Abstract
Caspases have functions other than in apoptosis. Here, we report that Caenorhabditis elegans CED-3 caspase regulates asymmetric cell division. Many of the 131 cells that are “programmed” to die during C. elegans development are the smaller daughter of a neuroblast that divides asymmetrically by size and fate. We have previously shown that CED-3 caspase is activated in such neuroblasts, and that before neuroblast division, a gradient of CED-3 caspase activity is formed in a ced-1 MEGF10 (multiple EGF-like domains 10)-dependent manner. This results in the nonrandom segregation of active CED-3 caspase or “apoptotic potential” into the smaller daughter. We now show that CED-3 caspase is necessary for the ability of neuroblasts to divide asymmetrically by size. In addition, we provide evidence that a pig-1 MELK (maternal embryonic leucine zipper kinase)-dependent reciprocal gradient of “mitotic potential” is formed in the QL.p neuroblast, and that CED-3 caspase antagonizes this mitotic potential. Based on these findings, we propose that CED-3 caspase plays a critical role in the asymmetric division by size and fate of neuroblasts, and that this contributes to the reproducibility and robustness with which the smaller daughter cell is produced and adopts the apoptotic fate. Finally, the function of CED-3 caspase in this context is dependent on its activation through the conserved egl-1 BH3-only, ced-9 Bcl-2, and ced-4 Apaf-1 pathway. In mammals, caspases affect various aspects of stem cell lineages. We speculate that the new nonapoptotic function of C. elegans CED-3 caspase in asymmetric neuroblast division is relevant to the function(s) of mammalian caspases in stem cells.
Keywords: caspase, nonapoptotic function, asymmetric cell division, neuroblasts, C. elegans, pig-1 MELK
DURING embryonic and postembryonic Caenorhabditis elegans development, 131 somatic cells reproducibly die (Sulston and Horvitz 1977; Sulston et al. 1983). Genetic screens resulted in the identification of four genes that can mutate to block most of these cell deaths and that define a conserved apoptotic cell death pathway (egl-1 BH3-only, ced-9 Bcl-2, ced-4 Apaf-1, and ced-3 caspase) (Horvitz 2003; Conradt et al. 2016). Interestingly, most of the cells that are programmed to die during development are generated through divisions that are asymmetric by fate and size, and that produce a smaller daughter that is programmed to die. The apoptotic death of the smaller daughter is triggered through the transcriptional upregulation (and, hence, increase in expression) in that cell of egl-1 BH3-only, which induces apoptosome formation, and the maturation and activation of the protease CED-3 caspase. Active CED-3 caspase cleaves specific substrates and thereby induces the killing, dismantling, and phagocytosis of the cell in a cell-autonomous manner. For example, CED-3 caspase cleaves and activates the lipid scramblase CED-8 Xkr8, which results in the exposure of the “eat-me signal” phosphatidylserine (PS) on the surface of the dying cell (Stanfield and Horvitz 2000; Suzuki et al. 2013). This signal is recognized by receptors on neighboring cells, namely CED-1 MEGF10 (multiple EGF-like domains 10), which leads to receptor clustering and the activation of two conserved parallel engulfment pathways in the engulfing cell (Zhou et al. 2001; Venegas and Zhou 2007). Recently, we demonstrated that active CED-3 caspase is already present in the mother of at least one cell programmed to die, the embryonic neurosecretory motor neuron (NSM) neuroblast, which divides to give rise to the larger NSM, which survives and differentiates into a serotonergic motor neuron, and the smaller NSM sister cell (NSMsc), which dies (Chakraborty et al. 2015; Lambie and Conradt 2016). Furthermore, this active CED-3 caspase causes the clustering and activation (in a ced-8 Xkr8- and PS-independent manner) of CED-1 MEGF10 and the two engulfment pathways in the two dorsal neighbors of the NSM neuroblast. This activation of the engulfment pathways in turn is necessary for the formation and/or maintenance of a gradient of CED-3 caspase activity in the NSM neuroblast, and the nonrandom segregation of active CED-3 caspase into the smaller NSMsc, where it promotes the robust and swift execution of apoptotic cell death (Chakraborty et al. 2015; Lambie and Conradt 2016).
The formation of a gradient of CED-3 caspase activity in the mother of a cell programmed to die has so far only been demonstrated in the embryonic NSM neuroblast lineage. For this reason, the generality of this phenomenon has so far been unclear. In addition, whether active CED-3 caspase plays a role in the mother other than promoting its own enrichment in one part of the cell, has been unknown. To address these questions, we examined the postembryonic QL.p neuroblast lineage. Our results support the notion that the formation of a gradient of CED-3 caspase activity is a general phenomenon. Furthermore, we provide evidence that the ced-3 caspase gene plays an active role in the asymmetric division of mothers. Specifically, we provide evidence that ced-3 caspase is required for their ability to divide asymmetrically by size and fate, and, hence, to produce the smaller daughter, which is programmed to die.
Materials and Methods
Strains and genetics
All C. elegans strains analyzed were maintained at 25° on Nematode Growth Medium, unless otherwise specified (Brenner 1974). The following mutations and transgenes were used in this study: LGI: ced-1(e1735) (Hedgecock et al. 1983); LGIII: ced-4(n1162) (Ellis and Horvitz 1986), ced-6(n1813) (Ellis et al. 1991), unc-119(ed3) (Maduro and Pilgrim 1995) and rdvIs1(Pegl‐17mCherry::his‐24, Pegl‐17myristoylated mCherry, Pegl‐17mig‐10::yfp) (Ou et al. 2010); LGIV: ced-2(n1994) (Ellis et al. 1991), ced-3(n717) (Ellis and Horvitz 1986), ced-3(n2433) (Shaham et al. 1999), ced-3(n2427) (Shaham et al. 1999), pig-1(gm344) (Cordes et al. 2006), and bcSi82 (Ptoe-2mKate2::tac-1) (this study); and LGV: egl-1(n3330) (Sherrard et al. 2017), bzIs190 (Pmec-4gfp) (M. Driscoll (Rutgers University), personal communication), ltIs44 (Ppie-1mCherry::phPLCδ) (Audhya et al. 2005), and enIs1 (Pced-1ced-1∆C::gfp) (Zhou et al. 2001). Additional transgenes used in this study are: bcIs133 (Ptoe-2gfp) (this study), bcEx1277 (Phyp7ced-1::gfp) (this study), and bcEx1334 (Ptoe-2ced-1::mKate2) (this study).
Cloning
pBC1565 (Ptoe-2gfp):
The toe-2 promoter (Ptoe-2) (2117 bp immediately upstream of the toe-2 start site) was amplified by PCR using N2 genomic DNA, and restriction sites for AgeI and SacI were introduced at the ends of the PCR product. Primers used to amplify Ptoe-2 were SacI Ptoe-2-F (5′-aaaaaGAGCTCttatctgtaccacaaattcc-3′) and AgeI Ptoe-2-R (5′-aaaaaACCGGTttttgacctgaggacatgatg-3′). The resulting PCR product and plasmid pBC1408 (Pces-2gfp::unc-54 3′UTR in pBluescript) were digested with AgeI and SacI (which drops out Pces-2 from pBC1408), and the PCR product cloned into the pBC1408 backbone using T4 ligation to obtain pBC1565 (Ptoe-2gfp).
pBC1591 (Phyp7gfp) and pBC1681 (Phyp7ced-1::gfp):
The promoter of gene Y37A1B.5 (Phyp7) (2910 bp immediately upstream of the Y37A1B.5 start site) (Hunt-Newbury et al. 2007) was amplified by PCR using N2 genomic DNA, and restriction sites for AgeI and SacI were introduced at the ends of the PCR product. Primers used to amplify Phyp7 were Phyp7-F (5′-aaaGAGCTCaaactttattagacgtcgcaattt-3′) and Phyp7-R (5′-aaaACCGGTtttggtttttgggatttttgatc-3′). The PCR product obtained (Phyp7) was used to replace Ptoe-2 in pBC1565 using restriction digest with AgeI and SacI, and T4 ligation to generate plasmid pBC1591 (Phyp7gfp). A ced-1 minigene was amplified from plasmid pZZ610 (Pced-1ced-1::gfp, Zhou et al. 2001) by PCR using the primers AgeI ced-1-F (5′-aaaACCGGTatgcgtctcattctccttgtgctac-3′) and ced-1-R (5′-tttttctaccggtacttgaattcct-3′). The resulting PCR product and pBC1591 were digested with AgeI, and the ced-1 minigene was inserted by T4 ligation into the linearized pBC1591 to obtain plasmid pBC1681 (Phyp7ced-1::gfp).
pBC1805 (Ptoe-2ced-1::mKate2):
pBC1805 was cloned by Gibson cloning (Gibson et al. 2009). Primers pBSKPtoe-2-F (5′-ATCCCCCGGGCTGCAGGAATTCGATTTATCTGTACCACAAATTCCTTG-3′) and ced-1Ptoe-2-R (5′-GAATGAGACGCATTTTTGACCTGAGGACATG-3′) were used to amplify Ptoe-2 using pBC1565 as a template. A ced-1 minigene was amplified in two parts: fragment 1 was amplified using primers Ptoe-2ced-1_fwd (5′-CCTCAGGTCAAAAATGCGTCTCATTCTCCTTG-3′) and ced-1_mini_(1)_rev (5′-ccgggtcacagttGGCTCCATTTTCACAGTC-3′), whereas fragment 2 was amplified using the primers ced-1_mini_(2)_fwd (5′-tgaaaatggagccAACTGTGACCCGGAACTC-3′) and ced-1_mini_(2)_rev (5′-ccttgatgagctcggaTTTTTCTACCGGTACTTGAATTC-3′). pZZ610 was used as a template for the amplification by PCR of both fragments (Zhou et al. 2001). mKate2::tbb-2 3′UTR was amplified from plasmid pEZ167 (Pmex-5fkbp12::mKate2::tbb-2 3′UTR; E. Zanin (LMU Munich), personal communication, Turek et al. 2013) as template with the help of the primers mK_tbb-2_fwd (5′-accggtagaaaaaTCCGAGCTCATCAAGGAG-3′) and mK_tbb-2_rev (5′-ggtcgacggtatcgataagcttgatCAATGAGACTTTTTTCTTGGC-3′). EcoRV-digested pBluescript II KS(+) was used as backbone and all the above fragments were assembled using Gibson assembly to obtain pBC1805 (Ptoe-2ced-1::mKate2).
pBC1807 (Ptoe-2mKate2::tac-1):
pBC1807 was generated by Gibson assembly (Gibson et al. 2009). Ptoe-2 was amplified using the primers NM1 (5′-gagctctggtaccctctagtcaaggTTATCTGTACCACAAATTCCTTG-3′) and NM2 (5′-tgagctcggacatTTTTGACCTGAGGACATG-3′), and pBC1565 as a template. mKate2::tac-1 was obtained by PCR amplification using plasmid TMD34 (Pmex-5mKate2::tac-1::tbb-2 3′UTR; T. Mikeladze-Dvali, (LMU Munich) personal communication) as a template, and the primers NM3 (5′-cctcaggtcaaaaATGTCCGAGCTCATCAAG-3′) and NM4 (5′-aattctacgaatgTTATGCATCCGTCGAAATAAC-3′). Similarly, pBC1565 was used as a template to amplify the unc-54 3′UTR using primers NM5 (5′-gacggatgcataaCATTCGTAGAATTCCAACTG-3′) and NM6 (5′-agtccgtaatacgactcacttaaggAAACAGTTATGTTTGGTATATTGG-3′). These fragments were introduced into StuI-digested pCFJ909 (Frøkjær-Jensen et al. 2014) using Gibson assembly to obtain pBC1807 (Ptoe-2mKate2::tac-1).
Microinjection
bcIs133 (Ptoe-2gfp):
pBC1565 (20 ng/μl) + pRF4 (80 ng/μl) was injected into the gonads of young N2 adult hermaphrodites and extrachromosomal array was integrated by UV irradiation to obtain bcIs133, which was 5× backcrossed with N2.
bcEx1277 (Phyp7ced-1::gfp):
pBC1681 (51 ng/μl) + IR101 (10 ng/μl) (Prps-0HygR::gpd-2/gpd-3::mCherry::unc-54 3′UTR, Radman et al. 2013) was injected into the gonads of young ced-1(e1735); ced-3(n2427); bcIs133 adult hermaphrodites to obtain ced-1(e1735); ced-3(n2427); bcIs133; bcEx1277.
bcEx1334 (Ptoe-2ced-1::mKate2):
bcEx1334 was generated by injecting pBC1805 (26 ng/μl) + pCFJ90 (Pmyo-2::mCherry::unc-54 3′UTR; Frøkjaer-Jensen et al. 2008) (2.6 ng/μl) + pBluescript II KS(+) (60 ng/μl) into the gonads of young ced-1(e1735); ced-3(n2427); bcIs133 adult hermaphrodites to obtain ced-1(e1735); ced-3(n2427); bcIs133; bcEx1334.
bcSi82 (Ptoe-2mKate2::tac-1):
bcSi82 was generated by miniMOS integration of pBC1807 (Ptoe-2mKate2::tac-1) into HT1593 [unc-119(ed3)] animals (Frøkjær-Jensen et al. 2014). First, 10 ng/μl of pBC1807 was injected into the gonads of HT1593 animals along with pCFJ601 (50 ng/μl), pGH8 (10 ng/μl), pCFJ90 (2.5 ng/μl), and pCFJ104 (5 ng/μl) (Frøkjær-Jensen et al. 2014). Worms were allowed to starve for 1 week, after which wild-type movers were examined for integration.
Extra PVM neurons
Posterior ventral mechanosensory (PVM) neurons were visualized using the transgene Pmec-4gfp (bzIs190), which labels all mechanosensory neurons (Mitani et al. 1993). Fourth larval stage (L4) larvae of the desired genotype were anesthetized in a drop of sodium azide solution (30 mM in M9 buffer) on a 2% agarose pad on a glass slide. A 100×/1.3 NA oil-immersion objective lens on a Zeiss ([Carl Zeiss], Thornwood, NY) Imager.M2 epifluorescence microscope was used to visualize PVM neurons.
QL.pp survival
QL.pp survival was determined using the transgene Ptoe-2gfp (bcIs133), which labels cells of the Q lineages (Gurling et al. 2014). In wild-type animals, QL.pp dies within ∼17 hr postegg laying during the first larval stage (L1 stage) (at 25°) (Sulston and Horvitz 1977). To avoid false positives due to delayed cell death, we used larvae of the second larval stage (L2 larvae) (25–30 hr postegg laying) for analysis. Larvae were anesthetized in a drop of levamisole solution (10 mM in M9 buffer) on a 2% agarose pad on a glass slide. Ptoe-2gfp was visualized with the help of a 100X/1.3 NA oil-immersion objective lens on a Zeiss Imager.M2 epifluorescence microscope. We only considered those worms for assessing QL.pp survival in which QL.pa had divided and its daughters had formed visible neurite extensions at the time of counting. Thus, we ensured that slow-growing strains were not analyzed at an earlier developmental time point. Importantly, upon failure of death, QL.aa, which is produced in the vicinity of QL.p, migrates toward the tail, and therefore away from QL.pp. As a result, it does not interfere with our determination of QL.pp survival.
Division of “undead” QL.pps
The division of undead QL.pps was determined using the transgene Ptoe-2gfp (bcIs133). L2 larvae were prepared similarly as for QL.pp survival. Animals were assessed after QL.pa had divided to form PVM and SDQL neurons with visible neurite outgrowths.
Live imaging of QL.p and QL.a divisions
The transgene Ptoe-2gfp (bcIs133) was used to identify QL.p and QL.a, and to analyze their asymmetric divisions. L1 larvae were immobilized in 1 μl polybead microsphere suspension (0.1 μm diameter, 2.5% w/v, catalog number 00876; Polysciences, Warrington, PA) on a 10% agarose pad (agarose was dissolved in 67% M9 buffer). A glass coverslip was placed on the agar pad and the empty space around the agarose pad underneath the coverslip was filled with paraffin oil to prevent dehydration. Image Z-stacks were acquired every 3 min with a 1-μm step size using a 63×/1.4 NA oil-immersion objective lens on the UltraVIEW VoX spinning disk microscope (Perkin Elmer [Perkin Elmer-Cetus], Norwalk, CT).
QL.p and QL.a daughter cell sizes
Q-lineage cells are relatively flat cells. As a result, single-plane cell areas provide a fair estimation of cell sizes (i.e., area is directly proportional to cell volume) (Cordes et al. 2006). Therefore, the image Z-stacks acquired of the daughters of QL.p and QL.a using the transgene Ptoe-2gfp (bcIs133) (see Live imaging of QL.p and QL.a divisions) were converged to obtain maximum-intensity Z-projections. Cell sizes were estimated by circumscribing the cells and measuring their areas with Fiji (Schindelin et al. 2012; Schneider et al. 2012).
Determination of QL.p cleavage furrow position
Images of QL.p undergoing cytokinesis were obtained from the movies generated to assess QL.p daughter cell sizes. The position of the cleavage furrow was determined by measuring its distance from the anterior periphery of QL.p and dividing it by the total length of the cell (Figure 5D). Distances were measured using Fiji (Schindelin et al. 2012; Schneider et al. 2012).
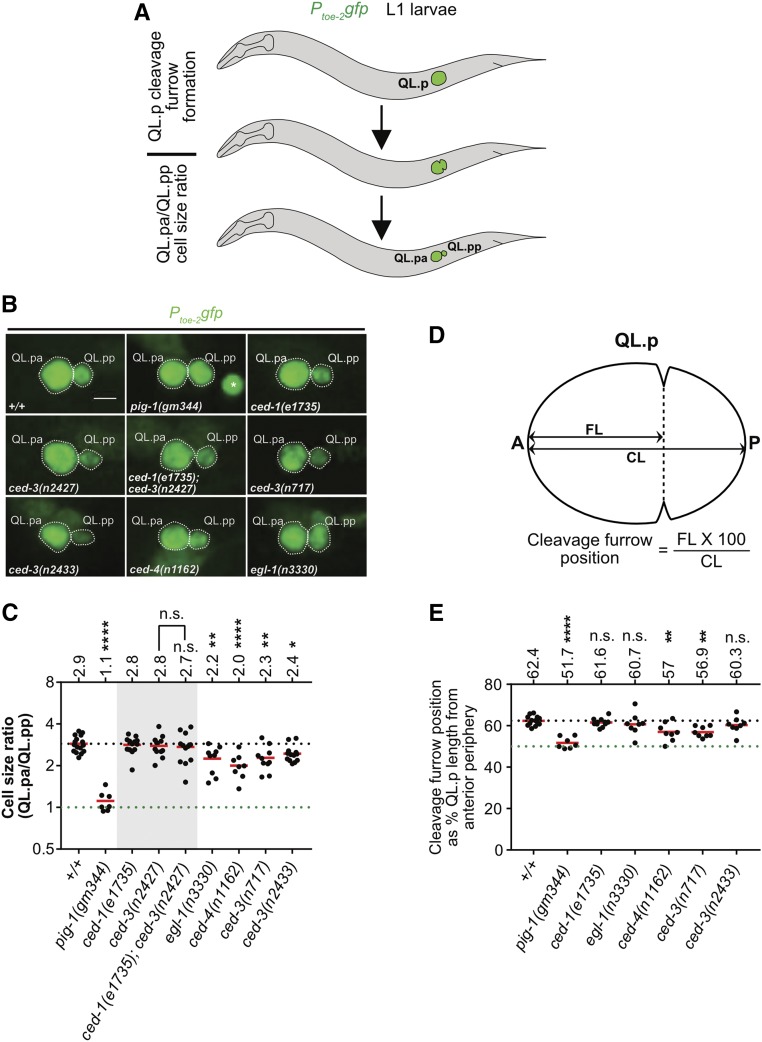
Figure 5.
Genetic requirements for the asymmetric division by size of QL.p. (A) Schematic representation of the assays. The division of QL.p was monitored by live imaging. QL.p division asymmetry was assessed at two stages during mitosis, specifically during and after cytokinesis. During cytokinesis, we determined the position of the cleavage furrow. The first time point upon the completion of cytokinesis was used to estimate daughter cell sizes based on maximum intensity projection images, as described in the Materials and Methods. QL.p and its daughter cells were visualized using the transgene Ptoe-2gfp (bcIs133). (B) Representative images of QL.p daughters immediately after QL.p cytokinesis in the indicated genotypes. Bar, 3 μm. In the image showing pig-1(gm344), the asterisk (*) marks QL.aa present in the vicinity of QL.pa and QL.pp. (C) Dot plot showing cell size ratios (QL.pa/QL.pp) in the indicated genotype. Each dot represents the cell size ratio for one QL.p. The horizontal red lines indicate the means for the respective genotypes, which are also mentioned on top for each genotype. The dotted black line represents the mean in wild-type (+/+). The dotted green line represents the ratio 1. (D) Schematic representation of the method used to determine the position of the cleavage furrow during QL.p cytokinesis. We measured the distance of the cleavage furrow from the anterior periphery of QL.p (FL), and divided it by QL.p length (CL) to obtain the cleavage furrow position. (E) Dot plot showing the position of the cleavage furrow during QL.p cytokinesis. Each dot represents the position in one QL.p. The horizontal red lines indicate the means for the respective genotypes, which are also mentioned at the top for each genotype. The dotted green line represents the position corresponding to 50% CL, and the dotted black line represents the mean for wild-type animals. Statistical test: one-way ANOVA followed by Benjamini and Hochberg multiple comparisons correction. ** P < 0.01, **** P < 0.0001, and n ≥ 7 QL.ps. Statistical test: one-way ANOVA followed by Benjamini and Hochberg multiple comparisons correction. * P < 0.05, ** P < 0.01, **** P, 0.0001, and n ≥ 7 QL.ps. n.s., not significant.
Localization studies for CED-1ΔC::GFP
CED-1 localization was analyzed using the transgene Pced-1ced-1∆C::gfp (enIs1) (Zhou et al. 2001). L1 larvae were anesthetized with levamisole (0.1 mM in M9 buffer), mounted on 3% agarose pads on glass slides, and examined using 63×/1.4 NA oil-immersion objectives on a Leica SP5 inverted confocal microscope. Q-lineage cells were identified with the help of the transgene Pegl‐17mCherry::his‐24, Pegl‐17myristoylated mCherry, Pegl‐17mig‐10::yfp (rdvIs1) (Ou et al. 2010).
TAC-1 ratio
The TAC-1 ratio was determined using the transgenes Ptoe-2mKate2::tac-1 (bcSi82) and Ptoe-2gfp (bcIs133). L1 larvae of the desired genotypes were grown at 20° and immobilized in 1 μl polybead microsphere suspension, and slides prepared as described earlier for live imaging of QL.p and QL.a divisions. Image Z-stacks were acquired every 5 min with a 0.5-μm step size using a 100×/1.4 NA oil-immersion objective lens on the UltraVIEW VoX spinning disk microscope (Perkin Elmer). The time point at which QL.p was at metaphase was used to determine the amount of TAC-1 associated with the two centrosomes. Quantification of the amount of centrosome-associated TAC-1 was performed as described previously (Chakraborty et al. 2015). A region of the same size on the slide but outside the animal was considered as background noise, and its intensity was subtracted from the measured intensities of centrosome-associated TAC-1.
NSM neuroblast daughter cell sizes
The NSM and NSMsc were identified using the transgene Ppie-1mCherry::phPLCδ (ltIs44), and their sizes determined as described (Chakraborty et al. 2015; Wei et al. 2017). All images were acquired using a Leica TCS SP5 II confocal microscope. All strains were incubated at 20° overnight before imaging.
Embryonic lethality
Five L4 larvae of each genotype were singled on seeded NGM plates. These were allowed to lay eggs for 30 hr at 20°. Two-days later, the numbers of larvae and dead eggs were determined.
Statistical analysis
Proportions were compared using Fisher’s Exact Test, and the obtained P-values were adjusted using the Benjamini and Hochberg test for multiple comparisons (Fisher 1935; Yoav 1995). Wherever applicable, data were tested for normal distribution using the D’Agostino and Pearson normality test (D’Agostino and Pearson 1993). When data were found to be distributed normally, the Student’s t-test or parametric one-way ANOVA were performed to determine statistical significance between groups assuming that the groups had unequal SD, and Tukey’s or the Benjamini and Hochberg multiple comparisons test was applied (Student 1908; Fisher 1921; Tukey 1949). When comparing the amounts of TAC-1 on the anterior and posterior centrosomes (where all posterior TAC-1 levels were set to 1), we used the Wilcoxon signed-rank test to determine if the groups were statistically different (Wilcoxon 1945). The Mann–Whitney test was used to compare the ratios of QL.a daughter cell sizes in wild-type animals with those in ced-3(n717) animals (Mann 1947).
Data availability
All reagents and strains generated for this study are available from the authors upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article and figures. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7058609.
Results
A ced-1 MEGF10-dependent, but pig-1 MELK-independent, gradient of CED-3 caspase activity is present in the postembryonic QL.p neuroblast
During the L1 stage of postembryonic C. elegans development, the neuroblast QL.p divides asymmetrically by size and fate to produce a larger anterior daughter, QL.pa, which survives, and a smaller posterior daughter, QL.pp, which is programmed to die (Sulston and Horvitz 1977; Cordes et al. 2006) (Figure 1, wild-type). The observation that defects in the asymmetric division of QL.p by size can affect the fate of its daughters (especially the fate of QL.pp) indicates that daughter cell size and daughter cell fate are functionally coupled (Cordes et al. 2006; Singhvi et al. 2011; Gurling et al. 2014; Teuliere et al. 2014; Teuliere and Garriga 2017). To determine whether a gradient of CED-3 caspase activity forms in QL.p prior to its division, we used a reporter for the protein TAC-1 (Ptoe-2mKate2::tac-1). TAC-1 is a component of the pericentriolar material and a substrate of CED-3 caspase (Bellanger and Gonczy 2003; Le Bot et al. 2003; Srayko et al. 2003; Chakraborty et al. 2015). We have previously shown that in the embryonic NSM neuroblast, the amount of TAC-1 associated with the centrosome that is inherited by the larger daughter, which survives, is greater (by 1.30-fold) than the amount of TAC-1 associated with the centrosome that is inherited by the smaller daughter, which is programmed to die (Chakraborty et al. 2015). This “TAC-1 asymmetry” is dependent on a functional ced-3 caspase gene as well as a CED-3 caspase cleavage site in the TAC-1 protein and, hence, reflects a gradient of CED-3 caspase activity along the cleavage axis of the NSM neuroblast. Furthermore, the establishment and/or maintenance of this gradient of CED-3 caspase activity in the NSM neuroblast is dependent on the two conserved C. elegans engulfment pathways (Chakraborty et al. 2015). We found that at QL.p metaphase, the amount of TAC-1 associated with the anterior centrosome is 1.25-fold greater than that associated with the posterior centrosome (Figure 2, A and B). Furthermore, this asymmetry is lost in the background of a strong loss-of-function (lf) mutation of ced-3 caspase, n717, or a strong lf mutation of the engulfment gene ced-1 MEGF10, e1735 (ratios of 0.97 and 0.99, respectively) (Figure 2, A and C). The gene pig-1 encodes a PAR-1-like kinase orthologous to mammalian MELK (maternal embryonic leucine zipper kinase) and is important for the asymmetric division of QL.p. Specifically, the loss of pig-1 MELK causes QL.p to divide symmetrically by size (Cordes et al. 2006) (see below). Interestingly, we found that TAC-1 asymmetry at QL.p metaphase is not affected by a strong lf mutation of pig-1 MELK, gm344 (ratio of 1.28) (Figure 2, A and C). Therefore, a ced-1 MEGF10-dependent gradient of CED-3 caspase activity is also formed in QL.p prior to its division. This gradient is along the anterior–posterior axis and presumably results in the nonrandom segregation of active CED-3 caspase into the smaller posterior daughter QL.pp, which is programmed to die. pig-1 MELK is not required for the formation of this CED-3 caspase activity gradient. However, because of its role in the asymmetric division of QL.p by size, its loss nevertheless probably affects the concentration of active CED-3 caspase in QL.pp (the concentration is probably less than that in wild-type; see below).
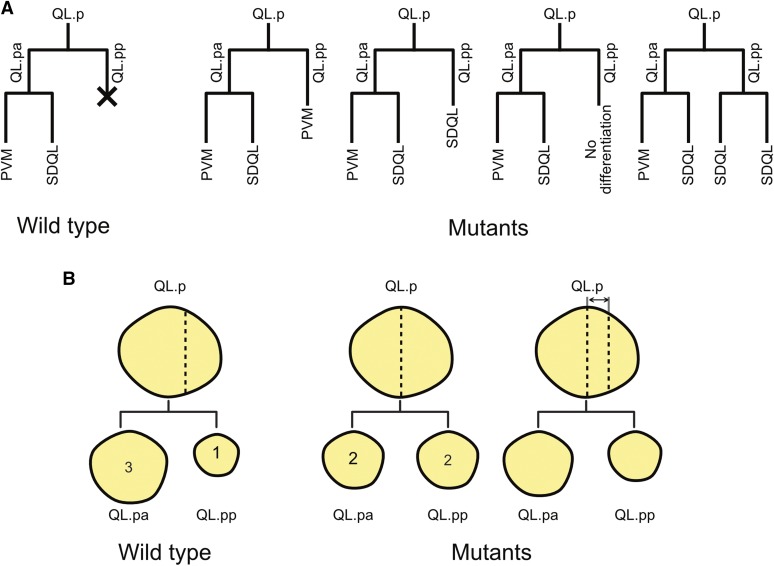
Figure 1.
The C. elegans QL.p neuroblast lineage. (A) Schematic representation of the postembryonic QL.p neuroblast lineage in wild-type (left) and mutant (right) worms. Vertical and horizontal lines represent individual cells and cell divisions, respectively. “X” denotes a cell death. (B) Schematic representation of the cell sizes of the QL.p neuroblast and its daughter cells in wild-type (left) and mutant (right) worms. In wild-type worms, QL.pa is nearly three times as large as its sister, QL.pp. However, in mutants with defects in the asymmetric division by size of QL.p, QL.pa and QL.pp can be of similar sizes.
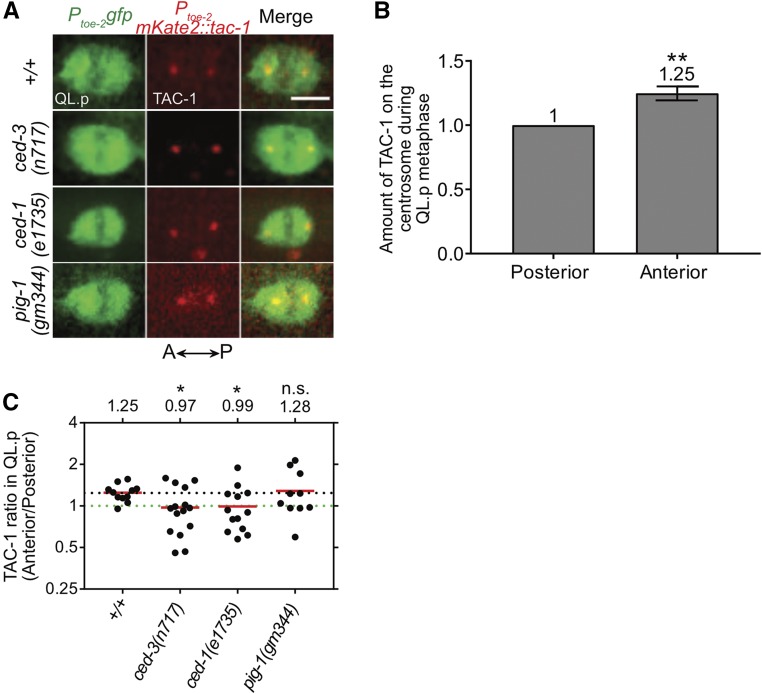
Figure 2.
A ced-1 MEGF10-dependent gradient of CED-3 caspase activity exists in QL.p at metaphase. (A) Representative images of centrosome-associated mKate2::TAC-1 in QL.p at metaphase in the indicated genotypes. Bar, 3 μm. QL.p was identified using Ptoe-2gfp (bcIs133) and TAC-1 was visualized using Ptoe-2mKate2::tac-1 (bcSi82). (B) Bar graph showing the comparison of total mKate2::TAC-1 signals associated with the anterior and posterior centrosomes during QL.p metaphase in wild-type animals. Total “Anterior” TAC-1 signal was normalized to the total “Posterior” TAC-1 signal, which was set to 1 for each animal. Wilcoxon signed-rank test was used to determine statistical significance. ** P < 0.01, n = 11 QL.ps. (C) Dot plot showing the spread of the TAC-1 ratios in QL.p (Anterior/Posterior) among animals of each genotype. The horizontal red lines indicate the mean for each genotype (also mentioned above the spread for each group). The dotted black line represents the mean for wild-type (+/+). The dotted green line represents the ratio 1. Each dot represents the TAC-1 ratio for one animal. Statistical significance was determined using the Student’s t-test and Benjamini and Hochberg multiple comparisons correction. * P < 0.05, n ≥ 10 QL.ps. n.s., not significant.
Consistent with the notion that ced-1 MEGF10 and the engulfment pathways play an instructive role in the establishment and/or maintenance of this gradient of CED-3 caspase activity, we found that an asymmetric contact exists between QL.p and a neighboring cell that exhibits detectable levels of CED-1 MEGF10 on its cell surface (Figure 3, Supplemental Material, Figure S1 and File S1). Specifically, QL.p is in contact with the syncytial cell hyp7, which is part of the hypodermis that covers large parts of the animal and which, after QL.p division, engulfs the QL.pp corpse (Sulston and Horvitz 1977). Using a CED-1 MEGF10 reporter (Pced-1ced-1∆C::gfp) (Zhou et al. 2001), we detected CED-1 MEGF10 on what appears to be the entire surface of hyp7 (Figure 3, Figure S1, and File S1). However, we found that while there is almost uniform contact between hyp7 and the lateral side of QL.p, there is an asymmetric contact between hyp7 and the medial side of QL.p. More specifically, the posterior, but not the anterior, part of QL.p’s medial side contacts hyp7. Therefore, a cell surface that contains detectable levels of CED-1 MEGF10 apposes the part of QL.p that will later form the smaller daughter, QL.pp, which is programmed to die, but not the part of QL.p that will later form the larger daughter, QL.pa, which survives (schematically represented in the drawing in the lower left hand corner of Figure 3; this drawing was compiled based on the images shown in Figure 3 and in Figure S1). We propose that this asymmetry in the presentation of CED-1 MEGF10 on apposing cell surfaces (and, hence, in the activation of CED-1 MEGF10 and the engulfment pathways) is critical for the establishment of a gradient of CED-3 caspase activity along the anterior–posterior axis of QL.p.
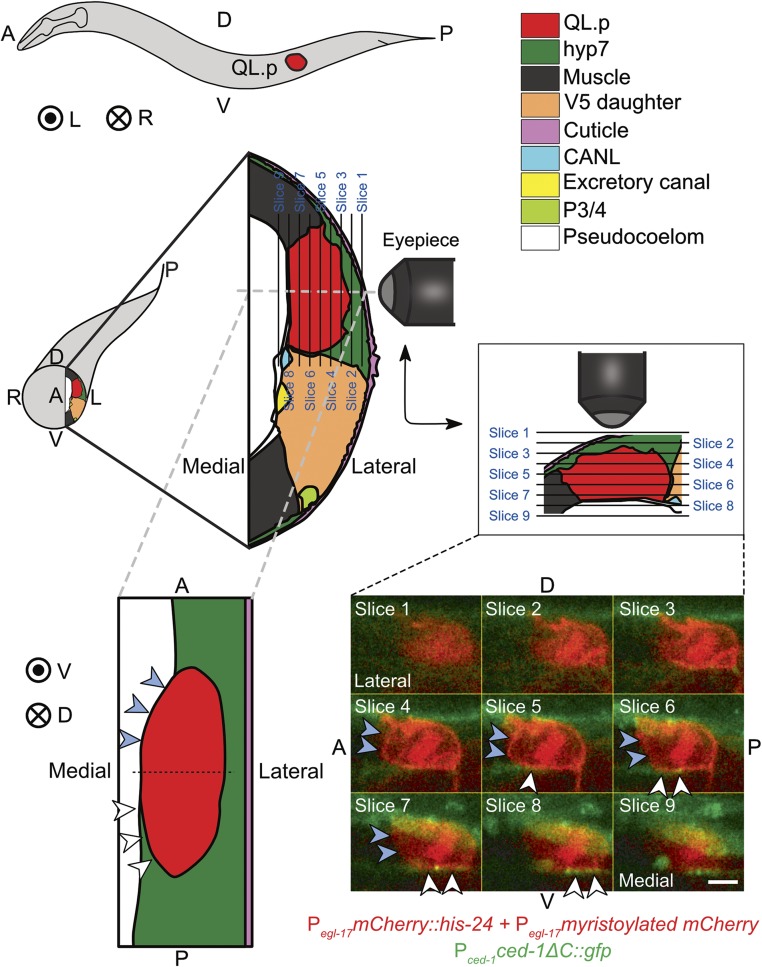
Figure 3.
Asymmetric presence of CED-1 MEGF10 around QL.p. Schematic of an L1 larva with the location of QL.p indicated is shown top-left. A: anterior, P: posterior, D: dorsal, V: ventral, L: left, and R: right. Concentric circles represent the direction projecting toward the reader, while the circle with a cross represents the direction projecting away from the reader. The schematic below the L1 larva represents a transverse section through the larva at the position where QL.p is localized. The section is enlarged in the adjoining image on the right. This and another section (bottom-left) shown were drawn based on fluorescence images and images of electron microscopy sections available on WormAtlas (http://www.wormatlas.org/). The key for cell colors is at the top-right. Larvae were imaged from the side, and individual image slices were acquired while gradually moving from the left to the middle of the worm. QL.p was labeled with the transgene Pegl-17mCherry::his‐24, Pegl-17myristoylated mCherry, Pegl-17mig‐10::yfp (rdvIs1) and hyp7 with the transgene Pced-1ced-1ΔC::gfp (enIs1) (see Materials and Methods for details). The diagram in the middle-right represents the orientation of the image planes, images for which have been shown at the bottom-right. In these images, white arrowheads indicate a contact between CED-1ΔC::GFP and QL.p, whereas blue arrowheads indicate the lack of contact. Bar, 2 μm. It can be seen in the images for slices 4–9 that CED-1ΔC::GFP is in contact with the posterior half of QL.p but not with its anterior part. The coronal section (bottom-left) has been created based on the fluorescence images (slices shown at the bottom-right) and the images of the electron microscopy sections. It can be seen in this diagram that while hyp7 almost entirely envelopes the posterior part of QL.p, it only forms contact with the left side of the anterior part.
The loss of ced-1 MEGF10 promotes the appearance of extra PVM neurons
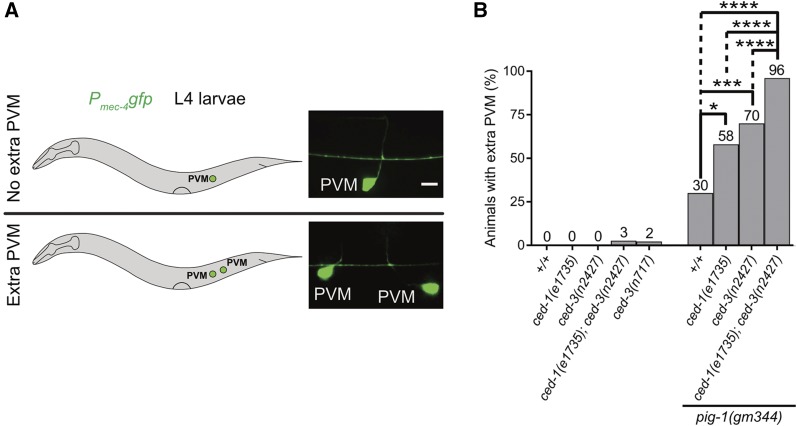
The larger daughter, QL.pa, not only survives but also divides to generate the neurons PVM and SDQL (Figure 1A, wild-type). To determine the possible function of the ced-1 MEGF10-dependent gradient of CED-3 caspase activity in QL.p, we analyzed the number of PVM neurons generated by this lineage. A block in apoptotic cell death prevents the death of QL.pp, and this can lead to the presence of two, rather than one, PVM neurons in L4 larvae (Figure 1A, Mutants). For example, using a PVM-specific reporter (Mitani et al. 1993), an extra PVM neuron is detected in 0% of wild-type animals but in 2% of ced-3(n717) animals (Chien et al. 2013) (Figure 4, A and B). In contrast, a compromised apoptotic cell death pathway, such as in animals homozygous for the weak ced-3 lf mutation n2427, does not lead to the presence of an extra PVM neuron. We found that in a ced-3(n2427), but not wild-type, background, ced-1(e1735) causes the presence of an extra PVM neuron in 3% of the animals (Figure 4B). We also analyzed ced-1(e1735) in the background of pig-1(gm344). In an otherwise wild-type background, the loss of pig-1 MELK causes the presence of an extra PVM neuron in 30% of the animals (Cordes et al. 2006) (Figure 4B). We found that ced-1(e1735) as well as ced-3(n2427) significantly enhanced this phenotype to 58 or 70%, respectively. Furthermore, we found that 96% of the triple-mutant animals had an extra PVM neuron. Therefore, the loss of ced-1 MEGF10 (and, hence, the loss of the gradient of active CED-3 caspase in QL.p) promotes the presence of extra PVM neurons in a background in which the apoptotic cell death pathway is compromised as well as in a background in which QL.p divides symmetrically. Furthermore, in a background in which QL.p divides symmetrically [i.e., in pig-1(gm344)], the enhancements of the extra PVM neuron phenotype, either through compromising the apoptotic cell death pathway or through the loss of ced-1 MEGF10, are additive.
Figure 4.
The loss of ced-1 MEGF10 enhances the extra PVM phenotype of ced-3(n2427) animals. (A) Schematic representation of assay and representative images of L4 larvae with no extra PVM neuron (top), or with an extra PVM neuron (bottom). Bar, 5 μm. PVM neurons were visualized using Pmec-4gfp (bzIs190) as reporter. (B) Bar graph showing the fraction of animals with extra PVM neurons for the indicated genotypes. Statistical test: Fisher’s exact test with Benjamini and Hochberg multiple comparisons correction. * P < 0.05, *** P < 0.001, **** P < 0.0001, and n > 50 animals.
ced-1 MEGF10 is not required for the asymmetric division of QL.p by size
The presence of an extra PVM neuron can be the result of a defect in the asymmetric division of QL.p by size, the asymmetric division of QL.p by fate, or a combination thereof. To determine which of these processes is affected by ced-1(e1735), we analyzed them individually. To analyze the asymmetric division of QL.p by size, we used a GFP reporter that is expressed in QL.p and its daughters (Ptoe-2gfp) (Gurling et al. 2014), and acquired image stacks of QL.p in developing L1 larvae every 3 min. The first stack acquired after the completion of QL.p cytokinesis was then used to estimate the sizes of the two daughter cells (Figure 5A) (Q-lineage cells are relatively flat cells. Hence, measurements of their areas of maximum intensity projection images provide reliable approximations of their cell sizes (Cordes et al. 2006)). In wild-type animals, QL.p divides asymmetrically by size to produce the larger QL.pa and the smaller QL.pp with a size ratio of QL.pa to QL.pp of ∼3.0 (Figure 1B, wild-type; Figure 5, B and C). As was reported previously (Cordes et al. 2006), in pig-1(gm344) animals, QL.p divides symmetrically to produce two cells of similar sizes with a size ratio of QL.pa to QL.pp of 1.1 (Figure 5, B and C). Using this assay, we did not observe a significant effect of ced-1(e1735) on the asymmetric division by size of QL.p in a wild-type or ced-3(n2427) background. Therefore, the loss of ced-1 MEGF10 does not promote the presence of additional PVM neurons by affecting the asymmetric division by size of QL.p.
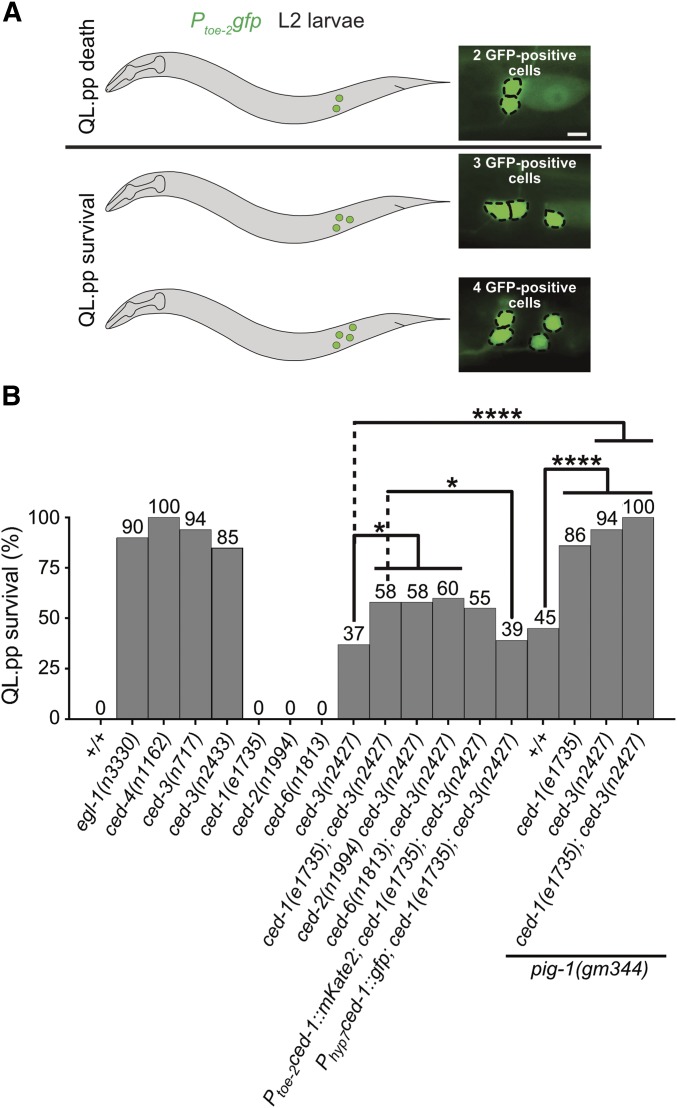
ced-1 MEGF10 promotes the apoptotic fate of QL.pp
QL.p divides asymmetrically by fate to produce QL.pa, which survives and divides to generate two neurons, PVM and SDQL (“mitotic fate”), and QL.pp, which is programmed to die by apoptosis (“apoptotic fate”) (Figure 1A). We tested whether the loss of ced-1 MEGF10 affects the ability of QL.pp to adopt the apoptotic fate. Using the Ptoe-2gfp reporter, we determined the fraction of QL.pps that inappropriately survive (regardless of whether they subsequently divided or not) (Figure 6A). In wild-type animals, QL.pp always dies. In contrast, in animals carrying strong lf mutations of egl-1 BH3-only, n3330, ced-4 Apaf-1, n1162, or ced-3 caspase, n717, QL.pp survives in almost all animals (Figure 6B). Furthermore, in animals homozygous for ced-3(n2427), 37% of QL.pps survive. We found that while ced-1(e1735) does not cause QL.pp survival in an otherwise wild-type background, it increases QL.pp survival in a ced-3(n2427) background to 58%. Similarly, lf mutations of ced-2 CrkII or ced-6 GULP (n1994 and n1813, respectively), two other engulfment genes, also enhance the ced-3(n2427) phenotype. To determine in which cell or cells the engulfment genes act to enhance QL.pp survival, we performed rescue experiments using promoters that drive transgene expression specifically in the Q lineage (toe-2 promoter) (Gurling et al. 2014) or in hyp7 (“hyp7 promoter”) (Hunt-Newbury et al. 2007). We found that the expression of a ced-1 minigene under the control of the hyp7 promoter, but not the toe-2 promoter, rescues the ced-1(e1735) phenotype (Figure 6B). Therefore, in a background in which apoptotic cell death is compromised, the loss of ced-1 MEGF10 or other engulfment genes increases the probability of QL.pp survival. Hence, the engulfment genes act to promote the apoptotic fate in QL.pp. Furthermore, in this context, ced-1 MEGF10, and most probably the other engulfment genes, act in hyp7 and therefore in a cell nonautonomous manner. Finally, based on our finding that a ced-1 MEGF10-dependent gradient of CED-3 caspase activity is established in QL.p before its division, we propose that ced-1 MEGF10 and the other engulfment genes promote the apoptotic fate of QL.pp by causing the nonrandom segregation of active CED-3 caspase into QL.pp.
Figure 6.
pig-1 MELK, ced-1 MEGF10, and ced-3 caspase interact to promote the apoptotic fate in QL.pp. (A) Schematic representation of assay and representative images of L2 larvae showing two, three, or four cells of the QL.p lineage. Bar, 3 μm. Cells were visualized using the transgene Ptoe-2gfp (bcIs133). For the purpose of this analysis, animals with three or four QL.p lineage cells were counted as an instance of “QL.pp survival,” and those with two QL.p lineage cells were counted as ones of “QL.pp death.” (B) Bar graph showing the fraction of animals in which QL.pp inappropriately survived for the indicated genotypes. Fisher’s exact test was used to determine statistical significance. Benjamini and Hochberg multiple comparisons correction was used to adjust individual P values. * P < 0.05, **** P < 0.0001, and n > 50 animals.
pig-1 MELK, ced-1 MEGF10, and ced-3 caspase interact to promote the apoptotic fate of QL.pp
We also determined the effect of ced-1(e1735) on the ability of QL.pp to adopt the apoptotic fate in a background in which QL.p divides symmetrically by size, i.e., in the pig-1(gm344) background. In pig-1(gm344) animals, 45% of QL.pps survive (Figure 6B). We found that the loss of ced-1 MEGF10 enhances QL.pp survival in this background to 86% (Figure 6B). Similarly, ced-3(n2427) enhances QL.pp survival in pig-1(gm344) animals from 45 to 94%. Finally, in the triple mutant, 100% of QL.pps inappropriately survive. The loss of pig-1 MELK does not affect the formation of the gradient of CED-3 caspase activity in QL.p (see above; Figure 2, A and C). However, since the size of QL.pp in pig-1(gm344) animals is increased by a factor of ∼2.0, the loss of pig-1 MELK probably results in a significant reduction of the concentration of active CED-3 caspase in QL.pp after QL.p division (since QL.p and its daughters are relatively flat cells, an increase by a factor of ∼2.0 in cell size results in an increase by a factor of ∼2.0 in cell volume as well; hence, the loss of pig-1 potentially could reduce the concentration of active CED-3 caspase by as much as twofold). Furthermore, the loss of pig-1 MELK may also affect the concentration in QL.pp of factors other than CED-3 caspase. For example, the loss of pig-1 MELK may cause a reduction in the concentration of other proapoptotic factors, such as transcriptional activators of egl-1 BH3-only. Combined with a reduction in the concentration of active CED-3 caspase, this may be sufficient to cause 45% of the QL.pps to inappropriately survive. Abolishing the gradient of CED-3 caspase activity in QL.p [i.e., ced-1(e1735)] or compromising the apoptotic cell death pathway [i.e., ced-3(n2427)] is expected to reduce the concentration of active CED-3 caspase in QL.pp in pig-1(gm344) animals even further, thereby increasing the fraction of animals in which QL.pp inappropriately survives. Finally, in ced-1(e1735); pig-1(gm344) ced-3(n2427) triple mutants, the concentration of active CED-3 caspase in QL.pp is reduced below the threshold required to execute the apoptotic fate in 100% of the animals.
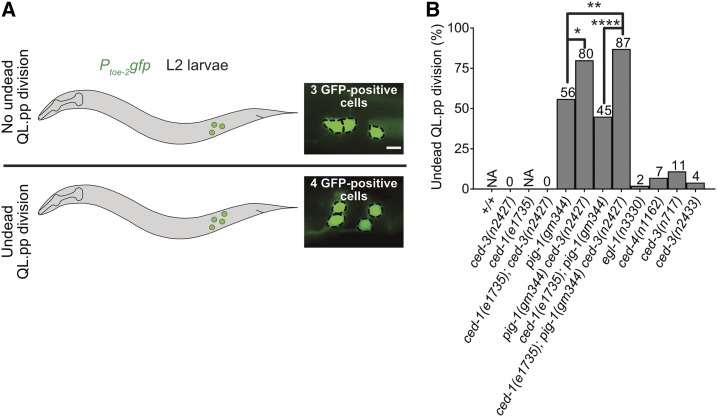
pig-1 MELK, but not ced-1 MEGF10, is required to restrict mitotic potential to QL.pa
Next, we asked whether the loss of ced-1 MEGF10 affects the ability of an undead QL.pp to inappropriately adopt the mitotic fate, i.e., the fate normally adopted by QL.pa. Specifically, using Ptoe-2gfp, we determined the fraction of undead QL.pps that divide (Figure 7A). Since in ced-1(e1735) animals QL.pp always dies, we addressed this question in the background of ced-3(n2427). In ced-3(n2427) or ced-1(e1735); ced-3(n2427) animals, 37 or 58% of QL.pps inappropriately survive, respectively. We found that none of the undead QL.pps divide in either of these genetic backgrounds (Figure 7B). In contrast, in pig-1(gm344) animals, in which 45% of QL.pps inappropriately survive, 56% of undead QL.pps divide. Therefore, the loss of pig-1 MELK, but not ced-1 MEGF10, causes undead QL.pps to inappropriately adopt the mitotic fate. Hence, pig-1 MELK, but not ced-1 MEGF10, is required to restrict the mitotic potential to QL.pa. Based on this observation, we propose that a pig-1 MELK-dependent, ced-1 MEGF10-independent gradient of mitotic potential exists in QL.p prior to its division. Furthermore, we propose that this gradient is along the anterior–posterior axis of QL.p reciprocal to the gradient of CED-3 caspase activity (with a higher concentration in the anterior rather than posterior part of QL.p) and that during QL.p division, this gradient results in the nonrandom segregation of mitotic potential into QL.pa, instead of QL.pp (Figure 9). According to this model, the loss of this gradient in pig-1(gm344) animals should result in the inappropriate presence of mitotic potential in QL.pp after QL.p division, and, hence, the ability of undead QL.pps to adopt the mitotic fate and divide. In support of this model, Garriga and co-workers found that while the loss of pig-1 MELK or strd-1 STRD1 causes similar defects in the asymmetric division by size of QL.p and QR.p, pig-1 mutants have a more penetrant “extra neuron phenotype,” most likely as a result of the division of more undead QL.pps and QR.pps (Chien et al. 2013). Based on this observation, the authors proposed that pig-1 may act in a strd-1-independent manner to affect the segregation of “fate determinants” during QL.p and QR.p division. Alternatively, the mitotic potential could be distributed throughout QL.p, and inherited into QL.pa and QL.pp proportional to cell size, but the small size of QL.pp (∼1/3 of QL.pa) may prevent its ability to divide. In that case, the loss of pig-1 MELK would increase by a factor of ∼2.0 both the size of QL.pp as well as the total amount of mitotic potential in QL.pp (with no change in its concentration), and this would lead to the inappropriate division of 56% of undead QL.pps. Finally, pig-1 MELK may also act in QL.pp to antagonize the activity of mitotic potential, and this could lead to the division of undead QL.pps in animals lacking pig-1.
Figure 7.
pig-1 MELK and ced-3 caspase interact to suppress the mitotic fate in QL.pp. (A) Schematic representation of assay and representative images of L2 larvae showing three or four QL.p lineage cells. Bar, 3 μm. Cells were visualized using the reporter Ptoe-2gfp (bcIs133). Only those animals, in which QL.pp failed to die were considered for this analysis. Animals with four QL.p lineage descendants were considered to have had an instance of “undead QL.pp division,” whereas the ones with three QL.p lineage cells were counted as an instance of “no undead QL.pp division.” (B) Bar graph showing the fraction of undead QL.pps that divided for the indicated genotypes. Statistical test: Fisher’s exact test with Benjamini and Hochberg multiple comparisons correction. * P < 0.05, ** P < 0.01, **** P < 0.0001, and n > 25 animals. NA, not applicable.
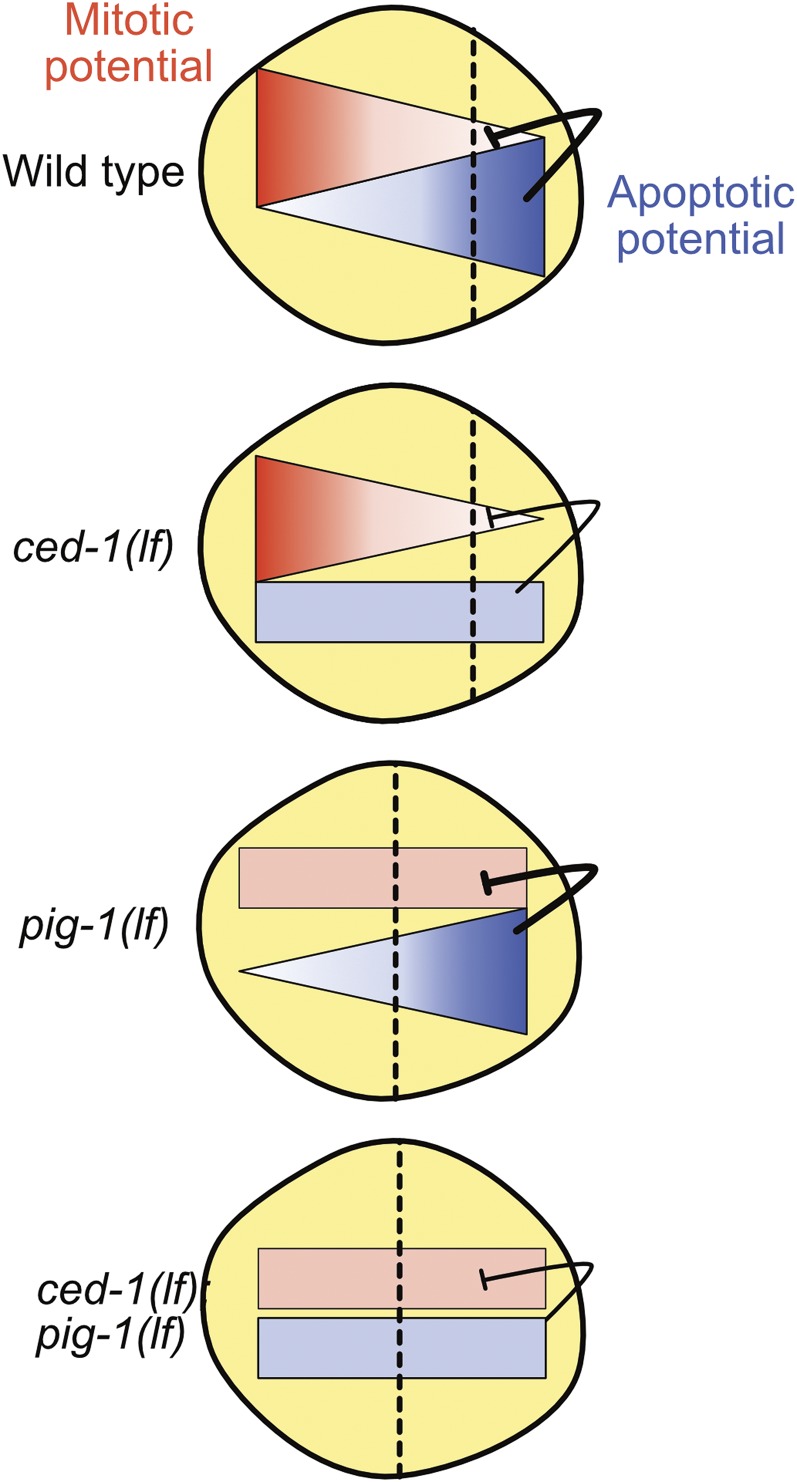
Figure 9.
Working model. Schematic representation of QL.p neuroblast at metaphase. We propose that reciprocal gradients of mitotic (red) and apoptotic (blue) potential are present in QL.p along the anterior–posterior axis in wild-type animals. The gradient of apoptotic potential is dependent on ced-1 MEGF10 function and the gradient of mitotic potential may be dependent on pig-1 MELK function. See text for details.
Surprisingly, we found that ced-3(n2427) significantly increases the fraction of undead QL.pps that divide in pig-1(gm344) animals (from 56 to 80%; Figure 7B). In contrast, ced-1(e1735) fails to do so in a pig-1(gm344) (56 vs. 45%) or pig-1(gm344) ced-3(n2427) (80 vs. 87%) background. Based on this, we propose that CED-3 caspase antagonizes the activity of the mitotic potential. Furthermore, the finding that compromising the apoptotic cell death pathway [i.e., ced-3(n2427)] but not abolishing the gradient of CED-3 caspase activity in QL.p [i.e., ced-1(e1735)] increases the fraction of undead QL.pps that divide in pig-1(gm344) animals, indicates that it is CED-3 caspase activity per se rather than a gradient of CED-3 caspase activity that is capable of antagonizing the activity of the mitotic potential. Possibly, the functional interaction between CED-3 caspase and the mitotic potential could occur in QL.p (before the formation of the gradient of CED-3 Caspase activity) and not in QL.pp after QL.p division. Alternatively, the interaction could take place in QL.pp; in this case, ced-3(n2427) but not ced-1(e1735) would reduce the level of CED-3 caspase activity below the threshold necessary to antagonize the activity of the mitotic potential. Finally, the observation that ced-3(n2427) enhances the fraction of undead QL.pps that divide in pig-1(gm344), but not in wild-type animals, provides additional support for the notion that a gradient of mitotic potential exists in QL.p and that during QL.p division, little or no mitotic potential is normally segregated into QL.pp.
egl-1 BH3-only, ced-4 Apaf-1, and ced-3 caspase promote the ability of QL.p to divide asymmetrically by size
Like ced-1 MEGF10, the genes egl-1 BH3-only, ced-4 Apaf-1, and ced-3 caspase are required for the formation of the gradient of CED-3 caspase activity in the NSM neuroblast and presumably also in QL.p. However, egl-1 BH3-only, ced-4 Apaf-1, and ced-3 caspase, but not ced-1 MEGF10, are required for the initial maturation and full activation of CED-3 caspase. We found that unlike ced-1(e1735), egl-1(n3330), ced-4(n1162), and ced-3(n717) affect the ability of undead QL.pps to divide. Specifically, we found that in an otherwise wild-type background, these mutations cause between 2 and 11% of undead QL.pps to divide (Figure 7B). We also tested the ced-3 mutation n2433, which is a missense mutation in the coding region of the ced-3 gene and leads to the synthesis of CED-3 protein that lacks protease activity. We found that, similarly, ced-3(n2433) causes 4% of undead QL.pps to divide. Hence, the loss of CED-3 caspase activity per se rather than the loss of a gradient of CED-3 caspase activity can result in the inappropriate presence of mitotic potential in undead QL.pps after QL.p division. This suggests that it is the loss of ced-3 caspase in QL.p rather than QL.pp that causes the inappropriate presence of mitotic potential in QL.pp. Therefore, we tested whether the loss of egl-1 BH3-only, ced-4 Apaf-1, or ced-3 caspase affects the asymmetric division of QL.p by size and, hence, the partitioning of mitotic potential during QL.p division. We found that rather than exhibiting a cell size ratio of QL.pa to QL.pp of ∼3.0, in these mutants, cell size ratios observed range from 2.4 to 2.0, indicating that the cleavage plane has shifted anteriorly resulting in larger QL.pps (Figure 5, B and C) [it has previously been reported that the loss of ced-4 affects the cell size ratio of QL.pa to QL.pp (Singhvi et al. 2011)]. To rule out that the changes in cell size ratios observed are a result of differences in the cell shape of undead QL.pps (and, hence, our ability to estimate QL.pp size rather than the asymmetry of the QL.p division), we determined the position of the cleavage furrow in QL.p in the different mutant backgrounds. To that end, we used the image stacks generated every 3 min of L1 larvae carrying the Ptoe-2gfp reporter that we generated to determine the cell size ratio of QL.pa to QL.pp (Figure 5A), and used the last stack before QL.p division to determine the position of the cleavage furrow as outlined in Figure 5D. Using this approach, we found that in wild-type animals, the cleavage furrow is positioned at 62.4% QL.p length and in pig-1(gm344) animals at 51.7% QL.p length (Figure 5E). Considering that the cell size ratio of QL.pa to QL.pp is 2.9 in the wild-type and 1.1 in pig-1(gm344) animals, it is likely that small changes in cleavage furrow position may result in significant changes in QL.pa to QL.pp cell size ratio. Next, we analyzed animals lacking egl-1 BH3-only, ced-4 Apaf-1, or ced-3 caspase, and found significant changes in cleavage furrow position in ced-4(n1162) and ced-3(n717) animals (57 and 56.9% of QL.p length, respectively). In addition, we found small changes in cleavage furrow position in egl-1(n3330) and ced-3(n2433), but not ced-1(e1735), animals. As mentioned above, small changes in cleavage furrow position may result in significant changes in QL.pa to QL.pp cell size ratio. The small changes in cleavage furrow position detected in egl-1(n3330) and ced-3(n2433) animals may therefore account for the significant changes in QL.pa to QL.pp cell size ratios observed in these animals (Figure 5, C and E).
Based on these observations, we conclude that the loss of egl-1 BH3-only, ced-4 Apaf-1, or ced-3 caspase compromises the ability of QL.p to divide asymmetrically by size. Therefore, CED-3 caspase contributes to the asymmetric division by size of QL.p. Furthermore, we propose that through the observed shift in cleavage plane toward the anterior, mitotic potential is segregated inappropriately into QL.pp during QL.p division, and that this accounts for the small fraction of undead QL.pps that divide in these mutants. Finally, the ability of CED-3 caspase to contribute to the asymmetric division by size of QL.p depends on its activation through egl-1 BH3-only and ced-4 Apaf-1, and, hence, the apoptotic cell death pathway.
The function of the apoptotic cell death pathway in asymmetric cell division by size is not restricted to the QL.p neuroblast
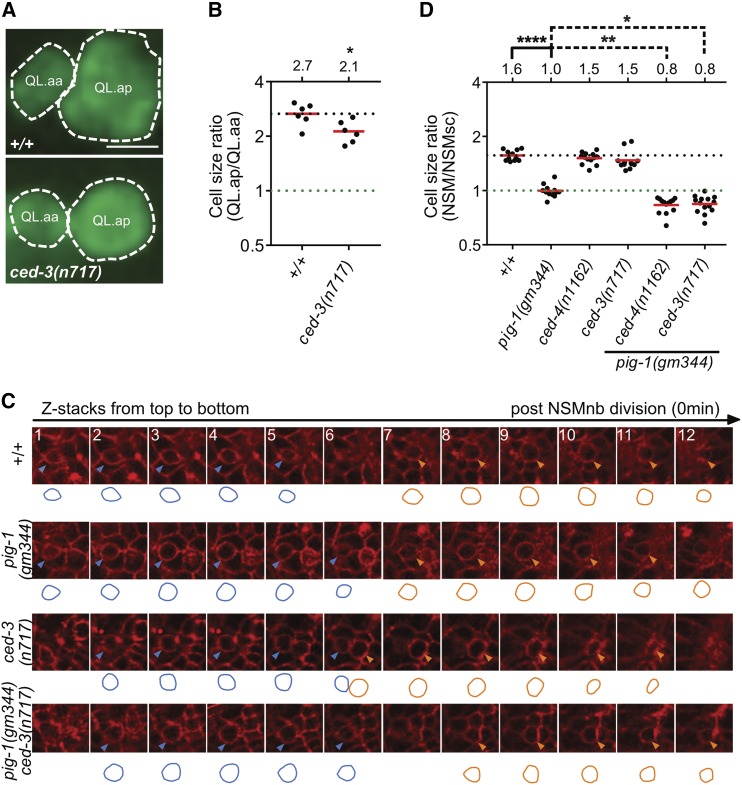
To determine whether the function of the apoptotic cell death pathway in asymmetric cell division by size is specific to the QL.p neuroblast, we analyzed the postembryonic QL.a neuroblast and the embryonic NSM neuroblast, both of which divide asymmetrically by size and fate to produce a smaller daughter that is programmed to die (Sulston et al. 1983; Hatzold and Conradt 2008). In wild-type animals, QL.a divides to generate a larger posterior daughter, QL.ap, and a smaller anterior daughter, QL.aa, with a cell size ratio of QL.ap to QL.aa of 2.67 (Ou et al. 2010) (Figure 8, A and B) (cells of the QL.a lineages are also relatively flat cells; therefore, cell size was determined as for QL.p daughters). We found that in ced-3(n717) animals, this ratio was reduced to 2.13. Furthermore, we observed an effect of the loss of ced-3 caspase on the asymmetric division by size of the NSM neuroblast; however, only in the background of pig-1(gm344). Specifically, the NSM neuroblast divides to produce a larger ventral daughter, the NSM, and a smaller dorsal daughter, the NSMsc, with a cell size ratio of NSM to NSMsc of 1.57 (Hatzold and Conradt 2008) (Figure 8, C and D) (since cells of the NSM neuroblast lineage are not flat cells, cell size was estimated as described in Figure 8C and in the Materials and Methods). As reported previously, in pig-1(gm344) animals, the NSM neuroblast divides symmetrically by size and generates two daughter cells of similar sizes, with a cell size ratio of NSM to NSMsc of 1.00 (Wei et al. 2017). In ced-3(n717) or ced-4(n1162) animals, the cell size ratio is not significantly different from that of wild-type animals (1.47 and 1.51, respectively). However, in a pig-1(gm344) background, the cell size ratio of ced-3(n717) or ced-4(n1162) animals is significantly reduced compared to that of pig-1(gm344) animals (0.84 and 0.83, respectively). A reduction in the ratio below 1.0 is indicative of a reversal of polarity. Indeed, in these double-mutant animals, the NSM is smaller than the NSMsc. In summary, these results indicate that the function of the apoptotic cell death pathway in asymmetric neuroblast division, at least by size, is not restricted to the QL.p neuroblast.
Figure 8.
The apoptotic pathway promotes asymmetric cell division in other neuroblast lineages. (A) Representative images and (B) dot plot showing relative sizes of QL.a daughter cells in wild-type and ced-3(n717) animals. Bar, 3 μm. Cells were visualized using the reporter Ptoe-2gfp (bcIs133). Statistical test: Mann–Whitney test. * P < 0.05, n = 6 QL.as. (C) Representative images showing NSM (Neurosecretory motor neuron) neuroblast (NSMnb) daughter cells visualized with Ppie-1mCherry::phplcδ (ltIs44). The circumferences of the daughter cells were traced and are shown below the respective images. Circumferences of NSM sister cells (NSMsc) are shown in blue and circumferences of NSM are shown in orange. (D) Dot blot showing cell size ratios (NSM/NSMsc) in the indicated genotypes. One-way ANOVA with Tukey’s multiple comparisons test was used to determine statistical significance. * P < 0.05, ** P < 0.01, **** P < 0.0001, n > 11 NSM neuroblasts.
Our finding that ced-4(n1162) and ced-3(n717) affect the asymmetric division of the NSM neuroblast in a pig-1(gm344), but not wild-type, background suggests that in certain lineages, the apoptotic cell death pathway may act in parallel to pig-1 MELK to affect cellular polarization. Therefore, we tested whether ced-3(n717) enhances embryonic lethality in pig-1(gm344) animals.
When grown at 20°, we observed 0% embryonic lethality among ced-3(n717) animals (n = 1413) and 8% embryonic lethality among pig-1(gm344) animals (n = 1514). However, in pig-1(gm344) ced-3(n717) double mutants, 43% of the animals arrested during embryonic development (n = 397). Therefore, the apoptotic cell death pathway may play a more general role in cellular polarization and asymmetric cell division, and, in this context, act in parallel to pig-1 MELK.
Discussion
The formation of reciprocal gradients of mitotic and apoptotic potential is critical for the asymmetric division of the QL.p neuroblast by fate
Based on results presented here, we propose that prior to QL.p division, reciprocal gradients of mitotic and apoptotic potential form in QL.p, and that these gradients result in the nonrandom segregation of mitotic potential into QL.pa and of apoptotic potential into QL.pp (Figure 9). We speculate that the mitotic potential in QL.pa promotes its ability to divide, and we provide evidence that the apoptotic potential in QL.pp promotes its ability to execute apoptotic cell death. Furthermore, while we know that apoptotic potential encompasses at least active CED-3 caspase, we do not know the molecular nature of the mitotic potential. However, we speculate that it encompasses factors involved in cell cycle control, and/or molecules or organelles required for energy production. We also provide evidence that distinct genetic pathways are responsible for the establishment and/or maintenance of these two gradients in QL.p. Specifically, the two conserved C. elegans engulfment pathways are necessary for the gradient of CED-3 caspase activity but not the gradient of mitotic potential. Conversely, a pig-1 MELK-dependent pathway is necessary for the gradient of mitotic potential but not the gradient of CED-3 caspase activity. How these distinct pathways control the two reciprocal gradients, and how these gradients are generated in the first place, remains to be determined. Interestingly, in the case of the gradient of CED-3 caspase activity, we found that there is a mechanistic difference in the symmetry-breaking event that orients this gradient along the dorsal–ventral axis in the NSM neuroblast and along the anterior–posterior axis in the QL.p neuroblast. In the case of the NSM neuroblast, all neighboring cells appear to present CED-1 MEGF10 on their cell surface; however, CED-1 MEGF10 clustering and activation specifically occurs on the cell surface of the two dorsal neighbors (Chakraborty et al. 2015). In contrast, in the case of QL.p, at least on the medial side, CED-1 MEGF10 is asymmetrically presented and, for this reason, results in CED-1 MEGF10 activation only on the posterior side. Finally, the results presented here indicate that the formation of a gradient of CED-3 caspase activity in a mother of a cell programmed to die is not specific to the NSM neuroblast lineage. Based on this, we speculate that it represents a general phenomenon of cell death lineages in developing C. elegans animals.
The apoptotic cell death pathway is required for the asymmetric division by size and fate of the QL.p neuroblast
The asymmetric division of mothers of cells programmed to die is critical for the correct fate of their daughters and, in particular, for the correct fate of the daughters programmed to die. Hence, it has been postulated that asymmetric cell division regulates the apoptotic cell death pathway (Hatzold and Conradt 2008; Teuliere and Garriga 2017). We now demonstrate that the apoptotic cell death pathway contributes to the ability of mothers of cells programmed to die to divide asymmetrically. Specifically, we present evidence that the apoptotic cell death pathway is required for the asymmetric division of QL.p by size and, hence, the correct sizes of the daughter cells QL.pa and QL.pp. In addition, we present evidence that the apoptotic cell death pathway is also required for the asymmetric division of QL.p by fate. Specifically, by controlling the sizes of QL.pa and QL.pp, the apoptotic cell death pathway indirectly influences the relative amounts of mitotic potential or apoptotic potential that are segregated into either QL.pa or QL.pp during QL.p division. In addition, the apoptotic cell death pathway is required for the activation of CED-3 caspase in QL.p, which we propose antagonizes the mitotic potential. Hence, the apoptotic cell death pathway controls the total amount of apoptotic and mitotic potential present in QL.p, and, therefore, the amounts that can be segregated into QL.pa and QL.pp. Based on these new findings, we now postulate that not only does asymmetric cell division regulate the apoptotic cell death pathway, but that in the context of cell death lineages, the apoptotic cell death pathway regulates asymmetric cell division. Furthermore, this regulation of asymmetric cell division through the apoptotic cell death pathway is necessary for the production of smaller daughters that are programmed to die.
A new nonapoptotic function of the C. elegans apoptotic cell death pathway in asymmetric neuroblast division
The C. elegans apoptotic cell death pathway, and CED-3 caspase in particular, has been implicated in a number of nonapoptotic processes, which range from aging and neuronal regeneration to the control of the expression of specific genes, such as the heterochronic gene lin-28 (Pinan-Lucarre et al. 2012; Weaver et al. 2014; Yee et al. 2014). Similarly, mammalian caspases have been shown to have various nonapoptotic functions during development (Nakajima and Kuranaga 2017). We now present evidence that the apoptotic cell death pathway is also involved in the asymmetric division of neuroblasts that generate a smaller daughter that is programmed to die. How CED-3 caspase affects the position of the cleavage plane in these neuroblasts remains to be determined. In C. elegans, the positioning of the cleavage plane is best understood in the one-cell embryo, which, like QL.p, divides asymmetrically by size and fate (Rose and Gonczy 2014; Wu and Griffin 2017). In general, the cleavage plane is perpendicular to and centered on the middle of the mitotic spindle. As a result of unequal dynein-mediated pulling forces emanating from the anterior and posterior poles (with more pulling forces emanating from the posterior pole), in one-cell embryos, the spindle is shifted posteriorly along the anterior–posterior axis. As a result, the cleavage plane is shifted posteriorly as well, generating a larger anterior cell, AB, and a smaller posterior cell, P1. It has been shown previously that the spindle is shifted posteriorly in QL.p as well (Ou et al. 2010), and we speculate that this is also caused by unequal dynein-mediated pulling forces. However, whereas the loss of pig-1 MELK causes QL.p to divide symmetrically rather than asymmetrically, it fails to do so in the one-cell embryo [however, the loss of pig-1 MELK does synergize with the loss of ani-1, which encodes one of two C. elegans anillin genes, to affect the position of the cleavage plane in one-cell embryos (Pacquelet et al. 2015)]. This suggests that the function of pig-1 MELK in the regulation of cleavage plane position differs between the one-cell embryo and the QL.p neuroblast lineage. Nevertheless, since the loss of ced-3 caspase causes a partial defect in the posterior shift of the cleavage plane in QL.p, ced-3 caspase may be necessary for the dynein-mediated pulling forces that emanate from the posterior pole. However, the finding that the loss of ced-3 caspase also affects the asymmetric division by size of the QL.a neuroblast suggests that dynein-mediated pulling forces may not be the target of CED-3 caspase activity in this context. Specifically, like QL.p, the neuroblast QL.a divides asymmetrically by size and fate to generate a larger daughter, which survives, and a smaller daughter, which dies (in contrast to QL.p, the smaller daughter of QL.a is the anterior rather than the posterior daughter). However, rather than through a shift of the mitotic spindle along the anterior–posterior axis, it has been suggested that two daughter cells of different sizes are generated from QL.a through asymmetric myosin-mediated contractile forces during QL.a division (Ou et al. 2010). Hence, ced-3 caspase may be required for general cellular polarization of QL.p and QL.a, and affect the position of the cleavage plane or myosin-mediated contractile forces indirectly. Consistent with the notion that ced-3 caspase may be required for general cellular polarization, we found that in the embryonic NSM neuroblast lineage, the loss of ced-3 caspase in a pig-1 MELK mutant background causes a reversal of polarity. Finally, the finding that the loss of ced-3 caspase enhances embryonic lethality in animals lacking pig-1 MELK function suggests that ced-3 caspase (and possibly the entire apoptotic cell death pathway) also functions in asymmetric cell division and cellular polarization in lineages other than cell death lineages, and that this may be crucial for normal animal development.
Interestingly, there is increasing evidence that mammalian caspases have functions in different types of embryonic and adult stem cells (Baena-Lopez et al. 2017). Hence, our finding that C. elegans CED-3 caspase plays a role in cellular polarization, and the asymmetric division by size and fate of neuroblasts may very well be relevant to these nonapoptotic functions of mammalian caspases in stem cell lineages.
Acknowledgments
The authors thank E. Lambie, E. Zanin, and B. Weaver for comments on the manuscript; G. Garriga, D. Hall, and members of the Conradt laboratory for constructive discussions; M. Bauer, N. Lebedeva, and M. Schwarz for excellent technical support; R. Sherrard for constructing plasmid pBC1805; Z. Zhou for plasmid pZZ610; T. Mikeladze-Dvali for plasmid TMD34; H. Bringmann for the mKate2 clone; Monica Driscoll for bzIs190; and E. Zanin for plasmid pEZ167. Some strains used in this study were provided by the Caenorhabditis Genetics Center (https://cbs.umn.edu/cgc/home), which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD-010440). N.M. was supported by a predoctoral fellowship from the Studienstiftung des deutschen Volkes (https://www.studienstiftung.de/) and H.W. was supported by a predoctoral fellowship from the China Scholarship Council (https://www.csc.edu.cn/). This work was supported by the Deutsche Forschungsgemeinschaft (Center for Integrated Protein Science Munich; EXC 114).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7058609.
Communicating editor: M. Sundaram
Literature Cited
- Audhya A., Hyndman F., McLeod I. X., Maddox A. S., Yates J. R., III, et al. , 2005. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J. Cell Biol. 171: 267–279. 10.1083/jcb.200506124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Lopez L. A., Arthurton L., Xu D. C., Galasso A., 2017. Non-apoptotic Caspase regulation of stem cell properties. Semin. Cell Dev. Biol. 82: 118–126. 10.1016/j.semcdb.2017.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger J. M., Gonczy P., 2003. TAC-1 and ZYG-9 form a complex that promotes microtubule assembly in C. elegans embryos. Curr. Biol. 13: 1488–1498. 10.1016/S0960-9822(03)00582-7 [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S., Lambie E. J., Bindu S., Mikeladze-Dvali T., Conradt B., 2015. Engulfment pathways promote programmed cell death by enhancing the unequal segregation of apoptotic potential. Nat. Commun. 6: 10126 10.1038/ncomms10126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien S. C., Brinkmann E. M., Teuliere J., Garriga G., 2013. Caenorhabditis elegans PIG-1/MELK acts in a conserved PAR-4/LKB1 polarity pathway to promote asymmetric neuroblast divisions. Genetics 193: 897–909. 10.1534/genetics.112.148106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt B., Wu Y. C., Xue D., 2016. Programmed cell death during Caenorhabditis elegans. Dev. Genet. 203: 1533–1562. 10.1534/genetics.115.186247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes S., Frank C. A., Garriga G., 2006. The C. elegans MELK ortholog PIG-1 regulates cell size asymmetry and daughter cell fate in asymmetric neuroblast divisions. Development 133: 2747–2756. 10.1242/dev.02447 [DOI] [PubMed] [Google Scholar]
- D’Agostino R. P., Pearson E. S., 1993. Tests for departure from normality. Empirical results for the distributions of b2 and √b1. Biometrika 60: 613–622. [Google Scholar]
- Ellis H. M., Horvitz H. R., 1986. Genetic control of programmed cell death in the nematode C. elegans. Cell 44: 817–829. 10.1016/0092-8674(86)90004-8 [DOI] [PubMed] [Google Scholar]
- Ellis R. E., Jacobson D. M., Horvitz H. R., 1991. Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics 129: 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., 1921. On the probable error of a coefficient of correlation deduced from a small sample. Metron 1: 3–32. [Google Scholar]
- Fisher R. A., 1935. The Design of Experiments. Macmillan, New York. [Google Scholar]
- Frøkjaer-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., et al. , 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40: 1375–1383. 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Sarov M., Taylor J., Flibotte S., et al. , 2014. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat. Methods 11: 529–534. 10.1038/nmeth.2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A., III, et al. , 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Gurling M., Talavera K., Garriga G., 2014. The DEP domain-containing protein TOE-2 promotes apoptosis in the Q lineage of C. elegans through two distinct mechanisms. Development 141: 2724–2734. 10.1242/dev.110486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzold J., Conradt B., 2008. Control of apoptosis by asymmetric cell division. PLoS Biol. 6: e84 10.1371/journal.pbio.0060084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock E. M., Sulston J. E., Thomson J. N., 1983. Mutations affecting programmed cell deaths in the nematode Caenorhabditis elegans. Science 220: 1277–1279. 10.1126/science.6857247 [DOI] [PubMed] [Google Scholar]
- Horvitz H. R., 2003. Nobel lecture. Worms, life and death. Biosci. Rep. 23: 239–303. 10.1023/B:BIRE.0000019187.19019.e6 [DOI] [PubMed] [Google Scholar]
- Hunt-Newbury R., Viveiros R., Johnsen R., Mah A., Anastas D., et al. , 2007. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 5: e237 10.1371/journal.pbio.0050237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambie E. J., Conradt B., 2016. Deadly dowry: how engulfment pathways promote cell killing. Cell Death Differ. 23: 553–554. 10.1038/cdd.2015.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bot N., Tsai M. C., Andrews R. K., Ahringer J., 2003. TAC-1, a regulator of microtubule length in the C. elegans embryo. Curr. Biol. 13: 1499–1505. 10.1016/S0960-9822(03)00577-3 [DOI] [PubMed] [Google Scholar]
- Maduro M., Pilgrim D., 1995. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics 141: 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann H. B. W., 1947. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 18: 50–60. 10.1214/aoms/1177730491 [DOI] [Google Scholar]
- Mitani S., Du H., Hall D. H., Driscoll M., Chalfie M., 1993. Combinatorial control of touch receptor neuron expression in Caenorhabditis elegans. Development 119: 773–783. [DOI] [PubMed] [Google Scholar]
- Nakajima Y. I., Kuranaga E., 2017. Caspase-dependent non-apoptotic processes in development. Cell Death Differ. 24: 1422–1430. 10.1038/cdd.2017.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G., Stuurman N., D’Ambrosio M., Vale R. D., 2010. Polarized myosin produces unequal-size daughters during asymmetric cell division. Science 330: 677–680. 10.1126/science.1196112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacquelet A., Uhart P., Tassan J. P., Michaux G., 2015. PAR-4 and anillin regulate myosin to coordinate spindle and furrow position during asymmetric division. J. Cell Biol. 210: 1085–1099. 10.1083/jcb.201503006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinan-Lucarre B., Gabel C. V., Reina C. P., Hulme S. E., Shevkoplyas S. S., et al. , 2012. The core apoptotic executioner proteins CED-3 and CED-4 promote initiation of neuronal regeneration in Caenorhabditis elegans. PLoS Biol. 10: e1001331 10.1371/journal.pbio.1001331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman I., Greiss S., Chin J. W., 2013. Efficient and rapid C. elegans transgenesis by bombardment and hygromycin B selection. PLoS One 8: e76019 10.1371/journal.pone.0076019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose L., Gonczy P., 2014. Polarity establishment, asymmetric division and segregation of fate determinants in early C. elegans embryos (December 30, 2014), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.30.2, http://www.wormbook.org. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., et al. , 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W., 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham S., Reddien P. W., Davies B., Horvitz H. R., 1999. Mutational analysis of the Caenorhabditis elegans cell-death gene ced-3. Genetics 153: 1655–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrard R., Luehr S., Holzkamp H., McJunkin K., Memar N., et al. , 2017. miRNAs cooperate in apoptosis regulation during C. elegans development. Genes Dev. 31: 209–222. 10.1101/gad.288555.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhvi A., Teuliere J., Talavera K., Cordes S., Ou G., et al. , 2011. The Arf GAP CNT-2 regulates the apoptotic fate in C. elegans asymmetric neuroblast divisions. Curr. Biol. 21: 948–954. 10.1016/j.cub.2011.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srayko M., Quintin S., Schwager A., Hyman A. A., 2003. Caenorhabditis elegans TAC-1 and ZYG-9 form a complex that is essential for long astral and spindle microtubules. Curr. Biol. 13: 1506–1511. 10.1016/S0960-9822(03)00597-9 [DOI] [PubMed] [Google Scholar]
- Stanfield G. M., Horvitz H. R., 2000. The ced-8 gene controls the timing of programmed cell deaths in C. elegans. Mol. Cell 5: 423–433. 10.1016/S1097-2765(00)80437-2 [DOI] [PubMed] [Google Scholar]
- Student1908. The probable error of a mean. Biometrika 6: 1–25. [Google Scholar]
- Sulston J. E., Horvitz H. R., 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156. 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., Thomson J. N., 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Suzuki J., Denning D. P., Imanishi E., Horvitz H. R., Nagata S., 2013. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 341: 403–406. 10.1126/science.1236758 [DOI] [PubMed] [Google Scholar]
- Teuliere J., Garriga G., 2017. Size matters: how C. elegans asymmetric divisions regulate apoptosis. Results Probl. Cell Differ. 61: 141–163. 10.1007/978-3-319-53150-2_6 [DOI] [PubMed] [Google Scholar]
- Teuliere J., Cordes S., Singhvi A., Talavera K., Garriga G., 2014. Asymmetric neuroblast divisions producing apoptotic cells require the cytohesin GRP-1 in Caenorhabditis elegans. Genetics 198: 229–247. 10.1534/genetics.114.167189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey J. W., 1949. Comparing individual means in the analysis of variance. Biometrics 5: 99–114. 10.2307/3001913 [DOI] [PubMed] [Google Scholar]
- Turek M., Lewandrowski I., Bringmann H., 2013. An AP2 transcription factor is required for a sleep-active neuron to induce sleep-like quiescence in C. elegans. Curr. Biol. 23: 2215–2223. 10.1016/j.cub.2013.09.028 [DOI] [PubMed] [Google Scholar]
- Venegas V., Zhou Z., 2007. Two alternative mechanisms that regulate the presentation of apoptotic cell engulfment signal in Caenorhabditis elegans. Mol. Biol. Cell 18: 3180–3192. 10.1091/mbc.e07-02-0138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver B. P., Zabinsky R., Weaver Y. M., Lee E. S., Xue D., et al. , 2014. CED-3 caspase acts with miRNAs to regulate non-apoptotic gene expression dynamics for robust development in C. elegans. Elife 3: e04265 10.7554/eLife.04265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Yan B., Gagneur J., Conradt B., 2017. Caenorhabditis elegans CES-1 snail represses pig-1 MELK expression to control asymmetric cell division. Genetics 206: 2069–2084. 10.1534/genetics.117.202754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcoxon F., 1945. Individual comparisons by ranking methods. Biom. Bull. 1: 80–83. 10.2307/3001968 [DOI] [Google Scholar]
- Wu Y., Griffin E. E., 2017. Regulation of cell polarity by PAR-1/MARK kinase. Curr. Top. Dev. Biol. 123: 365–397. 10.1016/bs.ctdb.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee C., Yang W., Hekimi S., 2014. The intrinsic apoptosis pathway mediates the pro-longevity response to mitochondrial ROS in C. elegans. Cell 157: 897–909. 10.1016/j.cell.2014.02.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoav B. H. Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Zhou Z., Hartwieg E., Horvitz H. R., 2001. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 104: 43–56. 10.1016/S0092-8674(01)00190-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All reagents and strains generated for this study are available from the authors upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article and figures. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7058609.