Abstract
Stem cells are considered as a multipotent regenerative source for diseased and dysfunctional tissues. Despite the promise of stem cells, the inherent capacity of stem cells to convert to tissue-specific lineages can present a major challenge to the use of stem cells for regenerative medicine. We hypothesized that epigenetic regulating molecules can modulate the stem cell’s developmental program, and thus potentially overcome the limited lineage differentiation that human stem cells exhibit based on the source and processing of stem cells. In this study, we screened a library of 84 small molecule pharmacological agents indicated in nucleosomal modification and identified a sub-set of specific molecules that influenced osteogenesis in human mesenchymal stem cells (hMSCs) while maintaining cell viability in-vitro. Pre-treatment with five candidate hits, Gemcitabine, Decitabine, I-CBP112, Chidamide, and SIRT1/2 inhibitor IV, maximally enhanced osteogenesis in-vitro. In contrast, five distinct molecules, 4-Iodo-SAHA, Scriptaid, AGK2, CI-amidine and Delphidine Chloride maximally inhibited osteogenesis. We then tested the role of these molecules on hMSCs derived from aged human donors and report that small epigenetic molecules, namely Gemcitabine and Chidamide, can significantly promote osteogenic differentiation by 5.9- and 2.3-fold, respectively. Taken together, this study demonstrates new applications of identified small molecule drugs for sensitively regulating the lineage plasticity fates of bone-marrow derived mesenchymal stem cells through modulating the epigenetic profile of the cells.
Introduction
There is a consensus in the scientific community about the potential of adult stem cell-based therapies and tissue regeneration1–3. Varied biochemical and biophysical cues have been employed to design efficacious cell-based therapies3–5. However, the ability to optimally harness the potential of adult human mesenchymal stem cells (hMSCs) and direct optimal cell phenotypic development for tissue formation continues to present a major challenge, which limits the translation of these technologies to the clinic. A major limitation is that the differentiation capabilities of MSCs deteriorate with age6,7 and with in-vitro passages8, thereby affecting their developmental potential and impairing the efficacy of cell therapy. The second major limitation is the poor stability of cell phenotypes9, which complicates the ability to accurately postulate the response of cells to engineered cues. Therefore, technologies that can enhance the potency of stem cells cultured in-vitro and modulate their sensitivity and stability to engineered cues, need to be developed to ensure a specific developmental fate of the cell and facilitate the advancement of cell-based therapies for tissue engineering applications.
Conventional regenerative tissue technologies have relied on extracellular signals (growth factors, small molecules and metabolic regulators) to accelerate lineage conversion and ameliorate age related MSC dysfunction10–12. While recent scientific evidence indicated that the epigenetic profile of the cell is a key determinant in guiding the developmental pathway of cells13,14, the role of epigenetic modifications in steering cell differentiation and the use of pharmacologic agents as epigenetic manipulators to optimize specific cell phenotypic development has not been explored. “Epigenetics” refers to the “non-genetically based cellular memory”, which involves heritable changes in gene expression that occur without alteration in DNA sequence. These changes can be a consequence of environmental factors or induced spontaneously, using two primary mechanisms of DNA methylation and covalent modification of histones15. The emerging field of epigenetics has thus far caught the interest of scientists globally by evidencing that the epigenetic markers influence gene expression and genome function, thereby directing DNA-based biological processes15,16. Recent studies have indicated the potential role of epigenetic modifiers such as trichostatin A, valproic acid and sodium butyrate in osteogenic differentiation17–19. Even so, the use of the many accessible pharmacologic agents as epigenetic manipulators and their application in optimizing specific cell phenotypic development has not been comprehensively realized.
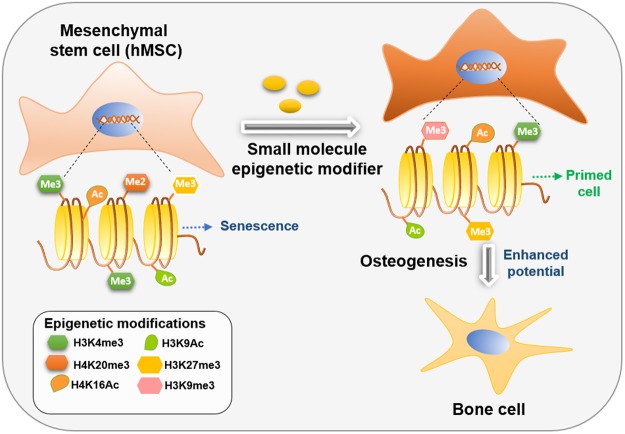
In this study, we systematically evaluated a library of pharmacological agents indicated in nucleosomal modification to identify specific compounds capable of modulating osteogenic differentiation (Fig. 1). 84 compounds capable of influencing the epigenetic profile of the cells and consequently the nucleosomal organization were screened (Table 1). The compounds included small molecules that modulate the activity of methyltransferases, demethylases, HATs, HDACs and acetylated lysine reader proteins. Top 10 compounds maximally enhancing or inhibiting osteogenesis in human mesenchymal stem cells (hMSCs) cultured in vitro, while maintaining cell viability, were identified and presented.
Figure 1.
Increasing differentiation potential of ex-vivo cultured stem cells through epigenetic modulation. In this study small molecules nucleosomal modifiers able to significantly increase osteogenic differentiation potential of hMSCs were identified.
Table 1.
List of all nucleosomal modifying drugs screened for modulating hMSC differentiation.
| List of drugs screened | ||
|---|---|---|
| (1) AGK2 | (29) Delphinidin chloride | (57) Oxamflatin |
| (2) CAY10433 | (30) ITF 2357 | (58) 2′,3′,5′-triacetyl-5-Azacytidine |
| (3) M 344 | (31) 3-Deazaneplanocin A | (59) Salermide |
| (4) HNHA | (32) Suramin (sodium salt) | (60) Mirin |
| (5) Octyl-α-ketoglutarate | (33) PFI-1 | (61) UNC1999 |
| (6) Cl-Amidine (trifluoroacetate salt) | (34) 5-Azacytidine | (62) Pimelic Diphenylamide 106 |
| (7) CAY10669 | (35) Chaetocin | (63) MS-275 |
| (8) HC Toxin | (36) Decitabine | (64) RG-108 |
| (9) JGB1741 | (37) (+)-JQ1 | (65) S-Adenosylhomocysteine |
| (10) Garcinol | (38) (—)-JQ1 | (66) UNC0224 |
| (11) MI-2 (hydrochloride) | (39) BSI-201 | (67) Chidamide |
| (12) Sinefungin | (40) IOX1 | (68) Pyroxamide |
| (13) Suberohydroxamic Acid | (41) MI-nc (hydrochloride) | (69) N-Oxalylglycine |
| (14) Valproic Acid | (42) Gemcitabine | (70) WDR5-0103 |
| (15) Resveratol | (43) Lomeguatrib | (71) AMI-1 (sodium salt) |
| (16) 3-amino Benzamide | (44) Daminozide | (72) UNC1215 |
| (17) 4-iodo-SAHA | (45) GSK-J1 (sodium salt) | (73) GSK 343 |
| (18) C646 | (46) GSKJ2 (sodium salt) | (74) Bromosporine |
| (19) Ellagic Acid | (47) GSK-J4 (hydrochloride) | (75) SIRT1/2 Inhibitor IV |
| 20) Scriptaid | 48) GSK-J5 (hydrochloride) | 76) I-CBP112 (hydrochloride) |
| (21) UNC0321 (trifluoroacetate salt) | (49) Tenovin-1 | (77) PFI-3 |
| (22) (-)-Neplanocin A | (50) Tenovin-6 | (78) 2,4-DPD |
| (23) F-Amidine (trifluoroacetate salt) | (51) BIX01294 (hydrochloride hydrate) | (79) DMOG |
| (24) UNC0638 | (52) Anacardic Acid | (80) Trichostatin A |
| (25) Phthalazinone pyrazole | (53) CAY10603 | (81) CAY10398 |
| (26) Isoliquiritigenin | (54) Splitomicin | (82) RSC-133 |
| (27) CCG-100602 | (55) CBHA | (83) Piceatannol |
| (28) Zebularine | (56) Oxamflatin | (84) CAY10591 |
In parallel, we also elucidated the cross-talk between the epigenetic effects and stem cell lineage phenotypes using single cell nucleosomal imaging and image informatics. To accomplish this, we performed high content image informatics to track and annotate cells using SC-35 as a surrogate marker20, which can be employed to profile changes the in-situ nucleosomal organization globally after exposure to small molecule modifiers.
SC-35 nuclear speckle domains constitute small nuclear ribonucleoprotein particles (snRNPs), spliceosomes, and transcription factors that mediate co-transcriptional modifications of RNA21,22. Recent body of work from our lab has shown that speckle factor SC-35 can be employed as an integrative surrogate marker to assess the effect of environmental factors (growth factors, topography, biomaterials) on MSC differentiation and parse the emergent hMSC phenotypes predictably within 72 hours of exposure to external modulating factors20,23. We believe that treatment with these small molecules modifies the epigenetic profile, which in turn influences the regulation of gene expression and consequently the SC-35 spatial organization. SC-35 can therefore be utilized as a universal surrogate marker to annotate the cells by mapping the resultant textural signatures, capturing minute variations in nucleosomal organization, post treatment with epigenetic manipulators. Therefore, this is the first study to demonstrate that osteogenic differentiation can be regulated through epigenetic modulation by small molecules (Fig. 1), and that high content image informatic of SC-35 spatial organization can be employed to parse the resultant variances in nucleosomal organization.
Results
Optimization of osteogenic differentiation by modulating nucleosomal organization through small molecule pharmacologic agents
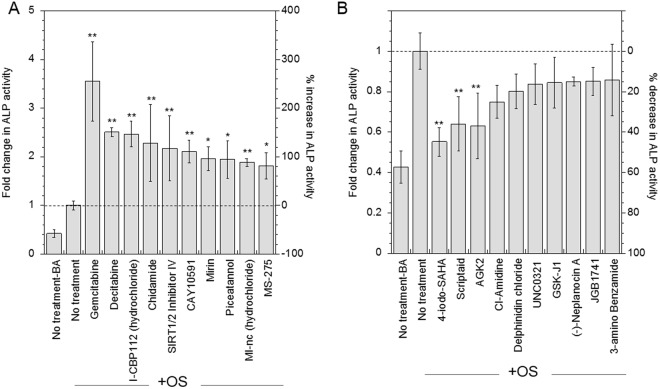
A screen of 84 small molecule drugs known to influence nucleosomal organization (Table 1) was applied to identify the drugs that significantly influence osteogenic differentiation of cultured hMSCs in vitro. Cells were pretreated with drugs for 24 hours and subsequently induced to differentiate using commonly used cocktail of differentiation factors (osteogenic medium). The effect of drug treatment on osteogenesis was assessed at 14 days using ALP assay (Supplemental Fig. 1). The top 10 drugs that most increased osteogenesis or decreased osteogenesis were identified (Fig. 2A,B). Their concentrations and chemical structures are listed in Supplemental Tables 1,2. The top 5 drugs that significantly increased (p < 0.05) osteogenesis included Gemcitabine, Decitabine, I-CBP112, Chidamide, and SIRT1/2 inhibitor IV. Gemcitabine maximally increased ALP activity by 3.5-fold, Decitabine and I-CBP112 increased ALP activity by 2.5-fold, and Chidamide and SIRT1/2 Inhibitor IV increased ALP activity by 2.3- and 2.2-fold, respectively (Fig. 2A).
Figure 2.
Small molecule nucleosomal modifiers influence osteogenic differentiation of hMSCs. The effect of treatment with pharmacological agents influencing the epigenetic profile of the cell on osteogenic differentiation was analyzed at Day 14 using ALP activity assay. Top 10 agents that significantly increased the ALP activity (A) and decreased ALP activity (B) were identified through screening. Statistical analysis was performed using the Dunnett Multiple Comparisons test, which compares all columns versus a control column (OS-no treatment). The symbols ** and * represent a significant change in ALP activity with respect to OS – no treatment condition to the level of p < 0.01 and p < 0.05, respectively.
The top 5 drugs that most decreased osteogenesis were 4-Iodo SAHA, Scriptaid, AGK2, CI-Amidine and Delphidine Chloride. Specifically, 4-Iodo-SAHA significantly inhibited ALP activity by 45%, followed by Scriptaid and AGK2, which inhibited ALP activity by at least 36% (Fig. 2B). Furthermore, there was no significant difference in ALP activity between cells cultured in basal media, and cells cultured in osteogenic media after pretreatment with 4-Iodo-SAHA, Scriptaid or AGK2, indicating that nucleosomal modifications by these drugs inhibit osteogenic differentiation completely even after induction with soluble factors.
The cells were also stained for ALP using Fast Blue RR salt staining (Supplemental Fig. 4), osteogenic differentiation marker RUNX2 using immune-staining and actin using phalloidin staining (Supplemental Fig. 5). The top 5 drugs identified to increase osteogenesis have more Fast Blue positive staining and RUNX2 expression, as compared to basal condition (BA), and to a level same or more than untreated cells cultured in osteogenic media (OS). Also, as observed from Supplemental Fig. 5, cells pre-treated with drugs identified to increase osteogenic differentiation, led to confluent mono-layers at 14-days, similar in morphology to untreated cells.
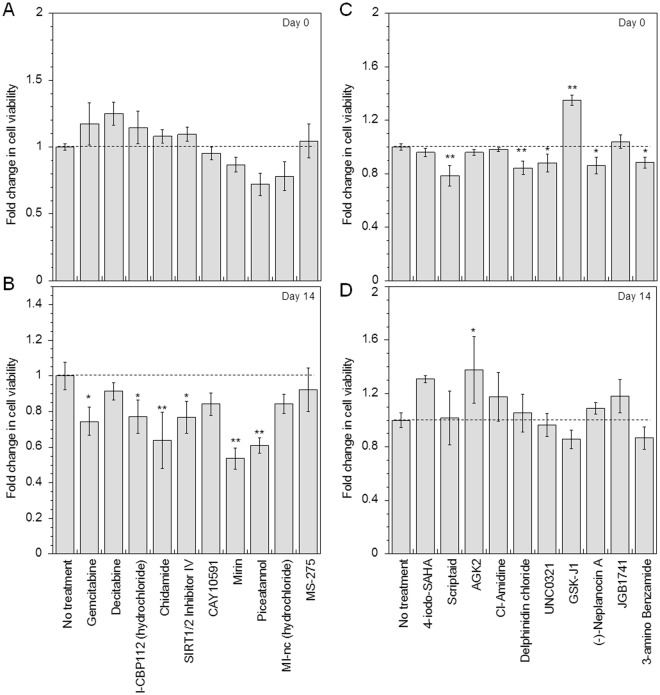
Effect of small molecule pharmacologic agents on cell viability
The effect of treatment with the library of pharmacologic agents on cell viability was assessed immediately post treatment (Day 0) and 14 days post treatment (Day 14) using MTS assay (Supplemental Fig. 2). Figure 3 shows the effect of the top 10 osteogenically sensitive drugs on the degree of cell viability at Day 0 and Day 14. Immediately post treatment (Day 0), >85% cell viability was maintained for all identified drugs, except Piceatannol, MI-nc and Scriptaid, which maintained at least 70% cell viability (Fig. 3A,C). On Day 14, cells that were pretreated with Decitabine, CAY10591, MI-nc and MS-275 maintained >80% cell viability (Fig. 3B). A significant decrease (at least p < 0.05) in cell viability was observed at 14 days with other compounds that increased osteogenesis including Gemcitabine, I-CBP112, SIRT1/2 Inhibitor IV, Chidamide, Mirin and Piceatannol (Fig. 3B). However, Gemcitabine, I-CBP112 and SIRT1/2 Inhibitor IV maintained at least 70% cell viability and Chidamide, and Piceatannol maintained at least 60% cell viability. On the other hand, at 14-day post treatment no significant differences in cell viability compared to untreated cells were observed for small molecules identified for inhibiting osteogenesis (Fig. 3D).
Figure 3.
Effect of identified small molecule nucleosomal modifying drugs on cell viability. Cell viability was analyzed immediately post treatment with drugs (Day 0) and at 14 days post drug treatment (Day 14) using MTS cell viability assay. (A) and (B) show the effect on cell viability for drugs that increase osteogenesis, at Day 0 and 14, respectively. (C) and (D) show influence on cell viability for drugs that increase or decrease osteogenesis, at Day 0 and 14, respectively. Statistical analysis was performed using the Dunnett Multiple Comparisons test, which compares all columns versus a control column (no treatment). The symbols **and *represent a significant change in cell viability with respect to no treatment condition to the level of p < 0.01 and p < 0.05, respectively.
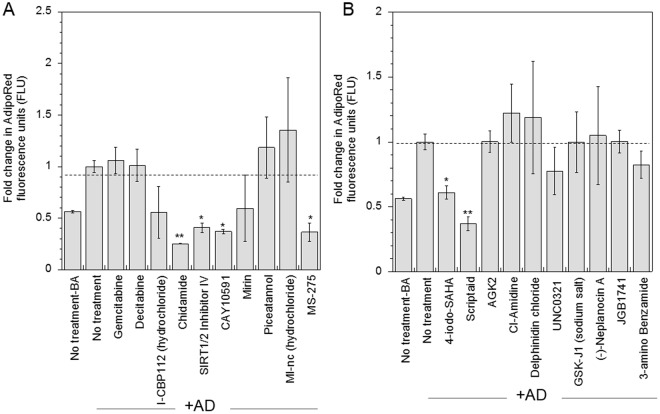
Specificity of identified compounds for osteogenesis versus adipogenesis
Next, we evaluated the specificity of the identified drugs in influencing the osteogenic versus adipogenic potential of hMSCs. The 10 drugs identified for significantly increasing osteogenesis did not similarly affect adipogenesis, and no increase in adipogenesis was observed. Interestingly, Chidamide, SIRT1/2 inhibitor IV, CAY10591 and MS-275 significantly decreased adipogenesis (at least p < 0.05) while Gemcitabine, Decitabine, I-CBP112, Mirin, Piceatannol and MI-nc did not significantly influence adipogenesis (Fig. 4A). Among the drugs identified for mediating inhibition of osteogenesis, 4-Iodo-SAHA and Scriptaid significantly inhibited adipogenesis as well (at least p < 0.05), while AGK2, CI-Amidine, Delphinidin Chloride, UNC0321, GSK-J1, Neplanocin A, JGB1741 and 3-Amino benzamide did not significantly alter adipogenesis (Fig. 4B).
Figure 4.
Effect of identified small molecule nucleosomal modifying drugs on adipogenesis. The specificity of identified drugs in influencing osteogenesis was evaluated by assessing the effect of drugs on adipogenesis. Cells treated with drugs identified to increase (A) and inhibit (B) osteogenesis were cultured in adipogenic differentiation media (AD) for 14 days and adipogenesis was evaluated using AdipoRed staining assay. Statistical analysis was performed using the Dunnett Multiple Comparisons test, which compares all columns versus a control column (AD-no treatment). The symbols ** and * represent a significant change in AdipoRed fluorescence units with respect to no treatment condition to the level of p < 0.01 and p < 0.05, respectively.
Profiling change in nucleosomal organization post treatment with identified compounds using a high content image informatics platform
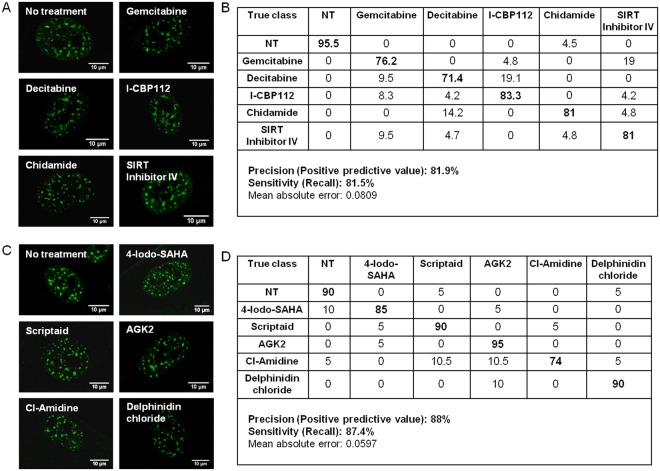
Previously, we demonstrated that speckle factor SC-35 can be used as a surrogate marker to classify cell-state in response to external cues during osteogenic differentiation20,23. Our next step was to test if the high content image informatics approach could be employed to profile and elucidate the influence of the drugs on nucleosomal organization globally using SC-35 organizational metrics. We applied this platform to track changes in nucleosomal organization induced via top 5 key drugs identified to either enhance or inhibit differentiation (Fig. 2A,B). By employing high content analysis to compute organizational metrics of speckle factor SC-35 and using J48 decision tree classification, untreated cells and cells treated with drugs identified to enhance osteogenesis, namely, Gemcitabine, Decitabine, I-CBP112, Chidamide and SIRT1/2 inhibitor IV, could be parsed with >80% precision and sensitivity, immediately post treatment (Fig. 5A,B). Likewise, untreated cells and cells treated with drugs identified to inhibit osteogenesis, namely, 4-Iodo-SAHA, Scriptaid, AGK2, CI-amidine and Delphinidin chloride could be parsed with > 85% precision and sensitivity based on variances in SC-35 spatial organization (Fig. 5C,D).
Figure 5.
Using SC-35 organizational metrics, resultant epigenetic cell-states can be parsed immediately post treatment. (A) and (B) show representative SC-35 images taken immediately post treatment and classification table for small molecules that increase osteogenesis. (C) and (D) show representative SC-35 images and classification table for small molecules that inhibit osteogenesis.
Variations in nucleosomal organization post drug treatment and in presence of osteogenic cues can be profiled using SC-35 metrics within 72 hours
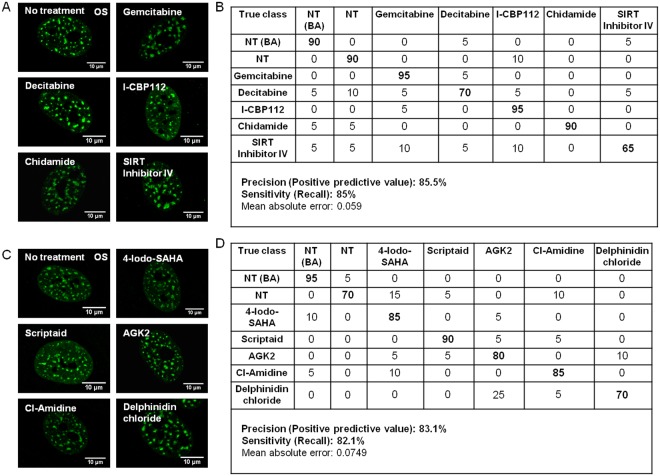
Next, we evaluated if cells pretreated with varied drugs and induced to differentiate using osteogenic cues could be profiled and parsed based on variations in nucleosomal organization present due to drug treatment. Our results indicate that using SC-35 organizational metrics, untreated cells and cells pre-treated with compounds capable of enhancing (Fig. 6A,B) or inhibiting osteogenesis (Fig. 6C,D) could be parsed with >80% precision and sensitivity in 72 hours post differentiation induction. Untreated cells or cells treated with pharmacological agents could also be differentiated from untreated cells cultured in basal condition with high fidelity (Fig. 6B,D). This indicates that the epigenetic modulations and lineage differentiation programs intersect within the nucleosome and can be forecast using SC35 organizational dynamics.
Figure 6.
Image informatics of SC-35 nucleosomal organization can effectively parse the effect of epigenetic drugs on cellular phenotypes (cells were compared with and without drug pretreatment). (A) and (C) Representative SC-35 images of cells cultured in osteogenic medium (OS) for 3 days post treatment with various pharmacological agents identified to enhance and inhibit osteogenesis, respectively. (B) and (D) Classification table.
Identified compounds enhance osteogenesis in hMSCs from an aged donor (>45 years) as well at a late passage number (>P7 hMSCs)
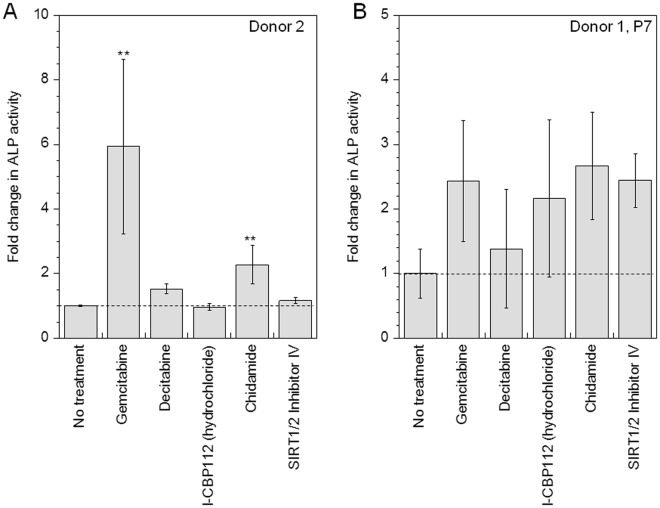
In order to test the hypothesis that epigenetic modifications can overcome regenerative tissue potential from aged donors, hMSCs derived from an aged donor (age: 46 years) were pre-treated with top 5 drugs identified to induce differentiation, namely Gemcitabine, Decitabine, I-CBP112, Chidamide and SIRT1/2 Inhibitor IV, and subsequently induced to differentiate. Osteogenic differentiation was assessed on Day 14 using ALP assay. It was observed, that even for cells from an aged donor, Gemcitabine and Chidamide significantly (p < 0.01) increased osteogenesis by 6- and 2.3- fold respectively, while Decitabine increased osteogenesis by 1.5-fold (Fig. 7A). These results indicate that these drugs effectively increase osteogenesis in hMSCs irrespective of donor age. On the other hand, I-CBP112 and SIRT Inhibitor IV did not modulate differentiation in hMSCs from an older donor.
Figure 7.
Identified drugs enhance osteogenesis in hMSCs from aged donor and at older passage number. (A) and (B) Influence of identified drugs on osteogenic differentiation was analyzed using ALP activity assay in cells from an older donor (age 46 years) and in cells at a late passage number (p7), respectively. Statistical analysis was performed using the Dunnett Multiple Comparisons test, which compares all columns versus a control column (no treatment). The symbol ** represent a significant change in ALP activity with respect to no treatment condition to the level of p < 0.01 and p < 0.05, respectively.
Next, we also evaluated if the identified drugs could enhance osteogenesis in cells at late passage number (P7). The identified drugs were observed to enhanced osteogenesis. (Fig. 7B). An increase in average ALP activity was observed for all five small molecules tested. However, statistical significant differences were not seen due to large error bars because of inherent variability and challenges faced in culturing hMSCs at a late passage number.
Discussion
Transplantation of bone tissues is often needed to repair bones damaged due to severe trauma or disease24,25. Conventional autografts, allografts and xenografts have many associated adverse effects (implant rejection, disease transfer) and complications (cost, surgical risks and injury at donor site)26,27. Consequently, there is heightened demand for an alternative bone repair treatment using biocompatible and functional engineered-bone tissue formed from autologous stem cells. Mesenchymal stem cells (MSCs) are widely believed to be an ideal cell source to engineer bone tissue given their role in natural bone development, due to their availability and lineage-tunability10,12,28. However, the application of adult mesenchymal stem cells remains limited primarily due to decrease in cell’s developmental potential caused by ex-vivo culture and constraints of the advancing age of prospective donors.
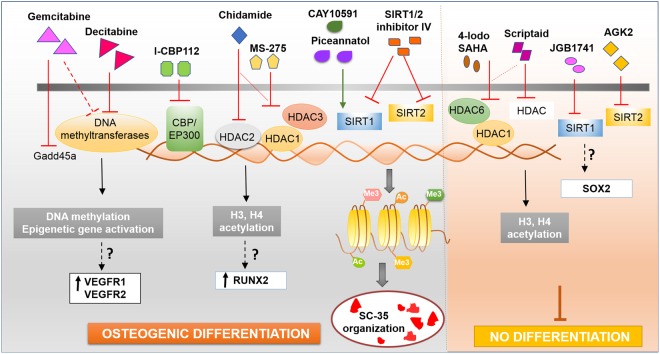
While the role of epigenetics and nucleosomal modifications in cancer progression and treatment has been widely investigated29, the role of epigenetics in healthy stem cells and cellular differentiation remains relatively underexplored. Recently Attema et al. and Zardo et al. indicated that epigenetic mechanisms contribute to controlling stem cell potency and fate of hematopoietic stem cells30,31. Epigenetic modification such as DNA methylation and bivalent histone modifications have been observed to be associated in lineage-affiliated genes and lymphoid-affiliated genes, respectively30. Even so, the role of epigenetic programing in regulating the therapeutic potential of mesenchymal stem cell (MSCs) has not been elucidated. Our results demonstrated that epigenetic modifications regulate MSC differentiation potential and can be modulated using small molecule nucleosomal modifiers to direct optimal differentiation. In this study, we systematically studied 84 compounds and identified molecules capable of significantly influencing osteogenesis through epigenetic modifications. The key top hits identified (Fig. 8) to significantly and maximally increase or inhibit osteogenesis have been discussed in detail below.
Figure 8.
Small molecules that sensitively modulate osteogenic differentiation through epigenetic modifications. The different small molecules influence gene activity, chromatin remodeling and the spatial SC-35 domain organization.
Our study demonstrated that pre-treatment of cells with Gemcitabine and Decitabine significantly increased osteogenesis in hMSCs by 3.5-fold and 2.5-fold, respectively, while maintaining cell proliferation (growth). Gemcitabine and Decitabine are both nucleoside analogs of cytosine and share structural similarities. Gemcitabine specifically inhibits the Growth Arrest and DNA Damage inducible protein 45 a (Gadd45a), a key mediator of active DNA demethylation32, while Decitabine inhibits DNA methyltransferases and causes hypomethylation of cytosine residues in the absence of any significant mutagenic effect33,34. Both drugs have been used to treat various cancers. A recent study comparing the effects of Gemcitabine with Decitabine demonstrated that Gemcitabine functionally inhibits and destabilizes DNA methyltransferases and reactivates epigenetically silenced genes having activity equivalent to decitabine at concentrations significantly lower than those achieved in the treatment of patients with solid tumors35. Interestingly, decitabine and Gemcitabine have been shown to induce both VEGFR1 and VEGFR2 in A549 (adenocarcinoma) cells35. We believe that Gemcitabine and Decitabine enhance osteogenesis in bone marrow derived hMSCs possibly through induction of critical proteins such as VEGFR1 and VEGFR2 via epigenetic modulation, which regulate osteogenic differentiation of hMSCs36,37. Furthermore, the differential regulation of hMSC differentiation by Gemcitabine and decitabine can be a resultant of different mechanisms of actions and potency of the drugs, as decitabine alters DNA CpG methylation while Gemcitabine reactivates (induces) epigenetically silenced genes through an alternate mechanism.
Beyond Gemcitabine and Decitabine, I-CBP112 was the next molecule identified to maximally increase osteogenesis. CBP (CREB (cAMP responsive element binding protein) binding protein (CREBBP)) and P300 (adenovirus E1A-associated 300 kDa protein) are two closely related histone acetyltransferases (HATs) and regulate gene-transcription. I-CBP112 is a selective inhibitor of CBP and EP300 and directly binds their bromodomains that mediate their binding to acetylated lysine residues on histones and other proteins38. While CREB binding protein (CBP) and P300 were shown to play distinct roles in hematopoietic stem cell renewal and differentiation39, this is the first study to indicate their role in regulating MSC differentiation.
Many histone deacetylase inhibitors (HDIs) are among the small molecules identified in this study to epigenetically influence osteogenesis. HDIs are a new category of drugs that inhibit histone deacetylases (HDACs) and have been shown to influence cell growth, cycle, and differentiation. HDACs are further classified into classes depending on their location, for example, Class I HDACs are found in the nucleus and Class II HDACs alternate between the nucleus and cytoplasm. After I-CBP112, Chidamide was the next molecule identified to maximally increase osteogenesis and is a benzamide HD1 that inhibits Class I HDAC1, HDAC2, HDAC3 as well as Class IIb HDAC1040,41. It inhibits epithelial-mesenchymal transition (EMT) in lung cancer cell lines and increases H3 acetylation levels41–43. Chidamide’s influence on osteogenic differentiation could possibly be through H3 acetylation based modifications, as H3 acetylation play a key role in maintaining the balance between genes associated with stem cell self-renewal and genes associated with osteogenic differentiation, for multipotent differentiation potential44, and increases the accessibility of osteocalcin promoter to the osteogenic transcription factors such as RUNX2. In addition to Chidamide, MS-275 is another HDI identified to significantly increase osteogenesis. MS-275 has been shown to preferentially inhibit HDAC1 over HDAC3, and does not inhibit HDAC845. Franci et al. recently showed that treatment with MS-275 modulated ESC fate by enhanced neural differentiation while preventing teratocarcinoma formation46. Our study demonstrates that priming MSCs with MS-275 treatment also modulates their differentiation.
Small molecules maximally inhibiting osteogenesis were also identified in this study. Interestingly, two of the five small molecule drugs found to inhibit MSC differentiation are also classified as HDIs, namely 4-iodo-SAHA and Scriptaid. 4-iodo-SAHA is a hydrophobic derivative of the HDI SAHA (Suberoylanilide Hydroxamic Acid), and is a Class I and Class II inhibitor that affects the activity of both HDAC1 and HDAC6, like SAHA47. SAHA has been shown to induce cell apoptosis in several cancer cell lines and is being applied to cancer therapy47–49. Duncan et al. recently showed that SAHA induced apoptosis in dental pulp cells (DPCs) at high concentration (5 µM), while lower concentrations (1 µM), maintained cell viability and regulated pupal reparative events50. While SAHA (branded as Vorinostat) has been previously shown to enhance osteogenic differentiation of hMSCs51, our results indicate that pre-treatment of cells with 4-Iodo-SAHA (1 µM), significantly inhibited both osteogenic and adipogenic differentiation, yet maintained cell viability. Similar results were observed for Scriptaid, which is also a HDAC inhibitor52 and has been studied in transformed cells52,53. Furthermore, we also tested Trichostatin A, a reversible inhibitor of class I, II, and IV histone deacetylases (HDACs) that has previously indicated in the likely enhancement of osteogenesis18,54. We observed that Trichostatin A did not influence hMCS differentiation (Supplemental Fig. 1). Taken together our results indicate that the varied class I and II HDAC inhibitors (Chidamide, 4-iodo-SAHA, Scriptaid, Trichostatin A, MS-275) have differential effect on hMSC differentiation as they have different mechanistic targets and are cell-type specific.
Another class of molecules that were among the top identified drugs was the SIRT1 and SIRT2 inhibitors and activators. SIRT1 and SIRT2 refers to sirtuin (silent mating type information regulation 2 homolog) 1 and 2 and are nicotinamide adenine dinucleotide (NAD)-dependent histone deacetylases (HDAC). They are considered Class III HDAC and differ from Class I and Class II as they require NAD+ for their deacetylase activity. SIRT1 and 2 have been shown to play a role in cancer55,56, aging57, differentiation58,59, and in regulating metabolic reprogramming and function of human pluripotent stem cells60. SIRT1/2 inhibitor IV, CAY10591 and Piceannatol were among the top identified drugs to enhance osteogenesis and led to >2-fold increase in ALP activity. SIRT1/2 inhibitor IV is a cell-permeable inhibitor of SIRT1 and 2, and their NAD+-dependent deacetylase activity in a substrate competitive manner, without effecting class I and II histone deacetylases61, while CAY10591 and Piceannatol are known to activate SIRT162,63. Recent studies have shown that SIRT1 influences multipotency of bone marrow derived stem cells by regulating SOX2 in the nucleus64, and activation of SIRT1 using resveratrol enhanced osteogenesis in cells64,65. Interestingly, our results indicate that combined inhibition of SIRT1 and 2 using SIRT1/2 inhibitor IV significantly increased osteogenic differentiation (p < 0.01). Our results also show that activation of SIRT1 using CAY1059162 and Piceannatol significantly increased osteogenesis (at least p < 0.05). Interestingly, while Piceannatol, a resveratrol analog66,67, increased ALP activity, treatment with trans-resveratrol, a compound known to inhibit cyclooxygenase 1 (COX-1)68 and activate SIRT169, did not influence osteogenesis in hMSCs (Supplemental Fig. 1). On the other hand, we also observed that specific inhibition of SIRT1 using JGB174170 and specific inhibition of SIRT2 (without effecting SIRT1 and 3), using AGK271, decreased osteogenesis without influencing adipogenesis or decreasing cell viability.
After identifying small molecules that influence osteogenic differentiation of hMCSs in-vitro through epigenetic modifications, we tested the effect of these molecules in mediating osteogenesis in aged hMSCs. Our results showed that Gemcitabine and Chidamide maximally increased osteogenic differentiation in hMSCs obtained from aged donors by 5.9- and 2.3- fold, respectively, and in hMSCs at late passage number by 2.4- and 2.6- fold respectively. Frazen et al. recently showed that epigenetic modifications are associated with senescence in hMSCs72. Our study has further demonstrated that age related loss of differentiation potential can be rescued through modulating epigenetic modifications by innovatively employing small molecules.
Current technologies like chromatin immunoprecipitation methods (e.g., CHIP) to elucidate the nucleosomal alterations in histone modifications are based on population averaged metrics. These methods are unable to catalog minute epigenetic variations in single cells that regulate the heterogeneity of cell-fate decisions73,74. These methods are also unable to profile the changes in in-situ nucleosomal organization elicited due to treatment by small molecules. Previously we have demonstrated that organizational metrics of speckle factor SC-35 are sensitive to signaling molecules mediating osteogenic differentiation in response to external cues and can be employed to parse emergent MSC phenotypes, within 72 hours of exposure20,23. Further, during hMSC differentiation, SC-35 domains co-localize with loci of active gene transcripts in the nucleus leading to increased expression23. SC-35 domain organization is therefore dynamic and regulated by nucleosomal organizational changes and gene transcription. In this study, our results demonstrate that exposure to the various small molecules produces distinct nucleosomal organizational profiles and that speckle factor SC-35 can also be employed as a surrogate marker to profile these in-situ variances in the nucleosomal organization. Using SC-35 organizational metrics and machine learning approaches, cells primed with different small molecules could be parsed with a precision of >85%. Furthermore, our results also indicate that SC-35 organization is highly sensitive (>85%) and modulated by underlying epigenetic modifications as they influence chromatic modeling and gene activation.
In summary, this study demonstrates that small molecules can be applied to modulate stem cell fate and direct cell development. In this study we have successfully identified novel indications for key small molecules to direct cell differentiation of hMSCs through epigenetic modulation. We have further shown that specific identified molecules also increase cellular differentiation in hMSCs obtained from an aged donor as well as in hMSCs at a late passage. Our results elucidate that the underlying epigenetic profile is a pivotal regulator of MSC potential and could afford an approach to design adult stem cell-based tissue engineering strategies. Further studies need to be conducted to decipher the mechanistic role of identified molecules in MSCs and develop strategies for implementing their applications in tissue engineering.
Methods
Cell culture
Human mesenchymal stem cells (hMSCs) were obtained from Texas A&M University (College Station, TX). Cells were cultured in a humidity-controlled environment under 5% CO2 and 37 °C and fed every 3 to 4 days with basal growth media (BA) consisting of Alpha Minimum Essential medium (αMEM) with L-glutamine (Life Technologies) supplemented with fetal bovine serum (10% v/v, Atlanta Biologicals) and penicillin-streptomycin (0.1% v/v, Life Technologies). Cells were received at passage 1 and used for up to 5 passages, for both young and old donors, unless specified otherwise as per experimental conditions. Osteogenic differentiation (OS) was induced by culturing hMSCs in BA media supplemented with 0.5 mM L-ascorbic acid-2-phosphate, 0.2 µM dexamethasone (dex), and 20 mM β-glycerophosphate. Adipogenic differentiation (AD) was induced with BA media supplemented with 1 µM dexamethasone, 50 µM indomethacin, 10 µg/ml insulin, and 100 µM 3-isobutyl-1-methyl-xanthine. Cells were allowed to adhere overnight in basal growth media, followed by a media change with appropriate induction media. All culture reagents were purchased from Sigma-Aldrich unless otherwise specified.
Treatment with small molecule pharmacological agents
An epigenetic screening library of small molecules (Cayman Chemicals) was utilized and 84 compounds from the library were screened to determine the molecules capable of influencing osteogenesis. The library included compounds that modulate the activity of methyltransferases, demethylases, HATs, HDACs and acetylated lysine reader proteins. To evaluate the effect of the small molecules, cells were seeded in basal medium overnight, and subsequently treated with pharmacological agents for 24 hours at concentrations recommended by the manufacturer. 24 hours post drug treatment, the medium was replaced with fresh medium of differentiation medium as per experimental conditions.
Immuno-fluorescence staining
hMSCs were seeded on eight-chamber glass slides (Nunc, Rochester, NY) at a density of 10,000 cells/cm2 and cultured in various conditions as required by the experiment. At desired time point for analysis, cells were fixed in 4% paraformaldehyde in PBS for 15 min, permeabilized with 0.1% Triton-X in PBS for 5 minutes, and blocked by incubating in blocking buffer (5% NGS and 1% BSA in PBS) for 1 hour at room temperature. For performing immunostaining for SC-35, samples were first incubated overnight with primary antibody (Abcam) in blocking buffer at a 1:500 ratio, followed by three 10-minute washes in blocking buffer. Next, samples were incubated with secondary antibody solution (Alexa Fluor; Invitrogen) in blocking buffer at a 1:250 ratio for 1–1.5 hours at room temperature, followed by three 10-minute washes with blocking buffer. All samples were counterstained with 5 μg/ml Hoechst (Sigma) in PBS and stored at 4 °C until imaging.
Differentiation assays
hMSCs were cultured for 14 days in differentiation induction medium prior to assessing differentiation. To analyze adipogenic differentiation, Adipo Red staining assay for staining intracellular triglycerides was performed on fixed cells as per manufacturer’s protocol (Lonza). For analyzing osteogenic differentiation, cells were either fixed or lysed at day 14. Alkaline phosphatase (ALP) was assessed by staining fixed cells using Fast Blue RR (Sigma) or by performing calorimetric assay ALP assay (Biovision Inc) on cell lysate as per manufacturer’s protocol. DNA content in cell lysate was determined by performing the PicoGreen assay (Life Technologies) as per manufacturer’s protocol. Analysis of ALP activity using calorimetric ALP assay was performed and represented after normalization by DNA content.
Assessing cell proliferation
The CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega) was performed to determine the cytotoxicity and proliferation of cells as per manufacturer’s protocol. Briefly, cells were cultured at a density of 10,000 cells/cm2 in a 96 well plate and treated with pharmacological agents. After treatment the media was replaced with fresh basal medium and cells were maintained in basal medium for up to 14 days. To assess cell viability, immediately post treatment and 14 days after treatment, Aqueous One Solution was added in each well to be assayed and incubated for 1 hour. The fluorescence was measured using a plate reader at 490 nm.
High content image analysis of nuclear speckle factor (SC-35) Organization
High content image analysis for SC-35 organization was performed using the similar methodology to the one described in Vega, S et al.20. Briefly, high resolution 1024 × 1024 images were first acquired for cells stained for speckle factor SC-35 using antibody staining and for DNA using Hoechst dye staining (as described above.), using 63x and 1.3 NA objective with a Leica TCS SP8 system (Leica Microsystems). To analyze the organization of SC-35, 26 texture-based Haralick texture features (13 mean and 13 standard deviation or range descriptors) were subsequently acquired for each cell using high content image analysis. First, images underwent intensity-based thresholding to create nuclear ROI masks for each nucleus in a given image, based on Hoechst DNA staining. Next, Haralick descriptors were obtained using a Matlab algorithm. A complete list of the calculated Haralick descriptors with their definitions is provided in Table S1. These descriptors are quantifiable measurements of texture features that represent the spatial organization of the SC-35 in the nucleus. The 26 descriptors were linearly reduced to a minimum number of eigenvectors that account for 95% variance of the data by performing principal component analysis (PCA) using the Weka (Waikato Environment for Knowledge Analysis) open source software (University of Waikato, New Zealand).
To illustrate differences between the various subpopulations, a predictive classification model was made using J48 decision tree analysis in the Weka software. J48 generated a C4.5 pruned decision tree, where tree pruning is used as a tool to correct for potential over fitting. The best performing classification tree was generated by using the experimental data as the training set. The quality of the tree is reported in terms of the percent of correctly classified instances, precision (positive predictive value), and recall (sensitivity). Briefly, Precision = TP/(TP + FP) and Recall = TP/(TP + FN). True positives (TP) are the number of instances correctly classified as belonging to the positive class. False positives (FP) are the number of instances incorrectly classified to the class. False negatives (FN) are the number of instances not classified to the class but belong to class. Precision is also defined as the number of instances that truly have class x among all those which are classified as class x.
Statistics
All statistical analysis was performed using the computer program Instat (GraphPad, San Diego, CA). Experiments were statistically analyzed using the Tukey-Kramer Multiple Comparisons test, which compares all pairs of columns, using a 95% confidence interval or using the Dunnett Multiple Comparisons test which compares all columns versus a control column.
Electronic supplementary material
Acknowledgements
The authors gratefully acknowledge funding from NIH EB001046 (RESBIO, Integrated Resources for Polymeric Biomaterials) and the NSF DGE 0801620 (IGERT on Integrated Science and Engineering of Stem Cells).
Author Contributions
A.D. and P.V.M. designed the experiments. A.D. and S.P. conducted the experiments for Figures 2–7, and supplemental data. D.S.G. designed and conducted experiments for supplemental Figures 4 and 5. A.D. analyzed and plotted the data and performed high content image informatics. A.D. and P.V.M. wrote the text and all authors reviewed the manuscript.
Data Availability Statement
The datasets generated during and/or analyzed in this study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-34511-7.
References
- 1.Chanda D, Kumar S, Ponnazhagan S. Therapeutic potential of adult bone marrow-derived mesenchymal stem cells in diseases of the skeleton. J Cell Biochem. 2010;111:249–257. doi: 10.1002/jcb.22701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caplan AI. Adult mesenchymal stem cells for tissue engineering versus regenerative medicine. J Cell Physiol. 2007;213:341–347. doi: 10.1002/jcp.21200. [DOI] [PubMed] [Google Scholar]
- 3.Hwang NS, Zhang C, Hwang YS, Varghese S. Mesenchymal stem cell differentiation and roles in regenerative medicine. Wiley Interdiscip Rev Syst Biol Med. 2009;1:97–106. doi: 10.1002/wsbm.26. [DOI] [PubMed] [Google Scholar]
- 4.Hiew VV, Simat SFB, Teoh PL. The Advancement of Biomaterials in Regulating Stem Cell Fate. Stem Cell Rev. 2017;1:43–57. doi: 10.1007/s12015-017-9764-y. [DOI] [PubMed] [Google Scholar]
- 5.Das RK, Zouani OF. A review of the effects of the cell environment physicochemical nanoarchitecture on stem cell commitment. Biomaterials. 2014;35:5278–5293. doi: 10.1016/j.biomaterials.2014.03.044. [DOI] [PubMed] [Google Scholar]
- 6.Maredziak M, Marycz K, Tomaszewski KA, Kornicka K, Henry BM. The Influence of Aging on the Regenerative Potential of Human Adipose Derived Mesenchymal Stem Cells. Stem Cells Int. 2016;2016:2152435. doi: 10.1155/2016/2152435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raggi C, Berardi AC. Mesenchymal stem cells, aging and regenerative medicine. Muscles Ligaments Tendons J. 2012;2:239–242. [PMC free article] [PubMed] [Google Scholar]
- 8.Kim M, Kim C, Choi YS, Park C, Suh Y. Age-related alterations in mesenchymal stem cells related to shift in differentiation from osteogenic to adipogenic potential: implication to age-associated bone diseases and defects. Mech Ageing Dev. 2012;133:215–225. doi: 10.1016/j.mad.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 9.Rosland GV, et al. Long-term cultures of bone marrow-derived human mesenchymal stem cells frequently undergo spontaneous malignant transformation. Cancer Res. 2009;69:5331–5339. doi: 10.1158/0008-5472.CAN-08-4630. [DOI] [PubMed] [Google Scholar]
- 10.Amini AR, Laurencin CT, Nukavarapu SP. Bone tissue engineering: recent advances and challenges. Crit Rev Biomed Eng. 2012;40:363–408. doi: 10.1615/CritRevBiomedEng.v40.i5.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sui BD, Hu CH, Zheng CX, Jin Y. Microenvironmental Views on Mesenchymal Stem Cell Differentiation in Aging. J Dent Res. 2016;95:1333–1340. doi: 10.1177/0022034516653589. [DOI] [PubMed] [Google Scholar]
- 12.Sima LE. Extracellular Signals for Guiding Mesenchymal Stem Cells Osteogenic Fate. Curr Stem Cell Res Ther. 2017;12:139–144. doi: 10.2174/1574888X10666151026114411. [DOI] [PubMed] [Google Scholar]
- 13.Lynch Patrick J., Thompson Elaine E., McGinnis Kathleen, Rovira Gonzalez Yazmin I., Lo Surdo Jessica, Bauer Steven R., Hursh Deborah A. Chromatin Changes at thePPAR-γ2Promoter During Bone Marrow-Derived Multipotent Stromal Cell Culture Correlate With Loss of Gene Activation Potential. STEM CELLS. 2015;33(7):2169–2181. doi: 10.1002/stem.1967. [DOI] [PubMed] [Google Scholar]
- 14.Noer A, Lindeman LC, Collas P. Histone H3 modifications associated with differentiation and long-term culture of mesenchymal adipose stem cells. Stem Cells Dev. 2009;18:725–736. doi: 10.1089/scd.2008.0189. [DOI] [PubMed] [Google Scholar]
- 15.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 16.Siggens L, Ekwall K. Epigenetics, chromatin and genome organization: recent advances from the ENCODE project. J Intern Med. 2014;276:201–214. doi: 10.1111/joim.12231. [DOI] [PubMed] [Google Scholar]
- 17.de Boer J, et al. Inhibition of histone acetylation as a tool in bone tissue engineering. Tissue Eng. 2006;12:2927–2937. doi: 10.1089/ten.2006.12.2927. [DOI] [PubMed] [Google Scholar]
- 18.Cho HH, et al. Induction of osteogenic differentiation of human mesenchymal stem cells by histone deacetylase inhibitors. J Cell Biochem. 2005;96:533–542. doi: 10.1002/jcb.20544. [DOI] [PubMed] [Google Scholar]
- 19.Lee S, et al. Histone deacetylase inhibitors decrease proliferation potential and multilineage differentiation capability of human mesenchymal stem cells. Cell Prolif. 2009;42:711–720. doi: 10.1111/j.1365-2184.2009.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vega SL, et al. Organizational metrics of interchromatin speckle factor domains: integrative classifier for stem cell adhesion & lineage signaling. Integr Biol (Camb) 2015;7:435–446. doi: 10.1039/C4IB00281D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shopland LS, Johnson CV, Byron M, McNeil J, Lawrence JB. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J Cell Biol. 2003;162:981–990. doi: 10.1083/jcb.200303131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szczerbal I, Bridger JM. Association of adipogenic genes with SC-35 domains during porcine adipogenesis. Chromosome Res. 2010;18:887–895. doi: 10.1007/s10577-010-9176-1. [DOI] [PubMed] [Google Scholar]
- 23.Dhaliwal A, et al. Profiling stem cell states in three-dimensional biomaterial niches using high content image informatics. Acta Biomater. 2016;45:98–109. doi: 10.1016/j.actbio.2016.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baroli B. From natural bone grafts to tissue engineering therapeutics: Brainstorming on pharmaceutical formulative requirements and challenges. J Pharm Sci. 2009;98:1317–1375. doi: 10.1002/jps.21528. [DOI] [PubMed] [Google Scholar]
- 25.Report. Bone Grafts and Substitutes Market by Product, by Application - Global Opportunity Analysis and Industry Forecast, Allied Market Research, 2014 – 2022 (2016).
- 26.Ebraheim NA, Elgafy H, Xu R. Bone-graft harvesting from iliac and fibular donor sites: techniques and complications. J Am Acad Orthop Surg. 2001;9:210–218. doi: 10.5435/00124635-200105000-00007. [DOI] [PubMed] [Google Scholar]
- 27.Banwart JC, Asher MA, Hassanein RS. Iliac crest bone graft harvest donor site morbidity. A statistical evaluation. Spine (Phila Pa 1976) 1995;20:1055–1060. doi: 10.1097/00007632-199505000-00012. [DOI] [PubMed] [Google Scholar]
- 28.Kobolak J, Dinnyes A, Memic A, Khademhosseini A, Mobasheri A. Mesenchymal stem cells: Identification, phenotypic characterization, biological properties and potential for regenerative medicine through biomaterial micro-engineering of their niche. Methods. 2016;99:62–68. doi: 10.1016/j.ymeth.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 29.Avgustinova A, Benitah SA. The epigenetics of tumour initiation: cancer stem cells and their chromatin. Curr Opin Genet Dev. 2016;36:8–15. doi: 10.1016/j.gde.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Attema JL, et al. Epigenetic characterization of hematopoietic stem cell differentiation using miniChIP and bisulfite sequencing analysis. Proc Natl Acad Sci USA. 2007;104:12371–12376. doi: 10.1073/pnas.0704468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zardo G, Cimino G, Nervi C. Epigenetic plasticity of chromatin in embryonic and hematopoietic stem/progenitor cells: therapeutic potential of cell reprogramming. Leukemia. 2008;22:1503–1518. doi: 10.1038/leu.2008.141. [DOI] [PubMed] [Google Scholar]
- 32.Schafer A, Schomacher L, Barreto G, Doderlein G, Niehrs C. Gemcitabine functions epigenetically by inhibiting repair mediated DNA demethylation. PLoS One. 2010;5:e14060. doi: 10.1371/journal.pone.0014060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 34.Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst. 2005;97:1498–1506. doi: 10.1093/jnci/dji311. [DOI] [PubMed] [Google Scholar]
- 35.Gray SG, et al. Gemcitabine reactivates epigenetically silenced genes and functions as a DNA methyltransferase inhibitor. Int J Mol Med. 2012;30:1505–1511. doi: 10.3892/ijmm.2012.1138. [DOI] [PubMed] [Google Scholar]
- 36.Berendsen AD, Olsen BR. How vascular endothelial growth factor-A (VEGF) regulates differentiation of mesenchymal stem cells. J Histochem Cytochem. 2014;62:103–108. doi: 10.1369/0022155413516347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berendsen AD, Olsen BR. Regulation of adipogenesis and osteogenesis in mesenchymal stem cells by vascular endothelial growth factor A. J Intern Med. 2015;277:674–680. doi: 10.1111/joim.12364. [DOI] [PubMed] [Google Scholar]
- 38.Popp TA, et al. Development of Selective CBP/P300 Benzoxazepine Bromodomain Inhibitors. J Med Chem. 2016;59:8889–8912. doi: 10.1021/acs.jmedchem.6b00774. [DOI] [PubMed] [Google Scholar]
- 39.Rebel VI, et al. Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc Natl Acad Sci USA. 2002;99:14789–14794. doi: 10.1073/pnas.232568499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu L, et al. Chidamide and 5-flurouracil show a synergistic antitumor effect on human colon cancer xenografts in nude mice. Neoplasma. 2016;63:193–200. doi: 10.4149/203_150422N214. [DOI] [PubMed] [Google Scholar]
- 41.Ning ZQ, et al. Chidamide (CS055/HBI-8000): a new histone deacetylase inhibitor of the benzamide class with antitumor activity and the ability to enhance immune cell-mediated tumor cell cytotoxicity. Cancer Chemother Pharmacol. 2012;69:901–909. doi: 10.1007/s00280-011-1766-x. [DOI] [PubMed] [Google Scholar]
- 42.Chang R, You J, Zhou Q. Research advance on mechanism and application of HATs and HDACs in epithelial-mesenchymal transition of lung cancer. Zhongguo Fei Ai Za Zhi. 2013;16:211–215. doi: 10.3779/j.issn.1009-3419.2013.04.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin SH, et al. Chidamide alleviates TGF-beta-induced epithelial-mesenchymal transition in lung cancer cell lines. Mol Biol Rep. 2016;43:687–695. doi: 10.1007/s11033-016-4005-z. [DOI] [PubMed] [Google Scholar]
- 44.Li Z, et al. Epigenetic dysregulation in mesenchymal stem cell aging and spontaneous differentiation. PLoS One. 2011;6:e20526. doi: 10.1371/journal.pone.0020526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu E, et al. Identification of novel isoform-selective inhibitors within class I histone deacetylases. J Pharmacol Exp Ther. 2003;307:720–728. doi: 10.1124/jpet.103.055541. [DOI] [PubMed] [Google Scholar]
- 46.Franci G, et al. The class I-specific HDAC inhibitor MS-275 modulates the differentiation potential of mouse embryonic stem cells. Biol Open. 2013;2:1070–1077. doi: 10.1242/bio.20135587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salmi-Smail C, et al. Modified cap group suberoylanilide hydroxamic acid histone deacetylase inhibitor derivatives reveal improved selective antileukemic activity. J Med Chem. 2010;53:3038–3047. doi: 10.1021/jm901358y. [DOI] [PubMed] [Google Scholar]
- 48.Komatsu N, et al. SAHA, a HDAC inhibitor, has profound anti-growth activity against non-small cell lung cancer cells. Oncol Rep. 2006;15:187–191. [PubMed] [Google Scholar]
- 49.Wang L, et al. Increased expression of histone deacetylaces (HDACs) and inhibition of prostate cancer growth and invasion by HDAC inhibitor SAHA. Am J Transl Res. 2009;1:62–71. [PMC free article] [PubMed] [Google Scholar]
- 50.Duncan HF, et al. The Histone-Deacetylase-Inhibitor Suberoylanilide Hydroxamic Acid Promotes Dental Pulp Repair Mechanisms Through Modulation of Matrix Metalloproteinase-13 Activity. J Cell Physiol. 2016;231:798–816. doi: 10.1002/jcp.25128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu S, et al. Effect of the HDAC inhibitor vorinostat on the osteogenic differentiation of mesenchymal stem cells in vitro and bone formation in vivo. Acta Pharmacol Sin. 2013;34:699–709. doi: 10.1038/aps.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su GH, Sohn TA, Ryu B, Kern SE. A novel histone deacetylase inhibitor identified by high-throughput transcriptional screening of a compound library. Cancer Res. 2000;60:3137–3142. [PubMed] [Google Scholar]
- 53.Keen JC, et al. A novel histone deacetylase inhibitor, scriptaid, enhances expression of functional estrogen receptor alpha (ER) in ER negative human breast cancer cells in combination with 5-aza 2′-deoxycytidine. Breast Cancer Res Treat. 2003;81:177–186. doi: 10.1023/A:1026146524737. [DOI] [PubMed] [Google Scholar]
- 54.Hu X, et al. Histone deacetylase inhibitor trichostatin A promotes the osteogenic differentiation of rat adipose-derived stem cells by altering the epigenetic modifications on Runx2 promoter in a BMP signaling-dependent manner. Stem Cells Dev. 2013;22:248–255. doi: 10.1089/scd.2012.0105. [DOI] [PubMed] [Google Scholar]
- 55.Cha YI, Kim HS. Emerging role of sirtuins on tumorigenesis: possible link between aging and cancer. BMB Rep. 2013;46:429–438. doi: 10.5483/BMBRep.2013.46.9.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeong SM, Haigis MC. Sirtuins in Cancer: a Balancing Act between Genome Stability and Metabolism. Mol Cells. 2015;38:750–758. doi: 10.14348/molcells.2015.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, Liang Y, Vanhoutte PM. SIRT1 and AMPK in regulating mammalian senescence: a critical review and a working model. FEBS Lett. 2011;585:986–994. doi: 10.1016/j.febslet.2010.11.047. [DOI] [PubMed] [Google Scholar]
- 58.Wang M, Yue Z, Paus R, Ramot Y. SIRT2 as a new player in epigenetic programming of keratinocyte differentiation and a candidate tumor suppressor. Exp Dermatol. 2014;23:636–638. doi: 10.1111/exd.12434. [DOI] [PubMed] [Google Scholar]
- 59.Hu B, et al. Repression of SIRT1 promotes the differentiation of mouse induced pluripotent stem cells into neural stem cells. Cell Mol Neurobiol. 2014;34:905–912. doi: 10.1007/s10571-014-0071-8. [DOI] [PubMed] [Google Scholar]
- 60.Cha Y, et al. Metabolic control of primed human pluripotent stem cell fate and function by the miR-200c-SIRT2 axis. Nat Cell Biol. 2017;19:445–456. doi: 10.1038/ncb3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heltweg B, et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 62.Nayagam VM, et al. SIRT1 modulating compounds from high-throughput screening as anti-inflammatory and insulin-sensitizing agents. J Biomol Screen. 2006;11:959–967. doi: 10.1177/1087057106294710. [DOI] [PubMed] [Google Scholar]
- 63.Kawakami S, et al. Piceatannol and its metabolite, isorhapontigenin, induce SIRT1 expression in THP-1 human monocytic cell line. Nutrients. 2014;6:4794–480. doi: 10.3390/nu6114794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoon DS, et al. SIRT1 directly regulates SOX2 to maintain self-renewal and multipotency in bone marrow-derived mesenchymal stem cells. Stem Cells. 2014;32:3219–3231. doi: 10.1002/stem.1811. [DOI] [PubMed] [Google Scholar]
- 65.Lee YM, et al. The role of sirtuin 1 in osteoblastic differentiation in human periodontal ligament cells. J Periodontal Res. 2011;46:712–721. doi: 10.1111/j.1600-0765.2011.01394.x. [DOI] [PubMed] [Google Scholar]
- 66.Potter GA, et al. The cancer preventative agent resveratrol is converted to the anticancer agent piceatannol by the cytochrome P450 enzyme CYP1B1. Br J Cancer. 2002;86:774–778. doi: 10.1038/sj.bjc.6600197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wolter F, Clausnitzer A, Akoglu B, Stein J. Piceatannol, a natural analog of resveratrol, inhibits progression through the S phase of the cell cycle in colorectal cancer cell lines. J Nutr. 2002;132:298–302. doi: 10.1093/jn/132.2.298. [DOI] [PubMed] [Google Scholar]
- 68.Jang M, et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- 69.Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 70.Kalle AM, et al. Inhibition of SIRT1 by a small molecule induces apoptosis in breast cancer cells. Biochem Biophys Res Commun. 2010;401:13–19. doi: 10.1016/j.bbrc.2010.08.118. [DOI] [PubMed] [Google Scholar]
- 71.Outeiro TF, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science. 2007;317:516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 72.Franzen J, Wagner W, Fernandez-Rebollo E. Epigenetic Modifications upon Senescence of Mesenchymal Stem Cells. Current Stem Cell Reports. 2016;2:248–254. doi: 10.1007/s40778-016-0051-7. [DOI] [Google Scholar]
- 73.Altschuler SJ, Wu LF. Cellular heterogeneity: do differences make a difference? Cell. 2010;141:559–563. doi: 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed in this study are available from the corresponding author on reasonable request.