Significance
Despite the initial success of therapeutic agents targeting the RAS/MAP kinase and PI3K/AKT/mTOR signalling networks in oncology, development of acquired resistance to such therapeutics represents a significant challenge in successful disease management. BCL-2–associated death promoter (BAD) is a common and core downstream molecule for both the RAS/MAP kinase and PI3K/AKT/mTOR pathways and regulates cancer cell survival. In its unphosphorylated state, BAD sequesters BCL-2, which results in BAK/BAX activation and apoptosis. Herein, we identified and characterized a small molecule which specifically inhibits BAD phosphorylation on Ser99. This molecule may be therapeutically useful, either alone or in combination, to delay or obviate the development of resistance to other therapeutic agents.
Keywords: BAD phosphorylation, AKT-PKB, NPB, carcinoma, Laplacian-modified naive Bayesian classifier
Abstract
Human BCL-2–associated death promoter (hBAD) is an apoptosis-regulatory protein mediating survival signals to carcinoma cells upon phosphorylation of Ser99, among other residues. Herein, we screened multiple small-molecule databases queried in a Laplacian-modified naive Bayesian-based cheminformatics platform and identified a Petasis reaction product as a site-specific inhibitor for hBAD phosphorylation. Based on apoptotic efficacy against mammary carcinoma cells, N-cyclopentyl-3-((4-(2,3-dichlorophenyl) piperazin-1-yl) (2-hydroxyphenyl) methyl) benzamide (NPB) was identified as a potential lead compound. In vitro biochemical analyses demonstrated that NPB inhibited the phosphorylation of hBAD specifically on Ser99. NPB was observed to exert this effect independently of AKT and other kinase activities despite the demonstration of AKT-mediated BAD-Ser99 phosphorylation. Using a structure-based bioinformatics platform, we observed that NPB exhibited predicted interactions with hBAD in silico and verified the same by direct binding kinetics. NPB reduced phosphorylation of BAD-Ser99 and enhanced caspase 3/7 activity with associated loss of cell viability in various human cancer cell lines derived from mammary, endometrial, ovarian, hepatocellular, colon, prostatic, and pancreatic carcinoma. Furthermore, by use of a xenograft model, it was observed that NPB, as a single agent, markedly diminished BAD phosphorylation in tumor tissue and significantly inhibited tumor growth. Similar doses of NPB utilized in acute toxicity studies in mice did not exhibit significant effects. Hence, we report a site-specific inhibitor of BAD phosphorylation with efficacy in tumor models.
The BCL-2–associated death promoter (BAD) plays a pivotal role in regulating apoptosis by interacting with BCL-2, BCL-xL, and BCL-w (1). Human BAD (hBAD) is phosphorylated at Ser75 (equivalent of the murine Ser112 residue) by p44/42 MAP kinase and at Ser99 (equivalent of the murine Ser136 residue) by AKT/p70S6K (2). Both serine residues are also phosphorylated by Pim family kinases and function to prevent apoptosis (3). Phosphorylation of BAD at either of these residues results in the loss of the ability of hBAD to heterodimerize with BCL-xL or BCL-2 (4). Phosphorylated BAD protein is heterodimerized with the 14-3-3 protein and is sequestered in the cytoplasm (5). This association can be reversed by dephosphorylation of BAD, following which it can dimerize with BCL-2, BCL-xL, or BCL-w, which allows activation of BAK/BAX and promotes the release of cytochrome C to the cytoplasm with the subsequent promotion of the intrinsic apoptotic pathway (6, 7). Independent of its phosphorylation status, BAD also interacts with TP53 and forms a BAD/TP53 complex at mitochondria to induce apoptosis (8). The critical involvement of BAD and its phosphorylation status in tumor initiation and disease progression has been documented (9). Phosphorylated BAD is also observed to be a predictive marker for drug response, chemosensitivity, and prognosis in various malignancies (9). For example, increased BAD phosphorylation has been reported to predict poor overall survival in ovarian cancer (10) and to be associated with resistance to cisplatin (9, 10). Phosphorylated BAD has been observed in more than 80% of the CD44+ cancer stem cell (CSC) population in breast cancer, and BAD phosphorylation has been reported to be essential for CSC survival (11). Furthermore, BAD phosphorylation is critically associated with acquired resistance to chemotherapeutic agents such as vemurafenib in melanoma (12) and other inhibitors of the RAS/MAP kinase and PI3K/AKT/mTOR pathways (9). Therefore, inhibition of BAD phosphorylation may be useful to improve clinical endpoints in oncology.
We performed virtual screening of 1.7 million compounds using a cheminformatics platform against a broad range of proteins and identified title compounds that were predicted to target BAD protein. The derivatives of the lead scaffold were synthesized using the Petasis reaction and were screened for their apoptotic efficacy in cancer cells. Among the derivatives, N-cyclopentyl-3-((4-(2,3-dichlorophenyl) piperazin-1-yl) (2-hydroxyphenyl) methyl) benzamide (NPB) was identified as a lead compound which inhibits site-specific (Ser99) phosphorylation of BAD. We have further validated the in vitro and in vivo inhibitory activity of NPB against BAD phosphorylation.
Results
A Cheminformatics Approach to Identify Ligands Targeting hBAD.
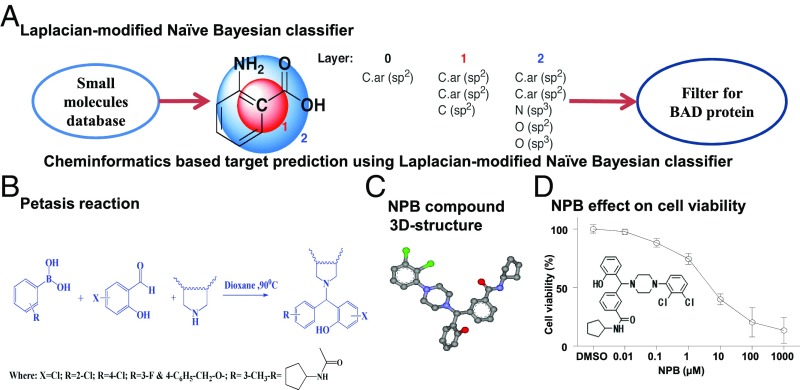
We screened 1.7 million compounds for predicted interaction with BAD using a unique in silico mapping of chemical space in the drug-discovery area (13). The structures of small molecules used in the study were retrieved from ZINC (14) and from traditional Chinese and Ayurvedic medicinal compound databases (15). A number of the title compounds were predicted to interact with BAD (Fig. 1A). We next utilized the Petasis reaction for the synthesis of derivatives of title compounds. The Petasis reaction is a three-component boronic Mannich-type reaction that contributes significantly to the advancement of organic transformations by providing greater flexibility, novelty, and efficiency (16). An antimalarial agent, Mefloquine, was previously generated using the Petasis reaction and is known to inhibit autophagy and induce cell death of various mammary carcinoma (MC) cells (17). With the goal of preparing a library of druggable small molecules, we used boronic acid as a potential nucleophilic species, salicylaldehyde and substituted piperazines to form the new C–C bond of the title compounds (Fig. 1B). The Petasis reaction proceeds via the formation of an iminium species, which reacts with the boronic acid to yield tertiary amines (SI Appendix, Supporting Information 1).
Fig. 1.
A cheminformatics approach to identify ligands that target hBAD. (A) Laplacian-modified naive Bayesian classifier algorithm analysis predicted a high-probability score for hBAD. (B) The Petasis reaction is a three-component boronic Mannich-type reaction. The method utilizes boronic acids as a potential nucleophilic species, salicylaldehyde, and substituted piperazines to form the new C–C bond of the formula I compound in which R is selected from group comprising chlorine, methyl, fluorine, and N-cyclopentylacetamido groups and X is selected from the group comprising chlorine and hydrogen or the tautomers, isomers, analogs, derivatives, or salts thereof, by the Petasis reaction. (C) The three-dimensional structure of NPB. (D) Dose-dependent effect of NPB (the chemical structure) on the viability of MCF7 cells measured using the AlamarBlue viability assay as described in Materials and Methods. Points are the mean of triplicate determinations; error bars indicate SD.
NPB Identified as a Lead Candidate Which Inhibits the Viability of Carcinoma-Derived Cells.
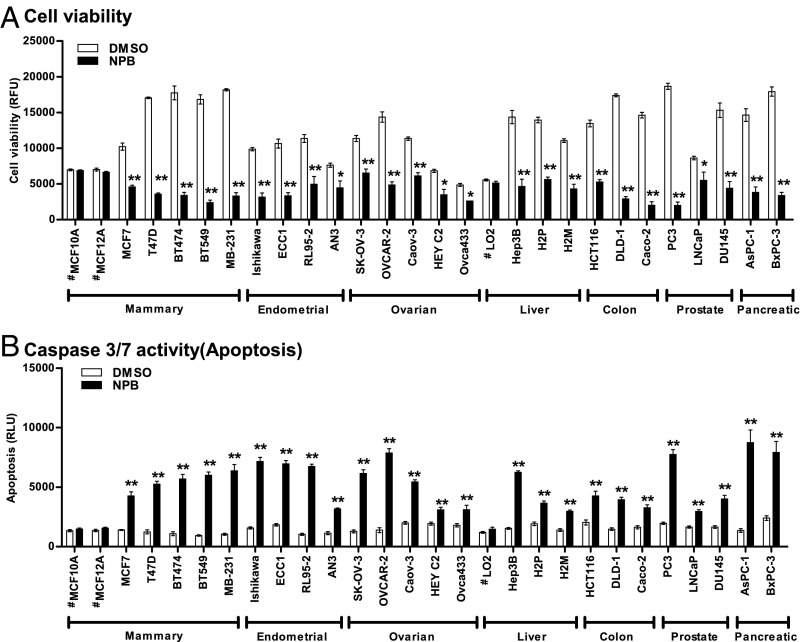
We first investigated the effect of newly synthesized small-molecule compounds against MCF7 cells using an AlamarBlue cell-viability assay. Among the series of small-molecule compounds, NPB (compound 4i) (Fig. 1C and SI Appendix, Supporting Information 5) was identified as a highly efficacious small-molecule compound reducing the viability of MCF7 cells compared with DMSO-treated cells (Fig. 1D). We next determined the IC50 of NPB in the various carcinoma-derived cell lines tabulated in Table 1. As normal cell controls, we also included MCF10A and MCF12A (mammary epithelial cells) and LO2 (hepatocytes) in the panel of cell lines. Next, we examined cell viability, cytotoxicity, and apoptotic cell death of the carcinoma cell lines after exposure to NPB or DMSO using the ApoTox-Glo Triplex Assay Kit. Exposure of normal hepatocytes or mammary epithelial cells to NPB did not produce substantial changes in cell viability, cytotoxicity, or apoptosis compared with DMSO-exposed cells. Exposure of carcinoma cells to NPB decreased cell viability significantly (P < 0.05) and increased caspase 3/7 activities (apoptosis) compared with DMSO-treated carcinoma cells (Fig. 2). A slight but significant cytotoxic effect of NPB was observed in a small number of the carcinoma cell lines compared with DMSO-exposed control cells specified in SI Appendix, Supporting Information 2A.
Table 1.
IC50 values of NPB in a panel of carcinoma cell lines
| Tissue | Cell line | NPB IC50* ± SD, µM |
| Mammary | MCF10A† | NV |
| MCF12A† | NV | |
| MCF7 | 6.5 ± 1.06 | |
| T47D | 7.24 ± 1.91 | |
| BT474 | 5.31 ± 2.04 | |
| BT549 | 4.88 ± 1.31 | |
| MDA-MB-231 | 6.94 ± 1.86 | |
| Endometrial | Ishikawa | 7.51 ± 2.08 |
| ECC1 | 2.61 ± 0.97 | |
| RL95-2 | 6.38 ± 1.85 | |
| AN3 | 11.37 ± 2.61 | |
| Ovarian | SK-OV-3 | 7.34 ± 2.03 |
| OVCAR-2 | 4.21 ± 1.74 | |
| Caov-3 | 3.95 ± 0.93 | |
| HEY C2 | 6.82 ± 1.94 | |
| Ovca433 | 9.79 ± 2.48 | |
| Hepatocellular | #LO2 | NV |
| Hep3B | 6.94 ± 1.07 | |
| H2P | 4.18 ± 0.83 | |
| H2M | 5.57 ± 2.61 | |
| Colon | HCT116 | 7.29 ± 2.02 |
| DLD-1 | 2.46 ± 0.91 | |
| Caco-2 | 3.08 ± 0.76 | |
| Prostate | PC3 | 3.77 ± 1.51 |
| LNCaP | 8.02 ± 3.3 | |
| DU145 | 6.99 ± 1.72 | |
| Pancreatic | AsPC-1 | 3.83 ± 1.14 |
| BxPC-3 | 7.25 ± 2.09 |
Cell viability was measured using the AlamarBlue cell viability assay. NV, no value.
IC50 values were calculated using GraphPad Prism software (version 5.0).
Normal cells.
Fig. 2.
NPB suppresses cell viability and promotes apoptosis in carcinoma cell lines. Effect of NPB (5 µM) on the viability of carcinoma cells including, mammary, endometrial, ovarian, liver, colon, prostate, and pancreatic carcinoma cell lines. (A) Cell viability and (B) caspase 3/7 activities were evaluated using the ApoTox-Glo Triplex Assay Kit as described in Materials and Methods. Statistical significance was assessed by an unpaired two-tailed Student’s t test using GraphPad Prism 5. The bars represent the mean of triplicate determinations; error bars indicate SD. **P < 0.001, *P < 0.05. #, nontransformed, immortalized epithelial cells; MB-231, MDA-MB-231; RFU, relative fluorescence units; RLU, relative luminescence units.
NPB Suppresses MC Cell Proliferation by Stimulating Apoptotic Cell Death.
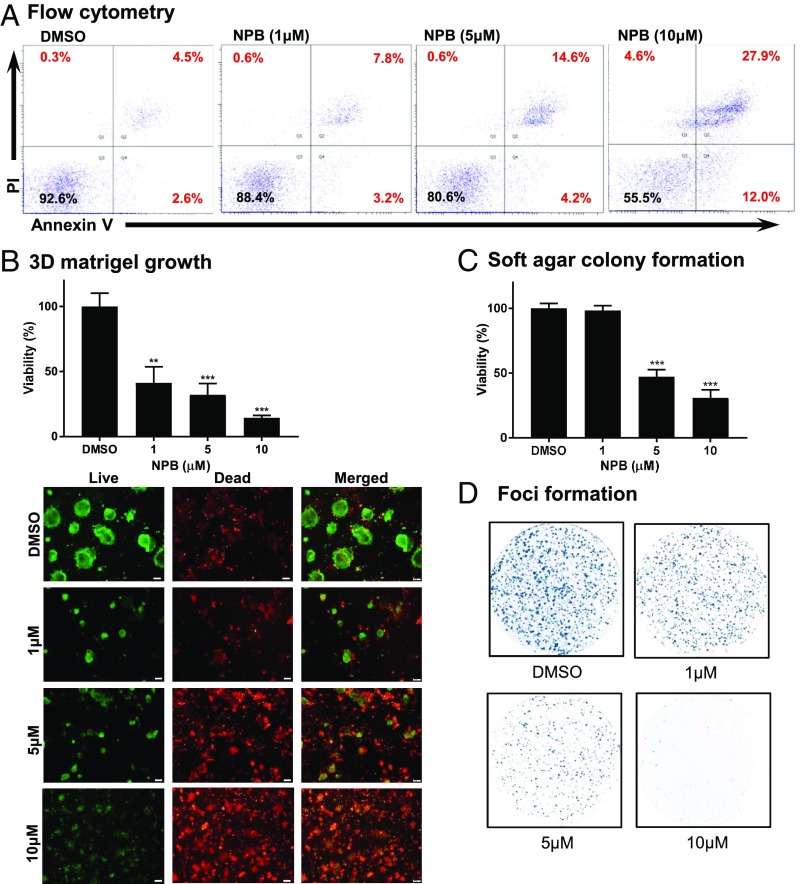
Using flow cytometry, we next examined whether NPB treatment of MCF7 cells promotes apoptotic cell death. NPB treatment of MCF7 cells significantly stimulated early (PI−, FITC-Annexin V+) and late (PI+, FITC-Annexin V+) apoptotic cell death compared with DMSO-exposed cells (Fig. 3A). Concomitantly, cell-cycle analysis of MCF7 cells after exposure to NPB demonstrated significantly increased cell populations in the sub-G1 phase along with increased numbers of cells in the G2M phase and reduced numbers of cells in the S phase compared with DMSO-exposed cells (SI Appendix, Supporting Information 2B). Subsequently, we also assessed the effect of NPB on pregrown colonies of MCF7 cells in 3D Matrigel. Our analyses demonstrated that NPB treatment significantly suppressed the growth of these cells as indicated by cell viability and live-cell calcein AM staining (Fig. 3B). NPB treatment of MCF7 cells also suppressed the growth of colonies pregrown in soft agar and also foci formation as compared with DMSO-exposed cells (Fig. 3 C and D). Thus, NPB treatment of MCF7 cells decreased cell proliferation and anchorage-independent growth by stimulating apoptotic cell death.
Fig. 3.
NPB stimulates apoptotic cell death in MCF7 cells. (A) Apoptotic cell death of MCF7 cells measured after treatment with 10 µM NPB using flow cytometry analysis. Annexin V-FITC staining is indicated on the x axis, and PI staining is indicated on the y axis. The lower left quadrants represent live cells, the lower right quadrants represent early apoptotic cells, the upper left quadrants represent necrotic cells, and the upper right quadrants display late apoptotic cells. Acquisition of Annexin V and PI data are presented as a percentage (%) in each quadrant. (B, Upper) Cell viability was determined using the AlamarBlue viability assay of colonies generated by MCF7 cells cultured for 14 d in 3D Matrigel after exposure to NPB or DMSO. (Lower) Microscopic visualization of calcein AM-stained colonies generated by MCF7 cells cultured in 3D Matrigel after exposure to NPB or DMSO. (C) Cell viability determined using the AlamarBlue viability assay of colonies generated by MCF7 cells cultured in soft agar after exposure to NPB or DMSO. (D) Crystal Violet staining of foci in colonies generated by MCF7 cells after exposure to NPB or DMSO. All assays were performed as described in Materials and Methods. Statistical significance was assessed by an unpaired two-tailed Student’s t test using GraphPad Prism 5. The bars represent the mean of triplicate determinations; error bars indicate SD. ***P < 0.0001, **P < 0.001.
In Silico and Surface Plasmon Resonance Analysis Predicts an Interaction of NPB Compound with the BAD Protein.
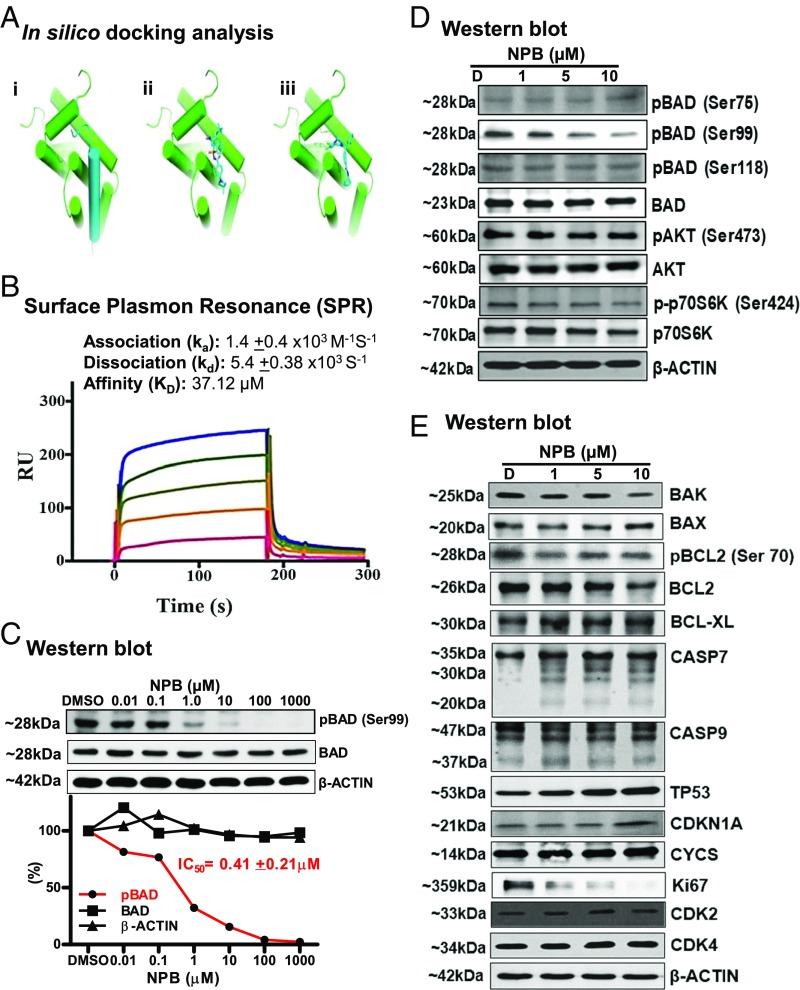
The Laplacian-modified naive Bayesian classifier algorithm analysis identified a high-probability score of NPB for hBAD interaction (SI Appendix, Supporting Information 3). In silico docking analyses were then performed for NPB, which was predicted to occupy the hydrophobic groove within the protein–protein interface of BCL-2/BAD. These observations were found to be in concordance with other known BCL-2 inhibitors (18). The dichlorophenyl moiety of NPB was predicted to occupy an additional hydrophobic side pocket within the BCL-2/BAD interface formed by the side chains of Leu97 (Leu94 in mouse), Trp144 (Trp141 in mouse), and Phe198 (Phe195 in mouse) in contrast to a BCL-2 inhibitor (Fig. 4A).
Fig. 4.
Cheminformatics and SPR analysis predict an interaction of NPB with BAD protein. (A) Docking studies of NPB toward the Bcl-2/BAD interface. (i) Modeled complex of Bcl-2 (green cartoon) and a 25-residue helical fragment of BAD (blue cartoon) based on the structures of Protein Data Bank ID code 2O22 and 1G5J (70, 74). (ii) NMR-derived structure of a small molecule [(4-(4,4-Dimethylpiperidin-1yl)-N-(4-(2-methyl-1-(phenylthio)-propan-2-ylamino)-3-nitrophenylsulfonyl) benzamide] in stick representation binding to a groove within the protein–protein interface of Bcl-2 (70). (iii) Predicted binding mode of NPB to Bcl-2: NPB occupies a binding region similar to that of a known Bcl-2 binder but additionally exploits a hydrophobic pocket formed by Leu94, Trp141, and Phe195 (in mouse) (shown as lines) in the BCL-2/BAD interface. (B) Sensorgrams obtained by SPR analysis of NPB with the BAD protein subunit. The BAD protein subunit was immobilized on the surface of a CM5 sensor chip. A solution of NPB at variable concentrations (20–100 µM) was injected to generate the binding responses (RU) recorded as a function of time (s). The results were analyzed using BIA evaluation 4.1. (C, Upper) WB analysis was used to assess the level of BAD phosphorylation at Ser99 in MCF7 cells after treatment with NPB. (Lower) IC50 of NPB calculated from the dose–response curves for BAD phosphorylation at Ser99, BAD, and β-actin using ImageJ software (NIH) (https://imagej.nih.gov/ij/). (D) WB analysis was used to assess the level of multiple proteins upstream of BAD in MCF7 cells after treatment with NPB. (E) WB analysis was used to assess the level of multiple proteins involved in cell survival and cell proliferation in MCF7 cells after treatment with NPB. β-Actin was used as an input control for cell lysate. The sizes of detected protein bands are shown on the left. For WB analysis, soluble whole-cell extracts (30 µg for C and 50 µg for D and E) were run on an SDS/PAGE gel and were immunoblotted as described in Materials and Methods.
We next performed surface plasmon resonance (SPR) measurement with immobilized BAD protein using NPB as the analyte. This potential interaction was analyzed using the BIAcore system, and the recombinant BAD was immobilized on a CM5 sensor chip. Various concentrations of NPB were injected onto the BAD-coated surface of the sensor chip, and the association and dissociation curves were recorded. The overlaid sensorgrams are shown in Fig. 4B and were analyzed collectively. The calculation of kinetic parameters for the interaction of NPB with BAD revealed an association rate constant (Ka) of 1.4 ± 0.4 × 103 M/s and a dissociation rate constant of 5.4 ± 0.38 × 103 s of binding affinity, which yielded dissociation equilibrium constants (Kds) of 37.12 µM (Fig. 4B). These kinetic parameters provide substantial evidence for the interaction of NPB with BAD protein.
NPB Inhibits Site-Specific Phosphorylation of BAD at Ser99 in Carcinoma Cells Independently of AKT Signaling.
Phosphorylation of hBAD at residues Ser75 (ser112 in mouse) and Ser99 (ser136 in mouse) is crucial in regulating the activity of the BCL-2 family of proteins (4). To further validate BAD as a target, we first analyzed the effect of NPB on phosphorylation of hBAD at Ser99 by Western blot (WB) analysis. Treatment of MCF7 cells with NPB produced a dose-dependent decrease in phosphorylation of hBAD at Ser99 without a significant change in total hBAD protein (Fig. 4D). The calculated IC50 for the inhibition of BAD phosphorylation at Ser99 by NPB was 0.41 ± 0.21 µM, using WB analysis (Fig. 4C).
To determine the possibility that NPB decreased hBAD phosphorylation at Ser99 by modulation of kinase activity, we assessed the effects of NPB on various kinases using a human phospho-kinase antibody array. No significant changes in kinase activity or phosphorylated substrates were observed in MCF7 cells exposed to NPB compared with DMSO-exposed cells despite NPB inhibition of hBAD phosphorylation at Ser99 in the same extract (SI Appendix, Supporting Information 4 A and B). Specifically, the phosphorylation of the direct AKT substrate [GSK-3 α/β (S21/S9)] (19) was not affected by NPB treatment, despite NPB inhibition of hBAD phosphorylation at Ser99 in the same cell extract (SI Appendix, Supporting Information 4B). Furthermore, we also confirmed that NPB had no effect on AKT kinase activity after cellular treatment with NPB or in the in vitro kinase assays (SI Appendix, Supporting Information 4C).
We analyzed the effects of NPB exposure on upstream kinases reported to phosphorylate hBAD at Ser99 (2, 20). BAD phosphorylation at Ser99 was decreased upon increasing NPB dosage; neither the levels of phosphorylated and total AKT nor the levels of phosphorylated and total p70S6K were altered by NPB, as indicated by WB analysis (Fig. 4D). Also, analysis of other BCL-2 family members (BAK, BAX, BCL-2, and BCL-xL) and cell-cycle markers (CDKN1A, CDK2, and CDK4) demonstrated no changes in protein levels. Concomitantly, however, the protein levels of apoptosis-related markers (CASP7, CASP9, and TP53) were increased, and the protein levels of Ki67 were decreased after exposure to NPB (Fig. 4E).
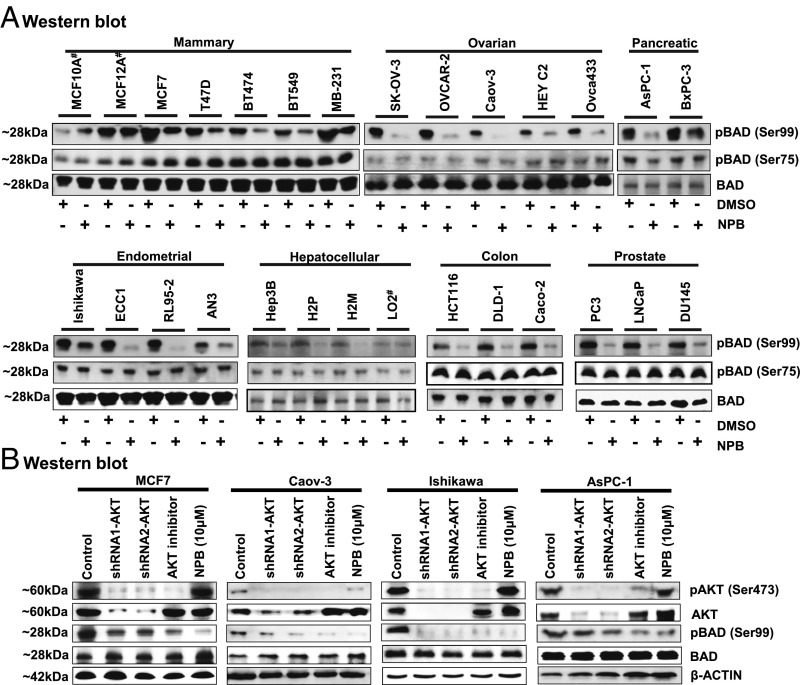
We next analyzed the effect of NPB on phosphorylation of hBAD at both Ser75 and Ser99 by WB analysis in 25 carcinoma cell lines derived from seven different types of cancer. It was observed that NPB inhibited the phosphorylation of BAD at the Ser99 site in all the tested carcinoma cell lines; however, NPB demonstrated no effect on the phosphorylation of hBAD at the Ser75 site in the same cells, indicating that NPB specifically inhibited phosphorylation of hBAD at Ser99 (Fig. 5A).
Fig. 5.
NPB specifically inhibits BAD phosphorylation at Ser99 in carcinoma cell lines independently of AKT signaling. (A) WB analysis was used to assess the levels of phosphorylated hBAD at Ser75 and Ser99 and BAD protein in a range of carcinoma cell lines, including mammary, ovarian, pancreatic, endometrial, hepatocellular, colon, and prostate cancer, after treatment with NPB (5 µM). Total BAD was used as an input control for cell lysate. (B) WB analysis was used to assess the levels of pBAD at Ser99, pAKT at Ser473, AKT, and BAD in MCF7, Caov-3, Ishikawa, and AsPC-1 cells. AKT inhibitor IV, and NPB (5 µM each) were used to treat cells. Depletion of AKT expression was achieved using transient transfection of shRNA (1 and 2) directed to the AKT transcript as described in Materials and Methods. β-Actin was used as an input control for cell lysate. For WB analysis, soluble whole-cell extracts were run on an SDS/PAGE gel and were immunoblotted as described in Materials and Methods. The sizes of detected protein bands in kDa are shown on the left. #, nontransformed immortalized cell line.
We next determined whether NPB inhibits the phosphorylation of hBAD at Ser99 by modulating AKT activity (as indicated by phosphorylation at Ser473) using WB analysis. We observed no change in the levels of pAKT or of total AKT protein after exposure of four different carcinoma cell lines to 10 µM NPB (Fig. 5B). However, all NPB-treated carcinoma cell lines exhibited inhibition of BAD phosphorylation at the Ser99 site with no change in the level of total BAD protein. Additionally, we examined BAD phosphorylation after depletion of AKT using two independent shRNAs targeting AKT expression or the inhibition of AKT activity with AKT inhibitor IV as a positive control in the different carcinoma cell lines. We observed that reduced expression of AKT in the carcinoma cell lines was associated with a concomitant decrease in pAKT levels at Ser474 and pBAD levels at Ser99 compared with control cells. Therefore, BAD phosphorylation at Ser99 is AKT dependent in all tested carcinoma cell lines, as previously reported by others (2, 4, 21–23). Hence, NPB inhibits the phosphorylation of BAD specifically at Ser99, independently of the upstream AKT-kinase.
siRNA-Mediated Depletion of BAD Expression Prevented the Effect of NPB in Carcinoma Cell Lines.
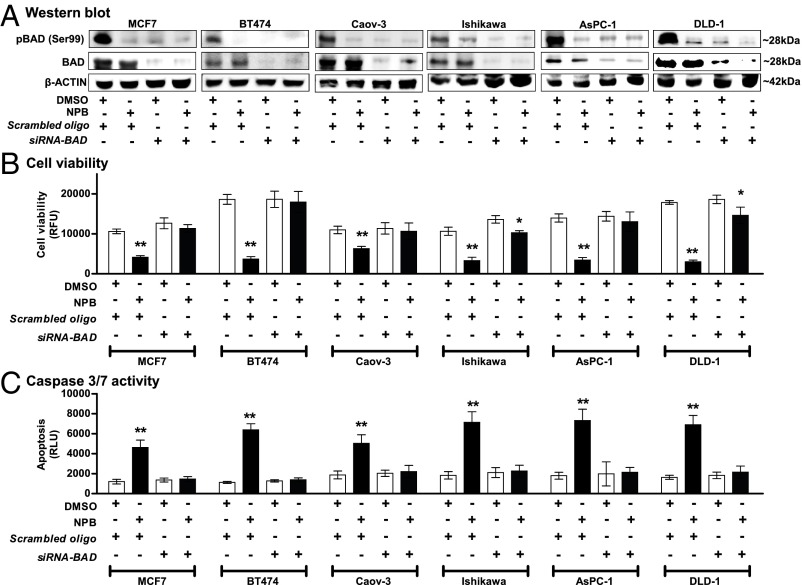
As previously described, the function of hBAD in apoptosis/cell survival is determined by its phosphorylation status, primarily on residues Ser75 and Ser99, with phosphorylated BAD promoting cell survival (9, 20, 24–26). Extensive experimental evidence in the literature clearly demonstrates that neither depletion nor forced expression of BAD (27–29) affects cell viability, whereas forced expression of BAD phosphorylation-deficient mutants (11, 30) results in loss of cell viability. Hence, the equilibrium between nonphosphorylated and phosphorylated BAD, rather than the level of BAD protein per se, apparently determines the cellular response. We therefore exploited this attribute of BAD to confirm the functional specificity of NPB directed to the BAD protein by examining the effect of NPB exposure after siRNA-mediated depletion of BAD expression in six carcinoma cell lines (Fig. 6A). Transient transfection of the different carcinoma cells with siRNA directed to the BAD transcript decreased BAD expression and also decreased levels of phospho-Ser99 BAD compared with levels in control cells (transfected with scrambled oligo), as observed by WB analysis (Fig. 6A). No significant change in cell viability or apoptosis was observed upon siRNA-mediated depletion of BAD in carcinoma cells (Fig. 6 B and C), as previously reported (25, 28, 31–34). As described above, NPB treatment of the control transfected carcinoma cell lines decreased BAD phosphorylation at Ser99 compared with vehicle-treated control cells. Concomitantly, exposure of the same carcinoma cell lines to NPB decreased cell viability and increased caspase 3/7 activity compared with vehicle-exposed cells. In contrast, NPB did not affect cell viability or caspase 3/7 activity in carcinoma cell lines with depleted expression of BAD (Fig. 6 B and C).
Fig. 6.
siRNA-mediated depletion of BAD expression prevents the effect of NPB in carcinoma cell lines. (A) WB analysis was used to assess the levels of pBAD at Ser99 activity and BAD protein in MCF7, BT474, Caov-3, Ishikawa, AsPC-1, and DLD-1 cells after treatment with 5 µM NPB. Depletion of BAD expression was achieved using transient transfection of siRNA directed to the BAD transcript. Soluble whole-cell extracts were run on an SDS/PAGE gel and were immunoblotted as described in Materials and Methods. β-Actin was used as input control. (B and C) Effects of NPB (5 µM) in MCF7, BT474, Caov-3, Ishikawa, AsPC-1, and DLD-1 cells. Cell viability (B) and caspase 3/7 activities (C) were evaluated using the ApoTox-Glo Triplex Assay Kit. All assays were performed as described in Materials and Methods. Statistical significance was assessed by an unpaired two-tailed Student’s t test (P < 0.05 was considered as significant) using GraphPad Prism 5. The bars represent the mean of triplicate determinations; error bars indicate SD. **P < 0.001, *P < 0.05. RFU, relative fluorescence units; RLU, relative luminescence units.
Pharmacological Features of NPB.
We determined the pharmacological features of NPB (SI Appendix, Supporting Information 5). The solubility of NPB was found to be 37.7 ± 3.2 μM at 24 h at 23 °C in universal buffer (pH 7.4) as determined by the filtration method using multiscreen plates. We next determined the pharmacokinetic (PK) characteristics of NPB via i.v. and oral administration in Wistar rats. Following a single 2-mg/kg i.v. dose, NPB showed multiexponential disposition with first-order kinetics, a moderate clearance of 1.2 L−1⋅h−1⋅kg−1 (37% of rat liver blood flow), a high volume of distribution at steady state (Vss) of 3.1 L/kg, and a moderately long terminal t1/2 of 6.1 h. Following a single oral dose of 10 mg/kg, NPB showed slow zero-order absorption (tmax = 4 h) characterized by a low phase up to 8 h postdose followed by an apparent monoexponential disposition. The maximum concentration (Cmax), area under the plasma concentration-time curve (AUC)last, and t1/2 were 73.4 ng/mL, 991 ng⋅h−1⋅mL−1, and 5.4 h, respectively. The absolute oral bioavailability was low (∼12%). We also performed acute toxicity analysis on mice to determine the suitability of NPB for in vivo use by i.p. injection of 5 or 20 mg/kg NPB. Various standard parameters of toxicity analyses did not exhibit any significant effects in the NPB-treated mice (SI Appendix, Supporting Information 6–8). However, the exposure of mouse mammary carcinoma cells (MMC) 4T1 and 67NR to NPB significantly inhibited Bad phosphorylation at Ser136 and decreased cell viability in a dose-dependent manner compared with the DMSO-treated control cells (SI Appendix, Supporting Information 9).
NPB Inhibits Phosphorylation of BAD at Ser99 in MC and Inhibits Tumor Growth.
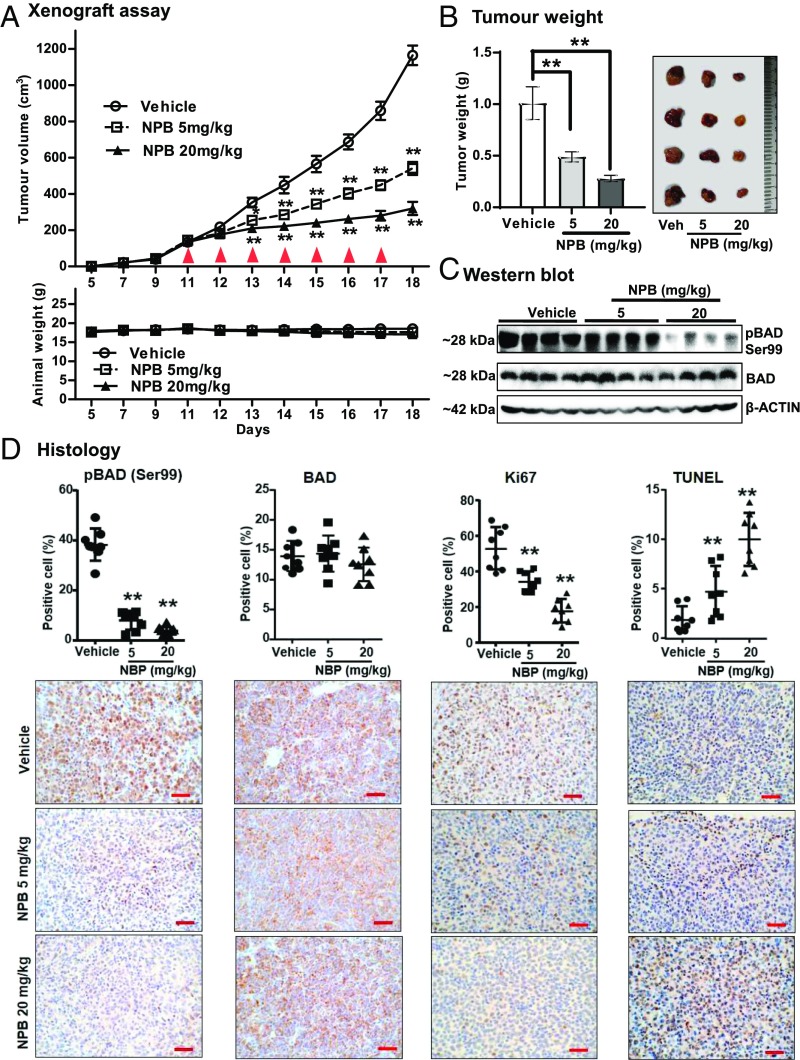
Finally, we examined the in vivo efficacy of NPB in a xenograft model (MCF7) of MC. Randomly grouped mice with preformed tumors (volume ∼150 cm3) were injected i.p. with vehicle or NPB at 5 mg/kg or 20 mg/kg. A significant reduction in tumor volume was observed in NPB-treated mice compared with their vehicle-treated counterparts (Fig. 7A). During this period, animal weight was not significantly different between the groups (Fig. 7A, Lower). However, the tumor weight of NPB-treated animals was reduced in a dose-dependent manner compared with vehicle-treated mice (Fig. 7B). We further analyzed the effect of NPB on the levels of hBAD phosphorylation at Ser99 in tumor tissue using WB analysis. NPB treatment significantly inhibited BAD phosphorylation at Ser99 in tumor tissues compared with control specimens, as demonstrated by WB analysis (Fig. 7C and SI Appendix, Supporting Information 9). No significant difference was observed in total levels of BAD protein between the NPB-treated and vehicle-treated tumors (SI Appendix, Supporting Information 10).
Fig. 7.
NPB inhibits BAD phosphorylation at Ser99 in mammary carcinoma and inhibits tumor growth. (A) Measurement of tumor volume in BALB/c-nu female mice as described in Materials and Methods. Animals (n = 5 in each group) were treated with vehicle, 5 mg/kg NPB, or 20 mg/kg NPB, and relative tumor burden was recorded. Animal weight was measured daily for the duration of the experiment. Red arrowheads indicate the days on which NPB was administered. (B, Left) Tumors were excised after the NPB treatment regime and weighed. (Right) Representative resected tumors. (C) WB of tumor tissue to determine levels of pBAD at Ser99 and BAD. Soluble whole-cell extracts were run on an SDS/PAGE gel and were immunoblotted as described in Materials and Methods. β-Actin was used as an input control. The sizes of detected protein bands in kDa are shown on the left. (D, Upper) Histological analyses of pBAD at Ser99, BAD, Ki67, and TUNEL staining. Statistical significance was assessed by an unpaired two-tailed Student’s t test (P < 0.05 was considered as significant) using GraphPad Prism 5. The points represent the means of triplicate experiments; error bars indicate SD. **P < 0.001. (Lower) Tumor tissue sections were immunolabeled with goat anti-pBAD (Ser136) polyclonal antibody (Santa Cruz Biotechnology), mouse anti-BAD monoclonal antibody (Santa Cruz Biotechnology), and anti-Ki67 antibody (ab15580; Abcam) and were stained with hematoxylin. (Scale bar, 100 μm.) Apoptotic DNA fragmentation was detected using the TUNEL Apoptosis Detection Kit (GenScript USA, Inc.) as described in Materials and Methods.
Histological analyses of tumor specimens resected from the animals treated with NPB showed significantly reduced pBAD at Ser99 compared with vehicle-treated tumors (Fig. 7D), whereas BAD protein was not significantly different between the groups. Animals treated with NPB exhibited a significantly decreased percentage of Ki67+ cells in tumors and significantly increased TUNEL positivity compared with vehicle-treated animals (Fig. 7D).
Discussion
The AKT/protein kinase B (PKB)-signaling pathway has a prominent role in promoting cell survival in most carcinomas, including hepatocellular, cervical, pancreatic, and mammary cancers (35–38). Multiple reports have demonstrated the persistent activation of AKT in carcinomas with resultant phosphorylation of BAD at Ser99 to promote cell survival (29, 35). Aberrant activation of AKT is positively associated with a poor prognosis in carcinomas and with resistance to endocrine therapy and chemotherapy in MC (39). Previous reports have also demonstrated that AKT inhibitors also decrease the phosphorylation of BAD at Ser99, and numerous AKT inhibitors, including ARQ 092, AZD5363, GSK690693, GDC-0068, and MK-2206, have been advanced to clinical trial and have shown encouraging results (40, 41). However, high degrees of homology in the ATP-binding domain between AKT, protein kinase A (PKA), and protein kinase C (PKC) and mutations in the AKT gene present possible significant obstacles for the clinical specificity and efficacy of AKT inhibitors (42). Some small-molecule inhibitors of BCL-2 proteins, such as ABT-199, ABT-263, ABT737, TW-37, and navitoclax, have also been reported (43). Single-agent activity of these compounds has observed in clinical trials for hematological malignancies. However, thus far, the response in solid tumors has been limited (44, 45). One explanation is that BCL-xL expression is increased more often than BCL-2 expression in solid tumors (43). This notion is exemplified in a recent report that investigated the response of colon carcinoma stem cell-like cells to a selective inhibitor of BCL-2 (ABT199), a BCL-2/BCL-xL dual inhibitor (ABT737), and a selective inhibitor of BCL-xL (WEHI-539). No efficacy of ABT199 was observed, but ABT737 and WEHI-539 both promoted apoptosis of the colon carcinoma cells utilized (43, 46). Compounds targeting BAD, such as NPB, would be useful in solid tumors regardless of whether a tumor was predominantly dependent on BCL-2 or BCL-xL, because both BCL-2 and BCL-xL are BAD dimerization partners (23, 43). Indeed, targeting BAD phosphorylation would prevent the alternate utilization of BCL-2 or BCL-xL as an escape mechanism promoting cellular resistance to therapeutics targeting either one individually. Furthermore, BAD appears to exert BCL-2 protein-independent functions by interacting with 14-3-3 (5, 47), by modulating apoptosis in a complex with p53 (8), by mediating PAK1- and RAF (including BRAF-V600E)-stimulated proliferation and survival (48, 49) and cytokine-stimulated proliferation mediated by CaMKII-γ (50). Regardless of the different sites of hBAD phosphorylation utilized for these different functions, inhibition of BAD phosphorylation at Ser99 by NPB would promote apoptosis and hence negate the other cellular functions of BAD. Also, by the use of clinico-genomic datasets, the BAD-mediated apoptotic pathway has been reported to be highly significantly associated with the development and progression of breast, ovarian, colon, and endometrial cancers (51). Our work herein has shown a broad apoptotic response of carcinoma cells originating from various organs to the inhibition of BAD phosphorylation at Ser99 by NPB. Hence, NPB inhibition of BAD function by diminished phosphorylation of Ser99 could be expected to provide a broadly efficacious therapeutic response in various solid tumors. Indeed, we demonstrated herein that NPB [at fold lower doses than those used for ABT-737 (52)] inhibited the growth of an MC xenograft model. Further use of various xenograft or spontaneous tumor models should assist in elucidating the potential clinical utility of NPB.
The paucity of single-agent efficacy of BCL-2 protein inhibitors such as ABT-737 or ABT-199 (52, 53) in solid tumors has prompted the examination of these compounds in combination therapy (54). NPB could be useful in all situations in which BCL-2 protein inhibitors are used in combination with other therapeutics, including tamoxifen treatment of breast cancer and treatment of other cancers with cytotoxic agents such as docetaxel (55). Also, NPB could be used as single-agent therapy in tumors in which high BAD expression or high BAD phosphorylation is associated with clinical outcome. Such examples would include ovarian cancer, in which BAD phosphorylation has been positively associated with dismal survival outcomes (10). Specific chemotherapeutics such as cisplatin have also been reported to stimulate BAD phosphorylation via PIM kinases, promoting cisplatin resistance (56), and NPB could prove useful as a combination therapy to improve therapeutic outcome. The next step in augmenting the potential clinical utility of NPB would be to design various analogs to improve the oral bioavailability while retaining activity.
No significant toxicities of NPB were observed herein. BCL-xL is critical for determining platelet lifespan (57). Therefore, the effective dose-limiting toxicity observed with the use of BCL-xL inhibitors such as ABT-263 was thrombocytopenia (43). Such toxicity was not observed with BCL-2–only inhibitors such as ABT-199. It could be postulated that inhibition of BAD phosphorylation, as a heterodimerizing partner of BCL-xL, may also result in thrombocytopenia. However, BAD gene-deleted mice mature to adulthood and are largely indistinguishable from control mice, except that their seminiferous tubules demonstrated prominent multinucleated giant cells (33) and ∼20% developed B cell lymphoma at 18 mo. Furthermore, mice containing a triple knock-in mutation of three functional serine residues in bad (Ser112, Ser136, and Ser155 in murine bad) displayed no gross abnormalities, although they demonstrated increased sensitivity to apoptotic stimuli (loss of growth factor-dependent lymphocyte survival) (23). NPB did not significantly alter lymphocyte or platelet number in the acute toxicity studies herein. Mice deficient for BAD or expressing the triple serine mutation described above also display fasting hyperglycemia (58). AKT inhibitors such as ARQ092 (59) also raise blood glucose levels clinically (which is successfully treated with metformin), as is consistent with AKT-dependent phosphorylation of BAD (60). Given the current interest in cancer cell dependence on glucose (61), it is noteworthy that glucose deprivation results in BAD dephosphorylation and BAD-mediated apoptosis (58). Hence, to date, there is no evidence that NPB inhibition of BAD phosphorylation at Ser99 will exhibit short-term toxicity.
In conclusion, NPB is a potent inhibitor of hBAD phosphorylation at Ser99. Further preclinical progression of this molecule is warranted.
Materials and Methods
Cell Lines and Reagents.
The human immortalized mammary epithelial cell lines MCF10A and MCF12A and the immortalized hepatocellular epithelial cell line LO2 were obtained from the American Type Culture Collection (ATCC) and were cultured per ATCC propagation instructions. MCF7, T47D, BT474, BT549, and MDA-MB-231 (denoted as “MB-231”); Ishikawa, ECC1, RL95-2 and AN3; Hep3B, H2P, and H2M; HCT116, DLD-1, and Caco-2; and LNCaP and DU145 cells were obtained from the ATCC. SK-OV-3, OVCAR-2, Caov-3, HEY C2, and Ovca433 cells were obtained from Ruby Huang’s laboratory at The Cancer Science Institute of Singapore (CSI), National University of Singapore (NUS). Pancreatic carcinoma cell lines were obtained from H. Phillip Koeffler’s laboratory at CSI, NUS. The MMC cell lines 4T1 and 67NR were obtained from ATCC. All carcinoma cell lines were cultured per ATCC propagation instructions. AKT inhibitor IV was purchased from Calbiochem. BAD directed stealth (sh)-RNA-BAD (shRNA-BAD1, 5′-GCUCCGCACCAUGAGUGACGAGUUU-3′ and shRNA-BAD2, 5′-AAACUCGUCACUCAUCCUCCGGAGC-3′) were purchased from Life Technologies. AKT-directed shRNA (shRNA-AKT1, 5′-CCGGCGCGTGACC ATGAACGAGTTTCTCGAGAAACTCGTTCATGGTCACGCGTTTTTG-3′ and shRNA2-AKT, 5′-CCGGGGACTACCTGCACTCGGAGAACTCGAGTTCTCCGAGTGCAGGTAG TCCTTTTTG-3′) were purchased from Life Technologies and were cloned into PLKO.1 vector (Sigma). Cells were transiently transfected with 20 nM AKT or BAD shRNA or with universal negative control (Invitrogen) using FuGENE HD (Promega) for 24 h, and further assays were performed. Alanine transaminase, aspartate transaminase, lactate dehydrogenase, creatine kinase, and blood urea nitrogen commercial kits were purchased from Agappe Diagnostics Ltd.
Cheminformatics Assay.
Using in silico chemogenomics approaches, we analyzed large set of small molecules retrieved from ZINC (14) and from a traditional Chinese and Ayurvedic medicinal compounds database (15). Initially, we employed the Laplacian-modified naive Bayesian classifier to rank hBAD as the target for the queried compounds (62). The classifier was trained on a large dataset obtained from ChEMBL (63). The hBAD protein target was normalized by comparing the predicted targets in the dataset used in this study with target predictions of a background dataset comprising 3,000 compounds in total from PubChem, GDB13, and ChEMBL; a probability of 0.05 or higher was considered “as predicted” in this case.
Synthesis of the Petasis Reaction Products.
The synthesis of the Petasis reaction products was performed as described previously (16). The desired product was obtained by separation using column chromatography. All compounds were characterized entirely using advanced techniques including IR, NMR, electrospray ionization-MS, and elemental analyses. The complete characterization of the tested library is provided in SI Appendix, Supporting Information 1.
Oncogenicity Assays.
The AlamarBlue viability assay, 3D Matrigel growth assay, soft agar colony-formation assay, and foci-formation assay were performed as previously described (64–66). Whole-cell viability, apoptosis, and cytotoxicity were assessed using the ApoTox-Glo Triplex Assay Kit (Promega) according to the manufacturer’s instructions (www.promega.com/resources/protocols/?utm_source=promega&utm_medium=Vanity&utm_campaign=promegahttp://www.promega.com/tbs/). Fluorescence and luminescence were determined using a Tecan microplate reader for fluorescence (Tecan). Phosphatidylserine treatment and cell death were assessed using the Annexin-V-FLUOS Staining Kit (Life Technologies) and propidium iodide (PI)-stained cells as described previously (65, 67). Samples were analyzed using a BD FACSAria cell sorter (BD Biosciences).
SPR Analysis.
Molecular interactions were analyzed based on SPR using a BIAcore 2000 system (BIAcore AB). hBAD recombinant protein (catalog no. MBS143012; MyBioSource) was immobilized on a sensor chip as described by the manufacturer’s protocol. For the interaction of BAD with NPB, various concentrations of NPB (20–100 µM) in the running buffer were injected onto the surface of the BAD-immobilized sensor chip with a flow rate of 15 µL/min per the manufacturer’s directions. The kinetic parameters were measured using BIA evaluation software 4.1 (BIAcore AB).
Immunoblot Analysis.
WB analysis was performed as previously described (18, 64) using the following antibodies: rabbit anti-pBAD (Ser112), goat anti-pBAD (Ser136), mouse anti-BAD, mouse anti-BAK, mouse anti-BAX, mouse anti–BCL-2, mouse anti–BCL-XL, mouse anti-CDKN1A, mouse anti-CDK2, rabbit anti-CDK4, mouse anti-CYCS, and mouse anti-TP53 from Santa Cruz Biotechnology; rabbit anti-caspase 7, mouse anti-caspase 9, and rabbit anti-Ki67 from Abcam; rabbit anti-p70S6K (Ser424), rabbit anti-70S6K, rabbit anti-pAKT (Ser473), rabbit anti-AKT, rabbit anti-pBAD (Ser155), and rabbit anti–pBCL-2 (Ser70) from Cell Signaling; and a mouse anti–β-actin (Santa Cruz Biotechnology).
Proteome Profiler Array and Akt Kinase Kit.
Phospho-kinases were detected using a Western blotting array (Proteome Profiler Human Phospho-Kinase Array Kit, ARY003B; R&D Systems) according to the manufacturer’s instructions. After cell extraction using lysis buffer provided by the kit, 200 μg of protein was added per sample. Spots were analyzed using ImageJ software (https://imagej.nih.gov/ij/). For significant changes reading, a statistical cutoff for mean pixel density was set to ≥25 present.
AKT kinase activity was detected using the Akt Kinase Activity Kit (Non-Radioactive) from Enzo Life Science (catalog no. ADI-EKS-400A) according to the manufacturer’s instructions (SI Appendix, Supporting Information 4C). The intensity of color was measured in a Tecan microplate reader at 450 nm.
Molecular Docking Analysis.
We employed computational docking to validate the binding mode of NPB and other compounds to the Bcl-2/BAD interface. We docked the series of compounds to the exposed Bcl-2/BAD protein–protein interface known to bind small molecules (68). We used protonate3D of MOE (69) to prepare the NMR-derived structure of a Bcl-2 small-molecule complex (70). Afterward, we docked the compound set using MOE’s default setting and the affinity dG scoring function, aiming at a reasonable description of hydrophobic interactions. We retained the highest-scoring pose for each compound out of 30 intermediate docking runs for later comparison.
In Vivo Tumor Studies.
In vivo studies were performed according to The Guide for the Care and Use of Laboratory Animals (71), approved by Laboratory Animal Ethics Committee, at Jinan University, Guangzhou. Briefly, 5- to 6-wk-old BALB/c-nu female mice were s.c. implanted in the scruff of the neck with 17β-estradiol pellets (Innovative Research of America) at 0.72 mg per pellet with a 60-d release. After 3 d mice were injected s.c. in the right flank with 100 µL of cell suspension (1 × 107 cells). Tumor growth was monitored by measuring the tumor size using calipers. About 10 d after implantation mice were randomized and divided into three groups (each group, n = 8) according to treatment, and were administered of vehicle or 200 µL NPB [dissolved in 5% DMSO, 50% PEG400, and 45% water (pH 5.0)] by i.p. injection every day for 7 d. The first group of mice was treated with vehicle, the second was treated with a 5-mg/kg dose of NPB, and the third was treated with a 20-mg/kg dose of NPB. Animal weight and tumor volumes were measured daily. After completion, tumors were excised, photographed, weighed, and fixed or stored in liquid nitrogen for later analysis. Histological analysis was performed as previously described (64, 72, 73).
Pharmacological Features and Acute Toxicity Methodology.
The kinetic solubility of NPB was determined using multiscreen filter plates from Millipore. This method gives the kinetic solubility of NPB. Briefly, a saturated solution of NPB was prepared in universal buffer (45 mM ethanolamine, 45 mM potassium dihydrogen phosphate, 45 mM potassium acetate dissolve in Milli-Q water, pH 7.4) (1% DMSO, vol/vol), agitated for a fixed period, and filtered. The concentration of the filtrate was determined by UV spectroscopy to give an estimate of solubility. PK studies were performed by TheraIndx Lifesciences Pvt. Ltd. The animal work followed ethical practices as laid down in the guidelines for animal care (registration no. 1852/PO/Rc/S/16/CPCSEA). Approval was obtained from the Institutional Animals Ethics Committee of the test facility of Theralndx Lifesciences Pvt. Ltd. (protocol no. IAEC/01/2016/024). Briefly, the i.v. and oral dose PK of NPB (2 and 10 mg/kg in the vehicle, respectively) was characterized in male Wistar rats. The animals were anesthetized before a cannula was inserted into the jugular vein 1 d before the PK study. The animals were fasted on the day before the study (SI Appendix, Supporting Information 6–8).
Statistical Analysis.
One-way ANOVA followed by Bonferroni’s posttest correction was used to analyze acute cytotoxicity. For in vitro assays, the statistical differences among subgroup analyses were compared using an unpaired two-tailed Student t test. All analyses were done using GraphPad Prism software (version 5.0). P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This research was supported by Council of Scientific and Industrial Research Grant 02(0291)17/EMR-II; Department of Biotechnology, Ministry of Science & Technology, India Grant BT/PR/8064/BID/7/441/2013; The Cancer Science Institute of Singapore through grants from the National Research Foundation and Ministry of Education of Singapore; National Medical Research Council of Singapore Grant R-713-000-163-511; and Shenzhen Development and Reform Commission Subject Construction Project (2017)1434. P.E.L. was supported by Chinese Academy of Sciences President’s International Fellowship Initiative Grant 2015VBA031.
Footnotes
Conflict of interest statement: V.P., C.D.M., S.R., V.S., B., K.S.R., and P.E.L. are listed as inventors on a provisional patent application based on components of this work.
This article is a PNAS Direct Submission. J.W.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804897115/-/DCSupplemental.
References
- 1.Doerflinger M, Glab JA, Puthalakath H. BH3-only proteins: A 20-year stock-take. FEBS J. 2015;282:1006–1016. doi: 10.1111/febs.13190. [DOI] [PubMed] [Google Scholar]
- 2.Hayakawa J, et al. Inhibition of BAD phosphorylation either at serine 112 via extracellular signal-regulated protein kinase cascade or at serine 136 via Akt cascade sensitizes human ovarian cancer cells to cisplatin. Cancer Res. 2000;60:5988–5994. [PubMed] [Google Scholar]
- 3.Macdonald A, et al. Pim kinases phosphorylate multiple sites on Bad and promote 14-3-3 binding and dissociation from Bcl-XL. BMC Cell Biol. 2006;7:1. doi: 10.1186/1471-2121-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang X, et al. Regulation of BAD phosphorylation at serine 112 by the Ras-mitogen-activated protein kinase pathway. Oncogene. 1999;18:6635–6640. doi: 10.1038/sj.onc.1203076. [DOI] [PubMed] [Google Scholar]
- 5.Masters SC, Yang H, Datta SR, Greenberg ME, Fu H. 14-3-3 inhibits Bad-induced cell death through interaction with serine-136. Mol Pharmacol. 2001;60:1325–1331. doi: 10.1124/mol.60.6.1325. [DOI] [PubMed] [Google Scholar]
- 6.Harada H, et al. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol Cell. 1999;3:413–422. doi: 10.1016/s1097-2765(00)80469-4. [DOI] [PubMed] [Google Scholar]
- 7.Li C, Zhang G, Zhao L, Ma Z, Chen H. Metabolic reprogramming in cancer cells: Glycolysis, glutaminolysis, and Bcl-2 proteins as novel therapeutic targets for cancer. World J Surg Oncol. 2016;14:15. doi: 10.1186/s12957-016-0769-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang P, Du W, Heese K, Wu M. The Bad guy cooperates with good cop p53: Bad is transcriptionally up-regulated by p53 and forms a Bad/p53 complex at the mitochondria to induce apoptosis. Mol Cell Biol. 2006;26:9071–9082. doi: 10.1128/MCB.01025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bui NL, et al. Bad phosphorylation as a target of inhibition in oncology. Cancer Lett. 2018;415:177–186. doi: 10.1016/j.canlet.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 10.Marchion DC, et al. BAD phosphorylation determines ovarian cancer chemo-sensitivity and patient survival. Clin Cancer Res. 2011;17:6356–6366. doi: 10.1158/1078-0432.CCR-11-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sastry KSR, et al. Targeting proapoptotic protein BAD inhibits survival and self-renewal of cancer stem cells. Cell Death Differ. 2014;21:1936–1949. doi: 10.1038/cdd.2014.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perna D, et al. BRAF inhibitor resistance mediated by the AKT pathway in an oncogenic BRAF mouse melanoma model. Proc Natl Acad Sci USA. 2015;112:E536–E545. doi: 10.1073/pnas.1418163112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider G. Automating drug discovery. Nat Rev Drug Discov. 2018;17:97–113. doi: 10.1038/nrd.2017.232. [DOI] [PubMed] [Google Scholar]
- 14.Irwin JJ, Shoichet BK. ZINC–A free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novick PA, Ortiz OF, Poelman J, Abdulhay AY, Pande VS. SWEETLEAD: An in silico database of approved drugs, regulated chemicals, and herbal isolates for computer-aided drug discovery. PLoS One. 2013;8:e79568. doi: 10.1371/journal.pone.0079568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petasis NA, Zavialov IA. A new and practical synthesis of alpha-amino acids from alkenyl boronic acids. J Am Chem Soc. 1997;119:445–446. [Google Scholar]
- 17.Sharma N, et al. Inhibition of autophagy and induction of breast cancer cell death by mefloquine, an antimalarial agent. Cancer Lett. 2012;326:143–154. doi: 10.1016/j.canlet.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 18.Mohan CD, et al. Development of a novel azaspirane that targets the Janus kinase-signal transducer and activator of transcription (STAT) pathway in hepatocellular carcinoma in vitro and in vivo. J Biol Chem. 2014;289:34296–34307. doi: 10.1074/jbc.M114.601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salas TR, et al. Alleviating the suppression of glycogen synthase kinase-3beta by Akt leads to the phosphorylation of cAMP-response element-binding protein and its transactivation in intact cell nuclei. J Biol Chem. 2003;278:41338–41346. doi: 10.1074/jbc.M302972200. [DOI] [PubMed] [Google Scholar]
- 20.Harada H, Andersen JS, Mann M, Terada N, Korsmeyer SJ. p70S6 kinase signals cell survival as well as growth, inactivating the pro-apoptotic molecule BAD. Proc Natl Acad Sci USA. 2001;98:9666–9670. doi: 10.1073/pnas.171301998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seow HF, et al. Immunohistochemical detection of phospho-Akt, phospho-BAD, HER2 and oestrogen receptors alpha and beta in Malaysian breast cancer patients. Pathol Oncol Res. 2010;16:239–248. doi: 10.1007/s12253-009-9216-3. [DOI] [PubMed] [Google Scholar]
- 22.Kanamori Y, et al. Correlation between loss of PTEN expression and Akt phosphorylation in endometrial carcinoma. Clin Cancer Res. 2001;7:892–895. [PubMed] [Google Scholar]
- 23.Datta SR, Brunet A, Greenberg ME. Cellular survival: A play in three Akts. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 24.Shamas-Din A, Kale J, Leber B, Andrews DW. Mechanisms of action of Bcl-2 family proteins. Cold Spring Harb Perspect Biol. 2013;5:a008714. doi: 10.1101/cshperspect.a008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datta SR, et al. Survival factor-mediated BAD phosphorylation raises the mitochondrial threshold for apoptosis. Dev Cell. 2002;3:631–643. doi: 10.1016/s1534-5807(02)00326-x. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, et al. Rapamycin induces Bad phosphorylation in association with its resistance to human lung cancer cells. Mol Cancer Ther. 2012;11:45–56. doi: 10.1158/1535-7163.MCT-11-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tran AT, Cortens JP, Du Q, Wilkins JA, Coombs KM. Influenza virus induces apoptosis via BAD-mediated mitochondrial dysregulation. J Virol. 2013;87:1049–1060. doi: 10.1128/JVI.02017-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmelzle T, et al. Functional role and oncogene-regulated expression of the BH3-only factor Bmf in mammary epithelial anoikis and morphogenesis. Proc Natl Acad Sci USA. 2007;104:3787–3792. doi: 10.1073/pnas.0700115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernando RI, Wimalasena J. Estradiol abrogates apoptosis in breast cancer cells through inactivation of BAD: Ras-dependent nongenomic pathways requiring signaling through ERK and Akt. Mol Biol Cell. 2004;15:3266–3284. doi: 10.1091/mbc.E03-11-0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruiz R, et al. The RhoJ-BAD signaling network: An Achilles’ heel for BRAF mutant melanomas. PLoS Genet. 2017;13:e1006913. doi: 10.1371/journal.pgen.1006913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boisvert-Adamo K, Aplin AE. Mutant B-RAF mediates resistance to anoikis via Bad and Bim. Oncogene. 2008;27:3301–3312. doi: 10.1038/sj.onc.1211003. [DOI] [PubMed] [Google Scholar]
- 32.Gilmore AP, et al. Activation of BAD by therapeutic inhibition of epidermal growth factor receptor and transactivation by insulin-like growth factor receptor. J Biol Chem. 2002;277:27643–27650. doi: 10.1074/jbc.M108863200. [DOI] [PubMed] [Google Scholar]
- 33.Ranger AM, et al. Bad-deficient mice develop diffuse large B cell lymphoma. Proc Natl Acad Sci USA. 2003;100:9324–9329. doi: 10.1073/pnas.1533446100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Letai A, et al. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 35.Ma Y, Qin H, Cui Y. MiR-34a targets GAS1 to promote cell proliferation and inhibit apoptosis in papillary thyroid carcinoma via PI3K/Akt/Bad pathway. Biochem Biophys Res Commun. 2013;441:958–963. doi: 10.1016/j.bbrc.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 36.Gu Z, et al. Polyunsaturated fatty acids affect the localization and signaling of PIP3/AKT in prostate cancer cells. Carcinogenesis. 2013;34:1968–1975. doi: 10.1093/carcin/bgt147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng SF, et al. Curcumin-loaded nanoparticles enhance apoptotic cell death of U2OS human osteosarcoma cells through the Akt-Bad signaling pathway. Int J Oncol. 2014;44:238–246. doi: 10.3892/ijo.2013.2175. [DOI] [PubMed] [Google Scholar]
- 38.Akl H, et al. HTLV-I infection of WE17/10 CD4+ cell line leads to progressive alteration of Ca2+ influx that eventually results in loss of CD7 expression and activation of an antiapoptotic pathway involving AKT and BAD which paves the way for malignant transformation. Leukemia. 2007;21:788–796, and erratum (2009) 23:2187. doi: 10.1038/sj.leu.2404585. [DOI] [PubMed] [Google Scholar]
- 39.Tokunaga E, et al. Activation of PI3K/Akt signaling and hormone resistance in breast cancer. Breast Cancer. 2006;13:137–144. doi: 10.2325/jbcs.13.137. [DOI] [PubMed] [Google Scholar]
- 40.Anonymous AKT inhibitors take steps forward. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-NB2013-053. [DOI] [Google Scholar]
- 41.Vivanco I, et al. A kinase-independent function of AKT promotes cancer cell survival. eLife. 2014;3:03751. doi: 10.7554/eLife.03751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meuillet EJ. Novel inhibitors of AKT: Assessment of a different approach targeting the pleckstrin homology domain. Curr Med Chem. 2011;18:2727–2742. doi: 10.2174/092986711796011292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogler M. Targeting BCL2-proteins for the treatment of solid tumours. Adv Med. 2014;2014:943648. doi: 10.1155/2014/943648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mustata G, et al. Development of small-molecule PUMA inhibitors for mitigating radiation-induced cell death. Curr Top Med Chem. 2011;11:281–290. doi: 10.2174/156802611794072641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masood A, Azmi AS, Mohammad RM. Small molecule inhibitors of bcl-2 family proteins for pancreatic cancer therapy. Cancers (Basel) 2011;3:1527–1549. doi: 10.3390/cancers3021527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colak S, et al. Decreased mitochondrial priming determines chemoresistance of colon cancer stem cells. Cell Death Differ. 2014;21:1170–1177. doi: 10.1038/cdd.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenquist M. 14-3-3 proteins in apoptosis. Braz J Med Biol Res. 2003;36:403–408. doi: 10.1590/s0100-879x2003000400001. [DOI] [PubMed] [Google Scholar]
- 48.Polzien L, Baljuls A, Albrecht M, Hekman M, Rapp UR. BAD contributes to RAF-mediated proliferation and cooperates with B-RAF-V600E in cancer signaling. J Biol Chem. 2011;286:17934–17944. doi: 10.1074/jbc.M110.177345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye DZ, Jin S, Zhuo Y, Field J. p21-activated kinase 1 (Pak1) phosphorylates BAD directly at serine 111 in vitro and indirectly through Raf-1 at serine 112. PLoS One. 2011;6:e27637. doi: 10.1371/journal.pone.0027637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hojabrpour P, Waissbluth I, Ghaffari M, Cox ME, Duronio V. CaMKII-γ mediates phosphorylation of BAD at Ser170 to regulate cytokine-dependent survival and proliferation. Biochem J. 2012;442:139–149. doi: 10.1042/BJ20111256. [DOI] [PubMed] [Google Scholar]
- 51.Stickles XB, et al. BAD-mediated apoptotic pathway is associated with human cancer development. Int J Mol Med. 2015;35:1081–1087. doi: 10.3892/ijmm.2015.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oakes SR, et al. Sensitization of BCL-2-expressing breast tumors to chemotherapy by the BH3 mimetic ABT-737. Proc Natl Acad Sci USA. 2012;109:2766–2771. doi: 10.1073/pnas.1104778108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaillant F, et al. Targeting BCL-2 with the BH3 mimetic ABT-199 in estrogen receptor-positive breast cancer. Cancer Cell. 2013;24:120–129. doi: 10.1016/j.ccr.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 54.Deng J, Letai A. Priming BCL-2 to kill: The combination therapy of tamoxifen and ABT-199 in ER+ breast cancer. Breast Cancer Res. 2013;15:317. doi: 10.1186/bcr3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamaki H, et al. Bcl-2 family inhibition sensitizes human prostate cancer cells to docetaxel and promotes unexpected apoptosis under caspase-9 inhibition. Oncotarget. 2014;5:11399–11412. doi: 10.18632/oncotarget.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Musiani D, et al. PIM2 kinase is induced by cisplatin in ovarian cancer cells and limits drug efficacy. J Proteome Res. 2014;13:4970–4982. doi: 10.1021/pr500651n. [DOI] [PubMed] [Google Scholar]
- 57.Mason KD, et al. Programmed anuclear cell death delimits platelet life span. Cell. 2007;128:1173–1186. doi: 10.1016/j.cell.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 58.Danial NN, et al. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature. 2003;424:952–956. doi: 10.1038/nature01825. [DOI] [PubMed] [Google Scholar]
- 59.Yu Y, et al. Targeting AKT1-E17K and the PI3K/AKT pathway with an allosteric AKT inhibitor, ARQ 092. PLoS One. 2015;10:e0140479. doi: 10.1371/journal.pone.0140479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saleh M, et al. 2013. First in human study with ARQ 092, a novel pan AKT-inhibitor: Results from the advanced solid tumor cohorts. American Association for Cancer Research Annual Meet LB-197.
- 61.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lowe R, Mussa HY, Nigsch F, Glen RC, Mitchell JB. Predicting the mechanism of phospholipidosis. J Cheminform. 2012;4:2. doi: 10.1186/1758-2946-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gaulton A, et al. ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pandey V, et al. Trefoil factor 3 promotes metastatic seeding and predicts poor survival outcome of patients with mammary carcinoma. Breast Cancer Res. 2014;16:429. doi: 10.1186/s13058-014-0429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sebastian A, et al. Novel adamantanyl-based thiadiazolyl pyrazoles targeting EGFR in triple-negative breast cancer. ACS Omega. 2016;1:1412–1424. doi: 10.1021/acsomega.6b00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pandey V, et al. Hypomethylation associated enhanced transcription of trefoil factor-3 mediates tamoxifen-stimulated oncogenicity of ER+ endometrial carcinoma cells. Oncotarget. 2017;8:77268–77291. doi: 10.18632/oncotarget.20461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sulaiman NBS, et al. An azaspirane derivative suppresses growth and induces apoptosis of ER-positive and ER-negative breast cancer cells through the modulation of JAK2/STAT3 signaling pathway. Int J Oncol. 2016;49:1221–1229. doi: 10.3892/ijo.2016.3615. [DOI] [PubMed] [Google Scholar]
- 68.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 69. Molecular Operating Environment (MOE) (2018) New in MOE 2013.08 (Chemical Computing Group ULC, Montreal), Version 2013.08.
- 70.Bruncko M, et al. Studies leading to potent, dual inhibitors of Bcl-2 and Bcl-xL. J Med Chem. 2007;50:641–662. doi: 10.1021/jm061152t. [DOI] [PubMed] [Google Scholar]
- 71.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
- 72.Wang XN, et al. Trefoil factor 3 as a novel biomarker to distinguish between adenocarcinoma and squamous cell carcinoma. Medicine (Baltimore) 2015;94:e860. doi: 10.1097/MD.0000000000000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.You ML, et al. Trefoil factor 3 mediation of oncogenicity and chemoresistance in hepatocellular carcinoma is AKT-BCL-2 dependent. Oncotarget. 2017;8:39323–39344. doi: 10.18632/oncotarget.16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Petros AM, et al. Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies. Protein Sci. 2000;9:2528–2534. doi: 10.1110/ps.9.12.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.