Abstract
Antagonistic yeasts suppress plant pathogenic fungi by various mechanisms, but their biocontrol efficacy also depends on the ability to compete and persist in the environment. The goal of the work presented here was to quantify the composition of synthetic yeast communities in order to determine the competitiveness of different species and identify promising candidates for plant protection. For this purpose, colony counting of distinct species and matrix‐assisted laser desorption ionization time‐of‐flight mass spectrometry (MALDI‐TOF MS; MALDI biotyping) were used to distinguish different yeast species and to quantify the composition of a synthetic community of six yeasts (Aureobasidium pullulans, Candida subhashii, Cyberlindnera sargentensis, Hanseniaspora sp., Metschnikowia pulcherrima and Pichia kluyveri) over time, on apples and in soil, and in different growth media. These studies revealed important characteristics that predispose the different species for particular applications. For example, the competitiveness and antagonistic activity of C. subhashii was strongly increased in the presence of N‐acetylglucosamin as the sole carbon source, M. pulcherrima and A. pullulans were the strongest competitors on apple, and C. sargentensis competed the best in soil microcosms. Based on these laboratory studies, M. pulcherrima and A. pullulans are promising candidates for biocontrol applications against fungal phyllosphere diseases, while C. sargentensis may hold potential for use against soilborne fungal pathogens. These results document the potential of MALDI‐TOF MS for the quantitative analysis of synthetic yeast communities and highlight the value of studying microorganisms with relevant functions in moderately complex, synthetic communities and natural substrates rather than as individual isolates.
Keywords: antagonist, interspecific competition, MALDI‐TOF, mass spectrometry, synthetic community, yeast
1. INTRODUCTION
Although antagonistic yeasts are an attractive, environmentally friendly solution for combatting plant pathogens, and despite the long history of research on such applications, they are not yet widely used (Droby, Wisniewski, Teixidó, Spadaro, & Jijakli, 2016). One of the main reasons for the lack of or limited success of such biological control measures is the discrepancy between the effect in defined laboratory experiments and that in complex, agricultural environments (Spadaro & Droby, 2016). In order to better understand and predict the biocontrol activity of a particular antagonist in different environmental systems and crops, new types of assays, more complex than assays in the laboratory and less laborious and costly than field trials, are thus necessary.
Quantifying the species composition of synthetic yeast communities over time and under different conditions can reveal important phenomena such as antagonism, competition or nutrient cross feeding and thus identify functions and processes that may affect the biocontrol activity of yeasts. Studying interspecific competition in synthetic communities also characterizes individual species and may identify functions that are only expressed and observable in communities and not in pure cultures. Simple, synthetic communities of visually distinct species can be easily quantified by counting colonies, while complex, natural communities can be studied by sequencing‐based (e.g. metabarcoding approaches, genetically tagging of individual strains) or large‐scale, cultivation‐based methods (e.g. culturomics) (Lagier et al., 2012, 2015; Lawrence et al., 2012; Tyc, Wolf, & Garbeva, 2015). Matrix‐assisted laser desorption ionization time‐of‐flight mass spectrometry (MALDI‐TOF MS; also called MALDI biotyping) is a convenient tool for the identification of individual microorganisms and can be used as a fast, cheap and reliable identification method in culturomics projects (Lagier et al., 2015).
It was the goal of this study to characterize and assess potential biocontrol yeasts. We employed synthetic communities of antagonistic yeasts to uncover the competitiveness of six species in different culture media, on the apple surface and in soil and thereby identified promising species for the application as biocontrol agents in soil or on apples. For this purpose, a fast and reliable MALDI‐based approach to quantitatively describe synthetic yeast communities was developed. We used custom‐made reference spectra and direct smears of cells from single yeast colonies, in combination with counting of unequivocally identified colonies to determine the temporal dynamics within synthetic communities of six antagonistic yeasts on apples, in soil and in different growth media. This combination of direct counting and MALDI‐based identification allowed the fast, reliable and quantitative analysis of this moderately complex, synthetic yeast community in liquid media in vitro, as well as in complex substrates such as the apple surface or soil.
Overall, this study revealed pronounced temporal changes in the species composition of a synthetic yeast community and identified growth conditions and substrates where different yeasts were most competitive. Such quantitative analyses of synthetic antagonist communities may thus provide important information for selecting and promoting the competitiveness of individual species or mixtures of antagonistic microorganisms for biocontrol applications in different crops and environments.
2. METHODS
2.1. Isolates and cultivation
All experiments were conducted with the yeast species Metschnikowia pulcherrima (isolate APC 1.2), Hanseniaspora sp. (APC 12.1), Cyberlindnera sargentensis (SHA 17.2), Aureobasidium pullulans (NBB 7.2.1), Candida subhashii (FGA 2.2) and Pichia kluyveri (APC 11.10B). These six isolates were previously characterized and identified as strong antagonists of a range of filamentous fungi (Hilber‐Bodmer, Schmid, Ahrens, & Freimoser, 2017). The soilborne fungal pathogen Thielaviopsis basicola (DCM 1603a; SH200142.07) was isolated from an infected carrot in Switzerland. All fungi were routinely maintained on Potato Dextrose Agar (PDA) plates (Becton, Dickinson and Company, Le Pont de Claix, France).
2.2. Preparation of synthetic yeast communities
Yeasts (less than 2 weeks old) were collected from a PDA plate, diluted in water, and the OD600 was determined. Since plating the same OD600 units (ODU) of the six yeasts resulted in different colony forming units (CFU) on agar plates for the six species, the ODUs of the six species were empirically adjusted in order to generate a synthetic community with comparable initial CFU for each species. For the synthetic community studied here, the following ODUs were mixed and diluted to a volume of 1 mL: 0.08 ODU A. pullulans, 0.1 ODU C. sargentensis, 0.3 ODU C. subhashii, 0.3 ODU Hanseniaspora sp., 0.1 ODU M. pulcherrima and 0.1 ODU P. kluyveri. In the different experiments, this solution contained 1–5 × 105 CFU/mL for each species. The species composition of the starting culture (day 0) was assessed by collecting 0.1 ODU of the mixture, preparing a 100‐fold dilution and plating 20 μL onto regular PDA plates (incubation at 22°C for 2–3 days). Each experiment was performed twice with four repetitions each and one representative example is shown.
2.3. Analysis of yeast communities in liquid medium
A 10 μL aliquot of the yeast mixture (see above) was added to 10 mL liquid medium. This starting culture was equally divided into four 13 mL round‐bottom tubes (Semadeni Plastics Market, Ostermundigen, Switzerland), and the cultures were kept on a shaking incubator (27°C, 250 rpm; Stuart orbital incubator S150, Staffordshire, UK). At each timepoint, the OD600 of each culture was determined, 0.1 ODU was collected, a 100‐fold dilution was prepared and 20 μL of the suspension was plated on PDA and incubated at 22°C for 2–3 days. Potato Dextrose Broth (PDB; BD Difco™, Becton, Dickinson and Company, Le Pont de Claix, France) as well as Yeast Nitrogen Base (YNB; Formedium™, Norfolk, UK) were used as the basal media. YNB medium was always supplemented with Complete Supplement Mixture (Formedium™, Norfolk, UK) and different carbon sources [glucose monohydrate, maltose monohydrate, melezitose monohydrate and N‐acetyl‐d‐glucosamine (NAG); all used at 10 g/L and obtained from Carl Roth GmbH, Karlsruhe, Germany].
2.4. Analysis of yeast communities on apple surfaces
Apples (Golden Delicious, grown under normal orcharding conditions) were sprayed with the synthetic yeast community (see above; OD600 = 0.1, ~10 mL per apple). The yeast suspension was sprayed with a spray diffusor onto each apple (at the top, middle and bottom and from three sides). Apples were stored in a plastic box in a climate cabinet (14:10 h light–dark cycle, 26/18°C during the day and night, respectively). After 1, 2 and 3 weeks, one slice of the apple skin was removed with a potato peeler, weighed, 10× the weight of peptone water (2 g/L; Carl Roth GmbH, Karlsruhe, Germany) was added, and yeasts were isolated by shaking for 20 min at 22°C and 250 rpm. The suspensions were diluted 10‐fold and 20 μL was plated on PDA. At each time point, four separate apples were analysed.
2.5. Analysis of yeast communities in soil microcosms
Sterile soil microcosms were prepared with 15 g of Multiplication Substrate Floradur® B Cutting (Floragard Vertriebs GmbH, Oldenburg, Germany) in Rotilabo®‐screw top glass bottles (Carl Roth GmbH, Karlsruhe, Germany). The microcosms were autoclaved twice at 121°C for 20 min. To each microcosm, 15 mL of water with 500 μL of the synthetic yeast community were added, the glass bottles were slightly shaken so that the soil was evenly wetted, and the soil microcosms were incubated at 22°C. After 2, 4 and 6 weeks, soil samples were removed, diluted 10‐fold with H2O, vigorously mixed and shaken for 20 min at 22°C and 250 rpm. The suspensions were diluted 500‐fold and 20 μL was plated on PDA.
2.6. Quantitative analysis of yeast communities
Individual yeast colonies were analysed after 2–3 days of incubation at 22°C. Aureobasidium pullulans (white to pale pink with a velvety, greyish fringe), M. pulcherrima (smooth‐red with reddish, diffusing halos) and P. kluyveri (white‐cream and farinose appearance, membranous texture; see also Westerdijk Fungal Biodiversity Institute, 2011) were directly identified and counted by eye. The colonies of C. sargentensis, C. subhashii and Hanseniaspora sp. were similar in appearance and therefore identified by MALDI‐TOF MS.
From each plate, a maximum of 24 yeast colonies per plate (from a randomly selected sector of the plate) were picked with sterile toothpicks and applied as thin films on a steel target plate (direct smear technique; Bruker Daltonics GmbH, Bremen, Germany). A 1 μL aliquot of matrix solution (α‐cyano‐4‐hydroxycinnamic acid, 10 mg/mL in acetonitrile–water–trifluoroacetic acid, 60:37.5:2.5; Sigma‐Aldrich Chemie GmbH, Steinheim, Germany; Merck KGaA, Darmstadt, Germany) was applied to the samples and co‐crystalized at room temperature (see also Rahi, Prakash, & Shouche, 2016). Data collection parameters were defined using flexControl software version 3.4 and spectra were acquired with a microflex LT MALDI‐TOF MS (Bruker Daltonics GmbH, Bremen, Germany). Except for an adjustment of the laser frequency to 50–60 Hz, previously established settings for the identification of fungi were used (Freimoser, Hilber‐Bodmer, Brunisholz, & Drissner, 2016). The spectra were compared to custom‐made reference spectra of the three yeast isolates (also generated by the direct smear method used here). According to the BioTyper software (Bruker Daltonics GmbH, Bremen, Germany), a score of ≥1.7 indicates the correct identification of the genus. Scores of ≥1.7 were thus defined as correct identifications because the three yeasts analysed by MALDI biotyping belonged to three different genera. Measurements that resulted in scores <1.7 were counted as ‘not identified’. In all cases, the second best match had a score of <1.7.
3. RESULTS
Six yeast isolates from the apple phyllosphere or soil (A. pullulans, C. sargentensis, C. subhashii, Hanseniaspora sp., M. pulcherrima and P. kluyveri), identified as overall strong antagonists of a broad range of different filamentous fungi (Hilber‐Bodmer et al., 2017), were used in the present study.
3.1. Temporal changes in the composition of a synthetic yeast community growing in PDB medium
Yeast species that are unequivocally identified based on their colony morphology can be counted by eye, but the colonies of most species closely resemble each other and thus cannot be counted. For this reason, MALDI biotyping of direct smears from single yeast colonies was used to identify those yeast species that cannot be distinguished based on their colony morphology. The species composition of a moderately complex, synthetic community of the six strongly antagonistic yeasts growing in PDB was followed over time (Figure 1). In the initial yeast mixture, at day 0, all six species were detected in comparable frequencies (black bars; 1.0–2.3 × 105 CFU/mL). After 1 day of growth, A. pullulans was not detected anymore and the C. sargentensis and P. kluyveri frequencies were strongly reduced, while C. subhashii, Hanseniaspora sp. and M. pulcherrima had increased in abundance. At day 3, the yeast community predominantly comprised C. subhashii and M. pulcherrima, while Hanseniaspora sp. constituted <5% of the community. After 7 days of growth, the community was dominated by M. pulcherrima, which constituted over 90% of all yeasts. As compared with the changes observed in the yeast community during the first 7 days, the species composition changed little thereafter (data not shown). Therefore, and since we were interested in an endpoint for the competitive success of the different yeasts under different conditions, all subsequent analyses in liquid media were evaluated at day 7.
Figure 1.
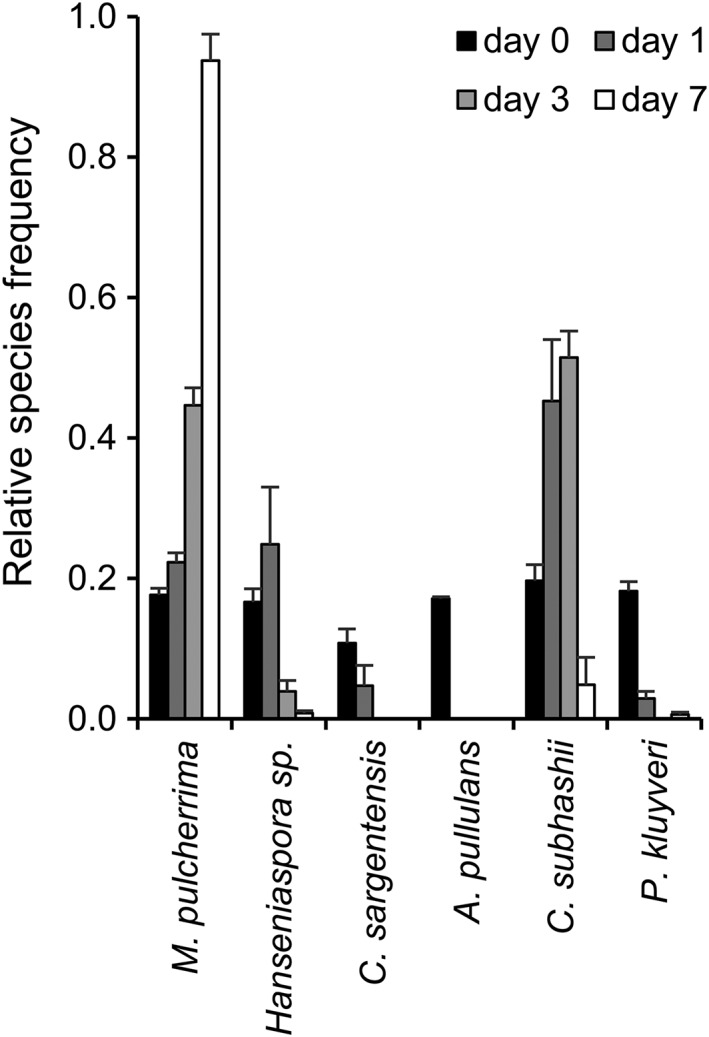
The composition of a synthetic yeast community growing in PDB medium changes over time. The synthetic mixture of six yeasts was grown in PDB for 7 days. Day 0 represents the initial yeast mixture that was used to inoculate the growth medium. The values represent the means of four replicates and standard errors are indicated. One example of at least two experiments is shown. The number of yeast colonies that were counted were 186 (day 0), 103 (day 1), 171 (day 3) and 803 (day 7, the community consisted almost exclusively of Metschnikowia pulcherrima, which could be identified and counted by eye; the total number of colonies that were counted at day 7 was thus higher). Overall,eight yeast colonies could not be identified
Aureobasidium pullulans, M. pulcherrima and P. kluyveri were identified based on their colony morphology and directly counted by eye. Direct smear MALDI biotyping was used to identify C. sargentensis, C. subhashii and Hanseniaspora sp. that cannot be distinguished based on their morphology on PDA plates. Our results clearly demonstrate the applicability and suitability of MALDI biotyping and direct counting for the quantitative analysis of a moderately complex, synthetic yeast community and indicated strong interspecific competition and vastly different competitiveness among the six strongly antagonistic yeasts studied here. In PDB medium, M. pulcherrima was the most competitive yeast.
3.2. Effect of different carbon sources on the competitiveness and antagonistic activity of the six yeast isolates
In an earlier study, the six antagonistic yeasts studied here exhibited marked differences with respect to the carbon sources that can be metabolized: Pichia kluyveri and Hanseniaspora sp. only grew well in the presence of glucose (where all yeasts grew well), and M. pulcherrima and C. subhashii were the only species that could utilize NAG (Hilber‐Bodmer et al., 2017). It was therefore expected that, depending on the carbon source provided, different yeasts would outcompete the other species within the synthetic community. In order to test this hypothesis, experiments with glucose, maltose, melezitose and NAG were performed in defined YNB (with ammonium sulphate) medium.
In contrast with PDB, where M. pulcherrima was the most competitive yeast (of the six isolates tested here; Figure 1), P. kluyveri outcompeted the other five species in defined YNB medium with glucose as the sole carbon source (Figure 2a; initial densities were 0.9–4.8 CFU/mL). Pichia kluyveri was also a strong competitor with maltose as the carbon source, but hardly detected if melezitose or NAG were provided as carbon sources. In YNB‐maltose medium, C. subhashii was the second most frequent species and constituted about 40% of the yeast community (after 7 days), while the other species were only rarely detected. With melezitose, C. subhashii (~40%) and M. pulcherrima (almost 60%) were most frequent and with NAG C. subhashii was the only yeast detected after 7 days.
Figure 2.
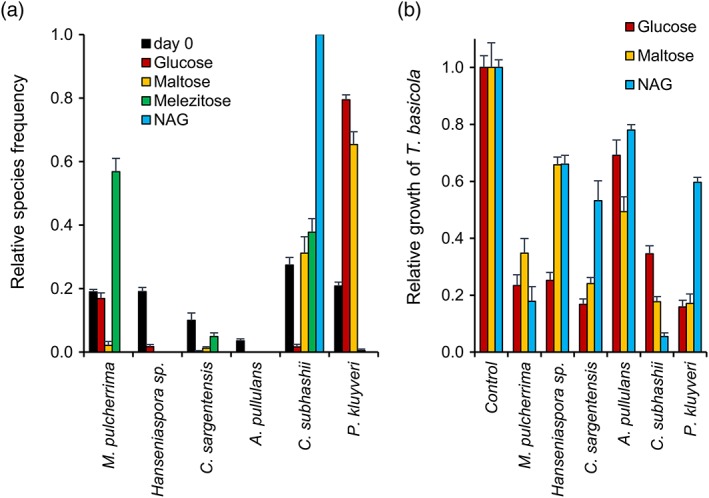
The competitive success (a) and antagonistic activity (b) of different yeasts depends on the carbon source that is provided in the growth medium. (a) A synthetic community of six yeast species was grown in YNB medium supplemented with different carbon‐sources (10 g/L). After 7 days of growth, the relative species frequency was determined. The numbers of yeast colonies that were analysed for each carbon source were 168 (day 0), 806 (glucose), 37 (maltose), 220 (melezitose) and 96 (NAG). Six colonies could not be identified. The values represent the means of four replicates and standard errors are indicated. One example of at least two experiments that resulted in similar outcomes is shown. (b) Binary competition assays of the six yeasts against the soilborne plant pathogen Thielaviopsis basicola were carried out on YNB medium with glucose, maltose, or NAG as carbon sources and otherwise performed as previously described (Hilber‐Bodmer et al., 2017). The mean relative growth (and standard error) of T. basicola in the presence of the six yeasts and on the control plate is shown (four replicates were used) [Colour figure can be viewed at http://wileyonlinelibrary.com]
To test if the competitiveness of the six yeasts in the different media correlated with their antagonistic activity under the respective conditions, binary competition assays of the six yeasts against a soilborne plant pathogen, T. basicola (DCM1603a; SH200142.07), were performed (Figure 2b). Under these conditions (YNB with the carbon sources glucose, maltose or NAG), A. pullulans was the least antagonistic of the five species. With NAG as the carbon source, M. pulcherrima and C. subhashii were most antagonistic against T. basicola, while Hanseniaspora sp., C. sargentensis and P. kluyveri were more strongly antagonistic with glucose as compared with the other sugars. These results reflected the observations made in the synthetic communities, which revealed strongly competitive behaviour of M. pulcherrima and C. subhashii in medium containing NAG, while P. kluyveri seemed to be more competitive with the sugars glucose and maltose. The weak competitiveness of A. pullulans in YNB‐base media also agreed with the low antagonistic activity under these conditions.
Overall, these results revealed strong effects of the different carbon sources on the competitiveness of the six yeasts studied here. Particularly noteworthy was the strong competitiveness of P. kluyveri in YNB medium, in particular as compared with PDB, and the complete exclusion of all five yeasts by C. subhashii in medium containing the carbon source NAG.
3.3. Changes in the composition of a synthetic yeast community on the apple surface and in soil
The competitiveness in a complex, natural environment is an important property for the success of a potential biocontrol agent. Therefore, we also determined the competitiveness of the six yeasts on the apple surface and in soil, where antagonistic yeasts could be employed as biocontrol agents against storage diseases or soilborne fungal pathogens, respectively.
In the initial yeast mixture, at day 0, all six species were clearly detectable in comparable frequencies (Figure 3; 2.0–4.6 × 105 CFU/mL). On the apple surface, only M. pulcherrima and A. pullulans increased in frequency over the 21 day duration of the experiment as compared with their initial occurrence (Figure 3a). Only these two yeasts were thus able to compete and efficiently colonize the apple surface. The relative frequency of the other four yeasts either declined (P. kluyveri, C. sargentensis, Hanseniaspora sp.) or reached undetectable levels after 14 days (C. subhashii). In soil, a complex yeast community of four species persisted throughout the 42 day duration of the experiment (Figure 3b). Only P. kluyveri and Hanseniaspora sp. were undetectable at days 28 and 42, but the other four yeasts persisted at frequencies between 10 and 30% (M. pulcherrima, C. subhashii) or increased to 40% of the synthetic yeast community at specific time points (C. sargentensis, A. pullulans).
Figure 3.
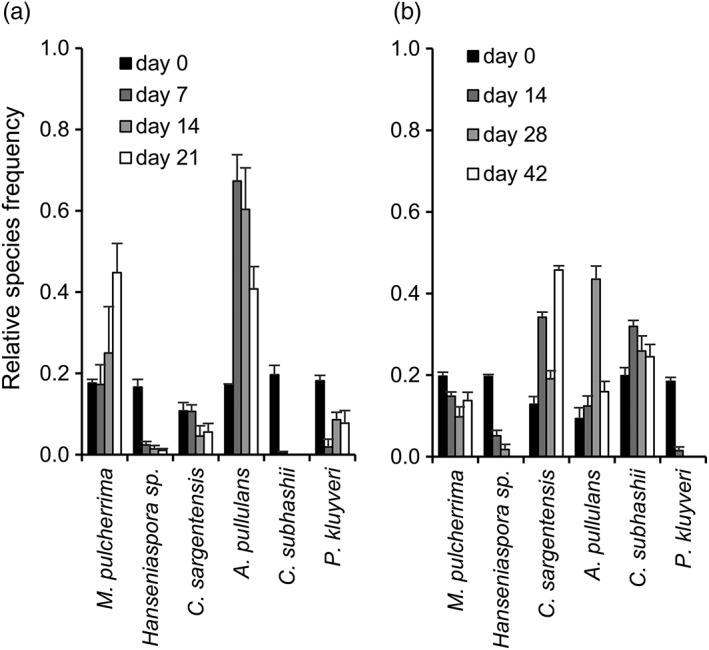
The competitive success of different yeast species on the apple surface (a) and in soil (b). The synthetic yeast community was sprayed on apples or inoculated into sterile soil and analysed at different time points (7–42 days after inoculation). (a) The total numbers of yeast colonies that were analysed for the apple experiment were 187 (day 0), 172 (day 7), 426 (day 14), and 584 (day 21). In total, six colonies could not be identified. (b) For the experiment in soil microcosms, 168 (day 0), 135 (day 14), 195 (day 28) and 129 (day 42) colonies were counted in total. Overall, 10 colonies could not be identified. All values represent the means of four replicates and standard errors are indicated. One example of at least two experiments with similar outcomes is shown
Overall, these experiments with apples and soil identified vastly different outcomes as compared with the results obtained from the in vitro studies and with each other. In particular, P. kluyveri, which was a strong competitor in YNB‐based media containing glucose or maltose, performed weakly in both complex substrates, while A. pullulans, a weak competitor against the other yeasts in liquid media, was competitive in soil and in particular on the apple surface.
4. DISCUSSION
Plant pathogenic, filamentous fungi threaten crop production worldwide and biocontrol agents are a desirable alternative and supplement to synthetic fungicides. As compared with chemical plant protection agents, biocontrol agents are less likely to drive the development of resistance owing to the many mechanisms by which they compete against pathogens (Janisiewicz & Korsten, 2002). Antagonistic yeasts are known to suppress pathogens via the production of toxic metabolites or lytic enzymes, by competition for space and nutrients or by biofilm formation (Bautista‐Rosales et al., 2014; Bautista‐Rosales, Calderon‐Santoyo, Servín‐Villegas, Ochoa‐Álvarez, & Ragazzo‐Sánchez, 2013; Spadaro & Droby, 2016). However, the success of a biocontrol agent depends not only on its mode of action under optimal conditions, but mostly on the ability to compete and persist in the environment where it is applied. Metabolic capabilities and nutritional factors can thus strongly affect the outcome of the antagonistic interactions mediating the biological control of plant pathogenic fungi (Blackburn, Shapiro‐Ilan, & Adams, 2016). The modes of action and metabolic capabilities of different species may also complement each other and lead to improved biocontrol efficacy of mixtures and communities as compared with individual isolates (Freimoser, Pelludat, & Remus‐Emsermann, 2016).
The six yeasts included in the synthetic community studied here were isolated from soil (A. pullulans, C. subhashii, C. sargentensis) or the apple phyllosphere (Hanseniaspora sp., M. pulcherrima, P. kluyveri; Hilber‐Bodmer et al., 2017). Despite its origin, the isolate NBB 7.2.1 of A. pullulans was a strong competitor on apple. At days 7 and 14, A. pullulans constituted over 60% of the total yeast population, which declined to 40% at day 21. Aureobasidium pullulans is a biotechnologically relevant and ubiquitous yeast (Prasongsuk, Lotrakul, Ali, Bankeeree, & Punnapayak, 2017; van Nieuwenhuijzen, 2014). However, the species seems well adapted to the phyllosphere and in particular to apples, which is also reflected by its use as a biocontrol agent against fireblight of apple trees and apple storage diseases (Castoria et al., 2001; Kunz & Haug, 2006; Leibinger, Breuker, Hahn, & Mendgen, 1997; Mari, Martini, Guidarelli, & Neri, 2012; Mari, Martini, Spadoni, Rouissi, & Bertolini, 2012; Mounir et al., 2007). The stronger competitiveness of our A. pullulans soil isolate on the apple surface, as compared with soil, may thus suggest that A. pullulans is adapted to the phyllosphere and reaches the soil compartment only accidentally via leaves and flowers falling to the ground and washing off (NBB 7.2.1 originates from a soil collected in an apple orchard). The second yeast that was able to compete on the apple surface was M. pulcherrima, which increased throughout the experiment and was slightly more abundant at day 21 than A. pullulans. The frequency of the other four yeasts was always lower than at the time of application (day 0). The strong competitiveness of M. pulcherrima and A. pullulans in the phyllosphere is documented by many studies. Aureobasidium pullulans and M. pulcherrima were regularly isolated from the leaf surface of 10 different species of trees in the Small Carpathian Mountains and the former was among the three most frequent species (Slavikova, Vadkertiova, & Vranova, 2007). Biodiversity studies on apple and pear surfaces in orchards in northern Italy frequently discovered A. pullulans and M. pulcherrima (Pelliccia et al., 2011) and a large field study of yeasts on apple, plum and pear found these two species regularly in flowers and on fruits (Vadkertiova, Molnarova, Vranova, & Slavikova, 2012). In contrast, on nectarines and plum, species of the genus Metschnikowia were only rarely or never found, while A. pullulans was frequently isolated (Janisiewicz, Jurick 2nd, Peter, Kurtzman, & Buyer, 2014; Janisiewicz, Kurtzman, & Buyer, 2010). Nevertheless, M. pulcherrima was considered an excellent candidate for the biological control of blue mould storage disease of apple (Janisiewicz, Tworkoski, & Kurtzman, 2001).
Of the species tested here, C. sargentensis, which was isolated from soil, was most competitive in our soil microcosms and constituted, at certain time points, up to 40% of the synthetic yeast community. Isolates of the Williopsis saturnus species complex, to which C. sargentensis belongs, have indeed been isolated from soils from all over the world, confirming the competitiveness in this environment (Naumova, Lee, Kondratieva, Sadykova, & Naumov, 2017). In addition to antagonistic activity against fungal pathogens, Williopsis complex isolates have been shown to promote plant growth, live endophytically and produce volatile sulphur compounds and antimycotic killer proteins (Goretti et al., 2009; Kurtzman, 1991; Kurtzman, Robnett, & Basehoar‐Powers, 2008; Minter, 2009; Nassar, El‐Tarabily, & Sivasithamparam, 2005; Tan, Lee, Seow, Ong, & Liu, 2012). Overall, these findings seem to confirm the potential of C. sargentensis as a plant protection agent against soilborne pathogens.
In the results presented here, P. kluyveri was more competitive in liquid YNB media (with glucose and maltose as carbon sources) as compared with PDB and complex substrates (e.g. apple or soil). Since the former medium contains large amounts of ammonium sulphate, in addition to all amino acids, it is possible that P. kluyveri is most efficient at utilizing ammonium as a nitrogen source and thus more competitive in this medium than the other yeasts. Alternatively, P. kluyveri may exhibit different competitive phenotypes owing to different metabolites that are secreted in YNB and PDB. Similarly, maltose may be more efficiently taken up or induce the synthesis of inhibitory compounds by P. kluyveri and to some extent C. subhashii, because these two yeasts constituted almost the entire community in the presence of this sugar. With the trisaccharide melezitose as the carbon source, M. pulcherrima and C. subhashii were the most competitive yeasts, while the other species were detected either in low frequency (C. sargentensis, P. kluyveri) or not at all (Hanseniaspora sp., A. pullulans). Interestingly, melezitose is found in honeydew and serves as an exogenous nutrient source in the phyllosphere that is rapidly consumed by phyllosphere yeasts, which thus prevents infections by foliar pathogens (Dik, Fokkema, & Van Pelt, 1991, 1992; Perez, French, Summy, Baines, & Little, 2009; Völkl, Woodring, Fischer, Lorenz, & Hoffmann, 1999). This may explain the competitiveness of the phyllosphere yeast M. pulcherrima in the medium containing melezitose as the sole carbon source. On the other hand, the weak competitiveness of the other yeasts isolated from the phyllosphere (Hanseniaspora sp., P. kluyveri), as well as the results obtained for the soil yeast C. subhashii, suggest that other reasons determine the specific preferences for different carbon sources. In the presence of the carbon source NAG, C. subhashii outcompeted the other five species and was the only yeast detectable after 7 days. Candida subhashii also inhibited the growth of T. basicola more strongly on agar plates containing NAG than on media supplemented with glucose or maltose. These findings may indicate, as suggested previously (Hilber‐Bodmer et al., 2017), that C. subhashii is adapted to grow on insect cuticle and/or fungal cell walls, of either dead or living organisms. Based on these findings, including NAG or chitin in a C. subhashii formulation may stimulate the antagonism and thus the biocontrol activity of this yeast. However, more detailed studies on the mechanisms underlying the differential competitive and antagonistic activities in different media are required to identify the most promising approaches for supporting and improving the biocontrol activity of such yeasts.
In this study, we have adopted MALDI‐TOF MS to develop an efficient, time‐saving method to quantify the yeast community composition in any growth medium or substrate at any given time point. Instead of preparing extracts for MALDI‐TOF MS, yeast cells were directly applied to the target plate (direct smear technique) in order to speed up sample preparation. Additional modifications, as compared with standard MALDI biotyping experiments, were custom‐made reference spectra (also obtained by direct smear) of only the yeasts to be identified and an increase in the laser frequency to 50–60 Hz, which reduced the scanning and identification times considerably. These adaptations led to significant time savings and an efficient protocol for the quantitative assessment of synthetic yeast communities that could be applied to other yeast species and microorganisms as well. For one experimenter it was possible to analyse four agar plates (e.g. the replicates of one condition/time point) in <2 h (including counting, picking colonies, drying of the matrix, MALDI‐TOF MS measurement). For defined, synthetic communities comprising few yeasts that cannot be distinguished by eye, as the one described here, MALDI biotyping by direct smear proved to be a powerful method. The identification threshold could be reduced because only genera had to be distinguished. The direct smear protocol is fast and reliable, and the costs per sample are negligible (for a review and comparison of different methods to identify fungal species see Drissner & Freimoser, 2017). Since individual colonies are identified, this method can be easily combined with counting of colonies/species that are recognized by eye. Other methods that have been used for fungal identifications, for example fatty acid analyses, Fourier‐transform infrared spectroscopy or sequence‐based analyses (sequencing of individual barcodes or metabarcoding), are more difficult to quantify and more costly and time‐consuming to perform and analyse (Botha & Kock, 1993; Drissner & Freimoser, 2017; Salman et al., 2010). However, direct smear MALDI biotyping is not applicable to natural, fungal communities, because filamentous fungi often require the preparation of crude extracts for MALDI biotyping (i.e. pure cultures are necessary) and the threshold for the identification of closely related species (of the same genus) by MALDI biotyping must be higher (usually ≥2), which would result in a larger number of unidentified isolates. For the quantitative analysis of complex, natural, fungal communities, culture‐based (‘culturomics’) or metabarcoding approaches, in combination with next‐generation sequencing, are thus more promising. Both approaches, culturomics and metabarcoding, have different advantages and shortcomings and rather complement each other, as they may not lead to the same quantitative and qualitative species composition of a particular sample (Drissner & Freimoser, 2017; Lagier et al., 2012, 2015).
In summary, we used MALDI‐TOF MS and colony counting to quantify the species composition of a synthetic yeast community over time, in different growth media, as well as on the apple surface and in soil. The results of these studies indicate a strongly increased competitiveness and antagonistic activity of C. subhashii in the presence of NAG as the sole carbon source and defined M. pulcherrima and A. pullulans as the strongest competitors on apple, while C. sargentensis competed the best in soil microcosms. Based on these laboratory studies M. pulcherrima and A. pullulans are promising candidates for biocontrol applications against fungal phyllosphere diseases and C. sargentensis should be further investigated with respect to its use against soilborne fungal pathogens. This study highlights the benefit of studying yeasts exhibiting relevant biological activities in moderately complex, synthetic communities and natural substrates, as compared with studying individual isolates. It may help speed up the time‐consuming analysis of different combinations of strains with promising antagonistic activities and to identify synergistic effects. Finally, this approach may present an avenue for performing and quantifying such experiments with other microorganisms and for other functions than antagonistic activity.
AUTHOR CONTRIBUTIONS
The authors declare that they have no conflict of interest.
ACKNOWLEDGEMENTS
Maja Hilber‐Bodmer is acknowledged for support and supervision in the laboratory. David Drissner and Maria Theresia Stergiou provided access to and supported MALDI‐TOF MS analyses. Christian Ahrens, Eduard Holliger and Inés Sumann made helpful suggestions for the manuscript and Daniel Feusi provided the apples for some experiments.
Gross S, Kunz L, Müller DC, Santos Kron A, Freimoser FM. Characterization of antagonistic yeasts for biocontrol applications on apples or in soil by quantitative analyses of synthetic yeast communities. Yeast. 2018;35:559–566. 10.1002/yea.3321
REFERENCES
- Bautista‐Rosales, P. U. , Calderon‐Santoyo, M. , Servín‐Villegas, R. , Ochoa‐Álvarez, N. A. , & Ragazzo‐Sánchez, J. A. (2013). Action mechanisms of the yeast Meyerozyma caribbica for the control of the phytopathogen Colletotrichum gloeosporioides in mangoes. Biological Control, 65, 293–301. [Google Scholar]
- Bautista‐Rosales, P. U. , Calderon‐Santoyo, M. , Servín‐Villegas, R. , Ochoa‐Álvarez, N. A. , Vázquez‐Juárez, R. , & Ragazzo‐Sánchez, J. A. (2014). Biocontrol action mechanisms of Cryptococcus laurentii on Colletotrichum gloeosporioides of mango. Crop Protection, 65, 194–201. [Google Scholar]
- Blackburn, D. , Shapiro‐Ilan, D. I. , & Adams, B. J. (2016). Biological control and nutrition: Food for thought. Biological Control, 97, 131–138. [Google Scholar]
- Botha, A. , & Kock, J. L. (1993). Application of fatty acid profiles in the identification of yeasts. International Journal of Food Microbiology, 19, 39–51. [DOI] [PubMed] [Google Scholar]
- Castoria, R. , de Curtis, F. , Lima, G. , Caputo, E. , Pacifico, S. , & de Cicco, V. (2001). Aureobasidium pullulans (LS30) an antagonist of postharvest pathogens of fruits: Study on its modes of action. Postharvest Biology and Technology, 22, 7–17. [Google Scholar]
- Dik, A. J. , Fokkema, N. J. , & Van Pelt, J. A. (1991). Consumption of aphid honeydew, a wheat yield reduction factor, by phyllosphere yeasts under field conditions. Netherlands Journal of Plant Pathology, 97, 209–232. [Google Scholar]
- Dik, A. J. , Fokkema, N. J. , & van Pelt, J. A. (1992). Influence of climatic and nutritional factors on yeast population dynamics in the phyllosphere of wheat. Microbial Ecology, 23, 41–52. [DOI] [PubMed] [Google Scholar]
- Drissner, D. , & Freimoser, F. M. (2017). MALDI‐TOF mass spectroscopy of yeasts and filamentous fungi for research and diagnostics in the agricultural value chain. Chemistry, Biology and Technology in Agriculture, 4, 13. [Google Scholar]
- Droby, S. , Wisniewski, M. , Teixidó, N. , Spadaro, D. , & Jijakli, M. H. (2016). The science, development, and commercialization of postharvest biocontrol products. Postharvest Biology and Technology, 122, 22–29. [Google Scholar]
- Freimoser, F. M. , Hilber‐Bodmer, M. , Brunisholz, R. , & Drissner, D. (2016). Direct identification of Monilinia brown rot fungi on infected fruits by matrix‐assisted laser desorption/ionization (MALDI) mass spectrometry. Chemistry, Biology and Technology in Agriculture, 3, 7. [Google Scholar]
- Freimoser, F. M. , Pelludat, C. , & Remus‐Emsermann, M. N. (2016). Tritagonist as a new term for uncharacterised microorganisms in environmental systems. ISME Journal, 10, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goretti, M. , Turchetti, B. , Buratta, M. , Branda, E. , Corazzi, L. , Vaughan‐Martini, A. , & Buzzini, P. (2009). In vitro antimycotic activity of a Williopsis saturnus killer protein against food spoilage yeasts. International Journal of Food Microbiology, 131, 178–182. [DOI] [PubMed] [Google Scholar]
- Hilber‐Bodmer, M. , Schmid, M. , Ahrens, C. H. , & Freimoser, F. M. (2017). Competition assays and physiological experiments of soil and phyllosphere yeasts identify Candida subhashii as a novel antagonist of filamentous fungi. BMC Microbiology, 17, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janisiewicz, W. J. , Jurick, W. M. 2nd , Peter, K. A. , Kurtzman, C. P. , & Buyer, J. S. (2014). Yeasts associated with plums and their potential for controlling brown rot after harvest. Yeast, 31, 207–218. [DOI] [PubMed] [Google Scholar]
- Janisiewicz, W. J. , & Korsten, L. (2002). Biological control of postharvest diseases of fruits. Annual Reviews in Phytopathology, 40, 411–441. [DOI] [PubMed] [Google Scholar]
- Janisiewicz, W. J. , Kurtzman, C. P. , & Buyer, J. S. (2010). Yeasts associated with nectarines and their potential for biological control of brown rot. Yeast, 27, 389–398. [DOI] [PubMed] [Google Scholar]
- Janisiewicz, W. J. , Tworkoski, T. J. , & Kurtzman, C. P. (2001). Biocontrol potential of Metchnikowia pulcherrima strains against blue mold of apple. Phytopathology, 91, 1098–1108. [DOI] [PubMed] [Google Scholar]
- Kunz, S. , & Haug, P. (2006). Development of a strategy for fire blight control in organic fruit growing In 12th International Conference on Cultivation Technique and Phytopathological Probelm in Organic Fruit‐Growing (pp. 113–117). Weinsberg. [Google Scholar]
- Kurtzman, C. P. (1991). DNA relatedness among saturn‐spored yeasts assigned to the genera Williopsis and Pichia . Antonie Van Leeuwenhoek, 60, 13–19. [DOI] [PubMed] [Google Scholar]
- Kurtzman, C. P. , Robnett, C. J. , & Basehoar‐Powers, E. (2008). Phylogenetic relationships among species of Pichia, Issatchenkia and Williopsis determined from multigene sequence analysis, and the proposal of Barnettozyma gen. nov., Lindnera gen. nov., & Wickerhamomyces gen. nov. FEMS Yeast Research, 8, 939–954. [DOI] [PubMed] [Google Scholar]
- Lagier, J. C. , Armougom, F. , Million, M. , Hugon, P. , Pagnier, I. , Robert, C. , … Raoult, D. (2012). Microbial culturomics: paradigm shift in the human gut microbiome study. Clinical and Microbiological Infections, 18, 1185–1193. [DOI] [PubMed] [Google Scholar]
- Lagier, J. C. , Hugon, P. , Khelaifia, S. , Fournier, P. E. , La Scola, B. , & Raoult, D. (2015). The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clinical Microbiology Reviews, 28, 237–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, D. , Fiegna, F. , Behrends, V. , Bundy, J. G. , Phillimore, A. B. , Bell, T. , & Barraclough, T. G. (2012). Species interactions alter evolutionary responses to a novel environment. PLoS Biology, 10, e1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibinger, W. , Breuker, B. , Hahn, M. , & Mendgen, K. (1997). Control of postharvest pathogens and colonization of the apple surface by antagonistic microorganisms in the field. Phytopathology, 87, 1103–1110. [DOI] [PubMed] [Google Scholar]
- Mari, M. , Martini, C. , Guidarelli, M. , & Neri, F. (2012). Postharvest biocontrol of Monilinia laxa, Monilinia fructicola and Monilinia fructigena on stone fruit by two Aureobasidium pullulans strains. Biological Control, 60, 132–140. [Google Scholar]
- Mari, M. , Martini, C. , Spadoni, A. , Rouissi, W. , & Bertolini, P. (2012). Biocontrol of apple postharvest decay by Aureobasidium pullulans . Postharvest Biology and Technology, 73, 56–62. [Google Scholar]
- Minter, D. W. (2009). Cyberlindnera, a replaement name for Lindnera Kurtzman et al., nom. illegit. Mycotaxon, 110, 473–476. [Google Scholar]
- Mounir, R. , Durieux, A. , Bodo, E. , Allard, C. , Simon, J. P. , Achbani, E. H. , … Jijakli, M. H. (2007). Production, formulation and antagonistic activity of the biocontrol like‐yeast Aureobasidium pullulans against Penicillium expansum . Biotechnology Letters, 29, 553–559. [DOI] [PubMed] [Google Scholar]
- Nassar, A. H. , El‐Tarabily, K. A. , & Sivasithamparam, K. (2005). Promotion of plant growth by an auxin‐producing isolate of the yeast Williopsis saturnus endophytic in maize (Zea mays L.) roots. Biology and Fertility of Soils, 42, 97–108. [Google Scholar]
- Naumova, E. S. , Lee, C.‐F. , Kondratieva, V. I. , Sadykova, A. Z. , & Naumov, G. I. (2017). Molecular genetic polymorphism of soil yeasts of the genus Williopsis from Taiwan Island. Russian Journal of Genetics, 53, 561–567. [Google Scholar]
- Pelliccia, C. , Antonielli, L. , Corte, L. , Bagnetti, A. , Fatichenti, F. , & Cardinali, G. (2011). Preliminary prospection of the yeast biodiversity on apple and pear surfaces from Northern Italy orchards. Annals of Microbiology, 61, 965–972. [Google Scholar]
- Perez, J. L. , French, J. V. , Summy, K. R. , Baines, A. D. , & Little, C. R. (2009). Fungal phyllosphere communities are altered by indirect interactions among trophic levels. Microbial Ecology, 57, 766–774. [DOI] [PubMed] [Google Scholar]
- Prasongsuk, S. , Lotrakul, P. , Ali, I. , Bankeeree, W. , & Punnapayak, H. (2017). The current status of Aureobasidium pullulans in biotechnology. Folia Microbiologia (Praha), 63, 129–140. [DOI] [PubMed] [Google Scholar]
- Rahi, P. , Prakash, O. , & Shouche, Y. S. (2016). Matrix‐assisted laser desorption/ionization time‐of‐flight mass‐spectrometry (MALDI‐TOF MS) based microbial identifications: Challenges and scopes for microbial ecologists. Frontiers in Microbiology, 7, 1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salman, A. , Tsror, L. , Pomerantz, A. , Moreh, R. , Mordechai, S. , & Huleihel, M. (2010). FTIR spectroscopy for detection and identification of fungal phytopathogenes. Spectroscopy, 24, 261–267. [Google Scholar]
- Slavikova, E. , Vadkertiova, R. , & Vranova, D. (2007). Yeasts colonizing the leaf surfaces. Journal of Basic Microbiology, 47, 344–350. [DOI] [PubMed] [Google Scholar]
- Spadaro, D. , & Droby, S. (2016). Development of biocontrol products for postharvest diseases of fruit: the importance of elucidating the mechanisms of action of yeast antagonists. Trends in Food Science and Technology, 47, 39–49. [Google Scholar]
- Tan, A. W. , Lee, P. R. , Seow, Y. X. , Ong, P. K. , & Liu, S. Q. (2012). Volatile sulphur compounds and pathways of L‐methionine catabolism in Williopsis yeasts. Applied Microbiology and Biotechnology, 95, 1011–1020. [DOI] [PubMed] [Google Scholar]
- Tyc, O. , Wolf, A. B. , & Garbeva, P. (2015). The effect of phylogenetically different bacteria on the fitness of Pseudomonas fluorescens in sand microcosms. PLoS One, 10, e0119838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadkertiova, R. , Molnarova, J. , Vranova, D. , & Slavikova, E. (2012). Yeasts and yeast‐like organisms associated with fruits and blossoms of different fruit trees. Canadian Journal of Microbiology, 58, 1344–1352. [DOI] [PubMed] [Google Scholar]
- van Nieuwenhuijzen, E. J. (2014). Aureobasidium In Tortorello M. L. (Ed.), Encyclopedia of food microbiology (2nd ed.) (pp. 105–109). Oxford: Academic Press. [Google Scholar]
- Völkl, W. , Woodring, J. , Fischer, M. , Lorenz, M. W. , & Hoffmann, K. H. (1999). Ant‐aphid mutualisms: The impact of honeydew production and honeydew sugar composition on ant preferences. Oecologia, 118, 483–491. [DOI] [PubMed] [Google Scholar]
- Westerdijk Fungal Biodiversity Institute (2011). Yeasts species and yeasts. Utrecht. Retrieved May 25, 2017 from http://www.westerdijkinstitute.nl/Collections/Biolomics.aspx?Table=Yeasts%202011 and http://www.westerdijkinstitute.nl/Collections/Biolomics.aspx?Table=Yeasts%20species


