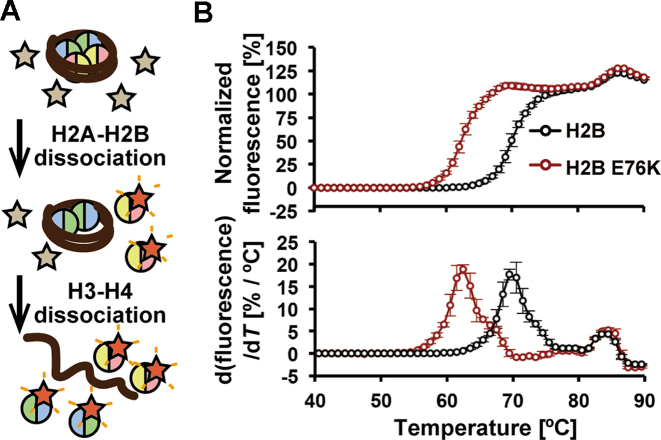
Figure 2.
The H2B E76K nucleosome is extremely unstable. (A) Schematic representation of the nucleosome thermal stability assay. In this assay, the histones that thermally dissociate from the nucleosome are detected with SYPRO Orange, as a fluorescent probe. In the first step, H2A-H2B dissociates from the nucleosome and is detected by the fluorescence of SYPRO Orange. In the second step, H3-H4 dissociates from the nucleosome and is detected in a similar manner. (B) Thermal stability assay with the H2B wild-type and H2B E76K nucleosomes. The upper panel shows the thermal stability curves of the H2B wild-type (black) and H2B E76K (red) nucleosomes. The bottom panel shows the differential values of the thermal stability curves presented in the upper panel. Means ± s.d. (n = 3) are shown.
Figure 6.
The H3.1 E97K mutation destabilizes the nucleosome and the histone complex. (A) Thermal stability assays of the H3.1 wild-type and H3.1 E97K nucleosomes. The upper panel shows the thermal stability curves of the H3.1 wild-type (black) and H3.1 E97K (red) nucleosomes. The bottom panel shows the differential values of the thermal stability curves presented in the upper panel. Means ± s.d. (n = 3) are shown. (B) Superdex 200 gel filtration chromatography. The red line indicates the elution profile of the H2A-H2B dimer and the H3.1 E97K-H4 tetramer. The black line indicates the elution profile of the H2A-H2B dimer and the H2A-H2B-H3-H4 complexes. (C and D) SDS-PAGE analyses of the elution fractions from the gel filtration chromatography shown in panel B. The experiments with wild-type H3.1 and H3.1 E97K are presented in panels C and D, respectively. The gels were stained with CBB. The full gel images of Figure 6C and D are shown in Supplementary Figure S13E and F, respectively. (E) Localization of GFP-H3.1 and GFP-H3.1 E97K in interphase and mitotic HeLa cells. Fluorescent (GFP) and bright-field (BF) images are shown with the merged images (merge). GFP-H3.1 E97K does not localize to the mitotic chromosomes. Bars, 10 μm. (F) Photobleaching assay. Bleaching a half of nucleus as in Figure 4 resulted in loss of fluorescence in GFP-H3.1 E97K because of a long bleaching period, during which free molecules are in and out of the bleached area. Therefore, a rapid imaging and bleaching assay was employed to evaluate the incorporation of GFP-H3.1 proteins. The relative intensity of bleached area was measured at ∼−2.0, −1.0, −0.0, 0.1, 0.3, 0.6, 1.0, 2.0, 3.0 and 4.0 s after bleaching. Averages with standard deviations are plotted (n = 17 and 15 for GFP-H3.1 and GFP-H3.1 E97K, respectively). The intensity of GFP-H3.1 E97K recovered to almost the original level within a few seconds, indicating that this H3.1 mutant is not stably incorporated into nucleosomes. A substantial recovery of wild-type H3.1 with a huge deviation was also observed. This high cell-to-cell variation of the rapidly recovered fraction likely reflects the amount of free GFP-H3.1 in cells at different cell cycle stages, as reported previously (32).
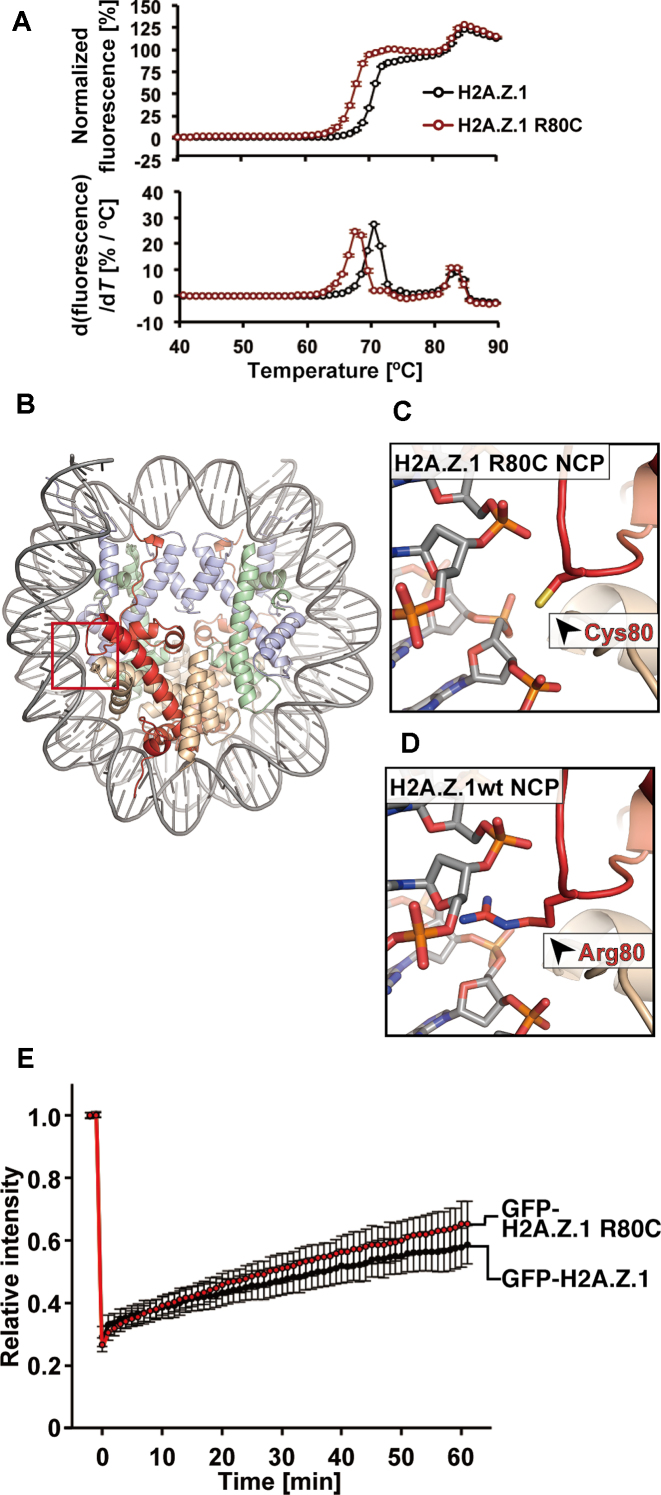
Figure 7.
The H2A.Z.1 R80C mutation destabilizes the nucleosome. (A) Thermal stability assay with the H2A.Z.1 wild-type and H2A.Z.1 R80C nucleosomes. The upper panel shows the thermal stability curves of the H2A.Z.1 wild-type (black) and H2A.Z.1 R80C (red) nucleosomes. The bottom panel shows the differential values of the thermal stability curves presented in the upper panel. Means ± s.d. (n = 3) are shown. (B) Overall crystal structure of the H2A.Z.1 R80C nucleosome. The H2A.Z.1 R80C, H2B, H3.1 and H4 molecules are colored red, orange, light blue and light green, respectively. (C) A close-up view around the Cys80 residue of H2A.Z.1 R80C in the H2A.Z.1 R80C nucleosome. (D) A close-up view around the Arg80 residue of H2A.Z.1 in the H2A.Z.1 wild-type nucleosome (PDB ID: 3WA9) (26). (E) Relative fluorescence intensity of the bleached area, normalized by the unbleached areas. Intensities were obtained from 2 min before photobleaching to 60 min after photobleaching. Averages with standard deviations are plotted (n = 24 and 20 for GFP-H2A.Z.1 and GFP-H2A.Z.1 R80C, respectively).
Nap1-mediated nucleosome formation assay
The purified H2A-H2B dimer or H2A-H2B E76K dimer (1.5, 3 and 4.5 pmol) and H3.1-H4 tetramer (0.75, 1.5 and 2.25 pmol) were incubated with 11.25 pmol of human Nap1 at 37°C for 15 min, in 3.7 μl of reaction solution. The pBluescript plasmid DNA containing eleven 5S repeats (5276 bp, 29 fmol), which was previously incubated with 1.7 units of calf thymus topoisomerase I (Takara) at 37°C for 60 min, was then added to the reaction mixture. The reactions were continued at 37°C for 60 min in 11 mM Tris–HCl buffer (pH 7.5), containing 84 mM NaCl, 0.43 mM MgCl2, 1.2 mM DTT, 0.73 mM EDTA, 1.4 mM 2-mercaptoethanol, 0.01 mM phenylmethylsulfonyl fluoride (PMSF) 0 and 1% glycerol (total volume: 4.7 μl). The reaction solutions were incubated with 2 μl of a proteinase K solution (1.67% sodium dodecyl sulphate (SDS) and 9.3 mg/ml proteinase K) for 15 min at room temperature, and the reactions were terminated. The DNA samples were then analyzed by 1% agarose gel electrophoresis in 1× Tris-acetate-EDTA (TAE) buffer, and were visualized by SYBR Gold staining (Invitrogen).
Gel filtration analysis
Purified H2A, H2B (or H2B E76K), H3.1 (or H3.1 E97K) and H4 were mixed and denatured in a 7 M guanidine hydrochloride solution, containing 20 mM Tris–HCl (pH 7.5) and 20 mM 2-mercaptoethanol, at 4°C for 60 min. In the gel filtration experiments, 1.3-fold excess amounts of H2A and H2B were used. The samples were then dialyzed against 10 mM Tris–HCl buffer (pH 7.5), containing 2 M sodium chloride, 1 mM EDTA and 5 mM 2-mercaptoethanol. After this dialysis step, the samples were subjected to gel filtration chromatography on a Superdex 200 pg 26/600 column (GE Healthcare). The peak fractions were analyzed by 18% SDS-PAGE with Coomassie Brilliant Blue (CBB) staining.
Fluorescence recovery after photobleaching (FRAP) analysis
The DNA fragment encoding green fluorescent protein (GFP)-fused H2B (HIST1H2BJ), H2B E76K, H3.1, or H3.1 E97K was inserted between the NheI and BamHI sites of the PB533A-2 vector (System Biosciences). The DNA fragment encoding GFP-fused H2A.Z.1 or H2A.Z.1 R80C was inserted between the NheI and BamHI sites of the PB510B-1 vector (System Biosciences). To establish the cells expressing GFP-H2B and GFP-H2B E76K, HeLa cells expressing proliferating cell nuclear antigen (PCNA)–mCherry were transfected with a Tn5-expression vector (System Biosciences) and either PB533A-2-GFP-H2B or PB533A-2-GFP-H2B E76K, using Lipofectamine 2000 (Thermo Fisher Scientific). To establish the cells expressing GFP-H3.1, GFP-H3.1 E97K, GFP-H2A.Z.1 and GFP-H2A.Z.1 R80C, HeLa cells were transfected with a Tn5-expression vector and either PB533A-2-GFP-H3.1, PB533A-2-GFP-H3.1 E97K, PB510B-1-GFP-H2A.Z.1 or PB510B-1-GFP-H2A.Z.1 R80C. HeLa cells were cultured as described previously (31). GFP-positive cells were sorted using a cell sorter, SH800Z (Sony). Fluorescence recovery after photobleaching (FRAP) was performed by the method described previously (31,32). For the cells expressing GFP-H2B, GFP-H2B E76K, GFP-H2A.Z.1 and GFP-H2A.Z.1 R80C, images were acquired every 1 min from 2 min before photobleaching to 60 min after photobleaching in the presence of 20 ng/ml cycloheximide (to monitor the exchange without the effect of newly synthesized protein molecules). For the cells expressing GFP-H3.1 or GFP-H3.1 E97K, images were acquired every 0.078–0.4 s, and the relative intensity was measured at ∼−2.0, −1.0, −0.0, 0.1, 0.3, 0.6, 1.0, 2.0, 3.0 and 4.0 s after bleaching. To obtain the relative intensities of the bleached area, the net intensities of the bleached areas (after background subtraction) were normalized to the net intensities of the unbleached areas in each time frame. The relative intensities in each time frame were then normalized to the intensity before bleaching. The half-lives of GFP-H2B and GFP-H2B E76K were calculated by fitting the FRAP curves of single cells to the exponential recovery curve, using Image J (1.47v). The box plot was generated using the BoxPlotR tool (http://shiny.chemgrid.org/boxplotr/).
Quantification of GFP-H2B or GFP-H2B E76K in HeLa cells
The whole cell extracts of the HeLa cells producing PCNA–mCherry and either GFP-H2B or GFP-H2B E76K were prepared to yield 7.5 × 106 cell equivalents per ml using 100 mM Tris–HCl (pH 6.8) buffer, containing 4% SDS, 20% glycerol and 0.2 M DTT. Three whole cell lysates (#1-#3) were independently prepared from HeLa cells producing GFP-H2B and GFP-H2B E76K. To estimate the amount of endogenous H2B, the whole cell lysates (5 × 104 cell equivalents) were analyzed by SDS-PAGE. The proteins were visualized by CBB staining. The calibration curve was obtained by the band intensities of purified recombinant H2B (50, 100, 200 and 400 ng), as the standard protein. To estimate the amount of GFP-H2B or GFP-H2B E76K, the whole cell lysates (3 × 104 cell equivalents) were analyzed by SDS-PAGE, followed by western blotting. The calibration curve was obtained by the band intensities of purified recombinant GFP-H2B (10, 20, 30 and 40 ng), as the standard protein. The GFP was detected by an ECL Western Blotting Detection System (GE Healthcare Biosciences), using a LAS4000 imager (Fujifilm). For the GFP detection, the anti-GFP rabbit polyclonal antibody (sc-8334, Santa Cruz Biotechnology) and the peroxidase-conjugated anti-rabbit IgG (NA9340, GE Healthcare Biosciences) were used as the primary and secondary antibodies, respectively. The band intensity was quantified using the Multi Gauge software (Fujifilm).
Composition of nucleosomes containing GFP-tagged histones in HeLa cells
The composition of nucleosomes containing GFP-tagged histones was analyzed as previously described (33). HeLa cells producing GFP-H2B, GFP-H2B E76K, GFP-H2A.Z.1 or GFP-H2A.Z.1 R80C were harvested and lysed, using a homogenizer. The insoluble fractions of the cell lysates were collected by centrifugation, and were digested with micrococcal nuclease (MNase, New England Biolabs) in 15 mM HEPES-NaOH (pH 7.4) buffer, containing 15 mM NaCl, 60 mM KCl, 340 mM sucrose, 1 mM CaCl2, 0.5 mM spermidine and 0.15 mM spermine, for 60 min at 30°C. After the MNase digestion, the insoluble fraction was collected by centrifugation and was suspended in a 500 mM NaCl solution containing 9 mM EDTA. The MNase-digested chromatin was then solubilized and those containing the GFP-fused histones were purified using GFP-Trap MA beads (ChromoTek). After the separation by SDS-PAGE and agarose gel electrophoresis, proteins and DNA were detected with CBB and GelRed (Biotium), respectively.
Colony formation assay
The DNA fragment encoding wild-type human H2B (HIST1H2BJ), H2B E76K, H3.1, H3.1 E97K, H2A.Z.1 or H2A.Z.1 R80C was inserted just downstream of the CAG promoter in pCAGGS, and the Internal Ribosome Entry Site (IRES) was placed downstream of each histone gene. The DNA fragment encoding GFP was then inserted downstream of IRES (pCAGGS-H2B E76K-IRES-GFP, pCAGGS-H2B-IRES-GFP, pCAGGS-H3.1-IRES-GFP, pCAGGS-H3.1 E97K-IRES-GFP, pCAGGS-H2A.Z.1-IRES-GFP or pCAGGS-H2A.Z.1 R80C-IRES-GFP). NIH3T3 cells were cultured in DMEM containing antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) and 10% Fetal bovine serum (FBS), and were transfected with these plasmids by the Nucleofector system (Lonza). At 40 h after transfection, about 500 cells were seeded onto 100-mm tissue culture dishes, and incubated for 10 days at 37°C, in a 5% CO2 atmosphere. After the incubation, the dishes were washed with PBS and stained with 4% Giemsa. Colonies containing over 50 cells were counted.
For the western blotting analysis, NIH3T3 cells were harvested at 48 h after transfection. The cells were washed with ice-cold PBS and lysed with RIPA buffer (10 mM Tris–HCl (pH 7.4), 150 mM sodium chloride, 5 mM EDTA, 1% Triton X-100 (Sigma-Aldrich), 1% sodium deoxycholate, 0.1% SDS and 0.2 mM PMSF). The cells were lysed by freezing and thawing, and after centrifugation, the supernatants were analyzed by western blotting. The GFP and β-actin were detected by an ECL Western Blotting Detection System (GE Healthcare). For the GFP detection, the anti-GFP rabbit polyclonal antibody (sc-8334, Santa Cruz Biotechnology) and the peroxidase-conjugated anti-rabbit IgG (NA934, GE Healthcare) were used as the primary and secondary antibodies, respectively. For the β-actin detection, the anti-β-actin mouse monoclonal antibody (013-24553, Wako) and the peroxidase-conjugated anti-mouse IgG (NA931, GE Healthcare) were used as the primary and secondary antibodies, respectively.
Cell cycle analysis
NIH3T3 cells (1.0 × 106) were transfected with pCAGGS-H2B E76K-IRES-GFP or pCAGGS-H2B-IRES-GFP by the Nucleofector system (Lonza), and were cultured in DMEM containing antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) and 10% FBS at 37°C, in a 5% CO2 atmosphere. At 40 h post-transfection, the cells were harvested and treated with 70% cold ethanol. After washing the cells with 1.5 ml of PBS, RNaseA (1.5 mg, SIGMA) was added, and the cells were incubated for 30 min at room temperature. After the RNaseA treatment, the cells were stained with propidium iodide (75 μg, SIGMA) for 30 min on ice. The propidium iodide intensity of each single cell was quantified by a flow cytometry system, BD FACS Calibur (BD Biosciences). The DNA content data for 100,000 cells were analyzed, and histograms were obtained using the FCS Express 6 software (De Novo Software).
Cell growth analysis
NIH3T3 cells (1.0 × 106) were transfected with pCAGGS-IRES-GFP, pCAGGS-H2B E76K-IRES-GFP, pCAGGS-H2B-IRES-GFP, pCAGGS-H3.1-IRES-GFP, pCAGGS-H3.1 E97K-IRES-GFP, pCAGGS-H2A.Z.1-IRES-GFP or pCAGGS-H2A.Z.1 R80C-IRES-GFP by the Nucleofector system (Lonza), and were cultured in DMEM containing antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) and 10% FBS at 37°C, in a 5% CO2 atmosphere. At 40 h post-transfection, the cells were harvested. The cells (2 × 104) were seeded onto 35-mm tissue culture dishes, and were incubated at 37°C, in a 5% CO2 atmosphere. The cells were harvested every 24 h for 7 days from independent dishes, and the cell numbers were manually counted.
RESULTS
The H2B E76K mutation changes the nucleosome structure at the interface between H2B and H4
Somatic missense mutations in canonical H2B genes have been reported in several types of cancer cells (13–15). We obtained the somatic mutation data for seventeen H2B genes (HIST1H2BB, HIST1H2BC, HIST1H2BD, HIST1H2BE, HIST1H2BF, HIST1H2BG, HIST1H2BH, HIST1H2BI, HIST1H2BJ, HIST1H2BK, HIST1H2BL, HIST1H2BM, HIST1H2BN, HIST1H2BO, HIST2H2BE, HIST2H2BF and HIST3H2BB) from the cBioPortal for Cancer Genomics database (34,35), and found that mutations of the H2B Glu76 residue occur with high frequency, implying that the H2B Glu76 mutations may be involved in cancer progression (Supplementary Figure S1). Among the H2B Glu76 mutations, the E76K mutation, in which the Glu76 residue is replaced by Lys, is the predominant substitution. If the H2B E76K mutation within one of the seventeen H2B genes is involved in cancer progression, then the presence of the H2B E76K mutant contaminating large amounts of wild-type H2B may dominantly induce the oncogenic transformation.
To study the effect of the H2B E76K mutation on the structure of the nucleosome, we first purified the H2B E76K mutant, and found that it was efficiently incorporated into nucleosomes (Supplementary Figure S2). The crystal structure of the H2B E76K nucleosome was then determined at 1.99 Å resolution (Figure 1A and Supplementary Table S1). For comparison, we also determined the canonical nucleosome structure containing wild-type H2B with a similar resolution, at 2.09 Å (Supplementary Table S1).
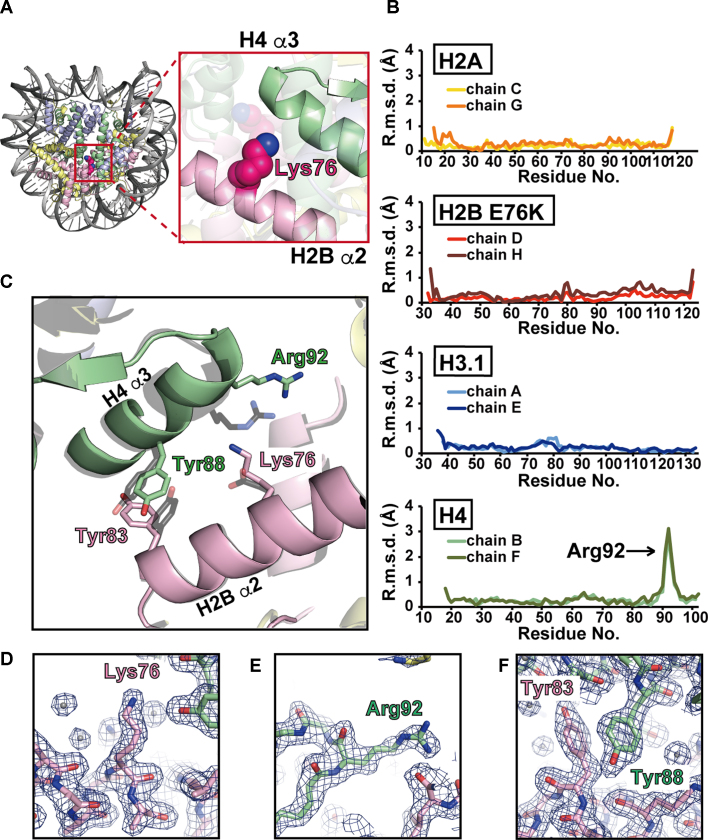
Figure 1.
Crystal structure of the H2B E76K nucleosome. (A) Left panel: overall structure of the H2B E76K nucleosome. The H2A, H2B E76K, H3.1 and H4 molecules are colored yellow, pink, blue and green, respectively. Right panel: close-up view around the Lys76 residue of H2B E76K. (B) The H2B E76K substitution significantly changed the backbone geometry around Arg92 of H4. The root mean square deviation (r.m.s.d.) values between the corresponding Cα atoms of the H2B E76K and wild-type H2B molecules in the nucleosomes are plotted against the amino acid residues. Two H2A molecules (chains C and G), two H2B molecules (chains D and H), two H3.1 molecules (chains A and E) and two H4 molecules (chains B and F) in the nucleosomes were independently compared. (C) The structures around the H2B Lys76 residue of the nucleosomes. The H4 and H2B molecules in the H2B E76K nucleosome are colored green and pink, respectively. The H4 and H2B molecules in the wild-type H2B nucleosome are colored gray. The structures of the H2B E76K and wild-type H2B nucleosomes are superimposed. (D–F) Close-up views of the Lys76 residue of H2B E76K (D), the Arg92 residue of H4 (E) and the Tyr83 residue of H2B E76K (F) in the H2B E76K nucleosome. Electron density maps are presented at the 1.0σ level.
We superimposed the H2B E76K nucleosome structure on the wild-type H2B nucleosome structure, and compared the backbone geometries of all of the histones. The rmsd values of the corresponding Cα atoms between the H2B E76K and wild-type nucleosomes were plotted (Figure 1B). Two histone peptides for H2A (chains C and G), H2B (chains D and H), H3.1 (chains A and E) and H4 (chains B and F) in the crystal structures were independently compared. We found that the H2B E76K mutation did not affect the backbone geometries of H2B, H2A and H3.1 in the nucleosome (Figure 1B, first, second and third rows). However, surprisingly, the H2B E76K substitution significantly changed the backbone geometry around Arg92 of H4 (Figure 1B, bottom row). In fact, in the H2B E76K nucleosome, the H2B Lys76 side chain caused electrostatic repulsion with the H4 Arg92 residue, and drastically changed the configuration of the H4 α3-helix (Figure 1C–E). Interestingly, the stacking interaction of the side chain moieties between the H2B Tyr83 and H4 Tyr88 residues was allosterically perturbed by the H2B E76K substitution (Figure 1C and F). Therefore, the H2B E76K mutation substantially changes the interaction between H2B and H4 within the nucleosome, and this modified H2B–H4 interaction may destabilize the H2B E76K nucleosome.
The H2B E76K nucleosome is unstable
We then performed a thermal stability assay (Figure 2A), in which the histones that thermally dissociate from the nucleosome are detected with SYPRO Orange, as a fluorescent probe (29,30). In the wild-type nucleosome, the two-step histone dissociation is detected as a bi-phasic thermal denaturation curve, in which the first and second peaks correspond to the H2A-H2B dissociation and the H3-H4 dissociation, respectively (Figure 2B). Intriguingly, in the H2B E76K nucleosome, the H2A-H2B E76K dissociation (first peak) occurred at a remarkably lower temperature (∼10°C difference), as compared to the H2A-H2B dissociation in the wild-type nucleosome (Figure 2B). In contrast, the H3-H4 dissociation (second peak) occurred at the same temperature between the H2B E76K and wild-type nucleosomes (Figure 2B). These results indicated that the H2A-H2B E76K complexes are weakly associated in the nucleosome.
The H2A-H2B E76K dimer does not bind to the H3-H4 complex without DNA
We then assessed the interaction between H2A-H2B and H3-H4 by gel filtration chromatography. As shown in Figure 3A (the chromatogram with the black line), wild-type H2A-H2B bound to H3-H4 in the absence of DNA, and efficiently formed the histone complex corresponding to the histone octamer or hexamer (Figure 3B). In contrast, the peak corresponding to the histone octamer or hexamer was not detected, when H2A-H2B E76K was incubated with H3-H4 (Figure 3A, the chromatogram with the red line, and Figure 3C). These results clearly showed that H2A-H2B E76K is defective in binding to H3-H4 without DNA. Therefore, the defective interaction between H2A-H2B E76K and H3-H4 may be responsible for the instability of the H2B E76K nucleosome.
Figure 3.
The H2A-H2B E76K dimer is defective in H3-H4 binding. (A) Superdex 200 gel filtration chromatography. The red line indicates the elution profile of the H2A-H2B E76K dimer and the H3-H4 tetramer. The black line indicates the elution profile of the H2A-H2B dimer and the H2A-H2B-H3-H4 complexes. (B and C) SDS-PAGE analyses of the elution fractions from the gel filtration chromatography shown in panel A. The experiments with wild-type H2B and H2B E76K are presented in panels B and C, respectively. The gels were stained with CBB. The full gel images of Figure 3B and C are shown in Supplementary Figures S13A and B, respectively.
H2B E76K is incorporated into chromatin and rapidly exchanged in living cells
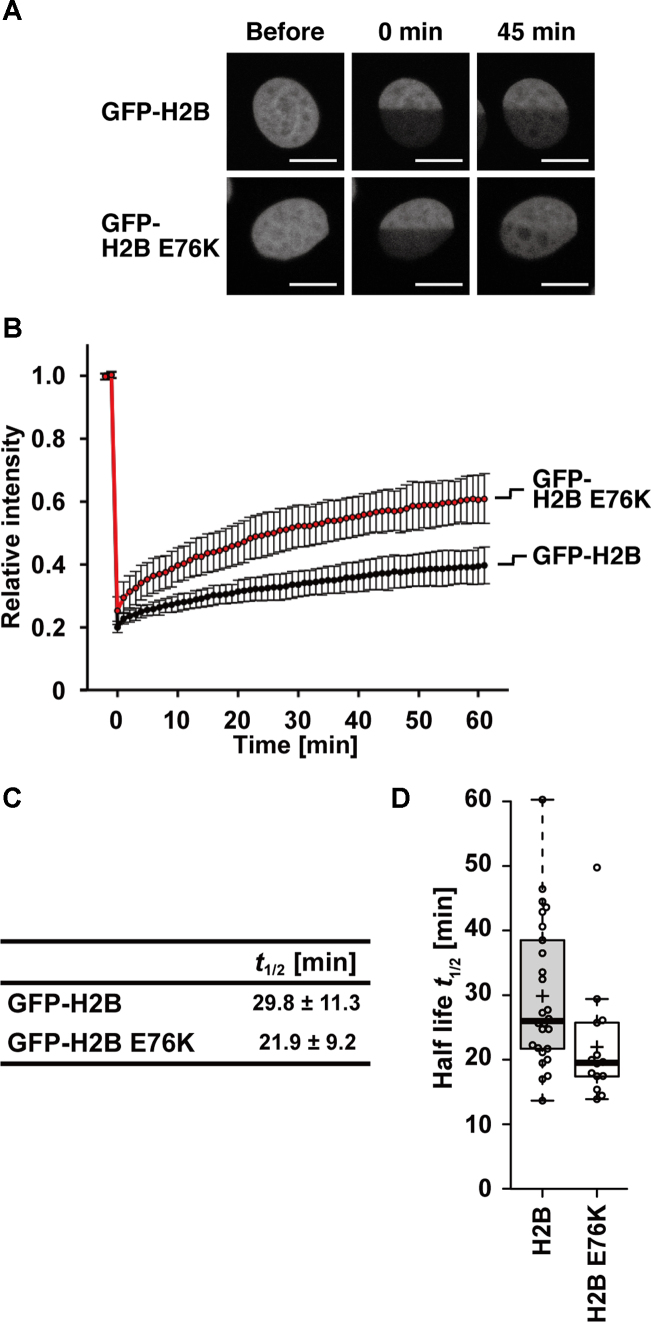
We next produced H2B E76K as a GFP-fusion protein in living cells, to compare its exchange rate with that of the wild-type H2B by FRAP (32). In stably expressing HeLa cells, the levels of GFP-H2B E76K and GFP-H2B were estimated to be ∼9% and ∼14% of the endogenous H2B, respectively (Supplementary Figure S3A and B). Similar to the wild-type H2B, GFP-H2B E76K was concentrated in interphase nuclei and in mitotic chromosomes (Supplementary Figure S3C). To assess whether GFP-H2B E76K is actually incorporated into nucleosomes in HeLa cells, we performed a micrococcal nuclease digestion followed by immunoprecipitation, using anti-GFP nanobody beads (Supplementary Figure S4A and B). GFP-H2B E76K was recovered in the nucleosome fraction, and the stoichiometry of endogenous H2B was lower in the nucleosomes containing GFP-H2B E76K, as in those containing GFP-H2B, suggesting that H2B E76K is properly incorporated into chromatin. To examine whether the H2B E76K mutant can be assembled into nucleosomes by a histone chaperone in vitro, we performed a nucleosome assembly assay with the prominent histone chaperone, Nap1 (Supplementary Figure S5A) (36). As expected, the H2B E76K mutant was incorporated into the nucleosomes; however, we reproducibly observed that the nucleosome formation with the H2B E76K mutant was subtly reduced, as compared to that with the wild-type H2B (Supplementary Figure S5B and C). We then performed FRAP by bleaching one-half of each nucleus (Figure 4A). The first point after bleaching was higher in H2B E76K than the wild type and the recovery kinetics was faster in the mutant (Figure 4B–D), indicating that H2B E76K is exchanged more rapidly than wild-type H2B. Thus, the nucleosome containing H2B E76K is also less stable than the wild-type H2B in living cells, in good agreement with the in vitro biochemical properties.
Figure 4.
Mobility of GFP-H2B E76K in living cells. (A) Representative fluorescence images of HeLa cell nuclei producing GFP-H2B or GFP-H2B E76K, before and after photobleaching. One-half of the nuclei were bleached using a 488-nm laser. Bars, 10 μm. (B) Relative fluorescence intensity of the bleached area, normalized by the unbleached areas. Intensities were obtained from 2 min before photobleaching to 60 min after photobleaching. Averages with standard deviations are plotted (n = 28 and 22 for GFP-H2B and GFP-H2B E76K respectively). (C) The half-lives of GFP-H2B and GFP-H2B E76K were estimated from the averages of FRAP curves of 26 and 14 cells for GFP-H2B and GFP-H2B E76K, respectively. (D) Box plots of the half-lives of GFP-H2B and GFP-H2B E76K FRAP curves.
Exogenous H2B E76K production affects colony formation ability of cells
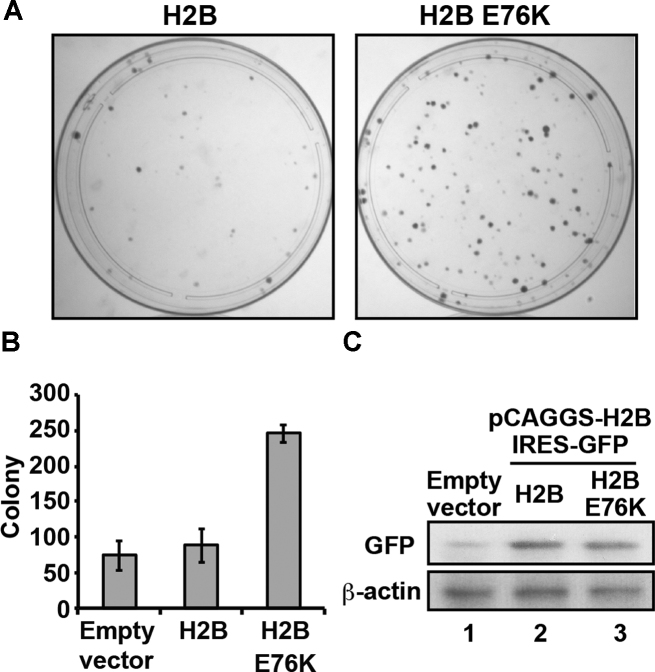
We then produced H2B E76K or wild-type H2B in NIH3T3 cells by transient transfection. At 10 days post-transfection, the colony number of the cells producing H2B E76K was higher than that of the cells producing wild-type H2B (Figure 5A and B). To test whether the H2B E76K mutation caused abnormal cell cycle progression, we measured the DNA content of an asynchronous population of the cells producing H2B E76K by flow cytometry, as previously reported for yeast histone mutants (37–39). The cells producing H2B E76K did not exhibit abnormal cell cycle progression, suggesting that this mutation did not drastically affect the cell-cycle progression (Supplementary Figure S6A). Consistently, the growth rate of the cells was not affected by the expression of the H2B E76K mutant (Supplementary Figure S6B). These results indicated that the production of the H2B E76K mutant promotes an oncogenic-like transformation, with reduced contact inhibition and enhanced colony formation ability, although the growth rate of the cells is not significantly affected. This may explain the dominant negative phenotype of the H2B E76K mutation in cancer cells. In this system, the mRNA encoding H2B E76K or wild-type H2B was expressed together with an mRNA encoding GFP as polycistronic genes, and stoichiometric amounts of exogenous H2B and GFP were produced in the cells. Therefore, the exogenous H2B production can be monitored as the GFP signal in cells. Similar amounts of GFP were detected in the cells expressing H2B E76K and wild-type H2B (Figure 5C), confirming that the production levels of H2B E76K and wild-type H2B were not substantially different in these cells.
Figure 5.
Exogenous H2B E76K production enhances malignant transformation of cells. (A) Colony formation assay with NIH3T3 cells expressing H2B or H2B E76K. Representative images are shown. Three independent experiments were performed, and similar results were obtained. (B) Graphic representation of the experiments shown in panel A. The colony numbers of cells expressing H2B or H2B E76K are plotted. Means ± s.d. (n = 3) are shown. (C) Western blotting analyses. The production of exogenous H2B and H2B E76K was detected by western blotting, using the α-GFP polyclonal antibody (upper panel). In this system, GFP was simultaneously produced with H2B or H2B E76K, from a polycistronic gene. For loading controls, endogenous β-actin was detected by western blotting using the anti-β-actin antibody (lower panel). The full images of the western blots are shown in Supplementary Figures S13C and D.
Somatic missense mutations, H3.1 E97K and H2A.Z.1 R80C, destabilize the nucleosome
It is intriguing to investigate whether the nucleosome-destabilizing character of the H2B E76K mutation is common in other canonical histone mutations. We investigated the somatic mutation data for canonical histone genes in the cBioPortal for Cancer Genomics database (34,35). We searched the somatic mutation data for ten H3 genes (HIST1H3A, HIST1H3B, HIST1H3C, HIST1H3D, HIST1H3E, HIST1H3F, HIST1H3G, HIST1H3H, HIST1H3I and HIST1H3J), and found that mutations of the H3 Glu97 residue occur with high frequency, similar to the H2B Glu76 mutations (Supplementary Figure S1). We prepared the H3 E97K mutant, and found that H3 E97K efficiently formed nucleosomes (Supplementary Figure S7). Our thermal stability assay revealed that the H3 E97K nucleosome exhibited a monophasic dissociation profile, and thus the H3 E97K-H4 dissociation may simultaneously occur with the H2A-H2B dissociation (Figure 6A). This indicated that the H3 E97K nucleosome was substantially unstable, as compared to the wild-type nucleosome. Interestingly, a gel filtration chromatographic analysis indicated that H3 E97K-H4 is defective in binding to H2A-H2B without DNA (Figure 6B–D). Our in vivo assays revealed that the H3 E97K mutant was minimally incorporated into chromosomes in cells (Figure 6E and F). However, the H3 E97K mutant slightly, but clearly, enhanced the colony formation ability of the cells (Supplementary Figure S8), although the growth rate of the cells was not affected (Supplementary Figure S9A). The H3 E97K mutant may compete with wild-type H3.1, and may perturb the proper function of H3.1. Further studies will be required to understand how the H3 E97K mutant enhances the colony formation ability of the cells.
To test whether the unstable nucleosome character by the cancer-associated mutations is common to histone variants, we searched the cBioPortal for the H2AFZ gene, a prominent histone variant (34,35). We found the H2A.Z.1 R80C mutant, in which the Arg80 residue is replaced by cysteine, as a cancer-associated somatic mutation (Supplementary Figure S1). We reconstituted the nucleosome containing the H2A.Z.1 R80C mutant (Supplementary Figure S10). The H2A.Z.1 R80C nucleosome migrated more slowly than the wild-type H2A.Z.1 nucleosome on a native polyacrylamide gel (Supplementary Figure S10A). A similar unusual gel mobility profile was observed for the H3 E97K nucleosome (Supplementary Figure S7A). These results suggest that, as in the H3 E97K nucleosome, the physical characteristics of the H2A.Z.1 R80C nucleosome are different from those of the wild-type H2A.Z.1 nucleosome. Consistent with this idea, the nucleosome containing H2A.Z.1 R80C was clearly unstable, as compared to the wild-type H2A.Z.1 nucleosome (Figure 7A). However, in contrast to the H3 E97K mutant, the H2A.Z.1 R80C mutant was incorporated into chromatin in cells (Figure 7E and Supplementary Figure S11). The mobility of the H2A.Z.1 R80C mutant was slightly faster than that of wild-type H2A.Z.1, but the H2A.Z.1 R80C production did not affect the cell growth and colony formation ability of the cells (Supplementary Figures S8 and 9). The crystal structure of the H2A.Z.1 R80C nucleosome revealed that the H2A.Z.1 Arg80 residue is located near the DNA backbone, and the local structure around the H2A.Z.1 R80 residue is changed by the R80C mutation (Figure 7B–D and Supplementary Figure S12). The perturbation of the local DNA–histone interaction induced by the H2A.Z.1 R80C mutation may be the structural basis for the nucleosome instability.
DISCUSSION
In the present study, we determined the crystal structure of the nucleosome containing the H2B E76K mutant at 1.99 Å resolution (Figure 1). This high-resolution analysis of the H2B E76K nucleosome structure revealed the detailed structural differences from the canonical nucleosome. We surprisingly found that only one amino acid substitution, at the H2B Glu76 residue, drastically perturbs the local histone interactions between the H2B α2 and H4 α3 helices within the nucleosome (Figure 1). Consistently, biochemical analyses revealed that the H2B E76K mutation reduces the interaction between H2A-H2B and H3-H4, and destabilizes the nucleosome (Figures 2 and 3).
The H2B Glu76 residue is frequently mutated in cancer cells (34,35). Therefore, the H2B E76K-mediated structural change may induce the oncogenic transformation in cells producing H2B E76K. We found that the H2B E76K mutant is efficiently incorporated into chromatin in cells (Figure 4; Supplementary Figures S3 and 4), and the production of the H2B E76K mutant transforms the cells (Figure 5). How does the nucleosome instability induced by the H2B E76K incorporation contribute to the oncogenic transformation of cells? One plausible explanation is that the unstable H2B E76K nucleosome may disturb the proper chromatin-mediated regulation of genomic DNA functions. This idea may be supported if other cancer-associated histone mutations also form unstable nucleosomes. In this context, we found that the cancer-associated R80C mutation of a major histone variant, H2A.Z.1, destabilizes the nucleosome (Figure 7A), although the colony formation ability of the cells producing H2A.Z.1 R80C seemed to be slightly enhanced (Supplementary Figure S8).
Another cancer-associated mutant of histone H3.1, H3.1 E97K, also destabilizes the nucleosome (Figure 6). The colony formation ability of the cells producing H3.1 E97K was enhanced (Supplementary Figure S8). However, the H3.1 E97K mutant may affect the cells by a different mechanism from that of the H2B E76K mutant, because it is not efficiently incorporated into chromatin in cells (Figure 6E and F). The H3.1 E97K mutant may inhibit the proper function of wild-type H3.1, probably by competing with interactions with the histone assembly complexes and/or histone chaperones, prior to the H3.1 deposition into the chromatin. Further studies are awaited to clarify this issue.
Mutations of the Lys27, Gly34 and Lys36 residues in the histone variant H3.3 have been reported in pediatric gliomas and chondroblastomas (7–9). The H3.3 Lys27, Gly34 and Lys36 residues, which are located in the N-terminal tail region, are highly conserved among histone variants, and are exposed to the solvent in the nucleosome (40,41). Therefore, these mutations may not directly affect the nucleosome structure and stability, and instead mainly function as binding sites for histone binding proteins, such as the histone modifiers and readers for post-translational modifications (10–12). In fact, our crystallographic analyses revealed that the mutations of these residues did not affect the structure of the nucleosome (data not shown). In contrast, we found that the H2B E76K and H2A.Z.1 R80C mutations substantially change the nucleosome structure and stability. In addition, our preliminary structural analysis of the nucleosome containing the H3.1 E97K mutation suggested that this nucleosome is structurally different from the wild-type nucleosome, as revealed by a small angle X-ray scattering analysis (data not shown). Therefore, the histone mutations found in the histone-fold domain may affect the cell fate in different pathways from the mutations in the N-terminal tail regions. The defective histone–histone or histone–DNA interaction inducing nucleosome instability may be a common characteristic for a certain group of cancer-associated histone mutations located in the histone-fold domain.
Recent evidence has suggested that specific chromatin structures, formed with certain histone modifications and variants, may play a central role in the epigenetic regulation of genomic DNA (10,17,18,42,43). The distributed incorporation of the H2A-H2B E76K and H2A.Z.1 R80C-H2B dimers may disturb such chromatin-mediated genomic DNA regulation. In general, nucleosomes negatively impact transcription (44,45). The disseminated H2B E76K and H2A.Z.1 R80C incorporation, which destabilizes the nucleosome, may reduce the nucleosome barrier during transcription processes throughout the genome. Such global de-suppression of transcription may induce the undesired expression of oncogenes, which are usually suppressed by chromatin, and potentially stimulate cancer progression.
DATA AVAILABILITY
The atomic coordinates of the H2B E76K nucleosome, H2A.Z.1 R80C nucleosome and the H2B wild-type nucleosome have been deposited in the Protein Data Bank, with the ID codes 5Y0D, 5Z30 and 5Y0C, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Ms Yukari Iikura (The University of Tokyo) for her assistance and Dr Masaaki Sugiyama (Kyoto University) for the X-ray scattering analysis. We thank the beamline scientists at the BL1A station of the Photon Factory and the BL41XU station of SPring-8 for their assistance with data collection. The synchrotron radiation experiments were performed with the approval of the Japan Synchrotron Radiation Research Institute (JASRI) [proposal nos. 2014A1042, 2014B1125 and 2015A1020] and the Photon Factory Program Advisory Committee [proposal nos. 2014G174 and 2014G556]. We also thank Sam-Yong Park (Yokohama City University) for advice about the structural refinement.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Waseda Research Institute for Science and Engineering; Research support programs of Waseda University; JSPS KAKENHI Grant [JP18H05534, JP25116002, JP17H01408 to H.Ku.; JP17K15043 to Y.A.; JP25116005 to H.Ki.; JP25116010 to Y.O.; JP16H01312 to S.T.; JP25116009 to M.H., JP16H01307 to T.I., in part]; JST CREST Grant [JPMJCR16G1 to H.Ku., H.Ki., Y.O., in part]; Platform Project for Supporting Drug Discovery and Life Science Research Japan Society for the Promotion of Science (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED [JP18am0101076 to H.Ku., in part]; Uehara Memorial Foundation (to H.Ku.). Funding for open access charge: Waseda University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J.. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997; 389:251–260. [DOI] [PubMed] [Google Scholar]
- 2. Tan S., Davey C.A.. Nucleosome structural studies. Curr. Opin. Struct. Biol. 2011; 21:128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y.. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004; 116:51–61. [DOI] [PubMed] [Google Scholar]
- 4. Marzluff W.F., Gongidi P., Woods K.R., Jin J., Maltais L.J.. The human and mouse replication-dependent histone genes. Genomics. 2002; 80:487–498. [PubMed] [Google Scholar]
- 5. Talbert P.B., Ahmad K., Almouzni G., Ausió J., Berger F., Bhalla P.L., Bonner W.M., Cande W.Z., Chadwick BP, Chan S.W. et al. A unified phylogeny-based nomenclature for histone variants. Epigenet. Chromatin. 2012; 5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Albig W., Kioschis P., Poustka A., Meergans K., Doenecke D.. Human histone gene organization: nonregular arrangement within a large cluster. Genomics. 1997; 40:314–322. [DOI] [PubMed] [Google Scholar]
- 7. Schwartzentruber J., Korshunov A., Liu X.Y., Jones D.T., Pfaff E., Jacob K., Sturm D., Fontebasso A.M., Quang D.A., Tönjes M. et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012; 482:226–231. [DOI] [PubMed] [Google Scholar]
- 8. Wu G., Broniscer A., McEachron T.A., Lu C., Paugh B.S., Becksfort J., Qu C., Ding L., Huether R., Parker M. et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012; 44:251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Behjati S., Tarpey P.S., Presneau N., Scheipl S., Pillay N., Van Loo P., Wedge D.C., Cooke S.L., Gundem G., Davies H. et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat. Genet. 2013; 45:1479–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kouzarides T. Chromatin modifications and their function. Cell. 2007; 128:693–705. [DOI] [PubMed] [Google Scholar]
- 11. Lewis P.W., Müller M.M., Koletsky M.S., Cordero F., Lin S., Banaszynski L.A., Garcia B.A., Muir T.W., Becher O.J., Allis C.D.. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013; 340:857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Herz H.M., Morgan M., Gao X., Jackson J., Rickels R., Swanson S.K., Florens L., Washburn M.P., Eissenberg J.C., Shilatifard A.. Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science. 2014; 345:1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nayak S.R., Harrington E., Boone D., Hartmaier R., Chen J., Pathiraja T.N., Cooper K.L., Fine J.L., Sanfilippo J., Davidson N.E. et al. A role for histone H2B variants in endocrine-resistant breast cancer. Horm. Cancer. 2015; 6:214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J.F., Wyczalkowski M.A. et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013; 502:333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Loudin M.G., Wang J., Leung H.C., Gurusiddappa S., Meyer J., Condos G., Morrison D., Tsimelzon A., Devidas M., Heerema N.A. et al. Genomic profiling in down syndrome acute lymphoblastic leukemia identifies histone gene deletions associated with altered methylation profiles. Leukemia. 2011; 25:1555–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Martincorena I., Campbell P.J.. Somatic mutation in cancer and normal cells. Science. 2015; 349:1483–1489. [DOI] [PubMed] [Google Scholar]
- 17. Maze I., Noh K.M., Soshnev A.A., Allis C.D.. Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat. Rev. Genet. 2014; 15:259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Venkatesh S., Workman J.L.. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 2015; 16:178–189. [DOI] [PubMed] [Google Scholar]
- 19. Tachiwana H., Kagawa W., Osakabe A., Kawaguchi K., Shiga T., Hayashi-Takanaka Y., Kimura H., Kurumizaka H.. Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:10454–10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tanaka Y., Tawaramoto-Sasanuma M., Kawaguchi S., Ohta T., Yoda K., Kurumizaka H., Yokoyama S.. Expression and purification of recombinant human histones. Methods. 2004; 33:3–11. [DOI] [PubMed] [Google Scholar]
- 21. Dyer P.N., Edayathumangalam R.S., White C.L., Bao Y., Chakravarthy S., Muthurajan U.M., Luger K.. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004; 375:23–44. [DOI] [PubMed] [Google Scholar]
- 22. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 23. Collaborative Computational Project Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994; 50:760–763. [DOI] [PubMed] [Google Scholar]
- 24. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsunaka Y., Kajimura N., Tate S., Morikawa K.. Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle. Nucleic Acids Res. 2005; 33:3424–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Horikoshi N., Sato K., Shimada K., Arimura Y., Osakabe A., Tachiwana H., Hayashi-Takanaka Y., Iwasaki W., Kagawa W., Harata M. et al. Structural polymorphism in the L1 loop regions of human H2A.Z.1 and H2A.Z.2. Acta Crystallogr. D Biol. Crystallogr. 2013; 69:2431–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 29. Taguchi H., Horikoshi N., Arimura Y., Kurumizaka H.. A method for evaluating nucleosome stability with a protein-binding fluorescent dye. Methods. 2014; 70:119–126. [DOI] [PubMed] [Google Scholar]
- 30. Arimura Y., Shirayama K., Horikoshi N., Fujita R., Taguchi H., Kagawa W., Fukagawa T., Almouzni G., Kurumizaka H.. Crystal structure and stable property of the cancer-associated heterotypic nucleosome containing CENP-A and H3.3. Sci. Rep. 2014; 4:7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Horikoshi N., Sato K., Shimada K., Arimura Y., Osakabe A., Tachiwana H., Hayashi-Takanaka Y., Iwasaki W., Kagawa W., Harata M. et al. Structural polymorphism in the L1 loop regions of human H2A.Z.1 and H2A.Z.2. Acta Crystallogr. D Biol. Crystallogr. 2013; 69:2431–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kimura H., Cook P.R.. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 2001; 153:1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kujirai T., Horikoshi N., Sato K., Maehara K., Machida S., Osakabe A., Kimura H., Ohkawa Y., Kurumizaka H.. Structure and function of human histone H3.Y nucleosome. Nucleic Acids Res. 2016; 44:6127–6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013; 6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Osakabe A., Tachiwana H., Matsunaga T., Shiga T., Nozawa R.S., Obuse C., Kurumizaka H.. Nucleosome formation activity of human somatic nuclear autoantigenic sperm protein (sNASP). J. Biol. Chem. 2010; 285:11913–11921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Baker S.P., Phillips J., Anderson S., Qiu Q., Shabanowitz J., Smith M.M., Yates J.R. 3rd, Hunt D.F., Grant P.A.. Histone H3 Thr 45 phosphorylation is a replication-associated post-translational modification in S. cerevisiae. Nat. Cell Biol. 2010; 12:294–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yu Y., Srinivasan M., Nakanishi S., Leatherwood J., Shilatifard A., Sternglanz R.. A conserved patch near the C terminus of histone H4 is required for genome stability in budding yeast. Mol. Cell. Biol. 2011; 31:2311–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chavez M.S., Scorgie J.K., Dennehey B.K., Noone S., Tyler J.K., Churchill M.E.. The conformational flexibility of the C-terminus of histone H4 promotes histone octamer and nucleosome stability and yeast viability. Epigenet. Chromatin. 2012; 5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kurumizaka H., Horikoshi N., Tachiwana H., Kagawa W.. Current progress on structural studies of nucleosomes containing histone H3 variants. Curr. Opin. Struct. Biol. 2013; 23:109–115. [DOI] [PubMed] [Google Scholar]
- 41. Koyama M., Kurumizaka H.. Structural diversity of the nucleosome. J. Biochem. 2018; 163:85–95. [DOI] [PubMed] [Google Scholar]
- 42. Margueron R., Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat. Rev. Genet. 2010; 11:285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vardabasso C., Hasson D., Ratnakumar K., Chung C.Y., Duarte L.F., Bernstein E.. Histone variants: emerging players in cancer biology. Cell Mol. Life Sci. 2013; 71:379–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bondarenko V.A., Steele L.M., Ujvári A., Gaykalova D.A., Kulaeva O.I., Polikanov Y.S., Luse D.S., Studitsky V.M.. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol. Cell. 2006; 24:469–479. [DOI] [PubMed] [Google Scholar]
- 45. Petesch S.J., Lis J.T.. Overcoming the nucleosome barrier during transcript elongation. Trends Genet. 2012; 28:285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates of the H2B E76K nucleosome, H2A.Z.1 R80C nucleosome and the H2B wild-type nucleosome have been deposited in the Protein Data Bank, with the ID codes 5Y0D, 5Z30 and 5Y0C, respectively.