Mallik et al. identify Xrp1 as a nuclear chromatin-binding protein involved in gene expression regulation that mediates phenotypes induced by loss of function of cabeza (caz), the Drosophila melanogaster orthologue of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) protein FUS. Knockdown of Xrp1 in motor neurons rescues phenotypes induced by ALS-mutant FUS.
Abstract
Cabeza (caz) is the single Drosophila melanogaster orthologue of the human FET proteins FUS, TAF15, and EWSR1, which have been implicated in amyotrophic lateral sclerosis (ALS) and frontotemporal dementia. In this study, we identified Xrp1, a nuclear chromatin-binding protein, as a key modifier of caz mutant phenotypes. Xrp1 expression was strongly up-regulated in caz mutants, and Xrp1 heterozygosity rescued their motor defects and life span. Interestingly, selective neuronal Xrp1 knockdown was sufficient to rescue, and neuronal Xrp1 overexpression phenocopied caz mutant phenotypes. The caz/Xrp1 genetic interaction depended on the functionality of the AT-hook DNA-binding domain in Xrp1, and the majority of Xrp1-interacting proteins are involved in gene expression regulation. Consistently, caz mutants displayed gene expression dysregulation, which was mitigated by Xrp1 heterozygosity. Finally, Xrp1 knockdown substantially rescued the motor deficits and life span of flies expressing ALS mutant FUS in motor neurons, implicating gene expression dysregulation in ALS-FUS pathogenesis.
Introduction
Amyotrophic lateral sclerosis (ALS) is an adult-onset neurodegenerative disorder characterized by motor neuron loss, leading to progressive muscle weakness and ultimately complete paralysis and death (Taylor et al., 2016). Mutations in several genes encoding RNA-binding proteins (RBPs) cause familial ALS (FALS), including TDP-43 (Gitcho et al., 2008; Kabashi et al., 2008; Sreedharan et al., 2008), FUS (Kwiatkowski et al., 2009; Vance et al., 2009), TAF15 (Couthouis et al., 2011), EWSR1 (Couthouis et al., 2012), hnRNPA1 and hnRNPA2B1 (Kim et al., 2013b), and matrin-3 (Johnson et al., 2014). Furthermore, TDP-43–positive inclusions are found in most sporadic ALS patients (Neumann et al., 2006; Taylor et al., 2016), and inclusions containing either TDP-43 or FUS are a pathological hallmark in ∼45% and ∼10% of patients with frontotemporal dementia (FTD), respectively (Ling et al., 2013). These findings implicated defects in RNA biogenesis in ALS and FTD pathogenesis.
Of the ALS-associated RBPs, FUS, EWSR1, and TAF15 (FET) proteins are highly homologous proteins that constitute the FET family (Schwartz et al., 2015). The FET proteins are DNA-binding proteins and RBPs involved in gene expression regulation, including transcription, mRNA splicing, and mRNA subcellular localization (Schwartz et al., 2015). Heterozygous mutations in FUS account for ∼5% of FALS (Ling et al., 2013), while mutations in TAF15 and EWSR1 are rare (Couthouis et al., 2011, 2012). Most ALS-associated mutations cluster in the nuclear localization signal of FUS, resulting in a shift from a predominantly nuclear to a more cytoplasmic localization, formation of cytoplasmic aggregates, and reduced nuclear FUS levels (Da Cruz and Cleveland, 2011). This suggests that loss of nuclear FUS function may contribute to ALS pathogenesis, although evidence from ALS-FUS mouse models indicates that ALS-FUS mutations also result in a novel toxic function that triggers motor neuron degeneration (Scekic-Zahirovic et al., 2016, 2017; Sharma et al., 2016). Moreover, in FTD with FUS pathology (FTLD-FUS), the three FET proteins are found in pathogenic inclusions, with reduced levels or complete loss of nuclear FET proteins in inclusion-bearing cells, indicating that loss of nuclear FET function may contribute to FTLD-FUS (Neumann et al., 2011; Davidson et al., 2013).
The Drosophila melanogaster gene cabeza (caz) encodes the single fly orthologue of the three human FET proteins. Accordingly, Caz is a predominantly nuclear RBP that contains the functional domains of the human FET proteins (Schwartz et al., 2015). When expressed in mammalian cells, Caz elicits down-regulation of FUS protein levels (Immanuel et al., 1995), while FUS expression in Drosophila rescues caz mutant phenotypes (Wang et al., 2011), indicating functional homology. We previously generated caz mutant animals, which exhibit pupal lethality because adult flies fail to eclose due to motor deficits (Frickenhaus et al., 2015). In this study, we performed a genetic screen to gain insight into the molecular mechanisms underlying caz mutant phenotypes. Exhaustive screening of ∼80% of the Drosophila genome identified Xrp1 as the only gene for which heterozygosity could rescue caz mutant phenotypes. Xrp1 encodes a protein containing an AT-hook DNA-binding domain often found in proteins involved in chromatin remodeling, transcriptional regulation, and DNA repair (Reeves, 2010). Xrp1 expression was increased in caz mutants, and neuron-selective knockdown of Xrp1 was sufficient to rescue caz mutant phenotypes. Importantly, the DNA-binding capacity of the AT-hook domain of Xrp1 was required to mediate caz mutant phenotypes, and caz mutants displayed substantial gene expression dysregulation, which was significantly mitigated by heterozygosity for Xrp1. Finally, Xrp1 knockdown in motor neurons rescued phenotypes induced by ALS mutant FUS expression, underscoring the potential relevance of our findings for human disease. Together, we propose that caz mutant phenotypes are mediated by up-regulation of Xrp1, leading to gene expression dysregulation and neuronal dysfunction.
Results
A genetic screen to identify suppressors of caz mutant phenotypes
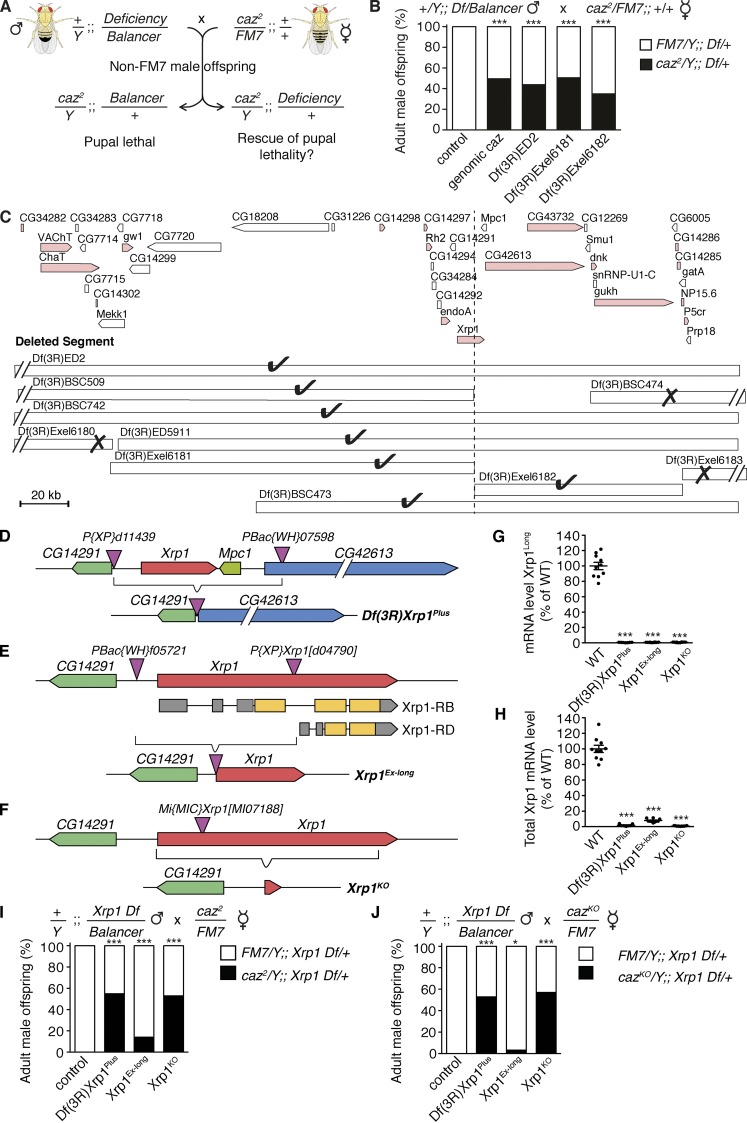
We previously generated two independent caz null alleles: (1) caz2, an imprecise excision allele, and (2) cazKO, generated by homologous recombination (Frickenhaus et al., 2015). Caz mutants die during the pupal stage due to motor incapability resulting in pharate adults failing to eclose from the pupal case. This phenotype was used to perform a dominant suppressor screen whereby males carrying chromosomal deficiencies were crossed to caz2 heterozygous females. Since caz is on the X chromosome, this approach allowed us to screen for genes on the second and third chromosomes for which hemizygosity would rescue the pupal lethality of caz2 males (Fig. 1 A). This screen yielded only a single deficiency that rescued caz2 pupal lethality, Df(3R)ED2 (Fig. 1 B).
Figure 1. .
Heterozygosity for Xrp1 rescues caz mutant pupal lethality. (A) Screening strategy to identify chromosomal deficiencies that rescue caz2 pupal lethality. (B) Frequency of adult male offspring from the indicated cross that is heterozygous for a genomic caz transgene or the indicated deficiencies. n > 128 per genotype. ***, P < 0.0001; χ2 test. (C) Genomic region uncovered by Df(3R)ED2. Pink indicates genes in the plus orientation; white indicates genes in the minus orientation. The different smaller deficiencies within this region which were tested for rescue of caz2 pupal lethality are shown. Check marks indicate deficiencies that rescue; X marks indicate deficiencies that do not rescue. (D–F) Xrp1 genomic locus showing the insertion sites of the transposable elements used to generate Xrp1 mutant alleles. In the Df(3R)Xrp1Plus allele (D), Xrp1, Mpc1, and the 5′ end of CG42613 are deleted. In the Xrp1Ex-long allele (E), the 5′ half of Xrp1 is deleted, predicted to abolish expression of the Xrp1Long isoform. The Xrp1Short isoform, encoded by Xrp1-RD, may still be expressed. In the Xrp1KO allele (F), the Xrp1 coding region is precisely deleted. (G and H) Xrp1 transcript levels in Xrp1 mutant lines relative to WT controls (100%) determined by qPCR using primers either selectively detecting Xrp1Long transcripts (G) or detecting all Xrp1 transcripts (H). n = 10. ***, P < 0.0001; one-way ANOVA. Mean ± SEM. (I and J) Frequency of adult male offspring from the indicated crosses that is heterozygous for the indicated Xrp1 allele. n > 87 per genotype. *, P < 0.05; ***, P < 0.0001; χ2 test.
Fine mapping using smaller overlapping deficiencies reduced the number of candidate genes to 11 (Fig. 1 C). As Df(3R)Exel6181 and Df(3R)Exel6182 are neighboring but nonoverlapping deficiencies that have a common break point in Xrp1, the fact that heterozygosity for either of these deficiencies rescued caz2 pupal lethality suggested that heterozygous loss of Xrp1 may mediate the rescue (Fig. 1, B and C).
Heterozygosity for Xrp1 rescues caz mutant pupal lethality
Xrp1 is predicted to encode seven alternative transcripts (Fig. S1 A), four of which can be translated into a 668-aa “long” isoform (Xrp1Long) and the remaining three into a 406-aa “short” isoform (Xrp1Short). We generated two Xrp1 deletion alleles: Df(3R)Xrp1Plus is a ∼25-kb deletion of Xrp1, Mpc1, and part of CG42613 (Fig. 1 D), whereas Xrp1Ex-long selectively deletes two thirds of Xrp1, expected to abolish expression of all Xrp1Long transcripts, but possibly leaving expression of a Xrp1Short transcript (Xrp1-RD) intact (Fig. 1 E). In addition, we used in vivo homologous recombination to generate the Xrp1KO allele, in which the entire Xrp1 coding region is precisely deleted (Figs. 1 F and S1 C).
All three Xrp1 mutant alleles were homozygous viable, and quantitative PCR (qPCR) using primers that selectively detect Xrp1Long revealed loss of Xrp1Long transcript in homozygous Df(3R)Xrp1Plus, Xrp1Ex-long, and Xrp1KO animals (Fig. 1 G). qPCR using primers detecting all Xrp1 isoforms revealed loss of Xrp1 transcript in Df(3R)Xrp1Plus and Xrp1KO flies, whereas in Xrp1Ex-long flies, some residual Xrp1 transcript could be detected (∼8% of WT levels), presumably reflecting expression of the short Xrp1-RD mRNA isoform (Fig. 1 H).
Crossing males heterozygous for either Df(3R)Xrp1Plus, Xrp1Ex-long, or Xrp1KO to caz2/FM7 or cazKO/FM7 females revealed that heterozygosity for Df(3R)Xrp1Plus or Xrp1KO rescued caz mutant pupal lethality to a similar extent as Xrp1 deficiencies and a genomic caz transgene, independent of the caz null allele used (Fig. 1, I and J). Heterozygosity for Xrp1Ex-long only partially rescued, presumably due to residual Xrp1 expression (Fig. 1, I and J). Furthermore, the mild rough eye phenotype previously reported in caz mutant adult escaper flies (Wang et al., 2011) was rescued by Xrp1 heterozygosity. While a genetic interaction was previously reported between caz and TBPH, the Drosophila homologue of TARDBP, encoding TDP-43 (Wang et al., 2011), Xrp1 heterozygosity could not rescue the adult eclosion defect of TBPH mutants (Fig. S2). Thus, loss of 50% of Xrp1 gene dosage is sufficient to rescue pupal lethality induced by loss of caz but not TBPH function.
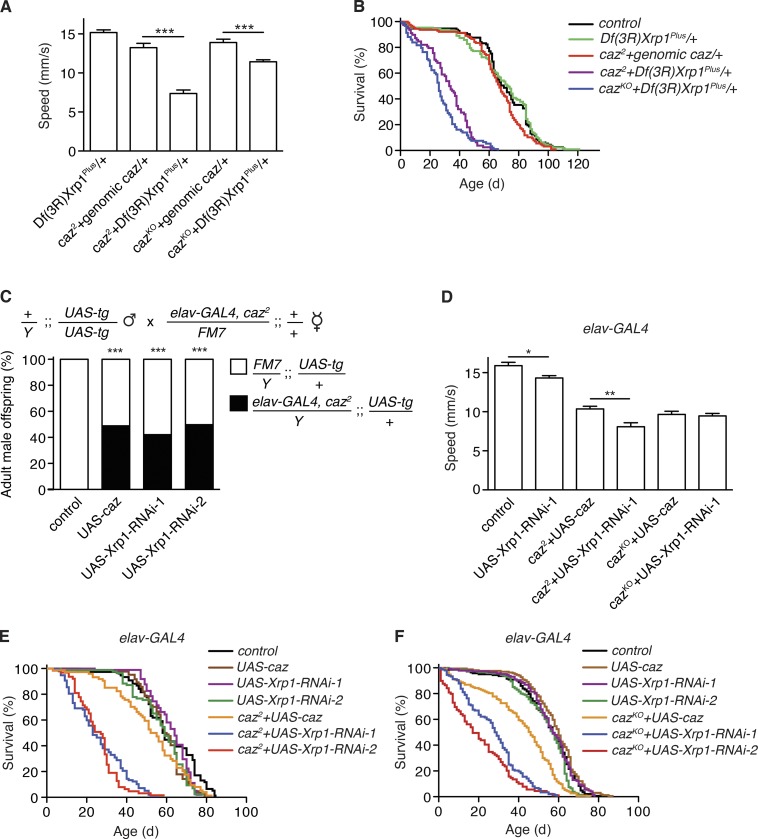
Xrp1 heterozygosity rescues motor deficits and life span of caz mutant flies
As caz mutant flies display motor defects (Wang et al., 2011; Frickenhaus et al., 2015), we evaluated motor performance of caz mutant Xrp1 heterozygous flies using an automated negative geotaxis climbing assay (Niehues et al., 2015). While we failed to obtain caz mutant adult escapers, Xrp1 heterozygous caz mutant flies managed to climb the wall of a test vial, although their climbing speed was still significantly reduced as compared with control animals (Fig. 2 A). Analysis of larval locomotion revealed that caz mutant third instar larvae display a significantly reduced crawling speed, which was fully rescued by heterozygosity for Xrp1 (Fig. S3 A).
Figure 2. .
Rescue of caz mutant phenotypes by Xrp1 heterozygosity or by selective knockdown of Xrp1 in neurons. (A) Average climbing speed in an automated negative geotaxis assay of heterozygous Df(3R)Xrp1Plus male flies (control) and caz mutant males rescued by genomic caz or heterozygosity for Xrp1. n > 100 per genotype. ***, P < 0.0005; Mann-Whitney test. Mean ± SEM. (B) Life span of WT (control), Df(3R)Xrp1Plus heterozygous, and caz mutant male flies rescued by genomic caz or heterozygosity for Xrp1. n = 78–127 per genotype. (C) Frequency of adult male offspring from the indicated cross that carries UAS-caz, UAS-Xrp1-RNAi, or no UAS transgene (control). n > 176 per genotype. ***, P < 0.0001; χ2 test. (D) Average climbing speed of adult male control (driver-only) flies, flies with neuronal (elav-GAL4) Xrp1 knockdown, and caz mutants rescued by neuronal Caz or neuronal Xrp1 knockdown. n > 100 per genotype. *, P < 0.05; **, P < 0.01; Mann-Whitney test. Mean ± SEM. (E and F) Life span of male flies selectively expressing caz or Xrp1-RNAi in neurons either in a WT, caz2 (E), or cazKO (F) background. n = 76–164 per genotype.
Evaluation of life span revealed that Xrp1 heterozygosity resulted in a substantial rescue of life span as compared with pupal lethality of caz2 and cazKO flies, although the life span of rescued caz mutants was still shorter than control flies (Fig. 2 B). Thus, heterozygosity for Xrp1 partially but substantially rescues caz mutant motor performance and life span.
Selective knockdown of Xrp1 in neurons rescues caz mutant phenotypes
Selective reintroduction of Caz in neurons was shown to rescue caz mutant phenotypes (Wang et al., 2011; Frickenhaus et al., 2015), while selective inactivation of caz in neurons resulted in severe motor deficits and reduced life span (Frickenhaus et al., 2015), indicating that loss of caz function in neurons is both necessary and sufficient to induce caz mutant phenotypes. We therefore evaluated whether selective knockdown of Xrp1 in neurons was sufficient to rescue caz mutant phenotypes despite the fact that the FlyAtlas and modENCODE databases report Xrp1 expression in all tissues throughout development and adult life. Two independent transgenic Xrp1-RNAi lines revealed that selective knockdown of Xrp1 in neurons (elav-GAL4) fully rescued caz mutant pupal lethality (Fig. 2 C) and partially rescued caz mutant motor performance for cazKO even to a similar extent as neuronal Caz reintroduction (Fig. 2 D). Importantly, selective knockdown of Xrp1 in motor neurons (D42-GAL4) was sufficient to fully rescue the reduced crawling speed of caz mutant larvae (Fig. S3 B), indicating that the larval locomotion deficit is attributable to dysfunction of motor neurons. Finally, the median life span of caz mutant flies with neuronal Xrp1 knockdown ranged from 30% to 50% of the life span of their respective controls (Fig. 2, E and F), comparable with the life span of Xrp1 heterozygous caz mutants. To evaluate the level of knockdown induced by the two Xrp1-RNAi lines, qPCR revealed residual Xrp1 transcript levels in the central nervous system (CNS) of actin5C-GAL4>UAS-Xrp1-RNAi third instar larvae of ∼7% and ∼11% of control levels for UAS-Xrp1-RNAi-1 and UAS-Xrp1-RNAi-2, respectively (Fig. S1 B). Furthermore, selective Xrp1 knockdown in glial cells did not rescue caz mutant pupal lethality (Fig. S4 A). Together, these findings demonstrate that selective down-regulation of Xrp1 in neurons is sufficient to rescue caz mutant phenotypes.
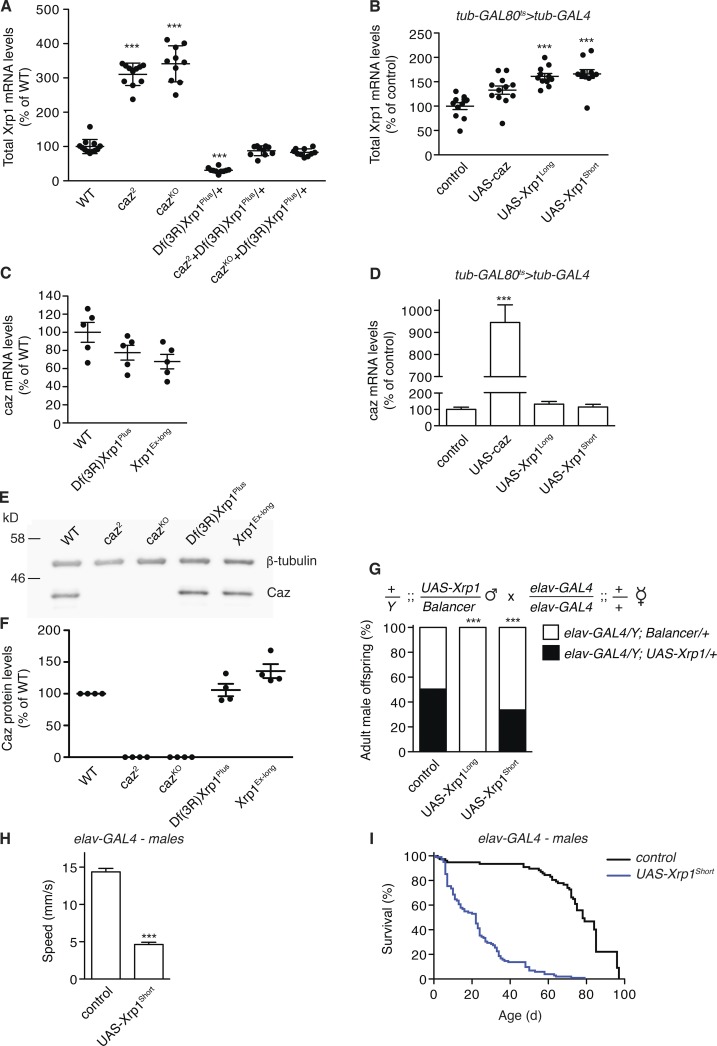
Increased Xrp1 expression mediates caz mutant phenotypes
Our finding that reducing Xrp1 expression rescues caz mutant phenotypes raised the possibility that these phenotypes are caused by up-regulation of Xrp1 expression, with particularly deleterious effects in neurons. Consistent with this hypothesis, qPCR revealed that Xrp1 mRNA levels are increased three- to fourfold in both CNS and body wall of caz mutants (Figs. 3 A and S4, B and C). Remarkably, in caz mutant Xrp1 heterozygotes, Xrp1 transcript levels were not significantly different from controls (Fig. 3 A). Thus, phenotypic rescue of caz mutants by Xrp1 heterozygosity is associated with normalization of Xrp1 expression levels. Vice versa, ubiquitous caz overexpression from the adult stage onwards did not reduce Xrp1 mRNA levels (Fig. 3 B). To evaluate whether Xrp1 gene dosage modifies caz expression levels, qPCR and Western blotting was performed on Xrp1 mutants and transgenic flies ubiquitously overexpressing Xrp1Long or Xrp1Short. These analyses revealed that Caz levels are not significantly changed in Xrp1 mutants (Fig. 3, C, E, and F) or upon Xrp1 overexpression (Fig. 3 D). Thus, loss of caz increases Xrp1 expression, but Caz overexpression does not down-regulate Xrp1, and alteration of Xrp1 levels has no effect on caz expression.
Figure 3. .
Xrp1 expression is up-regulated in caz mutants, and selective neuronal Xrp1 overexpression phenocopies caz mutant phenotypes. (A) Xrp1 transcript levels as determined by qPCR on CNS of WT, caz mutant, Xrp1 heterozygous, and caz mutant Xrp1 heterozygous larvae. n = 10. (B) Xrp1 transcript levels in heads of adult male flies that ubiquitously (tub-GAL4) overexpress Caz, Xrp1Long, Xrp1Short, or no transgene (control) from the adult stage onwards. n = 10. (C) Caz transcript levels in larval CNS of WT and two Xrp1 mutants. n = 10. (D) Caz transcript levels in heads of adult male flies that ubiquitously overexpress caz, Xrp1Long, Xrp1Short, or no transgene (control) from the adult stage onwards. n = 10. ***, P < 0.0001; one-way ANOVA. (E) Representative Western blot to evaluate Caz protein levels in larval CNS from WT, caz mutants, and Xrp1 mutants. β-tubulin was used as loading control. (F) Quantification of Caz protein levels relative to β-tubulin. n = 5. P = NS; one-way ANOVA. (G) Frequency of adult male offspring from the indicated cross. n > 111 per genotype. ***, P < 0.0005; χ2 test. (H) Average climbing speed of adult male flies selectively overexpressing Xrp1Short in neurons (elav-GAL4) as compared with driver-only controls. n > 100 per genotype. ***, P < 10−9; Mann-Whitney test. (I) Life span of male flies selectively overexpressing Xrp1Short in neurons (elav-GAL4) as compared with driver-only controls. n = 77–102. All graphs display mean ± SEM.
To further test the hypothesis that increased Xrp1 expression in neurons is a key mediator of caz mutant phenotypes, we evaluated the effect of selective Xrp1 overexpression in neurons of otherwise WT flies. Neuronal overexpression of either Xrp1Long or Xrp1Short induced developmental lethality, with a fraction of adult escapers emerging (Figs. 3 G and S4 D). These adult escapers displayed substantial motor performance deficits (Figs. 3 H and S4 E) and a significantly shortened life span (Figs. 3 I and S4 F). Thus, neuronal Xrp1 overexpression phenocopies caz mutant phenotypes. Together with the findings that Xrp1 heterozygosity or Xrp1 neuronal knockdown rescue caz mutant phenotypes, these results indicate that increased neuronal Xrp1 levels mediate caz mutant phenotypes.
Xrp1 is a nuclear protein that binds chromatin
The Xrp1 protein is predicted to contain two conserved DNA-binding domains in its C terminus: (1) an AT-hook motif consisting of nine amino acids centered on the invariant tripeptide glycine-arginine-proline (Reeves, 2010) and (2) a basic-region leucine zipper (bZIP) motif found in the bZIP family of transcription factors, which typically consists of a basic region of ∼20 aa that mediates sequence-specific DNA binding along with a leucine zipper, a sequence of 40–60 hydrophobic amino acids in which leucine occurs every seventh residue, which mediates dimerization (Vinson et al., 2002). Despite the fact that Xrp1 was reported to heterodimerize with the bZIP protein Irbp18 (Francis et al., 2016), coimmunoprecipitation experiments on extracts of Drosophila S2 cells cotransfected with N-terminal HA-tagged and Flag-tagged variants of either Xrp1Long or Xrp1Short indicated that Xrp1 does not homodimerize (Fig. S4, G and H).
As Ensembl and NCBI Blastp searches failed to identify a human Xrp1 orthologue, we used HHpred (Zimmermann et al., 2018), the most sensitive homology detection tool, to identify human Xrp1 homologues. This yielded human homologues for the C-terminal ∼150 aa of Xrp1 that contain the conserved DNA-binding domains (Table S1). Although these human proteins all contain a bZIP domain, they do not appear to be Xrp1 orthologues, as reciprocal searches against Drosophila proteins using HHpred with the “best hits” from human, a reliable method to detect orthologues whose sequence homology is not apparent with pairwise searches (Szklarczyk et al., 2012), did not uncover Xrp1. Furthermore, none of the human Xrp1 homologues contained an AT-hook motif, and in fact, none of the bZIP proteins in the SMART database (Letunic and Bork, 2018) contained an AT-hook motif.
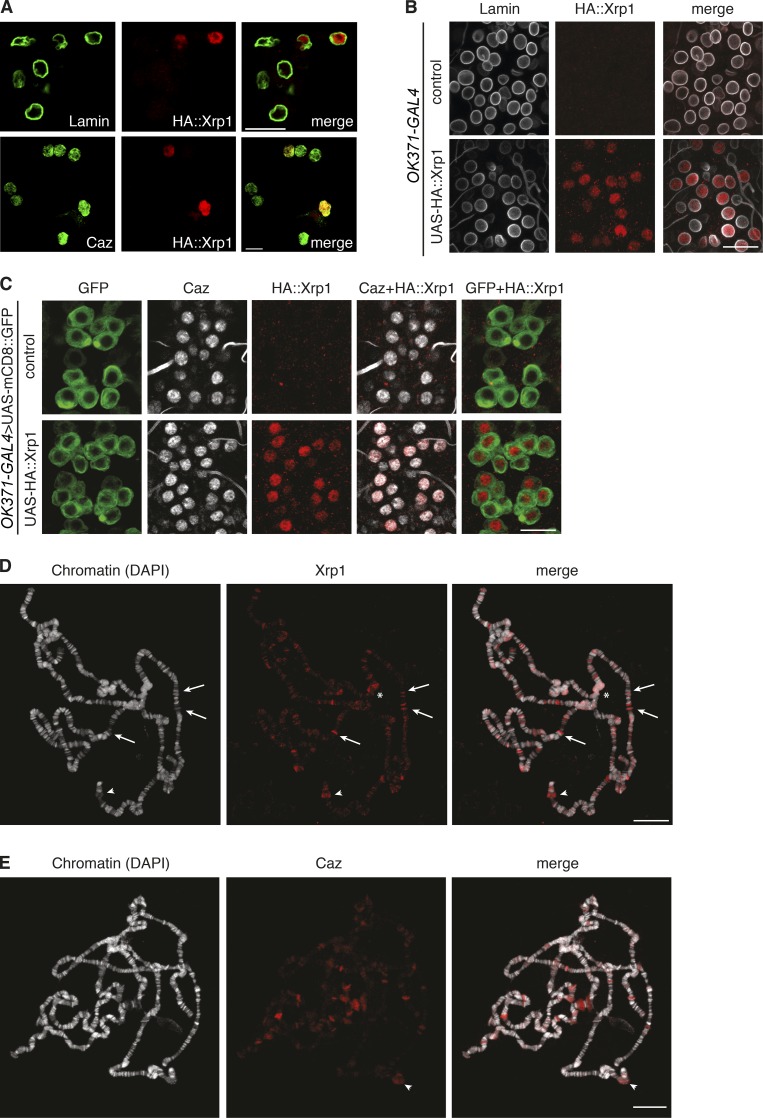
Consistent with the presence of two putative DNA-binding domains and its reported roles in protection against genotoxic stress and DNA repair (Brodsky et al., 2004; Akdemir et al., 2007; Francis et al., 2016), subcellular localization experiments revealed that Xrp1 is localized to the nucleus, where it colocalizes with Caz, both in Drosophila S2R+ cells and in motor neurons in vivo (Fig. 4, A–C). To evaluate whether Xrp1 binds chromatin, immunostaining for Xrp1 was performed on polytene chromosomes from larval salivary glands. Xrp1 was found to preferentially localize to euchromatic bands and to “puffs,” enlarged regions which indicate sites of active transcription (Fig. 4 D), suggesting a possible involvement of Xrp1 in regulation of gene expression. Furthermore, Xrp1 also localized to centromeric β-heterochromatin (Fig. 4 D). Caz prominently localized to puffs on polytene chromosomes (Fig. 4 E), and overall, Caz and Xrp1 displayed a distinct binding pattern with some overlap (e.g., arrowhead in Fig. 4, D and E).
Figure 4. .
Xrp1 is a nuclear protein that binds chromatin. (A) Immunostaining of Drosophila S2R+ cells expressing N-terminal HA-tagged Xrp1Short for lamin (labels the nuclear membrane), and the HA tag revealed Xrp1 localization to the nucleus (top). Immunostaining for Caz and HA::Xrp1 showed colocalization (bottom). (B) HA-tagged Xrp1 was selectively expressed in larval motor neurons (OK371-GAL4). Immunostaining for lamin and HA::Xrp1 revealed nuclear localization. Control animals are driver only. (C) HA-tagged Xrp1 was coexpressed with membrane-bound GFP in larval motor neurons. Immunolabeling for Caz and HA::Xrp1 showed colocalization of Caz and Xrp1. Control animals do not express HA::Xrp1. (D and E) Immunostaining of Xrp1 (D) or Caz (E) on polytene chromosomes. Chromatin was counterstained with DAPI. Xrp1 prominently localizes to euchromatic bands (arrows indicate examples), to puffs (arrowheads), and to centromeric β-heterochromatin (asterisks). Caz prominently localizes to puffs. Overall, Caz and Xrp1 display a distinct binding pattern, although there is some overlap, e.g., the puff region indicated by an arrowheads in D and E. Bars: 10 µm (A); 20 µm (B–E).
Xrp1-interacting proteins suggest a role in gene expression regulation
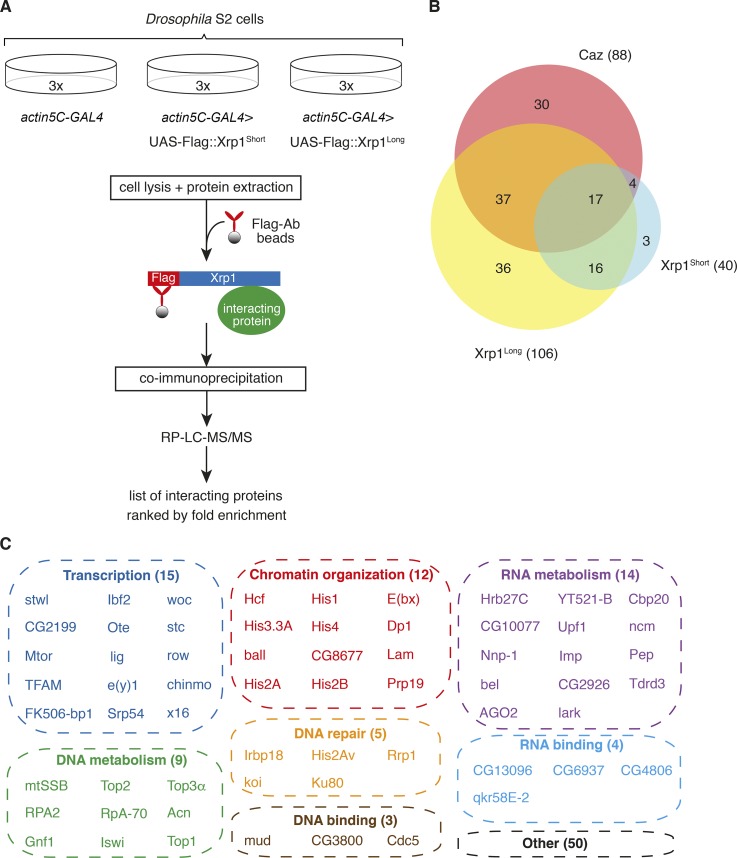
To obtain a comprehensive overview of the molecular processes in which Xrp1 may be involved, we immunoprecipitated N-terminal Flag-tagged Xrp1 (either short or long isoform) from Drosophila S2 cells and identified interacting proteins by mass spectrometry (MS; Fig. 5 A). 106 Xrp1long-interacting proteins and 40 Xrp1short-interacting proteins were identified (Tables S2 and S3). The substantially higher number of Xrp1long-interacting proteins is likely attributable to its 262 additional N-terminal amino acids. Importantly, of the 40 Xrp1short-interacting proteins, 33 were also identified as Xrp1long-interacting proteins (Fig. 5 B). In addition, consistent with heterodimer formation between Xrp1 and Irbp18 (Francis et al., 2016), Irbp18 was identified as an Xrp1-interacting protein (Fig. 5 C and Table S2). Remarkably, out of a total of 112 Xrp1-interacting proteins, 62 (55.4%) are involved in gene expression regulation or DNA/RNA metabolism, including regulation of transcription (15), chromatin organization (12), DNA metabolism (9), DNA repair (5), RNA metabolism (14), and DNA-binding proteins (3) and RBPs (4; Fig. 5 C).
Figure 5. .
Identification of Xrp1- and Caz-interacting proteins. (A) Approach used to identify Xrp1-interacting proteins. N-terminal Flag-tagged Xrp1 was expressed in Drosophila S2 cells. Coimmunoprecipitation followed by quantitative MS was used to identify Xrp1-interacting proteins, defined as proteins significantly enriched in Flag::Xrp1-expressing cells as compared with control cells. (B) Venn diagrams illustrating the substantial overlap between Xrp1Long-, Xrp1Short-, and Caz-interacting proteins. (C) The majority (55.4%) of Xrp1-interacting proteins are involved in gene expression regulation or DNA/RNA metabolism, including regulation of transcription, chromatin organization, DNA metabolism, DNA repair, RNA metabolism, and DNA- or RNA-binding proteins.
Following a similar experimental approach, we immunoprecipitated endogenous Caz from S2 cells and identified 88 Caz-interacting proteins (Table S4). Remarkably, 58 of these proteins (65.9%) were also identified as Xrp1-interacting proteins (Fig. 5 B), suggesting that Caz and Xrp1 are involved in similar molecular processes and/or commonly reside in protein complexes. The latter possibility is unlikely, as Caz did not coimmunoprecipitate with Xrp1 (Fig. S4 I).
The caz-Xrp1 genetic interaction depends on the functionality of the AT-hook domain of Xrp1
We next wanted to evaluate whether the rescue of caz mutant phenotypes by Xrp1 heterozygosity depends on the functionality of the AT-hook DNA-binding domain of Xrp1. We therefore evaluated whether a mild increase of Xrp1 levels in “rescued” Xrp1 heterozygous caz mutant flies would revert the rescue and result in pupal lethality, and if so, whether expression of Xrp1 with a subtle mutation in the AT-hook motif that precludes DNA binding would still revert the rescue.
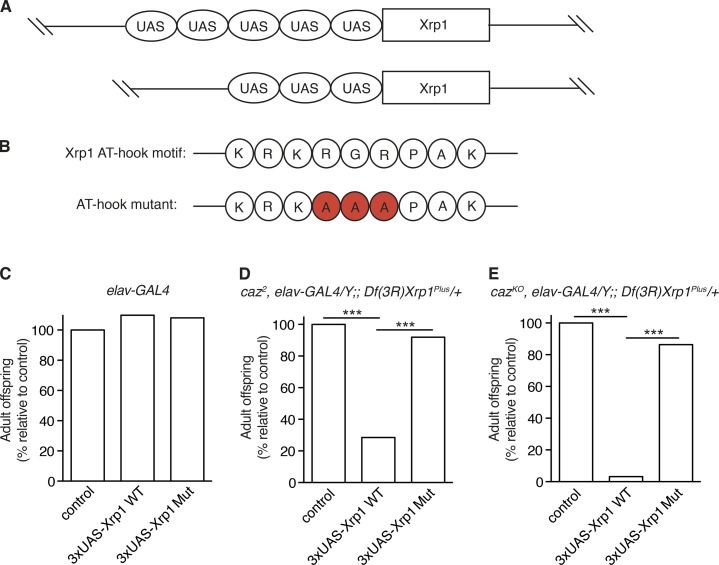
As neuron-selective expression of Xrp1 from the standard pUAST transgenesis vector induces phenotypes by itself (Fig. 3, G–I; and Fig. S4, D–F), we used a modified pUAST vector with only three UAS sites, known to result in lower transgene expression levels (Fig. 6 A; Pfeiffer et al., 2010). The resulting 3×UAS-Xrp1 lines did not induce developmental lethality when selectively expressed in neurons (elav-GAL4; Fig. 6 C). Neuronal Xrp1 expression from 3×UAS transgenes was nevertheless able to revert the rescue of caz mutant pupal lethality by Xrp1 heterozygosity (Fig. 6, D and E). We therefore inactivated the Xrp1 AT-hook domain in the 3×UAS constructs by mutagenizing the RGR triplet of the KRKRGRPAK motif to AAA, known to abolish AT-hook–mediated DNA binding (Fig. 6 B; Metcalf and Wassarman, 2006; Turlure et al., 2006; Baker et al., 2013). The subtle AT-hook mutation altered the binding pattern of Xrp1 on polytene chromosomes (Fig. S3 C) but did not reduce the stability of the Xrp1 protein and in fact increased Xrp1 protein level (Fig. S1, D–F). In spite of this, AT-hook–mutant UAS-Xrp1 transgenes were no longer able to revert the rescue of caz mutant pupal lethality by Xrp1 heterozygosity (Fig. 6, D and E). These data demonstrate that the genetic interaction between caz and Xrp1 is dependent on the functionality of the AT-hook DNA-binding motif in Xrp1.
Figure 6. .
Functionality of the AT-hook DNA-binding domain of Xrp1 is required to mediate caz mutant phenotypes. (A) Schematic representation of the Xrp1 transgene in the pUAST transgenesis vector, with five UAS GAL4-binding sites, and the pJFRC4 vector, with three UAS sites. (B) The Xrp1 AT-hook motif consists of nine amino acids including the invariant GRP triplet (top). To inactivate the DNA-binding capacity of the AT-hook motif, three amino acids essential for DNA binding (RGR) were mutagenized to alanine (bottom). (C) Panneuronal expression (elav-GAL4) of WT or AT-hook mutant (Mut) 3×UAS-Xrp1 transgenes (short isoform) does not induce developmental lethality. Adult offspring frequency relative to a driver-only control (100%) is shown. n > 376 per genotype. (D and E) Adult offspring frequency of caz2, elav-GAL4/Y;; Df(3R)Xrp1Plus/+ (D) or cazKO, elav-GAL4/Y;; Df(3R)Xrp1Plus/+ (E) males expressing 3×UAS-Xrp1 transgenes (WT or AT-hook mutant) or no transgene (control). n > 115 per genotype. ***, P < 0.0001; χ2 test.
Gene expression dysregulation in caz mutants is rescued by Xrp1 heterozygosity
FUS is known to be involved in transcriptional regulation and mRNA splicing (Schwartz et al., 2012, 2015; Tan et al., 2012; Yang et al., 2014), and knockdown or knockout of Fus in the mouse brain results in gene expression dysregulation (Ishigaki et al., 2012; Lagier-Tourenne et al., 2012; Scekic-Zahirovic et al., 2016). Furthermore, AT-hook proteins are often involved in gene expression regulation either as transcription factors or as chromatin architectural proteins (Reeves, 2010), and our results thus far indicate that Xrp1 is a nuclear chromatin-binding protein likely involved in gene expression regulation. We therefore hypothesized that loss of caz function may result in gene expression dysregulation, which could possibly be mitigated by heterozygosity for Xrp1.
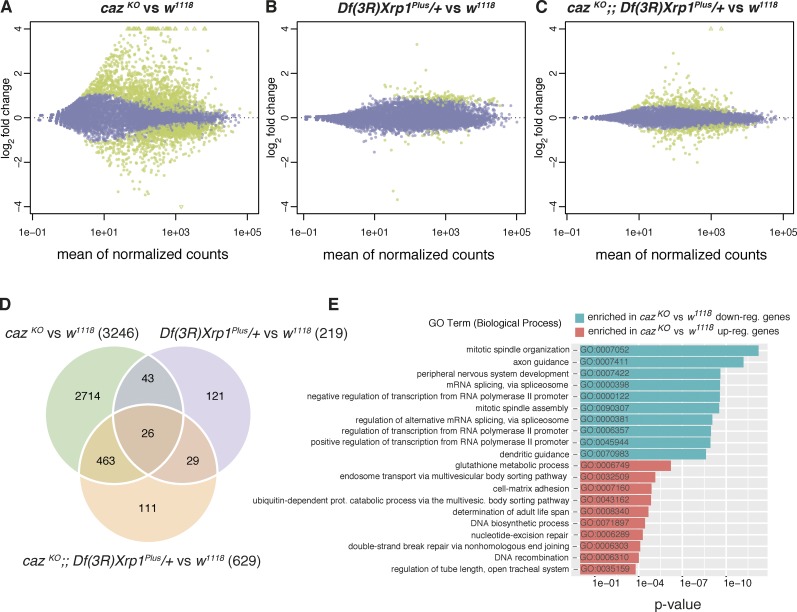
To test this hypothesis, we used RNA sequencing (RNA-seq) to evaluate mRNA expression levels in third instar larval CNS of cazKO and cazKO Xrp1 heterozygous animals as well as Xrp1 heterozygous and WT animals as controls. Principal component analysis and clustering of the samples discriminated the four genotypes from each other, with a certain degree of overlap between Xrp1 heterozygous and WT samples (Fig. S5, A and B). Differential gene expression analysis between mRNA levels from WT and cazKO identified 1,641 up-regulated and 1,605 down-regulated genes in caz mutants (FRD-adjusted P value <0.05; Fig. 7, A and D; and Table S5), indicating substantial gene expression dysregulation. Gene expression changes for caz and Xrp1 were consistent with the previously obtained qPCR data (Fig. S5 C). In contrast, comparison between WT and Xrp1 heterozygotes identified only 184 up-regulated and 30 down-regulated genes (Fig. 7, B and D). Most interestingly, in caz mutant Xrp1 heterozygous CNS, 315 up-regulated and 314 down-regulated genes were identified, with >90% of these displaying a less than twofold change (Fig. 7, C and D). Thus, heterozygosity for Xrp1 significantly mitigated gene expression dysregulation in caz mutant CNS. Principal component analysis confirmed the dramatic gene expression dysregulation in caz mutant CNS, which was significantly rescued in caz mutant Xrp1 heterozygous animals (Fig. S5 A).
Figure 7. .
Heterozygosity for Xrp1 mitigates gene expression dysregulation in caz mutant CNS. (A–C) MA plots displaying gene expression changes in cazKO versus w1118 (genetic background control; A), Df(3R)Xrp1Plus/+ versus w1118 (B), and cazKO;; Df(3R)Xrp1Plus/+ versus w1118 (C). X axes represent the mean of the normalized read counts per gene across all samples included in each comparison. Y axes represent the log2 fold change per gene resulting from each comparison. Green dots correspond with differentially expressed genes, with P < 0.05 adjusted for multiple testing. (D) Venn diagram representing the overlap between differentially expressed genes across the three comparisons. Numbers in parenthesis indicate the total number of differentially expressed genes in each comparison. (E) Top 10 enriched GO terms (Biological Process ontology) in the cazKO versus w1118 comparison for the set of up-regulated (red) and down-regulated genes (blue).
Up- or down-regulation of a panel of 19 genes was validated by qPCR (Fig. S5 E), and gene ontology (GO) analysis showed that transcripts whose expression was altered in caz mutant animals were enriched for genes involved in processes such as axon and dendrite guidance, peripheral nervous system development, regulation of transcription and mRNA splicing, DNA repair, and mitotic spindle organization and assembly (Fig. 7 E). GO analysis for molecular function revealed that transcripts with altered expression in caz mutants were enriched for mRNAs encoding DNA- and chromatin-binding proteins as well as mRBPs, along with transcription factor activity, among others (Fig. S5 D). Overall, this is in line with the known functions of Caz and its mammalian FET protein orthologues (Schwartz et al., 2015) and strikingly similar to results from RNA-seq experiments in Fus−/− mice (Scekic-Zahirovic et al., 2016). In conclusion, our transcriptome analysis revealed that gene expression dysregulation in caz mutant CNS is substantially mitigated by Xrp1 heterozygosity.
Phenotypes induced by motor neuron–selective expression of ALS mutant FUS are substantially mitigated by Xrp1 knockdown
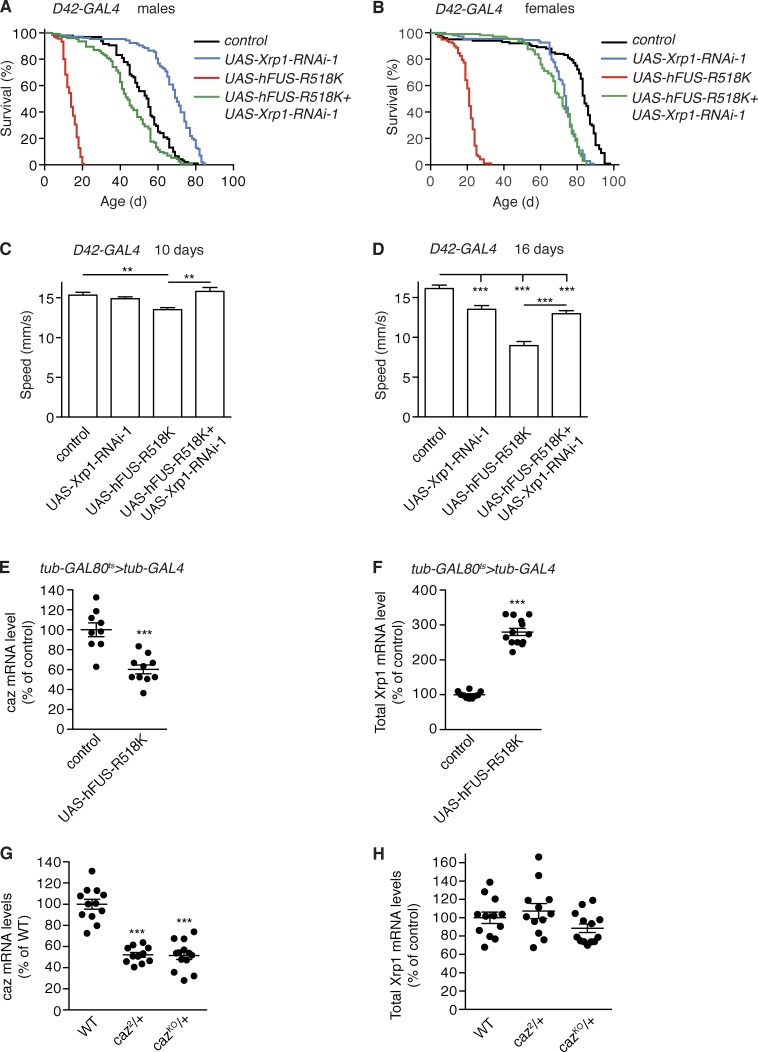
To evaluate the potential relevance of our findings for human ALS, we used a Drosophila ALS-FUS model. Selective expression of R518K mutant human FUS in motor neurons (D42-GAL4) yielded adult flies that developed progressive motor deficits and displayed a substantially shortened life span (Fig. 8, A–D). We therefore evaluated the effect of Xrp1 knockdown on motor behavior and life span of these flies. Coexpression of Xrp1-RNAi more than tripled the median life span of both male and female D42-GAL4>UAS-FUS-R518K flies (Fig. 8, A and B). At 10 d of age, FUS-R518K flies displayed a mild climbing defect (reduction in speed by ∼12%), which was fully rescued by motor neuron–selective Xrp1 knockdown (Fig. 8 C). At 16 d of age, FUS-R518K flies displayed a severe climbing defect (reduction in speed by ∼45%), which was rescued by knockdown of Xrp1 to a level that was not significantly different from D42-GAL4>UAS-Xrp1-RNAi control flies (Fig. 8 D). Thus, reduction of Xrp1 levels in motor neurons substantially rescues the motor deficits and shortened life span of a Drosophila ALS-FUS model.
Figure 8. .
Motor neuron–selective Xrp1 knockdown mitigates motor deficits and shortened life span induced by ALS mutant FUS expression. (A and B) Life span of control (driver only) flies and flies with motor neuron–selective (D42-GAL4) expression of human FUS-R518K, Xrp1-RNAi, or both transgenes. Data for male (A) and female (B) flies are shown. n > 75 per genotype. (C and D) Average climbing speed of adult female flies with motor neuron–selective (D42-GAL4) expression of human FUS-R518K, Xrp1-RNAi, or both transgenes versus driver-only controls. Flies were tested at 10 (C) and 16 (D) d of age. n > 100 per genotype. **, P < 0.01; ***, P < 0.005; Mann-Whitney test. (E and F) Transcript levels of caz (E) and Xrp1 (F) in heads of adult female flies 3 d after induction of ubiquitous FUS-R518K expression (tub-GAL4) versus driver-only controls. n = 9–11. ***, P < 0.0005; two-tailed unpaired t test. (G and H) Caz (G) and Xrp1 (H) transcript levels in the CNS of third instar female larvae heterozygous for caz2 or cazKO versus WT controls. n = 11–13. ***, P < 0.001; one-way ANOVA. All graphs display mean ± SEM.
To gain insight into the mechanism underlying this major phenotypic rescue, we evaluated the effect of FUS-R518K overexpression on caz and Xrp1 transcript levels. Ubiquitous overexpression of FUS-R518K in adult female flies moderately reduced caz transcript levels to ∼60% of control levels (Fig. 8 E). Strikingly, Xrp1 transcript levels were about threefold increased upon FUS-R518K expression (Fig. 8 F). This substantial increase in Xrp1 expression cannot be attributed to the moderate reduction in caz levels because Xrp1 transcript levels were not altered in caz heterozygous females in spite of a ∼50% reduction of caz transcript levels (Fig. 8, G and H). Thus, expression of ALS mutant FUS results in substantial up-regulation of Xrp1 expression independent of caz levels. The fact that Xrp1 knockdown substantially rescues phenotypes induced by ALS mutant human FUS indicates that these phenotypes are to a large extent mediated by increased Xrp1 expression.
Discussion
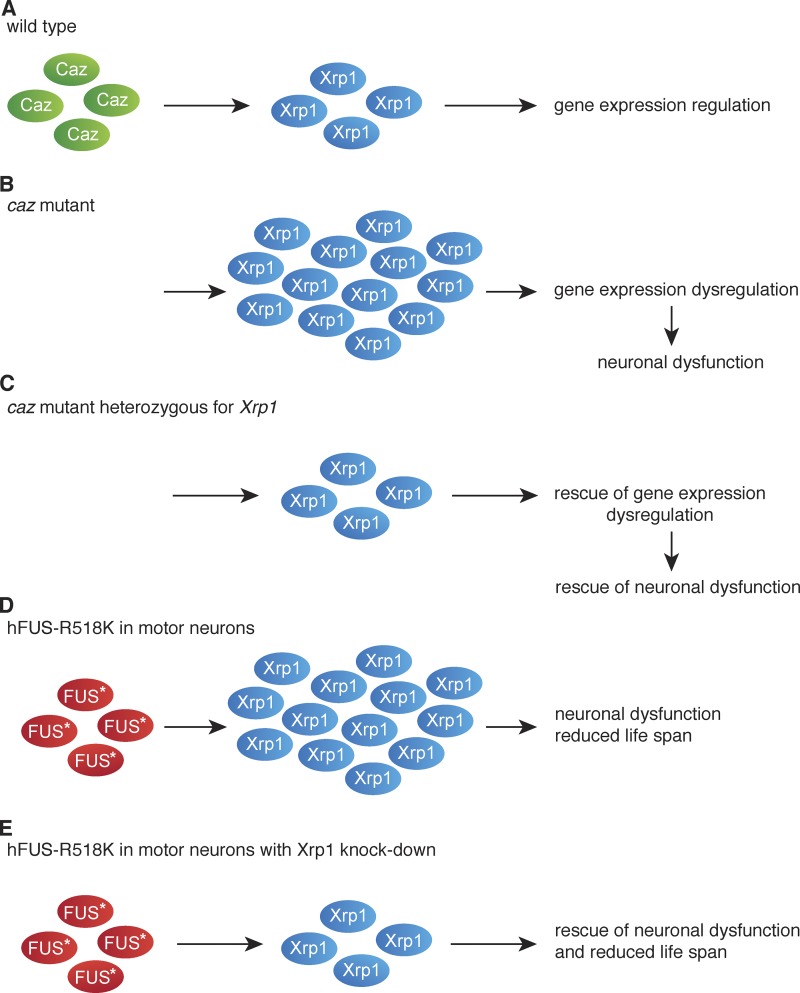
In this study, we identified Xrp1 as a genetic modifier of caz mutant phenotypes. Caz is the single Drosophila orthologue of the three human FET family proteins FUS, EWSR1, and TAF15 (Schwartz et al., 2015). Xrp1 expression was up-regulated by three- to fourfold in caz mutant animals, and heterozygosity for Xrp1 fully rescued the caz mutant eclosion defect and partially but substantially rescued adult motor performance and life span. Exhaustive genetic screening of the second and third chromosome, which together constitute ∼80% of the fly genome, identified Xrp1 as the only gene for which reduction of gene dosage by 50% could rescue caz mutant pupal lethality, indicating that Xrp1 is a key modifier of caz mutant phenotypes. Interestingly, in spite of the previously reported ubiquitous expression of Xrp1 (Tsurui-Nishimura et al., 2013) and the fact that Xrp1 expression was not only increased in the CNS but also in the body wall and presumably other tissues of caz mutants, neuron-selective knockdown of Xrp1 was sufficient to rescue caz mutant phenotypes, and selective neuronal overexpression of Xrp1 in otherwise WT animals phenocopied caz mutant phenotypes. This is consistent with the previously reported key function of Caz in neurons (Wang et al., 2011; Frickenhaus et al., 2015). Together, our data indicate that caz mutant phenotypes are largely mediated by increased Xrp1 expression, with particularly deleterious effects in neurons (Fig. 9).
Figure 9. .
Xrp1 is a key mediator of caz mutant phenotypes. (A) In WT animals, Caz controls Xrp1 levels, resulting in normal gene expression regulation. (B) Loss of caz function results in substantial up-regulation of Xrp1 expression, leading to gene expression dysregulation and neuronal dysfunction. (C) In caz mutant Xrp1 heterozygous animals, Xrp1 levels are normalized, resulting in rescue of gene expression dysregulation and neuronal dysfunction. (D) Expression of ALS mutant human FUS results in substantial up-regulation of Xrp1 expression, and motor neuron–selective expression induces neuronal dysfunction and reduced life span. (E) Simultaneous knockdown of Xrp1 in motor neurons expressing FUS-R518K rescues neuronal dysfunction and reduced life span.
Xrp1 has previously been implicated in protection against genotoxic stress and DNA damage repair (Brodsky et al., 2004; Akdemir et al., 2007; Francis et al., 2016). Consistently, a number of Xrp1-interacting proteins are involved in DNA repair (Fig. 5 C), and GO analysis of our RNA-seq data revealed enrichment for genes involved in DNA repair among genes up-regulated in caz mutant CNS (Fig. 7 E). This study revealed a novel role for Xrp1 in gene expression regulation as a substantial number of Xrp1-interacting proteins are involved in regulation of transcription, chromatin organization, and RNA metabolism (Fig. 5 C). Interestingly, neuronal overexpression of the long Xrp1 isoform induced significantly stronger phenotypes as compared with the short isoform (Fig. 3, G–I; and Fig. S4, D–F), despite insertion of the transgenes in the same genomic site and similar expression levels (Fig. 3 B). This is likely attributable to the additional N-terminal 262 aa of the long isoform, allowing the binding of substantially more interacting proteins (Fig. 5 B and Table S2), which may result in more pronounced gene expression dysregulation and stronger phenotypes.
Consistent with a key role of Xrp1 in gene expression regulation, significant gene expression dysregulation was found in caz mutant CNS, which was substantially mitigated by normalizing Xrp1 levels in caz mutants (Fig. 7). Importantly, of the 3,246 differentially expressed genes in caz mutants, only 489 are still significantly up- or down-regulated caz mutant Xrp1 heterozygotes (Fig. 7 D). The 2,757 genes that are significantly changed in caz mutants but not in caz mutant Xrp1 heterozygotes are likely direct or indirect targets of Xrp1, and up- or down-regulation of these genes in caz mutants may contribute to neuronal dysfunction. The novel function of Xrp1 in gene expression regulation is likely dependent on the capacity of Xrp1 to bind DNA, presumably mediated by two predicted DNA-binding domains in its C terminus: an AT-hook motif and a bZIP motif. Whereas the functionality of the predicted bZIP motif remains to be investigated, the AT-hook motif of Xrp1 conforms with the consensus sequence, consisting of nine amino acids centered on the invariant tripeptide glycine-arginine-proline (Reeves, 2010). The DNA-binding capacity of the Xrp1 AT-hook motif is likely required to mediate gene expression regulation and dysregulation, as the introduction of a subtle mutation in this motif demonstrated that its functionality is essential to mediate caz mutant phenotypes. Based on our findings, we propose a working model in which caz mutant phenotypes are mediated by increased Xrp1 expression, leading to gene expression dysregulation and neuronal dysfunction (Fig. 9).
Extensive bioinformatic searches did not reveal a clear one-to-one Xrp1 orthologue in mammals. However, we believe that it is highly likely that Xrp1 has functional homologues in mammals. Candidate functional homologues include 27 human genes encoding proteins predicted to contain at least one AT-hook motif (Table S6), including the Rett syndrome gene MECP2 (Amir et al., 1999). Interestingly, DNA binding mediated by the MeCp2 AT-hook domains has been implicated in the pathogenesis of Rett syndrome (Baker et al., 2013), and FUS was reported to bind the MECP2 promotor and positively regulate MECP2 transcription (Tan et al., 2012). In addition, FUS also binds MECP2 mRNA (Lagier-Tourenne et al., 2012), indicating that MECP2 is both a transcriptional and mRNA target of FUS. Furthermore, brains from Fus−/− mice or transgenic mice overexpressing ALS mutant FUS display up-regulation of Cbx2, Dot1l, Elf3, Prr12, and KMT2B (Scekic-Zahirovic et al., 2016; Shiihashi et al., 2016), and 17 of the 27 AT-hook genes are reported FUS RNA targets (Table S6). En route toward identification of human functional homologues of Xrp1, it will be particularly important to gain detailed molecular insight into how Xrp1 regulates gene expression. A first step could be the identification of the genomic binding sites of Xrp1 and its putative target genes. Furthermore, since Xrp1 does not have other predicted functional domains apart from the AT-hook and bZIP domains, it is tempting to speculate that Xrp1 regulates gene expression by recruiting other proteins that contain functional domains such as transactivation or histone-modifying domains to specific genomic sites. Several of the Xrp1-interacting proteins identified in this study contain such functional domains and have human orthologues (e.g., TAF9, ZMYM2, NFX1, HCFC1/2, VRK1, RSF1, and BPTF).
Interestingly, gene expression dysregulation was previously implicated in ALS and FTD pathogenesis. For instance, a significant enrichment in de novo mutations in the chromatin regulatory pathway in sporadic ALS patients was reported (Chesi et al., 2013). Furthermore, involvement of the three FET proteins in regulation of transcription and mRNA splicing is well established (Schwartz et al., 2015), and in fact, several other RBPs that have been implicated in ALS and FTD pathogenesis are also known to play important roles in gene expression regulation (Ling et al., 2013). These include TDP-43, involved in regulation of transcription and mRNA splicing (Buratti and Baralle, 2010), which mislocalizes to cytoplasmic inclusions with nuclear clearance in >95% of ALS and ∼45% of FTD patients (Ling et al., 2013). Furthermore, the ALS-causing expanded hexanucleotide repeat in C9orf72 may sequester RBPs, thus inducing gene expression dysregulation (Lee et al., 2013; Haeusler et al., 2014). The potential relevance of our findings for ALS-FUS pathogenesis is further indicated by the fact that knockdown of Xrp1 substantially rescues the motor deficits and shortened life span of flies that selectively express ALS mutant human FUS in motor neurons (Figs. 8 and 9). This may be explained by a substantial increase in Xrp1 expression induced by mutant FUS, which is not attributable to the moderate (∼40%) down-regulation of caz expression.
Finally, our findings may also be relevant for FTLD-FUS pathogenesis as this disease is characterized by pathological inclusions containing not only FUS but also TAF15 and EWS, with reduced levels or complete loss of nuclear FET proteins in inclusion-bearing neurons and glial cells (Neumann et al., 2011; Davidson et al., 2013). Thus, loss of FET protein function and consequent gene expression dysregulation may contribute to FTLD-FUS pathogenesis. Consistently, FUS knockout or knockdown in the hippocampus of mice induces behavioral aberrations related to FTD symptoms (Kino et al., 2015; Udagawa et al., 2015). In conclusion, our findings provide important novel insights into the molecular mechanisms by which loss of Caz, the Drosophila orthologue of human FET proteins, induces motor deficits and reduced life span, and they suggest that gene expression dysregulation may be involved in the pathogenesis of human FUSopathies.
Materials and methods
Drosophila genetics
Flies were housed in a temperature-controlled incubator with 12:12 h on/off light cycle at 25°C, and for some experiments, at 23°C (5×UAS-Xrp1 overexpression), in vials containing standard cornmeal medium. X chromosome–inserted elav-GAL4 (458; Bloomington Drosophila Stock Center [BDSC]) was used for panneuronal expression of UAS transgenes, OK371-GAL4 and D42-GAL4 were used for targeted expression in motor neurons, and tub-GAL4 was used for ubiquitous expression of UAS transgenes.
For the dominant suppressor screen, deficiencies covering the second and the third chromosome from the Bloomington Deficiency Kit were used as this kit provides maximal coverage (euchromatic coverage ≥ 97.5%) with a minimal number of deletions (Cook et al., 2012). Df/Balancer males were crossed to caz2/FM7 females to screen for the emergence of caz2/Y; Df/+ males in the offspring, which would indicate suppression of pupal lethality of caz2 males by hemizygosity for the deficiency. To narrow down the genomic region that is uncovered by Df(3R)ED2, the deficiency that mediated rescue of caz mutant pupal lethality, molecularly mapped smaller deficiency lines in this region were ordered from the BDSC. PCR genotyping of caz mutant males was used to exclude X chromosome nondisjunction in all experiments.
The UAS-Xrp1-RNAi lines used in this study were P[TRiP.HMS00053]attP2 (34521; BDSC; UAS-Xrp1-RNAi-1) and P[GD9476]v33010 obtained from the Vienna Drosophila Resource Center (UAS-Xrp1-RNAi-2). The UAS-FUS-R518K transgenic line was generated by and obtained from Lanson et al. (2011).
Generation of Xrp1 mutant lines
For generation of Xrp1 deletion lines, P[XP]d11439, P[XP]Xrp1d04790, PBac[WH]f07598, and PBac[WH]f05721 were obtained from the Drosophila Genetic Resource Center at the Kyoto Institute of Technology and used to isolate Xrp1 chromosomal deletions (Fig. 1, D and E) following the basic schemes outlined by Parks et al. (2004). The chromosomal deletions generated were verified by PCR with primers flanking the transposable element insertions (Table S7), followed by sequencing of the PCR fragments.
For generation of a clean Xrp1-null allele, in vivo homologous recombination was used to target the Xrp1 gene. Following a previously published strategy (Vilain et al., 2014), the presence of a Mi{MIC} transposon in the Xrp1 gene (Mi{MIC}Xrp1MI07118) was exploited for site-specific insertion of a targeting construct (Figs. 1 and S1 C). For the construction of a targeting vector, a fosmid (FlyFos clone number FF017187) containing the extended Xrp1 genomic region was used to PCR amplify left and right homology arms using the primers Xrp1_LHA_FW, Xrp1_LHA_REV, Xrp1_RHA_FW, and Xrp1_RHA_REV (Table S7). To minimize the chance of introducing mutations during PCR amplification, Phusion high-fidelity DNA polymerase (New England Biolabs) was used with only 20 cycles of PCR amplification. The obtained PCR products were subcloned in a Zero Blunt TOPO PCR cloning vector (Invitrogen) and sequence verified. The presence of a HindIII site in primer Xrp1_LHA_FW and an EcoRI site in primer Xrp1_LHA_REV was subsequently used to clone the left homology arm into pABC (Choi et al., 2009). Next, the presence of an EcoRI site in Xrp1_RHA_FW and a KpnI site in Xrp1_RHA_REV was used to clone the right homology arm into the pABC vector that already contained the left homology arm.
The obtained targeting vector was sequence verified and injected into Mi{MIC}Xrp1MI07118 embryos for site-specific integration of the targeting construct into the Mi{MIC} transposable element in the Xrp1 gene. Transgenic lines in which the transgenic construct was integrated into Mi{MIC}Xrp1MI07118 in the correct orientation were identified. These lines were subsequently crossed to a transgenic line that expresses I-SceI under the control of a heat-inducible promoter. Given the presence of an I-SceI restriction site in primer Xrp1_LHA_FW, this will induce a double-strand break adjacent to the left homology arm of the targeting construct, allowing for precise removal of the Xrp1 and Mi{MIC} sequences left of the targeting construct through homologous recombination (Fig. S1 C). Lines with successful homologous recombination were identified by PCR and sequencing of the obtained PCR fragments. Next, these lines were crossed to a transgenic line that expresses I-CreI under the control of a heat-inducible promotor. Given the presence of an I-CreI restriction site in primer Xrp1_RHA_REV, this will induce a double-strand break adjacent to the right homology arm of the targeting construct, allowing for precise removal of the Xrp1 and Mi{MIC} sequences right of the targeting construct through homologous recombination. Lines with successful homologous recombination were identified by PCR and sequencing of the obtained PCR fragments.
Generation of UAS-Xrp1 transgenic lines
For generation of UAS-Xrp1Long transgenic lines, RNA was extracted from WT flies and converted into cDNA, which was used as a template for PCR (primer sequences in Table S7) to amplify the transcript coding for the long Xrp1 isoform. Gold clone FI10013 containing the Xrp1Short cDNA was obtained from Kyoto Stock Center. Xrp1Long and Xrp1Short cDNAs were subsequently cloned into pUAST-attB using either NotI or EagI as well as XhoI restriction sites (Table S7). Site-directed PCR mutagenesis was used to generate AT-hook mutant versions of the long and short Xrp1 isoforms (mutagenesis primers are included in Table S7). WT and AT-hook mutant Xrp1 cDNAs were subsequently amplified by PCR using Phusion high-fidelity DNA polymerase (New England Biolabs) and primers containing XhoI and XbaI restriction sites (Table S7). The obtained PCR products were subcloned in a Zero Blunt TOPO PCR cloning vector (Invitrogen), and XhoI and XbaI were used to transfer the Xrp1 cDNAs to the pJFRC4 vector, which contains three UAS sites (Pfeiffer et al., 2010). UAS constructs were embryo injected following standard procedures. For each of the constructs, VK31 (on III) and VK37 (on II) genomic landing sites were used to avoid any influence of neighboring genomic sequences on transgene expression. As neuronal expression of 5×UAS-Xrp1 transgenes (elav-GAL4) in many cases resulted in developmental lethality with no adult escapers when raised at 25°C, experiments in which elav-GAL4 was used to drive expression of 5×UAS-Xrp1 transgenes were performed at 23°C.
Motor performance assay
For assaying mobility, flies were collected within 24 h after eclosion and divided into groups of 10 individuals. Motor performance of 3-, 10-, or 16-d-old flies was evaluated as described earlier (Frickenhaus et al., 2015; Niehues et al., 2015), and average climbing speed (mm/s) was determined and compared between genotypes. As female D42-GAL4>FUS-R518K flies lived longer than males, we studied the effect of Xrp1 knockdown on age-dependent motor deficits in female flies.
Larval locomotion was analyzed using the frustrated total internal reflection–based imaging method FIM (Risse et al., 2013, 2014, 2017). Batches of 15 third instar larvae were allowed to freely move for 3 min on a recording platform at RT. Tracking data were obtained using FIMTrack (http://fim.uni-muenster.de), and output files were analyzed with MatLab (MathWorks). In Fig. S3 B, larvae were sorted in a Petri dish with water for ∼5 min, and only male larvae were recorded.
Adult offspring frequency and life span analysis
For determination of adult offspring frequencies, appropriate crosses were set up, and the number of adult flies eclosing was counted for each genotype. For life span analysis, newly eclosed flies were collected and housed at a density of 10 flies per vial. At least 75–100 flies were tested for each genotype. The number of dead flies was counted every day, and the flies were transferred to fresh food vials every 2–3 d.
Real-time qPCR
Total RNA was extracted from 15–20 third instar larval brains or from four adult male flies per biological replicate using NucleoSpin RNA (Macherey-Nagel) according to the manufacturer’s instructions. Reverse transcription was performed on 1 µg RNA treated with gDNA Wipeout Buffer using the Quantitect Reverse Transcription kit (QIAGEN). Resulting cDNA samples were used as templates for real-time PCR assays performed on an ABI 7300 system (Applied Biosystems) with iTaq Universal SYBR Green supermix (Bio-Rad Laboratories). Primers used for quantitation of caz and Xrp1 transcript levels are listed in Table S7. Measurements were normalized to EifTuM and rp49 controls. Data were analyzed using the ΔΔCt calculation method. Experiments included no–reverse transcriptase controls for each template and no-template controls for each pair of primers.
Western blotting
For Western blots, protein extracts were made by homogenizing third instar larval CNS in extraction buffer (50 mM Tris/HCl, pH 7.4, 150 mM KCl, 0.25 M sucrose, 5 mM MgCl2, and 0.5% Triton X-100). Lysates of Drosophila S2 cells transfected with plasmids encoding actin5C-GAL4 alone or cotransfected with plasmids encoding N-terminal HA-tagged Xrp1Short or Xrp1Long were prepared in cell lysis buffer (20 mM Tris, 200 mM NaCl, 1 mM EDTA, and 0.5% NP-40 containing 1 U complete mini protease inhibitor cocktail [Roche]). Samples separated on 10% SDS-PAGE were electrotransferred onto polyvinylidene difluoride (EMD Millipore) for 45 min at 15 V. Blotted membranes were incubated overnight at 4°C with primary antibodies against Caz (mouse monoclonal 3F4; 1:30; Immanuel et al., 1995), HA epitope tag (mouse monoclonal HA.11; 1:1,000; Covance), and β-tubulin (mouse monoclonal E7; 1:700; Developmental Studies Hybridoma Bank). Immunoreactive proteins were visualized after incubation with anti-mouse IgG coupled to horseradish peroxidase (W402B; 1:2,500; Promega) for 1 h at RT. Blots were developed with enhanced chemiluminescence (GE Healthcare), and x-ray film images of chemiluminescence were developed and scanned. Densitometric quantification of images was performed with ImageJ/FIJI (National Institutes of Health).
Coimmunoprecipitation
For detection of potential homodimers of Xrp1Short and Xrp1Long, Drosophila S2 cells were either transfected with a plasmid encoding actin5C-GAL4 alone or cotransfected with plasmids encoding N-terminal HA- or Flag-tagged Xrp1Short or Xrp1Long constructs, all under UAS control. 48 h after transfection, protein lysates of cells expressing HA- or Flag-tagged Xrp1 were either directly used or combined in a 1:1 ratio. 5% of protein extracts were used for Western blotting, while the remaining 95% were added to anti-HA agarose beads for 24 h at 4°C. Immunoprecipitation of HA-tagged proteins was performed using an Anti-HA Immunoprecipitation Kit (Sigma-Aldrich), according to the manufacturer’s protocol.
To evaluate whether Caz and Xrp1 physically associate with each other, S2 cells expressing the actin5C-GAL4 plasmid alone (control) or in conjunction with the N-terminal HA-tagged Xrp1Short or Xrp1Long constructs were seeded at a density of 106 cells/ml in 1 ml Shields and Sang medium (Sigma-Aldrich) in 12-well plates 1 d before transfection. Transfection was performed using Fugene HD Transfection Reagent (Promega) according to manufacturer’s instructions. Cell lysates were prepared, cellular debris was pelleted by centrifugation at 12,000 g for 15 min and washed, and the protein-containing supernatant was incubated overnight at 4°C with 100 µl of either anti-Flag (10 µg; clone M2; F1804; Sigma-Aldrich) or anti-Caz (10 µg; 3F4; Immanuel et al., 1995) conjugated SureBeads Protein G Magnetic Beads (Bio-Rad Laboratories) according to the manufacturer’s instructions. Bound proteins were eluted by heating to 70°C for 10 min with 40 µl of 1× Laemmli buffer (Bio-Rad Laboratories).
Inputs, precipitates, and binding proteins were analyzed by SDS-PAGE and immunoblotting. The immunoblot analyses were performed using the following primary antibodies: anti-Flag (clone M2; F1804; 1:1,500; Sigma-Aldrich) and anti-HA (Mono HA.11; 1:1,000; Covance).
Liquid chromatography (LC)–tandem MS analysis
S2 cells were seeded at a density of 106 cells/ml in 3 ml Shields and Sang medium (Sigma-Aldrich) in six-well plates 1 d before transfection. Flag-tagged Xrp1Short or Xrp1Long plasmids were transfected, along with the actin5C-GAL4 construct. Three replicates were processed for each condition. After incubating the cells for 2 d with the transfection mixes, cells were collected and lysed in cell lysis buffer (described above). Cellular debris was cleared by centrifugation at 12,000 g for 15 min. To pull down Xrp1-interacting proteins, the protein-containing supernatant was applied to 100 µl anti-Flag (clone M2; F1804; Sigma Aldrich)–conjugated SureBeads Protein G Magnetic Beads (Bio-Rad Laboratories) according to the manufacturer’s instructions. Cell lysates were incubated with the antibody beads overnight at 4°C. Washed beads were resuspended in 4% SDS and 50 mM Tris, pH 7.5, and bound proteins were eluted by heating to 95°C for 10 min and then precipitated with a fourfold excess (vol/vol) of ice-cold acetone overnight to remove detergent and salts. Precipitated protein pellets were washed twice with 90% acetone, air dried, and then resuspended in 8 M urea and 50 mM Tris-HCl, pH 8.5, before in-solution digestion, first with endopeptidase LysC (1 µg/immunoprecipitation) for 3 h at 37°C, and then with trypsin overnight at 37°C (1.5 µg/immunoprecipitation). After acidification of the digest by addition of 1% formic acid (final concentration), peptides were desalted using Empore-C18 StageTips (Rappsilber et al., 2003) and stored at 4°C until further use. Prior to LC–tandem MS, peptides were eluted using 2 × 20 µl of 80% acetonitrile and 0.1% formic acid, and then they were dried in an Eppendorf concentrator to a volume of ∼2 µl and resuspended in 10 µl buffer A (0.1% acetic acid). 6 µl of this peptide solution was then analyzed by nanoscale reverse-phase chromatography using an EASY nLC 1200 (Thermo Fisher Scientific) as a high-performance LC pump and a Picofrit column (25 cm × 75 µm ID; New Objective) filled with C18 reverse-phase material (Reprosil pur C18-AQ; 1.9 µm; Dr. Maisch GmbH) that was online coupled via a Nanospray Flex electrospray ionization source (Thermo Fisher Scientific) to a QExactive HF mass spectrometer (Thermo Fisher Scientific). Peptides were separated at a flow rate of 300 nl/min using a gradient running from 3–35% B (80% acetonitrile and 0.1% formic acid) in 90 min, which was ramped up to 100% B in 5 min, where it was maintained for additional 10 min before reequilibration at starting conditions. Column temperature was maintained at 45°C with the help of a column oven (PRSO-V1; Sonation). The mass spectrometer was operated in data-dependent mode, acquiring full-scan spectra in profile mode at a resolution of 60,000 and an automatic gain control target value of 3 × 10−6 (scan range 300–1,650 m/z). Spray voltage was set to 2.1 kV. The 17 most intense ions were chosen for higher energy collisional dissociation with a resolution of 15,000 at m/z 200 and a target value of 10−5. The isolation window was set to 1.6 m/z, and the normalized collision energy to a value of 28. Dynamic exclusion was allowed and set to 20 s. Uncharged as well as singly charged compounds were excluded from the analysis as well as peptides with a charge state >6. Data were recorded with Xcalibur software (Thermo Fisher Scientific).
MS data analysis
Raw MS files were processed using the MaxQuant computational platform (version 1.5.3.8; Cox and Mann, 2008). The Andromeda search engine integrated into MaxQuant was used for the identification of peptides and proteins by querying a concatenated forward and reverse UniProt Drosophila database (UP000000803_7227.fasta; release 2015-12), including common laboratory contaminants. The search for precursor and fragment ions was performed allowing an initial mass deviation of 20 ppm and 0.5 D, respectively. Trypsin with full enzyme specificity was selected, and only peptides with a minimum length of seven amino acids were allowed. A maximum of two missed cleavages was allowed. Carbamidomethylation (Cys) was set as fixed modification, while oxidation (Met) and N-acetylation were defined as variable modifications. For protein and peptide identification, a minimum false discovery rate of 1% was required. Label-free quantification (LFQ) was based on the measurements of three independent biological replicates for each strain analyzed by the MaxQuant LFQ algorithm with the “match between runs” option turned on (Cox et al., 2014). Further data processing was performed using the bioinformatics module Perseus (version 1.5.6.0; Cox et al., 2011). Following initial filtering and grouping (actin; caz; XRP1-L; XRP1-s), LFQ values were log2 transformed, and only proteins were included in the analysis that were identified with at least three valid values in at least one of the four groups. Still-missing values (NaN) were replaced by imputation, simulating signals of low abundant proteins within the distribution of measured values. A width of 0.3 SD and a downshift of 1.8 SD were used for this purpose. To identify proteins that displayed significant differences between the groups, ANOVA testing was performed (P = 0.05). Fold enrichment was calculated based on LFQ intensity values. The MS proteomics data have been deposited to the public PRIDE repository (Vizcaíno et al., 2013) via the ProteomeXchange platform (http://proteomecentral.proteomexchange.org) with the dataset identifier PXD008417.
Immunocytochemistry and histochemistry
Drosophila S2R+ cells were seeded in a density of 4 × 105 cells/ml on Concanavalin A (Sigma-Aldrich)–treated circular microscope cover glasses (12 mm; VWR) in a 24-well cell culture dish. After 24 h at 25°C, cells were transfected with a mix containing the Fugene HD transfection reagent (Promega) and actin5C-GAL4 and UAS-HA::Xrp1Short plasmids. 48 h later, cells were fixed in 4% PFA for 15 min followed by two 5-min washes in DPBS (1×; Gibco) at RT. After permeabilization with DPBS (1×) and 0.5% Triton X-100, cells were washed twice with DPBS. Cells were blocked for 1 h in 2% BSA and 10% goat serum in DPBS, followed by overnight incubation at 4°C with primary antibodies against Caz (mouse monoclonal clone 3F4; 1:30; Immanuel et al., 1995), lamin (mouse monoclonal ADL67.10; 1:100; Developmental Studies Hybridoma Bank), and HA (rabbit polyclonal; 1:50; Santa Cruz Biotechnology) diluted in 2% BSA and 10% goat serum in DPBS. After two washes in DPBS, secondary goat anti-mouse and anti-rabbit antibodies (Alexa Fluor 488 and 568; 1:500) were applied for 2 h at RT, followed by three washes in DPBS and mounting on microscopy slide with Aqua-Poly/Mount (Polysciences, Inc.).
For subcellular localization of Xrp1 in motor neurons, brains/CNS from wandering third instar larvae expressing OK371-GAL4>UAS-mCD8::GFP alone (control) or in conjunction with UAS-Xrp1Short were dissected in PBS and fixed in 4% PFA for 30 min. Tissues were washed 3 × 10 min in PBS/0.2% Triton X-100 and blocked for 1 h at RT in 10% goat serum in PBS. Tissues were incubated with primary antibodies against Caz (mouse monoclonal clone 3F4; 1:30; Immanuel et al., 1995), lamin (mouse monoclonal ADL67.10; 1:100; Developmental Studies Hybridoma Bank), or HA (rabbit polyclonal; 1:50; Santa Cruz Biotechnology) overnight at 4°C with gentle agitation. Appropriate secondary antibodies conjugated either with Alexa Fluor 405, Alexa Fluor 488, or Alexa Fluor 568 (Molecular Probes) were used to detect the given primary antibody. All images were acquired using ZEN 2010 software on a Zeiss LSM700 laser scanning confocal microscope using an EC Plan neofluar 1.3 NA 40× oil-immersion objective.
Squash preparation of polytene chromosomes from larval salivary glands
For preparing polytene chromosome squashes, salivary glands of WT third instar larvae were dissected in PBS (1×) and transferred to 1% Triton X-100 for 30 s. Fixation was in 4% PFA (1 min) and in 45% acetic acid/4% PFA (2 min). The glands were then incubated in 45% acetic acid (1 min) and subsequently squashed in the same solution under a coverslip to get polytene spreads. After freezing the slides in liquid nitrogen, coverslips were flipped off with a sharp blade, and slides were stored in 90% ethanol.
For immunostaining, squash preparations were rehydrated twice for 5 min in PBS (1×). Immunostaining was performed following the Dangli and Bautz (1983) procedure using rabbit polyclonal anti-Xrp1 (1:50; Francis et al., 2016) followed by an Alexa Fluor 568–conjugated secondary antibody (1:500; Molecular Probes). The preparation was counterstained with DAPI and mounted in Vectashield antifade mounting medium (Vector Laboratories) for confocal microscopy. All images were acquired using ZEN 2010 software on a Zeiss LSM700 laser scanning confocal microscope using an EC Plan neofluar 1.3 NA 40× oil immersion objective.
RNA-seq and data analysis
For RNA-seq, total RNA was extracted from 15–20 brains dissected from wandering third instar male larvae using NucleoSpin RNA (Macherey-Nagel) according to the manufacturer’s instructions. After performing quality control checks, the RNA was sent to the Max Planck Genome Center, where cDNA libraries were prepared using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs) using standard procedures. The libraries were sequenced on an Illumina HiSeq 2500 instrument as 100-bp paired-end reads each according to the manufacturer’s standard protocols. Three biological replicates per genotype were sequenced, with an average of 8.6 million nonredundant read pairs uniquely mapped to the Drosophila genome.
Preprocessing filtering of the reads before alignment, e.g., quality- or adapter-trimming, was not necessary. The Drosophila reference genome was downloaded from Flybase. Revision 6.04 of the genome assembly and gene annotation was used for all analyses. We aligned reads to the reference transcriptome using the TopHat pipeline (version 2.0.14; Kim et al., 2013a) with Bowtie2 (version 2.2.5; Langmead and Salzberg, 2012) and the flags b2-very-sensitive and library-type = fr-firststrand. The mapped reads were assigned to genes using the HTseq-count script from the HTseq package (Anders et al., 2015). We used the intersection-nonempty mode to exclude ambiguous gene assignments. Aligned pairs with a mapping quality <10 were excluded, and rRNA genes were removed from the gene list for further analysis.
Differential gene expression analysis was performed using DeSeq2 (version 1.11; Love et al., 2014). All comparisons were performed in a pairwise manner, comparing samples of each genotype separately against the WT. We chose to disable filtering genes based on Cook’s distance for the whole analysis because we observed that the high biological variability of Xrp1 heterozygous animals lead to the exclusion of a large number of genes. Genes were called differentially expressed if the log2 fold change differed significantly from 0 with a false discovery rate–adjusted P value of <0.05. Expression levels for each annotated protein-coding gene were determined by the number of mapped reads per kilobase of exon per million mapped reads (RPKM).
Principal component analysis and hierarchical clustering of the global expression profile was performed on a variance-stabilized transformation of the read counts per gene using methods provided by the DeSeq2 R-package. For clustering, the distances between samples were calculated using the Manhattan distance metric. Based on the distance matrix, hierarchical clustering by complete linkage was performed using standard R functions.
GO-term enrichment analysis was done using the topGO package for R (version 2.22; Alexa et al., 2006). We extracted the sets of up- and down-regulated genes for each comparison from the DeSeq2 analysis and used the “weight01” algorithm in topGO in combination with Fisher’s exact test to check for enrichment of specific GO terms in these gene sets.
Statistical analysis
χ2 statistics were used to analyze offspring frequency data. For life span analysis, the log-rank test was used to test for statistical significance. Motor performance was analyzed using the Mann-Whitney U test to compare climbing speed of individual flies per genotype and per run. As all flies were tested in three independent runs, three P values were generated per genotype. These P values were combined using the Fisher’s combined probability test. To analyze larval locomotion data, Mann-Whitney rank-sum tests were performed with MatLab. One-way ANOVA with Bonferroni correction was used to analyze Caz and Xrp1 mRNA and protein levels as data displayed normal distribution and equal variance. All images were assembled in figure panels using the Adobe Illustrator CS5 software.
Online supplemental material
Fig. S1 shows generation and characterization of Xrp1 mutant and transgenic lines. Fig. S2 shows that heterozygosity for Xrp1 does not rescue the adult eclosion defect of TBPH mutant flies. Fig. S3 shows larval locomotion phenotypes and binding of WT or AT-hook mutant Xrp1 to polytene chromosomes. Figs. S4 shows characterization of the caz-Xrp1 genetic interaction. Fig. S5 shows that heterozygosity for Xrp1 mitigates gene expression dysregulation in caz mutant CNS. Table S1 lists human homologues of Xrp1. Tables S2, S3, and S4 list Xrp1long, Xrp1Short, and Caz-interacting proteins, respectively. Table S5 lists RNA-seq results. Table S6 lists human AT-hook genes. Table S7 lists oligonucleotide primers.
Supplementary Material
Acknowledgments
We thank B. McCabe (École Polytechnique Fédérale de Lausanne Brain Mind Institute, Lausanne, Switzerland), P. Verstreken (VIB–KU Leuven, Leuven, Belgium), F. Feiguin (International Centre for Genetic Engineering and Biotechnology, Trieste, Italy), and U. Pandey (University of Pittsburgh, Pittsburgh, PA) for fly lines, D.C. Rio (University of California, Berkeley, Berkeley, CA) for the Xrp1 antibody, S. Bogdan for technical advice on S2 cultures, M. Been for help with figure design, and F. Schmalbein, Y. Schmidt, and A. Nolte for excellent technical assistance.
This work is supported by funding to E. Storkebaum from the state North Rhine Westphalia, the Max Planck Society, The Bruno and Ilse Frick Foundation for ALS Research, the Minna-James-Heineman-Stiftung, the French Muscular Dystrophy Association, the US Muscular Dystrophy Association, the EU Joint Programme–Neurodegenerative Disease Research (grants ZonMW 733051075 [TransNeuro] and ZonMW 733051073 [LocalNMD]), and a European Research Council consolidator grant (ERC-2017-COG 770244). M. Mallik was supported by a Humboldt Foundation research fellowship, C.B. Hug by a fellowship from the International Max Planck Research School, and J. Bussmann by a fellowship from the Cell Dynamics and Disease (CEDAD) graduate school. Stocks were obtained from the BDSC (National Institutes of Health grant P40OD018537).
The authors declare no competing financial interests.
Author contributions: M. Mallik and E. Storkebaum designed the research; M. Mallik, M. Catinozzi, M. Wagner, L. Zhang, J. Bussmann, J. Bittern, S. Mersmann, H.C.A. Drexler, and E. Storkebaum performed the research; C.B. Hug, M.A. Huynen, and J.M. Vaquerizas performed bioinformatic analysis; M. Mallik, M. Catinozzi, L. Zhang, and E. Storkebaum analyzed data; all authors commented on the paper; M. Mallik, C.B. Hug, and E. Storkebaum made figures; M. Mallik and E. Storkebaum wrote the paper with contributions from C.B. Hug and J.M. Vaquerizas.
References
- Akdemir F., Christich A., Sogame N., Chapo J., and Abrams J.M.. 2007. p53 directs focused genomic responses in Drosophila. Oncogene. 26:5184–5193. 10.1038/sj.onc.1210328 [DOI] [PubMed] [Google Scholar]
- Alexa A., Rahnenführer J., and Lengauer T.. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 22:1600–1607. 10.1093/bioinformatics/btl140 [DOI] [PubMed] [Google Scholar]
- Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., and Zoghbi H.Y.. 1999. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23:185–188. 10.1038/13810 [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., and Huber W.. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S.A., Chen L., Wilkins A.D., Yu P., Lichtarge O., and Zoghbi H.Y.. 2013. An AT-hook domain in MeCP2 determines the clinical course of Rett syndrome and related disorders. Cell. 152:984–996. 10.1016/j.cell.2013.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky M.H., Weinert B.T., Tsang G., Rong Y.S., McGinnis N.M., Golic K.G., Rio D.C., and Rubin G.M.. 2004. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol. Cell. Biol. 24:1219–1231. 10.1128/MCB.24.3.1219-1231.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E., and Baralle F.E.. 2010. The multiple roles of TDP-43 in pre-mRNA processing and gene expression regulation. RNA Biol. 7:420–429. 10.4161/rna.7.4.12205 [DOI] [PubMed] [Google Scholar]
- Chesi A., Staahl B.T., Jovičić A., Couthouis J., Fasolino M., Raphael A.R., Yamazaki T., Elias L., Polak M., Kelly C., et al. . 2013. Exome sequencing to identify de novo mutations in sporadic ALS trios. Nat. Neurosci. 16:851–855. 10.1038/nn.3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C.M., Vilain S., Langen M., Van Kelst S., De Geest N., Yan J., Verstreken P., and Hassan B.A.. 2009. Conditional mutagenesis in Drosophila. Science. 324:54 10.1126/science.1168275 [DOI] [PubMed] [Google Scholar]
- Cook R.K., Christensen S.J., Deal J.A., Coburn R.A., Deal M.E., Gresens J.M., Kaufman T.C., and Cook K.R.. 2012. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 13:R21 10.1186/gb-2012-13-3-r21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J., Hart M.P., Shorter J., DeJesus-Hernandez M., Erion R., Oristano R., Liu A.X., Ramos D., Jethava N., Hosangadi D., et al. . 2011. A yeast functional screen predicts new candidate ALS disease genes. Proc. Natl. Acad. Sci. USA. 108:20881–20890. 10.1073/pnas.1109434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J., Hart M.P., Erion R., King O.D., Diaz Z., Nakaya T., Ibrahim F., Kim H.J., Mojsilovic-Petrovic J., Panossian S., et al. . 2012. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum. Mol. Genet. 21:2899–2911. 10.1093/hmg/dds116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., and Mann M.. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., and Mann M.. 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 13:2513–2526. 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Cruz S., and Cleveland D.W.. 2011. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr. Opin. Neurobiol. 21:904–919. 10.1016/j.conb.2011.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangli A., and Bautz E.K.. 1983. Differential distribution of nonhistone proteins from polytene chromosomes of Drosophila melanogaster after heat shock. Chromosoma. 88:201–207. 10.1007/BF00285621 [DOI] [PubMed] [Google Scholar]
- Davidson Y.S., Robinson A.C., Hu Q., Mishra M., Baborie A., Jaros E., Perry R.H., Cairns N.J., Richardson A., Gerhard A., et al. . 2013. Nuclear carrier and RNA-binding proteins in frontotemporal lobar degeneration associated with fused in sarcoma (FUS) pathological changes. Neuropathol. Appl. Neurobiol. 39:157–165. 10.1111/j.1365-2990.2012.01274.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis M.J., Roche S., Cho M.J., Beall E., Min B., Panganiban R.P., and Rio D.C.. 2016. Drosophila IRBP bZIP heterodimer binds P-element DNA and affects hybrid dysgenesis. Proc. Natl. Acad. Sci. USA. 113:13003–13008. 10.1073/pnas.1613508113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickenhaus M., Wagner M., Mallik M., Catinozzi M., and Storkebaum E.. 2015. Highly efficient cell-type-specific gene inactivation reveals a key function for the Drosophila FUS homolog cabeza in neurons. Sci. Rep. 5:9107 10.1038/srep09107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitcho M.A., Baloh R.H., Chakraverty S., Mayo K., Norton J.B., Levitch D., Hatanpaa K.J., White C.L. III, Bigio E.H., Caselli R., et al. . 2008. TDP-43 A315T mutation in familial motor neuron disease. Ann. Neurol. 63:535–538. 10.1002/ana.21344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler A.R., Donnelly C.J., Periz G., Simko E.A., Shaw P.G., Kim M.S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R., et al. . 2014. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 507:195–200. 10.1038/nature13124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immanuel D., Zinszner H., and Ron D.. 1995. Association of SARFH (sarcoma-associated RNA-binding fly homolog) with regions of chromatin transcribed by RNA polymerase II. Mol. Cell. Biol. 15:4562–4571. 10.1128/MCB.15.8.4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki S., Masuda A., Fujioka Y., Iguchi Y., Katsuno M., Shibata A., Urano F., Sobue G., and Ohno K.. 2012. Position-dependent FUS-RNA interactions regulate alternative splicing events and transcriptions. Sci. Rep. 2:529 10.1038/srep00529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.O., Pioro E.P., Boehringer A., Chia R., Feit H., Renton A.E., Pliner H.A., Abramzon Y., Marangi G., Winborn B.J., et al. ITALSGEN . 2014. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat. Neurosci. 17:664–666. 10.1038/nn.3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E., Valdmanis P.N., Dion P., Spiegelman D., McConkey B.J., Vande Velde C., Bouchard J.P., Lacomblez L., Pochigaeva K., Salachas F., et al. . 2008. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 40:572–574. 10.1038/ng.132 [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S.L.. 2013a TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Kim N.C., Wang Y.D., Scarborough E.A., Moore J., Diaz Z., MacLea K.S., Freibaum B., Li S., Molliex A., et al. . 2013b Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 495:467–473. 10.1038/nature11922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino Y., Washizu C., Kurosawa M., Yamada M., Miyazaki H., Akagi T., Hashikawa T., Doi H., Takumi T., Hicks G.G., et al. . 2015. FUS/TLS deficiency causes behavioral and pathological abnormalities distinct from amyotrophic lateral sclerosis. Acta Neuropathol. Commun. 3:24 10.1186/s40478-015-0202-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski T.J. Jr., Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T., et al. . 2009. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 323:1205–1208. 10.1126/science.1166066 [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C., Polymenidou M., Hutt K.R., Vu A.Q., Baughn M., Huelga S.C., Clutario K.M., Ling S.C., Liang T.Y., Mazur C., et al. . 2012. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat. Neurosci. 15:1488–1497. 10.1038/nn.3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanson N.A. Jr., Maltare A., King H., Smith R., Kim J.H., Taylor J.P., Lloyd T.E., and Pandey U.B.. 2011. A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum. Mol. Genet. 20:2510–2523. 10.1093/hmg/ddr150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.B., Chen H.J., Peres J.N., Gomez-Deza J., Attig J., Stalekar M., Troakes C., Nishimura A.L., Scotter E.L., Vance C., et al. . 2013. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Reports. 5:1178–1186. 10.1016/j.celrep.2013.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., and Bork P.. 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46(D1):D493–D496. 10.1093/nar/gkx922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling S.C., Polymenidou M., and Cleveland D.W.. 2013. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 79:416–438. 10.1016/j.neuron.2013.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf C.E., and Wassarman D.A.. 2006. DNA binding properties of TAF1 isoforms with two AT-hooks. J. Biol. Chem. 281:30015–30023. 10.1074/jbc.M606289200 [DOI] [PubMed] [Google Scholar]
- Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. . 2006. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 314:130–133. 10.1126/science.1134108 [DOI] [PubMed] [Google Scholar]
- Neumann M., Bentmann E., Dormann D., Jawaid A., DeJesus-Hernandez M., Ansorge O., Roeber S., Kretzschmar H.A., Munoz D.G., Kusaka H., et al. . 2011. FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain. 134:2595–2609. 10.1093/brain/awr201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehues S., Bussmann J., Steffes G., Erdmann I., Köhrer C., Sun L., Wagner M., Schäfer K., Wang G., Koerdt S.N., et al. . 2015. Impaired protein translation in Drosophila models for Charcot-Marie-Tooth neuropathy caused by mutant tRNA synthetases. Nat. Commun. 6:7520 10.1038/ncomms8520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks A.L., Cook K.R., Belvin M., Dompe N.A., Fawcett R., Huppert K., Tan L.R., Winter C.G., Bogart K.P., Deal J.E., et al. . 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36:288–292. 10.1038/ng1312 [DOI] [PubMed] [Google Scholar]
- Pfeiffer B.D., Ngo T.T., Hibbard K.L., Murphy C., Jenett A., Truman J.W., and Rubin G.M.. 2010. Refinement of tools for targeted gene expression in Drosophila. Genetics. 186:735–755. 10.1534/genetics.110.119917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Ishihama Y., and Mann M.. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75:663–670. 10.1021/ac026117i [DOI] [PubMed] [Google Scholar]
- Reeves R. 2010. Nuclear functions of the HMG proteins. Biochim. Biophys. Acta. 1799:3–14. 10.1016/j.bbagrm.2009.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risse B., Thomas S., Otto N., Löpmeier T., Valkov D., Jiang X., and Klämbt C.. 2013. FIM, a novel FTIR-based imaging method for high throughput locomotion analysis. PLoS One. 8:e53963 10.1371/journal.pone.0053963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risse B., Otto N., Berh D., Jiang X., and Klämbt C.. 2014. FIM imaging and FIMtrack: two new tools allowing high-throughput and cost effective locomotion analysis. J. Vis. Exp. 94:e52207 10.3791/52207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risse B., Berh D., Otto N., Klämbt C., and Jiang X.. 2017. FIMTrack: An open source tracking and locomotion analysis software for small animals. PLOS Comput. Biol. 13:e1005530 10.1371/journal.pcbi.1005530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scekic-Zahirovic J., Sendscheid O., El Oussini H., Jambeau M., Sun Y., Mersmann S., Wagner M., Dieterlé S., Sinniger J., Dirrig-Grosch S., et al. . 2016. Toxic gain of function from mutant FUS protein is crucial to trigger cell autonomous motor neuron loss. EMBO J. 35:1077–1097. 10.15252/embj.201592559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scekic-Zahirovic J., Oussini H.E., Mersmann S., Drenner K., Wagner M., Sun Y., Allmeroth K., Dieterlé S., Sinniger J., Dirrig-Grosch S., et al. . 2017. Motor neuron intrinsic and extrinsic mechanisms contribute to the pathogenesis of FUS-associated amyotrophic lateral sclerosis. Acta Neuropathol. 133:887–906. 10.1007/s00401-017-1687-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J.C., Ebmeier C.C., Podell E.R., Heimiller J., Taatjes D.J., and Cech T.R.. 2012. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev. 26:2690–2695. 10.1101/gad.204602.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J.C., Cech T.R., and Parker R.R.. 2015. Biochemical Properties and Biological Functions of FET Proteins. Annu. Rev. Biochem. 84:355–379. 10.1146/annurev-biochem-060614-034325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Lyashchenko A.K., Lu L., Nasrabady S.E., Elmaleh M., Mendelsohn M., Nemes A., Tapia J.C., Mentis G.Z., and Shneider N.A.. 2016. ALS-associated mutant FUS induces selective motor neuron degeneration through toxic gain of function. Nat. Commun. 7:10465 10.1038/ncomms10465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiihashi G., Ito D., Yagi T., Nihei Y., Ebine T., and Suzuki N.. 2016. Mislocated FUS is sufficient for gain-of-toxic-function amyotrophic lateral sclerosis phenotypes in mice. Brain. 139:2380–2394. 10.1093/brain/aww161 [DOI] [PubMed] [Google Scholar]
- Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J.C., Williams K.L., Buratti E., et al. . 2008. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 319:1668–1672. 10.1126/science.1154584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk R., Wanschers B.F., Cuypers T.D., Esseling J.J., Riemersma M., van den Brand M.A., Gloerich J., Lasonder E., van den Heuvel L.P., Nijtmans L.G., and Huynen M.A.. 2012. Iterative orthology prediction uncovers new mitochondrial proteins and identifies C12orf62 as the human ortholog of COX14, a protein involved in the assembly of cytochrome c oxidase. Genome Biol. 13:R12 10.1186/gb-2012-13-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A.Y., Riley T.R., Coady T., Bussemaker H.J., and Manley J.L.. 2012. TLS/FUS (translocated in liposarcoma/fused in sarcoma) regulates target gene transcription via single-stranded DNA response elements. Proc. Natl. Acad. Sci. USA. 109:6030–6035. 10.1073/pnas.1203028109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.P., Brown R.H. Jr., and Cleveland D.W.. 2016. Decoding ALS: from genes to mechanism. Nature. 539:197–206. 10.1038/nature20413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurui-Nishimura N., Nguyen T.Q., Katsuyama T., Minami T., Furuhashi H., Oshima Y., and Kurata S.. 2013. Ectopic antenna induction by overexpression of CG17836/Xrp1 encoding an AT-hook DNA binding motif protein in Drosophila. Biosci. Biotechnol. Biochem. 77:339–344. 10.1271/bbb.120756 [DOI] [PubMed] [Google Scholar]
- Turlure F., Maertens G., Rahman S., Cherepanov P., and Engelman A.. 2006. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 34:1653–1665. 10.1093/nar/gkl052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udagawa T., Fujioka Y., Tanaka M., Honda D., Yokoi S., Riku Y., Ibi D., Nagai T., Yamada K., Watanabe H., et al. . 2015. FUS regulates AMPA receptor function and FTLD/ALS-associated behaviour via GluA1 mRNA stabilization. Nat. Commun. 6:7098 10.1038/ncomms8098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C., Rogelj B., Hortobágyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., et al. . 2009. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 323:1208–1211. 10.1126/science.1165942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilain S., Vanhauwaert R., Maes I., Schoovaerts N., Zhou L., Soukup S., da Cunha R., Lauwers E., Fiers M., and Verstreken P.. 2014. Fast and efficient Drosophila melanogaster gene knock-ins using MiMIC transposons. G3 (Bethesda). 4:2381–2387. 10.1534/g3.114.014803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson C., Myakishev M., Acharya A., Mir A.A., Moll J.R., and Bonovich M.. 2002. Classification of human B-ZIP proteins based on dimerization properties. Mol. Cell. Biol. 22:6321–6335. 10.1128/MCB.22.18.6321-6335.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J.A., Côté R.G., Csordas A., Dianes J.A., Fabregat A., Foster J.M., Griss J., Alpi E., Birim M., Contell J., et al. . 2013. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41(D1):D1063–D1069. 10.1093/nar/gks1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.W., Brent J.R., Tomlinson A., Shneider N.A., and McCabe B.D.. 2011. The ALS-associated proteins FUS and TDP-43 function together to affect Drosophila locomotion and life span. J. Clin. Invest. 121:4118–4126. 10.1172/JCI57883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Gal J., Chen J., and Zhu H.. 2014. Self-assembled FUS binds active chromatin and regulates gene transcription. Proc. Natl. Acad. Sci. USA. 111:17809–17814. 10.1073/pnas.1414004111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann L., Stephens A., Nam S.Z., Rau D., Kubler J., Lozajic M., Gabler F., Soding J., Lupas A.N., and Alva V.. 2018. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 430:2237–2243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.