Significance
Fitness landscapes map the relationship between genotype and phenotype, and are a core tool for predicting evolutionary processes from the emergence of resistant pathogens to cancer. The topography of fitness landscapes is determined by the environment. However, populations can also dynamically modify their environment, for instance by releasing metabolites to it, and thus they may potentially deform their own adaptive landscape. Using a combination of genome-scale metabolic simulations and experiments with Escherichia coli strains from the Lenski laboratory Long-Term Evolution Experiment, we systematically and quantitatively characterize the deformability of an empirical fitness landscape. We show that fitness landscapes retain their power to forecast evolution over short mutational distances but environment building may attenuate this capacity over longer adaptive trajectories.
Keywords: fitness landscapes, eco-evolutionary feedbacks, ecologically mediated gene interactions, gene × environment × gene interactions, noncommutative epistasis
Abstract
A fitness landscape is a map between the genotype and its reproductive success in a given environment. The topography of fitness landscapes largely governs adaptive dynamics, constraining evolutionary trajectories and the predictability of evolution. Theory suggests that this topography can be deformed by mutations that produce substantial changes to the environment. Despite its importance, the deformability of fitness landscapes has not been systematically studied beyond abstract models, and little is known about its reach and consequences in empirical systems. Here we have systematically characterized the deformability of the genome-wide metabolic fitness landscape of the bacterium Escherichia coli. Deformability is quantified by the noncommutativity of epistatic interactions, which we experimentally demonstrate in mutant strains on the path to an evolutionary innovation. Our analysis shows that the deformation of fitness landscapes by metabolic mutations rarely affects evolutionary trajectories in the short range. However, mutations with large environmental effects produce long-range landscape deformations in distant regions of the genotype space that affect the fitness of later descendants. Our results therefore suggest that, even in situations in which mutations have strong environmental effects, fitness landscapes may retain their power to forecast evolution over small mutational distances despite the potential attenuation of that power over longer evolutionary trajectories. Our methods and results provide an avenue for integrating adaptive and eco-evolutionary dynamics with complex genetics and genomics.
When a new genotype appears in a population its reproductive success is largely governed by the environment. Although the environment is often thought of as an external driver of natural selection, it can also be shaped by the evolving population itself, for instance through its metabolic activity or through interactions with the abiotic habitat or other species (1–3). These population-driven environmental changes can in turn modify the fitness effects of future mutations, closing in an eco-evolutionary feedback loop (4). Eco-evolutionary feedbacks are well documented in natural (5) and experimental (6) populations, and at all scales of biological organization: from the cellular scale [e.g., in the evolution of cancer (7) and microbial populations (8)] to the organismal scale in animal (9) and plant evolution (10). Given the growing evidence that evolutionary and ecological processes, including niche construction, occur on similar timescales, there is a critical need to understand the genomic bases of these eco-evolutionary feedbacks (11).
The “map” between each genotype and its adaptive value in a given environment is known as the “fitness landscape” (12). Because populations actively modify their environment, new mutations can, in principle, have environmental as well as fitness effects. Thus, evolving populations may dynamically reshape (“deform”) the fitness landscapes on which they are adapting (13, 14). Although they are often used only metaphorically to depict or visualize adaptation, fitness landscapes are a major determinant of evolution. In particular, the topography of a fitness landscape (i.e., the location of fitness peaks and valleys and their connectivity) plays a pivotal role, as it governs the accessibility of evolutionary trajectories (15–17), the role of population structure on evolution (18), the degree of evolutionary convergence among populations (19), the expected role of drift, selection, and sex in the evolutionary process (20, 21), the discovery of evolutionary innovations (22), and the predictability of evolution (23), a subject of growing importance for the management of pathogens and cancer treatment (24–28). Given the fundamental role that fitness landscapes play in adaptation, if populations do indeed change the topography of their fitness landscapes as they evolve, it is imperative to understand precisely how. Do mutations that alter the environment generally also alter the fitness of all subsequent mutations or only a subset of them? If the latter, where are those deformations localized in the genotype space, and how strong are they? All these questions remain open, as the deformability (or “rubberness”) of fitness landscapes has never been systematically studied in empirical systems at the genomic scale.
Substantial experimental evidence suggests that microbial fitness landscapes are likely to exhibit deformability (29–33), making microbes an ideal system for addressing this issue. Microbial metabolism leads to large-scale environmental construction through the uptake and release of metabolites (30, 32). Which nutrients are taken up, which byproducts are released, and in what amounts, are all governed by the structure of the metabolic network and therefore by the genotype (34, 35). As a result, new mutations that change the metabolic network can also change the patterns of metabolic uptake and secretion, altering the environment and potentially also altering the fitness of future mutations (32).
Microbial physiology and growth can be explicitly simulated using genome-scale metabolic models (36–38). Due to their excellent predictive capabilities (36) and utility for easily and rapidly screening millions of genotypes, these models have been successfully used to systematically explore the genotype space (39). Recent advances in dynamic metabolic modeling make it possible to explicitly simulate the growth of microbial communities and their environmental feedbacks with evolution (40, 41), making genome-wide dynamic metabolic modeling of microbial genotypes a promising method to examine the deformability of fitness landscapes (Fig. 1A).
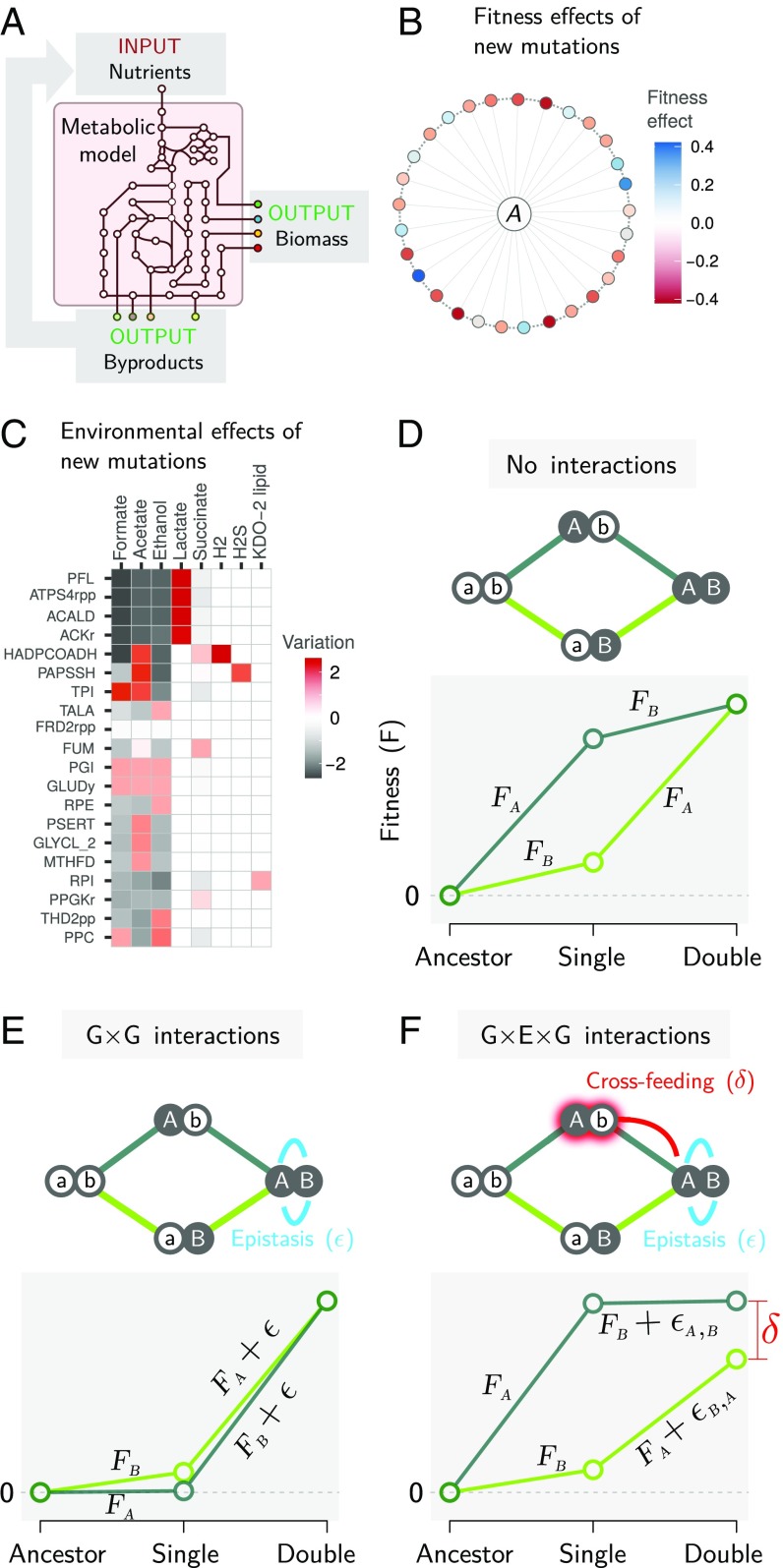
Fig. 1.
Measuring deformability in the E. coli metabolic fitness landscape. (A) Schematic depiction of dFBA simulations. Given an input in the form of nutrients, metabolic fluxes through an explicit and empirically curated metabolic model are optimized to maximize the biomass growth yield. The optimal metabolic fluxes produce metabolic byproducts that are released to the external environment, becoming part of future inputs. (B) A subset of genotypes differing from our E. coli metabolic model by a single mutation (an added or deleted reaction), colored according to their effect on fitness in competition with the ancestor (A). (C) Environmental effects of a subset of mutants expressed as the variation in the profile of secreted metabolites compared with the ancestral E. coli genotype (computed as log-modulus transformed difference in the amount of a given secreted molecule; Materials and Methods). Mutant labels are given in Biochemical Genetic and Genomic (BiGG) database notation. (D) Two loci fitness landscapes in the absence of gene–gene interactions in which the fitness effect of each mutation is the same in all genetic backgrounds. The fitness of each genotype was calculated in direct competition with its immediate ancestor. Mutations A and B correspond to the addition of GLYCL_2 (glycine cleavage system) and AIRCr (phosphoribosylaminoimidazole carboxylase), respectively. (E) Two-loci fitness landscapes with gene–gene interactions giving rise to epistasis (ε). Mutations A and B were SO3R (sulfite reductase) and PAPSSH (phosphoadenylylsulfatase), respectively, simulated in a constant environment. (F) Two-loci fitness landscapes in which one of the mutants transforms the environment, leading to cross-feeding toward the double mutant. Mutations A and B correspond to the addition of PAPSSH and HADPCOADH (3-hydroxyadipyl-CoA dehydrogenase). In addition to regular epistasis, this led to a noncommutative epistatic shift (δ = εA,B − εB,A).
Here, we first use metabolic modeling to show that the environmental effect of new mutations can make genetic interactions (or “epistasis”) noncommutative or dependent on the order in which mutations occur. We then use evolved strains from one of the populations in Lenski and coworkers’ (42, 43) Escherichia coli Long-Term Evolution Experiment (LTEE) to experimentally demonstrate the presence of noncommutative epistasis and quantitatively validate the predictive capabilities of our model. We then scale up our study to include tens of millions of genotypes from the metabolic genotype space. By systematically screening the in silico metabolic fitness landscape of E. coli, we are able to offer a precise view of how deformability by eco-evolutionary feedbacks plays out over short and long mutational distances.
Results
Noncommutative Epistasis Characterizes Fitness Landscape Deformability.
To investigate the effect of metabolic secretions on the fitness landscape, we used dynamic flux balance analysis (dFBA) to determine the distribution of fitness and environmental effects of new mutations in the local mutational neighborhood of a recently curated, genome-scale metabolic model of E. coli (36). Our screen included all possible single-addition and -deletion mutants (Materials and Methods), whose growth was simulated on anaerobic glucose medium until saturation was reached. Of all nonessential mutations, 147 (3.3%) affected growth rate either positively or negatively (Fig. 1B). All these mutations also altered the chemical composition of the environment (Materials and Methods; also see Fig. 1C for a representative subset and SI Appendix, Fig. S1 for the full set), and the magnitude of the environmental and fitness effects were strongly correlated (Pearson’s ρ = 0.61, P < 10−6) (SI Appendix, Fig. S2). This suggests that the extracellular environment will change as new mutations fix in the population, which could in turn alter the fitness effects of new mutations, thus deforming the fitness landscape.
We explored the extent to which this fitness landscape may be deformed by the effect of metabolic secretions using a dataset that consisted of ∼107 single and double mutants, representing the entire second-order metabolic mutational neighborhood of E. coli. The fitness of each mutant (M) was determined in competition with its immediate ancestor (A) as FM(A) = log([X′M/XM]/[X′A/XA]) (42, 44),where XA and XM represent the initial densities of ancestor and mutant and X′A and X′M represent their final respective densities after 10 h of competition (Materials and Methods). All competitions were performed at an initial mutant frequency of 0.01. Using this measure, the fitness effects of two mutations are expected to combine additively when they act independently (Fig. 1D). As shown in Fig. 1E, when two mutations without an environmental effect interact with one another, epistasis (ε) will cause the fitness of the double mutant to deviate from additivity. This is the usual definition of epistasis in the literature, which is invariant as to the order in which mutations occur (16). In contrast, when at least one of the single mutants has an environmental effect, the double mutant experiences a different extracellular environment depending on which of the two single mutants was its immediate ancestor. For example, a double mutant could cross-feed on the byproducts of one of its possible single-mutant ancestors but not on the byproducts of the other (Fig. 1F). The result is a gene-by-environment-by-gene (G × E × G) interaction in which the magnitude of epistasis may depend on the order in which mutations occur. In other words, epistasis becomes noncommutative. The value of that noncommutative fitness shift (δ) characterizes the deformation of a two-step mutational trajectory (Fig. 1F).
Noncommutative epistasis and fitness intransitivity are closely related but not identical concepts (SI Appendix, Fig. S3) (45). In its simplest, qualitative formulation, “intransitivity” refers to situations in which the fitness of three mutants (A, B, C) in pairwise competition are nonhierarchical (i.e., A invades B, B invades C, and C invades A). A less stringent, quantitative definition of intransitivity has been applied when the relative fitness between a mutant and its ancestor cannot be predicted by the sum of cumulative fitness gains along a mutational trajectory (45). This definition is close to but distinct from the concept of noncommutativity (Fig. 1). Noncommutativity quantifies the difference in cumulative fitness gains along two different mutational trajectories without regard for the fitness of the final point of the trajectory in competition with the original ancestor (SI Appendix, Fig. S3). Interestingly, noncommutativity and intransitivity are mathematically related to one another but must be estimated using independent experiments (SI Appendix, Fig. S3). Genotypes along an evolutionary trajectory usually compete with their immediate mutational ancestors rather than with their original ancestral strain (29). Therefore, noncommutativity is a suitable metric for characterizing fitness landscape deformability, while intransitivity can be more suitable for ecological questions, such as the possibility of coexistence of different genotypes (46, 47).
Deformability in the Path to an Evolutionary Innovation in E. coli.
To experimentally validate and assess the potential relevance of noncommutative epistasis as a metric of fitness landscape deformability, we studied two mutations on the path to the evolutionary innovation of strong aerobic growth on citrate (Cit++) in the Ara-3 population of the LTEE (43). The two principal mutations underlying this phenotype are known to have profound ecological consequences, suggesting that noncommutative epistasis may be present (Fig. 2A). The first mutation is a tandem amplification overlapping the citrate fermentation operon, cit, which occurred after 31,000 generations. This amplification caused aerobic expression of the CitT transporter, producing a weak citrate growth phenotype (Cit+) (48). CitT is an antiporter that imports citrate, present in large amounts in the LTEE DM25 growth medium, while exporting intracellular C4-dicarboxylate TCA intermediates, e.g., succinate and malate (34), thereby increasing their concentration in the extracellular environment. A subsequent mutation causes high-level, constitutive expression of DctA, a proton-driven dicarboxylic acid transporter. This mutation refines the Cit+ trait to Cit++ by allowing recovery of the C4-dicarboxylates released into the medium by both the progenitor and the double mutant itself during growth on citrate (Fig. 2A) (49). We reasoned that these mutations together enable the exploitation of environments built by progenitor strains, producing a stronger increase in fitness than expected in the absence of environmental construction (Fig. 2B). In contrast, had the DctA mutation occurred before the CitT-activating duplication, it would have conferred no fitness benefit and would not have produced any changes in the environment relative to the ancestor (Fig. 2B).
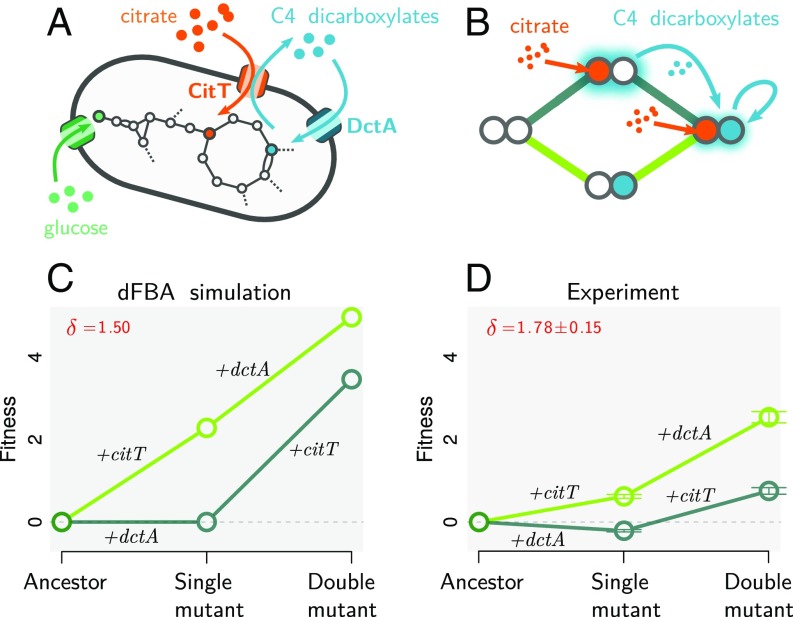
Fig. 2.
Noncommutative epistasis in the evolution of aerobic citrate use in E. coli. (A) Function of the two transporters involved in the innovation of strong aerobic growth on citrate (Cit++) in E. coli. CitT is an antiporter that exchanges extracellular citrate for internal C4-dicarboxylates (e.g., succinate, fumarate, and malate). DctA is a carboxylic acid transporter that imports C4-dicarboxylates from the extracellular space into the cytoplasm. (B) The two possible mutational trajectories leading to the Cit++ trait. If the mutation leading to expression of citT (+citT) occurs first, it will transform the environment leading to cross-feeding toward the double mutant. This should not occur if the dctA overexpression mutation (+dctA) occurs first. (C and D) Simulated (C) and experimentally measured (D) fitness landscapes in the DM25 medium used in the LTEE (Materials and Methods). Experimentally obtained values are reported as mean ± SEM (n = 10).
We tested this prediction by performing competitive fitness assays with different combinations of a spontaneous Cit– mutant and dctA– knockout strains derived from ZDB89, a 35,000-generation Cit++ clone that possesses both the DctA-activating and CitT-activating mutations (Materials and Methods). Competitions were carried out with equal volumes of each combination of competitors, and relative fitness was determined using colony counts obtained after 0 and 24 h of growth (42). In parallel, we used our dFBA model to simulate these competitions, relying solely on known parameters from the experiments and on published parameters pertaining to the physiology of E. coli [(Materials and Methods) 41, 50]. Confirming our expectations, the dFBA model predicts strong noncommutative epistasis (δ = 1.50) (Fig. 2C). This is confirmed by the experimental results (δ = 1.78 ± 0.15) (Fig. 2D). The agreement between the empirically calibrated computational model and the experiments is not only qualitative but also is quantitative: With no fitting parameters, dFBA is predictive of the outcome of the experimental pairwise competitions, explaining 52% of the variance in colony counts from all experiments (n = 120) (SI Appendix, Fig. S4).
Short-Range Deformability in E. coli Is Weak and Rare.
Although the above examples demonstrate the potential presence of fitness landscape deformability, its pervasiveness in empirical fitness landscapes remains unclear. To address this question, we screened the entire first- and second-order mutational neighborhood of E. coli using our computational model (Fig. 3A). In Fig. 3B we represent all pairs of mutations that exhibit deformability as nodes in a network that are connected if their noncommutative fitness shift (δ) is larger than 1% of the fitness effects (FMAX; also see SI Appendix, Fig. S5). These represent only a small subset (203/3,343, or 6.1%) of all epistatic interactions, which for the most part are not altered by the environmental effects of mutations.
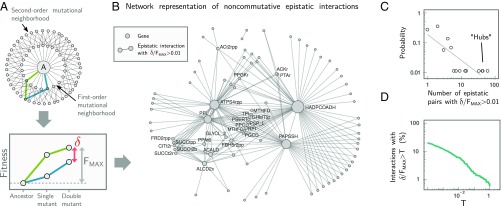
Fig. 3.
Short-range deformability in E. coli is rare, weak, and directional. (A) Systematic exploration of the second-order mutational neighborhood of E. coli. We exhaustively simulated every possible mutational trajectory starting from the ancestral (A) metabolism and ending in each double mutant. Noncommutative epistasis (δ) was measured for each pair of mutants and was normalized by FMAX, i.e., the maximal cumulative fitness effect of the double mutant: FMAX = max[ |Fi(A) + Fij(i)|, |Fj(A) + Fij(j)| ], where, e.g., Fx(y) denotes the fitness of mutant x when invading its immediate ancestor y at low frequency (Materials and Methods). (B) Network representation of all noncommutative epistatic pairs. Nodes represent mutations, and two nodes are joined by an edge if δ/FMAX >0.01 for that pair. Node labels (BiGG database notation) are shown for hubs (mutations with more than four noncommutative interactions). (C) Distribution of deformability for each gene in the network, measured as the number of other genes with which it has a noncommutative epistatic interaction. (D) Strength of all noncommutative epistatic interactions, i.e., the percentage of epistatic pairs with δ/FMAX > T as a function of T.
Noncommutative interactions also tend to be unevenly distributed: Most mutations do not deform the fitness of any other mutation, and only 15 (0.3%) of them deform the fitness of five or more other mutations (Fig. 3 B and C). These few highly connected hubs on the network tend to be the mutations with the strongest environmental effects (Pearson’s ρ = 0.79, P < 10−6) (SI Appendix, Fig. S5). Noncommutative epistasis also tends to be small in magnitude (Fig. 3D); only 1.6% (55/3,343) of epistatic pairs have a noncommutative epistatic shift larger than 10% of the total fitness increase (δ/FMAX > 0.1) (Fig. 3D). This reveals that the deformability of the local mutational neighborhood of the E. coli metabolic landscape is generally weak, rare, and highly anisotropic (i.e., nonhomogeneous), with deformations limited to localized directions in genotype space.
Long-Range Deformability of the E. coli Metabolic Fitness Landscape.
The low deformability of the local mutational neighborhood could be explained by the strong genetic similarity between the mutants and the ancestral genotype: Genotypically close descendants will rarely be able to use metabolites that are discarded by their immediate ancestors. By the same logic, one may predict that over longer mutational distances metabolic differences might accumulate that enable the use of extracellular metabolites that are left as a “legacy” by previous mutations. Thus, we hypothesize that changes to the extracellular environment produced by a given mutation will primarily deform the fitness landscape at distant positions on the genotype space.
To test this hypothesis, we set out to introduce a mutation with a strong environmental effect and measure the deformation it causes at different distances in the genotype space. We chose the ACKr (acetate kinase) mutation (the deletion of the acetate kinase gene), which as shown in Fig. 1C modifies the environment by releasing large amounts of lactate at the expense of lower secretions of formate, acetate, and ethanol. To quantify the deformation introduced by this mutation, we compared the fitness of thousands of genotypes at increasing mutational distances from the ancestor in competition with either the ancestor E. coli model (A) or the ACKr mutant (M) (Fig. 4A). The deformation introduced by M at genotype G is thus quantified by the parameter ΔFitness = |FG(M) − FG(A) − FM(A)| (Fig. 4B). Consistent with our hypothesis, and as shown in Fig. 4 B and C, we found that the fitness landscape deformation ΔFitness introduced by the ACKr mutation is negligible at short genotypic distances from it (e.g., 16 mutations or less), but it becomes stronger at longer distances. Fifteen other mutants in addition to ACKr were also tested, with similar results (SI Appendix, Fig. S7). Furthermore, by comparing the growth rate of thousands of genotypes in the environments constructed by A and M (noted by EA and EM, respectively), we found that increasingly distant genotypes become increasingly sensitive to the differences between the two environments (Fig. 4D and SI Appendix, Fig. S8). This provides an explanation for the observed pattern of fitness landscape deformation as a function of genotypic distance.
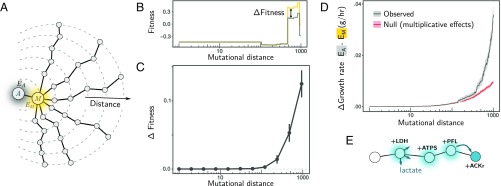
Fig. 4.
Long-range deformability of the E. coli metabolic fitness landscape. (A) We performed random walks (length = 1,000 mutations) in genotype space starting from an E. coli ancestor (A; gray) and first passing through a mutant (M; orange) with large environmental effect. (B and C) Fitness of mutants along these random walks was measured in competition with A (gray) in the environment it generates (EA), as well as in competition with M (orange) in the environment it generates (EM). In B we show the result for a single example of a random walk. Note that fitness in competition with M is shifted by the difference in fitness between M and A so all observed differences in fitness are due to deformation (for any genotype G, ΔFitness = |FG(A) − FG(M) − FM(A) |). (C) Average ΔFitness at increasing mutational distances from A in over n = 100 random walks (error bars represent SEM; n = 100). (D) Average difference (absolute value) in growth rate between environments EM and EA (in grams of dry cell weight × h−1) at varying genotype distances (gray line; shading represents SEM; n = 1,000). In red, we show the predicted difference in growth rates for a null model that assumes independent effects of mutations (Materials and Methods). (E) An example of an adaptive trajectory showing complex genetic interactions in a long-range deformation. The addition of LDH leads to the release of lactate as a by-product. Three additional mutations, ACKr, ATPS, and PFL, are required together for lactate to be used by a descendant genotype.
What are the genetic mechanisms underlying the long-range environmental effects of new mutations on growth rate? One possibility could be an increased probability of sampling mutations that produce a difference in growth rate between EA and EM. Alternatively, this effect could be caused by genetic interactions between two or more mutations that allow distant genotypes to use differently the resources secreted by A and M. To discriminate between these two possibilities, we compared the observed difference in growth rates (Fig. 4D, gray line) with the difference expected if mutations do not interact (Fig. 4D, red line) (Materials and Methods). As shown in Fig. 4D, the null model that only incorporates increased sampling of mutations at growing mutational distances (while assuming no interactions) vastly underestimates the observed growth difference between EA and EM and thus is insufficient to explain our results. This suggests that, although both mechanisms are present, interactions between mutations dominate the deformation of the fitness landscape at large mutational distances (see also SI Appendix, Fig. S8).
To mechanistically illustrate the role of complex genetic interactions in long-range landscape deformation, in Fig. 4E we show an adaptive trajectory in which a first mutation (lactate dehydrogenase; LDH) causes the release of lactate to the extracellular space. A complex metabolic innovation involving several reaction additions [ATP synthase (ATPS), pyruvate formate lyase (PFL), and ACKr] (51) is subsequently required to confer the ability to metabolize this lactate (Fig. 4E; see also SI Appendix, Fig. S8). Notably, lactate becomes metabolized only by the final genotype, which contains all three required mutations.
Discussion
Darwin (52) was perhaps the first to recognize that the environment experienced by an evolving population can also be shaped by the population itself. Long neglected, this concept was revived by Lewontin (1, 53), and has gained added momentum in recent years as the important role played by eco-evolutionary feedbacks in both ecology and evolution has become better appreciated (3, 4, 11). Due to technical limitations, experimental studies of eco-evolutionary feedbacks and the adaptive dynamics models that seek to explain them often lack explicit, genome-wide representations of the adaptive landscape, in particular with regard to complex traits and gene–gene interactions (11). The exact state of the environment, which is intrinsically complex and multidimensional, is also rarely measured experimentally or explicitly included in eco-evolutionary models. In return, genome-wide genotype fitness maps have largely ignored the effects of eco-evolutionary feedbacks, despite early abstract models of species coevolution, which introduced the idea of fitness landscape deformability (also referred to as “rubberness,” refs. 13 and 54), and the many examples of their importance in coevolutionary arms races and other forms of coevolution (55). This is particularly important in light of the argument, made by many authors, that the deformability of fitness landscapes (or its consequences, in the form of frequency-dependent selection) would erode their practical and conceptual utility (56–58).
Our work empirically addresses this latter argument. Encouragingly, our results show that fitness landscapes may retain their local properties in the presence of mutations that significantly alter the environment. By systematically mapping an empirical fitness landscape, we have found that ignoring deformability and assuming a rigid landscape is a good approximation over short genotypic distances. This is because closely related genotypes are unlikely to differ from one another in their physiological response to the built environment. In contrast, over longer mutational distances, fitness landscapes are likely to be affected by environmental construction, an effect that is shaped by complex genetic interactions. This suggests an ecologically mediated mechanism by which historical contingency may shape downstream evolution even in clonal populations. In summary, our work suggests that, depending on the scale at which they are examined, fitness landscapes can either behave as a fixed externally determined topography on which adaptation proceeds, or become a dynamic property of the populations adapting on them (57, 58).
One limitation of our study is the inability of our model to predict changes in the sign of the fitness effect of a new mutation. This is a common limitation of most FBA-based models (but see refs. 59 and 60), as they do not consider the potential costs of adding a new biochemical reaction, or of maintaining a flux through it. These costs can arise in microbial cells either through the cost of increasing genome size (61) or through the cost of expressing the enzymes required for the new reaction. Given the absence of such costs in our FBA model, a deletion can never provide any advantage, and an addition will never be detrimental. Costs have already been incorporated in nondynamic FBA models (59, 60), allowing the prediction of phenomena such as overflow metabolism. One can certainly imagine situations in which an addition that is detrimental due to its maintenance cost could become beneficial in the presence of the metabolic byproducts of its ancestor, leading to an ecologically mediated inversion of fitness effect (or “sign-δ”). Incorporating costs into a dynamic genome-scale modeling framework represents a promising future direction.
The idea that under frequency-dependent selection fitness landscapes change as populations move on them has been conceptually discussed and studied within the theoretical framework of adaptive dynamics (13, 14, 62). A solution to the problem of fitness landscape deformability was found in the formulation of the invasion fitness landscape, i.e., the map between the relative fitness S(x,y) of an invader with phenotype y against a resident phenotype x (62). In principle our results and methods might allow one to map an empirical invasion fitness landscape, at least locally. However, one would need to identify a scalar phenotype that can be mapped to the invasion success against a resident genotype in the environment this resident constructs. Under what conditions this is possible is an open question that lies beyond the scope of this study, but it poses an interesting future challenge.
In line with this discussion, our results indicate that simulating cellular adaptive dynamics with an explicit and biologically realistic genome-wide representation of the genotype–phenotype map is within reach. Such an approach will shed light into the role played by dynamic niche construction in cellular evolution. We believe that it will also create multiple opportunities to incorporate genomics into the study of eco-evolutionary dynamics and thus reveal the genetic, biochemical, and environmental constraints that simultaneously govern the ecology and evolution of cellular populations.
Materials and Methods
Detailed materials and methods regarding reconstruction of the prokaryotic genotype space, in silico simulations, fitness, environmental effects, and deformability measurements and experiments and simulations related to the LTEE can be found in SI Appendix. All data in this paper have been deposited in a public repository and can be accessed at https://github.com/vilacelestin/Bajicetal2018.
Supplementary Material
Acknowledgments
We thank Gunter Wagner, Steve Stearns, David Post, and members of the A.S. laboratory for helpful discussions and feedback on the manuscript; Daniel Segrè and Ilija Dukovski for helpful advice during the early implementation of the software COMETS [Computation of Microbial Ecosystems in Time and Space (COMETS)]; Maia Rowles, Kiyana Weatherspoon, and Brooke Sommerfeld for assistance in the construction of Ara-3–derived strains; Richard Lenski for use of LTEE strains and materials; Jonathan Fritzemeier and Martin Lercher for assistance with model curation; and two anonymous reviewers for their critical comments, which significantly improved the manuscript. This work was partially funded by Young Investigator Award RGY0077/2016 from the Human Frontier Science Program (to A.S.) and John Templeton Foundation Foundational Questions in Evolutionary Biology Grant FQEB RFP-12-13 (to Z.D.B.). The LTEE is supported in part by National Science Foundation Grant DEB-1019989.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All data in this paper have been deposited in GitHub, https://github.com/vilacelestin/Bajicetal2018.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808485115/-/DCSupplemental.
References
- 1.Lewontin RC. The organism as the subject and object of evolution. Scientia. 1983;77:65. [Google Scholar]
- 2.John Odling-Smee F, Laland KN, Feldman MW. Niche Construction: The Neglected Process in Evolution (MPB-37) Princeton Univ Press; Princeton: 2013. [Google Scholar]
- 3.Laland K, et al. Does evolutionary theory need a rethink? Nature. 2014;514:161–164. doi: 10.1038/514161a. [DOI] [PubMed] [Google Scholar]
- 4.Post DM, Palkovacs EP. Eco-evolutionary feedbacks in community and ecosystem ecology: Interactions between the ecological theatre and the evolutionary play. Philos Trans R Soc Lond B Biol Sci. 2009;364:1629–1640. doi: 10.1098/rstb.2009.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hendry AP. Eco-Evolutionary Dynamics. Princeton Univ Press; Princeton: 2016. [Google Scholar]
- 6.Jones LE, et al. Rapid contemporary evolution and clonal food web dynamics. Philos Trans R Soc Lond B Biol Sci. 2009;364:1579–1591. doi: 10.1098/rstb.2009.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basanta D, Anderson ARA. Homeostasis back and forth: An ecoevolutionary perspective of cancer. Cold Spring Harb Perspect Med. 2017;7:a028332. doi: 10.1101/cshperspect.a028332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez A, Gore J. Feedback between population and evolutionary dynamics determines the fate of social microbial populations. PLoS Biol. 2013;11:e1001547. doi: 10.1371/journal.pbio.1001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthews B, Aebischer T, Sullam KE, Lundsgaard-Hansen B, Seehausen O. Experimental evidence of an eco-evolutionary feedback during adaptive divergence. Curr Biol. 2016;26:483–489. doi: 10.1016/j.cub.2015.11.070. [DOI] [PubMed] [Google Scholar]
- 10.terHorst CP, Zee PC. Eco-evolutionary dynamics in plant–Soil feedbacks. Funct Ecol. 2016;30:1062–1072. [Google Scholar]
- 11.Rudman SM, et al. What genomic data can reveal about eco-evolutionary dynamics. Nat Ecol Evol. 2018;2:9–15. doi: 10.1038/s41559-017-0385-2. [DOI] [PubMed] [Google Scholar]
- 12.Wright S. Proceedings of the Sixth International Congress of Genetics. Vol 1. Genetics Society of America; Rockville, MD: 1932. The roles of mutation, inbreeding, crossbreeding and selection in evolution; pp. 356–366. [Google Scholar]
- 13.Kauffman SA, Johnsen S. Coevolution to the edge of chaos: Coupled fitness landscapes, poised states, and coevolutionary avalanches. J Theor Biol. 1991;149:467–505. doi: 10.1016/s0022-5193(05)80094-3. [DOI] [PubMed] [Google Scholar]
- 14.Watson RA, Ebner M. Eco-evolutionary dynamics on deformable fitness landscapes. In: Richter H, Engelbrecht A, editors. Recent Advances in the Theory and Application of Fitness Landscapes. Springer Berlin Heidelberg; Berlin: 2014. pp. 339–368. [Google Scholar]
- 15.Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 16.Poelwijk FJ, Kiviet DJ, Weinreich DM, Tans SJ. Empirical fitness landscapes reveal accessible evolutionary paths. Nature. 2007;445:383–386. doi: 10.1038/nature05451. [DOI] [PubMed] [Google Scholar]
- 17.Hartl DL. What can we learn from fitness landscapes? Curr Opin Microbiol. 2014;21:51–57. doi: 10.1016/j.mib.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nahum JR, et al. A tortoise-hare pattern seen in adapting structured and unstructured populations suggests a rugged fitness landscape in bacteria. Proc Natl Acad Sci USA. 2015;112:7530–7535. doi: 10.1073/pnas.1410631112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Cleve J, Weissman DB. Measuring ruggedness in fitness landscapes. Proc Natl Acad Sci USA. 2015;112:7345–7346. doi: 10.1073/pnas.1507916112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rozen DE, Habets MGJL, Handel A, de Visser JAGM. Heterogeneous adaptive trajectories of small populations on complex fitness landscapes. PLoS One. 2008;3:e1715. doi: 10.1371/journal.pone.0001715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moradigaravand D, Engelstädter J. The effect of bacterial recombination on adaptation on fitness landscapes with limited peak accessibility. PLoS Comput Biol. 2012;8:e1002735. doi: 10.1371/journal.pcbi.1002735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barve A, Wagner A. A latent capacity for evolutionary innovation through exaptation in metabolic systems. Nature. 2013;500:203–206. doi: 10.1038/nature12301. [DOI] [PubMed] [Google Scholar]
- 23.de Visser JAGM, Krug J. Empirical fitness landscapes and the predictability of evolution. Nat Rev Genet. 2014;15:480–490. doi: 10.1038/nrg3744. [DOI] [PubMed] [Google Scholar]
- 24.Barber LJ, Davies MN, Gerlinger M. Dissecting cancer evolution at the macro-heterogeneity and micro-heterogeneity scale. Curr Opin Genet Dev. 2015;30:1–6. doi: 10.1016/j.gde.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao B, Hemann MT, Lauffenburger DA. Modeling tumor clonal evolution for drug combinations design. Trends Cancer. 2016;2:144–158. doi: 10.1016/j.trecan.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luksza M, Lässig M. A predictive fitness model for influenza. Nature. 2014;507:57–61. doi: 10.1038/nature13087. [DOI] [PubMed] [Google Scholar]
- 27.Nourmohammad A, Held T, Lässig M. Universality and predictability in molecular quantitative genetics. Curr Opin Genet Dev. 2013;23:684–693. doi: 10.1016/j.gde.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Lässig M, Mustonen V, Walczak AM. Predicting evolution. Nat Ecol Evol. 2017;1:77. doi: 10.1038/s41559-017-0077. [DOI] [PubMed] [Google Scholar]
- 29.Paquin CE, Adams J. Relative fitness can decrease in evolving asexual populations of S. cerevisiae. Nature. 1983;306:368–370. doi: 10.1038/306368a0. [DOI] [PubMed] [Google Scholar]
- 30.Good BH, McDonald MJ, Barrick JE, Lenski RE, Desai MM. The dynamics of molecular evolution over 60,000 generations. Nature. 2017;551:45–50. doi: 10.1038/nature24287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Gac M, Doebeli M. Epistasis and frequency dependence influence the fitness of an adaptive mutation in a diversifying lineage. Mol Ecol. 2010;19:2430–2438. doi: 10.1111/j.1365-294X.2010.04664.x. [DOI] [PubMed] [Google Scholar]
- 32.Rosenzweig RF, Sharp RR, Treves DS, Adams J. Microbial evolution in a simple unstructured environment: Genetic differentiation in Escherichia coli. Genetics. 1994;137:903–917. doi: 10.1093/genetics/137.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friesen ML, Saxer G, Travisano M, Doebeli M. Experimental evidence for sympatric ecological diversification due to frequency-dependent competition in Escherichia coli. Evolution. 2004;58:245–260. [PubMed] [Google Scholar]
- 34.Quandt EM, et al. Fine-tuning citrate synthase flux potentiates and refines metabolic innovation in the Lenski evolution experiment. eLife. 2015;4:e09696. doi: 10.7554/eLife.09696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paczia N, et al. Extensive exometabolome analysis reveals extended overflow metabolism in various microorganisms. Microb Cell Fact. 2012;11:122. doi: 10.1186/1475-2859-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orth JD, et al. A comprehensive genome-scale reconstruction of Escherichia coli metabolism–2011. Mol Syst Biol. 2011;7:535. doi: 10.1038/msb.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis NE, et al. Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol Syst Biol. 2010;6:390. doi: 10.1038/msb.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Brien EJ, Monk JM, Palsson BO. Using genome-scale models to predict biological capabilities. Cell. 2015;161:971–987. doi: 10.1016/j.cell.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matias Rodrigues JF, Wagner A. Evolutionary plasticity and innovations in complex metabolic reaction networks. PLoS Comput Biol. 2009;5:e1000613. doi: 10.1371/journal.pcbi.1000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahadevan R, Edwards JS, Doyle FJ., 3rd Dynamic flux balance analysis of diauxic growth in Escherichia coli. Biophys J. 2002;83:1331–1340. doi: 10.1016/S0006-3495(02)73903-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harcombe WR, et al. Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep. 2014;7:1104–1115. doi: 10.1016/j.celrep.2014.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138:1315–1341. [Google Scholar]
- 43.Blount ZD, Borland CZ, Lenski RE. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci USA. 2008;105:7899–7906. doi: 10.1073/pnas.0803151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Travisano M, Lenski RE. Long-term experimental evolution in Escherichia coli. IV. Targets of selection and the specificity of adaptation. Genetics. 1996;143:15–26. doi: 10.1093/genetics/143.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Visser JAGM, Lenski RE. Long-term experimental evolution in Escherichia coli. XI. Rejection of non-transitive interactions as cause of declining rate of adaptation. BMC Evol Biol. 2002;2:19. doi: 10.1186/1471-2148-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kerr B, Riley MA, Feldman MW, Bohannan BJM. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- 47.Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 48.Blount ZD, Barrick JE, Davidson CJ, Lenski RE. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature. 2012;489:513–518. doi: 10.1038/nature11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quandt EM, Deatherage DE, Ellington AD, Georgiou G, Barrick JE. Recursive genomewide recombination and sequencing reveals a key refinement step in the evolution of a metabolic innovation in Escherichia coli. Proc Natl Acad Sci USA. 2014;111:2217–2222. doi: 10.1073/pnas.1314561111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gallet R, et al. The evolution of bacterial cell size: The internal diffusion-constraint hypothesis. ISME J. 2017;11:1559–1568. doi: 10.1038/ismej.2017.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pál C, Papp B. Evolution of complex adaptations in molecular systems. Nat Ecol Evol. 2017;1:1084–1092. doi: 10.1038/s41559-017-0228-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Darwin C. The Formation of Vegetable Mould Through the Action of Worms, with Observations on Their Habits. John Murray; London: 1881. [Google Scholar]
- 53.Lewontin RC. Adaptation. Sci Am. 1978;239:212–218, 220, 222 passim. doi: 10.1038/scientificamerican0978-212. [DOI] [PubMed] [Google Scholar]
- 54.Solé RV, Sardanyés J. Red Queen coevolution on fitness landscapes. In: Richter H, Engelbrecht A, editors. Recent Advances in the Theory and Application of Fitness Landscapes. Springer Berlin Heidelberg; Berlin: 2014. pp. 301–338. [Google Scholar]
- 55.Morran LT, Schmidt OG, Gelarden IA, Parrish RC, 2nd, Lively CM. Running with the Red Queen: Host-parasite coevolution selects for biparental sex. Science. 2011;333:216–218. doi: 10.1126/science.1206360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schuster P. A revival of the landscape paradigm: Large scale data harvesting provides access to fitness landscapes. Complexity. 2012;17:6–10. [Google Scholar]
- 57.Doebeli M, Ispolatov Y, Simon B. Towards a mechanistic foundation of evolutionary theory. eLife. 2017;6:e23804. doi: 10.7554/eLife.23804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moran PA. On the nonexistence of adaptive topographies. Ann Hum Genet. 1964;27:383–393. doi: 10.1111/j.1469-1809.1963.tb01535.x. [DOI] [PubMed] [Google Scholar]
- 59.Mori M, Hwa T, Martin OC, De Martino A, Marinari E. Constrained allocation flux balance analysis. PLoS Comput Biol. 2016;12:e1004913. doi: 10.1371/journal.pcbi.1004913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beg QK, et al. Intracellular crowding defines the mode and sequence of substrate uptake by Escherichia coli and constrains its metabolic activity. Proc Natl Acad Sci USA. 2007;104:12663–12668. doi: 10.1073/pnas.0609845104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giovannoni SJ, Cameron Thrash J, Temperton B. Implications of streamlining theory for microbial ecology. ISME J. 2014;8:1553–1565. doi: 10.1038/ismej.2014.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waxman D, Gavrilets S. 20 questions on adaptive dynamics. J Evol Biol. 2005;18:1139–1154. doi: 10.1111/j.1420-9101.2005.00948.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.