Abstract
Network analysis can provide insight into key organizational principles of brain structure and help identify structural changes associated with brain disease. Though static differences between diseased and healthy networks are well-characterized, the study of network dynamics, or how brain networks change over time, is increasingly central to understanding ongoing brain changes throughout disease. Accordingly, we present a short review of network models of spread, network dynamics and network degeneration. Borrowing from recent suggestions, we will divide this review into two processes by which brain networks can change: “Dynamics on networks,” which are functional and pathological consequences taking place atop a static structural brain network; and “dynamics of networks,” which constitutes a changing structural brain network. We focus on diffusion MRI-based structural or anatomic connectivity graphs. We address psychiatric disorders like schizophrenia; developmental disorders like epilepsy; stroke; Alzheimer’s disease and other neurodegenerative diseases.
Keywords: brain networks, connectomics, graph theory, diffusion tensor imaging, neurodegeneration, neurological disease, schizophrenia, neural networks, network neuroscience
1. Introduction
The brain is a large complex network whose processing elements communicate along neural projections. At various spatial scales the processing elements can be neurons, local circuits or large gyri, and the connections between them may be constituted by local dendritic and axonal arbors, all the way to long range bundles of axonal projections. Recent advances in graph-based network analyses make it possible to model, from a local to a global level, the structural wiring that supports the brain’s functional behavior (1–4). The mechanistic role of structural networks for shaping brain dynamics is a key rationale for mapping the human connectome (4). The emerging field of network neuroscience visualizes the brain network as a graph consisting of vertices representing regions and edges as connections between them. Edges can be constructed from a wide array of noninvasive human neuroimaging methodologies to represent both structural and functional networks.
Quickly following the birth of the connectome came pathoconnectomics, which focuses on mapping abnormal brain networks, with the overall goal of understanding brain disorders at the causal mechanistic level (5, 6). Disturbances in global and local network organization are well documented in psychiatric disorders like schizophrenia (SZ) (7–11), developmental disorders like epilepsy (12), stroke (13, 14), severe brain injury (15) as well as neurodegenerative diseases including Alzheimer’s disease (AD) (16), frontotemporal dementia (17) and amyotrophic lateral sclerosis (18). For reviews, see (6, 19–22).
Though static differences between diseased and healthy networks are well-characterized, ongoing brain changes throughout disease are less understood. Hence, this review will focus on a narrower set of concepts involving spread on networks rather than network statistics. This motivates the exploration of network dynamics, or how networks change over time (23). The mechanistic role of structural networks for shaping brain dynamics is a key rationale for mapping the human connectome (24). Here we review recent advances on how network dynamics induced by neurological diseases cause widespread impairment, with a specific focus on the analysis of diffusion MRI-based structural connectivity graphs.
Broadly, disease can affect the anatomic network in two possible ways: First, disease can cause malfunctioning of the nodes of the graph directly and cause either localized or widespread functional impairment by propagating the disease effect along neural connections to other areas. Second, some diseases primarily target the neural connections themselves, for instance via demyelination and axonal injury, leading to anomalous connectivity that then cause widespread information-processing impairments and aberrant function (22).
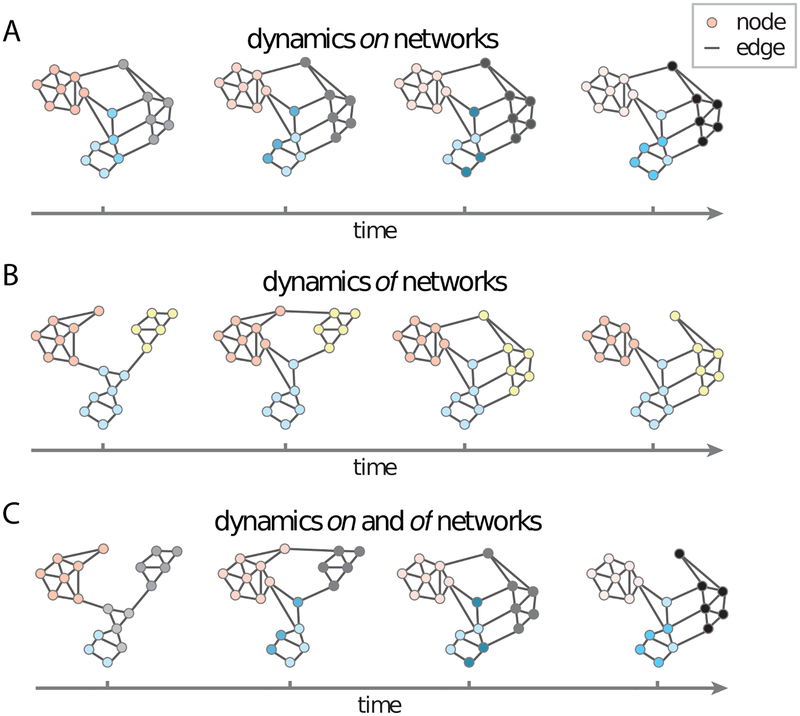
We will refer to these two modes respectively as disease dynamics on brain networks, and of brain networks. This rubric is borrowed from a recent discussion by Bassett and Sporns in 2017 (Figure 1). This review identifies how various diseases induce dynamics of and on networks, and extends the latter to include pathology transmission on networks, especially pertinent to emerging models of neurodegeneration. This of/on dichotomy is a slight over-simplification of network degeneration, since some diseases can have both kinds of effects (23). For example, AD exhibits dynamics both “of” and “on” networks (25, 26). Nonetheless, this framework provides a parsimonious and convenient understanding of network effects. Exceptions and caveats will be noted wherever applicable.
Figure 1.
Dynamic network models. Adapted (permission pending) from Bassett and Sporns, 2017. In the field of network science, two types of dynamic processes are studied in some detail: Dynamics on networks and dynamics of networks. (a) Dynamics on networks indicates that the activity (or some other property of interest) of nodes changes as a function of time. Here we illustrate decreasing activity (pink), increasing activity (gray) and changes in the pattern of activity (blue) over time in distinct network modules or communities. (b) Dynamics of networks indicates that the edges of the network themselves change either in their existence/absence or in their strength. Here we illustrate the coalescence of modules (blue and yellow), as well as the transfer of allegiance of a single region from one module (pink) to another (yellow) over time.
1.1. Current techniques of network construction, graph theory metrics and their limitations
A technical overview of current methods of constructing brain anatomic networks is contained in the supplemental information; this description is selective and brief rather than thorough, and the reader is pointed to several comprehensive review papers on this topic. Before discussing network spread, it is important to understand that network construction comes with several caveats, which have been reviewed comprehensively (27, 28). Briefly, tractography is known to yield false-positive and false-negative connections, where as a consequence, spurious tracts might be detected as plausible and genuine ones as invalid. Erroneous or biased conclusions can result from missing, partial or duplicate fibers (28). Similarly, graph theory metrics of structural networks suffer from pitfalls arising from lack of reliable approaches to edge thresholding, binarization and multiple comparisons (29, 30). These choices can affect the size, robustness and intrinsic organization of a network. Several network summary metrics (e.g., efficiency and path length) are non-linear and sensitive to such error because they are defined in respect to implied neighbors. These metrics are examined extensively in prior reviews, see (19–22).
2. Neurodegeneration: Dynamics ON or OF brain networks?
Nearly all neurodegenerative disorders are characterized by stereotypical patterns of disease progression measurable from longitudinal volumetric MRI analysis (e.g. FreeSurfer) (31). AD-associated atrophy, tau pathology, amyloid pathology and metabolic load all display highly stereotyped progression into brain circuits, from entorhinal cortex and hippocampus to temporal, parietal and eventually frontal regions (32, 33). This closely mirrors pathological tau spread first observed by Braak (34, 35). These converging results suggest that all relevant biomarkers of disease - atrophy, tau and amyloid – exhibit progression that follows fiber pathways rather than proximity (36–41). This section describes recent studies of network spread in neurodegeneration, with a focus on Alzheimer’s disease, the most prevalent degenerative disease.
2.1. Network degeneration implies dynamics of networks.
Conventionally, degeneration is thought to cause progressive disconnections of vulnerable fiber pathways via secondary Wallerian degeneration, loss of signaling, axonal retraction and post-synaptic dendrite retraction (36–38, 42), collectively referred to as network degeneration; for a review see (43). Affected brain regions send disordered information to connected regions, where they cause dysfunction and atrophy over time (42). Although neither Powell et al., 2018 (44) nor Oxtoby et al., 2017 (26) found significant alteration in global network statistics in AD, the latter, using a data-driven event-based model for sequencing the progression of AD patients, demonstrated that node-level measures of network hubs deteriorate. This deterioration causes progressive anatomical network disruption, not just neurodegeneration. A wider body of evidence suggests different dementias selectively target distinct intrinsic functional networks (33, 36, 42, 45) perhaps due to differential patterns insults governed by genetic, molecular, metabolic or oxidative factors (46). This is an example of dynamics of networks, since network topology is itself progressively altered. For a graph-theoretic exploration of progressive disconnection see (47), and for a comprehensive review see (22).
2.2. Networked spread of misfolded proteins: An example of dynamics on networks.
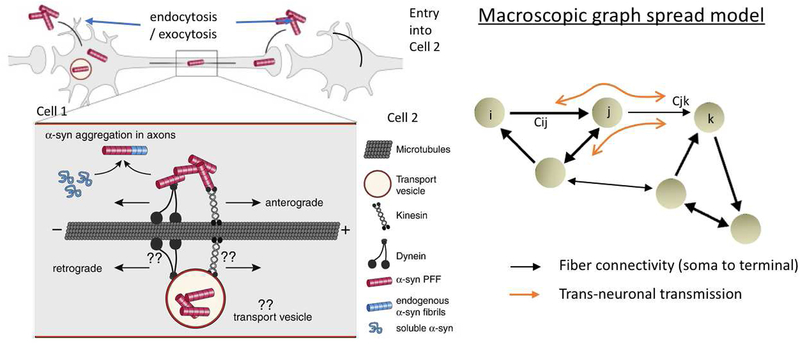
An alternate view has emerged, that instead of being primarily impaired in degeneration, the network serves mainly as a conduit for the transmission of disease – an example of dynamics on networks. Recently, it was shown (44) that despite measurable changes in integrity of specific fiber tracts, the overall structural network organization in AD is preserved, with similar graph metrics to controls. The authors state that the combined effect of edge thresholding, binarization and non-inclusion of subcortical regions might be responsible for global topological disturbances found in previous reports. Structural connectivity was found to mediate the relationships between regional atrophy, metabolism and amyloid in AD (15). This idea is underpinned by evidence that progression might occur via “prion-like” protein aggregation followed by trans-synaptic transmission of toxic proteins along neuronal pathways (48–54). Indeed, toxic tau and other proteins have the capacity to misfold, induce non-folded tau to adopt pathological conformations in a template-directed manner, travel across synapses into connecting neurons, and finally march throughout local and then long-range circuits, slowly ramifying across widespread brain circuits (48, 50, 52–54). In this manner pathology can spread along axonal pathways either anterogradely, or retrogradely toward the cell body, or both (52–54). The cellular processes involved in the transmission of synuclein, for example, were recently reviewed comprehensively by Bieri et al (55), and pictorially depicted in Figure 2A. In Figure 2B the graph analog of these cellular-level transneuronal transmission and transport processes is illustrated on the whole brain network.
Figure 2.
Modeling the process of pathology transmission between neurons using network spread. A: Pictorial example of transmission of α-syn between neurons. α-Syn fibrils can be internalized both in the dendrite/cell body compartment and in axons. α-Syn fibrils are actively transported along microtubules in both anterograde and retrograde direction, whether directly in the cytoplasm or in transport vesicles following endocytosis. Aggregation occurs via the growth of α-syn fibrils upon misfolding and recruitment of soluble endogenous α-syn proteins. Figure adapted with permission from Bieri, et al, 2018. B: Illustration of the whole brain connectivity network, with nodes representing brain regions, and connection strengths Cij representing fiber connectivity between regions i and j. On this network the cellular-level trans-neuronal transmission and transport processes depicted in panel A are modeled via macroscopic network spread shown in orange.
2.3. Models of trans-neuronal network propagation
The first aspect of the emerging model – template-driven protein aggregation - has benefitted from quantitative modeling using differential equations, mainly in the prion disease context. For a recent review see (56). These include the original heterodimer model (57), nucleated polymerization (NPM) (58) and Smoluchowski aggregation model (59, 60). The incorporation of classical spatial diffusion was proposed into prion aggregation models (61), into NPM model (62) and into truncated Smoluchowski’s equations (60). Models are listed and briefly described in Table 1.
Table 1.
Selected mathematical modeling papers on neurodegeneration (protein aggregation and network spread) highlighted in the current review. Please refer to section 2.3 for brief description of individual models listed below, and to the original papers for further information.
| Name (Citation) |
Description | Comments | Validation |
|---|---|---|---|
| Protein Aggregation Models | |||
| Heterodimer model (57) Prusiner et al. 1990 | Original landmark, template-driven heterodimer model of prion spread | Local protein aggregation, not network-based | In vitro |
| NPM (58) Masel et al. 1999 | Nucleated Polymerization Model: Monte Carlo discrete event simulations to model prion aggregation | Local protein aggregation, not network-based | In vivo (animal) |
| Smoluchowski Aggregation (60) Bersth et al. 2016 | Smoluchowski’s coagulation equation describes time evolution of concentration of polymers as they coagulate | Local protein aggregation, not network-based. Simultaneously modeled Amyloid cascade and prion hypotheses | X |
| Heterodimer model (61) Payne et al. 1998 | Prion aggregation described utilizing concentration dynamics of 2 strains of prions undergoing competition | Local protein aggregation, not network-based | In vivo (animal) |
| Matthaus (62) Matthaus et al. 2006 | Applied classic epidemic spread models of networks to recapitulate prion-like disease | Used synthetic lattice networks | In vitro |
| Network Spread Models | |||
| NDM (39) Raj et al. 2012 | Applied graph diffusion to model AD progression on large scale brain networks | Real human connectomes. Suggested spatially distinct “eigen-modes” which mediate network vulnerability | In vivo (Human neuroimaging). Validation limited to 2–4 year follow-up |
| NDM v gene expression (70) Acosta et al., 2018 | Compared graph diffusion against innate regional vulnerability (given by healthy regional gene expression) as predictors of the observed crosssectional atrophy patterns in AD | Found that NDM is a far better predictor of AD topography than is innate regional vulnerability | Validation on cross sectional regional atrophy data obtained from in vivo human MRI |
| ESM (25) Iturria-Medina et al. 2014 | Epidemic Spreading Model in AD interrelating structural connectivity and in vivo amyloid beta | Explicit Amyloid beta production/ clearance terms | In vivo (Human neuroimaging) validation on Amyloid PET |
| General Graph Theory Models | |||
| Graph Spread Koenigsberger et al., 2017 (71) Misic et al 2015 (72) | Models of graph spread, based on cooperative and competitive spreading dynamics on human connectomes | Model uses general principles of spread rather than biophysical modeling of pathology | In vivo (Human neuroimaging) |
The misfolded and aggregated proteins can travel along propagation pathways (63) via spatial gradient-driven processes, which would imply dependence on fiber length. However, it is not clear whether protein transmission occurs via passive diffusion, active axonal transport or other distance-independent processes – see reviews (64, 65). In the latter case, the overall density of fiber projections, captured via the connection strength measure used in graph theory, would assume higher importance than fiber distance-dependent spatial diffusion (see Figure 2B). Franziska Matthäus (62) implemented an epidemiological spread model on a simple lattice network topology, as did (66). The Network Diffusion Model (NDM) mathematically derived the behavior of protein transmission as a graph heat equation under a connectivity-driven rather than spatial- or fiber distance-driven mechanism (39). They obtained an analytical, closed-from solution for transneuronal propagation on whole brain structural connectivity networks. The macroscopic patterns predicted by the model, via spatially distinct Laplacian “eigen-modes,” were consistent with known patterns of atrophy in various dementias. An epidemic spreading model of network spread was given by (25), which included, in contrast to the NDM model, explicit production/clearance terms, and accessed via stochastic rather than analytical equations. The ESM model was successfully validated on PET Amyloid-ß patterns in 733 patients. Other network models were described in (67).
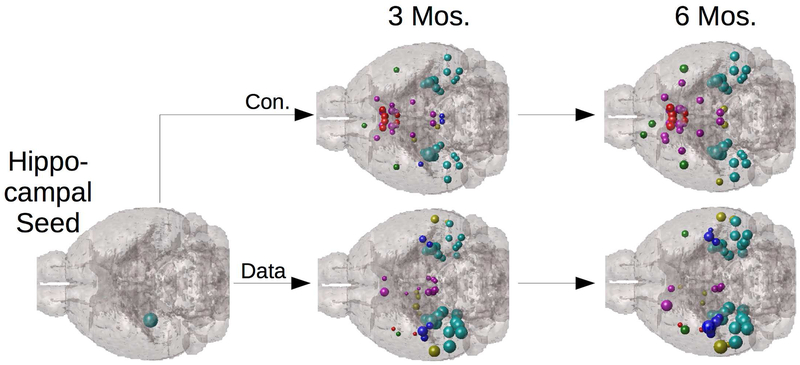
Although these network models were initially applied to human data, recently it was shown that network spread is equally effective at predicting the progression of tau pathology in transgenic mouse models (68), see Figure 3. This study showed using a series of statistical linear mixed models that the contribution of network connectivity far exceeds innate region-specific molecular factors governed by regional gene expression of AD-implicated genes. A caveat here is that network transmission models perform with higher accuracy for tau pathology than for beta amyloid, which appears to accommodate spatial diffusion with equal or higher plausibility, as shown recently (69). In similar vein, it was recently shown (70) via a series of mixed statistical models with both NDM and regional gene expression as predictors, only the former was a significant predictor of regional atrophy of human AD patient. This finding suggests innate properties of brain regions are insufficient to explain regional vulnerability to AD, whereas network transmission is a reliable predictor. Finally, general, high-level graph models of communication dynamics are also available; see (71). A graph model of cooperative spread illustrated that more central “hub” regions (those that are most well connected to the rest of the brain) in the network facilitate early spreading. Conversely, short paths (the set of graph connections required to connect any two nodes with the smallest sum of connection weights) accelerate communication cascades (72).
Figure 3.
Network Diffusion of Tau Pathology. Network diffusion model implemented on the mesoscale mouse connectome, starting from hippocampal seeding (top row) closely recapitulates the 3- and 6-month follow up pattern of tau proliferation in a transgenic AD mouse model (bottom row). Spheres represent mouse brain structures, color coded by lobe (cortical = cyan, limbic = red), and sphere size represents tau concentration. This kind of validation of network spread is critical since mouse data can directly measure both connectivity (via viral tracers) and tau pathology (via AT8 staining). Adapted with permission from Mezias et al., 2017.
Clinical utility.
By successfully formalizing qualitative neuropathological observations into quantitative network models, these emerging approaches can have wide applicability. The NDM was shown to have high ability to predict future disease patterns from the baseline scans of AD spectrum patients (40). However, this approach has yet to be replicated by others. Interestingly, these models can also be used to infer the likely regions of origin in individual cases. Seeding patterns from potentially multiple pathologic attacks over time were inferred by (73) using a sophisticated but expensive gradient descent algorithm. A simplified and less computationally demanding algorithm for the inference of likely initial seed patterns was proposed by (74) using the sparsity-preserving approach. Normally, model fitting is accomplished by minimizing a suitable cost function that is designed to penalize deviation from observed data. This method fails when the problem is ill-posed, whereby there is no unique minimum. The approach of (73) and (74) resolve this problem by adding a sparsity-preserving penalty, under the assumption that the desired initial pathology seeding pattern must be non-zero at only a small number of sites. Such penalty terms are widely used in the field of inverse problems, especially in image processing, in cases where there is a prior expectation that the solution be sparse.
2.4. Other degenerative diseases
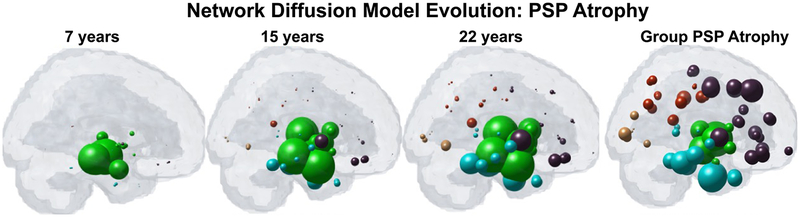
Pathological commonality and overlap is observed in various dementias, and specific biochemical properties of the pathologic entity may be inconsequential for the macroscopic and chronic clinical manifestation of disease (36, 45). Tau, A-beta and alpha-synuclein are present to varying degree in most degenerative diseases: AD, semantic dementia, Frontotemporal Dementia (FTD) (75), dementia with Lewy bodies (DLB) and posterior cortical atrophy (76). Furthermore, it was shown that resting-state functional connectivity of healthy brain regions is correlated with vulnerability to atrophy in five different neurodegenerative diseases (36, 45). Utilizing expression of a DNA binding protein, pTDP-43, from post-mortem brain tissue, Schmidt and colleagues constructed a network model to simulate pathology spread in Amyotrophic lateral sclerosis (ALS) and found that pTDP-43 in ALS may exhibit dynamics similar to amyloid and/or tau in dementia, following a topological pattern of spread along the brain’s anatomical pathways (77). In a rare neurodegenerative tauopathy called Progressive Supranuclear Palsy (PSP), pathology patterns were likewise recapitulated utilizing a structural connectome-based NDM (41); see Figure 4. This work also highlights the potential utility of using a directional connectome, as defined by the polarity of individual axonal projections (soma to axonal terminal or vice versa). Although not available from dMRI, a directional connectome can be inferred from tracer studies in mouse models (41). Alternatively, directionality of the connectome can be inferred from emerging methods involving metabolic activity mapping (78).
Figure 4.
Demonstration of NDM in predicting PSP atrophy. Top: Regional PSP group atrophy t-statistic. Middle: NDM predicted atrophy pattern from a seed placed at the hypothalamus. Bottom: snapshots of time-resolved NDM seeded at hypothalamus, showing progressive atrophy of subcortical and temporal areas. Spheres are color coded by lobe (green=subcortical, blue = temporal, brown = frontal, red = parietal, yellow = occipital). Spheres are sized according to regional atrophy. Reproduced with permission from (Pandya et al, 2017a).
Together, these network dynamic models provide a mechanical explanation of macroscopic archetypal patterns of regional specificity in various dementias, whereby diverse degenerative etiologies share a common network-based progression mechanism. Network dynamics might be sufficient to explain archetypal patterns of regional specificity in various dementias, with no particular need to invoke region- or tissue-specific factors of selective vulnerability. In this view the observed patterns of disease are simply a mechanical result of the way the disease moves around within the brain network (39).
3. Schizophrenia: Dynamics “on” or “of” networks?
There is evidence for schizophrenia (SZ) for being a disorder “on” as well as “of” networks. However, network models capable of simultaneously testing competing hypotheses have not yet been developed. The following section will review evidence for both hypotheses, highlighting the need for additional network models to be tested in psychiatric disease.
3.1. Support for dynamics “on” networks in SZ
Regional atrophy in SZ shows progressive waves of tissue loss over time (79–83), which might be mediated by connectivity – an example of dynamics “on” networks. Such topological changes may be the outcome of different growth processes and neurodevelopmental abnormalities in SZ that impact large multimodal cortical organization (84). Indeed, functional network models of SZ suggest changes in synaptic excitation and inhibition in schizophrenia disrupt delta rhythm-mediated cortico-cortical communication, while enhancing thalamocortical communication. The contrasting relationships between delta and higher frequencies in thalamus and cortex generate frequency mismatches in inter-regional connectivity, leading to a disruption in temporal communication between higher-order brain regions (85). Since these functional connectivity changes between regions take place atop a fixed structural network, they may be modeled as different modes of activity spread on the anatomic network, e.g. using theoretical network spread models (86, 87).
3.2. Support for dynamics “of” networks in SZ
Functional changes, such as cortical atrophy and EEG, are accompanied by extensive structural damage (88), which lends support to dynamics “of” networks hypotheses. Diffusion MRI studies report altered WM integrity in SZ (89–92) and graph theory analysis shows widespread disturbances in structural organization, as reflected by metrics such as path length, centrality, efficiency and clustering coefficient (8, 10, 93–96).
Age-dependent network alternations.
As in any developmental disorder, many of the network-level alterations in SZ have a strong age effect. Some studies suggest brain abnormalities exist early in the disease that lessen over time as the brain appears to normalize (97–101). However, age-related normalization is controversial, as other work suggests network integrity deteriorates throughout disease (102–105). There are differing perspectives on how the network changes in SZ as a joint function of disease, age and regional gene expression (83).
Powell et al. 2018 computationally modeled age-and genetic expression-related structural network changes in SZ (100). In a cross-sectional design, they found that control network topology degrades in a stereotyped linearly degrading manner, while corresponding patient networks experience a degree of post-onset compensatory rewiring. Younger adult patients (ages 20–37), show compromised topology, whereas older adult patients (ages 38–68) show linear network degradation and less pronounced network differences between patients and controls. This was presented as evidence of compensatory rewiring, reflective of the dynamic nature “of” brain networks.
4. Epilepsy: Utilizing network models to test competing hypotheses of dynamics
Epilepsy is a canonical example of neural dysfunction caused by the spread of hypersynchrony and hyperactivity on the anatomic network. Morphometric analysis using MRI indicates progressive extra-hippocampal and extra-temporal atrophy (106–111) in temporal lobe epilepsy (TLE), likely a result of seizures (112). Since damaged regions tend to be functionally and anatomically connected to seizure-prone structures (108, 113), this suggests a strong network effect confirmed by graph theory analysis (12, 110, 114) see reviews in (6, 115, 116).
Atrophy distribution in TLE may either be consequence of the propagation of epileptogenic activity (113, 117) resulting from excitotoxicity (12, 118, 119) (dynamics on networks); or due to a progressive deafferentation process followed by gradual and progressive neuronal loss in connected remote regions (dynamics of networks). Several mechanisms for deafferentation-induced atrophy are known (120, 121). Further, seizure spread may not be related to atrophy (122), while white matter fiber integrity was correlated with remote atrophy (120). To understand which of these models is more predictive of empirical atrophy patterns in TLE, Abdelnour and colleagues (123) modeled both processes using network spread, following prior theoretical work (86, 87). They found that while both models can reproduce the empirical atrophy distribution, the model corresponding to dynamics of networks, out-performs the hyperactivity spread model corresponding to dynamics on networks. However, further graph theoretic research incorporating EEG and MEG networks (124) is needed to resolve this matter. A good example of what is needed is provided by (125) where a detailed network of spiking neurons was used to simulate the spread of hyperexcitability. Apart from analytical models like network diffusion (86, 87), other graph theoretic models of the structure-function relationship are available on human neuroimaging data, for example (126).
5. Summary and Outlook
This mini review was aimed at surveying current approaches of modeling the network effect of neurodegenerative and psychiatric diseases, under the rubric of dynamics on versus of brain networks. This scheme is pedagogically useful but clearly has limitations. Many brain disorders, including those highlighted here, do not sort themselves neatly into one or the other box. Generally, any process that impairs neural systems will also secondarily alter the network itself, hence a strong interplay may be expected. Further, any model of network dynamics, whether of or on, cannot address disease etiology or pathophysiology; its value lies in showing that the macroscopic effect of network dynamics can largely explain the stereotyped patterns of disease irrespective of individual subjects’ and diseases’ etiologic factors. Nonetheless, this review suggests that a distinction between dynamics on and of networks is important in understanding how disease ramifies on brain networks. Too frequently, current graph theoretic research takes statistical or descriptive approaches aimed at demonstrating disease-induced alterations in network statistics but are unable to trace the underlying network dynamical processes that cause those changes. The value of network dynamics is that understanding the manner of network ramification can lead to clinical, diagnostic and therapeutic interventions. The field of network dynamics and network spread may well be the next big frontier in brain-related graph theory.
Supplementary Material
Acknowledgements
AR was supported in part by NIH grants R01NS092802 and R01 EB022717.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
Dr. Raj discloses his patent on a related topic, entitled: Methods and tools for analyzing brain images. USPTO patent number: 9563950. Dr. Raj reports no other biomedical financial interests or potential conflicts of interest. Dr. Powell reports no biomedical financial interests or potential conflicts of interest.
References
- 1.Hagmann P, Kurant M, Gigandet X, Thiran P, Wedeen VJ, Meuli R, Thiran J-P (2007): Mapping Human Whole-Brain Structural Networks with Diffusion MRI. (Sporns O, editor) PLoS One. 2: e597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iturria-Medina Y, Canales-Rodríguez EJ, Melie-García L, Valdés-Hernández PA, Martínez-Montes E, Alemán-Gómez Y, Sánchez-Bornot JM (2007): Characterizing brain anatomical connections using diffusion weighted MRI and graph theory. Neuroimage. 36: 645–60. [DOI] [PubMed] [Google Scholar]
- 3.Kaiser M, Hilgetag CC (2006): Nonoptimal component placement, but short processing paths, due to long-distance projections in neural systems. PLoS Comput Biol. 2: e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sporns O (2012): From simple graphs to the connectome: Networks in neuroimaging. Neuroimage. 62: 881–886. [DOI] [PubMed] [Google Scholar]
- 5.Rubinov M, Bullmore E (2013): Fledgling pathoconnectomics of psychiatric disorders. Trends Cogn Sci. 17: 641–647. [DOI] [PubMed] [Google Scholar]
- 6.Stam CJ (2014): Modern network science of neurological disorders. Nat Rev Neurosci. 15: 683–695. [DOI] [PubMed] [Google Scholar]
- 7.Skudlarski P, Schretlen DJ, Thaker GK, Stevens MC, Keshavan MS, Sweeney JA, et al. (2013): Diffusion tensor imaging white matter endophenotypes in patients with schizophrenia or psychotic bipolar disorder and their relatives. Am J Psychiatry. 170: 886–898. [DOI] [PubMed] [Google Scholar]
- 8.van den Heuvel MP, Mandl RC, Stam CJ, Kahn RS, Hulshoff Pol HE (2010): Aberrant frontal and temporal complex network structure in schizophrenia: a graph theoretical analysis. J Neurosci. 30: 15915–15926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q, Su T-P, Zhou Y, Chou K-H, Chen I-Y, Jiang T, Lin C-P (2012): Anatomical insights into disrupted small-world networks in schizophrenia. Neuroimage. 59: 1085–1093. [DOI] [PubMed] [Google Scholar]
- 10.Zalesky A, Fornito A, Seal ML, Cocchi L, Westin CF, Bullmore ET, et al. (2011): Disrupted axonal fiber connectivity in schizophrenia. Biol Psychiatry. 69: 80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang R, Wei Q, Kang Z, Zalesky A, Li M, Xu Y, et al. (2015): Disrupted brain anatomical connectivity in medication-naïve patients with first-episode schizophrenia. Brain Struct Funct. 220: 1145–1159. [DOI] [PubMed] [Google Scholar]
- 12.Bernhardt BC, Chen Z, He Y, Evans AC, Bernasconi N (2011): Graph-theoretical analysis reveals disrupted small-world organization of cortical thickness correlation networks in temporal lobe epilepsy. Cereb Cortex. 21: 2147–57. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Yu C, Chen H, Qin W, He Y, Fan F, et al. (2010): Dynamic functional reorganization of the motor execution network after stroke. Brain. 133: 1224–1238. [DOI] [PubMed] [Google Scholar]
- 14.Kuceyeski A, Kamel H, Navi BB, Raj A, Iadecola C (2014): Predicting future brain tissue loss from white matter connectivity disruption in ischemic stroke. Stroke. 45: 717–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pandya S, Kuceyeski A, Raj A (2016): The Brain’s Structural Connectome Mediates the Relationship between Regional Neuroimaging Biomarkers in Alzheimer’s Disease. (Hatami A, editor) J Alzheimer’s Dis. Preprint: 1–19. [DOI] [PubMed] [Google Scholar]
- 16.Lo C-Y, Wang P-N, Chou K-H, Wang J, He Y, Lin C-P (2010): Diffusion Tensor Tractography Reveals Abnormal Topological Organization in Structural Cortical Networks in Alzheimer’s Disease. J Neurosci. 30. doi: 10.1523/JNEUROSCI.4136-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuceyeski A, Zhang Y, Raj A (2012): Linking white matter integrity loss to associated cortical regions using structural connectivity information in Alzheimer’s disease and fronto-temporal dementia: the Loss in Connectivity (LoCo) score. Neuroimage. 61: 1311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verstraete E, Veldink JH, Mandl RC, van den Berg LH, van den Heuvel MP (2011): Impaired structural motor connectome in amyotrophic lateral sclerosis. PLoS One. 6: e24239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bassett DS, Bullmore E (2006): Small-World Brain Networks. Neurosci. 12: 512–523. [DOI] [PubMed] [Google Scholar]
- 20.Bullmore E, Sporns O (2009): Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 10: 186–198. [DOI] [PubMed] [Google Scholar]
- 21.Zalesky A, Fornito A, Harding IH, Cocchi L, Yücel M, Pantelis C, et al. (2010): Revisiting the Foundations of Network Analysis. Neuroimage. 20: 353–364. [Google Scholar]
- 22.Iturria-Medina Y (2013): Anatomical brain networks on the prediction of abnormal brain states. Brain Connect. 3: 1–21. [DOI] [PubMed] [Google Scholar]
- 23.Bassett DS, Sporns O (2017): Network neuroscience. Nat Neurosci. 20: 353–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sporns O (2012): Discovering the human connectome. MIT Press. [Google Scholar]
- 25.Iturria-Medina Y, Sotero RC, Toussaint PJ, Evans AC (2014): Epidemic spreading model to characterize misfolded proteins propagation in aging and associated neurodegenerative disorders. (Sporns O, editor) PLoS Comput Biol. 10: e1003956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oxtoby NP, Garbarino S, Firth NC, Warren JD, Schott JM, Alexander DC (2017): Data-Driven Sequence of Changes to Anatomical Brain Connectivity in Sporadic Alzheimer’s Disease. Front Neurol. 8: 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones DK, Cercignani M (2010): Twenty-five pitfalls in the analysis of diffusion MRI data. NMR Biomed. 23: 803–820. [DOI] [PubMed] [Google Scholar]
- 28.Daducci A, Dal Palú A, Descoteaux M, Thiran J-P (2016): Microstructure Informed Tractography: Pitfalls and Open Challenges. Front Neurosci. 10. doi: 10.3389/fnins.2016.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fornito A, Zalesky A, Breakspear M (2013): Graph analysis of the human connectome: Promise, progress, and pitfalls. Neuroimage. 80: 426–444. [DOI] [PubMed] [Google Scholar]
- 30.Drakesmith M, Caeyenberghs K, Dutt A, Lewis G, David AS, Jones DK (2015): Overcoming the effects of false positives and threshold bias in graph theoretical analyses of neuroimaging data. Neuroimage. 118: 313–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. (2002): Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 33: 341–55. [DOI] [PubMed] [Google Scholar]
- 32.Thompson PM, Hayashi KM, de Zubicaray G, Janke AL, Rose SE, Semple J, et al. (2003): Dynamics of gray matter loss in Alzheimer’s disease. J Neurosci. 23: 994–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buckner RL, Snyder AZ, Shannon BJ, LaRossa G, Sachs R, Fotenos AF, et al. (2005): Molecular, Structural, and Functional Characterization of Alzheimer’s Disease: Evidence for a Relationship between Default Activity, Amyloid, and Memory. J Neurosci. 25: 7709–7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braak H, Braak E (1991): Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82: 239–59. [DOI] [PubMed] [Google Scholar]
- 35.Braak H, de Vos RAI, Bohl J, Del Tredici K (2006): Gastric α-synuclein immunoreactive inclusions in Meissner’s and Auerbach’s plexuses in cases staged for Parkinson’s disease-related brain pathology. Neurosci Lett. 396: 67–72. [DOI] [PubMed] [Google Scholar]
- 36.Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD (2009): Neurodegenerative Diseases Target Large-Scale Human Brain Networks. Neuron. 62: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Englund E, Brun A, Alling C (1988): White matter changes in dementia of Alzheimer’s type. Biochemical and neuropathological correlates. Brain. 111 (Pt 6): 1425–39. [DOI] [PubMed] [Google Scholar]
- 38.Villain N, Desgranges B, Viader F, de la Sayette V, Mézenge F, Landeau B, et al. (2008): Relationships between hippocampal atrophy, white matter disruption, and gray matter hypometabolism in Alzheimer’s disease. J Neurosci. 28: 6174–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raj A, Kuceyeski A, Weiner M (2012): A Network Diffusion Model of Disease Progression in Dementia. Neuron. 73: 1204–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raj A, LoCastro E, Kuceyeski A, Tosun D, Relkin N, Weiner M (2015): Network Diffusion Model of Progression Predicts Longitudinal Patterns of Atrophy and Metabolism in Alzheimer’s Disease. Cell Rep. 10: 359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pandya S, Mezias C, Raj A (2017): Predictive Model of Spread of Progressive Supranuclear Palsy Using Directional Network Diffusion. Front Neurol. 8: 692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delbeuck X, Van der Linden M, Collette F (2003): Alzheimer’s disease as a disconnection syndrome? Neuropsychol Rev. 13: 79–92. [DOI] [PubMed] [Google Scholar]
- 43.Brier MR, Thomas JB, Ances BM (2014): Network dysfunction in Alzheimer’s disease: refining the disconnection hypothesis. Brain Connect. 4: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Powell F, Tosun D, Sadeghi R, Weiner M, Raj A (2018): Preserved Structural Network Organization Mediates Pathology Spread in Alzheimer’s Disease Spectrum Despite Loss of White Matter Tract Integrity. J Alzheimer’s Dis. 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou J, Gennatas E, Kramer J, Miller B, Seeley W, Bangaru S, et al. (2012): Predicting Regional Neurodegeneration from the Healthy Brain Functional Connectome. Neuron. 73: 1216–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saxena S, Caroni P (2011): Selective neuronal vulnerability in neurodegenerative diseases: from stressor thresholds to degeneration. Neuron. 71: 35–48. [DOI] [PubMed] [Google Scholar]
- 47.Drzezga A, Becker JA, Van Dijk KRA, Sreenivasan A, Talukdar T, Sullivan C, et al. (2011): Neuronal dysfunction and disconnection of cortical hubs in non-demented subjects with elevated amyloid burden. Brain. 134: 1635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jucker M, Walker L (2013): Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature. Retrieved June 12, 2017, from http://www.nature.com/nature/journal/v501/n7465/abs/nature12481.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frost B, Ollesch J, Wille H, Diamond MI (2009): Conformational diversity of wild-type Tau fibrils specified by templated conformation change. J Biol Chem. 284: 3546–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frost B, Diamond MI (2010): Prion-like mechanisms in neurodegenerative diseases. Nat Rev Neurosci. 11: 155–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palop JJ, Chin J, Mucke L (2006): A network dysfunction perspective on neurodegenerative diseases. Nature. 443: 768–773. [DOI] [PubMed] [Google Scholar]
- 52.Clavaguera F, Bolmont T, Crowther RA, Abramowski D, Frank S, Probst A, et al. (2009): Transmission and spreading of tauopathy in transgenic mouse brain. Nat Cell Biol. 11: 909–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu L, Drouet V, Wu JW, Witter MP, Small SA, Clelland C, Duff K (2012): Trans-synaptic spread of tau pathology in vivo. PLoS One. 7: e31302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iba M, McBride JD, Guo JL, Zhang B, Trojanowski JQ, Lee VM-Y (2015): Tau pathology spread in PS19 tau transgenic mice following locus coeruleus (LC) injections of synthetic tau fibrils is determined by the LC’s afferent and efferent connections. Acta Neuropathol. 130: 349–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bieri G, Gitler AD, Brahic M (2018): Internalization, axonal transport and release of fibrillar forms of alpha-synuclein. Neurobiol Dis. 109: 219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carbonell F, Iturria-Medina Y, Evans AC (2018): Mathematical Modeling of Protein Misfolding Mechanisms in Neurological Diseases: A Historical Overview. Front Neurol. 9: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prusiner SB, Scott M, Foster D, Pan KM, Groth D, Mirenda C, et al. (1990): Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 63: 673–86. [DOI] [PubMed] [Google Scholar]
- 58.Masel J, Jansen VA, Nowak MA (1999): Quantifying the kinetic parameters of prion replication. Biophys Chem. 77: 139–52. [DOI] [PubMed] [Google Scholar]
- 59.Achdou Y, Franchi B, Marcello N, Tesi MC (2013): A qualitative model for aggregation and diffusion of $ $\beta $ $ -amyloid in Alzheimer’s disease. J Math Biol. 67: 1369–1392. [DOI] [PubMed] [Google Scholar]
- 60.Bertsch M, Franchi B, Marcello N, Tesi MC, Tosin A (2016): Alzheimer’s disease: a mathematical model for onset and progression. Math Med Biol A J IMA. 34: 193–214. [DOI] [PubMed] [Google Scholar]
- 61.Payne RJ, Krakauer DC (1998): The spatial dynamics of prion disease. Proceedings Biol Sci. 265: 2341–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matthäus F (2006): Diffusion versus network models as descriptions for the spread of prion diseases in the brain. J Theor Biol. 240: 104–113. [DOI] [PubMed] [Google Scholar]
- 63.Warren JD, Rohrer JD, Schott JM, Fox NC, Hardy J, Rossor MN (2013): Molecular nexopathies: a new paradigm of neurodegenerative disease. Trends Neurosci. 36: 561–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brundin P, Melki R, Kopito R (2010): Prion-like transmission of protein aggregates in neurodegenerative diseases. Nat Rev Mol Cell Biol. Retrieved June 12, 2017, from http://www.nature.com/nrm/journal/v11/n4/abs/nrm2873.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clavaguera F, Hench J, Goedert M, Tolnay M (2015): Invited review: Prion-like transmission and spreading of tau pathology. Neuropathol Appl Neurobiol. 41: 47–58. [DOI] [PubMed] [Google Scholar]
- 66.Stumpf MP, Krakauer DC (2000): Mapping the parameters of prion-induced neuropathology. Proc Natl Acad Sci U S A. 97: 10573–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iturria-Medina Y, Carbonell FM, Sotero RC, Chouinard-Decorte F, Evans AC (2017): Multifactorial causal model of brain (dis)organization and therapeutic intervention: Application to Alzheimer’s disease. Neuroimage. 152: 60–77. [DOI] [PubMed] [Google Scholar]
- 68.Mezias C, LoCastro E, Xia C, Raj A (2017): Connectivity, not region-intrinsic properties, predicts regional vulnerability to progressive tau pathology in mouse models of disease. Acta Neuropathol Commun. 5: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mezias C, Raj A (2017): Analysis of Amyloid-β Pathology Spread in Mouse Models Suggests Spread Is Driven by Spatial Proximity, Not Connectivity. Front Neurol. 8: 653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Acosta D, Powell F, Zhao Y, Raj A (2018): Regional vulnerability in Alzheimer’s: The role of cell-autonomous and transneuronal processes. Alzheimer’s Dement. doi: 10.1016/j.jalz.2017.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Avena-Koenigsberger A, Misic B, Sporns O (2017): Communication dynamics in complex brain networks. Nat Rev Neurosci. 19: 17–33. [DOI] [PubMed] [Google Scholar]
- 72.Mišić B, Betzel RF, Nematzadeh A, Goñi J, Griffa A, Hagmann P, et al. (2015): Cooperative and Competitive Spreading Dynamics on the Human Connectome. Neuron. 86: 1518–1529. [DOI] [PubMed] [Google Scholar]
- 73.Hu C, Hua X, Ying J, Thompson PM, Fakhri GE, Li Q (2016): Localizing Sources of Brain Disease Progression with Network Diffusion Model. IEEE J Sel Top Signal Process. 10: 1214–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Torok J, Maia PD, Powell F, Pandya S, Raj A (2018): A method for inferring regional origins of neurodegeneration. Brain. 141: 863–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pereira JMS, Williams GB, Acosta-Cabronero J, Pengas G, Spillantini MG, Xuereb JH, et al. (2009): Atrophy patterns in histologic vs clinical groupings of frontotemporal lobar degeneration. Neurology. 72: 1653–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rabinovici GD, Jagust WJ (2009): Amyloid imaging in aging and dementia: testing the amyloid hypothesis in vivo. Behav Neurol. 21: 117–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schmidt R, de Reus MA, Scholtens LH, van den Berg LH, van den Heuvel MP (2016): Simulating disease propagation across white matter connectome reveals anatomical substrate for neuropathology staging in amyotrophic lateral sclerosis. Neuroimage. 124: 762–769. [DOI] [PubMed] [Google Scholar]
- 78.Neitzel J, Nuttall R, Sorg C (2018): Perspectives on How Human Simultaneous Multi-Modal Imaging Adds Directionality to Spread Models of Alzheimer’s Disease. Front Neurol. 9: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.DeLisi LE, Sakuma M, Tew W, Kushner M, Hoff AL, Grimson R (1997): Schizophrenia as a chronic active brain process: a study of progressive brain structural change subsequent to the onset of schizophrenia. Psychiatry Res Neuroimaging. 74: 129–140. [DOI] [PubMed] [Google Scholar]
- 80.Thompson PM, Vidal C, Giedd JN, Gochman P, Blumenthal J, Nicolson R, et al. (2001): Mapping adolescent brain change reveals dynamic wave of accelerated gray matter loss in very early-onset schizophrenia. Proc Natl Acad Sci U S A. 98: 11650–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van Haren NEM, Pol HEH, Schnack HG, Cahn W, Brans R, Carati I, et al. (2008): Progressive Brain Volume Loss in Schizophrenia Over the Course of the Illness: Evidence of Maturational Abnormalities in Early Adulthood. Biol Psychiatry. 63: 106–113. [DOI] [PubMed] [Google Scholar]
- 82.Bassett DS, Bullmore E, Verchinski BA, Mattay VS, Weinberger DR, Meyer-Lindenberg A (2008): Hierarchical organization of human cortical networks in health and schizophrenia. J Neurosci. 28: 9239–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gogtay N, Giedd JN, Lusk L, Hayashi KM, Greenstein D, Vaituzis AC, et al. (2004): Dynamic mapping of human cortical development during childhood through early adulthood. Proc Natl Acad Sci U S A. 101: 8174–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bassett DS, Bullmore E, Verchinski BA, Mattay VS, Weinberger DR, Meyer-Lindenberg A (2008): Hierarchical organization of human cortical networks in health and schizophrenia. J Neurosci. 28: 9239–9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hunt MJ, Kopell NJ, Traub RD, Whittington MA (2017): Aberrant Network Activity in Schizophrenia. Trends Neurosci. 40: 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Abdelnour F, Voss HU, Raj A (2014): Network diffusion accurately models the relationship between structural and functional brain connectivity networks. Neuroimage. 90: 335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abdelnour F, Raj A, Dayan M, Devinsky O, Thesen T (2015): Estimating function from structure in epileptics using graph diffusion model. 2015 IEEE 12th Int Symp Biomed Imaging. IEEE, pp 466–469. [Google Scholar]
- 88.Kubota M, Miyata J, Sasamoto A, Sugihara G, Yoshida H, Kawada R, et al. (2013): Thalamocortical disconnection in the orbitofrontal region associated with cortical thinning in schizophrenia. JAMA psychiatry. 70: 12–21. [DOI] [PubMed] [Google Scholar]
- 89.Kubicki M, Alvarado JL, Westin CF, Tate DF, Markant D, Terry DP, et al. (2011): Stochastic tractography study of Inferior Frontal Gyrus anatomical connectivity in schizophrenia. Neuroimage. 55: 1657–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kubicki M, Westin CF, Maier SE, Frumin M, Nestor PG, Salisbury DF, et al. (2002): Uncinate fasciculus findings in schizophrenia: a magnetic resonance diffusion tensor imaging study. Am J Psychiatry. 159: 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Price G, Cercignani M, Parker GJ, Altmann DR, Barnes TR, Barker GJ, et al. (2008): White matter tracts in first-episode psychosis: a DTI tractography study of the uncinate fasciculus. Neuroimage. 39: 949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Szeszko PR, Robinson DG, Ashtari M, Vogel J, Betensky J, Sevy S, et al. (2008): Clinical and neuropsychological correlates of white matter abnormalities in recent onset schizophrenia. Neuropsychopharmacology. 33: 976–984. [DOI] [PubMed] [Google Scholar]
- 93.Wang Q, Su TP, Zhou Y, Chou KH, Chen IY, Jiang T, Lin CP (2012): Anatomical insights into disrupted small-world networks in schizophrenia. Neuroimage. 59: 1085–1093. [DOI] [PubMed] [Google Scholar]
- 94.Fornito A, Zalesky A, Pantelis C, Bullmore ET (2012): Schizophrenia, neuroimaging and connectomics. Neuroimage. 62: 2296–2314. [DOI] [PubMed] [Google Scholar]
- 95.Nelson BG, Bassett DS, Camchong J, Bullmore ET, Lim KO (2017): Comparison of large-scale human brain functional and anatomical networks in schizophrenia. NeuroImage Clin. 15: 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Drakesmith M, Caeyenberghs K, Dutt A, Zammit S, Evans CJ, Reichenberg A, et al. (2015): Schizophrenia-like topological changes in the structural connectome of individuals with subclinical psychotic experiences. Hum Brain Mapp. 36: 2629–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kyriakopoulos M, Frangou S (2009): Recent diffusion tensor imaging findings in early stages of schizophrenia. Curr Opin Psychiatry. 22: 168–176. [DOI] [PubMed] [Google Scholar]
- 98.Karlsgodt KH, van Erp TG, Poldrack RA, Bearden CE, Nuechterlein KH, Cannon TD (2008): Diffusion tensor imaging of the superior longitudinal fasciculus and working memory in recent-onset schizophrenia. Biol Psychiatry. 63: 512–518. [DOI] [PubMed] [Google Scholar]
- 99.Pantelis C, Yücel M, Wood SJ, Velakoulis D, Sun D, Berger G, et al. (2005): Structural brain imaging evidence for multiple pathological processes at different stages of brain development in schizophrenia. Schizophr Bull. 31: 672–696. [DOI] [PubMed] [Google Scholar]
- 100.Powell F, LoCastro E, Acosta D, Ahmed M, O’Donoghue S, Forde N, et al. (2017): Age-Related Changes in Topological Degradation of White Matter Networks and Gene Expression in Chronic Schizophrenia. Brain Connect. 7: 574–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jones DK, Catani M, Pierpaoli C, Reeves SJ, Shergill SS, O’Sullivan M, et al. (2006): Age effects on diffusion tensor magnetic resonance imaging tractography measures of frontal cortex connections in schizophrenia. Hum Brain Mapp. 27: 230–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mori T, Ohnishi T, Hashimoto R, Nemoto K, Moriguchi Y, Noguchi H, et al. (2007): Progressive changes of white matter integrity in schizophrenia revealed by diffusion tensor imaging. Psychiatry Res. 154: 133–145. [DOI] [PubMed] [Google Scholar]
- 103.Friedman JI, Tang C, Carpenter D, Buchsbaum M, Schmeidler J, Flanagan L, et al. (2008): Diffusion Tensor Imaging Findings in First-Episode and Chronic Schizophrenia Patients. Am J Psychiatry. 165: 1024–1032. [DOI] [PubMed] [Google Scholar]
- 104.Cropley VL, Klauser P, Lenroot RK, Bruggemann J, Sundram S, Bousman C, et al. (2017): Accelerated Gray and White Matter Deterioration With Age in Schizophrenia. Am J Psychiatry. 174: 286–295. [DOI] [PubMed] [Google Scholar]
- 105.Schnack HG, van Haren NEM, Nieuwenhuis M, Hulshoff Pol HE, Cahn W, Kahn RS (2016): Accelerated Brain Aging in Schizophrenia: A Longitudinal Pattern Recognition Study. Am J Psychiatry. 173: 607–616. [DOI] [PubMed] [Google Scholar]
- 106.Keller SS, Wieshmann UC, Mackay CE, Denby CE, Webb J, Roberts N (2002): Voxel based morphometry of grey matter abnormalities in patients with medically intractable temporal lobe epilepsy: effects of side of seizure onset and epilepsy duration. J Neurol Neurosurg Psychiatry. 73: 648–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bernasconi N, Andermann F, Arnold DL, Bernasconi A (2003): Entorhinal cortex MRI assessment in temporal, extratemporal, and idiopathic generalized epilepsy. Epilepsia. 44: 1070–4. [DOI] [PubMed] [Google Scholar]
- 108.Bonilha L, Kobayashi E, Rorden C, Cendes F, Li LM (2003): Medial temporal lobe atrophy in patients with refractory temporal lobe epilepsy. J Neurol Neurosurg Psychiatry. 74: 1627–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mueller SG, Laxer KD, Barakos J, Cheong I, Garcia P, Weiner MW (2009): Widespread neocortical abnormalities in temporal lobe epilepsy with and without mesial sclerosis. Neuroimage. 46: 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Raj A, Mueller SG, Young K, Laxer KD, Weiner M (2010): Network-level analysis of cortical thickness of the epileptic brain. Neuroimage. 52: 1302–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bonilha L, Nesland T, Martz GU, Joseph JE, Spampinato MV, Edwards JC, Tabesh A (2012): Medial temporal lobe epilepsy is associated with neuronal fibre loss and paradoxical increase in structural connectivity of limbic structures. J Neurol Neurosurg Psychiatry. 83: 903–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bernhardt BC, Worsley KJ, Kim H, Evans AC, Bernasconi A, Bernasconi N (2009): Longitudinal and cross-sectional analysis of atrophy in pharmacoresistant temporal lobe epilepsy. Neurology. 72: 1747–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Riederer F, Lanzenberger R, Kaya M, Prayer D, Serles W, Baumgartner C (2008): Network atrophy in temporal lobe epilepsy: A voxel-based morphometry study. Neurology. 71: 419–425. [DOI] [PubMed] [Google Scholar]
- 114.Jiang W, Li J, Chen X, Ye W, Zheng J (2017): Disrupted Structural and Functional Networks and Their Correlation with Alertness in Right Temporal Lobe Epilepsy: A Graph Theory Study. Front Neurol. 8: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chiang S, Haneef Z (2014): Graph theory findings in the pathophysiology of temporal lobe epilepsy. Clin Neurophysiol. 125: 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bernhardt BC, Bonilha L, Gross DW (2015): Network analysis for a network disorder: The emerging role of graph theory in the study of epilepsy. Epilepsy Behav. 50: 162–170. [DOI] [PubMed] [Google Scholar]
- 117.Sutula TP, Hagen J, Pitkänen A (2003): Do epileptic seizures damage the brain? Curr Opin Neurol. 16: 189–95. [DOI] [PubMed] [Google Scholar]
- 118.Meldrum BS (1993): Excitotoxicity and selective neuronal loss in epilepsy. Brain Pathol. 3: 405–12. [DOI] [PubMed] [Google Scholar]
- 119.Mehta A, Prabhakar M, Kumar P, Deshmukh R, Sharma PL (2013): Excitotoxicity: bridge to various triggers in neurodegenerative disorders. Eur J Pharmacol. 698: 6–18. [DOI] [PubMed] [Google Scholar]
- 120.Bonilha L, Edwards JC, Kinsman SL, Morgan PS, Fridriksson J, Rorden C, et al. (2010): Extrahippocampal gray matter loss and hippocampal deafferentation in patients with temporal lobe epilepsy. Epilepsia. 51: 519–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Viscomi MT, Molinari M (2014): Remote neurodegeneration: multiple actors for one play. Mol Neurobiol. 50: 368–89. [DOI] [PubMed] [Google Scholar]
- 122.Liu RSN, Lemieux L, Bell GS, Hammers A, Sisodiya SM, Bartlett PA, et al. (2003): Progressive neocortical damage in epilepsy. Ann Neurol. 53: 312–324. [DOI] [PubMed] [Google Scholar]
- 123.Abdelnour F, Mueller S, Raj A (2015): Relating Cortical Atrophy in Temporal Lobe Epilepsy with Graph Diffusion-Based Network Models. PLoS Comput Biol. 11: e1004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Niso G, Carrasco S, Gudín M, Maestú F, Del-Pozo F, Pereda E (2015): What graph theory actually tells us about resting state interictal MEG epileptic activity. NeuroImage Clin. 8: 503–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dyhrfjeld-Johnsen J, Santhakumar V, Morgan RJ, Huerta R, Tsimring L, Soltesz I (2007): Topological determinants of epileptogenesis in large-scale structural and functional models of the dentate gyrus derived from experimental data. J Neurophysiol. 97: 1566–87. [DOI] [PubMed] [Google Scholar]
- 126.Wirsich J, Perry A, Ridley B, Proix T, Golos M, Bénar C, et al. (2016): Whole-brain analytic measures of network communication reveal increased structure-function correlation in right temporal lobe epilepsy. NeuroImage Clin. 11: 707–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.