Significance
Most mutations are subject to competitive exclusion: Their descendants will either take over the population or go extinct. In special cases, a mutant may evade competitive exclusion by exploiting a different ecological niche. Both types of mutations can be found in large microbial populations, yet little is known about how they interact. By generalizing consumer-resource theory to include heritable beneficial mutations, we show that interactions between diversification and competitive exclusion can produce dramatic departures from existing models of evolution or ecology alone. These results suggest that short-term evolutionary processes could play an important role in shaping the structure of microbial communities.
Keywords: resource competition, asexual evolution, coexistence
Abstract
Microbial communities can evade competitive exclusion by diversifying into distinct ecological niches. This spontaneous diversification often occurs amid a backdrop of directional selection on other microbial traits, where competitive exclusion would normally apply. Yet despite their empirical relevance, little is known about how diversification and directional selection combine to determine the ecological and evolutionary dynamics within a community. To address this gap, we introduce a simple, empirically motivated model of eco-evolutionary feedback based on the competition for substitutable resources. Individuals acquire heritable mutations that alter resource uptake rates, either by shifting metabolic effort between resources or by increasing the overall growth rate. While these constitutively beneficial mutations are trivially favored to invade, we show that the accumulated fitness differences can dramatically influence the ecological structure and evolutionary dynamics that emerge within the community. Competition between ecological diversification and ongoing fitness evolution leads to a state of diversification–selection balance, in which the number of extant ecotypes can be pinned below the maximum capacity of the ecosystem, while the ecotype frequencies and genealogies are constantly in flux. Interestingly, we find that fitness differences generate emergent selection pressures to shift metabolic effort toward resources with lower effective competition, even in saturated ecosystems. We argue that similar dynamical features should emerge in a wide range of models with a mixture of directional and diversifying selection.
Ecological diversification and competitive exclusion are opposing evolutionary forces. Conventional wisdom suggests that most new mutations are subject to competitive exclusion, while ecological diversification occurs only under highly specialized conditions (1). Recent empirical evidence from microbial, plant, and animal populations has started to challenge this assumption, suggesting that the breakdown of competitive exclusion is a more common and malleable process than is often assumed (2–4). Particularly striking examples have been observed in laboratory evolution experiments, in which primitive forms of ecology evolve from a single ancestor over years (5), months (6), and even days (7).
In the simplest cases, the population splits into a pair of lineages, or “ecotypes,” that stably coexist with each other due to frequency-dependent selection, leading to a breakdown of competitive exclusion (5, 6, 8–10). The mechanism of coexistence can often be traced to differences in resource utilization or to the accessibility of privileged spatial or temporal niches. Interestingly, these microbes rarely cease their evolution once ecological diversification has been achieved. Sequencing studies have shown that adaptive mutations continue to accumulate within each ecotype, even when population-wide fixations are rare (11–13). This additional evolution can cause the ecological equilibrium to wander over longer timescales, as observed in the shifting population frequencies of the two ecotypes (5, 13). In certain cases, these evolutionary perturbations can even drive one of the original lineages to extinction, either through the outright elimination of the niche (9) or by the invasion of individuals that mutate from the opposing ecotype (12).
Pairwise coexistence is the simplest form of community structure, but similar dynamics have been observed in more complex communities as well. Some laboratory experiments diversify into three or more ecotypes (7, 14, 15), and it is likely that previously undetected ecotypes may be present in existing experiments (13). Moreover, many natural microbial populations evolve in communities with tens or hundreds of ecotypes engaged in various degrees of competition and coexistence (16–18). Although the evolutionary dynamics within these communities are less well characterized, recent work suggests that similar short-term evolutionary processes can occur in these natural populations as well (19–21).
While the interactions between microbial adaptation and ecology are known to be important empirically, our theoretical understanding of this process remains limited in comparison. Early work in the field of adaptive dynamics (22) showed how ecological diversification emerges under very general models of frequency-dependent trait evolution, which are thought to describe the limiting behavior of a wide class of ecological interactions near the point of diversification. Numerous studies have also investigated the effects of evolution on ecological diversification and stability using computer simulations, in which the parameters of a particular ecological model are allowed to evolve over time (23–29). Yet while both approaches can reproduce some of the qualitative behaviors observed in experiments, it has been difficult to forge a more quantitative connection between these models and the large amount of molecular data that is now available.
One reason that quantitative comparisons have been difficult is that evolution also selects for other traits that are not directly involved in diversification. For example, natural selection always works to maintain essential cellular functions, and there may be a benefit to removing costly functions that are not needed in the current environment. As a result, mutations that influence an ecologically relevant phenotype like acetate metabolism might have to compete with constitutively beneficial mutations that are only tangentially related to metabolism [e.g., the loss of the yeast mating pathway (30)]. In some experiments, these constitutively beneficial mutations may even compose the bulk of the mutations that reach observable frequencies (12, 13, 31). Although many models exist for describing constitutively beneficial or deleterious mutations in the absence of ecology (32), we lack even a basic theoretical understanding of how they behave when they are linked to ecological phenotypes and vice versa. The absence of quantitative theoretical predictions makes it difficult to draw any inferences from the vast molecular data that are now available.
To start to bridge this gap, we introduce a simple, empirically motivated model that describes the interplay between ecological diversification and directional selection at a large number of linked loci. The ecological interactions derive from a well-studied class of consumer resource models (33–36), in which individuals compete for multiple substitutable resources (e.g., different carbon sources) in a well-mixed environment. We extend this ecological model to allow for heritable mutations in resource uptake rates, which can either divert metabolic effort between resources or increase the growth rate on all resources. The latter class of mutations provides a natural way to model adaptation at linked genomic loci.
Constitutively beneficial mutations might seem like an ecologically trivial addition to the model, since they are always favored to invade on short timescales. On longer timescales, however, we show that these accumulated fitness differences can dramatically influence both the ecological structure and the evolutionary dynamics that take place within the community. By focusing on the weak mutation limit, we derive analytical expressions for these dynamics in the two-resource case, and we show how our results extend to larger communities as well. These analytical results provide a general framework for integrating ecological and population-genetic processes in evolving microbial communities and suggest ways in which these processes might be inferred from time-resolved molecular data.
Evolutionary Model of Resource Competition
To investigate the interactions between ecological diversification and directional selection, we focus on a simple ecological model in which individuals compete for an assortment of externally supplied resources in a well-mixed, chemostat-like environment (Fig. 1). This resource-based model aims to capture some of the key ecological features observed in certain microbial evolution experiments (5, 8), as well as more complex ecosystems such as the gut microbiome (18), while remaining as analytically tractable as possible.
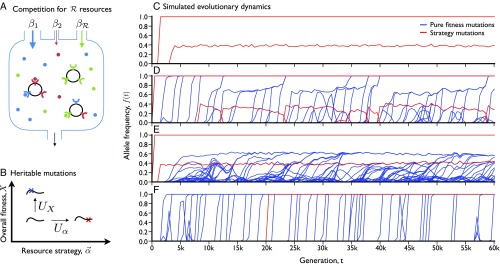
Fig. 1.
Ecological and evolutionary dynamics in a simplified consumer-resource model. (A) Schematic depiction of ecological dynamics. Substitutable resources are supplied to the chemostat at constant rates (), measured in units of biomass (). Cells import resources at genetically encoded rates, , which define a normalized resource strategy and overall fitness . (B) Schematic depiction of evolutionary dynamics. Mutations that alter resource strategies () occur at rate , while mutations that alter overall fitness () occur at rate . (C–F) Simulated ecological and evolutionary dynamics, starting from a clonal ancestor, in an environment with resources. C–F represent independent populations evolved under different sets of parameters, which differ only in the mutation rates and fitness benefits of pure fitness mutations (SI Appendix, section 5.1). Lines denote the population frequency trajectories of all mutations that reached frequency in at least one time point. Resource strategy mutations are shown in red, while pure fitness mutations are shown in blue.
In our idealized setting, individuals compete for substitutable resources, which are supplied by the environment at fixed rates (Fig. 1). Individuals are characterized by a resource utilization vector , which describes how well they can grow on each of the resources. We assume that the resource utilization phenotypes are constitutively expressed, so that we may neglect complicating factors like regulation. We find it useful to decompose the phenotype into a normalized portion and an overall magnitude . The components of represent the fractional effort devoted to growth on resource , so we refer to this quantity as the “resource strategy vector.” In contrast, the parameter resembles an environment-independent measure of “overall fitness,” an analogy that we make more precise below.
We assume that individuals reproduce asexually, so that the state of the ecosystem can be described by the number of individuals with a given resource strategy vector and fitness . Under suitable assumptions, the ecosystem can be described by the stochastic differential equation,
| [1] |
where is a fixed carrying capacity, is the relative frequency of strain , and is a stochastic contribution arising from genetic drift (SI Appendix, section 1.1). The state of the environment is encoded by the set of resource-specific mean fitnesses,
| [2] |
where denotes the fractional flux supplied by resource . Eq. 1 is an example of a more general and well-studied class of consumer-resource models introduced by refs. 33 and 37, whose ecological properties have been explored in several recent works (34–36).
However, the essential features of Eq. 1 are not limited to this consumer-resource framing. In SI Appendix, section 1.4, we argue that Eq. 1 captures the limiting behavior of a much larger class of models in the limit that and are both small compared with one. In this case, we can expand the exponential in Eq. 1 to obtain the lowest-order contribution,
| [3] |
When , we recover the standard Wright–Fisher model of population genetics (38), with its logistic growth function, . The parameter coincides with the standard measure of (log) fitness. In the presence of multiple resources, Eq. 3 can be regarded as the simplest generalization of the Wright–Fisher model that incorporates multiple fitness dimensions. From an ecological perspective, Eq. 3 can also be viewed as a special version of the Lotka–Volterra model that arises when the interactions between strains are mediated by intensive variables (the resource-specific mean fitnesses, ). This is an important simplification: Although there is no closed-form solution for when , Eq. 1 still possesses a convex Lyapunov function (SI Appendix, section 1.2), which implies that must approach a unique and stable equilibrium at long times.
The ecological model in Eq. 1 describes only the competition between a fixed set of strains. To incorporate evolution, we also allow for new strains to be created through the process of mutation. We show that it is useful to distinguish between two broad classes of mutations. The first class consists of mutations that alter an individual’s resource uptake strategy (“strategy mutations”). For simplicity, we assume that these mutations constitute a perfect tradeoff, so that the overall fitness remains unchanged (although we eventually relax this assumption below). We assume that strategy mutations occur at a per-genome rate and result in a new resource strategy drawn from some distribution . In addition to strategy mutations, we consider a second class of “pure fitness mutations,” which alter the overall fitness but leave the resource strategy unchanged. These mutations capture the effects of directional selection at a large number of other loci throughout the genome, which may be only tangentially related to the resource utilization strategy. We assume that these fitness mutations arise at a per-genome rate and that they increment by an amount drawn from the distribution of fitness effects, . For simplicity, we assume that there is no macroscopic epistasis for fitness (39), so that remains the same for all genetic backgrounds.
We note that this division into fitness and strategy mutations is neither exhaustive nor unambiguous. Some changes in resource strategy may also incur a fitness cost, and one can simulate a pure fitness mutation by shifting metabolic effort away from resources that are not present in the current environment (i.e., those with ). Nevertheless, considering these idealized cases as our fundamental axes will prove to be a useful conceptual tool, which provides additional insight into the behavior of our model.
For example, pure fitness mutations might seem like an ecologically trivial addition to the model, because they are always favored to invade. However, computer simulations show that these accumulated fitness differences can still have a dramatic influence on both the ecological structure and the evolutionary dynamics that arise in a given population. Fig. 1 depicts individual-based simulations of four populations, which are subject to the same environmental conditions and the same supply of strategy mutations, but have different values of and (SI Appendix, section 5.1). Depending on the supply of fitness mutations, the behaviors can include rapid diversification and stasis (Fig. 1C), unstable but continually renewed coexistence (Fig. 1D), stable coexistence and rapid within-clade evolution (Fig. 1E), or the permanent disruption of coexistence (Fig. 1F). In this way, the seemingly simple model in Eq. 1 can produce a diverse range of behaviors, which at least superficially resemble the complex dynamics observed in some microbial evolution experiments.
To understand these different behaviors and how they depend on the underlying parameters, we start by analyzing the simplest nontrivial scenario, in which the strains evolve in an environment with just two resources. In this case, the environmental supply rates and resource uptake strategies can be described by scalar parameters and , respectively. This case is already sufficient to elucidate many of the key qualitative behaviors and fundamental timescales involved, while maximizing analytical tractability. In Analysis, we extend this analysis to larger numbers of resources and comment on the additional features that are unique to this more complicated scenario.
Analysis
Selection for Ecosystem to Match Environment, Stable Coexistence.
We begin by considering the dynamics in the absence of fitness differences (, ). The ecological dynamics in this “neutral” scenario were recently described in ref. 34, and it will be useful to build on these results when we introduce fitness differences below.
We begin by considering a single strategy mutation that occurs in a clonal population of type , creating a new strain of type . The initial dynamics of this mutation can be described by a branching process with growth rate , also known as the “invasion fitness” (SI Appendix, section 2.1). In this case, the invasion fitness is given by
| [4] |
where is the difference between the mutant and wild-type uptake rates. The invasion fitness is positive whenever and have the same sign: If , then selection will favor mutations that increase , while if , selection will favor mutations that decrease . In this way, selection tries to tune the population uptake rate to match the environmental supply rate. If , then the invasion fitness vanishes for all further strategy mutants. This constitutes a marginal evolutionarily stable state (ESS). Using the definition of in Eq. 2, we can see that the universal dynamics in Eq. 3 correspond to a near-ESS limit, where the resource uptake rates remain close to . For the sake of generality, we focus on this limit for the remainder of the main text. The full expressions for the microscopic model in Eq. 1 are listed in SI Appendix. When and are both close to , Eq. 4 reduces to the quadratic form,
| [5] |
Since all mutations first arise in a single individual, many will be lost to genetic drift, even when their invasion fitness is positive. With probability , the mutant lineage will survive drift long enough to reach frequency and will then start to increase deterministically at rate . In sufficiently large populations, the transition to deterministic growth will occur long before the mutant starts to influence its own growth rate, so that the constant invasion fitness assumption is justified (SI Appendix, section 2.1).
At long times, the ecological dynamics will lead to one of two final states: Either the mutant will replace the wild type (competitive exclusion) or the two will coexist at some intermediate frequency (Fig. 2A). The latter scenario will occur if and only if the wild type can reinvade a population of mutants, which requires that the reciprocal invasion fitness, , is also positive. By examining this expression, we see that the mutant will outcompete the wild type if its strategy lies between and , while stable coexistence occurs when and span (i.e., or vice versa). When this condition for coexistence is met, the steady-state frequencies are determined by the linear equation,
| [6] |
whose solution is given by (34). In other words, the relative frequencies of the strains are inversely proportional to their distance from the environmental supply rate. According to Eq. 6, these frequencies are chosen such that the population-averaged uptake rate exactly balances the resource supply rate . This provides an intuitive explanation for the cause of coexistence: By maintaining the strains at intermediate frequencies, the population is able to match the environmental supply rate more closely than it could with either strain on its own.
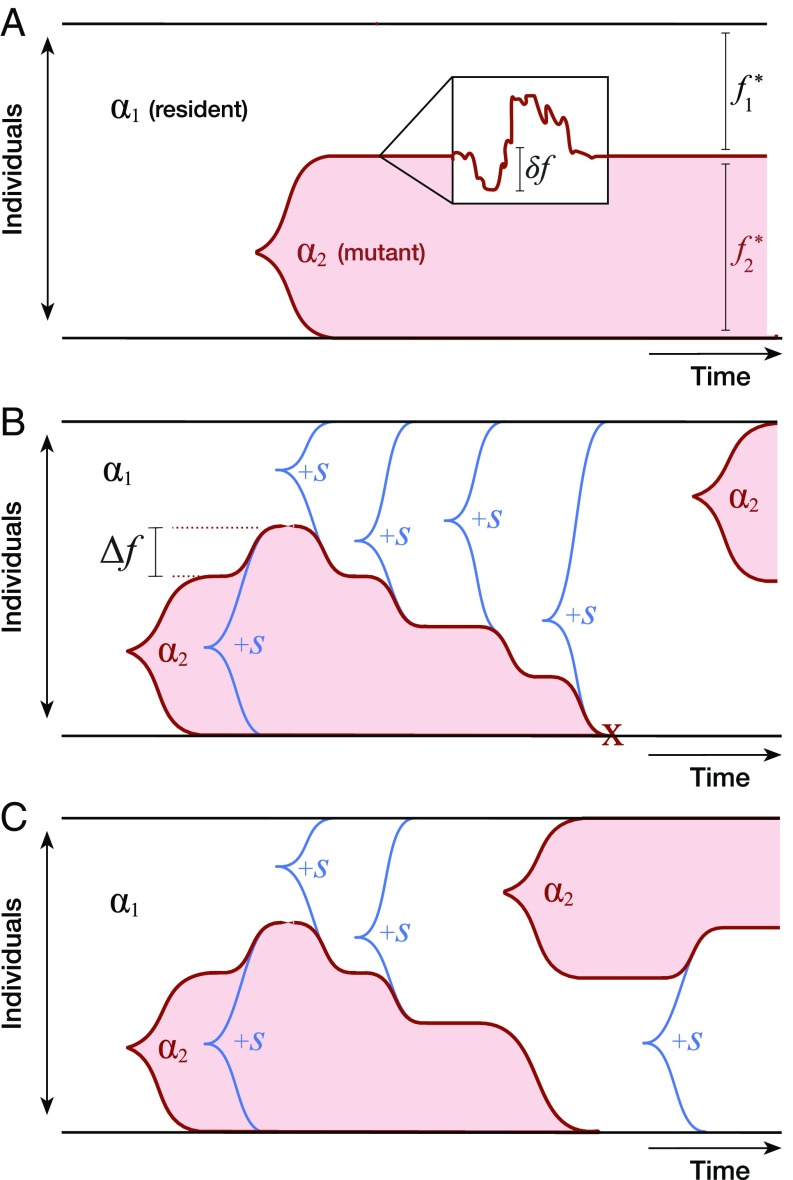
Fig. 2.
Schematic illustration of key eco-evolutionary processes in a two-resource ecosystem. (A) Ecological diversification from a clonal ancestor. In the absence of fitness mutations, strains coexist at a stable equilibrium () with fluctuations () controlled by genetic drift. Further strategy mutations are not favored to invade. (B) Pure fitness mutations that sweep within an ecotype shift the stable equilibrium by ; accumulated fitness differences can ultimately drive ecotypes to extinction. Further strategy mutations allow the winning clade to rediversify at a later time. (C) Occupied niches can also be invaded by strategy mutations that arise in fitter genetic backgrounds. In this case, the original ecotype lineage is driven to extinction while the ecological structure of the community is preserved.
Once this ecological equilibrium is attained, number fluctuations will continuously perturb the true frequency away from (Fig. 2A), subject to a linearized restoring fitness (SI Appendix, section 2.1). The restoring force is strong compared with genetic drift when , which leads to linearized fluctuations of order and a lifetime for the stable state that is exponentially long in . At this point, additional strategy mutants are subject to very weak selection pressures: Fluctuations will induce momentary invasion fitnesses of order (which can be large compared with ), but these fitness effects are quickly averaged to zero during the generations required for such a mutation to establish (SI Appendix, section 2.2). Thus, once the population diversifies to fill the two niches, the rate of evolution dramatically slows down (as in Fig. 1A), since the relevant timescales are controlled by genetic drift. In this way, a large-effect mutation can allow the ecosystem as a whole to reach an effective ESS, long before any of the constituent strains reach the ESS on their own.
Diversification Load.
We are now in a position to analyze how fitness alters the basic picture above. We begin by revisiting the invasion of a mutant strain in an initially clonal population, this time allowing for a fitness difference between the mutant and wild type. In this case, the new invasion fitness is given by a simple linear combination,
| [7] |
where is the invasion fitness for a pure strategy mutation from Eq. 4. This result describes, in quantitative terms, how selection balances its ecological preferences () with its desire to maximize fitness (). When the uptake rate of the resident population is far from the environmental supply rate [], the ecological selection pressures can be quite strong, with invasion fitnesses as high as . This implies that strongly deleterious mutations of order
| [8] |
can hitchhike to fixation when the population colonizes a new ecological niche (a form of “diversification load”).
Fitness Differences Perturb Ecological Equilibria.
In addition to shifting the invasion fitness of a new mutation, fitness differences can also alter the long-term ecological equilibrium between mutant and wild type in Eq. 6. In the extreme limit, this can disrupt the stable coexistence altogether. If the mutant is less fit than the wild type (), this will occur whenever is less than the maximum diversification load in Eq. 8. On the other hand, if , extinction will occur when the wild type no longer has positive invasion fitness or when exceeds a threshold
| [9] |
We note that the fitness differences in Eqs. 8 and 9 are lower than the values required for the mutant or wild type to dominate in all environmental conditions (SI Appendix, section 2.1). Instead, the fitness thresholds strongly depend on how the resource strategies differ from each other and how they differ from the environmental supply rate. When , even small fitness differences () can disrupt the stable ecology, while for , much larger fitness differences () can be tolerated.
When , the two strains continue to coexist, but their equilibrium frequency is no longer given by Eq. 6. In this case, the competing drive to maximize fitness means that selection will no longer favor an ecology that matches the environmental resource distribution, at least not perfectly. In SI Appendix, section 2.1, we show that the new equilibrium frequency is given by
| [10] |
where is the neutral ecological equilibrium from Eq. 6. From this expression, we can read off the typical fitness differences required to perturb from its present value. This fitness sensitivity is again determined by the distance between the two resource strategies. If , large fitness differences () are required to change the equilibrium frequency, while for , even very small fitness differences () can generate large changes in the equilibrium frequency.
Further Fitness Evolution and Diversification–Selection Balance.
Once the population achieves the stable ecology in Eq. 10, additional fitness mutations will occur in each strain with probability proportional to the equilibrium frequency . In our model, the invasion fitness of such a mutation is simply its fitness effect , independent of the ecological state of the population. With probability , this mutation will sweep through its parent clade, changing the fitness difference between the clades by and the equilibrium frequency by (Fig. 2B). In the linear regime of Eq. 10, the frequency and fitness changes are directly related,
| [11] |
where is the fitness scale that determines changes in equilibrium frequency. If , then the stable coexistence will be disrupted, and the mutant clade will take over the population. We refer to such a scenario as ecosystem collapse, since one of the niches is no longer occupied.
Similar behavior can occur when as well, except that now the ecosystem collapse occurs due to the cumulative effect of many pure fitness mutations. When the fitness mutations accumulate independently, this process can be described by an effective diffusion model
| [12] |
with a bias that reflects the higher probability of producing a mutation in a larger clade (SI Appendix, section 2.3). Eq. 12 superficially resembles the drift-induced perturbations at ecological equilibrium, except that the bias is now unstable rather than restoring. When , the mutation bias is weak, and the clade frequencies undergo a random walk . But after a time of order , the frequency differential grows large enough that the more prevalent clade will deterministically produce more beneficial mutations, so that it is destined for fixation. After a time of order , the fitness difference between the clades grows so large that the ecosystem finally collapses (Fig. 2B). This timescale sets an upper limit on the lifetime of the stable state when many fitness mutations are available.
Once the ecosystem collapses, there will be a strong selection pressure for the winning clade to rediversify through additional strategy mutations and restart this process from the beginning (Fig. 2B). To gain insight into these dynamics, we first consider the case where the resource strategies are controlled by a single genetic locus, with fixed phenotypes and , and mutations that alternate between the two states at rate . After an ecosystem collapse, Eq. 4 shows that the invasion fitness for the opposite strategy is given by , so the collapsed state will persist for a time of order , until the stable ecology is reestablished. If the two strategies are symmetric about , so that , the new stable state will persist for generations in the absence of additional strategy mutations, and the process will then repeat itself. The relative probability of observing the population in the collapsed () or saturated () states is therefore given by
| [13] |
This expression shows the minimum amount of strategy mutations, or the maximum amount of pure fitness mutations, that allow the population to maintain a saturated ecosystem. We refer to this dynamic steady state as “diversification–selection balance,” in analogy to mutation–selection balance in population genetics (40). Note that this balance crucially depends on the state of the ecosystem through . All else being equal, ecosystems with more similar resource uptake strategies will be disrupted more easily than those with a higher degree of specialization.
Invading Ecotypes Can Delay Ecosystem Collapse.
Strictly speaking, the derivation of Eq. 13 is valid only in the limit that , since we neglected mutations between and when both niches were filled. When (i.e., when the ecosystem spends an appreciable amount of time in the saturated state), we must also account for mutations between the two strategies that arise before the ecosystem collapses. Those mutations that arise in the less-fit clade will have little chance of invading. However, a mutation from the more-fit to the less-fit strategy will establish with probability , where is the current fitness difference between the two clades. If this mutation is successful, it will outcompete the resident lineage with the corresponding value of and reset the fitness difference to (Fig. 2C). In this way, invasion from one ecotype to another can significantly delay the process of ecosystem collapse, since it relieves the tension between fitness maximization () and selection to match the environment ().
To analyze this process, we note that successful invasion events will occur as an inhomogeneous Poisson process with rate , where and are again determined by the diffusion model in Eq. 12. This leads to a characteristic invasion timescale
| [14] |
which is derived in SI Appendix, section 2.3. Each of these regimes corresponds to a different intuitive picture of the dynamics. In the first case, strategy mutations are frequent compared with pure fitness mutations, and invasion occurs almost immediately after the first fitness mutation arises. In the second case, invasion occurs after multiple fitness mutations have accumulated, but when the frequencies of the clades still wander diffusively relative to each other []. In the third regime, invasion occurs after one of the clades has grown to a sufficiently large frequency that it would have deterministically led to ecosystem collapse. When the invading mutant establishes, it will therefore cause a rapid shift in the frequencies of the ecotypes as returns to .
Finally, when , strategy mutants are sufficiently rare that the ecosystem will typically collapse and rediversify before invasion can occur. This sets the region of validity of the diversification–selection balance in Eq. 13. Interestingly, Eq. 13 shows that collapse and rediversification can still dominate over invasion even when both niches are typically filled (). In this case, both the genealogical structure and the typical state of the ecosystem will resemble the invasion regime, but the historical record would contain a series of punctuated extinction and diversification events, interspersed with long periods of gradual fitness evolution.
Fitness Differences Create Opportunities for Ecological Tinkering.
Our derivation of Eqs. 12 and 14 assumed that the two ecotypes were fixed by the genetic architecture of the organism. Individuals could mutate between and , but mutations to other points in strategy space were forbidden. In the absence of fitness differences, we saw that selection for these additional strategy mutants is weak once both niches have been filled (), potentially justifying the single-locus assumption in terms of a priority effect. However, the previous analysis shows that there can be strong selection to switch strategies once fitness mutations accumulate, so it is also plausible that fitness differences could lead to selection for strategy mutations more generally.
To investigate these selection pressures, we consider a population that is currently described by the steady state in Eq. 10. We then consider strategy mutations that occur on the background of , altering its strategy to while leaving its fitness intact. The invasion fitness for such a mutation is given by
| [15] |
As anticipated, fitness differences create additional selection pressure for strategy mutations beyond the simple switching behavior considered above.
The direction of selection is determined by the sign of . In the background of the fitter clade (), selection favors mutations that increase the strategy in the direction of (a form of generalism), while simultaneously disfavoring mutations that lead to increased specialization (Fig. 3). The opposite behavior occurs in the less-fit background, with selection favoring mutations that increase the distance from , leading to increased specialization. Both behaviors have an intuitive explanation in terms of individuals preferring to allocate their metabolic energy toward the resource with the least-fit consumers, thereby minimizing the effective competition that they experience.
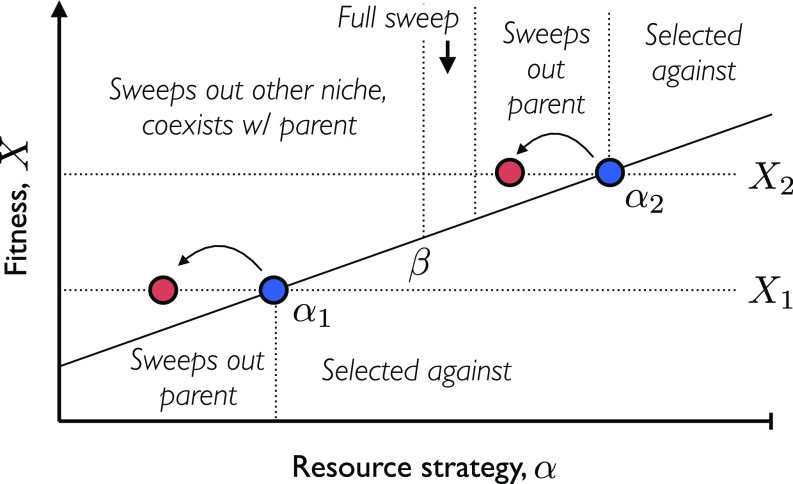
Fig. 3.
Invasion fitness landscape for additional strategy mutations in a two-resource ecosystem. The two resident ecotypes are illustrated by blue circles, while red circles denote mutant strains created by strategy mutations on one of the ecotype backgrounds. The solid black line denotes the effective mean fitness, , experienced by a given resource strategy. Strains with overall fitness () above this line are favored to invade, while others are selected against. If a mutant successfully invades, its effect on the ecosystem is indicated by the text.
Once a successful strategy mutation arises, it will sweep through part of the population and alter the ecological equilibrium (Fig. 3). Mutations in the less-fit clade are straightforward to analyze. Since these are always directed away from both and , these mutants will sweep through their parent clade and increase the equilibrium frequency according to Eq. 10. Successful mutations in the fitter clade have a wider range of outcomes, since these are always directed toward and . If , the mutant lineage will outcompete the less-fit strain and will stably coexist with its parent clade at an equilibrium frequency . On the other hand, if , the mutant lineage will always sweep through its parent clade . If is sufficiently close to , this will simply lead to an increase in frequency according to Eq. 10. However, if is close enough to that becomes less than the actual fitness difference, , then the mutant will sweep out both clades and lead to an ecosystem collapse and subsequent rediversification. Thus, in addition to creating a larger target for invasion events, these additional strategy mutations can also enhance the probability of ecosystem collapse. The balance between these competing tendencies will depend on the genetic architecture of the resource strategies, , which is poorly parameterized by existing data. A detailed exploration of the potential regimes remains an interesting topic for future work.
Beyond Pairwise Coexistence.
Our previous analysis focused on environments with only two substitutable resources, where at most two strains can coexist at equilibrium. In this case, the structure of the stable ecosystem was simple enough to admit a full analytical solution, which we could use to derive explicit predictions for many evolutionary quantities of interest. However, many microbial communities are found in environments with large numbers of potential resources and flexible gene pools that allow them to alter their resource uptake rates through horizontal gene transfer (41). It is therefore natural to ask how our results generalize to these more complicated environments as well. A full analysis of this case is beyond the scope of the present work, as there are even fewer constraints on the space of ecological and evolutionary parameters compared with the two-resource case. Nevertheless, it is still useful to know whether our qualitative results extend beyond and whether there are fundamentally new behaviors that arise only in higher dimensions.
For a general ecological equilibrium, a mutation that alters the phenotype of a resident strain from to will have an invasion fitness
| [16] |
where the resource-specific mean fitnesses in Eq. 2 must be evaluated at the equilibrium frequencies (SI Appendix, section 3.1). Increases in are favored when is lower than the “effort-averaged” for the other resources and vice versa. Thus, similar to the two-resource case, there is still a sense in which selection favors mutations that flow from high values of to lower values of , although there are now beneficial directions, , rather than just one.
The invasion fitness in Eq. 16 depends on the current community composition only through the intensive variables . In a “saturated” ecosystem, where the number of coexisting strains is equal to the number of resources, these can be directly obtained by a matrix inversion of Eq. 1,
| [17] |
where is the left inverse of . Thus, we see that in a saturated ecosystem, the are given by linear combinations of the strain fitnesses , justifying their interpretation as resource-specific mean fitnesses. Moreover, perturbation expansions of suggest that the prefactor is still inversely proportional to an effective distance between the strategies (SI Appendix, section 3.2), similar to the two-resource case in Eq. 15. We note that the equilibrium values of are conditionally independent of both the resource supply vector and the strain frequencies ; these quantities influence only through shaping the set of resource strategies that coexist at equilibrium. Thus, these saturated ecosystems dynamically adjust their composition to screen the internal selection pressures from the external environmental conditions. Similar findings were recently obtained for the neutral case [where (34)], as well as in certain community assembly processes in the limit (35, 36). Eq. 17 shows that this is a generic property that occurs whenever the number of surviving species is equal to the number of resources.
In this limit, the steady-state frequencies can be obtained from a similar matrix inversion,
| [18] |
which serves as the generalization of Eq. 10 for multiple resources (SI Appendix, section 3.2). As in the two-resource case, small fitness differences perturb the neutral ecological equilibrium via linear combinations of the strain fitnesses, with a prefactor that is inversely proportional to the square of the effective distance between the resource strategies.
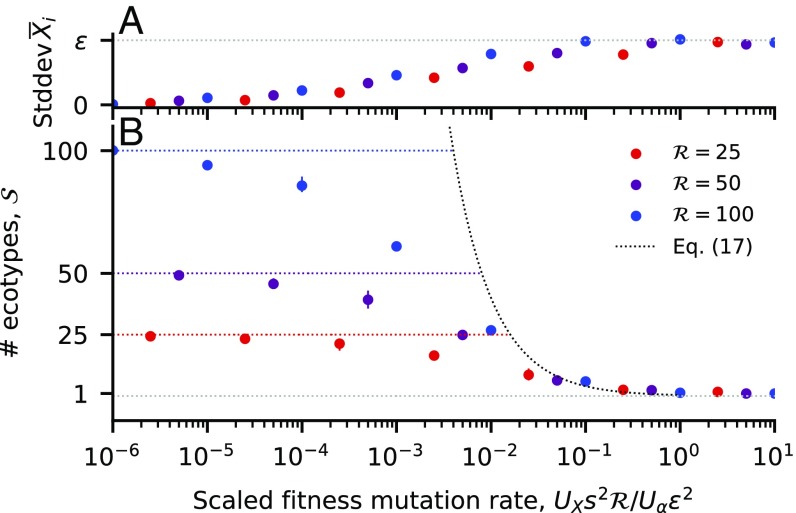
While the saturated case is particularly simple, we saw above that fitness mutations can drive the number of surviving species below this saturated value. In contrast to the two-resource case, these “unsaturated ecosystems” can now harbor multiple coexisting strains when , leading to a continuous generalization of the diversification–selection balance in Eq. 13. To investigate this effect, we performed computer simulations of a binary strategy space model, in which individuals can either use or not use a given resource, with mutations that toggle individual uptake rates on and off (SI Appendix, section 3.3). The results recapitulate the qualitative behavior observed for resources, in that a sufficiently high rate of pure fitness mutations can constrain the number of distinct strategies that are able to coexist (Fig. 4B). To compensate for the strong ecological selection pressures that can arise when , the populations are forced to evolve consortia of “generalist” strains such that is still close to , at least at lowest order (Fig. 4A).
Fig. 4.
Diversification–selection balance when . Circles depict the long-term steady state from simulations of a binary resource-use model in the strong-selection, weak mutation limit, with nearly uniform resource supply rates (SI Appendix, section 5.2). Each point denotes an average over multiple time points from three independent replicates; solid lines indicate the minimum and the maximum replicate. (A) The SD in across the resources. (B) The number of coexisting ecotypes. The colored dashed lines denote the maximum ecosystem capacity . The black dashed line depicts the scaling relation in Eq. 19 which applies for , with an prefactor of included for visualization.
In a nearly uniform environment [], simulations show that the steady-state ecosystem tends to be dominated by a generalist strain () and a collection of single loss-of-function variants () that recently descended from mutations in the generalist background (Fig. 5 and SI Appendix, Figs. S2–S4). This is reminiscent of a mutation-load argument (40), in which the preference for the generalist strain is balanced by the greater entropy of loss-of-function mutants. However, a key difference in this case is that the generalist strain is not actually favored by selection. By definition, all of the transient states in Fig. 5 are ecologically stable, while the preference for the generalist strain arises dynamically from the race to acquire pure fitness mutations.
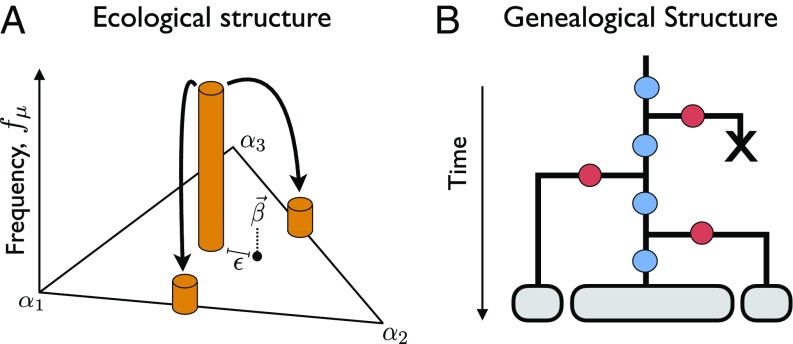
Fig. 5.
(A and B) Schematic of (A) ecological and (B) genealogical structure at the evolutionary steady state described in Eq. 19. In B, blue circles represent pure fitness mutations and red circles represent loss-of-function strategy mutations.
The dynamics of this process can be characterized analytically in the weak mutation limit, yielding a simple heuristic expression for the diversification–selection balance,
| [19] |
which is valid for (SI Appendix, section 3.3). The transition to the fully saturated state () requires an even more stringent condition, which implies that unsaturated ecosystems are obtained for a very broad parameter regime (Fig. 4B). In both cases, a larger number of substitutable resources will lead to a less diverse ecosystem at diversification–selection balance. This is ultimately due to the fact that the difference between generalists and single loss-of-function variants becomes increasingly small as .
This suggests that the relative frailty of the diversification–selection balance in Eq. 19 may be a pathological feature of the simple genetic architecture that we have assumed, in which fit generalist phenotypes are easily accessible. If we instead impose an upper limit on the number of resources that a strain can metabolize, heuristic calculations suggest that diversification–selection balance will be achieved for substantially higher values of , even for large (SI Appendix, section 3.4). In this case, the ecological and genealogical structures that are attained at this evolutionary steady state will be considerably more complex than the shallow star-shaped genealogies in Fig. 5. A detailed analysis of this steady state remains an interesting topic for future work.
Discussion
In microbial populations, primitive ecological interactions can evolve spontaneously over years (5), months (6), and even days (7). Yet this process rarely takes place in isolation. In rapidly evolving populations, diversifying selection must compete with directional selection acting on other loci throughout the genome. Here, we have introduced a simple mathematical framework to model the interactions between these two processes in asexually reproducing organisms.
The ecological interactions in our model emerge from the competition for substitutable resources (e.g., different carbon sources), according to a well-studied class of models from theoretical ecology (33–36). To incorporate evolution into this model, we assumed that individuals can acquire mutations that alter their resource uptake rates. We showed that it is useful to distinguish between two characteristic types of mutations: (i) strategy mutations, which divert metabolic effort from one resource to another, and (ii) fitness mutations, which increase the overall growth rate but leave the relative uptake rates unchanged. Strategy mutations enable ecological diversification, while fitness mutations capture the effects of directional selection at other genomic loci.
This classification scheme is best viewed as a conceptual tool, rather than a statement about the underlying biology. We have mostly focused on mutations with either a perfect tradeoff or a perfect benefit, but Eqs. 7 and 16 apply equally well in more realistic cases where a shift in resource strategy is accompanied by a change in the overall growth rate. These expressions can be used to predict when the costs of an opportunistic mutation will outweigh its ecological benefits or vice versa. Similarly, true fitness mutations (e.g., an increase in ATP efficiency) are assumed to be rare in nature, since they could have fixed in the population long ago. In practice, effective fitness mutations are more likely to correspond to strategy mutations whose tradeoffs are simply not exposed by the current environmental conditions. In this picture, the overall fitness can be viewed as an emergent trait that arises whenever we project high-dimensional cellular phenotypes onto a restricted set of axes (SI Appendix, section 4.3). The presence of effective fitness mutations may therefore be a more general phenomenon than their name would seem to imply.
The creation of new strains via mutation bears a superficial resemblance to immigration from a fixed species pool, which is the traditional scenario considered in theoretical ecology. However, this analogy is exact only in the absence of inheritance, when the phenotypes of nearby genotypes are uncorrelated from each other. In contrast, when the effects of mutations are heritable, we have seen that directional selection can produce dramatic departures from traditional ecological predictions.
Similar to immigration (36, 42), strategy mutations allow an initially clonal population to diversify into stably coexisting ecotypes, whose upper bound is set by the number of resources. Yet because fitness mutations are heritable, further evolution will lead to fitness differences between the clades, which can dynamically shift the ecological equilibrium over time and eventually drive less-fit clades to extinction. The mere observation that selection can disrupt coexistence is not surprising, since drug resistance or other harsh selection regimes provide striking examples of this effect. However, our quantitative analysis shows that this collapse can happen long before any clade is universally inferior to another and that it can result from the compound effect of many small-effect mutations that would not lead to extinction on their own. These results suggest that ongoing directional selection may have a larger impact on the structure of microbial communities than is often assumed. In particular, while previous ecological analyses suggest that the number of ecotypes should meet (35, 36) or even exceed (34) the number of resources, our results raise the possibility that they could also reside at a diversification–selection balance below the maximum capacity of the ecosystem.
In addition to their influence on coexistence, we also found that fitness differences accrued via directional selection will generate emergent selection pressures for continual evolution of the ecological phenotypes, even in a saturated ecosystem. While these internal selection pressures are reminiscent of the Red Queen effect (43), our quantitative analysis shows that they select for different phenotypes than in the standard predator–prey setting. In particular, less-fit clades do not experience increased selection pressure to narrow their fitness deficit by accumulating fitness mutations. Instead, selection favors mutations that divert metabolic effort toward resources with lower effective competition, even at the cost of widening the fitness deficit. Moreover, the direction of selection toward any given resource can shift dynamically as the fitness differences and resource uptake strategies evolve over time.
Most of our analysis focused on the strong-selection–weak-mutation regime, in which the current ecological equilibrium is attained before the next mutation occurs. In this limit, when the resource uptake strategies are sufficiently close to the supply rates, our model takes on a universal form that closely resembles traditional models of adaptive dynamics (22, 44). The key difference is that directional selection behaves as an additional trait dimension, which is effectively constrained to remain far from its optimum at all times (SI Appendix, Fig. S1 and section 4). Our results show that this simple broken symmetry can lead to dramatic deviations from the standard adaptive dynamics picture.
In contrast to adaptive dynamics, we also allow for mutations that have noninfinitesimal effects on resource uptake rates, which turn out to play a key role in controlling the dynamical behaviors that we observe. In practice, the genetic architectures of most ecological interactions remain poorly characterized empirically. In a few well-studied cases, ecological diversification can be traced to a single large-effect mutation (9, 45), while in others, a series of smaller mutations have been implicated (46). Our present analysis suggests potential ways to constrain this key parameter experimentally, either by analyzing fluctuations in ecotype frequencies on long timescales (13) or by measuring the joint distribution invasion fitness () and ecological perturbation () across a panel of engineered mutations (46).
Of course, the present work has focused on a highly simplified model, which omits many of the complicating factors expected in either natural or laboratory settings. A particularly important limitation is our focus on the weak mutation limit (). While analytically convenient, this assumption is violated by many of the laboratory experiments that motivated this study. In SI Appendix, section 2.3, we describe a preliminary extension of our results to the case where , which builds on the intuition gleaned from the weak mutation limit. However, a more thorough investigation of this regime is required to quantitatively fit the model to evolutionary measurements [e.g., in an approximate-likelihood framework (39)]. Future work will also be required to fully explore the effects of cross-feeding, time-varying environments, recombination, and other additions to our basic model [e.g., selection (47)]. We believe that our results provide a promising analytical framework in which to investigate these effects, which have mostly been confined to simulations so far.
It is also interesting to ask whether our results can be mapped onto more diverse modes of ecological interaction or whether there are other universality classes yet to be discovered. Since our model can be viewed as the simplest generalization of population genetics with multiple fitness axes, we hypothesize that it may capture the limiting behavior of a broader class of ecological interactions that are mediated by a small number of intensive variables. If so, its analytical tractability may offer a promising avenue for investigating the interactions between ecology and evolution more generally.
Supplementary Material
Acknowledgments
B.H.G. thanks Evgeni Frenkel and Ned Wingreen for discussions that inspired the development of the model. B.H.G. acknowledges support from the Miller Institute for Basic Research in Science at the University of California, Berkeley. O.H. acknowledges support from National Science Foundation Career Award 1555330, Simons Investigator Award 327934 from the Simons Foundation, and National Institutes of Health Grant R01GM115851. This research used resources of the National Energy Research Scientific Computing Center, a Department of Energy (DOE) Office of Science User Facility supported by the Office of Science of the US DOE under Contract DE-AC02-05CH11231.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807530115/-/DCSupplemental.
References
- 1.Maynard Smith J. The genetics of stasis and punctuation. Annu Rev Genet. 1983;17:11–25. doi: 10.1146/annurev.ge.17.120183.000303. [DOI] [PubMed] [Google Scholar]
- 2.Mallet J. Hybridization, ecological races and the nature of species: Empirical evidence for the ease of speciation. Philos Trans R Soc B Biol Sci. 2008;363:2971–2986. doi: 10.1098/rstb.2008.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nosil P, Harmon LJ, Seehausen O. Ecological explanations for (incomplete) speciation. Trends Ecol Evol. 2009;24:145–156. doi: 10.1016/j.tree.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Shapiro BJ, Leducq JB, Mallet J. What is speciation? PLoS Genet. 2016;12:e1005860. doi: 10.1371/journal.pgen.1005860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rozen DE, Lenski RE. Long-term experimental evolution in Escherichia coli. viii. Dynamics of a balanced polymorphism. Am Nat. 2000;155:24–35. doi: 10.1086/303299. [DOI] [PubMed] [Google Scholar]
- 6.Helling RB, Vargas CN, Adams J. Evolution of Escherichia coli during growth in a constant environment. Genetics. 1987;116:349–358. doi: 10.1093/genetics/116.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 8.Friesen ML, Saxer G, Travisano M, Doebeli M. Experimental evidence for sympatric ecological diversification due to frequency-dependent competition in Escherichia coli. Evolution. 2004;58:245–260. [PubMed] [Google Scholar]
- 9.Frenkel EM, et al. Crowded growth leads to the spontaneous evolution of semistable coexistence in laboratory yeast populations. Proc Natl Acad Sci USA. 2015;112:11306–11311. doi: 10.1073/pnas.1506184112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sousa A, et al. Recurrent reverse evolution maintains polymorphism after strong bottlenecks in commensal gut bacteria. Mol Biol Evol. 2017;34:2879–2892. doi: 10.1093/molbev/msx221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herron MD, Doebeli M. Parallel evolutionary dynamics of adaptive diversification in Escherichia coli. PLoS Biol. 2013;11:e1001490. doi: 10.1371/journal.pbio.1001490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Traverse CC, Mayo-Smith LM, Poltak SR, Cooper VS. Tangled bank of experimentally evolved Burkholderia biofilms reflects selection during chronic infections. Proc Natl Acad Sci USA. 2013;110:E250–E259. doi: 10.1073/pnas.1207025110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Good BH, McDonald MJ, Barrick JE, Lenski RE, Desai MM. The dynamics of molecular evolution over 60,000 generations. Nature. 2017;551:45–50. doi: 10.1038/nature24287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finkel SE, Kolter R. Evolution of microbial diversity during prolonged starvation. Proc Natl Acad Sci USA. 1999;96:4023–4027. doi: 10.1073/pnas.96.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poltak SR, Cooper VS. Ecological succession in long-term experimentally evolved biofilms produces synergistic communities. ISME J. 2011;5:369–378. doi: 10.1038/ismej.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torsvik V, Øvreås L. Microbial diversity and function in soil: From genes to ecosystems. Curr Opin Microbiol. 2002;5:240–245. doi: 10.1016/s1369-5274(02)00324-7. [DOI] [PubMed] [Google Scholar]
- 17.Kashtan N, et al. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science. 2014;344:416–420. doi: 10.1126/science.1248575. [DOI] [PubMed] [Google Scholar]
- 18.Foster KR, Schluter J, Coyte KZ, Rakoff-Nahoum S. The evolution of the host microbiome as an ecosystem on a leash. Nature. 2017;548:43–51. doi: 10.1038/nature23292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bendall ML, et al. Genome-wide selective sweeps and gene-specific sweeps in natural bacterial populations. ISME J. 2016;10:1589–1601. doi: 10.1038/ismej.2015.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao S, et al. 2017. Adaptive evolution within the gut microbiome of individual people. bioRxiv.
- 21.Garud NR, Good BH, Hallatschek O, Pollard KS. 2017. Evolutionary dynamics of bacteria in the gut microbiome within and across hosts. bioRxiv. [DOI] [PMC free article] [PubMed]
- 22.Geritz SA, et al. Evolutionarily singular strategies and the adaptive growth and branching of the evolutionary tree. Evol Ecol. 1998;12:35–57. [Google Scholar]
- 23.Dieckmann U, Marrow P, Law R. Evolutionary cycling in predator-prey interactions: Population dynamics and the red queen. J Theor Biol. 1995;176:91–102. doi: 10.1006/jtbi.1995.0179. [DOI] [PubMed] [Google Scholar]
- 24.Doebeli M. A model for the evolutionary dynamics of cross-feeding polymorphisms in microorganisms. Popul Ecol. 2002;44:59–70. [Google Scholar]
- 25.Kondoh M. Foraging adaptation and the relationship between food-web complexity and stability. Science. 2003;299:1388–1391. doi: 10.1126/science.1079154. [DOI] [PubMed] [Google Scholar]
- 26.Ackermann M, Doebeli M. Evolution of niche width and adaptive diversification. Evolution. 2004;58:2599–2612. doi: 10.1111/j.0014-3820.2004.tb01614.x. [DOI] [PubMed] [Google Scholar]
- 27.Ackland G, Gallagher I. Stabilization of large generalized Lotka-Volterra foodwebs by evolutionary feedback. Phys Rev Lett. 2004;93:158701. doi: 10.1103/PhysRevLett.93.158701. [DOI] [PubMed] [Google Scholar]
- 28.Shoresh N, Hegreness M, Kishony R. Evolution exacerbates the paradox of the plankton. Proc Natl Acad Sci USA. 2008;105:12365–12369. doi: 10.1073/pnas.0803032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vetsigian K. Diverse modes of eco-evolutionary dynamics in communities of antibiotic-producing microorganisms. Nat Ecol Evol. 2017;1:0189. [Google Scholar]
- 30.Lang GI, Murray AW, Botstein D. The cost of gene expression underlies a fitness trade-off in yeast. Proc Natl Acad Sci USA. 2009;106:5755–5760. doi: 10.1073/pnas.0901620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lang GI, et al. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature. 2013;500:571–574. doi: 10.1038/nature12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neher RA. Genetic draft, selective interference, and population genetics of rapid adaptation. Annu Rev Ecol Evol Syst. 2013;44:195–215. [Google Scholar]
- 33.Tilman D. Resource Competition and Community Structure. Princeton Univ Press; Princeton: 1982. [PubMed] [Google Scholar]
- 34.Posfai A, Taillefumier T, Wingreen NS. Metabolic trade-offs promote diversity in a model ecosystem. Phys Rev Lett. 2017;118:028103. doi: 10.1103/PhysRevLett.118.028103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tikhonov M, Monasson R. Collective phase in resource competition in a highly diverse ecosystem. Phys Rev Lett. 2017;118:048103. doi: 10.1103/PhysRevLett.118.048103. [DOI] [PubMed] [Google Scholar]
- 36.Advani M, Bunin G, Mehta P. Statistical physics of community ecology: A cavity solution to MacArthur’s consumer resource model. J Stat Mech Theor Exp. 2018;2018:033406. doi: 10.1088/1742-5468/aab04e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mac Arthur R. Species packing, and what competition minimizes. Proc Natl Acad Sci USA. 1969;64:1369–1371. doi: 10.1073/pnas.64.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ewens WJ. Mathematical Population Genetics. 2nd Ed Springer; New York: 2004. [Google Scholar]
- 39.Good BH, Desai MM. The impact of macroscopic epistasis on long-term evolutionary dynamics. Genetics. 2015;199:177–190. doi: 10.1534/genetics.114.172460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gillespie JH. Population Genetics: A Concise Guide. 2nd Ed Johns Hopkins Univ Press; Baltimore: 1998. [Google Scholar]
- 41.McInerney JO, McNally A, O’Connell MJ. Why prokaryotes have pangenomes. Nat Microbiol. 2017;2:17040. doi: 10.1038/nmicrobiol.2017.40. [DOI] [PubMed] [Google Scholar]
- 42.Tikhonov M, Monasson R. Innovation rather than improvement: A solvable high-dimensional model highlights the limitations of scalar fitness. J Stat Phys. 2017:74–104. [Google Scholar]
- 43.Van Valen L. A new evolutionary law. Evol Theor. 1973;1:1–30. [Google Scholar]
- 44.Doebeli M. Adaptive Diversification. Princeton Univ Press; Princeton: 2011. [Google Scholar]
- 45.McDonald MJ, Gehrig SM, Meintjes PL, Zhang XX, Rainey PB. Adaptive divergence in experimental populations of Pseudomonas fluorescens. iv. Genetic constraints guide evolutionary trajectories in a parallel adaptive radiation. Genetics. 2009;183:1041–1053. doi: 10.1534/genetics.109.107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plucain J, et al. Epistasis and allele specificity in the emergence of a stable polymorphism in Escherichia coli. Science. 2014;343:1366–1369. doi: 10.1126/science.1248688. [DOI] [PubMed] [Google Scholar]
- 47.MacArthur RH, Wilson EO. The Theory of Island Biogeography. Princeton Univ Press; Princeton: 1967. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.