Abstract
Small heat shock proteins (sHsps) function mainly as molecular chaperones that play vital roles in response to diverse stresses, especially high temperature. However, little is known about the molecular characteristics and evolutionary history of the sHsp family in Salix suchowensis, an important bioenergy woody plant. In this study, 35 non-redundant sHsp genes were identified in S. suchowensis, and they were divided into four subfamilies (C, CP, PX, and MT) based on their phylogenetic relationships and predicted subcellular localization. Though the gene structure and conserved motif were relatively conserved, the sequences of the Hsp20 domain were diversified. Eight paralogous pairs were identified in the Ssu-sHsp family, in which five pairs were generated by tandem duplication events. Ka/Ks analysis indicated that Ssu-sHsps had undergone purifying selection. The expression profiles analysis showed Ssu-Hsps tissue-specific expression patterns, and they were induced by at least one abiotic stress. The expression correlation between two paralogous pairs (Ssu-sHsp22.2-CV/23.0-CV and 23.8-MT/25.6-MT) were less than 0.6, indicating that they were divergent during the evolution. Various cis-acting elements related to stress responses, hormone or development, were detected in the promoter of Ssu-sHsps. Furthermore, the co-expression network revealed the potential mechanism of Ssu-sHsps under stress tolerance and development. These results provide a foundation for further functional research on the Ssu-sHsp gene family in S. suchowensis.
Keywords: small heat shock proteins (sHsp), phylogenetic analysis, gene structure, expression analysis, Salix suchowensis
1. Introduction
Plants are often exposed to complex environmental stimuli, so they inevitably suffer various stresses including extreme temperature, flooding, drought, and salinity, leading to restriction of their growth or even cause death. As a sessile organism, plants have evolved sophisticated mechanisms to accommodate these stresses over long lifespans [1]. Heat shock proteins (Hsps) function as molecular chaperones that are involved in the folding, localization, repairing, and degradation of protein molecules in all living organisms and play vital roles in response to diverse stresses [2].
Hsps can be divided into five major groups, including Hsp100, Hsp90, Hsp70, Hsp60, and small Hsp (sHsp)/Hsp20 according to their protein molecular weight [3]. The monomer molecular weight of sHsp ranges from 15 to 42 kDa. As molecular chaperones, sHsp perform functions in an ATP-independent manner through forming a complex with the denatured proteins to avoid protein degradation [4]. On the basis of the subcellular localization, sHsps are divided into five subfamilies: cytosol (C) subfamilies (CI-CVI) are located in the cytoplasm or nucleus; mitochondria (M) subfamilies (MTI and MTII) are located in the mitochondria; the endoplasmic reticulum (ER), chloroplast (CP), and peroxisome (PX) subfamilies are located in the endoplasmic reticulum, chloroplast, and peroxisome, respectively [5]. The essential characteristic of sHsp is the presence of a highly conserved 80–100 amino acid α-crystallin domain (ACD)/Hsp20 domain which are located in the C-terminal region. This domain is composed of a series of β-strand structures and can be divided into three parts, N-terminal consensus region I (CRI) with β2-β3-β4-β5, C-terminal consensus region II (CR II) with β7-β8-β9, and a hydrophobic region loop (β6-loop) that connects CRI and CRII [5].
To date, sHsps have been identified in various organisms from lower eukaryotes to higher plants and animals, two sHsps were identified in Saccharomyces cerevisiae [6], 10 sHsps in human [7], and 13 sHsps in zebrafish [8], while 19 sHsps in Arabidopsis thaliana [9], 23 in rice (Oryza sativa) [10], 27 in wheat (Triticum aestivum) [11], 35 in pepper (Capsicum annuum) [12], and 37 in Populus trichocarpa [13]. This indicates that the number of sHsps in the sessile plant species are higher than that in bacteria and animals. The expansion levels of the sHsp family is positively correlated with their diversification and adaptation to complicated stress conditions in high plants. In addition, the functions of several sHsps have been investigated based on genetic evidence. The expression of sHsp24.4 from Musa accuminata is induced under high temperature, and its overexpression can increase tolerance against heat stress in the transgenic tomato [14]. Heterologous expression of rice OsHsp26 enhances tolerance against oxidative and heat stresses possibly through protecting photosystem II in transgenic tall fescue [15]. Transgenic Arabidopsis plants with overexpression of poplar PtsHsp17.8 show higher tolerance to heat and salt stresses than control plants through increasing antioxidative enzyme activities [16]. After ectopically expressing NnHsp17.5 from Nelumbo nucifera in Arabidopsis, transgenic seeds exhibit increased superoxide dismutase activity after accelerated aging treatment [17]. In addition, the function of sHsp17.3 from Juglans regia [18], sHsp17.7 from Vaccinium corymbosum [19], and sHsp22.5 from Capsicum annuum [20] were proved to be involved in extreme temperature and salt stresses. All these studies have indicated that sHsps are involved in stress responses and play important roles in plant adaptation to various environments.
Willow (Salix) is a member of Salicaceae family and it is regarded as a promising economic plant that can be used as renewable biomass for bioenergy, because of its easily vegetative propagation, rapid growth, short-rotation, and substantial biomass yields [21]. As protective proteins, sHsps in willow have attracted attention for their potential roles against stresses. However, systematic research on the sHsp family in willow is still lacking. Salix suchowensis is a special shrub willow species that is found in the north of China [22]. The completion of the genome sequence of S. suchowensis make it possible to investigate the Ssu-sHsp gene family [23]. Nowadays, increasing public data (genomic, transcriptomic, or proteomic data) provide abundant information to help understand gene function, like sHsp in poplar [13], Arabidopsis thaliana [9], rice (Oryza sativa) [10], etc. In the present study, 35 genes encoding Ssu-sHsp proteins were identified in the S. suchowensis genome using bioinformatics methods. We analyzed their chromosomal localization, genes and protein structure, and duplication events. Subsequently, the expression profiles of all Ssu-sHsp genes were detected in various organs/tissues and under different abiotic stresses (heat, drought, cold, salt, and abscisic acid (ABA) treatment) to explore their potential functions in stress responses. The cis-acting elements and co-expression network were also analyzed to investigate the possible regulation mechanisms of Ssu-sHsps. Our analysis lays the foundation for the further characterization of Ssu-sHsp genes and genetic improvement of S. suchowensis resistance to stress conditions.
2. Results
2.1. Genome-Wide Identification and Phylogenetic Analysis of Ssu-sHsp in S. suchowensis
To identify sHsp genes in S. suchowensis, the protein sequences of 19 A. thaliana sHsp (AtsHsp) [9] and 37 P. trichocarpa sHsp (PtsHsp) [13] were used as a query to perform basic local alignment search tool algorithms (BLASTP) and Hidden Markov models (HMMs) search against the S. suchowensis genome database [23]. A total of 35 non-redundant Ssu-sHsp proteins were identified, and the information of open reading frame length, protein length, molecular weight (Mw), and isoelectric point (pI) are shown in Table 1. The length of proteins varied from 116 to 350 amino acids, the predicted pI ranged from 4.60 to 9.65, and the Mw from 13.3 to 38.7 kDa (Table 1 and Table S1).
Table 1.
The Ssu-sHsp gene family identified in S. suchowensis.
| Gene Name | Gene ID | Location (bp) | Coding Sequence (CDS) Length (bp) | Protein Length (aa) | Molecular Weight (MW) (kDa)/pI | Subfamily |
|---|---|---|---|---|---|---|
| Ssu-sHsp13.3-CI | willow_GLEAN_10013268 | chr19:6883846-6884193(−) | 348 | 116 | 13.3/8.92 | CI |
| Ssu-sHsp16.5-CI | willow_GLEAN_10019104 | chr01:18114468-18114899(−) | 432 | 144 | 16.5/5.81 | CI |
| Ssu-sHsp17.7A-CI | willow_GLEAN_10001930 | scaffold704:2227-2685(−) | 459 | 153 | 17.7/5.52 | CI |
| Ssu-sHsp17.7B-CI | willow_GLEAN_10001931 | scaffold704:5790-6248(−) | 459 | 153 | 17.7/5.5 | CI |
| Ssu-sHsp17.7C-CI | willow_GLEAN_10013270 | chr19:6871309-6871767(−) | 459 | 153 | 17.7/5.34 | CI |
| Ssu-sHsp17.8-CI | willow_GLEAN_10013271 | chr19:6867577-6868035(−) | 459 | 153 | 17.8/5.72 | CI |
| Ssu-sHsp17.9-CI | willow_GLEAN_10023007 | chr09:3274010-3274480(−) | 471 | 156 | 17.9/6.77 | CI |
| Ssu-sHsp18.0A-CI | willow_GLEAN_10015047 | chr01:12867685-12868164(−) | 480 | 159 | 18.0/5.82 | CI |
| Ssu-sHsp18.0B-CI | willow_GLEAN_10015393 | chr04:10868237-10868704(−) | 468 | 156 | 18.0/5.54 | CI |
| Ssu-sHsp18.1-CI | willow_GLEAN_10023005 | chr09:3270244-3270711(−) | 468 | 156 | 18.1/6.77 | CI |
| Ssu-sHsp18.2A-CI | willow_GLEAN_10001113 | scaffold03801:634-1107(+) | 474 | 158 | 18.2/5.55 | CI |
| Ssu-sHsp18.2B-CI | willow_GLEAN_10022937 | chr09:2679684-2680154(−) | 471 | 157 | 18.2/6.76 | CI |
| Ssu-sHsp18.3-CI | willow_GLEAN_10001192 | scaffold03321:1968-2444(−) | 477 | 159 | 18.3/5.51 | CI |
| Ssu-sHsp18.5-CI | willow_GLEAN_10019417 | chr10:11252074-11252562(+) | 489 | 163 | 18.5/5.57 | CI |
| Ssu-sHsp18.6-CI | willow_GLEAN_10024904 | chr08:2910419-2910907(−) | 489 | 163 | 18.6/6.77 | CI |
| Ssu-sHsp23.5-CI | willow_GLEAN_10013269 | chr19:6875448-6880158(−) | 618 | 206 | 23.5/5.77 | CI |
| Ssu-sHsp17.8A-CII | willow_GLEAN_10007257 | chr15:3576323-3576796(−) | 474 | 158 | 17.8/6.74 | CII |
| Ssu-sHsp17.8B-CII | willow_GLEAN_10026164 | chr06:13631015-13631488(+) | 474 | 158 | 17.8/6.32 | CII |
| Ssu-sHsp17.8-CIII | willow_GLEAN_10001482 | scaffold02057:529-1140(−) | 471 | 157 | 17.8/6.32 | CIII |
| Ssu-sHsp21.9-CIV | willow_GLEAN_10001528 | scaffold01874:2153-2906(−) | 576 | 191 | 21.9/9.59 | CIV |
| Ssu-sHsp23.1-CIV | willow_GLEAN_10008116 | chr13:3276222-3276989(−) | 615 | 204 | 23.1/9.65 | CIV |
| Ssu-sHsp28.0-CIV | willow_GLEAN_10023903 | chr09:8210641-8211561(−) | 762 | 253 | 28.0/7.8 | CIV |
| Ssu-sHsp38.2-CIV | willow_GLEAN_10015419 | chr04:11093447-11096554(−) | 1050 | 350 | 38.2/6.32 | CIV |
| Ssu-sHsp38.7-CIV | willow_GLEAN_10015420 | chr04:11103548-11105801(−) | 1053 | 350 | 38.7/8.43 | CIV |
| Ssu-sHsp22.2-CV | willow_GLEAN_10017670 | chr01:24155089-24155990(+) | 591 | 196 | 22.2/5.11 | CV |
| Ssu-sHsp23.0-CV | willow_GLEAN_10006909 | chr11:9175250-9176081(+) | 609 | 202 | 23.0/5.02 | CV |
| Ssu-sHsp26.3-CP | willow_GLEAN_10005204 | chr12:637010-638195(+) | 720 | 240 | 26.3/6.35 | CP |
| Ssu-sHsp26.5-CP | willow_GLEAN_10014198 | chr10:277029-277816(−) | 705 | 235 | 26.5/7.89 | CP |
| Ssu-sHsp23.8-MT | willow_GLEAN_10017874 | chr16:5313895-5314699(−) | 645 | 215 | 23.8/5.44 | MT |
| Ssu-sHsp25.6-MT | willow_GLEAN_10022135 | chr03:3924709-3926100(−) | 678 | 225 | 25.6/5.82 | MT |
| Ssu-sHsp15.9A-PX | willow_GLEAN_10001081 | scaffold04006:1618-2317(+) | 420 | 140 | 15.9/7.02 | PX |
| Ssu-sHsp15.9B-PX | willow_GLEAN_10018481 | chr04:3932697-3933402(+) | 426 | 141 | 15.9/6.2 | PX |
| Ssu-sHsp16.1-PX | willow_GLEAN_10002647 | chr17:7449402-7450136(+) | 426 | 142 | 16.1/6.92 | PX |
| Ssu-sHsp23.1 | willow_GLEAN_10011114 | chr15:5028375-5029436(+) | 609 | 202 | 23.1/4.6 | * |
| Ssu-sHsp31.8 | willow_GLEAN_10023904 | chr09:8208040-8209110(−) | 858 | 285 | 31.8/6.04 | * |
C, cytosol; CP, chloroplast; PX, peroxisome; MT, mitochondria; *, unclustered member.
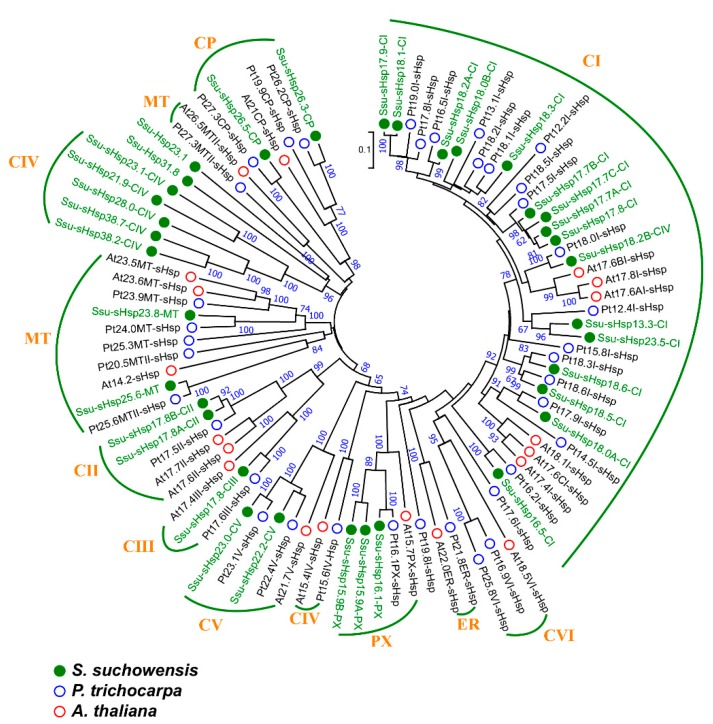
A phylogenetic tree was constructed using the full-length protein sequences of sHsp from S. suchowensis, A. thaliana, and P. trichocarpa to investigate their evolutionary relationship. In S. suchowensis, the sHsp family was mainly classified into four subfamilies, including C (cytosol), CP (chloroplast), PX (peroxisome), and MT (mitochondria), but lacked the endoplasmic reticulum (ER) subfamily in comparison with AtsHsps and PtsHsps [9,13]. The C subfamily of Ssu-sHsp, which had 26 members, was subdivided into five groups (CI-CV): 16 members in CI, two members in CII and CV, one member in CIII, and five members in CIV, respectively; the CP and MT subfamilies individually contained two members; three members were existed in PX subfamily (Table 1 and Figure 1). On the basis of their phylogenetic relationships, potential subcellular localization/group, and their Mw, we named the Ssu-sHsps the “Ssu-sHsp (Mw)-(group)”. For example, the Ssu-sHsp member willow_GLEAN_10019104 with Mw 16.5 kD belonging to the CI group was named Ssu-sHsp16.5-CI (Table 1 and Figure 1).
Figure 1.
Phylogenetic relationships of sHsp family members from Salix suchowensis, Populus trichocarpa, and Arabidopsis thaliana. Multiple alignment was performed using Clustal X2.1, and the phylogenetic tree was constructed using MEGA 6.06 by the neighbor-joining (NJ) method with 1000 bootstrap replicates. sHsp from S. suchowensis, P. trichocarpa, and A. thaliana were marked with green, blue, and red circles, respectively.
2.2. Gene Structure and Protein Domain Analysis of Ssu-sHsps
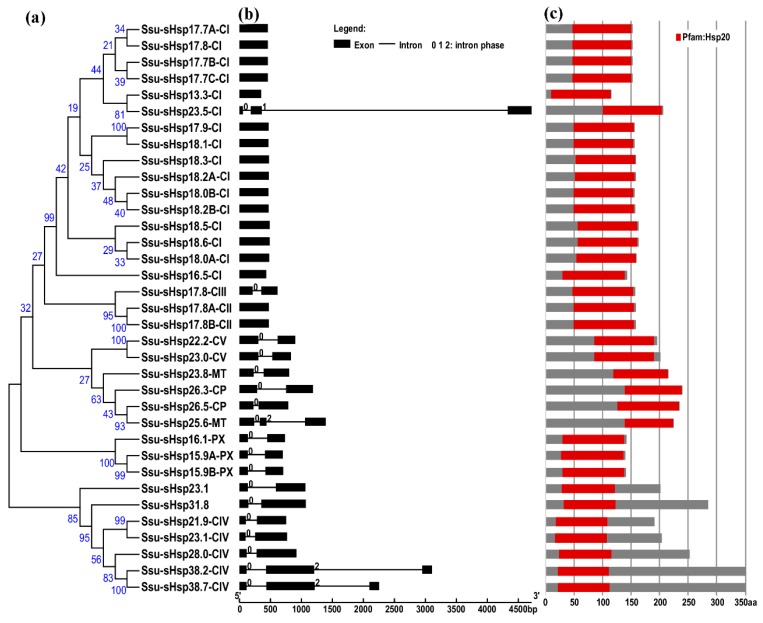
To gain further insight into the structural diversity of different groups of Ssu-sHsp genes, the simplified neighbor-joining (NJ) phylogenetic tree of their Hsp20 domains was constructed, as well as the analysis of gene structures and functional domain (Figure 2). As shown in Figure 2a, all of the Ssu-sHsp members still remained in the same subfamily as that used by the full-length protein sequences. All the members of CI (except Ssu-sHsp23.5-CI) and CII groups were intronless, four Ssu-sHsps (23.5-CI, 25.6-MT, 38.2-CIV and 38.7-CIV) were comprised of two introns, and the other 14 Ssu-sHsps contained one intron. While the Ssu-sHsps with two-introns showed phase 0 in the first intron, but the second intron of Ssu-sHsp23.5-CI showed phase 1 and others Ssu-sHsp (25.6-CP, 38.2-CIV and 38.7-CIV) showed phase 2 in the second intron. These results indicated that the members of the same subfamily shared similar gene structures and intron phases (Figure 1 and Figure 2).
Figure 2.
Phylogenetic relationships, gene structure, and protein structures of the sHsp genes in Salix suchowensis. (a) Multiple alignment of Hsp20 domain was executed using Clustal X2.1 and a phylogenetic tree was constructed using MEGA 6.06 by the neighbor-joining (NJ) method with 1000 bootstrap replicates. (b) The exon/intron structures of the Ssu-sHsps genes. Boxes represent exons and lines represent introns. The numbers indicate the splicing phases of the Ssu-sHsps: 0, phase 0; 1, phase 1; and 2, phase 2. (c) The protein structures of the Ssu-sHsp proteins. Red boxes represent the Hsp20 domain.
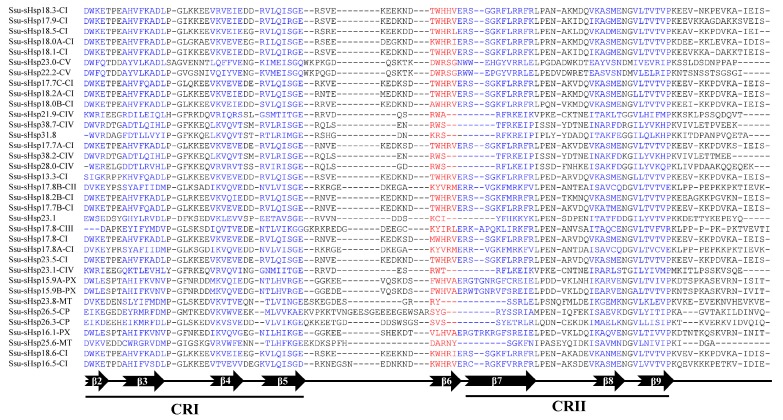
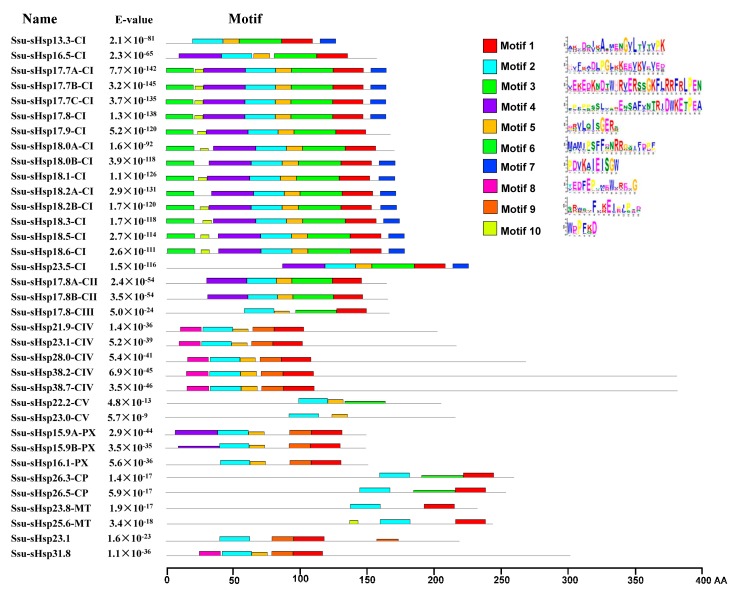
The analysis of the functional domain showed that all the Ssu-sHsp proteins have the Hsp20 domain, but their location was different and there existed two types: the Hsp20 domain of Ssu-sHsp23.1, Ssu-sHsp31.8, and the members in CIV subgroup were located near the N-terminal, while the Hsp20 domain of the others near the C-terminal (Figure 2c). Multiple sequence alignment was conducted to identify the variations in Hsp20 domains. As shown in Figure 3, the Hsp20 domain in most of Ssu-sHsps was composed by CRI, CRII, and β6-loop, which was consistent with the report in switchgrass [24] and pepper [12]. However, Ssu-sHsp17.8-CIII lacked the β2, and ten Ssu-sHsps (21.9-CIV, 23.1, 23.1-CIV, 26.3-CP, 26.5-CP, 23.8-MT, 28.0-CIV, 31.8, 38.2-CIV, and 38.7-CIV) lacked the β6-loop and contained incomplete β7 (Figure 3). After searching with the MEME program, ten conserved motifs were identified in Ssu-sHsps (Figure 4). Thereinto, the CRI contained the motifs 2, 4, and 5, while CRII contained motifs 1 and 3. However, only 19 of the 35 Ssu-sHsp proteins contained these five motifs, revealing the diversification of Hsp20 domain in S. suchowensis sHsps (Figure 4).
Figure 3.
Multiple sequence alignment of the Hsp20 domains of the Ssu-sHsp proteins.
Figure 4.
Distribution of conserved motifs in the Ssu-sHsp proteins. The motifs were identified by MEME. Different motifs are indicated by different colored number 1–10.
2.3. Expansion of the sHsp Family in S. suchowensis
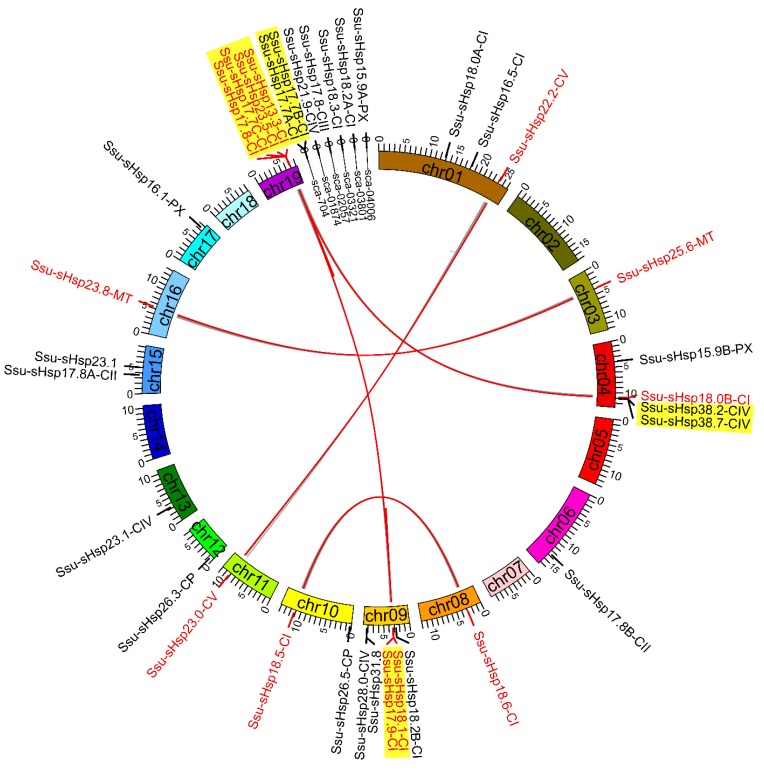
To analyze the expansion and genomic distribution of Ssu-sHsp genes, the chromosomal location and duplication events of the Ssu-sHsp family were analyzed based on the current genomic information from the S. suchowensis genome database. The 35 Ssu-sHsp genes were unevenly mapped onto 14 chromosomes and six scaffolds in the S. suchowensis genome (Figure 5). Chr09 contained the largest number of five Ssu-sHsps, followed by Chr04 and Chr19 that each contained four Ssu-sHsps (Figure 5 and Table 1). On the basis of the chromosomal location, phylogenetic relationships, and gene structures, eight paralogous pairs were identified in the Ssu-sHsp family (Table 2). Among of them, three paralogous pairs (Ssu-sHsp18.5-CI/18.6-CI, 22.2-CV/23.0-CV, 25.6-MT/23.8-MT) were generated by whole genome duplication (WGD) events and another five pairs (Ssu-sHsp17.7A-CI/17.7B-CI, 17.8-CI/17.7C-CI, 18.1-CI/17.9-CI, 23.5-CI/13.3-CI, 38.2-CIV/38.7-CIV) were generated by tandem duplication events (Figure 3 and Table 2). The substitution ratio of non-synonymous (Ka)/synonymous (Ks) was used to measure the selection pressures for duplicated gene pairs [25]. In our study, the Ka/Ks ratios of all the eight Ssu-sHsp paralogous pairs were less than 1, suggesting that these Ssu-sHsp paralogous pairs have evolved from purifying selection (Table 2).
Figure 5.
Chromosomal location and duplication event analyses of Ssu-sHsps in the S. suchowensis genome.
Table 2.
Divergence between paralogous Ssu-sHsp gene pairs in S. suchowensis.
| Gene 1 | Gene 2 | Ka | Ks | Ka/Ks | Duplication | Subfamily | p-Value (Fisher) |
|---|---|---|---|---|---|---|---|
| Ssu-sHsp17.7A-CI | Ssu-sHsp17.7B-CI | 0.0167 | 0.0676 | 0.2475 | T | CI | 4.9063 × 10−3 |
| Ssu-sHsp17.8-CI | Ssu-sHsp17.7C-CI | 0.0349 | 0.0612 | 0.5697 | T | CI | 1.4443 × 10−1 |
| Ssu-sHsp18.1-CI | Ssu-sHsp17.9-CI | 0.0061 | 0.0477 | 0.1271 | T | CI | 1.0528 × 10−3 |
| Ssu-sHsp23.5-CI | Ssu-sHsp13.3-CI | 0.0185 | 0.0485 | 0.3808 | T | CI | 5.1005 × 10−2 |
| Ssu-sHsp18.6-CI | Ssu-sHsp18.5-CI | 0.0711 | 0.4235 | 0.1678 | W | CI | 2.9675 × 10−17 |
| Ssu-sHsp38.2-CIV | Ssu-sHsp38.7-CIV | 0.1624 | 0.1997 | 0.8135 | T | CIV | 1.8343 × 10−1 |
| Ssu-sHsp22.2-CV | Ssu-sHsp23.0-CV | 0.1153 | 0.5552 | 0.2077 | W | CV | 1.6456 × 10−24 |
| Ssu-sHsp25.6-MT | Ssu-sHsp23.8-MT | 0.5873 | 0.9437 | 0.6223 | W | MT | 3.8210 × 10−17 |
2.4. Expression Patterns of Ssu-sHsps across Tissues and under Various Stresses
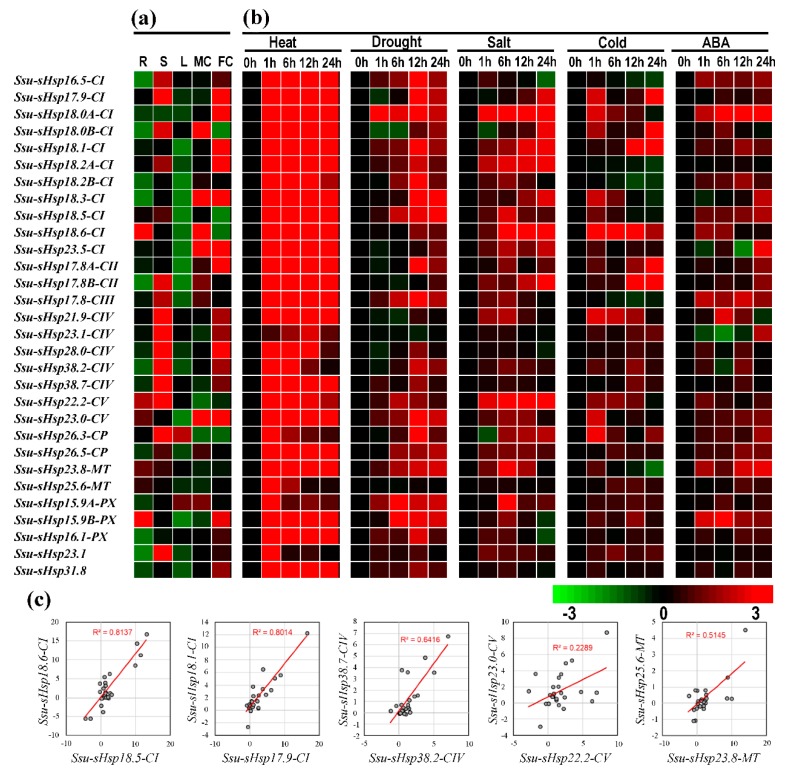
In order to gain the information about the spatial and temporal expression pattern of Ssu-sHsps, qRT-PCR was performed to explore the expression levels of Ssu-sHsps in five different tissues including root (R), stem (S), leaf (L), female catkin (FC), and male catkin (MC). As illustrated in Figure 6a, the majority of Ssu-sHsps have special tissue expression patterns. Four Ssu-sHsps (18.6-CI, 15.9B-PX, 23.8-MT, and 25.6-MT) were predominantly expressed in the root. Thirteen Ssu-sHsps (16.5-CI, 18.5-CI, 17.8B-CIII, 17.8-CIII, 21.9-CIV, 23.1-CIV, 28.0-CIV, 38.2-CIV, 38.7-CIV, 22.2-CV, 26.3-CP, 26.5-CP, and 23.1) were highly expressed in the stem. Thirteen Ssu-sHsps (18.0B-CI, 18.3-CI, 17.8B-CII, 15.9A-PX, 17.9-CI, 18.0A-CI, 18.1-CI, 18.2A-CI, 23.5-CI, 17.8A-CII, 23.0-CV, 16.1-PX, and 31.8) displayed relatively high expression levels in the reproductive organs (female catkin or male catkin). Notably, the expression levels of all the Ssu-sHsp genes were relatively low in the leaf across the five analyzed tissues (Figure 6a).
Figure 6.
Expression profiles of Ssu-sHsps by qRT-PCR. (a) Expression analysis of Ssu-sHsps in five tissues: root (R), stem (S), leaf (L), female catkin (FC), and male catkin (MC). The expression of Ssu-sHsp in different tissues were compared with the median value among these tissues. (b) Expression analysis of Ssu-sHsp genes under various abiotic stresses after treatments at 1, 6, 12, or 24 h under heat (37 °C), drought (20% w/v PEG 6000), salt (150 mM NaCl), cold (4 °C), and 100 µM abscisic acid (ABA) treatment. The different colors represent log2 transformed values compared with control (0 h). (c) Correlation of expression of five Ssu-sHsp paralogous pairs across various tissues and under stresses. The x- and y-axis indicate expression levels across various tissues and under stresses in each paralogous pair. For example, the x-axis represents Ssu-sHsp18.5-CI and y-axis represents Ssu-sHsp18.6-CI in pair Ssu-sHsp18.5-CI/Ssu-sHsp18.6-CI.
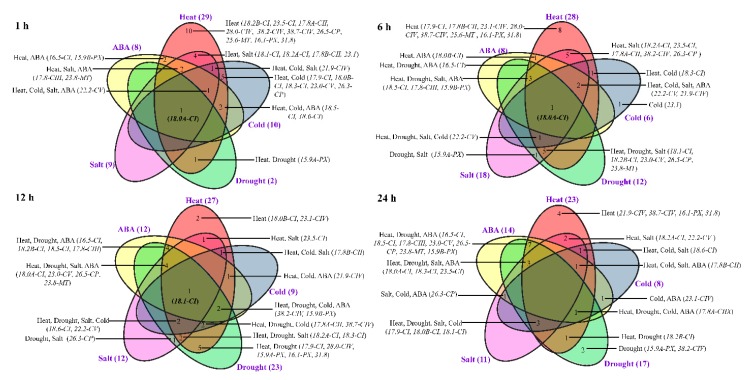
To investigate the responses of Ssu-sHsps to various environmental stresses, the expression profiles of Ssu-sHsps were detected using qRT-PCR under heat, drought, salt, cold, and ABA treatments. Among the detected 30 Ssu-sHsps, 29 Ssu-sHsps were promptly and dramatically up-regulated after heat treatments for 1 h (Figure 6b and Figure 7), and most of them maintained high expression levels until the end (24 h) of the heat stress (Figure 6b and Figure 7). Aside from heat stress, several Ssu-sHsps (e.g., Ssu-sHsp18.0A-CI, 18.1-CI, 18.2A-CI, 22.2-CV, and 15.9A-PX) were responsive to both drought and salt stresses. In contrast to the large number of Ssu-sHsps strongly induced by heat, drought, salt, and ABA stresses, only six, nine, and eight Ssu-sHsps were responsive to cold treatment at 6, 12 and 24 h, respectively. It was noted that Ssu-sHsp18.0A-CI were significantly responsive to all the tested treatments at 1 h and 6 h of treatment (Figure 6 and Figure 7).
Figure 7.
Different stress response model of Ssu-sHsp genes at different times after treatments.
Among the eight pairs, the expression of five Ssu-sHsp paralogous pairs (18.5-CI/18.6-CI, 17.9-CI/18.1-CI, 38.2-CIV/38.7-CIV, 22.2-CV/23.0-CV, and 23.8-MT/25.6-MT) were detected successfully. Only three Ssu-sHsp pairs (Ssu-sHsp18.5-CI/18.6-CI, 17.9-CI/18.1-CI, and 38.2-CIV/38.7-CIV) showed highly similar patterns with R2 > 0.6 across various tissues and stresses, while the other two detected paralogous pairs showed divergent expression (Figure 6c).
2.5. Characterization of Cis-acting Elements in the Promoter Regions of Ssu-sHsps
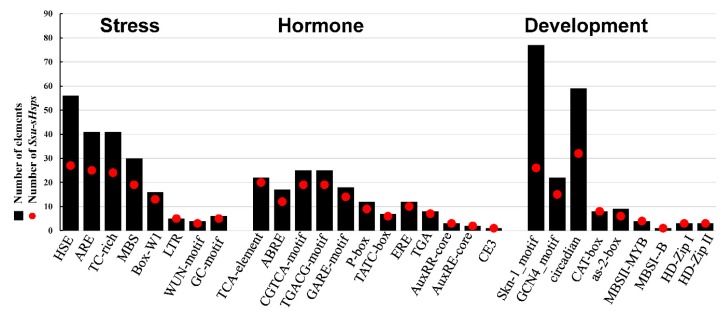
To further investigate the possible regulation mechanism of Ssu-sHsps in S. suchowensis, the cis-elements in the promoter regions of Ssu-sHsps were analyzed using the PlantCARE database. A series of stress-related (e.g., heat, drought, low temperature, and anaerobic induction), hormone-related (e.g., ABA, SA, GA, and MeJA), and development-related (e.g., endosperm expression, circadian control, meristem expression, and light response) cis-acting elements were identified (Figure 8 and Figure S1). In stress-related cis-acting elements, 77.1% of Ssu-sHsps (27 genes) had the HSE (heat responsive elements) which was the most abundant cis-acting element with its number reaching 56. There were 41 ARE (essential for the anaerobic induction) and 41 TC-rich repeats (stress responsiveness) that were located in 12 and 10 Ssu-sHsp promoters, respectively. In hormone-related cis-acting elements, TCA-element (salicylic acid responsiveness), ABRE (abscisic acid responsiveness), CGTCA-motif (MeJA-responsiveness), and GARE-motif (MeJA-responsiveness) were identified in the promoters of 20, 11, 19, and 19 Ssu-sHsps, respectively. Furthermore, large numbers of cis-acting elements related to endosperm expression (Skn-1) and circadian (GCN4_motif and circadian cis-acting element) were detected, implying that sHsp might be also involved in developmental processes in S. suchowensis.
Figure 8.
Various cis-acting elements in the promoter of Ssu-sHsp genes.
2.6. The Co-expression Network of Ssu-sHsps
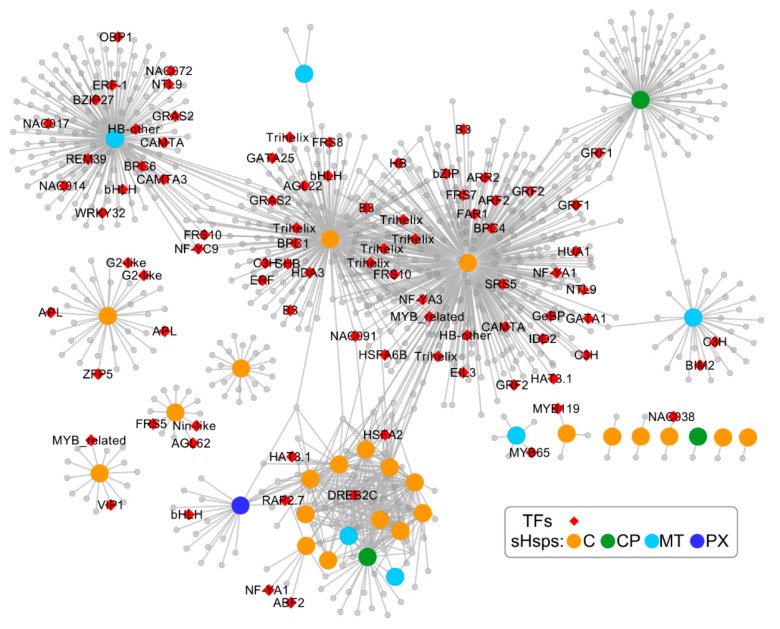
To further predict the relationship between Ssu-sHsp genes and other genes, a co-expression network of sHsp genes was constructed according to their orthologous in P. trichocarpa (Figure 9). The Ssu-sHsps lacked an ER subfamily compared with the PtsHsp family, and Ssu-sHsp did not correspond one-to-one with P. trichocarpa members (Figure 1). Thus, the ER members of PtsHsps were removed in the co-expression network, and the subfamily was used as a unit to study the co-expression relationship. In this network, 970, 235, 116, and 20 genes were co-expressed with sHsp in the C, CP, MT, and PX subfamily, respectively. Moreover, some of them shared the same co-expressed genes. It was noted that the network contained a transcription factor which related to stress response (e.g., HSFA6B, HSFA2, DREB2C), development (e.g., GRF, Floral homeotic protein HUA1, AGAMOUS-like 62), and hormone (e.g., ARF2, ARR2 and AREB3), and several of them were hub genes (Figure 9 and Table S2). This indicated that the sHsps in S. suchowensis might be function in stress tolerance or developmental processes through cooperation with specific transcription factors or other functional genes.
Figure 9.
Co-expression network of sHsp gene family. Orange, green, cyan, and blue nodes represent C, CP, MT, and PX subfamily members, respectively; red diamond nodes represent transcription factors; gray nodes are other co-expressed genes. The functional descriptions of these transcription factors are listed in Table S2.
3. Discussion
As ubiquitous molecular chaperones, sHsps are present in various organisms and play prominent roles in alleviating injuries from diverse abiotic and biotic stresses, especially in heat stress [26]. The features and functions of the sHsp family have been studied in many model plants (e.g., Arabidopsis, rice, and poplar) [9,10,13,27], however, very little is known regarding sHsp genes in S. suchowensis, which is regarded as a short-rotation woody biomass for bioenergy sources and biofuel.
In this study, 35 Ssu-sHsp genes were identified in the S. suchowensis genome, which is less than that in P. trichocarpa (37 PtsHsps), and this number is consistent with the relative genome size of these two specie (~425 Mb of S. suchowensis and ~485 Mb of P. trichocarpa) [13]. However, no ER-family member was identified, possibly because of the incomplete genome assembly of S. suchowensis. Gene duplication including WGD and tandem duplication events play a crucial role in expansions of the gene family during species evolution [28]. In the Ssu-sHsp gene family, eight paralogous pairs were identified, including three and five pairs which were generated by WGD and tandem duplication events, respectively, and contributing to the expansion of the CI (five pairs), CIV (one pair), CV (one pair), and MT (one pair) subgroups. Among of them, CI was the largest subgroup containing most expansion genes, which was consistent with reports in other plants [29]. The duplicated gene pairs shared high amino acid sequence identity and similar gene structures, but the expression patterns of duplicated gene pairs (Ssu-sHsp18.5-CI/18.6-CI, 22.2-CV/23.0-CV) were different across various tissues and under stresses, and they kept a lower correlation with R2 < 0.6 (Figure 6) indicating that their function might be divergent during the evolution.
The investigation of gene organization showed that 31 Ssu-sHsp genes (~88.5%) contained one or no intron and confirmed that sHsps have less introns in higher plants. Its reason might be that the length of sHsp itself is too short. It has been reported that the intron number likely affects the efficiency of transcription [30]. When comparing the relationship between the intron numbers and their expression patterns of Ssu-sHsps under stress conditions, we found less introns is helpful for Ssu-sHsps to prompt induction under stresses (Figure 6). Therefore, the gene structure might also play roles in controlling its transcriptional level under stress condition [31].
In the native state, sHsps always exist as multimeric complexes with variable numbers of subunits [5]. The Hsp20 domain is crucial for the formation of multimeric complexes that are important for their chaperone activity [4]. In our study, all the Ssu-sHsp proteins contained the Hsp20 domain, but multiple sequence alignments and motifs analysis showed that the sequences of Hsp20 were diverse among the Ssu-sHsp family and even parts of them lacked the β2-strand, β6-loop, or the β7-strand (Figure 3 and Figure 4). It is reported that the β2-strand is associated with structure dimerization [11,26]. Additionally, the β6-loop that links the β5-strand and β7-strand is related to dimer formation [32]. Thus, the Ssu-sHsp proteins lacking β2, β6, or β7 might be loss-of-function, or even leading to functional diversity which will need to be fully elucidated.
In the co-expression network, floral development-related genes (AGAMOUS-like 62, MADS-box, and Floral homeotic protein HUA1) were co-expressed with C subfamily members (Figure 9 and Table S2). In our study, 14 Ssu-sHsps had relatively high expression levels in reproductive organs, and ten of them belonged to the C subfamily (Figure 6a), in which most members were induced by at least one or two stresses (Figure 6b and Figure 7). During the reproductive growth, flowers are always sensitive to high temperature and other environmental stresses [33,34]. Therefore, it can be inferred that Ssu-sHsp with high expression levels in reproductive organs might be cooperating with the flower development-related genes in regulating flower development and maintaining the cellular homeostasis between stress and normal conditions. In addition, 13 Ssu-sHsps were detected as being highly expressed in the stem, a similar result is also reported in poplar [13]. The stem is the product of continuous division and differentiation of the cambium cells in the secondary growth. In addition, the activity of the cambium is susceptible to the external environment [35]. Considering the stem-specific expression patterns of Ssu-sHsps and their conserved function, they might be involved in adjustment stem development by protecting the protein folding which plays roles in maintaining the function of the cambium [16].
Stress response analysis showed that the transcript levels of Ssu-sHsps was generally induced by at least one tested stress, especially heat stress. The similar expression patterns of sHsps from different species in response to stress indicated that they might possess conservation function [9,10,13]. In order to analyze the potential regulatory mechanism, the cis-acting elements and the co-expression network of Ssu-sHsps were analyzed. As the binding sites of transcription factors, cis-acting elements in the promoter of the gene determine its expression patterns. In our study, a series of stress-, hormone-, and development-related cis-acting elements were detected in the promoter of Ssu-sHsps. HSE, the target site of Hsf, was abundantly present in the promoters of Ssu-sHsps (Figure 8). In our study, the Hsf family member (HsfA6B and HsfA2) were co-expressed with the C and CP subfamily sHsps (Figure 9). In Arabidopsis, HsfA6B participate in ABA-mediated salt and drought response and is required for heat stress resistance [36], and HsfA2 is regarded as a heat-inducible transactivator that plays a positive role in enhancing the heat tolerance by activating the expression of Hsp or sHsp [37]. In addition, DREB2C was also detected in our co-expressed network and co-expressed with sHsps from the C and MT subfamily (Figure 9). In Arabidopsis, DREB2C is responsive to heat stress by activating the expression of HsfA3 [38] and the Thermotolerance-related Phytocystatin 4 (AtCYS4) [39], and DREB2C is also involved in ABA signing by regulating the expression of the ABA responsive gene [40]. In our study, several members in the C subfamily were induced by ABA treatment (Figure 6 and Figure 7), this infers that HsfA6B, HsfA2, and DREB2C may directly or indirectly bind to the promoter region of Ssu-sHsps to activate their expression in the ABA dependent pathway. In addition, some other transcription factors and functional genes that are related to stress response and development were identified in the co-expression network (Figure 9 and Table S2). This suggests that Ssu-sHsps might be involved in stress tolerance and plant development through cooperating with these genes. Further verification will need to be performed in the future.
4. Materials and Methods
4.1. Identification of sHsp Genes in S. suchowensis Genome
The protein sequence of S. suchowensis was obtained from the S. suchowensis genome database (http://115.29.234.170/cgi-bin/gbrowse/gbrowse/Ssuchowensis4/) [23]. The sHsp protein sequences of A. thaliana and P. trichocarpa obtained from phytozome (https://phytozome.jgi.doe.gov) were used as query sequences for the BLASTP to identify sHsp sequences in the S. suchowensis genome, with a cutoff E-value < 10−10. An additional search strategy, Hsp20 domain (PF00011) was used as a query to identify sHsp proteins using the HMMER (v3.0) software with an E-value < 10−3. SMART program (http://smart.embl-heidelberg.de/) [41] and the InterPro databases (http://www.ebi.ac.uk/interpro/) [42] were used to screen and confirm the Hsp20 domain in the candidate sHsp proteins, and the proteins lacking Hsp20 domain were removed.
4.2. Sequence Analysis and Structural Characterization
The isoelectric points (pI) and molecular weight (Mw) of Ssu-sHsps were estimated using the compute pI/Mw tool from ExPASy (http://web.expasy.org/compute_pi). The online program gene structure display server (http://www.gsds.cbi.pku.edu.cn/) [43] was used to analyze the gene structure (exon-intron) of Ssu-sHsps. The motifs in Ssu-sHsps were investigated using the MEME 4.11.1 online program (http://meme.nbcr.net/meme/intro.html) [44]. The protein subcellular localization was predicted using WoLF PSORT (http://psort.hgc.jp/) [45].
4.3. Phylogenetic Analysis and Chromosomal Location
The sHsp protein sequences from S. suchowensis, A. thaliana, and P. trichocarpa were used for phylogenetic analysis. Clustal X 2.1 software [46] was used for the multiple alignment of sHsp proteins and unrooted phylogenetic trees were constructed using MEGA 6.06 software [47] with the neighbor-joining method. The information of the chromosomal location of all Ssu-sHsp genes was downloaded from Phytozome 12.1 and the chromosomal locations and duplication of the Ssu-sHsp genes were drawn using Circos software [48]. In the analysis of the duplication, the array of two genes located within a 100 kb region were regarded as duplicated genes. DnaSP 5 software (http://www.ub.edu/dnasp/) [49] was used to calculate the ratio between nonsynonymous (Ka) and synonymous nucleotide substitutions (Ks).
4.4. In Silico Analysis of Regulatory Elements in the Promoter Regions of the Ssu-sHsp Gene Family
The upstream regions (2.0 kb) of the translation initiation sites (ATG) of Ssu-sHsp genes were used as promoter fragments. In addition, the elements were located in the promoter sequences using the program PlantCARE online (http://bioinformatics. psb.ugent.be/webtools/plantcare/html/) [50].
4.5. Plant Materials, Growth Conditions, and Treatments
The different tissues of cultivated S. suchowensis, including root (R), stem (S), leaf (L), female catkin (FC), and male catkin (MC), were harvested from triennial S. suchowensis. Four-week-old clones of S. suchowensis were water cultured using Hoagland’s nutrient solution in a growth chamber under long-day conditions (16 h light/8 h dark) at 25 °C/22 °C (day/night). In order to investigate the expression profiles of Ssu-sHsps under abiotic stresses and hormone treatment, the clones were treated under 150 mM NaCl (for salt stress), 20% (w/v) polyethylene glycol (PEG 6000, for drought stress), 37 °C (for heat stress), 4 °C (for cold stress), or 100 µM abscisic acid (ABA), respectively. Samples were collected after five time periods (0, 1, 6, 12, and 24 h) of each treatment. Fully matured leaves from five individual clones were immediately harvested at each time period of different treatments, and then rapidly frozen in liquid nitrogen and stored at −80 °C for further analysis. The dosages of the abiotic stresses and hormone treatment were determined based on treatments in S. suchowensis [22]. Three biological replicates were performed in different pots for each treatment.
4.6. RNA Isolation and qRT-PCR
The RNA isolation, primer design, qRT-PCR reactions, and program were performed according to Li et al [51]. Because of the high similarity of Ssu-sHsp13.3-CI, Ssu-sHsp17.7A-CI, Ssu-sHsp17.7B-CI, Ssu-sHsp17.7C-CI, and Ssu-sHsp17.8-CI, their expression levels cannot be distinguished by qRT-PCR. On the basis of our previous research, EF1a (Elongation factor-1 alpha) was used as reference gene for different tissue types; HIS (Histone superfamily protein H3) and Actin7 for cold treatment and salt treatment, respectively; UBC (Ubiquitin-conjugating enzyme E2) both for heat and drought treatment [51]. The expression levels of Ssu-sHsps under abiotic stresses and hormone treatment were compared with control (0 h). In addition, the significantly induced genes were selected when the value of Log2 (fold change) ≥1. All the primers used in this study are listed in Table S3.
4.7. Integrated Network Analysis
The co-expression data of sHsps was obtained from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html). The co-expression network was constructed according to Tian et al [52].
5. Conclusions
On the basis of the S. suchowensis genome sequence, a total of 35 Ssu-sHsp genes were identified in this study. A comprehensive genome-wide analysis was performed to reveal phylogenetic relationship, gene organization, protein structure, chromosomal localization, co-expression network, and expression patterns across the different tissues and under stress conditions. According to the phylogenetic relationships and the predicted subcellular localization, the 35 Ssu-sHsps were classified into four subfamilies (C, CP, MT, and PX). A total of eight paralogous pairs were identified in the Ssu-sHsp gene family, five pairs were generated by tandem duplication and three pairs were generated by whole genome duplication, and all of them had undergone purifying selection during evolution. Multiple sequence alignment and motif analysis showed that the sequences of the Hsp20 domain were diverse among Ssu-sHsps. Expression profiling and co-expression network analyses revealed that Ssu-sHsps might be involved in plant stress tolerance and development through cooperating with a series of transcription factors and functional genes. The results provide a basis for comprehensively understanding the Ssu-sHsp gene family and will be beneficial to investigate the function of Ssu-sHsps.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/10/3246/s1.
Author Contributions
J.L. and J.Z. conceived and designed the research. J.L. and H.J. performed all the experiments. J.H, X.X., M.L. and Z.Y. contributed with valuable discussions. All authors read and approved the final manuscript.
Funding
This work was supported by the National Nonprofit Institute Research Grant of the Chinese Academy of Forestry (CAFYBB2017ZY008), (CAFYBB2018ZY001-9), the National Natural Science Foundation of China (31570669) and (31800570).
Conflicts of Interest
The authors declare that they have no conflicts of interests.
References
- 1.Zhu J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahuja I., Vos R.C.H.D., Bones A.M., Hall R.D. Plant molecular stress responses face climate change. Trends Plant Sci. 2010;15:664–674. doi: 10.1016/j.tplants.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Hu W., Hu G., Han B. Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Sci. 2009;176:583–590. doi: 10.1016/j.plantsci.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 4.De Jong W.W., Leunissen J.A., Voorter C.E. Evolution of the alpha-crystallin/small heat-shock protein family. Mol. Biol. Evol. 1992;10:103–126. doi: 10.1093/oxfordjournals.molbev.a039992. [DOI] [PubMed] [Google Scholar]
- 5.Van Montfort R.L., Basha E., Friedrich K.L., Slingsby C., Vierling E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat. Struct. Biol. 2001;8:1025–1030. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- 6.Bentley N.J., Fitch I.T., Tuite M.F. The small heat-shock protein Hsp26 of Saccharomyces cerevisiae assembles into a high molecular weight aggregate. Yeast. 1992;8:95–106. doi: 10.1002/yea.320080204. [DOI] [PubMed] [Google Scholar]
- 7.Kappé G., Franck E., Verschuure P., Boelens W.C., Leunissen J.A., de Jong W.W. The human genome encodes ten alpha-crystallin-related small heat shock proteins: HspB1-10. Cell Stress Chaperones. 2003;8:53–61. doi: 10.1379/1466-1268(2003)8<53:THGECS>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elicker K.S., Hutson L.D. Genome-wide analysis and expression profiling of the small heat shock proteins in zebrafish. Gene. 2007;403:60–69. doi: 10.1016/j.gene.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scharf K.D., Siddique M., Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing α-Crystallin domains (Acd Proteins) Cell Stress Chaperones. 2001;6:225–237. doi: 10.1379/1466-1268(2001)006<0225:TEFOAT>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkar N.K., Kim Y.K., Grover A. Rice sHsp genes: Genomic organization and expression profiling under stress and development. BMC Genom. 2009;10:393. doi: 10.1186/1471-2164-10-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandey B., Kaur A., Gupta O.P., Sharma I., Sharma P. Identification of HSP20 gene family in wheat and barley and their differential expression profiling under heat stress. Appl. Biochem. Biotech. 2015;175:2427–2446. doi: 10.1007/s12010-014-1420-2. [DOI] [PubMed] [Google Scholar]
- 12.Guo M., Liu J.H., Lu J.P., Zhai Y.F., Wang H., Gong Z.H., Wang S.B., Lu M.H. Genome-wide analysis of the CaHsp20 gene family in pepper: Comprehensive sequence and expression profile analysis under heat stress. Front. Plant Sci. 2015;6:806. doi: 10.3389/fpls.2015.00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J., Liu B., Li J., Zhang L., Wang Y., Zheng H., Lu M., Chen J. Hsf and Hsp gene families in Populus: Genome-wide identification, organization and correlated expression during development and in stress responses. BMC Genom. 2015;16:1. doi: 10.1186/s12864-015-1398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahesh U., Mamidala P., Rapolu S., Aragao F.J.L., Souza M.T., Rao P.J.M., Kirti P.B., Nanna R.S. Constitutive overexpression of small HSP24.4 gene in transgenic tomato conferring tolerance to high-temperature stress. Mol. Breeding. 2013;32:687–697. doi: 10.1007/s11032-013-9901-5. [DOI] [Google Scholar]
- 15.Kim K.H., Alam I., Kim Y.G., Sharmin S.A., Lee K.W., Lee S.H., Lee B.H. Overexpression of a chloroplast-localized small heat shock protein OsHsp26 confers enhanced tolerance against oxidative and heat stresses in tall fescue. Biotechnol Lett. 2012;34:371–377. doi: 10.1007/s10529-011-0769-3. [DOI] [PubMed] [Google Scholar]
- 16.Li J., Zhang J., Jia H., Li Y., Xu X., Wang L., Lu M. The Populus trichocarpa PtHsp17.8 involved in heat and salt stress tolerances. Plant Cell Rep. 2016;35:1587–1599. doi: 10.1007/s00299-016-1973-3. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y., Chen H., Chu P., Li Y., Tan B., Ding Y., Tsang E.W.T., Jiang L., Wu K., Huang S. NnHsp17.5, a cytosolic class II small heat shock protein gene from Nelumbo nucifera, contributes to seed germination vigor and seedling thermotolerance in transgenic Arabidopsis. Plant Cell Rep. 2012;31:379–389. doi: 10.1007/s00299-011-1173-0. [DOI] [PubMed] [Google Scholar]
- 18.Zhai M., Sun Y., Jia C., Peng S., Liu Z., Yang G. Over-expression of JrsHsp17.3 gene from Juglans regia confer the tolerance to abnormal temperature and NaCl stresses. J. Plant Biol. 2016;59:549–558. doi: 10.1007/s12374-015-0507-9. [DOI] [Google Scholar]
- 19.Shi W., Su S., Li B., Yang J., Gong Z., Hou Z. Molecular characterisation and expression analysis of a sHsp gene involved in heat shock treatment-induced chilling tolerance in highbush blueberry fruit. J. Hortic. Sci. Biotech. 2017;92:455–464. doi: 10.1080/14620316.2017.1304167. [DOI] [Google Scholar]
- 20.Li M., Ji L., Jia Z., Yang X., Meng Q., Guo S. Constitutive expression of CaHsp22.5 enhances chilling tolerance in transgenic tobacco by promoting the activity of antioxidative enzymes. Funct. Plant Biol. 2018;45:575–585. doi: 10.1071/FP17226. [DOI] [PubMed] [Google Scholar]
- 21.Hanley S.J., Karp A. Genetic strategies for dissecting complex traits in biomass willows (Salix spp.) Tree Physiol. 2014;34:1167–1180. doi: 10.1093/treephys/tpt089. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J., Li Y., Jia H., Li J., Huang J., Lu M., Hu J. The heat shock factor gene family in Salix suchowensis: A genome-wide survey and expression profiling during development and abiotic stresses. Front. Plant Sci. 2015;6:748. doi: 10.3389/fpls.2015.00748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai X., Hu Q., Cai Q., Feng K., Ye N., Tuskan G.A., Milne R., Chen Y., Wan Z., Wang Z., et al. The willow genome and divergent evolution from poplar after the common genome duplication. Cell Res. 2014;24:1274–1277. doi: 10.1038/cr.2014.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan H., Zhang A., Chen J., He X., Xu B., Xie G., Miao Z., Zhang X., Huang L. Genome-wide analysis of the PvHsp20 family in switchgrass: Motif, genomic organization, and identification of stress or developmental-related Hsp20s. Front. Plant Sci. 2017;8:1024. doi: 10.3389/fpls.2017.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch M., Conery J.S. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 26.Haslbeck M., Vierling E. A first line of stress defense: Small heat shock proteins and their function in protein homeostasis. J. Mol. Biol. 2015;427:1537–1548. doi: 10.1016/j.jmb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopes-Caitar V.S., Carvalho M.C.D., Darben L.M., Kuwahara M.K., Nepomuceno A.L., Dias W.P., Abdelnoor R.V., Marcelino-Guimarães F.C. Genome-wide analysis of the Hsp 20 gene family in soybean: Comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses. BMC Genom. 2013;14:577. doi: 10.1186/1471-2164-14-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrew B., Arvind M., Cannon S.B., Young N.D., Georgiana M. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waters E.R. The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 2013;64:391–403. doi: 10.1093/jxb/ers355. [DOI] [PubMed] [Google Scholar]
- 30.Jeffares D.C., Penkett C.J., Bähler J. Rapidly regulated genes are intron poor. Trends Genet. 2008;24:375–378. doi: 10.1016/j.tig.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 31.Mittal D., Chakrabarti S., Sarkar A., Singh A., Grover A. Heat shock factor gene family in rice: Genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol. Biochem. 2009;47:785–795. doi: 10.1016/j.plaphy.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 32.Montfort R.V., Slingsby C., Vierlingt E. Structure and function of the small heat shock protein/alpha-crystallin family of molecular chaperones. Adv. Protein Chem. 2001;59:105–156. doi: 10.1016/s0065-3233(01)59004-x. [DOI] [PubMed] [Google Scholar]
- 33.Saini H.S., Sedgley M., Aspinall D. Effect of heat stress during floral development on pollen tube growth and ovary anatomy in wheat (Triticum aestivum L.) Funct. Plant Biol. 1983;10:137–144. [Google Scholar]
- 34.Peet M.M., Sato S., Gardner R.G. Comparing heat stress effects on male-fertile and male-sterile tomatoes. Plant Cell Environ. 2010;21:225–231. doi: 10.1046/j.1365-3040.1998.00281.x. [DOI] [Google Scholar]
- 35.Begum S., Nakaba S., Yamagishi Y., Oribe Y., Funada R. Regulation of cambial activity in relation to environmental conditions: Understanding the role of temperature in wood formation of trees. Physiol. Plant. 2013;147:46–54. doi: 10.1111/j.1399-3054.2012.01663.x. [DOI] [PubMed] [Google Scholar]
- 36.Huang Y.C., Niu C.Y., Yang C.R., Jinn T.L. The heat stress factor HsfA6b connects ABA signaling and ABA-mediated heat responses. Plant Physiol. 2016;172:1182–1199. doi: 10.1104/pp.16.00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Charng Y., Liu H., Liu N., Chi W., Wang C., Chang S., Wang T. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 2007;143:251–262. doi: 10.1104/pp.106.091322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H., Jungeun H., Chanju L., Doolyi K., Sangyeol L., Chaeoh L. Arabidopsis DREB2C functions as a transcriptional activator of HsfA3 during the heat stress response. Biochem. Bioph. Res. Commun. 2010;401:238–244. doi: 10.1016/j.bbrc.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 39.Je J., Song C., Hwang J.E., Chung W.S., Lim C.O. DREB2C acts as a transcriptional activator of the thermo tolerance-related phytocystatin 4 (AtCYS4) gene. Transgenic Res. 2014;23:109–123. doi: 10.1007/s11248-013-9735-2. [DOI] [PubMed] [Google Scholar]
- 40.Lee S., Kang J., Park H., Kim M., Bae M., Choi H., Kim S. DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol. 2010;153:716–727. doi: 10.1104/pp.110.154617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letunic I., Doerks T., Bork P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43:257–260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell A., Chang H.Y., Daugherty L., Fraser M., Hunter S., Lopez R., Mcanulla C., Mcmenamin C., Nuka G., Pesseat S. The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 2015;43:213–221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu B., Jin J., Guo A.Y., Zhang H., Luo J., Gao G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bailey T.L., Johnson J., Grant C.E., Noble W.S. The MEME suite. Nucleic Acids Res. 2015;43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horton P., Park K.J., Obayashi T., Fujita N., Harada H., Adams-Collier C.J., Nakai K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007;35:585–587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larkin M.A., Blackshields G., Brown N.P., Chenna R., Mcgettigan P.A., Mcwilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 47.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Librado P., Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 50.Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Peer Y.V.D., Rouz P., Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J., Jia H., Han X., Zhang J., Sun P., Lu M., Hu J. Selection of reliable reference genes for gene expression analysis under abiotic stresses in the desert biomass willow, Salix psammophila. Front. Plant Sci. 2016;7:1505. doi: 10.3389/fpls.2016.01505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian F., Chang E., Yu L., Pei S., Hu J., Zhang J. Expression and integrated network analyses revealed functional divergence of NHX-type Na+/H+ exchanger genes in poplar. Sci. Rep. 2017;7:2607. doi: 10.1038/s41598-017-02894-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.