Abstract
The rat is the preferred model for toxicology studies, and it offers distinctive advantages over the mouse as a pre-clinical research model including larger sample size collection, lower rates of drug clearance, and relative ease of surgical manipulation. An immunodeficient rat would allow for larger tumor size development, prolonged dosing and drug efficacy studies, and preliminary toxicological testing and PK/PD studies in the same model animal. Here we created an immunodeficient rat with a functional deletion of the Rag2 gene, using genetically modified spermatogonial stem cells (SSCs). We targeted the Rag2 gene in rat SSCs with TALENs and transplanted these Rag2 deficient SSCs into sterile recipients. Offspring were genotyped and a founder with a 27bp deletion mutation was identified and bred to homozygosity to produce the Sprague-Dawley Rag2 - Rag2tm1Hera (SDR) knockout rat. We demonstrated that SDR rat lacks mature B and T cells. Furthermore, the SDR rat model was permissive to growth of human glioblastoma cell line subcutaneously resulting in successful growth of tumors. Additionally a human KRAS mutant non-small cell lung cancer cell line (H358), a patient derived high grade serous ovarian cancer cell line (OV81), and a patient derived recurrent endometrial cancer cell line (OV185) were transplanted subcutaneously to test the ability of the SDR rat to accommodate human xenografts from multiple tissue types. All human cancer cell lines showed efficient tumor uptake and growth kinetics indicating that the SDR rat is a viable host for a range of xenograft studies.
Keywords: SCID rat, xenograft, PDX, humanized rat, SCID mouse, ovarian cancer, Non-small cell lung cancer
Introduction
Preclinical research and drug discovery relies heavily on in vitro systems and animal models for safety and efficacy studies. However, in vitro and animal models don’t always accurately predict human metabolism and toxicity (1–5). As a result, there have been cases in which a drug was deemed safe or efficacious in rodent studies, but which failed in clinical trials in humans (6–9). Immunodeficient mouse models of human cancer have paved the way for studying cancer biology, genomics, effects on cancer growth kinetics, propensity for metastasis, and treatment response. A plethora of genetically immunodeficient mouse models, with varying immune phenotypes, exist for such studies(10). However, drug efficacy testing and downstream analysis such as pharmacokinetic (PK) / pharmacodynamic (PD) studies are limited because of inconsistent or poor tumor engraftment, high variability in tumor growth kinetics and limited tumor growth potential. As a result, a significantly large number of mice are used for drug efficacy screening in order to achieve a cohort of animals with tumors of similar size and similar tumor growth kinetics for treatment. We explored whether these cell lines might grow more consistently in a versatile in vivo model such as the immunodeficient rat.
The laboratory rat remains the favored species for toxicology research because of its relative physiological similarity to humans (11–14). The metabolism and pharmacokinetic properties of drugs in rats is similar to humans compared to mice. All toxicology and safety profiling of drugs is performed in rats while efficacy studies are conducted primarily in mice models due to a lack of appropriate SCID-rat models. Data quality for drug development would be much improved if all the relevant data sets are generated in the same model.
Due to the large size of the rats, tumors can be grown to nearly ten times the volume (or double the diameter) allowed in the mouse (15, 16). Rats have ten times the blood volume of mice. Therefore, rats can accommodate multiple blood samplings from the same test animal at different time points for blood cancer efficacy assessment, clinical pathology profiling, and pharmacokinetic sampling. Since the rat is the preferred model for toxicology and safety testing, a rat with human cancer would allow for a combination of chemotherapy efficacy, pharmacokinetic and preliminary toxicology testing all in one animal thereby greatly reducing the number of animals needed while improving the quality of data generated.
In order to generate cancer xenograft models or “humanize” a tissue in the rodent by replacing endogenous cells with human cells or ectopically transplanting human tissues, the animal must be immunodeficient to inhibit rejection of the xenogeneic cells. While many immunodeficient mouse models exist with differing capabilities for accepting human cells (10), very few rat models can engraft human cells (17, 18). The nude rat (RNU; NIH-Foxn1rnu) is one such model that is completely devoid of T cells but has limited capacity for human cell engraftment because it still has a normal repertoire of B and NK cells (19). Several studies have demonstrated that the nude mouse is superior in its ability to engraft and support the growth of human cancers compared to the nude rat and that there is an increased incidence of tumor regression in the nude rat, likely due to its age-dependent changes in immune competence (20–22). Data from studies with several different immunodeficient mouse models suggest that mice in which both B and T cells are absent have a better propensity for supporting human cell engraftment and growth (23–28). Therefore, we have created a genetically immunodeficient rat which completely lacks B cells and has a severely reduced population of mature T cells.
Although targeted genetic modification of the laboratory mouse has been possible since the first isolation of mouse embryonic stem cells (29, 30), targeted gene knockouts and knock-ins have not been described in rats until recently. New gene editing technologies such as TALENs and the CRISPR/Cas9 systems have now made targeted genetic modifications possible in the rat. We used Xanthomonas TALE Nuclease (XTN) to create a mutation in Rag2 (Recombination Activating Gene 2) which is critical for V(D)J recombination and its deletion disrupts maturation of B and T cells of the immune system (31, 32). Rat spermatogonial stem cells (SSCs) were targeted, which have recently been described as an alternative to genetic manipulation of embryos in rats (33). These modified SSCs can assimilate into the testes of sterile males and give rise to normal offspring, allowing germline transmission of the genetic modification of interest in one generation.
Here we report the generation of a Sprague-Dawley Rag2 knockout (SDR) rat characterized by a loss of mature B cells and severely reduced T cells compared with wild-type Sprague Dawley rats. We demonstrate that these immunodeficient rats are permissive for human cancer xenografts with high efficiency and desirable uniformity in tumor growth profiles. Our data suggest that the SDR rat may be a viable novel model in which to study human cancers and may also be useful for transplantation of various other human cells and tissues.
Materials and methods
Editing spermatogonial genome
All animal experiments were approved and met guidelines set forth by the University of Kentucky’s IACUC. Rat spermatogonial line SD-WT2 (Dr. Kent Hamra’s lab, UTSW) were propagated in Spermatogonial Medium (SG) and cryopreserved in SG freezing medium, as previously described (34). To generate Rag2 mutants, spermatogonia were expanded to passage 16 (from cryopreserved passage 13 stocks) in fresh SG medium before collecting for nucleofection with Rag2-XTNs. 10μg total DNA of Rag2-XTN plasmids was added to 3×106 SSCs suspended in 100μl Nucleofection Solution L (Amaxa), and subjected to nucleofection using settings A020 on the Nucleofector (Amaxa). After transfection, spermatogonia were subjected to 75μg/ml G418 treatment for 20 days following a 7-day recovery, and cryopreserved until transplantation.
Prior to transplantation, G418-selected spermatogonia were verified for carrying the desired mutation and correct karyotype. Genomic DNA was harvested from about 50,000 G418-selected spermatogonia and the targeted locus amplified. This pool of amplicons was TOPO cloned and a 96-well plate of clones was sequenced for analysis to confirm disruption of the Rag2 gene. Another cohort of nucleofected spermatogonia were sent to IDEXX BioResearch laboratories (Sacramento, CA) for karyotype analysis.
Immunocytochemistry
SD-WT2 spermatogonia at passage 16 were also subjected to immunocytochemistry to verify expression of spermatogonial stem cell marker Plzf (ZBTB16). Spermatogonia were plated on cover slips in a 24-well plate. 3T3 cells were plated on uncoated cover slips, MEF feeders on gelatin-coated cover slips, SSCs co-cultured with MEFs on laminin and gelatin coated cover slips. Plated cells were washed twice with SG medium and fixed with 4% PFA for 7min, and washed 3 times with PBS. They were then permeabilized with PBS plus 0.1%(v/v) TritonX-100 for 15min., washed 3 times with PBS. Blocking reagent (11096176001; Roche) dissolved in Maleic acid was applied to 2hrs, followed by a 20hr incubation at room temperature with 1μg/ml of mouse anti-Plzf (OP128L; Calbiochem) in blocking reagent. Slides were washed 3 times with TBST to remove unbound IgG and incubated with 4μg/ml Alexa Fluor 596 donkey anti-mouse IgG (A21203; Invitrogen), as the secondary antibody, in PBS containing 5μg/ml Hoecsht33342 for 40 min at RT. Slides were washed 3 times with TBST and finished with Fluoromount (F4680; Sigma) for observation under fluorescent microscope.
Transplantation of modified spermatogonia
G418-selected spermatogonia were thawed and transplanted within 3 hours of thawing into Busulfan-treated, Dazl-deficient male Sprague Dawley rats as previously described (35, 36). Briefly, Dazl-deficient males were injected intraperitoneally with 12mg/kg busulfan. Twelve days later, G418-resistant donor rat spermatogonia were thawed from cryo-preserved stock, resuspended in ice cold SG medium and loaded into injection needles at concentration 3X105 cells/50μl. The entire 50μl volume was injected into the seminiferous tubes of anesthetized rats by retrograde injection. Transplanted males were subsequently bred to wildtype females 70–81 days later.
Screening for SDR rats
Total genomic DNA was extracted from pups born from Dazl-deficient males transplanted with modified spermatogonia. The targeted Rag2 locus was PCR-amplified from the gDNA with Rag2_NHEJ-Fwd (GAGAAGGTGTCTTACGGTTCTATG) and Rag2_NHEJ-Rev (GCAGGCTTCAGTTTGAGATG) primers. The PCR product was purified and subjected to MseI digestion and the digestion product was analyzed via gel electrophoresis with a 1% agarose gel for disruption of the targeted MseI restriction site. PCR-amplicons that failed to be completely digested by MseI restriction enzyme was TOPO cloned and 6 clones sequenced to identify the sequence of the alleles in that sample.
FACS analysis of immune cells
To detect T, B, and NK cells in SDR rats, flow cytometric analysis was performed on splenocytes and thymocytes using a BD LSRII (BD Biosciences) flow cytometer. Spleen and thymus were collected in FACS buffer (BD Pharmingen 554656). The tissues were homogenized and passed through a 70μm cell strainer to remove clumps. Red blood cells were lysed by incubating with ACK Lysing Buffer (Quality Biological #118–156-721) for 10 minutes at room temperature. Cells were stained with fluorophore-labeled antibodies at a final concentration of 25μg/ml in 20μl volume for 20 minutes. Antibodies used were Goat anti-rat IgM-APC (Stem Cell Technologies #10215), FITC Mouse anti-rat CD45R (BD Pharmingen #561876), FITC Mouse Anti-Rat CD8b (BD Pharmingen #554973), APC Mouse Anti-Rat CD4 (BD Pharmingen #550057), and FITC Mouse Anti-Rat CD161a (BD Pharmingen #561781).
Transplantation of human cancer cell lines
Cell culture:
Human glioblastoma cell line U87MG Red FLuc (Perkin Elmer, BW124577) was a gift from Dr. Bjoern Bauer at the University of Kentucky, human NSCLC cell line H358 (bronchioalveolar carcinoma, mutant KRAS) was a gift from Dr. Goutham Narla at the Case Comprehensive Cancer Center, Cleveland, Ohio, human ovarian carcinoma cell line OV81 obtained from high grade serous ovarian cancer patient’s PDX tumor and human endometriod cancer cell line OV185 obtained from a recurrent endometriod patient’s PDX tumor was a gift from Dr. Analisa DiFeo at the Case Comprehensive Cancer Center, Cleveland, Ohio. U87MG cells were grown in Eagle’s Minimum Essential Medium with L-Glutamine (ATCC #30–2003), Sodium Pyruvate (Gibco #11360–070), FBS and Non-Essential Amino Acids (Gibco #11140–050). H358 cell line were cultured in Advanced RPMI 1640 Medium (Gibco #12633–012) supplemented with 10% fetal bovine serum (Atlanta Biologicals # S12450) and 1% penicillin and streptomycin solutions (Cat# 15140–122, Themofisher). OV81 and OV185 were grown in Dulbecco Modified Eagle Medium (Gibco #10566–016) supplemented with 10% fetal bovine serum (Atlanta Biologicals # S12450) and 1% penicillin and streptomycin solutions (Cat# 15140–122, Themofisher). All the cells were grown in a humidified incubator at 37° C with 5% CO2. Cell lines were tested for Mycoplasma using the MycoAlert Mycoplasma detection kit (Cat # LT07, Lonza). Experiments were performed within 6–8 cell passages after thaw.
Tumor xenografts:
For transplantation 1×106 U87MG cells, 1×106, 5×106 or 10×106 H358 cells, 2×106 OV81 cells and OV185 cells for each animal were resuspended in 250μl sterile 1xPBS (Gibco #14190–144). Immediately prior to injection, 250μl 10mg/ml Geltrex (Gibco #A14132–02) was added to the cell suspension for a final Geltrex concentration of 5mg/ml. The cell/Geltrex suspension was injected subcutaneously into the hindflank. Tumor diameter was measured using digital calipers (Fisher #14–648-17) 3 times a week. Tumor volume was calculated as (L × W2)/2 (37), where width and length were measured at the longest edges.
Immunohistochemistry of tumors
Tumors were excised and fixed in 10% neutral buffered formalin for 48 hours. Tissue was processed, paraffin embedded and sectioned by the University of Kentucky Imaging Core Facility. Standard 5um sections were collected. Sections were stained with Harris’ Hematoxylin (Sigma #HHS128) and Eosin (Sigma #318906) for basic histology. Human cells were visualized by staining with an antibody that recognizes a protein found in all human mitochondria (mouse anti-human mitochondria antibody, clone 113–1; EMD Millipore #1273). Chromogenic staining was performed by using biotinylated goat-anti mouse (Vector Labs #BA-9200), then Vectastain Elite ABC HRP Reagent, R.T.U. (Vector Labs #PK-7100) and developed using Pierce DAB substrate kit (ThermoFisher #34002). In some cases, sections were counterstained with Harris’ Hematoxylin to visualize nuclei. Images were taken using a Zeiss Palm Laser Microbeam Microscope in the University of Kentucky Imaging Core Facility.
Results
Rag2 knockout rat SSCs
XTNs ( Xanthomonas TALE Nucleases) (38, 39) were used to create a pool of spermatogonia in which about 3% of the cells contained disruptions in the Rag2 gene. Rag2 has a single coding exon. XTNs targeting a MseI restriction site near the start of this coding exon were nucleofected into SD-WT2 spermatogonial stem cells derived from wild-type Sprague-Dawley rat (Hsd: Sprague-Dawley SD; Harlan, Inc.). The location of the XTN binding sites and PCR primers for genotyping are shown in Fig. 1A. Total genomic DNA was extracted from transfected spermatogonia clusters. The targeted locus was PCR amplified, TOPO cloned, and sequenced to look for disruption of the Rag2 gene. Out of seventy six sequenced clones, two clones had deletions in the Rag2 gene (Fig. 1B). One clone contained a single nucleotide deletion and a second clone contained a two-nucleotide deletion. Six other clones contained point mutations within the targeted region.
Fig. 1.
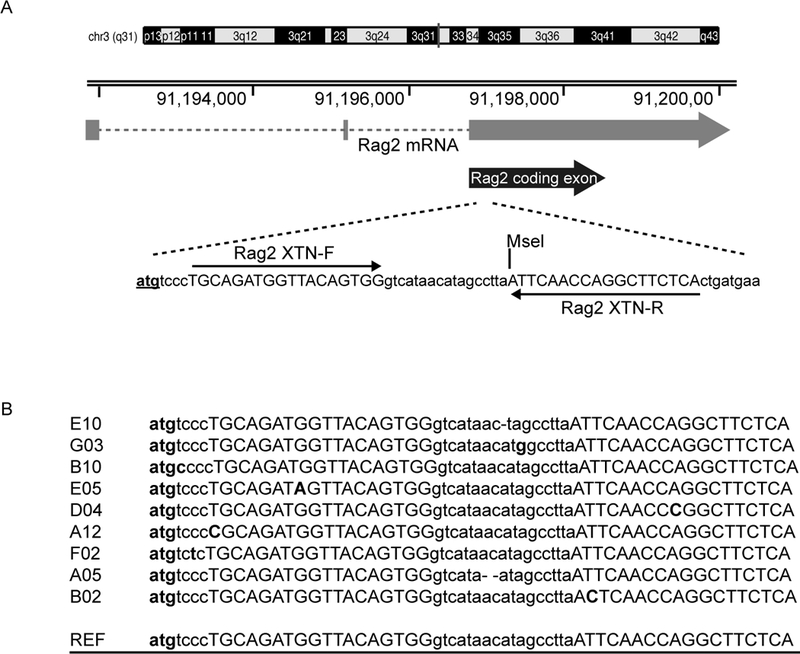
Disruption of Rag2 gene in rat SSCs. (A) The XTN pair targets early in the single coding exon of Rag2 gene. XTN binding sites are capitalized, the Rag2 start codon is shown in boldface font and underlined, the MseI site utilized for genotyping is marked. (B) Alignment of clones to the wild type reference sequence. Reference sequence is underlined, the XTN binding sites are shown in uppercase, mutations are shown in bold font and Rag2 start codon is in bold font and underlined. Clone E10 contains a single nucleotide deletion, clone A05 contains a two nucleotide deletion.
Genome modified spermatogonia express stemness marker
Spermatogonia were expanded until passage 16 (4.5 months in culture) for nucleofection to introduce the genetic mutations. To confirm that these spermatogonia maintained their stemness and chromosomal integrity, we analyzed the expression of ZBTB16 (PLZF), which is a marker for mammalian type A spermatogonia and has been shown to be critical for spermatogonial stem cell self-renewal (40, 41). MEFs did not express ZBTB16, whereas fresh SSCs did (Fig. 2A). SD-WT2 spermatogonia showed consistent and robust expression of ZBTB16 even after being expanded to passage 16 over a 5-month culture period (Fig. 2B). Spermatogonia at passage 16, were also sent to IDEXX for karyotype analysis, which verified that these SSCs did not contain any chromosomal anomalies (Supplementary Fig. S1A,B).
Fig. 2.
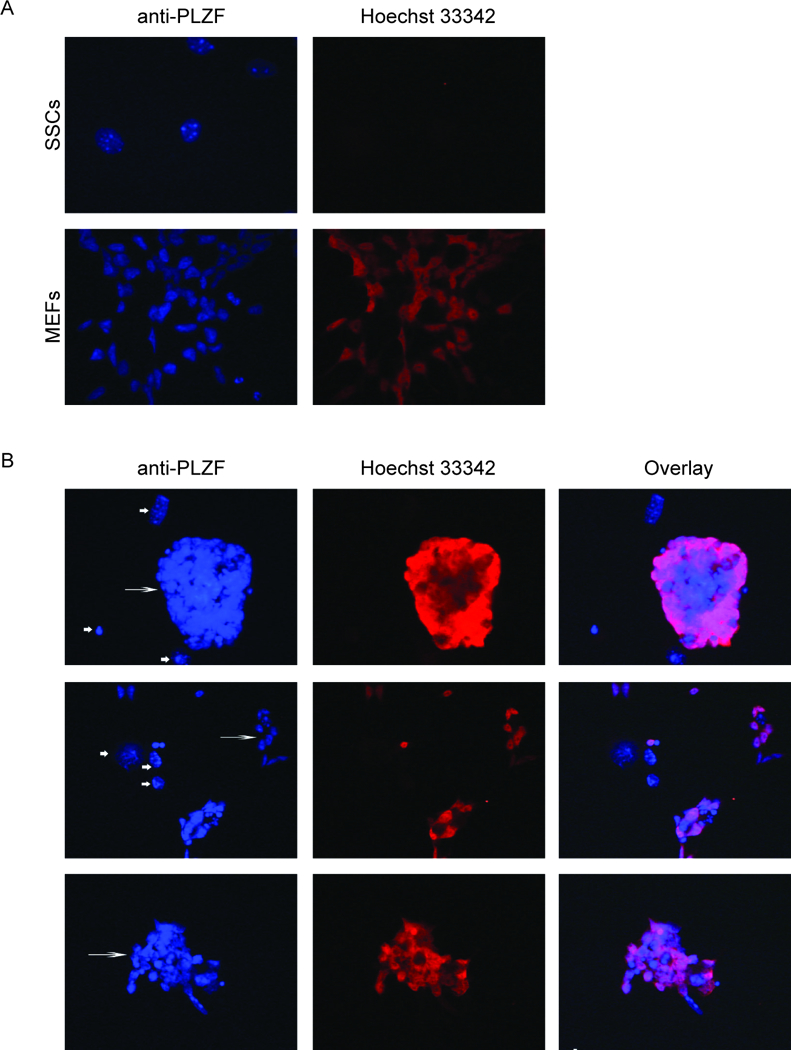
Expression of ZBTB16 (PLZF) in MEFs, fresh SSCs, and parental SD-WT2 SSCs kept in culture for 4.5 months (passage 16). Cells were stained with anti-PLZF antibody (red) and Hoechst 33342 (blue). (A) Fresh SSCs (passage 9) on laminin shows undifferentiated spermatogonia that express ZBTB16 (PLZF), whereas the MEFs do not. (B) Immunocytochemistry of SSC clusters at passage 16 on feeders. First column with Hoechst33342 nuclear staining shows SSCs (long arrow) and feeders (short arrow). Second column shows PLZF staining. Third column shows merged images of PLZF staining and Hoechst33342.
Sprague-Dawley Rag2 knockout rat (SDR Rat)
The pool of spermatogonia containing 3% modified spermatogonial stem cells were transplanted into seminiferous tubules (35, 36) of four busulfan-treated Dazl-deficient rats. At approximately eighty days post-transplantation, recipient males were paired with wild-type females. All modified SSCs were transplanted into recipient rats within three hours of their thawing. Three out of the four recipients had greater than 50% fill in at least one of their testes, with two also showing signs of bleeding during transplantation surgery. The recipient that had a high fill percentage and no bleeding during transplantation produced pups (Table 1). The recipient sired eight litters with a total of sixty pups. The pups were screened for loss of the targeted MseI site by subjecting the amplified locus to MseI digestion. As shown in Fig. 3A, one out of the sixty pups (2f) was identified to carry a disrupted allele. Sequencing analysis revealed this allele to be a 27bp deletion mutant. The allele was then bred to homozygosity to generate the SDR rat. Thymus and spleen were collected from SDR rat and western blot was performed to determine Rag2 protein expression using a Rag2 antibody. Because the SDR rat carries an in-frame 27bp deletion mutation in the Rag2 gene, western blot analysis showed similar expression of the Rag2 protein in the SDR and wild-type rats (Supplementary Fig. S2).
Table1. Production of SDR rats by implanting Rag2 SSCs into sterile males.
Four recipient males were transplanted with genetically modified SSCs. Only one recipient (526300) had no surgical complications and had good fill of testis with modified SSCs. Only that recipient produced pups when mated with wild type females.
| Animal ID |
Time from thaw to transplant |
Right testis (fill %) |
Left Testis (fill %) |
Surgery outcome |
Days to mating |
Days to 1st litter |
Total pups screened |
Rag2 pups |
|---|---|---|---|---|---|---|---|---|
| 526308 | 0.75 h | 20 | 10 | Good | 77 | n/a | n/a | n/a |
| 526300 | 1.6 h | 15 | 90 | Good | 81 | 142 | 60 | 1 |
| 526309 | 2 h | 60 | 100 | Survived, but was bleeding during surgery |
70 | n/a | n/a | n/a |
| 526310 | 2.75 h | Miss | 75 | Survived, but was bleeding during surgery |
70 | n/a | n/a | n/a |
Fig. 3.
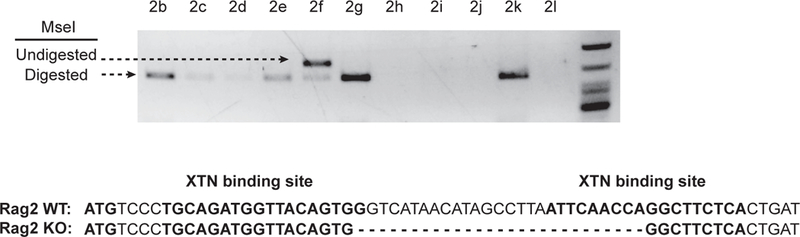
Genotyping pups for disruptions in Rag2 gene. The targeted locus was amplified from pups sired by the implanted male and subjected to MseI digestion. Animal 2f contained at least one allele that is resistant to MseI digestion (white arrow). The PCR product from animal 2f was TOPO cloned and sequenced which revealed this animal carries a mutant allele with a 27bp deletion.
SDR rat lacks mature B and T cell populations
Splenocytes and thymocytes were collected from age-matched wild-type and homozygous mutant animals and analyzed by flow cytometry to characterized the immune cell populations in the SDR rat. Homozygous mutant animals were essentially athymic with residual tissue equating to less than 10% the weight of thymus tissue in the wild-type. Mature T cells were identified by double staining for T cell antigens CD4 and CD8 (32, 42). The SDR thymocyte population contained only 7.55% CD4+/CD8+ cells and 81% CD4-/CD8- cells (Fig. 4A, right panel), drastically reduced from the wild-type thymocyte population, which consisted of 89.24% CD4+/CD8+ cells and less than 2% CD4-/CD8- cells (Fig. 4B, left panel).
Fig. 4.
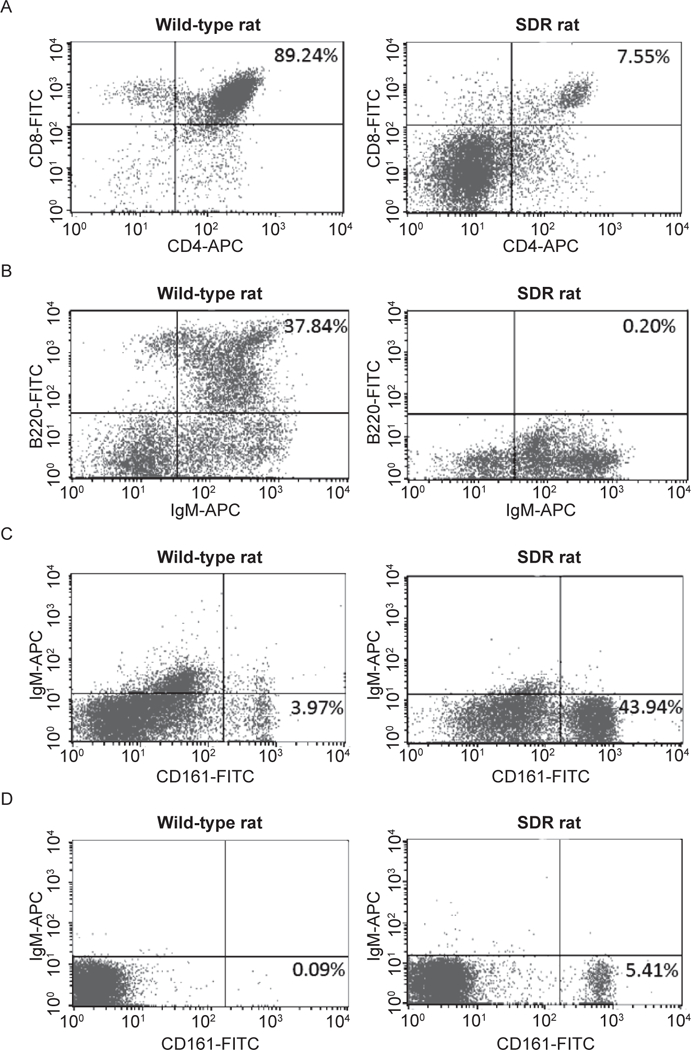
Immunophenotyping of the SDR rat. (A) SDR thymocytes contain only 7.55% CD4+/CD8+ mature T cells (right panel), compared to 89.24% in a wild-type control (left panel). The majority of thymocytes are CD4 and CD8 double negative in the SDR rat. (B) The SDR rat spleen contains no mature B cells as demonstrated by lack of B220+/IgM+ cells (right panel), whereas the wildtype spleen contains 37.84% B220+/IgM+ mature B cells (left panel). (C, D) SDR rat spleen (C) and thymus (D) have an increased NK cell population (43.94% and 5.41%, respectively) compared to only 3.97% in the wild-type spleen (C) and less than 1% in the wild-type thymus. Left panels: wild-type rat. Right panels: SDR rat.
Mature B cells were identified as double positive B220/IgM population of splenocytes (32, 43). The spleens from homozygous mutant animals were smaller compared to the wild-type spleen. In a Rag2 null genotype the B-cell receptor genes should not be capable of V(D)J recombination. Thus, as expected, the SDR rats had no B220/IgM double positive splenocytes (Fig. 4B, right panel).
Interestingly, the SDR rat had an increase in NK cells compared to the wild-type rat. Whereas the wild-type rat has 3.97% NK cells in the splenocytes and less than 1% NK cells in the thymocyte population (Fig. 4C, left panels), the SDR rat splenocytes and thymoctyes contained 43.94% and 5.41% NK cells, respectively (Fig. 4C, 4D, right panels). The increase in NK cells seen in our SDR rats is similar to that seen in a Rag1 knockout rat (17), a different Rag2 knockout rat (44), the Prkdc SCID rat which lacks mature B and T cells (45) and the SCID mouse. In addition, Rag2 null mice exhibit greater NK cell activity than their wild-type littermates (32). Although we do not know the mechanism resulting in the increased NK cells, these published data suggest this is a common phenomenon among immunodeficient animals lacking mature T and B cells.
Analysis of SDR whole blood demonstrated greatly diminished T and B cells compared to the wild-type rat (Supplementary Fig. S3A-C). The wild-type rat has a significant population of circulating CD4+, CD8+, and CD4/CD8 double positive T cell populations compared to the SDR rat (Supplementary Fig. S3A). The wild-type rat has over 20% B220/IgM double positive mature B cells, compared to 3.5% in the SDR rat (Supplementary Fig. S3B). Interesting, the SDR rat has slightly less circulating NK cells, unlike what is observed in the spleen (Supplementary Fig. S3C).
Subcutaneous human glioblastoma tumor growth
To determine if the SDR rat was permissive for human cancer xenotransplantation, we transplanted a human glioblastoma cell line (U87MG) subcutaneously into six SDR rats, two Rag2 heterozygous rats, and one wild-type rat. The cells were resuspended in Geltrex prior to inoculation to provide structural support for cell survival and growth. No tumors grew during the study period in the wildtype and heterozygous animals. All six SDR rats showed subcutaneous tumor growth (Fig. 5A), detectable as early as 10 days post-injection. All tumors reached the maximum allowable size of 40mm diameter by 49 days post-injection (Fig. 5B). The tumors all stained positive for the human mitochondrial protein (Fig. 5C). Tissue from a rat that had not been injected with human cells did not show positive staining with the antibody (Fig. 5C).
Fig. 5.
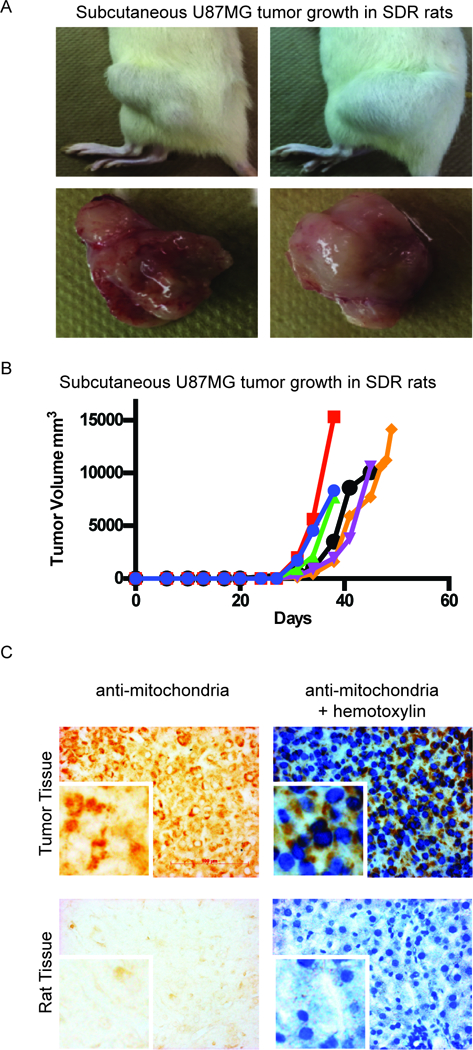
Subcutaneous growth of human glioblastoma U87MG cells in the SDR rat. 1×106 U87MG cells resuspended in Geltrex were injected subcutaneously into SDR rats. (A) Tumor growth in two different SDR animals with images of their excised tumors. (B) Tumor volume (mm3) over time. Each line represents tumor growth in an individual rat. (C) Immunohistochemistry of anti human-mitochondria in tumor tissue and rat tissue. Brown staining demonstrates peri-nuclear localization of human-mitochondria protein in a tumor section, with (right) and without (left) hematoxylin counterstain. 40x magnification. The antibody for human mitochondria protein does not show staining in tissue from a rat that was not injected with human cells (negative control). Right panel with hematoxylin counterstain; 40× magnification; Scale bar = 100μm.
SDR rat is a suitable model for human xenograft studies
In the second study, a KRAS mutant non-small cell lung cancer (NSCLC) cell line H358 was implanted into the SDR rat subcutaneously. Tumor growth was observed and compared to growth in nude (nu/nu) and NSG mice. 1×106, 5×106 or 1×107 were implanted in the SDR rat while 1×107 cells were implanted in the nude and NSG mice. The engraftment rate was 100% in the SDR rat for all three cell-densities implanted and growth rate was directly proportional to the amount of cells transplanted. In addition, the tumors grew much faster and more consistently in the SDR rat than in the mouse models (Fig. 6A, Supplementary Table S1).
Fig. 6.
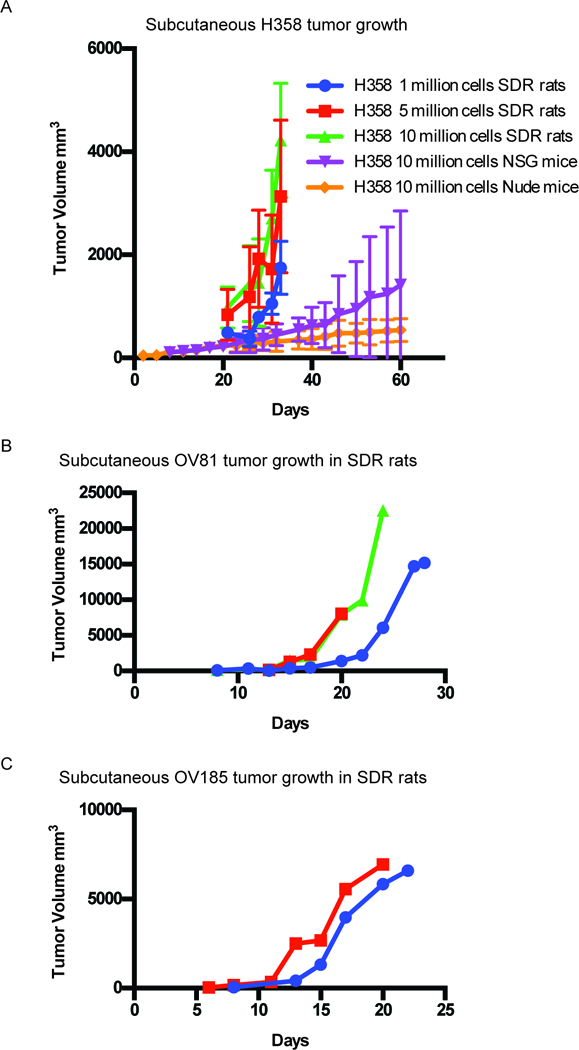
Subcutaneous tumor growth of NSCLC, ovarian and endometrial cells in SDR rats. (A) H358 cancer cells were transplanted subcutaneously in the SDR rat. Three groups of six rats received either 1×106, 5×106 or 1×107 cells in 5mg/ml Geltrex. In comparison, 1×107 H358 cells were injected in six nude and six NSG mice and growth was tracked for 60 days. Average tumor growth (mm3) over time. (B) 2×106 OV81 cells resuspended in 5mg/ml Geltrex were injected subcutaneously into three female SDR rats. Graph shows three individual rat tumor volumes over time (mm3). Each line represents tumor growth in an individual rat. (C) 2×106 OV185 cells resuspended in 5mg/ml Geltrex were injected subcutaneously into two female SDR rats. Graph shows two individual rat tumor volumes over time (mm3). Each line represents tumor growth in an individual rat.
In a pilot study performed to further explore the ability of the SDR rat to accommodate patient primary tumor derived cell lines, human ovarian cancer (OV81) (46) and human endometrial cancer (OV185) xenografts were established in the flanks of female SDR rats by implanting 2×106 cells of OV81 and OV185 in to three SDR rats each. Tumors implanted with OV81 cells showed 100% engraftment rate with rapid tumor uptake in as early as eight days and consistent growth kinetics (Fig. 6B). In SDR rats implanted with OV185 cells, the tumor engraftment rate was 66.7% (2/3 rats engrafted) with consistent growth kinetics and tumors detectable in 6–8 days (Fig. 6C). One rat did not engraft the tumor and we attribute this to a technical problem with the implantation.
Discussion
Despite its central role in toxicological, pharmacological, neurobehavioral and physiological studies, rats have lagged far behind the mouse as a genetic model (14).We have created a rat with a mutation in the Rag2 gene resulting in a complete lack of mature B cells and significantly reduced mature T cell population. Although targeting the Rag2 locus resulted in an in-frame deletion that does not alter Rag2 protein levels, the lack of mature B and T cells in the rats suggests that the deletion results in a non-functional protein. The mechanism has not been determined.
Several immunodeficient mouse strains exist, exhibiting a range of immune phenotypes, all with differing capabilities for accepting various human cell types for xenograft studies. No two immunodeficient mouse strains are alike. However, until recently, with the discovery of TALENs and CRISPR/Cas9 technology, the only immunodeficient rat strain in existence was the Nude rat (RNU; NIH-Foxn1rnu), which only lacks mature T cells. While the Nude rat has been useful for some human xenograft studies, there are far less human cancer cell lines with survival and growth data in the rat compared to the plethora of cell lines that have been able to be modeled in the mouse. Still, there are some human cancer cell lines that have not been successfully grown in any immunodeficient mouse model. It is possible that these cell lines will not be rejected in a different immunodeficient species, such as the rat.
Here we show proof of principle for human xenograft capabilities for our immunocompromised rat. SDR rat demonstrated efficient uptake and tumor growth kinetics for a wide range of human cancer types. For the NSCLC cell line H358, SDR rat demonstrated superior uptake rate than the NSG mouse – which is the gold standard for human xenograft models. Furthermore the H358 xenografts grew much more uniformly with far less variance in the SDR rat compared to the NSG mouse. In our pilot studies with PDX derived ovarian (OV81) and endometrial (OV185) cell lines, SDR rats also demonstrated a rapid tumor uptake and consistent growth kinetics, with faster growth to much larger tumor sizes than seen in mice.
While our data demonstrate that this SDR rat can accept human xenografts, it is possible that the increase in NK cells will impede engraftment of other human cell types. For this reason, we have created another genetically modified rat with depleted NK cells in addition to the loss of mature T and B cells. This rat strain will potentially support the growth of more human cell types. This model is currently being characterized and validated for its ability to support the growth of human cells.
Supplementary Material
Acknowledgements
We thank Dr. Bjoern Bauer at the University of Kentucky for the gift of the human glioblastoma cell. All tissue processing, embedding and sectioning was performed by the University of Kentucky Imaging Core Facility. Histology and immunohistochemistry imaging was done using equipment at the University of Kentucky Imaging Core Facility. Flow Cytometry analysis was performed by the University of Kentucky Flow Cytometry Core.
Financial support:
KSTC-18451217249: T. Jamling (PI, alias: T. Yeshi), F. Noto, W. Zhang, A. Arey, C. McClain, J. CrawfordR44GM099206: E. Ostertag (PI), T. Jamling.
Footnotes
Conflicts of interest
None.
References
- 1.Anderson S, Luffer-Atlas D, Knadler MP. Predicting circulating human metabolites: how good are we? Chem Res Toxicol. 2009;22(2):243–56. [DOI] [PubMed] [Google Scholar]
- 2.Leclercq L, Cuyckens F, Mannens GS, de Vries R, Timmerman P, Evans DC. Which human metabolites have we MIST? Retrospective analysis, practical aspects, and perspectives for metabolite identification and quantification in pharmaceutical development. Chem Res Toxicol. 2009;22(2):280–93. [DOI] [PubMed] [Google Scholar]
- 3.Walker D, Brady J, Dalvie D, Davis J, Dowty M, Duncan JN, et al. A holistic strategy for characterizing the safety of metabolites through drug discovery and development. Chem Res Toxicol. 2009;22(10):1653–62. [DOI] [PubMed] [Google Scholar]
- 4.Can Peltz G. ‘humanized’ mice improve drug development in the 21st century? Trends Pharmacol Sci. 2013;34(5):255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mak IW, Evaniew N, Ghert M. Lost in translation: animal models and clinical trials in cancer treatment. Am J Transl Res. 2014;6(2):114–8. [PMC free article] [PubMed] [Google Scholar]
- 6.Xu D, Peltz G. Can Humanized Mice Predict Drug “Behavior” in Humans? Annu Rev Pharmacol Toxicol. 2016;56:323–38. [DOI] [PubMed] [Google Scholar]
- 7.Fattinger K, Funk C, Pantze M, Weber C, Reichen J, Stieger B, et al. The endothelin antagonist bosentan inhibits the canalicular bile salt export pump: a potential mechanism for hepatic adverse reactions. Clin Pharmacol Ther. 2001;69(4):223–31. [DOI] [PubMed] [Google Scholar]
- 8.Manning F, Swartz M. Review of the Fialuridine (FIAU) Clinical Trials. Washington DC: Institute of Medicine (US); 1995. [PubMed] [Google Scholar]
- 9.McKenzie R, Fried MW, Sallie R, Conjeevaram H, Di Bisceglie AM, Park Y, et al. Hepatic failure and lactic acidosis due to fialuridine (FIAU), an investigational nucleoside analogue for chronic hepatitis B. N Engl J Med. 1995;333(17):1099–105. [DOI] [PubMed] [Google Scholar]
- 10.Shultz LD, Goodwin N, Ishikawa F, Hosur V, Lyons BL, Greiner DL. Human cancer growth and therapy in immunodeficient mouse models. Cold Spring Harb Protoc. 2014;2014(7):694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gad S Animal Models in Toxicology. 2 ed: CRC Press; 2006. [Google Scholar]
- 12.Jacob HJ. Functional genomics and rat models. Genome Res. 1999;9(11):1013–6. [DOI] [PubMed] [Google Scholar]
- 13.Iannaccone PM, Jacob HJ. Rats! Dis Model Mech. 2009;2(5–6):206–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazar J, Moreno C, Jacob HJ, Kwitek AE. Impact of genomics on research in the rat. Genome Res. 2005;15(12):1717–28. [DOI] [PubMed] [Google Scholar]
- 15.Mollard S, Mousseau Y, Baaj Y, Richard L, Cook-Moreau J, Monteil J, et al. How can grafted breast cancer models be optimized? Cancer Biol Ther. 2011;12(10):855–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nofiele JT, Cheng HL. Establishment of a lung metastatic breast tumor xenograft model in nude rats. PLoS One. 2014;9(5):e97950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuchida T, Zheng YW, Zhang RR, Takebe T, Ueno Y, Sekine K, et al. The development of humanized liver with Rag1 knockout rats. Transplant Proc. 2014;46(4):1191–3. [DOI] [PubMed] [Google Scholar]
- 18.Ménoret S, Fontanière S, Jantz D, Tesson L, Thinard R, Rémy S, et al. Generation of Rag1-knockout immunodeficient rats and mice using engineered meganucleases. FASEB J. 2013;27(2):703–11. [DOI] [PubMed] [Google Scholar]
- 19.Festing MF, May D, Connors TA, Lovell D, Sparrow S. An athymic nude mutation in the rat. Nature. 1978;274(5669):365–6. [DOI] [PubMed] [Google Scholar]
- 20.Colston MJ, Fieldsteel AH, Dawson PJ. Growth and regression of human tumor cell lines in congenitally athymic (rnu/rnu) rats. J Natl Cancer Inst. 1981;66(5):843–8. [PubMed] [Google Scholar]
- 21.Drewinko B, Moskwa P, Lotzovà E, Trujillo JM. Successful heterotransplantation of human colon cancer cells to athymic animals is related to tumor cell differentiation and growth kinetics and to host natural killer cell activity. Invasion Metastasis. 1986;6(2):69–82. [PubMed] [Google Scholar]
- 22.Maruo K, Ueyama Y, Kuwahara Y, Hioki K, Saito M, Nomura T, et al. Human tumour xenografts in athymic rats and their age dependence. Br J Cancer. 1982;45(5):786–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fowler JA, Mundy GR, Lwin ST, Lynch CC, Edwards CM. A murine model of myeloma that allows genetic manipulation of the host microenvironment. Dis Model Mech. 2009;2(11–12):604–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.J. B. Immunodeficient Mouse Models: An Overview. The Open Immunology Journal. 2009;2:79–85. [Google Scholar]
- 25.Holzapfel BM, Wagner F, Thibaudeau L, Levesque JP, Hutmacher DW. Concise review: humanized models of tumor immunology in the 21st century: convergence of cancer research and tissue engineering. Stem Cells. 2015;33(6):1696–704. [DOI] [PubMed] [Google Scholar]
- 26.Mazurier F, Fontanellas A, Salesse S, Taine L, Landriau S, Moreau-Gaudry F, et al. A novel immunodeficient mouse model--RAG2 × common cytokine receptor gamma chain double mutants--requiring exogenous cytokine administration for human hematopoietic stem cell engraftment. J Interferon Cytokine Res. 1999;19(5):533–41. [DOI] [PubMed] [Google Scholar]
- 27.van Rijn RS, Simonetti ER, Hagenbeek A, Hogenes MC, de Weger RA, Canninga-van Dijk MR, et al. A new xenograft model for graft-versus-host disease by intravenous transfer of human peripheral blood mononuclear cells in RAG2−/− gammac−/− double-mutant mice. Blood. 2003;102(7):2522–31. [DOI] [PubMed] [Google Scholar]
- 28.Ishikawa F, Yasukawa M, Lyons B, Yoshida S, Miyamoto T, Yoshimoto G, et al. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor {gamma} chain(null) mice. Blood. 2005;106(5):1565–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevens LC, Little CC. Spontaneous Testicular Teratomas in an Inbred Strain of Mice. Proc Natl Acad Sci U S A. 1954;40(11):1080–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51(3):503–12. [DOI] [PubMed] [Google Scholar]
- 31.Tasher D, Dalal I. The genetic basis of severe combined immunodeficiency and its variants. Appl Clin Genet. 2012;5:67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68(5):855–67. [DOI] [PubMed] [Google Scholar]
- 33.Chapman KM, Medrano GA, Jaichander P, Chaudhary J, Waits AE, Nobrega MA, et al. Targeted Germline Modifications in Rats Using CRISPR/Cas9 and Spermatogonial Stem Cells. Cell Rep. 2015;10(11):1828–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Z, Falciatori I, Molyneux LA, Richardson TE, Chapman KM, Hamra FK. Spermatogonial culture medium: an effective and efficient nutrient mixture for culturing rat spermatogonial stem cells. Biol Reprod. 2009;81(1):77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Izsvák Z, Fröhlich J, Grabundzija I, Shirley JR, Powell HM, Chapman KM, et al. Generating knockout rats by transposon mutagenesis in spermatogonial stem cells. Nat Methods. 2010;7(6):443–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivics Z, Izsvák Z, Chapman KM, Hamra FK. Sleeping Beauty transposon mutagenesis of the rat genome in spermatogonial stem cells. Methods. 2011;53(4):356–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol. 1989;24(3):148–54. [DOI] [PubMed] [Google Scholar]
- 38.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30(5):460–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reyon D, Maeder ML, Khayter C, Tsai SQ, Foley JE, Sander JD, et al. Engineering customized TALE nucleases (TALENs) and TALE transcription factors by fast ligation-based automatable solid-phase high-throughput (FLASH) assembly. Curr Protoc Mol Biol. 2013;Chapter 12:Unit 12.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, et al. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36(6):653–9. [DOI] [PubMed] [Google Scholar]
- 41.Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36(6):647–52. [DOI] [PubMed] [Google Scholar]
- 42.Bruno L, Res P, Dessing M, Cella M, Spits H. Identification of a committed T cell precursor population in adult human peripheral blood. J Exp Med. 1997;185(5):875–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kövesdi D, Bell SE, Turner M. The development of mature B lymphocytes requires the combined function of CD19 and the p110δ subunit of PI3K. Self Nonself. 2010;1(2):144–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Q, Fan C, Zhou S, Guo Y, Zuo Q, Ma J, et al. Bioluminescent imaging of vaccinia virus infection in immunocompetent and immunodeficient rats as a model for human smallpox. Sci Rep. 2015;5:11397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mashimo T, Takizawa A, Kobayashi J, Kunihiro Y, Yoshimi K, Ishida S, et al. Generation and characterization of severe combined immunodeficiency rats. Cell Rep. 2012;2(3):685–94. [DOI] [PubMed] [Google Scholar]
- 46.Nagaraj AB, Joseph P, Kovalenko O, Singh S, Armstrong A, Redline R, et al. Critical role of Wnt/β-catenin signaling in driving epithelial ovarian cancer platinum resistance. Oncotarget. 2015;6(27):23720–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


