Abstract
In the nematode Caenorhabditis elegans, the conserved LIN-41 RNA-binding protein is a translational repressor that coordinately controls oocyte growth and meiotic maturation. LIN-41 exerts these effects, at least in part, by preventing the premature activation of the cyclin-dependent kinase CDK-1. Here we investigate the mechanism by which LIN-41 is rapidly eliminated upon the onset of meiotic maturation. Elimination of LIN-41 requires the activities of CDK-1 and multiple SCF (Skp1, Cul1, and F-box protein)-type E3 ubiquitin ligase subunits, including the conserved substrate adaptor protein SEL-10/Fbw7/Cdc4, suggesting that LIN-41 is a target of ubiquitin-mediated protein degradation. Within the LIN-41 protein, two nonoverlapping regions, Deg-A and Deg-B, are individually necessary for LIN-41 degradation; both contain several potential phosphodegron sequences, and at least one of these sequences is required for LIN-41 degradation. Finally, Deg-A and Deg-B are sufficient, in combination, to mediate SEL-10-dependent degradation when transplanted into a different oocyte protein. Although LIN-41 is a potent inhibitor of protein translation and M phase entry, the failure to eliminate LIN-41 from early embryos does not result in the continued translational repression of LIN-41 oocyte messenger RNA targets. Based on these observations, we propose a model for the elimination of LIN-41 by the SEL-10 E3 ubiquitin ligase and suggest that LIN-41 is inactivated before it is degraded. Furthermore, we provide evidence that another RNA-binding protein, the GLD-1 tumor suppressor, is regulated similarly. Redundant mechanisms to extinguish translational repression by RNA-binding proteins may both control and provide robustness to irreversible developmental transitions, including meiotic maturation and the oocyte-to-embryo transition.
Keywords: oocyte meiotic maturation, oocyte-to-embryo transition, translational regulation, RNA-binding proteins, ubiquitin-mediated protein degradation
IN Caenorhabditis elegans, as in many animals, full-grown oocytes are transcriptionally quiescent and depend on a maternal load of protein and messenger RNA (mRNA) to complete their development. As a consequence, the dramatic cell cycle and developmental changes that occur during the transition from oogenesis to embryogenesis are driven by post-transcriptional mechanisms. Such mechanisms include protein phosphorylation, the elimination of maternally provided proteins or mRNAs, and the regulation of maternal mRNA translation [reviewed by Verlhac et al. (2010), Robertson and Lin (2015), Svoboda et al. (2017)]. The oocyte-to-embryo transition (OET) initiates when oocytes exit meiotic prophase and enter the first meiotic metaphase, a cell cycle and developmental event also known as meiotic resumption or meiotic maturation. The OET completes when zygotic gene transcription begins after fertilization in the early embryo.
Pioneering studies using amphibian oocytes established that oocyte meiotic maturation is initiated by the activation of maturation-promoting factor (MPF), in response to progesterone from the follicle cells [Masui and Markert 1971; reviewed by Masui (2001)]. The principal components of MPF are the cyclin-dependent kinase Cdk1 catalytic subunit and a cyclin B regulatory subunit [Dunphy et al. 1988; Gautier et al. 1988, 1990; Lohka et al. 1988; reviewed by Nurse (1990)]. In Xenopus, which represents the best-studied system from a biochemical standpoint, MPF activation involves the translation of multiple, apparently redundantly acting factors, including the c-mos protein kinase, B-type cyclins, and proteins that remain to be identified [Kobayashi et al. 1991; Minshull et al. 1991; Nebreda et al. 1995; Frank-Vaillant et al. 1999; Haccard and Jessus 2006a; reviewed by Haccard and Jessus (2006b)]. Once activated, MPF stimulates multiple positive feedback mechanisms, resulting in the activation of the CDC25 phosphatase, which removes the inhibitory CDK1 phosphorylations at Thr14 and Tyr15 catalyzed by the Wee1 or Myt1 kinases (Kornbluth et al. 1994; Mueller et al. 1995; Kumagai and Dunphy 1996). This regulatory mechanism generates the “switch-like” activation of MPF that promotes the rapid and irreversible cell cycle transition from prophase to metaphase [reviewed by O’Farrell (2001), Kishimoto (2015)].
MPF is the master regulator of cell cycle progression during oocyte meiotic maturation in C. elegans as in all examined species (Boxem et al. 1999; Burrows et al. 2006; van der Voet et al. 2009), yet MPF activation is regulated somewhat differently than in Xenopus. For example, the signal that triggers MPF activation for meiotic maturation in C. elegans is not progesterone, but rather the major sperm protein, an abundant cytoskeletal protein that is released from sperm (Miller et al. 2001; Kosinski et al. 2005). The latter control mechanism, which serves to link meiotic maturation and ovulation to sperm availability, likely evolved in gonochoristic predecessors of facultative hermaphroditic nematode species like C. elegans. The rate of meiotic maturation declines substantially as a C. elegans hermaphrodite utilizes its limited supply of sperm for self-fertilization but rapidly increases upon mating (Kosinski et al. 2005). When sperm are absent, as in mutant hermaphrodites that do not produce sperm (e.g., fog mutant females), oocytes arrest for prolonged periods and the rate of production and growth of new oocytes declines until insemination (McCarter et al. 1999; Wolke et al. 2007; Govindan et al. 2009). This serves to preserve metabolically costly oocytes when sperm are unavailable for fertilization. Thus, the molecular mechanisms that control MPF activation must be exquisitely fine-tuned for sperm sensing.
Another commonality between the C. elegans and Xenopus systems is that MPF activation depends on translational control mechanisms, although the details differ. In C. elegans, large ribonucleoprotein (RNP) complexes containing the tripartite motif (TRIM)-NHL (NCL-1, HT2A, and LIN-41) RNA-binding protein LIN-41 and the tristetraprolin/TIS11-related RNA-binding proteins OMA-1 and OMA-2 (referred to collectively as the OMA proteins) are major downstream targets of major sperm protein signaling (Spike et al. 2014a,b; Tsukamoto et al. 2017). LIN-41 is the chief determinant of the extended meiotic prophase of C. elegans oocytes (Spike et al. 2014a). In lin-41 null mutants, pachytene-stage oocytes cellularize prematurely, activate CDK-1, aberrantly disassemble the synaptonemal complex, and enter M phase precociously, causing sterility (Spike et al. 2014a; Tocchini et al. 2014; Matsuura et al. 2016). Premature CDK-1 activation causes lin-41 mutant oocytes to abnormally transcribe and express genes that are ordinarily expressed after the OET and restricted to differentiated cells (Allen et al. 2014; Spike et al. 2014a; Tocchini et al. 2014). In mammals, LIN-41/TRIM71 has been found to promote pluripotency through its activity as a translational repressor (Loedige et al. 2013, 2015; Worringer et al. 2014). In C. elegans, LIN-41 inhibits CDK-1 activation in part through the 3′-untranslated region (UTR)-mediated translational repression of the CDC-25.3 phosphatase (Spike et al. 2014a,b). By contrast, the OMA proteins are redundantly required for CDK-1 activation (Detwiler et al. 2001). In the absence of the OMA proteins, oocytes fail to undergo meiotic maturation despite the presence of sperm, resulting in sterility (Detwiler et al. 2001).
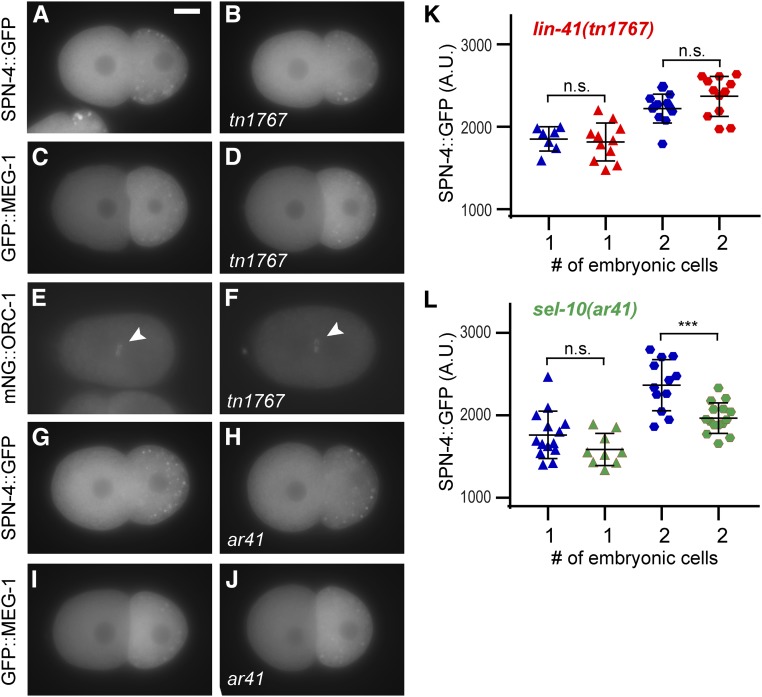
Genetic analysis suggests the OMA proteins promote meiotic maturation by inhibiting the function of LIN-41 in the most proximal oocyte. Two lines of molecular evidence are consistent with the idea that LIN-41 must be inactivated to promote meiotic maturation. First, LIN-41 is degraded upon the onset of meiotic maturation in response to CDK-1 activation (Spike et al. 2014a; Figure 1, A and B). Second, LIN-41 is a potent translational repressor, yet several of the mRNAs it associates with and represses are translated and coexpressed with LIN-41 prior to meiotic maturation in the −1 and −2 oocytes (Tsukamoto et al. 2017). These mRNAs include those encoding the RNA-binding protein SPN-4, which is required for development of the embryonic germline and the mesendoderm (Gomes et al. 2001), and MEG-1, which is a germplasm or P granule component needed for germline development (Leacock and Reinke 2008; Kapelle and Reinke 2011; Wang et al. 2014). By contrast, the OMA proteins are required for the translation of spn-4 and meg-1 transcripts in proximal oocytes, providing a molecular mechanism by which the OMA proteins might antagonize LIN-41 function (Tsukamoto et al. 2017).
Figure 1.
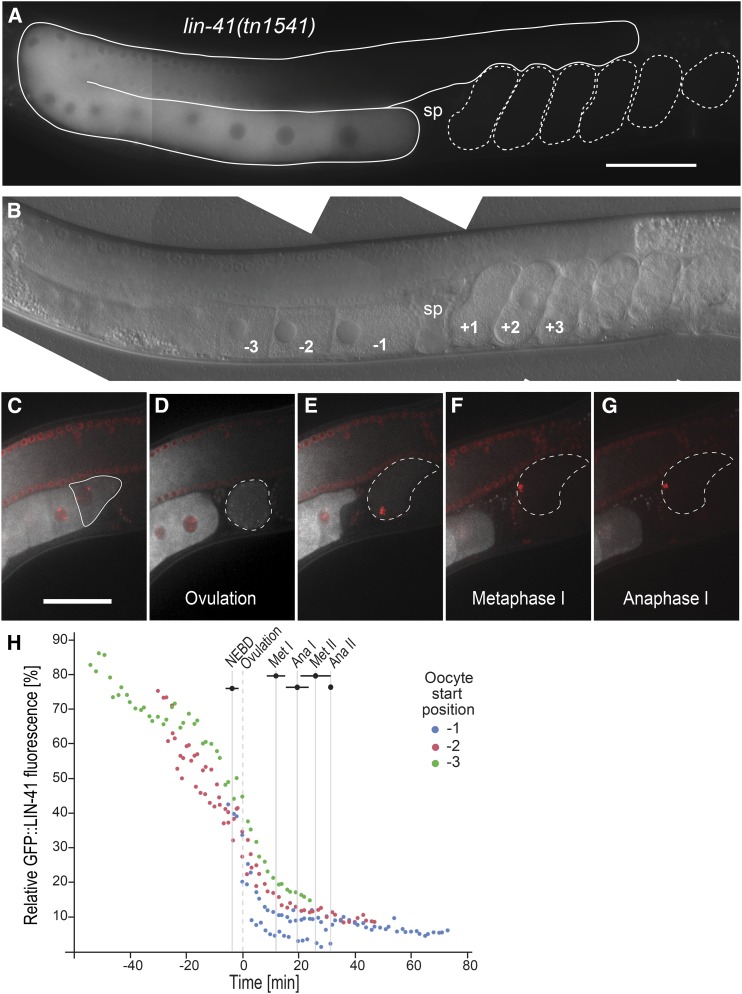
GFP::LIN-41 is eliminated during the first meiotic division. (A and B) Composite GFP (A) and DIC (B) images of a lin-41(tn1541[gfp::tev::s-tag::lin-41]) adult hermaphrodite. GFP::LIN-41 is apparent in the middle and proximal regions of the germline (solid outline, A), with reduced levels in the −1 oocyte immediately adjacent to the spermatheca (sp). The positions of some embryos (dashed outlines, A) and oocytes are indicated relative to the spermatheca in B; a fertilized embryo in the spermatheca would be at the zero position. These labels and naming conventions are used throughout. 100 ms GFP exposures; Bar, 50 μm. (C–G) Time-lapse images of GFP::LIN-41 (white) and mCHERRY::HISTONE-labeled chromosomes (red) were acquired in a living lin-41(tn1541); itIs37[pie-1p::mCherry:::H2B::pie-1 3′UTR, unc-119(+)] adult hermaphrodite by confocal microscopy. Images are shown for select time points (t) prior to meiotic maturation (C, t= −4.5 min), at ovulation (D, t = 0 min), and during the first meiotic division (E, t= +4 min; F, t = +11.8 min; G, t= +16.9 min) as an individual oocyte (C, solid outline) progresses from the −1 to the +1 position and through the OET (D–G, dashed outlines). Bar, 50 μm. Movie S1, worm 1, shows the complete time-lapse series from which the still images were taken. (H) Five oocytes were imaged as they progressed from the −1 position through meiotic divisions; the relative amount of background-corrected GFP::LIN-41 with respect to distal oocytes is shown on the graph at each time point. Three of the oocytes were also imaged at earlier stages as they moved from a more distal location [−2 oocyte (red) or −3 oocyte (green) position] into the −1 oocyte position (blue), as indicated. Timing on the x-axis is relative to ovulation (t = 0). Bars indicate the SD for different meiotic events. Ana, anaphase; Met, metaphase; NEBD, nuclear envelope breakdown.
Here we examine the mechanism by which LIN-41 is eliminated by the end of the first meiotic division. We identify two LIN-41 degradation domains, Deg-A and Deg-B, and a potential CDK-1 phosphorylation site within Deg-A, each of which is required for efficient degradation. Transplantation of both LIN-41 degradation domains into OMA-2 results in the premature degradation of the resulting fusion protein during meiosis. Furthermore, we find that a Skp, Cullin, and F-box (SCF) E3 ubiquitin ligase complex containing the substrate recognition subunit SEL-10 (referred to as SCFSEL-10) promotes the degradation of LIN-41 and likely functions through the newly identified degradation domains of LIN-41. SEL-10 is a highly conserved F-box protein important for cell cycle regulation in both yeast (cell division control protein 4; Cdc4) and humans (F-box and WD repeat domain protein; FBW7) [reviewed in Deshaies and Ferrell (2001), Welcker and Clurman (2008)]. In C. elegans, sel-10 does not have an essential mitotic cell cycle role but was identified as a negative regulator of LIN-12/Notch signaling (Sundaram and Greenwald 1993; Hubbard et al. 1997).
Intriguingly, we show that SEL-10 is also important for the degradation of the tumor suppressor protein GLD-1/STAR RNA-binding protein, which is required for oocyte differentiation and represses translation in oocytes (Francis et al. 1995a,b; Jones and Schedl 1995; Lee and Schedl 2001; Schumacher et al. 2005; Jungkamp et al. 2011; Wright et al. 2011; Farley and Ryder 2012; Scheckel et al. 2012; Doh et al. 2013). GLD-1 was independently identified as a target of SEL-10-mediated degradation by Kisielnicka et al. (2018) along with CPB-3, a cytoplasmic polyadenylation element-binding protein, which is also important for oocyte development (Boag et al. 2005; Hasegawa et al. 2006). GLD-1 and CPB-3 are degraded during meiotic prophase, as immature oocytes transition from pachytene to diplotene, considerably earlier than the degradation of LIN-41 during the OET. This variation in timing is due to contrasting mechanisms of signaling-mediated control; while the degradation of LIN-41 is regulated by activated CDK-1 (Spike et al. 2014a and this work), the degradation of GLD-1 and CPB-3 is regulated by the mitogen-activated protein (MAP) kinase MPK-1 (Kisielnicka et al. 2018 and this work). Contrary to a recent report (Bohnert and Kenyon 2017), we found that the SEL-10-dependent degradation of GLD-1 is not dependent on the presence of sperm in the gonad, contradicting a central aspect of the model proposed by Bohnert and Kenyon (2017) for the regulation of proteostasis renewal during oogenesis. A surprising finding of our study is that the ectopic expression of LIN-41 and GLD-1 in sel-10 mutants has only minor effects on fertility and the expression of mRNAs that are translationally repressed by either LIN-41 or GLD-1 during oogenesis. We suggest that the LIN-41 that persists in the embryos of sel-10 and certain lin-41 mutants is likely inactivated by additional post-transcriptional mechanisms that remain to be identified.
Materials and Methods
C. elegans strains and phenotypic analysis
Strains:
The genotypes of strains used in this study are reported in Supplemental Material, Table S1. The following mutations were used: LGI: mex-3(tn1753[gfp::3xflag::mex-3]), air-2(or207ts), unc-13(e1091), rrf-1(pk1417), gld-1(q485), lin-41(tn1487ts), lin-41(tn1541[gfp::tev::s-tag::lin-41], lin-41(tn1541tn1548), lin-41(tn1541tn1562), lin-41(tn1541tn1571), lin-41(tn1541tn1618), lin-41(tn1541tn1620), lin-41(tn1541tn1622), lin-41(tn1541tn1628), lin-41(tn1541tn1630), lin-41(tn1541tn1635), lin-41(tn1541tn1638), lin-41(tn1541tn1641), lin-41(tn1541tn1643), lin-41(tn1541tn1645), lin-41(tn1541tn1661), lin-41(tn1541tn1663), lin-41(tn1541tn1665), lin-41(tn1541tn1668), lin-41(tn1541tn1684), lin-41(tn1541tn1775), lin-41(tn1767), fog-3(q470), and lin-11(n566); LGIII: mpk-1(ga111ts), emb-30(tn377ts), cdk-1(ne2257ts), orc-1(tn1732[mng::3xflag::orc-1]), and cul-2(or209ts); LGIV: pgl-1(sam37[pgl-1R765S::mTagRFPT::3xflag) (kindly provided by Dustin Updike), cks-1(ne549ts), and oma-1(zu405te33). LGV: spn-4(tn1699[spn-4::gfp::3xflag]), oma-2(te51), oma-2(cp145[mng::3xflag::oma-2]), oma-2(tn1760[mng::3xflag::degA::oma-2]), oma-2(tn1764[mng::3xflag::degA::degB::oma-2]), lon-3(e2175), sel-10(ar41), sel-10(ok1632), him-5(e1490), and fog-2(oz40); and LGX: meg-1(tn1724[gfp::3xflag::meg-1]). The following rearrangements were used: hT2[bli-4(e937) let-?(q782) qIs48] (I; III) and nT1[qIs51] (IV; V). The following transgene insertions were used: axIs1498[pie-1p::gfp::gld-1::gld-1 3′UTR, unc-119(+)](Merritt et al. 2008), itIs37[pie-1p::mCherry::H2B::pie-1 3′UTR, unc-119(+)] IV (McNally et al. 2006), ozIs5[gld-1::gfp] I (kindly provided by Tim Schedl), ozIs2[gld-1::gfp] II (Schumacher et al. 2005), and pwIs116[rme-2p::rme-2::gfp::rme-2 3′UTR, unc-119(+)] (Balklava et al. 2007).
Phenotypic analysis:
Protein expression in living animals or embryos was analyzed using fusions of endogenous loci to green fluorescent protein (GFP; Chalfie et al. 1994) or mNeonGreen (mNG; Allele Biotechnology, San Diego, CA; Shaner et al. 2013). The persistence of GFP::LIN-41 was evaluated in embryos between the 1- and 12-cell stages located in the spermathecae and uteri of >100 animals. All mutant lin-41 and sel-10 alleles with persisting GFP::LIN-41 proteins were fully penetrant. Null mutations in lin-41 are sterile, with small, abnormal oocytes, and some hypomorphic alleles of lin-41 affect the production of high-quality oocytes (Slack et al. 2000; Spike et al. 2014a). Thus, GFP::LIN-41[Δ] was often examined in both heterozygotes (lin-41(tn1541Δ)/unc-13(e1091) lin-11(n566) genotypes) and homozygotes (lin-41(tn1541Δ) genotypes), particularly when the lin-41(tn1541Δ) homozygotes produced obviously small or abnormal oocytes or produced a significant number of dead embryos. All the sel-10(ar41) strains used also contain lon-3(e2175), a convenient cis-linked marker that encodes a cuticle collagen (Nyström et al. 2002; Suzuki et al. 2002). The lon-3(e2175) marker does not affect any of the gene expression patterns reported in this study and was used as a negative control where indicated.
Genome editing
Plasmids capable of expressing guide RNAs (gRNAs) that target the lin-41 gene were generated as described by Arribere et al. (2014) from the vector pRB1017 and sequence-specific oligonucleotides. We estimated the efficiency with which each lin-41 gRNA was able to target the lin-41(tn1541) locus by: (1) co-injecting a mixture of the gRNA plasmid (25 ng/μl), the pDD162 plasmid (Dickinson et al. 2013), which supplies the Cas9 enzyme (50 ng/μl), and a co-injection marker (myo-2p::Tdtomato, 4 ng/μl) into lin-41(tn1541) hermaphrodites; (2) culturing individual F1 progeny that expressed the co-injection marker (typically ≤10 F1s from each injected parent); and (3) determining the number of F1s that segregated F2 progeny with a Dpy lin-41 loss-of-function (lf) phenotype. File S1 reports the sequences and estimated efficiencies of the gRNAs we used to generate the lin-41 deletions and point mutations described in this work; most were relatively effective at targeting lin-41.
During the efficiency experiments for lin-41 gRNAs #10 and #11, we identified lin-41(tn1541tn1562) and lin-41(tn1541tn1571), respectively, as GFP::LIN-41-positive lin-41(lf) mutants that appeared to have relatively large deletions by PCR. All of the other deletions were generated in a targeted manner by co-injecting two or more lin-41 gRNA plasmids (25 ng/μl each), a single-strand oligonucleotide repair template (500 nM), the pDD162 plasmid (50 ng/μl), and a co-injection marker (myo-2p::Tdtomato, 4 ng/μl) into lin-41(tn1541) hermaphrodites. We used gRNAs on each side of the desired deletion that, in most cases, would not produce substrates for Cas9 digestion after the deletion event. Otherwise, silent mutations were included in the repair template to prevent recutting. To identify lin-41(tn1541) deletion mutants, we individually placed F1 worms expressing the co-injection marker on plates, allowed them to lay eggs, and then used PCR to screen pools of up to six F1 worms. Pools that appeared to be strongly positive for the desired deletion band were rescreened by PCR to identify F1 animals that had segregated candidate deletion mutants among their F2 progeny. Mutants were either allowed to become homozygous or were balanced using hT2[bli-4(e937) let-?(q782) qIs48] (I; III). Essentially the same method was used to generate amino acid substitutions in lin-41. However, in those experiments we used only one gRNA and repair events were identified using silent mutations that created restriction enzyme recognition sites in each repair template. Screening therefore consisted of PCR followed by a restriction enzyme digestion, and we only pooled two F1s in the initial round of screening so that the repair events would be easy to detect. All edited loci were validated by sequencing, and we were able to obtain multiple independent alleles for most targeted deletions and amino acid substitutions. Where possible, two alleles identical to the repair template (but derived from independently injected parents) were saved and assigned allele names. Other, typically imperfect, gene edits were also kept and given allele designations if they were informative or potentially useful. Additional information about all of these alleles and detailed genome-editing information, including gRNA, repair template, and PCR primer sequences, are provided in File S1.
oma-2(tn1760) and oma-2(tn1764) were created using the method described by Dickinson et al. (2015) to create oma-2(cp145). Indeed, we were careful to replicate oma-2(cp145) as closely as possible; we used the same gRNA plasmid (pDD223) and designed our repair templates to closely mimic pDD271, the repair template used to create oma-2(cp145). However, instead of using PCR to generate the 3′ homology arms of the repair templates, which contain sequences derived from both oma-2 and lin-41, we synthesized these sequences as gBlocks (Integrated DNA Technologies, Skokie, IL). We minimized the size and complexity of each gBlock by removing introns from the lin-41–encoding sequences. oma-2(tn1760) and oma-2(tn1764) were perfect matches to the desired repairs (repair templates pCS557 and pCS561, respectively). Gene edited alleles were outcrossed to the wild type before analyzing fertility and embryonic lethality. Specific genome-editing details are provided in File S1.
Microscopy
Movies of GFP::LIN-41, mNG::Deg-A::Deg-B::OMA-2, PGL-1::RFP, and mCHERRY::H2B during the OET were obtained using a Marianas 200 Microscopy Workstation (Intelligent Imaging Innovations, Denver, CO) built on an AxioObserver Z.1 stand (Carl Zeiss, Thornwood, NY), and driven by SlideBook 6.0 software (Intelligent Imaging Innovations). The imaging was performed using a 40× oil Carl Zeiss Plan-Apochromat objective lens (numerical aperture of 1.4) and an Evolve electron-multiplying charge-coupled device camera (Photometrics, Tucson, AZ). Fluorescence intensity quantification from time-lapse images was performed using ImageJ software. All of the other images were acquired on a Carl Zeiss motorized Axioplan 2 microscope with a 63× Plan-Apochromat (numerical aperture 1.4) objective lens, using a AxioCam MRm camera and AxioVision software, version 4.8.2.0 (Carl Zeiss). Image quantifications were performed using the interactive measurements tool in AxioVision to quantify the densitometric mean value of a circumscribed region. The average intensity of SPN-4::GFP fluorescence was measured in a ∼12 μm diameter circle in the anterior cytoplasm of one-cell and two-cell embryos to avoid the bright puncta of SPN-4::GFP in the posterior; these are likely P granules, as SPN-4 is known to associate with these nonmembrane-bound organelles in embryos (Ogura et al. 2003). The amount of diffusely cytoplasmic SPN-4::GFP appeared to be similar throughout the embryo during these early stages of embryogenesis and in each of the strains we analyzed. Likewise, the average intensity of GFP::MEX-3 and mNG::OMA-2 fluorescence was measured in a ∼10 μm diameter circle in the oocyte cytoplasm. Fluorescence was measured in the oocytes that expressed detectable levels of each fusion protein under our imaging conditions (100 and 120 ms for GFP::MEX-3 and mNG::OMA-2, respectively) and were large enough to fit a ∼10 μm diameter circle in the oocyte cytoplasm. GFP::MEX-3 was detected in four or five proximal oocytes in all strains, consistent with previous observations (Tsukamoto et al. 2017). mNG::OMA-2 was detected in five or six proximal oocytes in the sel-10(ar41) mutants and in seven or more proximal oocytes in the control strain.
RNA interference
Gene-specific RNA interference (RNAi) was performed by feeding C. elegans with double-strand RNA (dsRNA)-expressing Escherichia coli (Timmons and Fire 1998) at 22°, using the RNAi culture media described by Govindan et al. (2006). RNAi clones were obtained from Source BioScience (Nottingham, UK), and the identity of each RNAi clone was verified by DNA sequencing. Exposure to dsRNA-expressing E. coli was initiated during the fourth larval stage and GFP::LIN-41 was examined after 1 and 2 days. cdk-1(RNAi), skr-1(RNAi), and sel-10(RNAi) at least partially prevented the elimination of GFP::LIN-41 after 1 day, with completely penetrant phenotypic effects on day 2, while cul-1(RNAi) only prevented the elimination of GFP::LIN-41 after 2 days of RNAi treatment. All images of RNAi-treated animals were collected on day 2. cul-1, cul-2, and cul-3 are important for normal embryonic development. We observed highly penetrant embryonic lethality after treating animals with cul-1(RNAi) and cul-3(RNAi). However, the cul-2(RNAi) clone we used targets the cul-2 3′UTR and did not cause embryonic lethality. We therefore examined lin-41(tn1541); cul-2(or209ts) adults upshifted to 25° as stage 4 (L4) larvae to assess whether cul-2 is important for the elimination of GFP::LIN-41 from embryos. We observed that GFP::LIN-41 was eliminated normally from the dead embryos produced by cul-2(or209ts) parents at the restrictive temperature (n = 71). Postdauer lin-41(tn1541tn1618) animals [and parallel controls treated with cdk-1(RNAi)] were examined because strong loss-of-function lin-41 mutant animals are much healthier when they pass through the dauer stage (Spike et al. 2014a).
Western blots
Proteins were separated using NuPage 4–12% Bis-Tris gels or 3–8% Tris-Acetate gels (Invitrogen, Carlsbad, CA) and visualized after Western blotting. Blots were blocked with 5% nonfat dried milk. Primary antibodies used to detect proteins were affinity-purified rabbit anti-LIN-41 R214 (1:20,000 dilution) (Spike et al. 2014a), affinity-purified guinea pig anti-LIN-41 GP49E (1:4000 dilution) (Spike et al. 2014a), rabbit anti-GFP NB600-308 (1:4000 dilution; Novus Biologicals, Littleton, CO), and rabbit anti-GLD-1 (1:3000 dilution; kindly provided by Sarah Crittenden and Judith Kimble) (Jan et al. 1999). Secondary antibodies used for Western blots were peroxidase-conjugated donkey anti-guinea pig (1:40,000 dilution) (Jackson ImmunoResearch, West Grove, PA) and anti-rabbit (1:5000 dilution) (Thermo Scientific, Waltham, MA) antibodies. Detection was performed using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific).
Antibody staining
Dissected gonads stained with either the rabbit anti-phospho-histone H3 (Ser10) antibody (1:400 dilution; Millipore, Burlington, MA) or the rabbit anti-RME-2 antibody (1:50 dilution, kindly provided by Barth Grant) (Grant and Hirsh 1999) were fixed in 3% paraformaldehyde for 1 hr, as described (Rose et al. 1997). Dissected gonads stained with the rabbit anti-GLD-1 primary antibody (1:150 dilution; Jan et al. 1999) were fixed in 1% paraformaldehyde for 10 min with a 5-min postfix step in ice-cold methanol. Primary antibodies were detected using either Cy3-conjugated goat anti-rabbit or Alexa 488-conjugated donkey anti-rabbit secondary antibodies (1:500 dilutions; Jackson ImmunoResearch). For the strong loss-of-function lin-41(tn1541tn1618) mutant, gonads were dissected from postdauer animals on the first day of adulthood.
Data availability
All strains and newly created alleles (see File S1 and Table S1) are available upon request. The sequences of gRNAs, repair templates, PCR primers, lin-41 alleles, and oma-2 alleles are presented in File S1. Plasmids producing gRNAs and those containing repair templates for genome editing are available upon request. All Sanger sequencing files are available upon request. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7056557.
Results
GFP::LIN-41 is eliminated during the first meiotic division
Germline-expressed LIN-41 is restricted to oogenesis and required to prevent premature M phase entry and to promote growth of developing oocytes (Spike et al. 2014a). In the oogenic germlines of adult hermaphrodites, LIN-41 is expressed from midpachytene through subsequent stages of oocyte development, with a notable reduction in LIN-41 levels as oocytes initiate meiotic maturation at the end of oogenesis. Essentially the same pattern is seen in the oocytes of lin-41(tn1541[gfp::tev::s-tag::lin-41]) adult hermaphrodites; these animals carry a gfp-tagged allele of lin-41 and express only GFP-tagged LIN-41 (GFP::LIN-41), yet have essentially wild-type oocyte development and fertility (Figure 1, A and B; Spike et al. 2014a). GFP::LIN-41 is always visible in the oocyte immediately adjacent to the spermatheca (−1 oocyte), but is not detectable in most embryos, suggesting that GFP::LIN-41 is eliminated soon after meiotic maturation and ovulation (Figure 1, A and B; Spike et al. 2014a). To more precisely determine the stage at which GFP::LIN-41 is eliminated during the OET, we used time-lapse imaging to examine GFP::LIN-41 as oocytes proceed through meiotic maturation, are ovulated into and fertilized in the spermatheca, and complete their meiotic divisions (Movie S1 and Figure 1, C–H). We also imaged several oocytes as they moved into the −1 position from a slightly earlier developmental stage (−2 or −3 oocyte start position). Quantification of these images shows that GFP::LIN-41 levels decline slowly during the late stages of oogenesis and then drop dramatically after nuclear envelope breakdown and ovulation (Figure 1, D and H). As a result, GFP::LIN-41 is essentially undetectable well before the end of the first meiotic division (Figure 1, F and G). During meiotic maturation, the −1 oocyte undergoes a cortical cytoskeletal rearrangement prior to ovulation (McCarter et al. 1999). GFP::LIN-41 begins to localize to punctate structures in the oocyte cytoplasm during cortical rearrangement, at the onset of its dramatic disappearance (Movie S1). The nature of these punctate structures is unclear; however, most of them do not exhibit colocalization with PGL-1::RFP and therefore do not appear to be P granules (Figure S1, A–C). Collectively, these observations document that preexisting GFP::LIN-41 in the −1 oocyte is rapidly eliminated during meiosis I in a period of ∼10–15 min, likely as a result of protein degradation. lin-41 mRNA levels also decline during the OET (∼30-fold decrease in one-cell embryos relative to proximal oocytes; Stoeckius et al. 2014), suggesting that mRNA destabilization may prevent the resynthesis of LIN-41 after its elimination during meiosis.
Deg-A and Deg-B are required to eliminate GFP::LIN-41 from embryos
To identify the amino acid sequences of LIN-41 required for its elimination from early embryos, we generated a series of deletions in the coding region of the GFP::LIN-41-expressing lin-41(tn1541) gene using CRISPR-Cas9-based genome-editing approaches. Collectively, these deletions are predicted to remove 95% of the LIN-41 protein and disrupt all known structural domains of LIN-41 (Figure 2, A and B, and File S1). For each mutant, GFP::LIN-41[Δ] expression was examined to determine whether the deleted portion of LIN-41 is necessary for the elimination of GFP::LIN-41 from embryos (Figure 2, D–F, Figure S2, and Figure S3). This approach enabled us to determine that two nonoverlapping regions in the N-terminal third of LIN-41 are required for its elimination from embryos (Figure 2B). We will refer to these regions as the LIN-41 degradation domains Deg-A and Deg-B. Importantly, GFP::LIN-41[Δ] proteins with deletions affecting the Deg-A or Deg-B domains are expressed in proximal oocytes at comparable levels to the wild-type fusion protein (Figure 1 and Figure S2). However, the GFP::LIN-41[ΔDeg] proteins fail to be eliminated upon the onset of meiotic maturation. Sequences similar to Deg-A and Deg-B are found in Caenorhabditis nematodes but they appear to be rapidly evolving. In mammalian LIN-41/TRIM71, the region equivalent to Deg-B, which is located between the RING and B-box domains, is predicted to be intrinsically disordered like the C. elegans Deg domains (C. Spike and D. Greenstein, unpublished results).
Figure 2.
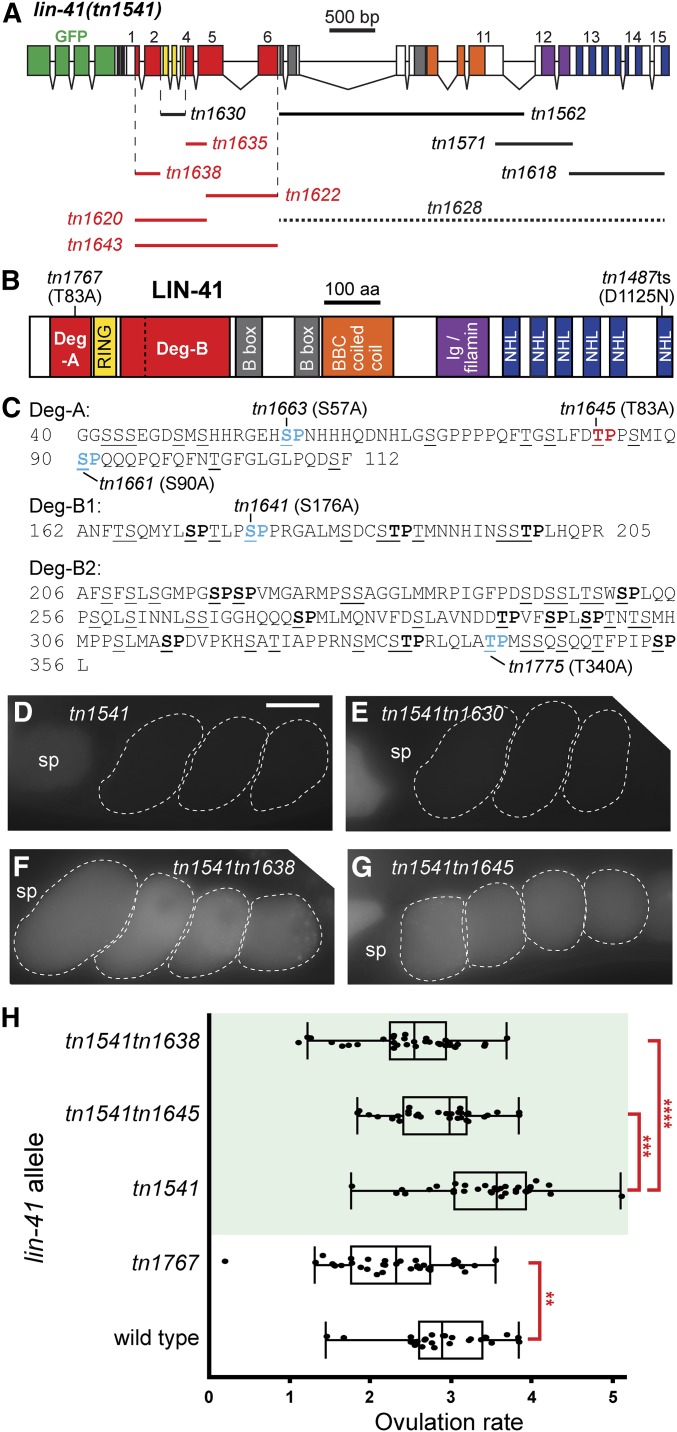
LIN-41 Deg domains are required for the elimination of GFP::LIN-41 upon the onset of meiotic maturation. (A) The exon–intron structure and deletion analysis of lin-41(tn1541). Colored boxes indicate exonic regions that encode GFP (green) or LIN-41 protein domains (see B). Deletions made in the context of lin-41(tn1541) are drawn as lines, labeled with a deletion-specific allele name, beneath the LIN-41-encoding exons and introns (exons labeled 1–15). GFP::LIN-41 can be detected in the germline of most deletion mutants (solid lines), with one exception (tn1628, dotted line). Deletions in red prevent the elimination of GFP::LIN-41 from early embryos. The vertical dashed lines delimit the beginning of Deg-A and the end of Deg-B, respectively. (B) The previously described [RING (yellow), B-box (gray), BBC (orange), Ig/filamin (purple), NHL (blue)] and newly identified [Deg (red)] protein domains of LIN-41. The vertical dashed line in B indicates the two parts of Deg-B, B1 and B2, which are individually removed in lin-41(tn1541tn1635) and lin-41(tn1541tn1622), respectively. The position of two lin-41 missense alleles (tn1767[T83A] and tn1487ts[D1125N]) generated in the endogenous lin-41 locus are indicated. (C) The amino acid sequences of Deg-A, Deg-B1, and Deg-B2. Many of the amino acids are serines and threonines (underlined) and some are potential targets of proline-directed serine/threonine [S/T] kinases (bold). The indicated alleles change an [S/T] residue to an alanine (colored and bold) in the context of lin-41(tn1541). The tn1645[T83A] mutation in Deg-A results in the persistence of GFP::LIN-41[T83A] in embryos (red), whereas the other changes do not (indicated in blue font). (D–G) GFP::LIN-41 is eliminated from the early embryos (dashed outlines) of lin-41(tn1541) (D, control) and lin-41(tn1541tn1630) (E, RING deleted) homozygous mutants but persists in the early embryos of lin-41(tn1541tn1638) (F, Deg-A deleted) and lin-41(tn1541tn1645) (G, LIN-41[T83A]) homozygous mutants. The position of the spermatheca (sp) is indicated, for reference. 100 ms GFP exposures; Bar, 20 μm. (H) The rate of ovulation is slightly reduced in mutants with a compromised LIN-41 Deg-A domain. Ovulation rate is expressed as the number of ovulations per gonad arm per hr and was measured in at least 25 day 2 adults. The three gfp-tagged alleles are in the top portion of the graph with green shading. Significance was determined using a one-way ANOVA test with Tukey’s post hoc test to compare the means; ** P < 0.01, *** P < 0.001, and **** P < 0.0001. itIs37[pie-1p::mCherry:::H2B::pie-1 3′UTR, unc-119(+)] was also present in each of the GFP::LIN-41-expressing strains; it is not expected to alter the ovulation rate.
The LIN-41 Deg-A domain is defined by the lin-41(tn1541tn1638) deletion allele. This deletion is predicted to affect GFP::LIN-41 by removing 73 amino acids on the N-terminal side of the LIN-41 RING domain (Figure 2C and File S1). lin-41(tn1541tn1638) is immediately adjacent to, but does not overlap, the lin-41(tn1541tn1630) deletion, which is predicted to affect GFP::LIN-41 by removing the LIN-41 RING finger domain (see File S1 for deleted residues). Consistent with previous amino acid substitution and transgenic rescue data (Tocchini et al. 2014), the RING domain is not required for the elimination of GFP::LIN-41 from embryos (Figure 2E and Figure S2). The LIN-41 Deg-B domain is defined by two contiguous, but nonoverlapping, deletions on the C-terminal side of the LIN-41 RING domain. The lin-41(tn1541tn1635) deletion is predicted to affect GFP::LIN-41 by removing 44 amino acids on the C-terminal side of the LIN-41 RING domain (Deg-B1) (Figure 2C and File S1). Compared to the Deg-A deletion mutant [lin-41(tn1541tn1638)], the Deg-B1 deletion mutant [lin-41(tn1541tn1635)] has a relatively low but detectable level of GFP::LIN-41[Δ] in early embryos (compare Figure S2, K and M). However, the lin-41(tn1541tn1622) deletion (which defines Deg-B2 and the remaining 151 amino acids of Deg-B; Figure 2C and File S1) has a robust defect in the elimination of GFP::LIN-41[Δ] from early embryos that is apparent in both heterozygous and homozygous deletion mutants (Figure S2, E and G). Finally, deletions predicted to affect GFP::LIN-41 by removing amino acids and structural domains C-terminal to Deg-B were able to eliminate GFP::LIN-41[Δ] from early embryos (Figure 2A and Figure S3). Interestingly, we found that C-terminal domains could only be removed individually or in small groups, as GFP::LIN-41[Δ] was not detectable when a majority of the C terminus was removed [lin-41(tn1541tn1628) deletion; Figure 2A and Figure S3, M–P].
LIN-41[T83] is required to eliminate LIN-41 from embryos
The results described above indicate that the elimination of GFP::LIN-41 does not depend on any of the previously described structural domains of LIN-41, but instead requires two new regulatory domains. Analysis of the amino acid sequences of Deg-A and Deg-B shows that each regulatory domain contains many possible phosphorylation sites (Figure 2C). Previously published results indicate that the elimination of GFP::LIN-41 from embryos also requires CDK-1 (Spike et al. 2014a), a highly conserved, proline-directed, serine/threonine kinase essential for M phase entry during oocyte meiotic maturation in C. elegans (Boxem et al. 1999). Thus, we hypothesized that LIN-41 might be a direct target of CDK-1 activity, and that phosphorylation of either Deg-A or Deg-B by CDK-1 could be sufficient to trigger the elimination of GFP::LIN-41 from embryos. Eighteen minimal CDK-1 consensus sequences ([S/T]P) are present in Deg-A and Deg-B (Figure 2C), but only a single site, found in Deg-B1, conforms to an expanded CDK1 consensus sequence ([S/T]PX[K/R]) (Ubersax et al. 2003). However, changing the potentially phosphorylated residue at this site to an alanine [S176A; e.g., lin-41(tn1541tn1641)] had no effect on the elimination of GFP::LIN-41 from embryos (Figure 2C and Figure S4A).
Consequently, we shifted our focus to Deg-A, which is relatively small and contains only three potential CDK-1 target sites, but strongly prevents the elimination of GFP::LIN-41 from embryos (Figure S2M). Each site in Deg-A was tested individually to see if it is required for the elimination of GFP::LIN-41 from embryos. Although the mutations S57A and S90A [e.g., lin-41(tn1541tn1663) and lin-41(tn1541tn1661), respectively] had no discernable effect (Figure S2C and Figure S4, C and E), the T83A mutation [e.g., lin-41(tn1541tn1645)] strongly prevented the elimination of GFP::LIN-41 from embryos, similar to the Deg-A deletion mutant (Figure 2G, Figure S2M, and Figure S4G). Time-lapse imaging of oocyte meiotic maturation, ovulation, and fertilization documents that the T83A mutation strongly abrogates the elimination of GFP::LIN-41[T83A] during meiosis I (Movie S2). During cortical rearrangement, GFP::LIN-41[T83A] localized partially to dynamic punctate structures like GFP::LIN-41 (compare Movie S1 and Movie S2); however, unlike the wild-type protein, puncta of GFP::LIN-41[T83A] were also observed during the meiotic divisions. Furthermore, GFP::LIN-41[T83A] persisted through multiple embryonic cleavage divisions and became at least partially associated with P granules by the two-cell stage (Figure S1, D–I and Movie S2). Importantly, total GFP::LIN-41[T83A] levels remain relatively constant throughout the corresponding time period of meiosis, during which GFP::LIN-41 is normally eliminated (compare Movie S1 and Movie S2). These results are consistent with the possibility that phosphorylation of LIN-41 by a proline-directed S/T kinase, such as CDK-1, promotes the rapid degradation of GFP::LIN-41 upon the onset of meiotic maturation. We next replaced T83 with a glutamic acid residue (T83E) [e.g., lin-41(tn1541tn1684)], which is negatively charged and might function as a phosphomimetic. However, T83E did not result in the premature elimination of GFP::LIN-41, as when CDK-1 is prematurely activated (Spike et al. 2014a). Instead, T83E prevented the elimination of GFP::LIN-41 from embryos, similar to T83A (Figure S4, G and I). This result is not unexpected because phosphorylation sites that function to recruit adapter proteins are often not recognized by binding partners after phosphomimetic substitution (Dephoure et al. 2013). In fact, phosphomimetic substitutions within recognition domains are known to be ineffective for Fbw7 substrates (Welcker et al. 2013), and this is a likely role for the function of T83 and the Deg domains of LIN-41, as we describe below.
A requirement for the extreme N terminus of LIN-41 (amino acids 1–39) with respect to the elimination of GFP::LIN-41 was not examined in the lin-41(tn1541) deletion analysis. Genetic analysis suggests that this region of LIN-41 is important for downregulating lin-41 function specifically in the male tail (Del Rio-Albrechtsen et al. 2006). Gain-of-function (gf) alleles that affect this part of LIN-41 have a defect in male tail tip retraction, while hermaphrodites appear overtly wild type. lin-41(tn1541) males also have a male tail tip retraction defect (Figure S5, A and B), suggesting that the GFP tag on the N terminus of LIN-41 disrupts this male-specific function. Furthermore, the amino acid change found in the lin-41(bx37gf) allele (G35R) does not affect the elimination of GFP::LIN-41 from early embryos [lin-41(tn1541tn1665); Figure S5, C and E]. For these reasons, we suspect that the extreme N terminus of LIN-41 is unlikely to be involved in the elimination of GFP::LIN-41 from early embryos. One possibility, however, might be that the N-terminal GFP tag on GFP::LIN-41 compromises a function that is required redundantly with Deg-A or Deg-B. To explore this possibility, we generated worms expressing LIN-41[T83A] [lin-41(tn1767)] and asked whether the untagged protein also persists in embryos. Using Western blots, we found that LIN-41 was undetectable in a lysate made from wild-type embryos, but that LIN-41[T83A] was clearly present in a lysate prepared from lin-41(tn1767) mutant embryos (Figure S6D). Thus, the T83A mutation abrogates the elimination of both LIN-41 and GFP::LIN-41 from embryos.
Functional requirements for individual LIN-41 domains
LIN-41 is a large protein with two well-defined domains that are proposed to have strikingly different activities. The first of these is actually the TRIM multidomain arrangement that contains RING, B-box, and coiled-coil domains; many TRIM proteins are thought to function as RING finger E3 ubiquitin ligases (Ikeda and Inoue 2012). The second functional domain is an RNA-binding domain composed of six NHL (NCL-1, HT2A, and LIN-41) repeats at the C terminus of LIN-41 (Slack and Ruvkun 1998; Loedige et al. 2015; Kumari et al. 2018). Forward and reverse genetic analyses strongly indicate that the NHL domain is important for both the germline and somatic functions of C. elegans LIN-41 (Slack et al. 2000; Spike et al. 2014a; Tocchini et al. 2014), consistent with the identification of LIN-41 as a translational repressor in both tissue types (Spike et al. 2014b; Aeschimann et al. 2017; Tsukamoto et al. 2017). By contrast, a deletion of the entire LIN-41 RING domain (Figure 2A), which confers in vitro E3 ligase catalytic activity to mouse LIN41 and other TRIM proteins (Rybak et al. 2009; Esposito et al. 2017), results in appreciable fertility (brood size of 210 ± 87; Table 1) and thus is nonessential for C. elegans oogenesis. As described below, the phenotypes seen in lin-41(tn1541) deletion mutants are consistent with prior observations and provide additional insights into the functions of LIN-41 protein domains.
Table 1. Fertility and fecundity of lin-41 alleles at 20°.
| Genotype | Predicted protein change | Fertile, %a | Brood sizeb | Dead embryos, %c |
|---|---|---|---|---|
| lin-41(tn1541) | N-terminal GFP | 100 (n = 68) | 316 ± 39d (n = 6) | 0.3 (n = 361) |
| lin-41(tn1541tn1618)e,f | Δ NHL (AA 819–1128) | 1.5 (n = 65) | 1 (n = 1) | ND |
| lin-41(tn1541tn1571)e,f | Δ Ig (AA 677–824) | 78.5 (n = 65) | 11 ± 12 (n = 9) | 57.1g (n = 35) |
| lin-41(tn1541tn1562)e,f | Δ B-box–coiled-coil (AA 356–707)h | 84 (n = 87) | 6 ± 3 (n = 17) | ND |
| lin-41(tn1541tn1643)e | Δ N-terminal (AA 40–356) | 66 (n = 90) | 6 ± 4 (n = 48) | 75.4g (n = 142) |
| lin-41(tn1541tn1620)e | Δ N-terminal (AA 40–205) | 97 (n = 67) | 39 ± 32 (n = 10) | 36.4g (n = 110) |
| lin-41(tn1541tn1622)e | Δ Deg-B2 (AA 206–356) | 100 (n = 65) | 33 ± 16 (n = 6) | 39.0g (n = 105) |
| lin-41(tn1541tn1635) | Δ Deg-B1 (AA 162–205) | 100 (n = 70) | 127 ± 108 (n = 10) | 2.9 (n = 105) |
| lin-41(tn1541tn1630) | Δ RING (AA 113–161) | 98.5 (n = 65) | 210 ± 87 (n = 12) | 2.8 (n = 144) |
| lin-41(tn1541tn1638) | Δ Deg-A (AA 40–112) | 100 (n = 70) | 217 ± 103 (n = 10) | 6.3 (n = 174) |
| lin-41(tn1541tn1645) | T83A | 100 (n = 70) | 251 ± 86 (n = 10) | 1.0 (n = 193) |
| lin-41(tn1767) | T83A | 98.3 (n = 120) | 313 ± 31 (n = 6) | 0.0 (n = 176) |
Fertile animals produced at least one viable offspring.
The average number of progeny that hatched from fertile animals ± SD.
The percent lethality among the embryos laid on day 1 of adulthood.
Essentially identical to the lin-41(tn1541) brood size previously reported in Spike et al. (2014a) (319 ± 28; n = 30).
The progeny of lin-41/hT2[qIs48] hermaphrodites.
These animals have a Dumpy (Dpy) body shape, as previously described for lin-41(lf) alleles (Slack et al. 2000).
Some of the embryos laid were small or otherwise appeared to be abnormal.
The minimum number of amino acids removed by tn1562. Assuming the use of an in-frame 5′ splice site in the 17-bp insertion, either one amino acid (L) or five amino acids (LSPLL) would replace amino acids 356–707.
TRIM (RING, B-box, and coiled-coil) domain:
Deletion of the RING finger in the context of GFP::LIN-41 (GFP::LIN-41[ΔRING]) results in only mild defects. Most lin-41(tn1541tn1630) animals are fertile and have a large number of progeny; no strong defects in oogenesis, embryonic development, or body shape are evident (Table 1 and Figure S2, I and J). We did note, however, that lin-41(tn1541tn1630) animals appear to be slightly sick and that they produce ∼33% fewer progeny than lin-41(tn1541) hermaphrodites (Table 1). Interestingly, deletion of the other two TRIM sub-domains (GFP::LIN-41[ΔB-box-CC]) causes a much stronger reduction in LIN-41 function. Most (84%) lin-41(tn1541tn1562) hermaphrodites are fertile, but produce very few progeny (6 ± 4) and have obvious defects in oogenesis as well as a Dumpy (Dpy) body shape (Table 1 and Figure S3, A and B). Thus, lin-41(tn1541tn1562) is clearly a hypomorphic allele of lin-41 that affects both its germline and somatic functions.
We note that lin-41(tn1541tn1562) might remove additional residues beyond the B-box–coiled-coil region because, unlike the other lin-41(tn1541Δ) mutants we created, lin-41(tn1541tn1562) is not a precise exon–exon fusion and requires a new in-frame splicing event to make a full-length protein (Figure 2A and File S1). However, the deletion in this mutant was accompanied by the insertion of a small sequence that includes two potential 5′ splice site consensus sequences; both are in-frame with the downstream exon. Furthermore, the relative size of GFP::LIN-41[ΔB-box-BBC] on SDS-PAGE Western blots is consistent with what we expect to see for the protein made by this particular deletion mutant (Figure S6B and File S1). This is also true for the other GFP::LIN-41[Δ] proteins detected using either anti-LIN-41 or anti-GFP antibodies (Figure S6, A–C).
IG/filamin domain:
IG/filamin (IG) domains are only found in a subset of TRIM-NHL proteins; structural analysis of this part of C. elegans LIN-41 suggests that it forms a classic IG-like domain fold (Tocchini et al. 2014). The IG domain has been proposed to function, along with the coiled-coil domain, as a binding platform for proteins that repress the translation of NHL-bound target mRNAs (Loedige et al. 2013). lin-41(ma104) is a hypomorphic allele that likely disrupts the structure and function of the LIN-41 IG domain (Tocchini et al. 2014). As previously reported (Spike et al. 2014b), outcrossed lin-41(ma104) mutant hermaphrodites have mild oocyte defects and a reduced, but still substantial, brood size of 181 progeny (n = 12). Deletion of the IG domain in the context of GFP::LIN-41 (GFP::LIN-41[ΔIG]) results in stronger defects. Most lin-41(tn1541tn1571) hermaphrodites are fertile, with a very low brood size (11 progeny) and obvious defects in oogenesis (Figure S3, C and D and Table 1). Both alleles also result in worms with an obviously Dpy body shape. Thus, despite the difference in brood size, the alleles that affect the IG domain are hypomorphic and reduce both the germline and somatic functions of lin-41. Indeed, it is potentially misleading to conclude that the relative severities of lin-41(ma104) and lin-41(tn1541tn1571) are meaningful, as LIN-41 function may be slightly compromised in the lin-41(tn1541) mutant despite its wild-type brood size (316 ± 39; Table 1; Spike et al. 2014a). For example, the introduction of the LIN-41[D1125N] amino acid change in the sixth NHL repeat (Figure 2B) that results in a temperature-sensitive (ts) phenotype in an otherwise wild-type LIN-41 protein [e.g., lin-41(tn1487(ts); 100% fertile at 15° (n = 224), average brood size of 104 (n = 64); Spike et al. 2014a] results in a stronger, but still hypomorphic, phenotype in a GFP::LIN-41 mutant background [e.g., lin-41(tn1541tn1548); 71% fertile at 15° (n = 21), average brood size of three (n = 15)].
The NHL domain:
Deletion of the C-terminal NHL domain in the context of GFP::LIN-41 (GFP::LIN-41[ΔNHL]) results in a strong loss-of-function lin-41 phenotype. Whereas lin-41(tn1541) hermaphrodites are fertile, with normal oocyte development and overall appearance, nearly all (98.5%) lin-41(tn1541tn1618) hermaphrodites are sterile and have a Dpy body shape (Table 1). Oogenesis is extremely abnormal in most animals (Figure S3, G and H), although lin-41(tn1541tn1618) hermaphrodites produce embryos on occasion (Figure S3, I and J and Table 1). The fact that deletion of the LIN-41 NHL domain does not result in 100% sterility is surprising because the lin-41(n2914) null mutation has never been observed to produce progeny. Thus, LIN-41 can exhibit some, albeit very low, biological function in the absence of the NHL domain. We suggest that this low-level function may be mediated through components of the LIN-41 RNP (Spike et al. 2014b; Tsukamoto et al. 2017). We confirmed that CDK-1 exhibits premature activation in lin-41(tn1541tn1618) mutants, as it does in lin-41(n2914) null mutants, by staining adult hermaphrodite germlines with an antibody specific to histone H3 phosphorylated on Serine 10 (pH3(S10)). This antibody stains the nucleoplasm and condensed chromosomes of wild-type diakinesis-stage oocytes as they prepare to enter M phase near the spermatheca (Hsu et al. 2000). Both M phase and anti-pH3(S10) staining occur prematurely in strong loss-of-function lin-41 mutants and are cdk-1-dependent (Spike et al. 2014a). As expected for a strong loss-of-function mutant, and consistent with the idea that premature M phase entry and CDK-1 activation occur prematurely, we detected pH3(S10)-positive condensed chromosomes in or near the loop region of the gonad, and just after the end of pachytene, in most lin-41(tn1541tn1618) oogenic germlines (n = 6/9). Interestingly, GFP::LIN-41[ΔNHL] forms abnormal aggregates in the oocytes of lin-41(tn1541tn1618) homozygotes; these aggregates are not seen in heterozygotes (Figure S3, G, I, and K). The reason for this aberrant pattern of localization is unknown, but GFP::LIN-41[ΔNHL] aggregation is also seen in lin-41(tn1541tn1618); cdk-1(RNAi) animals (n = 32), and therefore does not depend on the dysregulation of cdk-1 function that occurs during oogenesis in strong loss-of-function lin-41 mutants (Spike et al. 2014a). These aggregates may reflect abnormal biogenesis of LIN-41 RNPs in the absence of the NHL domain.
Meiotic degradation domains are nonessential:
We initially hypothesized that the deletion of LIN-41 degradation domains might result in a gain-of-function phenotype that would affect fertility or embryonic viability. However, lin-41(tn1541tn1643), a large deletion that removes Deg-A, the RING finger, and Deg-B in the context of GFP::LIN-41, behaves as a recessive hypomorph that preferentially affects germline function. Homozygous mutants do not have a strong Dpy phenotype, but do have an extremely small brood size and display obvious defects in oogenesis and embryogenesis (Figure S2, O and P and Table 1). In contrast, heterozygous lin-41(tn1541tn1643) mutants appear essentially normal (n = 20). This is also true for deletions that subdivide the large N-terminal region of LIN-41, such as lin-41(tn1541tn1620) and lin-41(tn1541tn1622) (Figure S2, A–H and Table 1). Indeed, even when homozygous, the relatively small Deg-A deletion [lin-41(tn1541tn1638)], which results in abundant GFP::LIN-41[ΔDeg-A] in early embryos, appears to have minimal consequences for GFP::LIN-41 function at 20° (Figure S2, M and N and Table 1). Likewise, animals expressing LIN-41[T83] and GFP::LIN-41[T83] appear essentially wild type; the latter have only a slightly reduced brood size relative to GFP::LIN-41-expressing controls (Figure S4, G and H and Table 1). Consequently, we decided to look carefully at the ovulation rates of the minimally affected LIN-41 Deg-A deletion [lin-41(tn1541tn1638)] and T83A point mutants [lin-41(tn1541tn1645) and lin-41(tn1767)]. Oocyte meiotic maturation is a rate-limiting step for hermaphrodite fertility and the ovulation rate approximates the rate of oocyte meiotic maturation (McCarter et al. 1999; Miller et al. 2001; Govindan et al. 2006). Importantly, several aspects of nuclear and cytoplasmic oocyte maturation occur prematurely in lin-41(lf) mutations (Spike et al. 2014a,b; Tsukamoto et al. 2017). Deg-A domain mutants exhibit mean ovulation rates that are significantly reduced relative to genotype-matched controls (Figure 2H). Interestingly, the mean ovulation rate of the lin-41(tn1541) control strain was elevated relative to wild-type animals (Figure 2H, 3.4 vs. 2.9 ovulations per gonad arm per hr). Together, these observations suggest: (1) lin-41(tn1541) might be a weak hypomorph that causes a slight increase in the oocyte maturation rate; and (2) Deg-A domain mutants cause the opposite phenotype, a reduced oocyte maturation rate. These changes in the rate of oocyte maturation are relatively modest, however, and our phenotypic analyses generally suggest that the elimination of LIN-41 from early embryos is not a critical control point for regulating LIN-41 function or activity levels in vivo.
LIN-41[Deg] domains are sufficient for degradation
OMA-1 and OMA-2 (OMA-1/2) are functionally redundant, cytoplasmic RNA binding proteins expressed in oocytes and early embryos (Detwiler et al. 2001) that copurify with LIN-41 RNP complexes (Spike et al. 2014b; Tsukamoto et al. 2017). OMA-1/2 levels remain high in one-cell embryos until the first mitotic division, when they are rapidly degraded (Lin 2003; Nishi and Lin 2005; Shirayama et al. 2006; Stitzel et al. 2006). The expression and subsequent elimination of OMA-2 can be easily visualized in oma-2(cp145) mutants (Dickinson et al. 2015; Figure 3A and Figure S7, A and B), which express an mNG-tagged form of OMA-2 that is largely functional in vivo [Table 2, compare oma-1(zu405te33); oma-2(cp145) to oma-1(zu405te33); oma-2(te51)]. We decided to test whether the LIN-41 Deg domains are sufficient to induce the premature degradation of mNG::OMA-2 during meiosis. Molecularly, we placed LIN-41 Deg domains between mNG and OMA-2 (Figure 3, A–C), as this is similar to their locations in GFP::LIN-41 (Figure 2, A and B) and no structural (e.g., X-ray crystallographic) data are available to aid the experimental design. We generated two new oma-2 alleles that also contain lin-41-encoded Deg domains and examined the pattern of mNG::Deg::OMA-2 accumulation prior to the first mitotic division (Figure 3 and Figure S7).
Figure 3.
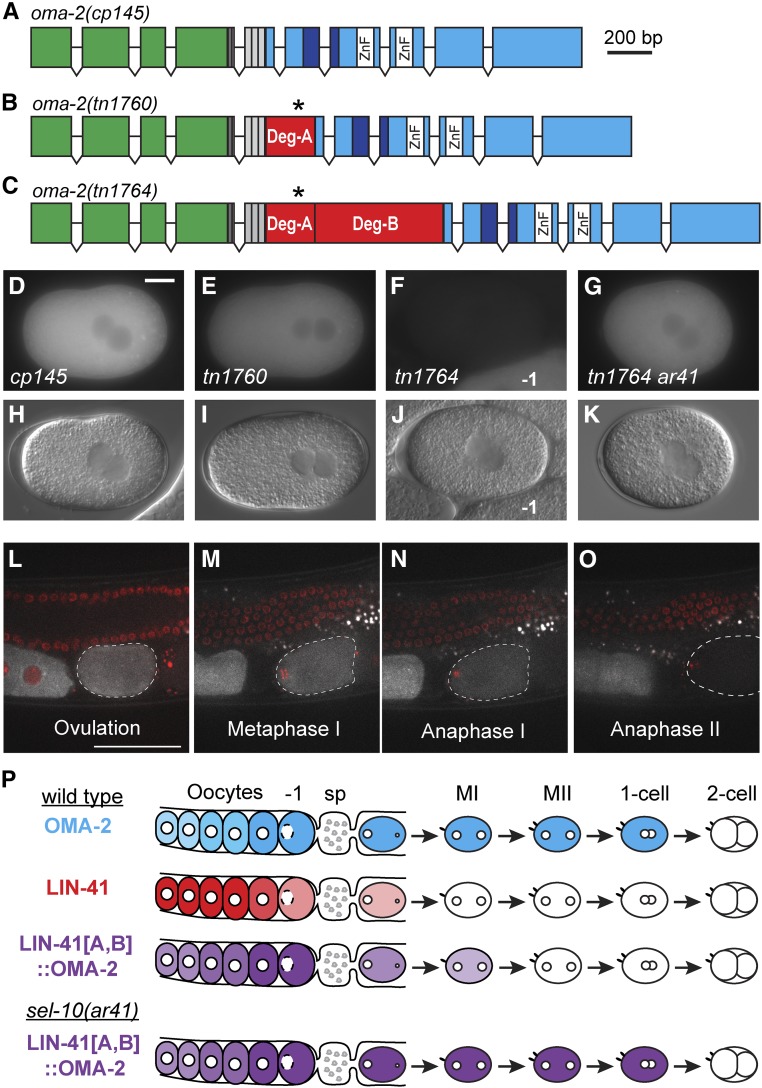
LIN-41 degradation domains when implanted into mNG::OMA-2 promote its rapid elimination during meiosis. (A–C) The exon–intron structures of oma-2(cp145[mng::tev::3xflag::oma-2]), oma-2(tn1760[mng::tev::3xflag::deg-a::oma-2]), and oma-2(tn1764[mng::tev::3xflag::deg-a::deg-b::oma-2]). Boxes represent exonic regions that encode mNeonGreen (green), the tobacco etch virus cleavage site (TEV; dark gray), FLAG epitope tags (light gray), LIN-41 Deg-A and Deg-B domains (red), the likely TAF-4 interaction domain of OMA-2 (dark blue), two OMA-2 CCCH zinc fingers (white), and other OMA-2 coding sequences (cyan). The position of LIN-41 T83 within the LIN-41 Deg-A domain is indicated by an asterisk. (D–K) GFP (D–G) and DIC (H–K) images of oma-2(cp145) (D and H), oma-2(tn1760) (E and I), oma-2(tn1764) (F and J), and oma-2(tn1764) lon-3(e2175) sel-10(ar41) (G and K) one-cell embryos at pronuclear meeting (E and I), or just slightly later, as the pronuclei begin a counterclockwise rotation (D, F–G, H, J, and K) prior to nuclear envelope breakdown and the first mitotic division. Part of a −1 oocyte is visible in F and J, and is indicated for reference. 150 ms GFP exposures; Bar, 10 μm. (L–O) Time-lapse images of mNG::Deg-A,B::OMA-2 (white) and mCHERRY::HISTONE-labeled chromosomes (red) were acquired in a living oma-2(tn1764); itIs37[pie-1p::mCherry:::H2B::pie-1 3′UTR, unc-119(+)] adult hermaphrodite by confocal microscopy. Images are shown for select time points (t) at ovulation (L, t = 0 min), during the first (M, t = +5 min; N, t = +10.5 min) and second meiotic divisions (O, t = +24.5 min) as an embryo (dashed outline) progresses through both meiotic divisions. See Movie S3 for the complete time-lapse sequence. Bar, 50 μm. (P) A visual summary of the dynamic expression patterns of mNG::OMA-2 (cyan), GFP::LIN-41 (red), and mNG::Deg-A,B::OMA-2 (purple). Oocytes are to the left and embryos are to the right of the spermatheca (sp). Meiotic embryos (MI, MII) have completed their respective divisions.
Table 2. Sterility and embryonic lethality in oma-2 and oma-1; oma-2 mutant strains at 20°.
| Genotype | Embryos laida | Dead embryos, % |
|---|---|---|
| oma-2(cp145) | 314 ± 48 (n = 6) | 0.6 (n = 1793) |
| oma-2(tn1760) | 306 ± 39 (n = 6) | 0.6 (n = 1835) |
| oma-2(tn1764) | 300 ± 45 (n = 6) | 2.1 (n = 1797) |
| oma-2(tn1764) lon-3(e2175) sel-10(ar41) | 288 ± 26 (n = 6) | 1.1 (n = 1727) |
| oma-1(zu405te33) | 261 ± 18 (n = 6) | 0.8 (n = 1568) |
| oma-1(zu405te33); oma-2(te51) M+Z–b | 0 (n > 21)c | NA |
| oma-1(zu405te33); oma-2(cp145) | 212 ± 29 (n = 12) | 12.3d (n = 2516) |
| oma-1(zu405te33); oma-2(tn1760) | 224 ± 35 (n = 6) | 60.3 (n = 1341) |
| oma-1(zu405te33); oma-2(tn1764) M+Z–b,e | 249 ± 29 (n = 5) | 100 (n > 1256) |
| oma-1(zu405te33); oma-2(tn1764) lon-3(e2175) sel-10(ar41) M+Z–b,f | 246 ± 32 (n = 6) | 89.4 (n = 1476) |
| oma-1(zu405te33); oma-2(tn1764) lon-3(e2175) sel-10(ar41) | 196 ± 56 (n = 6) | 84.9 (n = 1175) |
Average number of embryos laid per worm ± SD.
M+Z– animals were the progeny of nT1[qIs51] balancer-containing parents, which are heterozygous for both oma-1 and oma-2. All other animals were the progeny of parents of the listed genotype.
Sterile, with a defect in meiotic maturation as described by Detwiler et al. (2001).
Percent embryo lethality was variable among the 12 parents analyzed; it ranged between 6 and 35%.
These animals lay many eggs, none of which hatch (n = 30).
These animals lay many eggs, some of which hatch (n = 24).
We began by testing LIN-41 Deg-A, which contains T83, the potential CDK-1 target site required for the elimination of LIN-41. oma-2(tn1760) mutants express mNG::Deg-A::OMA-2 in oocytes and one-cell embryos. Similar to mNG::OMA-2, this protein is present in one-cell embryos just prior to the first mitotic division, but is eliminated from older embryos (Figure 3, D and E, and Figure S7, A, B, E, and F). However, we did note that the amount of mNG fluorescence in older one-cell pronuclear stage embryos was reduced 36% in oma-2(tn1760) embryos compared to oma-2(cp145) controls (compare Figure 3, D and E, and Figure S8A). This reduction might be caused by Deg-A-mediated destabilization of the mNG::OMA-2 fusion protein (see below), but is not equivalent to the rapid elimination of GFP::LIN-41 that occurs in meiosis I (Movie S1 and Figure 1). Because Deg-A on its own was not sufficient to trigger the rapid elimination of mNG::OMA-2 from one-cell embryos, we tested LIN-41 Deg-A and Deg-B together. oma-2(tn1764) mutants express mNG::Deg-A, Deg-B::OMA-2 in oocytes, but in one-cell embryos the amount of mNG fluorescence is substantially reduced or absent (Figure 3F and Figure S7, G and I). To more precisely determine the stage at which mNG::Deg-A, Deg-B::OMA-2 is eliminated, we used time-lapse imaging (Figure 3, L–O and Movie S3). Levels of this fusion protein drop by ∼50% during the first meiotic division (Figure 3, M and N) and become essentially undetectable before the end of the second meiotic division (Figure 3O). We conclude that Deg-A and Deg-B are sufficient in combination to trigger the rapid degradation of mNG::OMA-2 during meiosis, although this event is temporally delayed relative to GFP::LIN-41 (Figure 3P).
oma-1 and oma-2 share redundant functions during both oocyte and early embryo development (Detwiler et al. 2001; Guven-Ozkan et al. 2008). Double mutants carrying strong loss-of-function alleles [e.g., oma-1(zu405te33); oma-2(te51)] are sterile with a defect in meiotic maturation (Detwiler et al. 2001; Table 1). For the most part, the embryonic functions of oma-1/2 have been studied using conditions that reduce, but do not eliminate, OMA-1/2 function in embryos, such as double RNAi or reduction-of-function alleles that are incompletely sterile (Nishi and Lin 2005; Guven-Ozkan et al. 2008). In oma-1(zu405te33); oma-2(tn1764) double mutants, OMA-2 is expressed during oogenesis but eliminated prematurely from embryos. Consequently, these double mutants are very fertile but produce ∼249 progeny that die during embryogenesis (Table 2). Thus, as a novel allele that specifically reduces embryonic OMA-2, oma-2(tn1764) may be useful for studying the embryonic functions of oma-1/2. Our initial observations indicate that young oma-1(zu405te33); oma-2(tn1764) embryos exhibit cell division defects and ectopic cleavage furrows (Figure S8B); similar defects have been reported after oma-1/2(RNAi) depletion (Li et al. 2009).
When combined with oma-1(zu405te33), oma-2(tn1764) exhibits a stronger embryonic phenotype than either oma-2(cp145) or oma-2(tn1760). However, the severity of the oma-1(te33zu405); oma-2(tn1760) double-mutant phenotype relative to oma-1(te33zu405); oma-2(cp145) was surprising (Table 2; 60% vs. 12% embryonic lethality). One possibility for the increased severity of the oma-1(te33zu405); oma-2(tn1760) double-mutant embryonic phenotype might be the reduction in mNG::Deg-A::OMA-2 levels observed in oma-2(tn1760) pronuclear-stage embryos (Figure 3E and Figure S8A). We examined this more closely by crossing each mNG-tagged oma-2 allele into an emb-30(tn377ts) mutant background. emb-30 encodes a subunit of the Anaphase Promoting Complex (APC), and adult emb-30(tn377ts) hermaphrodites upshifted to a restrictive temperature (25°) produce one-cell embryos that arrest in the metaphase of the first meiotic division (Furuta et al. 2000). Arrest in meiotic metaphase does not prevent or delay the elimination of GFP::LIN-41, which is independent of APC function (Spike et al. 2014a). We observed that mNG::OMA-2 is turned over in arrested meiotic embryos, but could typically be seen in four embryos in the uterus of emb-30(tn377ts); oma-2(cp145) hermaphrodites after a 5–7 hr upshift to 25° (Figure S8C). In contrast, both of the LIN-41 Deg domain-containing OMA-2 proteins appeared to be less stable under the same conditions. mNG::Deg-A, Deg-B::OMA-2 was seen in 0–1 mNG-positive embryos and appeared to be the least stable (Figure S8E), as expected from our previous analysis (Figure 3 and Figure S7). mNG::Deg-A::OMA-2 was seen in two mNG-positive embryos and therefore appeared to be of intermediate stability (Figure S8D). Thus, although Deg-A is not sufficient for rapid elimination, it may reduce the stability of mNG::Deg-A::OMA-2 in meiotic embryos. The consequent reduction in protein levels could contribute to the enhanced severity of the oma-1(te33zu405); oma-2(tn1760) double-mutant embryonic phenotype. It is also possible that insertion of LIN-41 Deg-A at the N terminus of OMA-2 might perturb the nearby TAF-4-binding domain (Figure 3, A–C), which is critical for the function of OMA-2 in embryos (Guven-Ozkan et al. 2008). It is important to note that OMA-1 proteins with deletions in the TAF-4-binding domain are stable in one-cell embryos (Guven-Ozkan et al. 2008) and this is expected to be true for OMA-2. Therefore, we conclude that Deg-A by itself can confer destabilization activity to OMA-2, albeit less potent than that conferred by Deg-A and Deg-B in combination.
GFP::LIN-41 is eliminated from embryos by the SCFSEL-10 E3 ubiquitin ligase
Several different SCF-containing E3 ubiquitin ligase complexes promote protein degradation during meiosis and early embryogenesis in C. elegans (Peel et al. 2012; Du et al. 2015; Beard et al. 2016). We initially used RNAi to knock down the functions of each of the six cullins identified in the C. elegans genome (Kipreos et al. 1996; Nayak et al. 2002) to determine whether an SCF-type E3 ligase is involved in the elimination of GFP::LIN-41. In general, RNAi was initiated in lin-41(tn1541) hermaphrodites at the L4 larval stage and GFP::LIN-41 was examined in adults, 2 days after the initiation of the RNAi treatment at 22°. Because our cul-2(RNAi) clone was ineffective (see Materials and Methods), we also examined GFP::LIN-41 in cul-2(or209ts) mutant animals at the restrictive temperature (n = 71). Only the cul-1(RNAi)-treated animals produced multiple young embryos with faint GFP::LIN-41 (n = 12), suggesting that CUL-1 may be required to eliminate GFP::LIN-41 from embryos. rrf-1(pk1417) mutants are RNAi-defective in certain somatic cells, including the somatic gonad, but are sensitive to RNAi in the germline (Sijen et al. 2001; Kumsta and Hansen 2012). Treatment of rrf-1(pk1417) lin-41(tn1541) hermaphrodites with cul-1(RNAi) also resulted in the failure to eliminate GFP::LIN-41 from early embryos (n = 54; Figure 4C). Together, these results suggest that a germline-expressed CUL-1-containing SCF E3 ubiquitin ligase may eliminate GFP::LIN-41 from early embryos.
Figure 4.
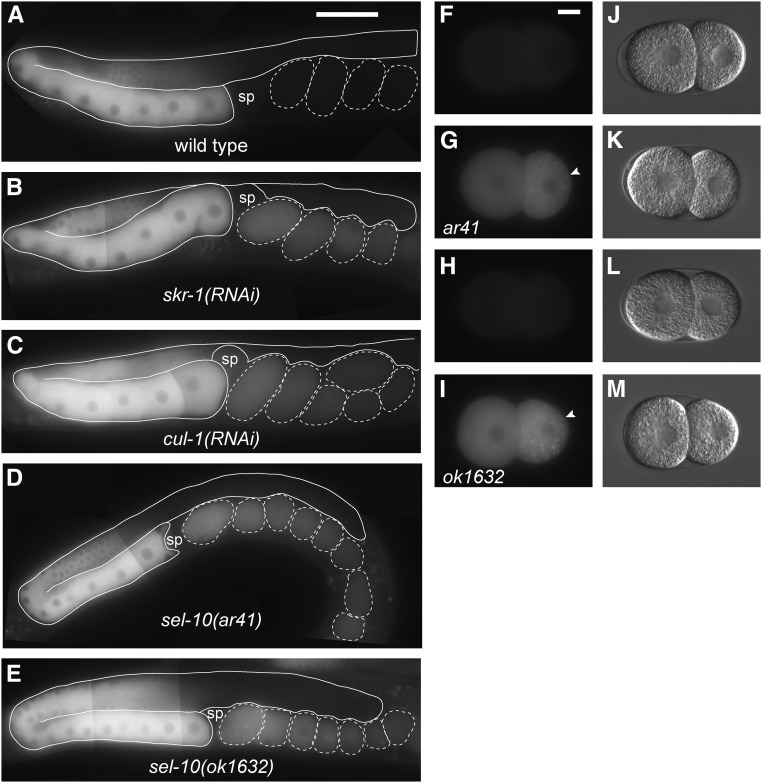
Subunits of the SCFSEL-10 E3 ubiquitin ligase are required for the elimination of GFP::LIN-41 from early embryos. (A–E) Composite images of GFP::LIN-41 in adult rrf-1(pk1417) lin-41(tn1541) hermaphrodites fed control RNAi bacteria (A), and adult hermaphrodites with reduced SCFSEL-10 E3 ubiquitin ligase activity (B–E): lin-41(tn1541); skr-1(RNAi) (B), rrf-1(pk1417) lin-41(tn1541); cul-1(RNAi) (C), lin-41(tn1541); lon-3(e2175) sel-10(ar41) (D), and lin-41(tn1541); sel-10(ok1632) (E); 100 ms GFP exposures, brightened slightly (and equivalently) to better visualize embryonic GFP::LIN-41; Bar, 50 μm. (F–M) Images of two-cell embryos removed from the uterus of hermaphrodites were imaged for GFP (F–I) and DIC (J–M); the genotypes were as follows: lin-41(tn1541); lon-3(e2175) (F and J), lin-41(tn1541); lon-3(e2175) sel-10(ar41) (G and K), lin-41(tn1541) (H and L), and lin-41(tn1541); sel-10(ok1632) (I and M). Arrowheads indicate a few of the GFP::LIN-41 aggregates in the posterior blastomeres of sel-10 mutant embryos, which likely correspond to P granules; 300 ms GFP exposures; Bar, 10 μm. sp, spermatheca.
In SCF-type E3 ligases, cullins interact with Skp1-related proteins. Twenty-one Skp1-related (skr) genes have been identified in C. elegans, and RNAi experiments suggest the closely related skr-1 and skr-2 genes function in the germline and early embryo (Nayak et al. 2002; Yamanaka et al. 2002; Shirayama et al. 2006; Fox et al. 2011; Mohammad et al. 2018). In addition, both SKR-1 and SKR-2 can interact with CUL-1 (Nayak et al. 2002; Yamanaka et al. 2002). We therefore examined whether skr-1(RNAi), which likely reduces the function of both skr-1 and skr-2, would prevent the elimination of GFP::LIN-41 from early embryos. lin-41(tn1541); skr-1(RNAi) animals produced embryos with defects in the elimination of GFP::LIN-41 2 days after RNAi treatment (n = 26; Figure 4B). Treatment of rrf-1(pk1417) lin-41(tn1541) animals with skr-1(RNAi) also prevented the elimination of GFP::LIN-41 from early embryos (n = 14). In addition, the rrf-1(pk1417) lin-41(tn1541) mutants treated with skr-1(RNAi) for 2 days at 22° exhibited defects in germline morphology and embryo production that are consistent with the phenotypes previously described after skr-1/2(RNAi) (Nayak et al. 2002).
At least three F-box-containing substrate recognition subunits, LIN-23, PROM-1, and SEL-10, are thought to function with either SKR-1 or SKR-2 and CUL-1 in the C. elegans germline or early embryos (Peel et al. 2012; Du et al. 2015; Kisielnicka et al. 2018; Mohammad et al. 2018). We used RNAi to knock down the activities of lin-23 and sel-10 as a first step toward the analysis of candidate F-box proteins. lin-23(RNAi) had no effect on the elimination of GFP::LIN-41 from rrf-1(pk1417) lin-41(tn1541) embryos (n = 52; Figure S9C). Consistent with this observation, mutations designed to prevent the phosphorylation of a possible β-TrCP/LIN-23-binding site near the amino terminus of LIN-41 (amino acids 32–38) also do not prevent the elimination of GFP::LIN-41 from embryos [lin-41(tn1541tn1668); Figure S5, D and E]. However, sel-10(RNAi) did prevent the elimination of GFP::LIN-41 from rrf-1(pk1417) lin-41(tn1541) embryos (n = 17). Similarly, the elimination of GFP::LIN-41 from young embryos is prevented by the strong loss-of-function mutations sel-10(ok1632) and sel-10(ar41) (Figure 4, D, E, G, and I). Whereas strong loss-of-function sel-10 mutant alleles abrogate the rapid elimination of GFP::LIN-41 that commences upon the onset of meiotic maturation, we note that the ∼twofold decline in GFP::LIN-41 levels that occurs as oocytes progress to the −1 position (Figure 1H) occurs independently of sel-10 (Figure 4, A, D, and E). Genetic and physical interactions indicate that SEL-10 and SKR-1 function together in C. elegans (Killian et al. 2008; Kisielnicka et al. 2018). Because our skr-1(RNAi) experiments are likely to also target skr-2 (Nayak et al. 2002), we are unable to parse out the relative roles of SKR-1 and SKR-2 at this time. Collectively, these observations suggest that a germline-expressed SCFSEL-10 E3 ubiquitin ligase containing SKR-1/2, CUL-1, and SEL-10 is likely involved in the elimination of GFP::LIN-41 from early embryos (Figure 5E).
Figure 5.
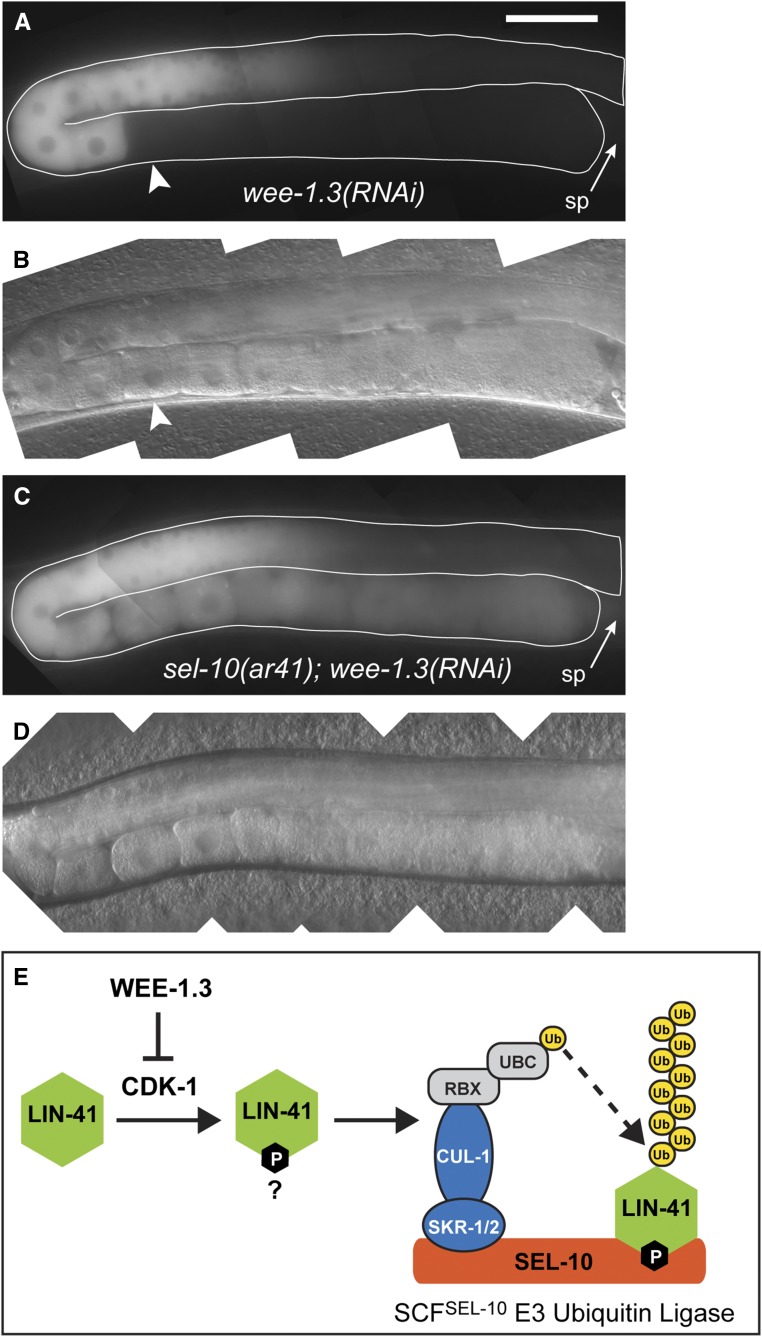
SEL-10 is required for the WEE-1.3–inhibited degradation of GFP::LIN-41. (A–D) Composite GFP (A and C) and DIC (B and D) images of lin-41(tn1541); lon-3(e2175); wee-1.3(RNAi) (A and B) and lin-41(tn1541); lon-3(e2175) sel-10(ar41); wee-1.3(RNAi) (C and D) animals. GFP::LIN-41 is prematurely eliminated from oocytes by wee-1.3(RNAi) (arrowhead), but persists in abnormal oocytes near the spermatheca (sp; arrow) in sel-10(ar41); wee-1.3(RNAi) animals (C and D), suggesting that SEL-10 is required for this process; 150 ms GFP exposures, brightened slightly; Bar, 50 μm. (E) A simple model for the elimination of LIN-41 (green) that incorporates the known molecular functions of WEE-1.3 kinase, cyclin-dependent kinase (CDK-1) and subunits of the SCFSEL-10 E3 ubiquitin ligase. In brief, we hypothesize that SEL-10 (orange) may recognize phosphorylated LIN-41 (green) and trigger its ubiquitin-mediated degradation in collaboration with the other SCF E3 ubiquitin ligase subunits, SKR-1/2 (blue), and CUL-1 (blue). CUL-1 orthologs bind RING finger proteins (RBX; gray), which recruit a ubiquitin-conjugating enzyme (UBC; gray) that catalyzes the transfer of ubiquitin (yellow) to protein substrates, such as LIN-41. Subsequent recruitment of polyubiquitinated substrates to the proteasome results in degradation (not shown). This model is consistent with the epistatic relationship between wee-1.3(RNAi) and sel-10(ar41) with respect to the elimination of GFP::LIN-41, but other models are also possible.
SEL-10 functions through LIN-41 degradation domains
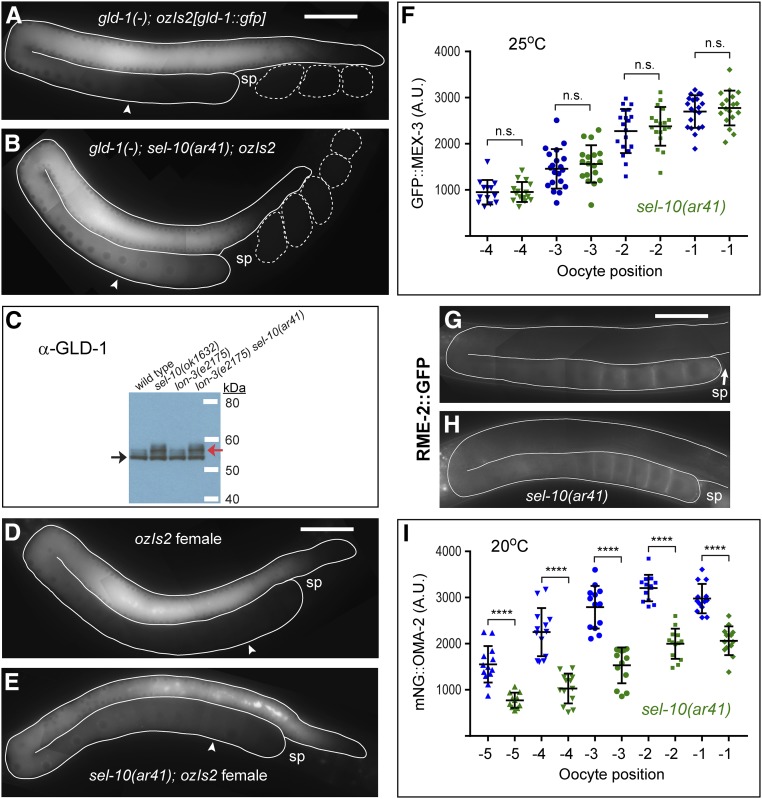
LIN-41 can be detected in sel-10(ok1632) mutant but not wild-type embryos by Western blot analysis (Figure S6D), indicating that endogenous and GFP-tagged LIN-41 behave similarly. We hypothesized that the Deg domains are likely used to target LIN-41 for degradation by SCFSEL-10. To test this hypothesis, we examined whether the premature elimination of mNG::Deg-A, Deg-B::OMA-2, which is mediated by the LIN-41 Deg domains, is prevented in sel-10 mutant embryos. Although mNG::Deg-A, Deg-B::OMA-2 is eliminated by the pronuclear stage in otherwise wild-type one-cell embryos, mNG::Deg-A, Deg-B::OMA-2 levels remain high in lon-3(e2175) sel-10(ar41) embryos at the same stage of embryonic development (Figure 3, F, G, J, K, and P and Figure S7, G, H, K, and L). These observations suggest that sel-10(ar41) should suppress the completely penetrant maternal-effect lethal phenotype exhibited by oma-1(zu405te33); oma-2(tn1764) mutants. Consistent with this expectation, oma-1(zu405te33); oma-2(tn1764) lon-3(e2175) sel-10(ar41) animals produce hatchlings and can be maintained as a homozygous strain (Table 2). However, sel-10(ar41) is a relatively weak suppressor of the oma-1(zu405te33); oma-2(tn1764) maternal-effect lethal mutant phenotype, since only 10–15% of the embryos produced by oma-1(zu405te33); oma-2(tn1764) lon-3(e2175) sel-10(ar41) animals hatch. This observation is consistent with the possibility described above that the LIN-41 Deg domains might perturb the TAF-4-binding function of OMA-2 in the one-cell embryo. Additionally, sel-10(ar41) mildly reduces mNG::OMA-2 accumulation in the germline, likely through effects on GLD-1 (see below).
Although the degradation of OMA-1, and presumably OMA-2, appears to be mediated by several SCF E3 ubiquitin ligases, SEL-10 has not been implicated in this process (Shirayama et al. 2006; Du et al. 2015). Indeed, mNG::OMA-2 is degraded at the expected time in oma-2(cp145) lon-3(e2175) sel-10(ar41) embryos (Figure S7, C and D). Likewise, mNG::Deg-A, Deg-B::OMA-2 levels only remain high until the end of the one-cell stage in oma-2(tn1764) lon-3(e2175) sel-10(ar41) embryos, when the degradation of OMA-2 is normally initiated (Figure S7, K and L). We conclude that SEL-10 is not required for the elimination of OMA-2 and likely functions through the LIN-41 Deg domains to promote the proteolytic degradation of LIN-41 and mNG::Deg-A, Deg-B::OMA-2 during meiosis.
SEL-10 is required for the CDK-1-dependent elimination of GFP::LIN-41
LIN-41 Deg domains contain potential Cdc4 phosphodegron sequences:
Substrate recognition subunits such as SEL-10 recognize their targets by binding to short linear sequence motifs called degrons (Lucas and Ciulli 2017). LIN-41 Deg domains were therefore examined for sequences similar to the SEL-10/Fbw7/Cdc4 degron consensus sequence ΦΦ[pT/pS]PXX[pT/pS/E/D], where Φ represents a hydrophobic amino acid. This degron is commonly referred to as a Cdc4 phosphodegron or CPD; it contains two essential residues, a phosphorylated residue that is typically a phosphothreonine, immediately followed by a proline (pTP) (Nash et al. 2001). SEL-10 appears to be recruited to its substrates via CPD sequences (de la Cova and Greenwald 2012). Residues surrounding LIN-41 T83, which is important for the elimination of LIN-41 from embryos, are poor matches to the CPD consensus sequence (sequence FDTPPSM, mismatches are underlined; Figure S10). The best match to a high-affinity CPD appears to be around residue T340 in the LIN-41 Deg-B2 domain (sequence LATPMSS; Figure S10), which is the only candidate Fbw7-binding site identified in LIN-41 using the Eukaryotic Linear Motif database (Gouw et al. 2018). However, changing T340 to an alanine (T340A) [e.g., lin-41(tn1541tn1775] has no effect on the elimination of GFP::LIN-41 from embryos (Figure 2C and Figure S4, K and L). Therefore, if SEL-10 binds directly to LIN-41 Deg domains it might recognize imperfect or lower-affinity degrons. We note that the SEL-10 ortholog Cdc4p utilizes multiple imperfect degrons to target the cell division protein Sic1p for degradation (Nash et al. 2001). Likewise, multiple weak degrons in an intrinsically disordered region of the c-Jun protein synergize to promote a high-affinity interaction with the SEL-10 ortholog Fbw7 (Csizmok et al. 2018). It seems plausible that SEL-10 might function similarly. Alternatively, the failure to eliminate LIN-41 from embryos could be an indirect consequence of the lack of SCFSEL-10. To begin to address this possibility, we sought to clarify the epistatic relationships between sel-10 and other factors involved in the elimination of GFP::LIN-41 from embryos.
SEL-10 functions downstream or in parallel to CDK-1:
CDK-1 was previously shown to be required for the elimination of GFP::LIN-41 (Spike et al. 2014a). Likewise, cdk-1(RNAi) on rrf-1(pk1417) lin-41(tn1541) hermaphrodites prevents the elimination of GFP::LIN-41 from embryos (n = 67; Figure S9, A and B). Therefore, germline-expressed CDK-1 likely promotes the elimination of GFP::LIN-41. CDK-1 is a conserved and essential cell cycle regulator required for M phase entry and progression during both meiotic and mitotic cell divisions (Boxem et al. 1999). Consequently, most cdk-1 alleles are sterile, precluding the examination of GFP::LIN-41 in cdk-1 mutant embryos. Two temperature-sensitive alleles of cdk-1 that produce oocytes have been described; both cause a later embryonic arrest phenotype than the one-cell meiotic arrest phenotype seen after cdk-1(RNAi) (Boxem et al. 1999; Shirayama et al. 2006). Furthermore, although both mutations alter residues in the T-loop/activation domain of CDK-1, neither cdk-1(ts) allele causes obvious cell cycle defects (Shirayama et al. 2006). We examined GFP::LIN-41 in cdk-1(ne2257ts) animals at the restrictive temperature and found that GFP::LIN-41 disappears normally from embryos (n = 57; Figure S9, E and F). Similarly, GFP::LIN-41 disappears normally in cks-1(ne549ts) mutant embryos (n = 33; Figure S9, G and H), which phenotypically resemble cdk-1(ne2257ts) embryos at the restrictive temperature (Shirayama et al. 2006). Thus, the subset of CDK-1 activities affected by cdk-1(ne2257ts) does not include either the elimination of GFP::LIN-41 or entry into meiotic M phase.
SEL-10/Fbw7/Cdc4p degrons are only activated after being phosphorylated by a proline-directed serine/threonine kinase such as CDK-1 (Koepp et al. 2001; Nash et al. 2001; Strohmaier et al. 2001; Welcker and Clurman 2008). Thus, prior phosphorylation by CDK-1 might be required for SEL-10 to directly interact with degrons in the LIN-41 Deg domains. cdk-1(RNAi) and sel-10(lf) cause the same phenotype with respect to GFP::LIN-41 degradation, precluding a direct analysis of their epistatic relationship. However, it is possible to examine this relationship indirectly through wee-1.3 (Burrows et al. 2006). GFP::LIN-41 is eliminated prematurely when wee-1.3 function is attenuated by RNAi (n = 21; Figure 5, A and B; Spike et al. 2014a). When the same experiment is performed in lon-3(e2175) sel-10(ar41) mutants, however, GFP::LIN-41 is not eliminated prematurely. Instead, GFP::LIN-41 persists, typically at reduced levels, in the proximal oocytes of lon-3(e2175) sel-10(ar41); wee-1.3(RNAi) animals (n = 51; Figure 5, C and D). Because CDK-1 is prematurely activated, wee-1.3(RNAi) oocytes mature prematurely and exhibit numerous defects (Burrows et al. 2006). Obvious oocyte abnormalities caused by strong wee-1.3(RNAi) are evident in sel-10(ar41) mutants, confirming that these animals are competent to respond to wee-1.3(RNAi) (Figure 5, compare B and D). We conclude that the epistatic relationship between wee-1.3 and sel-10 is consistent with the model shown in Figure 5E, which postulates that active CDK-1 promotes the phosphorylation of LIN-41 and its subsequent destruction by an SCFSEL-10 E3 ubiquitin ligase.
MPK-1 MAP kinase is dispensable for LIN-41 degradation:
Kinases other than CDK-1 might play a role in the SEL-10-mediated elimination of LIN-41. However, our attempts to identify additional kinases that affect the elimination of GFP::LIN-41 have so far been unsuccessful. For example, the MAP kinase MPK-1 is active in the late stages of oogenesis and is an important regulator of oocyte meiotic maturation (Lee et al. 2007). Furthermore, as a proline-directed serine/threonine kinase, MPK-1 could potentially phosphorylate CPDs in LIN-41 Deg domains. However, GFP::LIN-41 disappears normally in mpk-1(ga111ts) embryos at the restrictive temperature (n = 97; Figure S9, I–L). We found no evidence that GFP::LIN-41 degradation involves the AIR-2/Aurora B kinase, GSK-3/glycogen synthase kinase, CDK-2, PLK-1/polo-like kinase, or MBK-2/minibrain kinase (Figure S9; Spike et al. 2014a). Since LIN-41 functions to inhibit CDK-1 activation for M phase entry during meiotic maturation (Spike et al. 2014a), CDK-1 may be the chief effector kinase mediating feedback regulation of wild-type LIN-41.
Embryonic LIN-41 does not strongly inhibit the expression of mRNAs repressed by LIN-41
LIN-41 represses the translation of several different mRNAs during oogenesis (Spike et al. 2014b; Tsukamoto et al. 2017). Their protein products normally begin to accumulate in late oogenesis or early embryogenesis, and some are essential for normal development (Gomes et al. 2001; Leacock and Reinke 2008; Tsukamoto et al. 2017). We therefore anticipated that the failure to eliminate LIN-41 would result in the ectopic repression of these mRNAs, and that this, in turn, might result in embryo or oocyte abnormalities. However, the lin-41(tn1767) mutant, which fails to eliminate LIN-41[T83A] from early embryos, appears essentially wild type at 20° (Table 1). Similarly, sel-10(ok1632) and sel-10(ar41) mutants, which fail to eliminate LIN-41 from early embryos, produce large broods of progeny at 20° that are similar in size to genotype-matched controls (Table 3). Indeed, we only observed a moderate decrease in fertility when sel-10(ok1632) mutants were grown at an elevated temperature (25°; Table 3). We therefore decided to examine the amount of protein made by several LIN-41 target mRNAs (spn-4, meg-1, and orc-1 mRNAs, respectively) in strains that fail to eliminate LIN-41 from embryos, as this should provide a sensitive way to monitor LIN-41 translational repression activity. Protein expression was examined using fluorescence-tagged alleles of each gene; the proteins made by spn-4(tn1699[spn-4::gfp::3xflag]), meg-1(tn1724[gfp::3xflag::meg-1]), and orc-1(tn1732[mng::3xflag::orc-1) were previously shown to be ectopically or prematurely expressed in lin-41(lf) oocytes (Tsukamoto et al. 2017). As described below, only minor differences in protein expression were observed in lin-41(tn1767) and sel-10(ar41) embryos and oocytes (Figure 6 and Figure S11). Collectively, these observations suggest that the ectopic LIN-41 present in lin-41(tn1767) and sel-10(lf) embryos is largely ineffective at repressing translation.
Table 3. sel-10 mutant brood sizes at 20 and 25°.
| Genotype | Temperature | Brood size |
|---|---|---|
| Wild type | 20 | 304.0 ± 31.1 (n = 29) |
| Wild typea | 25 | 266.8 ± 39.0 (n = 19) |
| sel-10(ok1632)a | 20 | 258.3 ± 67.7 (n = 30) |
| sel-10(ok1632)a | 25 | 72.2 ± 34.0b (n = 30) |
| lin-41(tn1487ts); sel-10(ok1632)c | 20 | 3.2 ± 3.4 (n = 53) |
| lin-41(tn1487ts)d | 20 | 41.4 ± 23.8 (n = 36) |
| lon-3(e2175) | 20 | 294.5 ± 36.7 (n = 20) |
| lon-3(e2175) sel-10(ar41) | 20 | 280.2 ± 40.1 (n = 20) |
Newly fertilized embryos were collected at 15° and shifted to 25°.
Approximately 10.0 ± 5.3% of sel-10(ok1632) hermaphrodites (n = 3568) are infertile, exhibiting incompletely penetrant sterility or maternal-effect lethality when grown and examined for seven generations at 25°.
The progeny of sel-10(ok1632); lin-41(tn1487ts)/hT2[qIs48] hermaphrodites.
The progeny of lin-41(tn1487ts)/hT2(qIs48) hermaphrodites.
Figure 6.
Persisting LIN-41 or LIN-41[T83A] does not strongly inhibit the expression of LIN-41 targets of translational repression in young embryos. (A–J) Young embryos express similar levels of SPN-4::GFP (A, B, G, and H), GFP::MEG-1 (C, D, I, and J) and mNG::ORC-1 (arrowhead in E and F) when ectopic LIN-41[T83A] [B, D, and F; lin-41(tn1767) mutant embryos], ectopic LIN-41 [H and J; sel-10(ar41) mutant embryos] or normal (undetectable) levels of LIN-41 (A, C, E, G, and I) are present. Exposures were 100 ms for SPN-4::GFP, 200 ms for GFP::MEG-1, and 600 ms for mNG::ORC-1; Bar, 10 μm. (K and L) Quantification of the intensity of SPN-4::GFP expression in wild type, lin-41(tn1767), and sel-10(ar41) genetic backgrounds. Statistical significance was evaluated using an unpaired t-test. The plots show the mean and SD. (K) Comparison of spn-4(tn1699) and lin-41(tn1767); spn-4(tn1699) one- and two-cell embryos; no significant differences were seen (n.s.). (L) Comparison of spn-4(tn1699); lon-3(e2175) and spn-4(tn1699); lon-3(e2175) sel-10(ar41) one- and two-cell embryos. Levels appeared to be slightly lower in the sel-10(ar41) two-cell embryos (P < 0.001). Note that the slightly reduced level of SPN-4::GFP in H relative to G accurately illustrates the very modest magnitude of this difference in expression.
LIN-41 mediates 3′-UTR-dependent translational repression of spn-4, and spn-4 mRNA is the most abundant and enriched mRNA in LIN-41 RNPs (Tsukamoto et al. 2017). SPN-4::GFP is faint, but visible, in one or two proximal oocytes and rapidly accumulates during the OET (Tsukamoto et al. 2017). This pattern, and the amount of SPN-4::GFP in early embryos, is largely unaffected by lin-41(tn1767) and sel-10(ar41) at 20° (Figure 6, A, B, G, and H, and Figure S11, A, B, K, and L). Quantification of GFP fluorescence levels revealed no differences in SPN-4::GFP levels in lin-41(tn1767); spn-4(tn1699) one- and two-cell embryos and a slight reduction in SPN-4::GFP levels in spn-4(tn1699) lon-3(e2175) sel-10(ar41) two-cell embryos relative to age and genotype-matched controls (Figure 6, K and L). Finally, we also failed to identify any apparent differences in SPN-4::GFP accumulation or intensity in spn-4(tn1699) lon-3(e2175) sel-10(ar41) (n = 26) and spn-4(tn1699) lon-3(e2175) (n = 19) animals upshifted as L4 larvae to 25°.
GFP::MEG-1 expression is evident somewhat earlier in oogenesis than SPN-4::GFP and seems to accumulate more slowly. Again, the pattern and amount of GFP::MEG-1 was largely unaffected in lin-41(tn1767) and sel-10(ar41) mutants at 20° (Figure 6, C, D, I, and J, and Figure S11, C, D, G, and H). GFP::MEG-1 has a complex pattern of accumulation in the early embryo; it appears to be eliminated from somatic blastomeres and localizes, at least partially, to P granules (Figure 6C and Figure S11C), similar to what was previously described by Leacock and Reinke (2008) for endogenous MEG-1. Because of these complexities, we quantified GFP::MEG-1 levels in the cytoplasm of proximal oocytes instead of embryos. There were no differences in GFP::MEG-1 levels in lin-41(tn1767); meg-1(tn1724) animals and only a slight reduction in the −1 oocytes of lon-3(e2175) sel-10(ar41); meg-1(tn1724) animals relative to controls. Finally, we examined lon-3(e2175) sel-10(ar41); meg-1(tn1724) (n = 17) and lon-3(e2175); meg-1(tn1724) (n = 15) animals upshifted as L4 larvae to 25°, but again failed to identify any apparent differences in GFP::MEG-1 accumulation.
mNG::ORC-1 is not visibly expressed in oocytes but becomes increasingly evident in embryos as they develop (Tsukamoto et al. 2017). mNG::ORC-1 associates with chromatin at certain stages of the cell cycle (Sonneville et al. 2012), and is faintly visible in one-cell embryos during metaphase of the first mitotic division (Figure 6E). mNG::ORC-1 was only examined in lin-41(tn1767) mutants at 20°. As for SPN-4::GFP and GFP::MEG-1, the pattern and amount of mNG::ORC-1 was largely unaffected by lin-41(tn1767) (Figure 6, E and F and Figure S11, E and F). Most importantly, the small amount of mNG::ORC-1 visible in one-cell embryos was not obviously reduced in the lin-41(tn1767) background. Because LIN-41 is a potent translational repressor of spn-4, meg-1, and orc-1 (Tsukamoto et al. 2017), we conclude that a mechanism distinct from SCFSEL-10-mediated degradation antagonizes LIN-41 function to promote their expression during the late stages of oogenesis and the OET.
Because molecular tests failed to reveal an increase in LIN-41 activity in sel-10 mutants, we also tested the ability of a sel-10(ok1632) strong loss-of-function mutation to suppress the temperature-sensitive lin-41(tn1487ts) allele at a semipermissive temperature. This was found not to be the case; rather, sel-10(ok1632) enhanced the lin-41(tn1487ts) defects (Table 3). The basis for this enhancement is presently not clear. Taken together, these results indicate that the regulation of LIN-41 by sel-10 is nonessential.
SEL-10 promotes the elimination of GLD-1 from oocytes
GLD-1 is a translational repressor that, like LIN-41, controls and coordinates oocyte differentiation and cell cycle progression (Francis et al. 1995a,b; Jones et al. 1996). In gld-1(q485) null mutants, pachytene-stage oocytes re-enter the mitotic cell cycle and form a tumor (Francis et al. 1995a,b). GLD-1 also has redundant functions to inhibit the proliferative fate of germline progenitor cells and to promote their entry into the meiotic pathway of development during oogenesis and spermatogenesis, as well as a function to promote spermatogenesis in hermaphrodites (Francis et al. 1995a,b; Kadyk and Kimble 1998). GLD-1 is abundantly expressed during the early and middle stages of meiotic prophase, but eliminated from oocytes as they progress from late pachytene through diplotene and to diakinesis during the later stages of oocyte development (Jones et al. 1996). GLD-1 binds to, and represses the translation of, many mRNAs that are normally translated in oocytes (Lee and Schedl 2001, 2004; Schumacher et al. 2005; Jungkamp et al. 2011; Wright et al. 2011; Scheckel et al. 2012). Thus, it has generally been assumed that the elimination of GLD-1 from oocytes permits the translation of these mRNAs [reviewed in Lee and Schedl (2010)]. Although it occurs at an earlier stage of oocyte development, this model is analogous to what we originally hypothesized with respect to LIN-41. However, because the LIN-41 ectopically found in sel-10 loss-of-function embryos appears to be insufficient to sustain translational repression, it seems likely that the activity of LIN-41 is also regulated by a nonproteolytic mechanism. Given the similarities between LIN-41 and GLD-1, we wondered whether GLD-1 might also be regulated by both proteolytic and nonproteolytic mechanisms.
We investigated whether the elimination of GLD-1, like LIN-41, requires SEL-10. Surprisingly, we found that GLD-1::GFP and GLD-1 do indeed persist at elevated levels in the oocytes of sel-10(ar41) and sel-10(ok1632) mutants (Figure 7, A and B and Figure S12, A, B, D, and E), indicating that LIN-41 and GLD-1 may be regulated similarly. Indeed, while we were completing this work, the failure to eliminate GLD-1 in a timely fashion from sel-10(ok1632) mutant oocytes was independently discovered by Kisielnicka et al. (2018). Their results suggest that both GLD-1 and the cytoplasmic polyadenylation element-binding protein CPB-3 are likely degraded by essentially the same SCFSEL-10 E3 ubiquitin ligase (Kisielnicka et al. 2018) that regulates LIN-41 (this work). Consistent with this hypothesis, they observed that slow-migrating isoforms of GLD-1, which are likely phosphorylated (Jeong et al. 2011), accumulate in sel-10(ok1632) mutants. In agreement with this finding, we also observe an increase in the slow-migrating isoforms of GLD-1 in both sel-10(ok1632) and sel-10(ar41) mutants relative to controls (Figure 7C).
Figure 7.
GLD-1 persists at elevated levels in the oocytes of sel-10(ar41) mutants. (A and B) Composite images of GLD-1::GFP in gld-1(q485); lon-3(e2175); ozIs2[gld-1::gfp] (A) and gld-1(q485); lon-3(e2175) sel-10(ar41); ozIs2[gld-1::gfp] (B) adult hermaphrodites. GLD-1::GFP levels remain elevated in the proximal oocytes (e.g., −4 oocytes, arrowheads) of sel-10(ar41) animals (B) relative to controls (A); 17 ms GFP exposures, brightened slightly. (C) Slow-migrating forms of GLD-1 (red arrow) are more abundant in sel-10(lf) adult hermaphrodites than in sel-10(+) controls, where the fast-migrating form of GLD-1 (black arrow) predominates. (D and E) Composite images of GLD-1::GFP in fog-3(q470); lon-3(e2175); ozIs2[gld-1::gfp] (D) and fog-3(q470); lon-3(e2175) sel-10(ar41); ozIs2[gld-1::gfp] females (E). GLD-1::GFP levels are elevated in the proximal oocytes (e.g., −4 oocytes, arrowheads) of sel-10(ar41) females (B) relative to controls (A), although this is not as dramatic as in hermaphrodites. A somewhat longer GFP exposure (35 ms, brightened slightly) was needed than in A and B, likely due to the presence of endogenous GLD-1. (F) Quantification of the intensity of GFP::MEX-3 in the proximal oocytes of lon-3(e2175); mex-3(tn1753) and lon-3(e2175) sel-10(ar41); mex-3(tn1753) hermaphrodites at 25°. No significant differences were seen (n.s.). (G and H) Composite images of lon-3(e2175); pwIs116[rme-2p::rme-2::GFP::rme-2 3′UTR] (G) and lon-3(e2175) sel-10(ar41); pwIs116 [rme-2p::rme-2::GFP::rme-2 3′UTR] (H) hermaphrodites at 22°; 300 ms GFP exposures. Neither target of GLD-1 translational repression (MEX-3, RME-2) was strongly or even marginally reduced in expression in sel-10(ar41) oocytes. (I) Quantification of the intensity of mNG::OMA-2 in the proximal oocytes of oma-2(cp145) lon-3(e2175) and oma-2(cp145) lon-3(e2175) sel-10(ar41) hermaphrodites at 20°. Differences in expression were highly significant (**** P < 0.0001), but relatively modest in magnitude. For example, we measured a 37% reduction in average fluorescence in the −2 oocytes of sel-10(ar41) animals relative to the same oocytes in control animals. Statistical tests (F and I) employed an unpaired t-test. The plots show the mean and SD. All phenotypes were analyzed on the first day of adulthood. Bar, 50 μm (A, B, D, E, G, and H). sp, spermatheca.
It was recently proposed that sperm trigger the proteasome-dependent elimination of GLD-1 from oocytes such that a GFP::GLD-1 transgene (an N-terminal fusion) was expressed at higher levels in unmated females than in mated females or hermaphrodites (Bohnert and Kenyon 2017). Therefore we examined the localization of a rescuing GLD-1::GFP transgene (a C-terminal fusion) in both wild-type and sel-10(ar41) mutant females, which lack sperm. We used strong loss-of-function mutations in both fog-2 (Schedl and Kimble 1988) and fog-3 (Ellis and Kimble 1995) to feminize the germline. However, in our experiments, GLD-1::GFP did not persist at elevated levels in female oocytes; instead, it was eliminated from oocytes in both the presence and absence of sperm (Figure 7, A and D, and Figure S12, A and C). Likewise, endogenous GLD-1, detected with specific antibodies (Jan et al. 1999), also disappeared from oocytes in both hermaphrodites and females (Figure S12, D and F). However, GLD-1::GFP levels remained elevated in the oocytes of sel-10(ar41) mutant females (Figure 7E). Oocytes remain in the gonad for an extended period of time in the absence of sperm (McCarter et al. 1999). Indeed, we noticed that there seemed to be relatively less GLD-1::GFP in the sel-10(ar41) oocytes of females as compared to hermaphrodites, possibly as a result of sel-10-independent protein turnover. From these results, we conclude that the sel-10-dependent elimination of GLD-1::GFP is sperm-independent. Furthermore, the expression patterns of GLD-1::GFP, which rescues the gld-1(q485) null mutation to fertility (Schumacher et al. 2005; Figure 7A), and endogenous GLD-1 (Figure S12, D and F), fail to support the hypothesis that sperm trigger the elimination of GLD-1 from oocytes. We have tested the GFP::GLD-1 transgene (axIs1498[pie-1p::gfp::gld-1::gld-1 3′UTR, unc-119(+)]; Merritt et al. 2008) used by Bohnert and Kenyon (2017) to monitor GLD-1 expression in females; however, we found that it fails to rescue gld-1(q485) null mutants to fertility. Adult gld-1(q485); axIs1498 hermaphrodites are invariably sterile and exhibit a range of phenotypes from a tumorous phenotype that is equivalent to the null allele to the production of abnormal oocytes. We analyzed 192 progeny from gld-1(q485)/+; axIs1498 adult hermaphrodites; 53 (27.6%) were gld-1(q485); axIs1498 and were sterile, consistent with the inability of axIs1498 to provide wild-type gld-1 function. We conclude that the increased expression of GFP::GLD-1 observed in the oocytes of axIs1498 females (Bohnert and Kenyon 2017) is most likely a transgene expression artifact and does not reflect the expression and regulation of endogenous GLD-1.
Ectopic GLD-1 in proximal oocytes does not strongly inhibit the expression of mRNAs repressed by GLD-1
As for LIN-41, we examined whether the mRNA targets of GLD-1-mediated translational repression are ectopically repressed in sel-10(ar41) mutant oocytes. Studies of GLD-1 function in the proliferative vs. meiotic entry decision of germline progenitor cells demonstrate that GLP-1/Notch signaling functions to inhibit GLD-1 accumulation in the distal end of the germline (Hansen and Schedl 2013). When GLD-1 accumulates ectopically in the distal stem cell niche in glp-1 mutants, or double mutants affecting the Pumilio and FBF proteins FBF-1 and FBF-2, germline progenitor cells fail to proliferate and prematurely enter the meiotic pathway of development (Crittenden et al. 2002; Hansen et al. 2004). Thus, our initial expectation was that ectopic GLD-1 expression in proximal oocytes in strong sel-10 loss-of-function mutants might exert substantial effects on the repression of its mRNA targets. As for LIN-41, this proved not to be the case; only subtle or modest effects were observed, as described below.
GLD-1 binds the 3′-UTR of the spn-4 mRNA (Jungkamp et al. 2011), which we initially examined as a LIN-41 target, and GLD-1 appears to repress SPN-4 accumulation in the distal germline (Mootz et al. 2004). As described previously, SPN-4::GFP expression was not strongly affected by the sel-10(ar41) mutation (Figure S11, I–L) despite the ectopic expression of both GLD-1 and LIN-41 (Figure 4 and Figure 7). MEX-3 is expressed in proximal oocytes and also appears to be repressed by GLD-1 (Mootz et al. 2004; Jungkamp et al. 2011). We used the fluorescence-tagged mex-3(tn1753[gfp::3xflag::mex-3]) allele to quantitatively examine the expression of GFP::MEX-3 in these oocytes at both 20 and 25°. GFP::MEX-3 levels were not reduced in sel-10(ar41) oocytes at either temperature, but were generally very similar to the wild-type controls (Figure 7F, and Figure S12H). In addition, we examined the expression of the yolk receptor RME-2 (Grant and Hirsh 1999), a well-established target of GLD-1-mediated translational repression (Lee and Schedl 2001; Jungkamp et al. 2011; Wright et al. 2011). We began by examining the expression of RME-2::GFP from pwIs116[rme-2p::rme-2::GFP::rme-2 3′UTR] in oocytes at 22°, to prevent transgene silencing. Again, the levels of RME-2::GFP were similar in the proximal oocytes of sel-10(ar41) and wild-type controls (Figure 7, G and H). Likewise, similar levels of endogenous RME-2 were seen in sel-10(ok1632) and wild-type oocytes stained with anti-RME-2-specific antibodies (Figure S12, I and J). Finally, we examined the expression of OMA-2, another well-established target of GLD-1-mediated translational repression (Lee and Schedl 2004; Wright et al. 2011; Scheckel et al. 2012). We quantitatively compared the expression level of mNG::OMA-2 expression in the proximal oocytes of the wild type and sel-10(ar41) mutants and observed a modest reduction (∼30–50%) in mNG::OMA-2 expression levels in sel-10(ar41) mutants (Figure 7I). This result is agreement with the finding that an antibody that detects OMA-2 and its paralog OMA-1 exhibits a modest reduction in immunofluorescence staining (∼10–33%, depending on the region of the proximal gonad analyzed) in the sel-10(ok1632) strong loss-of-function mutant (Kisielnicka et al. 2018).
Collectively, these results suggest that the ectopic GLD-1 in sel-10 mutant oocytes is minimally effective at repressing translation of mRNA targets. The observation that some targets (e.g., spn-4, mex-3, and rme-2) might be unaffected by ectopic GLD-1, whereas others (e.g., oma-2) are modestly affected, is consistent with the observation that certain gld-1 mutant alleles disrupt binding and repression of some mRNA targets but not others (Schumacher et al. 2005). Furthermore, these observations are again consistent with the fact that sel-10 mutants are viable and fertile (Table 3), as the efficient repression of proteins such as SPN-4, MEX-3, and RME-2 during oogenesis should have negative consequences for embryonic development (Draper et al. 1996; Grant and Hirsh 1999; Gomes et al. 2001).
The SCFSEL-10-dependent degradation of LIN-41 and GLD-1 depend on different kinases
As described above, the SCFSEL-10-dependent degradation of LIN-41 depends on CDK-1, but not MPK-1 (Figure S9, I–L). Consequently, we examined the requirement of these kinases for the SCFSEL-10-dependent degradation of GLD-1. Whereas cdk-1(RNAi) or cdk-2(RNAi) had no effect on the accumulation of GLD-1::GFP in proximal oocytes (n = 14 and n = 23, respectively), we observed ectopic expression of GLD-1::GFP in the proximal oocytes of ozIs5[gld-1::gfp]; mpk-1(ga111ts) hermaphrodites at the nonpermissive temperature (Figure S13). Thus, although both GLD-1 and LIN-41 are regulated by SCFSEL-10-dependent degradation, the temporal and spatial control of their accumulation during oogenesis is differentially responsive to protein kinase signaling, befitting their individual biological functions in promoting oogenesis.
Discussion
Feedback regulation of LIN-41 and the spatial control of oocyte meiotic maturation
The oocytes of most sexually reproducing animals arrest in meiotic prophase for a prolonged period [reviewed by Huelgas-Morales and Greenstein (2017), Avilés-Pagán and Orr-Weaver (2018)]. This conserved arrest likely enables transcriptionally quiescent oocytes to grow by accumulating cellular organelles and cytoplasmic factors needed for embryonic development. Indeed, in C. elegans oocyte growth and meiotic maturation are coordinately controlled by LIN-41. In the absence of LIN-41 function, pachytene-stage oocytes abruptly cellularize, activate CDK-1, and enter M phase (Spike et al. 2014a). A salient feature of C. elegans oogenesis is that meiotic maturation is restricted to the oocyte in the most proximal position adjacent to the spermatheca. This restriction ensures that only fully grown oocytes undergo meiotic maturation when they are positioned to enter the spermatheca during ovulation so they can become fertilized. Genetic evidence suggests that OMA proteins function to inhibit LIN-41 to facilitate meiotic maturation of the most proximal oocyte (Spike et al. 2014a). Specifically, proximal oocytes fail to enter M phase in lin-41(ts); oma-1(null); oma-2(null) triple mutants; whereas pachytene stage oocytes prematurely enter M phase in lin-41(null); oma-1(null); oma-2(null) triple mutants (Spike et al. 2014a). Thus, the OMA proteins are absolutely required to spatially restrict M phase entry to the –1 oocyte, where they counteract the inhibitory activity of LIN-41. Consistent with this idea, molecular evidence suggests that LIN-41 is inactivated as a translational repressor in the final stages of oogenesis (Spike et al. 2014a; Tsukamoto et al. 2017), which precedes the elimination of LIN-41 upon the onset of meiotic maturation. Specifically, two targets of LIN-41-mediated translational repression, spn-4 and meg-1, are coexpressed with LIN-41 in the most proximal oocytes. The expression of spn-4 and meg-1 in proximal oocytes requires the function of the OMA proteins (Tsukamoto et al. 2017), consistent with the idea that the OMA proteins antagonize LIN-41 function in the late stages of oogenesis.
Here we show that SCFSEL-10 promotes the rapid ubiquitin-mediated degradation of LIN-41 that leads to its elimination during meiosis I. Analysis of sel-10 mutants indicates that the inactivation and degradation of LIN-41 are separable; the LIN-41 that accumulates in sel-10 mutants appears to be largely inactive as a translational repressor. However, we did note that several LIN-41 variants (LIN-41[T83A] and LIN-41[ΔDeg-A]), which are defective in the SCFSEL-10-mediated degradation, decrease the meiotic maturation rate. This finding is consistent with the idea that LIN-41 inhibits meiotic maturation and that SCFSEL-10-mediated degradation constitutes a nonessential component of the regulatory mechanism. The nature of the “primary” mechanism inactivating LIN-41 prior to its degradation is currently unknown but could act on LIN-41 directly or another component of the large RNP complex it associates with (Spike et al. 2014b; Tsukamoto et al. 2017).
LIN-41 and CDK-1 reciprocally inhibit each other’s activity. Thus, the primary inactivation mechanism might play a key role in tipping the balance between LIN-41 and CDK-1 to generate a spatially restricted all-or-none meiotic maturation response. Upon its activation, CDK-1 triggers meiotic maturation and promotes the SCFSEL-10-dependent elimination of LIN-41. The elimination of LIN-41 requires the Deg-A and Deg-B domains in the LIN-41 N-terminal region. The LIN-41 Deg-A and Deg-B domains are intrinsically disordered and contain sequences that might function as phosphodegrons. The SCFSEL-10-dependent elimination of LIN-41 is blocked by the T83A mutation affecting a potential CDK-1 phosphorylation site within the Deg-A domain, although whether this regulation is direct or indirect remains to be determined. We note one exception to the rule that CDK-1 activity promotes LIN-41 degradation. The lin-41(tn1541tn1618) mutation (Figure 2), which deletes the NHL domain, produces a strong loss-of-function lin-41 mutant phenotype in which pachytene-stage oocytes enter M phase precociously. Nonetheless, we observe that the GFP::LIN-41(ΔNHL) protein still accumulates in the proximal gonad, albeit in an aberrantly punctate pattern (Figure S3, G–J). Interestingly, in the presence of a wild-type LIN-41 protein, the GFP::LIN-41(ΔNHL) mutant protein accumulates normally and is subject to SCFSEL-10-dependent degradation on schedule. It may be that the accumulation of the GFP::LIN-41(ΔNHL) protein in a punctate pattern correlates with its inaccessibility to CDK-1-dependent regulation. Alternatively, the degradation of LIN-41 during meiotic maturation may depend on LIN-41 activity during pachytene, as could be the case if a component of the SCFSEL-10 degradation mechanism depends on lin-41 function for its synthesis or activity.
The Deg domains may function as a timer to ensure that CDK-1 activity reaches an optimal threshold to ensure the successful completion of the meiotic divisions prior to the initiation of LIN-41 degradation. If LIN-41 is eliminated too early, the fidelity of meiotic chromosome segregation may be compromised, as is observed in certain hypomorphic lin-41 mutant alleles (e.g., tn1487tn1515, tn1487tn1516, tn1487tn1536, and tn1487tn1539; Spike et al. 2014a). Thus, it will be important to elucidate the precise mechanisms by which the LIN-41 Deg domains link CDK-1 activity to SCFSEL-10-mediated degradation. The regulation of the G1/S phase transition in budding yeast provides a framework for thinking about this issue [Nash et al. 2001; Kõivomägi et al. 2011; Yang et al. 2013; reviewed by Hopkins et al. (2017)]. The cyclin-dependent kinase complex, Cdk1-Clb5/6, promotes the entry into S phase but is inhibited by binding to its inhibitor Sic1 (Nugroho and Mendenhall 1994; Schwob et al. 1994). Sic1 is a substrate of the Cdk1-Clb5/6 kinase, which phosphorylates Sic1 to promote SCFCdc4-mediated degradation (Feldman et al. 1997; Verma et al. 1997; Nash et al. 2001). The cyclin-dependent kinase Cdk1-Cln1/2 initiates the decision to enter S phase during G1 and is not inhibited by Sic1. Phosphorylation of Sic1 by Cdk1-Cln1/2, while not sufficient to trigger Sic1 degradation, primes Sic1 for multisite phosphorylation by Clb5/6. The Sic1 CPD sequences contain multiple sites for phosphorylation by both Cdk1-Clb5/6 and Cdk1-Cln1/2, which results in the switch-like destruction of Sic1. A failure to degrade Sic1 substantially delays the G1/S transition, whereas deletion of SIC1 causes DNA replication to initiate too early, resulting in genome instability (Nugroho and Mendenhall 1994; Cross et al. 2007). Further dissection of the mechanism by which the LIN-41 Deg domains function will illuminate whether analogous mechanisms are employed in a developmental context.
Ubiquitin-mediated protein degradation and the OET
Signaling pathways and downstream kinase activation coordinate the cell cycle and developmental events that underpin oocyte and early embryo development. In C. elegans the ERK MAP kinase signaling pathway and its effector kinase MPK-1 regulate pachytene progression and multiple aspects of oogenesis, including oocyte growth and specific events that occur during meiotic maturation [reviewed by Arur (2017)]. Consistent with these phenotypes, sustained activation of MPK-1 occurs during pachytene and in proximal oocytes (Lee et al. 2007). Likewise, in proximal oocytes activated cyclin-dependent kinase CDK-1 regulates an important aspect of oocyte meiotic maturation by promoting the transition from meiotic prophase to meiotic M phase, as we have described. Once activated, CDK-1 phosphorylates the DYRK mini-brain kinase MBK-2 as part of an intricate regulatory mechanism that permits MBK-2 activation near the end of the first meiotic division (Pellettieri et al. 2003; Stitzel et al. 2006, 2007; Cheng et al. 2009; Parry et al. 2009). These three kinases (MPK-1, CDK-1, and MBK-2) all function, at least in part, to promote the degradation of one or more RNA-binding proteins during oogenesis or the OET.
After meiosis, the OMA proteins are detectably phosphorylated by activated MBK-2 (Nishi and Lin 2005). Phosphorylation by MBK-2 promotes a direct physical interaction between the OMA proteins and the transcription factor TAF-4; this permits the sequestration of TAF-4 in the cytoplasm and prevents the premature onset of zygotic transcription (Guven-Ozkan et al. 2008). Furthermore, MBK-2-dependent phosphorylation primes the OMA proteins for phosphorylation by the glycogen synthase kinase GSK-3 and for degradation during the first mitotic division (Nishi and Lin 2005; Shirayama et al. 2006). In addition to MBK-2 and GSK-3, the degradation of the OMA proteins requires the normal activities of additional kinases, including CDK-1/Cyclin B3, and several proposed E3 ubiquitin ligases (Shirayama et al. 2006; Du et al. 2015). The failure to degrade OMA-1 and eliminate it from early embryos is deleterious (Lin 2003) and contributes to phenotypes exhibited by mutants that fail to degrade the OMA proteins (Shirayama et al. 2006). Indeed, the ectopic expression of OMA-1 in early embryos represses the translation of at least one mRNA target of the OMA proteins, zif-1 mRNA, but only when OMA-1 is not phosphorylated by MBK-2, as in the oma-1(zu405gf) mutant (Guven-Ozkan et al. 2010). Thus, the MBK-2-dependent phosphorylation of the OMA proteins not only primes these proteins for degradation but also inhibits their ability to function as translational repressors.
Likewise, the ability of GLD-1 to function as a translational repressor might be inhibited by MPK-1-dependent phosphorylation (Kisielnicka et al. 2018; this work). Since mpk-1 activity is also required for the elimination of GLD-1, MPK-1-dependent phosphorylation would coordinate the inactivation of GLD-1 as a translational repressor with GLD-1 degradation. Consistent with this hypothesis, MPK-1 promotes the phosphorylation of GLD-1 and promotes its SCFSEL-10-mediated degradation (Kisielnicka et al. 2018; this work). Furthermore, this hypothesis potentially explains why the ectopic GLD-1 expressed in sel-10 mutant oocytes is relatively ineffective at repressing the translation of multiple target mRNAs.
In sharp contrast to OMA-1 and GLD-1, our current understanding of the regulation of LIN-41 suggests that the inactivation of LIN-41 as a translational repressor is temporally and molecularly distinct from its degradation. Targets of LIN-41 translational repression such as spn-4 and meg-1 are actively translated prior to meiotic maturation and the CDK-1-dependent elimination of LIN-41. We have not yet determined that LIN-41 is phosphorylated by CDK-1 or any other kinase, as electrophoretic mobility changes are not reproducibly observed in sel-10 mutants using several standard gel systems (Figure S6D and C. Spike and D. Greenstein, unpublished results). Using the Phos-Tag gel system, which is useful for detecting phosphoproteins (Wang et al. 2014), the total LIN-41 from sel-10 mutant adults migrates more slowly; however, we are uncertain as to whether this is due to phosphorylation (C. Spike and D. Greenstein, unpublished results). The fact that spn-4 and meg-1 mRNAs are translated normally when cdk-1 function is attenuated by RNAi (Tsukamoto et al. 2017) suggests that the CDK-1 is not required to inactivate LIN-41 as a translational repressor. In addition, we show here that mutations affecting the function of the LIN-41 Deg domains do not exhibit gain-of-function phenotypes or substantially repress the translation of LIN-41 target mRNAs.
Multiple mechanisms regulate LIN-41 proteins
LIN-41 was first identified through its role in the heterochronic gene regulatory pathway that controls the timing of postembryonic cell divisions and cell fate decisions in somatic cells in C. elegans [Reinhart et al. 2000; Slack et al. 2000; reviewed by Rougvie and Moss (2013)]. In this capacity, LIN-41 functions to repress the translation of several transcription factors, including LIN-29, MAB-3, MAB-10, and DMD-3, which play key roles in specifying somatic cell fates during the L4 and adult stages (Reinhart et al. 2000; Harris and Horvitz 2011; Aeschimann et al. 2017). LIN-41 binds to the mRNAs of these genes and represses their translation during early larval stages (e.g., L1–L3) (Aeschimann et al. 2017). The let-7 microRNA promotes the switch from early larval stages to the L4 and adult stages by repressing translation of LIN-41 beginning in the L4 stage (Reinhart et al. 2000; Slack et al. 2000). This regulation is specific to the soma as the let-7(n2583ts) mutation does not increase the accumulation of LIN-41 in the oogenic germline (Spike et al. 2014a). It is not clear whether specific protein degradation mechanisms collaborate with Let-7-mediated regulation to ensure that LIN-41 does not perdure from the early larval stages into the L4 and adult stage in somatic cells. If such mechanisms exist, they are unlikely to depend solely on the Deg domains because lin-41 mutations affecting the Deg domains (e.g., tn1620, tn1622tn1635, tn1638, tn1643, and tn1645) do not phenocopy let-7 mutations or exhibit dominant somatic defects. Additionally, the Deg mutations do not confer an overt lin-41(lf) Dpy phenotype. Further, lin-41(tn1541tn1643[ΔDeg-A–RING–Deg-B]) L3-stage larvae do not exhibit precocious adult alae (n = 7; Ann Rougvie, personal communication) as is frequently observed in lin-41(lf) mutants (Slack et al. 2000). The Deg domains mediate LIN-41 degradation during the OET over short timescales (i.e., 10–15 min), whereas the larval stages last for hours. This difference may obviate a requirement for SCFSEL-10-mediated degradation of LIN-41 during the larval stages. Interestingly, several lin-41 gain-of-function alleles affecting the N-terminal 39 amino acid residues result in a defect in tip retraction during male tail development resulting in the production of a leptoderan (Lep) tail characteristic of other rhabditid nematode species (Del Rio-Albrechtsen et al. 2006). These lin-41(Lep) gain-of-function alleles do not affect LIN-41 degradation during the OET and thus define a site for LIN-41 regulation in somatic cells, which could involve proteolytic degradation.
LIN-41 is highly conserved. The mammalian ortholog LIN-41/TRIM71 is required for embryonic viability and neural tube closure in mice (Maller Schulman et al. 2008; Cuevas et al. 2015; Mitschka et al. 2015). LIN-41/TRIM71 was found to promote reprogramming of dermal fibroblasts to induced-pluripotent stem cells through the negative regulation of differentiation genes, including the transcription factor EGR1 (Worringer et al. 2014). Importantly, the let-7 microRNA inhibits reprogramming in part through the repression of LIN-41. Thus, the regulation of LIN-41 by Let-7 is a conserved regulatory module. By contrast, the Deg domains of C. elegans LIN-41 are not conserved at the amino acid sequence level in the mammalian orthologs and appear to be rapidly evolving in closely related rhabditid nematodes.
Developing systems must deploy mechanisms to extinguish translational repression mediated by RNA-binding proteins. Such mechanisms may function to promote translation of batteries of genes needed to drive developmental transitions. LIN-41-associated mRNAs include many key genes required for embryonic development (Tsukamoto et al. 2017). Thus, inactivation of LIN-41 likely plays a key role in shaping the proteome during the OET. The primary mechanism inactivating LIN-41 prior to its degradation, and its potential conservation in LIN-41 orthologs or members of the TRIM-NHL class of RNA-binding proteins, remain to be determined.
Acknowledgments
We are grateful to Swathi Arur, Sarah Crittenden, Claire de la Cova, Daniel Dickinson, Bob Goldstein, Barth Grant, Iva Greenwald, Judith Kimble, Tim Schedl, and Dustin Updike for providing strains or reagents. We thank G. W. Gant Luxton for the use of his spinning disc confocal microscope. We also thank WormBase for sequences and annotations. We thank Cynthia Kenyon for discussions on GLD-1 regulation. Ann Rougvie and Todd Starich provided helpful suggestions during the course of this work. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by grant P40OD010440 from the National Institutes of Health Office of Research Infrastructure Programs. This work was supported by National Institutes of Health grant GM57173 (to D.G.).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7056557.
Communicating editor: V. Reinke
Literature Cited
- Aeschimann F., Kumari P., Bartake H., Gaidatzis D., Xu L., et al. , 2017. LIN41 post-transcriptionally silences mRNAs by two distinct and position-dependent mechanisms. Mol. Cell 65: 476–489.e4. 10.1016/j.molcel.2016.12.010 [DOI] [PubMed] [Google Scholar]
- Allen A. K., Nesmith J. E., Golden A., 2014. An RNAi-based suppressor screen identifies interactors of the Myt1 ortholog of Caenorhabditis elegans. G3 (Bethesda) 4: 2329–2343. 10.1534/g3.114.013649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere J. A., Bell R. T., Fu B. X., Artiles K. L., Hartman P. S., et al. , 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arur S., 2017. Signaling-mediated regulation of meiotic prophase I and transition during oogenesis. Results Probl. Cell Differ. 59: 101–123. 10.1007/978-3-319-44820-6_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilés-Pagán E. E., Orr-Weaver T. L., 2018. Activating embryonic development in Drosophila. Semin. Cell Dev. Biol. (in press) 10.1016/j.semcdb.2018.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balklava Z., Pant S., Fares H., Grant B. D., 2007. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat. Cell Biol. 9: 1066–1073. 10.1038/ncb1627 [DOI] [PubMed] [Google Scholar]
- Beard S. M., Smit R. B., Chan B. G., Mains P. E., 2016. Regulation of the MEI-1/MEI-2 microtubule-severing Katanin complex in early Caenorhabditis elegans development. G3 (Bethesda) 6: 3257–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boag P. R., Nakamura A., Blackwell T. K., 2005. A conserved RNA-protein complex component involved in physiological germline apoptosis regulation in C. elegans. Development 132: 4975–4986. 10.1242/dev.02060 [DOI] [PubMed] [Google Scholar]
- Bohnert K. A., Kenyon C., 2017. A lysosomal switch triggers proteostasis renewal in the immortal C. elegans germ lineage. Nature 551: 629–633. 10.1038/nature24620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxem M., Srinivasan D. G., van den Heuvel S., 1999. The Caenorhabditis elegans gene ncc-1 encodes a cdc2-related kinase required for M phase in meiotic and mitotic cell divisions, but not for S phase. Development 126: 2227–2239. [DOI] [PubMed] [Google Scholar]
- Burrows A. E., Sceurman B. K., Kosinski M. E., Richie C. T., Sadler P. L., et al. , 2006. The C. elegans Myt1 ortholog is required for the proper timing of oocyte maturation. Development 133: 697–709. 10.1242/dev.02241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M., Tu Y., Euskirchen G., Ward W. W., Prasher D. C., 1994. Green fluorescent protein as a marker for gene expression. Science 263: 802–805. 10.1126/science.8303295 [DOI] [PubMed] [Google Scholar]
- Cheng K. C., Klancer R., Singson A., Seydoux G., 2009. Regulation of MBK-2/DYRK by CDK-1 and the pseudophosphatases EGG-4 and EGG-5 during the oocyte-to-embryo transition. Cell 139: 560–572. 10.1016/j.cell.2009.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden S. L., Bernstein D. S., Bachorik J. L., Thompson B. E., Gallegos M., et al. , 2002. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417: 660–663. 10.1038/nature754 [DOI] [PubMed] [Google Scholar]
- Cross F. R., Schroeder L., Bean J. M., 2007. Phosphorylation of the Sic1 inhibitor of B-type cyclins in Saccharomyces cerevisiae is not essential but contributes to cell cycle robustness. Genetics 176: 1541–1555. 10.1534/genetics.107.073494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csizmok V., Montecchio M., Lin H., Tyers M., Sunnerhagen M., et al. , 2018. Multivalent interactions with Fbw7 and Pin1 facilitate recognition of c-Jun by the SCFFbw7 ubiquitin ligase. Structure 26: 28–39.e2. 10.1016/j.str.2017.11.003 [DOI] [PubMed] [Google Scholar]
- Cuevas E., Rybak-Wolf A., Rhode A. M., Nguyen D. T. T., Wulczyn F. G., 2015. Lin41/Trim71 is essential for mouse development and specifically expressed in postnatal ependymal cells of the brain. Front. Cell Dev. Biol. 3: 20 10.3389/fcell.2015.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cova C., Greenwald I., 2012. SEL-10/Fbw7-dependent negative feedback regulation of LIN-45/Braf signaling in C. elegans via a conserved phosphodegron. Genes Dev. 26: 2524–2535. 10.1101/gad.203703.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rio-Albrechtsen T., Kiontke K., Chiou S. Y., Fitch D. H., 2006. Novel gain-of-function alleles demonstrate a role for the heterochronic gene lin-41 in C. elegans male tail tip morphogenesis. Dev. Biol. 297: 74–86. 10.1016/j.ydbio.2006.04.472 [DOI] [PubMed] [Google Scholar]
- Dephoure N., Gould K. L., Gygi S. P., Kellogg D. R., 2013. Mapping and analysis of phosphorylation sites: a quick guide for cell biologists. Mol. Biol. Cell 24: 535–542. 10.1091/mbc.e12-09-0677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R. J., Ferrell J. E., 2001. Multisite phosphorylation and the countdown to S phase. Cell 107: 819–822. 10.1016/S0092-8674(01)00620-1 [DOI] [PubMed] [Google Scholar]
- Detwiler M. R., Reuben M., Li X., Rogers E., Lin R., 2001. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev. Cell 1: 187–199. 10.1016/S1534-5807(01)00026-0 [DOI] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. 10.1038/nmeth.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Pani A. M., Heppert J. K., Higgins C. D., Goldstein B., 2015. Streamlined genome engineering with a self-excising drug selection cassette. Genetics 200: 1035–1049. 10.1534/genetics.115.178335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doh J. H., Jung Y., Reinke V., Lee M. H., 2013. C. elegans RNA-binding protein GLD-1 recognizes its multiple targets using sequence, context, and structural information to repress translation. Worm 2: e26548 10.4161/worm.26548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper B. W., Mello C. C., Bowerman B., Hardin J., Priess J. R., 1996. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell 87: 205–216. 10.1016/S0092-8674(00)81339-2 [DOI] [PubMed] [Google Scholar]
- Du Z., He F., Yu Z., Bowerman B., Bao Z., 2015. E3 ubiquitin ligases promote progression of differentiation during C. elegans embryogenesis. Dev. Biol. 398: 267–279. 10.1016/j.ydbio.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy W. G., Brizuela L., Beach D., Newport J., 1988. The Xenopus cdc2 protein is a component of MPF, a cytoplasmic regulator of mitosis. Cell 54: 423–431. 10.1016/0092-8674(88)90205-X [DOI] [PubMed] [Google Scholar]
- Ellis R. E., Kimble J., 1995. The fog-3 gene and regulation of cell fate in the germ line of Caenorhabditis elegans. Genetics 139: 561–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito D., Koliopoulos M. G., Rittinger K., 2017. Structural determinants of TRIM protein function. Biochem. Soc. Trans. 45: 183–191. 10.1042/BST20160325 [DOI] [PubMed] [Google Scholar]
- Farley B. M., Ryder S. P., 2012. POS-1 and GLD-1 repress glp-1 translation through a conserved binding-site cluster. Mol. Biol. Cell 23: 4473–4483. 10.1091/mbc.e12-03-0216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman R. M., Correll C. C., Kaplan K. B., Deshaies R. J., 1997. A complex of Cdc4p, Skp1p, and Cdc53/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91: 221–230. 10.1016/S0092-8674(00)80404-3 [DOI] [PubMed] [Google Scholar]
- Fox P. M., Vought V. E., Hanazawa M., Lee M. H., Maine E. M., et al. , 2011. Cyclin E and CDK-2 regulate proliferative cell fate and cell cycle progression in the C. elegans germline. Development 138: 2223–2234. 10.1242/dev.059535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R., Maine E., Schedl T., 1995a. Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics 139: 607–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R., Barton M. K., Kimble J., Schedl T., 1995b. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics 139: 579–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Vaillant M., Jessus C., Ozon R., Maller J. L., Haccard O., 1999. Two distinct mechanisms control the accumulation of cyclin B1 and Mos in Xenopus oocytes in response to progesterone. Mol. Biol. Cell 10: 3279–3288. 10.1091/mbc.10.10.3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta T., Tuck S., Kirchner J., Koch B., Auty R., et al. , 2000. EMB-30: an APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans. Mol. Biol. Cell 11: 1401–1419. 10.1091/mbc.11.4.1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier J., Norbury C., Lohka M., Nurse P., Maller J., 1988. Purified maturation-promoting factor contains the product of a Xenopus homolog of the fission yeast cell cycle control gene cdc2+. Cell 54: 433–439. 10.1016/0092-8674(88)90206-1 [DOI] [PubMed] [Google Scholar]
- Gautier J., Minshull J., Lohka M., Glotzer M., Hunt T., et al. , 1990. Cyclin is a component of maturation-promoting factor from Xenopus. Cell 60: 487–494. 10.1016/0092-8674(90)90599-A [DOI] [PubMed] [Google Scholar]
- Gomes J. E., Encalada S. E., Swan K. A., Shelton C. A., Carter J. C., et al. , 2001. The maternal gene spn-4 encodes a predicted RRM protein required for mitotic spindle orientation and cell fate patterning in early C. elegans embryos. Development 128: 4301–4314. [DOI] [PubMed] [Google Scholar]
- Gouw M., Michael S., Sámano-Sánchez H., Kumar M., Zeke A., et al. , 2018. The eukaryotic linear motif resource - 2018 update. Nucleic Acids Res. 46: D428–D434. 10.1093/nar/gkx1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan J. A., Cheng H., Harris J. E., Greenstein D., 2006. Galphao/i and Galphas signaling function in parallel with the MSP/Eph receptor to control meiotic diapause in C. elegans. Curr. Biol. 16: 1257–1268. 10.1016/j.cub.2006.05.020 [DOI] [PubMed] [Google Scholar]
- Govindan J. A., Nadarajan S., Kim S., Starich T. A., Greenstein D., 2009. Somatic cAMP signaling regulates MSP-dependent oocyte growth and meiotic maturation in C. elegans. Development 136: 2211–2221. 10.1242/dev.034595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B., Hirsh D., 1999. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell 10: 4311–4326. 10.1091/mbc.10.12.4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven-Ozkan T., Robertson S. M., Nishi Y., Lin R., 2010. zif-1 translational repression defines a second, mutually exclusive OMA function in germline transcriptional repression. Development 137: 3373–3382. 10.1242/dev.055327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven-Ozkan T., Nishi Y., Robertson S. M., Lin R., 2008. Global transcriptional repression in C. elegans germline precursors by regulated sequestration of TAF-4. Cell 135: 149–160. 10.1016/j.cell.2008.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haccard O., Jessus C., 2006a. Redundant pathways for Cdc2 activation in Xenopus oocyte: either cyclin B or Mos synthesis. EMBO Rep. 7: 321–325. 10.1038/sj.embor.7400611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haccard O., Jessus C., 2006b. Oocyte maturation, Mos and cyclins–a mater of synthesis: two functionally redundant ways to induce meiotic maturation. Cell Cycle 5: 1152–1159. 10.4161/cc.5.11.2800 [DOI] [PubMed] [Google Scholar]
- Hansen D., Schedl T., 2013. Stem cell proliferation vs. meiotic fate decision in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 71–99. 10.1007/978-1-4614-4015-4_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D., Wilson-Berry L., Dang T., Schedl T., 2004. Control of the proliferation vs. meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development 131: 93–104. 10.1242/dev.00916 [DOI] [PubMed] [Google Scholar]
- Harris D. T., Horvitz H. R., 2011. MAB-10/NAB acts with LIN-29/EGR to regulate terminal differentiation and the transition from larva to adult in C. elegans. Development 138: 4051–4062. 10.1242/dev.065417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa E., Karashima T., Sumiyoshi E., Yamamoto M., 2006. C. elegans CPB-3 interacts with DAZ-1 and functions in multiple steps in germline development. Dev. Biol. 295: 689–699. 10.1016/j.ydbio.2006.04.002 [DOI] [PubMed] [Google Scholar]
- Huelgas-Morales G., Greenstein D., 2017. Control of oocyte meiotic maturation in C. elegans. Semin. Cell Dev. Biol. (in press) 10.1016/j.semcdb.2017.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins M., Tyson J. J., Novák B., 2017. Cell-cycle transitions: a common role for stoichiometric inhibitors. Mol. Biol. Cell 28: 3437–3446. 10.1091/mbc.e17-06-0349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J. Y., Sun Z. W., Li X., Reuben M., Tatchell K., et al. , 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102: 279–291. 10.1016/S0092-8674(00)00034-9 [DOI] [PubMed] [Google Scholar]
- Hubbard E. J., Wu G., Kitajewski J., Greenwald I., 1997. sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes Dev. 11: 3182–3193. 10.1101/gad.11.23.3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K., Inoue S., 2012. TRIM proteins as RING finger E3 ubiquitin ligases. Adv. Exp. Med. Biol. 770: 27–37. 10.1007/978-1-4614-5398-7_3 [DOI] [PubMed] [Google Scholar]
- Jan E., Motzny C. K., Graves L. E., Goodwin E. B., 1999. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 18: 258–269. 10.1093/emboj/18.1.258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Verheyden J. M., Kimble J., 2011. Cyclin E and Cdk2 control GLD-1, the mitosis/meiosis decision, and germline stem cells in Caenorhabditis elegans. PLoS Genet. 7: e1001348 10.1371/journal.pgen.1001348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. R., Schedl T., 1995. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes Dev. 9: 1491–1504. 10.1101/gad.9.12.1491 [DOI] [PubMed] [Google Scholar]
- Jones A. R., Francis R., Schedl T., 1996. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 180: 165–183. 10.1006/dbio.1996.0293 [DOI] [PubMed] [Google Scholar]
- Jungkamp A. C., Stoeckius M., Mecenas D., Grün D., Mastrobuoni G., et al. , 2011. In vivo and transcriptome-wide identification of RNA binding protein target sites. Mol. Cell 44: 828–840. 10.1016/j.molcel.2011.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk L. C., Kimble J., 1998. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 125: 1803–1813. [DOI] [PubMed] [Google Scholar]
- Kapelle W. S., Reinke V., 2011. C. elegans meg-1 and meg-2 differentially interact with nanos family members to either promote or inhibit germ cell proliferation and survival. Genesis 49: 380–391. 10.1002/dvg.20726 [DOI] [PubMed] [Google Scholar]
- Killian D. J., Harvey E., Johnson P., Otori M., Mitani S., et al. , 2008. SKR-1, a homolog of Skp1 and a member of the SCF(SEL-10) complex, regulates sex-determination and LIN-12/Notch signaling in C. elegans. Dev. Biol. 322: 322–331. 10.1016/j.ydbio.2008.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos E. T., Lander L. E., Wing J. P., He W. W., Hedgecock E. M., 1996. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell 85: 829–839. 10.1016/S0092-8674(00)81267-2 [DOI] [PubMed] [Google Scholar]
- Kishimoto T., 2015. Entry into mitosis: a solution to the decades-long enigma of MPF. Chromosoma 124: 417–428. 10.1007/s00412-015-0508-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisielnicka E., Minasaki R., Eckmann C. R., 2018. MAPK signaling couples SCF-mediated degradation of translational regulators to oocyte meiotic progression. Proc. Natl. Acad. Sci. USA 115: E2772–E2781. 10.1073/pnas.1715439115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H., Minshull J., Ford C., Golsteyn R., Poon R., et al. , 1991. On the synthesis and destruction of A- and B-type cyclins during oogenesis and meiotic maturation in Xenopus laevis. J. Cell Biol. 114: 755–765. 10.1083/jcb.114.4.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp D. M., Schaefer L. K., Ye X., Keyomarsi K., Chu C., et al. , 2001. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294: 173–177. 10.1126/science.1065203 [DOI] [PubMed] [Google Scholar]
- Kõivomägi M., Valk E., Venta R., Iofik A., Lepiku M., et al. , 2011. Cascades of multisite phosphorylation control Sic1 destruction at the onset of S phase. Nature 480: 128–131. 10.1038/nature10560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornbluth S., Sebastian B., Hunter T., Newport J., 1994. Membrane localization of the kinase which phosphorylates p34cdc2 on threonine 14. Mol. Biol. Cell 5: 273–282. 10.1091/mbc.5.3.273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski M., McDonald K., Schwartz J., Yamamoto I., Greenstein D., 2005. C. elegans sperm bud vesicles to deliver a meiotic maturation signal to distant oocytes. Development 132: 3357–3369. 10.1242/dev.01916 [DOI] [PubMed] [Google Scholar]
- Kumagai A., Dunphy W. G., 1996. Purification and molecular cloning of Plx1, a Cdc25-regulatory kinase from Xenopus egg extracts. Science 273: 1377–1380. 10.1126/science.273.5280.1377 [DOI] [PubMed] [Google Scholar]
- Kumari P., Aeschimann F., Gaidatzis D., Keusch J. J., Ghosh P., et al. , 2018. Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition. Nat. Commun. 9: 1549 10.1038/s41467-018-03920-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumsta C., Hansen M., 2012. C. elegans rrf-1 mutations maintain RNAi efficiency in the soma in addition to the germline. PLoS One 7: e35428 10.1371/journal.pone.0035428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leacock S. W., Reinke V., 2008. MEG-1 and MEG-2 are embryo-specific P-granule components required for germline development in Caenorhabditis elegans. Genetics 178: 295–306. 10.1534/genetics.107.080218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Schedl T., 2001. Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes Dev. 15: 2408–2420. 10.1101/gad.915901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Schedl T., 2004. Translation repression by GLD-1 protects its mRNA targets from nonsense-mediated mRNA decay in C. elegans. Genes Dev. 18: 1047–1059. 10.1101/gad.1188404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Schedl T., 2010. C. elegans star proteins, GLD-1 and ASD-2, regulate specific RNA targets to control development. Adv. Exp. Med. Biol. 693: 106–122. 10.1007/978-1-4419-7005-3_8 [DOI] [PubMed] [Google Scholar]
- Lee M. H., Ohmachi M., Arur S., Nayak S., Francis R., et al. , 2007. Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics 177: 2039–2062. 10.1534/genetics.107.081356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., DeBella L. R., Guven-Ozkan T., Lin R., Rose L. S., 2009. An eIF4E-binding protein regulates katanin protein levels in C. elegans embryos. J. Cell Biol. 187: 33–42. 10.1083/jcb.200903003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., 2003. A gain-of-function mutation in oma-1, a C. elegans gene required for oocyte maturation, results in delayed degradation of maternal proteins and embryonic lethality. Dev. Biol. 258: 226–239. 10.1016/S0012-1606(03)00119-2 [DOI] [PubMed] [Google Scholar]
- Loedige I., Gaidatzis D., Sack R., Meister G., Filipowicz W., 2013. The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucleic Acids Res. 41: 518–532. 10.1093/nar/gks1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loedige I., Jakob L., Treiber T., Ray D., Stotz M., et al. , 2015. The crystal structure of the NHL domain in complex with RNA reveals the molecular basis of Drosophila brain-tumor-mediated gene regulation. Cell Rep. 13: 1206–1220. 10.1016/j.celrep.2015.09.068 [DOI] [PubMed] [Google Scholar]
- Lohka M. J., Hayes M. K., Maller J. L., 1988. Purification of maturation-promoting factor, an intracellular regulator of early mitotic events. Proc. Natl. Acad. Sci. USA 85: 3009–3013. 10.1073/pnas.85.9.3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas X., Ciulli A., 2017. Recognition of substrate degrons by E3 ubiquitin ligases and modulation by small-molecule mimicry strategies. Curr. Opin. Struct. Biol. 44: 101–110. 10.1016/j.sbi.2016.12.015 [DOI] [PubMed] [Google Scholar]
- Maller Schulman B. R., Liang X., Stahlhut C., DelConte C., Stefani G., et al. , 2008. The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle 7: 3935–3942. 10.4161/cc.7.24.7397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui Y., 2001. From oocyte maturation to the in vitro cell cycle: the history of discoveries of Maturation-Promoting Factor (MPF) and Cytostatic Factor (CSF). Differentiation 69: 1–17. 10.1046/j.1432-0436.2001.690101.x [DOI] [PubMed] [Google Scholar]
- Masui Y., Markert C. L., 1971. Cytoplasmic control of nuclear behavior during meiotic maturation of frog oocytes. J. Exp. Zool. 177: 129–145. 10.1002/jez.1401770202 [DOI] [PubMed] [Google Scholar]
- Matsuura R., Ashikawa T., Nozaki Y., Kitagawa D., 2016. LIN-41 inactivation leads to delayed centrosome elimination and abnormal chromosome behavior during female meiosis in Caenorhabditis elegans. Mol. Biol. Cell 27: 799–811. 10.1091/mbc.e15-10-0713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., Schedl T., 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 205: 111–128. 10.1006/dbio.1998.9109 [DOI] [PubMed] [Google Scholar]
- McNally K., Audhya A., Oegema K., McNally F. J., 2006. Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175: 881–891. 10.1083/jcb.200608117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C., Rasoloson D., Ko D., Seydoux G., 2008. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol. 18: 1476–1482. 10.1016/j.cub.2008.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. A., Nguyen V. Q., Lee M. H., Kosinski M., Schedl T., et al. , 2001. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science 291: 2144–2147. 10.1126/science.1057586 [DOI] [PubMed] [Google Scholar]
- Minshull J., Murray A., Colman A., Hunt T., 1991. Xenopus oocyte maturation does not require new cyclin synthesis. J. Cell Biol. 114: 767–772. 10.1083/jcb.114.4.767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitschka S., Ulas T., Goller T., Schneider K., Egert A., et al. , 2015. Co-existence of intact stemness and priming of neural differentiation programs in mES cells lacking Trim71. Sci. Rep. 5: 11126 10.1038/srep11126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad A., Vanden Broek K., Wang C., Daryabeigi A., Jantsch V., et al. , 2018. Initiation of meiotic development is controlled by three posttranscriptional pathways in Caenorhabditis elegans. Genetics 209: 1197–1224. 10.1534/genetics.118.300985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz D., Ho D. M., Hunter C. P., 2004. The STAR/Maxi-KH domain protein GLD-1 mediates a developmental switch in the translational control of C. elegans PAL-1. Development 131: 3263–3272. 10.1242/dev.01196 [DOI] [PubMed] [Google Scholar]
- Mueller P. R., Coleman T. R., Dunphy W. G., 1995. Cell cycle regulation of a Xenopus Wee1-like kinase. Mol Biol Cell 6: 119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash P., Tang X., Orlicky S., Chen Q., Gertler F. B., et al. , 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414: 514–521. 10.1038/35107009 [DOI] [PubMed] [Google Scholar]
- Nayak S., Santiago F. E., Jin H., Lin D., Schedl T., et al. , 2002. The Caenorhabditis elegans Skp1-related gene family: diverse functions in cell proliferation, morphogenesis, and meiosis. Curr. Biol. 12: 277–287. 10.1016/S0960-9822(02)00682-6 [DOI] [PubMed] [Google Scholar]
- Nebreda A. R., Gannon J. V., Hunt T., 1995. Newly synthesized protein(s) must associate with p34cdc2 to activate MAP kinase and MPF during progesterone-induced maturation of Xenopus oocytes. EMBO J. 14: 5597–5607. 10.1002/j.1460-2075.1995.tb00247.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi Y., Lin R., 2005. DYRK2 and GSK-3 phosphorylate and promote the timely degradation of OMA-1, a key regulator of the oocyte-to-embryo transition in C. elegans. Dev. Biol. 288: 139–149. 10.1016/j.ydbio.2005.09.053 [DOI] [PubMed] [Google Scholar]
- Nugroho T. T., Mendenhall M. D., 1994. An inhibitor of yeast cyclin-dependent protein kinase plays an important role in ensuring the genomic integrity of daughter cells. Mol. Cell. Biol. 14: 3320–3328. 10.1128/MCB.14.5.3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P., 1990. Universal control mechanism regulating onset of M-phase. Nature 344: 503–508. 10.1038/344503a0 [DOI] [PubMed] [Google Scholar]
- Nyström J., Shen Z. Z., Aili M., Flemming A. J., Leroi A., et al. , 2002. Increased or decreased levels of Caenorhabditis elegans lon-3, a gene encoding a collagen, cause reciprocal changes in body length. Genetics 161: 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Farrell P. H., 2001. Triggering the all-or-nothing switch into mitosis. Trends Cell Biol. 11: 512–519. 10.1016/S0962-8924(01)02142-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura K., Kishimoto N., Mitani S., Gengyo-Ando K., Kohara Y., 2003. Translational control of maternal glp-1 mRNA by POS-1 and its interacting protein SPN-4 in Caenorhabditis elegans. Development 130: 2495–2503. 10.1242/dev.00469 [DOI] [PubMed] [Google Scholar]
- Parry J. M., Velarde N. V., Lefkovith A. J., Zegarek M. H., Hang J. S., et al. , 2009. EGG-4 and EGG-5 link events of the oocyte-to-embryo transition with meiotic progression in C. elegans. Curr. Biol. 19: 1752–1757. 10.1016/j.cub.2009.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel N., Dougherty M., Goeres J., Liu Y., O’Connell K. F., 2012. The C. elegans F-box proteins LIN-23 and SEL-10 antagonize centrosome duplication by regulating ZYG-1 levels. J. Cell Sci. 125: 3535–3544. 10.1242/jcs.097105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J., Reinke V., Kim S. K., Seydoux G., 2003. Coordinate activation of maternal protein degradation during the egg-to-embryo transition in C. elegans. Dev. Cell 5: 451–462. 10.1016/S1534-5807(03)00231-4 [DOI] [PubMed] [Google Scholar]
- Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., et al. , 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. 10.1038/35002607 [DOI] [PubMed] [Google Scholar]
- Robertson S., Lin R., 2015. The maternal-to-zygotic transition in C. elegans. Curr. Top. Dev. Biol. 113: 1–42. 10.1016/bs.ctdb.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Rose K. L., Winfrey V. P., Hoffman L. H., Hall D. H., Furuta T., et al. , 1997. The POU gene ceh-18 promotes gonadal sheath cell differentiation and function required for meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 192: 59–77. 10.1006/dbio.1997.8728 [DOI] [PubMed] [Google Scholar]
- Rougvie A. E., Moss E. G., 2013. Developmental transitions in C. elegans larval stages. Curr. Top. Dev. Biol. 105: 153–180. 10.1016/B978-0-12-396968-2.00006-3 [DOI] [PubMed] [Google Scholar]
- Rybak A., Fuchs H., Hadian K., Smirnova L., Wulczyn E. A., et al. , 2009. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat. Cell Biol. 11: 1411–1420. 10.1038/ncb1987 [DOI] [PubMed] [Google Scholar]
- Scheckel C., Gaidatzis D., Wright J. E., Ciosk R., 2012. Genome-wide analysis of GLD-1-mediated mRNA regulation suggests a role in mRNA storage. PLoS Genet. 8: e1002742 10.1371/journal.pgen.1002742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl T., Kimble J., 1988. fog-2, a germline-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics 119: 43–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B., Hanazawa M., Lee M. H., Nayak S., Volkmann K., et al. , 2005. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell 120: 357–368. 10.1016/j.cell.2004.12.009 [DOI] [PubMed] [Google Scholar]
- Shaner N. C., Lambert G. G., Chammas A., Ni Y., Cranfill P. J., et al. , 2013. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10: 407–409. 10.1038/nmeth.2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Soto M. C., Ishidate T., Kim S., Nakamura K., et al. , 2006. The conserved kinases CDK-1, GSK-3, KIN-19, and MBK-2 promote OMA-1 destruction to regulate the oocyte-to-embryo transition in C. elegans. Curr. Biol. 16: 47–55. 10.1016/j.cub.2005.11.070 [DOI] [PubMed] [Google Scholar]
- Schwob E., Böhm T., Mendenhall M. D., Nasmyth K., 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisae. Cell 79: 233–244. 10.1016/0092-8674(94)90193-7 [DOI] [PubMed] [Google Scholar]
- Sijen T., Fleenor J., Simmer F., Thijssen K. L., Parrish S., et al. , 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107: 465–476. 10.1016/S0092-8674(01)00576-1 [DOI] [PubMed] [Google Scholar]
- Slack F. J., Ruvkun G., 1998. A novel repeat domain that is often associated with RING finger and B-box motifs. Trends Biochem. Sci. 23: 474–475. 10.1016/S0968-0004(98)01299-7 [DOI] [PubMed] [Google Scholar]
- Slack F. J., Basson M., Liu Z., Ambros V., Horvitz H. R., et al. , 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5: 659–669. 10.1016/S1097-2765(00)80245-2 [DOI] [PubMed] [Google Scholar]
- Sonneville R., Querenet M., Craig A., Gartner A., Blow J. J., 2012. The dynamics of replication licensing in live Caenorhabditis elegans embryos. J. Cell Biol. 196: 233–246. 10.1083/jcb.201110080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike C. A., Coetzee D., Eichten C., Wang X., Hansen D., et al. , 2014a. The TRIM-NHL protein LIN-41 and the OMA RNA-binding proteins antagonistically control the prophase-to-metaphase transition and growth of Caenorhabditis elegans oocytes. Genetics 198: 1535–1558. 10.1534/genetics.114.168831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike C. A., Coetzee D., Nishi Y., Guven-Ozkan T., Oldenbroek M., et al. , 2014b. Translational control of the oogenic program by components of OMA ribonucleoprotein particles in Caenorhabditis elegans. Genetics 198: 1513–1533. 10.1534/genetics.114.168823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitzel M. L., Pellettieri J., Seydoux G., 2006. The C. elegans DYRK kinase MBK-2 marks oocyte proteins for degradation in response to meiotic maturation. Curr. Biol. 16: 56–62. 10.1016/j.cub.2005.11.063 [DOI] [PubMed] [Google Scholar]
- Stitzel M. L., Cheng K. C., Seydoux G., 2007. Regulation of MBK-2/Dyrk kinase by dynamic cortical anchoring during the oocyte-to-zygote transition. Curr. Biol. 17: 1545–1554. 10.1016/j.cub.2007.08.049 [DOI] [PubMed] [Google Scholar]
- Stoeckius M., Grün D., Kirchner M., Ayoub S., Torti F., et al. , 2014. Global characterization of the oocyte-to-embryo transition in Caenorhabditis elegans uncovers a novel mRNA clearance mechanism. EMBO J. 33: 1751–1766. 10.15252/embj.201488769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohmaier H., Spruck C. H., Kaiser P., Won K.-A., Sangfelt O., et al. , 2001. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature 413: 316–322. 10.1038/35095076 [DOI] [PubMed] [Google Scholar]
- Sundaram M., Greenwald I., 1993. Suppressors of a lin-12 hypomorph define genes that interact with both lin-12 and glp-1 in Caenorhabditis elegans. Genetics 135: 765–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Morris G. A., Han M., Wood W. B., 2002. A cuticle collagen encoded by the lon-3 gene may be a target of TGF-beta signaling in determining Caenorhabditis elegans body shape. Genetics 162: 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P., Fulka H., Malik R., 2017. Clearance of parental products. Adv. Exp. Med. Biol. 953: 489–535. 10.1007/978-3-319-46095-6_10 [DOI] [PubMed] [Google Scholar]
- Timmons L., Fire A., 1998. Specific interference by ingested dsRNA. Nature 395: 854 10.1038/27579 [DOI] [PubMed] [Google Scholar]
- Tocchini C., Keusch J. J., Miller S. B., Finger S., Gut H., et al. , 2014. The TRIM-NHL protein LIN-41 controls the onset of developmental plasticity in Caenorhabditis elegans. PLoS Genet. 10: e1004533 10.1371/journal.pgen.1004533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T., Gearhart M. D., Spike C. A., Huelgas-Morales G., Mews M., et al. , 2017. LIN-41 and OMA ribonucleoprotein complexes mediate a translational repression-to-activation switch controlling oocyte meiotic maturation and the oocyte-to-embryo transition in Caenorhabditis elegans. Genetics 206: 2007–2039. 10.1534/genetics.117.203174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax J. A., Woodbury E. L., Quang P. N., Paraz M., Blethrow J. D., et al. , 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425: 859–864. 10.1038/nature02062 [DOI] [PubMed] [Google Scholar]
- van der Voet M., Lorson M. A., Srinivasan D. G., Bennett K. L., van den Heuvel S., 2009. C. elegans mitotic cyclins have distinct as well as overlapping functions in chromosome segregation. Cell Cycle 8: 4091–4102. 10.4161/cc.8.24.10171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlhac M. H., Terret M. E., Pintard L., 2010. Control of the oocyte-to-embryo transition by the ubiquitin-proteolytic system in mouse and C. elegans. Curr. Opin. Cell Biol. 22: 758–763. 10.1016/j.ceb.2010.09.003 [DOI] [PubMed] [Google Scholar]
- Verma R., Annan R. S., Huddleston M. J., Carr S. A., Reynard G., et al. , 1997. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278: 455–460. 10.1126/science.278.5337.455 [DOI] [PubMed] [Google Scholar]
- Wang J. T., Smith J., Chen B. C., Schmidt H., Rasoloson D., et al. , 2014. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife 3: e04591 10.7554/eLife.04591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M., Clurman B. E., 2008. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8: 83–93. 10.1038/nrc2290 [DOI] [PubMed] [Google Scholar]
- Welcker M., Larimore E. A., Swanger J., Bengoechea-Alonso M. T., Grim J. E., et al. , 2013. Fbw7 dimerization determines the specificity and robustness of substrate degradation. Genes Dev. 27: 2531–2536. 10.1101/gad.229195.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolke U., Jezuit E. A., Priess J. R., 2007. Actin-dependent cytoplasmic streaming in C. elegans oogenesis. Development 134: 2227–2236. 10.1242/dev.004952 [DOI] [PubMed] [Google Scholar]
- Wright J. E., Gaidatzis D., Senften M., Farley B. M., Westhof E., et al. , 2011. A quantitative RNA code for mRNA target selection by the germline fate determinant GLD-1. EMBO J. 30: 533–545. 10.1038/emboj.2010.334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worringer K. A., Rand T. A., Hayashi Y., Sami S., Takahashi K., et al. , 2014. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell 14: 40–52. 10.1016/j.stem.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka A., Yada M., Imaki H., Koga M., Ohshima Y., et al. , 2002. Multiple Skp1-related proteins in Caenorhabditis elegans: diverse patterns of interaction with Cullins and F-box proteins. Curr. Biol. 12: 267–275. 10.1016/S0960-9822(02)00657-7 [DOI] [PubMed] [Google Scholar]
- Yang X., Lau K.-Y., Sevim V., Tang C., 2013. Design principles of the yeast G1/S switch. PLoS Biol. 11: e1001673 10.1371/journal.pbio.1001673 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains and newly created alleles (see File S1 and Table S1) are available upon request. The sequences of gRNAs, repair templates, PCR primers, lin-41 alleles, and oma-2 alleles are presented in File S1. Plasmids producing gRNAs and those containing repair templates for genome editing are available upon request. All Sanger sequencing files are available upon request. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7056557.