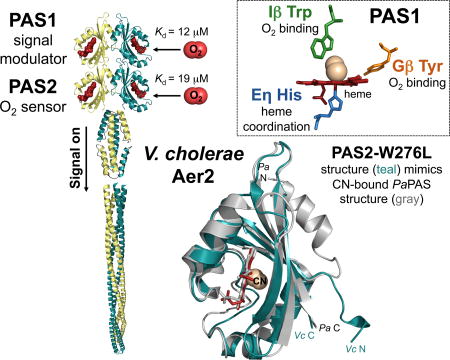
SUMMARY
The diarrheal pathogen Vibrio cholerae navigates complex environments using three chemosensory systems and 44–45 chemoreceptors. Chemosensory cluster II modulates chemotaxis, whereas clusters I and III have unknown functions. Ligands have been identified for only five V. cholerae chemoreceptors. Here we report that the cluster III receptor, VcAer2, binds and responds to O2. VcAer2 is an ortholog of Pseudomonas aeruginosa Aer2 (PaAer2), but differs in that VcAer2 has two, rather than one, N-terminal PAS domain. We have determined that both PAS1 and PAS2 form homodimers and bind penta-coordinate b-type heme via an Eη-His residue. Heme binding to PAS1 required the entire PAS core, but receptor function also required the N-terminal cap. PAS2 functioned as an O2-sensor [Kd(O2), 19 µM], utilizing the same Iβ Trp (W276) as PaAer2 to stabilize O2. The crystal structure of PAS2-W276L was similar to that of PaAer2-PAS, but resided in an active conformation mimicking the ligand-bound state, consistent with its signal-on phenotype. PAS1 also bound O2 [Kd(O2), 12 µM], although O2 binding was stabilized by either a Trp or Tyr residue. Moreover, PAS1 appeared to function as a signal modulator, regulating O2-mediated signaling from PAS2, and resulting in activation of the cluster III chemosensory pathway.
Keywords: Vibrio cholerae, chemoreceptor, PAS domain, heme, oxygen, signal transduction
Graphical abstract
INTRODUCTION
Vibrio cholerae, the diarrheal pathogen that causes cholera, is a highly motile bacterium that encodes 44 to 45 chemoreceptors and three distinct chemosensory systems (chemosensory clusters I, II and III) (Boin et al., 2004). Chemosensory cluster II expresses a membrane-bound array that modulates bacterial chemotaxis (Gosink et al., 2002, Hyakutake et al., 2005, Briegel et al., 2016). Cluster I functions in microaerobic environments (Hiremath et al., 2015), although its precise role, as well as the role of cluster III, is currently unknown. Cluster III (VCA1089-VCA1096) resides on chromosome II and encodes a complete set of chemosensory proteins (CheY4, CheA3, CheW3, CheW4, CheR3, CheD and CheB3), including a chemoreceptor (VCA1092, Mlp45) that we have named VcAer2 (Fig. 1A). This designation is based on its homology to the Pseudomonas aeruginosa Aer2 receptor (PaAer2), which we have previously characterized (Airola et al., 2010, Watts et al., 2011, Airola et al., 2013a, Airola et al., 2013b, Garcia et al., 2017). V. cholerae cluster III is also orthologous to the P. aeruginosa Che2 chemosensory cluster, but includes a second cheW gene (Fig. 1A). Notably, the expression of both of these chemosensory clusters i) occurs during stationary phase, ii) requires the alternative sigma factor RpoS, and iii) produces proteins that form polar clusters independent of chemotaxis clusters (Schuster et al., 2004, Guvener et al., 2006, Ringgaard et al., 2015). Cluster III is also expressed in cultures after growth arrest from conditions of carbon starvation or culture saturation (Ringgaard et al., 2015).
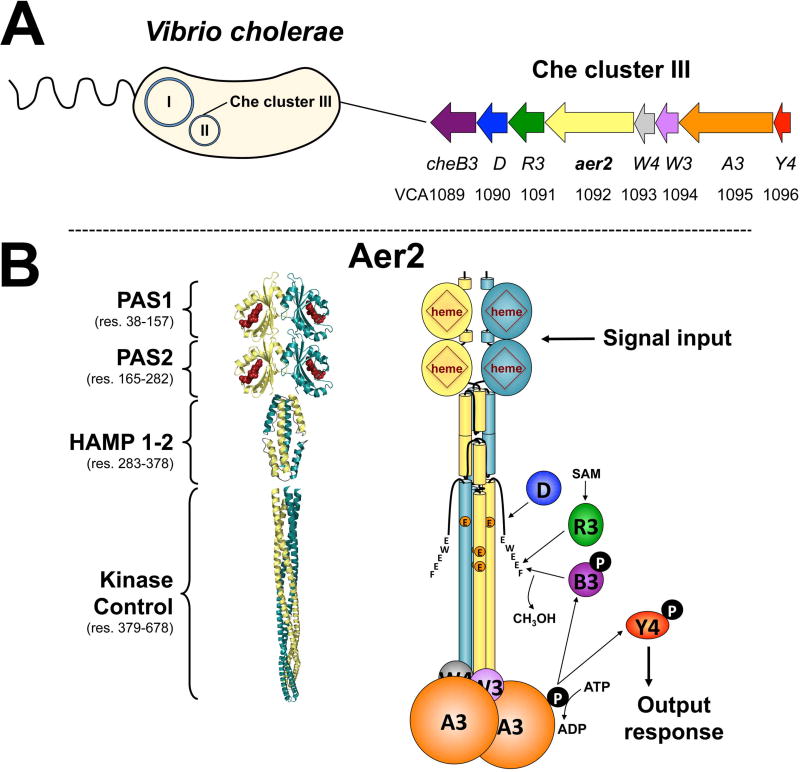
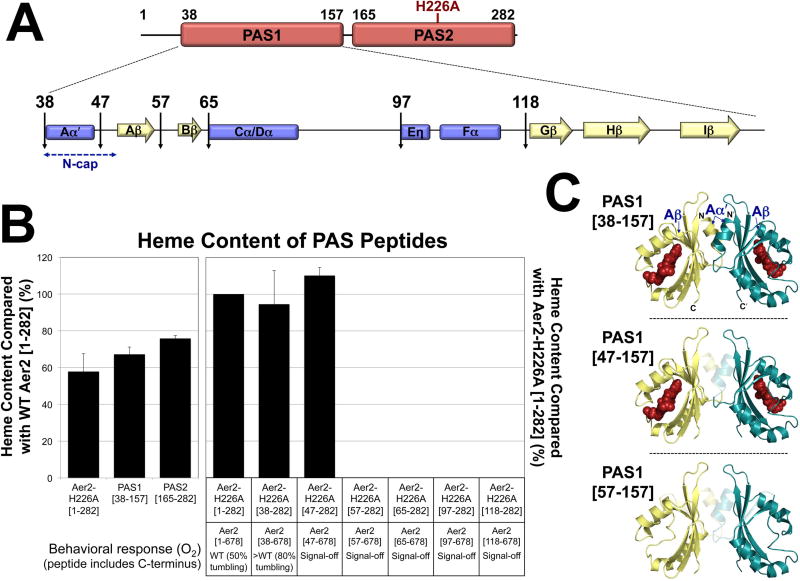
Fig. 1. V. cholerae chemosensory cluster III and the proposed structure of VcAer2.
A. V. cholerae chemosensory (Che) cluster III resides on chromosome II and encodes a complete chemosensory system. This includes a chemoreceptor (Aer2), a histidine kinase (CheA3), two coupling proteins (CheW3 and W4), a response regulator (CheY4), and three adaptation enzymes (CheR3, D and B3).
B. The proposed structure of a VcAer2 dimer (left) and the cluster III chemosensory pathway (right). VcAer2 is predicted to contain two N-terminal PAS domains (PAS1 and PAS2), a di-HAMP unit (HAMP 1–2), and a kinase control module that is typical of methyl-accepting chemoreceptors [containing three putative methylation sites: EEE, residues 414, 421 and 603, and a C-terminal pentapeptide sequence (EWEEF) for the binding of adaption enzymes]. In the left panel, PAS1 and PAS2 are modeled on the structure of P. aeruginosa Aer2 PAS [PDB code: 4HI4, (Airola et al., 2013a)], the di-HAMP unit is modeled on the structure of P. aeruginosa Aer2 HAMP 2–3 [PDB code: 3LNR, (Airola et al., 2010)], and the kinase control module is modeled on the structure of MCP1143C [PDB code: 2CH7, (Park et al., 2006)]. VcAer2 signaling is proposed to activate CheA3 autophosphorylation, which in turn phosphorylates CheY4, which purportedly regulates the response from the cluster III chemosensory system. VcAer2 signaling is modulated by the adaptation enzymes CheR3, D and B3, which bind the C-terminal pentapeptide EWEEF and/or the kinase control module to modify the methylation status of VcAer2. Abbreviation: SAM, S-adenosylmethionine.
The V. cholerae chemotaxis system (cluster II) mirrors that of a classic chemotaxis pathway, whereby chemoreceptor signaling activates the autophosphorylation of receptor-bound CheA2, which in turn phosphorylates the response regulator CheY3 (Boin et al., 2004). Phospho-CheY3 then binds to the flagellar motor protein, FliM (Biswas et al., 2013), changing the direction of flagellar rotation from counterclockwise to clockwise, and causing mono-flagellated V. cholerae to reverse its direction. In contrast to Cluster II, the Cluster III components CheA3, CheW3, and CheY4 do not support chemotaxis in V. cholerae (Gosink et al., 2002, Hyakutake et al., 2005, Selvaraj et al., 2015). In addition, CheY4 can’t bind to FliM because it lacks appropriate interacting residues (Dasgupta & Dattagupta, 2008, Biswas et al., 2013).
VcAer2 is predicted to be a soluble receptor containing a kinase control module with 51% homology to PaAer2, three potential methylation sites (EEE), and a C-terminal pentapeptide sequence (EWEEF) for binding adaptation enzymes like CheB3 and CheR3 (Fig. 1B). The principal difference between VcAer2 and PaAer2 is that VcAer2 is predicted to contain two, rather than one, N-terminal PAS (Per-ARNT-Sim) domains: PAS1 (res. 38–157) and PAS2 (res. 165–282) (Fig. 1B). PAS1 is situated in place of the three N-terminal HAMP domains of PaAer2, whereas PAS2 is positionally equivalent to the PaAer2 PAS sensing domain, PaPAS (Airola et al., 2010, Watts et al., 2011). By analogy to PaPAS, PAS2 may have a similar sensory role. PAS domains are common sensing and signaling domains in nature that maintain high structural conservation, even in instances of low sequence similarity. This conserved structure consists of an antiparallel β-sheet (containing strands Aβ, Bβ, Gβ, Hβ, and Iβ) surrounded by several α-helices (Cα, Dα, Eα, and Fα) (Moglich et al., 2009). Resolved structures of the PaPAS domain revealed several variations on the conserved PAS theme, including an extended Cα/Dα helix and a short 310 helix called Eη that replaces Eα (Sawai et al., 2012, Airola et al., 2013a). PaPAS binds penta-coordinate b-type heme via a His residue on Eη and functions as an O2 sensor (Garcia et al., 2017). Gas binding displaces a Leu residue on Hβ that occupies the ligand-binding site, causing an unorthodox Trp residue on Iβ to rotate ~90° to stabilize gas binding and initiate signaling (Airola et al., 2013a, Garcia et al., 2017). PaPAS shares 32% and 35% sequence identity with V. cholerae PAS1 and PAS2, respectively, whereas PAS1 and PAS2 share 38% sequence identity between themselves. However, both PAS1 and PAS2 contain Eη His residues that should support heme binding and Iβ Trp residues that should stabilize O2-binding (see Fig. 3A). The purpose of the current study is to gain greater insight into the function and role of Aer2-type chemoreceptors from different microorganisms with a different number of PAS domains. Since Aer2 receptors are likely to be the sole chemoreceptor holding their associated chemosensory clusters together [as is the case for PaAer2, (Guvener et al., 2006)], this may also shed light on the function of cluster III in V. cholerae.
RESULTS
V. cholerae Aer2 can hijack E. coli chemotaxis and mediate signaling in response to O2
The Aer2 ortholog from P. aeruginosa (PaAer2) does not direct chemotaxis in its native host but it can hijack the E. coli chemotaxis system and induce ~98% of cells to tumble (a repellent response) in the presence of O2, CO or NO (Watts et al., 2011). To determine if V. cholerae Aer2 (VcAer2) can similarly elicit E. coli responses, aer2 (VCA1092) was cloned from V. cholerae O1 JBK 70 genomic DNA and expressed from pProEXHTa in chemoreceptor-less E. coli BT3388, which is smooth biased (~2% tumbling). In E. coli, full-length VcAer2 (res. 1–678, 77.7 kDa including the His-tag) was stable (Fig. 2A) and soluble; it partitioned into the high-speed supernatant after spinning low-speed culture supernatants at 485 000 g for 1 hr. When VcAer2 was expressed in BT3388 under the same conditions previously used for PaAer2-mediated responses (200 µM IPTG induction for 45 min), it did not promote cell tumbling in air (20.9% O2). Induction with 1 mM IPTG for 45 min similarly did not elicit cell tumbling. VcAer2/BT3388 responses were not observed until cells had been induced with 200 µM IPTG for 2 hr. Under these conditions, ~30% of VcAer2/BT3388 cells tumbled in air. Adding 25 µg ml−1 of 5-aminolevulinic acid (ALA) during growth to accelerate heme synthesis increased the air tumbling response from ~30% to ~50% (Fig. S1A). Ten seconds after air was replaced with N2, the cells became smooth swimming (~2% of cells tumbled at any time, Fig. S1A). Removing the predicted PAS1 domain from VcAer2 (VcAer2 [165–678]) increased receptor stability (Figs. 2A and S2A), but abolished the O2 response. BT3388 cells expressing VcAer2 [165–678] swam smoothly and were non-responsive (signal-off) in both air and N2 (~2% of cells tumbled). Lastly we tested cells expressing full-length VcAer2 (induced with 200 µM or 1 mM IPTG, and with or without 25 µg ml−1 ALA) for a response to CO and NO. VcAer2-expressing cells did not respond to either CO or NO, unlike BT3388 cells expressing PaAer2 (Watts et al., 2011).
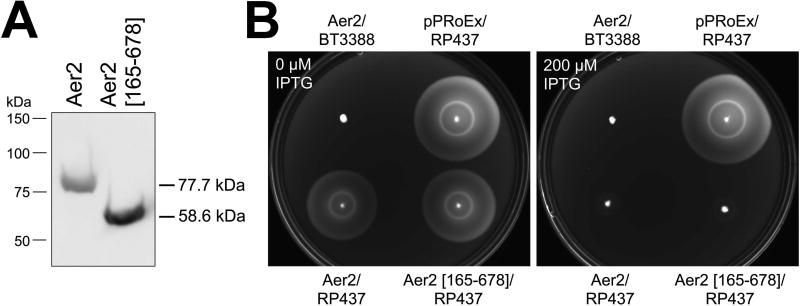
Fig. 2. Expression of VcAer2 in E. coli and its effect on E. coli chemotaxis.
A. Full-length VcAer2 and VcAer2 [165–678] expression in E. coli BT3388 after induction with 50 µM IPTG. Full-length VcAer2 is stably expressed in E. coli, although VcAer2 [165–678] (ΔPAS1) has a higher steady-state level (see Fig. S2A).
B. E. coli BT3388 and E. coli RP437 expressing full-length VcAer2 or VcAer2 [165–678] in tryptone soft agar with 0 or 200 µM IPTG. BT3388 lacks the five native chemoreceptors of E. coli, whereas RP437 is a WT E. coli chemotaxis strain. Plates were incubated at 30 °C for 9 h. VcAer2 does not induce chemotaxis ring formation in E. coli BT3388, but disrupts chemotaxis ring formation by WT E. coli chemoreceptors (the outer Tsr serine ring and the inner Tar aspartate ring). Adding 25 µg ml−1 ALA to the plates (to assist heme incorporation in VcAer2) made no difference to the appearance of the colonies compared to colonies in plates without ALA (not shown).
VcAer2/BT3388 cells had a repellent response to O2, and would therefore not be expected to exhibit aerotaxis in tryptone soft agar. When VcAer2/BT3388 cells were inoculated into tryptone soft agar, they did not elicit either aerotaxis or chemotaxis (with 0 to 1000 µM IPTG, and with or without 25 µg ml−1 ALA, Fig. 2B). However, VcAer2 did inhibit wild-type (WT) E. coli RP437 chemotaxis rings in tryptone soft agar after induction with at least 100 µM IPTG (Fig. 2B, 0 and 200 µM IPTG plates are shown). At the same induction levels, PAS1-less VcAer2 [165–678] also inhibited E. coli RP437 chemotaxis (Fig. 2B). Since BT3388 cells expressing VcAer2 [165–678] are smooth swimming, this inhibition of E. coli chemotaxis most likely results from titrating chemotaxis components away from native E. coli chemoreceptors.
The PAS1 and PAS2 domains of VcAer2 both bind b-type heme
VcAer2 contains two predicted PAS domains, PAS1 (res. 38–157) and PAS2 (res. 165–282) (Fig. 1B), which share ~30% sequence identity with PaPAS. PAS1 and PAS2 also have the Eη His that coordinates heme in PaPAS and the Iβ Trp that stabilizes O2 binding to PaPAS (Fig. 3A). When PAS1 [38–157] and PAS2 [165–282] were expressed in E. coli BT3388, both formed stable soluble peptides (Fig. S2C) that purified to ~98% apparent homogeneity on nickel-nitrilotriacetic acid (Ni-NTA) agarose (Fig. 3B). PAS1 and PAS2 both bound b-type heme and exhibited spectra similar to PaPAS [Fig. 3C, (Watts et al., 2011)]. In the deoxy-state, both PAS domains had penta-coordinate heme, judged by both a red-shifted Soret peak and a single broad band replacing the α/β-bands of ligand-bound PAS (Fig. 3C). O2 titrations yielded O2 dissociation constants [Kd(O2)] of 12 and 19 µM for PAS1 and PAS2, respectively, while CO titrations gave Kd(CO)’s of 7 and 5 µM, respectively (see Fig. S3A and C for representative O2 titrations). These affinities are similar to those previously determined for PaPAS [Kd(O2) of 16 µM and Kd(CO) of 2 µM, (Garcia et al., 2017)]. In addition, the O2 and CO affinities of PAS2 were not altered by the presence of the C-terminus in VcAer2 [165–678] (Kd(O2) of 20 µM and Kd(CO) of 2 µM). This indicates that the C-terminal HAMP and kinase control domains do not contribute to, or attenuate, gas-binding to PAS2.
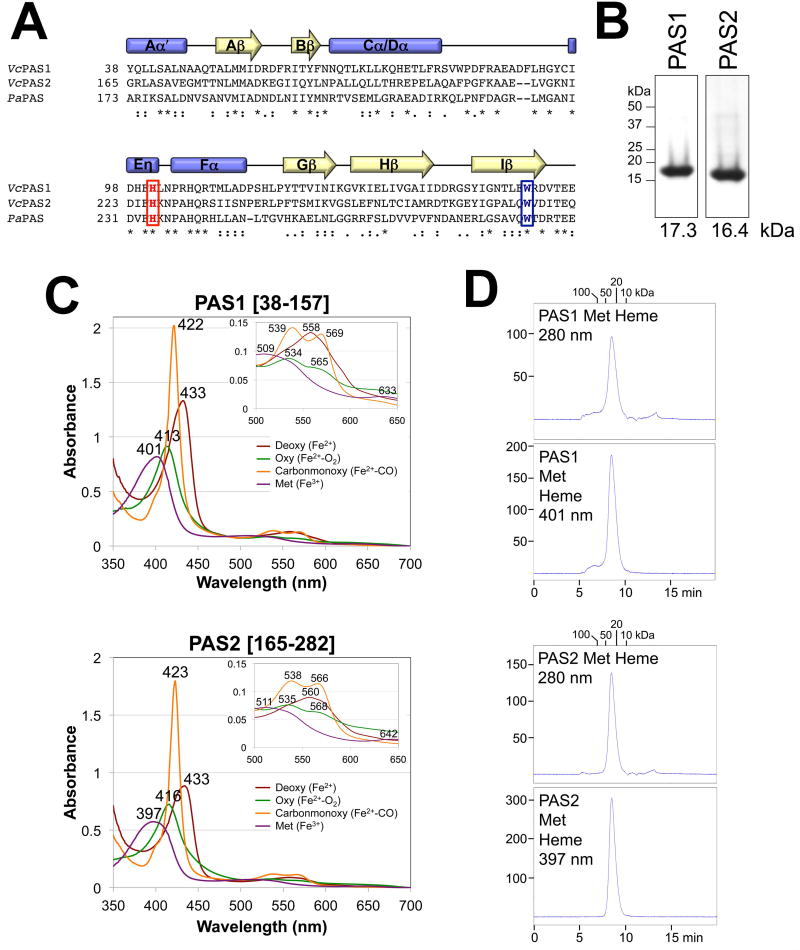
Fig. 3. Secondary structure, heme binding and oligomeric state of the PAS1 and PAS2 domains of VcAer2.
A. Sequence alignment of PAS1 and PAS2 from V. cholerae Aer2 and PaPAS from P. aeruginosa Aer2 as generated in ClustalW. The conserved His that coordinates heme in PaPAS, and the conserved Trp that stabilizes O2-binding to PaPAS, are highlighted red and blue, respectively. Secondary structure elements are based on the solved structures of PaPAS (Sawai et al., 2012, Airola et al., 2013a). Stars indicate conserved residues, colons indicate similar amino acids, and periods indicate amino acids with weakly similar properties.
B. Coomassie-stained SDS-PAGE of 5 µg of purified PAS1 [38–157] and PAS2 [165–282] peptides.
C. Absorption spectra of 10 µM purified PAS1 and PAS2 domains in the reduced (deoxy), oxidized (met), carbon monoxide-bound (carbonmonoxy) and oxygen-bound (oxy) states. The wavelength for each absorbance maximum is indicated. The insert shows an expanded view of peaks between 500 and 650 nm.
D. Elution profiles of isolated PAS1 and PAS2 peptides (200 µg) in their met-heme states during size-exclusion chromatography. Elution profiles are shown in arbitrary units at 280 nm to reveal total protein content (top panels) and at 401 nm (PAS1) or 397 nm (PAS2) to detect the elution of met-heme (bottom panels). The area under the peak for PAS2 is 97% the area of PAS1. Fractions were removed and analyzed by Western blotting; in all cases, PAS peptide co-eluted with the heme (not shown).
Using size exclusion chromatography, the isolated PaPAS domain was determined to be a compact monomer in both its met and deoxy heme states (Watts et al., 2011). In contrast, both V. cholerae PAS1 [38–157] and PAS2 [165–282] eluted as homodimers from the same TSKgel G2000SW column in their met heme state (Fig 3D, Mwapp of 30 and 33 kDa, respectively) and as more compact homodimers in their deoxy heme state (each with a Mwapp of 25 kDa). For PAS 1–2 [1–282] and PAS1 [1–164] peptides, both dimers (Mwapp of 75 and 43 kDa, respectively) and larger complexes (Mwapp of >200 kDa) were evident, suggesting that the N-terminal 37 amino acids contiguous to PAS1 promote protein aggregation (data not shown).
The Eη His coordinates heme in PAS1 and PAS2
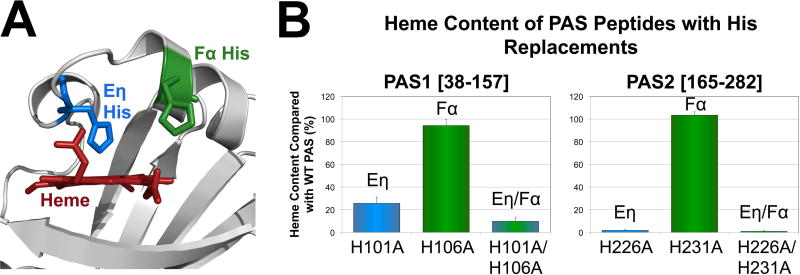
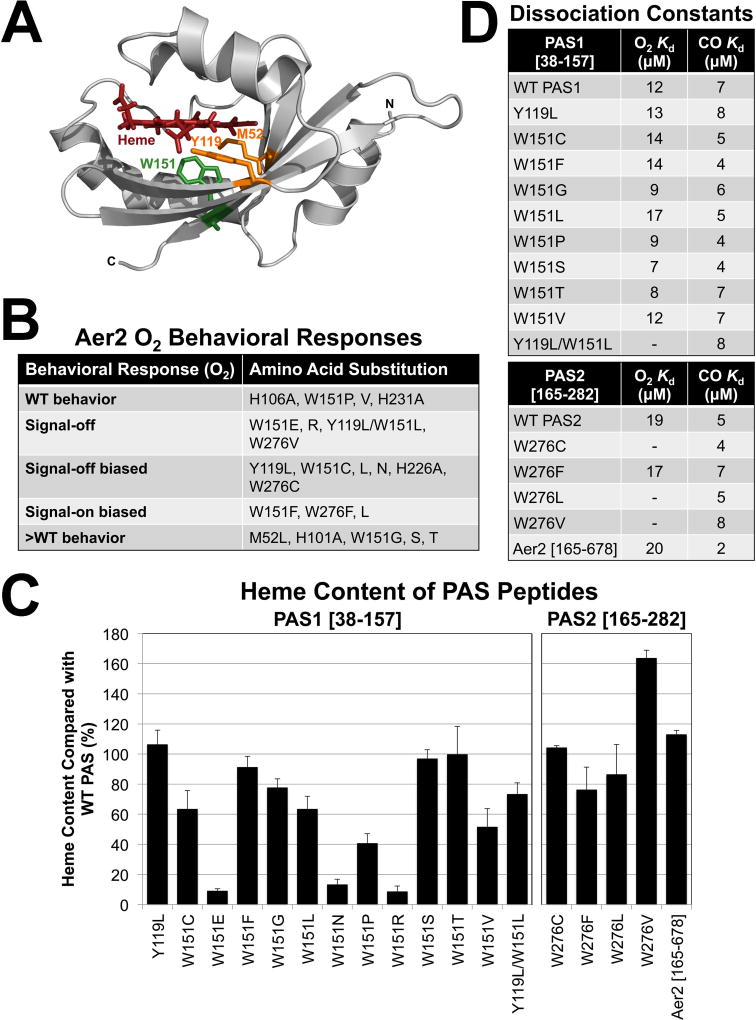
PAS domains often coordinate b-type heme via a His residue on Fα (Gilles-Gonzalez & Gonzalez, 2005, Kerby et al., 2008), whereas PaAer2 coordinates heme via a His residue on Eη (Sawai et al., 2012, Airola et al., 2013a, Garcia et al., 2017). The Eη and Fα His residues are conserved in PaPAS and both VcAer2 PAS domains (Figs. 3A and 4A). To test the contributions of each His to heme binding and function in PAS1 and PAS2, Eη, Fα, and Eη/Fα His to Ala mutants were created for PAS1 and PAS2 in both PAS peptides and full-length VcAer2. Similar to what was observed with PaPAS (Garcia et al., 2017), the Eη His mutants (PAS1-H101A and PAS2-H226A) showed substantial heme-binding defects (26% and 2% of WT PAS heme content, respectively), whereas the Fα His mutants (PAS1-H106A and PAS2-H231A) retained WT heme content (Fig. 4B). This confirms that the Eη His coordinates heme in both PAS1 and PAS2. However, 26% of PAS1-H101A molecules retained heme, whereas the dual His mutant PAS1-H101A/H106A retained only 10% heme (P < 0.05, Fig. 4B). This phenomenon was also observed for PaPAS (Garcia et al., 2017), and suggests that for PAS1, Fα-H106 may contribute to heme coordination in the absence of Eη-H101. Another possibility is that heme is retained in PAS1-H101A by hydrophobic interactions, and that the additional H106A replacement distorts the heme pocket to impact heme retention. The same conclusions could not be reached for PAS2 because the Eη His mutant, PAS2-H226A, retained only 2% heme.
Fig. 4. Heme coordination in the PAS1 and PAS2 domains of VcAer2.
A. Location of the Eη and Fα His side chains on the PAS2-W276L structure (see Fig. 7). The distance from the Fe atom to the Eη His NE2 atom is 2.5 Å, whereas the distance from the Fe atom to the Fα His NE2 atom is 10.5 Å.
B. Average heme content of PAS1 [38–157] and PAS2 [165–282] peptides with Eη and Fα His replacements, given as a percentage of WT PAS heme content, corrected for peptide concentration. Error bars represent standard deviations from two to three experiments. The heme content of PAS1-H101A is significantly different from the heme content of PAS1-H101A/H106A (P < 0.05).
To determine the effects of the His substitutions on VcAer2 function, the relevant mutations were introduced into pProEXHTa containing full-length Vcaer2. Each of the mutants had steady-state expression levels comparable with WT VcAer2 (Fig. S2A). In BT3388, the Fα His mutants VcAer2-H106A and VcAer2-H231A had WT responses (Fig. S1A). In contrast, VcAer2-H101A, whose PAS1 peptide retained 26% heme, had a more robust O2 response than did WT receptor, promoting ~95% cell tumbling in air compared with ~50% tumbling for WT VcAer2 (Fig. S1E). This response represents the aggregate output of both heme-bound and unbound receptors, but suggests that PAS1 may not be required for O2 sensing and instead functions to regulate O2-mediated signaling from PAS2. VcAer2-H226A, whose PAS2 peptide retained 2% heme, was signal-off biased, directing ~10% of BT3388 cells to tumble in air (Fig. S1C). All of the His mutants tested had WT smooth-swimming responses in response to O2 removal.
Heme binding requires the entire PAS1 core, but Aer2 function requires the PAS N-cap
PAS domain function often requires a short region N-terminal to the PAS core, called the PAS N-terminal cap (N-cap) (Kurokawa et al., 2004, Watts et al., 2006, Key et al., 2007). N-cap regions contain an Aα′ helix (res. 38–45 in PAS1) and a short loop that precedes the Aβ strand (Aβ begins the PAS1 core at res. 50, Fig. 3A). Both PaPAS structures contain part (Sawai et al., 2012) or all (Airola et al., 2013a) of the Aα′ helix, and heme is bound to the PAS core in both instances. To determine how much of the N-terminus is required for heme binding to PAS1, a series of N-terminal truncations were introduced into VcAer2-H226A [1–282] (Fig. 5A), and heme content was measured. H226A should prevent ~98% of heme binding to PAS2 (Fig. 4B), so that the heme content of VcAer2-H226A [1–282] (which contains both PAS1 and PAS2) should instead reflect the amount of heme bound to PAS1. Indeed, the amount of heme bound to VcAer2-H226A [1–282] was not significantly different from the amount of heme bound to PAS1 [38–157] alone (P = 0.3, Fig. 5B, left panel). The heme content of WT VcAer2 [1–282] was less than twice the heme content of VcAer2-H226A [1–282], PAS1 [38–157] or PAS2 [165–282] (Fig. 5B, left panel). This discrepancy suggests that it may be more difficult to incorporate two heme molecules into PAS1–2 than incorporate one heme molecule into either PAS1 or PAS2.
Fig. 5. Effects of N-terminal truncations on PAS1 heme binding and VcAer2 behavior.
A. Cartoon of PAS1-PAS2 (res. 1–282), the predicted secondary structure elements of PAS1, and locations of the N-terminal truncations.
B. Average heme content of PAS peptides, given as a percentage of the heme content in either WT VcAer2 [1–282] (left panel) or VcAer2-H226A [1–282] (right panel), corrected for peptide concentration. Error bars represent standard deviations from two to four experiments. The amount of heme bound to VcAer2-H226A [1–282] (i.e., heme bound to PAS1 in the presence of heme-less PAS2, left panel) is not significantly different from the amount of heme bound to PAS1 [38–157] (P = 0.3). The behavioral responses of BT3388 cells expressing VcAer2 mutants with N-terminal truncations are provided beneath the bar graph. The >WT mutant (VcAer2 [38–678]) exhibited ~80% tumbling in air compared with ~50% for WT VcAer2, whereas the signal-off mutants exhibited smooth-swimming behavior (2–5% tumbling) in both air and N2.
C. PAS1 dimer model based on the structure of P. aeruginosa Aer2 PAS (Airola et al., 2013a) showing the truncations that retained heme (top and center panels) and the shortest truncation that no longer retained heme (57–157, bottom panel). In each case, the translucent region represents the N-terminal residues (Aα′ helix, Aα-Aβ loop, and Aβ strand) that were sequentially deleted from PAS1.
Of the N-terminal truncation peptides, only VcAer2-H226A [38–282] and VcAer2-H226A [47–282] retained heme; here the heme content matched that of VcAer2-H226A [1–282] (Figs. 5B and C). Longer truncations abolished any measureable heme (Fig. 5B), even though the corresponding peptides were stably expressed (Fig. S2B). VcAer2-H226A [57–282], which lacked the PAS1 Aβ strand, contained the shortest truncation that eliminated heme binding (Figs. 5B and C). The N-cap was not required for heme binding, but it was required for function: VcAer2 [47–678] (with the N-cap removed) and all longer N-terminal truncations resulted in non-functional, signal-off mutants (Fig. 5B). In contrast, VcAer2 [38–678], which lacked the region N-terminal to the Aα′ helix, had a more robust O2 response than WT VcAer2 [1–678] (Figs. 5B and S1E). This finding suggests that the first 37 residues of VcAer2 are not necessary for Aer2 signaling, but that they assist PAS1 in regulating PAS2 signaling.
The conserved Iβ Trp stabilizes O2 binding to PAS2 whereas PAS1 uses either the Iβ Trp or a Gβ Tyr residue
In PaPAS, gas binding causes the Iβ Trp, W283, to rotate towards the ligand to stabilize gas binding and initiate signaling (Airola et al., 2013a, Garcia et al., 2017). This may be a canonical mechanism for O2-signaling in Aer2-type PAS domains, as the Iβ Trp is universally present (Garcia et al., 2017). In VcAer2, the Iβ Trp resides at W151 in PAS1 (Fig. 6A) and at W276 in PAS2 (Fig. 3A). Of these two PAS domains, PAS2 most closely resembles PaPAS in that it lies directly upstream of the C-terminal HAMP and kinase control domains of VcAer2 (Fig. 1B). However, in most other chemoreceptors and histidine kinases with tandem PAS-like domains, the second domain does not bind ligands (Zhang & Hendrickson, 2010, Glekas et al., 2012, Nishiyama et al., 2012, Liu et al., 2015, Nishiyama et al., 2016). A notable exception was recently described for the Helicobacter pylori TlpC receptor, where the first PAS-like domain binds ligand (Machuca et al., 2017). To determine whether W276 is required for O2-mediated signaling in PAS2, W276C, F, L and V [selected because the equivalent PaPAS substitutions retained heme, (Garcia et al., 2017)] were introduced into full-length VcAer2 and into the PAS2 [165–282] peptide. The mutants were then tested for their responses to, and affinities for, O2. Similar to PaAer2-W283L, VcAer2-W276L was a signal-on biased mutant that caused ~98% of BT3388 cells to tumble in air and had a 30 sec delayed smooth-swimming response in N2 (Figs. 6B and S1D). Like PaAer2-W283V, VcAer2-W276V was signal-off, whereas VcAer2-W276C was signal-off biased (~20% of cells tumbled in the presence of O2, Fig. S1C). Purified PAS2 peptides containing W276C, L and V retained heme (Fig. 6C) and bound CO, but none bound O2 (Fig. 6D). Instead, O2 rapidly oxidized each peptide from Fe(II) to Fe(III) heme (Fig. S3D), as was previously observed for PaPAS-W283C, L and V (Garcia et al., 2017). Thus, the isolated PAS2-W276L and PAS2-W276C peptides did not stably bind O2, even though the corresponding full-length receptors responded to O2 in vivo. This suggests that O2 binding is too transient to observe in vitro, but is sufficiently stable in vivo to generate partial behavioral responses. A similar scenario was observed for PaAer2-W283L (Garcia et al., 2017).
Fig. 6. VcAer2 PAS mutant behavior, heme content and gas-binding affinities.
A. The location of PAS1 residues M52, Y119 and W151 based on the structure of the P. aeruginosa Aer2 PAS domain (Airola et al., 2013a).
B. The behavior of full-length VcAer2 mutants in BT3388 compared with WT VcAer2 (which caused ~50% tumbling in air and ~2% tumbling in N2). Mutants with WT behavior exhibited 30–55% tumbling in air; signal-off mutants exhibited 2–5% tumbling in air; signal-off biased mutants exhibited 10–25% tumbling in air; >WT mutants exhibited 80–98% tumbling in air. All of these mutants exhibited ~2% tumbling in N2. In contrast, the signal-on-biased mutants caused 95–98% of cells to tumble in air, but had 20–50 sec delayed smooth-swimming (~2% tumbling, W151F and W276L) or incomplete smooth swimming (50–80% tumbling, W276F) responses in N2.
C. Average heme content of PAS peptides with amino acid substitutions, given as a percentage of WT PAS1 (left panel) or WT PAS2 (right panel) heme content, corrected for peptide concentration. VcAer2 [165–678] heme content is given as a percentage of full-length VcAer2 [1–678] heme content. Values below 15% indicate a substantial heme-binding defect. Error bars represent standard deviations from two to five experiments.
D. PAS peptide O2 and CO binding affinities. A dash indicates that O2-bound spectra were not observed, so no binding affinity was determined.
In contrast to the other W276 mutants, VcAer2-W276F was a signal-on biased mutant that retained heme (Figs. 6C and S1D), and bound O2 and CO with similar affinities to the WT PAS2 domain (Fig. 6D). The same finding was previously observed for PaAer2-W283F, which was the only Iβ Trp mutant that retained O2 binding in PaAer2 (Garcia et al., 2017). Overall, the PAS2-W276 mutants in this study produced comparable results to the corresponding PaPAS-W283 mutants, supporting the hypothesis that the PAS2 Iβ Trp (W276) stabilizes O2 binding in an analogous manner to PaPAS (W283).
To analyze the PAS1 domain, W151L was introduced into full-length VcAer2 and into the PAS1 [38–157] peptide and tested for its response to, and affinity for, O2. Unlike PAS2 and PaPAS, PAS1-W151L bound O2 and CO with WT affinities (Figs. 6D and S3B). In BT3388, VcAer2-W151L was a signal-off biased mutant that induced ~10% cell tumbling in air, and ceased tumbling in response to O2 removal like WT receptor (Figs. 6B and S1C). To determine if O2 binding was unique to PAS1-W151L, an additional 10 W151 mutants were created by site-directed random or specific mutagenesis. PAS1 [38–157] peptides containing W151E, W151N and W151R, had low heme binding (9–13% of WT heme content, Fig. 6C) and gas-binding affinities could not be determined. The seven remaining PAS1-W151 mutants (W151C, F, G, P, S, T and V) had at least 41% heme content compared with WT PAS1 (Fig. 6C) and bound O2 and CO with WT affinities (Fig. 6D). The corresponding full-length VcAer2-W151 mutants also responded to O2 with either signal-off or -on biased, WT or more robust (>WT) behavior (Figs. 6B and S1). Overall, these data suggest that W151 alone does not stabilize O2 binding to the PAS1 domain.
To find residues other than W151 that might stabilize O2-binding in the distal heme pocket of PAS1, polar residues within 10 Å of ligand bound heme were sought by comparing the sequence of PAS1 with the structure of PaPAS (Sawai et al., 2012). In the distal heme pocket, only W151, M52 (on Aβ) and Y119 (on Gβ) were potentially within range to contact a ligand bound to heme (Fig. 6A), so we created Leu mutants of M52 and Y119 in full-length VcAer2. In BT3388, VcAer2-M52L had a more robust O2 response than WT, whereas VcAer2-Y119L was signal-off biased (~25% of cells tumbled in the presence of O2, Fig. S1C). Analysis of the PAS1-Y119L peptide showed that it had WT heme content and WT O2 and CO affinities (Fig. 6). Thus, single residues with the potential to stabilize O2 were dispensable for O2 binding to PAS1. We next considered whether a combination of amino acids could provide O2-stabilizing interactions for PAS1, similar to what has been described for Ascaris hemoglobin [Tyr and Gln, (Yang et al., 1995)] and for soybean leghemoglobin [Tyr and His, (Kundu & Hargrove, 2003)]. We created a PAS1 peptide and full-length VcAer2 receptor containing dual Y119L and W151L substitutions. In BT3388, VcAer2-Y119L/W151L did not respond to O2 (it was signal-off), and PAS1-Y119L/W151L bound heme and CO, but it did not bind O2 (Fig. 6). This suggests that interactions from either Y119 or W151 stabilize O2 binding to PAS1.
Signal-on behavior is not dependent on Aer2 methylation
Robust PaAer2 responses in E. coli require Aer2 methylation by E. coli CheR (Watts et al., 2011). However, signal-on mutants of PaAer2 are signal-on irrespective of methylation status (Garcia et al., 2017). To determine if this is also true for VcAer2 mutants, mutants that were determined to have signal-on biased behavior or more robust signaling responses than WT (Figs. 5B and 6B) were expressed in E. coli UU2610, which lacks all E. coli chemoreceptors in addition to the adaptation enzymes CheR and CheB. In UU2610, the tumbling biases of cells expressing the three signal-on biased mutants (VcAer2-W151F, VcAer2-W276F or VcAer2-W276L) and the more robust response mutants VcAer2-W151G and VcAer2-W151T were unaffected by the lack of E. coli adaptation enzymes (Fig. S1F). This suggests that the signaling behavior induced by these receptors is primarily due to the PAS residue substitutions, rather than receptor methylation status. In contrast, the tumbling biases of cells expressing WT VcAer2, VcAer2 [38–678], VcAer2-M52L, VcAer2-H101A, and VcAer2-W151S were reduced by 20–40% in air (Fig. S1F), suggesting that methylation partly contributes to the signal-on biases observed for these receptors in air.
The structure of the PAS2 domain
The structure of the signal-on PAS2-W276L peptide with ferric [Fe(III)] heme was determined to 1.67 Å resolution (Table S1, PDB code: 6CEQ). PAS2-W276L maintained a conserved PAS fold with most features similar to those of PaPAS (Fig. 7A–B), including the PAS β-sheet consisting of Aβ, Bβ, Gβ, Hβ and Iβ and the extended Cα/Dα helix with a kink at R201 (A209 in PaPAS). Also like PaPAS, the Eα helix was distorted into an Eη helix. The Eη His residue H226 was 2.5 Å from the Fe atom and, as anticipated, served as the proximal heme ligand (Fig. 7C–D).
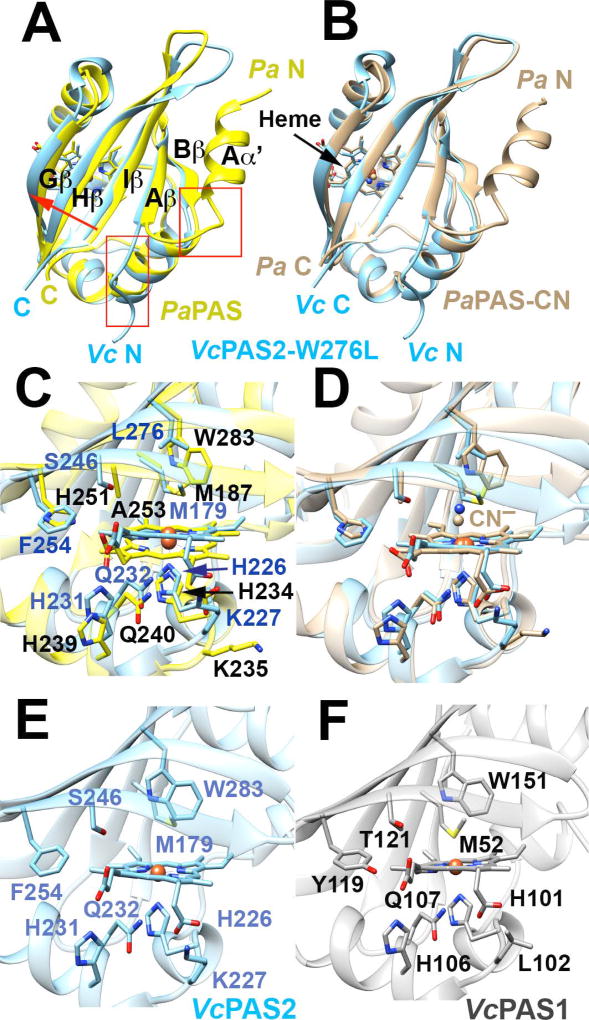
Fig. 7. The structure of PAS2-W276L and its relationship to PaPAS structures and WT PAS1 and PAS2 homology models.
A–B. Superposition of PAS2-W276L (A; PDB code: 6CEQ, blue) and PaPAS in the unliganded ferric-heme form (A; PDB code: 4HI4, yellow) and the CN−-bound form (B; PDB code: 3VOL, brown). The structures of PAS2-W276L and both PaPAS structures are similar, except that the Aα′ helix is dissociated and unstructured in PAS2-W276L (red boxes). However, PAS2-W276L is most similar to the ligand bound form of PaPAS, with the β-sheet shifted slightly relative to the ferric form of PaPAS (A; red arrow).
C–D. Superposition of the heme-binding pockets of PAS2-W276L (blue) and PaPAS in the unliganded ferric-heme form (C; yellow) and CN−-bound form (D; brown).
E–F. Comparison of the heme pockets of WT PAS2 (E) and WT PAS1 (F) homology models. Modeling of the WT PAS2 structure and PAS1 domain was accomplished in SWISS-MODEL using the PAS2-W276L structure as a template in both cases.
Despite the overall similarity between PAS2-W276L and PaPAS, there were also some notable structural differences. Whereas the N-cap of PaPAS forms a well defined Aα′ helix (Sawai et al., 2012, Airola et al., 2013a), that of PAS2-W276L continued the extended structure of the Aβ strand to end in a disordered region (Fig. 7A–B). Indeed, electron density for the first five residues of PAS2-W276L was not discernible. Sequence homology in the Aα′ region is relatively strong between PAS2-W276L and PaPAS, which suggests that other factors, including the W276L substitution and possibly crystal contacts, may influence the conformational difference in the N-cap. PAS2-W276L formed an antiparallel dimer in the crystal with β-β contacts that included the extended Aβ′, whereas PaPAS crystallizes as a parallel dimer whose subunit interface is formed from both the Aα′ helices and the β-sheets (Airola et al., 2013a). In addition to these differences in conformation and oligomerization, PAS2-W276L also contained an additional turn of 310 helix between the Fα helix and the Gβ strand that is not found in PaPAS.
Both PAS2 and PaPAS bind b-type hemes that associate into a hydrophobic pocket of non-polar side chains. For PaPAS, H251 on the Gβ strand hydrogen bonds with the 7-propionate of the heme (Sawai et al., 2012, Airola et al., 2013a). PAS2 contains Phe (F244) instead of His at this position, but in PAS2, H231 is shifted closer to the heme propionate than its counterpart, H239 in PaPAS (Fig. 7C–D) to provide a compensating interaction.
Consequences of Trp substitution in the distal ligand-binding pocket of PAS2
Although PAS2-W276L was crystalized in the ferric, ligand-free form, its overall structure was more similar to the structure of CN−-bound PaPAS than to the structure of unliganded ferric PaPAS. This similarity was especially true at the C-terminal β-strands (Gβ-Hβ-Iβ), which aligned well with those of CN−-bound PaPAS (Fig. 7B). In addition, the PAS2-W276L variant appeared to recapitulate the conformational changes of the CN−-bound species despite having an unoccupied distal pocket (Fig. 7A–D). The substitution of the W276 indole ring for the shorter Leu side chain promoted a similar movement of Gβ-Hβ-Iβ towards the heme carboxylates and the periphery of the protein, including a displaced Hβ Leu (Fig. 7A–D). In PAS2-W276L, these combined motions led to both a shift in the position of the Iβ strand and changes in the interactions between the PAS β-sheet and juxtaposed Aα′, which completely dissociated from the protein core. Importantly, the spatial arrangement of the Iβ strand is critical for signal transduction because it connects directly to the downstream HAMP domains. Furthermore, the Aα′ helix contributes to the proposed PAS-PAS interface in the full-length protein (Sawai et al., 2012, Airola et al., 2013a). Thus, the Iβ Trp indole buttresses the Gβ-Hβ-Iβ region against the porphyrin, and either its substitution to Leu, or reorientation due to ligand binding, elicits similar repositioning of the PAS β-sheet and subsequent repacking of Aα′. The signal-on-biased character of VcAer2-W276L is thus consistent with the similarity of the PAS2-W276L structure to that of the ligand-bound form of PaPAS.
PAS1 structural model
A homology model was created for the wild type PAS1 domain by threading the PAS1 sequence (res. 38–157) onto the PAS2-W276L structure. The heme-binding pockets of PAS1 and PAS2 showed high similarity for the proximal heme-ligating residue (H101 in PAS1 and H226 in PAS2) and distal ligand-coordinating residue (W151 in PAS1 and W276 in PAS2) (Fig. 7E–F). In both PAS domains the Fα His (H106 in PAS1 and H231 in PAS2) formed a hydrogen bond with the 7-propionate of the heme, as did an Fα Gln (Q107 in PAS1 and Q232 in PAS2) (Fig. 7 E–F). Some minor differences in the distal ligand-binding pocket included the change of F244 to Y119 in PAS1 (which contributes to O2 binding) and also S246 to T121 (Fig. 7E–F). Interestingly, given this high degree of homology, relatively conservative changes in ionizable residues produce a considerably more negative potential in the heme pocket for PAS1 compared to PAS2 (Fig. S4). The O2 affinity of heme proteins can depend on many factors including proximal ligand electronic effects, distal ligand hydrogen bonding, heme distortions and heme redox potential. Lower potential hemes (those in more negative environments) generally produce more stable O2 binding by favoring the ferric-superoxy state of the heme-liganded complex (Grinstaff et al., 1995). Hence, the more negative heme environment of PAS1 may contribute to stable oxy complexes in the absence of either Y119 or W151.
DISCUSSION
V. cholerae Aer2 is a dual-PAS heme O2 sensor
In this study we have shown that the V. cholerae cluster III chemoreceptor, VcAer2, directly senses O2 via two PAS-heme domains. This was shown in vitro by measuring the O2 binding affinity of each PAS domain, and in vivo in E. coli by demonstrating the O2-directed response of full-length VcAer2. In V. cholerae, cluster III proteins (Fig. 1) do not direct chemotaxis or aerotaxis (Gosink et al., 2002, Hyakutake et al., 2005, Dasgupta & Dattagupta, 2008, Biswas et al., 2013), but when VcAer2 was expressed in E. coli, it orchestrated a repellent response to O2. The ability to hijack (Fig. 2B) and control E. coli chemotaxis presumably stems from the 54% sequence homology between the kinase control module of VcAer2 (Fig. 1B) and the major E. coli chemoreceptor Tsr. When VcAer2 was expressed in E. coli BT3388, the most robust signaling response occurred when ALA, the first compound in the porphyrin synthesis pathway, was added to cultures during growth. This response was weaker than that observed for PaAer2: ~50% of WT VcAer2/BT3388 cells tumbled in air (with ALA) compared with ~98% for PaAer2/BT3388 [without ALA, (Watts et al., 2011)]. Cells ceased tumbling when air was removed, indicating that O2 had activated VcAer2 signaling. The lower tumbling bias of VcAer2/BT3388 versus PaAer2/BT3388 in air may reflect differences in predicted methylation sites, which, when methylated, promote the kinase-on state. VcAer2 has three methylation sites (EEE), while PaAer2 has four (QEEE) including a signal-inducing Gln that is not efficiently deamidated by E. coli CheB (Watts et al., 2011). Unlike PaAer2, VcAer2 did not respond to CO or NO even though CO bound to both PAS1 and PAS2 (Fig. 3C).
VcAer2 differs from PaAer2 in that it has two N-terminal PAS domains (Fig. 1B). PAS1 replaces the HAMP1–3 domains of PaAer2, and PAS2 is positionally equivalent to PaPAS (Airola et al., 2010, Watts et al., 2011). Our preliminary analysis suggests that this Aer2 architecture also exists in other microbes, e.g., in Shewanella oneidensis SO2123, where both predicted PAS domains contain the Eη His and Iβ Trp residues. The Aer2 ortholog from Vibrio vulnificus (Mcp III) is predicted to have three PAS domains and each PAS domain contains the conserved Eη His and Iβ Trp residues. In VcAer2, both PAS1 and PAS2 coordinated b-type heme via Eη-His and exhibited spectra and gas-binding affinities that were similar to PaPAS [(Garcia et al., 2017), Figs. 3, 4 and 6]. The O2 affinities of PAS1 and PAS2 (12 and 19 µM, respectively) were comparable to PaAer2 (16 µM) and to those of other O2-sensing PAS domains, e.g., E. coli DOS (13 µM) and Sinorhizobium meliloti FixL (31 µM) (Delgado-Nixon et al., 2000, Gilles-Gonzalez & Gonzalez, 2005, Garcia et al., 2017). It is probable that Aer2 homologs are all PAS heme-O2 sensors, regardless of their specific PAS-HAMP domain architecture.
PAS N-cap rearrangements and VcAer2 signaling
In VcAer2, the entire PAS core was required for heme binding to PAS1 (Fig. 5). The Aα′ helix of the PAS1 N-cap was not required for heme binding, but it was required for VcAer2 function (Fig. 5). PAS N-caps are functionally important, and in PaPAS and other PAS dimers, the Aα′ helix, along with the PAS β-sheet, stabilize the PAS dimer interface (Watts et al., 2006, Key et al., 2007, Moglich et al., 2009, Airola et al., 2013a). In solution, the isolated PAS1 and PAS2 peptides formed homodimers in both met heme (Fig. 3D) and deoxy heme states, yet homodimers were more compact in the deoxy, unliganded state. Previously, when the structure of the CN−-bound PaPAS monomer was superimposed on the ligand-free PaPAS dimer structure, collisions occurred between the N-caps and Hβ strands (Airola et al., 2013a). Incompatibility of the ligand-bound form with the ligand-free dimer suggested that ligand binding to the Aer2-PAS domain promotes PAS-PAS rearrangements at the dimer interface that involves the PAS N-cap. In the current study, removing Aα′ from PAS1 resulted in a signal-off phenotype (Fig. 5). This may indicate that PAS1 does not dimerize correctly in the absence of Aα′, and/or that rearrangements of the Aα′ helix are essential for VcAer2 signaling. In further support of this idea, it is striking that the structure of the PAS2-W276L signal-on variant was very different from that of PaPAS in the region of Aα′ despite strong sequence similarity between the two proteins. The dissociation and unfolding of Aα′ from the PAS2 β-sheet may well promote an activated conformation in keeping with the behavior of this variant. Structural changes in both N-cap and C-cap elements on the opposite side of the β-sheet from the ligand-binding pocket is a common feature of PAS domain signaling. A classic example involves restructuring of the Jα and Aα′ helices in LOV domain proteins (Harper et al., 2003, Zoltowski et al., 2007). The fact that the signal-on variant of PAS2 had a completely dissociated Aα′ helix is in keeping with this theme. The antiparallel dimer formed in the crystal of PAS2-W276L is likely only a consequence of favorable packing for this altered conformation, but the change in conformation for Aα′ is a strong indication that perturbations in the ligand-binding pocket are relayed through changes in the Hβ-Gβ-Iβ strands, where they are able to alter the conformation and thereby interactions of the N-cap. The structure of PAS2-W276L supports the view that ligand binding ultimately restructures the PAS-PAS interface in full-length Aer2 proteins.
The distinct roles of the PAS1 and PAS2 domains of VcAer2
The PAS1 and PAS2 domains of VcAer2 both bound O2 with similar affinities (Fig. 6D), but they stabilized O2 binding by different mechanisms. In PAS2, the Iβ Trp, W276, stabilized O2 binding in an analogous manner to the Iβ Trp, W283, from PaAer2 (Garcia et al., 2017). PAS2 peptides containing W276C, L and V were rapidly oxidized by O2 (Fig. S3D), whereas W276F bound O2 with WT affinity (Fig. 6D). This finding is similar to that of PaAer2, where W283F was the only Iβ Trp mutant that retained O2 binding. In that case O2 stabilization may have occurred via a solvent molecule in a manner similar to that shown for a Tyr to Phe replacement mutant of Mycobacterium tuberculosis DevS (Yukl et al., 2008). PAS1 was different; the Iβ Trp mutants PAS1-W151C, F, G, L, P, S, T and V all bound O2 with WT affinities (Fig. 6D), and their corresponding full-length mutants responded to O2 (Figs. 6B and S1). For PAS O2 sensors that bind b-type heme, only the Iβ Trp residue (in Aer2) and a Gβ Arg residue [in FixL, DOS and PDEA-1 (Gilles-Gonzalez & Gonzalez, 2005)] have been shown to H-bond to ligand in the distal heme pocket. In other heme proteins, His [e.g., vertebrate hemoglobin and myoglobin], Gln and Tyr [e.g., Ascaris hemoglobin (Kloek et al., 1994)] stabilize O2 binding. In soybean leghemoglobin, a combination of His and Tyr residues synergistically provide weak interactions with bound O2 (Kundu & Hargrove, 2003). The proximal heme pocket of leghemoglobin enhances Fe2+-O2 interactions and the weak distal pocket interactions facilitate faster O2 dissociation. In VcAer2, PAS1-Y119L and PAS1-W151L both had WT O2 and CO affinities, whereas PAS1-Y119L/W151L bound CO, but did not bind O2 (Fig. 6D). In leghemoglobin, removal of either the His or Tyr residues allowed the remaining side-chain to stabilize O2 to a larger extent than both in combination (Kundu & Hargrove, 2003). A similar scenario in VcAer2 might explain why the O2 affinity of PAS1 was not affected by individually removing Y119 or W151. Whether PAS1, like leghemoglobin, requires faster O2 dissociation is currently under investigation.
In PaPAS, gas binding displaces the Hβ Leu that occupies the ligand-binding site, eliciting the Iβ Trp to rotate towards the ligand. These movements stabilize gas binding and initiate conformational signaling (Airola et al., 2013a, Garcia et al., 2017). The Hβ Leu is also conserved in PAS1 and PAS2 (Fig. 3A) and presumably moves out of the ligand-binding site for O2 binding in both PAS domains. It is intriguing that the signal-on mutant W276L assumed an activated conformation with the Hβ Leu displaced in the absence of ligand. This finding suggests that, although the distal Hβ Leu must certainly move for the heme to bind ligand, ligand-induced rearrangements of the Iβ Trp may be the dominant factor in promoting the switch to the active conformation.
In most chemoreceptors or histidine kinases with tandem PAS-like domains, the second domain does not bind small ligands (Nishiyama et al., 2016, Machuca et al., 2017). This is clearly not the case for VcAer2, where PAS2 binds O2 and signals its binding. Our data suggest that the primary role of PAS1 is to regulate O2-mediated signaling from PAS2. For example, VcAer2-H101A, whose PAS1 peptide retained only 26% heme (Fig. 4B), had a more robust O2 response than did WT receptor. An additional four PAS1 mutants likewise had better than WT function (Figs. 6B and S1E). However, removing PAS1 altogether (VcAer2 [165–678]) resulted in a non-functional receptor, even though PAS2 presumably remained dimeric. The only PAS1 mutant in this study that did not bind O2 (PAS1-Y119L/W151L) was similarly non-functional. Thus, PAS1-heme appears tuned to regulate PAS2 function, and this function may require PAS1 to bind O2. Since the Iβ strand of PAS1 is fused directly to the N-cap of PAS2, regulatory effects could be directed through the PAS1 Iβ-PAS2 N-cap connection, or could involve more global changes, such as PAS domain rotations or association/dissociation (signal-off, deoxy heme/signal-on, ligand bound heme, respectively). PAS2-mediated signaling then results in activation of the cluster III chemosensory pathway (Fig. 1B), resulting in a cellular response.
EXPERIMENTAL PROCEDURES
Mutagenesis and cloning
The aer2 gene (VCA1092) was amplified from V. cholerae O1 JBK 70 (Kaper et al., 1984) genomic DNA using PfuUltra DNA polymerase (Agilent Technologies, Santa Clara, CA) and cloned into the NcoI and PstI sites of pProEXHTa to express VcAer2 (res. 1–678) with an N-terminal His6 tag. This construct was named pKGB1. To create PAS peptides or VcAer2 constructs with N-terminal truncations, DNA fragments were PCR amplified from pKGB1, pKGB1-derived plasmids, or from V. cholerae O1 JBK 70 genomic DNA and cloned into the NcoI and PstI sites of pProEXHTa or the NdeI and XhoI sites of pET28a. Site-directed mutagenesis was performed on pKGB1, pProEXHTa-PAS2 (for PAS2-W276F), or pET28a-PAS2 (for PAS2-W276L) using site-specific primers and PfuUltra II Fusion DNA polymerase (Agilent Technologies) or Phusion® DNA polymerase (New England Biolabs, Ipswich, MA). For site-directed random mutagenesis, primers containing an equimolar mix of all four nucleotides at the W151 codon were used to amplify pKGB1 with 66 °C annealing and 20 amplification cycles. Site-specific mutagenesis products were treated with DpnI (New England Biolabs) to remove template strands and then electroporated into E. coli. For all constructs, VcAer2 expression was induced with 600 µM IPTG and products of the correct size were confirmed by Western blotting with HisProbe™-HRP (Thermo Scientific, Rockford, IL). All protein masses in this manuscript are reported for proteins without heme. All mutations were confirmed by sequencing the entire aer2 coding sequence.
Bacterial strains
VcAer2 plasmids were expressed in E. coli BL21(DE3), the WT E. coli chemotaxis strain RP437 (Parkinson, 1978), and in the chemoreceptorless E. coli strains BT3388 [tar, tsr, trg, tap, aer (Yu et al., 2002)] and UU2610 [tar, tsr, trg, tap, aer, cheR, cheB (Zhou et al., 2011)].
Steady-state cellular VcAer2 levels
The steady-state cellular levels of the full-length VcAer2 mutants (res. 1–678) and N-terminal truncation mutants were compared with WT VcAer2 after inducing BT3388 cells with 50 µM IPTG (Fig. S2A). Aer2-W151E and Aer2-W151R were instead induced with 200 µM IPTG. The cellular levels of the PAS peptides were compared with PAS1–2 (res. 1–282, with or without H226A, Fig. S2B–C), WT PAS1 (res. 38–157, Fig. S2D), or WT PAS2 (res. 165–282, Fig. S2E) after inducing expression in BT3388 with 100 µM IPTG. PAS1-W151E, PAS1-W151N, and PAS1-W151R were instead induced with 200 µM IPTG. Samples were electrophoresed in duplicate and experiments were repeated on two to four separate occasions. Bands were visualized on HisProbe Western blots and quantified on a BioSpectrum® digital imager (UVP, Upland, CA).
Behavioral assays
BT3388 cells were grown at 30 °C in tryptone broth containing 0.5 µg ml−1 thiamine and 25 µg ml−1 5-aminolevulinic acid (Sigma-Aldrich) and induced at an OD600nm of 0.2–0.25 for 2 h with 200 µM IPTG. Cells were then placed in a gas perfusion chamber where the gas was toggled between air (20.9% O2) and N2, and cell behavior was analyzed (Rebbapragada et al., 1997, Taylor et al., 2007). Signal-off mutants (those that were smooth swimming in air and in N2) were retested after inducing expression with 1 mM IPTG for 2 h. Behavioral responses to O2 were repeated two or more times on at least two separate occasions. Estimation of percent tumbling was determined for all motile bacteria in a field of view at 800× magnification. To determine CO responses, BT3388 cells induced with 200 µM or 1 mM IPTG were perfused with N2 for 30 sec prior to perfusing with CO gas (>99% purity, Sigma-Aldrich, St. Louis, MO) for 10 sec. NO responses were assessed using the NO donor Proli NONOate as previously described (Watts et al., 2011). Swim plate responses were determined by inoculating RP437 or BT3388 cells into tryptone soft agar containing 0–1000 µM IPTG (Taylor et al., 2007) and incubating at 30 °C for 9–16 h.
Protein purification for heme and gas binding studies
BT3388 cells expressing full-length VcAer2, truncation mutants or PAS peptides were grown in LB broth, Lennox (5 g L−1 NaCl), containing 0.5 µg ml−1 thiamine and 25 µg ml−1 5-aminolevulinic acid to augment heme synthesis and incorporation. Protein expression was induced with 600 µM IPTG and proteins were purified on Ni-NTA agarose columns (Qiagen, Valencia, CA) as previously described (Garcia et al., 2017). In addition to wash buffers 1 and 2 (containing 50 mM Tris, pH 7.5, 100 mM NaCl and 20 mM or 50 mM imidazole, respectively), all PAS peptides except PAS2 peptides were also washed with wash buffer 3 (containing 100 mM imidazole). The concentration of the eluted proteins was determined in a BCA™ Protein Assay (Thermo Scientific) using BSA as a standard, and sample quality was assessed by SDS-PAGE (2.5 µg of each protein), followed by staining with Coomassie Brilliant Blue.
Heme binding
The proportion of heme bound to the WT PAS1 (res. 38–157) and PAS2 (res. 165–282) peptides was determined by using a pyridine hemochrome assay (Garcia et al., 2017). Results were used to standardize PAS-heme concentrations in ligand-binding assays. To determine the heme content of purified PAS peptides, the Soret height and purity of 10 µM imidazole-bound PAS peptides were compared with corresponding WT PAS peptides, as previously described (Garcia et al., 2017). Full-length VcAer2 and VcAer2 [165–678] were analyzed using 5 µM purified protein. Heme ratios below 15% indicated a substantial heme-binding defect from which gas affinity constants could not be determined.
Absorption spectra and gas binding affinities
The deoxy, oxy, carbonmonoxy and met heme spectra of 10 µM purified PAS1 (res. 38–157) and PAS2 (res. 165–282) were determined as previously described for PaAer2 (Watts et al., 2011). Dissociation constants for O2 (Kd(O2)) and CO (Kd(CO)) binding to PAS1, PAS2 and VcAer2 [165–678] peptides were estimated by linear interpolation of unliganded (Fe2+) and liganded (Fe2+-O2, Fe2+-CO) spectra, as previously described (Garcia et al., 2017). Experiments were repeated on two to eight occasions, from which average Kd’s were determined and rounded to the nearest whole number.
Solubility assays
VcAer2/BT3388 cells were grown, lysed and centrifuged at low (10 000 g) and then high (485 000 g) speed as per the protein purification method above. Low- and high-speed pellets were resuspended in buffer containing 50 mM Tris, pH 7.5, 100 mM NaCl and 10 mM imidazole. Equal volumes of the low- and high-speed supernatants and resuspended pellets were analyzed by SDS-PAGE and HisProbe Western blots.
Size-exclusion chromatography
Purified PAS peptides and protein standards [from Schwarz/Mann Biotech (Cleveland, OH) and Sigma-Aldrich] were filtered though 0.2 µM centrifugal filters (Millipore, Billerica, MA) and 200 µg of proteins (in 100 µl) were separated on a TSKgel G2000SW size-exclusion column (Tosoh Bioscience, King of Prussia, PA) in 50 mM NaPO4, pH 7.0, 300 mM NaCl and 0.02% NaN3 (TSK buffer), as previously described (Watts et al., 2011). Under these conditions, imidazole dissociated from the PAS peptides, leaving met heme. To analyze peptides containing deoxy heme, TSK buffer was perfused with N2 for at least one hour, before adding sodium dithionite to 2 mM and perfusing with N2 for an additional 15 min. The column was then equilibrated with this buffer for 15 min. Sodium dithionite grains were added to the protein samples before loading onto the column. The presence of deoxy heme was determined by monitoring the elution spectra. For all samples, 1 ml fractions were collected and analyzed after ammonium sulfate precipitation for the presence of PAS peptide on HisProbe Western blots.
Crystallization and data collection
PAS2-W276L was expressed in E. coli BL21(DE3) along with E. coli ferrochelatase to promote PAS-heme incorporation (Sudhamsu et al., 2010). Protein expression was induced with 400 µM IPTG at 37 °C for 16 hours. PAS2-W276L was purified on a Ni-NTA column and eluted in 10 mM Tris pH 8.0, 500 mM NaCl, 10% glycerol and 250 mM Imidazole pH 8.0. The eluted peptide was subjected to buffer exchange into 20 mM imidazole pH 8.0 and 100 mM NaCl before overnight digestion with thrombin (0.7 µg ml−1). The tag-free peptide was further purified using a Superdex 75 26–60 size exclusion column (GE Life Sciences, Pittsburgh, PA), and eluted in 20 mM imidazole pH 8.0 and 100 mM NaCl. Crystals of PAS2-W276L were grown by vapor diffusion upon mixing 1 µl of protein (12 mg ml−1) with 1 µl of well solution against a reservoir containing 2.6 M (NH4)2SO4, 0.1 M citric acid pH 5.5. A solution of NiCl2 (10 mM final concentration) was added directly to the protein-well solution mixture to influence crystallization and promote better diffraction. Diffraction data was collected at the Cornell High Energy Synchroton Source (CHESS) on the A1 beamline with an ADSC Quantum 210 CCD detector. Data were processed with HKL2000 (Otwinowski & Minor, 1997).
Structure determination and refinement
The structure of PAS2-W276L in the ligand-free form was determined by molecular replacement with PHENIX AutoMR using the CN−-bound PaPAS structure (PDB code: 3VOL) as a search model. The structure was built using Coot (Emsley et al., 2010), and refined in PHENIX (Adams et al., 2011) amid manual model building, minimization, B-factor refinement, and application of non-crystallographic symmetry (NCS) restraints. NCS restraints were removed in the later stages of refinement.
Homology modelling
Homology models of WT PAS1 (res. 38–157) and WT PAS2 were produced based on the PAS2-W276L structure using SWISS-MODEL (Arnold et al., 2006). A b-type heme molecule was positioned into the PAS1 homology model by superimposing the heme-containing PAS2-W276L structure. The same heme-ligation pattern was maintained as found in the PAS2-W276L structure.
Supplementary Material
ABBREVIATED SUMMARY.
In this study we show that V. cholerae Aer2 is an O2 receptor with two related but functionally divergent PAS-heme domains. PAS2 is an O2 sensor that stabilizes O2 binding via a conserved Trp residue, whereas PAS1 is a signal modulator that binds O2 to regulate signaling from PAS2. O2 binding to PAS1 required at least one of two heme pocket residues: the conserved Trp or a specific Tyr residue.
Acknowledgments
We thank Kahryl Bennett, Adwoa Wiafe, Andrew Hong, Magi Ishak Gabra, and Lana Haddad for constructing and testing several of the mutants in this study. This research was supported by laboratory start-up funds to K. Watts, the Loma Linda University Center for Health Disparities and Molecular Medicine (CHDMM) summer research program [National Institutes of Health (NIH) award number P20MD006988], the National Institute of General Medical Sciences (NIGMS) of the NIH award number R01GM108655 to K. Watts, and award number R35GM122535 to B. Crane. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health.
Footnotes
AUTHOR CONTRIBUTIONS
NS and BRC acquired, and TKC refined, the PAS2 structure; SGP and KJW acquired all remaining data; MSJ, BRC and KJW designed this study, interpreted the data, and wrote this manuscript.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Echols N, Headd JJ, Hung LW, Jain S, Kapral GJ, Kunstleve RWG, McCoy AJ, Moriarty NW, Oeffner RD, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airola MV, Huh D, Sukomon N, Widom J, Sircar R, Borbat PP, Freed JH, Watts KJ, Crane BR. Architecture of the soluble receptor Aer2 indicates an in-line mechanism for PAS and HAMP domain signaling. J Mol Biol. 2013a;425:886–901. doi: 10.1016/j.jmb.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airola MV, Sukomon N, Samanta D, Borbat PP, Freed JH, Watts KJ, Crane BR. HAMP domain conformers that propagate opposite signals in bacterial chemoreceptors. PLOS Biol. 2013b;11:e1001479. doi: 10.1371/journal.pbio.1001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airola MV, Watts KJ, Bilwes AM, Crane BR. Structure of concatenated HAMP domains provides a mechanism for signal transduction. Structure. 2010;18:436–448. doi: 10.1016/j.str.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Biswas M, Dey S, Khamrui S, Sen U, Dasgupta J. Conformational barrier of CheY3 and inability of CheY4 to bind FliM control the flagellar motor action in Vibrio cholerae. PLOS One. 2013;8:e73923. doi: 10.1371/journal.pone.0073923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boin MA, Austin MJ, Hase CC. Chemotaxis in Vibrio cholerae. FEMS Microbiol Lett. 2004;239:1–8. doi: 10.1016/j.femsle.2004.08.039. [DOI] [PubMed] [Google Scholar]
- Briegel A, Ortega DR, Mann P, Kjaer A, Ringgaard S, Jensen GJ. Chemotaxis cluster 1 proteins form cytoplasmic arrays in Vibrio cholerae and are stabilized by a double signaling domain receptor DosM. Proc Natl Acad Sci USA. 2016;113:10412–10417. doi: 10.1073/pnas.1604693113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta J, Dattagupta JK. Structural determinants of V. cholerae CheYs that discriminate them in FliM binding: comparative modeling and MD simulation studies. J Biomol Struct Dyn. 2008;25:495–503. doi: 10.1080/07391102.2008.10507196. [DOI] [PubMed] [Google Scholar]
- Delgado-Nixon VM, Gonzalez G, Gilles-Gonzalez MA. Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry. 2000;39:2685–2691. doi: 10.1021/bi991911s. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Orillard E, Johnson MS, Watts KJ. Gas sensing and signaling in the PAS-heme domain of the Pseudomonas aeruginosa Aer2 receptor. J Bacteriol. 2017;199:3–17. doi: 10.1128/JB.00003-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles-Gonzalez MA, Gonzalez G. Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J Inorg Biochem. 2005;99:1–22. doi: 10.1016/j.jinorgbio.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Glekas GD, Mulhern BJ, Kroc A, Duelfer KA, Lei V, Rao CV, Ordal GW. The Bacillus subtilis chemoreceptor McpC senses multiple ligands using two discrete mechanisms. J Biol Chem. 2012;287:39412–39418. doi: 10.1074/jbc.M112.413518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosink KK, Kobayashi R, Kawagishi I, Hase CC. Analyses of the roles of the three cheA homologs in chemotaxis of Vibrio cholerae. J Bacteriol. 2002;184:1767–1771. doi: 10.1128/JB.184.6.1767-1771.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinstaff MW, Hill MG, Bimbaum ER, P SW, Labinger JA, Gray HB. Structures, electronic properties, and oxidation-reduction reactivity of halogenated iron porphyrins. Inorg Chem. 1995;34:4896–4902. [Google Scholar]
- Guvener ZT, Tifrea DF, Harwood CS. Two different Pseudomonas aeruginosa chemosensory signal transduction complexes localize to cell poles and form and remould in stationary phase. Mol Microbiol. 2006;61:106–118. doi: 10.1111/j.1365-2958.2006.05218.x. [DOI] [PubMed] [Google Scholar]
- Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- Hiremath G, Hyakutake A, Yamamoto K, Ebisawa T, Nakamura T, Nishiyama S, Homma M, Kawagishi I. Hypoxia-induced localization of chemotaxis-related signaling proteins in Vibrio cholerae. Mol Microbiol. 2015;95:780–790. doi: 10.1111/mmi.12887. [DOI] [PubMed] [Google Scholar]
- Hyakutake A, Homma M, Austin MJ, Boin MA, Hase CC, Kawagishi I. Only one of the five CheY homologs in Vibrio cholerae directly switches flagellar rotation. J Bacteriol. 2005;187:8403–8410. doi: 10.1128/JB.187.24.8403-8410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper JB, Lockman H, Baldini MM, Levine MM. Recombinant nontoxinogenic Vibrio cholerae strains as attenuated cholera vaccine candidates. Nature. 1984;308:655–658. doi: 10.1038/308655a0. [DOI] [PubMed] [Google Scholar]
- Kerby RL, Youn H, Roberts GP. RcoM: a new single-component transcriptional regulator of CO metabolism in bacteria. J Bacteriol. 2008;190:3336–3343. doi: 10.1128/JB.00033-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key J, Hefti M, Purcell EB, Moffat K. Structure of the redox sensor domain of Azotobacter vinelandii NifL at atomic resolution: signaling, dimerization, and mechanism. Biochemistry. 2007;46:3614–3623. doi: 10.1021/bi0620407. [DOI] [PubMed] [Google Scholar]
- Kloek AP, Yang J, Mathews FS, Frieden C, Goldberg DE. The tyrosine B10 hydroxyl is crucial for oxygen avidity of Ascaris hemoglobin. J Biol Chem. 1994;269:2377–2379. [PubMed] [Google Scholar]
- Kundu S, Hargrove MS. Distal heme pocket regulation of ligand binding and stability in soybean leghemoglobin. Proteins. 2003;50:239–248. doi: 10.1002/prot.10277. [DOI] [PubMed] [Google Scholar]
- Kurokawa H, Lee DS, Watanabe M, Sagami I, Mikami B, Raman CS, Shimizu T. A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor. J Biol Chem. 2004;279:20186–20193. doi: 10.1074/jbc.M314199200. [DOI] [PubMed] [Google Scholar]
- Liu YC, Machuca MA, Beckham SA, Gunzburg MJ, Roujeinikova A. Structural basis for amino-acid recognition and transmembrane signalling by tandem Per-Arnt-Sim (tandem PAS) chemoreceptor sensory domains. Acta Crystallogr D Biol Crystallogr. 2015;71:2127–2136. doi: 10.1107/S139900471501384X. [DOI] [PubMed] [Google Scholar]
- Machuca MA, Johnson KS, Liu YC, Steer DL, Ottemann KM, Roujeinikova A. Helicobacter pylori chemoreceptor TlpC mediates chemotaxis to lactate. Sci Rep. 2017;7:14089. doi: 10.1038/s41598-017-14372-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama S, Suzuki D, Itoh Y, Suzuki K, Tajima H, Hyakutake A, Homma M, Butler-Wu SM, Camilli A, Kawagishi I. Mlp24 (McpX) of Vibrio cholerae implicated in pathogenicity functions as a chemoreceptor for multiple amino acids. Infect Immun. 2012;80:3170–3178. doi: 10.1128/IAI.00039-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama S, Takahashi Y, Yamamoto K, Suzuki D, Itoh Y, Sumita K, Uchida Y, Homma M, Imada K, Kawagishi I. Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants. Sci Rep. 2016;6:20866. doi: 10.1038/srep20866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Park SY, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- Parkinson JS. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J Bacteriol. 1978;135:45–53. doi: 10.1128/jb.135.1.45-53.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbapragada A, Johnson MS, Harding GP, Zuccarelli AJ, Fletcher HM, Zhulin IB, Taylor BL. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci USA. 1997;94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringgaard S, Hubbard T, Mandlik A, Davis BM, Waldor MK. RpoS and quorum sensing control expression and polar localization of Vibrio cholerae chemotaxis cluster III proteins in vitro and in vivo. Mol Microbiol. 2015;97:660–675. doi: 10.1111/mmi.13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai H, Sugimoto H, Shiro Y, Ishikawa H, Mizutani Y, Aono S. Structural basis for oxygen sensing and signal transduction of the heme-based sensor protein Aer2 from Pseudomonas aeruginosa. Chem Commun (Camb) 2012;48:6523–6525. doi: 10.1039/c2cc32549g. [DOI] [PubMed] [Google Scholar]
- Schuster M, Hawkins AC, Harwood CS, Greenberg EP. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol Microbiol. 2004;51:973–985. doi: 10.1046/j.1365-2958.2003.03886.x. [DOI] [PubMed] [Google Scholar]
- Selvaraj P, Gupta R, Peterson KM. The Vibrio cholerae ToxR regulon encodes host-specific chemotaxis proteins that function in intestinal colonization. SOJ Microbiol Infect Dis. 2015;3:1–5. doi: 10.15226/sojmid/3/3/00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhamsu J, Kabir M, Airola MV, Patel BA, Yeh SR, Rousseau DL, Crane BR. Co-expression of ferrochelatase allows for complete heme incorporation into recombinant proteins produced in E. coli. Protein Expr Purif. 2010;73:78–82. doi: 10.1016/j.pep.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL, Watts KJ, Johnson MS. Oxygen and redox sensing by two-component systems that regulate behavioral responses: behavioral assays and structural studies of Aer using in vivo disulfide cross-linking. Methods Enzymol. 2007;422:190–232. doi: 10.1016/S0076-6879(06)22010-X. [DOI] [PubMed] [Google Scholar]
- Watts KJ, Sommer K, Fry SL, Johnson MS, Taylor BL. Function of the N-terminal cap of the PAS domain in signaling by the aerotaxis receptor Aer. J Bacteriol. 2006;188:2154–2162. doi: 10.1128/JB.188.6.2154-2162.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts KJ, Taylor BL, Johnson MS. PAS/poly-HAMP signalling in Aer-2, a soluble haem-based sensor. Mol Microbiol. 2011;79:686–699. doi: 10.1111/j.1365-2958.2010.07477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Kloek AP, Goldberg DE, Mathews FS. The structure of Ascaris hemoglobin domain I at 2.2 A resolution: molecular features of oxygen avidity. Proc Natl Acad Sci USA. 1995;92:4224–4228. doi: 10.1073/pnas.92.10.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HS, Saw JH, Hou S, Larsen RW, Watts KJ, Johnson MS, Zimmer MA, Ordal GW, Taylor BL, Alam M. Aerotactic responses in bacteria to photoreleased oxygen. FEMS Microbiol Lett. 2002;217:237–242. doi: 10.1111/j.1574-6968.2002.tb11481.x. [DOI] [PubMed] [Google Scholar]
- Yukl ET, Ioanoviciu A, Nakano MM, de Montellano PR, Moenne-Loccoz P. A distal tyrosine residue is required for ligand discrimination in DevS from Mycobacterium tuberculosis. Biochemistry. 2008;47:12532–12539. doi: 10.1021/bi801234w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hendrickson WA. Structural characterization of the predominant family of histidine kinase sensor domains. J Mol Biol. 2010;400:335–353. doi: 10.1016/j.jmb.2010.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Ames P, Parkinson JS. Biphasic control logic of HAMP domain signalling in the Escherichia coli serine chemoreceptor. Mol Microbiol. 2011;80:596–611. doi: 10.1111/j.1365-2958.2011.07577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, Dunlap JC, Crane BR. Conformational switching in the fungal light sensor Vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.