Significance
The shielding strategy for self-resistance in antibiotic biosynthesis from bacteria has been known to employ reversible group-transfer reactions that usually do not directly act on the antibiotic pharmacophore. Typically, the phosphorylation-modified, glycosylation-modified, acetylation-modified, and prepeptide-modified prodrug need to be activated by removing the protection group during or following export and, thereby, employing the hydrolysis reaction as the final step. Herein, we discover an unprecedented oxidative activation and overoxidative inactivation of a matured prodrug; significantly, the oxidation reaction directly deals with the drug warhead and occurs outside the host cells. This cryptic self-resistance mechanism sheds light on antibiotic resistance and the complex biology of extracellular environment.
Keywords: self-protection, secreted enzyme, tetrahydroisoquinoline alkaloids, warhead, natural product biosynthesis
Abstract
Understanding how antibiotic-producing bacteria deal with highly reactive chemicals will ultimately guide therapeutic strategies to combat the increasing clinical resistance crisis. Here, we uncovered a distinctive self-defense strategy featured by a secreted oxidoreductase NapU to perform extracellularly oxidative activation and conditionally overoxidative inactivation of a matured prodrug in naphthyridinomycin (NDM) biosynthesis from Streptomyces lusitanus NRRL 8034. It was suggested that formation of NDM first involves a nonribosomal peptide synthetase assembly line to generate a prodrug. After exclusion and prodrug maturation, we identified a pharmacophore-inactivated intermediate, which required reactivation by NapU via oxidative C-H bond functionalization extracellularly to afford NDM. Beyond that, NapU could further oxidatively inactivate the NDM pharmacophore to avoid self-cytotoxicity if they coexist longer than necessary. This discovery represents an amalgamation of sophisticatedly temporal and spatial shielding mode conferring self-resistance in antibiotic biosynthesis from Gram-positive bacteria.
Antibiotic resistance has sparked extraordinary concerns because it is becoming one of the major crises for global health, which calls for not only the perpetual generation of new antibiotics but also detailed understanding of resistance mechanisms (1–3). In general, antibiotic modification, target modification or duplication, and antibiotic efflux are canonical strategies coevolved with the biosynthetic capabilities by antibiotic-producing microbes (4, 5). Similarly, such detoxification strategies have also been exploited by antibiotic-resistant strain of pathogenic bacteria presumptively via resistance gene transfer (6, 7). In rare cases, some special mechanisms, exemplified by self-sacrifice resistance proteins and a damaged base excision repair system, have been found in the biosynthesis of DNA-targeting antibiotics, calicheamicin and yatakemycin, respectively, for self-protection (8, 9). Recently, a prodrug activation strategy, in which an N-terminal capped prepeptide is biogenerated and undergoes a final, enzyme-hydrolytic cap-removing step during or following export, is identified in the biosynthesis of some nonribosomal peptides to avoid self-destruction of the producers (10). Therefore, elucidation of more underlying self-resistance mechanisms from the antibiotic biosynthetic gene clusters will not only direct the discovery of microbial natural products with biological activities (11–14), but also provide key clues for target identification and mode of action/resistance studies of these compounds (14–16).
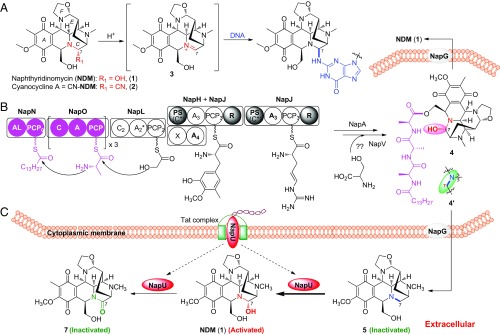
Naphthyridinomycin (NDM, 1) and cyanocycline (2, Fig. 1A), isolated from Streptomyces, belong to a family of tetrahydroisoquinoline (THIQ) alkaloids and exhibit strong antitumor and antimicrobial activities, extraordinarily effective against Gram-positive bacteria, even methicillin-resistant Staphylococcus aureus (17, 18). Their biological activities are originated from the highly functionalized hexacyclic framework, especially the labile hemiaminal functional group in C ring. The elimination of C-7 hydroxyl or cyano group results in formation of a highly reactive, electrophilic iminium species 3, which can efficiently alkylate DNA at the N-2 residue of guanine in the GC-rich region located in the minor groove (Fig. 1A) (18). Recently, biosynthetic studies of NDM have established a nonribosomal peptide synthetase (NRPS) platform that features not only using nonproteinogenic amino acid building blocks as precursors but also employing a leader peptide/prodrug activation mechanism (19–22). In this model, prodrug 4 was suggested, which underwent hydrolytic cap removal by a membrane protease NapG following export to yield the end product 1 (Fig. 1B). Preceding studies have assigned the functions of most genes in the biosynthetic gene cluster. However, napU, encoding a FAD-binding oxidoreductase, originally proposed as a tailoring enzyme, seems not necessary for the biosynthesis. Herein, we uncover a cryptically physiological role of this enzyme. It catalyzes an extracellular C-H bond functionalization for oxidative activation and overoxidative inactivation of the NDM matured prodrug, which constitutes a distinctively noncanonical self-resistance strategy to protect the producers from self-harm during the whole biosynthetic process.
Fig. 1.
Activation and biosynthesis of NDM. (A) Proposed mechanism of DNA alkylation. (B) Previously suggested biosynthetic pathway of NDM. (C) The revised pathway including extracellular activation and overoxidative inactivation catalyzed by NapU in NDM biosynthesis.
Results
Genetic Investigation of napU Revealing Its Essential Role in NDM Biosynthesis.
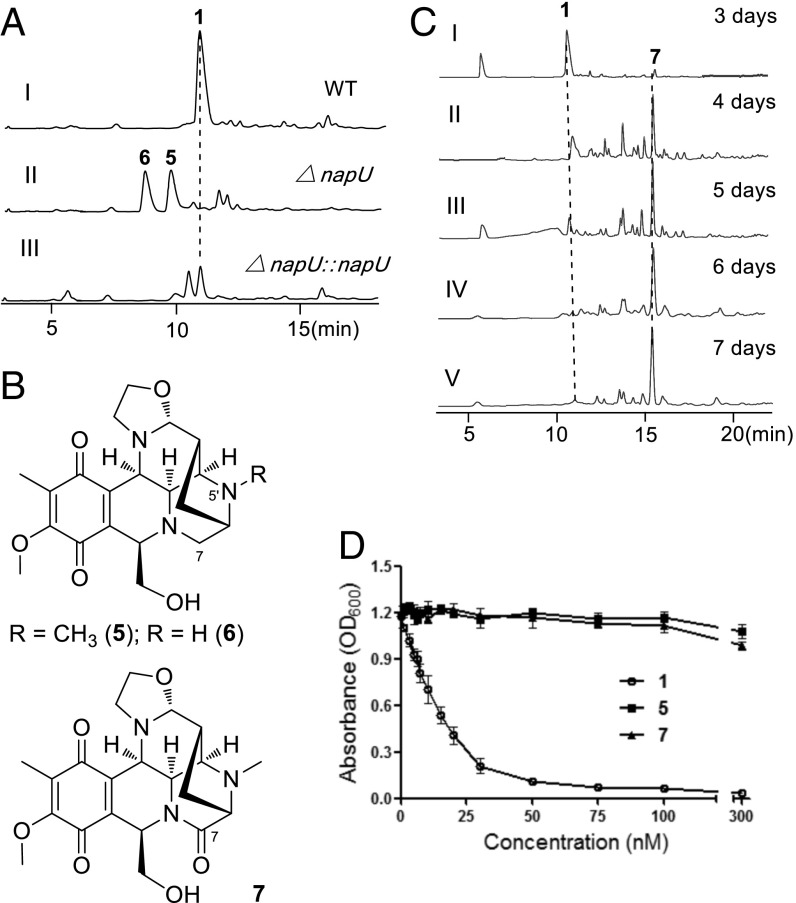
Our initial effort to explore the relationship between napU and NDM biosynthesis was first performed by inactivation of this gene in vivo (SI Appendix, Fig. S1). This gene replacement mutant strain did not produce NDM anymore but generated two metabolites showing an [M+H]+ ion with m/z of 402 (5) and 388 (6) respectively; and gene complementation of this mutant could restore the production of NDM (Fig. 2 A, II and III). Afterward, this mutant was further fermented, the compounds were isolated, and their structures were elucidated by NMR analysis and compared with NDM (SI Appendix, Figs. S2 and S3 and Table S3): The C-7 hydroxyl group of NDM is lost in compound 5; meanwhile, the N-methyl group is also missing in compound 6 (Fig. 2B).
Fig. 2.
Genetic characterization of napU in NDM biosynthesis. (A) Genetic investigation of napU in vivo. HPLC analysis with UV detected at 270 nm S. lusitanus NRRL 8034 (WT) (I); mutant S. lusitanus ΔnapU (II); complementary mutant S. lusitanus ΔnapU::napU (III). (B) Chemical structures of new NDM analogs. (C) Analysis of the metabolites produced by WT strain with different fermentation time. (D) Antibacterial activity of NDM (1) and new analogs (5 and 7) using E. coli DH5α as assay strain cultured in liquid media.
The isolation of C-7 dehydroxyl analogs from the ΔnapU mutant suggests that NapU is involved in NDM biosynthesis and possibly catalyzes the hydroxylation of C-7 using 5 as a substrate. Given that the hemiaminal structure is the key active pharmacophore, the disappearance of C-7 functional group likely means loss of the DNA alkylating activity. As expected, compound 5 shows no antibacterial activity when Escherichia coli is used as the test strain (Fig. 2D and SI Appendix, Fig. S4). Therefore, we surmise that the installation of a hydroxyl group at C-7 position is probably catalyzed by NapU, which may regenerate the pharmacophore and likely occurs as the final step in NDM biosynthesis.
Biochemical Characterization of NapU as a Secreted Flavoprotein.
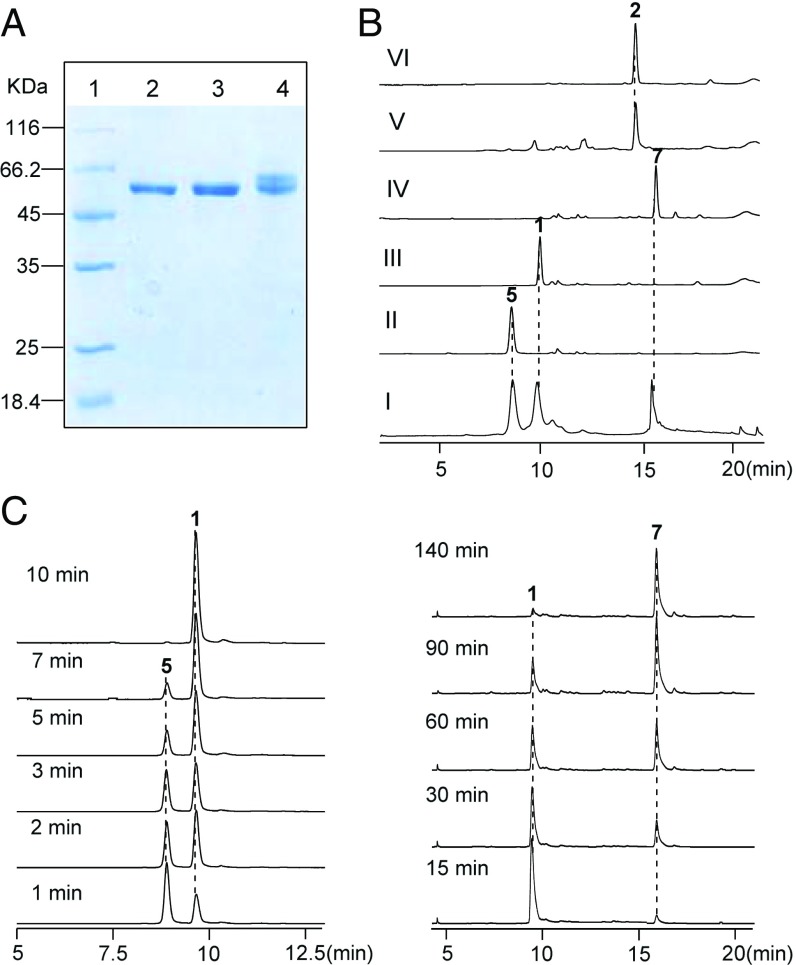
NapU bears conserved residues (i.e., the invariant His and Cys) for the covalent binding of FAD, as well as a berberine bridge enzyme domain (SI Appendix, Fig. S5). It exhibits high sequence similarity to TamL (46% identity) and AknOx (47% identity), which catalyze two-electron oxidation of hydroxyl to ketone and four-electron oxidation of rhodinose to l-aculose, respectively (23, 24). To further investigate the enzymatic function, the recombinant NapU was purified as a C-His8–tagged fusion protein from E. coli, which exhibits a yellow color and displays a UV-Vis absorption spectrum consistent with typical flavoproteins (SI Appendix, Fig. S6), but surprisingly affords two bands in SDS/PAGE analysis (Fig. 3A). Further bioinformatic analysis shows that NapU contains a 34-amino acid, N-terminal signal peptide sequence (SI Appendix, Fig. S5), which belongs to the twin-arginine translocation (Tat) signal peptides with conserved RR—AXA/G (25). Previous studies have established that the Tat system is a major and general route of protein export in S. coelicolor (26). Additionally, we recently identified a secreted oxidoreductase SfmCy2 bearing a similar Tat signal peptide, which catalyzes an oxidative deamination extracellularly in saframycin A biosynthesis from S. lavendulae (27). These collective evidences facilitate us to speculate that: (i) NapU might be a secreted oxidase to catalyze an extracellular hydroxylation reaction; (ii) the two bands in SDS/PAGE should be the full and N signal-cleaved variants. To verify these hypotheses, napU was cloned into a Streptomyces-E. coli shuttle vector under the control of thiostrepton-inducible promoter and then expressed in S. lividans 1326. After induction, the cultures were centrifuged to remove the cells and the resulting supernatants were incubated with Ni-NTA affinity resin for purifying the secreted C-terminal His-tagged fusion proteins, which indeed yields only one band by SDS/PAGE analysis (Fig. 3A). Importantly, the N-terminal amino acid analysis gave an A-G-K-N-T sequence (SI Appendix, Fig. S7), which unequivocally proved the signal peptide was actually cleaved between Ala-34 and Ala-35. We therefore expressed another truncated variant of this enzyme in E. coli by deletion of the N-terminal signal peptide sequence, the resulting protein indeed showed one band as the same as that purified from supernatants of S. lividans (Fig. 3A). Taken together, we conclude that NapU is a secreted flavoenzyme and likely conducts an extracellular reaction.
Fig. 3.
Biochemical characterization of FAD-binding oxidoreductase NapU in vitro. (A) SDS/PAGE (12%) analysis of the recombinant C-His8–tagged NapU. Lane 1, protein marker; lane 2, full length of NapU purified from S. lividans 1326; lane 3, truncated NapU by removing the N-terminal signal peptide purified from E. coli BL21(DE3); lane 4, full length of NapU purified from E. coli BL21(DE3). (B) HPLC analysis of NapU-catalyzed reaction: mixture of 1, 5, and 7 (I); control reaction with heat-inactivated NapU (II); full reaction for 10 min (III); reaction for 3 h (IV); derivation of reaction product (10 min) by KCN (V); standard of 2 (VI). (C) HPLC analysis of NapU-mediated reaction using 5 as substrate with different reaction time.
Biochemical Elucidation of NapU-Catalyzed Reactions.
With recombinant NapU in hand, we next set out to test the hydroxylation reaction using compound 5 as a substrate. We found that 5 could be efficiently converted into NDM (1) by LC-MS analysis, which was further confirmed by derivation using potassium cyanide to give 2 (Fig. 3B). Interestingly, this enzymatic reaction yielded another product 7 with the elongation of reaction time (Fig. 3B). Further investigation of the time-course reaction showed that NapU completely transformed 5 into 1 in 10 min; subsequently, this enzyme continued to catalyze another reaction using 1 as substrate to afford another product 7 with a decreased molecular mass by 2 Da less than 1 (Fig. 3C). Inspired by this observation, we are wondering whether the similar biotransformation occurs in vivo during the biosynthetic process. Indeed, the titer of 1 was decreased, and meantime 7 was increased with the extension of fermentation time; 1 was converted to 7 almost completely after the fermentation lasting up to 7 d (Fig. 2C). Despite instability, 7 was further isolated and elucidated by NMR and compared with compound 2 (SI Appendix, Fig. S8 and Table S3), in which the C-7 hydroxyl group is further oxidized into ketone group (Fig. 2B). Intriguingly, this further overoxidation by NapU suggests the pharmacophore is inactivated again. Subsequent biological activity assay indeed showed that compound 7 also lost its antibacterial activity compared with 1 (Fig. 2D and SI Appendix, Fig. S4). Enzymatic kinetics analyses indicated that NapU exhibits higher catalytic efficiency for the conversion of 5 to 1 (kcat/KM = 1.43 × 104 M−1·s−1) than that of 1 to 7 (kcat/KM = 36.48 M−1·s−1) as well as higher substrate affinity toward 5 [KM (5) = 48.89 ± 6.02 μM, KM (1) = 82.23 ± 5.06 μM] (SI Appendix, Fig. S9). These kinetic properties suggest that NapU functions as a biosynthetic role to catalyze the oxidation of 5 to 1, as well as an antibiotic resistance protein to perform detoxification by overoxidation of 1 when accumulating too much before diffusing in case of self-harmness.
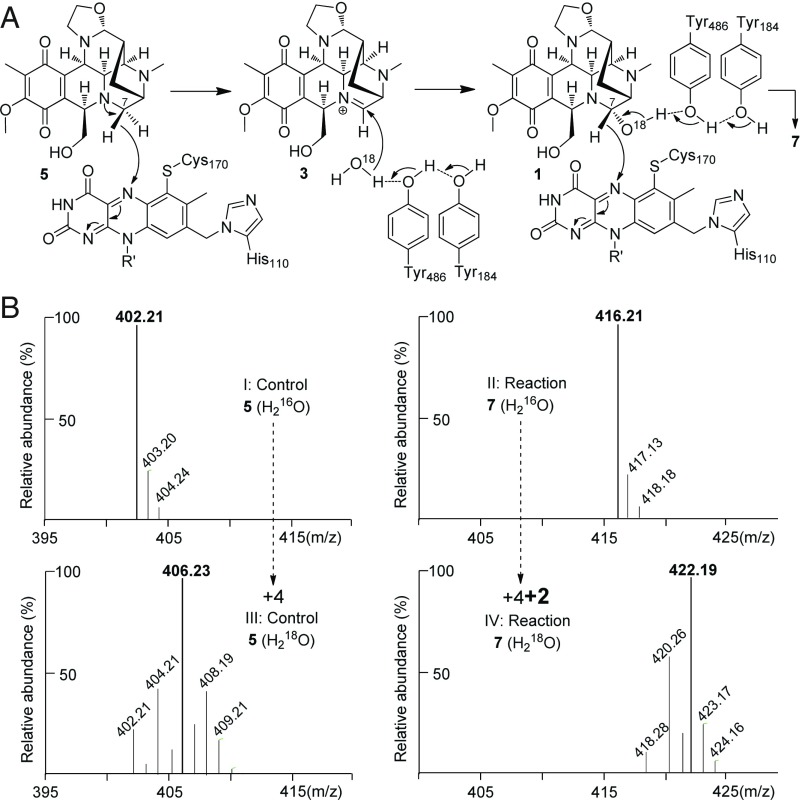
Collectively, we now have illustrated that NapU not only catalyzes the hydroxylation of 5 to give NDM, but it also could further mediate a four-electron oxidation of 5 into 7 (Fig. 1C). The first two-electron oxidative C-H bond functionalization step possibly proceeds through the reactive iminium intermediate 3. Therefore, following the well-studied TamL and AknOx (23, 24), we proposed an enzymatic mechanism: hydride transfer from C-7 to the covalent 8α-histidyl-6-S-cysteinyl-FAD results in formation of intermediate 3; subsequently, H2O as a nucleophilic reagent, which may be activated by Tyr184 and Tyr486 (multiple sequence alignment shows they may serve as active sites, see SI Appendix, Fig. S10), attacks the iminium group to form 1; another round of oxidation affords the ketone compound 7 (Fig. 4A). The subsequent site-directed mutation studies confirmed that these amino acid residues are critical for the enzymatic activity: Either of the mutant H110A or C170A, both of which are conserved for FAD covalent binding, completely lost the biological activity, while the Y184F or Y486F mutant dramatically reduced reaction efficiency of both steps (SI Appendix, Fig. S10). Then we performed this enzyme reaction in H218O buffer. The final enzymatic product 7 exhibited an additional increase in molecular mass by 2 Da compared with that formed in H216O buffer (Fig. 4B). Therefore, these data provided validation that the ketone oxygen atom originates from H2O, which supports our proposed mechanism.
Fig. 4.
Enzymatic mechanism investigation of NapU-catalyzed reaction. (A) Proposed mechanism. (B) MS analysis of enzymatic products in H216O (I, control; II, reaction) and H218O (III, control; IV, reaction) after 3 h.
Extension of the Biosynthetic Pathway and Establishment of the Cryptic Self-Resistance Model.
The biosynthetic gene cluster encodes a homolog of major facilitator subfamily transporter (NapR4) and a UV-repair protein NapR1, both of which were assumed to confer self-resistance (20). Herein, combining genetic characterization and biological investigation, we discovered a cryptic self-defense strategy during the biosynthesis of NDM in S. lusitanus NRRL 8034. This specific self-defense strategy was evolved by integrating with the pathway to ensure the survival of S. lusitanus during NDM biosynthesis, which includes a key tailoring step via oxidative reactivation of a warhead-blocked intermediate occurring outside the cells. A fully modified and harmless prodrug 4′ is first generated by NRPSs, followed by tailoring enzymes in the cytoplasm. Next, 4′ is pumped out of cells and maturated through the hydrolysis of the ester bond to remove the N-terminal peptides by a membrane-located peptidase NapG to yield 5. Finally, the matured but still inactivated intermediate 5 will be reactivated via extracellularly oxidative C-H bond functionalization catalyzed by secreted oxidase NapU to afford NDM (Fig. 1C). Once the activated antibiotic NDM accumulated long enough with NapU before it is effectively diffused far away from the host cell, the enzyme will catalyze an additional oxidation of the hemiaminal pharmacophore into an inactive lactam derivative to avoid self-toxicity (Fig. 1C).
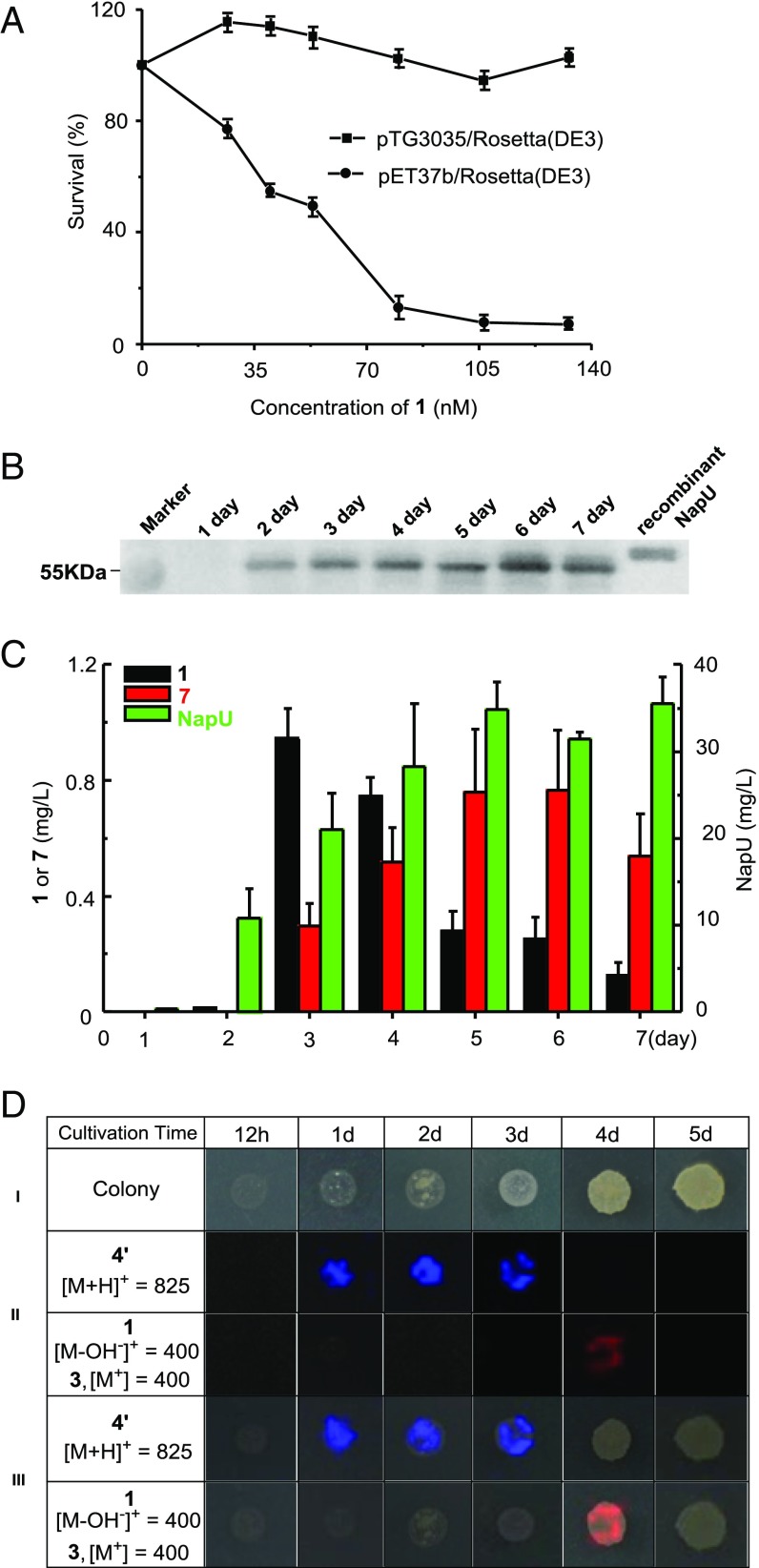
To support this proposed model, we next tested the effect of NapU against 1 by heterologous expression of napU in E. coli Rosetta (DE3) strains. The bacterial growth curve indicated that in vivo NapU indeed confers better resistance to NDM (Fig. 5A). Subsequently, we decided to probe whether the host-producing NapU is secreted as an extracellular protein (predicted size: 55.0 kDa) in a physiological condition using polyclonal antibody against the recombinant truncated NapU (56.8 kDa). Western blot analysis of the supernatants shows that NapU was obviously produced from day 2 to day 7 under the fermentation conditions (Fig. 5B). The supernatants were also collected and analyzed by proteomic approach, which further confirmed that NapU is secreted out of cells with the yield increased from day 1 to day 5 (Fig. 5C). Moreover, the relationship between the production of extracellular NapU and compounds 1 with 7 were monitored and summarized in Fig. 5C. Compound 1 was produced slightly on the second day and the yield reached its maximum value on the third day, then decreased from day 3 to day 7. The yield of compound 7 increased continuously from day 3 to day 6 and decreased in day 7 because of its instability. These data illuminated that the biotransformation in vivo is consistent with that in vitro, and the timing of 1/7 production is well corresponding to the appearance of NapU extracellularly. Additionally, we explored the in situ production of these related metabolites by MALDI-TOF imaging MS (28–30). We interrogated S. lusitanus colonies and monitored the compounds 4′ and 1/3 over a time from 12 h to 5 d, albeit the production on solid medium could not be detected by HPLC analysis. The visualization of the chemical output strikingly showed that the prodrug 4′ is generated during the early growth phase, and this intermediate disappears with the emergence of NDM by day 4 (Fig. 5D). Due to low production and instability of 1 and 7, we could not detect their signals in day 5. The temporal and spatial distribution of the related intermediates is consistent with the proposed model (Fig. 1C) in which the biosynthetic intermediate is indeed inactivated by deficiency of warhead and the end product is reactivated finally.
Fig. 5.
The function relationship between NapU and the NDM relative metabolites in physiological conditions. (A) The effect of NapU against 1 using E. coli Rosetta (DE3) harboring the expression plasmid pTG3035 or an empty vector as test strains at 18 °C for 72 h. (B) Western blot for detecting natural NapU secreted in fermental supernatants. (C) Time-course for production of compounds 1, 7, and secreted protein NapU. Data are mean ± SD; n = 3 independent experiments. (D) Time-course production of NDM-relevant metabolites detected by MALDI-imaging MS. (I) Photograph of the microorganisms grown on ISP-2 in a Petri dish (diameter 90 mm). (II) Spatial and temporal distribution of the m/z signals of compound 4′ and 1/3. In these images, the most abundant ions are shown with a 1-Da window. (III) Overlapped images of the chemical signal and microorganism colony growth.
Discussion
A particularly significant feature of this self-resistance mechanism in NDM biosynthesis is the space-time shielding mode employing oxidation reaction to handle a pharmacophore. In fact, the cellular or subcellular trafficking models have been occasionally adopted to biosynthesize natural products with different biological activity in multicellular eukaryotes, such as plants and filamentous fungi (31, 32). In those cases, this trafficking highway that compartmentalizes the biosynthetic steps not only contributes to avoiding self-harm but also guarantees to deliver specific compounds at the right time to the right tissue for physiological requirement. However, the similar space-time shielding pattern is seldom observed in bacteria. In the biosynthesis of some aminoglycoside and macrolide antibiotics, phosphorylation and glycosylation of compounds have been evaluated for host cell immunity, and final products are activated during or following export by dephosphorylation and deglycosylation via hydrolysis (5, 33). These simple metabolic shielding strategies are basically similar to the recently characterized prodrug maturation pathway involved in some NRPS systems: The prodrug bearing a protection group is employed to prevent binding to the responding cellular target inside the cell. Moreover, these modifications are limited by group-transfer reactions and have never touched the warhead. In addition, an enzyme-hydrolytic reaction is used to remove the protection group during or after secretion, which is the last step for the pathway. In the NDM pathway, the producer has evolved a sophisticated space-time shielding mode through adopting an extracellular oxidation reaction catalyzed by a secreted oxidoreductase. Aside from atypical enzymatic steps beyond known group-transfer reactions, this self-resistance mechanism is distinctive from other bacterial systems. Overall, this space-time shielding pattern is highly hybrid with the prodrug NRPS system, while the prepeptide seems essential to the NRPS-mediated reactions and export, and not only contributes to the self-resistance. Additionally, the prodrug maturation via hydrolysis during or following export is just one of the tailoring steps, which produce a fully modified, but still inactive, intermediate out of the cell remaining to be further activated. Finally, the extracellularly oxidative reaction not only reinstalls the warhead back to the intermediate to afford the final active antibiotic, but also contributes to balance the concentration of antibiotic around the host cell. More importantly, the comprehensive, multilevel self-resistance system is consummately integrated with the biosynthetic steps spatially, which might minimize the potential damage caused by the endogenous antibiotics and accurately control the efficiency of biosynthetic machinery.
In summary, we have discovered a distinctive self-resistance in NDM biosynthesis representing an unprecedented amalgamation of prodrug mechanism and space-time shielding mode in Streptomyces. With the increasing demand to access new antibiotics and understand the antibiotic resistance, the biosynthetic studies in antibiotic-producing microorganisms provide a reservoir for not only upgrading or mining antibiotics but also discovering physiological antagonistic effects or self-resistance mechanisms. Given the fact that THIQ antibiotics alkylate DNA via the similar electrophilic iminium species (18), it will be interesting to investigate whether such a resistance mechanism is also employed in the biosynthesis of other members. Additionally, considering the potentially clinical application of this family of antibiotics (22, 34), understanding of the activation-inactivation mechanism will be critical for dealing with the potentially clinical resistance problems.
Materials and Methods
General materials, gene inactivation/complementation, fermentation, production and analysis of NDM related intermediates, protein expression, and purification are all following previous methods (19, 20, 27) except using different plasmids, strains (SI Appendix, Table S1), and PCR primers (SI Appendix, Table S2). Additional SI includes three tables (SI Appendix, Tables S1–S3) and 10 figures (SI Appendix, Figs. S1–S10).
Purification of Compound 5 and 6 from the ΔnapU Mutant.
Four liters of fermentation broth of S. lusitanus ΔnapU was filtered and extracted with methylene chloride (4 L × 3 times), and the combined methylene chloride extracts were evaporated under reduced pressure to afford the crude extract. The crude extract was subjected to separation by column chromatography (silica gel). Compound 5 (20 mg) and a crude fraction were thus obtained by eluting with MeOH/EtOAc gradient (0–30% MeOH in EA). Further separating the crude fraction by using preparative TLC under the elution condition of Acetone/Methol = 3:1 resulted in the purification of Compound 6 (15 mg). Compound 5 was isolated as an orange red amorphous powder: 1H and 13C NMR data (in methanol-d4, SI Appendix, Table S3). HR-ESI-MS (positive mode) m/z calculated for C21H27N3O5: 402.2023 [M+H]+, found 402.2018. Compound 6 was also isolated as an orange red amorphous powder: 1H and 13C NMR data (in chloroform-d3, SI Appendix, Table S3). HR-ESI-MS m/z for C20H25N3O5: calculated 388.1867 [M+H]+, found 388.1868.
Enzymatic Reaction and Purification of Compound 7.
Standard assays were carried out in 50 μL of reaction mixture containing 50 mM NaH2PO4-NaOH (pH 8.5), 1.0 μM NapU, and 100 μM substrate 5. For production of 7, 150 mL of reaction containing 200 μM 5 and 10 μM NapU was incubated at 30 °C for 3 h, monitored by HPLC. The mixture was added to MeOH (300 mL) and subsequently centrifuged at 4,000 × g for 20 min to remove the enzyme. The supernatant was evaporated and then extracted with CH2Cl2 (200 mL × 3). The combined organic phase was evaporated and purified by preparative TLC (CHCl3/MeOH = 10:1) resulted in the purification of compound 7 (3 mg). It was a bright yellow amorphous powder: 1H and 13C NMR data (in methanol-d4, SI Appendix, Table S3). HR-ESI-MS m/z for C21H25N2O6: calculated 416.1816 [M+H]+, found 416.1811.
Bacterial Inhibition Assay of Compounds.
The solutions containing either 100 μM, 10 μM, or 1 μM compound 1 (fresh prepared by NapU-catalyzed reaction using 5 as substrate), 5, and 7 were prepared. Two microliters of solutions were dropped onto solid LB agar plate applied with 400 μL of E. coli DH5α liquid culture grown overnight from a single colony, respectively. The plate was incubated overnight at 37 °C, and the zones of inhibition were subsequently observed. Further, the solutions were applied to test tubes to a final concentration of 1, 3, 5, 7, 10, 15, 20, 30, 50, 75, 100, or 300 nM. These tubes contained 3 mL of liquid LB and 1% (v%v) of E. coli DH5α liquid culture grown overnight from a single colony. The tubes were incubated 6 h at 37 °C. Growth in tubes containing compound 1 or 5 or 7 was compared with control (no antibiotic). To test the effect of NapU against 1 in vivo, the E. coli Rosetta (DE3) cells were transferred to a 18 °C incubator for another 12 h after induction by IPTG. Then, the culture was inoculated to several test tubes, which individually contained 3 mL of LB with 100 μM IPTG. Different concentrations of 1 was added into test tubes.
Western Blot Analysis of Natural NapU.
The truncated NapU with the C-terminal His-tagged fusion was delivered to ABclonal Technology Company for preparing the rabbit polyclonal antibody as primary antibody. One hundred milliliters of the fermental supernatant was concentrated to 1 mL. One microliter of the concentrated solution was separated by SDS/PAGE, transferred to a nitrocellulose membrane and then blocked with 5% nonfat milk in TBST (100 mM Tris⋅HCl, 150 mM NaCl, 0.1% Tween 20, pH 7.4) buffer for 1 h at room temperature (RT). The membrane was incubated with primary antibody at 1:500 dilution in dilution buffer (ABclonal Technology Company) for 1 h, and after washing for five times with TBST for a total of 1 h, the secondary antibody conjugated with horseradish peroxidase at 1:5,000 dilution for 1 h at RT and washed for five times with TBST. The blots were visualized with the Thermo Scientific ECL Western blotting detection reagents.
MALDI-TOF Imaging MS Analysis of Production of NDM-Related Compounds.
The IMS approach directly measure microbial inoculum on solid media using MALDI-TOF imaging mass spectrometer. ITO-coated glass slide was sterilized in 1% NaClO for 1 h and buried with 17 mL of ISP-2 media in 90-mm Petri dish. The plates were inoculated with a 10-μL spore suspension (∼108) of S. lusitanus NRRL 8034 and incubated at 25 °C. The slides were removed from the plates at 12 h and 1–6 d each day, then vacuum dried at 37 °C for 1 h. Five milliliters of α-cyano-4-hydroxycinnamic acid dissolved in methanol (10 mg/mL;; Sigma-Aldrich) was evenly sprayed on top of the slide to form uniform layers of the matrix. The sample was subjected to Autoflex Bruker Daltonics MALDI-TOF MS for imaging MS acquisition with scan range from 400 to 2,500 m/z, and the data were analyzed using the FlexImaging 2.0 software. The sample was run in positive reflection or linear mode, with 800-μm laser intervals in XY.
Supplementary Material
Acknowledgments
This work was supported in part by National Natural Science Foundation of China Grants 31330003, 21750002, and 21621002; Chinese Academy of Sciences Grant XDB20000000; and K. C. Wong Education Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800502115/-/DCSupplemental.
References
- 1.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wright GD. Molecular mechanisms of antibiotic resistance. Chem Commun (Camb) 2011;47:4055–4061. doi: 10.1039/c0cc05111j. [DOI] [PubMed] [Google Scholar]
- 3.Walsh CT, Wencewicz TA. Prospects for new antibiotics: AA molecule-centered perspective. J Antibiot (Tokyo) 2014;67:7–22. doi: 10.1038/ja.2013.49. [DOI] [PubMed] [Google Scholar]
- 4.Hopwood DA. How do antibiotic-producing bacteria ensure their self-resistance before antibiotic biosynthesis incapacitates them? Mol Microbiol. 2007;63:937–940. doi: 10.1111/j.1365-2958.2006.05584.x. [DOI] [PubMed] [Google Scholar]
- 5.Cundliffe E, Demain AL. Avoidance of suicide in antibiotic-producing microbes. J Ind Microbiol Biotechnol. 2010;37:643–672. doi: 10.1007/s10295-010-0721-x. [DOI] [PubMed] [Google Scholar]
- 6.D’Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 7.Forsberg KJ, et al. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biggins JB, Onwueme KC, Thorson JS. Resistance to enediyne antitumor antibiotics by CalC self-sacrifice. Science. 2003;301:1537–1541. doi: 10.1126/science.1086695. [DOI] [PubMed] [Google Scholar]
- 9.Xu H, et al. Self-resistance to an antitumor antibiotic: AA DNA glycosylase triggers the base-excision repair system in yatakemycin biosynthesis. Angew Chem Int Ed Engl. 2012;51:10532–10536. doi: 10.1002/anie.201204109. [DOI] [PubMed] [Google Scholar]
- 10.Reimer D, Bode HB. A natural prodrug activation mechanism in the biosynthesis of nonribosomal peptides. Nat Prod Rep. 2014;31:154–159. doi: 10.1039/c3np70081j. [DOI] [PubMed] [Google Scholar]
- 11.Thaker MN, et al. Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat Biotechnol. 2013;31:922–927. doi: 10.1038/nbt.2685. [DOI] [PubMed] [Google Scholar]
- 12.Thaker MN, Waglechner N, Wright GD. Antibiotic resistance-mediated isolation of scaffold-specific natural product producers. Nat Protoc. 2014;9:1469–1479. doi: 10.1038/nprot.2014.093. [DOI] [PubMed] [Google Scholar]
- 13.Tang X, et al. Identification of thiotetronic acid antibiotic biosynthetic pathways by target-directed genome mining. ACS Chem Biol. 2015;10:2841–2849. doi: 10.1021/acschembio.5b00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan Y, et al. Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action. Nature. 2018;559:415–418. doi: 10.1038/s41586-018-0319-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kling A, et al. Antibiotics. Targeting DnaN for tuberculosis therapy using novel griselimycins. Science. 2015;348:1106–1112. doi: 10.1126/science.aaa4690. [DOI] [PubMed] [Google Scholar]
- 16.Johnston CW, et al. Assembly and clustering of natural antibiotics guides target identification. Nat Chem Biol. 2016;12:233–239. doi: 10.1038/nchembio.2018. [DOI] [PubMed] [Google Scholar]
- 17.Bernan VS, et al. Bioxalomycins, new antibiotics produced by the marine Streptomyces sp. LL-31F508: TTaxonomy and fermentation. J Antibiot (Tokyo) 1994;47:1417–1424. doi: 10.7164/antibiotics.47.1417. [DOI] [PubMed] [Google Scholar]
- 18.Scott JD, Williams RM. Chemistry and biology of the tetrahydroisoquinoline antitumor antibiotics. Chem Rev. 2002;102:1669–1730. doi: 10.1021/cr010212u. [DOI] [PubMed] [Google Scholar]
- 19.Peng C, et al. Hijacking a hydroxyethyl unit from a central metabolic ketose into a nonribosomal peptide assembly line. Proc Natl Acad Sci USA. 2012;109:8540–8545. doi: 10.1073/pnas.1204232109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pu J-Y, et al. Naphthyridinomycin biosynthesis revealing the use of leader peptide to guide nonribosomal peptide assembly. Org Lett. 2013;15:3674–3677. doi: 10.1021/ol401549y. [DOI] [PubMed] [Google Scholar]
- 21.Hiratsuka T, et al. Core assembly mechanism of quinocarcin/SF-1739: BBimodular complex nonribosomal peptide synthetases for sequential mannich-type reactions. Chem Biol. 2013;20:1523–1535. doi: 10.1016/j.chembiol.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Tang G-L, Tang M-C, Song L-Q, Zhang Y. Biosynthesis of tetrahydroisoquinoline antibiotics. Curr Top Med Chem. 2016;16:1717–1726. doi: 10.2174/1568026616666151012112329. [DOI] [PubMed] [Google Scholar]
- 23.Carlson JC, et al. Tirandamycin biosynthesis is mediated by co-dependent oxidative enzymes. Nat Chem. 2011;3:628–633. doi: 10.1038/nchem.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexeev I, Sultana A, Mäntsälä P, Niemi J, Schneider G. Aclacinomycin oxidoreductase (AknOx) from the biosynthetic pathway of the antibiotic aclacinomycin is an unusual flavoenzyme with a dual active site. Proc Natl Acad Sci USA. 2007;104:6170–6175. doi: 10.1073/pnas.0700579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer T, Berks BC. The twin-arginine translocation (Tat) protein export pathway. Nat Rev Microbiol. 2012;10:483–496. doi: 10.1038/nrmicro2814. [DOI] [PubMed] [Google Scholar]
- 26.Widdick DA, et al. The twin-arginine translocation pathway is a major route of protein export in Streptomyces coelicolor. Proc Natl Acad Sci USA. 2006;103:17927–17932. doi: 10.1073/pnas.0607025103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song L-Q, et al. Catalysis of extracellular deamination by a FAD-linked oxidoreductase after prodrug maturation in the biosynthesis of saframycin A. Angew Chem Int Ed Engl. 2017;56:9116–9120. doi: 10.1002/anie.201704726. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y-L, Xu Y, Straight P, Dorrestein PC. Translating metabolic exchange with imaging mass spectrometry. Nat Chem Biol. 2009;5:885–887. doi: 10.1038/nchembio.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. MALDI imaging mass spectrometry: MMolecular snapshots of biochemical systems. Nat Methods. 2007;4:828–833. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 30.Calligaris D, et al. MALDI mass spectrometry imaging analysis of pituitary adenomas for near-real-time tumor delineation. Proc Natl Acad Sci USA. 2015;112:9978–9983. doi: 10.1073/pnas.1423101112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Luca V, Salim V, Thamm A, Masada SA, Yu F. Making iridoids/secoiridoids and monoterpenoid indole alkaloids: PProgress on pathway elucidation. Curr Opin Plant Biol. 2014;19:35–42. doi: 10.1016/j.pbi.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 32.Keller NP. Translating biosynthetic gene clusters into fungal armor and weaponry. Nat Chem Biol. 2015;11:671–677. doi: 10.1038/nchembio.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wakimoto T, et al. Calyculin biogenesis from a pyrophosphate protoxin produced by a sponge symbiont. Nat Chem Biol. 2014;10:648–655. doi: 10.1038/nchembio.1573. [DOI] [PubMed] [Google Scholar]
- 34.Le VH, Inai M, Williams RM, Kan T. Ecteinascidins. A review of the chemistry, biology and clinical utility of potent tetrahydroisoquinoline antitumor antibiotics. Nat Prod Rep. 2015;32:328–347. doi: 10.1039/c4np00051j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.