Biologists have long appreciated natural variation in the nematode phylum. The development of Caenorhabditis elegans as a model organism has provided a rich set of specific genetic and cellular mechanisms that have been subjected to...
Keywords: C. elegans, connectome, developmental systems drift, embryo, evolution, gene regulatory network, sex determination, sperm, vulva, WormBook
Abstract
Since the earliest days of research on nematodes, scientists have noted the developmental and morphological variation that exists within and between species. As various cellular and developmental processes were revealed through intense focus on Caenorhabditis elegans, these comparative studies have expanded. Within the genus Caenorhabditis, they include characterization of intraspecific polymorphisms and comparisons of distinct species, all generally amenable to the same laboratory culture methods and supported by robust genomic and experimental tools. The C. elegans paradigm has also motivated studies with more distantly related nematodes and animals. Combined with improved phylogenies, this work has led to important insights about the evolution of nematode development. First, while many aspects of C. elegans development are representative of Caenorhabditis, and of terrestrial nematodes more generally, others vary in ways both obvious and cryptic. Second, the system has revealed several clear examples of developmental flexibility in achieving a particular trait. This includes developmental system drift, in which the developmental control of homologous traits has diverged in different lineages, and cases of convergent evolution. Overall, the wealth of information and experimental techniques developed in C. elegans is being leveraged to make nematodes a powerful system for evolutionary cellular and developmental biology.
THE small, laboratory-friendly nematodes of the genus Caenorhabditis were first developed as a system for genetic analysis of animal development by a few early champions. One of the first experimental studies on C. elegans was performed by Japanese American Hikokuro Honda, who found that sperm determine the sex of progeny, and discovered that oocyte meiosis is not completed until after fertilization (Honda 1925). Two decades later, the French biologist Victor Nigon and his American colleague Ellsworth Dougherty greatly extended this work (Nigon 1943; Dougherty and Nigon 1949; Ferris and Hieb 2015; Nigon and Félix 2017), aided by improvements in culture methodology by Briggs (1946). These workers set the stage for Sydney Brenner’s breakthroughs with C. elegans (Brenner 1974, 2009). Along with French biologist Emile Maupas, who first described C. elegans (Maupas 1900), all of these early researchers were struck by the fact that, within a stereotypical body form, evolutionary variation in habitat choice, feeding strategy, reproductive mode, behavior, and anatomical details are rampant. Thus, research focusing on C. elegans was always complemented by the work of other nematologists working in other groups, such as other nematodes in the order Rhabditida (Figure 1) (Sudhaus 1976). It can therefore be fairly said that questions of biodiversity, the evolution of developmental processes, and their connections to ecology were very much lingering over the field even in the earliest days. The authors of this review represent examples of contemporary biologists who share their predecessors’ fascination with the evolution of nematode development. Trained in the C. elegans paradigm, we and others take particular delight in gazing outward across the phylogeny, always on the lookout for new phenomena and explanations for how they evolved.
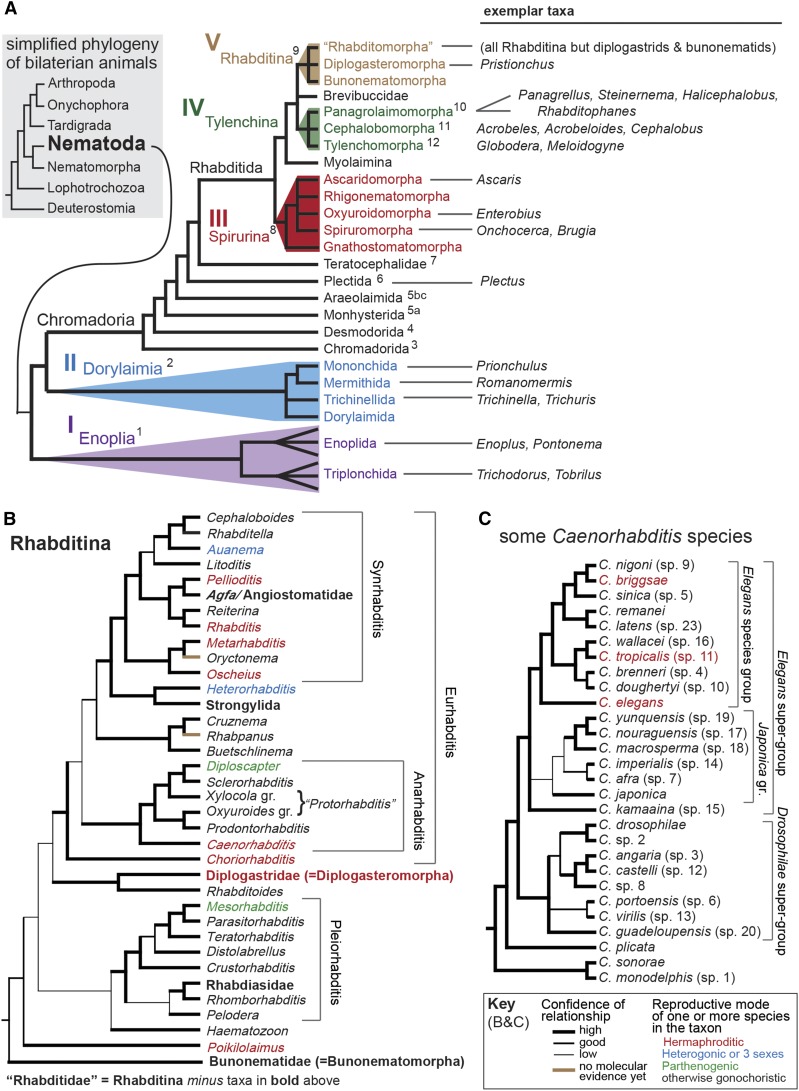
Figure 1.
Phylogenies of phylum Nematoda, suborder Rhabditina, and genus Caenorhabditis, based on molecular data. (A) Inset shows the phylogenetic position of Nematoda within a very simplified phylogeny of bilaterian animals. Recent molecular studies place Nematoda together with its sister group Nematomorpha as the closest relatives of Panarthopoda (Arthropoda, Onychophora, Tardigrada) in a clade often called Ecdysozoa (Giribet 2016; Giribet and Edgecombe 2017). The phylogeny of Nematoda has been derived mainly from ribosomal RNA (rRNA) genes and contains several well-defined clades: clades I–V (De Ley and Blaxter 2004; De Ley 2006) designated in like-colored roman numerals, taxon names, and polygons; and clades 1–12 designated in black superscripts to corresponding taxon names (Holterman et al. 2008; van Megen et al. 2009). Some taxa have been left out here for simplicity. Taxa other than Rhabditina that are mentioned in this review are listed at the right. Adapted with permission from Blaxter (2011) and Kiontke and Fitch (2013). Taxa in quotation marks are paraphyletic: “Rhabditomorpha” includes all Rhabditina except Diplogasteromorpha and Bunonematomorpha. (B) Phylogeny of Rhabditina (clade V), almost entirely based on molecular data from rRNA and other loci (Kiontke et al. 2007; Ross et al. 2010; Kanzaki et al. 2017). Thickness of the lineages, as indicated in the key at lower right, indicates the approximate level of confidence estimated from statistical tests. The systematics of “Rhabditidae” was recently revised (Sudhaus 2011) based almost entirely on the molecular phylogeny (Kiontke et al. 2007) with some consideration of morphological characters to place taxa only known from literature descriptions (brown lineages). A few, mostly monotypic taxa of uncertain position are not shown. Four named suprageneric clades are shown with brackets. Despite being paraphyletic, “Rhabditidae” is a useful taxon because it includes many free-living (rarely parasitic) species with fairly similar Bauplan and excludes three specialized parasitic taxa (Angiostomatidae/Agfa, Strongylida, Rhabdiasidae) and Diplogastridae, a clade of species morphologically distinguished from “Rhabditidae” that have undergone an extensive adaptive radiation. Pristionchus pacificus and its relatives are included in the Diplogastridae. The “Rhabditidae” sister taxa to each of these special groups provide important resources for investigating the evolutionary origins of parasitism and other specializations that have resulted in adaptive radiations. Colored fonts indicate taxa in which reproductive mode has evolved from gonochorism to hermaphroditism, heterogonism or parthenogenesis (see key at lower right). Taxon names in bold font are at higher levels than the genera otherwise depicted. For more complete information, see RhabditinaDB at rhabditina.org. (C) Phylogeny for some Caenorhabditis species as inferred by molecular data from rRNA and several other loci (Kiontke et al. 2011). Due to the rapid rate of discovery, species are provisionally designated with numbers (sp. n) until names can be attached to these species units (Félix et al. 2014). Only 28 of the ∼50 known species are shown here; however, this phylogeny shows all the major known clades (demarcated here as “species groups”). Several Caenorhabditis species are only known from morphological descriptions and not included here. Hermaphroditic species are indicated in red font; other species are gonochoristic.
Unique Attributes of the Caenorhabditis System
Caenorhabditis offers an attractive set of attributes for evolutionary developmental biology (EDB, or “evo-devo”). First, it presents a highly simplified and stereotyped developmental system. Worms are transparent and have a small number of somatic cells formed by a predictable lineage (Sulston and Horvitz 1977; Kimble and Hirsh 1979; Sulston et al. 1983). This allows one to homologize and compare developmental processes at the resolution of individual cells (Zhao et al. 2008). Nevertheless, the major tissues of larger, more complex animals (e.g., muscles, integument, nerves, sensory cells, renal, digestive and reproductive organs, and immune cells) are present (see www.wormatlas.org). While zoologists of the past believed the simple anatomy of nematodes represented a primitive state, molecular phylogenetics (Figure 1) have generally supported the membership of the phylum Nematoda in the Ecdysozoan superphylum of protostomes (Giribet and Edgecombe 2017). This implies that nematodes’ often miniature bodies are actually highly derived and highly specialized. An alternative interpretation to the C. elegans body, therefore, is that it is a sophisticated, “microchip animal” that evolved from a larger progenitor. Along the way, some ancestral regulators of animal development have been shed or modified. For example, Caenorhabditis have fewer Hox genes than other nematodes or more distantly related animals (Aboobaker and Blaxter 2003), and the hedgehog signaling pathway has both diverged in its roles (Bürglin and Kuwabara 2006; Soloviev et al. 2011) and been co-opted to form the core of the global sex determination pathway (Zarkower 2006). Simultaneously, proteins implicated in chemosensation, such as rhodopsin-related G protein-coupled receptors, have been amplified and diversified (Bargmann 2006).
A striking variable distinguishing some nematodes, such as Caenorhabditis, Pristionchus, and some other clade V taxa relates to sexual mode. Although the ancestral gonochoristic (male-female), obligately outcrossing mode is retained by most species, several have evolved a self-fertile hermaphrodite (Kiontke et al. 2011) (Figure 1). Males persist at greatly reduced frequencies, creating an androdioecious mating system. Androdioecy is rare in both animals and plants (Pannell 2002; Weeks et al. 2006), but because it makes genetic manipulations simpler and faster, selfing species like C. elegans and C. briggsae (and peas!) have always been favored by experimental biologists. A major area of research reviewed below involves comparisons between close relatives with different sexual modes.
Despite its fame for exhibiting “invariant” development, C. elegans also offers one of the best-characterized examples of an adaptive phenotypic plasticity: the formation of the dauer larva. This resistant variant of the third larval stage is triggered by crowding or starvation in the previous stage (Albert et al. 1981), which, in turn, alters pheromones and nutritional status. These cues are then translated into differential states of signaling pathways and circulating hormones (Fielenbach and Antebi 2008). Because the dauer larva appears to be a universal dispersal form for both free-living and parasitic terrestrial nematodes (Crook 2014), the cues that induce its development and the attributes it possesses are likely to vary with ecological niche. Some first examples of this variation are reviewed below.
C. elegans has also enjoyed early and intense attention to the characterization of its genome and its relation to various processes. It was the first animal species to have a complete sequence assembly (C. elegans Sequencing Consortium 1998), and this quickly became a handmaiden to gene-focused EDB (e.g., Kuwabara and Shah 1994; Haag and Kimble 2000). Interest in examining interspecies variation led to a collection of genome assemblies from other Caenorhabditis species (Stein et al., 2003; Hillier et al. 2007; Ross et al. 2011; Fierst et al. 2015; see also http://www.nematodes.org/nematodegenomes/index.php/Main_Page). This work is ongoing on an ever-larger scale, driven by both the discovery of many new species (Kiontke et al. 2011; Barrière and Félix 2014; Huang et al. 2014; Ferrari et al. 2017; Slos et al. 2017) and advances in sequencing technology (see caenorhabditis.org). Note that genome sequencing and annotation have been completed, or are in progress, for all of the Caenorhabditis species shown in Figure 1C except for C. sonorae, which has been refractory to reisolation. Within species, the genomes of many genetically distinct isolates from around the world are also being characterized (Cutter et al. 2006; Rockman and Kruglyak 2009; Dey et al. 2012; Thomas et al. 2015; Cook et al. 2016). This presents a rich resource with which to examine standing variation in molecules and processes (e.g., Cook et al. 2017).
Perhaps not surprisingly, evolutionary studies of Caenorhabditis grew as comparative offshoots of the major topics of C. elegans research. Essentially, once an aspect of the development of C. elegans came to be understood in some detail, several obvious questions followed quickly: Is that general? If it is general, can it help us understand natural variation in form? If it is not general, how did it evolve? Sometimes the reverse line of questioning, starting with an appreciation of variation in a particular feature, has also sparked more in-depth work in C. elegans itself. In this fashion, EDB using nematodes has focused on these topics:
zygotic mitosis and founder cell specification.
embryonic cell lineage.
developmental regulation of gene expression.
neuroanatomy.
sex determination.
germ cell development.
spermatogenesis and sexual behavior.
vulva and somatic gonad development.
nongonadal somatic sexual dimorphism and male development, e.g., the tail.
dauer formation.
Below, we summarize key findings from C. elegans research on the above developmental processes, and discuss the evolutionary studies that they have enabled. While the latter would, in principle, include many studies of deeply diverged nematodes and other phyla, we emphasize here the more recent evolution revealed by comparisons within Rhabditida (Clade V, Figure 1). Finally, we attempt to distill the major insights that have emerged from nematode EDB.
Findings
Zygotic mitosis
The first embryonic divisions of the C. elegans embryo have been extensively described (Rose and Gönczy 2014) (Figure 2). Briefly, oocytes are blocked in prophase of meiosis I and unpolarized. At fertilization, the sperm brings in the paternal DNA and a pair of centrioles. These centrioles rapidly recruit pericentriolar material, which locally destabilizes the cortical actomyosin contractility, leading to the asymmetric repartition of the PAR polarity proteins. At fertilization, the anteroposterior (AP) axis of the cell is thus established, and the sperm entry site defines the posterior side of the cell. In response to the PAR polarity, cytoplasmic proteins localize asymmetrically in the cell, and the mitotic spindle that is initially centrally located becomes posteriorly positioned along the AP axis during anaphase (Figure 2). This asymmetric displacement comes with impressive transverse oscillations of the spindle—the manifestation of excess forces pulling on posterior astral microtubules. Because the cell cleavage plane is perpendicular to the spindle, two daughter cells of unequal size and asymmetric fate are formed, the posterior cell P1 being smaller than the anterior cell AB. This stereotyped asymmetric division has become a model to study oriented cell division because of the exquisite spatiotemporal resolution of events during this first cell cycle and because of the strong conservation of molecules involved across phyla (Neumuller and Knoblich 2009). At each subsequent division, a similar asymmetric cell division is reproduced in the P lineage, ultimately giving birth to the founder cell of the germline, the P4 cell.
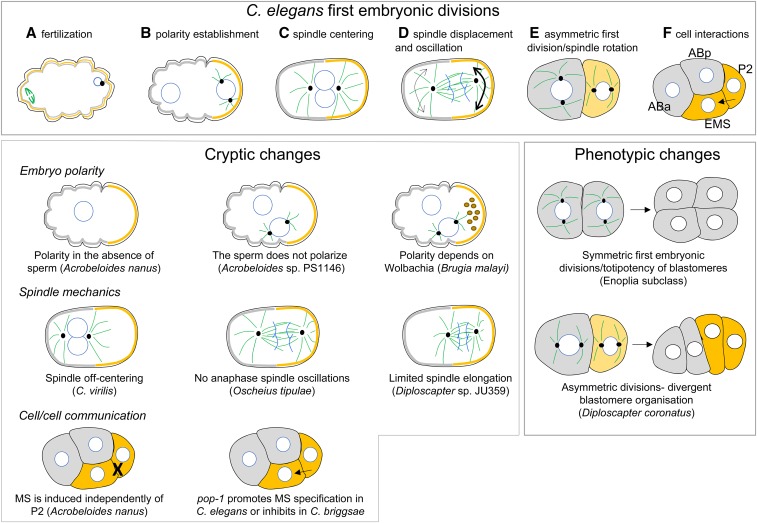
Figure 2.
First embryonic cell divisions in C. elegans and variations in other species. Top panel (A–F) schematic representation of the two first cell division of the C. elegans embryo. Microtubules are shown in green, centrosomes are represented by black dots and nuclei by white circles. Polarity proteins are shown in gray and yellow. (A) Initially, the oocyte is unpolarized. After the sperm entry (on the right), female meiosis resumes (spindle on the left). (B) After fertilization, polarity proteins are asymmetrically localized and the sperm entry site defines the posterior pole of the cell on the right (B). In response to polarity, the mitotic spindle (D) and cell fate determinants (E) are asymmetrically localized. During the second cell division, spindle orientation is different between the two cells (E), giving rise to a rhomboid organization of blastomeres at the four-cell stage (F). At this stage, the P2 cell sends a Wnt signal to EMS. Phenotypic changes: timing of cell divisions and cell orientations can vary between species leading to different cellular contacts and blastomeres organization. Cryptic changes: among species that have similar embryonic cell divisions than C. elegans, evolutionary changes are found in the polarization of the embryo, the positioning of the first mitotic spindle or in cell/cell communication.
At the second cell cycle, while the founder cell AB divides symmetrically to generate ABa and ABp, P1 gives rise to the small P2 cell and EMS. During this division, the mitotic spindle of P1 rotates along the AP axis of the embryo and becomes perpendicular to the spindle of AB. This leads to a rhomboid organization of the four first blastomeres, which is essential for the subsequent cellular interactions (Figure 2). Indeed, at the four-cell stage, P2 sends a Wnt signal to EMS, which then divides asymmetrically to give rise to the founder cell of the intestine (the E cell) and the founder cell of the mesoderm (MS). In the absence of P2 or Wnt signaling, EMS gives rise to two MS cells. Through Notch/Delta signaling, P2 also induces different fate acquisition in ABp compared to ABa. P2 next divides to give the founder cell C and P3, which divides again to give the founder cells D and P4. These cell divisions thus rapidly produce the six key founder cells of C. elegans embryos (Sulston et al. 1983).
The embryos of most nematodes, in particular free-living forms, can easily develop ex-utero. The first cell divisions are easy to monitor under slide and coverslip because cells are large and transparent and the pace of cell divisions is relatively fast. These properties allowed the analysis of the early steps of embryogenesis in very diverse nematode species, starting with the founding work of T. Boveri on Ascaris megalocephala (=Parascaris equorum) (Maderspacher 2008) and followed by Nigon and others in the early twentieth century (Nigon and Félix 2017).
Among the long list of free-living and parasitic species that have been observed since then, only species from Enoplia (Clade I, Figure 1A) undergo a series of symmetric embryonic first divisions, with late specification of cellular identity (Malakhov 1994; Schulze and Schierenberg 2008, 2009, 2011). In all the other Chromadoria species so far observed, the first embryonic division is asymmetric, giving rise to two unequally sized, asymmetrically fated daughter cells (Brauchle et al. 2009; Schulze and Schierenberg 2011; Landmann et al. 2014; Calderón-Urrea et al. 2016). Thus, as in C. elegans, the polarity of the embryo is already established during the first mitosis in all these species. However, in Acrobeloides sp. PS1146 (Cephalobomorpha), the sperm entry site does not correlate with the posterior side of the embryo, in contrast to C. elegans (Goldstein et al. 1998). There is also an absence of cytoplasmic movements toward the site of sperm entry, further suggesting that the sperm is not the polarity cue in this species. In a study of 16 other free-living and parasitic nematodes, a clade that includes Acrobeloides does not show any sign of cytoplasmic flow, while the other groups resemble C. elegans (Goldstein et al. 1998) (Figure 2). Thus, embryonic early polarity can be established independently of the sperm centrosomes in many nematode species. Parthenogenesis has emerged several times in the group of species for which polarity is independent of the sperm, leading to the hypothesis that the ability to polarize the embryo independently of sperm might have been a preadaptation to the emergence of parthenogenesis (Goldstein et al. 1998). Such a transition state—to sperm-independent polarization in gonochoristic species—is, however, not a prerequisite for the emergence of parthenogenesis, because parthenogenesis is also found in the Diploscapter genus within the “Rhabditidae” (Figure 1B) (Fradin et al. 2017)—a paraphyletic family composed mainly of gonochoristic species that use sperm as a polarity cue.
The origin of the polarity cue in the absence of sperm has been investigated (Lahl et al. 2006). Because, in some experimental conditions, the female meiotic spindle can promote PAR asymmetric localization in C. elegans embryos (Wallenfang and Seydoux 2000), one tempting hypothesis is that the female meiotic spindle becomes the polarity cue in parthenogenetic species. However, this hypothesis can be ruled out, as the position of the posterior pole does not correlate with the position of the polar bodies in Acrobeloides nanus and in Diploscapter coronatus (Lahl et al. 2009). In Acrobeloides, the anterior side of the embryos always faces the vulva, suggesting that the orientation of the oocytes within the gonadal tract provides a polarity cue. In contrast, there is no correlation between embryo orientation within the uterus and the position of the posterior pole in Diploscapter, suggesting that polarity in these species is established randomly. Because polarization relies on the local destablilization of the actomyosin network in C. elegans, one could imagine that spontaneous self-organization of the actomyosin cortex triggers symmetry breaking to define the anterior–posterior axis of the embryo in Diploscapter.
Early embryo polarization is also observed in the parasitic nematode Brugia malayi (Spirurina) (Landmann et al. 2014). In this species, a microtubule-organizing center is found in oocytes prior to fertilization at the future posterior side of the cell, opposite to the location of the female meiotic spindle, suggesting a microtubule-based mechanism of polarization from a maternal origin. Wolbachia endosymbionts are found enriched at the posterior side of the one-cell embryo and in the P1 cell after the first division in this species; their removal leads to polarity defects in two-cell embryos (Landmann et al. 2014). Whether Wolbachia are required for the initiation of polarity or its maintenance remains to be determined, but this example nicely illustrates the diversity of mechanisms that exist to establish the first embryonic polarity axis of nematode embryos during the first cell cycle.
In C. elegans, the early polarization of the embryo after fertilization can be easily scored by a series of cortical contractions following the reorganization of the actomyosin network. Similarly, in response to the asymmetric localization of PAR proteins, microtubule force generators produce movements of the nuclei and the spindle that are extremely stereotyped. In one study, the first two embryonic divisions of 34 rhabditids were scored, uncovering a large degree of variability in these subcellular phenomena (Brauchle et al. 2009). Farhadifar et al. (2015) analyzed the first embryonic cell division of 42 different rhabditid species and of natural isolates and mutation accumulation lines of C. elegans. Spindle length appears to be constrained by stabilizing selection on cell and embryo size, with the two linked in C. elegans by a linear scaling relationship. However, the observed variations in spindle movements could not be explained by evolutionary changes in cell size between species (Valfort et al. 2018). Moreover, traits associated with spindle movements combined in ways contrasting with the expectation based on C. elegans studies, suggesting that mechanical optimization of the mitotic spindle differs between species despite a conserved output phenotype: the asymmetry of division.
The origin of differences in spindle positioning between C. elegans and its congener C. briggsae have been explored (Riche et al. 2013). In C. briggsae, at the onset of mitosis, the spindle is anteriorly shifted in contrast to a central position found in C. elegans. During anaphase, the spindle is pulled posteriorly in both species. However, this movement is accompanied by much-reduced transverse spindle oscillations in C. briggsae compared to C. elegans. These phenotypes were attributable to the GPR-1/2 proteins—components of the cortical force generator complex. While two recently duplicated genes gpr-1 and gpr-2 are found in the genome of C. elegans, C. briggsae has only one gpr-2 gene. This difference in gene copy number correlated with a lower expression level in C. briggsae compared to C. elegans but also with a different spatio-temporal regulation.. Thus, the processes that produce a conserved and essential cellular feature, asymmetric spindle position, are distinct. This represents a case of what has been dubbed developmental system drift (DSD; True and Haag 2001) or phenogenetic drift (Weiss and Fullerton 2000) at the earliest stages of embryonic development.
Postzygotic cell lineage and founder cell specification
Although descriptions of early embryogenesis in Enoplia and Dorylaimia (Clades I and II; Figure 1) remain scarce because species of these clades are difficult to maintain in laboratory conditions (Schulze and Schierenberg 2011), what is known suggests that a striking diversity of mechanisms for early-development evolved early in the phylum. In Enoplus brevis (Enoplia, Clade I) the first embryonic divisions are symmetric and body axes are not specified during the first cell divisions. Moreover, except for the endoderm (E) lineage, no founder cells are identified and cells become determined later “en bloc” (Schulze and Schierenberg 2011). In Pontonema vulgare, the spatial arrangements of the blast cells producing specific lineages can also vary substantially among embryos (Malakhov 1994; Voronov 1999), also suggestive of “regulative” development. In another representative of Enoplia, Tobrilus, a blastocoel is even observed with a canonical gastrulation—a feature that was unexpected in this phylum of pseudocoelomate worms. However, anteroposterior polarity is established at the four-cell stage, and three founder cells for the germline, the pharynx, and the intestine are found (Schierenberg 2005; Schulze and Schierenberg 2011). Results obtained in Prionchulus punctatus (Mononchida) are contradictory. On the one hand, laser ablation of half of the embryo does not prevent the development of a normal fertile adult (Borgonie et al. 2000). On the other hand, there are five founder cells (E, pharynx, D, C, and P), suggesting an early specification of cellular identities (Schulze and Schierenberg 2011). While Romanomermis culicivorax (Dorylaimia, a.k.a. Clade II) has six founder cells like C. elegans, tissues are formed by rings of cells, reminiscent of a segmentation process (Schulze and Schierenberg 2008, 2009). These species present extremely divergent early embryonic development, making it difficult to infer the ancestral pattern of development in nematodes. Nevertheless, because it is shared with outgroup phyla, the absence of deterministic lineage was most likely an ancestral character associated with slow embryogenesis.
On the other hand, the more derived Chromadoria are characterized by a fast embryonic development with largely deterministic lineages (Malakhov 1994; Schulze and Schierenberg 2011). Plectus species (Plectida) seem to have an intermediate way to specify cell types between Enoplia (no early founder cells) and Rhabditina (six founder cells established by the 16-cell stage): while the P lineage is clearly specified, the AB lineage is highly variable, leading to variable cell–cell contacts from one embryo to the other (Schulze et al. 2012). Within the Chromadoria, the early lineages of many species resemble C. elegans. Early examples came from work with the Clade IV species Panagrellus redivivus, Turbatrix aceti, and Aphelencoides blastophthorus imbedded in the seminal description of the C. elegans embryonic lineages (Sulston et al. 1983). Later studies examined fellow Clade V taxa, such as C. briggsae (Zhao et al. 2008), Litoditis marina (a.k.a. Pellioditis marina) (Houthoofd et al. 2003), Pristionchus pacificus (Vangestel et al. 2008), and Oscheius shamimi (Tahseen and Nisa 2006). Other species within the Tylenchina a.k.a. Clade IV have also been described, such as Rhabditophanes (Houthoofd et al. 2008), Halicephalobus (Borgonie et al. 2000), as well as from the more distantly related Spirurina a.k.a. Clade III (Ascaris; Boveri 1899). Nevertheless, in a detailed analysis of 70 different species from 19 different nematode families within Chromadoria, differences were found in the spatial and temporal organization of the founder cells (Dolinski et al. 2001). First, AB and P1 divide at the same rate as C. elegans (synchrony), or at different rates (asynchrony) as in Acrobeloides, in which all the P divisions take place before the first division of AB. It has been previously proposed that such timely separation of soma and germline divisions would ensure proper germline identity (Schlicht and Schierenberg 1991). Yet, in species for which AB and P1 divide in the same generation, a delay in cell divisions can exist, such as in C. elegans, where AB divides 2 min before P1, or in Diploscapter and Poikilolaimus oxycercus, where P1 divides first (Brauchle et al. 2009). Moreover, species have either a rhomboid organization of blastomeres as in C. elegans, or a linear arrangement at the four-cell stage when both AB and P1 spindles rotate to align along the AP axis. Such linear organization is found in Diploscapter and some “Protorhabditis” species (Dolinski et al. 2001; Brauchle et al. 2009; Lahl et al. 2009; Fradin et al. 2017) or in Meloidogyne (Dolinski et al. 2001; Calderón-Urrea et al. 2016) (Figure 2).
In species with linear arrangement of the early blastomeres, the question of lineage specification remains open. In C. elegans, ABp fate is induced by P2 via Notch signaling (Mello et al. 1994; Mickey et al. 1996). The linear arrangement in the four-cell embryo means that this signaling must occur in a different way, if it occurs at all (Brauchle et al. 2009). Also, in Diploscapter coronatus, and some other species of the “Protorhabditis” group, P2 has already divided into C and P3 at the time of EMS and ABp division (Lahl et al. 2009; Fradin et al. 2017). Moreover, the orientation of C and P3 is random, at least in Diploscapter coronatus. Thus, in only 50% of embryos does ABp contact C while EMS contacts P3. Despite these random contacts, ABp and ABa have a distinct lineage, suggesting that ABp specification is independent of an induction by either C or P3. Whether EMS requires an inductive signal by a neighboring cell or is cell-autonomous remains to be determined. Importantly, removal of EMS leads to an absence of intestinal cells, demonstrating an absence of multipotency, as in C. elegans (Lahl et al. 2009). In striking contrast, in Acrobeloides nanus, where cellular contacts at the four-cell stage are similar to C. elegans, the absence of P2 does not prevent gut specification (Figure 2). Rather, any cell at the three-cell stage can give rise to intestinal cells after ablation of the others. Similarly, if AB is ablated, EMS takes over and C becomes EMS. Thus, in this species, multipotency and hierarchy of transformations is observed, despite an early segregation of the lineage in wild-type embryos (Wiegner and Schierenberg 1998, 1999). Unexpectedly, in the distantly related Plectus, the situation resembles C. elegans, since an induction of EMS by P2 is necessary to specify the intestine (Schulze et al. 2012). Therefore, many different solutions and reversals are found over the course of nematode evolution to specify cellular identities during early embryogenesis.
Interestingly, even within Caenorhabditis, differences in gut specification have been revealed at the molecular level, despite conservation of cellular interactions and blastomere specification (Coroian et al. 2006; Lin et al. 2009). Upon Wnt signaling by P2, the transcription factors SKN-1 and POP-1 act to specify E and MS identity. While POP-1 has a positive contribution to MS specification in C. elegans, it represses the MS fate in C. briggsae. In an interesting twist to the story, MED-1,2, two GATA transcription factors that act downstream of SKN-1, evolved in the lineage to C. elegans and are not present in C. briggsae. One model for the co-option of these new factors is via a transitional feed-forward architecture in which SKN-1 acts both through and independently of MED-1,2 (Maduro 2009). Given the highly conserved cell lineages in the two species (Zhao et al. 2008), such an opposite role for a key signaling pathway is an unexpected case of DSD.
The above results demonstrate that—despite a very constrained body plan—early steps of embryogenesis vary considerably between nematodes. The molecular signature of such diversity in the early steps of embryogenesis was explored in five different species within Caenorhabditis (Levin et al. 2012). Embryos from 10 different morphological stages were collected, from four-cell stage embryos to L1 larvae, and their transcriptomes were analyzed. Despite species-specific developmental timing, embryos from specific stages showed a similar pattern of gene expression across species, suggesting the existence of conserved “milestones” in development. Importantly, at midembryogenesis, corresponding to ventral enclosure, transcriptomes from different species were the least divergent. Moreover, genes that were activated at this stage showed enrichment in crucial functions such as patterning by Hox genes or locomotion. These results led to the proposition (Levin et al. 2012) that for nematodes, ventral enclosure represents a key, body plan-defining point in development, the so-called phylotypic stage (Slack et al. 1993; Richardson et al. 1998). Transcriptome profiles throughout embryonic development were also performed in 20 mutation accumulation lines of C. elegans, in which the effect of selection is largely abolished. For all developmental stages, except ventral enclosure, variation in gene expression was much higher in the MA lines. This result strongly suggests that gene expression during ventral enclosure is highly conserved because of stabilizing selection (Zalts and Yanai 2017). Regardless of whether or not there is a phylotypic stage, these results do support an hourglass model (Raff 1996), in which nematode development shows the greatest diversity prior to or after a conserved point midway through embryogenesis.
Developmental regulation of gene expression
The variation in global embryonic gene expression described above indicates that the transcriptional controls acting on each gene evolve readily. Several studies have examined this process at the level of individual genes. One early focus was on lin-48, which encodes a transcription factor related to Drosophila ovo. lin-48 is expressed in the developing excretory duct cell in C. elegans, but is not in C. briggsae. Using reporter transgenes, Wang and Chamberlin (2002) found that only the combination of C. elegans regulatory sequences with a C. elegans host supported lin-48 excretory cell expression, suggesting the difference between species was due to changes in both cis-regulatory sequences and trans-acting factors. At least four C. elegans-specific sequences contribute to the former. Further, the absence of lin-48 expression in C. briggsae correlates with a more anterior location of the excretory duct cell—a shift also seen in lin-48 loss-of-function mutants in C. elegans. A subsequent study (Wang and Chamberlin 2004) found that C. elegans lin-48 recently gained a binding site for the bZip transcription factor CES-2 that is necessary in C. elegans for both strong excretory cell expression and anterior excretory duct cell location. Forcing expression of LIN-48 in the C. briggsae excretory duct cell is sufficient for anterior location. Thus, the gain of a novel regulatory linkage during evolution altered both lin-48 expression and morphology. In addition, enhancers that mediate the conserved hindgut expression of lin-48, which are bound by EGL-38, have diverged between C. elegans and C. briggsae (Wang et al. 2004).
Gene regulatory evolution has also been examined in subsets of homologous neurons conserved across Caenorhabditis. Barrière et al. (2012) focused on the GABAergic cell marker unc-47. Though expressed in identical ways in C. elegans and C. briggsae, cross-species reporter transgenes produced additional, ectopic sites of expression. Further experiments revealed that coordinated evolution between cis and trans factors has occurred in each lineage. A subsequent study (Barrière and Ruvinsky 2014) expanded the neuronal genes analyzed to seven (unc-25, unc-46, unc-47, oig-1, acr-14, gpa-5, and mod-5) and the species to five (C. elegans, C. briggsae, C. remanei, C. brenneri, and C. japonica). Again, while regulatory regions from non-elegans species generally drive expression in the expected C. elegans cells, ectopic expression and/or cell-specific lack of expression is seen in nearly all cases. Interestingly, ectopic expression of cross-species transgenes is much more common, suggesting that the repressive mode of regulation evolves faster than the activating mode. Similar reporters based on homologs from the much more distantly related parasites Meloidogyne, Brugia, and Trichinella (Figure 1) showed that conserved patterns of expression can be driven by sequences that are essentially unalignable (Gordon et al. 2015).
The above studies show that changes in cis-regulatory sequences evolve rapidly. They can sometimes have developmental effects, but more often remain phenotypically cryptic. This is likely due to the action of stabilizing selection, which mandates an outcome, but not a mechanism. This allows compensatory evolution (or apparently compensatory, see Haag 2007) to proceed unchecked, accelerated by directional selection on other loci that share trans-regulators (Johnson and Porter 2007). Over time complex dependencies between distinct promoter regions form (Ludwig et al. 2000).
Neuronal development
Of the 957 somatic cells of the C. elegans hermaphrodite, 302 are neurons, with another 56 providing support (Chalfie and White 1988). Males have over 100 additional neurons and glia, mostly with mating-related roles. Pioneering work of John White and his colleagues determined the full connectome of the hermaphrodite (White et al. 1986), and 25 years later a full description of the male posterior nervous system completed the picture (Jarrell et al. 2012). A large body of literature has described normal and perturbed nervous system development in C. elegans as well (Hobert 2010; Cherra and Jin 2015; Shaham 2015). Such a wealth of information about this one species, as with other topics explored here, begs the question of conservation. Are all Caenorhabditis nematodes put together this way? How about more distantly related nematodes? The earliest comparisons were with the larger, distantly related parasite, Ascaris (e.g., Sulston et al. 1975; Walrond et al. 1985; Niebur and Erdos 1993; Holden-Dye and Walker 1994), and revealed a surprising fine-scale congruence of neurons over a large evolutionary distance (Schafer 2016).
Perhaps not surprising given their overtly similar anatomy, homologous neurons are produced in C. briggsae from a congruent embryonic cell lineage (Zhao et al. 2008). The more distantly related P. pacificus shares all 20 of the pharyngeal neurons, despite substantial divergence in feeding strategies (Bumbarger et al. 2013). Interestingly, however, these homologous pharyngeal neurons are connected in substantially different ways. The cell lineages producing them have yet to be determined in P. pacificus, but even if they differ somewhat, the nervous system appears to evolve novel connections far faster than novel neurons. That finding presents an interesting parallel to work on the evolution of gene regulatory networks (GRNs; Peter and Davidson 2011). In both cases, homologous components (either neurons or genes) evolve distinct regulatory connections to other components. How neural development is modified to produce novel connections is an important area for future research.
Sex determination
Sex determination was one of the first aspects of C. elegans development to be tackled using forward genetic approaches (Hodgkin and Brenner 1977; Hodgkin 2002). X chromosome dosage had long been known to be the ultimate regulator of sexual fate (Nigon 1951). The discovery of a genetic pathway linking X dosage to cell fate (Hodgkin 1986) was subsequently confirmed by molecular cloning of the genes (reviewed by Zarkower 2006). It soon became apparent, however, that this pathway did not resemble those that link chromosomes to sexual fate in Drosophila or mammals (Cline and Meyer 1996; Eggers et al. 2014). The cloning of C. elegans mab-3 revealed the first widely conserved sex-specifier, the DM family of transcription factors (Raymond et al. 1998; Zarkower 2001). Thus, the disparity in sex determination mechanisms among different phyla is not due to wholly independent origins of sexual dimorphism, but rather to rapid divergence of sex determination pathways, most likely upstream and downstream of conserved DM factors (Haag and Doty 2005; Kopp 2012). This realization provided further motivation to examine the evolution of sex determination over shorter time scales.
The first comparisons of sex determination genes within Caenorhabditis focused on the “core pathway” that regulates dimorphism body-wide (Figure 3), starting with the identification of C. briggsae homologs of the genes tra-2 (Kuwabara and Shah 1994; Kuwabara 1996) and tra-1 (de Bono and Hodgkin 1996). These early studies revealed rapid sequence evolution but conserved functions in the promotion of female somatic development. Similar results were subsequently reported for the male-promoting xol-1, her-1, fem-2, and fem-3 (Hansen and Pilgrim 1998; Streit et al. 1999; Haag et al. 2002; Luz et al. 2003) and the male-promoting tra-3 (Kelleher et al. 2008). XOL-1 and FEM-3 are particularly divergent, with only 22 and 38% amino acid sequence identity between their C. elegans and C. briggsae orthologs, respectively. For FEM-3, its overall rapid divergence is mirrored by the region of the C-terminal domain of TRA-2 with which it interacts (Haag and Kimble 2000). In three species tested, the interaction between conspecific TRA-2 and FEM-3 partners was conserved, but interspecies pairings invariably failed (Haag et al. 2002). Less complete interspecies incompatibility was observed for the FEM-2-FEM-3 interaction (Stothard and Pilgrim 2006). Another interaction, between a C-terminal domain of TRA-2 and TRA-1, has been documented in both C. elegans and C. briggsae (Lum et al. 2000; Wang and Kimble 2001). These results indicate that, contrary to the conventional wisdom of molecular biology, even protein domains of critical importance can evolve rapidly. This may be especially true if the only role of a sequence is to interact with one other partner (i.e., there is no pleiotropy at the molecular level). Abundant polymorphisms that do not disrupt interaction are observed in C. remanei TRA-2 and FEM-3 (Haag and Ackerman 2005). A population model suggests such variants can allow rapid coevolution by reducing the deleterious effects of other changes that would reduce fitness on their own (Haag and Molla 2005).
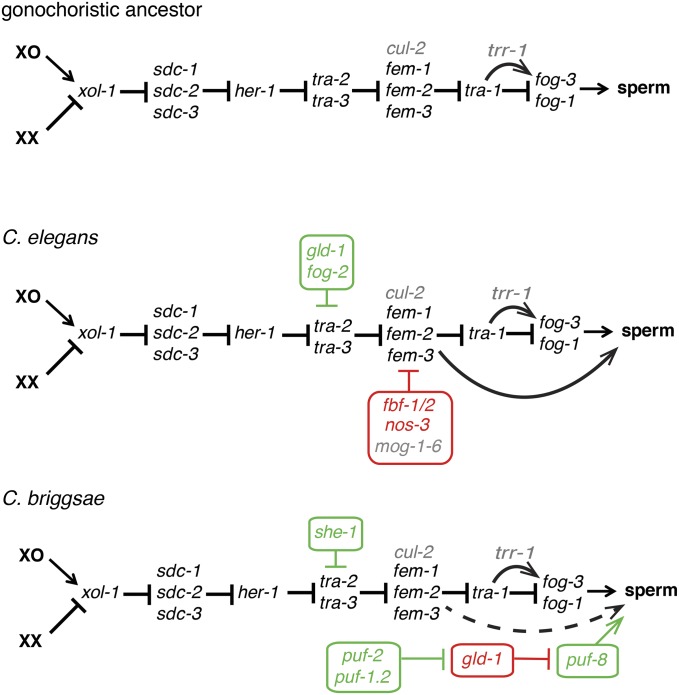
Figure 3.
Convergent evolution of self-fertility via distinct changes alterations of germline sex determination. The core body-wide sex determination pathway (black), which acts in all dimorphic tissues, is shared with outcrossing relatives (top). Upstream factors that sense X dosage and regulate both sex determination and dosage compensation (xol-1 and the sdc genes), are not depicted here for simplicity. The XX hermaphrodites of C. elegans (middle) and C. briggsae (bottom) both produce sperm in an otherwise female body by germline-specific modification of sex determination. Germline-specific factors that promote sperm production in each are indicated in green, while those limiting it are in red. Note that in the C. briggsae case, the influence of she-1 on tra-2 is indirect, and the action of the pathway consisting of puf-2, puf-1.2, gld-1, and puf-8 has not yet been placed along the global pathway, and is thus conservatively depicted as a parallel pathway. Pleiotropic accessory factors with important roles in sexual fate are indicated in gray. The alternative functions of homologous genes and the role of species-specific genes in both hermaphrodites are particularly noteworthy. The arrows connecting fem genes directly to sperm fate in C. elegans depicts how loss of any of the fem genes phenotypically feminizes tra-1 germ cells without loss of fog-3 expression (Chen and Ellis 2000). In C. briggsae, a similar result is found for fem-3, but not fem-2, and the effect is to convert the mostly male tra-1 germ line to a consistent hermaphroditic (rather than female) pattern (Hill and Haag 2009). For this reason, the equivalent arrow is dashed.
In addition to rapid ortholog sequence evolution, C. briggsae is apparently lacking a clear ortholog of sea-1, an autosomal regulator of xol-1, the upstream-most “master regulator” of sexual fate (E.S.H., unpublished data). Thus, over the roughly 20 MY since C. elegans and C. briggsae diverged (Cutter 2008), their global sex determination pathways have undergone rapid sequence evolution and coevolution of conserved genes, and have begun to exhibit gene-level pathway incongruence. The existence of a highly diverged tra-1 homolog in the more distantly related P. pacificus (Pires-daSilva and Sommer 2004) suggests that key aspects of the core sex determination pathway nevertheless remain after substantially longer periods of divergence.
Self-fertile hermaphrodites have evolved at least three times within Caenorhabditis (Kiontke et al. 2004, 2011) (Figure 1 and Figure 3). This novel strategy is enabled by production of sperm in the XX ovary, making germline sex determination an obvious topic of interest for EDB. Before examining that, however, it is worth noting that, unlike Drosophila and mammals, the somatic niches for germline stem cells are very similar (if not identical) in male and female Caenorhabditis (Kimble and Hirsh 1979; Kimble and White 1981; Milloz et al. 2008), and a male somatic gonad is not required to support the differentiation of spermatocytes (Graham and Kimble 1993; Graham et al. 1993). Further, the C. elegans hermaphrodite does not express HER-1, a secreted protein that specifies male fate in XO animals, even in the L4 stage when sperm are produced (Trent et al. 1991; Perry et al. 1993). Self-fertility thus represents a cell-autonomous change in sexual fate. Extensive mutagenesis screens for XX animals with germline-specific sexual transformations (e.g., the masculinization of germline, or Mog, and feminization of germline, or Fog phenotypes) have identified cis-regulatory elements in core sex-determination gene mRNAs that are sites of negative regulation by germline RNA-binding proteins [RBPs, reviewed by Zanetti and Puoti (2013)]. The reconfiguration of RBP-target mRNA networks thus appears to be the key to XX spermatogenesis, distinguishing it from other phenotypic novelties that are rooted in changes in transcription factors and their target genes (Carroll 2008).
What were the changes that allowed XX spermatogenesis to evolve, and how similar are they in selfing species that evolved convergently? Examination of conserved global sex-determiners in the hermaphroditic C. briggsae and the outcrossing C. remanei revealed identical roles for the female-promoting tra-1, tra-2, and tra-3 (de Bono and Hodgkin 1996; Kuwabara 1996; Haag and Kimble 2000; Kelleher et al. 2008), and the male-promoting her-1 (Streit et al. 1999). In contrast, while RNAi knockdown of Cbr-fem-2 and Cbr-fem-3 function could feminize the germ cells of C. briggsae males, it had no effect on hermaphrodites (Haag et al. 2002; Stothard et al. 2002). The dispensability of the C. briggsae fem genes for hermaphrodite spermatogenesis was subsequently confirmed by deletion mutations and exhaustive tra-2(ts) suppressor screens (Hill et al. 2006). These results suggested that regulatory mechanisms that allow C. briggsae spermatogenesis act downstream of the fem genes. Cbr-fem-3; Cbr-tra-1 double mutants have the perfect male soma characteristic of Cbr-tra-1 mutants, but a well-regulated hermaphrodite germline, as in Cbr-fem-3 mutants (Hill and Haag 2009). This indicates that, as in C. elegans (Hodgkin 1986), the fem mutations are epistatic to tra-1 in the germ line. Interestingly, in both species fog-3 expression, which is controlled by tra-1, and thus by the fem genes, remains high in tra-1 mutants even when the germline is feminized by simultaneous loss of one or more fem genes (Chen and Ellis 2000; Hill and Haag 2009). This indicates that the fem genes act in multiple places near the terminus of the germline sex determination pathway. The degree of identity of these sites of control between the two selfing species, and the extent to which they were present in their gonochoristic ancestors, remains to be determined.
How conserved are the germline-specific sex determination factors known from C. elegans? The promoter of sperm fate fog-3 is conserved and plays a similar role across the genus (Chen et al. 2001). Clear orthologs of fog-1 also exist in all Caenorhabditis species (Cho et al. 2004), but their loss-of-function phenotypes have not yet been reported. GLD-1, the RBP that binds the tra-2 3′ UTR (Jan et al. 1999), is also conserved across species (Nayak et al. 2005; Beadell and Haag 2014). However, C. briggsae GLD-1 is a repressor of sperm fate, rather than an enabler, Cbr-tra-2 lacks the duplicated motifs that recruit GLD-1 in C. elegans, and gld-1 has no apparent role in sex determination in any male-female species (Nayak et al. 2005; Beadell et al. 2011). GLD-1 thus appears to have been co-opted into sex determination independently, and to opposite effect, in C. elegans and C. briggsae. This may have occurred because of its simple RNA target motif (Ryder et al. 2004) and conserved expression in early meiotic germ cells (Jones et al. 1996; Nayak et al. 2005). FOG-2, an F-box protein cofactor for GLD-1 in C. elegans that is essential for XX (but not male) spermatogenesis (Schedl and Kimble 1988; Clifford et al. 2000) is a recent gene duplicate that is found only in this species (Nayak et al. 2005).
The gld-1 mRNA is itself subject to translational repression via its own 3′ UTR by the PUF family RBP FBF (Crittenden et al. 2006). The PUF family is somewhat dynamic in Caenorhabditis, such that in C. briggsae there are not strict orthologs of FBF. However, both biochemical and genetic studies indicate that the three paralogs of the PUF-2 subfamily (Cbr-puf-2, Cbr-puf-1.1, and Cbr-puf-1.2) represent the C. briggsae equivalents (Liu et al. 2012). Given the opposite roles of C. briggsae and C. elegans gld-1 in sex determination, it is not surprising that simultaneous RNAi knockdown of Cbr-puf-2 and Cbr-puf-1.2 function feminizes the germ line, rather than masculinizes as does C. elegans fbf(RNAi). Surprisingly, however, complete elimination of Cbr-puf-2 activity alone (via a deletion mutation) leads to a fully penetrant larval arrest. Subsequent studies revealed this was due to a defect in pharyngeal development, apparently related to the brief expression of Cbr-puf-2 in three pharyngeal muscle cells (Liu and Haag 2014). This suggests that the PUF protein family may spin off paralogs as they acquire novel roles outside of the germ line, an example of the neofunctionalization process thought to favor retention of otherwise redundant gene copies (Lynch et al. 2001).
The above studies revealed evolutionary variation through reverse-genetic targeting of conserved genes. Another fruitful approach has been to conduct unbiased forward screens for germline-specific feminizers in C. briggsae (reviewed by Ellis 2017). For example, alleles of Cbr-gld-1 emerged from screens for Mog hermaphrodites (Beadell et al. 2011). Similarly, screens for fog-2-like mutations conferring hermaphrodite-specific germline feminization led to the discovery of she-1 (Guo et al. 2009). Like FOG-2, SHE-1 is an F-box protein that depends upon tra-2 for its function. However, there is no indication that it directly regulates tra-2, nor that it interacts with GLD-1. Its exact role in enabling XX spermatogenesis thus remains a subject for future work.
Another novel factor required for sperm development of both sexes of C. briggsae is encoded by trr-1 (Guo et al. 2013). This component of the Tip60 histone acetyl transferase complex is conserved across Caenorhabditis, but loss of trr-1 alone is incapable of causing similar feminization of the C. elegans germ line. Cbr-trr-1 mutations enhance the incomplete somatic masculinization of Cbr-tra-2, and, in the germ line, help activate fog-3 expression, suggesting that TRR-1 promotes male development. However, the effect on fog-3 is dependent upon the presence of tra-1. This suggests that, as for Gli and its other hedgehog pathway transcription factor homologs, TRA-1 has both activating and repressing effects on target genes, with TRR-1 being important for the former. A previously unknown role of the C. elegans trr-1 ortholog in promoting male development can be revealed through enhancement of weak fem alleles (Guo et al. 2013). These results are consistent with existence of separate and conserved tra-2/fem (repressor) and tra-1/trr-1 (activator) branches of the sex determination pathway. Though apparently conserved in both C. elegans and C. briggsae, their relative importance is reversed. The case of trr-1 also shows how use of a second “satellite model” organism can shed important light on cryptic evolution underlying conserved phenotypes of the more widely studied species.
The impact of trr-1 described above, as well as related work in C. elegans (Grote and Conradt 2006) suggest that chromatin regulators may be frequent contributors to sexual regulation via modulation of TRA-1 function. Chen et al. (2014) thus pursued possible roles for the nucleosome remodeling factor (NURF) complex in C. briggsae. Using the TALEN-based genome editing methods they had developed (Wei et al. 2014a), they discovered that, while complete loss of Cbr-isw-1 and Cbr-nurf-1 were sterile, hypomorphic mutations were sometimes Fog, and RNAi knockdown of either gene increased the penetrance of this. Surprisingly, however, the feminizing impact is not observed in C. elegans, or in the outcrossing C. nigoni and C. remanei. The NURF complex thus appears to be uniquely important in C. briggsae, and likely represents another component of the species-specific regulation that each hermaphrodite evolved to produce sperm transiently.
Germ cell proliferation
The proliferation of germ cells at the distal tip of the C. elegans gonad is directed by a somatic niche, comprised of the many finger-like projections of a single distal tip cell (DTC, Hall et al. 1999; Byrd et al. 2014). The DTC stimulates mitotic proliferation of germline stem cells via Notch signaling (Kimble and Hirsh 1979; Kimble and White 1981; Austin and Kimble 1987; Cinquin et al. 2010). As proliferation pushes stem cells out of the DTC niche, they undergo a final mitosis and then enter meiosis. No further mitoses are normally observed in either sex, and there is no evidence for a mostly quiescent, or “label-retaining” subpopulation of stem cells (Crittenden et al. 2006). In addition, for all Caenorhabditis species that are self-fertile, spermatocytes are found only during the L4 larval stage and (depending on species) the first few hours of adulthood as defined by the final molt. Thus, sperm are of a finite number established prior to ovulation, and when sperm are exhausted reproduction ceases unless mating with a male occurs. Recent studies in other nematode groups have revealed significant deviations from these aspects of Caenorhabditis germ cell proliferation.
The recently described genus Auanema (Kanzaki et al. 2017) has presented several unexpected aspects of germline development. Though similar to Caenorhabditis in overall form and habitat, and within the same family, “Rhabditidae” (Kiontke and Fitch 2005), at least three Auanema species (A. rhodensis, A. freiburgenesis, and A. viguieri) exhibit a reproductive polyphenism in the development of XX individuals, such that those that develop directly via a normal L3 larva mature into females, while those produced from dauer larvae (L3d) develop as selfing hermaphrodites (Félix 2004; Kanzaki et al. 2017). This presents another convergently evolved self-fertile taxon, which has now been examined in some detail. Among their unexpected features, hermaphrodite spermatocytes are not specified briefly in the L4 stage, as in Caenorhabditis, but instead are continuously replenished via coherent populations of spermatagonia (McCaig et al. 2017). These form elongated cysts that proliferate mitotically far from the distal stem cell niche, and undergo meiosis and spermatogenesis adjacent to oocytes. Other surprising features of Auanema germline biology are described below.
More distant relatives of Caenorhabditis are the mammalian filarial parasites (onchocercids, Spiruromorpha, Figure 1), such as Brugia malayi, the causative agent of human filariasis. These parasites have a radically different life history from the bacteriovores in “Rhabditidae” discussed thus far. A female Brugia adult can lay over 1000 embryos per day, and sustain this rate for over 5 years (Taylor et al. 2010)—a reproductive output three orders of magnitude greater than that of C. elegans. In addition, they and many of their relatives have harbored Wolbachia bacteria as obligatory symbionts for millions of years (McLaren et al. 1975; Bandi et al. 1998; Taylor et al. 1999). Importantly, curing these nematodes of Wolbachia with antibiotics adversely affects them without harming their mammalian host (Bosshardt et al. 1993). In Onchocerca ochengi, a parasite of livestock, tetracycline treatment kills adults (Langworthy et al. 2000). In cured Brugia malayi and B. pahangi, females produce inviable embryos that die via extensive apoptosis, while males retain normal fertility (Bandi et al. 1999; Landmann et al. 2011). This inviability is likely caused by the requirement for Wolbachia in proper polarization of the first zygotic cell division, as noted earlier (see section Zygotic mitosis). A subsequent study (Foray et al. 2018) revealed that the Wolbachia symbiont and the Brugia female have coevolved to jointly support oocyte proliferation. The dynamics of this proliferation differ markedly from that of Caenorhabditis, in that it occurs predominantly in a zone proximal to the distal stem cell niche, with the most distal cells represent a quiescent population (Foray et al. 2018). Loss of Wolbachia stimulates ectopic proliferation in the distal zone, with the effect of exhausting the quiescent pool. It thus appears that Wolbachia has become such an integral part of the female germline development that the nematodes can no longer prosper without it. What, if anything, the nematode hosts derive from the symbiosis is another mystery.
In addition to the presence of the Wolbachia symbiont, the somatic niche for germline stem cells differs between Caenorhabditis and Brugia (Foray et al. 2018). Ablation of the DTC in Brugia is not sufficient to eliminate germline proliferation, as it is in Caenorhabditis. Nevertheless, broad treatment with inhibitors of Notch signaling reduce proliferation. These results suggest that the somatic niche in Brugia is similar to that of Caenorhabditis, but on a larger scale. This finding is consistent with the ongoing anatomical (Rundell and Leander 2010) and genomic (Aboobaker and Blaxter 2003) miniaturization of nematodes that accompanied their invasion of tiny meiofaunal habitats.
Spermatogenesis
Compared to the wild variety seen in other phyla (Lüpold and Pitnick 2018), the peculiar amoeboid sperm of nematodes are notably constant in their major sperm protein (MSP)-based motility and overall shape. However, this outward constancy masks tremendous variation that impacts organismally important traits, such as sex ratio, self-fertility, sexual selection, and resistance to cross-species mating. Some of these variables are described below.
Just within Caenorhabditis, sperm can differ in volume as much as 50-fold between species (Vielle et al. 2016). Sperm size is correlated with competitive ability within species (LaMunyon and Ward 1998). In selfing species, male sperm are consistently larger than those of hermaphrodites, in part because of somatic gonad effects (Baldi et al. 2011). However, male sperm of outcrossing species are generally larger than those of males from selfing species (LaMunyon and Ward 1999; Hill and L’Hernault 2001). Further, conditions that select for the most competitive sperm also increase sperm size (LaMunyon and Ward 2002). These correlations indicate that postcopulatory sexual selection and its relaxation in selfing species is a major force that shapes sperm development. They also suggest a simple effect of sperm size on competitive ability, yet interspecies matings reveal a more complex relationship. Males of outcrossing species frequently suppress self-fertility in hermaphrodites, and tend to have larger sperm. However, across a matrix of many pair-wise crosses, the extent of this effect is not correlated with difference in the sperm size of the two species (Ting et al. 2014). This suggests that other factors contribute to competitiveness. A likely candidate is the sperm proteome, which can be much larger in outcrossing species (Thomas et al. 2012b; Yin et al. 2018).
Several lines of evidence have revealed that male-expressed genes are disproportionately lost as part of widespread genome shrinkage in self-fertile lineages (Thomas et al. 2012b; Fierst et al. 2015). One case that has been investigated functionally is that of the MSS family of sperm surface glycoproteins. Yin et al. (2018) found that mss genes are found in nearly all outcrossing Caenorhabditis, but are missing in all self-fertile species. MSS proteins are both necessary (in C. remanei) and sufficient (when restored to C. briggsae) for optimal sperm competition. The increased success in siring cross-progeny that an mss+ transgene confers to C. briggsae males (Yin et al. 2018) may provide an important clue about its independent loss. With greater suppression of selfing comes a greater fraction of male progeny. The reproductive assurance and lack of inbreeding depression of selfing species (Dolgin et al. 2008), combined with the small, transient habitats they favor likely create conditions that select for lower male frequency via interdemic selection. This is reminiscent of the local mate competition scenario of Hamilton (1967). Loss of mss may provide a way to reduce male frequency without complete loss of outcrossing, which is likely needed at some level (Morran et al. 2009a,b).
Beyond competiveness, the sperm of some nematodes exhibit oddities that lead to unexpected sex ratios, as in the heterogonic sheep parasite Strongyloides papillosus. Adults in a host are always parthenogenic females. Many of their XX progeny develop directly into infective larvae, creating a simple asexual life cycle. However, females can also produce sexual XX female and XO male progeny, which mate outside the host and produce outcrossed infective larvae. How does a parthenogenic XX female produce a male without mating? Albertson et al. (1979) had suggested that one X chromosome (present as part of an X-autosome fusion in this species) may be lost in some diploid oocytes via chromosomal diminution. Using molecular markers and heroic crosses through sheep, Nemetschke et al. (2010) found clear support for this hypothesis. Which of the two X chromosomes is lost appears to be random, but some mechanism must prevent both from being lost.
Auanema rhodensis presents another interesting sperm-mediated sex ratio anomaly. Though males are XO and females XX, cross progeny are <2% male (Félix 2004). Examination of male spermatogenesis provided an explanation (Shakes et al. 2011). As spermatocytes proceed through meiosis I, the two X chromosomes are not paired, as in C. elegans. This produces secondary spermatocytes with one X chromatid. When these divide, the spermatid possessing the X attracts nearly all of the organelles required for sperm function (mitochondria, membranous organelles, and MSP), while the nullo-X chromosome set ends up in a residual body incapable of supporting spermiogenesis. As a result, nearly all spermatozoa capable of fertilizating an oocyte are X-bearing, which, in turn, produces extremely female-biased broods. Interestingly, matings between male and free-living female Strongyloides papilillosus also produce all-female broods, but it is not yet known whether the mechanism is the same as that described for A. rhodensis (Streit et al. 1999). A. rhodensis hermaphrodite morphs employ yet another non-Mendelian mechanism of X chromosome segregation during spermatogenesis, as the functional self-sperm contain two X chromosomes (Tandonnet et al. 2018). This is coupled with loss of both oocyte X chromosomes to the first polar body. As a result, self-progeny are always XX, but crosses between XX hermaphrodites and males yield exclusively male progeny. These dynamics are another strong indication that selection on sex ratio can push the evolution of sperm attributes, in this case via unexpected meiotic novelties.
Though it is obvious that self-fertility depends upon XX spermatogenesis, the final step of sperm development—spermiogenesis or activation—plays another important role in its evolution. In male nematodes, spermatids are stored in an inactive state in the seminal vesicle, and are not activated to become motile spermatozoa until exposure to factors during their passage through the vas deferens activates two parallel pathways (Ellis and Stanfield 2014). One of these pathways is composed of SPE-8 and associated sperm proteins, which responds to a signal from the vas deferens (Nishimura and L’Hernault 2010) that may be zinc cations (Liu et al. 2013). The other is mediated by the seminal protease TRY-5 and its inhibitor, SWM-1 (Stanfield and Villeneuve 2006; Smith and Stanfield 2011). The requirement for activators expressed in the male somatic gonad presents a problem for would-be selfing hermaphrodites, which must evolve male-independent sperm auto-activation.
C. elegans spe-8 group mutants exhibit hermaphrodite-specific activation defects, suggesting that only the spe-8 pathway is used to achieve auto-activation. The independent origins of selfing in C. briggsae and C. tropicalis raise the question of whether convergently evolved hermaphrodites used the identical means to achieve sperm auto-activation. Wei et al. (2014b) found that genes of both male sperm activation pathways are conserved across the genus. Knockout mutants in multiple spe-8 group genes cause self-sterility in C. briggsae hermaphrodites (but not males), suggesting parallel co-options of the same sperm activation pathway. However, loss of spe-8 group homologs had no effect on C. tropicalis hermaphrodites, but try-5 mutant hermaphrodites were self-sterile. This indicates that, in the C. tropicalis lineage, the alternative pathway evolved to enable auto-activation (i.e., convergence). Surprisingly, C. tropicalis males are also rendered sterile upon loss of only try-5, indicating that the two pathways are no longer redundant in this species.
Given the need for both XX spermatogenesis and sperm auto-activation, how did self-fertility ever evolve? In an elegant experiment, Baldi et al. (2009) simulated this transformation using the gonochoristic C. remanei. Partial loss of Cre-tra-2 function with RNA interference creates XX pseudohermaphrodites that produce sperm (Haag and Kimble 2000), but these sperm are not active and the animals are not self-fertile. However, mating with males is sufficient to activate these sperm and allow production of selfed progeny. Moreover, when Cre-swm-1 is also knocked down, pseudohermaphrodite self sperm spontaneously activate and sire self-progeny (Baldi et al. 2009). Because there is no known role for TRY-5 protease in females, this ability of Cre-swm-1(RNAi) to activate these XX sperm is surprising. Examination of sex-specific transcriptome data (Thomas et al. 2012a) reveals that C. remanei swm-1 is abundantly and comparably expressed in both females and males, while try-5 is highly male-biased. One possibility is that the low level of TRY-5 expression in C. remanei females is sufficient to activate self sperm when SWM-1 is eliminated. Alternatively, knockdown of tra-2 may elevate TRY-5 levels to a point that potentiates loss of swm-1. In either case, simultaneous modification of sex determination and sperm activation factors is sufficient to allow rudimentary selfing.
The above experiments suggest a two-step model for the evolution of self-fertility (Figure 4). In the first phase, a germline-specific change in the regulation of the sex determination (discussed above) could have produced a small population of XX spermatids. By virtue of developing in a female body, these were initially inactive, and also smaller than male sperm (Baldi et al. 2011). However, their trans-activation could be achieved via seminal fluid from conspecific (or closely related) males, as shown by Baldi et al. (2009). Evolving in a population of gonochoristic conspecifics, such mates would likely be readily available to the incipient selfer. This would produce a mixed-paternity brood with an XX-skewed sex ratio, which has two potential consequences. First, selfed progeny would retain the maternal genotype that promotes XX spermatogenesis, which is likely a recessive trait (Woodruff et al. 2010). Second, the XX-biased broods may be adaptive if population sizes shrink and local mate competition conditions set in (see above). Such partial selfing would have the further benefit of allowing recessive deleterious mutations to be gradually purged (Garcia-Dorado 2012). Eventually, a greater degree of selfing would become well tolerated. In the final phase, hermaphrodites could have evolved sperm auto-activation by upregulating expression of one of the spermiogenesis signals in their otherwise female reproductive tract. Baldi et al. (2009) also suggested that the capacity for XX sperm autoactivation may have evolved as a neutral polymorphism first, allowing subsequent changes in sex determination that enable spermatogenesis to achieve great impact without mating. In either case, an autonomous selfer would enjoy reproductive assurance at low density without accompanying inbreeding depression, allowing them access to habitats that would be marginal for obligate outcrossers.
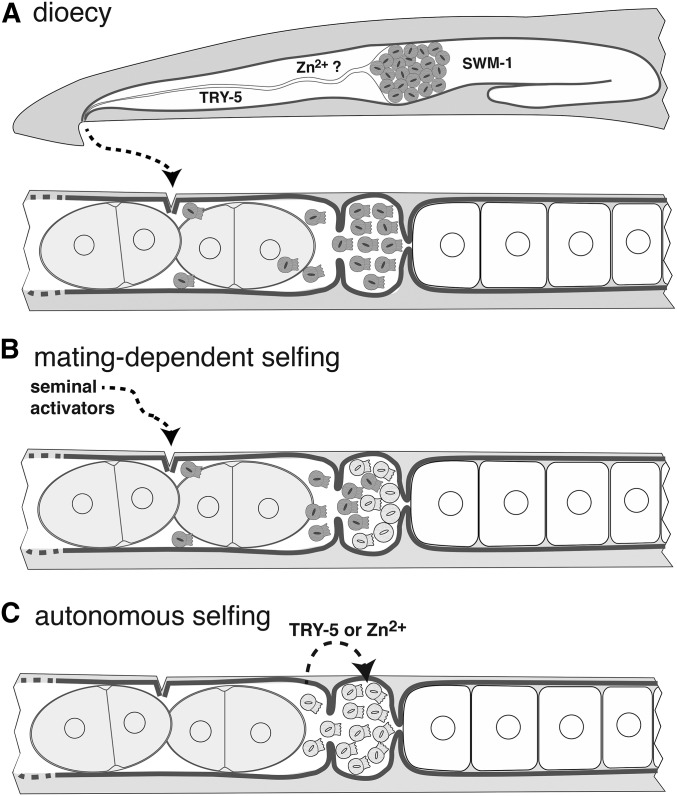
Figure 4.
Scenario for the evolution of self-fertility in Caenorhabditis. (A) In the gonochoristic/dioecious Caenorhabditis ancestor, males (top) store gametes as inactive spermatids in the seminal vesicle, maintained in this state by the protease inhibitor SWM-1. Upon mating and ejaculation, male spermatids (gray) pass through the glandular vas deferens, where they encounter active TRY-5 protease and the signal for the spe-8 pathway, which may be zinc ions. Once inside the female (bottom), they are activated and migrate from the uterus to the spermatheca, where they await ovulation and a chance to fertilize an egg. (B) A hypothetical first step to self-fertility is a change in germline sex determination that allows the production of some self-spermatids (light circles). These cannot activate on their own, but, after mating and transfer of some male seminal fluid, they are activated in trans (light spermatozoa). (C) In the second step, hermaphrodites evolve the ability to activate self-spermatids autonomously, by increasing the level of active TRY-5 protease (as in C. tropicalis) or the signal for the spe-8 pathway (in C. elegans and C. briggsae).
Vulva development
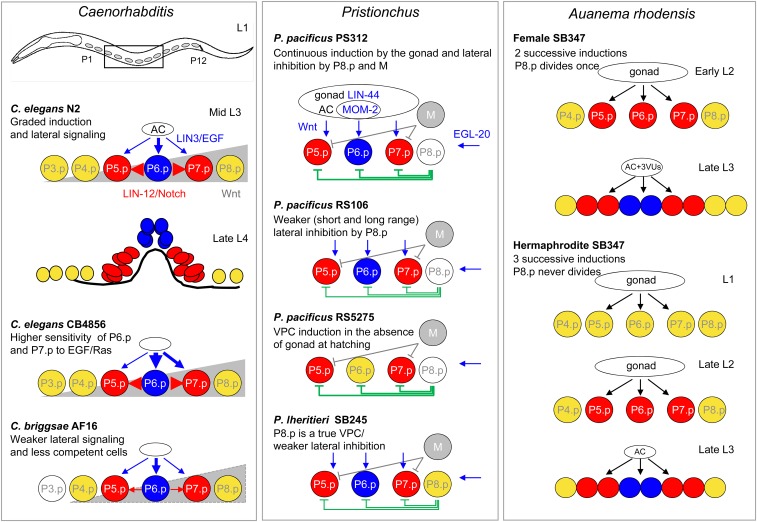
The vulva is an opening in the center of the C. elegans hermaphrodite (but can be in different positions in other taxa) that serves for copulation and egg laying through its direct connection with the uterus. Because of extensive work on the vulva in C. elegans, it has also become an important evo-devo model and is a primary exemplar of DSD. The vulva is a simple organ that originates from a handful of ventral epidermal cells during the larval stages (Figure 5). Cellular division and organogenesis can be tracked by differential interference contrast (DIC) microscopy. Moreover, C. elegans mutants with abnormal vulvae remain fertile, which has allowed vulva development to be explored in exquisite detail (Sternberg 2005; Gupta et al. 2012). While the morphology of the adult vulva can be a slit or a round pore depending on the species (Kiontke et al. 2007), the fate patterns of the vulva precursor cells (VPCs) are quite conserved between species. Are the VPCs specified similarly in all species? Where does the induction signal come from, and is it always the same molecular signal? Studies that have posed these questions have revealed an impressive diversity of cryptic changes (i.e., changes in mechanism without changes in phenotype), between both closely and distantly related species, and even strains of the same species. The field has also benefited from the establishment of other selfing species, P. pacificus and Oscheius tipulae, as genetically tractable systems to explore changes in vulval development over large evolutionary distances (Félix 2006; Sommer 2006). Cryptic genetic changes have been deciphered further by exploring different species of the same genus and even different natural isolates of the same species in the three genera, Caenorhabditis, Oscheius and Pristionchus.
Figure 5.
Variations in vulval development in Caenorhabditis, Pristionchus, and Auanema. Left panel: schematic representation of vulva development in C. elegans, and some cryptic variations found within Caenorhabditis. From ventral epidermal cells, six competent cells, P3.p to P8.p are defined in C. elegans. During the L3 larval stage, VPCs are specified and induced by the combined action of a graded EGF signal from the anchor cell (AC), a lateral Notch signal between the most central cells and a Wnt gradient emanating from the posterior of the body (gray wedge). Blue cells adopt a primary fate and divide to form the center of the vulva in late L4. Red cells have a secondary fate and form the lateral part of the vulva. Yellow cells form the vulva only if blue or red cells are absent. The respective contributions of the EGF and Notch pathways vary quantitatively (shown by arrows of different size) among Caenorhabditis species and even among strains of the same species. In C. briggsae, reduction in Wnt signaling (compared to C. elegans) is responsible for the lack of competency of P3.p. This could be due to truncation of the Wnt gradient (depicted here), or because of reduced sensitivity of P3.p to an identical gradient. Middle panel: Schematic representation of vulva development in P. pacificus, and variations found within Pristionchus. In P. pacificus, the VPCs are induced by redundant Wnt signaling signals sent by the gonad and the AC. The M cell, as well as the P8.p cell (which is only partially competent), send lateral inhibitory signals to prevent the adoption of 1° Cell fate by P5.p and P7.p. Within Pristionchus, cryptic quantitative changes in the signaling pathways are observed. In particular, the extent of lateral inhibition by P8.p varies frequently between and within species. Right panel: example of changes in vulva development between morphs of the same species is shown for Auanema rhodensis SB347. In this species, three sexes coexist because female larvae that go through the dauer stage become self-fertile hermaphrodite adults. This plasticity is accompanied by changes in vulva formation between females and hermaphrodites, in the number of inductive signaling steps from the gonad that are required to specify the Pn.p cells, as well as in the number of divisions of P8.p.
Twelve ventral epidermal cells (P1.p to P12.p) originate during the L1 stage (Figure 5): P1.p being the most anterior cell and P12.p the most posterior cell (Sulston and Horvitz 1977). In C. elegans, during the L3 stage, and upon signaling from a specialized cell of the uterus called the anchor cell (AC), the central cells P5.p to P7.p divide to give rise to 22 cells that will fuse in late L4 in concentric circles to form the vulva. The pattern of division of each cell is highly reproducible and reflects the fate of the cells. While the most central cell P6.p adopts the 1° fate (“inner” vulval cells that will detach from the cuticle and involute), P5.p and P7.p adopt the 2° fate (“outer” vulval cells that remain connected to the cuticle to anchor the vulva). P3.p, P4.p and P8.p daughter cells adopt a 3° (nonvulval) fate, because they do not form the vulva in wild type animals. All of these P(5–7).p cells form a vulval competence group, as any of these cells can replace an ablated cell and become vulval. All the other Pn.p cells are incapable of forming the vulva, even upon ablation of P3.p to P8.p (Sternberg and Horvitz 1986). This vulval competence group is defined by expression of two Hox proteins, with LIN-39 in central cells promoting competence and MAB-5 repressing it in more posterior Pn.p cells (Clandinin et al. 1997). The AC sends a LIN-3/EGF signal that acts as a morphogen on the VPCs (Hill and Sternberg 1992). Closest proximity to the signal determines the 1° fate (generally P6.p); P5.p and P7.p receive a lower dose and adopt the 2° fate. Activation of the EGF/Ras signaling pathway in P6.p activates the Notch/Delta pathway (Sternberg 1988). This leads to the inhibition of the 1° fate in P5.p and P7.p through lateral inhibition and to the activation of the 2° fate in these same cells. The Wnt pathway is also involved in vulval specification, as loss of negative regulators of Wnt causes mote than three VPCs to be induced (Gleason et al. 2002). Conversely, VPCs adopt a 3° fate or fuse with the hypodermis in the absence of positive regulators of the Wnt pathway (Eisenmann and Kim 2000).
Variation in the position of the vulva:
C. elegans has two gonadal arms extending from the center of the animal, with a central uterus and vulva derived from the central epidermal Pn.p cells. Some species have a single gonadal arm (monodelphy), which extends anteriorly. The evolution of monodelphy per se will not be covered here (but see Félix 1999). In most cases, monodelphy is accompanied by a posterior shift of the uterus and the vulva. In the monodelphic species P. redivivus, the vulva forms at 60% of body length because of a posterior displacement of the central Pn.p cells and because the vulva is centered in between P6.p and P7.p (Sternberg and Horvitz 1982). Within “Rhabditidae,” monodelphy and a posterior vulva are derived and evolved several times (Kiontke et al. 2007). In the three posterior-vulva species of Cruznema, Mesorhabditis, and Teratorhabditis that have been analyzed, again only the central Pn.p cells (P5.p to P7.p) are competent, and they migrate posteriorly (Sommer and Sternberg 1994). However, mechanisms of vulva induction differ between species. The developing gonad induces the VPCs in Cruznema, but is not required to induce the VPCs in Mesorhabditis and Teratorhabditis. Establishment of the competence group by LIN-39 is conserved in P. pacificus (Eizinger and Sommer 1997) and O. tipulae (Louvet-Vallee et al. 2003). HOX specification represents a constraint on specifying which cells can form the vulva; to make a posterior vulva, this constraint has been overcome in at least four independent lineages in Rhabditida by a similar mechanism, i.e., posterior migration of the vulva cells (Kiontke et al. 2007).
Variation in the number and fate of VPCs:
Large variations are found in the size of the competence group, in the number of divisions of competent cells, as well as in the fate of the noncompetent Pn.p cells (for an evolutionary synthesis of most of these differences among species in Rhabditida, see Kiontke et al. 2007). While the competence group includes P3.p up to P10.p in P. redivivus (Sternberg and Horvitz 1982), much larger than the number of cells that are induced, in Rhabditophanes and Strongyloides ratti the competence group is restricted to the cells that form the vulva (Félix et al. 2000a). The size of the competence group can also vary between closely related species. For instance, P3.p is competent in C. elegans but not in C. briggsae or in some other Caenorhabditis species (Pénigault and Félix 2011). Overexpression of Wnt in C. briggsae is sufficient to induce the division of P3.p, while downregulation in C. elegans prevents the division of P3.p (Pénigault and Félix 2011). Because several Wnt ligands are expressed in a gradient from posterior to anterior in the C. elegans body (Gleason et al. 2006), one possible explanation is that the lack of competency of P3.p in C. briggsae could be due to a shorter Wnt gradient in C. briggsae compared to C. elegans (Figure 5; Pénigault and Félix 2011). It is also possible that, in C. briggsae, P3.p is less sensitive to Wnt signals. Within Pristionchus, P8.p is partially competent in P. pacificus (Sommer 1997), but is a true VPC in P. lheritieri (Srinivasan et al. 2001).
Within the competence group, the pattern of cell division is also variable. The numbers of cells that form the vulva vary between 16 cells in O. tipulae, to 34 cells in Rhabditoides regina (Sommer and Sternberg 1995). Within Caenorhabditis, 22 cells form the vulva in all species that have been observed (Félix 2007). However, the division pattern of the competent cell P3.p is highly variable, between and within Caenorhabditis species (Delattre and Félix 2001; Félix 2007; Pénigault and Félix 2011). Similarly, the pattern of P4.p and P8.p division shows a high degree of intraspecies and interspecies variation in Oscheius. The fate of the cells that are not competent to form the vulva also vary. Pn.p cells that do not express LIN-39 fuse with the hypodermis in C. elegans and in O. tipulae (Louvet-Vallee et al. 2003), while the expression of LIN-39 prevents cell death in P. pacificus (Eizinger and Sommer 1997). The restriction of the competence group by cell death is a derived character that is observed several times in the phylogeny, yet reversals of this restriction are very rare, constituting an interesting evolutionary bias (Sommer and Sternberg 1996; Kiontke et al. 2007). In Turbatrix aceti, as in three other Panagrolaimomorpha species, the survival of the VPCs depends on a signal emanating from the gonad during the L2 stage, in contrast to P. pacificus (Sternberg and Horvitz 1982; Félix and Sternberg 1997, 1998).
Variation in the mechanisms of VPC induction:
Laser ablation of the AC or the gonad has been performed in a wide range of species. These experiments have revealed an impressive diversity of induction mechanisms, apparently evolved from an ancestral two-step induction signal from the gonad (Kiontke et al. 2007). Most surprisingly, systematic characterization of Caenorhabditis and Pristionchus has uncovered cryptic genetic changes (e.g., changes in the contributions of different signaling mechanisms, in competence level and even genetic variation affecting the requirement for induction) between closely related species and even between strains of the same species (Srinivasan et al. 2001; Félix 2007; Zauner and Sommer 2007; Milloz et al. 2008; Kienle and Sommer 2013).
In Clades IV and V (Figure 1), the VPCs can be induced independently of the gonad, as in Brevibucca, or require continuous or possibly consecutive signals from the gonad, as in Halicephalobus sp. (Félix et al. 2000a). As shown above, Mesorhabditis and Teratorhabditis also do not rely on the gonad for induction (Sommer and Sternberg 1994), while two consecutive signals from the AC are required in O. tipulae and Rhabditella axei (Félix and Sternberg 1997). In O. tipulae, the early and late induction signals depend on the activity of MEK kinase—a component of the Ras pathway involved in C. elegans late-only induction (Dichtel-Danjoy and Félix 2004b). One possible evolutionary scenario is that a heterochronic shift occurred in the Caenorhabditis lineage with regard to both the requirement for, and expression of, the homologous induction event (Kiontke et al. 2007).
In P. pacificus, a continuous 10-hr induction from several cells of the somatic gonad is required to induce the VPCs (Sigrist and Sommer 1999), seemingly comparable to the two-step induction of O. tipulae (Kiontke et al. 2007). As in C. elegans (Gleason et al. 2006), simultaneous inactivation of several Wnt ligands and receptors leads to Vulvaless phenotypes in P. pacificus (Zheng et al. 2005; Tian et al. 2008; Wang and Sommer 2011). However, after inactivation of Ppa-bar-1/β-catenin (the Wnt signal transducer), VPCs do not die of apoptosis but adopt a 3° fate, similar to the phenotype obtained after gonad ablation (Tian et al. 2008). Further, Wnt ligands MOM-2 and LIN-44 are expressed in the AC before the division of VPCs and in the central cells of the somatic gonad, respectively (Tian et al. 2008). This suggests that Wnt signals comprise the gonadal signal that induces formation of the vulva in P. pacificus (Figure 5), whereas they are primarily involved in establishing VPC competence to respond to that signal in Caenorhabditis. The involvement of LIN-3 and its downstream cascade has not been demonstrated in P. pacificus vulval induction, raising the possibility that a secondary, largely redundant Wnt pathway in one species (C. elegans) could be central to the homologous process in another (P. pacificus). Interestingly, P8.p and the mesoblast M cell are both responsible for the lateral inhibition that prevents too many VPCs from adopting a 1° fate (Jungblut and Sommer 2000). Thus, P8.p inhibits the induction of VPCs even though it is not a VPC itself. Of note, Wnt signaling is also used in P. pacificus to shape the distinct “pretzel” morphology of the somatic gonad (Rudel et al. 2008). Selection on either gonad shape or vulva induction would thus target a pleiotropic Wnt module, with potential consequences (overt or cryptic) for the other trait.
Variation of the induction mechanism of VPCs is also found between species belonging to the same genus or even between strains of the same species. For instance, a single late induction from the AC is required for VPC divisions in Panagrolaimus sp. PS1579 as in C. elegans, while early and continuous (or possibly two consecutive) signals from the gonad are necessary in another Panagrolaimus species, P. sp. PS1732 (Félix et al. 2000). Within Pristionchus, the system of induction found in the laboratory strain P. pacificus PS312 is not widely conserved. For instance, in P. lheritieri and P. maupasi and even different strains of P. pacificus, some VPCs are induced even when the gonad is ablated just after hatching, and the extent of the lateral inhibition exerted by P8.p on the VPCs can vary (Srinivasan et al. 2001; Zauner and Sommer 2007). Mapping of the quantitative trait locus (QTL) responsible for the differences in the gonad-independent induction of VPCs between P. pacificus strains revealed a new role for the Notch ligand apx-1/Delta (Kienle and Sommer 2013). In many wild strains, absence of a binding site for the HAIRY transcription factor in the cis-regulatory region of apx-1 leads to its expression in P6.p and confers a gonad-independent induction of this cell, while in the laboratory strain PS312, apx-1 is not expressed in the VPCs, which thus require the gonad for induction (Kienle and Sommer 2013).
While the pattern of division of the VPCs is very conserved among Caenorhabditis species, cryptic changes in the mechanism of induction were revealed by ablation of the AC or overexpression of the LIN-3/EGF inductive signal (Félix 2007). Early ablation of the AC leads to adoption of the 3° fate for all VPCs in all species. However, ablation of the AC during patterning, i.e., mid-L3 stage, has different outcomes depending on the species or strain within a species (Félix 2007; Milloz et al. 2008). For instance, in C. remanei, the VPCs adopt a 2°3°2° pattern, suggesting that, in contrast to C. elegans, a low level of induction from the AC is sufficient for P6.p to induce its neighboring cells, but not enough for its own fate acquisition. In C. briggsae, the same experiment leads to a 2°2°2° pattern. Moreover, mild overexpression of LIN-3 in C. briggsae generates adjacent 1° cells, a phenotype that is obtained in C. elegans only after strong overexpression of LIN-3 (Katz et al. 1995). Thus, lateral inhibition from P6.p on adjacent Pn.p cells can be overcome easily in C. briggsae and less so in C. elegans. Nevertheless, the LIN-12/Notch pathway is still involved in lateral inhibition in C. briggsae and LIN-3/EGF acts in a dose-dependent manner on VPC fate specification (Félix 2007). However, inactivation of genes of the vulva specification pathways in C. elegans and C. briggsae often leads to different phenotypes, revealing a difference in the respective contributions of these pathways (Rudel and Kimble 2001; Sharanya et al. 2012, 2015; Mahalak et al. 2017). Thus, evolutionary changes in the patterning of the vulva are not necessarily due to rewiring of the signaling pathways, but can also be attributed to quantitative changes of the same network of signaling pathways (Haag and True 2007). Modeling of the vulva induction pathways confirms that quantitative tuning of the same network parameters could account for the different vulva patterning obtained experimentally among Caenorhabditis species (Hoyos et al. 2011).
Other experiments provided indirect evidence of cryptic genetic changes between species or between strains of the same species. One approach is mutagenesis screens for vulva defects. These yielded a different spectrum of mutations depending on the species. In O. tipulae, although 50,000 gametes were mutagenized, only a handful of hypo- and hyper-induction mutants were isolated. The far larger category was represented by mutations that affect the number of cell division of VPCs, but not their specification, while this category of mutants was very rarely found in C. elegans. These results may indicate a rewiring of the system, depending on the species. Alternatively, higher pleiotropy of the genes involved in vulval patterning in O. tipulae could lead to embryonic death or sterility, thus preventing the detection of specific classes of mutants (Dichtel et al. 2001; Louvet-Vallee et al. 2003; Dichtel-Danjoy and Félix 2004a). Similarly, screens in C. briggsae (Sharanya et al. 2012, 2015) failed to identify mutants lacking VPC induction, and mapping of mutants indicates there are novel players relative to the C. elegans paradigm.
An alternative approach is to characterize the spectrum of vulval defects in mutation accumulation lines. Phenotypes observed in C. elegans are quantitatively different from those obtained in C. briggsae (Braendle et al. 2010). Although the division of the VPCs is highly reproducible within a strain, developmental errors arise at low frequency (∼1%; Braendle and Félix 2008). Interestingly, the frequency and type of errors differ between closely related species or strains of the same species (Zauner and Sommer 2007; Braendle and Félix 2008). Introgression of a mutant allele in different strains of C. elegans also revealed intraspecific cryptic changes. For instance, the impact of different alleles of the Ras pathway on vulva induction vary, depending on the genetic background (Milloz et al. 2008). The background factors that distinguish C. elegans natural isolates were next explored by introgressing an allele of the EGF receptor let-23 and performing QTL mapping. This revealed that C. elegans N2 harbors a mutation in the conserved acetyltransferase NATH-10 that is mainly responsible for the difference in expressivity of the let-23 allele. Because this nath-10 allele also confers high fitness on the laboratory strain N2 compared to others, this experiment demonstrated that cryptic genetic changes can accumulate in the genomes by indirect selection and pleiotropic effects (Duveau and Félix 2012). Even seemingly constant features of the signaling network are subject to DSD at the molecular level. For example, while expression of lin-3/EGF remained constant between Caenorhabditis species and O. tipulae, the cis-regulatory elements that underlie it have been substantially reconfigured, with elements required for expression in one species completely missing in the other (Barkoulas et al. 2016).
Last, but not least, the development of the vulva has been shown to vary between female and hermaphrodite morphs of A. rhodensis (Félix 2004; Kanzaki et al. 2017). While both females and hermaphrodites have a competence group formed by P(4-8).p, P8.p divides in females but not in hermaphrodites. Most strikingly, while three successive gonadal inductions are necessary to form a vulva in hermaphrodites, two rounds of induction are sufficient in females. Although the molecular basis of such a switch remains unknown, this example illustrates that vulva induction can go through different routes even for animals from the same genotype.
The comparative work on vulva development reviewed above has initiated a virtuous cycle, in which interesting differences between C. elegans and its relatives have been appreciated directly, and also motivated further research in C. elegans. For example, the observation of an intrinsic difference among Pn.p cells in Mesorhabditis (Sommer and Sternberg 1994) led to re-evaluation of the differential competence of cells of the “equivalence” group in C. elegans (Clandinin et al. 1997). Similarly, C. briggsae pry-1 mutants are multi-vulva (like their C. elegans counterparts), but also frequently show a failure of P7.p induction. This led to the discovery of a similar, albeit weakly penetrant, defect in C. elegans pry-1 mutants (Seetharaman et al. 2010).
Male tail
Outwardly at least, nematodes appear to vary little with regard to morphology, especially compared to animals with appendages, like arthropods. One clear exception is the male copulatory apparatus, or “male tail.” Because of the abundant variation in male tail morphology, it has long been used as an important tool (along with the feeding apparatus) for nematode morphological systematics (Chitwood and Chitwood 1974; Sudhaus 1976; Andrássy 1983, 1984; Fagerholm 1991; Sudhaus and Fitch 2001; Sudhaus and Fürst von Lieven 2003; Sudhaus 2011; Ragsdale et al. 2015). In the context of the C. elegans model system, additional “satellite” model species and the phylogeny of related species, male tail morphological variation also provides much material for studying the developmental-genetic basis of morphological evolution.
Much of the evolutionary developmental work on the male tail has involved rhabditid nematodes, on which this review is primarily focused. Mapping male tail character states onto the rhabditid phylogeny shows that some characters have evolved uniquely, or nearly uniquely, in some clades (i.e., are “apomorphic”) and are thus important for systematics (Sudhaus 2011). On the other hand, several characters have evolved repeatedly (i.e., are “homoplastic”). In many ways, these latter characters are the more interesting for EDB, since such repeated evolution provides the potential to address questions about bias or constraints of the developmental-genetic system on evolutionary trajectories.
These male tail structures all play some role in the series of stereotypical behaviors involved in copulation (Sudhaus 1976; Loer and Kenyon 1993; Liu and Sternberg 1995; Barr and Garcia 2006; Koo et al. 2011; Sherlekar and Lints 2014). External structures that help the male sense contact with a hermaphrodite and determine correct orientation and body position include the genital papillae. In many species, these sensilla are arrayed within a cuticular “bursa velum” or “fan,” which in different species exist in different sizes and shapes or can be absent altogether. The mechanosensory genital papillae (called “rays” when they form finger-like extensions in the fan) occur in different positional patterns in different species. Other structures include the precloacal papilla and any associated structure (e.g., the “hook” in C. elegans), the chemosensory phasmids (which exist in both sexes but with some sexual dimorphism, and are found in different positions relative to the rays in different species), and the tail tip (which undergoes male-specific morphogenesis in some species, like C. elegans, but is sexually monomorphic in other species). Internal structures include the sclerotic spicules and the gubernaculum. The spicules are inserted into the vulva, providing a means of anchorage and sperm delivery; the gubernaculum covers the roof of the proctodeum and provides a shield that guides the spicules during their protraction. Both of these structures also show marked morphological variation among different groups of rhabditid species (Sudhaus and Fitch 2001; Kiontke et al. 2011).
Evolutionary developmental studies have focused primarily on variation in the patterning of genital papillae/rays and tail tip morphogenesis (Sternberg and Horvitz 1982; Fitch and Emmons 1995; Fitch 1997, 2000; Baird 2001; Sudhaus and Fitch 2001; Sudhaus and Fürst von Lieven 2003; Baird et al. 2005; Kiontke and Fitch 2005). For C. elegans, much progress has been made in elucidating the ultrastructural anatomy, neural connectivity, development, and genetics underlying these structures (Emmons 2005, 2014). Briefly, the overall form of the male tail is a result of morphogenetic “retractions” that begin in the second half of the L4 stage (Figure 6). The first indication of this process occurs as the tail tip cells detach from the L4 cuticle, become rounded, fuse, and migrate a short distance anteriorly. This is followed by the retraction of more anterior hypodermal cells. Because the inner and outer layers of the adult cuticle in the area of the fan are not connected, the outer layer folds and flattens in the wake of the retractions of the cells that are covered by the inner cuticle layer. As the tips of the rays have fixed points of attachment to the outer cuticle, they are drawn out into finger-like projections during the retractions of surrounding tissue, and sandwiched between the dorsal and ventral layers of outer cuticle that form the fan.
Figure 6.
Male tail development in C. elegans and male tails of some other species. Top left: the left-side cell lineages giving rise to the Rn.p “tail seam” hypodermal cell, the three cells of each ray rn (with designations v1–v7, ad and pd used for comparing ray homologs across species), and the phasmid socket cells (see text). These lineages are produced from bilateral pairs of V5, V6 and T blast cells, shown in the L1 larva (Sulston et al. 1980). Red lines represent apical boundaries of cells as would be visualized by immunostaining with MH27 or AJM-1::GFP. Inset: canonical ray sublineage in which an Rn neuroblast produces an Rn.p hypodermal cell (part of the “tail seam”), two ray neurons RnA and RnB, a ray structural cell Rnst and a programmed cell death (“x”). Below the cell lineage: arrangements of these cells in the left lateral hypodermis right after their origins at early L4, and at mid-L4 after the RnA and RnB neurons have sunk a little below the surface. Tail tip cells hyp(8–11) and phasmid socket are also depicted. At the mid-L4 stage, the tail tip cells fuse and some of the Rn.p cells fuse together (leading to absence of adherens junctions separating those cells) and begin to change shape (Fitch and Emmons 1995). DIC photomicrographs at the right: tail morphogenesis, left side view. The first visual sign of morphogenesis occurs when the tail tip cells separate from the pointed L4 cuticle, round up and retract anteriorly at mid-L4. At late L4, because the tips of the rays are attached to the adult outer cuticle (beneath the L4 cuticle), the rays are formed as the rest of the body retracts and the fan folds flat around them. The fully formed adult emerges after the pointed-tailed L4 cuticle is molted off. These events do not occur in hermaphrodites/females, which retain the pointed shape of the larval tails. Bottom: left side views of adult male tails of C. elegans and four other species: Pelodera strongyloides, Metarhabditis blumi, Rhabditella axei, and Panagrellus redivivus. Outlines of the body and fan (if any) are depicted as gray lines, the internal left spicule (or fused left-right spicule in P. strongyloides) and gubernaculum are in brown, and the rays are outlined in black and labeled using the ray homolog designations. Also shown is the position of the phasmid. The tail tip cells retract to make the independently evolved peloderan tails of C. elegans and P. strongyloides, and do not retract in R. axei and M. blumi (derived, “apomorphic” leptoderan) and P. redivivus (ancestrally, “plesiomorphic” leptoderan) (see references cited in text).
The cell lineages that produce the nine rays on each side originate from left-right pairs of the three most posterior blast cells of the lateral “seam:” V5, V6 and T (Figure 6) (Sulston et al. 1980; Emmons 2014). The posterior branch of the T blast cell lineage produces phasmid socket cells that hold the phasmid neurons in place; the anterior branch gives rise to the three most posterior rays. V5 gives rise to the most anterior ray and V6 produces the lineages of the other five rays. Each sublineage that produces a ray is stereotypic: for each ray rn (n = 1–9) an Rn blast cell divides at the end of L3 to produce a posterior Rn.p hypodermal or “tail seam” cell and an anterior blast cell. Then, at the beginning of the L4 stage, this blast cell produces four granddaughters, one of which dies and three of which become the ray components, the RnA and RnB neurons and the glial-like Rnst “structural cell.” The structural cell holds the ray in place and forms a clearly visible papilla on the surface of the tail before the morphogenetic retractions reveal the rays.
The ray cell sublineages are determined by the proneural lin-32/atonal gene, which also acts in combination with the lin-44/Wnt pathway to pattern asymmetry within the sublineage (Zhao and Emmons 1995; Portman and Emmons 2000, 2004). Male-specificity provided via DM-domain genes mab-3, mab-23, and dmd-3 (homologous to Drosophila dsx and human DMRT) is required for these lineages as well (Shen and Hodgkin 1988; Yi et al. 2000; Lints and Emmons 2002; Ross et al. 2005; Siehr et al. 2011). Despite identical sublineage patterns, each bilateral pair of rays has a different identity from every other pair in terms of its AP position, whether its terminus opens on the dorsal or ventral surface of the fan, and what type of neurotransmitters, connectivities, and behavioral proclivities are associated with it (Loer and Kenyon 1993; Chow and Emmons 1994; Liu and Sternberg 1995; Chamberlin and Thomas 2000; Lints et al. 2004; Sherlekar and Lints 2014; Serrano-Saiz et al. 2017b). These identities are patterned along the AP axis by HOX genes (mab-5/Antp and egl-5/AbdB) and their Polycomb- and trithorax-group regulators (sop-2, lin-49, lin-59) (Chow and Emmons 1994; Salser and Kenyon 1996; Chamberlin and Thomas 2000; Lints et al. 2004; Zhang et al. 2004). Identity of the dopaminergic rays that open dorsally on the fan requires signaling by a TGFβ morphogen, DBL-1 (Savage et al. 1996; Krishna et al. 1999; Lints and Emmons 1999; Morita et al. 1999; Suzuki et al. 1999; Wong et al. 2010; Siehr et al. 2011). Further differentiation among ray identities is provided by temporal differences in HOX expression during the ray lineages (Ferreira et al. 1999) and by additional factors, including VAB-3/Pax6 (Baird et al. 1991; Zhang and Emmons 1995), ephrins, and semaphorins (Roy et al. 2000; Hahn and Emmons 2003), and the probable chromatin modifier VAB-3 (Chow et al. 1995; Ho et al. 2001). These patterning systems act combinatorially to establish individual ray identities (Lints et al. 2004); perturbations to these systems result in ray losses/gains or changes in ray positions, including complete homeotic transformations—different rays adopting the same identity cluster together and can fuse into a single ray (Baird et al. 1991; Chow and Emmons 1994).
Evolution of ray pattern:
Rays provide a good example of how developmental analysis can provide information about organ homologies between different species, a fundamental step to any evolutionary reconstruction. In P. redivivus—a member of suborder Tylenchina and thus an outgroup representative relative to the rhabditids—the genital papillae are generated by Rn sublineages identical to those in C. elegans (Figure 6) (Sternberg and Horvitz 1982). The main difference is that V5 does not produce a ray lineage, and V6 only produces rays homologous to C. elegans rays r3–r6; the T lineage produces the same three rays as in C. elegans. The spatial pattern of ray cell origins in the L4 hypodermis (Figure 6) is also highly conserved, allowing the ray homologies among species to be traced (Fitch 1997, 2000). Some rules have emerged, allowing ray homologies to be inferred without having to follow development in each species: (1) the rays homologous to rays r5 and r7 of C. elegans are always dorsal (labeled “ad” and “pd,” the anterior and posterior dorsal rays respectively; Figure 6), and (2) the seven other rays homologous to C. elegans r1–r4, r6, r8 and r9 (relabeled “v1-v7”; Figure 6) are almost always arranged in that order ventral to the two dorsal rays (Fitch 1997, 2000; Sudhaus and Fitch 2001; Sudhaus and Fürst von Lieven 2003).
Whereas the pattern of ray and Rn.p cell origins in the L4 hypodermis is highly conserved across Clade V (and likely even Clade IV, to which Panagrellus belongs), the ray structural cell tips then migrate to species-specific positions, tending to be at junctions between Rn.p cells (Fitch and Emmons 1995). This planar array of structural cells prefigures the species-specific pattern of rays in the adult tail. The two dorsal rays can thus be in very different positions relative to the ventral (v1–v7) rays, which can also cluster together in different groups in different species. The ability to identify the ray homologies allows evolutionary changes in ray positioning to be reconstructed on the phylogeny (Fitch 1997).
The ability to homologize rays also allows identification of which rays are missing in species with fewer than nine ray pairs. For example, the R8 cell stopped dividing in the lineage leading to the Metarhabditis clade, resulting in a loss of v6 (homologous to C. elegans ray r8) (Figure 6) (Fitch and Emmons 1995; Fitch 1997). It has been noted that this ray is particularly susceptible to loss in mutants with altered activity of the proneural factor LIN-32/Achaete-Scute (Zhao and Emmons 1995; Fitch 1997). Thus, lin-32 or its regulatory pathway are good candidate loci in which variation could cause such an evolutionary change in development and morphology and Metarhabditis would be a good group in which to test this hypothesis in future work.
Other C. elegans male tail mutants suggest candidate loci for evolutionary change. For example, in C. elegans and its close relatives, the first two rays are located anterior of the cloaca, ray v3 is positioned at the cloaca and the other rays are clustered in triplets posterior of the cloaca. In C. briggsae, ray v3 moved posterior of the cloaca and is frequently found to be fused with ray v4 (Nigon and Dougherty 1949; Friedman et al. 1977), as if the v3 identity were transformed partially or fully to a v4 identity (Fitch 1997; Baird 2001; Baird et al. 2005). This phenotype is mimicked by several C. elegans mutations, including mutations in HOX genes mab-5/Antp and egl-5/AbdB or in genes that regulate HOX genes (Chow and Emmons 1994; Chamberlin and Thomas 2000; Toker et al. 2003; Lints et al. 2004; Zhang et al. 2004; Baird et al. 2005). The AP position of ray v3 in the fan coincides with the border between mab-5 and egl-5 expression domains (Ferreira et al. 1999). Thus, variation in HOX genes or their regulators are likely to underlie evolutionary changes in ray pattern, a hypothesis that could be tested further, e.g., by CRISPR editing experiments in future research.
Although the posterior placement of ray v3 is canonical for C. briggsae, there is considerable strain-specific variation with respect to the frequency of the derived vs. ancestral v3 positions (Baird 2001; Baird et al. 2005). Using recombinant inbred lines (RILs) between these different strains, it has been shown that as few as two loci—the C. briggsae HOX genes mab-5 and egl-5 or closely linked loci—are sufficient to explain the posterior v3 localization, but that at least two additional loci are involved, one of which only affects the frequency of v3+v4 fusion (Baird et al. 2005). A similar RIL approach revealed cryptic genetic variation, i.e., transgressive variation in v3 position, likely due to epistatic interactions between different alleles at different loci from different C. elegans strains (Guess 2008). Such intraspecific genetic variation has the potential for leading to the types of evolutionary differences in ray pattern that we now observe between species.
Evolution of phasmid position:
Several rhabditid species have been described as having ten bilateral pairs of rays, but one of these is actually the phasmid, which is positioned anterior (instead of posterior) to the posterior-most three rays (Fitch and Emmons 1995; Kiontke and Sudhaus 2000; Sudhaus and Fitch 2001). Because the phasmid tip is usually attached to the outer cuticle that makes the fan, the phasmid is drawn out like the rays, and was often mistaken as a 10th ray in original species descriptions. Evolutionary switches between “anterior” and “posterior” positions of phasmids occurred independently in at least three lineages in Clade V: in the clade including Haematozoon + Pleiorhabditis (e.g., Pelodera strongyloides; Figure 6), in the lineage to Cruznema, and in Diplogastridae; Figure 1 and Figure 6; Fitch and Emmons 1995; Kiontke and Sudhaus 2000). These apparently “saltational” changes in phasmid positions have been hypothesized to be due to a simple developmental change: i.e., a reversal in T blast cell division polarity (Fitch 1997; Kiontke and Sudhaus 2000). In C. elegans, the three most posterior rays derive from T.a and the phasmid sockets derive from T.p, the anterior and posterior daughters of the T blast cell, respectively (Figure 6) (Sulston and Horvitz 1977; Sulston et al. 1980). Reversal of the T division, as occurs in mutants of the lin-44/Wnt signaling pathway (Herman and Horvitz 1994; Herman et al. 1995), would place phasmids in the anterior position. Similar cell division polarity differences have been observed in other nematode species comparisons (Sternberg and Horvitz 1981, 1982). However, an alternative hypothesis is that the polarity of the T division has not changed and there have been subsequent migrations of ray or phasmid precursors along the AP axis. These hypotheses are currently being tested.
Male-specific tail tip morphogenesis and its evolution:
In C. elegans, the four cells (hyp8-11) that constitute the tip of the tail in both sexes originate during embryogenesis and form their tapered, pointed shape during elongation (Hall and Altun 2008). This pointed shape is maintained throughout development in both sexes and into the adult stage of the hermaphrodite. In L4 males, however, these cells fuse, round up, and migrate inwardly and anteriorly (Nguyen et al. 1999). An associated sex-shared neuron is also extensively remodeled (Serrano-Saiz et al. 2017a). As a result of these processes, the tail tip of the adult male is rounded, or “peloderan” (Gk. “bowl” + “skin”). It is noteworthy that this is a case of sexual dimorphism at the level of homologous, sex-shared cells.
Tail tip morphogenesis has changed repeatedly during the evolution of rhabditid nematodes (Sudhaus and Fitch 2001; Kiontke and Fitch 2005). Besides peloderan species like C. elegans (and M. blumi and P. strongyloides; Figure 6), there are “leptoderan” (Gk. “narrow” + “skin”) species in which tail tip retraction does not occur in males, and in which the tail tips (but not the rest of the tails) are thus sexually monomorphic. In leptoderan males, the pointed tail tips nearly always stick out behind the fan (e.g., Rhabditella axei; Figure 6). Within rhabditids, species with peloderan tails have evolved from leptoderan ancestors and vice versa several times (Kiontke and Fitch 2005). Such repeated evolution provides an opportunity to explore the extent to which evolutionary trajectories are constrained by genetic architecture, “developmental constraints” or other biases in the production of morphological variation (Funk and Brooks 1990; Harvey and Pagel 1991; Brooks 1996; Kiontke et al. 2007; Gompel and Prud’homme 2009).
DMD-3 is a transcription factor required and sufficient for initiation of male tail tip morphogenesis, as well as the remodeling of associated neurons (Mason et al. 2008; Serrano-Saiz et al. 2017a). Mutants of dmd-3 generate males with leptoderan tails. DMD-3 appears to be at the center of a “bow-tie” gene-regulatory network, in which it integrates temporal (the heterochronic pathway), spatial (HOX genes), sexual and other cues and coordinates downstream processes associated with cell fusion, vesicular trafficking, and regulation of cytoskeletal architecture (Nelson et al. 2011). Such DMRT factors have been repeatedly recruited for the production of male-specific features (Kopp 2012); whether or not DMD-3 has been recruited in the repeated evolution of tail tip sexual dimorphism is a focus of current studies.
An obvious question with regard to male tail variation is whether or not a particular feature of an organism is an adaptation crafted by natural selection. Though fundamental, this can be difficult to test. The “comparative method” tests for phylogenetic correlations between traits that indicate if one trait is dependent on another; repeated, homoplastic evolutionary events provide the power to test such correlations (Funk and Brooks 1990; Harvey and Pagel 1991; Brooks 1996). One hypothesis is that the shape of the male tail is an adaptation to mating behavior or mating position (Sudhaus 1976; Fitch 2000). For example, according to the most parsimonious reconstruction of trait evolution, ancestral rhabditids had no fan and were leptoderan. A fan then arose independently in the Pleiorhabditis and Eurhabditis clades (also in diplogastrids) and was subsequently lost or greatly reduced several times independently (Fitch 2000; Sudhaus and Fitch 2001; Sudhaus and Fürst von Lieven 2003). Males with broad fans (often but not always peloderan) tend to use their tails sort of like suction cups and mate in a “parallel” body position relative to the female, whereas leptoderan males with reduced or no fans mate in a “spiral” fashion, wrapping around the female’s body (Sudhaus 1976). Consistent with selection imposing interdependency on these characters, there is a significant correlation between retaining a fan and retaining parallel mating (Fitch 2000). It is conceivable that some mating positions may be more favorable in some ecological environments than others; e.g., spiral mating may provide stability in fluid environments, whereas parallel positions may be more efficient on solid substrates. Essentially nothing is known about the natural conditions of living for most of these species—an open field for future investigations.
Although this selectionist explanation seems reasonable for overall morphological differences in male tails, other aspects of male tail variation may have alternative explanations. For example, there is considerable redundancy among different rays regarding their functions in mating behavior, probably to ensure robustness and efficiency of mating success (Liu and Sternberg 1995; Koo et al. 2011; Sherlekar and Lints 2014). Variations in ray position might therefore contribute little, if any, advantage to mating success, but might instead be due to pleiotropic effects of selection on AP patterning of other parts of the body. Alternatively, variation in ray position may have little to do with selection at all, and instead be due to the fixation of particular variants by genetic drift. Whatever the ultimate cause of this variation, the male tail holds great promise to uncover proximate mechanisms underlying developmental changes important for morphological evolution.
Dauer formation and phenotypic plasticity
Though C. elegans is famous for having invariant embryonic development, it also presents one of the best characterized examples of developmental plasticity. In response to crowding (via a pheromone), starvation (via reduction of insulin and TGF-β signaling), and/or heat stress during the second larval stage (L2), an alternative form of the L3 larva develops that is highly resistant to subsequent stresses. The dauer is crucial across nematode diversity for dispersal and survival of adverse conditions (Perry and Wharton 2011), and, in Caenorhabditis, differs from the reproductive L3 in many ways (Androwski et al. 2017). We refer readers interested in the details of dauer regulation in C. elegans to other reviews (Hu 2007; Fielenbach and Antebi 2008), and focus here on conservation and variation across the nematodes.
A key role of insulin signaling in the transition from dauer to postdauer reproductive development is conserved in diverse nematodes. For example, pharmacological inhibition of PI3 kinase (AGE-1 in C. elegans) blocks dauer exit in the hookwoork Ancyclostoma (Brand and Hawdon 2004) and the related strongylid Nippostrongylus (Huang et al. 2010). Similarly, loss-of-function mutations in the DAF-16/FOXO homolog of Pristionchus pacificus block dauer entry (Ogawa et al. 2011). It is thus likely that a canonical insulin pathway regulated the entry and exit from the dauer, including its infective variant that enables parasitic life cycles (Crook 2014), in an ancestor to Rhabditida (Sudhaus 2010).
Beyond serving as a trait that may facilitate parasitism, dauer formation is tied to other variable phenotypes. As noted above, Auanema XX larvae develop into either selfing hermaphrodites or obligately outcrossing females, depending upon whether they pass through dauer or not. Because application of dafachronic acid blocks both dauer formation and hermaphrodite development (Chaudhuri et al. 2011), it appears that control of this sexual mode polyphenism is downstream of the same DAF-12 nuclear hormone receptor that integrates various sensory inputs to the dauer decision. Thus, a pre-existing switch mechanism has been co-opted to regulate a new trait: gonad development. A similar co-option appears to have evolved with regard to the mouth form of Pristionchus pacificus. This species exhibits a polyphenism, in which adults show either a narrow-mouthed (stenostomatous) bacteriovore or a wide-mouthed (eurystomatous) predator-omnivore morphology. The latter is likely to be adaptive when bacteria become limiting (Serobyan et al. 2014), and thus would be expected to form under dauer-inducing conditions. Indeed, starvation or application of crowding pheromones greatly increase the frequency of the eurystomatous form (Bento et al. 2010). Consistent with dauer signaling co-option, loss of P. pacificus daf-12 activity via mutations or application of exogenous dafachronic acid ligands greatly reduces formation of both dauer larvae and the eurystomatous form. Interestingly, though these perturbations block dauer formation (Ogawa et al. 2009), they do not eliminate the eurystomatous form completely, suggesting that other factors also play a role. In line with this, forward genetic screens have identified other genes that have no obvious relationship to C. elegans dauer formation with stronger effects (Ragsdale et al. 2013; Kieninger et al. 2016).
Discussion
The findings above concern a wide range of developmental processes, from the earliest embryonic divisions to adult reproduction, and were produced by applying the developmental genetic tools of C. elegans to a growing list of other nematodes (Table 1). From the details of these processes and how they vary, several principles can be inferred that are likely to be general for the evolution of animal development. Some of these ideas are incorporated into the review above, but some of the most salient are the following:
Table 1. Status of methods for developmental genetics in various nematode species.
Methods for perturbing or measuring gene activity in C. elegans (left column) have been employed successfully in a number of other free-living nematodes. The reference given is generally the first published example, but others often exist.
No published studies, but C. remanei dpy and unc mutants (isolated by K.L. Chow lab at Hong Kong University of Science and Technology) are available from the Caenorhabditis Genetics Center (https://cgc.umn.edu).
Susceptible only via gonadal injection.
An accurate, well-resolved phylogeny is essential for EDB
A phylogeny provides the framework for interspecific comparisons. For example, it is required for testing if compared genes, cells, developmental processes or traits in different species are homologous or independently evolved (nonhomologous). It allows directionality of evolutionary changes to be inferred (what was ancestral or derived), and how many times such changes occurred repeatedly. A phylogeny is required for tests of correlated evolution in different traits. Finally, a phylogeny allows informed selection of species for further research that are representative of the phylogenetic diversity. A good phylogenetic framework for nematodes, particularly for Rhabditida, and, especially, for Caenorhabditis, is now available, although improvements to resolution (e.g., using multilocus or whole-genome data) and species representation continue.
Many genes, pathways, and cells are highly conserved
Though variation was emphasized here, the very ability to describe the nature of that variation hinges upon recognition of homologous genes, pathways, and cells (which hinges upon a good phylogeny). For example, we are only able to say that gld-1 has opposite roles in hermaphrodite spermatogenesis, because unambiguous gld-1 orthologs exist in the species compared. Similarly, conserved developmental origins provide evidence for the homology of VPC and Rn cells and early blastomeres, supporting the many inferences of evolutionary changes made in the development of the vulva, male tail, and embryo.
Early events are surprisingly evolvable
Though the final outcome of nematode embryogenesis is predictably vermiform, there appear to be many ways to begin that all lead to this shape. Early variation appears to funnel into a more constrained “phylotypic stage,” with relatively constant morphogenetic processes and gene expression. This early variation is often apparent in the zygote and first embryonic cell cycle, indicating that the evolution of development can emerge from cell-level behaviors (polarity, lineage, signaling, etc.). Nematodes thus represent an excellent system for evolutionary cell biology and its interface with development. Similar variation in the earliest postfertilization events is seen in insects (El-Sherif et al. 2012) and vertebrates (Hasley et al. 2017). The same principle applies to the cellular differences underlying the evolution of sex determination, such as meiotic modifications that produce a nullo-X germ cell or an XO parthenogenic male via chromosome diminution.
Gene duplication allows partition and gain of gene functions
Gene duplication and divergence has long been recognized as an important phenomenon in evolution (Ono 1972), and nematodes provide excellent examples. Both fog-2 and she-1, lineage-specific genes that allow hermaphrodite spermatogenesis discussed above, are duplicated and diverged members of the large F-box protein family. Novel features crucial to the role of she-1 have yet to be determined, but for fog-2 it appears that acquisition of a C-terminal GLD-1-binding domain was a key event that allowed its co-option into the sex determination pathway (Nayak et al. 2005). Gene duplication and divergence can also underlie DSD, as exemplified by C. briggsae puf-2. Though Cbr-PUF-2 shares the same RNA-binding properties as its closest paralogs, it alone is required for pharyngeal development (Liu and Haag 2014). This is true even as the pharynx itself has not changed in any obvious way.
Genetic network architecture may influence evolutionary trajectories
Certain nodes in the GRNs, such as central nodes of “bow-tie” or “hour-glass” networks (Nelson et al. 2011), may be optimally positioned for evolutionary changes to produce targeted effects on particular traits (Kopp 2009). Examples include the DM-domain genes that coordinate the production of many male-specific traits in nematodes, flies, and other animals (Kopp 2012). Regulators of such genes (e.g., signaling or HOX patterning modules) as well as effectors (e.g., cytoskeletal components and other machinery involved in cellular morphogenesis) may themselves be too pleiotropic (and thus constrained) to be effective targets for specific evolutionary change. Regulatory elements of DM-domain genes themselves, however, could conceivably be more pliable and effective agents of evolutionary change (Kopp 2012). GLD-1 may represent a similar “sweet spot” or “hot-spot,” as indicated by its independent co-option in germline sex determination. As another example, the HOX specification of VPC fates appears to constrain which Pn.p cells can become vulval precursors; this constraint is overcome in posterior-vulva species by posteriad migration of the VPCs.
On the other hand, EDB research on the vulva has demonstrated that quantitative changes to the regulation of even very pleiotropic genes, such as those comprising the Wnt and Ras signaling modules, are surprisingly evolvable. There seem to be near-infinite combinations of signaling factors, targets, and quantitative variants in the strength of their interaction that can all produce a functional vulva. This has some bearing on the oft-discussed role of constraints in the evolution of development (Brakefield 2006; Vermeij 2015). Nematode EDB shows that for many traits we do not yet know how many paths within developmental network space are available to build a phenotype, and thus how many potential routes there may be to an adaptive variant. As a result, even factors that appear to constitute constraints at present may instead reflect insufficient sampling.
Pervasive DSD and its implications for research
Another major theme emerging from the above studies is the astonishing ubiquity of DSD. At the level of developmental gene regulation, rapid evolution of cis-regulatory sequences occurs, but often remains cryptic due to the action of stabilizing selection. Because stabilizing selection mandates an outcome, but not a mechanism, compensatory evolution [or apparently compensatory, see Haag (2007)] proceeds unchecked. This process can be accelerated by directional selection on other loci that share trans-regulators (Johnson and Porter 2007), and generates complex dependencies between distinct promoter regions (Ludwig et al. 2000). The facilitation of DSD by pleiotropy is expected at higher levels of organization; selection on one output of a pleiotropic locus is likely to also change its (cryptic) contribution to a second output not under selection. Here, we have seen such examples of DSD as the different contributions of different signaling modules to vulval induction, the conservation of core components and interactions in the sex determination pathway despite rapid evolution at protein–protein recognition domains, and transgressive variation underlying conserved ray positions in the male tail. Other studies have provided further evidence of DSD from genome-scale comparisons of gene function. For example, systematic RNAi knockdown of essential genes in wild isolates of C. elegans (Paaby et al. 2015), and of their C. briggsae orthologs (Verster et al. 2014) revealed many instances of distinct phenotypes in both cases that could not be explained by knockdown efficacy. Cryptic genetic changes underlying canalized developmental processes are thus apparently rampant.
In hindsight, the choice of species for initial characterization of a developmental process has a large influence on the path of research. For example, selection of P. pacificus for the first genetic analyses of vulva development would have led to focus on the Wnt pathway rather than Notch and EGF, while choosing Diploscapter for the oocyte-to-embryo transition would have led to a complete different picture of cellular interactions and embryo polarization.
Microevolution is reflected in macroevolution
Cryptic genetic variation in the development of a conserved phenotypic output abounds in nematodes, and variation between species is mirrored by variation within them. This is true both qualitatively and quantitatively, providing strong support for the existence of biases in the introduction of variation (mutation and its impact on networks), and their influence on interspecies divergence. The interaction between these biases and natural selection offer a more complete view of evolutionary causation (Stoltzfus 2006). Intraspecific variation also suggests that using a “representative strain” may sometimes lead to a false sense of species divergence. This is particularly true concerning the laboratory strain of C. elegans, which has adapted to laboratory conditions and fixed mutations that improve fitness but indirectly affect developmental processes (McGrath et al. 2009, 2011; Duveau and Félix 2012; Andersen et al. 2014).
Nematodes like Caenorhabditis offer the ability to connect specific developmental processes with the general phenomenon of cryptic variation. Beyond being interesting, such connections are fundamental to understanding the genetic architecture of non-Mendelian disease, the response to selection in agriculture, the resistance of pathogens and cancer cells to drugs, and the mechanisms that underlie cellular and organismal homeostasis (Gibson and Dworkin 2004). For example, susceptibility to the topoisomerase-targeting chemotherapy agent etoposide varies among C. elegans isolates, and much of this can be explained by a single amino acid polymorphism that appears to be neutral in the absence of the drug (Zdraljevic et al. 2017). Amazingly, the same polymorphism distinguishes topoisomerase paralogs in humans as well.
Repeated evolution involves both reproducible co-options and idiosyncratic components
As pointed out by others (Kopp 2009), repeated evolution (phylogenetic replication) in a clade of experimentally tractable species (a “metamodel”) provides the opportunity to look for more general principles in the evolution of developmental-genetic systems. The research summarized above abundantly demonstrates the utility of nematodes in such a research program. For example, the convergent evolution of self-fertility has shown how specific conserved genes (e.g., tra-2 and gld-1) or the same gene classes (e.g., those encoding F-box proteins like fog-2 and she-1) are repeatedly co-opted (parallel evolution). Another example is the involvement of spe-8 in the activation of hermaphrodite sperm in two independent lineages of Caenorhabditis. At the same time, repeated evolution can involve idiosyncratic solutions (convergent evolution). Sticking with self-fertility, we see that the sperm activation program deployed in hermaphrodites can vary, as does the precise role of GLD-1 in germline sex determination. Evolutionary biases may also be revealed; e.g., once programmed cell death evolves as the mechanism to restrict the VPC group, reversal is rare. Given the advantages of nematodes for EDB, we can look forward to many more discoveries.
Acknowledgments
We thank our many colleagues in the nematode EDB community, whose enthusiasm, openness, and cooperation has greatly accelerated the pace of discovery. Among them, three anonymous reviewers and our associate editor (M.A.F.) provided a large number of helpful comments. We also gratefully acknowledge the various funding agencies that have supported the work described here (in the laboratories of the authors and more generally) over the last two decades.
Footnotes
Communicating editor: M.-A. Félix
Literature Cited
- Aboobaker A. A., Blaxter M. L., 2003. Hox gene loss during dynamic evolution of the nematode cluster. Curr. Biol. 13: 37–40. 10.1016/S0960-9822(02)01399-4 [DOI] [PubMed] [Google Scholar]
- Albert P. S., Brown S. J., Riddle D. L., 1981. Sensory control of dauer larva formation in Caenorhabditis elegans. J. Comp. Neurol. 198: 435–451. 10.1002/cne.901980305 [DOI] [PubMed] [Google Scholar]
- Albertson D. G., Nwaorgu O. C., Sulston J. E., 1979. Chromatin diminution and a chromosomal mechanism of sexual differentiation in Strongyloides papillosus. Chromosoma 75: 75–87. 10.1007/BF00330626 [DOI] [PubMed] [Google Scholar]
- Andersen E. C., Bloom J. S., Gerke J. P., Kruglyak L., 2014. A variant in the neuropeptide receptor npr-1 is a major determinant of Caenorhabditis elegans growth and physiology. PLoS Genet. 10: e1004156 (erratum: PLoS Genet. 10: e1004316) 10.1371/journal.pgen.1004156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrássy I., 1983. A Taxonomic Review of the Suborder Rha bditina (Nematoda: Secernentia). L’Office de la Recherche Scientifique et Technique Outre-Mer, Paris. [Google Scholar]
- Andrássy I., 1984. Klasse Nematoda (Ordnungen Monhysterida, Desmoscolecida, Araeolaimida, Chromadorida, Rhabditida). Gustav Fischer Verlag, Stuttgart. [Google Scholar]
- Androwski R. J., Flatt K. M., Schroeder N. E., 2017. Phenotypic plasticity and remodeling in the stress-induced Caenorhabditis elegans dauer. Wiley Interdiscip. Rev. Dev. Biol. 6: null. 10.1002/wdev.278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin J., Kimble J., 1987. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 51: 589–599. 10.1016/0092-8674(87)90128-0 [DOI] [PubMed] [Google Scholar]
- Baird S., 2001. Strain-specific variation in the pattern of caudal papillae in Caenorhabditis briggsae (Nematoda: Rhabditidae); implication for species identification. Nematology 3: 373–376. 10.1163/156854101317020295 [DOI] [Google Scholar]
- Baird S. E., Fitch D. H., Kassem I. A., Emmons S. W., 1991. Pattern formation in the nematode epidermis: determination of the arrangement of peripheral sense organs in the C. elegans male tail. Development 113: 515–526. [DOI] [PubMed] [Google Scholar]
- Baird S. E., Davidson C. R., Bohrer J. C., 2005. The genetics of ray pattern variation in Caenorhabditis briggsae. BMC Evol. Biol. 5: 3 10.1186/1471-2148-5-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi C., Cho S., Ellis R. E., 2009. Mutations in two independent pathways are sufficient to create hermaphroditic nematodes. Science 326: 1002–1005. 10.1126/science.1176013 [DOI] [PubMed] [Google Scholar]
- Baldi C., Viviano J., Ellis R. E., 2011. A bias caused by ectopic development produces sexually dimorphic sperm in nematodes. Curr. Biol. 21: 1416–1420. 10.1016/j.cub.2011.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandi C., Anderson T. J., Genchi C., Blaxter M. L., 1998. Phylogeny of Wolbachia in filarial nematodes. Proc. Biol. Sci. 265: 2407–2413. 10.1098/rspb.1998.0591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandi C., McCall J. W., Genchi C., Corona S., Venco L., et al. , 1999. Effects of tetracycline on the filarial worms Brugia pahangi and Dirofilaria immitis and their bacterial endosymbionts Wolbachia. Int. J. Parasitol. 29: 357–364. 10.1016/S0020-7519(98)00200-8 [DOI] [PubMed] [Google Scholar]
- Bargmann C. I., 2006. Comparative chemosensation from receptors to ecology. Nature 444: 295–301. 10.1038/nature05402 [DOI] [PubMed] [Google Scholar]
- Barkoulas M., Vargas Velazquez A. M., Peluffo A. E., Félix M. A., 2016. Evolution of new cis-regulatory motifs required for cell-specific gene expression in Caenorhabditis. PLoS Genet. 12: e1006278 10.1371/journal.pgen.1006278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr M. M., Garcia L. R., 2006. Male mating behavior (June 19, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.78.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrière A., Félix M. A., 2014. Isolation of C. elegans and related nematodes (July 17, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.115.1, http://www.wormbook.org. 10.1895/wormbook.1.115.2 [DOI] [Google Scholar]
- Barrière A., Ruvinsky I., 2014. Pervasive divergence of transcriptional gene regulation in Caenorhabditis nematodes. PLoS Genet. 10: e1004435 10.1371/journal.pgen.1004435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrière A., Gordon K. L., Ruvinsky I., 2012. Coevolution within and between regulatory loci can preserve promoter function despite evolutionary rate acceleration. PLoS Genet. 8: e1002961 10.1371/journal.pgen.1002961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadell A. V., Haag E. S., 2014. Evolutionary dynamics of GLD-1-mRNA complexes in Caenorhabditis nematodes. Genome Biol. Evol. 7: 314–335. 10.1093/gbe/evu272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadell A. V., Liu Q., Johnson D. M., Haag E. S., 2011. Independent recruitments of a translational regulator in the evolution of self-fertile nematodes. Proc. Natl. Acad. Sci. USA 108: 19672–19677. 10.1073/pnas.1108068108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento G., Ogawa A., Sommer R. J., 2010. Co-option of the hormone-signalling module dafachronic acid-DAF-12 in nematode evolution. Nature 466: 494–497. 10.1038/nature09164 [DOI] [PubMed] [Google Scholar]
- Besnard F., Koutsovoulos G., Dieudonne S., Blaxter M., Félix M. A., 2017. Toward universal forward genetics: using a draft genome sequence of the nematode Oscheius tipulae to identify mutations affecting vulva development. Genetics 206: 1747–1761. 10.1534/genetics.117.203521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M., 2011. Nematodes: the worm and its relatives. PLoS Biol. 9: e1001050 (erratum: PLoS Biol. 9) 10.1371/journal.pbio.1001050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgonie G., Jacobsen K., Coomans A., 2000. Embryonic lineage evolution in nematodes. Nematology 2: 65–69. 10.1163/156854100508908 [DOI] [Google Scholar]
- Bosshardt S. C., McCall J. W., Coleman S. U., Jones K. L., Petit T. A., et al. , 1993. Prophylactic activity of tetracycline against Brugia pahangi infection in jirds (Meriones unguiculatus). J. Parasitol. 79: 775–777. 10.2307/3283620 [DOI] [PubMed] [Google Scholar]
- Boveri T., 1899. Die Entwickelung von Ascaris megalocephala mit besonderer Rücksicht auf die Kernverhältnisse, pp. 383–430 in Festschrift für C. v. Kupffer. Gustav Fischer Verlag, Jena, Germany. [Google Scholar]
- Braendle C., Félix M. A., 2008. Plasticity and errors of a robust developmental system in different environments. Dev. Cell 15: 714–724. 10.1016/j.devcel.2008.09.011 [DOI] [PubMed] [Google Scholar]
- Braendle C., Baer C. F., Félix M. A., 2010. Bias and evolution of the mutationally accessible phenotypic space in a developmental system. PLoS Genet. 6: e1000877 10.1371/journal.pgen.1000877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakefield P. M., 2006. Evo-devo and constraints on selection. Trends Ecol. Evol. 21: 362–368. 10.1016/j.tree.2006.05.001 [DOI] [PubMed] [Google Scholar]
- Brand A., Hawdon J. M., 2004. Phosphoinositide-3-OH-kinase inhibitor LY294002 prevents activation of Ancylostoma caninum and Ancylostoma ceylanicum third-stage infective larvae. Int. J. Parasitol. 34: 909–914. 10.1016/j.ijpara.2004.04.003 [DOI] [PubMed] [Google Scholar]
- Brauchle M., Kiontke K., MacMenamin P., Fitch D. H., Piano F., 2009. Evolution of early embryogenesis in rhabditid nematodes. Dev. Biol. 335: 253–262. 10.1016/j.ydbio.2009.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 2009. In the beginning was the worm. Genetics 182: 413–415. 10.1534/genetics.109.104976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs M., 1946. Culture Methods for a Free-Living Soil Nematode, pp. 63 Stanford University, Palo Alto, CA. [Google Scholar]
- Brooks D. R., 1996. Explanations of homoplasy at different levels of biological organization, in Homoplasy: The Recurrence of Similarity in Evolution, edited by Sanderson M. J., Hufford L. Academic Press, San Diego: 10.1016/B978-012618030-5/50003-4 [DOI] [Google Scholar]
- Bumbarger D. J., Riebesell M., Rodelsperger C., Sommer R. J., 2013. System-wide rewiring underlies behavioral differences in predatory and bacterial-feeding nematodes. Cell 152: 109–119. 10.1016/j.cell.2012.12.013 [DOI] [PubMed] [Google Scholar]
- Bürglin T. R., Kuwabara P. E., 2006. Homologs of the Hh signalling network in C. elegans (January 28, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.76.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd D. T., Knobel K., Affeldt K., Crittenden S. L., Kimble J., 2014. A DTC niche plexus surrounds the germline stem cell pool in Caenorhabditis elegans. PLoS One 9: e88372 10.1371/journal.pone.0088372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón-Urrea A., Vanholme B., Vangestel S., Kane S. M., Bahaji A., et al. , 2016. Early development of the root-knot nematode Meloidogyne incognita. BMC Dev. Biol. 16: 10 10.1186/s12861-016-0109-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. B., 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36. 10.1016/j.cell.2008.06.030 [DOI] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium, 1998 Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018 [corrigenda: Science 283: 35 (1999)]; [corrigenda: Science 283: 2103 (1999)]; [corrigenda: Science 285: 1493 (1999)]. [DOI] [PubMed]
- Chalfie M., White J. G., 1988. The nervous system, pp. 337–391 in The Nematode Caenorhabditis elegans, edited by Wood W. B. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Chamberlin H. M., Thomas J. H., 2000. The bromodomain protein LIN-49 and trithorax-related protein LIN-59 affect development and gene expression in Caenorhabditis elegans. Development 127: 713–723. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J., Kache V., Pires-daSilva A., 2011. Regulation of sexual plasticity in a nematode that produces males, females, and hermaphrodites. Curr. Biol. 21: 1548–1551. 10.1016/j.cub.2011.08.009 [DOI] [PubMed] [Google Scholar]
- Chen P., Ellis R. E., 2000. TRA-1A regulates transcription of fog-3, which controls germ cell fate in C. elegans. Development 127: 3119–3129. [DOI] [PubMed] [Google Scholar]
- Chen P., Cho S., Jin S., Ellis R., 2001. Specification of germ cell fates by FOG-3 has been conserved during nematode evolution. Genetics 158: 1513–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Shen Y., Ellis R. E., 2014. Dependence of the sperm/oocyte decision on the nucleosome remodeling factor complex was acquired during recent Caenorhabditis briggsae evolution. Mol. Biol. Evol. 31: 2573–2585. 10.1093/molbev/msu198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherra S. J., III, Jin Y., 2015. Advances in synapse formation: forging connections in the worm. Wiley Interdiscip. Rev. Dev. Biol. 4: 85–97. 10.1002/wdev.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood B. G., Chitwood M. B., 1974. Introduction to Nematology. University Park Press, Baltimore. [Google Scholar]
- Cho S., Jin S. W., Cohen A., Ellis R. E., 2004. A phylogeny of Caenorhabditis reveals frequent loss of introns during nematode evolution. Genome Res. 14: 1207–1220. 10.1101/gr.2639304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow K. L., Emmons S. W., 1994. HOM-C/Hox genes and four interacting loci determine the morphogenetic properties of single cells in the nematode male tail. Development 120: 2579–2592. [DOI] [PubMed] [Google Scholar]
- Chow K. L., Hall D. H., Emmons S. W., 1995. The mab-21 gene of Caenorhabditis elegans encodes a novel protein required for choice of alternate cell fates. Development 121: 3615–3626. [DOI] [PubMed] [Google Scholar]
- Cinkornpumin J. K., Hong R. L., 2011. RNAi mediated gene knockdown and transgenesis by microinjection in the necromenic nematode Pristionchus pacificus. J. Vis. Exp. 56: e3270 10.3791/3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinquin O., Crittenden S. L., Morgan D. E., Kimble J., 2010. Progression from a stem cell-like state to early differentiation in the C. elegans germ line. Proc. Natl. Acad. Sci. USA 107: 2048–2053. 10.1073/pnas.0912704107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clandinin T. R., Katz W. S., Sternberg P. W., 1997. Caenorhabditis elegans HOM-C genes regulate the response of vulval precursor cells to inductive signal. Dev. Biol. 182: 150–161. 10.1006/dbio.1996.8471 [DOI] [PubMed] [Google Scholar]
- Clifford R., Lee M., Nayak S., Ohmachi M., Giorgini F., et al. , 2000. FOG-2, a novel F-box-containing protein, associates with the GLD-1 RNA-binding protein and directs male sex determination in the C. elegans hermaphrodite germline. Development 127: 5265–5276. [DOI] [PubMed] [Google Scholar]
- Cline T. W., Meyer B. J., 1996. Vive la différence: males vs. females in flies vs. worms. Annu. Rev. Genet. 30: 637–702. 10.1146/annurev.genet.30.1.637 [DOI] [PubMed] [Google Scholar]
- Cook D. E., Zdraljevic S., Tanny R. E., Seo B., Riccardi D. D., et al. , 2016. The genetic basis of natural variation in Caenorhabditis elegans telomere length. Genetics 204: 371–383. 10.1534/genetics.116.191148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D. E., Zdraljevic S., Roberts J. P., Andersen E. C., 2017. CeNDR, the Caenorhabditis elegans natural diversity resource. Nucleic Acids Res. 45: D650–D657. 10.1093/nar/gkw893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coroian C., Broitman-Maduro G., Maduro M. F., 2006. Med-type GATA factors and the evolution of mesendoderm specification in nematodes. Dev. Biol. 289: 444–455. 10.1016/j.ydbio.2005.10.024 [DOI] [PubMed] [Google Scholar]
- Crittenden S. L., Leonhard K. A., Byrd D. T., Kimble J., 2006. Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol. Biol. Cell 17: 3051–3061. 10.1091/mbc.e06-03-0170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook M., 2014. The dauer hypothesis and the evolution of parasitism: 20 years on and still going strong. Int. J. Parasitol. 44: 1–8. 10.1016/j.ijpara.2013.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter A. D., 2008. Divergence times in Caenorhabditis and Drosophila, inferred from direct estimates of the neutral mutation rate. Mol. Biol. Evol. 25: 778–786. 10.1093/molbev/msn024 [DOI] [PubMed] [Google Scholar]
- Cutter A. D., Félix M. A., Barriere A., Charlesworth D., 2006. Patterns of nucleotide polymorphism distinguish temperate and tropical wild isolates of Caenorhabditis briggsae. Genetics 173: 2021–2031. 10.1534/genetics.106.058651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bono M., Hodgkin J., 1996. Evolution of sex determination in Caenorhabditis: unusually high divergence of tra-1 and its functional consequences. Genetics 144: 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre M., Félix M. A., 2001. Polymorphism and evolution of vulval precursor cell lineages within two nematode genera, Caenorhabditis and Oscheius. Curr. Biol. 11: 631–643. 10.1016/S0960-9822(01)00202-0 [DOI] [PubMed] [Google Scholar]
- De Ley P., 2006. A quick tour of nematode diversity and the backbone of nematode phylogeny (January 25, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.41.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ley, P., and M. L. Blaxter, 2004 A new system for Nematoda: combining morphological characters with molecular trees, and translating clades into ranks and taxa. In Cook R, Hunt DJ, editors, Nematology Monographs and Perspectives. Vol. 2. Leiden: E.J. Brill. 2004. p. 633–653. [Google Scholar]
- Dey A., Jeon Y., Wang G. X., Cutter A. D., 2012. Global population genetic structure of Caenorhabditis remanei reveals incipient speciation. Genetics 191: 1257–1269. 10.1534/genetics.112.140418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtel M.-L., Louvet-Vallée S., Viney M. E., Félix M.-A., Sternberg P. W., 2001. Control of vulval cell division number in the nematode Oscheius/Dolichorhabditis sp. CEW1. Genetics 157: 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtel-Danjoy M. L., Félix M. A., 2004a. Phenotypic neighborhood and micro-evolvability. Trends Genet. 20: 268–276. 10.1016/j.tig.2004.03.010 [DOI] [PubMed] [Google Scholar]
- Dichtel-Danjoy M. L., Félix M. A., 2004b. The two steps of vulval induction in Oscheius tipulae CEW1 recruit common regulators including a MEK kinase. Dev. Biol. 265: 113–126. 10.1016/j.ydbio.2003.09.010 [DOI] [PubMed] [Google Scholar]
- Dolgin E. S., Félix M. A., Cutter A. D., 2008. Hakuna Nematoda: genetic and phenotypic diversity in African isolates of Caenorhabditis elegans and C. briggsae. Heredity (Edinb) 100: 304–315. 10.1038/sj.hdy.6801079 [DOI] [PubMed] [Google Scholar]
- Dolinski C., Baldwin J. G., Thomas W. K., 2001. Comparative survey of early embryogenesis of Secernentea (Nematoda), with phylogenetic implications. Can. J. Zool. 79: 82–94. 10.1139/z00-179 [DOI] [Google Scholar]
- Dougherty E. C., Nigon V., 1949. A new species of the free-living nematode genus Rhabditis of interest in comparative physiology and genetics. J. Parasitol. 35: 11. [Google Scholar]
- Dufourcq P., Chanal P., Vicaire S., Camut E., Quintin S., et al. , 1999. lir-2, lir-1 and lin-26 encode a new class of zinc-finger proteins and are organized in two overlapping operons both in Caenorhabditis elegans and in Caenorhabditis briggsae. Genetics 152: 221–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duveau F., Félix M. A., 2012. Role of pleiotropy in the evolution of a cryptic developmental variation in Caenorhabditis elegans. PLoS Biol. 10: e1001230 10.1371/journal.pbio.1001230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers S., Ohnesorg T., Sinclair A., 2014. Genetic regulation of mammalian gonad development. Nat. Rev. Endocrinol. 10: 673–683. 10.1038/nrendo.2014.163 [DOI] [PubMed] [Google Scholar]
- Eisenmann D. M., Kim S. K., 2000. Protruding vulva mutants identify novel loci and Wnt signaling factors that function during Caenorhabditis elegans vulva development. Genetics 156: 1097–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eizinger A., Sommer R. J., 1997. The homeotic gene lin-39 and the evolution of nematode epidermal cell fates. Science 278: 452–455. 10.1126/science.278.5337.452 [DOI] [PubMed] [Google Scholar]
- Ellis R. E., 2017. “The persistence of memory”-Hermaphroditism in nematodes. Mol. Reprod. Dev. 84: 144–157. 10.1002/mrd.22668 [DOI] [PubMed] [Google Scholar]
- Ellis R. E., Stanfield G. M., 2014. The regulation of spermatogenesis and sperm function in nematodes. Semin. Cell Dev. Biol. 29: 17–30. 10.1016/j.semcdb.2014.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sherif E., Lynch J. A., Brown S. J., 2012. Comparisons of the embryonic development of Drosophila, Nasonia, and Tribolium. Wiley Interdiscip. Rev. Dev. Biol. 1: 16–39. 10.1002/wdev.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons S. W., 2005. Male development (November 10, 2005), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.33.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons S. W., 2014. The development of sexual dimorphism: studies of the Caenorhabditis elegans male. Wiley Interdiscip. Rev. Dev. Biol. 3: 239–262. 10.1002/wdev.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerholm H. P., 1991. Systematic implications of male caudal morphology in ascaridoid nematode parasites. Syst. Parasitol. 19: 215–229. 10.1007/BF00011888 [DOI] [Google Scholar]
- Farhadifar R., Baer C. F., Valfort A. C., Andersen E. C., Muller-Reichert T., et al. , 2015. Scaling, selection, and evolutionary dynamics of the mitotic spindle. Curr. Biol. 25: 732–740. 10.1016/j.cub.2014.12.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M. A., 1999. Evolution of developmental mechanisms in nematodes. J. Exp. Zool. 285: 3–18. [DOI] [PubMed] [Google Scholar]
- Félix M. A., 2004. Alternative morphs and plasticity of vulval development in a rhabditid nematode species. Dev. Genes Evol. 214: 55–63. 10.1007/s00427-003-0376-y [DOI] [PubMed] [Google Scholar]
- Félix M. A., 2006. Oscheius tipulae (August 16, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.119.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M. A., 2007. Cryptic quantitative evolution of the vulva intercellular signaling network in Caenorhabditis. Curr. Biol. 17: 103–114. 10.1016/j.cub.2006.12.024 [DOI] [PubMed] [Google Scholar]
- Félix M. A., Sternberg P. W., 1997. Two nested gonadal inductions of the vulva in nematodes. Development 124: 253–259. [DOI] [PubMed] [Google Scholar]
- Félix M. A., Sternberg P. W., 1998. A gonad-derived survival signal for vulval precursor cells in two nematode species. Curr. Biol. 8: 287–290. 10.1016/S0960-9822(98)70111-3 [DOI] [PubMed] [Google Scholar]
- Félix M. A., De Ley P., Sommer R. J., Frisse L., Nadler S. A., et al. , 2000a. Evolution of vulva development in the Cephalobina (Nematoda). Dev. Biol. 221: 68–86. 10.1006/dbio.2000.9665 [DOI] [PubMed] [Google Scholar]
- Félix M. A., Delattre M., Dichtel M. L., 2000b. Comparative developmental studies using Oscheius/Dolichorhabditis sp. CEW1 (Rhabditidae). Nematology 2: 89–98. 10.1163/156854100508809 [DOI] [Google Scholar]
- Félix M. A., Braendle C., Cutter A. D., 2014. A streamlined system for species diagnosis in Caenorhabditis (Nematoda: Rhabditidae) with name designations for 15 distinct biological species. PLoS One 9: e94723 [corrigenda: PLoS One 10: e0118327 (2015)] 10.1371/journal.pone.0094723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari C., Salle R., Callemeyn-Torre N., Jovelin R., Cutter A. D., et al. , 2017. Ephemeral-habitat colonization and neotropical species richness of Caenorhabditis nematodes. BMC Ecol. 17: 43 10.1186/s12898-017-0150-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira H. B., Zhang Y., Zhao C., Emmons S. W., 1999. Patterning of Caenorhabditis elegans posterior structures by the Abdominal-B homolog, egl-5. Dev. Biol. 207: 215–228. 10.1006/dbio.1998.9124 [DOI] [PubMed] [Google Scholar]
- Ferris H., Hieb W. F., 2015. Ellsworth C. Dougherty: a pioneer in the selection of Caenorhabditis elegans as a model organism. Genetics 200: 991–1002. 10.1534/genetics.115.178913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N., Antebi A., 2008. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 22: 2149–2165. 10.1101/gad.1701508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierst J. L., Willis J. H., Thomas C. G., Wang W., Reynolds R. M., et al. , 2015. Reproductive mode and the evolution of genome size and structure in Caenorhabditis nematodes. PLoS Genet. 11: e1005323 (erratum: PLoS Genet. 11: e1005497) 10.1371/journal.pgen.1005323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch D. H., 1997. Evolution of male tail development in rhabditid nematodes related to Caenorhabditis elegans. Syst. Biol. 46: 145–179. 10.1093/sysbio/46.1.145 [DOI] [PubMed] [Google Scholar]
- Fitch D. H. A., 2000. Evolution of “Rhabditidae” and the male tail. J. Nematol. 32: 235–244. [PMC free article] [PubMed] [Google Scholar]
- Fitch D. H. A., Emmons S. W., 1995. Variable cell positions and cell contacts underlie morphological evolution of the rays in the male tails of nematodes related to Caenorhabditis elegans. Dev. Biol. 170: 564–582. 10.1006/dbio.1995.1237 [DOI] [PubMed] [Google Scholar]
- Foray V., Pérez-Jiménez M. M., Fattouh N., Landmann F., 2018. Wolbachia control stem cell behavior and stimulate germline proliferation in filarial nematodes. Dev. Cell 45: 198–211.e3. 10.1016/j.devcel.2018.03.017 [DOI] [PubMed] [Google Scholar]
- Fradin H., Kiontke K., Zegar C., Gutwein M., Lucas J., et al. , 2017. Genome architecture and evolution of a unichromosomal asexual nematode. Curr. Biol. 27: 2928–2939.e6. 10.1016/j.cub.2017.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman P. A., Platzer E. G., Eby J. E., 1977. Species differentiation in Caenorhabditis briggsae and Caenorhabditis elegans. J. Nematol. 9: 197–203. [PMC free article] [PubMed] [Google Scholar]
- Funk V. A., Brooks D. R., 1990. Phylogenetic Systematics as the Basis for Comparative Biology. Smithsonian Institution Press, Washington, DC: 10.5962/bhl.title.123293 [DOI] [Google Scholar]
- Garcia-Dorado A., 2012. Understanding and predicting the fitness decline of shrunk populations: inbreeding, purging, mutation, and standard selection. Genetics 190: 1461–1476. 10.1534/genetics.111.135541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldziler B., Chatterjee I., Kadandale P., Putiri E., Patel R., et al. , 2006. A comparative study of sperm morphology, cytology and activation in Caenorhabditis elegans, Caenorhabditis remanei and Caenorhabditis briggsae. Dev. Genes Evol. 216: 198–208. 10.1007/s00427-005-0045-4 [DOI] [PubMed] [Google Scholar]
- Gibson G., Dworkin I., 2004. Uncovering cryptic genetic variation. Nat. Rev. Genet. 5: 681–690. 10.1038/nrg1426 [DOI] [PubMed] [Google Scholar]
- Giribet G., 2016. Genomics and the animal tree of life: conflicts and future prospects. Zool. Scr. 45: 14–21. 10.1111/zsc.12215 [DOI] [Google Scholar]
- Giribet G., Edgecombe G. D., 2017. Current understanding of Ecdysozoa and its internal phylogenetic relationships. Integr. Comp. Biol. 57: 455–466. 10.1093/icb/icx072 [DOI] [PubMed] [Google Scholar]
- Gleason J. E., Korswagen H. C., Eisenmann D. M., 2002. Activation of Wnt signaling bypasses the requirement for RTK/Ras signaling during C. elegans vulval induction. Genes Dev. 16: 1281–1290. 10.1101/gad.981602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason J. E., Szyleyko E. A., Eisenmann D. M., 2006. Multiple redundant Wnt signaling components function in two processes during C. elegans vulval development. Dev. Biol. 298: 442–457. 10.1016/j.ydbio.2006.06.050 [DOI] [PubMed] [Google Scholar]
- Goldstein B., Frisse L. M., Thomas W. K., 1998. Embryonic axis specification in nematodes: evolution of the first step in development. Curr. Biol. 8: 157–160. 10.1016/S0960-9822(98)70062-4 [DOI] [PubMed] [Google Scholar]
- Gompel N., Prud’homme B., 2009. The causes of repeated genetic evolution. Dev. Biol. 332: 36–47. 10.1016/j.ydbio.2009.04.040 [DOI] [PubMed] [Google Scholar]
- Gordon K. L., Arthur R. K., Ruvinsky I., 2015. Phylum-level conservation of regulatory information in nematodes despite extensive non-coding sequence divergence. PLoS Genet. 11: e1005268 10.1371/journal.pgen.1005268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham P., Kimble J., 1993. The mog-1 gene is required for the switch from spermatogenesis to oogenesis in Caenorhabditis elegans. Genetics 133: 919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham P. L., Schedl T., Kimble J., 1993. More mog genes that influence the switch from spermatogenesis to oogenesis in the hermaphrodite germ line of Caenorhabditis elegans. Dev. Genet. 14: 471–484. 10.1002/dvg.1020140608 [DOI] [PubMed] [Google Scholar]
- Grote P., Conradt B., 2006. The PLZF-like protein TRA-4 cooperates with the Gli-like transcription factor TRA-1 to promote female development in C. elegans. Dev. Cell 11: 561–573. 10.1016/j.devcel.2006.07.015 [DOI] [PubMed] [Google Scholar]
- Guess, A. J., 2008 QTL analysis of ray pattern in Caenorhabditis elegans recombinant inbred lines, M.S. Thesis in Biological Sciences. Wright State University, Dayton, OH. [Google Scholar]
- Guo Y., Lang S., Ellis R. E., 2009. Independent recruitment of F box genes to regulate hermaphrodite development during nematode evolution. Curr. Biol. 19: 1853–1860. 10.1016/j.cub.2009.09.042 [DOI] [PubMed] [Google Scholar]
- Guo Y., Chen X., Ellis R. E., 2013. Evolutionary change within a bipotential switch shaped the sperm/oocyte decision in hermaphroditic nematodes. PLoS Genet. 9: e1003850 10.1371/journal.pgen.1003850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta B. P., Hanna-Rose W., Sternberg P. W., 2012. Morphogenesis of the vulva and the vulval-uterine connection (November 30, 2012), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.152.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez A., Sommer R. J., 2007. Functional diversification of the nematode mbd2/3 gene between Pristionchus pacificus and Caenorhabditis elegans. BMC Genet. 8: 57 10.1186/1471-2156-8-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag E. S., 2007. Compensatory vs. pseudocompensatory evolution in molecular and developmental interactions. Genetica 129: 45–55. 10.1007/s10709-006-0032-3 [DOI] [PubMed] [Google Scholar]
- Haag E. S., Ackerman A. D., 2005. Intraspecific variation in fem-3 and tra-2, two rapidly coevolving nematode sex-determining genes. Gene 349: 35–42. 10.1016/j.gene.2004.12.051 [DOI] [PubMed] [Google Scholar]
- Haag E. S., Doty A. V., 2005. Sex determination across evolution: connecting the dots. PLoS Biol. 3: e21 10.1371/journal.pbio.0030021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag E. S., Kimble J., 2000. Regulatory elements required for development of Caenorhabditis elegans hermaphrodites are conserved in the tra-2 homologue of C. remanei, a male/female sister species. Genetics 155: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag E. S., Molla M. N., 2005. Compensatory evolution of interacting gene products through multifunctional intermediates. Evolution Int J Org Evolution 59: 1620–1632. 10.1111/j.0014-3820.2005.tb01813.x [DOI] [PubMed] [Google Scholar]
- Haag E. S., True J. R., 2007. Evolution and development: anchors away! Curr. Biol. 17: R172–R174. 10.1016/j.cub.2007.01.015 [DOI] [PubMed] [Google Scholar]
- Haag E. S., Wang S., Kimble J., 2002. Rapid coevolution of the nematode sex-determining genes fem-3 and tra-2. Curr. Biol. 12: 2035–2041. 10.1016/S0960-9822(02)01333-7 [DOI] [PubMed] [Google Scholar]
- Hahn A. C., Emmons S. W., 2003. The roles of an ephrin and a semaphorin in patterning cell-cell contacts in C. elegans sensory organ development. Dev. Biol. 256: 379–388. 10.1016/S0012-1606(02)00129-X [DOI] [PubMed] [Google Scholar]
- Hall D. H., Altun Z. F., 2008. C. elegans Atlas. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- Hall D. H., Winfrey V. P., Blaeuer G., Hoffman L. H., Furuta T., et al. , 1999. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: relations between the germ line and soma. Dev. Biol. 212: 101–123. 10.1006/dbio.1999.9356 [DOI] [PubMed] [Google Scholar]
- Hamilton W. D., 1967. Extraordinary sex ratios. Science 156: 477–488. 10.1126/science.156.3774.477 [DOI] [PubMed] [Google Scholar]
- Hansen D., Pilgrim D., 1998. Molecular evolution of a sex determination protein. FEM-2 (pp2c) in Caenorhabditis. Genetics 149: 1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey P. H., Pagel M. D., 1991. The Comparative Method in Evolutionary Biology. Oxford University Press, Oxford. [Google Scholar]
- Hasley A., Chavez S., Danilchik M., Wühr M., Pelegri F., 2017. Vertebrate embryonic cleavage pattern determination. Adv. Exp. Med. Biol. 953: 117–171. 10.1007/978-3-319-46095-6_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman M. A., Horvitz H. R., 1994. The Caenorhabditis elegans gene lin-44 controls the polarity of asymmetric cell divisions. Development 120: 1035–1047. [DOI] [PubMed] [Google Scholar]
- Herman M. A., Vassilieva L. L., Horvitz H. R., Shaw J. E., Herman R. K., 1995. The C. elegans gene lin-44, which controls the polarity of certain asymmetric cell divisions, encodes a Wnt protein and acts cell nonautonomously. Cell 83: 101–110. 10.1016/0092-8674(95)90238-4 [DOI] [PubMed] [Google Scholar]
- Hill K. L., L’Hernault S. W., 2001. Analyses of reproductive interactions that occur after heterospecific matings within the genus Caenorhabditis. Dev. Biol. 232: 105–114. 10.1006/dbio.2000.0136 [DOI] [PubMed] [Google Scholar]
- Hill R. C., Haag E. S., 2009. A sensitized genetic background reveals evolution near the terminus of the Caenorhabditis germline sex determination pathway. Evol. Dev. 11: 333–342. 10.1111/j.1525-142X.2009.00340.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill R. C., de Carvalho C. E., Salogiannis J., Schlager B., Pilgrim D., et al. , 2006. Genetic flexibility in the convergent evolution of hermaphroditism in Caenorhabditis nematodes. Dev. Cell 10: 531–538. 10.1016/j.devcel.2006.02.002 [DOI] [PubMed] [Google Scholar]
- Hill R. J., Sternberg P. W., 1992. The gene lin-3 encodes an inductive signal for vulval development in C. elegans. Nature 358: 470–476. 10.1038/358470a0 [DOI] [PubMed] [Google Scholar]
- Hillier L. W., Miller R. D., Baird S. E., Chinwalla A., Fulton L. A., et al. , 2007. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 5: e167 10.1371/journal.pbio.0050167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S. H., So G. M., Chow K. L., 2001. Postembryonic expression of Caenorhabditis elegans mab-21 and its requirement in sensory ray differentiation. Dev. Dyn. 221: 422–430. 10.1002/dvdy.1161 [DOI] [PubMed] [Google Scholar]
- Hobert O., 2010. Neurogenesis in the nematode Caenorhabditis elegans (October 4, 2010), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.12.2, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., 1986. Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics 114: 15–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., 2002. One lucky XX male: isolation of the first Caenorhabditis elegans sex-determination mutants. Genetics 162: 1501–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J. A., Brenner S., 1977. Mutations causing transformation of sexual phenotype in the nematode Caenorhabditis elegans. Genetics 86: 275–287. [PMC free article] [PubMed] [Google Scholar]
- Holden-Dye L., Walker R. J., 1994. Characterization of identifiable neurones in the head ganglia of the parasitic nematode Ascaris suum: a comparison with central neurones of Caenorhabditis elegans. Parasitology 108: 81–87. 10.1017/S0031182000078550 [DOI] [PubMed] [Google Scholar]
- Holterman M., Holovachov O., van den Elsen S., van Megen H., Bongers T., et al. , 2008. Small subunit ribosomal DNA-based phylogeny of basal Chromadoria (Nematoda) suggests that transitions from marine to terrestrial habitats (and vice versa) require relatively simple adaptations. Mol. Phylogenet. Evol. 48: 758–763. 10.1016/j.ympev.2008.04.033 [DOI] [PubMed] [Google Scholar]
- Honda H., 1925. Experimental and cytological studies on bisexual and hermaphrodite free-living nematodes, with special reference to problems of sex. J. Morph. Phys. 40: 191–233. 10.1002/jmor.1050400202 [DOI] [Google Scholar]
- Houthoofd W., Jacobsen K., Mertens C., Vangestel S., Coomans A., et al. , 2003. Embryonic cell lineage of the marine nematode Pellioditis marina. Dev. Biol. 258: 57–69. 10.1016/S0012-1606(03)00101-5 [DOI] [PubMed] [Google Scholar]
- Houthoofd W., Willems M., Jacobsen K., Coomans A., Borgonie G., 2008. The embryonic cell lineage of the nematode Rhabditophanes sp. Int. J. Dev. Biol. 52: 963–967. 10.1387/ijdb.072404wh [DOI] [PubMed] [Google Scholar]
- Hoyos E., Kim K., Milloz J., Barkoulas M., Penigault J. B., et al. , 2011. Quantitative variation in autocrine signaling and pathway crosstalk in the Caenorhabditis vulval network. Curr. Biol. 21: 527–538. 10.1016/j.cub.2011.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, P. J., 2007 Dauer (August 08, 2007), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.144.1, http://www.wormbook.org.
- Huang R. E., Ren X., Qiu Y., Zhao Z., 2014. Description of Caenorhabditis sinica sp. n. (Nematoda: Rhabditidae), a nematode species used in comparative biology for C. elegans. PLoS One 9: e110957 10.1371/journal.pone.0110957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. C., Chan D. T., Smyth D. J., Ball G., Gounaris K., et al. , 2010. Activation of Nippostrongylus brasiliensis infective larvae is regulated by a pathway distinct from the hookworm Ancylostoma caninum. Int. J. Parasitol. 40: 1619–1628. 10.1016/j.ijpara.2010.06.004 [DOI] [PubMed] [Google Scholar]
- Jan E., Motzny C. K., Graves L. E., Goodwin E. B., 1999. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 18: 258–269. 10.1093/emboj/18.1.258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell T. A., Wang Y., Bloniarz A. E., Brittin C. A., Xu M., et al. , 2012. The connectome of a decision-making neural network. Science 337: 437–444. 10.1126/science.1221762 [DOI] [PubMed] [Google Scholar]
- Johnson N. A., Porter A. H., 2007. Evolution of branched regulatory genetic pathways: directional selection on pleiotropic loci accelerates developmental system drift. Genetica 129: 57–70. 10.1007/s10709-006-0033-2 [DOI] [PubMed] [Google Scholar]
- Jones A. R., Francis R., Schedl T., 1996. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 180: 165–183. 10.1006/dbio.1996.0293 [DOI] [PubMed] [Google Scholar]
- Jud M., Razelun J., Bickel J., Czerwinski M., Schisa J. A., 2007. Conservation of large foci formation in arrested oocytes of Caenorhabditis nematodes. Dev. Genes Evol. 217: 221–226. 10.1007/s00427-006-0130-3 [DOI] [PubMed] [Google Scholar]
- Jungblut B., Sommer R. J., 2000. Novel cell-cell interactions during vulva development in Pristionchus pacificus. Development 127: 3295–3303. [DOI] [PubMed] [Google Scholar]
- Kanzaki N., Kiontke K., Tanaka R., Hirooka Y., Schwarz A., et al. , 2017. Description of two three-gendered nematode species in the new genus Auanema (Rhabditina) that are models for reproductive mode evolution. Sci. Rep. 7: 11135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz W. S., Hill R. J., Clandinin T. R., Sternberg P. W., 1995. Different levels of the C. elegans growth factor LIN-3 promote distinct vulval precursor fates. Cell 82: 297–307. 10.1016/0092-8674(95)90317-8 [DOI] [PubMed] [Google Scholar]
- Kelleher D. F., de Carvalho C. E., Doty A. V., Layton M., Cheng A. T., et al. , 2008. Comparative genetics of sex determination: masculinizing mutations in Caenorhabditis briggsae. Genetics 178: 1415–1429. 10.1534/genetics.107.073668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieninger M. R., Ivers N. A., Rodelsperger C., Markov G. V., Sommer R. J., et al. , 2016. The nuclear hormone receptor NHR-40 acts downstream of the sulfatase EUD-1 as part of a developmental plasticity switch in Pristionchus. Curr. Biol. 26: 2174–2179. 10.1016/j.cub.2016.06.018 [DOI] [PubMed] [Google Scholar]
- Kienle S., Sommer R. J., 2013. Cryptic variation in vulva development by cis-regulatory evolution of a HAIRY-binding site. Nat. Commun. 4: 1714 10.1038/ncomms2711 [DOI] [PubMed] [Google Scholar]
- Kimble J., Hirsh D., 1979. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 70: 396–417. 10.1016/0012-1606(79)90035-6 [DOI] [PubMed] [Google Scholar]
- Kimble J. E., White J. G., 1981. On the control of germ cell development in Caenorhabditis elegans. Dev. Biol. 81: 208–219. 10.1016/0012-1606(81)90284-0 [DOI] [PubMed] [Google Scholar]
- Kiontke, K., and D. H. Fitch, 2005 The phylogenetic relationships of Caenorhabditis and other rhabditids (August 11, 2005), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.11.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K., Fitch D. H. A., 2013. Nematodes. Curr. Biol. 23: R862–R864. 10.1016/j.cub.2013.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K., Sudhaus W., 2000. Phasmids in male Rhabditida and other secernentean nematodes. J. Nematode Morphol. Syst. 3: 1–37. [Google Scholar]
- Kiontke K., Gavin N. P., Raynes Y., Roehrig C., Piano F., et al. , 2004. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc. Natl. Acad. Sci. USA 101: 9003–9008. 10.1073/pnas.0403094101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K., Barrière A., Kolotuev I., Podbilewicz B., Sommer R. J., et al. , 2007. Trends, stasis and drift in the evolution of nematode vulva development. Curr. Biol. 17: 1925–1937. 10.1016/j.cub.2007.10.061 [DOI] [PubMed] [Google Scholar]
- Kiontke K. C., Félix M. A., Ailion M., Rockman M. V., Braendle C., et al. , 2011. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol. Biol. 11: 339 10.1186/1471-2148-11-339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo P. K., Bian X., Sherlekar A. L., Bunkers M. R., Lints R., 2011. The robustness of Caenorhabditis elegans male mating behavior depends on the distributed properties of ray sensory neurons and their output through core and male-specific targets. J. Neurosci. 31: 7497–7510. 10.1523/JNEUROSCI.6153-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A., 2009. Metamodels and phylogenetic replication: a systematic approach to the evolution of developmental pathways. Evolution 63: 2771–2789. 10.1111/j.1558-5646.2009.00761.x [DOI] [PubMed] [Google Scholar]
- Kopp A., 2012. Dmrt genes in the development and evolution of sexual dimorphism. Trends Genet. 28: 175–184. 10.1016/j.tig.2012.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S., Maduzia L. L., Padgett R. W., 1999. Specificity of TGFbeta signaling is conferred by distinct type I receptors and their associated SMAD proteins in Caenorhabditis elegans. Development 126: 251–260. [DOI] [PubMed] [Google Scholar]
- Kuwabara P. E., 1996. Interspecies comparison reveals evolution of control regions in the nematode sex-determining gene tra-2. Genetics 144: 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara P. E., Shah S., 1994. Cloning by synteny: identifying C. briggsae homologues of C. elegans genes. Nucleic Acids Res. 22: 4414–4418. 10.1093/nar/22.21.4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahl V., Sadler B., Schierenberg E., 2006. Egg development in parthenogenetic nematodes: variations in meiosis and axis formation. Int. J. Dev. Biol. 50: 393–397. 10.1387/ijdb.052030vl [DOI] [PubMed] [Google Scholar]
- Lahl V., Schulze J., Schierenberg E., 2009. Differences in embryonic pattern formation between Caenorhabditis elegans and its close parthenogenetic relative Diploscapter coronatus. Int. J. Dev. Biol. 53: 507–515. 10.1387/ijdb.082718vl [DOI] [PubMed] [Google Scholar]
- LaMunyon C. W., Ward S., 1998. Larger sperm outcompete smaller sperm in the nematode Caenorhabditis elegans. Proc. Biol. Sci. 265: 1997–2002. 10.1098/rspb.1998.0531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMunyon C. W., Ward S., 1999. Evolution of sperm size in nematodes: sperm competition favours larger sperm. Proc. Biol. Sci. 266: 263–267. 10.1098/rspb.1999.0631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMunyon C. W., Ward S., 2002. Evolution of larger sperm in response to experimentally increased sperm competition in Caenorhabditis elegans. Proc. Biol. Sci. 269: 1125–1128. 10.1098/rspb.2002.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmann F., Voronin D., Sullivan W., Taylor M. J., 2011. Anti-filarial activity of antibiotic therapy is due to extensive apoptosis after Wolbachia depletion from filarial nematodes. PLoS Pathog. 7: e1002351 10.1371/journal.ppat.1002351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmann F., Foster J. M., Michalski M. L., Slatko B. E., Sullivan W., 2014. Co-evolution between an endosymbiont and Its nematode host: wolbachia asymmetric posterior localization and AP polarity establishment. PLoS Negl. Trop. Dis. 8: e3096 10.1371/journal.pntd.0003096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langworthy N. G., Renz A., Mackenstedt U., Henkle-Duhrsen K., de Bronsvoort M. B., et al. , 2000. Macrofilaricidal activity of tetracycline against the filarial nematode Onchocerca ochengi: elimination of Wolbachia precedes worm death and suggests a dependent relationship. Proc. Biol. Sci. 267: 1063–1069. 10.1098/rspb.2000.1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T. S., Yang F. J., Lo Y. H., Chang T. C., Hsu J. C., et al. , 2017. Non-Mendelian assortment of homologous autosomes of different sizes in males is the ancestral state in the Caenorhabditis lineage. Sci. Rep. 7: 12819 10.1038/s41598-017-13215-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M., Hashimshony T., Wagner F., Yanai I., 2012. Developmental milestones punctuate gene expression in the Caenorhabditis embryo. Dev. Cell 22: 1101–1108. 10.1016/j.devcel.2012.04.004 [DOI] [PubMed] [Google Scholar]
- Lin K. T., Broitman-Maduro G., Hung W. W., Cervantes S., Maduro M. F., 2009. Knockdown of SKN-1 and the Wnt effector TCF/POP-1 reveals differences in endomesoderm specification in C. briggsae as compared with C. elegans. Dev. Biol. 325: 296–306. 10.1016/j.ydbio.2008.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lints R., Emmons S. W., 1999. Patterning of dopaminergic neurotransmitter identity among Caenorhabditis elegans ray sensory neurons by a TGFbeta family signaling pathway and a Hox gene. Development 126: 5819–5831. [DOI] [PubMed] [Google Scholar]
- Lints R., Emmons S. W., 2002. Regulation of sex-specific differentiation and mating behavior in C. elegans by a new member of the DM domain transcription factor family. Genes Dev. 16: 2390–2402. 10.1101/gad.1012602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lints R., Jia L., Kim K., Li C., Emmons S. W., 2004. Axial patterning of C. elegans male sensilla identities by selector genes. Dev. Biol. 269: 137–151. 10.1016/j.ydbio.2004.01.021 [DOI] [PubMed] [Google Scholar]
- Liu K. S., Sternberg P. W., 1995. Sensory regulation of male mating behavior in Caenorhabditis elegans. Neuron 14: 79–89. 10.1016/0896-6273(95)90242-2 [DOI] [PubMed] [Google Scholar]
- Liu Q., Haag E. S., 2014. Evolutionarily dynamic roles of a PUF RNA-binding protein in the somatic development of Caenorhabditis briggsae. J. Exp. Zoolog. B Mol. Dev. Evol. 322: 129–141. 10.1002/jez.b.22550 [DOI] [PubMed] [Google Scholar]
- Liu Q., Stumpf C., Thomas C., Wickens M., Haag E. S., 2012. Context-dependent function of a conserved translational regulatory module. Development 139: 1509–1521. 10.1242/dev.070128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Chen L., Shang Y., Huang P., Miao L., 2013. The micronutrient element zinc modulates sperm activation through the SPE-8 pathway in Caenorhabditis elegans. Development 140: 2103–2107. 10.1242/dev.091025 [DOI] [PubMed] [Google Scholar]
- Lo T. W., Pickle C. S., Lin S., Ralston E. J., Gurling M., et al. , 2013. Precise and heritable genome editing in evolutionarily diverse nematodes using TALENs and CRISPR/Cas9 to engineer insertions and deletions. Genetics 195: 331–348. 10.1534/genetics.113.155382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loer C. M., Kenyon C. J., 1993. Serotonin-deficient mutants and male mating behavior in the nematode Caenorhabditis elegans. J. Neurosci. 13: 5407–5417. 10.1523/JNEUROSCI.13-12-05407.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvet-Vallee S., Kolotuev I., Podbilewicz B., Félix M. A., 2003. Control of vulval competence and centering in the nematode Oscheius sp. 1 CEW1. Genetics 163: 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M. Z., Bergman C., Patel N. H., Kreitman M., 2000. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403: 564–567. 10.1038/35000615 [DOI] [PubMed] [Google Scholar]
- Lum D., Kuwabara P., Zarkower D., Spence A., 2000. Direct protein-protein interaction between the intracellular domain of TRA-2 and the transcription factor TRA-1A modulates feminizing activity in C. elegans. Genes Dev. 14: 3153–3165. [PMC free article] [PubMed] [Google Scholar]
- Lüpold S., Pitnick S., 2018. Sperm form and function: what do we know about the role of sexual selection? Reproduction 155: R229–R243. 10.1530/REP-17-0536 [DOI] [PubMed] [Google Scholar]
- Luz J. G., Hassig C. A., Pickle C., Godzik A., Meyer B. J., et al. , 2003. XOL-1, primary determinant of sexual fate in C. elegans, is a GHMP kinase family member and a structural prototype for a class of developmental regulators. Genes Dev. 17: 977–990. 10.1101/gad.1082303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., O’Hely M., Walsh B., Force A., 2001. The probability of preservation of a newly arisen gene duplicate. Genetics 159: 1789–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maderspacher F., 2008. Theodor Boveri and the natural experiment. Curr. Biol. 18: R279–R286. 10.1016/j.cub.2008.02.061 [DOI] [PubMed] [Google Scholar]
- Maduro M. F., 2009. Structure and evolution of the C. elegans embryonic endomesoderm network. Biochim. Biophys. Acta 1789: 250–260. 10.1016/j.bbagrm.2008.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalak K. K., Jama A. M., Billups S. J., Dawes A. T., Chamberlin H. M., 2017. Differing roles for sur-2/MED23 in C. elegans and C. briggsae vulval development. Dev. Genes Evol. 227: 213–218. 10.1007/s00427-017-0577-4 [DOI] [PubMed] [Google Scholar]
- Malakhov V. V., 1994. Nematodes—Structure, Development, Classification and Phylogeny. Smithsonian Institution Press, Washington, DC. [Google Scholar]
- Mason D. A., Rabinowitz J. S., Portman D. S., 2008. dmd-3, a doublesex-related gene regulated by tra-1, governs sex-specific morphogenesis in C. elegans. Development 135: 2373–2382. 10.1242/dev.017046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupas E., 1900. Modes et formes de reproduction des nématodes. Arch. Zool. Exp. 8: 463–624. [Google Scholar]
- McCaig C. M., Lin X., Farrell M., Rehain-Bell K., Shakes D. C., 2017. Germ cell cysts and simultaneous sperm and oocyte production in a hermaphroditic nematode. Dev. Biol. 430: 362–373. 10.1016/j.ydbio.2017.08.010 [DOI] [PubMed] [Google Scholar]
- McGrath P. T., Rockman M. V., Zimmer M., Jang H., Macosko E. Z., et al. , 2009. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron 61: 692–699. 10.1016/j.neuron.2009.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath P. T., Xu Y., Ailion M., Garrison J. L., Butcher R. A., et al. , 2011. Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes. Nature 477: 321–325. 10.1038/nature10378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren D. J., Worms M. J., Laurence B. R., Simpson M. G., 1975. Micro-organisms in filarial larvae (Nematoda). Trans. R. Soc. Trop. Med. Hyg. 69: 509–514. 10.1016/0035-9203(75)90110-8 [DOI] [PubMed] [Google Scholar]
- Mello C. C., Draper B. W., Priess J. R., 1994. The maternal genes apx-1 and glp-1 and establishment of dorsal-ventral polarity in the early C. elegans embryo. Cell 77: 95–106. 10.1016/0092-8674(94)90238-0 [DOI] [PubMed] [Google Scholar]
- Mickey K. M., Mello C. C., Montgomery M. K., Fire A., Priess J. R., 1996. An inductive interaction in 4-cell stage C. elegans embryos involves APX-1 expression in the signalling cell. Development 122: 1791–1798. [DOI] [PubMed] [Google Scholar]
- Milloz J., Duveau F., Nuez I., Félix M. A., 2008. Intraspecific evolution of the intercellular signaling network underlying a robust developmental system. Genes Dev. 22: 3064–3075. 10.1101/gad.495308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Chow K. L., Ueno N., 1999. Regulation of body length and male tail ray pattern formation of Caenorhabditis elegans by a member of TGF-beta family. Development 126: 1337–1347. [DOI] [PubMed] [Google Scholar]
- Morran L. T., Cappy B. J., Anderson J. L., Phillips P. C., 2009a Sexual partners for the stressed: facultative outcrossing in the self-fertilizing nematode Caenorhabditis elegans. Evolution 63: 1473–1482. 10.1111/j.1558-5646.2009.00652.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran L. T., Parmenter M. D., Phillips P. C., 2009b Mutation load and rapid adaptation favour outcrossing over self-fertilization. Nature 462: 350–352. 10.1038/nature08496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namai S., Sugimoto A., 2018. Transgenesis by microparticle bombardment for live imaging of fluorescent proteins in Pristionchus pacificus germline and early embryos. Dev. Genes Evol. 228: 75–82. 10.1007/s00427-018-0605-z [DOI] [PubMed] [Google Scholar]
- Nayak S., Goree J., Schedl T., 2005. fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biol. 3: e6 10.1371/journal.pbio.0030006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. D., Zhou E., Kiontke K., Fradin H., Maldonado G., et al. , 2011. A bow-tie genetic architecture for morphogenesis suggested by a genome-wide RNAi screen in Caenorhabditis elegans. PLoS Genet. 7: e1002010 10.1371/journal.pgen.1002010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemetschke L., Eberhardt A. G., Hertzberg H., Streit A., 2010. Genetics, chromatin diminution, and sex chromosome evolution in the parasitic nematode genus Strongyloides. Curr. Biol. 20: 1687–1696. 10.1016/j.cub.2010.08.014 [DOI] [PubMed] [Google Scholar]
- Neumuller R. A., Knoblich J. A., 2009. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 23: 2675–2699. 10.1101/gad.1850809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C. Q., Hall D. H., Yang Y., Fitch D. H., 1999. Morphogenesis of the Caenorhabditis elegans male tail tip. Dev. Biol. 207: 86–106. 10.1006/dbio.1998.9173 [DOI] [PubMed] [Google Scholar]
- Niebur E., Erdos P., 1993. Theory of the locomotion of nematodes: control of the somatic motor neurons by interneurons. Math. Biosci. 118: 51–82. 10.1016/0025-5564(93)90033-7 [DOI] [PubMed] [Google Scholar]
- Nigon V., 1943. Le déterminisme du sexe chez un nématode libre hermaphrodite (Rhabditis elegans Maupas). C R Soc Biol 137: 40–41. [Google Scholar]
- Nigon V., 1951. Polyploide experimentale chez un nematode libre, Rhabditis elegans Maupas. Bull. Biol. Fr. Belg. 85: 187–225. [Google Scholar]
- Nigon V., Dougherty E. C., 1949. Reproductive patterns and attempts at reciprocal crossing of Rhabditis elegans Maupas, 1900, and Rhabditis briggsae Dougherty and Nigon, 1949 (Nematoda, Rhabditidae). J. Exp. Zool. 112: 485–503. 10.1002/jez.1401120307 [DOI] [PubMed] [Google Scholar]
- Nigon V., Dougherty E. C., 1950. A dwarf mutation in a nematode; a morphological mutant of Rhabditis briggsae, a free-living soil nematode. J. Hered. 41: 103–109. 10.1093/oxfordjournals.jhered.a106095 [DOI] [PubMed] [Google Scholar]
- Nigon V. M., Félix M. A., 2017. History of research on C. elegans and other free-living nematodes as model organisms (September 07, 2017), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.181.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura H., L’Hernault S. W., 2010. Spermatogenesis-defective (spe) mutants of the nematode Caenorhabditis elegans provide clues to solve the puzzle of male germline functions during reproduction. Dev. Dyn. 239: 1502–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuez I., Félix M. A., 2012. Evolution of susceptibility to ingested double-stranded RNAs in Caenorhabditis nematodes. PLoS One 7: e29811 10.1371/journal.pone.0029811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa A., Streit A., Antebi A., Sommer R. J., 2009. A conserved endocrine mechanism controls the formation of dauer and infective larvae in nematodes. Curr. Biol. 19: 67–71. 10.1016/j.cub.2008.11.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa A., Bento G., Bartelmes G., Dieterich C., Sommer R. J., 2011. Pristionchus pacificus daf-16 is essential for dauer formation but dispensable for mouth form dimorphism. Development 138: 1281–1284. 10.1242/dev.058909 [DOI] [PubMed] [Google Scholar]
- Ono S., 1972. Gene duplication, mutation load, and mammalian genetic regulatory systems. J. Med. Genet. 9: 254–263. 10.1136/jmg.9.3.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby A. B., White A. G., Riccardi D. D., Gunsalus K. C., Piano F., et al. , 2015. Wild worm embryogenesis harbors ubiquitous polygenic modifier variation. eLife 4: e09178 10.7554/eLife.09178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannell J. R., 2002. The evolution and maintenance of androdioeecy. Annu. Rev. Ecol. Syst. 33: 397–425. 10.1146/annurev.ecolsys.33.010802.150419 [DOI] [Google Scholar]
- Pénigault J. B., Félix M. A., 2011. Evolution of a system sensitive to stochastic noise: P3.p cell fate in Caenorhabditis. Dev. Biol. 357: 419–427. 10.1016/j.ydbio.2011.05.675 [DOI] [PubMed] [Google Scholar]
- Perry M. D., Li W., Trent C., Robertson B., Fire A., et al. , 1993. Molecular characterization of the her-1 gene suggests a direct role in cell signaling during Caenorhabditis elegans sex determination. Genes Dev. 7: 216–228. 10.1101/gad.7.2.216 [DOI] [PubMed] [Google Scholar]
- Perry R., Wharton D. (Editors), 2011. Molecular and Physiological Basis of Nematode Survival. CABI, Wallingford, UK: 10.1079/9781845936877.0000 [DOI] [Google Scholar]
- Peter I. S., Davidson E. H., 2011. Evolution of gene regulatory networks controlling body plan development. Cell 144: 970–985. 10.1016/j.cell.2011.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires-daSilva A., Sommer R. J., 2004. Conservation of the global sex determination gene tra-1 in distantly related nematodes. Genes Dev. 18: 1198–1208. 10.1101/gad.293504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portman D. S., Emmons S. W., 2000. The basic helix-loop-helix transcription factors LIN-32 and HLH-2 function together in multiple steps of a C. elegans neuronal sublineage. Development 127: 5415–5426. [DOI] [PubMed] [Google Scholar]
- Portman D. S., Emmons S. W., 2004. Identification of C. elegans sensory ray genes using whole-genome expression profiling. Dev. Biol. 270: 499–512. 10.1016/j.ydbio.2004.02.020 [DOI] [PubMed] [Google Scholar]
- Raff R. A., 1996. The Shape of Life: Genes, Development, and the Evolution of Animal Form. University of Chicago Press, Chicago. [Google Scholar]
- Ragsdale E. J., Muller M. R., Rodelsperger C., Sommer R. J., 2013. A developmental switch coupled to the evolution of plasticity acts through a sulfatase. Cell 155: 922–933. 10.1016/j.cell.2013.09.054 [DOI] [PubMed] [Google Scholar]
- Ragsdale E. J., Kanzaki N., Herrmann M., 2015. Taxonomy and natural history: the genus Pristionchus, pp. 77–120 in Pristionchus Pacificus—a Nematode Model for Comparative and Evolutionary Biology, edited by Sommer R. J. Brill, Leiden: 10.1163/9789004260306_005 [DOI] [Google Scholar]
- Raymond C. S., Shamu C. E., Shen M. M., Seifert K. J., Hirsch B., et al. , 1998. Evidence for evolutionary conservation of sex-determining genes. Nature 391: 691–695. 10.1038/35618 [DOI] [PubMed] [Google Scholar]
- Richardson M. K., Minelli A., Coates M., Hanken J., 1998. Phylotypic stage theory. Trends Ecol. Evol. 13: 158 10.1016/S0169-5347(98)01340-8 [DOI] [PubMed] [Google Scholar]
- Riche S., Zouak M., Argoul F., Arneodo A., Pecreaux J., et al. , 2013. Evolutionary comparisons reveal a positional switch for spindle pole oscillations in Caenorhabditis embryos. J. Cell Biol. 201: 653–662. 10.1083/jcb.201210110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., Kruglyak L., 2009. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 5: e1000419 10.1371/journal.pgen.1000419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose L., Gönczy P., 2014. Polarity establishment, asymmetric division and segregation of fate determinants in early C. elegans embryos (December 30, 2014), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.30.2, http://www.wormbook.org. [DOI] [PubMed] [Google Scholar]
- Ross J. A., Koboldt D. C., Staisch J. E., Chamberlin H. M., Gupta B. P., et al. , 2011. Caenorhabditis briggsae recombinant inbred line genotypes reveal inter-strain incompatibility and the evolution of recombination. PLoS Genet. 7: e1002174 10.1371/journal.pgen.1002174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. L., Ivanova E. S., Spiridonov S. E., Waeyenberge L., Moens M., et al. , 2010. Molecular phylogeny of slug-parasitic nematodes inferred from 18S rRNA gene sequences. Mol. Phylogenet. Evol. 55: 738–743. 10.1016/j.ympev.2010.01.026 [DOI] [PubMed] [Google Scholar]
- Ross J. M., Kalis A. K., Murphy M. W., Zarkower D., 2005. The DM domain protein MAB-3 promotes sex-specific neurogenesis in C. elegans by regulating bHLH proteins. Dev. Cell 8: 881–892. 10.1016/j.devcel.2005.03.017 [DOI] [PubMed] [Google Scholar]
- Roy P. J., Zheng H., Warren C. E., Culotti J. G., 2000. mab-20 encodes Semaphorin-2a and is required to prevent ectopic cell contacts during epidermal morphogenesis in Caenorhabditis elegans. Development 127: 755–767. [DOI] [PubMed] [Google Scholar]
- Rudel D., Kimble J., 2001. Conservation of glp-1 regulation and function in nematodes. Genetics 157: 639–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel D., Tian H., Sommer R. J., 2008. Wnt signaling in Pristionchus pacificus gonadal arm extension and the evolution of organ shape. Proc. Natl. Acad. Sci. USA 105: 10826–10831. 10.1073/pnas.0800597105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundell R. J., Leander B. S., 2010. Masters of miniaturization: convergent evolution among interstitial eukaryotes. BioEssays 32: 430–437. 10.1002/bies.200900116 [DOI] [PubMed] [Google Scholar]
- Ryder S. P., Frater L., Abramovitz D., Goodwin E., Williamson J., 2004. RNA target specificity of the STAR/GSG domain posttranscriptional regulatory protein GLD-1. Nat. Struct. Mol. Biol. 11: 20–28. 10.1038/nsmb706 [DOI] [PubMed] [Google Scholar]
- Salser S. J., Kenyon C., 1996. A C. elegans Hox gene switches on, off, on and off again to regulate proliferation, differentiation and morphogenesis. Development 122: 1651–1661. [DOI] [PubMed] [Google Scholar]
- Savage C., Das P., Finelli A. L., Townsend S. R., Sun C. Y., et al. , 1996. Caenorhabditis elegans genes sma-2, sma-3, and sma-4 define a conserved family of transforming growth factor beta pathway components. Proc. Natl. Acad. Sci. USA 93: 790–794. 10.1073/pnas.93.2.790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer W., 2016. Nematode nervous systems. Curr. Biol. 26: R955–R959. 10.1016/j.cub.2016.07.044 [DOI] [PubMed] [Google Scholar]
- Schedl T., Kimble J., 1988. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics 119: 43–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierenberg E., 2005. Unusual cleavage and gastrulation in a freshwater nematode: developmental and phylogenetic implications. Dev. Genes Evol. 215: 103–108. 10.1007/s00427-004-0454-9 [DOI] [PubMed] [Google Scholar]
- Schlager B., Wang X., Braach G., Sommer R. J., 2009. Molecular cloning of a dominant roller mutant and establishment of DNA-mediated transformation in the nematode Pristionchus pacificus. Genesis 47: 300–304. 10.1002/dvg.20499 [DOI] [PubMed] [Google Scholar]
- Schlicht P., Schierenberg E., 1991. Altered establishment of cell lineages in the Caenorhabditis elegans embryo after suppression of the first cleavage supports a concentration-dependent decision mechanism. Rouxs Arch. Dev. Biol. 199: 437–448. 10.1007/BF01705780 [DOI] [PubMed] [Google Scholar]
- Schulze J., Schierenberg E., 2008. Cellular pattern formation, establishment of polarity and segregation of colored cytoplasm in embryos of the nematode Romanomermis culicivorax. Dev. Biol. 315: 426–436. 10.1016/j.ydbio.2007.12.043 [DOI] [PubMed] [Google Scholar]
- Schulze J., Schierenberg E., 2009. Embryogenesis of Romanomermis culicivorax: an alternative way to construct a nematode. Dev. Biol. 334: 10–21. 10.1016/j.ydbio.2009.06.009 [DOI] [PubMed] [Google Scholar]
- Schulze J., Schierenberg E., 2011. Evolution of embryonic development in nematodes. Evodevo 2: 18 10.1186/2041-9139-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze J., Houthoofd W., Uenk J., Vangestel S., Schierenberg E., 2012. Plectus - a stepping stone in embryonic cell lineage evolution of nematodes. Evodevo 3: 13 10.1186/2041-9139-3-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetharaman A., Cumbo P., Bojanala N., Gupta B. P., 2010. Conserved mechanism of Wnt signaling function in the specification of vulval precursor fates in C. elegans and C. briggsae. Dev. Biol. 346: 128–139. 10.1016/j.ydbio.2010.07.003 [DOI] [PubMed] [Google Scholar]
- Semple J. I., Garcia-Verdugo R., Lehner B., 2010. Rapid selection of transgenic C. elegans using antibiotic resistance. Nat. Methods 7: 725–727. 10.1038/nmeth.1495 [DOI] [PubMed] [Google Scholar]
- Semple J. I., Biondini L., Lehner B., 2012. Generating transgenic nematodes by bombardment and antibiotic selection. Nat. Methods 9: 118–119. 10.1038/nmeth.1864 [DOI] [PubMed] [Google Scholar]
- Serobyan V., Ragsdale E. J., Sommer R. J., 2014. Adaptive value of a predatory mouth-form in a dimorphic nematode. Proc. Biol. Sci. 281: 20141334 10.1098/rspb.2014.1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E., Oren-Suissa M., Bayer E. A., Hobert O., 2017a Sexually dimorphic differentiation of a C. elegans hub neuron is cell-autonomously controlled by a conserved transcription factor. Curr. Biol. 27: 199–209. 10.1016/j.cub.2016.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E., Pereira L., Gendrel M., Aghayeva U., Battacharya A., et al. , 2017b A Neurotransmitter atlas of the Caenorhabditis elegans male nervous system reveals sexually dimorphic neurotransmitter usage. Genetics 206: 1251–1269. 10.1534/genetics.117.202127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham S., 2015. Glial development and function in the nervous system of Caenorhabditis elegans. Cold Spring Harb. Perspect. Biol. 7: a020578 10.1101/cshperspect.a020578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakes D. C., Neva B. J., Huynh H., Chaudhuri J., Pires-Dasilva A., 2011. Asymmetric spermatocyte division as a mechanism for controlling sex ratios. Nat. Commun. 2: 157 10.1038/ncomms1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharanya D., Thillainathan B., Marri S., Bojanala N., Taylor J., et al. , 2012. Genetic control of vulval development in Caenorhabditis briggsae. G3 (Bethesda) 2: 1625–1641. 10.1534/g3.112.004598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharanya D., Fillis C. J., Kim J., Zitnik E. M., Jr., Ward K. A., et al. , 2015. Mutations in Caenorhabditis briggsae identify new genes important for limiting the response to EGF signaling during vulval development. Evol. Dev. 17: 34–48. 10.1111/ede.12105 [DOI] [PubMed] [Google Scholar]
- Shen M. M., Hodgkin J., 1988. mab-3, a gene required for sex-specific yolk protein expression and a male-specific lineage in C. elegans. Cell 54: 1019–1031. 10.1016/0092-8674(88)90117-1 [DOI] [PubMed] [Google Scholar]
- Sherlekar A. L., Lints R., 2014. Nematode tango milonguero - the C. elegans male’s search for the hermaphrodite vulva. Semin. Cell Dev. Biol. 33: 34–41. 10.1016/j.semcdb.2014.05.009 [DOI] [PubMed] [Google Scholar]
- Siehr M. S., Koo P. K., Sherlekar A. L., Bian X., Bunkers M. R., et al. , 2011. Multiple doublesex-related genes specify critical cell fates in a C. elegans male neural circuit. PLoS One 6: e26811 10.1371/journal.pone.0026811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist C. B., Sommer R. J., 1999. Vulva formation in Pristionchus pacificus relies on continuous gonadal induction. Dev. Genes Evol. 209: 451–459. 10.1007/s004270050278 [DOI] [PubMed] [Google Scholar]
- Slack J. M., Holland P. W., Graham C. F., 1993. The zootype and the phylotypic stage. Nature 361: 490–492. 10.1038/361490a0 [DOI] [PubMed] [Google Scholar]
- Slos D., Sudhaus W., Stevens L., Bert W., Blaxter M., 2017. Caenorhabditis monodelphis sp. n.: defining the stem morphology and genomics of the genus Caenorhabditis. BMC Zoology 2: 4. [Google Scholar]
- Smith J. R., Stanfield G. M., 2011. TRY-5 is a sperm-activating protease in Caenorhabditis elegans seminal fluid. PLoS Genet. 7: e1002375 10.1371/journal.pgen.1002375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soloviev A., Gallagher J., Marnef A., Kuwabara P. E., 2011. C. elegans patched-3 is an essential gene implicated in osmoregulation and requiring an intact permease transporter domain. Dev. Biol. 351: 242–253. 10.1016/j.ydbio.2010.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer R. J., 1997. Evolutionary changes of developmental mechanisms in the absence of cell lineage alterations during vulva formation in the Diplogastridae (Nematoda). Development 124: 243–251. [DOI] [PubMed] [Google Scholar]
- Sommer R. J., 2006. Pristionchus pacificus (August 14, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.102.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer R. J., Sternberg P. W., 1994. Changes of induction and competence during the evolution of vulva development in nematodes. Science 265: 114–118. 10.1126/science.8016644 [DOI] [PubMed] [Google Scholar]
- Sommer R. J., Sternberg P. W., 1995. Evolution of cell lineage and pattern formation in the vulval equivalence group of rhabditid nematodes. Dev. Biol. 167: 61–74. 10.1006/dbio.1995.1007 [DOI] [PubMed] [Google Scholar]
- Sommer R. J., Sternberg P. W., 1996. Apoptosis and change of competence limit the size of the vulva equivalence group in Pristionchus pacificus: a genetic analysis. Curr. Biol. 6: 52–59. 10.1016/S0960-9822(02)00421-9 [DOI] [PubMed] [Google Scholar]
- Srinivasan J., Pires-daSilva A., Gutierrez A., Zheng M., Jungblut B., et al. , 2001. Microevolutionary analysis of the nematode genus Pristionchus suggests a recent evolution of redundant developmental mechanisms during vulva formation. Evol. Dev. 3: 229–240. 10.1046/j.1525-142x.2001.003004229.x [DOI] [PubMed] [Google Scholar]
- Stanfield G. M., Villeneuve A. M., 2006. Regulation of sperm activation by SWM-1 is required for reproductive success of C. elegans males. Curr. Biol. 16: 252–263. 10.1016/j.cub.2005.12.041 [DOI] [PubMed] [Google Scholar]
- Stein L. D., Bao Z., Blasiar D., Blumenthalm T., Brent M. R., Chenm N., et al. 2003. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol 1: e45 10.1371/journal.pbio.0000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg P. W., 1988. Lateral inhibition during vulval induction in Caenorhabditis elegans. Nature 335: 551–554. 10.1038/335551a0 [DOI] [PubMed] [Google Scholar]
- Sternberg P. W., 2005. Vulval development (June, 25 2005), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.6.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg P. W., Horvitz H. R., 1981. Gonadal cell lineages of the nematode Panagrellus redivivus and implications for evolution by the modification of cell lineage. Dev. Biol. 88: 147–166. 10.1016/0012-1606(81)90226-8 [DOI] [PubMed] [Google Scholar]
- Sternberg P. W., Horvitz H. R., 1982. Postembryonic nongonadal cell lineages of the nematode Panagrellus redivivus: description and comparison with those of Caenorhabditis elegans. Dev. Biol. 93: 181–205. 10.1016/0012-1606(82)90251-2 [DOI] [PubMed] [Google Scholar]
- Sternberg P. W., Horvitz H. R., 1986. Pattern formation during vulval development in C. elegans. Cell 44: 761–772. 10.1016/0092-8674(86)90842-1 [DOI] [PubMed] [Google Scholar]
- Stoltzfus A., 2006. Mutationism and the dual causation of evolutionary change. Evol. Dev. 8: 304–317. 10.1111/j.1525-142X.2006.00101.x [DOI] [PubMed] [Google Scholar]
- Stothard P., Pilgrim D., 2006. Conspecific and interspecific interactions between the FEM-2 and the FEM-3 sex-determining proteins despite rapid sequence divergence. J. Mol. Evol. 62: 281–291. 10.1007/s00239-005-0084-5 [DOI] [PubMed] [Google Scholar]
- Stothard P., Hansen D., Pilgrim D., 2002. Evolution of the PP2C family in Caenorhabditis: rapid divergence of the sex-determining protein FEM-2. J. Mol. Evol. 54: 267–282. 10.1007/s0023901-0008-y [DOI] [PubMed] [Google Scholar]
- Streit A., Li W., Robertson B., Schein J., Kamal I., et al. , 1999. Homologs of the Caenorhabditis elegans masculinizing gene her-1 in C. briggsae and the filarial parasite Brugia malayi. Genetics 152: 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhaus W., 1976. Vergleichende Untersuchungen zur Phylogenie, Systematik, Ökologie, Biologie und Ethologie der Rhabditidae (Nematoda). Zoologica 43: 1–299. [Google Scholar]
- Sudhaus W., 2010. Preadaptive plateau in Rhabditida (Nematoda) allowed the repeated evolution of zooparasites, with an outlook on evolution of life cycles within Spiroascarida. Palaeodiversity 3: 117–130. [Google Scholar]
- Sudhaus W., 2011. Phylogenetic systematisation and catalogue of paraphyletic “Rhabditidae” (Secernentea, Nematoda). J. Nematode Morphol. Syst. 14: 113–178. [Google Scholar]
- Sudhaus W., Fitch D. H. A., 2001. Comparative studies on the phylogeny and systematics of the Rhabditidae (Nematoda). J. Nematol. 33: 1–70. [PMC free article] [PubMed] [Google Scholar]
- Sudhaus W., Fürst von Lieven A., 2003. A phylogenetic classification and catalogue of the Diplogastridae (Secernentea, Nematoda). J. Nematode Morphol. Syst. 6: 43–90. [Google Scholar]
- Sulston J., Dew M., Brenner S., 1975. Dopaminergic neurons in the nematode Caenorhabditis elegans. J. Comp. Neurol. 163: 215–226. 10.1002/cne.901630207 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R., 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156. 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Albertson D. G., Thomson J. N., 1980. The Caenorhabditis elegans male: postembryonic development of nongonadal structures. Dev. Biol. 78: 542–576. 10.1016/0012-1606(80)90352-8 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., Thomson J. N., 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Yandell M. D., Roy P. J., Krishna S., Savage-Dunn C., et al. , 1999. A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development 126: 241–250. [DOI] [PubMed] [Google Scholar]
- Tahseen Q., Nisa S., 2006. Embryology and gonad development in Oscheius shamimi sp. n. (Nematoda: Rhabditida). Nematology 8: 211–221. 10.1163/156854106777998665 [DOI] [Google Scholar]
- Tandonnet S., Farrell M. C., Koutsovoulos G. D., Blaxter M. L., Parihar M., et al. , 2018. Sex- and gamete-specific patterns of X chromosome segregation in a trioecious nematode. Curr. Biol. 28: 93–99.e3. 10.1016/j.cub.2017.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. J., Bilo K., Cross H. F., Archer J. P., Underwood A. P., 1999. 16S rDNA phylogeny and ultrastructural characterization of Wolbachia intracellular bacteria of the filarial nematodes Brugia malayi, B. pahangi, and Wuchereria bancrofti. Exp. Parasitol. 91: 356–361. 10.1006/expr.1998.4383 [DOI] [PubMed] [Google Scholar]
- Taylor M. J., Hoerauf A., Bockarie M., 2010. Lymphatic filariasis and onchocerciasis. Lancet 376: 1175–1185. 10.1016/S0140-6736(10)60586-7 [DOI] [PubMed] [Google Scholar]
- Thomas C. G., Li R., Smith H. E., Woodruff G. C., Oliver B., et al. , 2012a Simplification and desexualization of gene expression in self-fertile nematodes. Curr. Biol. 22: 2167–2172. 10.1016/j.cub.2012.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. G., Woodruff G. C., Haag E. S., 2012b Causes and consequences of the evolution of reproductive mode in Caenorhabditis nematodes. Trends Genet. 28: 213–220. 10.1016/j.tig.2012.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. G., Wang W., Jovelin R., Ghosh R., Lomasko T., et al. , 2015. Full-genome evolutionary histories of selfing, splitting, and selection in Caenorhabditis. Genome Res. 25: 667–678. 10.1101/gr.187237.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H., Schlager B., Xiao H., Sommer R. J., 2008. Wnt signaling induces vulva development in the nematode Pristionchus pacificus. Curr. Biol. 18: 142–146. 10.1016/j.cub.2007.12.048 [DOI] [PubMed] [Google Scholar]
- Ting J. J., Woodruff G. C., Leung G., Shin N. R., Cutter A. D., et al. , 2014. Intense sperm-mediated sexual conflict promotes reproductive isolation in Caenorhabditis nematodes. PLoS Biol. 12: e1001915 10.1371/journal.pbio.1001915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker A. S., Teng Y., Ferreira H. B., Emmons S. W., Chalfie M., 2003. The Caenorhabditis elegans spalt-like gene sem-4 restricts touch cell fate by repressing the selector Hox gene egl-5 and the effector gene mec-3. Development 130: 3831–3840. 10.1242/dev.00398 [DOI] [PubMed] [Google Scholar]
- Trent C., Purnell B., Gavinski S., Hageman J., Chamblin C., et al. , 1991. Sex-specific transcriptional regulation of the C. elegans sex-determining gene her-1. Mech. Dev. 34: 43–55. 10.1016/0925-4773(91)90090-S [DOI] [PubMed] [Google Scholar]
- True J. R., Haag E. S., 2001. Developmental system drift and flexibility in evolutionary trajectories. Evol. Dev. 3: 109–119. 10.1046/j.1525-142x.2001.003002109.x [DOI] [PubMed] [Google Scholar]
- Valfort A. C., Launay C., Sémon M., Delattre M., 2018. Evolution of mitotic spindle behavior during the first asymmetric embryonic division of nematodes. PLoS Biol. 16: e2005099 10.1371/journal.pbio.2005099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Megen H., van den Elsen S., Holterman M., Karssen G., Mooyman P., et al. , 2009. A phylogenetic tree of nematodes based on about 1200 full-length small subunit ribosomal DNA sequences. Nematology 11: 927–950. 10.1163/156854109X456862 [DOI] [Google Scholar]
- Vangestel S., Houthoofd W., Bert W., Borgonie G., 2008. The early embryonic development of the satellite organism Pristionchus pacificus: differences and similarities with Caenorhabditis elegans. Nematology 10: 301–312. 10.1163/156854108783900267 [DOI] [Google Scholar]
- Vargas-Velazquez A. M., Besnard F., Félix M. A., 2018. Necessity and contingency in developmental genetic screens: LIN-3, Wnt and semaphorin pathways in vulval induction of the nematode Oscheius tipulae. bioRxiv. 10.1101/383729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeij G. J., 2015. Forbidden phenotypes and the limits of evolution. Interface Focus 5: 20150028 10.1098/rsfs.2015.0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verster A. J., Ramani A. K., McKay S. J., Fraser A. G., 2014. Comparative RNAi screens in C. elegans and C. briggsae reveal the impact of developmental system drift on gene function. PLoS Genet. 10: e1004077 10.1371/journal.pgen.1004077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle A., Callemeyn-Torre N., Gimond C., Poullet N., Gray J. C., et al. , 2016. Convergent evolution of sperm gigantism and the developmental origins of sperm size variability in Caenorhabditis nematodes. Evolution 70: 2485–2503. 10.1111/evo.13043 [DOI] [PubMed] [Google Scholar]
- Voronov D. A., 1999. The embryonic development of Pontonema vulgare (Enoplida: Oncholaimidae) with a discussion of nematode phylogeny. Russ. J. Nematol. 7: 105–114. [Google Scholar]
- Wallenfang M. R., Seydoux G., 2000. Polarization of the anterior-posterior axis of C. elegans is a microtubule-directed process. Nature 408: 89–92. 10.1038/35040562 [DOI] [PubMed] [Google Scholar]
- Walrond J. P., Kass I. S., Stretton A. O., Donmoyer J. E., 1985. Identification of excitatory and inhibitory motoneurons in the nematode Ascaris by electrophysiological techniques. J. Neurosci. 5: 1–8. 10.1523/JNEUROSCI.05-01-00001.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Kimble J., 2001. The TRA-1 transcription factor binds TRA-2 to regulate sexual fates in Caenorhabditis elegans. EMBO J. 20: 1363–1372. 10.1093/emboj/20.6.1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chamberlin H., 2002. Multiple regulatory changes contribute to the evolution of the Caenorhabditis lin-48 ovo gene. Genes Dev. 16: 2345–2349. 10.1101/gad.996302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chamberlin H. M., 2004. Evolutionary innovation of the excretory system in Caenorhabditis elegans. Nat. Genet. 36: 231–232. 10.1038/ng1301 [DOI] [PubMed] [Google Scholar]
- Wang X., Sommer R. J., 2011. Antagonism of LIN-17/Frizzled and LIN-18/Ryk in nematode vulva induction reveals evolutionary alterations in core developmental pathways. PLoS Biol. 9: e1001110 10.1371/journal.pbio.1001110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Greenberg J. F., Chamberlin H. M., 2004. Evolution of regulatory elements producing a conserved gene expression pattern in Caenorhabditis. Evol. Dev. 6: 237–245. 10.1111/j.1525-142X.2004.04029.x [DOI] [PubMed] [Google Scholar]
- Weeks S. C., Benvenuto C., Reed S. K., 2006. When males and hermaphrodites coexist: a review of androdioecy in animals. Integr. Comp. Biol. 46: 449–464. 10.1093/icb/icj048 [DOI] [PubMed] [Google Scholar]
- Wei Q., Shen Y., Chen X., Shifman Y., Ellis R. E., 2014a Rapid creation of forward-genetics tools for C. briggsae using TALENs: lessons for nonmodel organisms. Mol. Biol. Evol. 31: 468–473. 10.1093/molbev/mst213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q., Zhao Y., Guo Y., Stomel J., Stires R., et al. , 2014b Co-option of alternate sperm activation programs in the evolution of self-fertile nematodes. Nat. Commun. 5: 5888 10.1038/ncomms6888 [DOI] [PubMed] [Google Scholar]
- Weiss K., Fullerton S., 2000. Phenogenetic drift and the evolution of genotype-phenotype relationships. Theor. Popul. Biol. 57: 187–195. 10.1006/tpbi.2000.1460 [DOI] [PubMed] [Google Scholar]
- White J. G., Southgate E., Thomson J. N., Brenner S., 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314: 1–340. 10.1098/rstb.1986.0056 [DOI] [PubMed] [Google Scholar]
- Wiegner O., Schierenberg E., 1998. Specification of gut cell fate differs significantly between the nematodes Acrobeloides nanus and Caenorhabditis elegans. Dev. Biol. 204: 3–14. 10.1006/dbio.1998.9054 [DOI] [PubMed] [Google Scholar]
- Wiegner O., Schierenberg E., 1999. Regulative development in a nematode embryo: a hierarchy of cell fate transformations. Dev. Biol. 215: 1–12. 10.1006/dbio.1999.9423 [DOI] [PubMed] [Google Scholar]
- Winston W. M., Sutherlin M., Wright A. J., Feinberg E. H., Hunter C. P., 2007. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc. Natl. Acad. Sci. USA 104: 10565–10570. 10.1073/pnas.0611282104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter A. D., Keskiaho K., Kukkola L., McCormack G., Félix M. A., et al. , 2007. Differences in collagen prolyl 4-hydroxylase assembly between two Caenorhabditis nematode species despite high amino acid sequence identity of the enzyme subunits. Matrix Biol. 26: 382–395. 10.1016/j.matbio.2007.01.008 [DOI] [PubMed] [Google Scholar]
- Witte H., Moreno E., Rödelsperger C., Kim J., Kim J. S., et al. , 2015. Gene inactivation using the CRISPR/Cas9 system in the nematode Pristionchus pacificus. Dev. Genes Evol. 225: 55–62. 10.1007/s00427-014-0486-8 [DOI] [PubMed] [Google Scholar]
- Wong Y. F., Sheng Q., Chung J. W., Chan J. K., Chow K. L., 2010. mab-31 and the TGF-beta pathway act in the ray lineage to pattern C. elegans male sensory rays. BMC Dev. Biol. 10: 82 10.1186/1471-213X-10-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A. J., Lo T. W., Zeitler B., Pickle C. S., Ralston E. J., et al. , 2011. Targeted genome editing across species using ZFNs and TALENs. Science 333: 307 10.1126/science.1207773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff G. C., Eke O., Baird S. E., Félix M. A., Haag E. S., 2010. Insights into species divergence and the evolution of hermaphroditism from fertile interspecies hybrids of Caenorhabditis nematodes. Genetics 186: 997–1012. 10.1534/genetics.110.120550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W., Ross J. M., Zarkower D., 2000. Mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development 127: 4469–4480. [DOI] [PubMed] [Google Scholar]
- Yin D., Schwarz E. M., Thomas C. G., Felde R. L., Korf I. F., et al. , 2018. Rapid genome shrinkage in a self-fertile nematode reveals sperm competition proteins. Science 359: 55–61. 10.1126/science.aao0827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalts H., Yanai I., 2017. Developmental constraints shape the evolution of the nematode mid-developmental transition. Nature Ecol. Evol. 1: 113 10.1038/s41559-017-0113 [DOI] [PubMed] [Google Scholar]
- Zanetti S., Puoti A., 2013. Sex determination in the Caenorhabditis elegans germline. Adv. Exp. Med. Biol. 757: 41–69. 10.1007/978-1-4614-4015-4_3 [DOI] [PubMed] [Google Scholar]
- Zarkower D., 2001. Establishing sexual dimorphism: conservation amidst diversity? Nat. Rev. Genet. 2: 175–185. 10.1038/35056032 [DOI] [PubMed] [Google Scholar]
- Zarkower, D., 2006 Somatic sex determination (February 10, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.84.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauner H., Sommer R. J., 2007. Evolution of robustness in the signaling network of Pristionchus vulva development. Proc. Natl. Acad. Sci. USA 104: 10086–10091. 10.1073/pnas.0610799104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdraljevic S., Strand C., Seidel H. S., Cook D. E., Doench J. G., et al. , 2017. Natural variation in a single amino acid substitution underlies physiological responses to topoisomerase II poisons. PLoS Genet. 13: e1006891 10.1371/journal.pgen.1006891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Smolen G. A., Palmer R., Christoforou A., van den Heuvel S., et al. , 2004. SUMO modification is required for in vivo Hox gene regulation by the Caenorhabditis elegans polycomb group protein SOP-2. Nat. Genet. 36: 507–511. 10.1038/ng1336 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Emmons S. W., 1995. Specification of sense-organ identity by a Caenorhabditis elegans Pax-6 homologue. Nature 377: 55–59. 10.1038/377055a0 [DOI] [PubMed] [Google Scholar]
- Zhao C., Emmons S. W., 1995. A transcription factor controlling development of peripheral sense organs in C. elegans. Nature 373: 74–78. 10.1038/373074a0 [DOI] [PubMed] [Google Scholar]
- Zhao Z., Boyle T. J., Bao Z., Murray J. I., Mericle B., et al. , 2008. Comparative analysis of embryonic cell lineage between Caenorhabditis briggsae and Caenorhabditis elegans. Dev. Biol. 314: 93–99. 10.1016/j.ydbio.2007.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Flibotte S., Murray J. I., Blick D., Boyle T. J., et al. , 2010. New tools for investigating the comparative biology of Caenorhabditis briggsae and C. elegans. Genetics 184: 853–863. 10.1534/genetics.109.110270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Messerschmidt D., Jungblut B., Sommer R. J., 2005. Conservation and diversification of Wnt signaling function during the evolution of nematode vulva development. Nat. Genet. 37: 300–304. 10.1038/ng1512 [DOI] [PubMed] [Google Scholar]