Abstract
The nuclear pore complex (NPC) mediates all macromolecular transport across the nuclear envelope. In higher eukaryotes that have an open mitosis, NPCs assemble at two points in the cell cycle: during nuclear assembly in late mitosis and during nuclear growth in interphase. How the NPC, the largest nonpolymeric protein complex in eukaryotic cells, self‐assembles inside cells remained unclear. Recent studies have started to uncover the assembly process, and evidence has been accumulating that postmitotic and interphase NPC assembly use fundamentally different mechanisms; the duration, structural intermediates, and regulation by molecular players are different and different types of membrane deformation are involved. In this Review, we summarize the current understanding of these two modes of NPC assembly and discuss the structural and regulatory steps that might drive the assembly processes. We furthermore integrate understanding of NPC assembly with the mechanisms for rapid nuclear growth in embryos and, finally, speculate on the evolutionary origin of the NPC implied by the presence of two distinct assembly mechanisms.
Keywords: cell cycle, mitosis, nuclear assembly
Abbreviations
AL, annulate lamellae
CDK1, cyclin‐dependent kinase 1
cNLS, classical nuclear localization signal
EM, electron microscopy
ER, endoplasmic reticulum
IBB, importin‐β‐binding
INM, inner nuclear membrane
LBR, lamin B receptor
LINC, linker of nucleoskeleton and cytoskeleton
NE, nuclear envelope
NPC, nuclear pore complex
ONM, outer nuclear membrane
SNARE, soluble N‐ethylmaleimide‐sensitive factor activating protein receptor
Eukaryotic cells are characterized by distinct nuclear and cytoplasmic compartments which are separated by the nuclear envelope (NE). The NE is penetrated by nuclear pore complexes (NPCs) which allow exchange of macromolecules between these two compartments. Proteins destined for nuclear function, such as histones and transcriptional factors, are imported through the NPC from the cytoplasm where they are synthesized. At the same time, mRNA, tRNA, and ribosomal subunits are exported through the NPC from the nucleus. The selective macromolecular transport across the NE is an integral part of eukaryotic cell functions, and the dysfunction of the NPC has been associated with many diseases, such as cancer, disorders of immune, and nervous system 1.
The NE consists of two lipid bilayers (outer and inner nuclear membrane; ONM and INM). The ONM is facing the cytoplasm and is continuous with endoplasmic reticulum (ER), whereas the INM is facing the nucleoplasm and interacts with chromatin and the nuclear lamina via integral membrane proteins. The INM proteins are known to be involved in chromosome organization and gene expression and play important roles especially in cell differentiation and development 2, 3. The linker of nucleoskeleton and cytoskeleton (LINC) complex forms a bridge connecting the ONM and the INM in the perinuclear space, which stabilizes the NE against cytoplasmic forces and facilitates nuclear positioning 4.
The NPC is the sole gate spanning the ONM and INM and is the largest nonpolymeric protein complex in eukaryotic cells with an estimated mass of 60 MDa in yeast and 120 MDa in vertebrates. Despite the rather large difference in mass, the unique overall architecture and transport mechanism of the NPC are well conserved between species 5, 6. Unlike protein transport through single‐membrane gates in the ER or mitochondria, the NPC can mediate transport of single proteins and very large complexes without unfolding their structure through its very large transport channel and unique transport selectivity mechanism 7, 8. To stabilize such a large channel, the NPC has an eight‐fold rotationally symmetric architecture which consists of several biochemically and ultra‐structurally defined substructures: the cytoplasmic filaments, the cytoplasmic and nuclear rings, the inner pore ring, the central transporter region, and the nuclear basket 9, 10, 11. Individual NPCs are composed of a total of 500–1000 polypeptides, comprising multiple copies (8–64) of around 30 different proteins termed nucleoporins (Nups) 12, 13. Scaffold Nups have been reported to be stably associated with the NPC with almost no turnover during the cell cycle, whereas some Nups localizing at the NPC periphery have been shown to interact transiently with the NPC 14, 15, 16.
In metazoan cells which undergo open mitosis, the NPCs disassemble and the NE breaks down at mitosis onset, and the cytoplasm and the nucleoplasm mix. NPCs and the NE reassemble during mitotic exit (called postmitotic assembly) (Fig. 1), and their rapid reformation is essential for establishing a functional nucleus in the daughter cell. The NPC also assembles during nuclear growth in interphase (called interphase assembly) and the number of the NPCs is almost doubled at the end of G2 phase for the next round of cell division in proliferating cells (Fig. 1). Over the last decade, it has been revealed that the postmitotic and interphase NPC assembly mechanisms are fundamentally different 4, 17, 18, 19. During mitotic exit, a large number of NPCs assemble within a few minutes concomitant with nuclear membrane sealing 20, 21, 22, 23, 24, whereas in interphase NPC assembly, it is sporadic, takes about an hour, and requires deformation and fusion of the intact double‐membraned nuclear boundary 25, 26, 27. Here, we will review what is known on the assembly mechanisms and discuss what drives them and how they are regulated. The two distinct assembly mechanisms provide very interesting connections to rapid nuclear growth in embryos and have fundamental implications to rationalize the evolutionary origin of the NPC.
Figure 1.
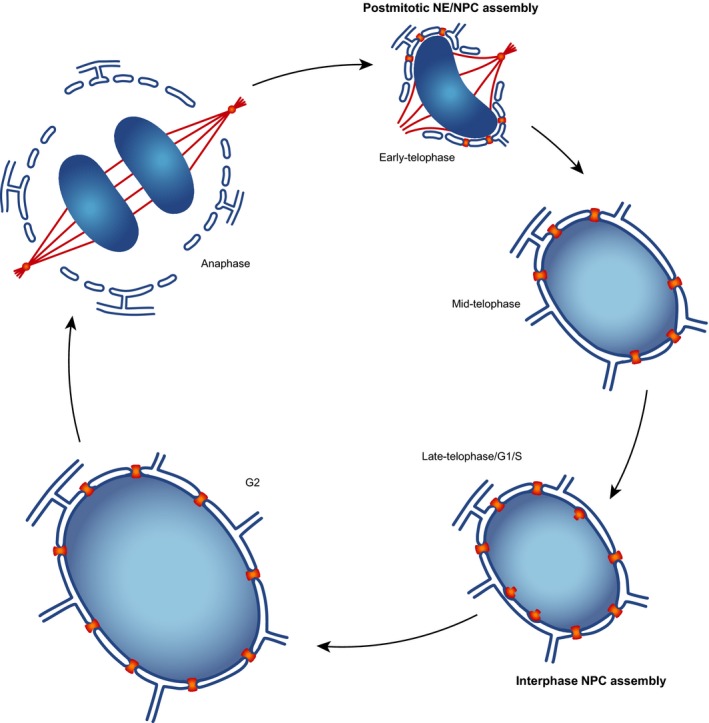
Nuclear pore assembly during cell cycle. During anaphase and telophase, the nuclear membranes that are absorbed in the endoplasmic reticulum (ER, blue) at metaphase assemble on chromatin (blue gradient) together with the nuclear pore complexes (NPCs, orange). The nuclear envelope (NE) assembly is delayed in the chromosome regions next to the spindle pole and the central spindle area (called ‘core region’) due to dense spindle microtubules (red lines) on the DNA surface. During nuclear growth after telophase, the NPCs assemble de novo into the double membrane barrier of the NE.
Postmitotic NE and NPC assembly
Regulation of mitotic exit
At the onset of mitosis, mitotic kinases such as Cyclin‐dependent kinase 1 (CDK1)/Cyclin B phosphorylate Nups, lamins, and INM proteins, which triggers the disassembly of the NPC and the nuclear lamina and the release of chromatin from the NE. For NPC disassembly, Nup98 has been shown to play a key role. Kinetic observations in live cells have shown that Nup98 is among the first Nups that dissociates from the NPC in prometaphase 21, 28, and the mitotic phosphorylation of Nup98 is required for its release from the NPC and disruption of the NPC permeability barrier 29. Since Nup98 interacts with inner pore ring components Nups205, 155, and 53, a subcomplex linked to both the transmembrane Nup NDC1 and the central channel subcomplex consisting of Nups62, 58, and 54 30, 31, 32, 33; Nup98 can be speculated to function as an important linker that connects the inner ring, central channel, and the nuclear membrane. Its disengagement from the NPC by mitotic phosphorylation would thus be an efficient way to trigger NPC disassembly. In addition to Nup98, Nup53, the Nup107‐160 complex, and transmembrane Nups NDC1, Pom121 and GP210 are also known to be phosphorylated at mitosis onset. However, how their phosphorylation modulates their functions remains to be studied 18, 34. During mitotic exit, the phosphorylation events that drive NPC disassembly are reverted due to the inactivation of mitotic kinases and the activation of phosphatases such as protein phosphatase 1 (PP1), which allows the NE to rebind to chromatin and Nups as well as lamins to reassemble 34, 35.
The assembly of the NE and the NPC is also regulated by small GTPase Ran. The GTP‐bound form of Ran (RanGTP) is enriched on mitotic chromosomes as a consequence of the activity of chromatin‐bound Ran guanine‐exchange factor RCC1 36. Nuclear transport receptors importin β1 and β2 bind to key mitotic proteins such as spindle assembly factors as well as lamin B, lamin B receptor (LBR) and Nups including Nups358, 214, 153, 98, and 62, the Nup107‐160 complex and ELYS, preventing them from interacting with other proteins 37, 38, 39, 40. The binding of RanGTP to the importins allosterically reduces their binding affinity to these proteins 41, allowing them to dissociate from importins in the vicinity of mitotic chromatin. The interaction of ELYS, a Nup specific to the nuclear ring of the NPC and an essential player in postmitotic NPC assembly (discussed in the following sections 2.3 and 2.4) 42, 43, 44, with mitotic chromatin has been shown to be regulated positively by RanGTP and negatively by importin β1 45, 46. In general, RanGTP displacement of importins spatially guides both NE and NPC assembly during mitotic exit by allowing the formation of key protein complexes in the proximity of chromatin while it remains inhibited in the cytosol.
Mechanism of nuclear membrane coverage of chromosomes during mitotic exit
Upon nuclear disassembly, INM proteins are detached from chromatin by mitotic phosphorylation and are absorbed in the mitotic ER together with the nuclear membranes 20, 22, 47. Until anaphase, the mitotic ER is largely excluded from chromosomes and the central spindle area by the active mechanism of ER clearance from chromatin by microtubule‐binding ER proteins REEP3/4 as well as the inhibition of microtubule‐binding competence of ER proteins STIM1 and CLIMP63 by mitotic phosphorylation 48, 49, 50. During chromosome segregation in anaphase, the masses of separated sister chromatids move to the opposite poles of the spindle and while the ‘core’ of these masses initially remains shielded from membranes by the persisting central spindle and kinetochore microtubules, their periphery (or ‘noncore’) starts to get in contact with the ER membranes (Fig. 1, early telophase).
How exactly the NE reforms from mitotic ER had been unclear and a matter of debate, mainly due to the resolution limitation of live cell fluorescence microscopy that is necessary for capturing the dynamic shaping of subdiffraction ER cisternae and tubules, as well as the difficulty to preserve the native membrane structure in in vitro assays. The structure of the ER in interphase and mitosis has been studied systematically in different cell types by electron microscopy (EM) 51. It has revealed that the ER undergoes structural reorganization from interphase to mitosis from sheet morphology to more fenestrated and tubular forms. It has also shown that the architecture of mitotic ER varies in different cell types: while mitotic HeLa, Huh‐7, NRK‐52E, and Vero cells contain multiple layers of fenestrated ER sheets, CHO‐K1 cells have a more extensively tubulated network 51. These differences may depend on the abundance of ER membranes as well as of ER‐shaping proteins, including reticulons, GTPases atlastins, and the protein lunapark 52, 53. More recently, a time course of EM reconstructions of entire HeLa cells from early to late anaphase by correlative live imaging with large volume EM could capture the dynamic process of covering chromosomes with ER‐derived nuclear membranes at nanometer resolution 24. This study showed that the NE forms from highly fenestrated ER sheets that contain a large number of discontinuities whose diameter shrinks as NE assembly proceeds. NE reformation is very fast, and most of chromosome surface is covered by the membrane within 2–4 min 24. Although nuclear membranes have been thought to be supplied by lateral expansion of ER after initial attachment to the chromatin surface 35, 54 (Fig. 2, lateral expansion model), the new EM data make it equally possible that multiple layers of ER sheets are stacked on top of each other on the chromosome surface during telophase (Fig. 2, cisternal stacking model), especially in the periphery of the chromosome mass where ER sheets may become locally packed by the ingressing cleavage furrow. Lateral expansion and cisternal stacking are not mutually exclusive mechanisms for the sealing of chromosomes by nuclear membrane and current evidence would indeed suggest that both occur during rapid assembly of the NE during mitotic exit.
Figure 2.
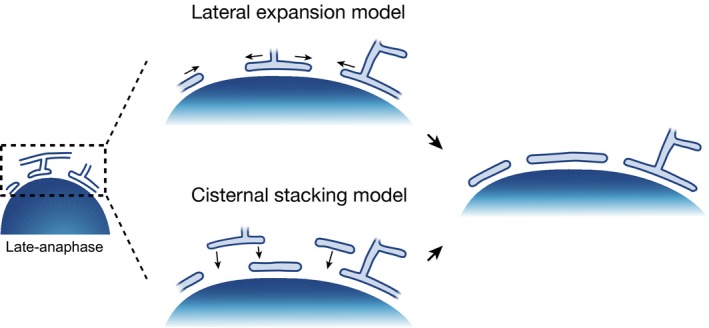
Models for the NE assembly during chromosome segregation in anaphase. The nuclear membranes will be supplied by the lateral expansion of the ER (blue) on the chromatin surface (blue gradient) and/or by the cisternal stacking of the ER.
What drives membrane coverage of chromosome during mitotic exit? ER‐chromatin contacts are initiated at the peripheral edge of the chromosome. When the ER membranes contact the chromosome surface, INM proteins including LBR, emerin, and Sun2 which can bind to DNA and/or lamin are locally enriched via lateral diffusion through the ER and retention at the membrane‐chromatin contact region 20, 22, 47, 55, 56. By contrast, NE assembly is delayed in the chromosome regions next to the spindle pole and the central spindle area due to dense spindle microtubules on the DNA surface (Fig. 1, early telophase). This core region is specifically enriched with emerin, LAP‐2β and lamin A, and the chromatin‐binding protein BAF plays a key role in establishing this transient chromosome subdomain in mid/late anaphase 55, 57. Recent studies have shown that members of endosomal sorting complex required for transport‐III (ESCRT‐III) complex and microtubule severing ATPase spastin localize where the reforming NE starts to enclose microtubules in the core region, and function cooperatively in microtubule disassembly to allow NE sealing from the periphery across the core domain in telophase 58, 59. Other membrane fusion mediators, SNARE (soluble N‐ethylmaleimide‐sensitive factor activating protein receptor) proteins and the atlastin family GTPases have been shown to be required to form the closed NE in vitro, and may also contribute to the NE sealing in intact cells 60, 61, 62. In general, the driving force for nuclear membrane coverage appears to be the multiple interactions between INM proteins with chromatin that become activated by postmitotic dephosphorylation and/or RanGTP‐induced release from importins, as well as the progressive accessibility of the chromosome surface for ER membranes made possible by retraction and active removal of spindle microtubules. It is very likely that homotypic fusion of ER membranes during cisternal stacking and/or lateral expansion further promotes nuclear membrane sealing.
Mechanism of NPC assembly in the reforming nuclear membrane
The coordinated reassembly of NPCs concomitant with the nuclear membrane sealing around chromosomes is essential to establish a transport‐competent nucleus after mitosis. Studies using in vitro assembled nuclei with Xenopus egg extracts have defined a step‐wise process of the Nup recruitment to the DNA surface 30, 63, 64, 65, 66. In vitro, the nuclear ring component ELYS first binds to chromatin in mid‐anaphase and recruits the nuclear and cytoplasmic ring components Nup107‐160 complex (also called Y‐shaped complex), which in turn allows the recruitment of transmembrane Nups Ndc1 and Pom121 as well as the membrane‐associated Nup53. An inner ring component Nup155 is then recruited through the interaction with Nup53, which is required for recruiting the other inner ring components Nups205, 188, and 93, and the subsequent recruitment of the central channel component Nup62 complex 30, 63, 64, 65, 66. The assembly order of Nups has also been studied by immunostaining of fixed human cells at different cell‐cycle stages 45, 67, 68. They have shown that ELYS, the Nup107‐160 complex, and the nucleoplasmic Nup153 are recruited in late anaphase and that the cytoplasmic Nup214 and the nuclear basket component TPR are recruited later at telophase. In addition to these studies, live cell imaging in both rat and human cells has directly visualized the dynamic process of Nup recruitment to the NE and shown that postmitotic NPC assembly indeed proceeds by sequential addition of Nups in an overall similar order to the one proposed by the in vitro and immuno‐localization studies 20, 21, 22, 23, 24.
While the order of postmitotic Nup recruitment to the chromosome surface is well defined with few exceptions, which structural intermediates are formed in order to build a mature NPC remained mysterious for a long time. The first suggestions for what such assembly intermediates (so called prepores) might look like came from scanning EM. Here, the outer surface of nuclei assembled in vitro using Xenopus egg extracts treated with inhibitors or in nuclei isolated from cycling Drosophila embryos were observed by scanning EM 69, 70, and different sized intermediates were observed 70, but it remained difficult to correlate them with the dynamic molecular progression, which starts on the nuclear side, which remained invisible in these studies. More recently it became possible to observe the three‐dimensional (3D) ultrastructure of precisely staged assembling NPCs in human cells by directly correlating live imaging with high‐resolution electron tomography 24. This approach has revealed that the first prepores form in small membrane holes about half the size of mature NPCs. Such prepores contained dense material in the center of the holes as well as nuclear ring‐like structures underneath them. The small membrane hole then dilated as the inner ring, and the cytoplasmic ring formed and the density of central channel increased 24. The molecular and structural progression of postmitotic NPC assembly described in this section is summarized in Fig. 3A.
Figure 3.

Two distinct assembly mechanisms of the NPC. (A) Postmitotic NPC assembly proceeds by a radial dilation of small membrane openings 24. The initial prepore contains dense material in the center of the membrane hole as well as the nuclear ring. It dilates radially and obtains active transport competence during the inner ring and the cytoplasmic ring complex assembly. The central channel continues to maturate even after the membrane dilation stops, and the NPC gradually obtains permeability barrier function against small proteins. Molecular players involved in individual steps of the assembly are indicated below. Possible molecular requirements are shown in gray. ONM, outer nuclear membrane; INM, inner nuclear membrane. (B) Interphase NPC assembly proceeds by an inside‐out extrusion of the nuclear membrane 27 . The intermediate is a dome‐shaped evagination of the INM, that contains the nuclear ring structure underneath the INM from the beginning and grows in size vertically and laterally until it fuses with the flat ONM. Possible molecular players in individual steps of the assembly are shown as in (A).
Possible molecular mechanisms for postmitotic NPC assembly
First of all, what are the molecular triggers for postmitotic NPC assembly? In vitro and in vivo studies have revealed that artificial tethering RCC1 and ELYS to DNA in Xenopus egg extracts or tethering ELYS to the surface of nucleosome‐depleted chromosomes in mouse zygotes is sufficient to initiate NPC assembly 71, 72. Thus, the minimal requirements for starting NPC assembly appear to be RanGTP promoted release of assembly factors, very likely early recruited Nups from importins, and ELYS localization at the nuclear periphery. Interestingly, ELYS localization to chromatin requires demethylation of histone H3 by the Lysine‐specific demethylase LSD1 in vitro, but the effect of LSD1 is mild in human cells 73, and to which extent histone modifications regulate NPC assembly in vivo remains to be examined further.
Recent EM observations of assembling NEs and NPCs have shown that the first density containing membrane discontinuities were small NE holes about the half the size of mature NPCs 24. Interestingly, similarly sized small holes were already present in the ER cisternae when they first made contact with chromosomes, suggesting that postmitotic NPC assembly uses pre‐existing shrinking ER fenestrations, rather than relying on de novo membrane fusion after chromosomes are sealed with a continuous double membrane 24. How then is the localization of ELYS on the chromosome surface coordinated with the positions of these small prepore holes in the ER as it contacts chromosomes? It is possible that ELYS localization precedes membrane contacts and seeds local Nup complexes that stop the fusion of the shrinking fenestrae in the ER‐derived nuclear membranes locally, to initiate NPC assembly. Consistent with this idea, the initial prepores contain not only nuclear ring‐like density (ELYS is a nuclear ring component) but also dense material in the center of membrane holes, which may be additional Nups recruited by ELYS and/or the proteins localizing at the curvature of the small hole (Fig. 3A). At this initial stage of NPC assembly in mid‐anaphase only, nuclear ring components have been reported to be recruited on the NE 20, 21, 23, 67. Possible components of the central dense material include Ndc1, Nup53, and Nup155, as well as reticulon and DP1/Yop1p family proteins that localize at the tip of ER membranes, which have a similar membrane curvature 56, 74. These proteins could stall the membrane hole shrinkage at the site of NPC assembly as defined by a local accumulation of ELYS and then recruit additional Nups.
Once initiated, prepores grow in size until they reach the full diameter of a mature NPC. What drives this prepore dilation? Since inner ring formation and hole dilation occur at the same time with similar kinetics 24, the recruitment of the inner ring components and their self‐assembly into eight regular subunits might be the driving force for pore dilation. In addition, the components of the central channel are being recruited to the pore following the inner ring assembly 21. The central channel components Nups62, 58, and 54 carry intrinsically disordered domains rich in phenylalanine‐glycine (FG) motifs and have been suggested to form a hydrophobic meshwork through FG‐FG interactions 7, 8, 75. A high concentration of intrinsically disordered domains in a small membrane hole may trigger additional self‐assembly steps that may further contribute to prepore dilation.
When do the assembling NPCs become transport competent? Live imaging studies have demonstrated that the nucleus obtains active nuclear import competence before all the Nups are recruited. Proteins containing classical nuclear localization signal (cNLS) or the importin β‐binding (IBB) domain of importin alpha start to be imported into the nucleus at early telophase, when the cytoplasmic Nup214, the cytoplasmic filament component Nup358, and the nuclear basket component TPR are not yet accumulated on the NE 20, 21, 23, 27, 68 (Fig. 3A), suggesting that a core NPC composed of nuclear and cytoplasmic rings, inner ring, and central channel but without cytoplasmic or nucleoplasmic filaments can already mediate nuclear import. Interestingly, live imaging studies using a photoswitchable protein have shown that although the nucleus very rapidly re‐establishes full active transport competence within 10 min after anaphase onset, it remains permeable for passive diffusion of small proteins for more than 1 h 23, 76. Since the NE sealing is completed until 10 min after anaphase onset 24, the gradual re‐establishment of the NE permeability barrier must be mediated by changes to the NPC. Indeed, EM observation has shown that newly assembled NPCs at 10 min after anaphase onset contain less central channel density than mature NPCs in interphase 24, suggesting the recruitment of additional proteins, potentially including importin β 77, 78, 79, to the central channel is needed to establish the permeability barrier.
Interphase NPC assembly
Inside‐out membrane extrusion mechanism
In contrast to postmitotic NPC assembly, NPC assembly during interphase occurs in continuous double‐lipid NE bilayers and thus requires an insertion into and fusion of the double nuclear membrane. Interphase NPC assembly starts in telophase when nuclear membrane sealing is completed and the postmitotic nucleus dynamically grows its surface to accommodate nuclear import and chromosome decompaction. Interphase assembly had been less‐well understood compared with postmitotic assembly, largely due to the experimental challenge of capturing the sporadic assembly events and the need to distinguish a few newly assembling from many already‐formed NPCs. Molecular players for interphase assembly have so far been identified by examining the increase of bulk NPC numbers after postmitotic assembly is completed. The studies using human cells and reconstituted nuclei with Xenopus egg extracts have identified molecular requirements including (a) reticulon and DP1/Yop1p family proteins 80, (b) the C‐terminal domain of Nup53 that has membrane deforming activity in vitro 66, (c) the membrane curvature‐sensing domain of Nup133 81, (d) the INM protein Sun1 82, (e) the targeting of Pom121 to the INM 83, and (f) the import of Nup153 into the nucleus to recruit the Nup107‐160 complex to the INM 84 (Fig. 3B). Interestingly, several of the above molecular requirements (b)–(f) have been shown to be specific for interphase assembly, but dispensable for postmitotic NPC assembly, suggesting that interphase and postmitotic assembly processes are regulated by molecularly distinct mechanisms.
High‐resolution live cell microscopy has directly visualized the assembly process at a single‐NPC resolution 25, 26. These studies have demonstrated that interphase NPC assembly is an order of magnitude slower than postmitotic assembly and that the order of the recruited components is different; in interphase assembly Pom121 is recruited earlier than the Nup107‐160 complex whereas this order is inverted in postmitotic assembly 21, 23, 26. Recently, interphase NPC assembly intermediates have been captured in situ in human and rat cells by EM tomography 27. This has revealed that the interphase assembly proceeds asymmetrically, starting with an unusual asymmetric de novo fusion event between INM and ONM. Interphase assembly proceeds by an inside‐out evagination of the INM that grows in diameter and depth until it fuses with the flat ONM (Fig. 3B). The dome‐shaped INM evagination is driven by a growing mushroom‐shaped density, supported by an eight‐fold symmetric nuclear ring structure that underlies the INM from the beginning of membrane bending (Fig. 3B). Only few ONM/INM fusion intermediates have been captured compared with other intermediates 27, suggesting that the fusion step itself is very short‐lived and that the mushroom‐shaped assembly intermediate must undergo rapid and drastic structural rearrangements to transform into a mature NPC.
Similar INM deformations have been observed in yeast mutants lacking several nucleoporins, ER proteins, and the AAA‐ATPase VPS4. These have been interpreted as malformed NPCs due to defects of nucleoporin quality control or errors in NPC assembly, or pleiotropic consequences of transport defects 85, 86, 87, 88, 89, 90. Since yeast undergoes a closed mitosis and all new NPCs assemble in a continuous NE, it can be speculated that the NPCs assemble by an inside‐out INM evagination similar to mammalian interphase NPC assembly. However, NPC assembly intermediates have rarely been observed in wild‐type yeast cells. The reason may be that in the small nuclei of yeast only about 70 new pores form during one whole cell cycle reducing the chance to capture assembly intermediates. In addition, ‘interphase’ NPC assembly in yeast could be faster than in mammalian cells, due to the lack of nuclear lamina. Understanding how the NPC assembles in wild‐type cells in yeast will be necessary in order to interpret the existing mutant phenotypes that involve nuclear membrane and nuclear pore deformations. It is worth mentioning that although recently reported nuclear egress structures transporting ribonucleoproteins and viruses sometimes also show INM deformations 91, 92, these structures are much larger (around 200 nm) and clearly distinct from NPC assembly intermediates.
Possible molecular mechanisms for interphase NPC assembly
What initiates interphase NPC assembly? Since membrane association of Nups153 and 53, as well as INM targeting of Pom121 and Sun1, have been shown to be required for the assembly 66, 82, 83, 84, 93, the initial cue may be the binding of at least one of these proteins to the INM. Some of the Nups including Nup53 and yeast nuclear basket proteins Nup1 and Nup60 have been shown to be capable of membrane deformation and could thus be candidates that drive the evagination of the INM 66, 93. Nup153 can recruit the Nup107‐160 complex to the INM, which enables to recruit additional Nups to the site of assembly. Importantly, interphase NPC assembly does not happen in close proximity to already‐formed NPCs 27 but maintains a constant density in the growing nuclear surface, suggesting that mechanisms exist to regulate the spacing of NPCs in the NE.
What drives the growth of membrane evagination and what triggers fusion? Sec13 and Seh1 are nuclear pore components and also the components of the late endosomal SEA (Seh1‐associated) protein complex in yeast 94 or in the case of Sec13 components of the COPII complex 95. They are thought to be involved in membrane budding, and might thus play an analogous role in INM evagination during interphase NPC assembly. Additional Nups such as the Nup93 complex and the Nup62 complex will be recruited to the site of assembling NPCs via the interaction with Nup53, and this Nup accumulation and subcomplex self‐assembly might generate the mechanical force needed for INM deformation, potentially helped by connections to the nuclear ring already underlying the INM. In addition to Nups, the ESCRT machinery, which mediates diverse membrane remodeling events 96, is likely to be involved either at the fusion step between the ONM and the INM during regular NPC assembly, or alternatively to seal membrane fusions that do not succeed in NPC assembly, but would locally rupture the double nuclear membrane. ESCRT‐III proteins and ATPase VPS4 genetically interact with the Nups (including Nups of the cytoplasmic, nuclear and inner rings, as well as transmembrane Nups) in yeast, and yeast cells lacking a transmembrane Nup Pom152 and one of these proteins accumulate malformed NPCs in the deformed INM 89. AAA‐ATPase Torsin family proteins might also contribute to the membrane evagination and/or fusion steps. Although no direct Torsin ortholog has been found in yeast, its knockout in human cells results in the NE blebs whose neck is enriched with Nups, which could represent stalled evagination intermediates 97.
As described above, the molecular mechanism for interphase NPC assembly remains poorly understood (Fig. 3B), largely due to the sporadic nature of the assembly, which has so far precluded a precise kinetic mapping of the sequence of molecular events. Recently, systematic EM observations of the NE in cells at different cell‐cycle stages has shown that interphase NPC assembly happens relatively synchronously during the first hour of G1, when the nuclear surface grows rapidly following postmitotic nuclear membrane sealing 27. Importantly, the assembly process during the rapid nuclear expansion in G1 has been suggested to be similar to the homeostatic NPC assembly later during interphase, based on the fact that the assembly intermediates are also constantly observed even in later interphase and that the abundance of them quantitatively explains the increase in mature NPCs during cell cycle 27. Since the former core region of the G1 nucleus is largely devoid of postmitotically assembled NPCs, the interphase assembly process can be examined more easily in this region. Taking advantage of this, the core region has indeed been monitored by live cell imaging and super‐resolution microscopy, and the accumulation kinetics and single‐NPC localization of Nups107 and 358 in interphase NPC assembly have been determined 27. In the future, extending such approaches to all Nups and integrating different imaging modalities that allow to determine the dynamic molecular choreography and the dramatically changing protein architecture are likely to be a powerful approach to unravel the mystery of the interphase NPC assembly mechanism.
Significance and implications of two distinct NPC assembly processes
Ensuring fast nuclear assembly process after mitosis
During mitotic exit many NPCs reassemble very rapidly and synchronously within 5 min in the reforming NE 20, 21, 23, 24, whereas NPC assembly into the interphase nucleus occurs only sporadically and requires close to 1 h 25, 26, 27. Why is NPC assembly so much faster after mitosis than during interphase? First of all, the mitotic cell contains a high concentration of ‘assembly ready’ NPC subcomplexes, already synthesized in the mother cell and disassembled by mitotic phosphorylation. This stockpile of building blocks becomes permissive for assembly synchronously by the reduction in mitotic kinase activity 34. By contrast, during interphase, the soluble reservoir of Nups inherited from the mother cell has been consumed, and Nups have to be newly synthesized, limiting the amount of building blocks available for homeostatic NPC assembly. A second fundamental difference is the topology of the membrane into which the new NPC channels are built. In the mitotic cell, the ER sheets that form the nuclear membrane are highly fenestrated and contain a large number of small membrane discontinuities on the surface of the chromosomes. NPC assembly would, therefore, not require a new local membrane fusion event, but could use a pre‐existing hole. Interphase NPC assembly by contrast requires a de novo local fusion event, and it appears that the formation of the nuclear ring and growing mushroom‐shaped density that drives the membrane evagination consumes most of the time required for interphase assembly. Membrane fusion could, thus, be a rate limiting step for the interphase assembly mechanism. Ubiquitously available building blocks and a membrane surface full of holes may thus be the two main factors that enable the high efficiency of postmitotic NPC assembly, that is, so crucial to rapidly establish a functional nucleus after mitosis.
Implications for understanding rapid nuclear growth in embryos
Annulate lamellae (ALs) are cytoplasmic stacked sheets of ER membranes penetrated by nuclear pores containing most Nups and are highly abundant in germ cells and early embryos, which stockpile large amounts of maternal proteins for later development. ALs have been thought to be a repository of maternally provided Nups 98, 99, 100, but how they would be incorporated into embryonic nuclei remained unclear. In rapidly cycling early Drosophila embryos, for example, the size of the nucleus doubles within an interphase that lasts only 10 min, in principle not leaving enough time to assemble a sufficient number of new NPCs using the slow mechanism of interphase assembly found in mammalian cells 27. Very interestingly, a recent study could show that embryonic Drosophila nuclei grow by incorporating pore‐perforated ER sheets ‘en bloc’ into their NE, thus inserting large membrane patches that already contain pores 101. Such merging of AL into a growing NE thus provides a mechanism for rapidly expanding the nuclear surface with a source of membrane already containing pre‐assembled NPCs. This mechanism, thus, formally does not involve de novo NPC assembly and seems ideally suited to support the rapid nuclear growth requirements of the short cell cycles in early embryogenesis.
Implications for the evolutionary origin of the NPC
Two theories have been discussed to explain the evolution of eukaryotic endomembrane compartmentalization, which involve the ancestral function of the NPC or its subcomplexes 102, 103, 104 (Fig. 4). The ‘Outside‐in’ model proposes that the surface of early eukaryotic cells invaginated, creating a cytoplasmic membrane network that eventually would have surrounded the cellular DNA, thus giving rise to the NE (Fig. 4A). By contrast the ‘inside‐out’ model proposes that ancestral eukaryotes evaginated their surface, so that membrane filopodia protruded through their protective glycocalyx to engulf extracellular material and thereby conquer new space, effectively leaving the original cell as the nucleus with the DNA behind and creating the cytoplasm de novo (Fig. 4B). In the outside‐in model, the original function of the NPC would have been to stabilize regions of high curvature in the internalized membrane cisternae and prevent their fusion around DNA to keep gates of communication and transport for macromolecules open. In the inside‐out model, the original function of the NPC would have been to actively promote membrane evaginations that penetrate the protective glycocalyx, but do so in a controlled manner without rupturing it and therefore would have had to constrain the neck diameter of the protrusion. It has been suggested that the assembly of protein complexes may recapitulate their molecular evolution 105, and it is, therefore, interesting to examine our knowledge about NPC assembly in the light of these models. Interestingly, the two different NPC assembly mechanisms we have discussed above are consistent with aspects of both models. The postmitotic NPC assembly process is reminiscent of the outside‐in model (‘stop membrane fusion and keep the gate open’), while the interphase NPC assembly process exhibits key features proposed by the inside‐out model (a controlled evagination of membrane). Although these models appear opposed to each other, it is plausible that both the membrane invagination and controlled membrane extrusion would have been advantageous for early eukaryotic cells. It is tempting to speculate that fusion stopping and membrane deforming activity may thus have initially been separate molecular modules, which became combined only later during eukaryotic evolution, when controlled transport through the gate became an advantage. This might explain why the same molecular machine can be made in two different ways.
Figure 4.

Two theories for the evolution of the eukaryotic endomembrane system. (A) ‘Outside‐in’ model. The surface of ancestral eukaryotic cells invaginated and branched progressively into a cytoplasmic membrane network. The function of NPC components (pink) would have been a passive stabilization of regions with high membrane curvature to prevent closure of the membrane cisternae around DNA (dark and light blue dots). (B) ‘Inside‐out’ model. Membrane filopodia protruded progressively through the glycocalyx to engulf extracellular material and thereby create new cytoplasm while the original cell becomes the nucleus. The function of NPC components would have been to actively promote membrane evaginations. Modified from 103. Arrows indicate direction of cytoplasmic flow.
Conclusions and perspectives
Recent conceptual advances in understanding the two distinct mechanisms of NPC assembly put us in a position to re‐interpret existing genetic and biochemical data and to propose and in the future test the molecular mechanisms regulating the dynamic assembly. Rapid conditional molecular perturbations will be needed to identify molecular players specifically regulating postmitotic or interphase NPC assembly, since scoring phenotypes after perturbations over one cell cycle makes it difficult to separate defects in postmitotic assembly from interphase assembly and vice versa. Once we are in a position to enrich the pores in physiological assembly states by defined molecular perturbations, high‐resolution analysis by super‐resolution microscopy and cryo‐EM combined with single particle averaging will allow us to reveal the molecular architecture and eventually atomic structure of the assembly intermediates. In addition, computational modeling based on biophysical and structural parameters will be necessary to understand what drives the progressive membrane deformation and how the large scale structural rearrangement of individual Nups and their subcomplexes powers NPC assembly. Knowing how the largest nonpolymeric protein complex assembles will lead us to an understanding of general principles of how multi‐protein complexes self‐assemble inside cells and provide important implications for how early eukaryotes evolved subcellular compartmentalization.
Acknowledgements
We thank Tabea Miriam Rauscher for help with figure illustration. This work was supported, in part, by grants from the German Research Council to JE (DFG EL 246/3‐2 within the priority program SPP1175), the Baden‐Württemberg Stiftung to JE, as well as by the European Molecular Biology Laboratory. SO was additionally supported by the EMBL Interdisciplinary Postdoc Programme (EIPOD) under Marie Curie Actions COFUND and a JSPS fellowship (The Japan Society for the Promotion of Science, postdoctoral fellowship for research abroad).
Edited by Wilhelm Just
Contributor Information
Shotaro Otsuka, Email: shotaro.otsuka@embl.de.
Jan Ellenberg, Email: jan.ellenberg@embl.de.
References
- 1. Jamali T, Jamali Y, Mehrbod M and Mofrad MR (2011) Nuclear pore complex: biochemistry and biophysics of nucleocytoplasmic transport in health and disease. Int Rev Cell Mol Biol 287, 233–286. [DOI] [PubMed] [Google Scholar]
- 2. Gomez‐Cavazos JS and Hetzer MW (2012) Outfits for different occasions: tissue‐specific roles of Nuclear Envelope proteins. Curr Opin Cell Biol 24, 775–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amendola M and van Steensel B (2014) Mechanisms and dynamics of nuclear lamina‐genome interactions. Curr Opin Cell Biol 28, 61–68. [DOI] [PubMed] [Google Scholar]
- 4. Ungricht R and Kutay U (2017) Mechanisms and functions of nuclear envelope remodelling. Nat Rev Mol Cell Biol 18, 229–245. [DOI] [PubMed] [Google Scholar]
- 5. Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y and Chait BT (2000) The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol 148, 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT and Matunis MJ (2002) Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 158, 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stanley GJ, Fassati A and Hoogenboom BW (2017) Biomechanics of the transport barrier in the nuclear pore complex. Semin Cell Dev Biol 68, 42–51. [DOI] [PubMed] [Google Scholar]
- 8. Sakiyama Y, Panatala R and Lim RYH (2017) Structural dynamics of the nuclear pore complex. Semin Cell Dev Biol 68, 27–33. [DOI] [PubMed] [Google Scholar]
- 9. Knockenhauer KE and Schwartz TU (2016) The nuclear pore complex as a flexible and dynamic gate. Cell 164, 1162–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoelz A, Glavy JS and Beck M (2016) Toward the atomic structure of the nuclear pore complex: when top down meets bottom up. Nat Struct Mol Biol 23, 624–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beck M and Hurt E (2017) The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Biol 18, 73–89. [DOI] [PubMed] [Google Scholar]
- 12. Grossman E, Medalia O and Zwerger M (2012) Functional architecture of the nuclear pore complex. Annu Rev Biophys 41, 557–584. [DOI] [PubMed] [Google Scholar]
- 13. Ori A, Banterle N, Iskar M, Andres‐Pons A, Escher C, Khanh Bui H, Sparks L, Solis‐Mezarino V, Rinner O, Bork P et al (2013) Cell type‐specific nuclear pores: a case in point for context‐dependent stoichiometry of molecular machines. Mol Syst Biol 9, 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rabut G, Doye V and Ellenberg J (2004) Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol 6, 1114–1121. [DOI] [PubMed] [Google Scholar]
- 15. D'Angelo MA, Raices M, Panowski SH and Hetzer MW (2009) Age‐dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 136, 284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W and Selbach M (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342. [DOI] [PubMed] [Google Scholar]
- 17. Rabut G, Lenart P and Ellenberg J (2004) Dynamics of nuclear pore complex organization through the cell cycle. Curr Opin Cell Biol 16, 314–321. [DOI] [PubMed] [Google Scholar]
- 18. Weberruss M and Antonin W (2016) Perforating the nuclear boundary ‐ how nuclear pore complexes assemble. J Cell Sci 129, 4439–4447. [DOI] [PubMed] [Google Scholar]
- 19. LaJoie D and Ullman KS (2017) Coordinated events of nuclear assembly. Curr Opin Cell Biol 46, 39–45. [DOI] [PubMed] [Google Scholar]
- 20. Haraguchi T, Koujin T, Hayakawa T, Kaneda T, Tsutsumi C, Imamoto N, Akazawa C, Sukegawa J, Yoneda Y and Hiraoka Y (2000) Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J Cell Sci 113, 779–794. [DOI] [PubMed] [Google Scholar]
- 21. Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L and Ellenberg J (2008) Systematic kinetic analysis of mitotic dis‐ and reassembly of the nuclear pore in living cells. J Cell Biol 180, 857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu L, Ladinsky MS and Kirchhausen T (2011) Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly. J Cell Biol 194, 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Otsuka S, Szymborska A and Ellenberg J (2014) Imaging the assembly, structure, and function of the nuclear pore inside cells. Methods Cell Biol 122, 219–238. [DOI] [PubMed] [Google Scholar]
- 24. Otsuka S, Steyer AM, Schorb M, Hériché JK, Hossain MJ, Sethi S, Kueblbeck M, Schwab Y, Beck M and Ellenberg J (2017) Postmitotic nuclear pore assembly proceeds by radial dilation of small ER membrane openings. bioRxiv, 10.1101/141150. [DOI] [PubMed] [Google Scholar]
- 25. D'Angelo MA, Anderson DJ, Richard E and Hetzer MW (2006) Nuclear pores form de novo from both sides of the nuclear envelope. Science 312, 440–443. [DOI] [PubMed] [Google Scholar]
- 26. Dultz E and Ellenberg J (2010) Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J Cell Biol 191, 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Otsuka S, Bui KH, Schorb M, Hossain MJ, Politi AZ, Koch B, Eltsov M, Beck M and Ellenberg J (2016) Nuclear pore assembly proceeds by an inside‐out extrusion of the nuclear envelope. eLife 5, e19071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lenart P, Rabut G, Daigle N, Hand AR, Terasaki M and Ellenberg J (2003) Nuclear envelope breakdown in starfish oocytes proceeds by partial NPC disassembly followed by a rapidly spreading fenestration of nuclear membranes. J Cell Biol 160, 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, Aebersold R, Antonin W and Kutay U (2011) Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell 144, 539–550. [DOI] [PubMed] [Google Scholar]
- 30. Eisenhardt N, Redolfi J and Antonin W (2014) Interaction of Nup53 with Ndc1 and Nup155 is required for nuclear pore complex assembly. J Cell Sci 127, 908–921. [DOI] [PubMed] [Google Scholar]
- 31. Fischer J, Teimer R, Amlacher S, Kunze R and Hurt E (2015) Linker Nups connect the nuclear pore complex inner ring with the outer ring and transport channel. Nat Struct Mol Biol 22, 774–781. [DOI] [PubMed] [Google Scholar]
- 32. Kosinski J, Mosalaganti S, von Appen A, Teimer R, DiGuilio AL, Wan W, Bui KH, Hagen WJ, Briggs JA, Glavy JS et al (2016) Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science 352, 363–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin DH, Stuwe T, Schilbach S, Rundlet EJ, Perriches T, Mobbs G, Fan Y, Thierbach K, Huber FM, Collins LN et al (2016) Architecture of the symmetric core of the nuclear pore. Science 352, aaf1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Champion L, Linder MI and Kutay U (2017) Cellular reorganization during mitotic entry. Trends Cell Biol 27, 26–41. [DOI] [PubMed] [Google Scholar]
- 35. Schellhaus AK, De Magistris P and Antonin W (2015) Nuclear reformation at the end of mitosis. J Mol Biol 128, 3466–3477. [DOI] [PubMed] [Google Scholar]
- 36. Hetzer M, Bilbao‐Cortes D, Walther TC, Gruss OJ and Mattaj IW (2000) GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol Cell 5, 1013–1024. [DOI] [PubMed] [Google Scholar]
- 37. Ma Y, Cai S, Lv Q, Jiang Q, Zhang Q, Sodmergen, Zhai Z and Zhang C (2007) Lamin B receptor plays a role in stimulating nuclear envelope production and targeting membrane vesicles to chromatin during nuclear envelope assembly through direct interaction with importin beta. J Cell Sci 120, 520–530. [DOI] [PubMed] [Google Scholar]
- 38. Peters R (2009) Translocation through the nuclear pore: Kaps pave the way. BioEssays 31, 466–477. [DOI] [PubMed] [Google Scholar]
- 39. Bernis C, Swift‐Taylor B, Nord M, Carmona S, Chook YM and Forbes DJ (2014) Transportin acts to regulate mitotic assembly events by target binding rather than Ran sequestration. Mol Biol Cell 25, 992–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Forbes DJ, Travesa A, Nord MS and Bernis C (2015) Nuclear transport factors: global regulation of mitosis. Curr Opin Cell Biol 35, 78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Christie M, Chang CW, Rona G, Smith KM, Stewart AG, Takeda AA, Fontes MR, Stewart M, Vertessy BG, Forwood JK et al (2016) Structural biology and regulation of protein import into the nucleus. J Mol Biol 428, 2060–2090. [DOI] [PubMed] [Google Scholar]
- 42. von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P et al (2015) In situ structural analysis of the human nuclear pore complex. Nature 526, 140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gomez‐Saldivar G, Fernandez A, Hirano Y, Mauro M, Lai A, Ayuso C, Haraguchi T, Hiraoka Y, Piano F and Askjaer P (2016) Identification of conserved MEL‐28/ELYS domains with essential roles in nuclear assembly and chromosome segregation. PLoS Genet 12, e1006131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hattersley N, Cheerambathur D, Moyle M, Stefanutti M, Richardson A, Lee KY, Dumont J, Oegema K and Desai A (2016) A nucleoporin docks protein phosphatase 1 to direct meiotic chromosome segregation and nuclear assembly. Dev Cell 38, 463–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Franz C, Walczak R, Yavuz S, Santarella R, Gentzel M, Askjaer P, Galy V, Hetzer M, Mattaj IW and Antonin W (2007) MEL‐28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep 8, 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rotem A, Gruber R, Shorer H, Shaulov L, Klein E and Harel A (2009) Importin beta regulates the seeding of chromatin with initiation sites for nuclear pore assembly. Mol Biol Cell 20, 4031–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ and Lippincott‐Schwartz J (1997) Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol 138, 1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vedrenne C, Klopfenstein DR and Hauri HP (2005) Phosphorylation controls CLIMP‐63‐mediated anchoring of the endoplasmic reticulum to microtubules. Mol Biol Cell 16, 1928–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smyth JT, Beg AM, Wu S, Putney JW Jr and Rusan NM (2012) Phosphoregulation of STIM1 leads to exclusion of the endoplasmic reticulum from the mitotic spindle. Curr Biol 22, 1487–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schlaitz AL, Thompson J, Wong CC, Yates JR III and Heald R (2013) REEP3/4 ensure endoplasmic reticulum clearance from metaphase chromatin and proper nuclear envelope architecture. Dev Cell 26, 315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Puhka M, Joensuu M, Vihinen H, Belevich I and Jokitalo E (2012) Progressive sheet‐to‐tubule transformation is a general mechanism for endoplasmic reticulum partitioning in dividing mammalian cells. Mol Biol Cell 23, 2424–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang S, Tukachinsky H, Romano FB and Rapoport TA (2016) Cooperation of the ER‐shaping proteins atlastin, lunapark, and reticulons to generate a tubular membrane network. eLife 5, e18605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Powers RE, Wang S, Liu TY and Rapoport TA (2017) Reconstitution of the tubular endoplasmic reticulum network with purified components. Nature 543, 257–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wandke C and Kutay U (2013) Enclosing chromatin: reassembly of the nucleus after open mitosis. Cell 152, 1222–1225. [DOI] [PubMed] [Google Scholar]
- 55. Haraguchi T, Kojidani T, Koujin T, Shimi T, Osakada H, Mori C, Yamamoto A and Hiraoka Y (2008) Live cell imaging and electron microscopy reveal dynamic processes of BAF‐directed nuclear envelope assembly. J Cell Sci 121, 2540–2554. [DOI] [PubMed] [Google Scholar]
- 56. Anderson DJ and Hetzer MW (2008) Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J Cell Biol 182, 911–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Haraguchi T, Koujin T, Segura‐Totten M, Lee KK, Matsuoka Y, Yoneda Y, Wilson KL and Hiraoka Y (2001) BAF is required for emerin assembly into the reforming nuclear envelope. J Cell Sci 114, 4575–4585. [DOI] [PubMed] [Google Scholar]
- 58. Olmos Y, Hodgson L, Mantell J, Verkade P and Carlton JG (2015) ESCRT‐III controls nuclear envelope reformation. Nature 522, 236–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vietri M, Schink KO, Campsteijn C, Wegner CS, Schultz SW, Christ L, Thoresen SB, Brech A, Raiborg C and Stenmark H (2015) Spastin and ESCRT‐III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature 522, 231–235. [DOI] [PubMed] [Google Scholar]
- 60. Baur T, Ramadan K, Schlundt A, Kartenbeck J and Meyer HH (2007) NSF‐ and SNARE‐mediated membrane fusion is required for nuclear envelope formation and completion of nuclear pore complex assembly in Xenopus laevis egg extracts. J Cell Sci 120, 2895–2903. [DOI] [PubMed] [Google Scholar]
- 61. Orso G, Pendin D, Liu S, Tosetto J, Moss TJ, Faust JE, Micaroni M, Egorova A, Martinuzzi A, McNew JA et al (2009) Homotypic fusion of ER membranes requires the dynamin‐like GTPase atlastin. Nature 460, 978–983. [DOI] [PubMed] [Google Scholar]
- 62. Wang S, Romano FB, Field CM, Mitchison TJ and Rapoport TA (2013) Multiple mechanisms determine ER network morphology during the cell cycle in Xenopus egg extracts. J Cell Biol 203, 801–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Antonin W, Franz C, Haselmann U, Antony C and Mattaj IW (2005) The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol Cell 17, 83–92. [DOI] [PubMed] [Google Scholar]
- 64. Mansfeld J, Guttinger S, Hawryluk‐Gara LA, Pante N, Mall M, Galy V, Haselmann U, Muhlhausser P, Wozniak RW, Mattaj IW et al (2006) The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol Cell 22, 93–103. [DOI] [PubMed] [Google Scholar]
- 65. Rasala BA, Ramos C, Harel A and Forbes DJ (2008) Capture of AT‐rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol Biol Cell 19, 3982–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vollmer B, Schooley A, Sachdev R, Eisenhardt N, Schneider AM, Sieverding C, Madlung J, Gerken U, Macek B and Antonin W (2012) Dimerization and direct membrane interaction of Nup53 contribute to nuclear pore complex assembly. EMBO J 31, 4072–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bodoor K, Shaikh S, Salina D, Raharjo WH, Bastos R, Lohka M and Burke B (1999) Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J Cell Sci 112, 2253–2264. [DOI] [PubMed] [Google Scholar]
- 68. Hase ME and Cordes VC (2003) Direct interaction with Nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol Biol Cell 14, 1923–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Goldberg MW, Wiese C, Allen TD and Wilson KL (1997) Dimples, pores, star‐rings, and thin rings on growing nuclear envelopes: evidence for structural intermediates in nuclear pore complex assembly. J Cell Sci 110, 409–420. [DOI] [PubMed] [Google Scholar]
- 70. Kiseleva E, Rutherford S, Cotter LM, Allen TD and Goldberg MW (2001) Steps of nuclear pore complex disassembly and reassembly during mitosis in early Drosophila embryos. J Cell Sci 114, 3607–3618. [DOI] [PubMed] [Google Scholar]
- 71. Zierhut C, Jenness C, Kimura H and Funabiki H (2014) Nucleosomal regulation of chromatin composition and nuclear assembly revealed by histone depletion. Nat Struct Mol Biol 21, 617–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Inoue A and Zhang Y (2014) Nucleosome assembly is required for nuclear pore complex assembly in mouse zygotes. Nat Struct Mol Biol 21, 609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Schooley A, Moreno‐Andres D, De Magistris P, Vollmer B and Antonin W (2015) The lysine demethylase LSD1 is required for nuclear envelope formation at the end of mitosis. J Cell Sci 128, 3466–3477. [DOI] [PubMed] [Google Scholar]
- 74. Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM and Rapoport TA (2010) Mechanisms determining the morphology of the peripheral ER. Cell 143, 774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Frey S, Richter RP and Gorlich D (2006) FG‐rich repeats of nuclear pore proteins form a three‐dimensional meshwork with hydrogel‐like properties. Science 314, 815–817. [DOI] [PubMed] [Google Scholar]
- 76. Dultz E, Huet S and Ellenberg J (2009) Formation of the nuclear envelope permeability barrier studied by sequential photoswitching and flux analysis. Biophys J 97, 1891–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yang W and Musser SM (2006) Nuclear import time and transport efficiency depend on importin beta concentration. J Cell Biol 174, 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lowe AR, Tang JH, Yassif J, Graf M, Huang WY, Groves JT, Weis K and Liphardt JT (2015) Importin‐beta modulates the permeability of the nuclear pore complex in a Ran‐dependent manner. eLife 4, e04052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kapinos LE, Huang B, Rencurel C and Lim RYH (2017) Karyopherins regulate nuclear pore complex barrier and transport function. J Cell Biol 216, 3609–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dawson TR, Lazarus MD, Hetzer MW and Wente SR (2009) ER membrane‐bending proteins are necessary for de novo nuclear pore formation. J Cell Biol 184, 659–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Doucet CM, Talamas JA and Hetzer MW (2010) Cell cycle‐dependent differences in nuclear pore complex assembly in metazoa. Cell 141, 1030–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Talamas JA and Hetzer MW (2011) POM121 and Sun1 play a role in early steps of interphase NPC assembly. J Cell Biol 194, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Funakoshi T, Clever M, Watanabe A and Imamoto N (2011) Localization of Pom121 to the inner nuclear membrane is required for an early step of interphase nuclear pore complex assembly. Mol Biol Cell 22, 1058–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Vollmer B, Lorenz M, Moreno‐Andres D, Bodenhofer M, De Magistris P, Astrinidis SA, Schooley A, Flotenmeyer M, Leptihn S and Antonin W (2015) Nup153 recruits the Nup107‐160 complex to the inner nuclear membrane for interphasic nuclear pore complex assembly. Dev Cell 33, 717–728. [DOI] [PubMed] [Google Scholar]
- 85. Wente SR and Blobel G (1993) A temperature‐sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J Cell Biol 123, 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Murphy R, Watkins JL and Wente SR (1996) GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol Biol Cell 7, 1921–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Scarcelli JJ, Hodge CA and Cole CN (2007) The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes. J Cell Biol 178, 799–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Makio T, Stanton LH, Lin CC, Goldfarb DS, Weis K and Wozniak RW (2009) The nucleoporins Nup170p and Nup157p are essential for nuclear pore complex assembly. J Cell Biol 185, 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Chadrin A, Hess B, San Roman M, Gatti X, Lombard B, Loew D, Barral Y, Palancade B and Doye V (2010) Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. J Cell Biol 189, 795–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Webster BM, Colombi P, Jager J and Lusk CP (2014) Surveillance of nuclear pore complex assembly by ESCRT‐III/Vps4. Cell 159, 388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, Li Y, Ataman B, Koon A, Chang YT, Li Q et al (2012) Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell 149, 832–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Mettenleiter TC, Muller F, Granzow H and Klupp BG (2013) The way out: what we know and do not know about herpesvirus nuclear egress. Cell Microbiol 15, 170–178. [DOI] [PubMed] [Google Scholar]
- 93. Meszaros N, Cibulka J, Mendiburo MJ, Romanauska A, Schneider M and Kohler A (2015) Nuclear pore basket proteins are tethered to the nuclear envelope and can regulate membrane curvature. Dev Cell 33, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Dokudovskaya S, Waharte F, Schlessinger A, Pieper U, Devos DP, Cristea IM, Williams R, Salamero J, Chait BT, Sali A et al (2011) A conserved coatomer‐related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol Cell Proteomics 10 (M110), 006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hsia KC, Stavropoulos P, Blobel G and Hoelz A (2007) Architecture of a coat for the nuclear pore membrane. Cell 131, 1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Olmos Y and Carlton JG (2016) The ESCRT machinery: new roles at new holes. Curr Opin Cell Biol 38, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Laudermilch E, Tsai PL, Graham M, Turner E, Zhao C and Schlieker C (2016) Dissecting Torsin/cofactor function at the nuclear envelope: a genetic study. Mol Biol Cell 27, 3964–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Soupart P and Strong PA (1974) Ultrastructural observations on human oocytes fertilized in vitro . Fertil Steril 25, 11–44. [PubMed] [Google Scholar]
- 99. Spindler M and Hemleben C (1982) Formation and possible function of annulate lamellae in a planktic foraminifer. J Ultrastruct Res 81, 341–350. [DOI] [PubMed] [Google Scholar]
- 100. Lenart P and Ellenberg J (2003) Nuclear envelope dynamics in oocytes: from germinal vesicle breakdown to mitosis. Curr Opin Cell Biol 15, 88–95. [DOI] [PubMed] [Google Scholar]
- 101. Hampoelz B, Mackmull MT, Machado P, Ronchi P, Bui KH, Schieber N, Santarella‐Mellwig R, Necakov A, Andres‐Pons A, Philippe JM et al (2016) Pre‐assembled nuclear pores insert into the nuclear envelope during early development. Cell 166, 664–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A and Rout MP (2004) Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol 2, e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Baum DA and Baum B (2014) An inside‐out origin for the eukaryotic cell. BMC Biol 12, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Rout MP and Field MC (2017) The Evolution of organellar coat complexes and organization of the eukaryotic cell. Annu Rev Biochem 86, 637–657. [DOI] [PubMed] [Google Scholar]
- 105. Kirschner M and Gerhart J (1998) Evolvability. Proc Nati Acad Sci 95, 8420–8427. [DOI] [PMC free article] [PubMed] [Google Scholar]


