ABSTRACT
Tissue growth needs to be properly controlled for organs to reach their correct size and shape, but the mechanisms that control growth during normal development are not fully understood. We report here that the activity of the Hippo signaling transcriptional activator Yorkie gradually decreases in the central region of the developing Drosophila wing disc. Spatial and temporal changes in Yorkie activity can be explained by changes in cytoskeletal tension and biomechanical regulators of Hippo signaling. These changes in cellular biomechanics correlate with changes in cell density, and experimental manipulations of cell density are sufficient to alter biomechanical Hippo signaling and Yorkie activity. We also relate the pattern of Yorkie activity in older discs to patterns of cell proliferation. Our results establish that spatial and temporal patterns of Hippo signaling occur during wing development, that these patterns depend upon cell-density modulated tissue mechanics and that they contribute to the regulation of wing cell proliferation.
KEY WORDS: Hippo, Stress, Biomechanics, Yorkie, Organ growth
Summary: Temporal and spatial patterns of Hippo signaling seen during Drosophila wing development are related to changes in mechanical stress, resulting from alterations in cell density and cytoskeletal tension.
INTRODUCTION
How organs achieve their correct size and shape remains a key unanswered question in developmental biology (Hariharan, 2015). Observations that mechanical stress can regulate cell proliferation and regulate signaling pathways such as Hippo that influence growth, suggest that mechanical stress could influence growth during development (Eder et al., 2017; Irvine and Shraiman, 2017; LeGoff and Lecuit, 2016). However, whether and how mechanical stress actually contributes to the normal control of growth in developing tissues remains controversial and poorly understood. Here, we describe evidence that increases in cell density that occur during Drosophila wing development regulate a biomechanical Hippo signaling pathway to influence patterns of growth in the developing wing imaginal disc.
Hippo signaling is a conserved signal transduction network that regulates growth and cell fate (Misra and Irvine, 2018; Yu et al., 2015). The central core of this pathway includes a pair of kinases, Hippo and Warts (Wts), which act in sequence to phosphorylate the transcriptional co-activator Yorkie (Yki) (Huang et al., 2005). Phosphorylation of Yki by Wts promotes cytoplasmic localization of Yki, thus reducing Yki-dependent transcription and growth (Dong et al., 2007; Oh and Irvine, 2008). Hippo signaling is regulated by diverse upstream inputs, many of which are dependent upon the local physical environment of cells (reviewed by Meng et al., 2016; Sun and Irvine, 2016). One such input is cytoskeletal tension experienced at adherens junctions, which in Drosophila can be mediated through the tension-dependent recruitment of the Ajuba LIM protein (Jub) to adherens junctions, and the Jub-dependent recruitment and inhibition of Wts (Rauskolb et al., 2014). This mechanism for tension-dependent regulation of Hippo signaling, which we refer to as the Jub biomechanical pathway, is conserved in mammalian cells (Dutta et al., 2017; Ibar et al., 2018).
Initial studies that have demonstrated the influences of mechanical stress on Hippo signaling were based largely on physical, genetic or pharmacological manipulations of cytoskeletal tension (Sun and Irvine, 2016). More recently, we observed that genetically altering growth rates in Drosophila imaginal discs could lead to compression of faster growing cells, and the consequent downregulation of Jub and Wts recruitment to junctions and downregulation of Yki activity (Pan et al., 2016). Related phenomena occur in cultured mammalian cell models, in which cells that are cultured at high density have low YAP (YAP1) activity, whereas cells that are cultured at low density, or are physically stretched, have high YAP activity (Aragona et al., 2013; Benham-Pyle et al., 2015; Zhao et al., 2007). Cell density-dependent effects on YAP correlate with tension-dependent recruitment of LATS proteins to adherens junctions (Dutta et al., 2017; Ibar et al., 2018). These effects of cell crowding provide experimental support and a molecular mechanism for ‘mechanical feedback’, a homeostatic mechanism for maintaining uniform or conformal growth rates in developing tissues (Irvine and Shraiman, 2017; Shraiman, 2005). However, the extent to which differences in Hippo signaling normally occur during development and their potential relationship to variations in mechanical stress have remained poorly understood.
The Drosophila wing imaginal disc has been used extensively as a model for in vivo studies of growth regulation (Hariharan, 2015; Irvine and Harvey, 2015). During larval stages, the wing disc grows from 30-50 cells to 30,000-50,000 cells (Martin et al., 2009; Milan et al., 1996; Worley et al., 2013), with the growth rate gradually declining throughout larval development (Mao et al., 2013; Martin et al., 2009; Wartlick et al., 2011). Hippo signaling is required for normal growth of Drosophila wing discs, as mutations in genes such as wts that increase Yki activity lead to wing overgrowth, whereas loss of Yki activity reduces wing growth (Hariharan, 2015; Irvine and Harvey, 2015). However, variations in Hippo signaling during normal wing development have not been described.
The wing has also been used as a model to study tissue mechanics (Landsberg et al., 2009; LeGoff et al., 2013; Major and Irvine, 2006; Mao et al., 2013; Nienhaus et al., 2009), but how or whether stresses in the wing are linked to Hippo signaling and growth remains unknown. To elucidate spatial and temporal patterns of Hippo signaling, and their potential relationship to mechanical stress, we undertook a detailed analysis of Yki activity, and components of the Jub biomechanical pathway, in wing discs throughout the third larval instar. We identified dynamic changes in Yki activity that occur during wing development and we show that they correlate with changes in components of the Jub biomechanical pathway, with changes in myosin accumulation and with changes in cell density. Experimental manipulations of tension and cell density confirm this linkage, identifying changes in mechanical stress as a contributor to patterns of Yki in the developing wing.
RESULTS
Dynamics of Yki activity during wing disc development
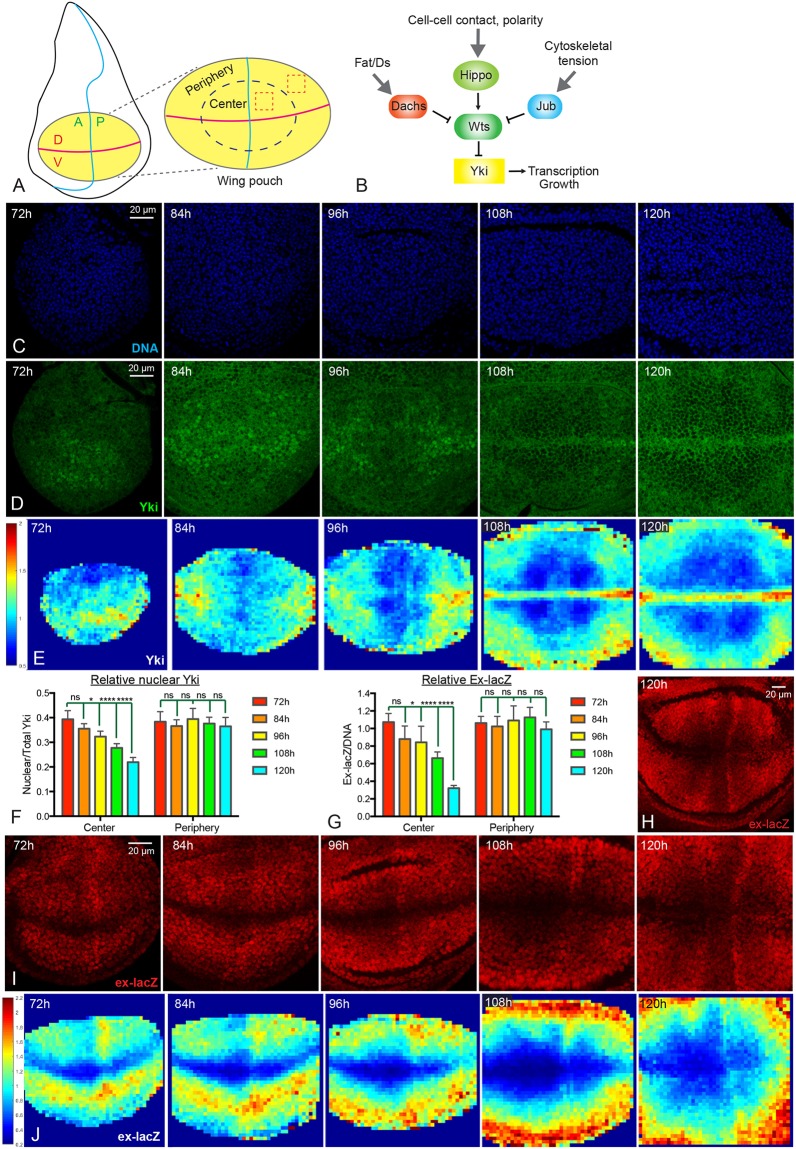
To investigate patterns of Yki activity throughout normal wing growth, we examined Yki protein localization within developing wing discs that were dissected from third instar larvae of different ages [72 h, 84 h, 96 h, 108 h and 120 h after egg laying (AEL)]. Hippo signaling regulates Yki by controlling its localization, and the relative fraction of nuclear Yki is an indicator of Yki activity (Dong et al., 2007; Oh and Irvine, 2008; Zhang et al., 2008). We focused on the region of the disc referred to as the wing pouch (Fig. 1A), which gives rise to the adult wing and is demarcated by folds that form in the disc. Visual inspection of wing discs of different ages revealed that in older wing discs (108 h and 120 h AEL) Yki is predominantly cytoplasmic within the wing pouch, consistent with initial descriptions of Yki localization (Dong et al., 2007; Oh and Irvine, 2008). In contrast, in younger wing discs (72 h and 84 h AEL), Yki is predominately nuclear within the wing pouch (Fig. 1D). There is also a spatial pattern of Yki localization, which changes over time. At 72 h AEL Yki is nuclear in most of the wing pouch, but around the periphery of the wing pouch Yki is distributed between both cytoplasm and nucleus. As wing discs age, nuclear Yki levels decline throughout the center of the wing pouch, such that by 108 and 120 h AEL, Yki is predominantly cytoplasmic in the center of the wing pouch, except along the dorsal-ventral (D-V) compartment boundary. Around the periphery of the wing pouch, however, relative levels of nuclear Yki decline very little during third instar. Thus, in older wing discs (108 h and 120 h AEL) nuclear Yki levels are mostly low in the center and relatively higher around the periphery (Fig. 1D).
Fig. 1.
Dynamics of Yki activity during wing disc development. (A) Schematic of the Drosophila wing disc. The yellow ellipse shows the wing pouch region, which is enlarged on the right. The horizontal magenta line indicates the D-V compartment boundary; the vertical blue line indicates the A-P compartment boundary. The dashed blue ellipse is drawn approximately halfway between the center and edge of the pouch, separating it into central and peripheral regions. Examples of cuboids used for quantification are indicated by dashed red boxes. (B) Simplified schematic of the Hippo pathway which, in the wing disc, is regulated by Fat/Ds signaling through Dachs, by cell contact and cell polarity through Hippo and Wts, and by cytoskeletal tension through Jub. (C,D) Wing discs at 72 h, 84 h, 96 h, 108 h and 120 h AEL stained for DNA (blue, C) and Yki (green, D). Images were captured under identical conditions. (E) Heat maps of relative nuclear Yki intensity in wing discs from 72 to 120 h AEL. In this and subsequent figures a scale for the heat maps is indicated on the left. Number of wing discs used for analysis: 72 h, n=6; 84 h, n=5; 96 h, n=5; 108 h, n=5; 120 h, n=6. (F,G) Histograms showing relative Yki levels (F, nucleus/total) and relative ex-lacZ levels (G) in the central and peripheral regions of wing discs at different stages. Error bars indicate 95% confidence interval. For Yki, 72 h, n=11; 84 h, n=14; 96 h, n=13; 108 h, n=12; 120 h, n=12. For ex-lacZ, n=12 for each time point. Statistical comparisons are shown relative to the 72 h timepoint. (H,I) Wing discs at 72 h, 84 h, 96 h, 108 h and 120 h AEL stained for the expression of ex-lacZ (red). Images were taken under the same conditions. H is a lower magnification image of ex-lacZ at 120 h AEL. (J) Heat maps of relative nuclear ex-lacZ intensity of wing discs from 72 to 120 h AEL. Number of wing discs used for analysis: 72 h, n=5; 84 h, n=5; 96 h, n=5; 108 h, n=5; 120 h, n=5. ns, P>0.05; *P≤0.05; ****P≤0.0001. Scale bars: 20 µm.
We used two approaches to quantify relative nuclear Yki levels. First, we divided the wing pouch into central and peripheral regions, taking half the distance between the center and edges of the pouch as the border between center and periphery (Fig. 1A). The ratio of nuclear (defined by DNA staining) to total Yki was then measured within equally sized cuboids in each central and peripheral quadrant. Cells along the compartment boundaries were excluded from this analysis because of their distinct gene expression profiles and biophysical properties compared with other wing cells. In a second approach, we divided wing disc confocal stacks into 4 µm squares in the xy plane, and then calculated the relative nuclear Yki levels within three-dimensional (3D) cuboids defined by each square to generate a quantitative intensity map of nuclear Yki. Intensity maps from several discs were averaged and relative nuclear Yki levels displayed on a red (high) to blue (low) heat map. These quantitative analyses confirmed the visual impression of declining nuclear Yki levels in the center of the wing pouch throughout third instar larval growth, resulting in a spatial profile in older wing discs with nuclear Yki lower in the center of the wing pouch than around the periphery, except along the D-V boundary (Fig. 1E,F).
To confirm that these changes in Yki localization reflect changes in Yki activity, we examined and quantified the expression of two direct targets of Yki, expanded (ex) and Diap1. Consistent with the dynamics of Yki localization, relative levels of ex transcription, monitored using a nuclear-localized ex-lacZ reporter, gradually decrease in the center of the wing pouch from 72 to 120 h AEL, but remain similar at the peripheral wing pouch (Fig. 1G-J). Hence in older wing discs (108 h and 120 h AEL), ex-lacZ expression is lower in the center than at the periphery, as has been noted previously (Fletcher et al., 2015). Relative Diap1 intensity also declines in the central wing pouch during third instar, resulting in a spatial pattern in older wing discs in which Diap1 levels are lower in the center, except along the compartment boundaries (Fig. S1). Taken together, these observations confirm the finding that Yki activity declines in the center of the wing pouch during larval development, leading to spatial differences in Yki activity between central and peripheral cells.
Dynamics of Jub biomechanical signaling during wing disc development
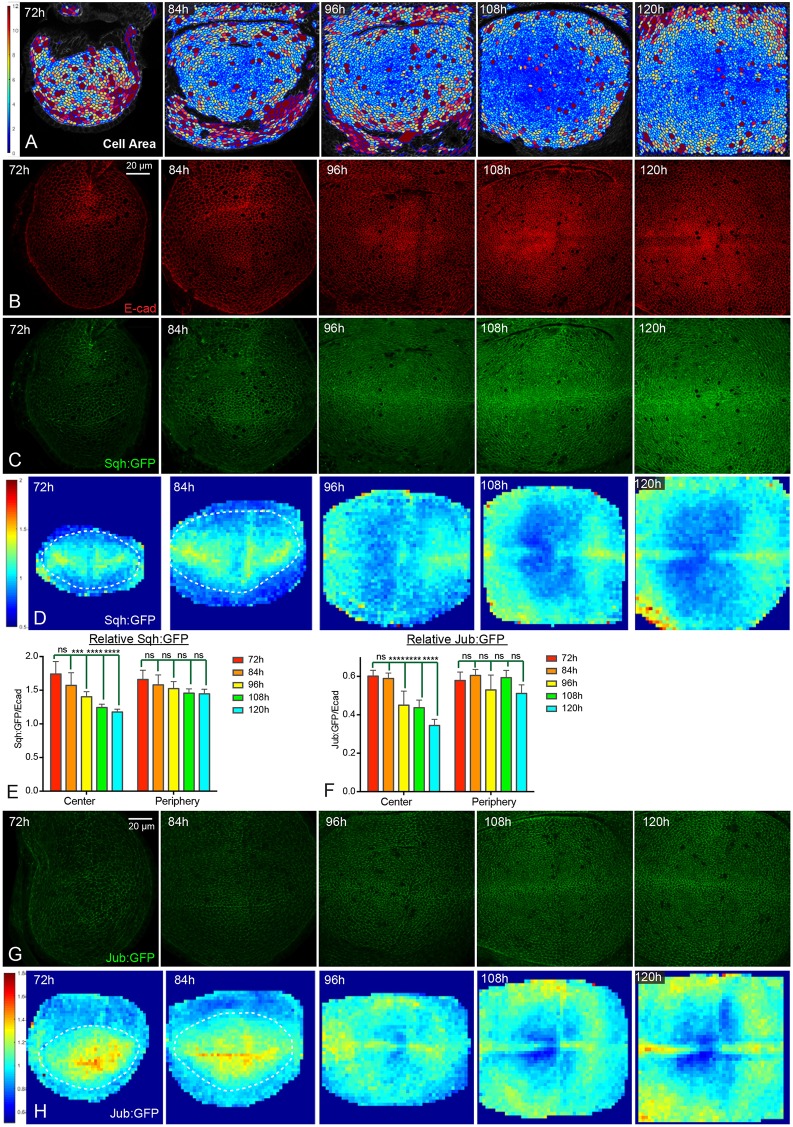
The temporal and spatial pattern of Yki activity is reminiscent of changes in cell shape and cytoskeletal tension that occur during wing development. For example, the recoil velocity of cell junctions after laser ablation can provide an indication of relative tension, and such experiments have revealed higher tension in younger wing discs compared with older wing discs, and within older wing discs, higher tension in peripheral regions compared with central regions (LeGoff et al., 2013; Mao et al., 2013; Rauskolb et al., 2014). Changes in tension are thought to stem, at least in part, from crowding of cells as the disc grows, and examination of apical cell areas indicates that cells are more crowded in the center of older wing discs than they are in the periphery of the wing pouch, or in younger discs (Aegerter-Wilmsen et al., 2012; LeGoff et al., 2013; Mao et al., 2013; Rauskolb et al., 2014) (Fig. 2A).
Fig. 2.
Dynamics of Jub biomechanical signaling during wing disc development. (A) Cell area color maps of wing discs at 72 h, 84 h, 96 h, 108 h and 120 h AEL. Cell areas are shown from 0 µm2 (blue) to 12 µm2 (violet). Cells larger than 12 µm2 have the same color as the 12 µm2 ones. (B,C) Wing discs stained for E-cad (red, B) and expressing Sqh:GFP (green, C) at 72 h, 84 h, 96 h, 108 h and 120 h AEL. Images were taken under the same conditions. (D) Heat map of relative junctional Sqh:GFP intensity of wing discs from 72 h to 120 h AEL. The wing pouch is demarcated with dashed lines for 72 h and 84 h as the wing pouch does not occupy the entire field analyzed here. Number of wing discs used for analysis: 72 h, n=6; 84 h, n=5; 96 h, n=5; 108 h, n=5; 120 h, n=5. (E,F) Histograms showing relative junctional Sqh:GFP levels (E) and relative junctional Jub:GFP levels (F) in the central and peripheral regions of wing discs at different stages. Error bars indicate 95% confidence interval. For Sqh:GFP, n=14 (72 h, 84 h or 108 h) or 15 (96 h or 120 h). For Jub:GFP n=12 (72 h, 84 h or 96 h), 13 (108 h) or 14 (120 h). Statistical comparisons are shown relative to the 72 h timepoint. (G) Wing discs expressing Jub:GFP (green) at 72, 84 h, 96 h, 108 h and 120 h AEL. Images were taken under the same conditions. (H) Heat map of relative junctional Jub:GFP intensity of wing discs from 72 h to 120 h AEL (red, high; blue, low). The wing pouch is demarcated with dashed lines for 72 and 84 h as the wing pouch does not occupy the entire field analyzed here. Number of wing discs used for analysis: 72 h, n=6; 84 h, n=5; 96 h, n=5; 108 h, n=6; 120 h, n=5. ns, P>0.05; ***P≤0.001; ****P≤0.0001. Scale bars: 20 µm.
To further investigate the relationship between developmental patterns of Yki activity and patterns of cytoskeletal tension, we examined the levels of Non-muscle myosin II (myosin) using Myosin light chain (Spaghetti squash, Sqh) or heavy chain (Zipper, Zip) GFP fusion proteins. Levels of myosin along cell-cell junctions (which were defined by staining for E-cadherin, E-cad) were quantified and compared with the levels of E-cad. Overall levels of both Sqh:GFP and E-cad appear to gradually increase within the wing pouch from 72 h to 120 h AEL (Fig. 2B,C, Fig. S2D-G). However, when junctional Sqh:GFP levels are quantified and normalized to E-cad levels, the relative myosin per junction (Sqh:GFP/E-cad ratio) declines over time (Fig. 2E). Moreover, junctional Sqh:GFP declines to a greater extent in the center of the wing pouch than in the periphery, such that in older wing discs (108 and 120 h AEL) junctional myosin is generally lower in the center than at the periphery, except along compartment boundaries (Fig. 2D,E). These spatial and temporal patterns of junctional myosin are consistent with examinations of apical cell areas and measurements of junctional tension (Aegerter-Wilmsen et al., 2012; LeGoff et al., 2013; Mao et al., 2013; Rauskolb et al., 2014) (Fig. 2A). We also note that the cellular distribution of apical myosin differs between younger and older wing discs. At 72 h AEL, apical myosin is predominantly junctional, whereas at 120 h AEL apical myosin is predominantly junctional in peripheral regions, but both junctional and medial in central regions (Fig. S2A-C).
The spatial and temporal patterns of junctional myosin during wing disc development correlate with the pattern of Yki activity. Tension at adherens junctions promotes Yki activity by recruiting a Jub-Wts complex that inhibits Wts activity (Rauskolb et al., 2014). To investigate whether the spatial and temporal patterns of Yki and myosin activity might be connected through tension-dependent regulation of Jub and Wts, we analyzed their localization throughout the third instar. This revealed that junctional Jub and Wts intensities, relative to E-cad, decline in the center of the wing pouch, whereas they remain similar in the periphery of the wing pouch (Fig. 2F-H, Fig. S2D-J). This matches the spatial and temporal dynamics of junctional Sqh:GFP localization and results in a spatial pattern in older wing discs (108 h and 120 h AEL) with lower levels of junctional Jub and Wts relative to E-cad in the center of the wing pouch, except along the D-V boundary.
Temporal differences in response to altered tension
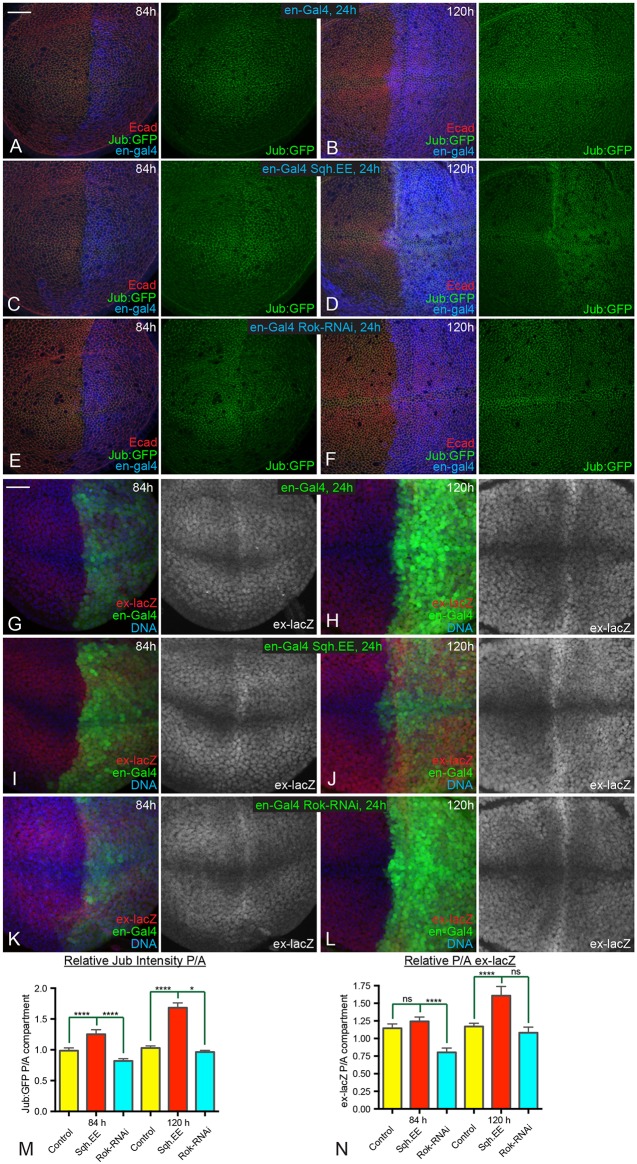
The spatial and temporal correlations among patterns of junctional myosin, Jub, Wts and Yki activity imply that cytoskeletal tension contributes to patterns of Yki activity during development, and that this occurs through the Jub biomechanical pathway. To test this, we compared the consequences of a 24 h change in cytoskeletal tension within younger (84 h) versus older (120 h) wing discs. We reasoned that if tension is normally higher in younger discs, then younger discs should be more sensitive than older discs to decreases in tension, whereas older discs should be more sensitive than younger discs to increases in tension. Tension was decreased by expressing a UAS-RNAi transgene targeting Rho-associated kinase (Rok), or increased by expressing an activated form of myosin light chain (Sqh.EE) under UAS control. These transgenes were expressed in the posterior half of the developing wing disc by placing them under control of en-Gal4, enabling anterior cells to be used as an internal control. Expression of UAS transgenes was made conditional by including a temperature-sensitive allele of Gal80 (Gal80ts).
Increasing tension for 24 h was sufficient to increase recruitment of Jub to adherens junctions, but the increase in Jub was greater in older wing discs than it was in younger wing discs (Fig. 3C,D,M). Conversely, decreasing tension for 24 h was sufficient to decrease recruitment of Jub to adherens junctions, but the decrease in Jub was greater in younger wing discs than it was in older wing discs (Fig. 3E,F,M). These differences between younger and older wing discs are consistent with inferences that junctional tension decreases in wing disc cells at older stages. Similar differential responses to altered tension were observed when we examined the Yki target ex-lacZ. Increasing tension significantly augmented ex-lacZ expression at 120 h AEL, particularly within the central wing pouch, whereas ex-lacZ expression at 84 h AEL was not significantly increased (Fig. 3I,J,N). Decreasing the tension substantially reduced ex-lacZ expression at 84 h AEL, but at 120 h AEL the decrease was not significant (Fig. 3K,L,N). These results further support the conclusion that spatial and temporal differences in tension occur during wing disc development, and that they influence the Jub biomechanical pathway and Yki activity.
Fig. 3.
Temporal differences in response to altered tension. (A-F) Wing discs expressing en-Gal4 UAS-dcr2 tub-Gal80ts Jub:GFP (green) in controls (A,B), UAS-Sqh.EE (C,D) and UAS-Rok-RNAi (E,F) stained for E-cad (red) and Dcr2 (blue). Larvae were cultured at 18°C, shifted to the restrictive temperature for Gal80ts (29°C) for 24 h before dissection at ages corresponding to 84 h (A,C,E) or 120 h (B,D,F) AEL as indicated. (G-L) Wing discs expressing en-Gal4 UAS-GFP (green) UAS-dcr2 tub-Gal80ts ex-lacZ in controls (G,H), UAS-Sqh.EE (I,J) and UAS-Rok-RNAi (K,L) stained for expression of ex-lacZ (red/white) and DNA (blue). Larvae were cultured at 18°C, shifted to the restrictive temperature for Gal80ts (29°C) for 24 h before dissection at ages corresponding to 84 h (G,I,K) or 120 h (H,J,L) AEL as indicated. (M,N) Histograms showing relative posterior to anterior (P/A) Jub:GFP (M) and ex-lacZ (N) intensities, normalized to E-cad or DNA, respectively, for the indicated genotypes at 84 h and 120 h AEL. Error bars indicate 95% confidence interval. n=12 for all time points. Statistical comparisons are shown relative to the control. ns, P>0.05; *P≤0.05; ****P≤0.0001. Scale bars: 20 µm.
Temporal differences in response to elevated Wts
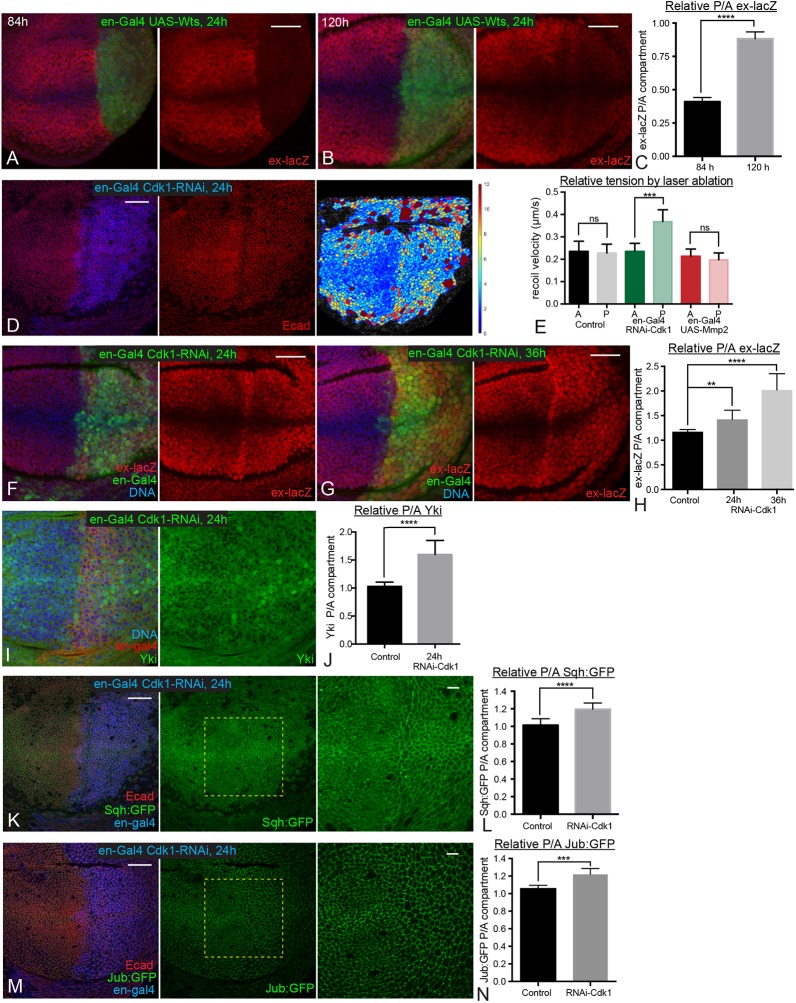
Another implication of our observation of temporal differences in tension-dependent Jub regulation, is that the Jub biomechanical pathway contributes more to promoting Yki activity in younger discs than it does in older discs. As Jub promotes Yki activity by inhibiting Wts (Das Thakur et al., 2010; Rauskolb et al., 2014; Sun et al., 2015), we reasoned that younger wing discs might therefore be more sensitive to increases in Wts levels than older wing discs. To test this hypothesis, we induced Wts overexpression for 24 h in posterior cells using the conditional system described above. Remarkably, this led to a strong reduction in ex-lacZ expression in 84 h AEL discs, but only a weak reduction in ex-lacZ expression in 120 h AEL discs (Fig. 4A-C). This observation further supports the conclusion that differences in cytoskeletal tension, acting through the Jub biomechanical pathway, modulate patterns of Yki activity during development.
Fig. 4.
Temporal differences in response to elevated Wts or reduction of cell density. (A,B) Wing discs expressing en-Gal4 UAS-GFP tub-Gal80ts UAS-Wts stained for DNA (blue) and ex-lacZ (red). Larvae were cultured at 18°C, shifted to the restrictive temperature for Gal80ts (29°C) for 24 h before dissection and dissected at ages corresponding to 84 h (A) and 120 h (B) AEL. (C) Histogram showing quantitation of relative posterior to anterior (P/A) ex-lacZ intensity for the wing discs at 84 h and 120 h AEL with Wts overexpression. n=14 at 84 h and 12 at 120 h. (D) Wing disc expressing en-Gal4 UAS-dcr2 tub-Gal80ts UAS-Cdk1-RNAi stained for Dcr2 (blue) and E-cad (red). Larvae were cultured at 18°C, shifted to the restrictive temperature for Gal80ts (29°C) for 24 h before dissection at an age corresponding to 108 h AEL. The rightmost panel is a cell area color map (blue to violet, 0 µm2 to 12 µm2). (E) Average recoil velocities after laser cutting of cell junctions in anterior (A) or posterior (P) compartments of wing discs from control animals, or those expressing cdk1 RNAi or Mmp2 in posterior cells. n=17 (control), 19 (cdk1 RNAi) or 19 (UAS-Mmp2). (F,G) Wing discs expressing en-Gal4 UAS-GFP UAS-dcr2 tub-Gal80ts UAS-Cdk1-RNAi ex-lacZ stained for expression of ex-lacZ (red) and DNA (blue). Larvae were cultured at 18°C, shifted to the restrictive temperature for Gal80ts (29°C) for 24 h (F) or 36 h (G) before dissection. (I) Wing disc expressing en-Gal4 UAS-dcr2 tub-Gal80ts UAS-Cdk1-RNAi stained for DNA (blue) and Yki (green). Larvae were cultured at 18°C, shifted to the restrictive temperature for Gal80ts (29°C) for 24 h before dissection. (K,M) Wing discs expressing en-Gal4 UAS-dcr2 tub-Gal80ts UAS-Cdk1-RNAi Sqh:GFP (K) or Jub:GFP (M) stained for Dcr2 (blue) and E-cad (red). Larvae were cultured at 18°C and shifted to the restrictive temperature for Gal80ts (29°C) for 24 h before dissection. Pictures on the right show higher magnification of boxed area in central panel. (H,J,L,N) Histograms showing relative posterior to anterior (P/A) ex-lacZ (H), nuclear Yki (J), Sqh:GFP (L) and Jub:GFP (N) intensity, normalized to DNA (H,J) or E-cad (L,N) for wing discs of the indicated genotypes. n=12 (ex-lacZ, Sqh:GFP and Jub:GFP) or 22 (Yki). Error bars indicate 95% confidence interval. ns, P>0.05; **P≤0.01; ***P≤0.001; ****P≤0.0001. Scale bars: 20 µm.
Influence of cell density on cytoskeletal tension and Hippo signaling
The decreasing junctional tension in the central wing pouch as wing discs age correlates with increasing cell density, as does the difference in tension between central and peripheral regions of late third instar wing discs (Aegerter-Wilmsen et al., 2012; LeGoff et al., 2013; Mao et al., 2013; Rauskolb et al., 2014). There is both theoretical and experimental support for the hypothesis that cytoskeletal tension decreases as cells become more crowded (Ibar et al., 2018; Noll et al., 2017; Pan et al., 2016; Puliafito et al., 2012). To directly test the relationship between crowding and junctional tension in the developing wing, and the influence of cellular crowding on the Jub biomechanical pathway during wing development, we assessed the consequences of experimentally altering cell density.
To decrease cell density, we took advantage of observations that knockdown of Cyclin-dependent kinase 1 (Cdk1) leads to both lower cell density and higher Yki activity (Montes and Morata, 2017). To determine whether this increase in Yki activity could be due to elevated junctional tension within the lower density cells, we examined Sqh:GFP and Jub:GFP in wing discs 24 h after en-Gal4-driven RNAi knockdown of Cdk1, at a stage roughly equivalent to 108 h AEL. This 24 h induction of cdk1 RNAi reduced cell density (Fig. 4D) and also increased ex-lacZ expression and nuclear localization of Yki (Fig. 4F-J), consistent with observations of Montes and Morata (2017). Examination of Sqh:GFP revealed increased levels of junctional myosin (Fig. 4K,L). A difference in junctional tension between anterior (control) cells and posterior (cdk1 RNAi) cells was further confirmed by measuring the recoil velocity of cell junctions after laser cutting (Fig. 4E). Examination of Jub:GFP confirmed that the increased junctional myosin and tension are associated with increased junctional recruitment of Jub (Fig. 4M,N). Therefore, reducing cell density by the knockdown of Cdk1 is sufficient to enhance junctional tension and Jub recruitment to junctions, which could account for the increased Yki activity that was observed.
To increase cell density, we used Minute mutations, which are mutations that affect ribosome function and cause a dominant slow growth phenotype (Marygold et al., 2007). In heterozygous Minute/+ animals, making a whole wing compartment wild type (+/+) through compartment-specific mitotic recombination that is induced by localized expression of Flipase, can lead to wing discs with a faster growing wild-type compartment in an otherwise slow-growing Minute heterozygous animal (Martin and Morata, 2006). In developing discs, this can result in posterior +/+ compartments with a higher cell density than anterior Minute/+ compartments, because of their faster growth (Fig. 5A-C). These wild-type posterior compartments also exhibit lower myosin intensity, lower Jub recruitment to junctions and lower Yki activity, compared with the Minute heterozygous cells in the anterior compartment (Fig. 5E-L). These effects appear to be more pronounced in the central wing pouch, in which cell density is highest. Although there is also some cell death in these discs, it is mostly localized near the anterior-posterior (A-P) compartment boundary (Fig. 5D) and so cannot account for the compartment-wide differences in myosin, Jub and Yki. These observations further support the conclusion that cell density regulates Yki activity during wing development and that it does so through influences on junctional tension.
Fig. 5.
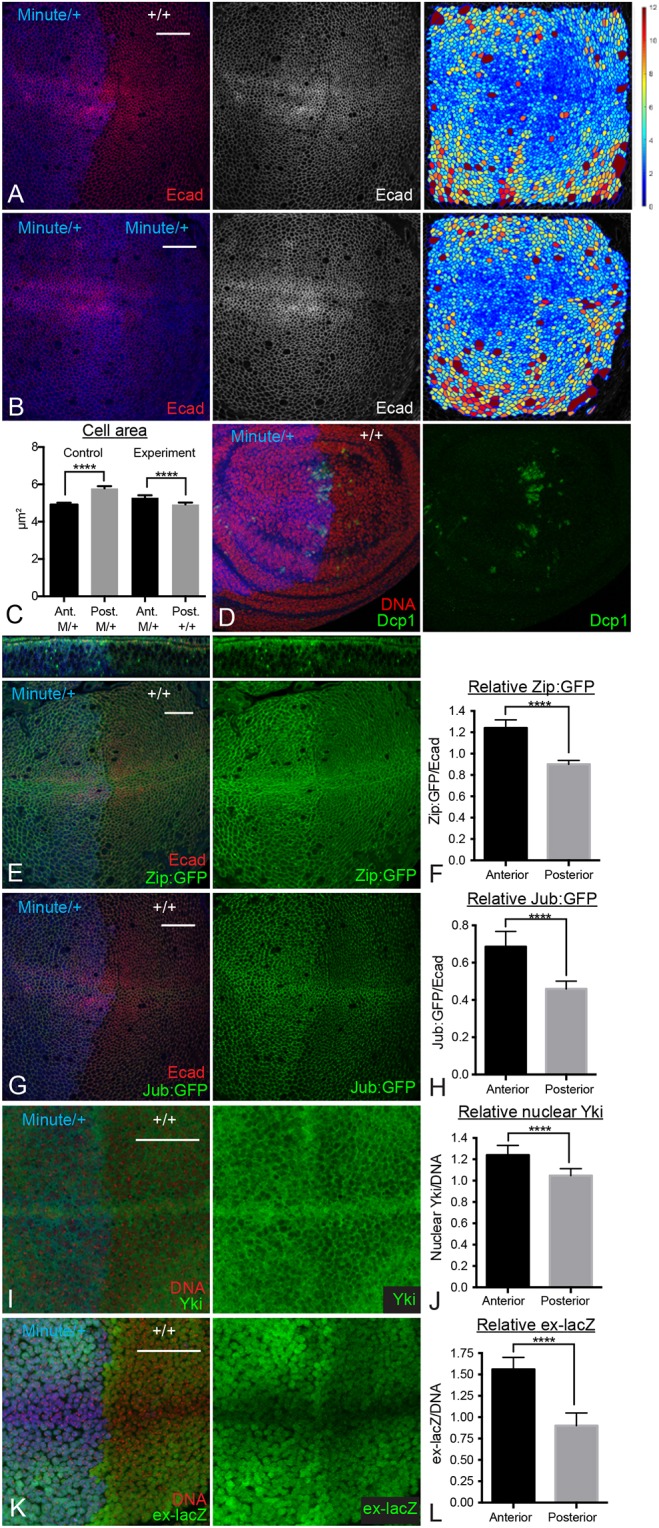
Increasing cell density reduces cytoskeletal tension and Yki activity. (A,D,E,G,I,K) Wing discs with posterior compartment composed of faster-growing wild-type (+/+) cells and the anterior compartment composed of slower-growing Minute (Rps17)/+ cells, at 132 h AEL, which corresponds to ∼108 h AEL in the wild type. The anterior compartment is marked by staining of β-galactosidase (A,B,D,E,G,I; blue) or His:RFP (K; blue). Wing discs were also stained for E-cad (red/white) or DNA (red). (A) Wing disc stained for E-cad (red/white), panel on the right shows a cell area color map (blue to violet, 0 µm2 to 12 µm2). (D) Staining for cleaved Dcp1 (green) reveals modest amounts of cell death, similar between the two compartments, except for some clusters of cell death near the A-P compartment boundary. (E) Wing disc showing Zip:GFP (green). Upper panels show a z slice through the disc. Junctional Zip:GFP decreased in the posterior compartment. (G,I,K) Wing discs showing Jub:GFP (G) or stained for Yki (I) or ex-lacZ (K). (B) Control Minute/+ wing disc stained for E-cad (red/white), panel on the right shows a cell area color map (blue to violet, 0 µm2 to 12 µm2). (C) Quantitation of average cell areas throughout the wing pouch of anterior and posterior compartments in control (M/+ posterior compartment) and experimental (+/+ posterior compartment) wing discs. (F,H,J,L) Histograms showing anterior and posterior relative levels of junctional Zip:GFP (F), junctional Jub:GFP (H), nuclear Yki (J) or ex-lacZ (L), normalized to E-cad (F,H) or DNA (J,L) in Minute/+ vs +/+ wing discs. n=12 for all experiments. Error bars indicate 95% confidence interval. ****P≤0.0001. Scale bars: 20 µm.
Degrading basement membrane does not increase cytoskeletal tension
Our observations, which confirm a key role for tissue mechanics in modulating growth during development, contrast with a recent paper claiming to show the opposite (Ma et al., 2017). That study was based on experimental manipulations of basement membrane, which can influence cell density, but did not actually include any experimental assessments of tissue mechanics. To resolve the potential discrepancy between that study and ours, we examined wing discs in which basement membrane was degraded by expression of Matrix metalloprotease 2 (Mmp2). Consistent with previous observations (Ma et al., 2017; Pastor-Pareja and Xu, 2011), we found that expression of Mmp2 for 18 h or 24 h to degrade basement membrane altered cell shape, leading to flatter cells with larger apical areas, but did not significantly affect Yki activity (Fig. 6). However, examination of Sqh:GFP and Jub:GFP, as well as laser cutting of cell junctions, revealed that this degradation of basement membrane did not significantly affect junctional tension (Figs 4E and 6C-F). Thus, although disrupting basement membrane leads to flatter cells with larger apical areas, it does not increase junctional tension, which explains why Yki activity is not increased despite the reduced cell density.
Fig. 6.
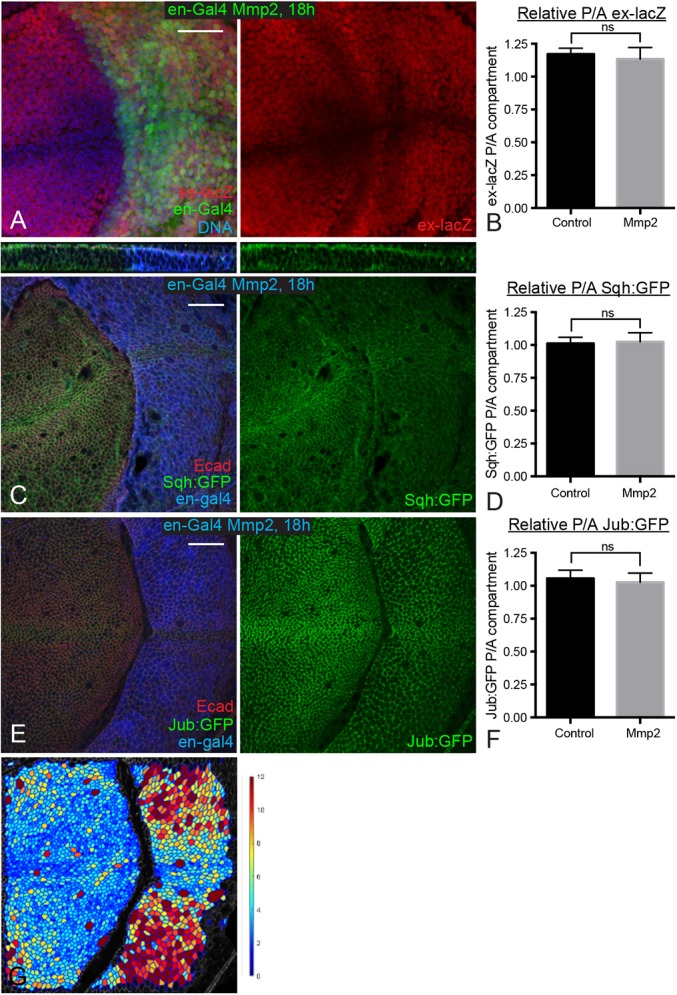
Degrading basement membrane does not increase cytoskeletal tension. (A) Wing disc expressing en-Gal4 UAS-GFP (green) UAS-Dcr2 tub-Gal80ts UAS-Mmp2 ex-lacZ stained for ex-lacZ (red) and DNA (blue). Larvae were cultured at 18°C, shifted to the restrictive temperature for Gal80ts (29°C) for 18 h before dissection. (C,E) Wing discs expressing en-Gal4 UAS-dcr2 tub-Gal80ts UAS-Mmp2 and Sqh:GFP (C) or Jub:GFP (E) were stained for Dcr2 (blue) and E-cad (red). Larvae were cultured at 18°C, shifted to the restrictive temperature for Gal80ts (29°C) for 18 h before dissection. The top panel in C shows a z section through the disc. (B,D,F) Histograms showing relative posterior to anterior (P/A) ex-lacZ (B), Sqh:GFP (D) and Jub:GFP (F) intensity for wing discs of the indicated genotypes, normalized to DNA (B) or E-cad (D,F). n=12 for all experiments. Error bars indicate 95% confidence interval. (G) A cell area color map of the disc shown in panel E shows quantitation of cell area, blue to violet, 0 µm2 to 12 µm2. ns, P>0.05. Scale bars: 20 µm.
Patterns of cell proliferation during wing disc development
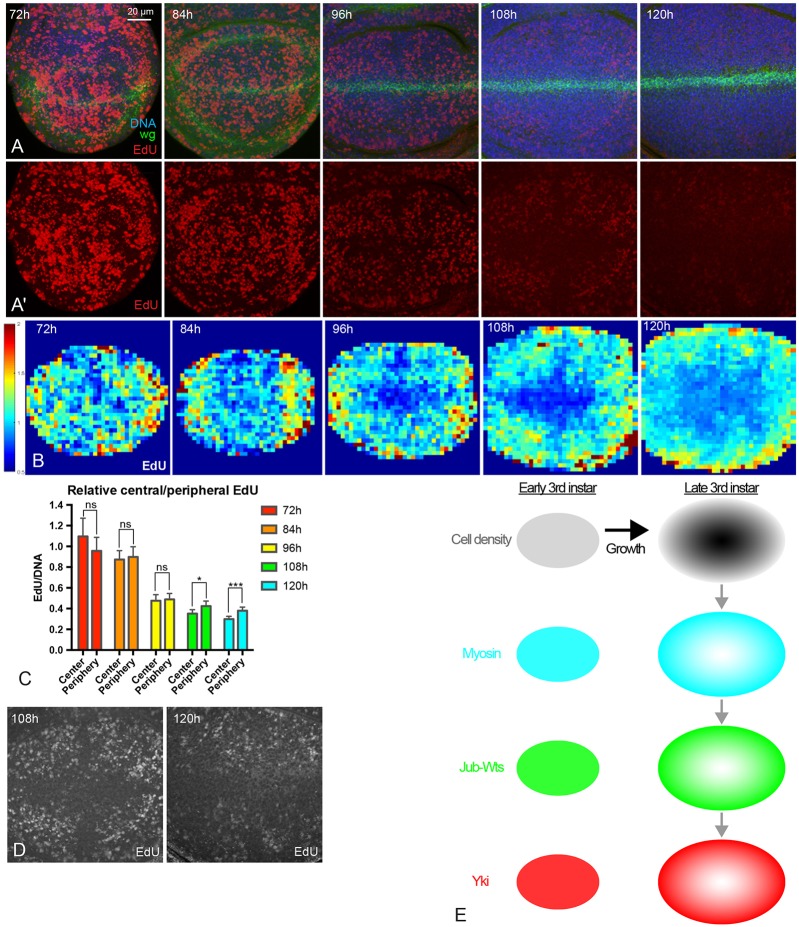
To investigate potential contributions of the spatial and temporal patterns of Yki activity that we identified to patterns of cell proliferation, we labeled proliferating cells by 5-ethynyl-2′-deoxyuridine (EdU) incorporation at 72 h, 84 h, 96 h, 108 h and 120 h AEL. We then quantified the EdU signal, normalized to total DNA labeling. EdU staining performed under identical conditions shows much lower rates of incorporation in older discs than in younger discs (Fig. 7A-D), which is consistent with studies showing that rates of growth and cell proliferation decline throughout the third larval instar (Mao et al., 2013; Martin et al., 2009; Wartlick et al., 2011). Notably, older discs (108 h and 120 h AEL) exhibit a spatial pattern of EdU labeling that appears to be largely similar to (except along the compartment boundaries, which were excluded from our quantitation) the pattern of Yki activity – higher in the proximal wing pouch, and lower in the central wing pouch. This observation suggests that the spatial pattern of Yki activity contributes to the spatial pattern of proliferation, consistent with its well-established role in promoting growth within the wing disc. We also directly confirmed effects of Yki on EdU labeling by reducing Yki levels with a 24 h induction of Yki RNAi, and by activating Yki with a 24 h induction of the activated Yki isoform YkiS168A (Fig. S3). This further supports the inference that spatial correlations between Yki activity and EdU labeling reflect a contribution of patterns of Yki activity in the wing disc to patterns of cell proliferation.
Fig. 7.
Patterns of cell proliferation during wing disc development. (A,A′) Wing discs at 72 h, 84 h, 96 h, 108 h and 120 h AEL stained for DNA (blue), Wg (green) and EdU (red). Pictures were taken under the same conditions. A′ shows only the EdU stain from the images in A. (B) Heat maps of relative EdU intensity of wing discs from 72 h to 120 h AEL (red, high; blue, low). Number of wing discs used for analysis: 72 h, n=5; 84 h, n=6; 96 h, n=5; 108 h, n=5; 120 h, n=5. (C) Histogram showing relative EdU intensity in the central and peripheral regions of wing discs at different stages. n=12 to 15 discs/timepoint. Error bars indicate 95% confidence interval. ns, P>0.05; *P<0.05; ***P<0.001. (D) Images with enhanced brightness to illustrate the spatial pattern of EdU (white) at 108 h and 120 h AEL. (E) Summary model of results. Growth of the wing disc leads to higher cell density in the center of the wing pouch (dark gray). Higher cell density reduces myosin levels and cytoskeletal tension, which reduces formation of Jub-Wts complexes at junctions, which reduces activity of Yki by decreasing inhibition of Wts by Jub. This results in lower Yki activity in the center of the wing pouch, except along compartment boundaries (not shown). Scale bar: 20 µm.
DISCUSSION
Hippo signaling was identified as a pathway that regulates organ growth, because mutations in pathway components can cause overgrowth or undergrowth phenotypes (Irvine and Harvey, 2015; Yu et al., 2015). However, whether Hippo signaling is normally modulated during Drosophila development to instruct temporal or spatial patterns of growth has remained unclear. Here, we have established that Hippo signaling is dynamically regulated during wing disc development, that this regulation occurs in response to changes in mechanical stress and that patterns of Yki activity correlate with patterns of cell proliferation.
Our results link the biomechanical modulation of Hippo signaling during wing development to increases in cell density as the wing disc grows. This increased cell density correlates with reduced junctional tension, reduced junctional recruitment of Jub and Wts, and reduced Yki activity. Moreover, our experimental manipulations of tension and of cell density confirm a functional connection between cells being more crowded and reduced Jub biomechanical signaling. This connection between growth-induced cell crowding and reduced Yki activity is consistent with observations of clones of cells that are genetically induced to overgrow surrounding cells (Pan et al., 2016), and with observations of density-dependent regulation of YAP activity and cell proliferation in cultured mammalian cells (Aragona et al., 2013; Benham-Pyle et al., 2015; Dutta et al., 2017; Ibar et al., 2018; Puliafito et al., 2012; Zhao et al., 2007).
Our observations contrast with a recent claim that cell density and shape do not influence Yki activity in the developing wing (Ma et al., 2017). However, that study was based on reducing or eliminating contact between cells and the extracellular matrix (ECM) (e.g. by degradation of ECM). Based on studies in cultured cells, the disruption of contact between cells and the ECM would be expected to decrease rather than to increase cytoskeletal tension within cells, because cytoskeletal tension is affected by cytoskeletal connections to cell-cell and cell-matrix attachments (Panciera et al., 2017). Both experimental and theoretical analyses of cell crowding, in both Drosophila wing discs and cultured mammalian cells, have emphasized that a key aspect of the response to growth-modulated cell density is its influence on cytoskeletal tension (Ibar et al., 2018; Irvine and Shraiman, 2017; Noll et al., 2017; Pan et al., 2016; Puliafito et al., 2012). Thus, although the flatter cells that are characteristic of young wing discs, or cells that have been treated with Mmp2, might superficially appear to be similar, they are flat for very different reasons, and these reasons have distinct effects on cytoskeletal tension. This difference, together with the crucial contribution of junctional tension to Hippo pathway regulation (Rauskolb et al., 2014), can explain why Mmp2 degradation does not generate the increased Yki activity that is characteristic of low-density cells. Similarly, treatment of cells with a Rok inhibitor increases apical cell area, making cells appear less crowded, but it decreases Yki activity because the influence of cytoskeletal tension predominates over any hypothesized influence of cell shape (Rauskolb et al., 2014). The fact that Yki activity correlates with cytoskeletal tension rather than with apical area under these experimental conditions also argues against speculation that Yki activity is regulated in the wing by a dilution of junctional regulators (Fletcher et al., 2018, 2015).
Our examination of EdU labeling suggests that reduced Yki activity contributes to reduced cell proliferation towards the center of late third instar wing discs. Cell proliferation has been described as evenly distributed throughout the wing disc, with two notable exceptions: a zone of non-proliferating cells (ZNP) along the D-V compartment boundary of late third instar wing discs (O'Brochta and Bryant, 1987) and a slight elevation of cell proliferation towards the center of second and early third instar wing discs (Mao et al., 2013). We observed slightly elevated EdU labeling towards the center of the wing pouch in 72 h AEL discs, but the difference between center and periphery was not significant in our assays. Measuring clone area, Mao et al. (2013) described faster growth in the center of the wing that ends around 72 h AEL, so their observations are not inconsistent with ours. Our discovery of lower proliferation towards the center of late third instar discs appears to be more surprising because it has not been described previously. However, proliferation rates decline throughout the entire disc at late third instar, which could make it difficult to detect local differences that are relatively small compared with the overall temporal decline in proliferation rates. In addition, using assays that lack single-cell resolution (e.g. measures of clone size), it could be difficult to disentangle the decrease in proliferation near the center of the wing disc from the cessation of proliferation throughout the ZNP.
Cell proliferation and transcription of Yki target genes is low along the D-V boundary, even though cytoskeletal tension and nuclear Yki levels are high. This has been linked to an inhibition of Yki activity by Notch activity in the wing, most likely due to activation of Vestigial (Vg) expression (Djiane et al., 2014). Vg partners with Scalloped (Sd) to regulate downstream genes (Halder et al., 1998; Simmonds et al., 1998). Two other proteins that can partner with Sd, Tgi and E2f1, have also been reported to inhibit Yki-dependent gene expression (Koontz et al., 2013; Zhang et al., 2017). Thus, a simple model for the ZNP, and for the generally low expression of Yki target genes despite high cytoskeletal tension and nuclear Yki, is that Yki activity is suppressed here by competition from Vg for Sd binding (Djiane et al., 2014).
Whereas our observations support a contribution of reduced Yki activity to the relative reduction in proliferation in the center of the wing, Yki activity appears to change relatively little in the proximal wing, even though proliferation rates decrease significantly. Thus, changes in Yki activity cannot fully explain the decrease in proliferation rates throughout the wing disc. Instead, we infer that other factors must also contribute to the general decline in growth rates as discs age. These could include changes in the patterning and response to local growth factors such as Dpp, as well as systemic effects such as changes in ecdysone signaling (reviewed by Hariharan, 2015; Irvine and Harvey, 2015).
MATERIALS AND METHODS
Drosophila culture
Unless otherwise indicated, crosses were performed at 25°C. To obtain wing discs at different stages, eggs were laid for 2 to 4 h and larvae were dissected at 72 h, 84 h, 96 h, 108 h and 120 h AEL. Protein or gene localization and expression was monitored using previously characterized transgenes: ex-lacZ (Hamaratoglu et al., 2006), jub:GFP (Sabino et al., 2011), wts:GFP (Rauskolb et al., 2014), zip:GFP and sqh:GFP (Royou et al., 2004).
To manipulate gene expression in the posterior compartment, en-gal4 tub-Gal80ts; UAS-dcr2 flies (created in house) were crossed with UAS-Rok-RNAi (Vienna Drosophila Resource Center, vdrc104675), UAS-Sqh.EE (Winter et al., 2001), UAS-Cdk1-RNAi (Bloomington Drosophila Stock Center, 36117), UAS-YkiS168A:V5 (Oh and Irvine, 2009), UAS-wts:Myc (Sun and Irvine, 2011) or UAS-Mmp2 (Bloomington Drosophila Stock Center, 58706) flies. Crosses were first maintained at 18°C and then shifted to 29°C for the indicated times (18, 24 or 36 h). For collecting wing discs at young (equivalent to 84 h AEL at 25°C) or old (equivalent to 120 h AEL at 25°C) stages for experiments involving UAS-Rok-RNAi, UAS-Sqh.EE, UAS-YkiS168A:V5, or UAS-wts:Myc, eggs were laid for 6 h and larvae were first kept at 18°C for 120 h (young) or 192 h (old), and then shifted to 29°C for 24 h before dissection. This protocol yielded discs similar in size to wild-type wing discs maintained at 25°C for 84 or 120 h AEL. For UAS-Cdk1-RNAi and UAS-Mmp2 we kept larvae at 18°C for ∼156 h before shifting, which yielded discs roughly equivalent to 108 h AEL at 25°C.
To study the effect of increasing cell density in the Minute/+ animals, y w; en-Gal4 UAS-Flp; FRT80B flies were crossed with w; Rps174 arm-lacZ FRT80B/TM6B (Bloomington Drosophila Stock Center, 6358) or with w; Rps174 His:RFP FRT80B/TM6B flies. Genotypes are y w; en-Gal4 UAS-Flp/Zip:GFP; Rps174 arm-lacZ FRT80B/FRT80B (Fig. 5A,E), y w; en-Gal4 UAS-Flp/Jub:GFP; Rps174 arm-lacZ FRT80B/FRT80B (Fig. 5G), y w; en-Gal4 UAS-Flp/+; Rps174 arm-lacZ FRT80B/FRT80B (Fig. 5D,I) and y w; en-Gal4 UAS-Flp/ex-lacZ; Rps174 His:RFP FRT80B/FRT80B (Fig. 5K). Larvae were dissected at 132 h AEL, which, because of the developmental delay in Minute/+ animals, yields discs that are roughly equivalent to 108 h AEL in size and cell density.
Histology and imaging
For most experiments wing discs were fixed in 4% paraformaldehyde for 15 min at room temperature. Wts:GFP discs were fixed for 8 min, and Sqh:GFP or Zip:GFP discs were fixed for 12 min. Primary antibodies used were rabbit anti-Yki (1:400; Oh and Irvine, 2008), mouse anti-β-galactosidase (1:200, Developmental Studies Hybridoma Bank, JIE7-c), mouse anti-Wg (1:800, Developmental Studies Hybridoma Bank, 4D4-c), mouse anti-Diap1 (1:200, B. Hay, California Institute of Technology, USA), rabbit anti-Dcp1 (1:800, Cell Signaling Technology, 9578S) and rat anti-E-cad (1:400, Developmental Studies Hybridoma Bank, DCAD2-c). Secondary antibodies were used at a 1:133 dilution, and included anti-rat Alexa Fluor 647 (Jackson ImmunoResearch, 712-605-153), anti-rabbit Alexa Flour 647 (Jackson ImmunoResearch, 711-605-152), anti-mouse Alexa Fluor 647 (Jackson ImmunoResearch, 715-605-151), anti-mouse Cy3 (Jackson ImmunoResearch, 715-165-151), anti-rabbit Cy3 (Jackson ImmunoResearch, 711-165-152), anti-rat Cy3 (Jackson ImmunoResearch, 712-165-153), anti-mouse Alexa Fluor 488 (Invitrogen, A21202) and anti-rabbit Alexa Flour 488 (Invitrogen, A21206). DNA was stained using Hoechst (Invitrogen). Confocal images were captured on a Leica SP8 microscope.
For EdU labeling, larvae were dissected in Ringer's solution and anterior halves were immediately placed in 250 μl room temperature wing disc medium 1 (WM1) (Zartman et al., 2013). An equal volume of 20 µM EdU (Click-iT EdU Alexa Fluor 555 Imaging Kit, Invitrogen, C10638) in WM1 was added for a final concentration of 10 µM EdU and samples were incubated for 10 min at room temperature. Tissue was then fixed for 15 min with 4% paraformaldehyde in PBS. Subsequent standard antibody staining protocol using mouse anti-Wg was then followed by 30 min EdU detection using 1.2 µl Alexa Fluor 555 Azide per 500 μl Click-iT reaction cocktail. Afterwards, tissues were treated with Hoechst and then washed with PBT (phosphate buffered saline, 1% bovine serum albumin, 0.1% Triton X-100). Wing discs were removed and mounted on a slide in Vectashield (Vector Laboratories, H-1000).
Live imaging and laser cutting of cell junctions
Live imaging and laser ablation experiments were performed as previously described (Pan et al., 2016). en-Gal4 UAS-RFP/CyO; tub-Gal80ts UAS-dcr2/TM6B flies were crossed with Ubi-Ecad:GFP or UAS-Cdk1-RNAi; Ubi-Ecad:GFP/TM6B or UAS-Mmp2; Ubi-Ecad:GFP/TM6B flies. Crosses were first maintained at 18°C and then shifted to 29°C for the indicated times. Wing discs were cultured in WM1 media in a 4-well chambered coverglass (Nunc Lab-Tek II, Fisher Scientific, 12-565-17) coated with poly-lysine. Discs were imaged every 0.2 s on a Perkin Elmer Ultraview spinning disc confocal microscope, and ablation of junctions was achieved using a Micropoint pulsed laser (Andor Technology) tuned to 365 nm. Paired cutting of junctions, one in the anterior compartment and another in the posterior compartment at a similar location, were performed and compared in the wing discs. The displacement of vertices for the first second after ablation was used to calculate the velocities.
Image processing and quantitative image analysis
To compensate for aberrations due to the curvature of wing disc and signals from the peripodial epithelium, we used the Matlab toolbox ImSAnE (Heemskerk and Streichan, 2015) to detect and isolate a slice of the disc epithelium that surrounds the adherens junctions, using E-cad as a reference, as described previously (Pan et al., 2016). The Sqh:GFP, Zip:GFP, Jub:GFP and Wts:GFP images were created using ImSAnE.
Quantification of Sqh:GFP, Zip:GFP, Jub:GFP and Wts:GFP intensity was performed as previously described (Rauskolb et al., 2014) using Volocity (Perkin Elmer) software. In brief, E-cad was used as a junctional reference to define the volumes to be quantified, and the relative Sqh:GFP, Zip:GFP, Jub:GFP or Wts:GFP to E-cad intensities were compared. Quantification of Yki, ex-lacZ and EdU levels was performed similarly, except we quantified and compared their relative nuclear intensities (Yki, ex-lacZ or EdU to DNA ratio), using DNA staining to define nuclei. To compare relative nuclear Yki at different developmental stages, nuclear Yki versus total Yki ratio was calculated for the comparison. To quantify the signals at central and peripheral regions of the wing pouch, we took half the distance between the center and edges of the pouch as the border between center and periphery (Fig. 1A). Equally sized cuboids in each central and peripheral quadrant were measured. To compare signals between the anterior and posterior compartment of wing discs, equally sized cuboids in the anterior and posterior compartment at similar proximal-distal regions were measured.
To make the cell area color map, confocal stacks were first processed by ImSAnE to make 2D surface images. The processed images were then segmented using ilastik software and the segmented pictures were used to perform cell area analysis with a custom Matlab script (github.com/idse/epitheliumAnalysis). The fluorescence intensity heat maps were generated using a custom Matlab script (github.com/alegoth/Wing-disc-Intensity-Analysis). In brief, the channel used to normalize (either E-cad or DNA) serves as a 3D mask to keep only the relevant pixels in the channel for the analysis. The center of the wing disc (AP-DV boundaries intersections) is manually picked for each image. We then split the picture into blocks of a given xy size (4×4 µm), starting from the center. We measured the average intensity per pixel of each channel. And the intensity of both the reference channel and the channel of interest is normalized over their respective average intensity. The ratio of the channel of interest over the reference channel is then determined. The ratio of each position is stored in a matrix for each image. To average several discs, only matrices of the same xy size blocks were used. The center of the disc serves as a reference point; smaller matrices were expanded to correspond to the size of the biggest matrix and filled with NaN (Not-a-Number). We determined the minimum number of values required (usually two) to average the ratio for a given position. This means that the edges of the average disk are composed of the same minimum number of values, this corresponds to the n given for each experiment. Finally, signals from several wing discs were averaged and represented by the heat map.
Variability in mean ratios is presented using 95% confidence intervals, determined using GraphPad Prism7 software. Statistical comparisons between these mean ratios were performed using the Student's t-test (for pair-wise comparisons) or one-way ANOVA with Tukey's correction (for multiple comparisons) on the log of the ratios, using GraphPad Prism7 software. Statistical markers used were ns=P>0.05, *P≤0.05, **P≤0.01, ***P≤0.001, ****P≤0.0001.
Supplementary Material
Acknowledgements
We thank B. Hay, G. Morata, T. Xu, J. P. Vincent, J. Zallen, the Developmental Studies Hybridoma Bank, the Vienna Drosophila Resource Center and the Bloomington Drosophila Stock Center for antibodies and Drosophila stocks. We thank Idse Heemskerk for providing the cell area color map Matlab script.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: Y.P., C.R., K.D.I.; Methodology: Y.P., H.A.; Software: H.A.; Investigation: Y.P., C.R.; Resources: K.D.I.; Writing - original draft: Y.P., K.D.I.; Writing - review & editing: K.D.I.; Supervision: K.D.I.; Project administration: K.D.I.; Funding acquisition: K.D.I.
Funding
This research was supported by the National Institutes of Health grant R01 GM078620 to K.D.I. and a Busch graduate fellowship to Y.P. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.165712.supplemental
References
- Aegerter-Wilmsen T., Heimlicher M. B., Smith A. C., de Reuille P. B., Smith R. S., Aegerter C. M. and Basler K. (2012). Integrating force-sensing and signaling pathways in a model for the regulation of wing imaginal disc size. Development 139, 3221-3231. 10.1242/dev.082800 [DOI] [PubMed] [Google Scholar]
- Aragona M., Panciera T., Manfrin A., Giulitti S., Michielin F., Elvassore N., Dupont S. and Piccolo S. (2013). A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154, 1047-1059. 10.1016/j.cell.2013.07.042 [DOI] [PubMed] [Google Scholar]
- Benham-Pyle B. W., Pruitt B. L. and Nelson W. J. (2015). Cell adhesion. Mechanical strain induces E-cadherin-dependent Yap1 and β-catenin activation to drive cell cycle entry. Science 348, 1024-1027. 10.1126/science.aaa4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Thakur M., Feng Y., Jagannathan R., Seppa M. J., Skeath J. B. and Longmore G. D. (2010). Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr. Biol. 20, 657-662. 10.1016/j.cub.2010.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djiane A., Zaessinger S., Babaoğlan A. B. and Bray S. J. (2014). Notch inhibits yorkie activity in Drosophila wing discs. PLoS ONE 9, e106211 10.1371/journal.pone.0106211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S. A., Gayyed M. F., Anders R. A., Maitra A. and Pan D. (2007). Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120-1133. 10.1016/j.cell.2007.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S., Mana-Capelli S., Paramasivam M., Dasgupta I., Cirka H., Billiar K. and McCollum D. (2017). TRIP6 inhibits Hippo signaling in response to tension at adherens junctions. EMBO Rep. 19, 337-350. 10.15252/embr.201744777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder D., Aegerter C. and Basler K. (2017). Forces controlling organ growth and size. Mech. Dev. 144, 53-61. 10.1016/j.mod.2016.11.005 [DOI] [PubMed] [Google Scholar]
- Fletcher G. C., Elbediwy A., Khanal I., Ribeiro P. S., Tapon N. and Thompson B. J. (2015). The Spectrin cytoskeleton regulates the Hippo signalling pathway. EMBO J. 34, 940-954. 10.15252/embj.201489642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher G. C., Diaz-de-la-Loza M.-D.-C., Borreguero-Muñoz N., Holder M., Aguilar-Aragon M. and Thompson B. J. (2018). Mechanical strain regulates the Hippo pathway in Drosophila. Development 145, dev159467 10.1242/dev.159467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G., Polaczyk P., Kraus M. E., Hudson A., Kim J., Laughon A. and Carroll S. (1998). The Vestigial and Scalloped proteins act together to directly regulate wing-specific gene expression in Drosophila. Genes Dev. 12, 3900-3909. 10.1101/gad.12.24.3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F., Willecke M., Kango-Singh M., Nolo R., Hyun E., Tao C., Jafar-Nejad H. and Halder G. (2006). The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8, 27-36. 10.1038/ncb1339 [DOI] [PubMed] [Google Scholar]
- Hariharan I. K. (2015). Organ size control: lessons from Drosophila. Dev. Cell 34, 255-265. 10.1016/j.devcel.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemskerk I. and Streichan S. J. (2015). Tissue cartography: compressing bio-image data by dimensional reduction. Nat. Methods 12, 1139-1142. 10.1038/nmeth.3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. B., Wu S., Barrera J., Matthews K. and Pan D. J. (2005). The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122, 421-434. 10.1016/j.cell.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Ibar C., Kirichenko E., Keepers B., Enners E., Fleisch K. and Irvine K. D. (2018). Tension-dependent regulation of mammalian Hippo signaling through LIMD1. J. Cell Sci. 131, jcs214700 10.1242/jcs.214700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine K. D. and Harvey K. F. (2015). Control of organ growth by patterning and hippo signaling in Drosophila. Cold Spring Harb. Perspect. Biol. 7, a019224 10.1101/cshperspect.a019224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine K. D. and Shraiman B. I. (2017). Mechanical control of growth: ideas, facts and challenges. Development 144, 4238-4248. 10.1242/dev.151902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koontz L. M., Liu-Chittenden Y., Yin F., Zheng Y., Yu J., Huang B., Chen Q., Wu S. and Pan D. (2013). The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev. Cell 25, 388-401. 10.1016/j.devcel.2013.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsberg K. P., Farhadifar R., Ranft J., Umetsu D., Widmann T. J., Bittig T., Said A., Jülicher F. and Dahmann C. (2009). Increased cell bond tension governs cell sorting at the Drosophila anteroposterior compartment boundary. Curr. Biol. 19, 1950-1955. 10.1016/j.cub.2009.10.021 [DOI] [PubMed] [Google Scholar]
- LeGoff L. and Lecuit T. (2016). Mechanical forces and growth in animal tissues. Cold Spring Harb. Perspect. Biol. 8, a019232 10.1101/cshperspect.a019232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeGoff L., Rouault H. and Lecuit T. (2013). A global pattern of mechanical stress polarizes cell divisions and cell shape in the growing Drosophila wing disc. Development 140, 4051-4059. 10.1242/dev.090878 [DOI] [PubMed] [Google Scholar]
- Ma M., Cao X., Dai J. and Pastor-Pareja J. C. (2017). Basement membrane manipulation in Drosophila wing discs affects Dpp retention but not growth mechanoregulation. Dev. Cell 42, 97-106.e4. 10.1016/j.devcel.2017.06.004 [DOI] [PubMed] [Google Scholar]
- Major R. J. and Irvine K. D. (2006). Localization and requirement for Myosin II at the dorsal-ventral compartment boundary of the Drosophila wing. Dev. Dyn. 235, 3051-3058. 10.1002/dvdy.20966 [DOI] [PubMed] [Google Scholar]
- Mao Y., Tournier A. L., Hoppe A., Kester L., Thompson B. J. and Tapon N. (2013). Differential proliferation rates generate patterns of mechanical tension that orient tissue growth. EMBO J. 32, 2790-2803. 10.1038/emboj.2013.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F. A. and Morata G. (2006). Compartments and the control of growth in the Drosophila wing imaginal disc. Development 133, 4421-4426. 10.1242/dev.02618 [DOI] [PubMed] [Google Scholar]
- Martin F. A., Herrera S. C. and Morata G. (2009). Cell competition, growth and size control in the Drosophila wing imaginal disc. Development 136, 3747-3756. 10.1242/dev.038406 [DOI] [PubMed] [Google Scholar]
- Marygold S. J., Roote J., Reuter G., Lambertsson A., Ashburner M., Millburn G. H., Harrison P. M., Yu Z., Kenmochi N., Kaufman T. C. et al. (2007). The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 8, R216 10.1186/gb-2007-8-10-r216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z. P., Moroishi T. and Guan K.-L. (2016). Mechanisms of Hippo pathway regulation. Gene Dev. 30, 1-17. 10.1101/gad.274027.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan M., Campuzano S. and Garcia-Bellido A. (1996). Cell cycling and patterned cell proliferation in the wing primordium of Drosophila. Proc. Natl. Acad. Sci. USA 93, 640-645. 10.1073/pnas.93.2.640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra J. R. and Irvine K. D. (2018). The Hippo signaling network and its biological functions. Annu. Rev. Genet. 52, (in press). 10.1146/annurev-genet-120417-031621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes A. J. and Morata G. (2017). Homeostatic response to blocking cell division in Drosophila imaginal discs: role of the Fat/Dachsous (Ft/Ds) pathway. Dev. Biol. 424, 113-123. 10.1016/j.ydbio.2017.03.009 [DOI] [PubMed] [Google Scholar]
- Nienhaus U., Aegerter-Wilmsen T. and Aegerter C. M. (2009). Determination of mechanical stress distribution in Drosophila wing discs using photoelasticity. Mech. Dev. 126, 942-949. 10.1016/j.mod.2009.09.002 [DOI] [PubMed] [Google Scholar]
- Noll N., Mani M., Heemskerk I., Streichan S. and Shraiman B. I. (2017). Active tension network model suggests an exotic mechanical state realized in epithelial tissues. Nat. Phys. 13, 1221-1226. 10.1038/nphys4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brochta D. A. and Bryant P. J. (1987). Distribution of S-phase cells during the regeneration of Drosophila imaginal wing discs. Dev. Biol. 119, 137-142. 10.1016/0012-1606(87)90215-6 [DOI] [PubMed] [Google Scholar]
- Oh H. and Irvine K. D. (2008). In vivo regulation of Yorkie phosphorylation and localization. Development 135, 1081-1088. 10.1242/dev.015255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H. and Irvine K. D. (2009). In vivo analysis of Yorkie phosphorylation sites. Oncogene 28, 1916-1927. 10.1038/onc.2009.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Heemskerk I., Ibar C., Shraiman B. I. and Irvine K. D. (2016). Differential growth triggers mechanical feedback that elevates Hippo signaling. Proc. Natl. Acad. Sci. USA 113, E6974-E6983. 10.1073/pnas.1615012113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panciera T., Azzolin L., Cordenonsi M. and Piccolo S. (2017). Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol. 18, 758-770. 10.1038/nrm.2017.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor-Pareja J. C. and Xu T. (2011). Shaping cells and organs in Drosophila by opposing roles of fat body-secreted collagen IV and Perlecan. Dev. Cell 21, 245-256. 10.1016/j.devcel.2011.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puliafito A., Hufnagel L., Neveu P., Streichan S., Sigal A., Fygenson D. K. and Shraiman B. I. (2012). Collective and single cell behavior in epithelial contact inhibition. Proc. Natl. Acad. Sci. USA 109, 739-744. 10.1073/pnas.1007809109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C., Sun S., Sun G., Pan Y. and Irvine K. D. (2014). Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell 158, 143-156. 10.1016/j.cell.2014.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royou A., Field C., Sisson J. C., Sullivan W. and Karess R. (2004). Reassessing the role and dynamics of nonmuscle myosin II during furrow formation in early Drosophila embryos. Mol. Biol. Cell 15, 838-850. 10.1091/mbc.e03-06-0440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino D., Brown N. H. and Basto R. (2011). Drosophila Ajuba is not an Aurora-A activator but is required to maintain Aurora-A at the centrosome. J. Cell Sci. 124, 1156-1166. 10.1242/jcs.076711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shraiman B. I. (2005). Mechanical feedback as a possible regulator of tissue growth. Proc. Natl. Acad. Sci. USA 102, 3318-3323. 10.1073/pnas.0404782102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds A. J., Liu X., Soanes K. H., Krause H. M., Irvine K. D. and Bell J. B. (1998). Molecular interactions between Vestigial and Scalloped promote wing formation in Drosophila. Genes Dev. 12, 3815-3820. 10.1101/gad.12.24.3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G. and Irvine K. D. (2011). Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Dev. Biol. 350, 139-151. 10.1016/j.ydbio.2010.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S. and Irvine K. D. (2016). Cellular organization and cytoskeletal regulation of the Hippo signaling network. Trends Cell Biol. 26, 694-704. 10.1016/j.tcb.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Reddy B. V. V. G. and Irvine K. D. (2015). Localization of Hippo signalling complexes and Warts activation in vivo. Nat. Commun. 6, 8402 10.1038/ncomms9402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartlick O., Mumcu P., Kicheva A., Bittig T., Seum C., Julicher F. and Gonzalez-Gaitan M. (2011). Dynamics of Dpp signaling and proliferation control. Science 331, 1154-1159. 10.1126/science.1200037 [DOI] [PubMed] [Google Scholar]
- Winter C. G., Wang B., Ballew A., Royou A., Karess R., Axelrod J. D. and Luo L. (2001). Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell 105, 81-91. 10.1016/S0092-8674(01)00298-7 [DOI] [PubMed] [Google Scholar]
- Worley M. I., Setiawan L. and Hariharan I. K. (2013). TIE-DYE: a combinatorial marking system to visualize and genetically manipulate clones during development in Drosophila melanogaster. Development 140, 3275-3284. 10.1242/dev.096057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.-X., Zhao B. and Guan K.-L. (2015). Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163, 811-828. 10.1016/j.cell.2015.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zartman J., Restrepo S. and Basler K. (2013). A high-throughput template for optimizing Drosophila organ culture with response-surface methods. Development 140, 667-674. 10.1242/dev.088872 [DOI] [PubMed] [Google Scholar]
- Zhang L., Ren F., Zhang Q., Chen Y., Wang B. and Jiang J. (2008). The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell 14, 377-387. 10.1016/j.devcel.2008.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Pei C., Wang X., Xiang J., Sun B. F., Cheng Y., Qi X., Marchetti M., Xu J. W., Sun Y. P. et al. (2017). A balance of Yki/Sd activator and E2F1/Sd repressor complexes controls cell survival and affects organ size. Dev. Cell 43, 603-617.e605. 10.1016/j.devcel.2017.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Wei X., Li W., Udan R. S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L. et al. (2007). Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747-2761. 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.