Abstract
Background
The familial hyperkalemic hypertension (FHHt) cullin 3 (CUL3) mutant does not degrade WNK kinases normally, thereby leading to thiazide-sensitive Na-Cl cotransporter (NCC) activation. CUL3 mutant (CUL3Δ9) does not bind normally to the COP9 signalosome (CSN), a deneddylase involved in regulating cullin-RING ligases. CUL3Δ9 also caused increased degradation of the CUL3-WNK substrate adaptor kelch-like 3 (KLHL3). Here, we sought to determine how defective CSN action contributes to the CUL3Δ9 phenotype.
Methods
The Pax8/LC1 mouse system was used to generate mice in which the catalytically active CSN subunit, Jab1, was deleted only along the nephron, after full development (KS-Jab1−/−).
Results
Western blot analysis demonstrated that Jab1 deletion increased the abundance of neddylated CUL3. Moreover, total CUL3 expression was reduced, suggesting decreased CUL3 stability. KLHL3 was almost completely absent in KS-Jab1−/− mice. Conversely, the protein abundances of WNK1, WNK4, and SPAK kinases were substantially higher. Activation of WNK4, SPAK, and OSR1 was indicated by higher phosphorylated protein levels and translocation of the proteins into puncta, as observed by immunofluorescence. The ratio of phosphorylated NCC to total NCC was also higher. Surprisingly, NCC protein abundance was low, likely contributing to hypokalemia and Na+ and K+ wasting. Additionally, long-term Jab1 deletion resulted in kidney damage.
Conclusions
Together, the results indicate that deficient CSN binding contributes importantly to the FHHt phenotype. Although defective CUL3Δ9-faciliated WNK4 degradation likely contributes, dominant effects on KLHL3 may be a second factor that is necessary for the phenotype.
Keywords: renal hypertension, Na transport, distal tubule, Cell & Transport Physiology
The thiazide-sensitive Na-Cl cotransporter (NCC) in the distal convoluted tubule (DCT) of the kidney is modulated through the WNK-SPAK/OSR1 kinase pathway. WNKs (with-no-lysine kinases) phosphorylate intermediary kinases serine/threonine protein kinase 39 (SPAK; STK39) and oxidative stress-response 1 (OxSR1 or OSR1; OXSR1), which then directly phosphorylate NCC, activating the cotransporter.1,2 Increased WNK activity causes the disease familial hyperkalemic hypertension (FHHt), also known as pseudohypoaldosteronism type 2 or Gordon syndrome.3 FHHt is characterized by hypertension, hyperkalemia, and metabolic acidosis, which can be treated with thiazide diuretics.4 The disease is caused by mutations in WNK1 and WNK4,5 and also mutations in the cullin-RING ligase (CRL) proteins kelch-like 3 (KLHL3)6 and cullin 3 (CUL3).7 Although the effects of CUL3 mutations have been studied extensively, a consensus understanding of the mechanisms by which they cause human disease has not emerged.
All mutations causing FHHt lead to accumulation of WNK proteins. WNK4, KLHL3, and CUL3 mutations all increase WNK abundance by impairing WNK protein degradation,8–12 via CRLs. CRLs are modular protein complexes containing one of eight cullin proteins (CUL1, 2, 3, 4A, 4B, 5, 7, and 9),13 bound to a RING protein, which transfers ubiquitin to a substrate targeting it to the proteasome for degradation. The substrate does not typically interact directly with the cullin but instead through an adaptor protein and substrate receptor. In the case of CUL3, however, the substrate adaptor and receptor are merged into a single protein.14 For simplicity we will refer to CRL adaptors and receptors as substrate adaptors. In humans, hundreds of substrate adaptors have been identified, each able to bind to one or a few specific proteins.15 WNKs interact with CUL3 via the substrate adaptor KLHL3.16
Before ubiquitylation can occur the CRL must first be activated. Activation occurs via neddylation, a process in which a NEDD8 (neuronal precursor cell expressed developmentally downregulated protein 8) protein is covalently attached to the cullin.17,18 Neddylation increases flexibility of the cullin-RING structure, which closes the gap between the ubiquitin-charged E2 (ubiquitin-conjugating enzyme) and the substrate.19 In vivo, neddylation activates the CRL, but normal function can only be maintained through subsequent NEDD8 removal, as hyperneddylation paradoxically impairs substrate degradation.20 NEDD8 removal occurs via attachment of the multisubunit deneddylase COP9 signalosome (CSN).21 Deneddylation prevents overactivation of the CRL, which can lead to aberrant ubiquitylation targeting the substrate adaptor and/or the cullin protein itself.22
The CSN is one of the three Proteasome-COP9 signalosome-Initiation factor 3 (PCI) domain-containing complexes and consists of eight subunits.23 The subunits CSN1–4, and CSN7–8 make up a horseshoe-shaped ring structure and each contain a PCI domain, whereas the core subunits CSN5 and CSN6 are Mpr1-Pad1-N-terminal (MPN) domain-containing proteins. Unlike CSN6, CSN5 contains a JAB1/MPN/Mov34 motif conferring Zn2+-dependent metalloprotease activity. Although CSN5 (also known as JAB1 or jun activation domain-binding protein-1) is the sole isopeptidase, all subunits are required for enzymatic activity.24 In mice, genetic deletion of CSN subunits 2, 3, 5, 6, or 8 is embryonic lethal.25–29 Inhibition of any subunit, either in vitro30–34 or in vivo,33,35 leads to decreased expression of substrate adaptor proteins. Two groups have shown that the CUL3 FHHt mutant (CUL3Δ9) has impaired binding to the CSN11,36; our group found that CUL3Δ9 also targets KLHL3 for degradation. The loss of interaction with the CSN could account for enhanced KLHL3 degradation, perhaps explaining the disease phenotype, as we postulated previously.37
We recently proposed, on the basis of work primarily in cells, that CUL3Δ9 exhibits both gain- and loss-of-function features, with regard to specific substrates and substrate adaptors.36 As CSN’s catalytic activity is contained within the JAB1 subunit,21 and CUL3Δ9 is hyperneddylated in cultured cells,11,37 we postulated that hyperneddylation contributes to substrate accumulation and substrate adaptor instability. Here, we generated mice in which JAB1 could be deleted from kidney-tubule cells (KS-Jab1−/−) to test the importance of the CSN-binding deficiency in disease pathogenesis, in vivo.
Methods
Antibodies
Antibodies used are described in Supplemental Table 1.
Animals
Animal studies were approved by the Oregon Health and Science University Institutional Animal Care and Use Committee (protocol IP000286). The Pax8-rtTA/LC1 system38,39 was used to generate inducible renal epithelia-specific Jab−/− mice. Jab1flx/flx mice were generated by R.P.40 and generously provided by Dr. Guang Zhou. These mice developed normally, were phenotypically similar to wild-type (WT) mice, and had normal JAB1 expression. Jab1flx/flx mice were crossed with Pax8/LC1 mice to generate Jab1flx/flx-Pax/LC1 mice. Doxycycline was administered at 2 mg/ml with 5% sucrose in drinking water for 3 weeks to generate renal epithelia-specific Jab1−/− mice (referred to as KS-Jab1−/−). Heterozygous Jab1 mice were generated by giving doxycycline to Jab1flx/− mice (referred to as KS-Jab1+/−). Some KS-Jab1+/− mice were fed a high sodium (6%), low potassium diet (0%). Control mice were littermates that received 5% sucrose in drinking water. Both males and female were used for experiments except for BP measurement, in which only male mice were used, and all mice were on a C57Bl/6J background.
The mice were returned to regular drinking water for 3 weeks before they were placed in metabolic cages for 24-hour urine collection and analysis of food and water consumption. To prevent urine contamination, mice were fed a gel diet. Mice were acclimated in the metabolic cages for 3 days before analysis. Urine K+ and Na+ were determined by flame photometry. Plasma Mg2+ and urinary Ca2+ was analyzed using colorimetric assay kits (Pointe Scientific, Canton, MI).To determine the effects of long-term deletion of Jab1, mice were returned to regular drinking water for 9 weeks before kidneys were harvested.
Immunohistochemistry
For hematoxylin and eosin staining, mice were anesthetized, kidneys were removed and placed in a 10% formalin solution (Thermo Fischer Scientific, Waltham, MA) for 24 hours. The kidneys were then transferred to a 70% ethanol solution for 72 hours. The Oregon Health and Science University Histopathology Core Facility embedded the kidney in paraffin and performed hematoxylin and eosin staining.
Immunofluorescence
Mice were anesthetized with a ketamine/xylazine/acepromazine cocktail and kidneys were perfusion-fixed with retrograde abdominal aortic perfusion of 3% paraformaldehyde in PBS (pH 7.4). Kidneys were removed, dissected, and cryopreserved in 800 mOsm sucrose in PBS overnight before embedding in Tissue-Tek O.C.T. compound (Sakura Finetek, Torrance, CA). Slides were prepared by cutting 5 mm sections and were stored at −80°C until use. Immunofluorescence staining was prepared as follows. Slides were incubated with 0.5% Triton-X 100 in PBS for 30 minutes. Sections were then blocked with 5% milk in PBS for 30 minutes, followed by incubation with primary antibody, diluted in blocking buffer, for 1 hour at room temperature or overnight at 4°C (details are provided in Supplemental Table 1). Sections were incubated with fluorescent dye–conjugated secondary antibody, diluted in blocking buffer, for 1 hour at room temperature before being mounted with Prolong Diamond Antifade Mountant with DAPI (Invitrogen, Carlsbad, CA). Images were captured using a Zeiss AXIO Imager M2 microscope and AxioVision software. Image processing was completed using ImageJ software (National Institutes of Health).
Western Blotting
Kidneys were harvested and flash frozen in liquid nitrogen and stored at −80°C. Whole kidney was homogenized in buffer containing enzyme inhibitors with 1 mM dithiothreitol and 1 mM PMSF. Protein samples were separated by electrophoresis on 4%–15% Criterion TGX stain-free gels (Bio-Rad Laboratories, Hercules, CA) and transferred to Immobilon-P polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA) using the Trans-Blot Turbo transfer system (Bio-Rad Laboratories). Stain-free imaging was used as a total protein loading control (Supplemental Figure 1). Membranes were blocked with 5% nonfat milk in PBS-Tween for 1 hour at room temperature, before incubation with primary antibody in blocking buffer for 1 hour at room temperature or overnight at 4°C (details are provided in Supplemental Table 1). KLHL3 antibody (Proteintech, Rosemont, IL) was incubated in Can Get Signal (TOYOBO, Osaka, Japan). Appropriate horseradish peroxidase–conjugated secondary antibody in blocking buffer was added to membranes for 1 hour at room temperature. Membranes were developed using enhanced chemiluminescence, Western Lightning Plus–ECL (Perkin Elmer, Waltham, MA), and proteins were visualized using PXi digital imaging system (Syngene, Frederick, MA).
Quantitative PCR
Kidneys were harvested and RNA was preserved by treating with RNAlater RNA Stabilization Reagent (Qiagen, Hilden, Germany) overnight at 4°C. Kidney tissue was then homogenized in PBS and immediately transferred to QIAzol lysis reagent, where it was further homogenized by running the sample through a 20 gauge blunt cannula. Total RNA was isolated using RNeasy Plus Universal Mini kit (Qiagen). The total RNA (1000 ng) was reverse transcribed using a High-Capacity RNA-to-cDNA kit (Applied Biosystems, Foster City, CA). The resulting complementary DNA was diluted 1:10 and real-time quantitative PCR was performed on a QuantStudio 7 Flex Real-Time PCR System using TaqMan Universal Master Mix II (Applied Biosystems). The primers used were TaqMan Gene Expression Assays ID Mm00490213_m1 for NCC (Slc12a3) and Mm03928990_g1 for 18S (Rn18s) containing FAM dye. mRNA expression levels were calculated with the comparative threshold cycle method (2−ΔΔCt) and 18S mRNA was used for normalization.
Blood Analysis
Whole blood was collected via cardiac puncture from mice under isoflurane anesthesia and transferred to heparin-containing tubes. The blood was analyzed via an iSTAT analyzer (Abbott, Princeton, NJ) using Chem8+ cartridges. Aldosterone was measured by ELISA (IBL-America, Minneapolis, MN) from the plasma by centrifuging whole blood at 2000×g for 10 minutes at 4°C.
BP Measurement
Radio telemetry was used to measure BP in male mice only, using PA-C10 transmitters (Data Sciences International, St. Paul, MN). Mice were anesthetized with isoflurane and transmitters were implanted in the left carotid artery. Measurements were recorded for 10 seconds every 10 minutes. Mean of the hourly averages were used to calculate mean systolic BP.
Cell Culture, Plasmids, and Transfections
HEK293 cells were maintained in DMEM supplemented with 10% FBS, 25 mM HEPES, 100 U/ml penicillin, and 100 µg/ml streptomycin. Cells were transiently transfected using lipofectamine 2000 (Ambion, Foster City, CA; Invitrogen). Constructs were made by amplifying FLAG-CUL3 WT DNA using Phusion Hot Start II DNA Polymerase (Thermo Fisher Scientific, Boston, MA) with the appropriate primers, purified with the PureLink PCR Purification Kit (Invitrogen) and properly digested. The products were then extracted using the UltraClean GelSpin DNA Extraction Kit (MoBio Laboratories, Inc., Carlsbad, CA) and ligated into mammalian plasmids with T4 DNA Ligase (New England BioLabs, Ipswich, MA). Ligated constructs were transformed using DH5α competent cells (Thermo Fisher Scientific) and plasmid DNA was purified with either the QIAprep Spin Miniprep Kit or HiSpeed Plasmid Midi Kit (Qiagen). Sanger sequencing was performed for all constructs. For small interfering RNA (siRNA) experiments, either 40 nM of COPS5 siRNA (Ambion) or control siRNA was transfected along with DNA plasmids. Cells were harvested at 36 hours post-transfection. Transfected cells were harvested in 0.5% Triton X-100 in PBS cell lysis buffer containing enzyme inhibitors. All experiments were performed in triplicate, with each well representing a unique transfection. Western blotting was performed as explained above.
Statistical Analyses
Data are presented as individual values as well as mean±SEM. Differences between two groups were determined using two-tailed unpaired t test. Differences between multiple groups were determined using one-way ANOVA with Tukey post hoc analysis. A P-value of <0.05 was considered significant. Statistical analyses were performed using GraphPad Prism 7 software (GraphPad Software, San Diego, CA).
Results
CSN Is Abundant in the Distal Nephron
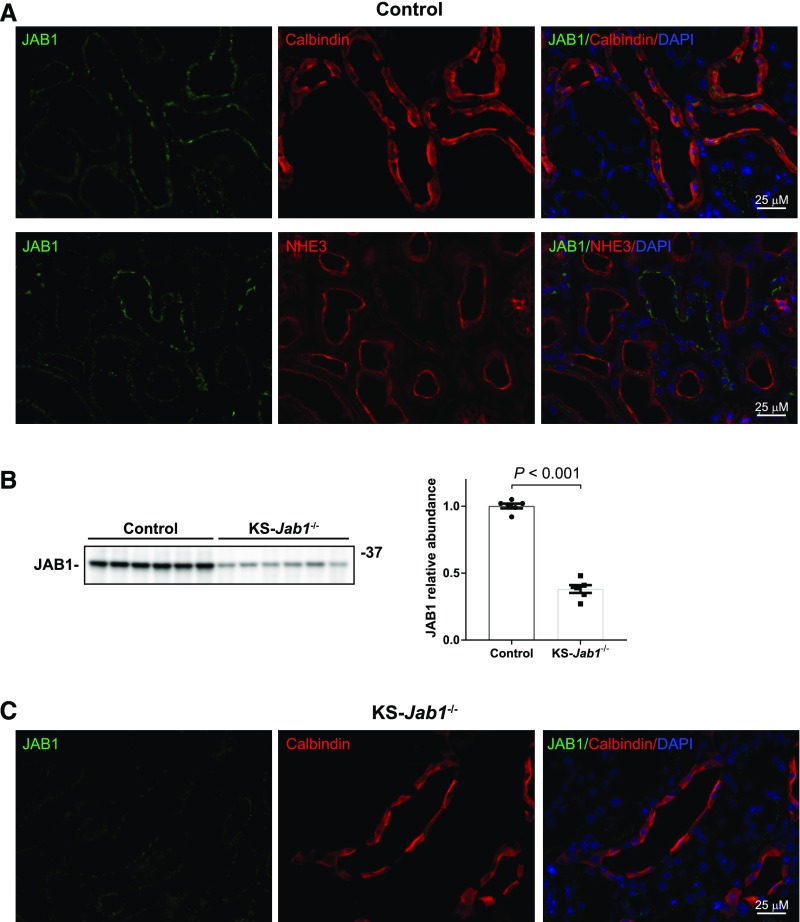
Immunofluorescence staining of mouse kidney sections showed that JAB1 is most highly expressed in segments that also express calbindin (Figure 1A), a marker for the distal nephron. Expression of JAB1 was most prominent in apical and subapical regions. In contrast, JAB1 was not abundant in proximal tubules, as indicated by costaining with Na-H exchanger 3.
Figure 1.
JAB1 expression is abundant in the distal nephron. (A) Immunofluorescence staining of kidney cortex sections in control mice. Top, JAB1 localization was examined in the distal nephron by costaining with the distal nephron marker calbindin. The staining intensity of JAB1 was higher in calbindin-positive cells. Bottom, JAB1 localization was examined in the proximal tubule by costaining with the Na-H exchanger 3 (NHE3). JAB1 did not colocalize with NHE3-positive tubules. (B and C) Doxycycline-induced Jab1 deletion. Doxycycline or vehicle control was administered to Jab1flx/flx Pax8/LC1 mice for 3 weeks. After 3 weeks of recovery, JAB1 protein abundance and localization was measured by Western blot (B) and immunofluorescence (C). Using whole kidney lysates, immunoblotting for JAB1 showed 62% knockdown in KS-Jab1−/− mice compared with controls. JAB1 staining was reduced in KS-Jab1−/− mice. Data represent individual values as well as mean±SEM relative to controls. Statistical differences were examined using two-tailed unpaired t test.
Disruption of the CSN along the Nephron Increases Both Neddylation and Degradation of CUL3
To examine the role of the CSN in kidney in vivo, we crossed Jab1flx/flx mice with Pax8/LC1. Homozygotic Jab1flx/flx and Pax8/LC1-positive mice were then administered doxycycline for 3 weeks before a 3-week recovery period. Protein analysis by Western blot showed a 62% knockdown of JAB1 in whole kidney lysates (Figure 1B). Although the Pax8/LC1 system is not 100% efficient, the residual JAB1 protein probably reflects expression in nontubular cells. Examination of JAB1 by immunofluorescence staining demonstrated lower staining intensity in the distal nephron of kidney sections, as determined by costaining with calbindin (Figure 1C). We have called these mice kidney-specific Jab1 knockout mice (KS-Jab1−/− mice).
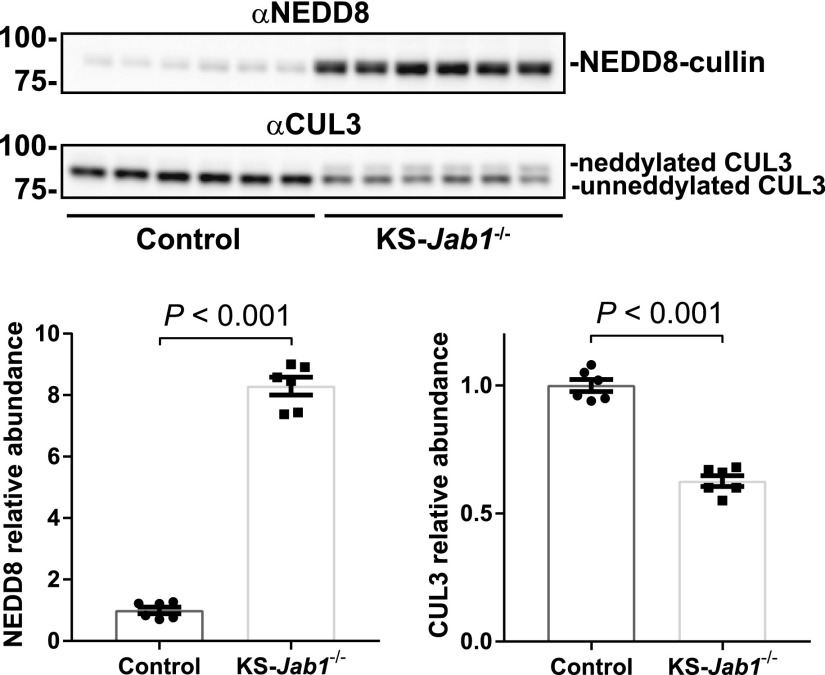
Inhibition of the CSN should block deneddylation, therefore increasing the amount of neddylated cullin. To test this, we immunoblotted for NEDD8. Western blot analysis showed a distinct band at approximately 85 kDa, which is the same as the molecular mass of cullins (Figure 2). The NEDD8-cullin band was strikingly more abundant in KS-Jab1−/− mice compared with controls. To confirm enhanced CUL3 neddylation we examined CUL3 protein. Although immunoblotting for CUL3 revealed two bands in the in KS-Jab1−/− mice, the higher molecular mass band, representing the neddylated form of CUL3, was absent in control mice. Additionally, quantitative analysis of total CUL3 showed a 37% reduction in protein abundance in KS-Jab1−/− mice compared with controls. The results indicate that most of the CUL3 protein under normal conditions is in the unneddylated state. Inhibition of the CSN causes hyperneddylation of CUL3, which also destabilizes the protein.
Figure 2.
Nephron-specific Jab1 deletion increases neddylation and degradation of CUL3. Western blot of whole kidney lysates from control and KS-Jab1−/− mice. Left, immunoblotting was performed with antibodies against NEDD8 and CUL3. Immunoblotting for NEDD8 showed increased protein abundance in KS-Jab1−/− mice compared with controls. Immunoblotting for CUL3 showed a neddylated (top) and unneddylated (bottom) band. The neddylated CUL3 band showed a higher abundance in KS-Jab1−/− mice compared with controls. Additionally, total CUL3 protein abundance was lower in KS-Jab1−/− mice compared with controls. Right, quantitative analysis of NEDD8 and CUL3 protein abundance. Data represent individual values as well as mean±SEM relative to controls. Statistical differences were examined using two-tailed unpaired t test.
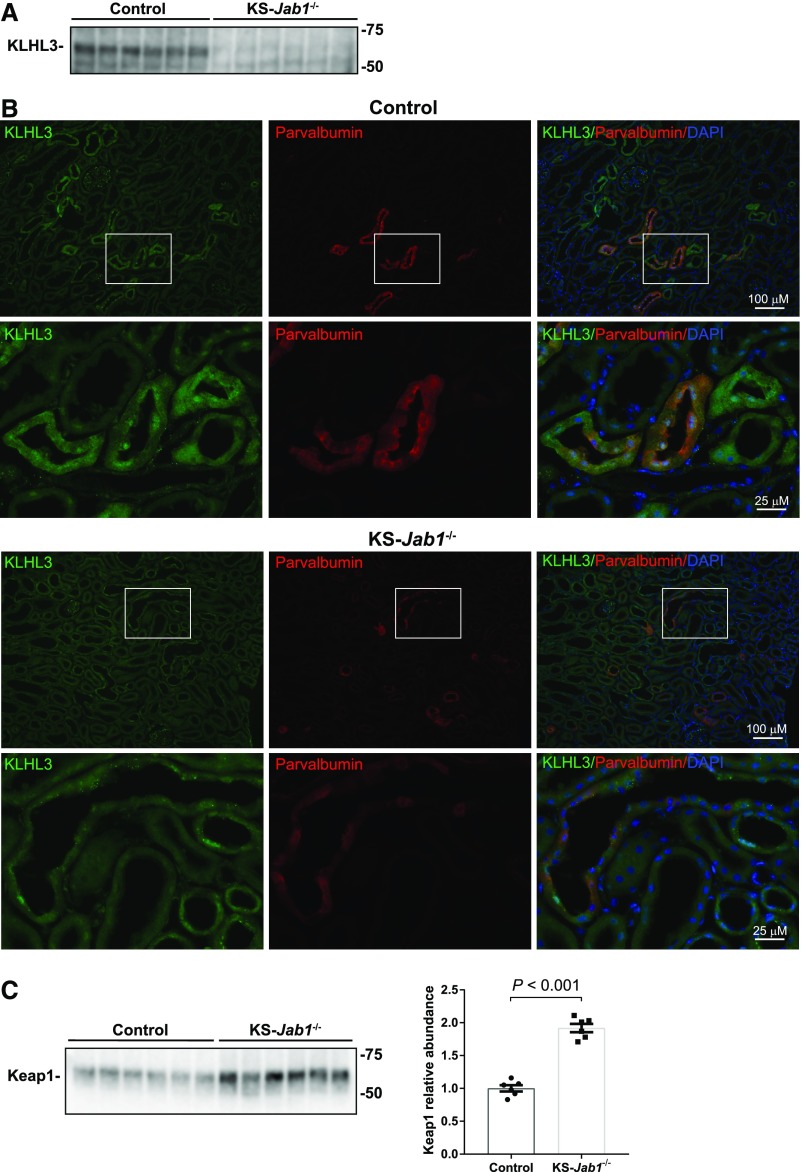
Disruption of CSN along the Nephron Reduces KLHL3 Protein Abundance
To examine the effect of CSN inhibition on KLHL3 protein abundance in vivo, we used KS-Jab1−/− mice and studied them using a commercial KLHL3 antibody that has been validated with KLHL3 knockout mice.41 Other KLHL3 antibodies validated in this way have proven nonspecific.42 KLHL3 protein abundance was substantially diminished, as shown by Western blot (Figure 3A) and immunofluorescence (Figure 3B). Costaining with the DCT1 marker parvalbumin in control mice demonstrated high levels of KLHL3 protein localized in the DCT1. Additionally, KLHL3 colocalized with calbindin (data not shown) indicating expression in the distal nephron, consistent with our previous work.37 Because CSN inhibition has been shown to differentially affect substrate adaptors, we examined the effect of Jab1 deletion on another CUL3 substrate adaptor Keap1 (kelch-like ECH-associated protein 1). Keap1 protein abundance was increased in KS-Jab1−/− mice (Figure 3C).
Figure 3.
Nephron-specific Jab1 deletion reduces KLHL3 protein abundance. (A and C) Western blot of whole kidney lysates from control and KS-Jab1−/− mice. (A) Immunoblotting was performed with antibodies against KLHL3. KLHL3 protein abundance was almost completely diminished in KS-Jab1−/− mice compared with controls. (B) Immunofluorescence staining of kidney cortex sections from control and KS-Jab1−/− mice. KLHL3 expression was examined in kidney cortex by costaining with the DCT1 marker parvalbumin. In control mice, KLHL3 staining intensity was most prominent in tubules expressing parvalbumin. KS-Jab1−/− mice showed lower levels of KLHL3 staining compared with controls. (C) Immunoblotting was performed with antibodies against Keap1. KS-Jab1−/− mice showed higher Keap1 protein levels compared with control mice. The graph shows the quantitative analysis of Keap1 protein abundance. Data represent individual values as well as mean±SEM relative to controls. Statistical differences were examined using two-tailed unpaired t test.
Disruption of CSN along the Nephron Activates the WNK-SPAK/OSR1 Pathway
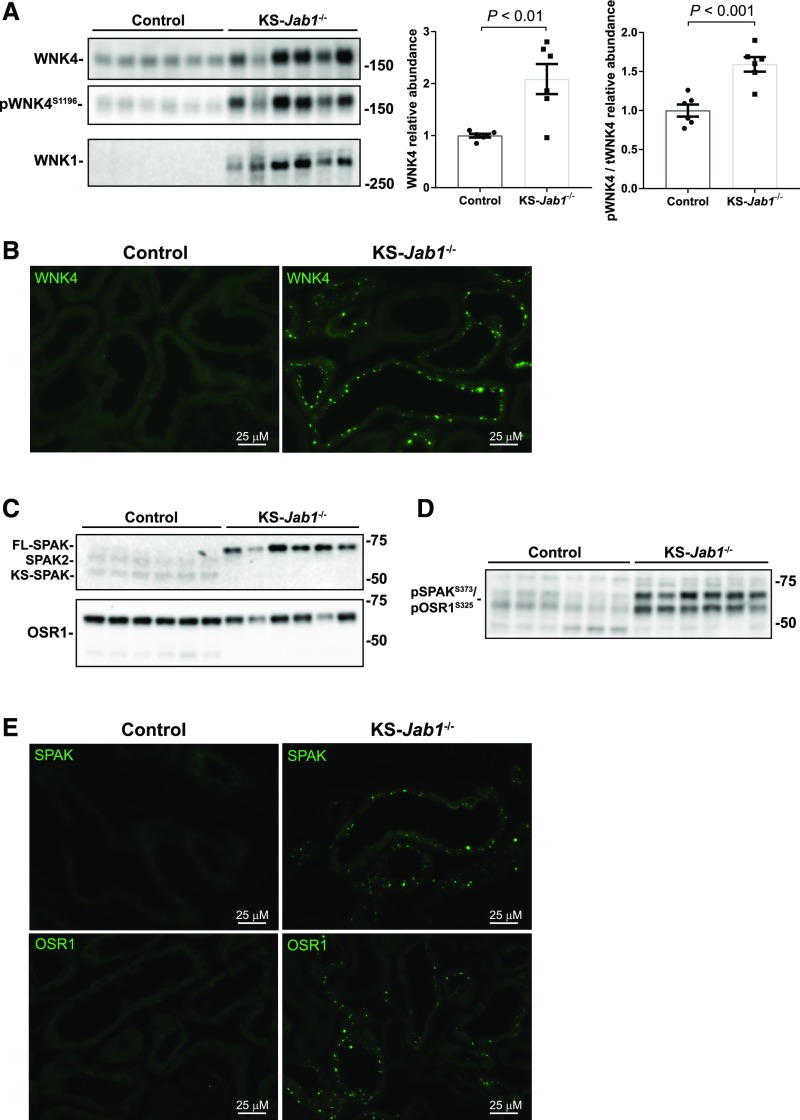
We next studied the effects of CSN inhibition on WNK/SPAK abundance. Here, KS-Jab1−/− mice had an increase in the total protein abundance of WNK1 and WNK4 (Figure 4A). Phosphorylation of WNK4 at S1196 was also higher in KS-Jab1−/− mice. Furthermore, the ratio of phosphorylated WNK4 to total WNK4 was increased. Immunofluorescence staining of WNK4 showed increased intensity and translocalization into puncta (Figure 4B) in the distal nephron, as identified by costaining with calbindin (Supplemental Figure 2). This punctate formation has been observed in CUL3Δ9 mice,11,43 and under conditions in which the protein is activated.44
Figure 4.
Nephron-specific Jab1 deletion activates the WNK-SPAK/OSR1 pathway. (A, C, and D) Western blot of whole kidney lysates from control and KS-Jab1−/− mice. (A) Left, immunoblotting was performed with antibodies against WNK4, phosphorylated WNK4 at serine 1196 (pWNK4S1196), and WNK1. WNK4, pWNK4S1196, and WNK1 protein abundance was higher in KS-Jab1−/− mice compared with controls. Right, quantitative analysis of total WNK4 protein abundance and the ratio of pWNK4S1196 to total WNK4 (pWNK4/tWNK4). Total WNK4 abundance and pWNK4/tWNK4 was higher in KS-Jab1−/− mice compared with controls. Data represent individual values as well as mean±SEM relative to controls. Statistical differences were examined using two-tailed unpaired t test. (B and E) Immunofluorescence staining of kidney cortex sections from control and KS-Jab1−/− mice. (B) Immunofluorescence staining was performed with antibodies against WNK4. There was low abundance of WNK4 staining in control mice relative to KS-Jab1−/− mice. Staining of KS-Jab1−/− mice kidney sections showed WNK4 protein translocated into puncta. (C) Immunoblotting was performed with antibodies against SPAK and OSR1. Protein abundance of full-length SPAK was higher, whereas SPAK2 and KS-SPAK were lower in KS-Jab1−/− mice compared with controls. (D) Immunoblotting was performed with antibodies against pSPAK/pOSR1 at serine 373 for SPAK and serine 325 for OSR1. Phosphorylation levels were higher in KS-Jab1−/− mice compared with controls. (E) Immunofluorescence staining was performed with antibodies against SPAK and OSR1. There was low abundance of SPAK (top) and OSR1 (bottom) staining in control mice relative to KS-Jab1−/− mice. Staining of KS-Jab1−/− mice kidney sections showed SPAK/OSR1 protein translocated into puncta.
The intermediary kinases SPAK and OSR1 link WNKs to NCC, and were examined by Western blot and immunofluorescence. Western blotting for SPAK showed an increase in the full-length SPAK, with lower abundance of the shorter forms, SPAK, SPAK2, and KS-SPAK in KS-Jab1−/− mice (Figure 4C). Additionally, immunoblotting for phosphorylated SPAK and OSR1 using an antibody that is specific for phospho-SPAK at serine 373 and phospho-OSR1 at serine 325 showed an increase in phosphorylated protein (Figure 4D). Immunofluorescence staining of SPAK and OSR1 showed a pattern similar to WNK4, with control mice displaying low staining intensity and KS-Jab1−/− mice showing staining localized into puncta (Figure 4E). Puncta formation was localized to distal nephron cells as shown by costaining with calbindin (Supplemental Figure 2). The results demonstrate that the WNK-SPAK/OSR1 pathway is activated in KS-Jab1−/− mice.
The results here also demonstrate a distal nephron-specific phenotype. WNK4 is expressed in both the DCT and thick ascending limb (TAL).45 Although SPAK and OSR1 are also expressed in these nephron segments, SPAK is the main activator of NCC in the DCT and OSR1 is the main activator of the Na-K-2Cl cotransporter (NKCC2) in the TAL.46–48 SPAK and OSR1 immunofluorescence showed punctate formation in the distal nephron, whereas there was no change in expression in the TAL (Supplemental Figure 2). Furthermore, total and phosphorylated NKCC2 protein levels were unchanged in KS-Jab1−/− mice (Supplemental Figure 3), indicating that the effects of KS-Jab1−/− mice on the WNK-SPAK/OSR1 pathway are localized to the distal nephron.
Disruption of CSN along the Nephron Reduces NCC Abundance
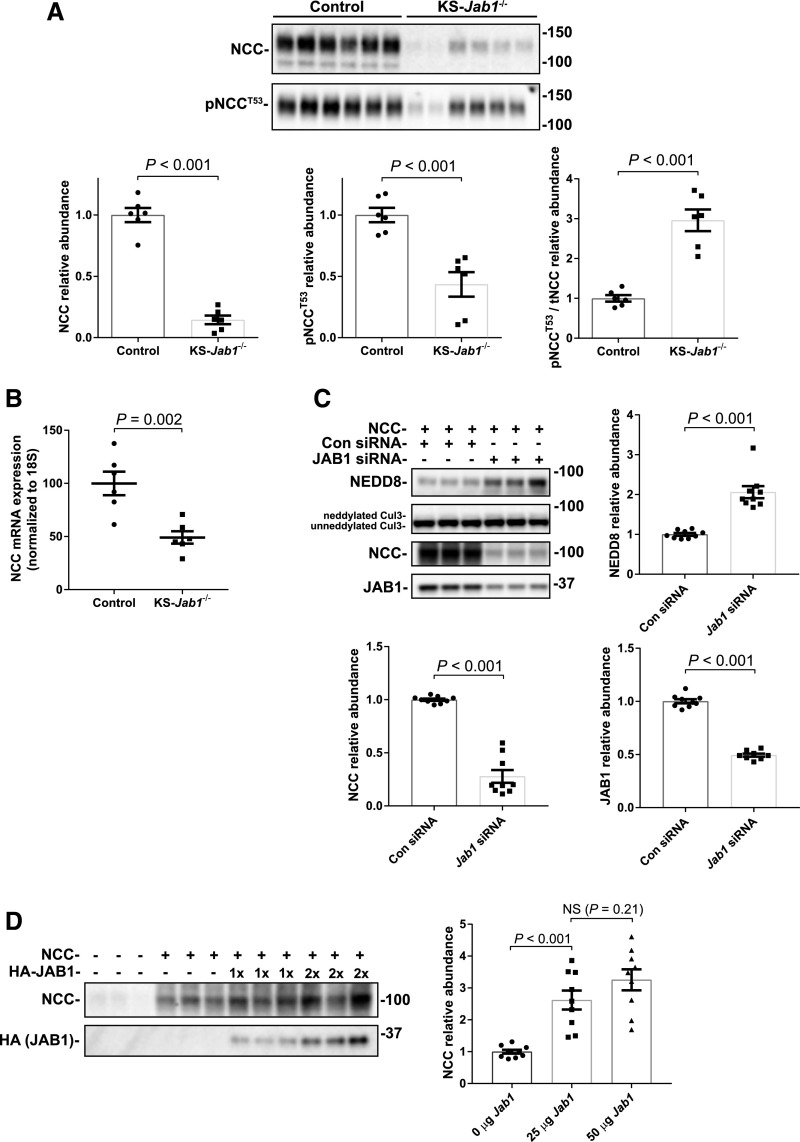
Because KS-Jab1−/− mice demonstrated activation of the WNK-SPAK pathway, it might be suspected that NCC protein phosphorylation would also be higher in these mice. In contrast, both total NCC (tNCC) and phosphorylated NCC (pNCC) at T53 were dramatically lower than in control (Figure 5A). Although both total and activated (phosphorylated) forms of NCC were low, there was a three-fold increase in the ratio of pNCC to tNCC in KS-Jab1−/− mice compared with controls. This suggests that, although the WNK-SPAK pathway may be functioning to activate NCC, a second process, either increased NCC degradation or decreased NCC synthesis must be present. Evidence for destructive effects outside of the DCT is the low aquaporin 2 abundance, as shown by Western blot (Supplemental Figure 3). To test whether NCC synthesis was decreased, we performed quantitative RT-PCR. Although NCC proteins levels were lower by approximately 85%, NCC (Slc12a3 gene) mRNA levels were decreased by approximately 50% in KS-Jab1−/− mice compared with controls (Figure 5B). This indicates that some, but not all, of the decreased NCC abundance may be due to effects on transcription.
Figure 5.
JAB1 dysfunction reduces NCC expression. (A) Top, Western blot of whole kidney lysates from control and KS-Jab1−/− mice. Immunoblotting was performed with antibodies against NCC or pNCC at threonine 53 (pNCCT53). Both NCC and pNCCT53 were lower in KS-Jab1−/− mice compared with controls. Bottom, quantitative analysis of NCC and pNCCT53 protein abundance. Both NCC and pNCCT53 protein abundance were lower in KS-Jab1−/− mice compared with controls. The ratio of pNCCT53 to tNCC was approximately three-fold higher in KS-Jab1−/− mice compared with controls. (B) Quantitative RT-PCR for the expression of NCC (Slc12a3 gene) in the kidney. The results were normalized to 18S mRNA expression. NCC mRNA expression was decreased in KS-Jab1−/− mice compared with controls. (C) NCC was cotransfected into HEK293 cells with either Jab1 siRNA or control siRNA. Top, Protein abundance was examined by immunoblotting using antibodies against NEDD8, CUL3, NCC, and JAB1. Bottom, quantitative analysis of NEDD8, NCC, and JAB1 protein abundance. Jab1 siRNA decreased JAB1 and NCC protein abundance and caused an increase in cullin neddylation as shown by increase in NEDD8 abundance at the molecular mass of cullins, and by an increase in the neddylated CUL3 band. (D) NCC was cotransfected into HEK293 cells together with increasing amounts of HA-tagged JAB1. Left, Immunoblotting for NCC showed an increase in protein abundance when cotransfected with 25 µg of JAB1, and trended higher when cotransfected with 50 µg of JAB1. Right, quantitative analysis of NCC protein. Data represent individual values as well as mean±SEM relative to controls. Statistical differences were examined using two-tailed unpaired t test (A–C) or by one-way ANOVA with Tukey post hoc analysis (D).
Changes in JAB1 Expression Affect NCC Abundance in HEK293 Cells
To determine whether the decrease in NCC abundance in KS-Jab1−/− mice is directly caused by Jab1 deletion and not from systemic effects, we examined the effects of JAB1 expression on NCC abundance in vitro using HEK293 cells. First, to mimic the KS-Jab1−/− mice we inhibited endogenous JAB1 expression by using siRNA in cells overexpressing NCC (Figure 5C). As shown previously,36 a reduction in JAB1 protein caused an increased in cullin neddylation compared with control siRNA. NCC abundance was dramatically reduced in cells transfected with Jab1 siRNA compared with cells containing control siRNA. Next, to see if the opposite result could be achieved, we transfected NCC together with overexpressed JAB1 (Figure 5D). Cells overexpressing JAB1 caused NCC abundance to increase compared with cells without transfected JAB1. Doubling the amount of JAB1 transfected into cells caused NCC abundance to trend higher; however, the result was not significant. Together the results indicate that JAB1 affects NCC through a WNK-independent pathway.
Disruption of CSN along the Nephron Causes Hypokalemia and Electrolyte Loss
KS-Jab1−/− mice were hypokalemic, hyperchloremic, and acidemic (Table 1). Additionally, the knockout mice exhibited Na+ and K+ wasting with hematocrits that tended to be higher, suggesting mild extracellular fluid volume contraction. The plasma aldosterone concentration in KS-Jab1−/− mice was twice the value of control mice (Table 1). Furthermore, telemetric analysis of BP showed no difference between KS-Jab1−/− and control mice (Supplemental Figure 4, Table 1).
Table 1.
Plasma and urine electrolytes and renal function
| Control | KS-Jab1−/− | Significance | P-Value | |
|---|---|---|---|---|
| Plasma | ||||
| Na+, mM | 144.8±0.75 | 144.3±0.76 | NS | 0.65 |
| K+, mM | 3.55±0.06 | 3.20±0.07 | a | 0.004 |
| Mg2+ (mg/dl) | 1.84±0.10 | 1.76±0.11 | NS | 0.58 |
| Cl− (mM) | 112.7±1.26 | 117.5±1.67 | b | 0.04 |
| iCa2+ (mM) | 1.24±0.02 | 1.26±0.02 | NS | 0.55 |
| TCO2 (mM) | 18.0±0.58 | 11.8±0.87 | c | <0.001 |
| BUN (mg/dl) | 28.7±1.15 | 94.5±13.41 | c | <0.001 |
| Glucose (mg/dl) | 325.5±14.3 | 288.5±17.8 | NS | 0.14 |
| Hct (% PCV) | 36.7±0.2 | 39.2±1.3 | NS | 0.09 |
| Hb (g/dl) | 12.5±0.08 | 13.3±0.44 | NS | 0.08 |
| AnGap (mM) | 18.8±0.7 | 19.3±0.5 | NS | 0.57 |
| Aldosterone (pg/ml) | 466±45.4 | 1031±117.1 | a | 0.001 |
| Urine | ||||
| UV (ml/d) | 4.71±0.50 | 9.41±1.60 | b | 0.02 |
| Na+ clearance (ml/d per gram body wt) | 0.088±0.006 | 0.135±0.013 | a | 0.009 |
| K+ clearance (ml/d per gram body wt) | 3.68±0.28 | 4.90±0.40 | b | 0.03 |
| Ca2+ (mg/dl) | 2.15±0.63 | 2.64±0.44 | NS | 0.53 |
| Systolic BP (mm Hg) | 128.7±1.1 | 126.3±7.5 | NS | 0.76 |
| Kidney weight/body wt (mg/g) | 10.71±0.39 | 15.69±1.03 | a | 0.002 |
Data are presented as mean±SEM. Systolic BP measure is the mean of 1 hour average values during three dark periods. For systolic blood pressure n=5. For all other measurements n=6. iCa2+, ionized calcium; TCO2, total carbon dioxide; Hct, hematocrit; PCV, packed cell volume; Hb, hemoglobin; AnGap, anion gap; UV, urine volume.
P<0.01, two-tailed unpaired t test.
P<0.05, two-tailed unpaired t test.
P<0.001, two-tailed unpaired t test.
Heterozygous Jab1 Mice on a High Sodium, Low Potassium Diet Have an Exaggerated Effect on WNK-NCC Pathway
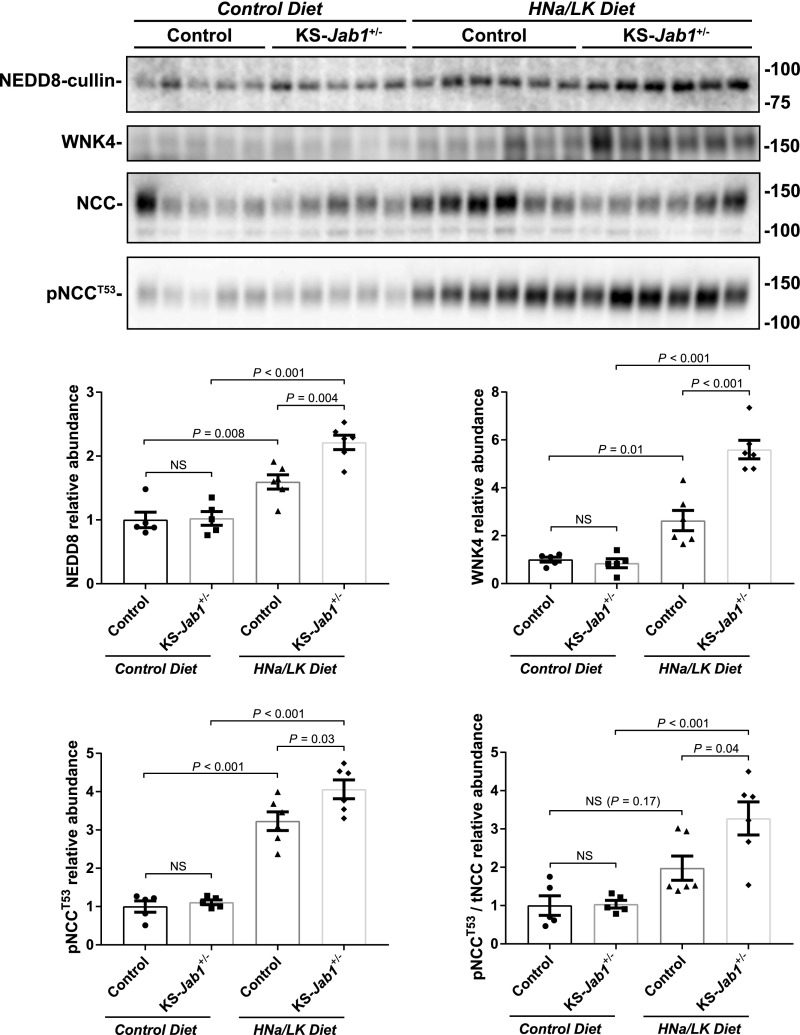
Although the WNK-SPAK/OSR1 pathway is activated in KS-Jab1−/− mice, the depletion of the extracellular fluid volume or hypokalemia might have contributed to the phenotype. To determine whether WNKs are activated by a compensatory physiologic response, we examined mice with heterozygous expression of Jab1 (KS-Jab1+/−). Mice expressing one Jab1 allele showed no phenotypic differences on control diet (data not shown). Additionally, plasma and urine electrolyte levels were similar between KS-Jab1+/− mice and control mice when place on a high sodium, low potassium diet. However, the high sodium, low potassium diet caused a more abundant increase in neddylated cullin, WNK4, and pNCC protein in KS-Jab1+/− mice compared with control mice (Figure 6). Thus, although the hypokalemia, volume depletion, and high plasma aldosterone levels may cause an increase in WNK4 activity in KS-Jab1−/− mice, the results indicate that JAB1 deficiency does contribute to the activation of the WNK-SPAK/OSR1 pathway, which is most likely caused by the substantial decrease in KLHL3.
Figure 6.
A high sodium, low potassium diet exaggerates the activation of the WNK-NCC pathway in heterozygous Jab1 mice. Mice containing only one floxed Jab1 allele were treated with doxycycline, similar to KS-Jab1−/− mice, to generate heterozygous Jab1 mice (KS-Jab1+/−). Top, protein was analyzed by Western blot with whole kidney lysate for NEDD8, WNK4, NCC, and pNCCT53 in mice on control and high sodium, low potassium diet (HNa/LK). Bottom, quantitative analysis of protein abundance. KS-Jab1+/− mice on control diet showed no differences in protein levels compared with control mice. Placing control mice on a HNa/LK diet caused an increase in neddylated cullin, WNK4, NCC, and pNCCT53 compared with control diet. When KS-Jab1+/− mice were fed a HNa/LK diet, neddylated cullin, WNK4, and pNCCT53 protein levels were further increased compared with control mice and the ratio of pNCCT53 to tNCC was significantly higher. Data represent individual values as well as mean±SEM relative to controls. Statistical differences were examined by one-way ANOVA with Tukey post hoc analysis.
Chronic Disruption of the CSN in Kidney Leads to Tubule Damage
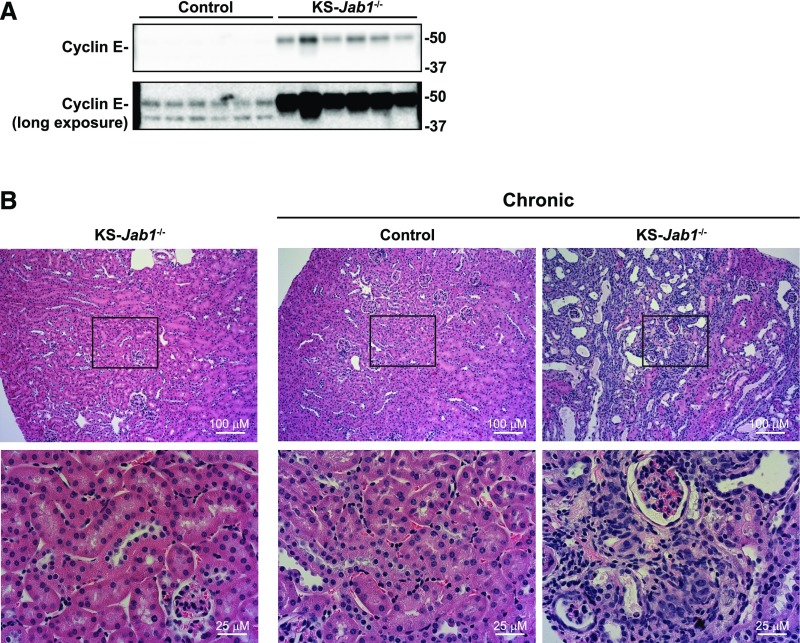
CUL3 also facilitates degradation of cyclin-dependent kinases, including cyclin E, a protein involved in cell cycle regulation.49 Our laboratory has previously shown that chronic kidney-tubule specific Cul3 deletion led to increased cyclin E, and histologic analysis showed tubulointerstitial inflammation and fibrosis.37 Similar to chronic Cul3 deletion, cyclin E abundance was greater in kidneys from KS-Jab1−/− mice than in controls (Figure 7A). Kidneys from KS-Jab1−/− mice were heavier and appeared discolored (Table 1), although histologic analysis of kidney sections did not show tubule damage (Figure 7B). However, when the period after Jab1 deletion was longer (9 weeks after doxycycline treatment), kidney damage was present, with increased nuclear staining and dilated tubules.
Figure 7.
Chronic nephron-specific Jab1 deletion induces kidney tubule damage. (A) Immunoblotting was performed on whole kidney lysates from control and KS-Jab1−/− mice using antibodies against cyclin E. Cyclin E abundance was higher in KS-Jab1−/− mice. (B) Doxycycline or vehicle control was administered to Jab1fl/fl Pax8/LC1 mice for 3 weeks. Mice were either allowed to recover the normal 3 weeks postdoxycycline treatment, or were euthanized after 9 weeks of recovery (chronic). Kidneys were harvested for immunohistochemistry and hematoxylin and eosin staining was performed with kidney sections after paraffin embedding. Mice with chronic deletion of Jab1 developed kidney tubule damage as observed by increased nuclear staining and dilated tubules.
Discussion
In cultured cells, the protein produced by the disease-causing mutant CUL3Δ9 does not associate normally with the CSN,11,36 is hyperneddylated,11,36 and degrades KLHL3 anomalously.37 On the basis of these findings, we postulated that the resulting low KLHL3 abundance impairs WNK kinase degradation, contributing to the phenotype of FHHt.37 Although this hypothesis has proved controversial,11 genetic deletion of KLHL3 does lead to a mouse phenotype that resembles FHHt,41 and recent results confirm that low KLHL3 abundance in mice carrying the pathogenic CUL3Δ9 mutation contributes to the phenotype.42 Here, we tested the hypothesis that defective CSN action contributes to the FHHt phenotype mice by deleting a component of the key enzyme involved in removing NEDD8 from the cullin proteins. The results confirm a role for deficient deneddylation for the disease process, but also provide novel insight into factors regulating NCC abundance, in vivo.
To disturb cullin deneddylation, we deleted CSN5, also known as JAB1, in kidney tubules; this deletion renders CSN ineffective. As expected, CUL3, and likely other cullins, was hyperneddylated in kidney tissue from KS-Jab1−/− mice. As predicted, the results also show low KLHL3 abundance, high WNK4 abundance, and a higher ratio of pNCC to tNCC, compared with control mice. Surprisingly, however, tNCC abundance was drastically reduced by approximately 85%, and the mice had polyuria, salt wasting, and hypokalemia. Although these features resemble Gitelman syndrome, the mice were also hyperchloremic and acidemic, indicating a mixed phenotype. The results suggest tubule dysfunction throughout the distal nephron, and more closely resemble another mouse model recently characterized by our laboratory, namely KS-Cul3−/− mice.
As shown previously,37 KS-Cul3−/− mice also develop polyuria, salt and water wasting, and hypokalemia, with increased WNK abundance and an increased ratio of pNCC to tNCC. Similar to KS-Jab1−/− mice, chronic Cul3 deletion also led to increased hematocrit and aldosterone levels, a lower abundance of aquaporin 2, and later to renal inflammation and fibrosis. One factor that likely contributed to the kidney damage is the increased abundance of cyclin E in both KS-Cul3−/− and KS-Jab1−/− mice; activation of this cyclin-dependent kinase favors cell proliferation and potential damage.50 Unlike KS-Jab1−/− mice, however, KS-Cul3−/− mice are alkalemic and hypochloremic, and tNCC abundance is unchanged.
An interesting feature of the KS-Jab1−/− mice is that CUL3 abundance is low. This likely reflects that, as reported previously,51 neddylated cullins are unstable, and suggests that hyperneddylation contributes substantially to the low abundance of CUL3Δ9 protein that has been described in models of CUL3Δ9-related FHHt.11,43 Interestingly, the KS-Jab1−/− mice also exhibited lower tNCC abundance despite the elevated WNK kinase abundance. This may reflect a previously unreported direct effect of JAB1 disruption on NCC (as suggested by our results in HEK293 cells), which would not be present in KS-Cul3−/− mice.
CSN disruption did not result in monotonic effects on all CUL3 BTB-Kelch substrate adaptors, as the effects on KLHL3 and Keap1 were different. This selectivity of substrate adaptor degradation by CSN inhibition has been shown in multiple in vitro and in vivo studies.30–35 A current model for CSN function is that it competitively binds to the neddylated-CRL when substrates are low and prevents erroneous autoubiquitylation of substrate adaptors and the cullin scaffold protein.33,52 Thus, hyperneddylation can disrupt regulated degradation of substrates by abnormally targeting its substrate adaptor. The results here are consistent with previous work in cultured cells using CSN inhibitors; KLHL3 was degraded36 and Keap1 abundance increased.53 Furthermore, CUL3Δ9 showed a similar effect on the substrate adaptors, both in vitro36,37 and in vivo.42 Because CUL3Δ9 also has an impaired interaction with the CSN, the results are consistent with decreased CSN activity as a partial mechanism for the disease. Additionally, the selectiveness of CSN inactivation to decrease KLHL3 but not Keap1 abundance may explain why the disease phenotype is only observed in the kidney and smooth muscle.54 Although CUL3 is expressed ubiquitously, KLHL3 is expressed most abundantly in the kidney and brain.41 Thus, the effects of KLHL3 degradation by CUL3Δ9 would only be observed in these tissues.
We propose that decreased KLHL3 is a second “hit” required for FHHt to occur. As noted above, CUL3Δ9 is unstable and its abundance in vivo is quite low.11,43 Yet, CUL3Δ9-FHHt is an autosomal dominant disease, meaning that the second WT CUL3 allele remains. Ferdaus et al.43 recently showed that CUL3 haploinsufficiency itself does not recapitulate the disease phenotype. The decreased abundance of CUL3 may, therefore, represent a “first factor” in the disease pathogenesis. When Ferdaus et al. introduced CUL3Δ9 on a haploinsufficient background, the disease phenotype was reproduced. This indicates clearly that a dominant effect of CUL3Δ9 is also essential, as a second hit. Previous results suggest that the second consequence of CUL3Δ9 involves enhanced KLHL3 degradation, leading to KLHL3 deficiency. In a cell culture model that expressed both WT and CUL3Δ9, depletion of KLHL3, driven by CUL3Δ9-mediated degradation, prevented the remaining WT CUL3 from effectively degrading WNK4 (it exerted a dominant-negative effect).36 The results here show that the KLHL3 degradation from CUL3Δ9 may be caused by its impaired interaction with the CSN.
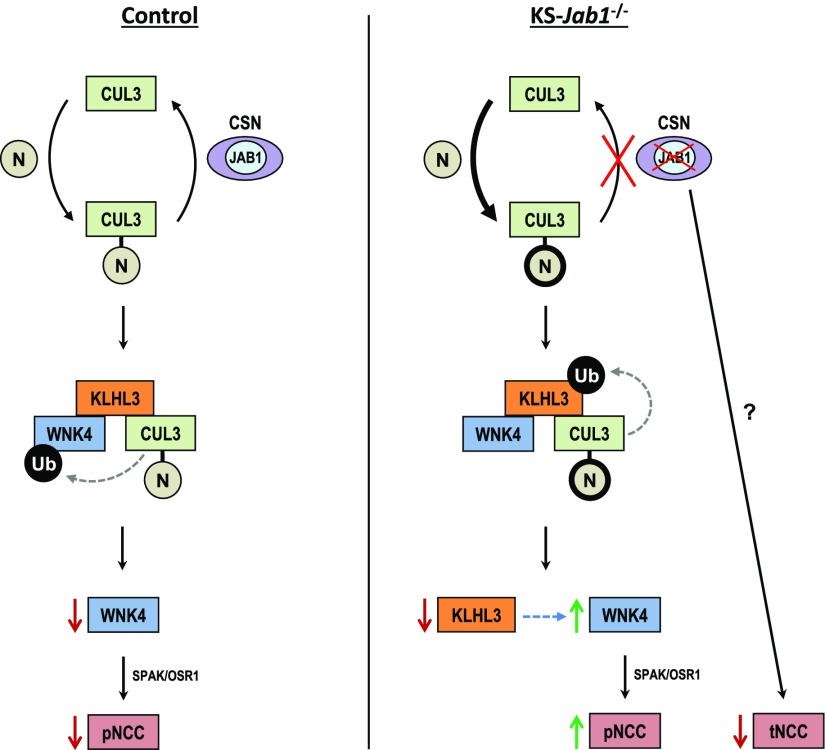
Thus, our results show that JAB1 dysfunction in vivo causes a mixed phenotype that in some respects resembles KS-Cul3−/− mice, with polyuria, salt wasting, hypokalemia, and kidney tubule damage. The KS-Jab1−/− mice also have unique features, including activation of the WNK-SPAK pathway with decreased NCC abundance (Figure 8). Furthermore, the low KLHL3 abundance suggests that an important effect of the CUL3Δ9 mutation is to disrupt CUL3 binding to the CSN, thus enhancing cullin neddylation, activating KLHL3 degradation, and thereby contributing to the disease. The results reveal unexpected complexity in the effects of CRL modulation of kidney structure and function. The dissociation between WNK kinase abundance and NCC activity observed here suggests that small molecule strategies to recapitulate this effect in humans might be useful to treat hypertension or edema.
Figure 8.
Simplified schematic of the effects of nephron-specific Jab1 deletion on the CUL3-KLHL3-WNK pathway. Cullin-RING ubiquitin ligases cycle between an “inactive” unneddylated and an “active” neddylated state. Left, when in homeostasis, NEDD8 (N) is removed from CUL3 by CSN and the substrate, WNK4, is ubiquitylated and degraded via interaction with CUL3 through the adaptor KLHL3, preventing NCC phosphorylation. Right, in KS-Jab1−/− mice, disturbed interaction of CUL3 with the CSN causes more neddylation of CUL3 and abnormal ubiquitylation and degradation of KLHL3, leading to an increase in WNK4 abundance. Phosphorylation of NCC is increased through downstream kinases SPAK/OSR1; however, deletion of Jab1 causes a decrease in NCC abundance that is independent from the WNK-SPAK pathway.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank Dr. Guang Zhou from Case Western Reserve University for providing the Jab1flx/flx mice, and Dr. Jim McCormick and Dr. Jeffrey Singer for the helpful discussions, as well as Lauren Miller for breeding and genotyping the mice. We thank the Oregon Health and Science University Histopathology Core Facility for performing hematoxylin and eosin staining.
C-L.Y., D.H.E., and R.J.C. conceived the study. R.J.C. performed most of the experiments. J.S., C.A.C., J.W.N., and B.D.K.G. helped with some of the experiments. R.P. generated Jab1flx/flx mice. R.J.C., C-L.Y., and D.H.E. analyzed data. R.J.C. and D.H.E. wrote the paper. R.J.C., C-L.Y., and D.H.E. edited the paper.
This work was supported by National Institutes of Health (NIH) grants R01 DK51496 (D.H.E. and C-L.Y.) and T32 DK067864 (D.H.E.), as well as by the merit review grant 1I01BX002228-01A1 from the Department of Veteran Affairs (D.H.E.), and by American Heart Association grant 16POST3064003 and NIH grant F32 DK112531 (R.J.C.).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2018030333/-/DCSupplemental.
References
- 1.Pacheco-Alvarez D, Cristóbal PS, Meade P, Moreno E, Vazquez N, Muñoz E, et al.: The Na+:Cl- cotransporter is activated and phosphorylated at the amino-terminal domain upon intracellular chloride depletion. J Biol Chem 281: 28755–28763, 2006. [DOI] [PubMed] [Google Scholar]
- 2.Vitari AC, Deak M, Morrice NA, Alessi DR: The WNK1 and WNK4 protein kinases that are mutated in Gordon’s hypertension syndrome phosphorylate and activate SPAK and OSR1 protein kinases. Biochem J 391: 17–24, 2005. 16083423 [Google Scholar]
- 3.Pathare G, Hoenderop JG, Bindels RJ, San-Cristobal P: A molecular update on pseudohypoaldosteronism type II. Am J Physiol Renal Physiol 305: F1513–F1520, 2013. [DOI] [PubMed] [Google Scholar]
- 4.Mayan H, Vered I, Mouallem M, Tzadok-Witkon M, Pauzner R, Farfel Z: Pseudohypoaldosteronism type II: Marked sensitivity to thiazides, hypercalciuria, normomagnesemia, and low bone mineral density. J Clin Endocrinol Metab 87: 3248–3254, 2002. Available at: http://www.ncbi.nlm.nih.gov/pubmed/12107233 [DOI] [PubMed] [Google Scholar]
- 5.Wilson FH, Disse-Nicodème S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, et al.: Human hypertension caused by mutations in WNK kinases. Science 293: 1107–1112, 2001. [DOI] [PubMed] [Google Scholar]
- 6.Susa K, Sohara E, Rai T, Zeniya M, Mori Y, Mori T, et al.: Impaired degradation of WNK1 and WNK4 kinases causes PHAII in mutant KLHL3 knock-in mice. Hum Mol Genet 23: 5052–5060, 2014. [DOI] [PubMed] [Google Scholar]
- 7.Boyden LM, Choi M, Choate KA, Nelson-Williams CJ, Farhi A, Toka HR, et al.: Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature 482: 98–102, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohta A, Schumacher F-R, Mehellou Y, Johnson C, Knebel A, Macartney TJ, et al.: The CUL3-KLHL3 E3 ligase complex mutated in Gordon’s hypertension syndrome interacts with and ubiquitylates WNK isoforms: Disease-causing mutations in KLHL3 and WNK4 disrupt interaction. Biochem J 451: 111–122, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shibata S, Zhang J, Puthumana J, Stone KL, Lifton RP: Kelch-like 3 and Cullin 3 regulate electrolyte homeostasis via ubiquitination and degradation of WNK4. Proc Natl Acad Sci U S A 110: 7838–7843, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wakabayashi M, Mori T, Isobe K, Sohara E, Susa K, Araki Y, et al.: Impaired KLHL3-mediated ubiquitination of WNK4 causes human hypertension. Cell Reports 3: 858–868, 2013. [DOI] [PubMed] [Google Scholar]
- 11.Schumacher FR, Siew K, Zhang J, Johnson C, Wood N, Cleary SE, et al.: Characterisation of the Cullin-3 mutation that causes a severe form of familial hypertension and hyperkalaemia. EMBO Mol Med 7: 1285–1306, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sohara E, Uchida S: Kelch-like 3/Cullin 3 ubiquitin ligase complex and WNK signaling in salt-sensitive hypertension and electrolyte disorder. Nephrol Dial Transplant 31: 1417–1424, 2016. [DOI] [PubMed] [Google Scholar]
- 13.Chen HY, Chen RH: Cullin 3 ubiquitin ligases in cancer biology: Functions and therapeutic implications. Front Oncol 6: 113, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wimuttisuk W, West M, Davidge B, Yu K, Salomon A, Singer JD: Novel Cul3 binding proteins function to remodel E3 ligase complexes. BMC Cell Biol 15: 28, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petroski MD, Deshaies RJ: Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6: 9–20, 2005. [DOI] [PubMed] [Google Scholar]
- 16.Wu G, Peng JB: Disease-causing mutations in KLHL3 impair its effect on WNK4 degradation. FEBS Lett 587: 1717–1722, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saha A, Deshaies RJ: Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell 32: 21–31, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K: Nedd8 on cullin: Building an expressway to protein destruction. Oncogene 23: 1985–1997, 2004. [DOI] [PubMed] [Google Scholar]
- 19.Boh BK, Smith PG, Hagen T: Neddylation-induced conformational control regulates cullin RING ligase activity in vivo. J Mol Biol 409: 136–145, 2011. [DOI] [PubMed] [Google Scholar]
- 20.Pintard L, Kurz T, Glaser S, Willis JH, Peter M, Bowerman B: Neddylation and deneddylation of CUL-3 is required to target MEI-1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr Biol 13: 911–921, 2003. [DOI] [PubMed] [Google Scholar]
- 21.Chung D, Dellaire G: The role of the COP9 signalosome and neddylation in DNA damage signaling and repair. Biomolecules 5: 2388–2416, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolf DA, Zhou C, Wee S: The COP9 signalosome: An assembly and maintenance platform for cullin ubiquitin ligases? Nat Cell Biol 5: 1029–1033, 2003 [DOI] [PubMed] [Google Scholar]
- 23.Wei N, Deng XW: The COP9 signalosome. Annu Rev Cell Dev Biol 19: 261–286, 2003. [DOI] [PubMed] [Google Scholar]
- 24.Sharon M, Mao H, Boeri Erba E, Stephens E, Zheng N, Robinson CV: Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure 17: 31–40, 2009. [DOI] [PubMed] [Google Scholar]
- 25.Yan J, Walz K, Nakamura H, Carattini-Rivera S, Zhao Q, Vogel H, et al.: COP9 signalosome subunit 3 is essential for maintenance of cell proliferation in the mouse embryonic epiblast. Mol Cell Biol 23: 6798–6808, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lykke-Andersen K, Schaefer L, Menon S, Deng XW, Miller JB, Wei N: Disruption of the COP9 signalosome Csn2 subunit in mice causes deficient cell proliferation, accumulation of p53 and cyclin E, and early embryonic death. Mol Cell Biol 23: 6790–6797, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomoda K, Yoneda-Kato N, Fukumoto A, Yamanaka S, Kato JY: Multiple functions of Jab1 are required for early embryonic development and growth potential in mice. J Biol Chem 279: 43013–43018, 2004. [DOI] [PubMed] [Google Scholar]
- 28.Menon S, Chi H, Zhang H, Deng XW, Flavell RA, Wei N: COP9 signalosome subunit 8 is essential for peripheral T cell homeostasis and antigen receptor-induced entry into the cell cycle from quiescence. Nat Immunol 8: 1236–1245, 2007. [DOI] [PubMed] [Google Scholar]
- 29.Zhao R, Yeung SC, Chen J, Iwakuma T, Su CH, Chen B, et al.: Subunit 6 of the COP9 signalosome promotes tumorigenesis in mice through stabilization of MDM2 and is upregulated in human cancers. J Clin Invest 121: 851–865, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su H, Huang W, Wang X: The COP9 signalosome negatively regulates proteasome proteolytic function and is essential to transcription. Int J Biochem Cell Biol 41: 615–624, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Denti S, Fernandez-Sanchez ME, Rogge L, Bianchi E: The COP9 signalosome regulates Skp2 levels and proliferation of human cells. J Biol Chem 281: 32188–32196, 2006. [DOI] [PubMed] [Google Scholar]
- 32.He Q, Cheng P, He Q, Liu Y: The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev 19: 1518–1531, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wee S, Geyer RK, Toda T, Wolf DA: CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat Cell Biol 7: 387–391, 2005. [DOI] [PubMed] [Google Scholar]
- 34.Cope GA, Deshaies RJ: Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem 7: 1, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su H, Li J, Menon S, Liu J, Kumarapeli AR, Wei N, et al.: Perturbation of cullin deneddylation via conditional Csn8 ablation impairs the ubiquitin-proteasome system and causes cardiomyocyte necrosis and dilated cardiomyopathy in mice. Circ Res 108: 40–50, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cornelius RJ, Zhang C, Erspamer KJ, Agbor LN, Sigmund CD, Singer JD, et al. : Dual gain and loss of cullin 3 function mediates familial hyperkalemic hypertension [published online ahead of print June 13, 2018]. Am J Physiol Renal Physiol [DOI] [PMC free article] [PubMed]
- 37.McCormick JA, Yang CL, Zhang C, Davidge B, Blankenstein KI, Terker AS, et al.: Hyperkalemic hypertension-associated cullin 3 promotes WNK signaling by degrading KLHL3. J Clin Invest 124: 4723–4736, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Traykova-Brauch M, Schönig K, Greiner O, Miloud T, Jauch A, Bode M, et al.: An efficient and versatile system for acute and chronic modulation of renal tubular function in transgenic mice. Nat Med 14: 979–984, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schönig K, Schwenk F, Rajewsky K, Bujard H: Stringent doxycycline dependent control of CRE recombinase in vivo. Nucleic Acids Res 30: e134, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panattoni M, Sanvito F, Basso V, Doglioni C, Casorati G, Montini E, et al.: Targeted inactivation of the COP9 signalosome impairs multiple stages of T cell development. J Exp Med 205: 465–477, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sasaki E, Susa K, Mori T, Isobe K, Araki Y, Inoue Y, et al. : KLHL3 knockout mice reveal the physiological role of KLHL3 and the pathophysiology of Pseudohypoaldosteronism Type II caused by mutant KLHL3. Mol Cell Biol 37: e00508–e00516, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida S, Araki Y, Mori T, Sasaki E, Kasagi Y, Isobe K, et al.: Decreased KLHL3 expression is involved in the pathogenesis of pseudohypoaldosteronism type II caused by cullin 3 mutation in vivo [published online ahead of print June 5, 2018]. Clin Exp Nephrol, 2018. 10.1007/s10157-018-1593-z [DOI] [PubMed] [Google Scholar]
- 43.Ferdaus MZ, Miller LN, Agbor LN, Saritas T, Singer JD, Sigmund CD, et al.: Mutant Cullin 3 causes familial hyperkalemic hypertension via dominant effects. JCI Insight 2: e96700, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terker AS, Zhang C, McCormick JA, Lazelle RA, Zhang C, Meermeier NP, et al.: Potassium modulates electrolyte balance and blood pressure through effects on distal cell voltage and chloride. Cell Metab 21: 39–50, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Reilly M, Marshall E, Macgillivray T, Mittal M, Xue W, Kenyon CJ, et al.: Dietary electrolyte-driven responses in the renal WNK kinase pathway in vivo. J Am Soc Nephrol 17: 2402–2413, 2006. [DOI] [PubMed] [Google Scholar]
- 46.McCormick JA, Mutig K, Nelson JH, Saritas T, Hoorn EJ, Yang CL, et al.: A SPAK isoform switch modulates renal salt transport and blood pressure. Cell Metab 14: 352–364, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saritas T, Borschewski A, McCormick JA, Paliege A, Dathe C, Uchida S, et al.: SPAK differentially mediates vasopressin effects on sodium cotransporters. J Am Soc Nephrol 24: 407–418, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grimm PR, Taneja TK, Liu J, Coleman R, Chen YY, Delpire E, et al.: SPAK isoforms and OSR1 regulate sodium-chloride co-transporters in a nephron-specific manner. J Biol Chem 287: 37673–37690, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singer JD, Gurian-West M, Clurman B, Roberts JM: Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev 13: 2375–2387, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kossatz U, Breuhahn K, Wolf B, Hardtke-Wolenski M, Wilkens L, Steinemann D, et al.: The cyclin E regulator cullin 3 prevents mouse hepatic progenitor cells from becoming tumor-initiating cells. J Clin Invest 120: 3820–3833, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu JT, Lin HC, Hu YC, Chien CT: Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat Cell Biol 7: 1014–1020, 2005. [DOI] [PubMed] [Google Scholar]
- 52.Wang X, Martin DS: The COP9 signalosome and cullin-RING ligases in the heart. Am J Cardiovasc Dis 5: 1–18, 2015. [PMC free article] [PubMed] [Google Scholar]
- 53.Schlierf A, Altmann E, Quancard J, Jefferson AB, Assenberg R, Renatus M, et al.: Targeted inhibition of the COP9 signalosome for treatment of cancer. Nat Commun 7: 13166, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Agbor LN, Ibeawuchi SC, Hu C, Wu J, Davis DR, Keen HL, et al.: Cullin-3 mutation causes arterial stiffness and hypertension through a vascular smooth muscle mechanism. JCI Insight 1: e91015, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.