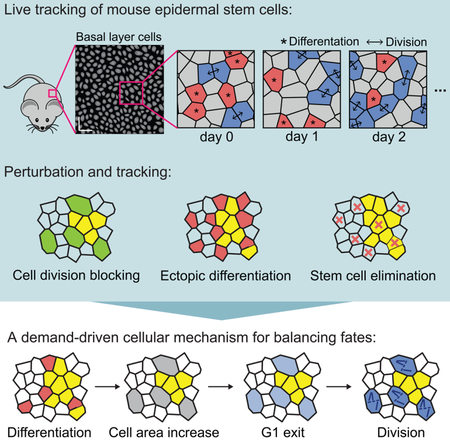
Summary:
Maintenance of adult tissues depends on sustained activity of resident stem cell populations, yet the mechanisms that regulate stem cell self-renewal during homeostasis remain largely unknown. Using an imaging and tracking approach that captures all epidermal stem cell activity in large regions of living mice, we show that self-renewal is locally coordinated with epidermal differentiation, with a lag time of one to two days. In both homeostasis and upon experimental perturbation, we find that differentiation of a single stem cell is followed by division of a direct neighbor, but not vice versa. Finally, we show that exit from the stem cell compartment is sufficient to drive neighboring stem cell self-renewal. Together, these findings establish that epidermal stem cell self-renewal is not the constitutive driver of homeostasis. Instead it is precisely tuned to tissue demand and responds directly to neighbor cell differentiation.
Blurb
By capturing all stem cell activity in large regions of mouse epidermis, Mesa, Kawaguchi, Cockburn and colleagues report that stem cell self-renewal is induced by the differentiation of neighbors. This study identifies the physiological factors that drive stem cell self-renewal, expanding the current understanding of epidermal homeostasis and regeneration.
Graphical abstract
INTRODUCTION
Maintenance of adult tissues depends on sustained activity of resident stem cell populations (Morrison & Spradling, 2008); (Simons & Clevers, 2011). An essential property of these stem cells is their ability to self-renew in order to preserve the size of the stem cell pool over time. However, the cellular mechanisms that regulate this homeostatic self-renewal remain poorly understood.
It remains generally unclear how stem cell self-renewal is regulated in the context of continual cell turnover (e.g. differentiation, cell death, etc.) in order to buffer against excess or insufficient cell divisions, such as in cancer or degenerative diseases, respectively. Work from epithelial tissues ranging from cultured cells to the developing mouse and zebrafish epidermis suggests that proliferation drives the delamination of nearby cells through a density-dependent mechanism (Eisenhoffer, et al., 2012; Marinari et al., 2017; Miroshnikova, et al., 2018). This coordination of behaviors is thought to maintain stem cell numbers and local density over time, allowing constitutive stem cell divisions to be compensated by the later exit of neighboring cells via delamination. We do not know whether this relationship between self-renewal and differentiation also occurs in fully-developed adult tissues. The ability to investigate this question depends on the tracking of co-existing stem cells as they execute both differentiation and self-renewal behaviors. However, to date this type of simultaneous, high-resolution spatiotemporal mapping of stem cell fates has not been possible in a live adult mammal.
The mouse skin epithelium offers a well-studied regenerative system in which to investigate the regulation of stem cell fates. Epidermal stem cells reside in an underlying basal layer, where they either self-renew within this compartment or differentiate by delaminating upward to contribute to the watertight barrier of the skin (Gonzales & Fuchs, 2017; Simpson, et al., 2011; Solanas & Benitah, 2013). Existing strategies to study these cell events have relied on clonal lineage tracing, which has provided fundamental insights into the self-renewal potential of epidermal stem cells, but has not addressed the factors that control self-renewal (Clayton, et al., 2007; Doupe, et al., 2010; Lim, et al., 2013; Mascre, et al., 2012; Rompolas, et al., 2016; Roy, et al., 2016; Sada, et al., 2016; Sanchez-Danes, et al., 2016). Collectively, these studies have shown that epidermal stem cells are equipotent, meaning they are equally capable to undergo self-renewal or terminal differentiation (Clayton, et al., 2007; Doupe, et al., 2010, Lim, et al., 2013; Mascre, et al., 2012; Rompolas, et al., 2016). Despite these advances in delineating stem cell potential, we still fail to understand the physiological cues of self-renewal in the context of other fate decisions taking place in neighboring stem cells, as well as how these cues ensure a precise balance of stem cell activity.
Here, we sought to directly interrogate epidermal stem cell self-renewal in relation to other cell fate decisions taking place in the surrounding tissue. We used an innovative imaging approach to map the timing and location of all self-renewal and differentiation events taking place in large epidermal regions. By combining spatiotemporal mapping of cell fates with newly developed statistical analysis, we find that cell fate choices are locally coordinated, with a lag time of one to two days. Surprisingly, and in contrast to the developing epidermis (Miroshnikova et al., 2018), we show that during homeostasis, differentiation of epithelial stem cells from the basal epidermal layer leads to neighboring cell size increase, cell cycle progression and ultimately cell division. Finally, we show that experimentally induced exit from the stem cell compartment is sufficient to drive local stem cell self-renewal. Altogether, this study identifies differentiation as the homeostatic driver of stem cell self-renewal in the mammalian epidermis.
RESULTS
Epidermal homeostasis is balanced through coupled, opposing fates of neighboring stem cells
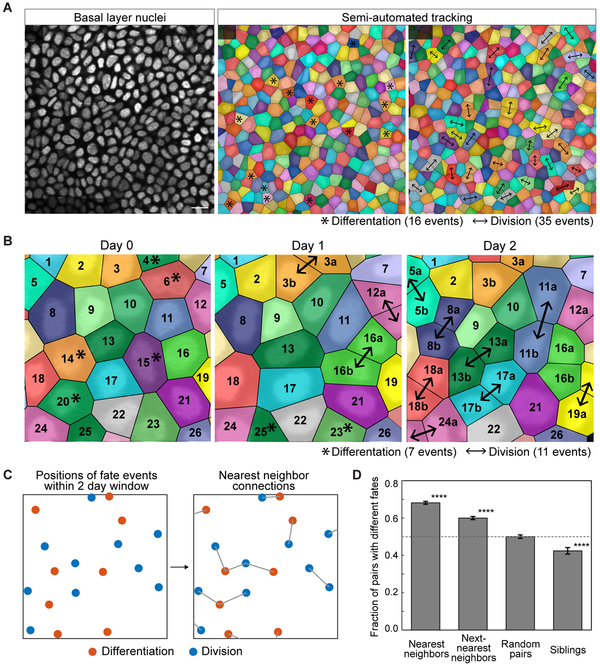
To begin to interrogate homeostatic regulation of stem cell self-renewal, we examined whether this behavior was occurring randomly with relation to surrounding stem cell activity. To address this question, we needed the ability to track every cell fate decision and evaluate it in the context of all other stem cell behaviors taking place around it. We therefore developed a spatiotemporal map of all division and differentiation events ocurring within a large region of tissue and over a long period of time. Building upon the live imaging approaches developed in our lab (Rompolas et al. 2012; Pineda et al 2015), we devised a whole-tissue tracking approach based on serial revisits of the same epidermal tissue over time (Figure 1A). Using mice expressing epidermal cortical and nuclear fluorescent markers (K14-actinGFP; (Vaezi, et al., 2002) and K14-H2BCerulean, respectively), we revisited the same epidermal regions every 12 hours for seven days (Figures S1A and S1B). Unlike previous efforts that only followed selected cells (Clayton, et al., 2007; Doupé, et al., 2010; Lim, et al., 2013; Mascré, et al., 2012; Rompolas, et al., 2016; Roy, et al., 2016; Sada, et al., 2016; Sánchez-Danés, et al.,2016), this approach for the first time accounts for the activity of every cell in a field of view, and thus provides a complete record of activity in homeostasis. Combining this imaging with the development of a semi-automated tracking method, we recorded 1527 divisions and 1540 differentiation events, representing a comprehensive record of all epidermal cell fate decisions across six regions from five mice (Figure 1A and Movie S1).
Figure 1. Basal fates are locally coordinated.
(A) Representative images showing the alignment, identification and tracking of individual basal cell behaviors over 12 hours. The regions of color represent the voronoi diagram calculated from the nuclear positions. Scale bar indicates 10 μm. Asterisks indicate that a cell will differentiate in the following 12 hours; arrows indicate that a cell has divided in the previous 12 hours.
(B) Representative tracking of basal cell fates over two days.
(C) Schematic of nearest neighbor fate analysis. Coordinates of the positions of divisions and differentiations were collected in 2 day time windows, and each of these positions were assigned a nearest neighbor fate decision point.
(D) Quantification of the fraction of pairs that execute opposite fates over a 2 day window when the pairs are nearest neighbors, next nearest neighbors, random pairs, or siblings. (**** represents p < 0.0001 obtained by a permutation test)
With this data, we were able to test whether stem cells might control their rates of self-renewal in response to differentiation of neighboring stem cells, or vice versa. We reasoned that the record of division and differentiation events would show if such control occurs rapidly, leading to instantaneous fate balance, or slowly, leading to a lag in fate balance. It would also reveal if stem cells respond directly to the fate of neighboring cells, or indirectly to the integrated activity of many stem cells. Finally, such data could also reveal if cell fate probabilities are ‘hard-wired’ in epidermal tissue (e.g. as odds of a coin flip), as previously proposed (Clayton et al., 2007; Simons and Clevers 2011). In such a case, each stem cell would commit to its fate in an autonomous manner with division and differentiation remaining only balanced over large fields of cells by the law of large numbers.
We first determined whether stem cells were compensating for the activity of their neighbors at all, and if so, how quickly they did this. For this, we focused on the extent to which division and differentiation behaviors were imbalanced over time. Although we observed both behaviors occurring in each imaged region and at each 12 hour time point, the number of cells dividing and differentiating over a given 12-hour period varied widely. Moreover, there was a significant discrepancy between the number of cells performing each activity between timepoints (Figures 1A and 1B), indicating the absence of any fast or instantaneous compensatory response of stem cells over this short interval. However, when we extended our analysis to a window of 7 days, we found that division and differentiation events occurred in nearly equal numbers, suggesting that stem cells might be responding to the activity of their neighbors over this longer period of time. Significantly, the imbalance between division and differentiation events in small regions of the tissue (22.5 μm × 22.5 μm) was significantly closer to zero than would be expected if each stem cell were autonomously deciding to divide or differentiate (standard deviation about zero of 2.9 events versus 5.7 events, p<0.0001, see STAR Methods for detail). This ruled out the possibility that stem cell fates are autonomous in the epidermis, as previously proposed and modeled by us and others (Klein and Simons 2011; Simons and Clevers 2011; Clayton et al. 2007; Doupe et al 2009; Lim et al. 2010; Rompolas et al. 2016). Instead, they appear to invoke mechanisms to actively balance their fates over time.
We then asked whether stem cells were responding to their local neighbors or to a broad neighborhood, by scoring how often a cell behavior (division or differentiation) is close to another behavior which is the same or the opposite. Interestingly, we found that within a two day window, nearest neighbor pairs of fate decision points were more likely to be opposite fates than would be expected by random chance (Figure 1C,D) (p<0.0001, permutation test). Correspondingly, direct neighbors that execute the same fates were found less frequently than expected. Such coordination became weaker when looking at next-nearest neighbors, but still significant. Consistent with our previous work (Rompolas et al., 2016), the fates of sibling pairs tended to be the same rather than the opposite. Together, these observations suggested that compensatory coordination between stem cell fates, which is key to balancing cell numbers over long periods of time, were occurring between neighboring pairs rather than between sibling pairs.
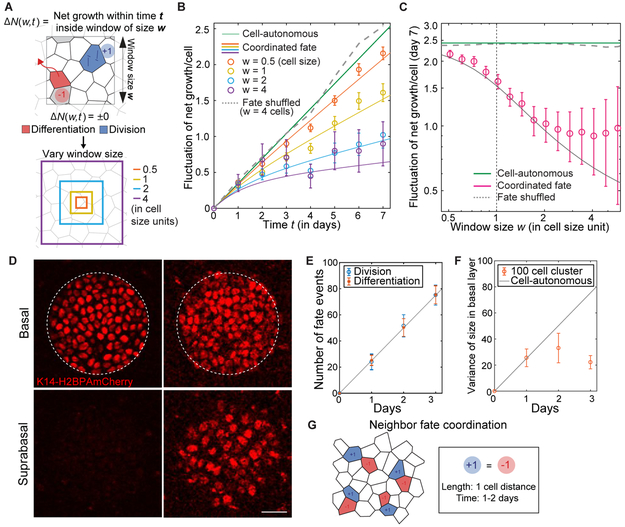
To precisely quantify the timing and spatial extent over which stem cells sense and respond to activity in the tissue, we devised a quantitative test that reveals whether the imbalance between the number of division and differentiation events differs from a null ‘non-response’ hypothesis, over any given time interval and any area of tissue. The methodology of this test defines the imbalance between the number of division and differentiation events as a function of window size w and time t, ΔN(w,t) (Figure 2A). We quantified ΔN(w,t) at many different spatial positions and over time to calculate its fluctuation (or Var[ΔN(w,t)]), and then compared this fluctuation to a null hypothesis randomizing the location of all division and differentiation events (Figures S2A-C). Over a short time window (one day), and after randomizing the location of all division and differentiation events, all cell fate choices appear to lack coordination as expected (Figures 2B, 2C S2D, and S2E). In contrast, at a 4-cell diameter (w = 4 cells, 27 μm), the variance of ΔN(w,t) departs clearly from random at later times (> two days), indicating that stem cells are coordinating their behaviors within their local neighborhood (Figure 2B and 2C). Strikingly, even when considering smaller distances w and time periods of less than three days, the variance of ΔN(w,t) remains distinct from random (Figure 2B and 2C). Fitting Var[ΔN(w,t)] to a generalized stochastic model with a coordination length scale / and timescale τ, we found that the time scale of coordination was approximately one to two days, while its length scale corresponded to the size of a single cell (~ 4 μm; Figures 2B, 2C, S2C and S2D; see STAR Methods for detail of the analysis). Thus, homeostatic self-renewal and differentiation are balanced by stem cells counteracting the fate of their neighbors, with a lag time of 1-2 days.
Figure 2. Coordination occurs between nearest epidermal neighbors.
(A) Schematic of net growth fluctuation analysis.
(B) Time dependence of net growth fluctuation. Data is from two 90 μm × 90 μm regions from one mouse; 507 divisions and 515 differentiations total. Solid lines indicate a model with the coordination time scale τ = 1.8 days and length scale l = 3.9 μm. Error bars represent s.e.m. See STAR Methods and Methods S1 for details of analysis. See Figure S2D for analyses from two additional mice.
(C) Window size dependence of net growth fluctuation. Using same data as (B). Error bars represent s.e.m. See Figure S2E for analyses from two additional mice.
(D) Representative images of a single 50 μm circle of K14H2B-PAmCherry labelled basal and suprabasal cells at 0 and 3 days post-labeling. Scale bar indicates 25 μm.
(E) Quantification of differentiation and division events in labeled circles (n = 34 circles from 2 mice). Differentiation events were quantified by counting the number of suprabasal H2B-PAmCherry positive cells (K14actin-GFP signal was used to help with scoring; see Figure S2G); division events were determined by comparing number of differentiation events with the number of labeled basal cells. Error bars represent s.e.m. Solid line is the least squares fit.
(F) Quantification of variance in labeled basal cells per circle (n = 34 circles from 2 mice) compared to the fluctuation predicted from a situation in which no local coordination occurs. Solid line is from rate obtained by fitting in (E) and assuming independent stochastic dynamics.
(G) Schematic of the length and time scale of fate coordination in epidermal stem cells.
To test this local coordination of behaviors, we used a second method to mark and follow geometrically defined regions in the basal layer over time. We used a recently described photo-activatable fluorescent reporter (K14H2B-PAmCherry) (Rompolas, et al., 2016) to label circular regions (50 μm in radius, 108 cells on average) designed to capture multiple local neighborhoods of stem cells and their first-and second-degree neighbors, and followed them for three consecutive days (Figures 2D and S2F). By counting the cells in the basal and suprabasal layers at each time point, we obtained the rates of division and differentiation as 0.23 events per day per basal layer cell (Figure 2E), consistent with rates obtained from previous tracking methods (Rompolas, et al., 2016). As a quantitative measure of behavioral coordination, we calculated the variance in the number of labeled basal cells per patch. In the absence of any local fate coordination within labeled patches, each basal cell should undergo independent stochastic dynamics as previously proposed (Clayton et al., 2007), leading to a linear increase in variance of approximately 80 cells2 after 3 days (Figure 2F). Yet strikingly, we observed very little variation in the number of cells contained within each circle after three days (Figures 2F; variance of 22 cells2). This clear deviation from the behaviors predicted by independent stochastic dynamics confirms that stem cells are indeed responding by self-renewing and/or differentiating to counteract local changes in cell number (Figure 2G).
Homeostatic self-renewal follows differentiation of immediate basal neighbors
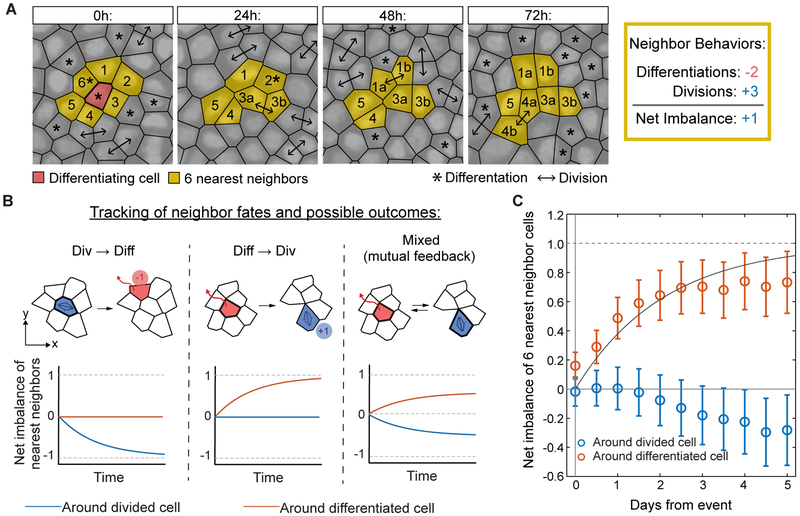
Thus far, we have demonstrated that neighboring stem cells locally balance self-renewal and differentiation rates, however it remained unclear if this relationship was a result of divisions driving local differentiation, as has been observed in the developing epidermis (Miroshnikova et al., 2018). To test for this scenario in the adult basal layer, we focused on individual cells that either divided or differentiated, and followed subsequent behaviors of the cells directly surrounding them (Figure 3A). To overcome the noise from simultaneous and highly frequent divisions and differentiations in the tissue (Figure 1A and Movie S1), we needed to average over hundreds of fate choice events to determine the overall imbalance of fates taking place between neighbors. If cell division leads to the differentiation of a neighbor, there should be a net imbalance of one extra differentiation event in the cells directly surrounding the original division, while the fates of neighbors surrounding a differentiating cell should, on average, be perfectly balanced (Figure 3B). These expectations reverse if differentiation leads to neighbor division, while a mixed scenario should lead to neighbor cell fate imbalances following both types of events (Figure 3B). Strikingly, the net imbalance across all dividing and differentiating cells showed a clear and unexpected unidirectional bias, with one excess stem cell division among cells directly neighboring a differentiation event, but no reciprocal compensation around dividing cells (Figures 3C and S2F). Our results thus indicate that in contrast to the embryonic scenario, homeostatic self-renewal in the adult epidermis is driven by neighboring stem cell differentiation events and not vice versa.
Figure 3. Stem cell self-renewal follows neighbor differentiation.
(A) Representative example of neighbor fate tracking following a differentiation event. The six cells in closest proximity to a differentiation event were followed to determine the imbalance in their fates over time. In this case, 2 direct neighbors differentiated (−2) and 3 neighbors divided (+3). We define difference between differentiation and division events, in this case +1 (1 more division than differentiation), as the “net imbalance”.
(B) Schematic of different possible neighbor fate imbalance scenarios. Depending on the temporal ordering of fates in neighboring cells, distinct patterns of imbalance surrounding divisions, differentiations or both are expected.
(C)Quantification of neighbor fate net imbalance corresponds to a model in which division follows differentiation within a short time scale. Data is from two 90 μm × 90 μm regions from one mouse; 524 divisions and 529 differentiations total. Solid line: , where τ = 1.8 days. Error bars represent s.e.m. See Figure S2F for analysis from two additional mice.
To functionally test this ordering of events, we reasoned that if differentiation is indeed a driver of self-renewal, manipulations that cause ectopic differentiation events should lead to a later compensatory increase in division. To test this hypothesis, we employed epidermal tape stripping, a method known to promote epidermal differentiation events through the removal of the outermost terminally differentiated layers of the skin (Potten, et al., 2000). Although this approach has long been known to also increase stem cell division (Pinkus, 1951), whether this wave of proliferation occurs subsequent to altered differentiation rates is not known. Thus, we sought to test our model by using groups of K14H2B-PAmCherry labeled cells to interrogate the sequence of events that occurs directly following tape stripping (Figure 4A). Tracking of labeled cells revealed a significant increase in differentiation rates within 12 hours of the procedure when compared to unperturbed control cells (Figure 4B). Notably, while tape stripping also increased the abundance of mitotic figures in the tissue as expected, this effect occurred subsequent to the rise in differentiation (Figure 4C). We next predicted that if differentiation is upstream and independent of self-renewal, blocking proliferation should not interfere with differentiation rates, leading to a rapid drop in basal cell density. By inhibiting cell division via either Mitomycin C (MMC) or Demecolcine treatment, we found that indeed, cell density progressively decreases in the basal layer (Figures 4D, S3A and S3B). Labeling and tracking cells with a photo-activatable fluorescent reporter (UBC-PAGFP) demonstrated that the number of cells differentiating out of the basal layer remained constant over multiple days of inhibition (Figures 4E and S3C), indicating that proliferation is not required to drive differentiation. Together, our experimental perturbations are entirely explained by the behaviors inferred from non-perturbative homeostatic imaging. They demonstrate an unexpected principle of homeostatic stem cell coordination, in which cell divisions follow differentiation to balance stem cell numbers over time (Figure 4F).
Figure 4. Differentiation is upstream and independent of self-renewal.
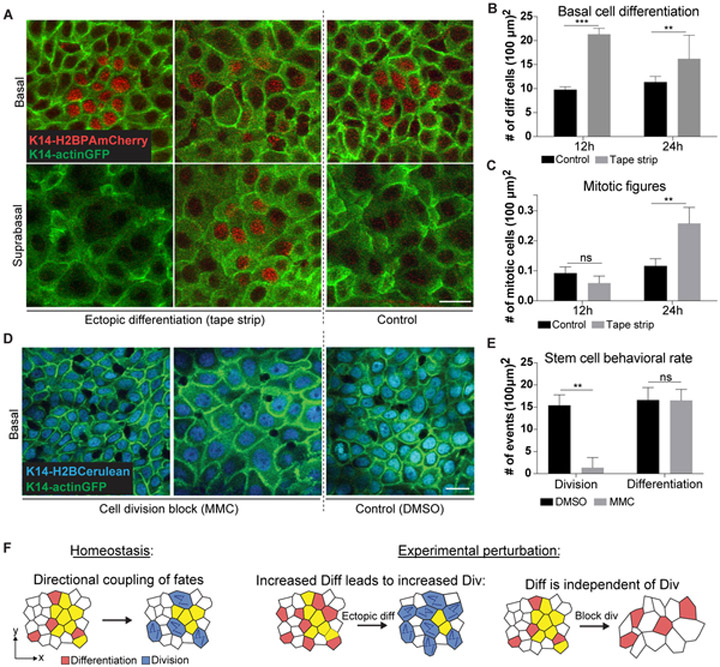
(A) Representative images of H2B-PAmCherry labeled basal and suprabasal cells after tape stripping to induce differentiation in mouse ear. Scale bar indicates 10 μm.
(B) Quantification of differentiation events by H2B-PAmCherry tracking after tape stripping in the mouse ear (n = 9 regions from 3 mice for each condition; error bars represent s.d.).
(C) Quantification of mitotic cells after tape stripping in the mouse ear. Mitotic events were scored based on cell morphology using K14-actinGFP signal (n = 9 regions from 3 mice for each condition; error bars represent s.d.).
(D) Representative images of basal cell density during Mitomycin C (MMC) induced cell division block in mouse ear. Scale bar indicates 10 μm.
(E) Quantification of basal cell division and differentiation events by PAGFP tracking after MMC treatment in the mouse ear (n = 9 regions from 3 mice for each condition; error bars represent s.d. See also Figure S3C).
(F) Schematic of directional fate coupling in homeostasis and after experimental perturbation.
Basal layer exit drives neighboring stem cell S phase entry and subsequent division
We next sought to understand the cellular mechanisms that lead to stem cell selfrenewal following local differentiation events. To this end, we first set out to resolve how cell cycle progression is affected by neighboring differentiation. We employed a Fucci-G1 reporter (mKO2-hCdt1(30/120), Sakaue-Sawano, et al., 2008) contained in our dataset, which specifically accumulates fluorescence during G1 of the cell cycle and drops this signal upon entry into S/G2 (Figures S4A and S4B). Our current dataset as well as previous work (Rompolas, et al., 2016), have demonstrated that epidermal stem cells can vary widely in their lifetimes, taking as few as 12 hours or as long as seven days between birth and their eventual division. Despite this variability, our Fucci reporter revealed that progression of the cell cycle through S/G2/M occurred for a predictable duration of approximately 12 hours (Figure S4C), demonstrating that it is the length of G1 that differs between cells of different lifetimes. Therefore, we hypothesized that a cell’s window of responsiveness to neighbor differentiation may occur during G1.
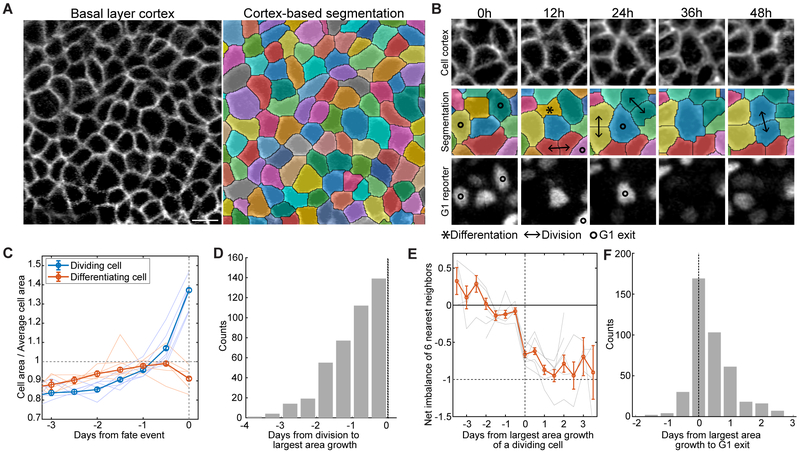
We next aimed to identify changes in the responding cells that occur prior to their G1 exit. We hypothesized that delamination of cells out of the basal layer allows neighbors to expand into newly freed space, resulting in area changes that promote G1 exit. To test this, we utilized the cortical K14-actinGFP signal in our imaged tissue to segment and measure the areas of each cell present in our seven-day timecourse (Figures 5A and 5B and Movies S2 and S3). We found that cells that eventually divide undergo significant area growth that starts as early as two days prior to division (Figures 5C and 5D). Moreover, a cell’s period of largest growth occurs concurrently with a neighboring differentiation event (Figure 5E). Finally, this period of largest growth is largely simultaneous with or precedes G1 exit (93% of cases, Figures 5B and 5F and Movie S3). Together, these data support a model in which a stem cell differentiation event provides space in the basal layer that drives S/G2 entry and later division of a directly neighboring stem cell (Figure 6D).
Figure 5. Cell enlargement precedes self-renewal.
(A) Representative images showing the segmentation of basal cells based on cortical K14-actinGFP signal. Scale bar indicates 10 μm.
(B) Representative example of an individual basal cell increasing in area concurrent with a neighbor differentiation, losing G1 reporter (mKO2-hCdt1(30/120)) signal, and dividing over the course of 48 hours.
(C) Quantification of average cell area prior to division and differentiation events. Thin lines correspond to 6 different regions from 5 mice (total of n = 1527 dividing cells and n = 1540 differentiating cells, error bars = s.e.m.).
(D) Histogram of the time elapsed between a cell’s period of largest area growth and its later division event (4 regions from 3 mice, n= 411 dividing cells).
(E) Quantification of fate imbalance in six nearest neighbors of dividing cells, expressed relative to the dividing cells’ period of largest area growth (6 regions from 5 mice, n = 1481 cells, error bars = s.e.m.).
(F) Histogram of the time elapsed between a cell’s period of largest area growth and its G1 exit (4 regions from 3 mice, n = 411 cells.).
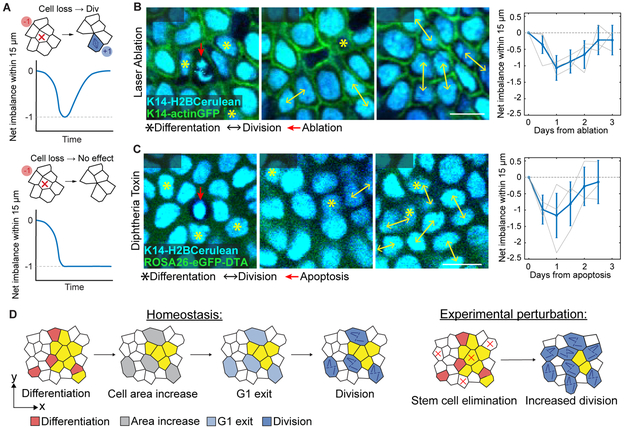
Figure 6. Elimination from the basal layer is sufficient to drive self-renewal.
(A) Expected net imbalance around an eliminated cell. If cell elimination is sufficient to drive local division, the net loss of the stem cell will be compensated by a later division. If elimination does not trigger local division, there will be net loss of a single cell that will not recover.
(B) Representative example of a single cell laser ablation in the basal layer, showing gradual elimination of the targeted cell (red arrow) along with compensatory division of neighboring cells. Quantification of net imbalance within a 15 μm radius of targeted ablations (n = 60 ablations from 3 mice), shows an initial trend towards cell loss followed by compensatory division and a return to balanced cell fates.
(C) Representative example of a DTA-induced apoptosis event in the basal layer (dying cell lacks cytoplasmic eGFP, indicating recombination of the ROSA26-eGFP-DTA allele to allow DTA expression), showing elimination of the dying cell (red arrow) along with compensatory division of neighboring cells. Quantification of net imbalance within 15 μm distance from single dying cells (n = 30 cells from 3 mice), shows an initial trend towards cell loss followed by compensatory division and a return to balanced cell fates.
(D) Schematic of relationship between cell area, cell cycle progression and division in homeostasis and after experimentally-induced basal cell elimination.
To test whether a local decrease of cell density in the basal layer by delaminating cells is sufficient to drive neighbor division, we devised a method to replicate these spatial changes. We used sub-micron scale laser ablations to precisely remove individual stem cells from the basal layer within approximately 24-36 hours (Figure S4D). To quantify the effect of this laser cell removal, we tracked the fate choice events in neighboring stem cells. If cell loss and consequent area increase is sufficient to drive compensatory proliferation, there should be a net excess of one neighboring division. If the spatial changes that follow cell removal have no effect on division, this compensation should not be observed (Figure 6A), suggesting that differentiating cells use alternative cues to induce neighbor cell division. As expected, we found that the net imbalance of cell fates within these small regions was significantly biased toward loss of basal cells up until 1.5 days post-ablation, reflecting the effective removal of cells (Figure 6B). Strikingly, however, the net imbalance returned to a neutral level by 2.5 days post-ablation as a result of compensatory neighboring cell divisions (Figures 6B), demonstrating similar compensatory self-renewal kinetics to our homeostatic tracking experiments.
While our ablation approach provides the advantage of selecting the specific cells we wish to ablate, it may also lead to unwanted damage or cause a local inflammatory response that could affect stem cell division rates. Although we saw no detectable difference in the number of immune cells present in the epidermis in the hours and days following ablation (Figures S4E and S4F and Movie S4), we additionally utilized a genetic model of Diptheria toxin (DTA) driven, non-inflammatory apoptosis (K14CreER combined with ROSA26-eGFP-DTA) (Ivanova et al., 2005). Notably, upon induction of DTA in low numbers of sparsely distributed basal cells (indicated by loss of cytoplasmic eGFP signal), we observed recovery of fate balance in neighboring cells with the same time kinetics as we observed following laser ablation (Figures 6C and S4G). Thus, forced removal of stem cells triggers neighboring cell divisions, demonstrating that exit from the basal layer itself is sufficient to drive stem cell self-renewal (Figure 6D).
DISCUSSION
Here, we describe a mechanism that coordinates stem cell self-renewal with differentiation in a live adult mammal. We provide direct evidence that the adult epidermis is maintained by local and rapid balancing of stem cell activity. Unexpectedly, we find that stem cell self-renewal does not drive differentiation during homeostasis, but rather that self-renewal is driven by neighboring differentiation events. Finally, we find this homeostatic process to be mediated by cell expansion-induced cell cycle progression following neighbor differentiation.
One notable distinction between stem cell behavior in adult epidermis versus what has been described during development is the direction in which coordination takes place. During embryogenesis, proliferation appears to be an upstream event that leads to local crowding and eventual compensatory cell delamination (Eisenhoffer, et al., 2012; Marinari et al., 2012; Miroshnikova, et al., 2018), while division in the adult context occurs as a downstream result of basal cell differentiation. This apparent discrepancy may represent the differing demands of developing versus homeostatic tissue. As a result of the need for constant growth and expansion during embryogenesis, progenitor cells might be primed to divide constitutively until the tissue growth is complete. Once a tissue structure has been established, the constant demand to replace lost cells may instead predominate, leading to a coordination strategy centered around stem cell differentiation. Given that our findings establish differentiation as a central driver of homeostatic behaviors, understanding the factors that cause selected stem cells to exit the basal layer will remain an important subject for future work. The unidirectional coupling we observe in the adult also poses intriguing questions: how does the tissue switch from the embryonic program to the adult homeostatic program? How does the stem cell compartment cope with situations where proliferation rates are increased, for instance due to oncogenic mutations? Further work will be required to determine whether the relationship we observe in homeostatic epidermis also predominates under pathological conditions, or whether other forms of feedback such as proliferation-driven delamination may instead emerge.
Notably, it appears that the majority of stem cells at any given time are in G1, remaining poised and competent to respond to external cues in a highly flexible manner. Yet, our observation that sibling cells tend to execute the same behavior may suggest that a stem cell’s age, or perhaps length time spent in G1, may also contribute toward biasing a cell to a particular fate. The specific molecular pathways linking cell area to G1 exit will also be an important subject for future study. Potential candidates include the Hippo and Wnt signaling pathways as well as mechanosensitive ion channels such as Piezo 1, all of which have been shown to sense and transduce cellular geometry and/or mechanical properties into cell cycle progression (Panciera, et al., 2017; Benham-Pyle, et al., 2015; Benham-Pyle, et al., 2016; Gudipaty, et al., 2017). Because of its central role in epidermal homeostasis, it is possible that fate coordination is mediated by multiple, redundantly acting molecular cues serving to reinforce the process. Additionally, it is far from clear how the sensing of cell loss is strongly localized to the neighboring cells, as opposed to a density or pressure-controlled feedback mechanism (Shraiman, 2005; Eisenhoffer, et al., 2012; Gudipaty, et al., 2017) which should have a larger length scale compared to the size of individual cells.
In conclusion, we have identified a behavioral coupling mechanism between neighboring epidermal stem cells that is mediated by local changes in cell area. The coordination we have uncovered indicates that instead of a cell-autonomous program of self-renewal, it is the constant production of differentiated cells that stimulates compensatory divisions in the epidermal stem cell pool. This study serves as an important step towards understanding the basic principles underlying stem cell fate coordination during normal tissue homeostasis as well as the potential loss of it in pathological states.
STAR Methods:
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Valentina Greco (Valentina.Greco@yale.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
K14-H2BPAmCherry, K14-H2BCherry and K14-H2BCerulean mice were generated by the Yale Transgenic Facility. K14-actinGFP mice were obtained from E. Fuchs. mKO2-hCdt1(30/120) (Fucci G1-reporter) mice were obtained from Shangqin Guo. UBC-PA-GFP, ROSA26-eGFP-DTA, K14CreER and CAG::H2BGFP mice were obtained from Jackson Laboratories. All experiments were performed on mice 1-2 months of age. Mice were not mated or used in previous procedures before experimentaion. Animals of both sexes were used for experiments. All procedures involving animal subjects were performed under the approval of the Institutional Animal Care and Use Committee (IACUC) of the Yale School of Medicine. Five mice were housed per cage, with a 12 hr light/dark cycle, and were provided food and water ad libitum.
METHOD DETAILS
Experimental treatment of mice
All imaging and experimental manipulation was performed on non-hairy mouse plantar (hind paw) skin except data shown in Figures 4A-E. Preparation of skin for intravital imaging was performed as described. Briefly, mice were anesthetized with IP injection of ketamine/xylazine (15 mg/ml and 1 mg/ml, respectively in PBS). After marking the area to be imaged for subsequent identification with a micro-tattoo, mice were returned to their housing facility. For subsequent revisits, the same mice were processed again with injectable anesthesia. The plantar epidermal regions were briefly cleaned with PBS pH 7.2, mounted on a custom-made stage and a glass coverslip was placed directly against the skin. Anesthesia was maintained throughout the course of the experiment with vaporized isoflurane delivered by a nose cone. Manipulations shown in Figures 4A-E were performed on ear skin which was shaved using an electric shaver and depilatory cream (Nair) at least 3 days prior to imaging, and then processed as described above. To induce sparse DTA-mediated cell death, mice received an intraperitoneal injection of Tamoxifen (10 mg/kg body weight in corn oil) 24h before imaging. All experiments were performed on at least 3 mice with the exception of the data shown Figure 2D-F, which was derived from 34 separate regions from 2 mice. Animal studies were not performed in a blinded fashion. Animals were assigned randomly to experimental groups.
Topical drug treatment
To inhibit cell proliferation in the epidermis, Mitomycin C (MMC) and Demecolcine were both delivered topically by applying it to ear skin. MMC was dissolved in a 15 mg/ml stock solution in dimethyl sulfoxide (DMSO), while Demecolcine was dissolved in a 25 mg/ml stock solution in the DMSO. The stock solution was diluted 100 times in 100% petroleum jelly (Vaseline; final concentration is 150 μg/ml). One hundred micrograms of either working concentration were spread evenly on the skin area daily. A mixture of 100% DMSO in petroleum jelly (1:100) was used as a vehicle control. In the analyses of the treated regions, the stars in Figures 4B, 4C, 4E, and S3B represent the statistical significance (**,***,****, and n.s. representing p < 0.01,0.001.0.0001, and nonsignificant, respectively).
Tape stripping
Tape stripping was performed by applying standard colored lab tape (Fisher Scientific) to the surface of the ear skin and removing it in ten sequential repetitions.
In vivo imaging
Image stacks were acquired with a LaVision TriM Scope II (LaVision Biotec, Germany) microscope equipped with both Chameleon Vision II and Discovery (Coherent, USA) 2-Photon lasers. For collection of serial optical sections, a laser beam (940nm for GFP/Cerulean and 1100nm for mCherry, respectively) was focused through a 20X or 40X water immersion lens (Zeiss W-Plan-APOCHROMAT, N.A. 1.0; Zeiss W-LD C-APOCHROMAT, N.A. 1.1 Zeiss) and scanned with a field of view of 0.5 mm × 0.5 mm at 600Hz. z-stacks were acquired in 1 μm steps for a ~40-80 μm range, covering the entire thickness of the epidermis. Cell tracking analysis was performed by re-visiting the same area of the epidermis in separate imaging experiments, as described in Image Analysis. A micro-tattoo was introduced in addition to using inherent landmarks of the skin to navigate back to the original region; including the vasculature and distinctive clustering of hair follicles.
Photo-activation
Photo-activation in K14-H2BPAmCherry and UBC-PA-GFP mice was carried out with the same optics as used for acquisition. An 810 nm laser beam was used to scan the target area (10–500 mm2). Activation of the PA-mCherry was achieved using 3% laser power for 2 min. For the K14-H2BPAmCherry experiments described in Figure 2, n = 34 circles from 2 mice were used for analyses. Activation of the PA-GFP shown in Figure S3 was achieved using 3% laser power for 30 sec.
Laser ablation
Laser ablation was carried out with the same optics as for acquisition. An 810nm laser beam was used to scan the target area (< 1 μm2 within the nucleus of a chosen cell) and ablation was achieved using 10-15% laser power for ~5-10 seconds. To look for signs of immune cell recruitment following ablation, 10-15 evenly distributed ablations were performed per 300 μm × 300 μm field of view in CAG::H2BGFP mice, followed by time-lapse recording of serial optical sections at 5 minute intervals. As a positive control, a much larger area of laser damage (~20 μm × 30 μm region scanned with 30% laser power) was created before time-lapse imaging.
QUANTIFICATION AND STATISTICAL ANALYSIS
Image analysis
Images which included the suprabasal layer, basal layer, and the extracellular matrix were obtained as large tiled image stacks at roughly the same positions every 12 hours for 7 days (mice 1-3) and 4 days (mice 4,5) guided by the micro-tattoo. We first manually aligned the images over the time course in Imaris (Bitplane) by using data from all three channels: K14-actinGFP, K14-H2BCerulean, and Fucci G1.
For the cell-tracking analysis performed by MATLAB scripts, we first cropped out regions with size 115 μm × 115 μm or 153 μm × 153 μm, which typically included more than 300 and 600 basal layer cells, respectively. To correct for the difference of height positions of the basal layer within the 3D images, we first Gaussian blurred the signal from the K14-actinGFP channel spatially in the xy-plane (width 4 um) to create a 3D mask representing the region covering the whole epidermis (Figure S1A). We then defined the height of the interface between the epidermis and the dermis from the 3D mask, and subtracted this height from the original 3D data to level the basal layer position.
From the height-corrected 3D images, we took a single z-position containing the nucleus of all the basal layer cells (Figure S1B), and performed cell tracking in the twodimensional data over the time-course. For this, we first manually corrected the shifts in the images to minimize the overall depositions of cells, and then ran an automatic cell tracking algorithm based on the positions of local maxima in the H2B-Cerulean channel. The algorithm assigned each cell (represented by the local maxima, calculated after Gaussian blurring the image with width 1 μm) to the closest cell in the previous time frame. Tracked cells were frequently lost or were associated with more than one cell in the subsequent time frame, which indicated cell differentiation (i.e., delamination from the basal layer) and cell division, respectively. After manually correcting the errors in the tracking with guide from the height-corrected 3D images in all three channels, the script outputted the positions of the local maxima in the H2B-Cerulean channel and the lineages of the cells present in the basal layer at each time point (Figure 1A).
For the fluctuation analysis (Figures 2A-C and S2A-E) and fate imbalance analysis (Figure 3 and S2F), we used the 7 day track data from mouse 1-3. Results from mouse 1 (n = 2 regions) are presented in Figures 2B, 2C, and 3C. Results from mouse 2 and 3 (n = 1 region for each) are presented in Figure S2D-F.
Quantification of cell area and G1 reporter signal level
At each time frame, we segmented the cells using the marker-controlled watershed algorithm. We first made a basal layer cortex image (Figure 5A) by subtracting the H2B-Cerulean channel from the Actin-GFP channel with appropriate scaling (set manually). We then imposed the cell position pixels (local maxima of H2B-Cerulean channel) as the local minima in the cell periphery image and applied the watershed transform for segmentation. Segmented regions were associated to cells (Figure 5B) and the size of the regions were recorded as the cell areas.
For the Fucci G1-reporter channel (mice 1, 4, and 5), we first z-score normalized the signal using all pixels at each time frame to account for the temporal fluctuation. Using this corrected signal, we calculated the signal per area of the cells at each time point using the segmented cell areas. We subtracted the background signal, defined as the average of the 10 cells with smallest signal per area at each time point.
Figure S4B shows the traces of the signal/area values for dividing cells (after subtracting background). Not all the cells had strong Fucci signal, and many of them had a low signal throughout their lifetime. For this figure we therefore filtered cells based on the maximum level of signal/area (threshold included 218 out of 509 dividing cells across two regions in mouse 1 and the same threshold used for mouse 4 and 5).
To show that after G1 exit cells divide within 24 hours, we defined the timing of G1 exit as the time where a cell experienced the largest fold change decrease in the signal/area value. We then tested that G1 exist, as defined, happens right before or 1 frame before the cell division. This analysis was restricted to the filtered cells as defined above.
Fluctuation test for uncoordinated fate choice
The fluctuation test (Fig. 2B) identifies whether fate coordination occurs over a time scale t and lengthscale w by comparing an observed test statistic Var[ΔN(w,t)], as defined in the text, with its value after breaking all coordination by spatially shuffling the location of division and differentiation events at each time point. This is done through the following steps for each lengthscale w:
Randomly choose 4096 positions of a window with size w within each region. To avoid tracking errors that occur near image boundaries, analyses focused on fate events that occur sufficiently far from the edge (central region with size 90 μm × 90 μm).
Calculate the time course ΔN(w, t) for each of the 4096 sampled windows
Calculate the variance Var [ΔN(w,t)] for each w and t over the 4096 positions.
Correct for a bias resulting from window overlap: Var[ΔN(w,t)](Corrected) = Var[ΔN(w,t)]/(1 – 0[L,w]), with 0[L,w] derived in Methods S1, Note 1.
Repeat steps 1-4 with randomized data. The final ‘shuffled’ curves in Figures 2B, 2C, S2D and S2E are the average of 128 independent randomizations of the data set.
The p-value of the fluctuation test corresponds to the fraction of randomized trials for which Var[ΔN(w,t)] is smaller than the empirical value. We calculated a p-value for w=22.5 μm and t=7 days, restricting to a total of 64 imaging regions (16 each from 4 regions, where 2 regions were from mouse 1 and 1 region each from mouse 2 and 3). The calculated value of p<0.0001 reported in the text associates with an empirical test statistic of Var[ΔN(w,t)]1/2 = 2.9 events, which was not observed in 10,000 randomizations of the positions of division and differentiation events (mean random test statistics Var[ΔN(w,t)]1/2 =5.7 events).
Extraction of timescales and lengthscales
In Figures 2B, 2C, S2D, and S2E, we show the best fit of a generalized stochastic model to the data of Var[ΔN(w,t)], obtained by the least-squares method while using an event rate of 㥋 = 9.3 × 103 events day−1 mm−2 for the first mouse (Figures 2B and 2C), Λ = 1.2 × 104 events day−1 mm−2 for the second mouse, and Λ = 1.1 × 104 events day−1 mm−2 for the third mouse (Figures S2D and S2E). The formula fit to the data is derived in Note 2 of Methods S1 [see Eq. (4) in Note 2]. The best fit parameters were τ = 1.8 days and l = 3.9 μm (Figures 2B and 2C), τ = 0.9 day and l = 5.2 μm, and τ = 1.2 days and l = 5.0 μm (Figures S2D and S2E) for the first, second, and third mouse, respectively.
Forward tracking of neighbor fate imbalance
In Figures 3C and S2F, we plotted the background-detrended net imbalance around differentiation and division events, Idiff(t,K) and Idiv(t,K), as functions of time t and considering K = 6 nearest neighbors, with formulae as defined in Methods S1, Note 3 [see Eqs. (12-13) in Note 3]. Error bars indicate fluctuations (s.e.m.) in INN(i,t,K) and B(t1,t2), as defined in Eqs. (10-11) in Note 3. We also plotted the expected theory line 1 – e−t/τ assuming a stochastic process for the compensation, where τ = 1.8 days (mouse 1), τ = 0.9 days (mouse 2), and τ = 1.2 days (mouse 3) were the best fit of the fate-coordination time scale from the fluctuation analyses (Figures 2B, 2C, S2D, and S2E).
Area growth analysis
In Figure S4, we plotted the cell area calculated from the Actin-GFP-based segmentation of dividing and differentiating cells, Adiv(t) and Adiff(t), and we indicated the time frame of largest cell area growth, tAG(i). Formulae for Adiv(t), Adiff(t) and tAG(i) are derived in Methods S1, Note 4.
Backward tracking of neighbor fate imbalance
In Figure 5E, we plotted the background-detrended net imbalance, IB(t,K), as a function of t with K = 6 for separate regions and the total mean of all regions. The errors come from the fluctuations (s.e.m) of IBNN(i,t,K) and BG(t1,t2). Formulae for IB(t,K), IBNN(i,t,K) and BG(t1,t2) are given in Methods S1, Note 5.
Data and software availability
All MATLAB codes and the data that support the findings of this study are available from the lead author (V.G.) upon request.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dulbecco’s Phosphate Buffered Saline (PBS) | Thermo Fisher | Cat#14190144 |
| Mitomycin C (MMC) | RPI | Cat#M92010 |
| Demecolcine | Sigma-Aldrich | Cat#D7385 |
| Dimethylsulfoxide (DMSO) | R&D | Cat#AB03091 |
| Tamoxifen | Sigma-Aldrich | Cat#T5648 |
| Experimental Models: Organisms/Strains | ||
| Mouse: K14-H2BPAmCherry | Rompolas et al., 2016 | N/A |
| Mouse: K14-actinGFP | Vaezi et al., 2012 | N/A |
| Mouse: K14-H2BCerulean | This paper | N/A |
| Mouse: mKO2-hCdt1(30/120) | Sakaue-Sawano, et al., 2008 | N/A |
| Mouse: K14-H2BmCherry | Mesa et al., 2015 | N/A |
| Mouse: B6.Cg-Ptprca Tg(UBC-PA-GFP) 1Mnz/J | The Jackson Laboratory | JAX: 022486 |
| Mouse: Gt(ROSA)26Sortm1 (D1A)Jpmb/J | The Jackson Laboratory | JAX: 006331 |
| Mouse: T g(KRT 14-cre/ERT)20Efu/J | The Jackson Laboratory | JAX: 005107 |
| Mouse: B6.Cg-Tg(HIST1 H2B/EGFP)1 Pa/J | The Jackson Laboratory | JAX: 006069 |
| Software and Algorithms | ||
| Fiji (ImageJ) | https://fiji.sc/ | N/A |
| MATLAB scripts | This paper | Available upon request |
Highlights.
Neighboring epidermal stem cells actively balance their fates over time
Stem cell division is triggered by demand for differentiated cells
Differentiation drives division by neighbor cell enlargement followed by S/G2 entry
Both forced and natural epidermal stem cell loss drive cell divisions
Acknowledgments:
We thank Lucy O’Brien and Sara Wickstrom for advice on the manuscript, Elaine Fuchs for K14-actinGFP mice, and Panteleimon Rompolas for CAG::H2B-EGFP timelapse data. This work is supported by The New York Stem Cell Foundation, The Edward Mallinckrodt, Jr. Foundation, the Glenn Foundation for Medical Research, the HHMI Scholar award, and the National Institute of Arthritis and Musculoskeletal and Skin Disease (NIAMS), NIH, grants no. 2R01AR063663-06A1; 1R01AR072668-01; 5R01AR067755-02. K.R.M was supported by the NIH Predoctoral Program in Cellular and Molecular Biology (grant T32GM007223). The content is the responsibility of the authors and does not necessarily represent the official views of the NIH. K.R.M was an NSF Graduate Research Fellow. V.G. is a New York Stem Cell Foundation Robertson Investigator and HHMI Scholar. A.M.K. is supported by a Career Award at the Scientific Interface from the Burroughs Wellcome Fund, and an Edward Mallinckrodt Jr. Foundation Grant. K.K. acknowledges Grants-in-Aid for JSPS Fellows (28-908). K.C. was supported by a Canadian Institutes of Health Research Postdoctoral Fellowship.
Footnotes
Declaration of Interests: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Benham-Pyle BW, Pruitt BL and Nelson WJ (2015). Mechanical strain induces E-cadherin--dependent Yap1 and β-catenin activation to drive cell cycle entry. Science, 348, 1024–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benham-Pyle BW, Sim JY, Hard KC, Pruitt BL and Nelson WJ (2016). Increasing β-catenin/Wnt3A activity levels drive mechanical strain-induced cell cycle progression through mitosis. Elife, 5, e19799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton E, Doupé D, Klein AM, Winton DJ, Simons BD and Jones PH (2007). A single type of progenitor cell maintains normal epidermis. Nature, 446, 185–189. [DOI] [PubMed] [Google Scholar]
- Doupe D, Klein AM, Simons BD and Jones H (2010). The ordered architecture of murine ear epidermis is maintained by progenitor cells with random fate. Dev. Cell, 18, 317–323. [DOI] [PubMed] [Google Scholar]
- Eisenhoffer GT, Loftus PD, Yoshigi M, Otsuna H, Chien CB, Morcos PA and Rosenblatt J (2012). Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature, 484, 546–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales KAU and Fuchs E (2017). Skin and Its Regenerative Powers: An Alliance between Stem Cells and Their Niche. Developmental Cell, 43, 387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipaty SA, Loftus PD, Redd MJ, Edes K, Davey CF, Krishnegowda V and Rosenblatt J (2017). Mechanical stretch triggers rapid epithelial cell division through Piezo1. Nature, 543, 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch T, Rothoeft T, Teig N, Bauer JW, Pellegrini G, De Rosa L, Scaglione D, Reichelt J, Klausegger A, et al. (2017). Regeneration of the entire human epidermis using transgenic stem cells. Nature, 551, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova A, Signore M, Caro N, Greene NDE, Copp AJ and Martinez-Barbera JP (2005). In vivo genetic ablation by Cre-mediated expression of diphtheria toxin fragment A. Genesis, 43, 129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM and Simons BD, 2011. Universal patterns of stem cell fate in cycling adult tissues. Development, 138, 3103–3111. [DOI] [PubMed] [Google Scholar]
- Lim X, Tan SH, Koh WLC, Chau RMW, Yan KS, Kuo CJ, van Amerongen R, Klein AM and Nusse R (2013). Interfollicular epidermal stem cells self-renew via autocrine Wnt signaling. Science, 342, 1226–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinari E, Mehonic A, Curran S, Gale J, Duke T and Baum B (2012). Live-cell delamination counterbalances epithelial growth to limit tissue overcrowding. Nature, 484, 542–545. [DOI] [PubMed] [Google Scholar]
- Mascré G, Dekoninck S, Drogat B, Youssef KK, Brohee S, Sotiropolou PA, Simons BD and Blanpain C (2012). Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature, 489, 257–262. [DOI] [PubMed] [Google Scholar]
- Miroshnikova YA, Le HQ, Schneider D, Thalheim T, Rubsam M, Bremicker N, Polleuz J, Kamprad N, Tarantola M, Wang I, et al. (2018). Adhesion forces and cortical tension couple cell proliferation and differentiation to drive epidermal stratification. Nat. Cell Biol, 20, 69. [DOI] [PubMed] [Google Scholar]
- Morrison SJ and Spradling AC (2008). Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell, 132, 598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panciera T, Azzolin L, Cordenonsi M and Piccolo S (2017). Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol, 18, 758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda C, Park S, Mesa KM, Wolfel M, Gonzalez DG, Haberman AM, Rompolas PM and Greco V (2015). Intravital imaging of hair follicle regeneration in the mouse. Nature Methods, 10, 1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkus H (1951). Examination of the epidermis by the strip method of removing horny layers: I. Observations on thickness of the horny layer, and on mitotic activity after stripping. J. Investig. Dermatol, 16, 383–386. [DOI] [PubMed] [Google Scholar]
- Potten CS, Barthel D, Li YQ, Ohlrich R, Matthe B and Loeffler M (2000). Proliferation in murine epidermis after minor mechanical stimulation Part 1. Sustained increase in keratinocyte production and migration. Cell Prolif., 33, 231–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Deschene ER, Zito G, Gonzalez DG, Saotome I, Haberman AM and Greco V (2012). Live imaging of stem cell and progeny behaviour in physiological hair follicle regeneration. Nature, 487, 496–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Mesa KR and Greco V (2016). Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science, 352, 1471–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy E, Neufeld Z, Cerone L, Wong HY, Hodgson S, Livet J and Khosrotehrani K (2016). Bimodal behaviour of interfollicular epidermal progenitors regulated by hair follicle position and cycling. EMBO J, 35, 2658–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sada A, Jacob F, Leung E, Wang S, White BS, Shalloway D and Tumbar T (2016). Defining the cellular lineage hierarchy in the interfollicular epidermis of adult skin. Nat. Cell Biol, 18, 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. (2008). Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell, 132, 487–498. [DOI] [PubMed] [Google Scholar]
- Sánchez-Danés A, Hannezo E, Larsimont J, Liagre M, Youssef KK, Simons BD and Blanpain C (2016). Defining the clonal dynamics leading to mouse skin tumour initiation. Nature, 536, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shraiman BI (2005). Mechanical feedback as a possible regulator of tissue growth. Proc. Natl. Acad. Sci, 102, 3318–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons BD and Clevers H (2011). Strategies for homeostatic stem cell self-renewal in adult tissues. Cell, 145, 851–862. [DOI] [PubMed] [Google Scholar]
- Simpson CL, Patel DM and Green KJ (2011). Deconstructing the skin: cytoarchitectural determinants of epidermal morphogenesis. Nat. Rev. Mol. Cell Biol, 12, 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solanas G and Benitah SA (2013). Regenerating the skin: a task for the heterogeneous stem cell pool and surrounding niche. Nat. Rev. Mol. Cell Biol,, 14, 737. [DOI] [PubMed] [Google Scholar]
- Vaezi A, Bauer C, Vasioukhin V and Fuchs E (2002). Actin cable dynamics and Rho/Rock orchestrate a polarized cytoskeletal architecture in the early steps of assembling a stratified epithelium. Dev. Cell, 3, 367–381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.