Significance
While most mammalian genes are expressed from both chromosomal copies, many autosomal genes randomly express only one allele in a given cell, resulting in somatic cellular mosaicism. To better understand the mechanisms, developmental aspects, and evolution of autosomal monoallelic expression (MAE), we used nucleotide polymorphism differences between hybrid mice to analyze MAE of clonal neural stem cell lines as they differentiated to astrocytes. We found that genes showing MAE are highly enriched among developmental stage-specific genes. Genes showing strong skewed expression are similarly enriched. We also found evidence suggestive of balancing selection not just for genes with MAE but also, for developmental stage-specific genes.
Keywords: evolution, Gstm5, overdominance, differentiation, schizophrenia
Abstract
Cellular mosaicism due to monoallelic autosomal expression (MAE), with cell selection during development, is becoming increasingly recognized as prevalent in mammals, leading to interest in understanding its extent and mechanism(s). We report here use of clonal cell lines derived from the CNS of adult female hybrid (C57BL/6 X JF1) mice to characterize MAE as neural stem cells (nscs) differentiate to astrocyte-like cells (asls). We found that different subsets of genes show MAE in the two populations of cells; in each case, there is strong enrichment for genes specific to the respective developmental state. Genes that exhibit MAE are 22% of nsc-specific genes and 26% of asl-specific genes. Moreover, the promoters of genes with MAE have reduced CpG dinucleotides but increased CpG differences between the two parental mouse strains. Extending the study of variability to wild populations of mice, we found evidence for balancing selection as a contributing force in evolution of those genes showing developmental specificity (i.e., expressed in either nsc or asl), not just for genes showing MAE. Furthermore, we found that genes showing skewed allelic expression (SKE) were similarly enriched among cell type-specific genes and also showed a heightened probability of balancing selection. Thus, developmental stage-specific genes and genes with MAE or SKE seem to make up overlapping classes subject to selection for increased diversity. The implications of these results for development and evolution are discussed in the context of a model with stochastic epigenetic modifications taking place only during a relatively brief developmental window.
Somatic cellular mosaicism is becoming increasingly recognized as a significant aspect of mammalian development, phenotypic variation, and disease (1–5). Epigenetic as well as genetic mosaicism is prevalent in the CNS, where it has implications for both normal function and neuropsychiatric disorders, especially since there can be cell selection during development (1). X chromosome inactivation (XCI) is a well-known example of an epigenetic process resulting in monoallelic expression, with resulting cellular mosaicism due to random selection and inactivation early in differentiation of most genes on one X chromosome (2). In any given cell, choice of either the maternal or paternal X chromosome is random, but after the decision is made, it is mitotically inherited with great stability. More recently, evidence has been accumulating that many autosomal genes also show monoallelic expression and that genes with monoallelic autosomal expression (MAE) are enriched for those that affect membrane proteins and development (3, 5). There are about 100 imprinted autosomal genes with expression that is determined by parental source (that is, their expression depends on whether they are paternally or maternally derived) (6). Even more genes show random MAE, where choice of allele is apparently stochastically determined (2, 4, 7, 8). Since mitotically persistent random MAE and skewed expression are likely to impact development as well as genetic inheritance and evolution, it is important to understand their extent and mechanism(s). Despite this, random MAE’s mechanism and evolutionary impetus remain largely unexplored. There have been a number of recent studies on MAE in humans and mice (reviewed in refs. 2 and 3), but significant puzzles remain. How is MAE affected by developmental changes? How does MAE affect development? How are alleles showing MAE regulated differently, although in the same nucleus? Are there specific sequences or patterns correlated with MAE? Does MAE have a special role in evolution? Even less is known about skewed allelic expression (SKE): selective mechanisms acting on MAE genes also are likely to be operating on SKE genes.
For several mouse genes, it is now known that stochastic epigenetic variability, sometimes influenced by the environment, can result in more than one phenotype from the same genotype (9–11). The best-studied example is mice carrying the agouti viable yellow gene: they are sometimes normal with agouti coat color, and they are sometimes yellow and obese. It is known that the insertion of a retroposon potentially provides an alternate promoter and that DNA methylation of this promoter correlates with phenotype (12). Methylation in the very early embryo is incomplete and stochastic, not always silencing the promoter. At or soon after implantation, the methylation state of the retroelement is locked. The key point here is that the retroelement introduces epigenetic variability dependent on stochastic DNA methylation during a developmental window. Theoretical modeling, based on somatically heritable stochastic epigenetic modifications taking place only during specific developmental windows of opportunity (10, 11), led to the surprising prediction (10) that genes associated with stochastic epigenetic modification can show overdominance (i.e., higher relative fitness of heterozygotes relative to either homozygote). Overdominance is associated with balancing selection (i.e., the selective process by which genetic polymorphisms are maintained in the gene pool). Loci associated with balancing selection have been recently reported for humans (13, 14) and Drosophila (15). Classical examples include sickle cell anemia, HLA (16), and olfactory receptor loci (17). It recently has been reported that some human genes with MAE, identified by a chromatin modification signature, show excessive variability and selection for maintenance of heterozygosity (balancing selection) (18). Do mouse genes exhibiting MAE similarly show increased variability and balancing selection? We report here evidence in support of this possibility for genes with MAE and for cell type-specific genes. We also report on changes in genes exhibiting MAE during development. Most of the genes showing MAE or skewed expression in astrocyte-like cells (asls) are in membrane-associated or cell-to-cell signaling categories. We have found increased sequence diversity in genes with MAE or SKE and in cell type-specific/developmental genes. This diversity may not only affect development but also, may provide some evolutionary advantage, perhaps related to the establishment of individual cell identities.
Results
Experimental Overview.
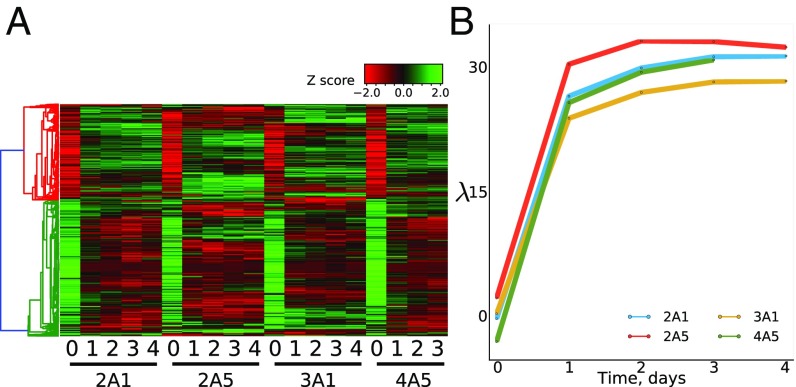
We performed transcriptome-wide profiling of four previously described F1(B6 X JF1) clonal cell lines (2A1, 2A5, 3A1, and 4A5) as they underwent in vitro differentiation along the astrocytic pathway (4). Cells were sampled at day 0 (before the start of differentiation) and at 1–4 d thereafter. A heat map describing changes in expression for each cell line is shown in Fig. 1A. Remarkably, most changes are visible by day 1 of differentiation, becoming more pronounced by days 3 and 4. Principal component analysis verified that about 85% of developmental changes occur by the first day after induction of differentiation (Fig. 1B). The astrocytic cells show considerable overlap in gene expression with astrocytes from adult mice (19) (Dataset S1).
Fig. 1.
(A) Heat map showing expression changes during in vitro differentiation of nscs to astrocytes. Results are shown for each cell line and day of differentiation. (B) Principal component analysis. In this constructed space, the component mainly associated with differentiation is shown to rise to near maximal levels by day 1 after induction of differentiation. Dataset S1 has a complete list of genes used in our study and their expression levels.
Establishment of a JF1 genomic SNP library allowed us to analyze transcriptome-wide allele-specific expression. As previously described, a cutoff of fragments per killobase per million mapped reads (fpkm) 3 and a pooled SNP depth of coverage 10 allowed us to analyze ∼10,000 genes in the four cell lines (8).
Definitions for Computational Analysis.
We define neural stem cells (s) and the s derived from them. refers to the cells before induction of differentiation (day 0), whereas refers to days 1–4 postinduction. The probability of each gene being expressed from the C57BL/6 (B6) allele was computed as the simple ratio between the SNP counts assigned to the B6 haplotype and the total number of SNP counts observed at the locus (details are in ref. 8). To measure the bias in expression for one allele with respect to the other, we define the monoallelic skew variable () for each gene as
| [1] |
where is bound between and is close to 0 for genes with biallelic expression, whereas it approaches 0.5 for genes with MAE.
It is important to note that for a gene to be considered within the set, we use strict criteria; it must have a large and statistically significant value of 0.35 and be both monoallelically expressed at fpkm 3 and biallelically expressed at 3 in at least one cell line at one stage. These criteria minimize contributions from statistical noise due to low expression and from apparent MAE due to erroneous SNP calls; they also provide assurance that both alleles have the potential to be expressed at similar levels. X-linked genes provide a control (8) but are excluded from analysis of MAE. We then define and as sets that contain genes displaying monoallelic expression in or , respectively. Analogously, we define and for the biallelically expressed genes.
nsc and asl—Specific Genes Preferentially Show MAE.
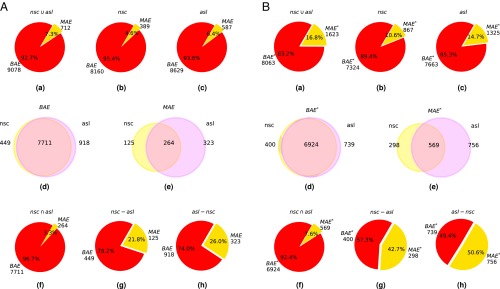
Overall, we found 712 genes (7.3%) showing MAE and 9,078 genes showing biallelic autosomal expression (BAE) (Fig. 2). Considering the two developmental stages independently, we observed the following: : is equal to 389 (4.6%), and is equal to 8,160 (95.4%); : is equal to 587 (6.4%), and is equal to 8,629 (93.6%) (Fig. 2 A, b and c and B, b and c). It has been reported for both mice and humans (2) that MAE is enriched in cell type-specific genes. To investigate this in our system, we divided the and sets into three subsets: those that belong exclusively to or and those that belong to both developmental stages (Fig. 2 A, d and e and B, d and e; Materials and Methods has a formal definition of the set, and Dataset S2 has gene lists). We then investigated if the set is enriched in cell type-specific genes [i.e., genes expressed in only one developmental stage ( and ) compared with genes that are in common ()]. The results in Fig. 2 A, f–h clearly show that the fraction of genes with MAE changes in different subsets: from 3.3% in the case of genes that are in common in the two developmental stages to 21.8% and 26.0% for genes present only in and , respectively. These highly statistically significant observations (P value ) clearly show that the probability of MAE is strongly increased and can be a high percentage in genes that are specific to a particular cell type.
Fig. 2.
Distribution of autosomal genes showing monoallelic or biallelic expression (MAE or BAE) in undifferentiated and . In A, is defined as 0.35; in B, is defined as 0.25. (a–c) Counts and percentages of () and () in the union set of (), , and , respectively. (d and e) The distribution of () and (), respectively, in and . (f) Counts and percentages of () and () in the intersection set of (; i.e., genes that are expressed in both and ). (g and h) Counts and percentages of () and () for genes that are expressed only in or , respectively.
Differences in Allele-Specific Expression Between nsc and asl.
Gene ontology (GO) analysis (20) was performed to explore the differences in the types of genes expressed among and . Genes showing MAE specifically associated with or or in common in both cell line are listed in Dataset S2. Genes enriched in include mostly those involved in cell division and DNA replication, while genes in cells are enriched for those involved in differentiated activity of neural cells, such as ion channels, transporters, and cell surface components (SI Appendix). Ontology analysis confirmed the enrichment of genes involved in cell adhesion and cell–cell signaling as well as extracellular matrix organization and calcium ion binding.
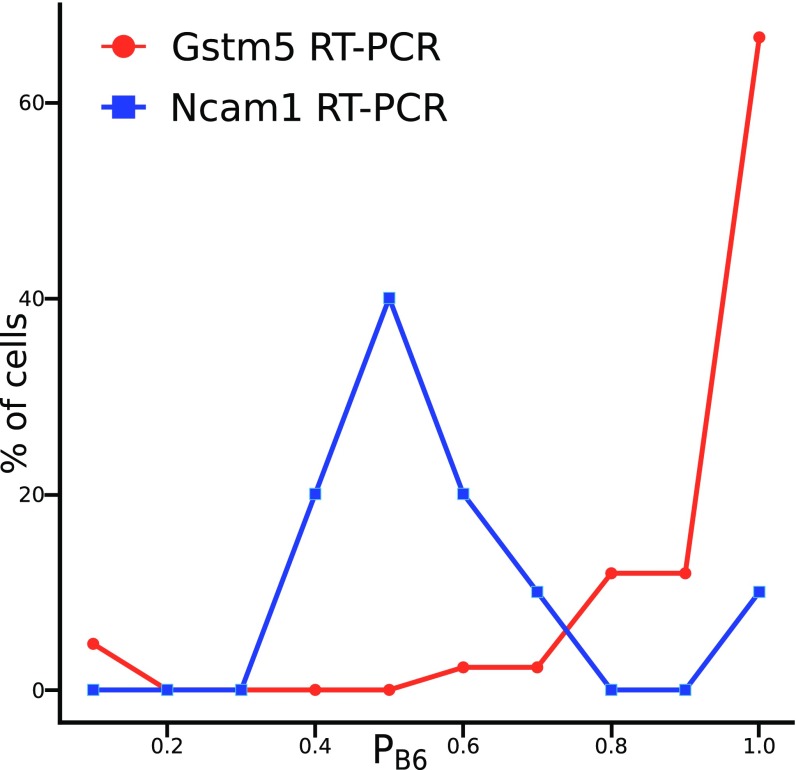
Our results confirmed previous findings of monoallelic expression of members of both the annexin and glutathione transferase (Gst) gene families and (8). For the latter gene family, we were able to confirm monoallelic expression of Gstm5 in individual hippocampal cells of postnatal mice (Fig. 3). In addition, a number of known imprinted genes show the expected pattern of reciprocal expression in lines 2A1, 3A1, and 4A5 (paternal JF1 allele) vs. 2A5 (paternal B6 allele): , , , , , , , , , , , and . Examining the remaining genes with MAE, we noted a small category that increases somewhat in expression as they transition from BAE in to MAE in astrocytes, including and pituitary tumor-transforming 1 (). codes for a brain-specific glycolytic enzyme (21). is involved in mitosis and has tumorigenic properties (22).
Fig. 3.
Allele-specific expression of Gstm5 and Ncam1 in individual hippocampal cells of postnatal day 7 B6 X JF1 hybrid mice. The x axis shows the fractional expression of the B6 allele. Gstm5 assay, n = 42 cells; Ncam1 assay, n = 25 cells.
Another interesting category includes genes that show a relatively low level of monoallelic expression in (or no expression at all) but are induced to high levels of MAE in astrocytes (Dataset S2). Some of these genes contribute to inflammation, such as oncostatin M (), a required gene for glioblastoma development (23), and chitinase-3-like protein 1 (; also termed Chil1) (24). is of particular interest, as it has been implicated as part of the known link between the immune system and schizophrenia (reviewed in ref. 25). Others are extracellular or membrane components, a group enriched among astrocytic genes as described above: these include (a Zinc carboxypeptidase) (26), integrin alpha chain 7 () (27), and transmembrane 4 superfamily member 1 (), a member of the tetraspanin family (www.ncbi.nlm.nih.gov/gene/17112). MAE of , and was independently confirmed by qRT-PCR (Dataset S3).
Dynamic Changes.
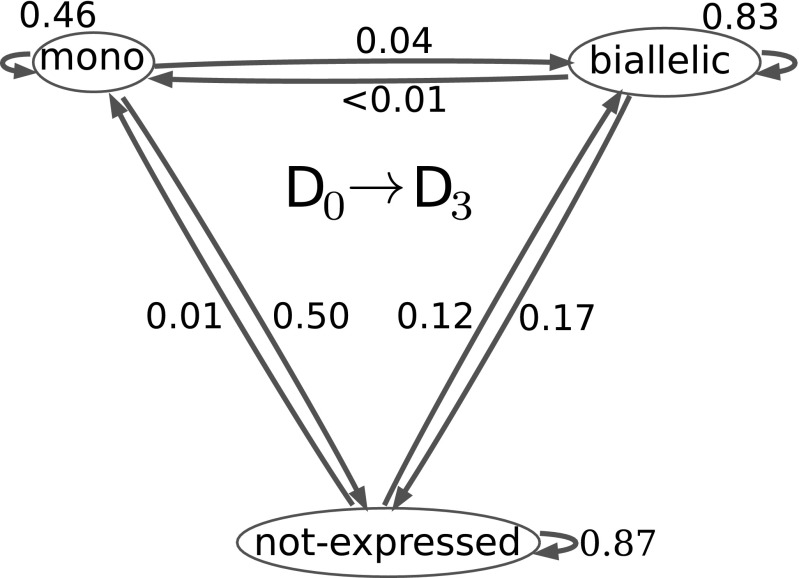
We next examined the dynamics of changes in allele-specific expression for each gene during differentiation. The transition of each gene is considered from a given cell line to the same cell line after differentiation to at day 3. Comparing day 0 with day 3, it can be seen (Fig. 4) that approximately one-half (0.46) of genes that display MAE in nsc continue to do so after differentiation to . The frequency of transition from BAE to MAE is 0.001 and to loss of expression is 0.17. The cases of BAE in cells are unchanged in most cases (0.83) from the state, while a small percentage transitions from lack of expression to MAE (0.01). Although the percentage is small, it should be emphasized that 44% of MAE in asl is from previously silent genes (SI Appendix), and a high percentage (26%) of asl-specific genes shows MAE when 0.35 (at least 85% expression from one allele) (Fig. 2 A, g and h and B, g and h). If minimum value is reduced to 0.25 (at least 75% from one allele), then even more asl-specific genes show MAE (50.6%) (Fig. 2 B, g and h). It is important to note that the differences between cell lines have persisted through numerous mitoses, consistent with other studies and with the hypothesis that stochastically variable epigenetic states, either preexisting in the or established during differentiation, can then become fixed and mitotically persistent.
Fig. 4.
Probability of change to (), where S is one of three possible states of expression: monoallelic, biallelic, or not expressed. refers to before induction of differentiation, and refers to after 3 d of differentiation.
Genes with MAE and Genes Expressed in Either nsc or asl Show Decreased CpG Frequency and Increased CpG Variability.
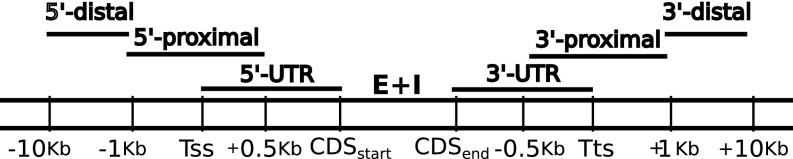
To explore possible sequence differences between the genes showing monoallelic vs. biallelic expression, we defined various regions (Fig. 5). For example, we defined the region from 1 kb upstream to 0.5 kb downstream of the transcription starting site as 5′ proximal. Given the well-known role of CpG dinucleotides in the epigenetic control of gene expression by DNA methylation, we also checked the distribution of , where o/e equals observed/expected. Similarly, we considered all other possible dinucleotides ().
Fig. 5.
Gene neighborhood scheme. E + I is the coding portion of exons plus introns, and and represent the Coding DNA Sequences starting and end points, respectively.
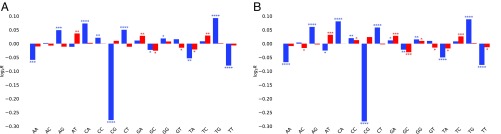
We first found that there is a large, statistically significant difference between MAE and BAE in the distribution of dinucleotides in the vicinity of the 5′-proximal region (Fig. 6A). We then found a similar difference for genes that are cell type specific (either asl or nsc) and those that are expressed by both cell types (Fig. 6B). In particular, shows the largest observed difference, with cell type-specific genes characterized by a lower than those expressed in common. Other dinucleotides show less pronounced but significant differences (Fig. 6)
Fig. 6.
Comparison (R) of the mean observed to expected dinucleotide distribution for (A) genes with MAE vs. BAE and (B) genes expressed specifically in nsc or asl vs. genes expressed in both cell types. Results are shown for the 5′-proximal (blue) and 3′-proximal (red) regions (Fig. 5 shows gene neighborhoods). *P < 10−2; **P < 10−5; ***P < 10−10; ****P < 10−15 (Mann–Whitney U rank test).
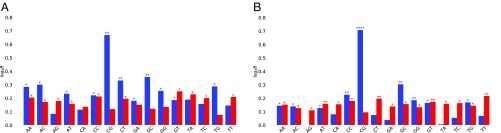
We also checked the differences in dinucleotide variability () defined as the number of dinucleotide changes per dinucleotide count between the two parental mouse strains, B6 and JF1. We then compared the distribution of in MAE vs. BAE genes and cell type-specific genes vs. those expressed in common. Overall variability was found to be significantly greater for genes that display MAE (Fig. 7), predominately at CpG dinucleotides, but again, the difference was found to be due mainly to differences between cell type-specific genes and those expressed in common (Fig. 7B). We conclude that cell type-specific genes (and genes with MAE) show lower at the promoter and more variability at CpGs between B6 and JF1. Taken together, our observations suggest that particular evolutionary mechanisms may be involved for genes displaying cell type-specific expression, with genes showing MAE or skewed expression as an overlapping group. This possibility is explored in the following section.
Fig. 7.
Comparison () of the mean dinucleotide variability distribution for (A) genes with MAE vs. BAE and (B) genes expressed specifically in nsc or asl vs. genes expressed in both cell types. Results are shown for the 5′-proximal (blue) and 3′-proximal (red) regions (Fig. 5 shows gene neighborhoods). *P value < 10−2; **P < 10−5; ****P < 10−15 (Mann–Whitney U rank test).
MAE and Cell Type-Specific Genes Show Increased Variability and Balancing Selection.
We first investigated the role of balancing selection in the evolution of the genes that we identified as showing MAE. The possibility of such a role has been previously suggested theoretically (10, 11), and it received some experimental corroboration recently in human (18). Using data from 28 individuals from four natural populations of mice of Mus musculus domesticus (28), we performed a genome-wide scan for regions with an excess of SNPs with intermediate frequencies, which is a signature of balancing selection (i.e., selection for heterozygosity) (29). To investigate the possibility of balancing selection, we used Tajima’s D, which results from an equation in widespread use (29) to detect evidence of balancing selection in natural populations. Briefly, under the infinite site model assumption, Tajima showed that the quantity should vanish under conditions of neutral evolution. A positive value of the statistic results from an increase in the intermediate frequencies of polymorphic nucleotides and suggests that a sequence or a genomic region is subject to balancing selection or another type of diversifying selection. A similar pattern can also be observed if the sampled population was formed from a recent admixture of two different populations (29). Negative values are associated with an excess of rare alleles, purifying selection, and/or expanding populations.
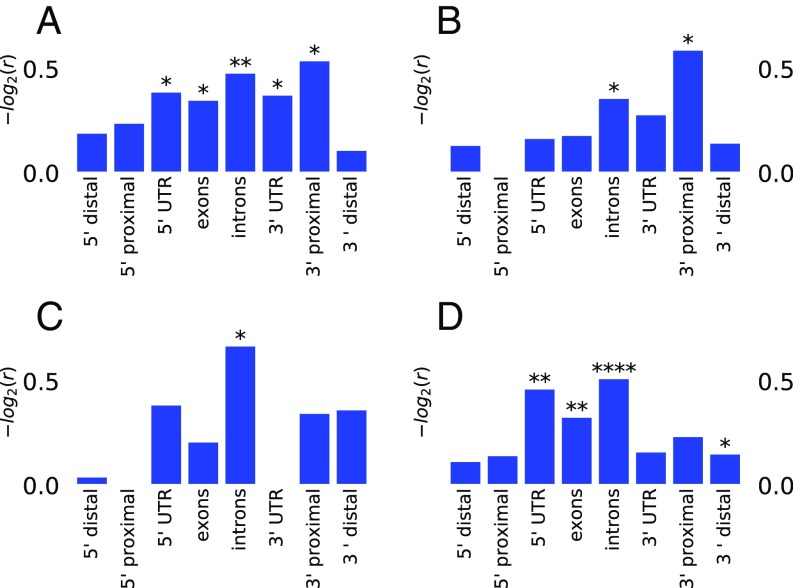
For each gene analyzed, we delineated six gene neighborhoods encompassing the 5′-distal to the 3′-distal region as defined in Fig. 5. We then computed the statistic for 1-kb windows in each neighborhood and recorded the largest value of observed in each window. As for the previous analysis, we divided the active genes into two sets, and BAE. We then considered as putative regions under balancing selection Tajima’s D statistic peaks with values of D larger than the 95th percentile. We used the Fisher exact test to assess whether the number of putative peaks under balancing selection in a given neighborhood differed significantly between the MAE and BAE sets. We observed that the MAE set is associated with peaks with a larger D statistic than the BAE set in introns, exons, and 3′-proximal regions (Fig. 8A). Because genes showing MAE are enriched among cell type-specific genes (Fig. 2 A, g and h and B, g and h), we next compared the Tajima’s D among genes showing monoallelic or biallelic expression in or only vs. genes expressed in both cell types. The results are reported in Fig. 8 B–D. When we compared cell type-specific MAE and BAE genes, we observed that the differences were marginal, with the only remaining significant difference being for the 3′-proximal region and introns (P value <0.05) (Fig. 8B). When we compared genes showing biallelic expression, we again observed a higher Tajima’s D for cell type-specific genes, especially at introns (Fig. 8D). We conclude that the increased variability is a property of astrocyte- or neuronal pathway-specific genes, not just genes showing MAE.
Fig. 8.
Balancing selection characterizes both MAE genes and cell type-specific genes (expressed in either nsc or asl but not in both). The Fisher test was performed, comparing the top 95th percentile Tajima’s D (calculated from natural mouse populations) observed in different gene neighborhoods as defined in Fig. 5 and Material and Methods. r is the odds ratio of the number of top 95th percentile Tajima’s D observed in different groups. *P < 0.05; **P < 10−3; ****P < 10−10 (Fisher test). (A) MAE vs. BAE. (B) MAE vs. BAE for cell type-specific genes. (C) MAE: cell type-specific genes vs. both cell types. (D) All expression (MAE or BAE): cell type-specific vs. genes expressed in both cell types.
Properties of Genes Showing SKE.
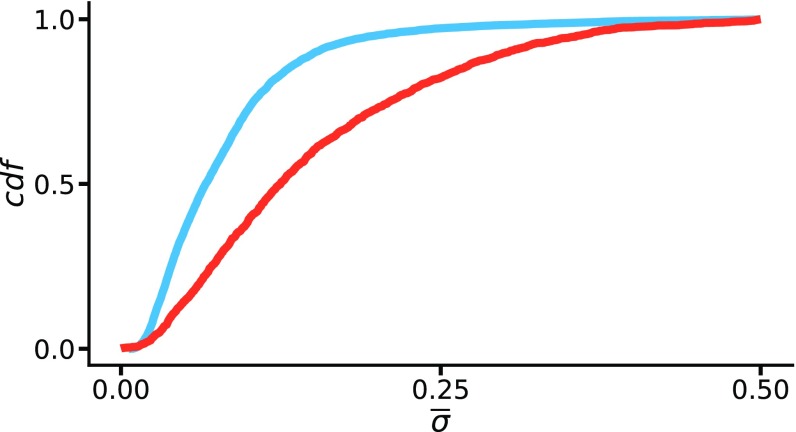
In earlier sections, we described as a monoallelic skew variable and considered the MAE set as including only cases where was 0.35. However, as shown in Fig. 9, the expression bias represented by is a smooth continuous function. We, therefore, were interested to see if some of the properties observed for MAE vs. BAE genes are a more general property of genes with skewed expression. We asked whether the enrichment for monoallelic expression observed for cell type-specific genes is only the tip of the iceberg (distributionally speaking) of the more general property of expression bias. To explore this possibility, we computed the average value of for each gene and divided the genes into two groups: those expressed in only one cell type ( or ) and those expressed in both cell types. The results shown in Fig. 9 clearly show a distributional difference between the two groups, with biased genes (high value) more likely present among cell type-specific genes (Wilcoxon rank sum test ). We further investigated whether the distributional difference in observed between the two groups of genes was still present if genes showing the most stringently described MAE were removed from the analysis. To do this, we removed the 712 genes defined as showing MAE (Fig. 2) from both groups of genes and tested whether the cell type-specific genes remained more biased in gene expression compared with the genes expressed in common. The results are reported in SI Appendix and show that cell type-specific genes are skewed toward higher , even if the MAE gene set is removed (Wilcoxon rank sum test ). Following a similar strategy, we performed GO analysis (20) to see if the ontology categories typically enriched for monoallelic expression also persist for genes showing moderately skewed expression. The results of the analysis of GO-ranked genes (reported in SI Appendix) show that skewed genes (high ) are enriched for categories, such as “extracellular region,” “integral component of the membrane,” “cell adhesion,” and “sensory perception,” just as are genes showing MAE. This category remains enriched even if the most skewed genes are removed from the GO ranked list (SI Appendix). This suggests that skewed expression may provide some selective advantage for genes involved in development and communication between cells, at least for the nervous system.
Fig. 9.
Developmental genes are associated with SKE expression. The cumulative density distribution is shown for the variable mean of (Eq. 1) for genes expressed in both cell types (blue) or only one cell type (red).
Discussion
Nag et al. (5) reported cell type-specific preferences for MAE in mouse lymphoblastoid cell lines using chromatin “signatures” to identify genes with MAE and suggested a link between MAE and cell identity. Here, we report direct measurements of monoallelic expression in mouse nscs and in vitro-generated astrocytes. The use of clonal cells derived from a hybrid of two distantly related mouse subspecies (J1 and B6) has allowed us to distinguish alleles of 10,000 genes and follow their ratio of expression during in vitro differentiation of nsc to asl cells (Fig. 2). Our results confirm and extend previous studies in mouse and human (5, 7, 8), which have established that (i) a subset of autosomal genes can be categorized as displaying MAE; (ii) the expression state of a given gene, MAE, BAE, or SKE, is persistent in cell culture; and (iii) the MAE category is enriched for cell type-specific genes, many of which are membrane-associated genes involved in cell–cell signaling and development. We find that 26% of astrocyte-specific genes show MAE using a strict criterion (0.85% expression of a given allele; 0.35). Use of a more relaxed criterion (0.75%; 0.25) reveals SKE for an even higher percentage of asl-specific genes (56%) (Fig. 2B).
Several reports have suggested that MAE is important for normal development as well as disease (3, 30–34). Chess (2), who first found that odorant genes show MAE, has reviewed random MAE and XCI and suggested that MAE and resulting cellular mosaicism have important implications, not only for development but also for evolution (2). We (10, 11) and others (18) have made similar suggestions.
It is widely known that many genotypes display incomplete penetrance (i.e., variable phenotypes). A dramatic example is the observation of incomplete concordance among identical twins for major human genetic disorders, such as schizophrenia and bipolar disorder (35). A very recent and relevant example is bistable epigenetic obesity in mice and humans (9). It also is well known that XCI takes place in the early embryo at about the time of implantation, and after it is established, it is very stable; that is, monoallelic expression resulting from stochastically random XCI during a developmental window is somatically heritable (2). Somatic heritability is due to epigenetic marks, which include DNA methylation, laid down during a developmental window. Reasoning from XCI and the prevalence of variable phenotypes, we proposed a stochastic epigenetic modification mechanism and did quantitative mathematical modeling (10, 11). The key assumptions of the stochastic epigenetic modification model are (i) stochastic and incomplete epigenetic marking during a relatively short developmental window in the early embryo, (ii) high-fidelity somatic heritability after the window of variability is closed, and (iii) the DNA sequence of cis-located elements, which can be due to retroposons or other mutations, affects the probability () of epigenetic marking of the cis-located gene that is subject to selection. Another assumption is that dosage compensation is absent or incomplete, and this has been confirmed for most genes with MAE (8, 36). A significant finding reported here is that skewed expression rather than strict MAE is quite prevalent. With respect to the stochastic epigenetic modification model, it is not critical whether or not at the single-cell level there is complete silencing. The fundamental assumption is that the ratio of expression between the maternal and paternal alleles follows a stochastic rule during the window of opportunity in early development, becoming fixed when the window closes. Quantitative analytical and computational modeling of stochastic epigenetic modification (11) showed that the fixation of genes in a population could be greatly accelerated by stochastic epigenetic modification. Another unanticipated result of our modeling was that overdominance is predicted for reasonable values of (e.g., 0.75). This means that the heterozygote has a selective advantage and will be maintained in the population, increasing genetic diversity.
Whether or not balancing selection is widespread in mammals has been controversial due to lack of convincing evidence for more than just a handful of genes. However, by analyzing SNPs in two human populations, Andrés et al. (13) obtained strong evidence of excess variation and balancing selection for at least 60 human genes. In addition, the laboratory of Gimelbrant and coworkers (7, 18) analyzed nucleotide diversity in genes having a chromatin signature suggestive of MAE. They concluded that human MAE genes contribute disproportionately to genetic diversity and are strongly enriched among those genes identified by Andrés et al. (13) as showing balancing selection. We confirm for mouse the findings of Savova et al. (18) for human that genes with MAE are more variable than genes with BAE. Moreover, we find that nsc or asl cell type-specific genes are also more variable than genes expressed in both cell types, overlapping with the set of genes. It remains to be determined by future work whether genes specific to the neuronal lineage are more variable than genes in other cell lineages.
Eckersley-Maslin et al. (36) found that overall DNA methylation was no different between genes showing MAE vs. BAE in embryonic stem cells or neuroprogenitor cells, but they did not analyze CpGs or other dinucleotide frequencies. A potential link of MAE to DNA methylation is suggested by a study using the MAUD assay: it showed that a set of genes with a distinct DNA methylation pattern in mouse brain was enriched for genes with MAE in nscs and also for genes showing lineage-specific differentiation within the developing CNS (34).
Savova et al. (18) have reported that, when MAE is compared with BAE, there is an increase in the density of CpGs in the gene body and in particular, in the coding portion of the genes. When we analyzed the 5′-proximal region, we observed that, in contrast to the gene body, Cp is lower for mouse genes that show MAE vs. those that show BAE, and there is increased dinucleotide variability, especially at CpGs (Fig. 6). This difference persists if housekeeping genes are removed from the analysis (SI Appendix). This finding led us to extend our variability analysis to different mouse strains by using Tajima’s D statistic, which yields a positive value for gene regions under balancing selection (29). We found that the gene body (coding region plus introns) and 3′-proximal regions (but not promoters and 5′-proximal regions) of the MAE genes show a significant positive Tajima’s D statistic compared with BAE genes (Fig. 8A). Housekeeping genes are known to have G + C (GC)- and CpG-rich promoters, with expression levels correlated with GC richness (37). Highly expressed housekeeping genes are also known to be under purifying selection (38), which would yield a negative Tajima’s D statistic. We, therefore, searched for sites with low Tajima’s D statistic in BAE or housekeeping genes and found no enrichment compared with MAE genes. This is in agreement with Savova et al. (18), who found that purifying selection similarly affects MAE and BAE genes. Therefore, the relative increase in Tajima’s D statistic in MAE genes is not likely due to a decrease in D score of BAE genes, and it is suggestive of balancing selection. We next found that the difference seen between MAE and BAE genes was mainly due to differences between cell type-specific genes and those genes expressed in both cell types (Fig. 8D). We conclude that there is an important, previously unappreciated difference in the variability and Tajima’s D statistic in the coding regions and introns of cell type-specific genes compared with those expressed by both cell types (including housekeeping genes). Cell type-specific genes are enriched for developmentally important, cell–cell, and cell–environment sensing genes. It is these genes, not just MAE genes, that our data suggest have a positive Tajima’s D statistic and are likely to be under balancing selection, resulting in increased genetic diversity.
We also addressed the question of whether SKE is subject to similar selection pressure as MAE. Our analysis, shown in Fig. 9, suggests that it is, since cell type-specific genes are skewed toward higher allele-specific expression bias, even when genes meeting our strict definition of MAE are removed (SI Appendix). Genes showing SKE were also found to be similarly enriched for the same GO categories as cell type-specific genes (SI Appendix). We suggest that skewed expression, resulting from stochastic epigenetic modification during a short developmental window, is evolutionarily important for giving each cell a unique cellular identity, with MAE genes being extreme examples of SKE genes.
Materials and Methods
Single-cell RT-PCR.
All mouse procedures were approved by the Institutional Animal Care and Use Committee at City of Hope (American Association for Laboratory Animal Care approval 000720). After euthanasia of postnatal (7.5 d) B6 × JF1 mice and dissection of hippocampi, the Worthington Papain Dissociation System was used to isolate neurons. Cells were diluted to one cell per 2 L and distributed to 48-well plates. Wells containing single cells were identified microscopically. After addition of Tri-zol and ethanol (50 L each), RNA was transferred to a Zymo spin collection column (Direct-zol RNA Microprep Kit; Zymo). Each RNA sample was eluted with 12 L of water and added to a Bioneer Accupower Dual-Hotstart RT-PCR pellet. After addition of primers, the mixture was transferred to 0.65-mL thin-walled Eppendorf tubes, and RT-PCR was performed for 35 cycles. The percentage B6 expression was determined as previously described (8). Ncam1 and Gstm5 cDNA primers are listed in SI Appendix.
High-Throughput Sequencing.
Clonal nsc lines were cultured and differentiated, and RNA was prepared for high-throughput (Illumina) sequencing as previously described (4, 8). A JF1 genomic SNP library was also created by use of Illumina sequencing. Briefly, short reads from JF1 DNA-seq were mapped to the mouse genome (GRCm38/mm10) using BWA (v. 0.7.5a) with default settings. Pileup of the sequences was created by samtools (v.0.1.19). The Genome Analysis Toolkit (v.1.4-17) was used with a minimum phred-scaled confidence threshold of calling a variant at 30 and other parameters at default settings. The JF1 DNA SNPs were further filtered with depth of coverage 5 and quality 30 with consensus probability 0.9. Although we have obtained JF1 DNA SNPS for the whole genome, monoallelic expression was assessed using JF1 SNPs located in the exons of Refseq genes. Short reads from RNA-seq were mapped to the mouse genome using TopHat (v.2.014). We allowed the reads to have a maximum of one hit in the genome, and reads with more than seven mismatches were discarded. The inner distance between mate pairs was set at 200 bp, and other parameters were set at default. Expression of Refseq genes was measured as fragments per kilobase of transcript per million mapped reads and was calculated using Cufflinks (v.2.2.1) with default settings.
During the course of this study, another JF1 genomic SNP library was independently published (39).
Mathematical Definitions.
Monoallelic expression.
The probability for each gene to be expressed from the B6 allele is
| [2] |
where is the total number of SNP counts observed at locus and and are the numbers of SNP counts assigned to the or haplotype, respectively (ref. 8 has details).
Moreover, for each gene , the exact binomial test was used to check the null hypothesis , and the probability of rejecting the hypothesis that expression was biallelic was computed. Finally, , obtained from RNA-seq experiments, was used as a measure of the level of expression of each gene. Defining as the set of all of the samples, a gene shows MAE for a sample if . We define the set as the set of all genes with in at least one sample and defined as genes that are expressed (i.e., with 3) but that do not display MAE in any sample. Finally, we define and as the sets of genes that show in or , respectively, but not both. Similarly, and are genes that are expressed (i.e., with 3) in or , respectively, but that do not display MAE in any sample. For cSNP-seq analysis,
| [3] |
where is the set of all autosomal expressed genes in our experiments; and are the sets of genes showing autosomal monoallelic and biallelic expressions, respectively; and cell type can be or . Formally,
| [4] |
| [5] |
and
| [6] |
Analogously, we can define , , and for (Fig. 2). We define as not expressed (with and ) if its expression cannot be classified as monoallelic () or biallelic (); formally,
| [7] |
where is the set of all genes in our experiments with 3 in at least one sample . The results are shown in Fig. 4, where each data point (i.e., gene, cell line, and developmental state) is considered independently.
Observed/expected dinucleotide frequency.
| [8] |
with equal to the frequency, in the region of interest, of the dinucleotide , respectively (Fig. 6)
Tajima’s D statistic.
| [9] |
where is the average number of pairwise mismatches observed in a set of sequences, is the number of segregating sites in a sample of sequences, , and is the operator for the variance.
Supplementary Material
Acknowledgments
A.D.R. holds the Samuel Rahbar Chair for Diabetes and Drug Discovery.
Footnotes
The authors declare no conflict of interest.
Data deposition: The genomic JF1 SNPs reported in this paper have been deposited in the European Nucleotide Archive, https://www.ebi.ac.uk/ena (accession no. ERZ772629).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808652115/-/DCSupplemental.
References
- 1.McConnell MJ, et al. Intersection of diverse neuronal genomes and neuropsychiatric disease: The brain somatic mosaicism network. Science. 2017;356:eaal1641. doi: 10.1126/science.aal1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chess A. Monoallelic gene expression in mammals. Annu Rev Genet. 2016;50:317–327. doi: 10.1146/annurev-genet-120215-035120. [DOI] [PubMed] [Google Scholar]
- 3.Gendrel A, Marion-Poll L, Katoh K, Heard E. Random monoallelic expression of genes on autosomes: Parallels with x-chromosome inactivation. Semin Cell Dev Biol. 2016;56:100–110. doi: 10.1016/j.semcdb.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Wang J, Valo Z, Smith D, Singer-Sam J. Monoallelic expression of multiple genes in the cns. PLoS One. 2007;2:e1293. doi: 10.1371/journal.pone.0001293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nag A, Vigneau S, Savova V, Zwemer L, Gimelbrant A. Chromatin signature identifies monoallelic gene expression across mammalian cell types. G3 (Bethesda) 2015;5:1713–1720. doi: 10.1534/g3.115.018853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21:457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Gimelbrant A, Hutchinson J, Thompson B, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 8.Li S, et al. Transcriptome-wide survey of mouse cns-derived cells reveals monoallelic expression within novel gene families. PLoS One. 2012;7:e31751. doi: 10.1371/journal.pone.0031751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalgaard K, et al. Trim28 haploinsufficiency triggers bi-stable epigenetic obesity. Cell. 2016;164:353–364. doi: 10.1016/j.cell.2015.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Branciamore S, Rodin AS, Gogoshin G, Riggs AD. Epigenetics and evolution: Transposons and the stochastic epigenetic modification model. AIMS Genet. 2015;2:148–162. [Google Scholar]
- 11.Branciamore S, Rodin AS, Riggs AD, Rodin SN. Enhanced evolution by stochastically variable modification of epigenetic marks in the early embryo. Proc Natl Acad Sci USA. 2014;111:6353–6358. doi: 10.1073/pnas.1402585111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolinoy DC, Weinhouse C, Jones TR, Rozek LS, Jirtle RL. Variable histone modifications at the avy metastable epiallele. Epigenetics. 2010;5:637–644. doi: 10.4161/epi.5.7.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andrés AM, et al. Targets of balancing selection in the human genome. Mol Biol Evol. 2009;26:2755–2764. doi: 10.1093/molbev/msp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leffler EM, et al. Multiple instances of ancient balancing selection shared between humans and chimpanzees. Science. 2013;339:1578–1782. doi: 10.1126/science.1234070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Croze M, et al. A genome-wide scan for genes under balancing selection in drosophila melanogaster. BMC Evol Biol. 2017;17:15. doi: 10.1186/s12862-016-0857-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedrick PW. Population genetics of malaria resistance in humans. Heredity. 2011;107:283–304. doi: 10.1038/hdy.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alonso S, López S, Izagirre N, de la Rúa C. Overdominance in the human genome and olfactory receptor activity. Mol Biol Evol. 2008;25:997–1001. doi: 10.1093/molbev/msn049. [DOI] [PubMed] [Google Scholar]
- 18.Savova V, et al. Genes with monoallelic expression contribute disproportionately to genetic diversity in humans. Nat Genet. 2016;48:231–237. doi: 10.1038/ng.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cahoy J, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beissbarth T, Speed TP. Gostat: Find statistically overrepresented gene ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 21.Paolella G, Buono P, Mancini F, Izzo P, Salvatore F. Structure and expression of mouse aldolase genes. brain-specific aldolase c amino acid sequence is closely related to aldolase a. Eur J Biochem. 1986;156:229–235. doi: 10.1111/j.1432-1033.1986.tb09572.x. [DOI] [PubMed] [Google Scholar]
- 22.Vlotides G, Eigler T, Melmed S. Pituitary tumor-transforming gene: Physiology and implications for tumorigenesis. Endocr Rev. 2007;28:165–186. doi: 10.1210/er.2006-0042. [DOI] [PubMed] [Google Scholar]
- 23.Jahani-Asl A, et al. Control of glioblastoma tumorigenesis by feed-forward cytokine signaling. Nat Neurosci. 2016;19:798–806. doi: 10.1038/nn.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiley C, et al. Role for mammalian chitinase 3-like protein 1 in traumatic brain injury. Neuropathology. 2015;35:95–106. doi: 10.1111/neup.12158. [DOI] [PubMed] [Google Scholar]
- 25.Horvath S, Mirnics K. Immune system disturbances in schizophrenia. Biol Psychiatry. 2014;75:316–323. doi: 10.1016/j.biopsych.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lei Y, Xin X, Morgan D, Pintar J, Fricker L. Identification of mouse cpx-1, a novel member of the metallocarboxypeptidase gene family with highest similarity to cpx-2. DNA Cell Biol. 1999;18:175–185. doi: 10.1089/104454999315565. [DOI] [PubMed] [Google Scholar]
- 27.Burkin D, Kaufman S. The alpha7beta1 integrin in muscle development and disease. Cell Tissue Res. 1999;296:183–190. doi: 10.1007/s004410051279. [DOI] [PubMed] [Google Scholar]
- 28.Harr B, et al. Genomic resources for wild populations of the house mouse, mus musculus and its close relative mus spretus. Sci Data. 2016;3:160075. doi: 10.1038/sdata.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartl DL, Clark AG. In: Principles of Population Genetics. 2nd Ed Hartl DL, Clark AG, editors. Sinauer Associates; Sunderland, MA: 1989. [Google Scholar]
- 30.Jeffries AR, et al. Random or stochastic monoallelic expressed genes are enriched for neurodevelopmental disorder candidate genes. PLoS One. 2013;8:e85093. doi: 10.1371/journal.pone.0085093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Do C, et al. Mechanisms and disease associations of haplotype-dependent allele-specific dna methylation. Am J Hum Genet. 2016;98:934–955. doi: 10.1016/j.ajhg.2016.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eckersley-Maslin M, Spector D. Random monoallelic expression: Regulating gene expression one allele at a time. Trends Genet. 2014;30:237–44. doi: 10.1016/j.tig.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zwemer LM, et al. Autosomal monoallelic expression in the mouse. Genome Biol. 2012;13:R10. doi: 10.1186/gb-2012-13-2-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, et al. Dual DNA methylation patterns in the cns reveal developmentally poised chromatin and monoallelic expression of critical genes. PLoS One. 2010;5:e13843. doi: 10.1371/journal.pone.0013843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torrey EF, Bowler AE, Taylor EH, Gottesman II. Schizophrenia and Manic-Depressive Disorder: The Biological Roots of Mental Illness as Revealed by the Landmark Study of Identical Twins. Vol 41. Basic Books; New York: 1994. pp. 424–425. [Google Scholar]
- 36.Eckersley-Maslin M, et al. Random monoallelic gene expression increases upon embryonic stem cell differentiation. Dev Cel. 2014;28:351–365. doi: 10.1016/j.devcel.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu J, He F, Hu S, Yu J. On the nature of human housekeeping genes. Trends Genet. 2008;24:481–484. doi: 10.1016/j.tig.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 38.Zhang L, Li WH. Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Mol Biol Evol. 2004;21:236–239. doi: 10.1093/molbev/msh010. [DOI] [PubMed] [Google Scholar]
- 39.Takada T, et al. The ancestor of extant Japanese fancy mice contributed to the mosaic genomes of classical inbred strains. Genome Res. 2013;23:1329–1338. doi: 10.1101/gr.156497.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.