Summary
Proline-rich antimicrobial peptides (PrAMPs) internalize into susceptible bacteria using specific transporters and interfere with protein synthesis and folding. To date, mammalian PrAMPs have so far only been identified in artiodactyls. Since cetaceans are co-phyletic with artiodactyls, we mined the genome of the bottlenose dolphin Tursiops truncates, leading to the identification of two PrAMPs, Tur1A and Tur1B. Tur1A, which is orthologous to the bovine PrAMP Bac7, is internalized into E. coli without damaging the membranes using the inner membrane transporters SbmA and YjiL/MdM. Furthermore, like Bac7, Tur1A also inhibits bacterial protein synthesis by binding to the ribosome and blocking the transition from the initiation to the elongation phase. By contrast, Tur1B is a poor inhibitor of protein synthesis and may utilize another mechanism of action. An X-ray structure of Tur1A bound within the ribosomal exit tunnel provides a basis to develop these peptides as novel antimicrobial agents.
Keywords: antibiotic, Cathelicidin, Bac7, dolphin, PrAMP, ribosome, proline-rich antimicrobial peptide, Tur1A, Tur1B
Introduction
Most antimicrobial peptides (AMPs), especially α-helical ones, mainly kill bacteria by disrupting the bacterial cell membrane. This mechanism of action makes such AMPs potent broad-spectrum antimicrobials, but with the drawback that they exhibit some toxicity towards host cells (Jenssen et al., 2006). By contrast, proline-rich antimicrobial peptides (PrAMPs) prevalently kill some Gram-negative bacteria without perturbing the cell membrane. PrAMPs utilize bacterial inner membrane proteins to translocate into the cytoplasm where they inhibit intracellular targets, presumably explaining their lower cytotoxicity (Scocchi et al., 2011). Until now, PrAMPs have been identified in some arthropods and mammals, but as evolutionarily unrelated AMPs (Graf et al., 2017; Otvos, 2002; Scocchi et al., 2011). However, within a specific class of animals, it is possible to group the PrAMPs into evolutionarily related families of peptides. In insects, for example, there are the apidaecins, isolated from members of the Apidea and Vespoidea superfamilies (Casteels et al., 1994). Similarly, pyrrochoricin (Cociancich et al., 1994), metalnikowins (Chernysh et al., 1996) and the oncocins (Knappe et al., 2010; Schneider and Dorn, 2001) were identified as members of PrAMPs from the Hemiptera order. In mammals, PrAMPs discovered within different animals from the Artiodactyla order can be grouped together as homologues of the same cathelicidin-derived peptides. For example, the proline-rich Bac5 and Bac7 were isolated from distinct members of the Bovidae family, such as Bos taurus (cow) (Gennaro et al., 1989), Capra hircus (goat) and Ovis aries (sheep) ((Huttner et al., 1998; Shamova et al., 1999), reviewed by (Graf et al., 2017; Scocchi et al., 2011)).
Distinct and unrelated PrAMPs do not necessarily display high sequence similarity, but rather appear to be related to each other by a generally high content of proline and arginine residues that are often arranged in short motifs repeated many times through-out the peptide sequence (e.g. - PPXR- in Bac5 and -PRPX- in Bac7) (Graf et al., 2017; Scocchi et al., 2011). Additionally, most of the PrAMPs characterized to date also display a similar uptake mechanism as well as inhibitory properties on bacterial growth (Graf et al., 2017). The inner-membrane protein SbmA appears to be the principle PrAMP transporter in E. coli (Mattiuzzo et al., 2007), while the MdtM/YjiL multi-drug resistance transporter seems to play an accessory role (Krizsan et al., 2015). Once in the bacterial cytosol, PrAMPs interfere with protein synthesis and folding (Graf et al., 2017; Scocchi et al., 2011). Both mammalian and invertebrate PrAMPs have been shown to inhibit protein synthesis by interacting with the ribosome (Krizsan et al., 2014; Mardirossian et al., 2014). Despite their diverse sequences, all characterized PrAMPs bind to an overlapping site within the ribosomal exit tunnel and inhibit translation by either blocking the transition from initiation to the elongation phase (Gagnon et al., 2016; Roy et al., 2015; Seefeldt et al., 2016; Seefeldt et al., 2015) or preventing the dissociation of the release factors during translation termination (Florin et al., 2017). Although PrAMPs also bind to and inhibit the activity of the bacterial chaperone DnaK (Otvos et al., 2000; Scocchi et al., 2009), this is not sufficient, per se, to kill bacteria (Krizsan et al., 2014; Scocchi et al., 2009). Inhibition of protein folding is therefore not the main mode of action of PrAMPs.
In mammals, all described PrAMPs belong so far to the cathelicidin family (Scocchi et al., 2011), one of the main families of vertebrate host defence peptides and a prime example of diversity among AMPs. Cathelicidins are characterized by the presence of the cathelin-like domain (CLD), a large, conserved pro-region of uncertain function (encoded by the first three exons), and a highly variable AMP located at the C-terminus (encoded by the fourth exon) (Figure 1) (Zanetti, 2005). Following the secretion of the pro-peptide into the extracellular medium or into the phagosomes of neutrophils, the active form of the AMP is produced upon proteolytic cleavage (Tomasinsig and Zanetti, 2005). Most vertebrate animals express only one or few cathelicidins, mostly comprising peptides that adopt helical conformations (Xhindoli et al., 2016). Artiodactyl species are an exception since they express numerous cathelicidins, which are comprised of AMPs with a diverse array of structures (α-helices, disulfide-stabilised β-hairpins or extended peptides rich in particular residues, such as tryptophan or proline) (Tossi et al., 2017). The presence of cathelicidin-derived PrAMPs in artiodactyls (Tossi et al., 2017) suggests that they may also be present in Cetacea since they are co-phyletic within the unique order of Cetartiodactyla (O'Leary and Gatesy, 2008; Spaulding et al., 2009). Here, we mined the available genome sequences of the cetacean Tursiops truncatus (the bottlenose dolphin), as well as physically probing its gDNA for homologues of known cathelicidin PrAMPs. This led to the identification of two previously unknown PrAMPs, which we termed Tur1A and Tur1B. The Tur1A and Tur1B peptides were synthesized and characterized for their antimicrobial activity, mode of entry into the bacterial cell and capacity to inhibit protein synthesis on ribosomes.
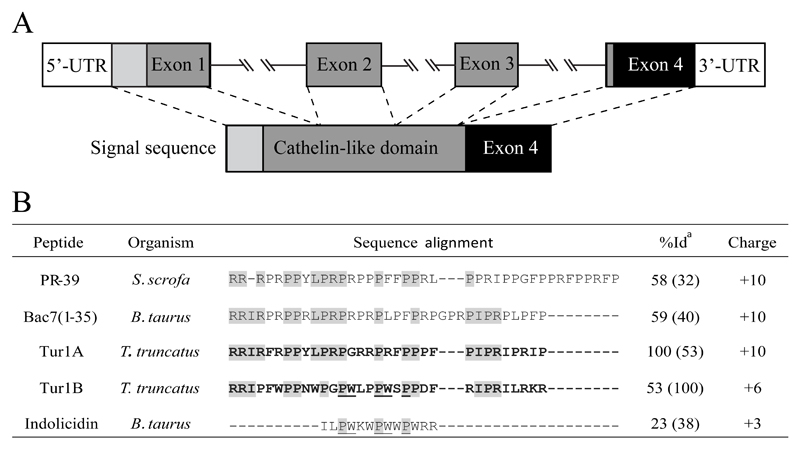
Figure 1. Gene structure and peptide sequences of dolphin PrAMPs Tur1A and Tur1B.
(A) Cathelicidin gene structure with PrAMP encoded in Exon 4. (B) Peptide sequences of dolphin Tur1A and Tur1B compared with orthologues from bovine Bac7(1-35) and porcine PR-39, as well as bovine indolicidin. a %Id indicates identity relative to the Tur1A or Tur1B sequences as determined using Clustal Omega. Conserved residues among PrAMPs are shaded grey. Conserved residues between Tur1B and indolicidin are underlined.
Results
Identification of potential PrAMPs in the bottlenose dolphin
To date, all mammalian PrAMPs identified in artiodactyls belong to the cathelicidin family. Since cetaceans are co-phyletic with artiodactyls, we hypothesized that cathelicidin-related PrAMPs could also exist in cetaceans. To investigate this, we searched the available genome sequence of the cetacean Tursiops truncatus (bottlenose dolphin) using the bovine and pig cathelicidin PrAMP sequences as queries. This led to the identification of a 32 residue proline-rich peptide with an overall charge of +10, which we termed Tur1A (Figure 1B). Analogous with other mammalian cathelicidin, the Tur1A peptide was also encoded within the fourth exon of the gene and preceded by three exons encoding the cathelin-like domain (Figure 1A). The sequence and conserved gene organisation suggested that Tur1A was indeed a bona fide PrAMP, orthologous to the cathelicidin Bac7.
In an attempt to directly validate the sequence, specific primers were used to selectively amplify the 4th exon of the gene from gDNA fragments extracted from Tursiops truncatus tissue. Unexpectedly, when sequenced, the amplified DNA encoded a sequence of what was apparently a paralogous PrAMP, which we termed Tur1B. Tur1B shares 53% identity with Tur1A, but has a lower overall charge (+6) and contains a number of tryptophan (W) residues (Figure 1B). While the EST database confirmed only the expression of Tur1B (GenBank: GT116023), the recently available Sequence Read Archive (SRA) database [bioproject PRJNA313464; (Morey et al., 2016)] validated the presence of both the tur1A and tur1B sequences in Tursiops truncatus. This bioproject provides an RNA-Seq with the hit frequency suggesting that tur1A is more extensively expressed than tur1B. These findings indicate that the bottlenose dolphin contains at least two distinct PrAMPs, Tur1A and Tur1B. Tur1A has high sequence similarity (58-59%) and similar charge with the fully active, 35-residue N-terminal regions of the bovine PrAMP Bac7 and porcine PR-39 (Figure 1B). The homology with PrAMPs such as Bac7 was lower for Tur1B, which instead shows similarity in sequence and charge with the bovine Trp-rich Indolicidin (38% identity based on a 13 residue stretch). Indolicidin is reported to internalize into the bacterial cytoplasm via a self-promoted uptake mechanism (Hsu et al., 2005; Shagaghi et al., 2016), where it is suspected to selectively inhibit DNA synthesis (Ghosh et al., 2014).
Antimicrobial activity of Tur1A and Tur1B
It has been previously shown that SbmA is the major PrAMP transporter, facilitating uptake of Bac7 fragments into the bacterial cytoplasm (Guida et al., 2015; Mattiuzzo et al., 2007), but that at higher concentrations of Bac7, the accessory transporter YjiL/MdtM also contributes to uptake (Krizsan et al., 2015). The antimicrobial activity of Tur1A and Tur1B was therefore assessed against E. coli strains lacking SbmA (ΔSbmA), YjiL (ΔyjiL) or lacking both SbmA and YjiL (ΔSbmAΔyjiL) (Krizsan et al., 2015), and compared with activity against the parental E. coli strain BW25113 (Table 1). Tur1A had a comparable minimal inhibitory concentration (MIC) to Bac7(1-35) against E. coli BW25113 (MICs of 1.2 μM and 2.8 μM, respectively), whereas Tur1B was less active (MIC of 7 μM). Furthermore, Tur1A seemed to be less dependent on the SbmA transporter than Bac7(1-35) since the ΔsbmA mutant showed little change in the MIC, whereas the MIC increased more evidently for Bac7(1-35). Knocking-out the yjiL gene had little effect on the MIC for either peptide, whereas the double ΔsbmA/ΔyjiL deletion mutant exhibited a marked increase (5-fold) in MIC for both Tur1A and Bac7(1-35). The activity of Tur1B, while generally less potent, did not seem to depend on the presence of either transporter. These data suggest that: (i) Tur1A uses an assisted internalization mode similar to other PrAMPs, such as for example Bac7, (ii) Tur1A can use either the SbmA or Yjil/MdtM transporters at concentrations close to the MIC (1-2 μM), whereas Bac7(1-35) seems to use preferentially SbmA at lower concentrations and Yjil/MdtM only at higher concentrations (8 μM), and (iii) Tur1B activity does not seem to depend on PrAMP transporters, suggesting it may have a different mechanism of action and/or entry.
Table 1. Minimal inhibitory concentrations (MIC) for Tur1A, Tur1B, Bac7(1-35) and Tur1A fragments against E. coli BW25113 strains.
| MIC (μM) on E. coli BW25113 strains | ||||
|---|---|---|---|---|
| Peptide | wild type | ΔsbmA | ΔyjiL | ΔsbmA / ΔyjiL |
| Tur1A | 1.2 (±0.4) | 2.3 (±1.3) | 1.9 (±1.1) | 6.4 (±2.2) |
| Tur1B | 7 (±1.9) | 4 | 8 | 8 |
| Bac7(1-35) | 2.8 (±1.0) | 6.3 (±2.6) | 1.8 (±0.5) | 14 (±3.7) |
| Tur1A(1-16) | 4 | nd | nd | nd |
| Tur1A(8-24) | >64 | nd | nd | nd |
| Tur1A(16-32) | >64 | nd | nd | nd |
Data represent the average and standard deviation (±) calculated after three independent experiments performed as internal duplicates (n=6). nd, not determined.
Internalization of Tur1A into E. coli cells
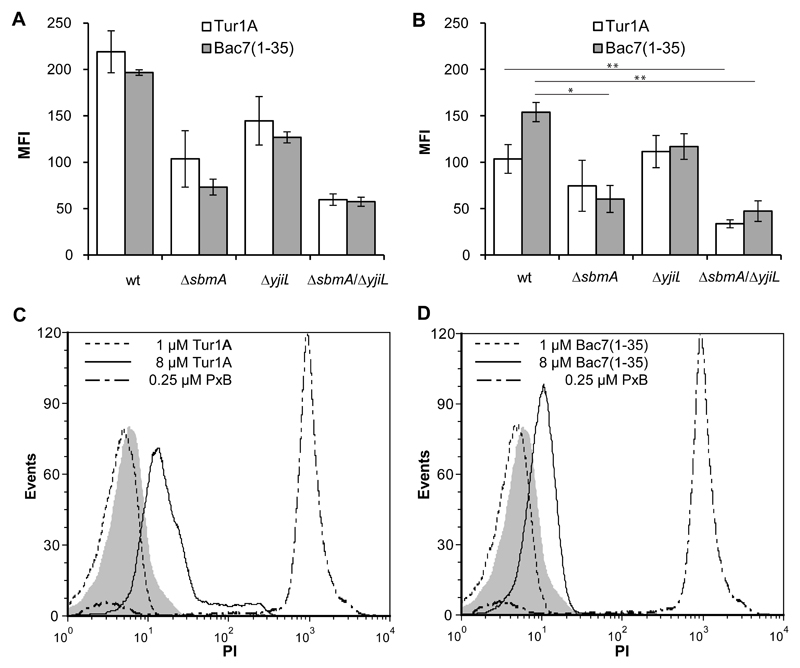
In order to better correlate the uptake of Tur1A with the presence of a specific transport system, internalization of boron-dipyrromethene (BODIPY)-labelled peptides into E. coli cells was evaluated by cytofluorimetric analysis (Figure 2A and B). It should be noted that for these experiments cells were treated with a quite low peptide concentration (0.1 μM). Cells were extensively washed before the total fluorescence was determined (Figure 2A). Because the total fluorescence comprises both tightly surface bound as well as internalized peptide, measurements were also made after washing with trypan blue, which quenches the BOPIDY (BY) fluorescence derived by the surface bound peptide and thus allows the amount of internalized peptide to be evaluated (Figure 2B). In the wildtype E. coli BW25113, the large difference between the Tur1A(Cys33)-BY fluorescence in the absence and presence of the trypan blue (Figure 2A and 2B) suggests a strong surface binding capacity of this peptide (Figure 2A). This probably explains the slower internalization of the peptide in comparison to the Bac7(1-35)(Cys36)-BY (Figure 2B). Nevertheless, the uptake of Tur1A(Cys33)-BY into E. coli BW25113 was efficient, even at very low concentrations. The absence of SbmA or YjiL did not significantly change the Tur1A(Cys33)-BY uptake, consistent with the unchanged MIC values (see Table 1). By contrast, uptake was significantly affected in the ΔsbmA/ΔyjiL double knock-out (Figure 2B) where corresponding increases in the MIC values were also observed (Table 1). Curiously, for both peptides, the presence of the transporters correlates not only with higher levels of internalized fluorescent peptide, but also with the increased amount of surface bound fluorescence. Propidium iodide (PI)-uptake assays carried out in parallel indicated however that no membrane permeabilization occurred under these conditions (not shown).
Figure 2. Flow-cytometry evaluation of Tur1A internalization into E. coli cells and membrane interaction/permeabilization.
The fluorescence intensity (MFI) of wildtype BW25113 (wt) and mutant ΔsbmA, ΔyjiL and ΔsbmA/ΔyjiL E. coli cells exposed to fluorescent derivatives of Tur1A or Bac7(1-35) is shown. Bacterial cells (1×106 CFU/ml) were incubated with 0.1 μM of each peptide for 10 min, extensively washed, and analyzed by flow cytometry (A) without or (B) with incubation with 1 mg/ml of the trypan-blue quencher for 10 min at 37°C. Data are expressed as the average MFI with a standard deviation for three independent experiments. Flow cytometric analysis of propidium iodide-uptake in E. coli BW25113 cells after 15 min treatment with (C) 1 μM and 8 μM Tur1A or (D) 1 μM and 8 μM Bac7(1-35) or 0.25 μM PolymixinB (PxB). The fluorescence of the untreated cells is indicated by the grey histogram. The reported data are representative of at least three independent experiments with comparable results (see Figure S1). PxB was used only once, as a comparison. 104 cells (events) were read for each measurement. Statistical significance: Student-Newman-Keuls Multiple Comparisons Test, ANOVA (*=p < 0.05; **= p < 0.01).
The effect of Tur1A on permeabilization of E. coli BW25113 membranes was characterized by monitoring the PI-uptake after 15 min of incubation with Tur1A (Figure 2C) or Bac7(1-35) (Figure 2D) at concentrations of 1 μM or 8 μM, respectively (i.e. the MIC values in the presence or absence of transporters, as seen in Table 1). The membranolytic antibiotic polymyxin B (PxB) was used at 0.25 μM as positive control for membrane permeabilization (>90% PI-positive cells). The level of membrane permeabilization for both Tur1A and Bac7(1-35) at 1 μM was very low (<3% of damaged cells). Since this is within the range of the MIC, this is consistent with the notion that Tur1A, like Bac7(1-35), inhibits bacteria using an intracellular mechanism rather than via inducing membrane damage. For comparison, almost all cells become permeabilized in the presence of 0.25 μM PxB. However, we note that by increasing the peptide concentration to 8 μM, deleterious effects on membrane integrity were observed for Tur1A, as previously reported for Bac7(1-35) on Salmonella enterica typhimurium (Podda et al., 2006). Specifically, at 8 μM, Bac7(1-35) damaged the membranes of ~50% of cells, whereas Tur1A damaged ~70% cells. By prolonging the incubation time (to 60 min), or by increasing Tur1A concentration (to 16 μM), permeabilization could be increased (but never to 100%), suggesting a time and concentration dependency (data not shown). However, for these longer times it is difficult to distinguish between a primary membranolytic effect and the disruption that occurs following bacterial death. Taken together, these results confirm that the antimicrobial activity of Tur1A against E. coli is principally correlated with its internalization into the cells rather than via membrane lysis, but that nevertheless at higher concentrations or incubation times, Tur1A, like Bac7(1-35), can induce membrane damage.
Inhibition of in vitro protein synthesis using E. coli lysates
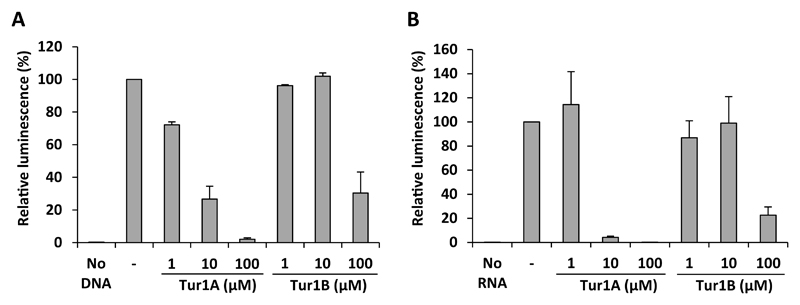
Since the PrAMP Bac7(1-35), which is homologous to the Tur1A and Tur1B peptides, is known to inhibit bacterial protein synthesis (Mardirossian et al., 2014; Seefeldt et al., 2016), we investigated whether the Tur1A and Tur1B peptides also have inhibitory activity in E. coli lysate-based in vitro coupled transcription/translation assays (Figure 3A). The effect of the Tur1A and Tur1B peptides was assessed by monitoring the luminescence resulting from translation of the firefly luciferase reporter gene. As seen in Figure 3A, increasing concentrations of Tur1A efficiently reduced the luminescence in a dose-dependent manner, with an estimated IC50 <5 μM. By contrast, Tur1B was less effective with no influence on the luminescence observed at concentrations up to 10 μM, and a partial (70%) inhibition of protein synthesis observed only at 100 μM. This suggests that Tur1B is a worse translation inhibitor than Tur1A, and that transcription-translation may not be the main target for Tur1B.
Figure 3. Effect of Tur1A and Tur1B on in vitro translation reactions.
(A) Effect of Tur1A and Tur1B on E. coli in vitro coupled transcription/translation assays and (B) on E. coli in vitro translation assay using mRNA template. Assays were performed in the absence (-) or presence of increasing concentrations of peptides (1 μM, 10 μM or 100 μM). The luminescence resulting from reactions performed in the absence of peptide was normalized to 100%. Reactions lacking DNA (No DNA) or RNA (No RNA) template were performed as negative controls. The error bars represent the standard deviation of the mean from three independent experiments.
To distinguish between effects on transcription and translation, we also assessed the direct effect on the bacterial translation machinery (Figure 3B). To do this, the in vitro translation assays were repeated but using transcribed mRNA template encoding the firefly luciferase, rather than DNA template as was used for the coupled assays. As seen in Figure 3B, similar results were obtained when mRNA template replaced the DNA template, namely, that Tur1A was an excellent inhibitor at concentrations over 10 μM and that the inhibition of luminescence was only observed for Tur1B at 100 μM. These findings led us to conclude that Tur1A acts directly on the translation apparatus, rather than on transcription, as was observed previously for Bac7 (Mardirossian et al., 2014), whereas Tur1B only effects translation at high concentrations and is therefore likely to have another target and/or mechanism of action.
Tur1A prevents the transition from initiation to elongation of translation
To understand which step of the protein synthesis was inhibited by Tur1A, toeprinting assays were performed using a fully reconstituted E. coli in vitro translation system (Shimizu et al., 2001), as described previously for other PrAMPs (Gagnon et al., 2016; Seefeldt et al., 2016; Seefeldt et al., 2015). The toeprinting assay uses reverse transcription to monitor the position of ribosomes on a reporter mRNA (Hartz et al., 1988). As shown in Figure 4, in the absence of antibiotic or peptide, ribosomes initiate and translate the mRNA until becoming stalled on three consecutive prolines (PPP) due to the absence of elongation factor EF-P in the system (Starosta et al., 2014). In the presence of the antibiotic thiostrepton (ThS), ribosomes can initiate at the AUG start codon but cannot translate further due to the inhibitory effect of the drug on elongation factors (Wilson, 2009). This is seen in the toeprint gel by an increase in the band corresponding to ribosomes stuck at the AUG codon and the loss of the band corresponding to ribosomes stalled at the PPP-motif. By contrast, the antibiotic edeine (Ede) prevents initiation complex formation by blocking fMet-tRNA binding to the 30S subunit (Dinos et al., 2004) and therefore represents a control for background reverse transcription stops. The reverse transcription stops, presumably due to secondary structure, are observed between the AUG and PPP codons in all reactions (Figure 4). For comparison, the PrAMP Myticalin A5 (MytA5) was also tested in the assay since this peptide inhibits T7 RNA polymerase but does not significantly affect bacterial protein synthesis (Leoni et al., 2017). As expected, increasing concentrations of MytA5 lead to a reduction and, at higher concentrations a complete loss, of all toeprint bands including the full-length mRNA. Like ThS, increasing concentrations of Tur1A led to the decrease in ribosomes stalled at the downstream polyproline sequence (PPP), as well as a concomitant appearance of toeprint signal corresponding to ribosomes stalled at the AUG start codon. At very high concentrations (100 μM), a reduction in the AUG toeprint signal was observed, which may result from drop-off of the fMet-tRNA and disassembly of the ribosome initiation complex (Figure 4), as was observed previously for high concentrations of the PrAMPs Bac7(1-16) and Bac7(1-35) (Seefeldt et al., 2016). In contrast to Tur1A, concentrations up to 100 μM of Tur1B did not lead to a complete loss of the toeprint signal at the PPP-motif, indicating that ribosomes can still elongate in the presence of the Tur1B peptide. However, a slight reduction in the signal intensity of the PPP-band, coupled with the appearance of weak toeprints at the AUG start codon at 10 μM and 100 μM suggest that Tur1B can inhibit translation at higher concentrations, consistent with the results obtained using the in vitro translation assays (Figure 3).
Figure 4. Toe-print analysis of Tur1A and Tur1B effect on translation.
Fluorescence scan of a polyacrylamide gel analysis of toeprinting reactions performed in the absence (-) or presence of 100 μM thiostrepton (ThS), 1-100 μM Tur1A, Tur1B, Myticalin A5 (MytA5) and 50 μM edeine (Ede). Toeprint signals corresponding to ribosomes stalled at the AUG start codon or at the polyproline stretch (PPP) are indicated with arrows, as is the reverse transcription product of the full-length mRNA (FL) and bands resulting from reverse transcription stops at secondary structure (SS). Sequencing lanes (C and A) are included for reference with corresponding nucleotide and peptide sequences for these regions. This gel is representative of three independent experiments (see Figure S2).
Tur1A inhibits protein synthesis on Thermus thermophilus ribosomes
Previous structural studies investigating the ribosome binding site of PrAMPs, such as oncocin and Bac7, utilized ribosome crystals from the thermophilic bacteria T. thermophilus (Gagnon et al., 2016; Roy et al., 2015; Seefeldt et al., 2016; Seefeldt et al., 2015), however, it was not demonstrated that such PrAMPs actually display inhibitory activity against T. thermophilus translation. To address this, we established a T. thermophilus lysate-based in vitro translation assay based on an S12 lysate protocol that has been successfully used in the past for E. coli (Kim et al., 2006) Huter et al., 2017) and Bacillus subtilis (Sohmen et al., 2015). Using the T. thermophilus lysate-based in vitro translation with firefly luciferase as a reporter it was possible to assess whether Tur1A can inhibit translation on T. thermophilus ribosomes. The Tur1B was not further tested since its inhibiting activity towards translation was low (see above Figure 3). As shown in Figure 5, the Tur1A peptide inhibited translation on T. thermophilus ribosomes very efficiently, with 80% inhibition observed even at 1 μM concentration of Tur1A. This indicates that structural studies using T. thermophilus ribosomes represent a valid model system for investigating the binding site of PrAMPs, such as Tur1A, on other bacterial ribosomes.
Figure 5. Effect of Tur1A on in vitro translation assays using T. thermophilus extracts.
Translation reactions were performed in the absence (-) or presence of increasing concentrations of Tur1A. The luminescence resulting from reactions performed in the absence of peptides was normalized to 100%. Reactions lacking RNA (No RNA) template were performed as negative controls. The error bars represent the standard deviation of the mean from three independent experiments.
The binding site of Tur1A on T. thermophilus ribosome
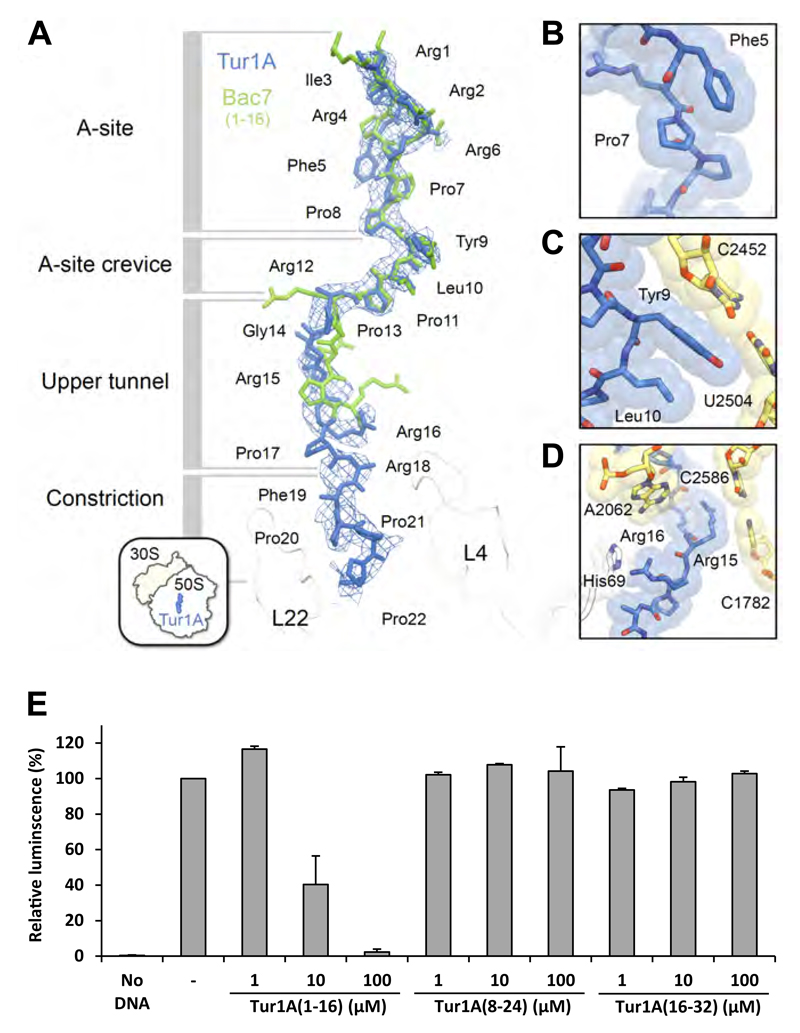
We determined the structure of Tur1A bound to the T. thermophilus 70S ribosome at 3.3 Å resolution from X-ray diffraction data collected using a single co-crystal of a ternary complex between T. thermophilus 70S, Tur1A and YfiA, a protein that is used to lock the head of the 30S subunit in order to improve crystal-to-crystal reproducibility (Table S1) (Polikanov et al., 2014). A minimally biased Fo-Fc difference map calculated after refinement of a model of an empty T. thermophilus 70S ribosome showed clear density for residues 1-16 of Tur1A, with weaker density visible for residues 17-22. This made it possible to build a complete model of Tur1A(1-16) and to trace the backbone for the remainder of the peptide. As in earlier structures of insect and mammalian PrAMPs bound to T. thermophilus 70S (Gagnon et al., 2016; Roy et al., 2015; Seefeldt et al., 2016; Seefeldt et al., 2015), Tur1A(1-16) binds to the exit tunnel in a reversed orientation relative to a nascent polypeptide chain (Figure 6A), blocking the binding site for the A-site tRNA and a significant portion of the ribosomal exit tunnel. Although its structure is very similar to that of the mammalian Bac7(1-16), the side chains of Arg1 and Arg4 of Tur1A showed no noticeable density, suggesting that these residues are more flexible in the case of Tur1A. Due to their strong structural resemblance to Bac7(1-16), the first 16 residues of Tur1A make similar contacts with the ribosome. A few differences are (i) in Tur1A, Phe5 stacks upon Pro7 (Figure 6B), whereas this is not possible in Bac7 because residue 5 is a proline, (ii) in Tur1A, Tyr9 stacks upon the basepair formed by nucleotides C2452 and U2504 of the 23S rRNA that comprise the ribosomal tunnel (Figure 6C). In Bac7, Arg9 makes an analogous stacking interaction with the C2452-U2504 base pair (Gagnon et al., 2016; Seefeldt et al., 2016), (iii) in Tur1A, Arg15 stacks upon basepair C2586-C1782 of the 23S rRNA (Figure 6D), whereas in Bac7, residue 15 is a proline, and (iv) in Tur1A, Arg16 stacks against the side chain of residue His69 of ribosomal protein L4 at the tunnel constriction (Figure 6D), whereas in Bac7, Arg16 stacks against A2062 (Gagnon et al., 2016; Seefeldt et al., 2016). Residues 17-22 of Tur1A extend towards the exit of the nascent polypeptide tunnel, but do not appear to make any specific contacts with ribosomal components, in agreement with their weaker electron density. As with other known PrAMPs, the structural data therefore indicate that Tur1A is likely to inhibit translation by interfering with the binding of aminoacyl-tRNA to the A-site.
Figure 6. Binding site of Tur1A on the ribosome.
(A) The structure of Tur1A(1-22) (blue) is superimposed with that of Bac7(1-16) (green) (Seefeldt et al., 2016) and density contoured at +2.5 σ of a minimally-biased Fo-Fc map shows the location of the Tur1A peptide (blue mesh). The density was trimmed using the carve function in Pymol, with a 3 Å cutoff. The various sections of the nascent polypeptide exit tunnel are labelled, as well as ribosomal proteins L4 and L22 (white). (B-D) detailed view of (B) Phe5 stacking on Pro7 of Tur1A, (C) Tyr9 of Tur1A stacking on the C2452-U2504 basepair of the 23S rRNA, and (D) Arg15 and Arg16 of Tur1A stacking on basepair C2586-C1782 of the 23S rRNA and His69 of L4, respectively. (E) Luciferase activity after in vitro E. coli coupled transcription/translation assays performed in presence of Tur1A(1-16), Tur1A(8-24) or Tur1A(16-32). Reactions were performed in the absence (-) or presence of increasing concentrations of peptides. The luminescence resulting of reactions performed in the absence of peptides was normalized to 100%. Reactions lacking DNA template (No DNA) were performed as negative controls. The error bars represent the standard deviation of the mean from two independent experiments.
The N-terminal region is critical for the inhibitory activity of Tur1A
The structural data suggest that the N-terminal region (residues 1-16) is critical for binding of Tur1A to the ribosome. To evaluate this, we synthesized three overlapping 16-residue Tur1A fragments, Tur1A(1-16), Tur1A(8-24) and Tur1A(16-32), and analysed their activity on living bacteria (Table 1). The MIC assays were performed against E. coli BW25113 and revealed that Tur1A(1-16) retained activity (MIC = 4μM), indicating that the C-terminal region is indeed dispensable for Tur1A antimicrobial activity. By contrast, both the Tur1A(8-24) and Tur1A(16-32) exhibited no antimicrobial activity, confirming that the N-terminal region is critical for the inhibitory activity of Tur1A. These findings are consistent with previous analysis of the related Bac7 orthologue, where the Bac7(1-16) fragment was also identified as being the shortest active fragment (Benincasa et al., 2004). Nevertheless, the absence of antimicrobial activity of the Tur1A(8-24) and Tur1A(16-32) fragments may be due to lack of uptake, rather than inability to bind to ribosomes. To distinguish between these scenarios, the three Tur1A fragments were analysed for their inhibitory activity in an E. coli in vitro coupled transcription/translation assay (Figure 6E). Similar to the MIC results using whole cells, only the Tur1A(1-16) fragment inhibited in vitro protein synthesis to any extent, whereas the Tur1A(8-24) and Tur1A(16-32) fragments were completely inactive, even at concentrations as high as 100 μM (Figure 6E). This indicates that the inactivity of the N-terminally deleted Tur1A peptides is mainly due to their inability to bind to the ribosome. Deletion of the N-terminus removes the RRIR sequence of Tur1A (Figure 1), which is also conserved and essential in the Bac7 PrAMP (Benincasa et al., 2004).
Effect of ribosomal RNA and protein mutations on PrAMP resistance
Tur1A binds the ribosome inside the exit-tunnel, which is also the ribosomal binding site of the antibiotic erythromycin. Previously, it was demonstrated that mutations in the ribosome that confer resistance to erythromycin also confer cross-resistance to PrAMPs, such as oncocin and apidaecin (Gagnon et al., 2016, Florin et al., 2017). Therefore, we determined the MIC of Tur1A against E. coli erythromycin-resistant strains bearing mutations within the ribosomal exit tunnel, namely, A2503C, A2059G or A2059C in the 23S rRNA and alterations in ribosomal proteins L4 (K63E) and L22 (Δ82MRK84) (Table S2). As controls, we also determined the MIC in the presence of Bac7(1-35), the apidaecin derivative Api137 and erythromycin. As expected, all the strains bearing rRNA mutations or ribosomal protein alterations had increased MIC against erythromycin compared to the wildtype strains (Table S2). Ribosomal protein alterations in L4 and L22 led to a modest increase in MIC for Api137, as reported previously (Florin et al., 2017). By contrast, there was no significant change in the MIC in the presence of Bac7(1-35), Tur1A or Tur1B (Table S2). While mutations of A2503 and A2059 have been reported to confer resistance to oncocin derivative Onc112 as well as Api137 (Gagnon et al. 2016; Florin et al., 2017), no resistance was observed with Bac7(1-35) (Gagnon et al., 2016). Curiously, we observed that while A2059C mutations confer resistance to Api137, A2059G did not, whereas the A2503G tested previously (Florin et al., 2017) and the A2503C tested here (Table S2), both conferred resistance to Api137. By contrast, we observed no increase in the MIC in the presence of Tur1A and Tur1B (Table S2), consistent with their high sequence similarity with Bac7 (Figure 1).
Discussion
Here we have identified two PrAMPs from the bottlenose dolphin Tursiops truncatus, which we termed Tur1A and Tur1B. Tur1A is highly similar to the PrAMPs from Bos taurus Bac7 and from Sus scrofa PR-39, sharing 58% and 59% sequence identity, respectively (Figure 1). According to our findings, Tur1A is the only PrAMP that is conserved across the order of Cetartiodactyla. This suggests that the Tur1A/Bac7/PR-39 family could be the most ancient family of mammalian PrAMPs, dating back approximately 60 million years ago, as calculated by TimeTree (Hedges et al., 2015), to before the evolutionary split between dolphins, cows and pigs. Consistent with their high sequence identity, the mechanism of action and uptake of Tur1A is also similar to Bac7 and PR-39. Like Bac7, Tur1A is also taken up by the bacterial cell using the SbmA and YjiL/MdtM transporters and mainly inhibits bacterial growth by targeting the ribosome, rather than via membrane permeabilization. However, unlike Bac7, the isolated role of both these transporter does not affect significantly the uptake of the peptide, indicating that Tur1A exploits the combined effect of these transporters to enter the bacterial cell.
The inhibition of protein synthesis in vitro by Tur1A has been demonstrated not only in E. coli, but also in T. thermophilus lysates, thereby confirming the validity of using T. thermophilus 70S ribosomes for the structural analysis. The binding site of Tur1A overlaps significantly with that observed previously for Bac7 (Gagnon et al., 2016; Seefeldt et al., 2016) (Figure 6), and Tur1A exhibits the same mechanism of action as Bac7 to prevent the transition from the initiation to the elongation phase of translation (Figure 4). However, slight sequence deviations between Tur1A and Bac7 result in subtle differences in the binding mode and interaction with the components of the ribosomal peptide exit tunnel (Figure 6). Such detailed insights will be important for understanding the sensitivity of particular regions of the PrAMPs to sequence variation. Sequence variations are also likely to explain why Tur1A was slightly more permeabilizing towards bacterial membranes than Bac7 (Figure 2). A detailed mutagenesis analysis exchanging residues between Tur1A and Bac7 could be used to identify which sequence determinants are critical for uptake and/or ribosome interaction, and thus provide a structure-activity relationship upon which to base further rational design of synthetic PrAMPs for clinical applications.
Although the partial sequence homology suggests that Tur1B may be evolutionarily related to the Tur1A/Bac7/PR-39 family of PrAMPs, Tur1B is significantly divergent so as to have only a modest inhibitory effect on translation (Figures 3 and 4). Instead, the presence of four tryptophan residues within Tur1B and its lower charge, imparts characteristics on this peptide that are similar to indolicidin, a short bovine proline-tryptophan rich AMP (Selsted et al., 1992). Tur1B almost appears to be intermediate between the proline-arginine rich AMPs and the proline-tryptophan rich AMPs, therefore further studies into Tur1B could provide additional evolutionary insight into this latter group of peptides. Indeed, as observed for indolicidin (Mattiuzzo et al., 2007), the transporter SbmA does not play a significant role in the mode of action of Tur1B. While the details of the mode of action of indolicidin are still under discussion, permeabilization of bacterial membrane appears to be an important part of its moderate antimicrobial activity (Falla et al., 1996). Therefore, we think it is likely that Tur1B also acts mainly on the bacterial membrane. However, further studies will be necessary to address this directly.
In conclusion, our study has extended our knowledge on the distribution among animals of PrAMPs and cathelicidins, providing insight into their evolution. We demonstrate that sequence variations within the cathelicidin family of PrAMPs, as evident by comparing Tur1A with Bac7 and PR-39 sequences, leads to subtle alterations in the binding mode and interaction with the ribosome but retains the potent activity and distinct mechanism of action. In contrast, further sequence variation as observed in the Tur1B peptide leads to a peptide with a completely unrelated properties and mechanism of action. We believe such structure-activity relationships will be critical for development of this class of AMPs as valid lead compounds to combat the ever-increasing emergence of multi-drug resistant bacteria.
Star Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial Strains | ||
| Escherichia coli K12 BW25115 | Keio collection (Baba et al., 2006) | N/A |
| Escherichia coli K12 BW25113ΔsbmA | Keio collection (Baba et al., 2006) | Strain number JW0368-1 |
| Escherichia coli K12 BW25113ΔyjiL | Keio collection (Baba et al., 2006) | Strain number JW5785-1 |
| Escherichia coli K12 BW25113ΔyjiL/ΔsbmA | (Krizsan et al., 2015) | N/A |
| Escherichia coli N281 | (Wittmann et al., 1973) | N/A |
| Escherichia coli N282 | (Wittmann et al., 1973) | N/A |
| Escherichia coli AB301 | (Bouck and Adelberg, 1970) | N/A |
| Escherichia coli SQ110ΔtolC | (Florin et al., 2017) | N/A |
| Escherichia coli SQ110ΔtolC A2059C | (Florin et al., 2017) | N/A |
| Escherichia coli SQ110ΔtolC A2059G | (Florin et al., 2017) | N/A |
| Escherichia coli SQ110ΔtolC A2305C | (Florin et al., 2017) | N/A |
| Thermus thermophilus HB8 | DSMZ | Catalog # DSM-579 |
| BL21 Star | Thermofisher | Catalog # C601003 |
| Biological Samples | ||
| Tursiops truncates muscle tissue | Mediterranean Marine Mammal Tissue Bank | N/A |
| tRNA from Escherichia coli MRE600 | Roche | Catalog # 10109550001 |
| Thermus thermophilus 70S ribosomes | This study | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| YfiA protein | This study | N/A |
| Tur1A | This study | N/A |
| Tur1B | This study | N/A |
| Tur1A(Cys33)-BY | This study | N/A |
| Bac7(Cys36)-BY | This study | N/A |
| Myticalin A5 | (Leoni et al., 2017) | N/A |
| Tur1A(1-16) | This study | N/A |
| Tur1A(8-24) | This study | N/A |
| Tur1A(16-32) | This study | N/A |
| Kanamycin | Sigma | Catalog # 60615 |
| Erythromycin | Sigma | Catalog # E6376 |
| Spectinomycin | Sigma | Catalog # S4014 |
| Thiostrepton | Sigma | Catalog # T8902 |
| Edeine | N/A | N/A |
| Fmoc-Arg(Pbf)-NovaSyn® TGA | Novabiochem | Catalog # 856042 |
| Trifluoroacetic acid (TFA) | Sigma | Catalog # 91700 |
| 3,6-dioxa-1,8-octanedithiol (DODT) | Sigma | Catalog # 465178 |
| Thioanisole | Sigma | Catalog # 88470 |
| Triisopropylsilane (TIPS) | Sigma | Catalog # 233781 |
| N,N'-diisopropylcarbodiimide (DIC) | Fluca | N/A |
| N-methylimidazole | VWR | Catalog # AAA12575-22 |
| Dimethylformamide (DMF) | VWR | Catalog # BDH1117-4LG |
| Piperidine | Acros Organics | Catalog # P/3520 |
| BDP FL maleimide | Lumiprobe | Catalog # 21480 |
| N-Methyl-2-Pyrrolidinone | VWR | Catalog # CA71007-814 |
| 1-hydroxybenzotriazole hydrate (hOBt) | Sigma | Catalog # 54802 |
| Prpidium iodide | Sigma | Catalog # P4170 |
| RNasin | Promega | Catalog # N2511 |
| rNTPs | Sigma | Catalog # 27-2025-01 |
| PEG-8000 | Sigma | Catalog # 1546605 |
| AMV reverse transcriptase | NEB | Catalog # M0277 |
| Hemo Klen Taq | NEB | Catalog # M0332 |
| ddNTPs | Sigma | Catalog # GE27-2045-01 |
| 2-methyl-2,4-pentanediol (MPD) | Sigma | Catalog # 000000008208191000 |
| Critical Commercial Assays | ||
| E.Z.N.A.® Mollusc DNA Kit | Omega Bio-Tek | Catalog # D3373 |
| Perfectprep® Gel Clean-up Kit | Eppendorf | N/A |
| RTS™ 100 Escherichia coli HY | Biotech Rabbit | Catalog # BR1400101 |
| Luciferase Assay System | Promega | Catalog # E1500 |
| PURExpress in vitro transcription/translation kit | NEB | Catalog # E6800S |
| QIAquick Nucleotide Removal Kit | Qiagen | Catalog # 28304 |
| Deposited Data | ||
| The crystal structure of the Tur1A-70S complex | This study | PDB ID 6FKR (https://www.rcsb.org/structure/6fkr) |
| Oligonucleotides | ||
| Tur1A FW 5’-CTTGTGACCCTGGGA-3’ | Eurofins Genomics | N/A |
| Tur1A RV 5’-ATAACTTCCTCCAGGCTTCA-3’ | Eurofins Genomics | N/A |
| NV-1 5’GGTTATAATGAATTTTGCTTATTAAC-3’ | Thermo-Fisher | N/A |
| Recombinant DNA | ||
| H-ns-PPP | This study | N/A |
| pGS21A | (Polikanov et al., 2014) | N/A |
| 2XermCL_S10K | (Arenz et al., 2016) | N/A |
| Software and Algorithms | ||
| Genomics Workbench 5.1 (CLCbio) | QIAGEN bioinformatics | https://www.qiagenbioinformatics.com/products/clc-genomics-workbench/ |
| FCS Express3 software | De Novo Software | https://www.denovosoftware.com/site/DownloadLanding.shtml |
| XDS | (Kabsch, 2010) | http://xds.mpimf-heidelberg.mpg.de/html_doc/downloading.html |
| Phenix | (Adams et al., 2010) | https://www.phenix-online.org/download |
| Coot | (Emsley and Cowtan, 2004) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Other | ||
| Hitachi 3130 Genetic Analyzer | Applied Biosystems | N/A |
| Biotage Initiator+ Alstra | Biotage | Catalog # 356017 |
| Kinetex, C18, 2.6 μm, 100 Å, 50x4,6 mm | Phenomenex | Catalog # 00A-4462-E0 00 |
| Esquire 4000 | Bruker Daltonics | N/A |
| NanoDrop2000 | Thermo Fisher Scientific | Catalog # ND-2000 |
| Jupiter™, C18, 5 μm, 300 Å, 100x10 mm | Phenomenex | Catalog # 00G-4053-E0 |
| MultiPep RSi | Intavis | N/A |
| Shim-pack VP-ODS column (120 Å, 150x4.6 mm,) | Shimadzu | N/A |
| Cytomics FC 500 | Beckman-Coulter | N/A |
| Tecan Infinite M1000 | Tecan | N/A |
| M-110L Microfluidizer Processor | Microfluidics | N/A |
| Typhoon FLA9500 | GE Healthcare | Catalog # 29187191 |
| Protino Ni-NTA agarose beads | Macherey-Nagel | Catalog # 745400 |
| Superdex HiLoad S75 16/600 | GE Healthcare | Catalog # 28989333 |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Daniel N. Wilson (daniel.wilson@chemie.uni-hamburg.de).
Experimental Model and Subject Details
E. coli strain and growth conditions
The E. coli strain BL21STAR was grown at 37°C in Luria-Bertani broth with shaking (200 rpm). All the other E. coli strains were grown at 37°C in Müller-Hinton broth with shaking (140 rpm). The E. coli strains BW25113, N281, N282 and AB301 required no antibiotics. The E. coli strains BW25113ΔsbmA and BW25113ΔyjiL required 50 μg/ml kanamycin, BW25113ΔyjiL/ΔsbmA required 50 μg/ml kanamycin and 15 μg/ml tetracycline. The E. coli strains SQ110ΔtolC, SQ110ΔtolC A2059C, SQ110ΔtolC A2059G, SQ110ΔtolC A2305C required 25 μg/ml kanamycin and 50 μg/ml spectinomycin. The T. thermophilus strain HB8 was grown in 1×YT medium without antibiotics at 70°C.
The E. coli strains BW25113, BW25113ΔsbmA::Kmr and BW25113ΔyjiL::Kmr are part of the KEIO collection (Baba et al., 2006). The double mutant E. coli BW25113ΔsbmA/ΔyjiL::KmrTetr (Krizsan et al., 2015) was generously provided by Prof. Ralf Hoffmann, University of Leipzig, Germany. The E. coli strains AB301 with its mutants N281(mutation on L22) and N282(mutation on L4), the SQ110ΔtolC::Kmr and its mutants SQ110ΔtolC::KmrSprA2059C, SQ110ΔtolC::KmrSprA2059G, SQ110ΔtolC::KmrSprA2305C were generously provided by Profs Alexander Mankin and Nora Vasquez-Laslop, University of Illinois, Chicago, USA.
Methods Details
Peptide identification
The Tursiops truncatus genome sequence is available as part of the Mammalian Genome Project (Lindblad-Toh et al., 2011), however, it was determined by first generation Sanger sequencing and has a relatively low coverage (2.6-fold), so it is only considered partial. Blasting the bovine and porcine PrAMP sequences led to the identification of a possible orthologue in both the Ensemble turTru1 scaffold 36647 and GeneScaffold 2343, which we termed Tur1A. The tur1A gene sequence encoding Tur1A was not present in Genebank, and the low coverage of turTru1 raised the question of the certainty of the identification. Thus, to validate the identification, an attempt was made to selectively amplify and sequence the corresponding gene fragment from gDNA purified from bottlenose dolphin tissue, at the Sequencing Facility of the Applied and Comparative Genomics group (University of Trieste). Frozen muscle tissue samples were obtained from the Mediterranean Marine Mammal Tissue Bank, at the Dept. Veterinary Experimental Sciences, Univ. of Padova, and DNA was extracted using the E.Z.N.A.® Mollusc DNA Kit (Omega Bio-Tek), following the provided protocol. Amplification primers were designed using Genomics Workbench 5.1 (CLCbio), based on the DNA sequence 5’ and 3’ to the 4th exon encoding Tur1A. The forward primer, 5’-CTTGTGACCCTGGGA-3’, was based on a sequence in the intron preceding the fourth exon, the reverse primer 5’-ATAACTTCCTCCAGGCTTCA-3’ was based on a sequence from the 3’-UTR. The PCR product was purified on a 2% agarose gel showing a major band of the expected size (470 bp), which was excised and extracted using the Perfectprep® Gel Clean-up kit (Eppendorf). Sanger sequencing was carried out on a Hitachi 3130 Genetic Analyzer (Applied Biosystems) using the same forward and reverse primers, but resulted in a different sequence than expected and was therefore named tur1B, encoding the Tur1B peptide. Since the direct sequencing had not confirmed the presence of tur1A, suggesting instead the presence of a paralogous sequence, the EST database was blasted with both sequences to determine if they were expressed. The EST database confirmed the expression of Tur1B only (GenBank: GT116023). On the other hand, the more recent Sequence Read Archive database bioproject PRJNA313464 (Morey et al., 2016) confirmed the presence of both the tur1A and tur1B sequences in Tursiops truncatus.
Peptide synthesis
Tur1A, Tur1B, the orthologous bovine peptide fragment Bac7(1-35) and the mussel peptide Myticalin A5 were synthesised on a Biotage Initiator+ Alstra automated microwave synthesizer using Fmoc chemistry. All peptides were synthesized on a 0.1 mmole scale using Fmoc-Pro-TGA or Fmoc-Arg(Pbf)-TGA resins (Novabiochem/Merck) as appropriate, and single couplings with a 6-fold amino acid excess at 75°C. Peptides were cleaved with a version of Reagent K (85% TFA, 6% DODT, 3% thioanisole, 2% phenol, 2% triisopropylsilane, 2% water), subsequently purified by preparative RP-HPLC (Phenomenex Kinetex, C18, 2.6 μm, 100 Å, 50x4,6 mm) using a 15-45% H2O to CH3CN gradient (0.05% trifluoroacetate (TFA)) and confirmed by ESI-MS (Bruker Esquire 4000) [Tur1A calculated MW=3972.9, measured MW=3972.3; Tur1B calculated MW=4032.8, measured MW=4033.2; Bac7(1-35) calculated MW=4201.2, measured MW=4207.1; MW calculated with Peptide Companion, Coshi Soft]). Tur1A and Tur1B stock solutions were prepared in deionized water from purified peptides that had been lyophilized three times from a 10 mM HCl solution to remove TFA. The concentrations were estimated by using a Nanodrop 2000 based on the extinction coefficients at 214 nm as described by (Kuipers and Gruppen, 2007), and based on the absorption of Tyr at 280 nm for Tur1A (ε = 1450 M-1cm-1) or of Trp at 280 nm for Tur1B (ε = 5500 M-1cm-1).
Fluorescently labelled Tur1A(Cys33)-BY and Bac7(1-35)(Cys36)-BY were prepared in the same manner, but adding a C-terminal Cys residue to each sequence by using Fmoc-Cys(Trt)-2-chlorotrityl chloride resin (Novabiochem/Merck). After cleavage, peptides were reacted with BODIPY-FL [N-(2-aminoethyl)maleimide] (1 eq. peptide/10 eq. dye) in 30% CH3CN, 10 mM sodium phosphate buffer at pH 7.4, as described previously (Mattiuzzo et al., 2007). The labelled peptides were purified by reverse-phase high performance liquid chromatography (RP-HPLC) on a Phenomenex semi-preparative column (Jupiter™, C18, 5 μm, 300 Å, 100x10 mm) with a linear gradient from 10-30% of CH3CN in 40 min, and the correct sequence verified by ESI-MS [Tur1A(Cys33)-BY calculated MW=4489.8, measured MW=4489.9; Bac7(1-35)(Cys36)-BY calculated MW=4310.3, measured MW=4310.7]. After lyophilisation from 10 mM HCl, the concentration of labelled peptide stock solution was determined by spectrophotometric determination of BODIPY (ε504 = 79000 M-1 cm-1 in MeOH) (Invitrogen Molecular Probes Handbook, section 2.2).
Tur1A fragments were synthesized by automated solid-phase peptide synthesis (SPPS) on a Whatman 50 cellulose membrane using a MultiPep RSI peptide synthesizer/pipetting robot (Intavis), the manual synthesis protocol was described in (Hilpert et al., 2007). Briefly, before robot synthesis, functionalisation of the cellulose membrane (10 cm x 15 cm) was carried out by overnight incubation in 0.2 M Fmoc-Gly-OH (Aldrich), 0.24 M N,N'-diisopropylcarbodiimide (DIC, Fluka) and 0.4 M N-methylimidazole (NMI, Aldrich) in dimethylformamide (DMF, VWR). Functionalisation was followed by Glycine deprotection in 20% piperidine (v/v, Acros Organics) in DMF (20 min + 10 min). Peptide synthesis at discrete spots addressed by the robot was performed using 9-fluorenyl-methoxycarbonyl/tert-butyl (Fmoc/tBu) strategy. Fmoc amino acids [Bachem, 0.5 M stock solutions in N-methyl-2-pyrrolidone (NMP, VWR)] were pre-activated with equimolar quantities of 1-hydroxybenzotriazole hydrate (HOBt, Aldrich) and DIC (both 1.1 M stock solutions in NMP) and assembled in double coupling procedure (2x10 min) per cycle to ensure higher coupling efficiency at each amino acid position. Spotting volumes of 0.8 μl for the first cycle and 0.9 μl for the following cycles were used. After amino acid coupling cycle, unreacted residues were capped applying a 5 min acetic anhydride treatment (5% v/v in DMF, Fluka). Subsequent Fmoc cleavage was achieved using 20% (v/v) piperidine in DMF (2x 5 min). Final cleavage of amino acid side-chain protecting groups was carried out with 25 ml of 90% TFA (Acros Organics), 3% triisopropylsilane (TIPS, Acros Organics) and 2% water in dichloromethane (DCM, Acros Organics) for 30 min followed by a 120 min treatment with 25 ml of 50% TFA, 3% TIPS and 2% water in DCM. Peptide amides were cleaved from the solid support by incubating the membrane in a saturated ammonia gas atmosphere overnight. An internally standardised control peptide, and individually chosen peptides from that synthesis, were used to determine SPOT synthesis yield and quality. Individual SPOTs were punched-out with a one-hole-puncher, transferred into a sterile 96-well round-bottomed polypropylene non-treated microtiter plate and dissolved overnight in 200 μl of autoclaved water. Peptide solution were quantified with a NanoDrop ND1000 spectrophotometry at 280 nm. Dissolved peptides were further analysed by analytical RP-HPLC on a Shim-pack VP-ODS column (120 Å, 150x4.6 mm, Shimadzu) using a LC2010AHT system (Shimadzu). The binary solvent system contained 0.1% (v/v) TFA in H2O (HPLC-grade, VWR, solvent A) and 0.1% (v/v) TFA in acetonitrile (HPLC-grade, VWR, solvent B). A linear gradient of 5% to 70% solv B in 32.5 min with an initial 3 min isocratic equilibration was used at a flow rate of 1 ml/min. The purity of the crude control peptides were between 37% and 68%. The remaining spots were then excised from the membrane, placed in microtiter plates and the cleaved peptides resuspended overnight in 200 μl sterile water, lyophilised and re-lyophilized from 200 μl 10 mM HCl solution to remove TFA. The final stock solutions, obtained by resuspending the pellets in 50 μl of sterile water, were quantified spectrophotometrically using a Nanodrop 2000 as described above.
Minimum inhibitory concentration determination
Bacterial cultures were grown overnight to the stationary phase, diluted in fresh Mueller-Hinton broth (MHB) and incubated to an OD600 ≈ 0.3 at 37°C under agitation (if required, in the presence of antibiotic) and diluted to 5 × 105 colony forming units (CFU)/ml in medium. Antimicrobial agents, diluted in MHB to a concentration of 128 μM, were added to the first wells of a round-bottom microtiter plate and then serially two-fold diluted with MHB into successive wells in a final volume of 50 μl. Subsequently, 50 μl of the bacterial suspension was added to each well, to a final load of 2.5 × 104 CFU/well and reducing by half the antibacterial compound concentration in each well. The plate was sealed to minimise evaporation and incubated overnight at 37°C. The MIC was calculated as the lowest compound concentration inhibiting visible bacterial growth.
Flow cytometry
The integrity of bacterial cell membranes was assessed by measuring the Propidium iodide (PI) uptake via flow cytometric assays, performed with a Cytomics FC 500 (Beckman-Coulter), acquiring 104 bacterial cells for each measurement. as described previously (Benincasa et al., 2009; Guida et al., 2015). Briefly, mid-log phase bacterial cultures, diluted to 1 × 106 CFU/ml in MHB, were incubated at 37°C for different times with increasing concentrations of peptides, in the presence of propidium iodide (PI) at a final concentration of 10 μg/ml. Membrane damage was assessed in terms of the % of PI positive cells. For evaluation of peptide uptake, bacterial cultures in mid-log phase were diluted to 1 × 106 CFU/ml in MHB and incubated at 37°C for different times with different concentrations of BOPIDY (BY) fluorescently marked peptides [Tur1A(Cys)33-BY or Bac7(1-35)(Cys)36-BY] and analysed as described previously (Benincasa et al., 2009; Guida et al., 2015)). Analyses were carried out after extensive washing only, or after washing and addition of Trypan Blue (TB) quenching the fluorescence of the peptide bound to the bacterial surface (Benincasa et al., 2009). Data analysis was performed with the FCS Express3 software (De Novo Software). Data are expressed as the mean ± standard deviation (S.D.).
In vitro transcription/translation and translation in E. coli
For in vitro transcription/translation assays, the RTS™ 100 Escherichia coli HY (Biotech Rabbit) was used for testing all samples. 0.1 μl of RNase inhibitor (RNasin®, 20-40 U/μl, Promega) and 1 μl of peptide solution were added to 5 μl of RTS reaction mix (containing either mRNA or DNA template encoding the Photinus pyralis luciferase protein), with a final peptide concentration of 1 μM, 10 μM or 100 μM as required. After incubation for 1 h at 30°C with shaking (750 rpm), 2 μl of each reaction were mixed with 8 μl kanamycin (50 mg/ml) to stop the process and 40 μl of Luciferase assay substrate solution (Promega) and then transferred into a white 96-well, flat bottom microtiter plate (Greiner). In positive controls, nuclease-free water was added instead of peptide solution. Negative controls contained nuclease-free water instead of both the peptide solution and mRNA/DNA template. The activity of the reporter protein was assessed and quantified using a Tecan Infinite M1000 plate reader. Relative values were calculated as a percentage of the positive control. The in vitro translation assay was performed using the PURExpress System (NEB). Reactions were performed according to the manual at 37°C for up to half an hour. Luminescence was determined as described above for the in vitro transcription/translation assay
In vitro translation on Thermus thermophilus ribosomes
The in vitro translation on T. thermophilus ribosomes was performed using an S12 lysate, prepared based on protocol described for E. coli (Kim et al., 2006) Huter et al., 2017) with minor variations. T. thermophilus strain HB8 (DSM-579) were grown in 1 x YT medium to an OD600 ≈ 0.6 at 70°C, centrifuged at 5000 x g at 4°C for 15’ and washed three times with Buffer A (10 mM Tris-acetate buffer (pH 8.2), 14 mM MgOAc, 60 mM KOAc, 1 mM DTT, 6 mM β-mercaptoethanol). The cell pellet was flash-frozen in liquid nitrogen and stored at -80°C until use. The pellet was thawed, resuspended in Buffer A and cells were lysed by three passages through an M-110L Microfluidizer Processor (Microfluidics) at >15000 psi. Cell debris was removed by centrifuging at 12000 x g for 10 min at 4°C. The cleared supernatant was aliquoted, flash frozen in liquid nitrogen and stored at -80°C. To perform the in vitro translation, 6.75 μL of cell lysate and 1 μL of peptide solution (to a final concentration of 1 μM, 10 μM or 100 μM) were added to a reaction mixture consisting of 240 mM HEPES-KOH, pH 8.0, 0.6 mM PEG8000, 60 mM glucose, 4.4 μg tRNA mix, 1.2 mM ATP, 1.2 mM GTP, 0.85 μg folinic acid, 1 mM DTT, 90 mM potassium glutamate; 80 mM ammonium acetate, 20 mM K2HPO4, 1.8 mM of each amino acid, 12.6 mM magnesium acetate and 200 ng of custom made firefly luciferase mRNA, to a final volume of 25 μL. Samples were incubated at 30°C (due to heat sensitivity of the reporter Fluc protein) for 1 h with shaking (550 rpm) and 8 μL from each sample were then mixed with 2 μL kanamycin (50 mg/ml) to block further translation, and 40 μL of Luciferase assay substrate (Promega) in the wells of a white 96-well flat bottom microtiter plate (Greiner). The luminescence was measured using a Tecan Infinite M1000 plate reader. Relative values were determined with respect to positive control in the absence of peptide and defined as 100%.
Toe-printing assay
For toe-printing assays, the PURExpress in vitro transcription/translation kit (NEB) was used with an H-ns-PPP template, comprising the N-terminal 1-36 nucleotides of E. coli hns (Uniprot-P0ACF8) gene but modified such that residues 20-22 are replaced by three proline residues (5’-ATTAATTACGACTCACTATAGGGATATAAGGAGGAAAACATATGAGCGAAGCACTTAA AATTCTGAACAACCTGCGTACTCTTCGTGCGCAGGCAATTCCGCCGCCGCTTGAAACG CTGGAAGAAATGCTGGAAAAATTAGAAGTTGTTGTTTAAGTGATAGAATTCTATCGTTAATAAGCAAAATTCATTATAACC-3’, start-, PRO3- and stop-codons respectively are in underlined bold). Reactions were set up by mixing 2 μL Solution A, 1.5 μL Solution B, 1 μL (0.5 pmol) of H-ns-PPP template 0.1 μL of RNasin® (20-40 U/μl, Promega), 1 μL of peptide (to a final concentration of 1 μM, 10 μM or 100 μM) or 1 μL of antibiotic (to a final concentration of 100 μM for thiostrepton or 50 μM for edeine) in PCR tubes. The control contained only nuclease-free water instead of peptide or antibiotic. Samples were incubated for 15 min at 37°C for transcription/translation, under agitation (550 rpm), cooled on ice for 5 min and then equilibrated at RT for 2 min. 1 μL (2 pmol) of Alexa647-labelled NV-1 toe-print primer (5’-GGTTATAATGAATTTTGCTTATTAAC-3’) was then added to each reaction and samples were incubated for 5 min at 37°C. For the reverse transcription, 0.5 μl of AMV reverse transcriptase (NEB), 0.1 μl dNTP mix (10 mM) and 0.4 μl Pure System Buffer were added to each reaction and samples were incubated for 20 min at 37°C. To stop the reaction and degrade RNA, 1 μl of 5M NaOH was added to each reaction and samples were incubated for 15 min at 37°C. After neutralizing with 0.7 μl HCl 25% (v/v) and 20 μl of toe-print resuspension buffer, samples were purified using the QIAquick Nucleotide Removal Kit (Qiagen), adding to samples 200 μl of PN1 buffer and following the supplier’s instructions. DNA was eluted using 80 μl of RNase-free water, dried in a vacuum centrifuge and re-suspended in 4 μl of formamide-loading dye. Samples were heated for 5 min at 95°C, then separated by electrophoresis on a 6% polyacrylamide gel (19:1 acrylamide: bisacrylamide) containing 7 M urea, at 2000 V. Gels were scanned using a Typhoon FLA9500 imaging system (GE Healthcare). Sequencing was carried out by mixing 1 μL (0.5-0.8 pmol) of DNA template, 5 μL of Sequencing buffer, 9 μL of nuclease-free water, 1 μL (10 pmol) of Alexa647-labelled NV-1 toe-print primer and 1 μL of Hemo Klen Taq polymerase. To 4 μL aliquots of this mix were then respectively added 2 μL of ddATP, ddTTP, ddGTP or ddCTP, and the mixtures incubated in a thermocycler [2 sec 95°C, 30 x (30 sec 95°C, 30 sec 42°C, 1 min 70°C), 1 min 70°C, 8°C storage]. Samples were then heated and loaded on a gel as indicated for the toe-print assay.
Purification of T. thermophilus 70S ribosomes
T. thermophilus 70S ribosomes were purified as described previously (Selmer et al., 2006) and resuspended in buffer containing 5 mM HEPES-KOH, pH 7.5, 50 mM KCl, 10 mM NH4Cl, and 10 mM Mg(CH3COO)2 to yield a final concentration of 26-32 mg/mL. For storage, T. thermophilus 70S ribosomes were flash frozen in liquid nitrogen and kept at -80 °C.
Purification of YfiA
YfiA was expressed from a pGS21A expression vector in BL21 Star RARE cells as described previously (Polikanov et al., 2014). The culture was grown at 37°C (220 rpm) to an OD600 of 0.6 and induced with 1 mM IPTG for 4 hours. Cells were harvested and lysed with a French press three times at 15000 psi in 20 mM Tris-HCl pH 7.6, 100 mM NH4Cl, 10 mM MgCl2 and 1 mM β-mercaptoethanol. The lysate was centrifuged at 50000 x g for 1 hour and the YfiA protein was purified on a Ni-NTA matrix using a linear gradient from 0-1 M imidazole in 20 mM Tris-HCl pH 7.6, 100 mM NH4Cl, 10 mM MgCl2 and 1 mM β-mercaptoethanol. Following size exclusion chromatography using a Superdex 75 column (GE healthcare) in 20 mM Tris-HCl pH 7.6, 100 mM NH4Cl, 10 mM MgCl2, 1 mM β-mercaptoethanol, YfiA was concentrated to 87 mg/mL and flash frozen in liquid nitrogen.
T. thermophilus 70S-YfiA-Tur1A Complex formation
A ternary complex was formed by first incubating a mixture of 5 μM T. thermophilus 70S ribosomes and 50 μM YfiA at 37°C for 10 min, followed by the addition of 50 μM Tur1A and further incubation at room temperature for at least 15 min. The complex was then centrifuged briefly before use for crystallization. The final sample buffer prior to crystallization contained 5 mM HEPES-KOH, pH 7.6, 50 mM KCl, 10 mM NH4Cl and 10 mM Mg(CH3COO)2.
Crystallization of the T. thermophilus 70S-YfiA-Tur1A complex
Published conditions were used as a starting point for screening crystallization conditions by vapor diffusion in sitting-drop trays at 20°C (Polikanov et al., 2014; Selmer et al., 2006). Crystallization drops consisted of 3 μl of ternary T. thermophilus 70S-YfiA-Tur1A complex and 3-4 μl of reservoir solution containing 100 mM Tris-HCl, pH 7.6, 2.9% (v/v) PEG 20,000, 7-10% (v/v) 2-methyl-2,4-pentanediol (MPD) and 175 mM L-arginine. Crystals appeared within 2-3 days and grew to approx. 1000 × 100 × 100 μm within 7-8 days. For cryoprotection, the concentration of MPD was increased in a stepwise manner to yield a final concentration of 40% (v/v). The ionic composition during cryoprotection was 100 mM Tris-HCl, pH 7.6, 2.9% (v/v) PEG 20000, 50 mM KCl, 10 mM NH4Cl and 10 mM Mg(CH3COO)2. Back soaking of the peptide was prevented by including 50 μM Tur1A in the final cryoprotection solution. Following overnight incubation at 20°C, crystals were flash frozen in a nitrogen cryostream at 90 K for subsequent data collection.
Data collection and processing
Diffraction data were collected at beamline ID23-1 of the European Synchrotron Radiation Facility (ESRF) in Grenoble, France. A complete dataset was obtained by merging 0.1° oscillation data collected at 100 K with a wavelength of 0.97625 Å from multiple regions of the same crystal. Initial data processing, including integration and scaling, was performed with XDS (Kabsch, 2010). The data collected could be indexed in the P212121 space group, with unit-cell dimensions around 210 Å × 450 Å × 625 Å and an asymmetric unit containing two copies of the T. thermophilus 70S ribosome.
Model building and refinement
A high-resolution structure of the apo T. thermophilus 70S ribosome crystallized in the same space group and similar unit cell as the crystal used in this study (PDB 4Y4O) was used directly for rigid body refinement in Phenix (Adams et al., 2010). Rigid bodies comprised four domains from the small 30S subunit (head, body, spur and helix h44) and three domains from the large 50S subunit (body, L1 stalk and the N-terminus of ribosomal protein L9). Restrained crystallographic refinement consisting of multiple cycles of positional and individual B-factor refinement was then carried out using the Phenix package. Non-crystallographic symmetry restraints between the two copies of the T. thermophilus 70S ribosome in the asymmetric unit were also applied during refinement. After confirming that density corresponding to the Tur1A peptide was visible inside the exit tunnel in a minimally biased Fo−Fc map, a model of Tur1A was built manually in Coot (Emsley and Cowtan, 2004). The sidechains of Arg1 and Arg4 of Tur1A showed no noticeable density in the original Fo-Fc map or in the 2Fo-Fc map obtained after complete refinement of the structure and therefore were not included in the final model. Further refinement and model validation were carried out in Phenix and on the MolProbity server (Chen et al., 2010), respectively.
Quantification and Statistical Analysis
In vitro data analysis
Data are presented as mean values +/- the standard deviation (SD) calculated from independent experiments using the software Excel (Microsoft). The number of experimental and technical replicates for each experiment is also described in each individual figure legend. The statistical significance has been calculated using the Student-Newman-Keuls Multiple Comparisons Test, ANOVA (*=p < 0.05; **= p < 0.01), with the software GraphPad Instat 3. For flow-cytometry experiments, data analysis was performed with the FCS Express3 software (De Novo Software).
Supplementary Material
Significance.
The discovery of the two proline-rich antimicrobial peptides (PrAMPs) Tur1A and Tur1B in dolphins indicates that these peptides also exist amongst cetaceans. Previously, evidence of PrAMPs was reported only in some terrestrial mammals. Tur1A was shown to kill bacteria by inhibiting protein synthesis, specifically preventing the transition from initiation to elongation. This reiterates the mode of action described for other PrAMPs, such as Bac7 and oncocin, previously identified in cows and insects. This also provides information on the evolutionary convergence of Tur1A with some insect PrAMPs, and on its evolutionary relationship with some bovine PrAMPs. By contrast, Tur1B displays a low inhibitory effect on protein synthesis. Instead, Tur1B appears to be an intermediate between a ribosome-targeting PrAMP and other proline-rich peptides that act mainly by permeabilizing the bacterial membrane, thus offering interesting hints on the relationship between these two groups of PrAMPs. Lastly, both the Tur peptides, but especially Tur1A, have a potent antibacterial activity to be exploited in the fight against the increasing prevalence of antibiotic-resistant pathogens.
Acknowledgments
M.M. acknowledges the “Talents3” fellowship program, from the Operative Regional Programme of the European Social Fund 2014-2020 of the Autonomous Region of Friuli Venezia Giulia. C.A.I has received funding for this project from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 724040). Data were collected at the European Synchrotron Radiation Facility (ESRF) and we thank the staff at beamline ID-23 for their support during data collection. The research in D.N.W. group is supported by grants from the Deutsche Forschungs gemeinschaft (DFG). A.T. acknowledges the program Finanziamento di Ateneo per la Ricerca Scientifica 2017 (FRA 2017) of the University of Trieste. S.H. acknowledges his fellowship from the William Harvey International Translational Research Academy (WHRI-ACADEMY). We thank the Applied and Comparative Genomics group at the Department of Life Sciences, University of Trieste, for helping to obtain the Tur1B sequence K.H. acknowledges the start-up grant from St. Georges University of London.
Footnotes
Data and Software Availability
Accession numbers
The atomic coordinates for the Tur1A-70S complex have been deposited in the PDB with the accession number 6FKR.
Authors Contribution
M.M., N.P., M.B., S.G., S.H., P.H., C.M. performed the experiments. M.M., D.N.W., N.P., C.A.I., M.B., S.G., A.T. designed the experiments and analysed data. M.M., D.N.W., A.T., K.H. wrote the paper. D.N.W. supervised the whole project.
Declaration of Interest
Kai Hilpert is also founder and director of TiKa diagnostics Ltd and Sven Hofmann is an employee of this company.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benincasa M, Pacor S, Gennaro R, Scocchi M. Rapid and reliable detection of antimicrobial peptide penetration into gram-negative bacteria based on fluorescence quenching. Antimicrob Agents Chemother. 2009;53:3501–3504. doi: 10.1128/AAC.01620-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benincasa M, Scocchi M, Podda E, Skerlavaj B, Dolzani L, Gennaro R. Antimicrobial activity of Bac7 fragments against drug-resistant clinical isolates. Peptides. 2004;25:2055–2061. doi: 10.1016/j.peptides.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Casteels P, Romagnolo J, Castle M, Casteels-Josson K, Erdjument-Bromage H, Tempst P. Biodiversity of apidaecin-type peptide antibiotics. Prospects of manipulating the antibacterial spectrum and combating acquired resistance. J Biol Chem. 1994;269:26107–26115. [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernysh S, Cociancich S, Briand JP, Hetru C, Bulet P. The inducible antibacterial peptides of the hemipteran insect Palomena prasina: Identification of a unique family of proline-rich peptides and of a novel insect defensin. J Insect Physiol. 1996;42:81–89. [Google Scholar]
- Cociancich S, Dupont A, Hegy G, Lanot R, Holder F, Hetru C, Hoffmann JA, Bulet P. Novel inducible antibacterial peptides from a hemipteran insect, the sap-sucking bug Pyrrhocoris apterus. Biochem J. 1994;300(Pt 2):567–575. doi: 10.1042/bj3000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinos G, Wilson DN, Teraoka Y, Szaflarski W, Fucini P, Kalpaxis D, Nierhaus KH. Dissecting the ribosomal inhibition mechanisms of edeine and pactamycin: the universally conserved residues G693 and C795 regulate P-site RNA binding. Mol Cell. 2004;13:113–124. doi: 10.1016/s1097-2765(04)00002-4. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Falla TJ, Karunaratne DN, Hancock REW. Mode of action of the antimicrobial peptide indolicidin. Journal of Biological Chemistry. 1996;271:19298–19303. doi: 10.1074/jbc.271.32.19298. [DOI] [PubMed] [Google Scholar]
- Florin T, Maracci C, Graf M, Karki P, Klepacki D, Berninghausen O, Beckmann R, Vazquez-Laslop N, Wilson DN, Rodnina MV, et al. An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome. Nat Struct Mol Biol. 2017 doi: 10.1038/nsmb.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon MG, Roy RN, Lomakin IB, Florin T, Mankin AS, Steitz TA. Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition. Nucleic Acids Res. 2016;44:2439–2450. doi: 10.1093/nar/gkw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennaro R, Skerlavaj B, Romeo D. Purification, composition, and activity of two bactenecins, antibacterial peptides of bovine neutrophils. Infect Immun. 1989;57:3142–3146. doi: 10.1128/iai.57.10.3142-3146.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Kar RK, Jana J, Saha A, Jana B, Krishnamoorthy J, Kumar D, Ghosh S, Chatterjee S, Bhunia A. Indolicidin targets duplex DNA: structural and mechanistic insight through a combination of spectroscopy and microscopy. ChemMedChem. 2014;9:2052–2058. doi: 10.1002/cmdc.201402215. [DOI] [PubMed] [Google Scholar]
- Graf M, Mardirossian M, Nguyen F, Seefeldt AC, Guichard G, Scocchi M, Innis CA, Wilson DN. Proline-rich antimicrobial peptides targeting protein synthesis. Nat Prod Rep. 2017 doi: 10.1039/c7np00020k. [DOI] [PubMed] [Google Scholar]
- Guida F, Benincasa M, Zahariev S, Scocchi M, Berti F, Gennaro R, Tossi A. Effect of size and N-terminal residue characteristics on bacterial cell penetration and antibacterial activity of the proline-rich peptide Bac7. J Med Chem. 2015;58:1195–1204. doi: 10.1021/jm501367p. [DOI] [PubMed] [Google Scholar]
- Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. Tree of Life Reveals Clock-Like Speciation and Diversification. Mol Biol Evol. 2015;32:835–845. doi: 10.1093/molbev/msv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilpert K, Winkler DF, Hancock RE. Peptide arrays on cellulose support: SPOT synthesis, a time and cost efficient method for synthesis of large numbers of peptides in a parallel and addressable fashion. Nat Protoc. 2007;2:1333–1349. doi: 10.1038/nprot.2007.160. [DOI] [PubMed] [Google Scholar]
- Hsu CH, Chen C, Jou ML, Lee AY, Lin YC, Yu YP, Huang WT, Wu SH. Structural and DNA-binding studies on the bovine antimicrobial peptide, indolicidin: evidence for multiple conformations involved in binding to membranes and DNA. Nucleic Acids Res. 2005;33:4053–4064. doi: 10.1093/nar/gki725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttner KM, Lambeth MR, Burkin HR, Burkin DJ, Broad TE. Localization and genomic organization of sheep antimicrobial peptide genes. Gene. 1998;206:85–91. doi: 10.1016/s0378-1119(97)00569-6. [DOI] [PubMed] [Google Scholar]
- Jenssen H, Hamill P, Hancock RE. Peptide antimicrobial agents. Clin Microbiol Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Keum JW, Oh IS, Choi CY, Park CG, Kim DM. Simple procedures for the construction of a robust and cost-effective cell-free protein synthesis system. J Biotechnol. 2006;126:554–561. doi: 10.1016/j.jbiotec.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Knappe D, Piantavigna S, Hansen A, Mechler A, Binas A, Nolte O, Martin LL, Hoffmann R. Oncocin (VDKPPYLPRPRPPRRIYNR-NH2): a novel antibacterial peptide optimized against gram-negative human pathogens. J Med Chem. 2010;53:5240–5247. doi: 10.1021/jm100378b. [DOI] [PubMed] [Google Scholar]
- Krizsan A, Knappe D, Hoffmann R. Influence of the yjiL-mdtM Gene Cluster on the Antibacterial Activity of Proline-Rich Antimicrobial Peptides Overcoming Escherichia coli Resistance Induced by the Missing SbmA Transporter System. Antimicrob Agents Chemother. 2015;59:5992–5998. doi: 10.1128/AAC.01307-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizsan A, Volke D, Weinert S, Strater N, Knappe D, Hoffmann R. Insect-derived proline-rich antimicrobial peptides kill bacteria by inhibiting bacterial protein translation at the 70S ribosome. Angew Chem Int Ed Engl. 2014;53:12236–12239. doi: 10.1002/anie.201407145. [DOI] [PubMed] [Google Scholar]
- Kuipers BJH, Gruppen H. Prediction of molar extinction coefficients of proteins and peptides using UV absorption of the constituent amino acids at 214 nm to enable quantitative reverse phase high-performance liquid chromatography-mass spectrometry analysis. J Agr Food Chem. 2007;55:5445–5451. doi: 10.1021/jf070337l. [DOI] [PubMed] [Google Scholar]
- Leoni G, De Poli A, Mardirossian M, Gambato S, Florian F, Venier P, Wilson DN, Tossi A, Pallavicini A, Gerdol M. Myticalins: A Novel Multigenic Family of Linear, Cationic Antimicrobial Peptides from Marine Mussels (Mytilus spp.) Mar Drugs. 2017;15 doi: 10.3390/md15080261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardirossian M, Grzela R, Giglione C, Meinnel T, Gennaro R, Mergaert P, Scocchi M. The host antimicrobial peptide Bac71-35 binds to bacterial ribosomal proteins and inhibits protein synthesis. Chem Biol. 2014;21:1639–1647. doi: 10.1016/j.chembiol.2014.10.009. [DOI] [PubMed] [Google Scholar]
- Mattiuzzo M, Bandiera A, Gennaro R, Benincasa M, Pacor S, Antcheva N, Scocchi M. Role of the Escherichia coli SbmA in the antimicrobial activity of proline-rich peptides. Mol Microbiol. 2007;66:151–163. doi: 10.1111/j.1365-2958.2007.05903.x. [DOI] [PubMed] [Google Scholar]
- Morey JS, Neely MG, Lunardi D, Anderson PE, Schwacke LH, Campbell M, Van Dolah FM. RNA-Seq analysis of seasonal and individual variation in blood transcriptomes of healthy managed bottlenose dolphins. BMC Genomics. 2016;17:720. doi: 10.1186/s12864-016-3020-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary MA, Gatesy J. Impact of increased character sampling on the phylogeny of Cetartiodactyla (mammalia): combined analysis including fossils. Cladistics. 2008;24:397–442. doi: 10.1111/j.1096-0031.2007.00187.x. [DOI] [PubMed] [Google Scholar]
- Otvos L., Jr The short proline-rich antibacterial peptide family. Cell Mol Life Sci. 2002;59:1138–1150. doi: 10.1007/s00018-002-8493-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otvos L, Jr, O I, Rogers ME, Consolvo PJ, Condie BA, Lovas S, Bulet P, Blaszczyk-Thurin M. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry. 2000;39:14150–14159. doi: 10.1021/bi0012843. [DOI] [PubMed] [Google Scholar]
- Podda E, Benincasa M, Pacor S, Micali F, Mattiuzzo M, Gennaro R, Scocchi M. Dual mode of action of Bac7, a proline-rich antibacterial peptide. Biochim Biophys Acta. 2006;1760:1732–1740. doi: 10.1016/j.bbagen.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Polikanov YS, Steitz TA, Innis CA. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat Struct Mol Biol. 2014;21:787–793. doi: 10.1038/nsmb.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy RN, Lomakin IB, Gagnon MG, Steitz TA. The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin. Nat Struct Mol Biol. 2015;22:466–469. doi: 10.1038/nsmb.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Dorn A. Differential infectivity of two Pseudomonas species and the immune response in the milkweed bug, Oncopeltus fasciatus (Insecta: Hemiptera) J Invertebr Pathol. 2001;78:135–140. doi: 10.1006/jipa.2001.5054. [DOI] [PubMed] [Google Scholar]
- Scocchi M, Lüthy C, Decarli P, Mignogna G, Christen P, Gennaro R. The Proline-rich Antibacterial Peptide Bac7 Binds to and Inhibits in vitro the Molecular Chaperone DnaK. International Journal of Peptide Research and Therapeutics. 2009;15:147–155. [Google Scholar]
- Scocchi M, Tossi A, Gennaro R. Proline-rich antimicrobial peptides: converging to a non-lytic mechanism of action. Cell Mol Life Sci. 2011;68:2317–2330. doi: 10.1007/s00018-011-0721-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seefeldt AC, Graf M, Perebaskine N, Nguyen F, Arenz S, Mardirossian M, Scocchi M, Wilson DN, Innis CA. Structure of the mammalian antimicrobial peptide Bac7(1-16) bound within the exit tunnel of a bacterial ribosome. Nucleic Acids Res. 2016;44:2429–2438. doi: 10.1093/nar/gkv1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seefeldt AC, Nguyen F, Antunes S, Perebaskine N, Graf M, Arenz S, Inampudi KK, Douat C, Guichard G, Wilson DN, et al. The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex. Nat Struct Mol Biol. 2015;22:470–475. doi: 10.1038/nsmb.3034. [DOI] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Selsted ME, Novotny MJ, Morris WL, Tang YQ, Smith W, Cullor JS. Indolicidin, a Novel Bactericidal Tridecapeptide Amide from Neutrophils. Journal of Biological Chemistry. 1992;267:4292–4295. [PubMed] [Google Scholar]
- Shagaghi N, Palombo EA, Clayton AH, Bhave M. Archetypal tryptophan-rich antimicrobial peptides: properties and applications. World J Microbiol Biotechnol. 2016;32:31. doi: 10.1007/s11274-015-1986-z. [DOI] [PubMed] [Google Scholar]
- Shamova O, Brogden KA, Zhao C, Nguyen T, Kokryakov VN, Lehrer RI. Purification and properties of proline-rich antimicrobial peptides from sheep and goat leukocytes. Infect Immun. 1999;67:4106–4111. doi: 10.1128/iai.67.8.4106-4111.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- Sohmen D, Chiba S, Shimokawa-Chiba N, Innis CA, Berninghausen O, Beckmann R, Ito K, Wilson DN. Structure of the Bacillus subtilis 70S ribosome reveals the basis for species-specific stalling. Nat Commun. 2015;6 doi: 10.1038/ncomms7941. 6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaulding M, O'Leary MA, Gatesy J. Relationships of Cetacea (Artiodactyla) among mammals: increased taxon sampling alters interpretations of key fossils and character evolution. PLoS One. 2009;4:e7062. doi: 10.1371/journal.pone.0007062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starosta AL, Lassak J, Peil L, Atkinson GC, Virumae K, Tenson T, Remme J, Jung K, Wilson DN. Translational stalling at polyproline stretches is modulated by the sequence context upstream of the stall site. Nucleic Acids Research. 2014;42:10711–10719. doi: 10.1093/nar/gku768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasinsig L, Zanetti M. The cathelicidins--structure, function and evolution. Curr Protein Pept Sci. 2005;6:23–34. doi: 10.2174/1389203053027520. [DOI] [PubMed] [Google Scholar]
- Tossi A, D'este F, Skerlavaj B, Gennaro R. Antimicrobial Peptides: Discovery, Design and Novel Therapeutic Strategies. CABI; 2017. Structural and Functional Diversity of Cathelicidins. [Google Scholar]
- Wilson DN. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol. 2009;44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- Xhindoli D, Pacor S, Benincasa M, Scocchi M, Gennaro R, Tossi A. The human cathelicidin LL-37--A pore-forming antibacterial peptide and host-cell modulator. Biochim Biophys Acta. 2016;1858:546–566. doi: 10.1016/j.bbamem.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Zanetti M. The role of cathelicidins in the innate host defenses of mammals. Curr Issues Mol Biol. 2005;7:179–196. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.