Abstract
It is clear that RhoA activates the DRF (diaphanous-related formin) mDia2 by disrupting the molecular interaction between the DAD (diaphanous autoregulatory domain) and the DID (diaphanous inhibitory domain). Previous studies indicate that a basic motif within the DAD contributes to mDia2 auto-inhibition, and results shown in the present study suggest these residues bind a conserved acidic region within the DID. Furthermore, we demonstrate that mDia2 is phosphorylated by ROCK (Rho-kinase) at two conserved residues (Thr1061 and Ser1070) just C-terminal to the DAD basic region. Phosphomimetic mutations to these residues in the context of the full-length molecule enhanced mDia2 activity as measured by increased actin polymerization, SRF (serum response factor)-dependent smooth muscle-specific gene transcription, and nuclear localization of myocardin-related transcription factor B. Biochemical and functional data indicate that the T1061E/S1070E mutation significantly inhibited the ability of DAD to interact with DID and enhanced mDia2 activation by RhoA. Taken together, the results of the present study indicate that ROCK-dependent phosphorylation of the mDia2 DAD is an important determinant of mDia2 activity and that this signalling mechanism affects actin polymerization and smooth muscle cell-specific gene expression.
Keywords: mDia2, RhoA, Rho-kinase (ROCK), diaphanous-related formin, serum response factor, smooth muscle
INTRODUCTION
The precise regulation of actin polymerization by the Rho family of small GTPases is essential for many important cellular functions, including adhesion, migration, polarity, cytokinesis and contraction. When bound to GTP, Rho, Rac and Cdc42 (cell division cycle 42) interact with a variety of effector molecules to induce the formation of stress fibres, lamellipodia and filopodia respectively. Of these effectors, the DRFs (diaphanous-related formins) mDia1, mDia2, mDia3 and FHOD1 [FH (formin homology) domain-containing protein 1] have received considerable attention because these proteins strongly catalyse actin nucleation and actin filament elongation and are regulated by specific interactions with the Rho GTPases. The DRFs are defined by the presence of two highly conserved FH domains (FH1 and FH2) that catalyse actin nucleation, a C-terminal DAD (diaphanous autoregulatory domain), an N-terminal DID (diaphanous inhibitory domain), and an N-terminal GBD (GTPase-binding domain) (see [1] and [2] for reviews). DRFs are maintained in an inactive conformation by a molecular interaction between DID and DAD (Figure 1A) [3,4]. Since the GBD- and DID-binding pockets slightly overlap, high-affinity binding of an activated GTPase to the GBD disrupts the DAD–DID interaction to expose the catalytically active FH1/FH2 domains [5,6]. Although the precise mechanism by which the DRFs stimulate actin polymerization is not fully understood, FH2-mediated actin filament elongation occurs at the barbed end and is enhanced by profilin binding to the FH1 domain.
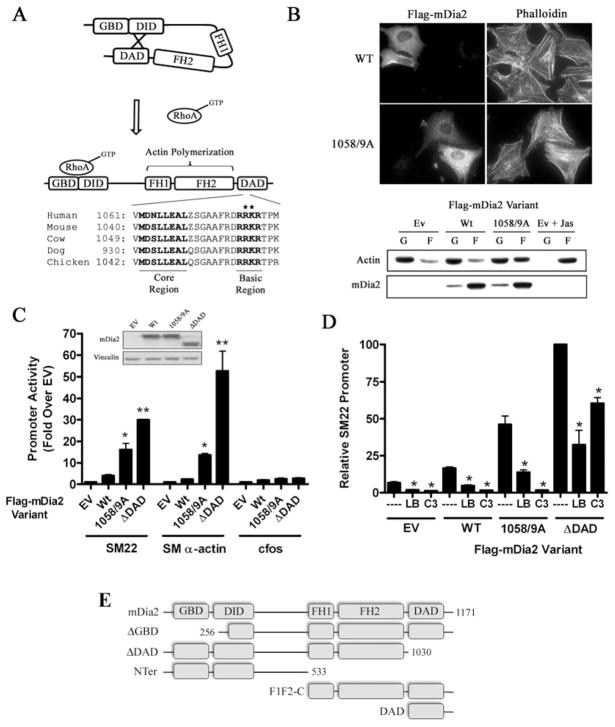
Figure 1. RhoA-dependent activation of mDia2 is enhanced by DAD basic region mutations.
(A) Schematic diagram of mDia2 activation by RhoA and DAD domain sequence conservation. Note: all sequence information presented in the manuscript was obtained from the NCBI database. *Targeted alanine mutations within the basic region. (B) HeLa cells were transfected with the indicated FLAG–mDia2 variant and then serum starved for 24 h. Actin polymerization was visualized by phalloidin staining (top) and actin sedimentation assays (bottom). In brief, F-actin (F) was separated from G-actin (G) by high-speed centrifugation. Actin levels in each fraction were then measured by Western blot analysis. The actin-stabilizing drug jasplakinolide (Jas, 1 μM) was used as a positive control. (C) Full-length FLAG–mDia2 variants were transfected into 10T1/2 cells along with luciferase reporter constructs driven by the SM22, SM α-actin or c-Fos promoters. Luciferase activity was analysed at 24 h and is expressed as the fold change compared with cells transfected with empty expression vector. mDia2 variant expression levels were determined by Western blot analysis and are shown in the inset. *P < 0.05 compared with wild-type, **P < 0.05 compared with R1058A/K1059A (1058/9A). (D) 10T1/2 cells were transfected with SM22–luciferase and the indicated FLAG–mDia2 variant. Some cells were co-transfected with C3 transferase or treated with 10 μM latrunculin B (LB) for 6 h before luciferase determinations. Luciferase activities were normalized to that obtained upon expression of mDia2ΔDAD alone set to 100 %. *P < 0.05 compared with control. (E) Schematic diagram of the mDia2 truncation variants used in the present studies. EV/Ev, empty vector; Wt/WT, wild-type.
In addition to its traditional role in controlling actin polymerization, RhoA signalling increases the expression of a variety of muscle differentiation and cytoskeletal genes by activating the SRF (serum response factor) (see [7] for a review). This effect is mediated by an actin polymerization-dependent depletion of G-actin (globular actin) pools that promotes the nuclear accumulation of the SRF co-factors, MRTF-A (myocardin-related transcription factor-A)/MKL-1 and MRTF-B/MKL-2 [8]. We have previously shown that RhoA/MRTF-A signalling regulates many SMC [SM (smooth muscle) cell] differentiation marker genes (i.e. SM α-actin, SM22, and SM myosin heavy chain) [9,10], and that the RhoA effectors mDia1 and mDia2 play critical roles in this process [11]. Importantly, MRTF-A- and MRTF-B-knockout mice exhibit defects in SMC differentiation marker gene expression, supporting a role for RhoA/MRTF signalling in the regulation of vascular development and perhaps in the progression of vascular disease [12–15]. Furthermore, a recent study by Medjkane et al. [16] demonstrated that MRTF-A was required for breast cancer cell metastasis because it regulates the expression of genes required for cell migration and invasion.
Given the importance of the DRFs in regulating actin-dependent cellular processes, it will be critical to identify the molecular mechanisms that regulate their activities. Since most studies have focused on DRF activation by the small GTPases, it is unclear whether post-translational modifications or additional proteins play a significant role. Interestingly, a polybasic region C-terminal to the core DAD domain is highly conserved in all four DRF members. Although not absolutely required for the DID–DAD interaction, studies suggest that it may contribute to DRF auto-inhibition [17,18]. Moreover, Takeya et al. [19] have demonstrated that several serine/threonine residues near the basic domain of FHOD1 are targets of ROCK (Rho-kinase), and that phosphorylation of these residues inhibits the DID–DAD interaction to fully activate FHOD1. Recent studies in our laboratory suggest that FHOD1 is phosphorylated by ROCK in SMCs and that this signalling mechanism regulates SMC-specific gene expression [20]. Interestingly, mDia2 contains two highly conserved potential phosphorylation sites, Thr1061 and Ser1070, just C-terminal to the DAD basic region, but whether mDia2 activity is regulated by a similar mechanism is currently unknown. The goals of the present study were to more fully characterize the electrostatic interactions by which the mDia2 basic domain stabilizes the DID–DAD interaction, to test whether phosphorylation of Thr1061 and/or Ser1070 plays a significant role in mDia2 activation, and if so, to determine the physiological consequences of this mechanism.
EXPERIMENTAL
Plasmids and reagents
The mDia2 and ROCK1 expression plasmids were gifts from Shuh Narumiya (Department of Pharmacology, Kyoto University, Kyoto, Japan) [19,21]. C3 transferase was a gift from Keith Burridge (Cell and Developmental Biology, University of North Carolina, Chapel Hill School of Medicine, NC, U.S.A.) [22]. The mDia2 ΔGBD (amino acids 256–1172), ΔDAD (1–1030), NTer (1–533), DAD (1030–1171) and F1F2-C (533–1172) fragments were generated by PCR and sub-cloned into pcDNA3.1 and pEGFP expression plasmids (Clonetech) and the pGEX-4T1 vector (Amersham Biosciences). The Ser1070 and Thr1061 alanine or glutamate mutations were made by the QuikChange® site-directed mutagenesis kit (Stratagene). Antibodies used were as follows: anti-ROCK1, anti-actin and anti-Myc (Cell Signaling Technology), M2-FLAG (Sigma), anti-GFP (green fluorescent protein; BioDesign) and mDia2 (gift from Henry Higgs, Department of Biochemistry, Dartmouth Medical School, Hanover, NH, U.S.A.). Chemical reagents used were as follows: Y-27632, calyculin A and latrunculin B (Calbiochem); jasplakinolide (EMD chemicals) and okadaic acid (MP Biomedicals).
Transient transfections and reporter gene assays
The maintenance and transfection of multipotential 10T1/2 cells were performed as described previously [11]. In brief, cells were maintained in 48-well plates in 10 % serum and were transfected 24 h after plating at 70–80 % confluency using the transfection reagent TransIT-LT1 (Mirus), following the manufacturer’s protocol. Luciferase assays (Promega) were conducted 24 h following transfection. The SM22, SM α-actin and c-Fos promoters have been described previously [23,24].
GST (glutathione transferase)-fusion protein expression
GST-fusion protein expression was induced in BL-21 bacteria by 100 μM isopropyl β-D-thiogalactopyranoside for 18 h at room temperature (22 °C). Following bacterial lysis, GST-fusion proteins were purified using glutathione–Sepharose beads (Sigma).
In vitro and in vivo kinase assays
In vitro ROCK assays were performed as described previously [25]. In brief, ROCK1Δ3 expressed in, and immunoprecipitated from, Cos-7 cells was incubated for 30 min at 30 °C in 25 μl of kinase buffer containing 250 μM ATP, 10 μCi of [γ-32P]ATP and 1 μg of GST–mDia2 DAD fusion protein. The ROCK inhibitor Y-27632 (10 μM) was added to some reactions prior to kinase addition. For in vivo 32P phosphate labelling, Cos-7 cells were transfected with either wild-type or phosphorylation mutants of FLAG-tagged mDia2 ± Myc–ROCK1Δ3. Cells were pre-incubated with phosphate-free medium for 3 h and then labelled with 1 mCi/ml [32P]Pi for 2 h. Some cells were treated with okadaic acid (1 μM) for 1 h before harvest. mDia2 was then immunoprecipitated using M2-FLAG antibody conjugated to agarose beads (Sigma) and analysed by autoradiography and Western blotting.
Phosphorylation mobility shifts
HeLa cells were treated with Y-27632 (or vehicle) for 1 h and then with calyculin A (or vehicle) for an additional 1 h. RIPA (50 mM Hepes, pH 7.2, 150 mM NaCl, 2 mM EDTA, 0.1%Nonidet P-40, 0.05% sodium deoxycholate and 0.5% Triton X-100) lysates were run on SDS/PAGE (7 % gel) and subjected to Western blot analysis. For SAP (shrimp alkaline phosphatase) (Roche) treatments, 20 μg of total protein was incubated with 5 units of SAP for 30 min at 37 °C prior to loading.
Immunoprecipitation and protein expression
Cells were lysed in buffer A (50 mM Tris/HCl, pH 7.2, 150 mM NaCl, 0.2 mM EDTA, 1 % Nonidet P40 and 0.5 % Triton X-100) and cleared by centrifugation for 15 min at 20 000 g at 4 °C. For co-immunoprecipitations, lysates were incubated with the appropriate Sepharose-conjugated antibody at 4 °C for 3 h and beads were washed three times with 1 ml of buffer A. Immunoprecipitants were separated by SDS/PAGE and transferred on to nitrocellulose membranes for Western blotting.
Actin sedimentation assays
Transfected HeLa cells were serum-starved for 48 h and then subjected to the actin sedimentation assay as described previously [20]. In brief, after centrifugation of cell lysates at 50 000 rev./min in a Beckman TLA-100.3 for 1 h and resuspension in equal amounts of sample buffer, levels of G-actin in the supernatant and F-actin (filamentous actin) in the pellet were measured by Western blotting.
Immunoflourescence
HeLa or 10T1/2 cells were plated and transfected in four-well chamber slides, and maintained overnight in medium containing 10 % serum. Cells were serum-starved for 16 h, fixed in 3.7 % paraformaldehyde/PBS for 20 min, and permeabilized in 0.5 % Triton X-100/PBS for 3 min. The M2-FLAG antibody (Sigma) was diluted 1:500 in 20 % goat serum/3 % BSA in PBS and incubated with the fixed cells for 2 h. Texas Red- or FITC-(Jackson ImmunoResearch) conjugated secondary antibodies were used at 1:1000, whereas Alexa Fluor® 555 phalloidin (Molecular Probes) and DAPI (4′,6-diamidino-2-phenylindole; Molecular Probes) were used at 1:100 and 1 nM respectively.
RESULTS
A DID acidic domain contributes to mDia2 auto-inhibition
The auto-inhibitory interaction between the amphipathic DAD core motif (see Figure 1A) and the hydrophobic DID binding pocket has been well characterized by a variety of structural and functional analyses [1,5,26]. Interestingly, additional studies on mDia2 and FHOD1 suggest that a string of basic amino acids just C-terminal to the core DAD may also be important [17,18]. However, due to a lack of crystal structure data on the basic domain and an inability to identify the acidic DID residues with which it interacts, its precise role in regulating mDia2 activity is not completely clear.
To better characterize the molecular interactions that regulate mDia2 auto-inhibition, we performed structure–function analyses using well-established assays of mDia2 activity. As shown in Figure 1(B), expression of FLAG-tagged full-length mDia2 failed to enhance stress fibre formation, as measured by phalloidin staining, or actin polymerization, as measured by actin sedimentation assays. These results reflect the auto-inhibited state of the full-length molecule. In contrast, expression of a full-length mDia2 variant containing a double alanine mutation within the basic domain (R1058A/K1059A) enhanced actin polymerization in both assays, confirming this domain’s role in mDia2 auto-inhibition. We have previously shown that activation of mDia2 strongly stimulated SRF-dependent SMC-specific gene transcription in multi-potential 10T1/2 cells [11]. As expected, the R1058A/K1059A mDia2 variant significantly activated the SM α-actin and SM22 promoters in this model but did not activate c-Fos, an SRF-dependent promoter not regulated by actin polymerization (Figure 1C). Importantly, R1058A/K1059A-dependent promoter activation was not as strong as that observed with an mDia2 variant lacking the entire DAD domain (see the schematic diagram in Figure 1E) and was completely inhibited by co-expression of C3 transferase (Figure 1D). These results strongly suggest that basic domain mutations do not render mDia2 constitutively active, but instead enhance RhoA-mediated mDia2 activity. The ability of latrunculin B to inhibit the transcriptional effects of the R1058A/K1059A mDia2 variant indicates that this response is mediated by actin polymerization.
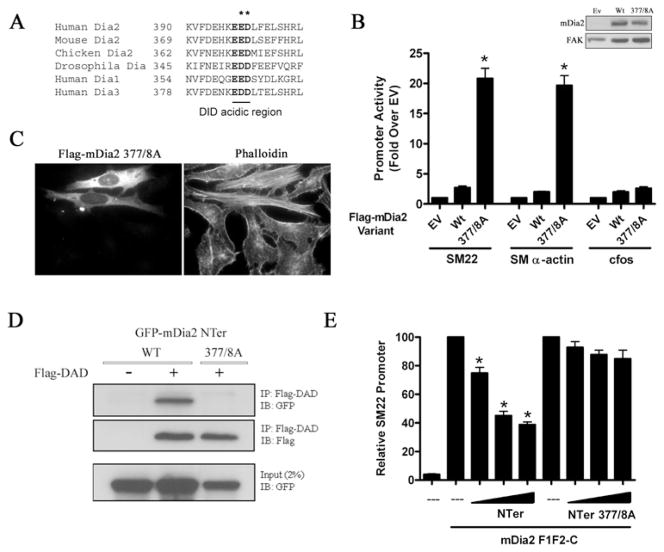
Given the sensitivity of the SRF/MRTF-dependent transcription assay, we used this model to screen the DID domain for residues that may interact with the DAD basic region. The DID-binding pocket consists of five armadillo repeats with all five β-helices making contact with the hydrophobic face of the amphipathic DAD helix [5]. Although the basic domain did not show sufficient order to be placed into the DID–DAD crystal structure, it probably extends C-terminally from the core DAD sequence towards the α5B helix of the DID. On the basis of sequence conservation (between species and other DRF family members; see Figure 2A) and positioning within the α5B helix, we hypothesized that a trio of acidic residues (Glu376, Glu377 and Asp378) were important for the DID–DAD interaction. To test this directly, we generated a E377A/D378A mutation in full-length mDia2 and found that this variant enhanced SMC-specific promoter activity by 20-fold (Figure 2B) and enhanced phalloidin staining when expressed in cells (Figure 2C). Importantly, co-immunoprecipitation assays (with N-terminal mDia2 fragments) clearly demonstrated that the E377A/D378A mutation inhibited the DID–DAD interaction (Figure 2D). As an alternative measure of DID–DAD binding, we used an assay originally described by Copeland et al. [27] that is based upon the ability of the N-terminal half of mDia2 (see Figure 1E) to inhibit the transcriptional activity of the C-terminal half of mDia2 even when these fragments are expressed as separate molecules. As expected, expression of the wild-type N-terminal fragment attenuated the activation of the SM22 promoter by the C-terminal fragment, whereas the N-terminal fragment containing the E377A/D378A double mutation had no significant effect in this model (Figure 2E).
Figure 2. An acidic DID region contributes to mDia2 auto-regulation.
(A) Conservation of an acidic region within the mDia2 DID α5B helix. (B) 10T1/2 cells were transfected with wild-type (Wt) or E377A/D378A (377/8A) mDia2 along with luciferase constructs driven by the SM22, SM α-actin or c-Fos promoter. *P < 0.05 compared with wild-type. Ev, empty vector. (C) HeLa cells were transfected with E377A/D378A mDia2, serum-starved for 24 h, and stained with phalloidin. (D) FLAG–mDia2 DAD was immunoprecipitated (IP) from Cos-7 cells co-expressing the indicated GFP-tagged N-terminal mDia2 fragment. Immunoprecipitants were probed with anti-GFP or anti-FLAG antibodies. IB, immunoblot. (E) 10T1/2 cells were transfected with the SM22 luciferase promoter, mDia2 F1F2-C, and increasing amounts of the indicated N-terminal mDia2 fragment. Luciferase activities were normalized to that obtained upon expression of mDia2F1F2-C alone set to 100 %. *P < 0.05 compared with control. FAK, focal adhesion kinase.
The mDia2 DAD domain is phosphorylated by ROCK
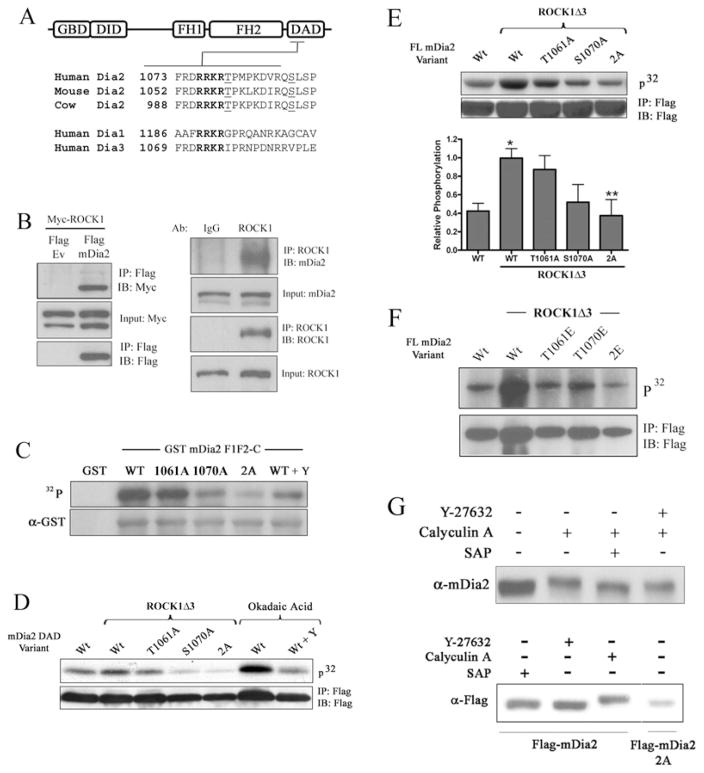
Our results described above suggested that an electrophilic interaction between the DAD basic and DID acidic domains regulates mDia2 auto-inhibition. Given the presence of two highly conserved potential phosphorylation sites, Thr1061 and Ser1070, just C-terminal to the DAD basic region (Figure 3A), we hypothesized that phosphorylation of these residues would inhibit the basic region’s contributions to the DID–DAD interaction. Each of these residues lies within a consensus sequence (RXS/T) that is targeted by basic domain kinases such as PKA (protein kinase A), PKG (protein kinase G) and ROCK [28]. However, on the basis of the demonstration that ROCK phosphorylated similar sequences in FHOD1 and LIM kinase [19,25], we postulated that mDia2 was a substrate for this enzyme. Indeed, we detected an interaction between endogenous ROCK1 and mDia2 in HeLa cells by co-immunoprecipitation (Figure 3B), strongly supporting this idea. In vitro kinase assays demonstrated that constitutively active ROCK1Δ3 immunoprecipitated from Cos-7 cells strongly phosphorylated a GST-fusion protein containing the C-terminal half of mDia2 (Figure 3C). Importantly, 32P incorporation was partially inhibited by single T1061A or S1070A mutations and almost completely inhibited by the double alanine mutation (2A; T1061A/S1070A). DAD phosphorylation in these in vitro assays was also attenuated by the ROCK inhibitor Y-27632.
Figure 3. The mDia2 DAD domain was phosphorylated by ROCK.
(A) Schematic diagram of phosphorylation site conservation near the mDia2 DAD basic region. Note that similar sites are not found in Dia1 or Dia3. (B) Left-hand panel, FLAG–mDia2 and Myc–ROCK1 were co-expressed in HeLa cells. FLAG immunoprecipitates (IP) were run on an SDS/PAGE gel, transferred on to nitrocellulose, and probed for Myc. Input controls represent 10 % of immunoprecipitated fractions. Ev, empty vector. Right-hand panel, endogenous ROCK1 immunoprecipitates were subjected to Western blotting (IB) for mDia2. (C) GST–mDia2 DAD variants containing the indicated phosphorylation mutations were incubated in vitro with constitutively active ROCKΔ3 and 10 μCi of [γ-32P]ATP. After removing unincorporated 32P, reactions were run on an SDS/PAGE gel and exposed to film. The ROCK inhibitor Y-27632 (Y; 10 μM) was added to some reactions. (D) Cos-7 cells were transfected with the indicated mDia2 DAD variant along with ROCK1Δ3. Following [32P]Pi labelling for 2 h, the mDia2 peptides were immunoprecipitated and analysed by autoradiography. Okadaic acid was added to some cells ± pretreatment with Y-27632 (10 μM). (E) Cos-7 cells were transfected with the indicated full-length mDia2 variant along with ROCK1Δ3. Three independent phosphorylation experiments were quantified by densitometry. *P < 0.05 compared with wild-type minus ROCK1Δ3, **P < 0.05 compared with wild-type plus ROCK1Δ3. (F) Same as in (E), except that cells were transfected with the indicated glutamate variants of mDia2. (G) HeLa cells (top) or HeLa cells transfected with the wild-type or DAD 2A mDia2 variant (bottom) were treated with Y-27632 (or vehicle) for 1 h and then with calyculin A (or vehicle) for an additional 1 h. RIPA lysates were run on an SDS/PAGE gel and subjected to Western blotting analysis using anti-mDia2 or anti-FLAG antibodies. Some lysates were treated with SAP before loading. FL, full-length; WT/Wt, wild-type.
As shown in Figure 3(D), nearly identical results were observed in vivo when DAD phosphorylation was measured in Cos-7 cells also overexpressing ROCK1Δ3. Interestingly, even in the absence of ROCK1Δ3, phosphorylation of the mDia2 DAD peptide was dramatically up-regulated in cells pre-treated with okadaic acid, indicating that DAD phosphorylation was maintained at low levels by phosphatase activity. Importantly, addition of the ROCK inhibitor Y-27632 prior to and during okadaic acid treatment significantly reduced DAD phosphorylation, providing strong evidence that the mDia2 DAD is a target for ROCK. Expression of ROCK1Δ3 also increased phosphorylation of full-length mDia2, and in excellent agreement with our in vitro experiments, this increase was partially reduced by the single T1061A and S1070A mutations, and completely eliminated by the double alanine mutation (Figure 3E). Interestingly, when the phosphomimetic glutamate mutations were used in these assays, the single T1061E mutation dramatically inhibited mDia2 phosphorylation (Figure 3F), perhaps suggesting that the relatively minor effect of the single T1061A mutation was due to compensatory phosphorylation of the Ser1070 site. We also found that the mobility of endogenous mDia2 was decreased by treatment of cells with the phosphatase inhibitor calyculin A, and that this mobility shift was prevented by pre-treatment of cells with Y-27632, incubation of lysates with SAP or mutation of both phosphorylation sites (Figure 3G). Additional in vivo phosphorylation experiments indicated that mDia2 was not a substrate for PKA, PKG or MAPK (mitogen-activated protein kinase; see Supplementary Figure S1 at http://www.BiochemJ.org/bj/439/bj4390057add.htm and results not shown).
Thr1061/Ser1070 phosphorylation inhibited DAD binding to DID
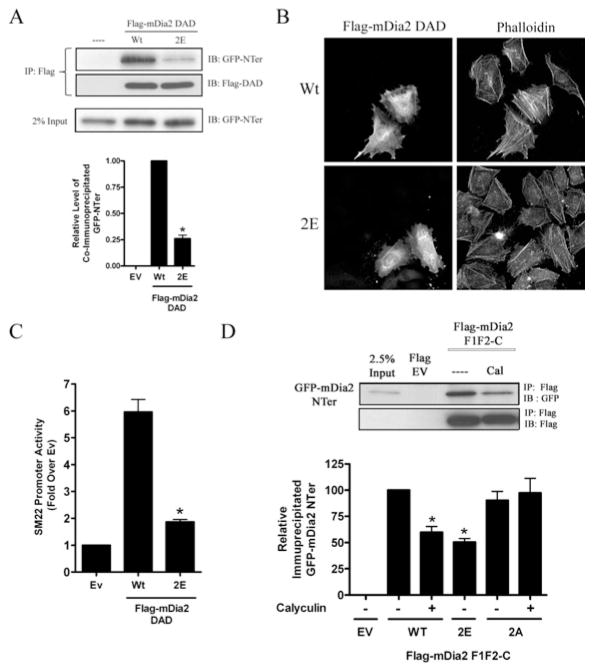
To test whether phosphorylation of Thr1061 and Ser1070 could interfere with the basic region’s role in stabilizing the mDia2 DID–DAD interaction, we generated a phosphomimetic variant of the mDia2 DAD that contained glutamate substitutions at both residues (2E; T1061E/S1070E) and performed co-immunoprecipitation assays with the wild-type and DAD 2E variants. As shown in Figure 4(A), wild-type DAD efficiently precipitated the N-terminal mDia2 fragment, but this interaction was significantly reduced by the DAD 2E mutation. Another method used to study DID–DAD interactions is based upon the ability of exogenously expressed DAD peptides to stimulate endogenous mDia activity by competitively interfering with the DID–DAD interaction [11,17]. As expected, expression of wild-type FLAG–mDia2 DAD enhanced phalloidin staining (Figure 4B) and SM22 promoter activity (Figure 4C). In contrast, expression of the DAD 2E variant had little, if any, effect in these models, indicating that the DAD 2E variant does not bind to DID as effectively as wild-type. To directly determine whether phosphorylation reduced the DID–DAD interaction in vivo, we performed co-immunoprecipitations in calyculin-treated cells that exhibit strong DAD phosphorylation. As shown in Figure 4(D), calyculin treatment resulted in a significant decrease in the interaction between the N- and C-terminal mDia2 fragments. Importantly, this decrease was completely reversed by the DAD 2A mutation, but was mimicked by the DAD 2E mutation in the absence of calyculin. These results clearly support a model in which phosphorylation of the mDia2 DAD inhibits it association with the DID.
Figure 4. Thr1061/Ser1070 phosphorylation inhibited DAD binding to DID.
(A) The DAD peptides, wild-type (Wt) or 2E, were immunoprecipitated (IP) from Cos-7 cells co-expressing an N-terminal fragment of mDia2–GFP (GFP-NTer). Quantification of three co-immunoprecipitations experiments is shown. *P < 0.05 compared with wild-type. (B) HeLa cells were transfected with wild-type or DAD 2E, serum-starved for 24 h, and stained with phalloidin. (C) 10T1/2 cells were transfected with wild-type or DAD 2E along with SM22–luciferase. *P < 0.05 compared with wild-type. (D) Lysates from non-treated and calyculin-treated HeLa cells expressing the indicated FLAG-tagged F1F2-C mDia2 fragment were mixed with lysates from HeLa cells expressing an N-terminal fragment of mDia2 fused to GFP. Following immunoprecipitation with anti-FLAG antibody, co-immunoprecipitation of the N-terminal mDia2 fragment was determined by Western blotting for GFP. Three separate experiments were quantified and levels of co-immunoprecipitation were normalized to that obtained with the wild-type F1F2-C fragment from untreated cells set to 100 %. *P < 0.05 compared with wild-type. EV/Ev, empty vector; IB, immunoblot
Phosphorylation of Thr1061/Ser1070 enhanced mDia2 activity
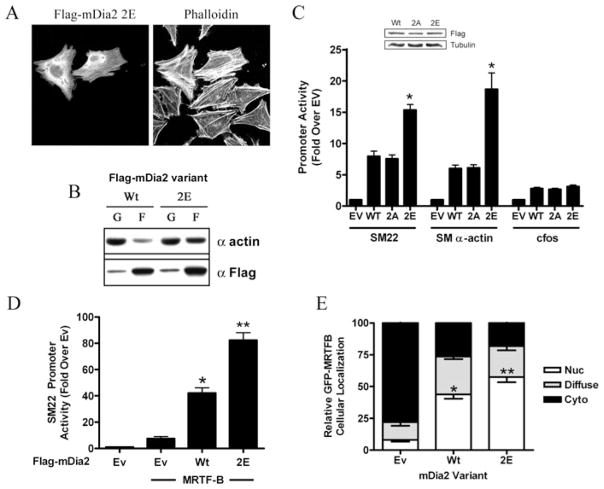
To begin to examine the consequences of Thr1061/Ser1070 phosphorylation on the function of full-length mDia2, we tested the effects of the DAD 2E phosphomimetic on actin polymerization. In contrast with wild-type mDia2, the DAD 2E mDia2 variant enhanced phalloidin staining (Figure 5A) and the amount of F-actin measured in actin sedimentation assays (Figure 5B), suggesting at least partial mDia2 activation by this mutation. We next examined the effect of the DAD 2E and phospho-deficient (DAD 2A) variants on mDia2’s ability to stimulate SMC-specific gene transcription. As shown in Figure 5(C), the DAD 2E variant activated SMC-specific promoter activity more strongly than wild-type, even though these molecules were expressed at nearly identical levels. In contrast, the DAD 2A mutation had little effect on mDia2’s ability to stimulate the SM22 promoter further, suggesting that basal levels of Thr1061/Ser1070 phosphorylation in 10T1/2 cells are relatively low. Expression of the mDia2 DAD 2E variant significantly increased the activity (Figure 5D) and nuclear accumulation (Figure 5E) of MRTF-B, providing additional evidence that phosphorylation of the mDia2 DAD enhances mDia2-mediated actin polymerization.
Figure 5. Phosphorylation of Thr1061/Ser1070 enhances mDia2 activity.
HeLa cells expressing full-length wild-type or DAD 2E mDia2 were serum-starved for 24 h. Cells were then fixed and stained with phalloidin (A) or subjected to actin sedimentation assays (B). (C) 10T1/2 cells were transfected with the indicated full-length mDia2 phosphorylation variant along with luciferase constructs driven by the SM22, SM α-actin or c-Fos promoter. *P < 0.05 compared with wild-type. (D) 10T1/2 cells were co-transfected with the SM22–luciferase, MRTF-B and the indicated FLAG–mDia2 variant. (E) 10T1/2 cells were transfected with GFP–MRTF-B and the full-length wild-type or DAD 2E mDia2 variant. Following serum starvation for 24 h, GFP–MRTF-B subcellular localization was scored as nuclear (Nuc), cytoplasmic (Cyto) or diffuse. At least 50 cells per group from five separate experiments were counted. *P < 0.05 compared with empty vector; **P < 0.05 compared with wild-type. EV/Ev, empty vector; F, F-actin; G, G-actin; WT/Wt, wild-type.
Phosphorylation enhances RhoA-mediated activation of mDia2
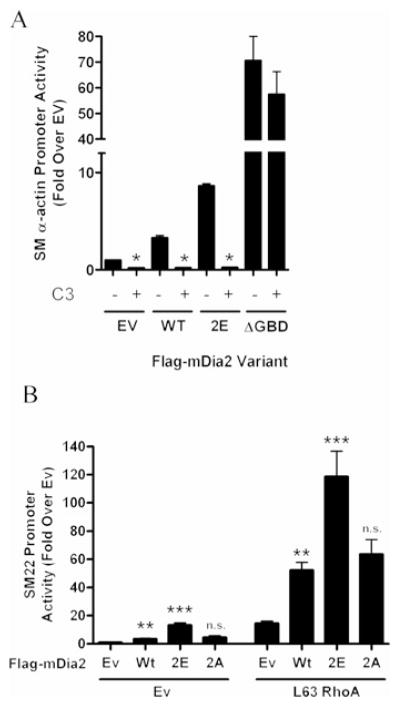
It has been suggested that GTPase binding to the GBD is not sufficient to fully activate DRF-dependent actin polymerization and that an additional, as yet undescribed signal, may also be required [29]. Results shown in Figure 6(A) demonstrate that the activity of the mDia2 phosphomimetic, unlike that of mDia2ΔGBD, was strongly inhibited by co-transfection of the RhoA inhibitor C3 transferase. When coupled with the observation that these mutations do not result in full mDia2 activation, we hypothesized that elimination of this stabilizing interaction renders mDia2 more sensitive to activation by RhoA. In support of this, the DAD 2E variant was synergistically activated by L63RhoA to a much greater extent than wild-type mDia2 or the DAD 2A variant (Figure 6B).
Figure 6. DAD phosphorylation sensitizes mDia2 to activation by RhoA.
(A) 10T1/2 cells were transfected with SM22–luciferase and the indicated mDia2 variant ± C3 transferase. *P < 0.05 compared with without C3. (B) 10T1/2 cells were transfected with SM22–luciferase and the indicated mDia2 variant ± constitutively active L63RhoA; **P < 0.05 compared with empty vector; ***P < 0.05 compared with wild-type. EV/Ev, empty vector; n.s., not significantly different from wild-type; WT/Wt, wild-type.
DISCUSSION
It is clear that mDia2 activity is inhibited by the DID–DAD interaction and that RhoA binding displaces DAD from the DID-binding pocket to expose the catalytically active FH2 domain. Virtually nothing is known about additional signalling mechanisms that regulate mDia2 activity, and the goal of the present study was to examine the effects of phosphorylation on this process. Our results demonstrate that the mDia2 DAD is phosphorylated at Thr1061 and Ser1070 by ROCK. These modifications weaken the DID–DAD interaction, sensitizing the phosphorylated form to activation by RhoA. Direct crosstalk between these two RhoA effectors should have important implications on a number of physiological processes that involve RhoA-dependent regulation of actin polymerization.
The core DAD sequence MDSLLEAL is critically important for the DID–DAD interaction, and crystal structure analyses of mDia1 have demonstrated that this region forms an amphipathic helix that binds tightly to a hydrophobic pocket on the DID surface [5]. Mutation analyses by Wallar et al. [17] implicated the basic region (RRKR) N-terminal to the core domain in mDia2 regulation. These authors demonstrated that single glutamate substitutions at any one of these residues resulted in a significant reduction in DAD affinity for DID and prevented exogenously expressed DAD from activating endogenous mDia. Our demonstration that the T1061E/S107E phosphomimetic enhanced mDia2 activity in the context of the full-length molecule and inhibited the effects of overexpressed DAD peptides fits well with these results. Moreover, Takeya et al. [19] demonstrated that FHOD1 was phosphorylated by ROCK at several residues C-terminal to the core DAD and that phosphorylation inhibited the interaction of the DAD with the putative FHOD1 DID. Similar basic sequences are found in mDia1 and mDia3, but only mDia2 and FHOD1 have conserved consensus ROCK phosphorylation sites near this domain, suggesting differential regulation of the DRFs by this mechanism. It is certainly possible that Thr1061 and/or Ser1070 are targeted by additional kinases, but our results from the present study would suggest that any such phosphorylation would enhance RhoA-dependent mDia2 activation.
The identification of conserved acidic residues in DID that potentially interact with the basic domain (amino acids 376–378) extends our understanding of the molecular interactions that regulate DID–DAD binding and mDia2 activity. During the completion of the present study, Lammers et al. [30] used isothermal titration calorimetry to demonstrate that the corresponding acidic residues of mDia1 (see Figure 1C) were important for DID–DAD binding, strongly supporting a role for this acidic region in DRF auto-inhibition. It will certainly be important to further characterize this region by additional structural studies.
Our results from the present study also suggest that mDia2 phosphorylation is maintained at relatively low levels by the type 1 or 2 phosphatases that are targets of okadaic acid and calyculin A. These findings are intriguing, since phosphatases have previously been shown to regulate several RhoGTPase effectors (see [31] for a review). For example, two PP2A (protein phosphatase 2A)-related phosphatases (POPX1 and POPX2) inhibited lamellipodia and membrane ruffle formation by inactivating the Cdc42/Rac effector PAK (p21-activated kinase) [32]. Interestingly, POPX2 was also identified as an mDia1-binding partner and inhibitor of SRF transcription and MRTF-A nuclear translocation [33]. However, phosphorylation of mDia1 has not yet been reported, and it is unclear whether phosphatase activity was required for this effect.
Although difficult to determine at present, the timing of DAD phosphorylation during the mDia2 activation cycle could have important implications on the regulation of mDia2 activity. For example, since mDia2 activation is essentially determined by competitive binding of RhoA and DAD to the GBD/DID domain, a phosphorylation-mediated weakening of the DID–DAD interaction might favour RhoA binding and initial activation. Alternatively, DAD phosphorylation that occurred subsequent to RhoA binding could prevent the re-association of the DID–DAD complex, leading to prolonged mDia2 activity. In support of the latter mechanism, we observed that introducing an mDia2 N-terminal DID fragment (amino acids 1–533) into our in vitro reaction inhibited phosphorylation of the C-terminal DAD fragment (see Supplementary Figure S2 at http://www.BiochemJ.org/bj/439/bj4390057add.htm), suggesting that the auto-inhibited conformation limits ROCK’s access to the DAD domain.
The precise physiological significance of mDia2 activation by ROCK is not completely clear. However, given that this mechanism occurs downstream of RhoA, we hypothesize that it allows for more dynamic temporal or spatial control of actin polymerization. This could be particularly relevant at the leading edge of migrating cells where precise actin polymerization is required for cell extension. In addition, since mDia2 activity has been shown to be targeted to filopodia by Cdc42 and Rif [34–36], it will be important to test the effects of phosphorylation on the extent and specificity of mDia2 activation by these small GTPases. ROCK and mDia2 activity are clearly important for SMC-specific gene expression [11], and it is likely that this mechanism contributes to the regulation of SMC differentiation. ROCK is also a major regulator of SMC contractility, and this mechanism could help ensure that levels of actin contractile fibres are sufficient to maintain correct SMC tone.
In summary, the results from the present study indicate that ROCK regulates mDia2 activity by phosphorylating the DAD at conserved residues near the basic region. This phosphorylation interferes with the intramolecular DID–DAD interaction and facilitates the activation of mDia2 by RhoA, resulting in increased actin polymerization. Given the importance of mDia2 in a number of physiological processes (especially in SMC), and the fact that this pathway links two important RhoA effectors, it will be critical to further characterize this signalling mechanism.
Supplementary Material
Acknowledgments
FUNDING
This work was supported by the National Institutes of Health [grant number HL-070953 (to C.P.M.)] and the American Heart Association [grant number 07152340U (to D.P.S.)].
Abbreviations used
- Cdc42
cell division cycle 42
- DAD
diaphanous autoregulatory domain
- DAD 2A
DAD T1061A/S1070A
- DAD 2E
DAD T1061E/S1070E
- DID
diaphanous inhibitory domain
- DRF
diaphanous-related formin
- F-actin
filamentous actin
- FH
formin homology
- FHOD1
formin homology domain-containing protein 1
- G-actin
globular actin
- GBD
GTPase-binding domain
- GFP
green fluorescent protein
- GST
glutathione transferase
- MRTF
myocardin-related transcription factor
- PKA
protein kinase A
- PKG
protein kinase G
- ROCK
Rho-kinase
- SAP
shrimp alkaline phosphatase
- SMC
smooth muscle cell
- SRF
serum response factor
Footnotes
AUTHOR CONTRIBUTION
Dean Staus conceived, designed and performed most experiments and wrote the majority of the paper. Joan Taylor helped conceive and design the research, and critically revised the paper. Christopher Mack helped conceive, design and perform the research, and helped write and revise the paper.
References
- 1.Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci. 2005;30:342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 2.Wallar BJ, Alberts AS. The formins: active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 2003;13:435–446. doi: 10.1016/s0962-8924(03)00153-3. [DOI] [PubMed] [Google Scholar]
- 3.Rose R, Weyand M, Lammers M, Ishizaki T, Ahmadian MR, Wittinghofer A. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature. 2005;435:513–518. doi: 10.1038/nature03604. [DOI] [PubMed] [Google Scholar]
- 4.Copeland JW, Copeland SJ, Treisman R. Homo-oligomerization is essential for F-actin assembly by the formin family FH2 domain. J Biol Chem. 2004;279:50250–50256. doi: 10.1074/jbc.M404429200. [DOI] [PubMed] [Google Scholar]
- 5.Nezami AG, Poy F, Eck MJ. Structure of the autoinhibitory switch in formin mDia1. Structure. 2006;14:257–263. doi: 10.1016/j.str.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Otomo T, Otomo C, Tomchick DR, Machius M, Rosen MK. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol Cell. 2005;18:273–281. doi: 10.1016/j.molcel.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Posern G, Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 9.Lockman K, Hinson JS, Medlin MD, Morris D, Taylor JM, Mack CP. Sphingosine 1-phosphate stimulates smooth muscle cell differentiation and proliferation by activating separate serum response factor co-factors. J Biol Chem. 2004;279:42422–42430. doi: 10.1074/jbc.M405432200. [DOI] [PubMed] [Google Scholar]
- 10.Hinson JS, Medlin MD, Lockman K, Taylor JM, Mack CP. Smooth muscle cell-specific transcription is regulated by nuclear localization of the myocardin-related transcription factors. Am J Physiol Heart Circ Physiol. 2007;292:H1170–H1180. doi: 10.1152/ajpheart.00864.2006. [DOI] [PubMed] [Google Scholar]
- 11.Staus DP, Blaker AL, Taylor JM, Mack CP. Diaphanous 1 and 2 regulate smooth muscle cell differentiation by activating the myocardin-related transcription factors. Arterioscler Thromb Vasc Biol. 2007;27:478–486. doi: 10.1161/01.ATV.0000255559.77687.c1. [DOI] [PubMed] [Google Scholar]
- 12.Oh J, Richardson JA, Olson EN. Requirement of myocardin-related transcription factor-B for remodeling of branchial arch arteries and smooth muscle differentiation. Proc Natl Acad Sci USA. 2005;102:15122–15127. doi: 10.1073/pnas.0507346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S, Chang S, Qi X, Richardson JA, Olson EN. Requirement of a myocardin-related transcription factor for development of mammary myoepithelial cells. Mol Cell Biol. 2006;26:5797–5808. doi: 10.1128/MCB.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Zhu X, Chen M, Cheng L, Zhou D, Lu MM, Du K, Epstein JA, Parmacek MS. Myocardin-related transcription factor B is required in cardiac neural crest for smooth muscle differentiation and cardiovascular development. Proc Natl Acad Sci USA. 2005;102:8916–8921. doi: 10.1073/pnas.0503741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Y, Boyd K, Xu W, Ma J, Jackson CW, Fu A, Shillingford JM, Robinson GW, Hennighausen L, Hitzler JK, et al. Acute myeloid leukemia-associated Mkl1 (Mrtf-a) is a key regulator of mammary gland function. Mol Cell Biol. 2006;26:5809–5826. doi: 10.1128/MCB.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat Cell Biol. 2009;11:257–268. doi: 10.1038/ncb1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallar BJ, Stropich BN, Schoenherr JA, Holman HA, Kitchen SM, Alberts AS. The basic region of the diaphanous-autoregulatory domain (DAD) is required for autoregulatory interactions with the diaphanous-related formin inhibitory domain. J Biol Chem. 2006;281:4300–4307. doi: 10.1074/jbc.M510277200. [DOI] [PubMed] [Google Scholar]
- 18.Schonichen A, Alexander M, Gasteier JE, Cuesta FE, Fackler OT, Geyer M. Biochemical characterization of the diaphanous autoregulatory interaction in the formin homology protein FHOD1. J Biol Chem. 2006;281:5084–5093. doi: 10.1074/jbc.M509226200. [DOI] [PubMed] [Google Scholar]
- 19.Takeya R, Taniguchi K, Narumiya S, Sumimoto H. The mammalian formin FHOD1 is activated through phosphorylation by ROCK and mediates thrombin-induced stress fibre formation in endothelial cells. EMBO J. 2008;27:618–628. doi: 10.1038/emboj.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staus DP, Blaker AL, Medlin MD, Taylor JM, Mack CP. Formin homology domain-containing protein 1 regulates smooth muscle cell phenotype. Arterioscler Thromb Vasc Biol. 2011;31:360–367. doi: 10.1161/ATVBAHA.110.212993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miki T, Okawa K, Sekimoto T, Yoneda Y, Watanabe S, Ishizaki T, Narumiya S. mDia2 shuttles between the nucleus and the cytoplasm through the importin-α/β-and CRM1-mediated nuclear transport mechanism. J Biol Chem. 2009;284:5753–5762. doi: 10.1074/jbc.M806191200. [DOI] [PubMed] [Google Scholar]
- 22.Worthylake RA, Lemoine S, Watson JM, Burridge K. RhoA is required for monocyte tail retraction during transendothelial migration. J Cell Biol. 2001;154:147–160. doi: 10.1083/jcb.200103048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mack CP, Somlyo AV, Hautmann M, Somlyo AP, Owens GK. Smooth muscle differentiation marker gene expression is regulated by RhoA-mediated actin polymerization. J Biol Chem. 2001;276:341–347. doi: 10.1074/jbc.M005505200. [DOI] [PubMed] [Google Scholar]
- 24.Mack CP, Thompson MM, Lawrenz-Smith S, Owens GK. Smooth muscle α-actin CArG elements coordinate formation of a smooth muscle cell-selective, serum response factor-containing activation complex. Circ Res. 2000;86:221–232. doi: 10.1161/01.res.86.2.221. [DOI] [PubMed] [Google Scholar]
- 25.Sumi T, Matsumoto K, Nakamura T. Specific activation of LIM kinase 2 via phosphorylation of threonine 505 by ROCK, a Rho-dependent protein kinase. J Biol Chem. 2001;276:670–676. doi: 10.1074/jbc.M007074200. [DOI] [PubMed] [Google Scholar]
- 26.Faix J, Grosse R. Staying in shape with formins. Dev Cell. 2006;10:693–706. doi: 10.1016/j.devcel.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 27.Copeland SJ, Green BJ, Burchat S, Papalia GA, Banner D, Copeland JW. The diaphanous inhibitory domain/diaphanous autoregulatory domain interaction is able to mediate heterodimerization between mDia1 and mDia2. J Biol Chem. 2007;282:30120–30130. doi: 10.1074/jbc.M703834200. [DOI] [PubMed] [Google Scholar]
- 28.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 29.Li F, Higgs HN. Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J Biol Chem. 2005;280:6986–6992. doi: 10.1074/jbc.M411605200. [DOI] [PubMed] [Google Scholar]
- 30.Lammers M, Meyer S, Kuhlmann D, Wittinghofer A. Specificity of interactions between mDia isoforms and Rho proteins. J Biol Chem. 2008;283:35236–35246. doi: 10.1074/jbc.M805634200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsen M, Tremblay ML, Yamada KM. Phosphatases in cell-matrix adhesion and migration. Nat Rev Mol Cell Biol. 2003;4:700–711. doi: 10.1038/nrm1199. [DOI] [PubMed] [Google Scholar]
- 32.Koh CG, Tan EJ, Manser E, Lim L. The p21-activated kinase PAK is negatively regulated by POPX1 and POPX2, a pair of serine/threonine phosphatases of the PP2C family. Curr Biol. 2002;12:317–321. doi: 10.1016/s0960-9822(02)00652-8. [DOI] [PubMed] [Google Scholar]
- 33.Xie Y, Tan EJ, Wee S, Manser E, Lim L, Koh CG. Functional interactions between phosphatase POPX2 and mDia modulate RhoA pathways. J Cell Sci. 2008;121:514–521. doi: 10.1242/jcs.013557. [DOI] [PubMed] [Google Scholar]
- 34.Westendorf JJ. The formin/diaphanous-related protein, FHOS, interacts with Rac1 and activates transcription from the serum response element. J Biol Chem. 2001;276:46453–46459. doi: 10.1074/jbc.M105162200. [DOI] [PubMed] [Google Scholar]
- 35.Gasteier JE, Madrid R, Krautkramer E, Schroder S, Muranyi W, Benichou S, Fackler OT. Activation of the Rac-binding partner FHOD1 induces actin stress fibers via a ROCK-dependent mechanism. J Biol Chem. 2003;278:38902–38912. doi: 10.1074/jbc.M306229200. [DOI] [PubMed] [Google Scholar]
- 36.Yasuda S, Oceguera-Yanez F, Kato T, Okamoto M, Yonemura S, Terada Y, Ishizaki T, Narumiya S. Cdc42 and mDia3 regulate microtubule attachment to kinetochores. Nature. 2004;428:767–771. doi: 10.1038/nature02452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.