Abstract
Plant myosin XI acts as a motive force for cytoplasmic streaming through interacting with actin filaments within the cell. Arabidopsis thaliana (At) has 13 genes belonging to the myosin XI family. Previous reverse genetic approaches suggest that At myosin XIs are partially redundant, but are functionally diverse for their specific tasks within the plant. However, the tissue-specific expression and enzymatic properties of myosin XIs have to date been poorly understood, primarily because of the difficulty in cloning and expressing large myosin XI genes and proteins. In this study, we cloned full-length cDNAs and promoter regions for all 13 At myosin XIs and identified tissue-specific expression (using promoter–reporter assays) and motile and enzymatic activities (using in vitro assays). In general, myosins belonging to the same class have similar velocities and ATPase activities. However, the velocities and ATPase activities of the 13 At myosin XIs are significantly different and are classified broadly into three groups based on velocity (high group, medium group and low group). Interestingly, the velocity groups appear roughly correlated with the tissue-specific expression patterns. Generally, ubiquitously expressed At myosin XIs belong to the medium-velocity group, pollen-specific At myosin XIs belong to the high-velocity group and only one At myosin XI (XI-I) is classified as belonging to the low-velocity group. In this study, we demonstrated the diversity of the 13 myosin XIs in Arabidopsis at the molecular and tissue levels. Our results indicate that myosin XIs in higher plants have distinct motile and enzymatic activities adapted for their specific roles.
Keywords: Arabidopsis thaliana, ATPase activity, Myosin XI, Tissue-specific expression, Velocity
Introduction
Myosin is a motor protein that converts chemical energy liberated by ATP hydrolysis into directed movement on actin filaments. Phylogenetic analyses of myosin motor domain sequences have shown that there are at least 79 myosin classes throughout all eukaryotes (Kollmar and Muhlhausen 2017). Among these myosins, plants possess only two classes of plant-specific myosins (classes VIII and XI). In angiosperms, more than a dozen isoforms of class XI myosin are present. For example, Arabidopsis thaliana, Oryza sativa and Brachypodium distachyon have 13, 12 and nine myosin XI genes, respectively (Reddy 2001, Jiang and Ramachandran 2004, Peremyslov et�al. 2011). Considering that the moss Physcomitrella patens has only two myosin XI genes (Vidali et�al. 2010), plant myosin XI isoforms would appear to have diverged and duplicated as land plants have evolved.
While the functions of myosin XI have been investigated to some extent, they are not yet fully understood (Tominaga et�al. 2012, Tominaga et�al. 2013, Tominaga and Ito 2015). Previously, the biological functions of myosin XIs have been investigated using knockout (KO) mutants of Arabidopsis thaliana (At) myosin XIs [XI-1 (MYA1), XI-2 (MYA2), XI-A, XI-B, XI-C, XI-D, XI-E, XI-F, XI-G, XI-H, XI-I, XI-J and XI-K]. Single and multiple At myosin XI KOs (among xi-1, xi-2, xi-b, xi-i and xi-k) have revealed that these myosin XIs are responsible for movement of organelles, including the endoplasmic reticulum (ER), Golgi apparatus, peroxisomes and mitochondria (Peremyslov et�al. 2008, Prokhnevsky et�al. 2008, Peremyslov et�al. 2010, Ueda et�al. 2010); further, it has been demonstrated in triple and quadruple At myosin XI KOs (among xi-1, xi-2, xi-b, xi-i and xi-k) that these myosin XIs are important in plant development (Prokhnevsky et�al. 2008, Peremyslov et�al. 2010, Ojangu et�al. 2012). Single xi-k KOs display a decreased abundance of fast and directional moving P-bodies (Steffens et�al. 2014), while single xi-i KOs show defects in nuclear movement and shape (Tamura et�al. 2013). A double At myosin XI KO (xi-f and xi-k) exhibits hypergravitropism and hyperphototropism in stems in response to gravity and light (Okamoto et�al. 2015). Another double At myosin XI KO (xi-c and xi-e) displays defective pollen tube growth (Madison et�al. 2015). A quadruple At myosin XI KO (xi-1, xi-2, xi-i and xi-k) was shown to have increased disease susceptibility to fungal pathogens (Yang et�al. 2014). These KO studies suggest that At myosin XIs are partially redundant but are functionally diverse for their specific tasks in various plant developmental processes. The discovery of a diverse array of adaptor proteins linking myosin XI to organelles or vesicles may contribute to this diversity (Hashimoto et�al. 2008, Peremyslov et�al. 2013, Tamura et�al. 2013, Steffens et�al. 2014, Kurth et�al. 2017). In order to understand the biological functions of diversified myosin XIs, it is essential to investigate the specific properties from the molecular level to the tissue level of each myosin XI. Expression profiles of At myosin XIs have been reported in the form of heat maps generated from public DNA microarray data (Peremyslov et�al. 2011). Moreover, the motile and enzymatic activity have been studied in a few myosin XIs (including At myosin XI-1, At myosin XI-I, a Chara myosin XI and a tobacco myosin XI) (Yokota et�al. 1999, Ito et�al. 2003b, Tominaga et�al. 2003, Hachikubo et�al. 2007, Ito et�al. 2007, Ito et�al. 2009, Diensthuber et�al. 2015). However, knowledge of the tissue-specific expression, and enzymatic properties of myosin XIs have to date been poorly understood, primarily because of the difficulty in cloning and expressing large myosin XI genes and proteins (approximately 4.5 kb for cDNA and 10 kb for genomic DNA). In this study, we cloned for the first time the full-length cDNAs and promoter regions of all 13 At myosin XIs. We then performed comprehensive analyses including: (i) direct observation of tissue-specific expression of At myosin XIs in plants; and (ii) measurement of motile and enzymatic activity of each At myosin XI.
Results
Constructs
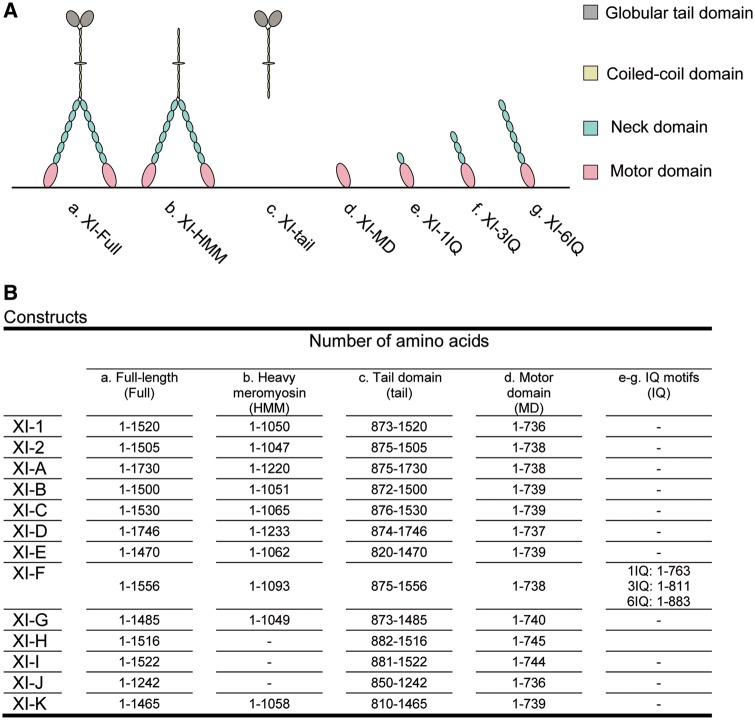
Arabidopsis thaliana possesses 13 genes encoding class XI myosins [At myosins XI-1 (MYA1), XI-2 (MYA2), XI-A, XI-B, XI-C, XI-D, XI-E, XI-F, XI-G, XI-H, XI-I, XI-J and XI-K] (Reddy and Day 2001). We found that the observed sequences of several myosin XIs (XI-C, -D, -E and -G) differed from the predicted sequences registered in TAIR (observed sequences have been submitted to DDBJ/EMBL/GenBank, see footnotes). These sequence differences were due to variations in splicing patterns (Supplementary Data S1). We did not observe any splice variants in different tissues or in cells cloned in this study (except for XI-E, which was only cloned from 30-day-old flowers) (Supplementary Table S1). A schematic diagram of At myosin XI deduced from its amino acid sequence is displayed in Fig.�1. In general, domain presence and composition were consistent across At myosin XIs and comprised a motor domain (MD), a neck domain with six IQ (isoleucine–glutamine) calmodulin-binding motifs, a coiled-coil region and a globular tail domain (GTD) (Tominaga and Nakano 2012, Tominaga and Ito 2015). It has been hypothesized that the long coiled-coil region allows for dimer formation, and that calmodulin or calmodulin-like proteins bind to each IQ motif as light chains, constituting a long lever arm.
Fig. 1.
Schematic diagrams showing the general morphology of At myosin XIs (A) deduced from amino acid sequences (B) of the constructs used in this study. (a) Full-length At myosin XI (XI-Full). (b) At myosin XI heavy meromyosin (XI-HMM) without the globular tail domain. (c) At myosin XI tail domain (XI-tail). (d) At myosin XI motor domain (XI-I MD). (e) At myosin XI motor domain with one IQ motif (XI-1IQ). (f) At myosin XI motor domain with three IQ motifs (XI-3IQ). (g) At myosin XI motor domain with six IQ motifs (XI-6IQ).
In this study, we generated seven types of recombinant At myosin XI constructs: full-length (XI-Full); heavy meromyosin which is truncated and lacks GTDs (XI-HMM); tail domain (XI-tail); motor domain (XI-MD); and motor domain with IQ motifs (XI-1IQ, -3IQ and -6IQ) (Fig.�1).
Promoter–GUS assays
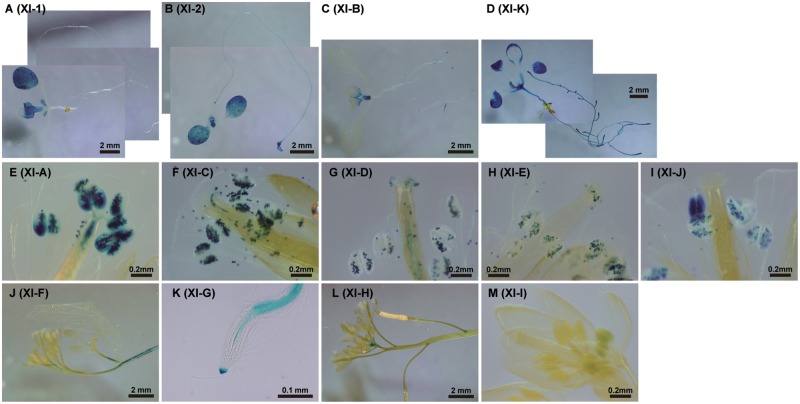
To determine the tissue-specific expression of At myosin XIs, we created transgenic Arabidopsis lines that expressed β-glucuronidase (GUS) fusion proteins under the transcriptional control of individual At myosin XI native promoters. GUS staining patterns were confirmed by observing three independent transgenic Arabidopsis lines. At myosin XIs were classified into three categories, according to the GUS expression pattern (Fig.�2; Table�1). Four At myosin XIs showed a ‘ubiquitous’ staining pattern which stained many plant tissues widely (XI-1, -2, -B and -K). These myosin XIs have been hypothesized to be responsible for organelle movement and for the generation of cytoplasmic streaming (Avisar et�al. 2008, Peremyslov et�al. 2008, Prokhnevsky et�al. 2008, Sparkes et�al. 2008, Avisar et�al. 2009, Peremyslov et�al. 2010). The GUS signal in the XI-K promoter lines was present from the root to the aerial regions (other than pollen) (Fig.�3). Although weaker than XI-K, the GUS signal of XI-2 was ubiquitously expressed, including in pollen. In the first rosette leaf, we commonly observed a decrease in GUS signal with leaf growth (Supplementary Fig. S1). The GUS signals of XI-1 and XI-B were weaker and more partial than they were for XI-2 and XI-K, with the XI-1 promoter lines displaying relatively strong GUS signal in the cotyledon and stipule (Supplementary Fig. S2), while XI-B expression was relatively strong in the ovule, pollen, shoot meristem, stipule and trichomes (Supplementary Fig. S3). Five At myosin XIs exhibited GUS staining patterns that were ‘pollen specific’ (XI-A, -C, -D, -E and -J) (Supplementary Figs. S4–S8). Four At myosin XIs exhibited ‘other’ patterns (XI-F, -G, -H and -I) (Supplementary Figs. S9–S12). The GUS signal of the XI-F promoter lines was observed specifically around the stele in the shoot, leaf and root (Supplementary Fig. S9). XI-G promoter GUS expression was predominantly observed in the stele of the root and the root cap (Supplementary Fig. S10). The GUS expression of XI-H was weakly observed in the style, node and stipule (Supplementary Fig. S11). GUS expression under the XI-I promoter was only observed slightly in the anthers (Supplementary Fig. S12). All data are summarized in Table�1.
Fig. 2.
Promoter–GUS assays of At myosin XI expression. The cDNA for β-glucuronidase (GUS) was ligated in-frame to the promoter regions and first exons of myosin XIs (XI-Pro), and introduced into Arabidopsis. (A) XI-1pro–GUS; 7-day-old seedling. (B) XI-2pro–GUS; 7-day-old seedling. (C) XI-Bpro–GUS; 7-day-old seedling. (D) XI-Kpro–GUS; 7-day-old seedling. (E) XI-Apro–GUS; flower of a 30-day-old plant. (F) XI-Cpro–GUS; flower of a 30-day-old plant. (G) XI-Dpro–GUS; flower of a 30-day-old plant. (H) XI-Epro–GUS; flower of a 30-day-old plant. (I) XI-Jpro–GUS; flower of a 30-day-old plant. (J) XI-Fpro–GUS; shoots and flowers of a 30-day-old plant. (K) XI-Gpro–GUS; root of a 7-day-old seedling. (L) XI-Hpro–GUS; shoots and flowers of a 30-day-old plant. (M) XI-Ipro–GUS; flower of a 30-day-old plant.
Table 1.
Promoter–GUS assays
| GUS activity | |||||
|---|---|---|---|---|---|
| Aerial | Root | ||||
| Reproductive | Shoot | Leaf | |||
| Ubiquitous | |||||
| XI-1 | + Style |
|
|
||
| XI-2 |
|
+++ Shoot Meristem |
|
|
|
| XI-B |
|
|
|
|
|
| XI-K |
|
+++ Shoot Meristem |
|
|
|
| Pollen | |||||
| XI-A | +++ Pollen | ||||
| XI-C | +++ Pollen | ||||
| XI-D | +++ Pollen | ||||
| XI-E | +++ Pollen | ||||
| XI-J | +++ Pollen | ||||
| Others | |||||
| XI-F | +++ Stele | ++ First leaf (midrib) | +++ Stele | ||
| XI-G |
|
|
|
||
| XI-H | ++ Style | ++ Node | ++ Stipule | ||
| XI-I | + Anther | ||||
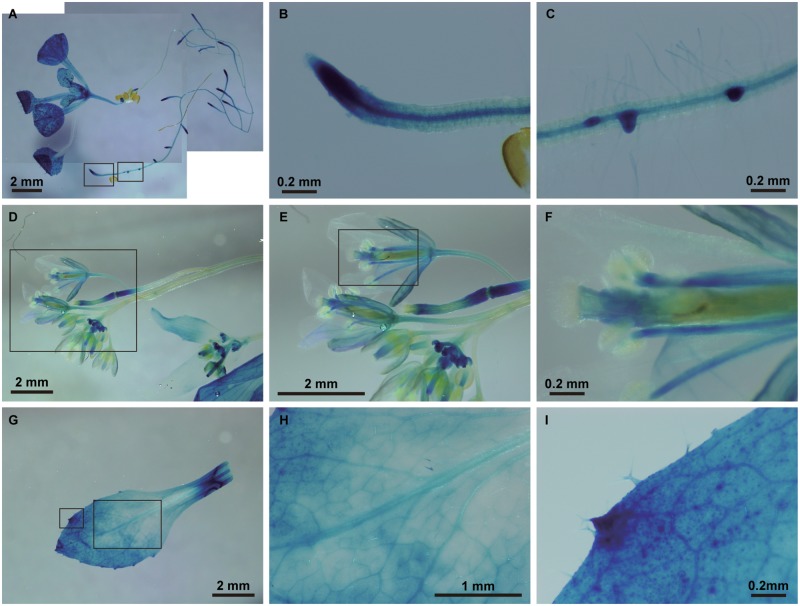
Fig. 3.
Promoter–GUS assay of At myosin XI-K expression. (A) A 7-day-old seedling. (B) Axial root tip of a 7-day-old seedling. (C) Lateral root tip of a 7-day-old seedling. (D) Shoot and flowers of a 30-day-old plant. (E) Magnified flowers from the region surrounded by a square in (D). (F) Magnified style and anther from the region surrounded by a square in (E). (G) First leaf of a 30-day-old plant. (H) Magnified first leaf from the region surrounded by the right hand square in (G). (I) Magnified trichomes from the region surrounded by the left-hand square in (G).
Velocities of At myosin XI-MDs
For measurements of the enzymatic activities and motile velocities of myosin XIs, constructs were expressed using a baculovirus transfer system in insect cells (High Five cells or Sf9 cells). We purified the recombinant myosin XIs using both nickel-affinity and Flag-affinity resin, as previously reported (Haraguchi et�al. 2016). In our previous work, we expressed and purified recombinant At myosin XI-1, -2 and -I using a baculovirus system. Of the three At myosin XIs, only XI-2 was shown to bind calmodulin at all IQ motifs as light chains. To avoid measurement errors due to the binding inability of light chains, we used XI-MD to measure the motile and enzymatic activity of all At myosin XIs in vitro.
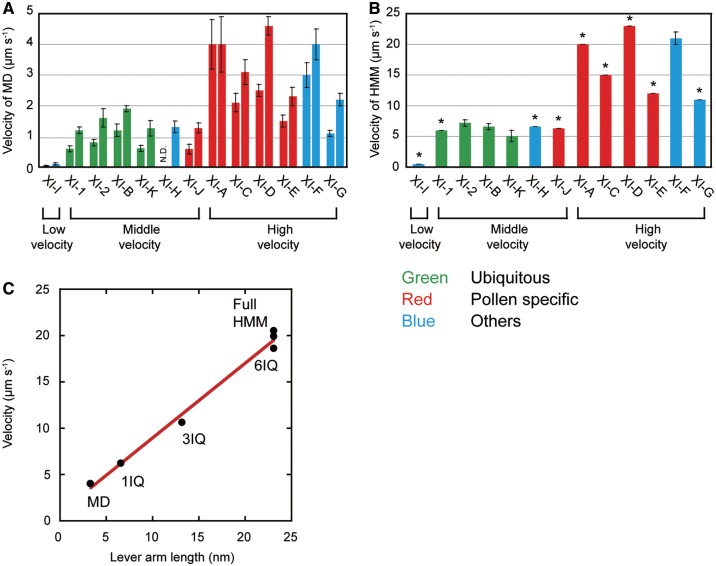
The sliding velocities of actin filaments on XI-MDs were measured at 25�C using an antibody-based version of the in vitro motility assay (Ito et�al. 2007, Ito et�al. 2009, Haraguchi et�al. 2014, Haraguchi et�al. 2016). Fig.�4A shows actin sliding velocities of XI-MDs under two different conditions: 150 mM KCl (a physiological ionic strength, right bars); and 25 mM KCl (the ionic strength used for standard biochemical assays, left bars). Previous studies have found that myosins belonging to the same class have similar velocities (El-Mezgueldi and Bagshaw 2008). However, our findings differ from the previously published work. The velocities of 13 At myosin XI MDs differed significantly, even though all 13 belong to the same myosin XI class. The 13 At myosin XIs were divided into three groups, according to their velocities: low velocity (XI-I); medium velocity (XI-1, -2, -B, -K, -J and -H); and high velocity (XI-A, -C, -D, -E, -F and -G) (Fig.�4A;Supplementary Table S3). Interestingly, this velocity grouping roughly corresponded to the expression patterns within the different plant tissues observed in our GUS assays. All four ubiquitous At myosin XIs (XI-1, -2, -B and -K) have a known role as motive forces of cytoplasmic streaming and belong to the medium-velocity group. Four of five pollen-specific At myosin XIs (XI-A, -C, -D and -E) belong to the high-velocity group. The velocities of the other At myosin XIs vary. XI-F, which is predominantly expressed in stele, belongs to the high-velocity group. Only XI-I, which is primarily located at the nuclear membrane, belongs to the low-velocity group, as has been previously reported (Haraguchi et�al. 2016).
Fig. 4.
The velocity of various At myosin XI constructs measured by in vitro motility assay. (A) Velocity of the At myosin XI motor domains (XI-MDs) at 25 mM KCl (left bar) and 150 mM KCl (right bar). (B) Velocity of the HMMs at 150 mM KCl. Asterisks show the calculated velocity from the velocity of the XI-MD and lever arm length: the velocities of HMMs were calculated by multi plying the velocities of MDs by 5. (C) The relationship between velocity and lever arm length of XI-F-MD, -1Q, -3IQ, -Full and -HMM.
Velocities of At myosin XI-HMM
The XI-HMMs of At myosin XIs were co-expressed with Arabidopsis or mouse calmodulin in insect cells because calmodulin acts as a light chain for many unconventional myosins including plant myosin XI (At myosin XI-2 and tobacco 175 kDa myosin) (Tominaga et�al. 2012, Tominaga et�al. 2013). During purification, exogenous calmodulin (1 �M) was added to solutions to prevent the dissociation of calmodulin from IQ motifs. However, the XI-HMMs of seven At myosin XIs (XI-1, -A, -C, -D, -E, -G and -I) did not result in smooth movement of the actin filaments even when adding excessive amounts of exogenous calmodulin (60 �M) to the buffer in the in vitro motility assays. We believe that it is likely that these results show that some IQ motifs of these At myosin XIs do not bind to calmodulin as light chains, because the velocities of myosins without light chains at the IQ motifs are very slow (Lowey et�al. 1993). We expect that specific light chains other than calmodulin would bind to IQ motifs of these myosin XIs in Arabidopsis cells. In contrast, XI-HMM of four At myosin XIs (XI-2, -B, -F and -K) moved actin smoothly. The velocities of these four XI-HMMs were 4- to 5-fold higher than were the velocities of XI-MDs (Fig.�4A, B;Supplementary Table S3). According to the prevailing model of myosin movement, known as the ‘swinging lever arm model’, myosin velocity is almost proportional to the length of the lever arm (Uyeda et�al. 1996, Spudich 2001). The ratio between the lever arm length of XI-HMM and XI-MD is 6.6-fold (23 nm/3.5 nm) (Ito et�al. 2007). This 6.6:1 ratio is similar to the ratio between the velocity of XI-HMM and XI-MD. This correlation strongly suggests that calmodulin binds to all IQ motifs of XI-2, -B and -F. The actual velocities of ubiquitous At myosins XI-2-, -B and -K-HMM were 5–7 �m s−1. This value is similar to the cytoplasmic streaming velocity in epidermal cells of Arabidopsis (Ueda et�al. 2010, Tominaga et�al. 2013).
We further studied the relationship between the lever arm length and the actin sliding velocities using several constructs of XI-F. We selected constructs so as to vary lever arm length in our experiments (Fig.�1, XI-MD, -1IQ, -3IQ, -6IQ, -HMM and -Full). As the lever arm length increased, the velocities increased linearly (Fig.�4C). These results are consistent with the swinging lever arm model and suggest that calmodulin binds to all IQ motifs of XI-F as light chains. The velocities of XI-F-Full, -HMM and -6IQ were 20 �m s−1 at 25�C. This velocity is about 3-fold higher than the velocity of ubiquitous At myosin XIs (5–7 �m s−1).
As reported in Fig.�4, the velocities of XI-HMM and -Full were about 5-fold higher than that of XI-MD, where we anticipated light chains to bind all IQ motifs. The velocities of remaining XI-Fulls can be estimated from the velocity of each XI-MD. The velocities of HMMs were calculated by multiplying the velocities of MDs by five (Fig.�4B, bars marked with *). This estimate suggests that the velocities of pollen-specific XI-A and -D were >20 μm s−1.
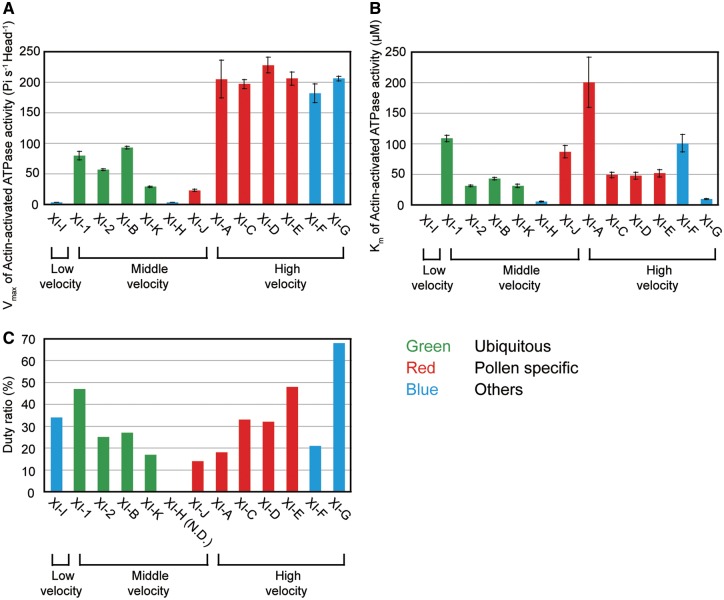
ATPase activities
Actin-activated ATPase activities of the MD for all 13 At myosin XIs were measured at standard ionic strength (approximately 50 mM) and temperature (25�C) (Ito et�al. 2003a, Ito et�al. 2007). Plots of ATPase activity against actin concentration were found to fit the Michaelis–Menten type curve. Using this curve, we determined the maximum rate of ATP turnover (Vmax) and the actin concentration at which the ATPase rate reached half of its maximum (Km). The Vmax values of four of five pollen-specific At myosin XIs, XI-A, -C, -D and -E, were much higher than the Vmax values of ubiquitous At myosin XIs (XI-1, -2, -B and -K). This relationship is similar to our findings with regards to velocities (Fig.�4A, B). Vmax values of At myosin XI-F, which is specifically expressed in the stele, were also higher than those Vmax values in ubiquitous At myosin XIs (Fig.�5A;Supplementary Table S3). At this ionic strength (approximately 50 mM), actin motility by XI-H-MD was not observed and the ATPase activity was extremely low. However, actin motility by XI-H-MD was observed in the presence of 150 mM KCl, suggesting that XI-H-MD may be denatured at low ionic strengths.
Fig. 5.
Enzymatic properties of At myosin XI-MDs. (A) Vmax of actin-activated ATPase activity for XI-MDs at 25 mM KCl. (B) Km of the actin-activated ATPase activity of XI-MDs at 25 mM KCl. (C) Duty ratio of the At myosin XI motor domains (XI-MDs). The duty ratio is estimated from the Vmax values of actin-activated ATPase activities and the actin sliding velocities of the MD (see Materials and Methods).
The Km values of XI-G, -H and -I were much lower than those of other At myosin XIs, indicating higher actin affinities of these At myosin XIs. In contrast, the Km value of XI-A was much higher than it was for other At myosin XIs, indicating that XI-A has a lower affinity for actin (Fig.�5B). At myosin XIs with high Km values (low affinity for actin) might be tuned to function optimally at the intracellular region with high actin concentration, namely the region where actin filaments are bundled. At myosin XIs with high Km values (low affinity for actin) might function to keep velocity high with low ATP consumption. In contrast, myosin XIs with low Km values (e.g. XI-I) may act as a tether to keep in contact with actin cables (Haraguchi et�al. 2016).
Duty ratio
The duty ratio is defined as the fraction of the ATPase cycle time that the myosin motor attaches to the actin filament. We estimated the duty ratio from the Vmax values of actin-activated ATPase activities and the actin sliding velocities of the MD (see the Materials and Methods). The duty ratio of all At myosin XI MDs, except XI-G MD, was <50% (Fig.�5C). Because the duty ratio of MD is required to be >50% (although commonly over 70%) for processivity, At myosin XIs other than XI-G are considered non-processive motors.
Discussion
While expression levels of At myosin XIs have been measured with public microarrays (Peremyslov et�al. 2011, Sparkes 2011), we took a different approach to assess plant-wide expression patterns. When tagged to the myosin XI promoter, GUS expression patterns cluster At myosin XIs into three groups (Fig.�2; Table�1). Four At myosin XIs (XI-1, -2, -B and -K) were expressed throughout the plant and were therefore categorized as ‘ubiquitous’. KO experiments suggested that these ubiquitous At myosin XIs are responsible for organelle movement and for the generation of cytoplasmic streaming (Tominaga and Ito 2015). Among the four ubiquitous At myosin XIs, GUS signals of XI-2 and XI-K were stronger than those of XI-1 and XI-B, corresponding to previous results showing that XI-2 and XI-K are major motive forces of cytoplasmic streaming (Peremyslov et�al. 2008, Prokhnevsky et�al. 2008, Peremyslov et�al. 2010). Interestingly, XI-2 expression was observed in pollen, but XI-K was not, which implies a role shared between XI-2 and XI-K with respect to tissue expression. GUS signals of XI-2 decreased with the progression of leaf development, although a similar pattern was not observed for XI-K (data not shown). Compared with XI-2 and XI-K, GUS signals of XI-1 and XI-B were weaker and only partial, supporting the suggestion that they only play a minor role in cytoplasmic streaming and plant development (Peremyslov et�al. 2008, Prokhnevsky et�al. 2008, Peremyslov et�al. 2010).
Of the 13 At myosin XIs, we found that five (XI-A, -C, -D, -E and -J) were specifically expressed and two (XI-2 and -B) were partially expressed in pollen. We believe that this indicates that the diversification of myosin XIs may be essential for the evolution of reproductive organs of angiosperms.
GUS expression patterns of the remaining four At myosin XIs (XI-F, -G, -H and -I) were inconsistent, and were therefore categorized as ‘other’. XI-F was expressed specifically in steles of the shoot, leaf and root, suggesting some involvement of material transport between sink and source organs. GUS signals of XI-G were strong at the root cap. GUS signals of XI-I were the weakest of all 13 At myosin XIs. Considering that the velocity of XI-I is an order of magnitude lower than that of other At myosin XIs (Haraguchi et�al. 2016), it may act as a regulator with a small number of molecules. Our results from promoter–GUS assays demonstrate that microarrays are insufficient to uncover the differences of At myosin XI expression.
Our results also suggest a need for standardization in the measurement of At myosin XI expression, as expression patterns were dependent on the method. For example, XI-2 expression in pollen was detected by promoter–GUS assays, although it was missed in previous microarray experiments (Peremyslov et�al. 2011, Sparkes 2011). XI-2 showed low GUS activity in root hairs, although the xi-2 mutant has clear root hair defects (Peremyslov et�al. 2008). The microarray data suggest high expression levels of XI-H and XI-I in most vegetative tissues, but they showed very low GUS activity in our experiments. Both XI-C and XI-E expression levels in pollen were relatively low in the microarray data, although xi-c xi-e mutants have clear pollen tube defects (Madison et�al. 2015). These differences reflect the strengths and weaknesses of each method. We believe that GUS expression profiles reflect only the promoter activity, and do not measure steady-state protein levels. In the present study, we defined the genomic sequence 3 kb upstream of the first codon as ‘the promoter’. However, there are many cases where essential promoter elements are also found downstream of the translation start site. On the other hand, expression profiles from public data often vary depending on sampling and detection.
Phylogenetic analyses of MD sequences revealed at least 79 classes of myosins (Kollmar and Muhlhausen 2017). In general, the motor properties are similar within the same myosin class (El-Mezgueldi and Bagshaw 2008). The velocities and actin-activated ATPase activities of myosin XIs studied thus far were found to be relatively high among the 79 myosin classes. However, we reported recently that the velocity and actin-activated ATPase activity of At myosin XI-I are very low compared with other myosin XIs measured thus far (Haraguchi et�al. 2016). This implies that there is a diversity of motor activity among myosin XIs. In this study, we conducted a comprehensive analysis of motor activities for all 13 At myosin XIs. The velocities of ubiquitous At myosin XIs (XI-1, -2, -B and -K) were between 5 and 7 �m s−1 (Fig.�4B). It has been observed that the velocities of purified myosin XI in vitro were higher than those of cytoplasmic streaming in cells (Tominaga et al. 2013). We speculate that this discrepancy between in vitro and cytoplasmic streaming velocities arises as a result of physical obstruction by cytoplasmic components (e.g. organelles and structural proteins), and as a result of competition with other slower myosin molecules (e.g. slower myosin XI-I) within the cells. In contrast, the velocities of most pollen-specific At myosin XIs (XI-A, -C, -D and -E) were about 2- to 3-fold higher (12–23 �m s−1) than were those of ubiquitous At myosin XIs. In epidermal cells of Arabidopsis, the velocities of peroxisome (maximum velocity: 3.25 �m s−1, Mano et�al. 2002) and Golgi (maximum velocity: 3.5 �m s−1, Akkerman et�al. 2011) were slightly slower than the velocity of ubiquitous At myosin XIs measured in vitro. In pollen tubes of Arabidopsis, the maximum velocities of peroxisome and Golgi were reported to be 6.6 and 5.9 �m s−1, respectively (Madison et�al. 2015); these velocities in pollen tubes were about twice as high as those in epidermal cells of Arabidopsis. It is likely that fast cytoplasmic streaming in pollen tubes is generated by these pollen-specific At myosin XIs. In addition, the velocity of the stele-specific At myosin XI (XI-F) was 20 �m s−1, which is also higher than those of ubiquitous At myosin XIs. A recent study by Okamoto et�al. (2015) has reported that XI-F is specifically expressed in fiber cells of the stem, and is also responsible for plant straightening. In these cells, vigorous cytoplasmic streaming was generated at a velocity of 8.6 �m s−1, which is 2-fold faster than the velocity of vigorous cytoplasmic streaming in epidermal cells. Plant straightening was disturbed by simultaneous KO of XI-F and ubiquitous XI-K. Cytoplasmic streaming velocity in fiber cells (8.6 �m s−1) was reduced to 8.2, 7.3 and 1.7 �m s−1 in the xi-k KO, xi-f KO and xi-f xi-k double KOs, respectively. These compound decreases in cytoplasmic streaming velocity suggest that they are a redundant motive force. Furthermore, fiber cells can reach a length of 1 mm. It is possible that higher velocity At myosin XIs are necessary for effective material transport in long cells, such as pollen or fiber cells.
Traditionally, the time myosin motors spend in the strong binding state (ts) is generally measured by kinetic analyses using stopped-flow apparatus. However, it is difficult to measure ts of fast myosins such as fast skeletal myosin II and myosin XI, because elementary processes constituting ts of these fast myosins at normal temperature (25�C) are too fast. Instead, the ts values of fast myosins were measured at multiple temperatures near 0 to 10�C, and the ts values at 25�C were estimated by extrapolating these values on an Arrhenius plot (Millar and Geeves 1983, Ito et�al. 2007). Because of its large amounts of protein and the long time required, it is impractical to measure the ts of the 13 At myosin XIs using this method in our study. Therefore, herein the duty ratios of At myosin XIs were estimated by using the Vmax values of actin-activated ATPase activities and the actin sliding velocities of the MD. The estimated duty ratio of all At myosin XI MDs except for XI-G MD are <50% (Fig.�5C), suggesting that most At myosin XIs are non-processive.
In our investigation, we examined the diversity of 13 myosin XIs in Arabidopsis at the molecular and tissue level, as predicted previously (Tominaga and Nakano 2012). Phylogenetic analysis of plant myosins indicated that At myosin XIs split into five lineages (Avisar et�al. 2008, Peremyslov et�al. 2011, Tominaga and Nakano 2012). However, we could not find any clear correlation between these lineages and the groupings generated in this study. Animals use various motor proteins (many classes of kinesin and myosin and cytoplasmic dynein) that have diverse abilities (e.g. velocity, directionality and cargo selectivity) for intracellular transport. In contrast, plants maintain intracellular transport by primarily using plant-specific myosin XIs. Considering that the moss P. patens has only two myosin XIs (Vidali et�al. 2010), the increased numbers of myosin XIs with diverse functions in angiosperms may have occurred during the evolution of land plants in order to sustain their highly complicated biological systems.
Materials and Methods
Constructs
Full-length At myosin XIs were cloned using PCR in two steps from total RNA. Total RNA was purified from 7-day-old Arabidopsis seedlings, 30-day-old Arabidopsis flowers and 7-day-old Arabidopsis cultured cells (Alex) using the RNeasy Plant Mini Kit (QIAGEN). First, myosin XI cDNAs including coding sequences (CDS), were amplified by PCR using forward and reverse primers (5'UTR-F and 3'UTR-R) designed 50–100 bp upstream from the predicted start codon and 50–100 bp downstream from the predicted stop codons, respectively (cDNAs successfully amplified from total RNA, along with the primers used, are described in Supplementary Table S2). Amplified cDNAs were then cloned into the pENTR-D-TOPO plasmid (Life Technologies) and the At myosin XI coding regions were sequenced. Secondly, full-length CDS regions of myosin XIs were amplified using forward primers (F) beginning from the start codon, and reverse primers (R) ending at the stop codon, according to the determined sequence (Supplementary Table S2). CDS regions were then cloned into the pENTR-D-TOPO plasmid.
For promoter–GUS assays, 3 kb of the 5′-flanking sequence upstream, including the first exon (determined from sequencing as described above) of myosin XIs (Supplementary Table S2), were amplified and cloned into pGWB533 (Nakagawa et�al. 2007), as described previously (Haraguchi et�al. 2014).
For measurements of enzymatic and motile activities, baculovirus transfer vectors of the At myosin XI MDs (pFastBac XI-1-, -2-, -A-, -B-, -C-, -D-, -E-, -F-, -G-, -H-, -I-, -J- and -K-MD) were used. At myosin XI HMMs, other than XI-1, -A and -C (pFastBac XI-2-, -B-, -D-, -E-, -F-, -G-, -H-, -I- and -K-HMM), At myosin XI full-length (Full) (pFastBac XI-F- and -J-Full) and the At myosin XI MD + IQ motif (IQ) (pFastBac XI-F-1IQ, -3IQ and -6IQ) were generated as follows: At myosin XI cDNAs were mutated to create an NcoI site at the 5' end of nucleotide 1 and at an AgeI site at the 3' end of the MD, HMM, Full, 3IQ and 6IQ (Supplementary Table S2). These were ligated to the NcoI–AgeI fragment of the pFastBac CCM MD (Ito et�al. 2007). Baculovirus transfer vectors for HMM of At myosin XI-1, A and C (pFastBac XI-1-, -A- and -C-HMM) were generated as follows: At myosin XI cDNAs were mutated to create restriction enzyme annealing sites for the pFastBac CCM MD at both the 5' end of nucleotide 1 of the At myosin XI coding regions and the 3' end of HMM (Supplementary Table S2). These were ligated to the NcoI–AgeI fragment of the pFastBac CCM MD using the In-Fusion cloning kit (TAKARA). The resultant constructs contained an N-terminal tag (MDYKDDDDKRSMNSRPA) consisting of the FLAG tag (DYKDDDDK), the amino acid residues of MD, 1IQ, 3IQ, HMM or Full, and a C-terminal tag (GGGEQKLISEEDLHHHHHHHH) containing a flexible linker (GGG), a Myc-epitope sequence (EQKLISEEDL) and a (His) 8 tag.
ATPase activities and in vitro actin gliding assays
Steady-state ATPase activity and in vitro actin gliding assays were measured at 25�C, as previously described (Ito et�al. 2007, Haraguchi et�al. 2014, Haraguchi et�al. 2016).
Estimation of the duty ratio
The duty ratio is the fraction of the ATPase cycle time that the myosin motor spends in the strong binding state (ts), and it is expressed as ts divided by the total ATPase cycle time (tc). tc is the inverse of the Vmax value of the ATPase activity. ts is the stroke size divided by the actin sliding velocity (Uyeda et�al. 1991). The stroke size is nearly equal to the lever arm length (Ruff et�al. 2001, Veigel et�al. 2002) and the lever arm length of MD is about 3.5 nm of the lever arm length of the MD. Therefore, ts is 3.5 nm divided by the actin sliding velocity of the MD. Thus, the duty ratio (ts/tc ) is estimated to be (3.5 nm divided by the actin sliding velocity of the MD)�(Vmax value of the ATPase activity of the MD).
Funding
This work was supported by the Japan Society for the Promotion of Science [KAKENHI 24658002, 26440131 and 15H01309 (to K.I.), 20001009, 23770060 and 25221103 (to M.T.)] and the Japan Science and Technology Agency, ALCA [JPMJAL1401 (to K.I., T.H., Z.D. and M.T.)].
Supplementary Material
Acknowledgments
The authors would like to thank Dr. K. Yoshino for assistance and suggestions, and A. Kimura, N. Shoji, K. Miyauchi, A. Sato, T. Suwa, S. Ohya, T. Wakisaka, C. Tsutsui and R. Umeda for technical assistance with the experiments.
Disclosures
The authors have no conflicts of interest to declare.
Glossary
Abbreviations
- GTD
globular tail domain
- GUS
β-glucuronidase
- HMM
heavy meromyosin
- IQ
isoleucine–glutamine
- KO
knockout
- MD
motor domain
Footnotes
References
- Akkerman M., Overdijk E.J., Schel J.H., Emons A.M., Ketelaar T. (2011) Golgi body motility in the plant cell cortex correlates with actin cytoskeleton organization. Plant Cell Physiol. 52: 1844–1855. [DOI] [PubMed] [Google Scholar]
- Avisar D., Abu-Abied M., Belausov E., Sadot E., Hawes C., Sparkes I.A. (2009) A comparative study of the involvement of 17 Arabidopsis myosin family members on the motility of Golgi and other organelles. Plant Physiol. 150: 700–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D., Prokhnevsky A.I., Makarova K.S., Koonin E.V., Dolja V.V. (2008) Myosin XI-K Is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol. 146: 1098–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diensthuber R.P., Tominaga M., Preller M., Hartmann F.K., Orii H., Chizhov I., et al. (2015) Kinetic mechanism of Nicotiana tabacum myosin-11 defines a new type of a processive motor. FASEB J. 29: 81–94. [DOI] [PubMed] [Google Scholar]
- El-Mezgueldi M., Bagshaw C.R. (2008) The myosin family: biochemical and kinetic properties In Myosins. Proteins and Cell Regulation, Vol. 7. pp. 55–93. Springer, Dordrecht. [Google Scholar]
- Hachikubo Y., Ito K., Schiefelbein J., Manstein D.J., Yamamoto K. (2007) Enzymatic activity and motility of recombinant Arabidopsis myosin XI, MYA1. Plant Cell Physiol. 48: 886–891. [DOI] [PubMed] [Google Scholar]
- Haraguchi T., Tominaga M., Matsumoto R., Sato K., Nakano A., Yamamoto K., et al. (2014) Molecular characterization and subcellular localization of Arabidopsis Class VIII myosin, ATM1. J. Biol. Chem. 289: 12343–12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T., Tominaga M., Nakano A., Yamamoto K., Ito K. (2016) Myosin XI-I is mechanically and enzymatically unique among class-XI myosins in Arabidopsis. Plant Cell Physiol. 57: 1732–1743. [DOI] [PubMed] [Google Scholar]
- Hashimoto K., Igarashi H., Mano S., Takenaka C., Shiina T., Yamaguchi M., et al. (2008) An isoform of Arabidopsis myosin XI interacts with small GTPases in its C-terminal tail region. J. Exp. Bot. 59: 3523–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Ikebe M., Kashiyama T., Mogami T., Kon T., Yamamoto K. (2007) Kinetic mechanism of the fastest motor protein, Chara myosin. J. Biol. Chem. 282: 19534–19545. [DOI] [PubMed] [Google Scholar]
- Ito K., Kashiyama T., Shimada K., Yamaguchi A., Awata J., Hachikubo Y., et al. (2003a) Recombinant motor domain constructs of Chara corallina myosin display fast motility and high ATPase activity. Biochem. Biophys. Res. Commun. 312: 958–964. [DOI] [PubMed] [Google Scholar]
- Ito K., Uyeda T.Q., Suzuki Y., Sutoh K., Yamamoto K. (2003b) Requirement of domain–domain interaction for conformational change and functional ATP hydrolysis in myosin. J. Biol. Chem. 278: 31049–31057. [DOI] [PubMed] [Google Scholar]
- Ito K., Yamaguchi Y., Yanase K., Ichikawa Y., Yamamoto K. (2009) Unique charge distribution in surface loops confers high velocity on the fast motor protein Chara myosin. Proc. Natl. Acad. Sci. USA 106: 21585–21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Ramachandran S. (2004) Identification and molecular characterization of myosin gene family in Oryza sativa genome. Plant Cell Physiol. 45: 590–599. [DOI] [PubMed] [Google Scholar]
- Kollmar M., Muhlhausen S. (2017) Myosin repertoire expansion coincides with eukaryotic diversification in the Mesoproterozoic era. BMC Evol. Biol. 17: 211.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth E.G., Peremyslov V.V., Turner H.L., Makarova K.S., Iranzo J., Mekhedov S.L., et al. (2017) Myosin-driven transport network in plants. Proc. Natl. Acad. Sci. USA 114: E1385–E1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowey S., Waller G.S., Trybus K.M. (1993) Skeletal muscle myosin light chains are essential for physiological speeds of shortening. Nature 365: 454–456. [DOI] [PubMed] [Google Scholar]
- Madison S.L., Buchanan M.L., Glass J.D., McClain T.F., Park E., Nebenfuhr A. (2015) Class XI myosins move specific organelles in pollen tubes and are required for normal fertility and pollen tube growth in Arabidopsis. Plant Physiol. 169: 1946–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano S., Nakamori C., Hayashi M., Kato A., Kondo M., Nishimura M. (2002) Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: dynamic morphology and actin-dependent movement. Plant Cell Physiol. 43: 331–341. [DOI] [PubMed] [Google Scholar]
- Millar N.C., Geeves M.A. (1983) The limiting rate of the ATP-mediated dissociation of actin from rabbit skeletal muscle myosin subfragment 1. FEBS Lett. 160: 141–148. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Suzuki T., Murata S., Nakamura S., Hino T., Maeo K., et al. (2007) Improved gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71: 2095–2100. [DOI] [PubMed] [Google Scholar]
- Ojangu E.L., Tanner K., Pata P., Jarve K., Holweg C.L., Truve E., et al. (2012) Myosins XI-K, XI-1, and XI-2 are required for development of pavement cells, trichomes, and stigmatic papillae in Arabidopsis. BMC Plant Biol. 12: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Ueda H., Shimada T., Tamura K., Kato T., Tasaka M., et al. (2015) Regulation of organ straightening and plant posture by an actin–myosin XI cytoskeleton. Nat. Plants 1: 15031.. [DOI] [PubMed] [Google Scholar]
- Peremyslov V.V., Mockler T.C., Filichkin S.A., Fox S.E., Jaiswal P., Makarova K.S., et al. (2011) Expression, splicing, and evolution of the myosin gene family in plants. Plant Physiol. 155: 1191–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov V.V., Morgun E.A., Kurth E.G., Makarova K.S., Koonin E.V., Dolja V.V. (2013) Identification of myosin XI receptors in Arabidopsis defines a distinct class of transport vesicles. Plant Cell 25: 3022–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov V.V., Prokhnevsky A.I., Avisar D., Dolja V.V. (2008) Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis. Plant Physiol. 146: 1109–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov V.V., Prokhnevsky A.I., Dolja V.V. (2010) Class XI myosins are required for development, cell expansion, and F-actin organization in Arabidopsis. Plant Cell 22: 1883–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhnevsky A.I., Peremyslov V.V., Dolja V.V. (2008) Overlapping functions of the four class XI myosins in Arabidopsis growth, root hair elongation, and organelle motility. Proc. Natl. Acad. Sci. USA 105: 19744–19749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A.S. (2001) Molecular motors and their functions in plants. Int. Rev. Cytol. 204: 97–178. [DOI] [PubMed] [Google Scholar]
- Reddy A.S., Day I.S. (2001) Analysis of the myosins encoded in the recently completed Arabidopsis thaliana genome sequence. Genome Biol. 2: RESEARCH0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff C., Furch M., Brenner B., Manstein D.J., Meyhofer E. (2001) Single-molecule tracking of myosins with genetically engineered amplifier domains. Nat. Struct. Biol. 8: 226–229. [DOI] [PubMed] [Google Scholar]
- Sparkes I. (2011) Recent advances in understanding plant myosin function: life in the fast lane. Mol. Plant 4: 805–812. [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Teanby N.A., Hawes C. (2008) Truncated myosin XI tail fusions inhibit peroxisome, Golgi, and mitochondrial movement in tobacco leaf epidermal cells: a genetic tool for the next generation. J. Exp. Bot. 59: 2499–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J.A. (2001) The myosin swinging cross-bridge model. Nat. Rev. Mol. Cell Biol. 2: 387–392. [DOI] [PubMed] [Google Scholar]
- Steffens A., Jaegle B., Tresch A., Hulskamp M., Jakoby M. (2014) Processing-body movement in Arabidopsis depends on an interaction between myosins and DECAPPING PROTEIN1. Plant Physiol. 164: 1879–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Iwabuchi K., Fukao Y., Kondo M., Okamoto K., Ueda H., et al. (2013) Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Curr. Biol. 23: 1776–1781. [DOI] [PubMed] [Google Scholar]
- Tominaga M., Ito K. (2015) The molecular mechanism and physiological role of cytoplasmic streaming. Curr. Opin. Plant Biol. 27: 104–110. [DOI] [PubMed] [Google Scholar]
- Tominaga M., Kimura A., Yokota E., Haraguchi T., Shimmen T., Yamamoto K., et al. (2013) Cytoplasmic streaming velocity as a plant size determinant. Dev. Cell 27: 345–352. [DOI] [PubMed] [Google Scholar]
- Tominaga M., Kojima H., Yokota E., Nakamori R., Anson M., Shimmen T., et al. (2012) Calcium-induced mechanical change in the neck domain alters the activity of plant myosin XI. J. Biol. Chem. 287: 30711–30718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga M., Kojima H., Yokota E., Orii H., Nakamori R., Katayama E., et al. (2003) Higher plant myosin XI moves processively on actin with 35 nm steps at high velocity. EMBO J. 22: 1263–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga M., Nakano A. (2012) Plant-specific myosin XI, a molecular perspective. Front. Plant Sci. 3: 211.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H., Yokota E., Kutsuna N., Shimada T., Tamura K., Shimmen T., et al. (2010) Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc. Natl. Acad. Sci. USA 107: 6894–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda T.Q.P., Abramson P.D., Spudich J.A. (1996) The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc. Natl. Acad. Sci. USA 93: 4459–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda T.Q., Warrick H.M., Kron S.J., Spudich J.A. (1991) Quantized velocities at low myosin densities in an in vitro motility assay. Nature 352: 307–311. [DOI] [PubMed] [Google Scholar]
- Veigel C., Wang F., Bartoo M.L., Sellers J.R., Molloy J.E. (2002) The gated gait of the processive molecular motor, myosin V. Nat. Cell Biol. 4: 59–65. [DOI] [PubMed] [Google Scholar]
- Vidali L., Burkart G.M., Augustine R.C., Kerdavid E., Tuzel E., Bezanilla M. (2010) Myosin XI is essential for tip growth in Physcomitrella patens. Plant Cell 22: 1868–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Qin L., Liu G., Peremyslov V.V., Dolja V.V., Wei Y. (2014) Myosins XI modulate host cellular responses and penetration resistance to fungal pathogens. Proc. Natl. Acad. Sci. USA 111: 13996–14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota E., Muto S., Shimmen T. (1999) Inhibitory regulation of higher-plant myosin by Ca2+ ions. Plant Physiol. 119: 231–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.