Abstract
Emerging evidence indicates that epigenetic mechanisms like DNA methylation directly contribute to metabolic regulation. For example, we previously demonstrated that de novo DNA methyltransferase Dnmt3a plays a causal role in the development of adipocyte insulin resistance. Recent studies suggest that DNA demethylation plays an important role in the developmental process of adipocytes. However, little is known about whether DNA demethylase ten-eleven translocation (TET) proteins regulate the metabolic functions of adipocytes.
METHODS:
The expression of Tet genes was assessed in the fractionated adipocytes of chow- and high fat diet–fed C57/Bl6 mice using qPCR and western blotting. The effect of Tet2 gain- or loss-of-function in fully mature 3T3-L1 adipocytes in the presence/absence of Rosiglitazone (Rosi) and TNF-α on insulin sensitivity was using the insulin-stimulated glucose uptake and insulin signaling assays. Gene expression and DNA methylation analyses of PPARγ target genes was performed in the same setting. In addition, PPARγ reporter assays, co-immunoprecipitation assays, PPARγ ChIP-PCR analyses were performed.
RESULTS:
We found that adipose expression of TET2, alone among its family members, was significantly reduced in diet-induced insulin resistance. TET2 gain-of-function was sufficient to promote insulin sensitivity while loss-of-function was necessary to facilitate insulin sensitization in response to the PPARγ agonist Rosiglitazone (Rosi) in cultured adipocytes. Consistent with this, TET2 was required for Rosi-dependent gene activation of certain PPARγ targets accompanied by changes in DNA demethylation at the promoter regions. Furthermore, TET2 was necessary to sustain PPARγ binding to target loci upon activation with Rosi via physical interaction with PPARγ.
CONCLUSIONS:
Our data demonstrate that TET2 works as an epigenetic regulator of Rosi-mediated insulin sensitization and transcriptional regulation in adipocytes.
Keywords: insulin sensitivity, adipocytes, epigenetics, DNA demethylation, TET proteins
Introduction
Insulin resistance is a key underlying feature of type 2 diabetes and a frequent complication in multiple clinical conditions including obesity, aging, and cardiovascular disease. Extraordinary efforts have gone into defining the mechanisms that underlie insulin resistance, with most of the attention focused on insulin signal transduction and other mitochondrial and cytosolic pathways such as ER stress [1-5]. The data supporting the involvement of these non-mutually exclusive pathways are convincing, but there is still a great deal of uncertainty about the mechanisms by which cells and organisms become insulin resistant.
Environmental factors, such as diet and aging, are important risk factors for metabolic disorders. The intricate interaction between genes and the environment is mediated by epigenetic mechanisms, suggesting they play a major role in the etiopathogenesis of metabolic dysregulation. DNA methylation is an epigenetic mark involving the covalent transfer of a methyl group to the C-5 position (5mC) of cytosine by DNA methyltransferases (Dnmts) [1]. Mounting evidence points to a role for DNA methylation in the pathogenesis of metabolic disorders: 1) obesity in agouti mice is associated with reduced DNA methylation at the regulatory region of the agouti gene, leading to its ectopic expression during development [2]; 2) changes in DNA methylation at key metabolic genes, such as Cox7a1 [3], PPARGC1a [4], IGF-2 [5], and Pomc [6], associate with various metabolic insults including those from aging, obesity, anorexia, and prenatal exposure to famine; 3) recent genome-wide profiling studies have identified distinct global DNA methylation patterns that associate with obesity and diabetes [7-9]; 4) altered DNA methylation is linked to the transgenerational passage of metabolic disorders [9-11]; and 5) both pharmacological and genetic studies have shown that certain DNMTs directly contribute to adipose insulin resistance [10, 11].
DNA methylation was long thought to be a static epigenetic mark, but recent evidence demonstrates that it undergoes dynamic remodeling [12], which involves the ten-eleven translocation (TET) protein family acting as DNA demethylases. TET proteins (TET1, 2, and 3) oxidize 5mC to hydroxymethylcytosine (5hmC), which is then converted to unmethylated cytosine (5C) through base excision repair (BER) and thymidine DNA glycosylase (TDG). In contrast to DNA methylation, TET-mediated DNA demethylation associates with increased enhancer activity and transcription factor occupancy in different biological contexts. For example, 5mC enrichment at distal enhancers is inversely correlated with enhancer activity [13-15], whereas 5hmC enrichment is positively correlated with enhancer activity and Foxp3 binding in immune cells [16-18]. A recent large-scale functional association analysis from GWAS data identified a genetic variant within TET2 (rs9884482) that associates with fasting insulin level, suggesting an impact on insulin resistance [19]. Furthermore, recent global profiling studies have demonstrated that 5hmC colocalizes with PPARγ binding at enhancers in 3T3-L1 adipocytes [20], and PPARγ-positive nuclei sorted from visceral adipose tissue from healthy humans is strongly co-enriched with 5hmC [21]. Together, these studies suggest the presence of functional and physical interactions between TET proteins and PPARγ.
Here, we found that adipocyte expression of Tet2 is downregulated in obesity. Our gain- and loss-of-function studies in 3T3-L1 adipocytes found that TET2 was required for Rosiglitazone (Rosi)-mediated insulin sensitization. Consistent with this, Tet2-knockdown adipocytes had decreased expression of some of the key PPARγ target genes important for insulin sensitivity in response to Rosi. TET2-dependent gene regulation of PPARγ targets involved site-selective DNA demethylation at the promoter regions. Mechanistically, we show that TET2 and PPARγ physically interact and that TET2 is required to maintain DNA binding of PPARγ upon ligand activation.
Methods
Cell culture:
3T3-L1 preadipocytes were obtained from ATCC and maintained and differentiated as described [22]. These cells had the ability to differentiate and were confirmed to be mycoplasma negative. To generate lentivirus particles, lentiviral constructs were cotransfected with pMD2.G- and psPAX2-expressing plasmids into 293T cells. After 48 hr, the virus-containing supernatant was collected, filtered through 0.45-mm filters, and added to mature 3T3-L1 adipocytes for 24 hr along with 8 mg/ml polybrene. Immortalized human preadipocyte cell line was cultured as described [23]. Transduction efficiency was determined by comparing to cells transduced in parallel with a GFP-expressing lentivirus.
3H-2-DG assay:
Mature 3T3-L1 adipocytes were incubated in serum-free DMEM for 4–6 hr. Cells were then washed three times with KRH buffer (12 mM HEPES, pH 7.4, 121 mM NaCl, 5 mM KCl, 0.33 mM CaCl2, and 1.2 mM MgSO4) and incubated for 20 min in KRH buffer in the absence or presence of 50 nM insulin. Cells were treated with 2-deoxy-d-[2,6-3H]-glucose (0.33 mCi/ml) for another 10 min. Glucose uptake was stopped quickly by three rapid washes with KRH buffer containing 200 mM glucose and 10 mM cytochalasin B on ice. Cells were solubilized in 0.1% SDS for 30 min, and radioactivity was measured by liquid scintillation counting.
Antibodies:
Antibodies were purchased from Cell Signaling Technology (PPARγ, Cat: 2443S, Lot: 4; Akt, Cat: 9272; Lot: 22; pAkt [S473], Cat: 9271, Lot: 12; IRS-1, Cat: 2382; Lot: 4), from Thermo Fisher Scientific (β-Actin: MA5-14739; pIRS1[pY612]: Cat: 44–816G, Lot: 1100477B), from Proteintech Group (TET2, Cat: 21207-1-AP, Lot: 00047680), Millipore (Ser82 PPARγ), from Santa Cruz Biotechnology (c-Myc, Cat: SC-40, Lot: F1917).
RNA extraction and quantitative PCR:
Total RNA was extracted from cells or tissues using TRIzol reagent according to the manufacturer’s instructions. cDNA was reverse-transcribed from 1 μg of RNA using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Quantitative PCR (qPCR) was performed with SYBR Green qPCR Master Mix (Bioneer) using a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Primer sequences are listed in Supplementary Table 1. The relative amount of mRNA normalized to 36B4 was calculated using the delta-delta method [24].
Plasmids:
Tet2-CD (#79554) and Tet2-HxD (#79611) cDNA was purchased from Addgene, PCR amplified, and subcloned into pCDH vector with the XbaI and EcoRI sites. sgRNAs that targeted Tet2 were cloned into lentiCRISPR v2 vector. Hairpin and sgRNA sequences are shown in Supplementary Table 1.
hMeDIP-qPCR:
Genomic DNA was sheared using a Covaris S220 to an average of 200–800 bp. Two micrograms of denatured DNA was incubated with 2 mg of anti-5-hydroxymethylcytidine antibody (EpiGentek) in IP buffer (10 mM Na-Phosphate pH 7.0, 0.14 M NaCl, 0.05% Triton X-100) for 2 hr at 4°C. Antibody-bound DNA was collected with 20 ml of Dynabeads anti-mouse IgG (Invitrogen Dynal AS) for 1 hr at 4°C on a rotating wheel and successively washed five times with washing buffer (0.1% SDS, 1 Triton X-100, 2 mM EDTA, 20 mM Tris-HCI pH 8.1, 150 mM NaCl) and twice with TE (10 mM TrisCl, 1 mM EDTA pH 8.0). DNA was recovered in 125 ml of digestion buffer (50 mM Tris pH 8.0, 10 mM EDTA, 0.5% SDS, 35 mg proteinase K) and incubated for 3 hr at 65°C. Recovered DNA was used for qPCR analysis. Primers for MeDIP-qPCR studies are listed in Supplementary Table 1. All data were normalized to input.
ChIP-qPCR:
Cells were crosslinked with 1% formaldehyde for 10 min at room temperature. Cross-linked chromatin was sonicated using an S220 Ultrasonicator (Covaris) to generate DNA fragments of ~150–600 bp. Inputs were taken from cleared lysates, and the rest were rotated O/N at 4°C with PPARγ (Cell Signaling Technology, 2443S) and IgG antibodies to precipitate protein:DNA complexes. An aliquot of 20 μl of pre-washed Dynabeads Protein G was added per IP and rotated 1 hr at 4°C. Beads were successively washed in low-salt RIPA buffer (20 mM Tris-HCl [pH 8.0], 1 mM EDTA, 1% Triton x-100, 0.1% SDS, 140 mM NaCl, 0.1% Na deoxycholate), high-salt RIPA buffer (20 mM Tris-HCl [pH 8.0], 1 mM EDTA, 1% Triton x-100, 0.1% SDS, 500 mM NaCl, 0.1% Na deoxycholate), LiCl buffer (250 mM LiCl, 0.5% NP40, 0.5% Na deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.0]) and TE buffer (10 mM Tris-HCl [pH 8.0] and 1 mM EDTA). Each reaction was then incubated in digestion buffer (50 mM Tris-HCl [pH 8.0], 1 mM EDTA, 100 mM NaCl, 0.5% SDS, proteinase K) for a minimum of 4 hr at 65°C to reverse cross-links. DNA was recovered using a phenol-chloroform extraction. Real-time qPCR primers are listed in Supplementary Table 1. All data were corrected for multiple hypothesis testing. All data were normalized to input.
Co-immunoprecipitation:
Co-immunoprecipitation (Co-IP) was performed according to a modified protocol as previously described[22]. Briefly, 293T cells were transfected with various DNA constructs using Lipofectamine 3000 (Invitrogen). A day after transfection, cells were lysed with Triton X lysis buffer (Boston Bio Products) containing 50 mM Tris-HCL, pH 7.4, 150 mM NaCl 1% Triton X-100, 5 mM EDTA, plus protease inhibitors (complete, Roche). 500 mg of protein was incubated with the appropriate antibodies overnight. The next day, protein A/G Dynabeads (Thermo Fisher Scientific) were added and incubated for 1 hr, washed with lysis buffer five times and PBS once. Beads were eluted with non-reducing SDS/PAGE loading buffer and subjected to SDS/PAGE and Western blotting.
Western blot analysis:
Whole-cell protein lysates were prepared according to the manufacturer’s protocol using RIPA lysis buffer and protease inhibitor cocktail. 20–30 μg of protein was resolved using 4–20% Tris-glycine gradient gels and transferred to PVDF membrane. After blocking with 5% non-fat dried milk in PBSTween (0.25%), membranes were incubated with the appropriate primary antibodies against FLAG (Sigma), MYC, PPARγ (Cell signaling technology), or TET2 (Proteintech Group). Anti-goat and anti-mouse or rabbit IgG–peroxidase conjugate (Sigma) were used to detect primary antibodies.
Luciferase reporter assays:
Reporter plasmids were co-transfected with expression vectors of Tet2-CD, Tet2-HxD, and PPARγ into 293T cells using a Profection kit (Promega Corporation). The Renilla reference plasmid was co-transfected to normalize transfection efficiency, and the mass of transfected plasmids was balanced with empty vector (pCDH-EGFP). 48 hr after transfection, cells were harvested and measured using the Dual-Luciferase Reporter Assay System (Promega Corporation).
Results
Tet2 expression is enriched in adipocytes, and adipocyte Tet2 expression is diminished in diet-induced obesity.
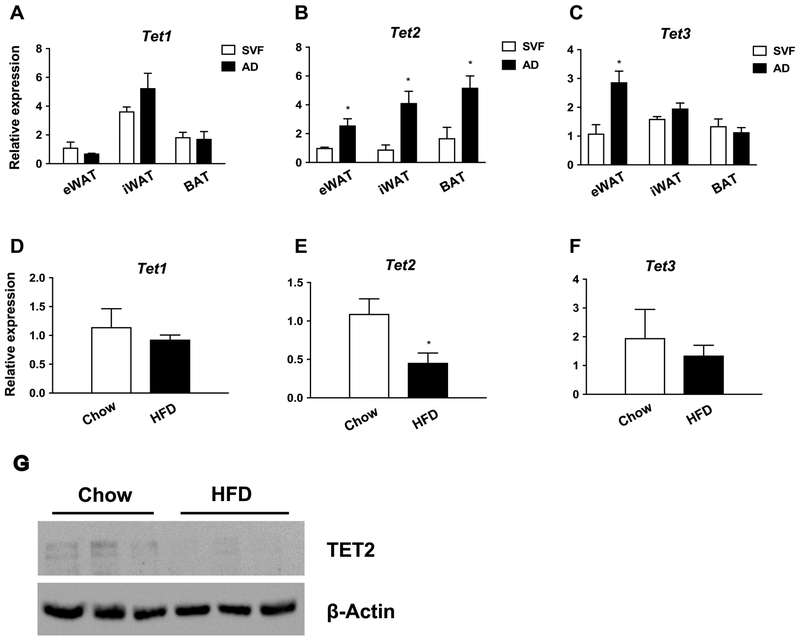
To explore the potential role of TET proteins in adipocytes, we assessed their relative expression between the adipose and stromal-vascular fraction (SVF), which contains preadipocytes (Fig. 1, Supplemental Fig. 1). Overall, Tet3 expression was highest, followed by Tet2 and Tet1, in all three tested adipose depots and in both fractions (Supplemental Fig. 1). However, only Tet2 was expressed more in adipocytes than in SVF (Fig. 1A-C). Moreover, both mRNA and protein expression of Tet2 in adipocytes was significantly reduced in a model of diet-induced obesity (DIO) (Fig. 1E, G). This implies a regulatory role for TET2 in the metabolic function of mature adipocytes.
Figure 1. Tet2 expression is enriched in adipocytes, and adipocyte Tet2 levels are reduced in obesity.
(A-C) Tet expression was measured by q-PCR in the stromal vascular fraction (SVF) and adipose fraction (AD) of the eWAT, iWAT, and BAT of chow-fed C57BI/6 mice (n= 3, p < 0.05, Student’s t-test, mean ± s.e.m.); (D-F) Tet expression in fractionated adipocytes from chow- and high fat diet–fed (HFD) C57Bl/6 mice (n = 3, p < 0.05, Student’s t-test, mean ± s.e.m.); (G) Western blot showing TET2 protein levels in fractionated adipocytes from chow vs. HFD mice. eWAT: epididymal WAT; iWAT: inguinal WAT; BAT: brown adipose tissue.
TET2 plays a necessary and sufficient role for Rosi-mediated insulin sensitization in cultured adipocytes.
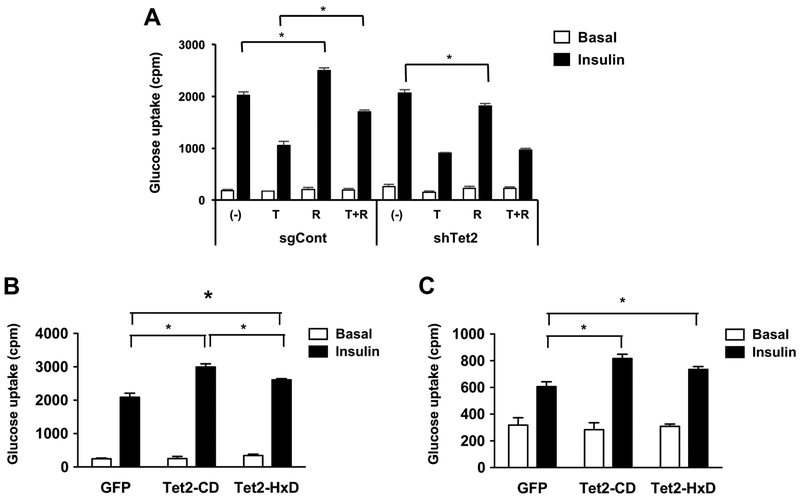
To investigate the potential role of TET2 in the regulation of insulin sensitivity, we performed a 2-deoxyglucose uptake assay in fully mature 3T3-L1 adipocytes that were lentivirally transduced with single-guide RNAs specific for Tet2 and an empty vector (Supplemental Fig. 2). In control adipocytes, TNF-α, a mediator of insulin resistance, potently inhibited insulin-stimulated glucose uptake, whereas the TZD compound Rosiglitazone (Rosi) enhanced it (Fig. 2A). Tet2*knockdown adipocytes had a similar basal insulin sensitivity, and as expected, TNF-α caused insulin resistance similar to control cells (Fig. 2A). However, Rosi largely failed to improve insulin sensitivity in the presence and absence of TNF-α (Fig. 2A). This result suggests that TET2 plays a necessary role in Rosi-mediated improvement of insulin sensitivity in cultured adipocytes.
Figure 2. TET2 is necessary and sufficient to facilitate Rosi-mediated insulin sensitization in vitro.
(A) Mature 3T3-L1 adipocytes at day 8 were lentivirally transduced with single-guide RNA specific for Tet2 (sgTet2) or an empty plasmid vector (sgCont). Two days after transduction, cells were treated with TNF (4 ng/ml), Rosi (100 nM), or both for 2 additional days and assessed for insulin-stimulated glucose uptake (3H-2-DG assay, n = 6, P < 0.05). (B, C) Basal and insulin-stimulated glucose uptake in mature 3T3-L1 adipocytes (B) and immortalized human adipocytes (C) transduced with lentivirus expressing the catalytic domain of wild-type (Tet2-CD) or mutated (Tet2-HxD) Tet2 or GFP (n = 6, p < 0.05, Student’s t-test, mean ± s.e.m.).
Next, we tested whether TET2 gain-of-function is sufficient to improve insulin sensitivity in adipocytes. Overexpressing the catalytic domain of the wild-type allele (Tet2-CD) was sufficient to increase insulin-stimulated glucose uptake (Fig. 2B). Previous studies report a catalytic and non-catalytic function for Tet2; therefore, we asked whether its DNA demethylase activity is critical for facilitating insulin sensitivity by overexpressing the catalytically inactive allele Tet2-HxD [25], which largely lacks DNA demethylase activity (Supplemental Fig. 3). To our surprise, Tet2-HxD still promoted insulin-stimulated glucose uptake, albeit to a lesser extent than wild type (Fig. 2B). To test whether the role of TET2 is conserved in human adipocytes, we performed gain-of-function study using immortalized human preadipocytes [23], Similar to 3T3-L1 adipocytes, overexpression of both TET2-CD and HxD significantly increased insulin-stimulated glucose uptake (Fig. 2C).
Together, these results indicate that TET2 facilitates insulin sensitivity both in a DNA demethylase-dependent and -independent manner. Despite a clear phenotypic difference in insulin-stimulated glucose uptake between these models of TET2 gain- and loss-of-function, there was a marginal difference in insulin signal transduction in both settings (Supplemental Fig. 4).
TET2 facilitates the transcriptional activity of PPARγ in both a catalytic-dependent and independent manner.
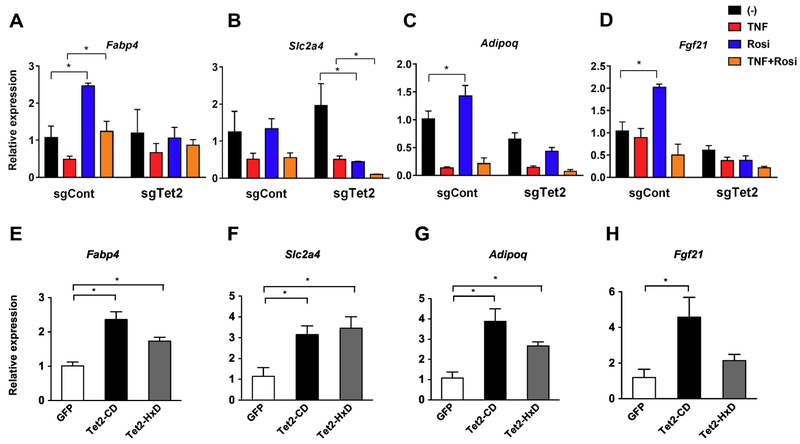
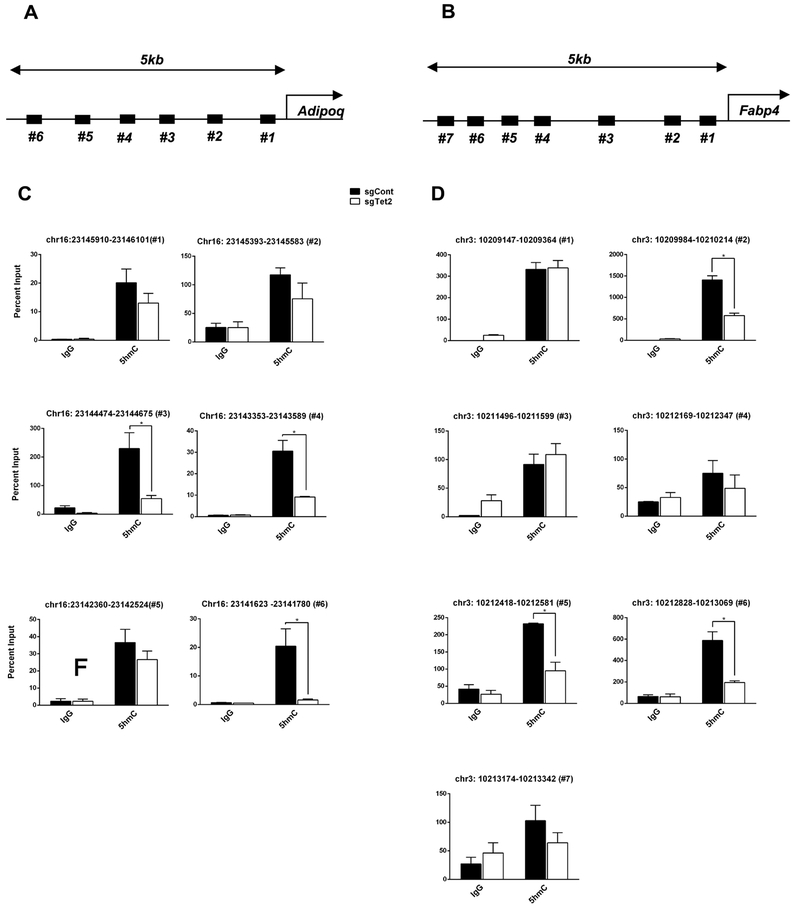
TET-dependent DNA demethylation associates with transcriptional regulation in several biological contexts including adipogenesis [26-29]. To determine the role of TET2 in Rosi-mediated transcriptional regulation, we measured a set of PPARγ target genes including those important to the regulation of insulin sensitivity [46]. Consistent with the insulin-stimulated glucose uptake assay, Tet2-knockdown adipocytes had reduced ability to acutely stimulate the transcription of PPARγ targets in response to Rosi, with and without TNF-α (Fig. 3). For example, Rosi-induced expression of Fabp4, Adipoq, and Fgf21 was not seen in Tet2-knockdown adipocytes (Fig. 3A-D). Of note, this was not due to the loss of PPARγ expression (Supplemental Fig. 5). Conversely, the expression of these PPARγ target genes was significantly increased by the overexpression of Tet2-CD and less potently by Tet2-HxD (Fig. 3E-H). Consistent with these expression patterns, we observed reduced DNA demethylation activity at several CpG-rich regions of the Adipoq and Fabp4 promoters upon TET2 loss-of-function (Fig 4).
Figure 3. TET2 is required for Rosi-dependent gene regulation of PPARγ targets.
Mature 3T3-L1 adipocytes were transduced with Tet2-knockdown (A-D) or -overexpressor (E-H) lentiviruses. Two days after transduction, cells were treated with TNF (4 ng/ml), Rosi (1 μM), or both for 4 hours. Gene expression of a set of PPARγ target genes was determined by q-PCR (n = 3, p < 0.05, Student’s t-test, mean ± s.e.m.).
Figure 4. TET2 loss-of-function leads to reduced DNA demethylation at the promoter regions of Adipoq and Fabp4.
Schematic showing the tested regions for hMeDIP-qPCR at Adipoq (A) and Fabp4 (C). Target regions were chosen using MethFinder (http://www.urogene.org/methprimer) on regions proximal (5 kb) to the Adipoq or Fabp4 transcriptional start site. (B, D) Genomic DNA extracted from mature 3T3-L1 adipocytes transduced with Tet2-overexpressor or -knockdown lentiviruses was subjected to hMeDIP-qPCR using anti-5hmC and IgG antibodies, (n = 3, p < 0.05, Student’s t-test, mean ± s.e.m.).
Tet2 promotes PPARγ-driven transcriptional activity through a physical interaction.
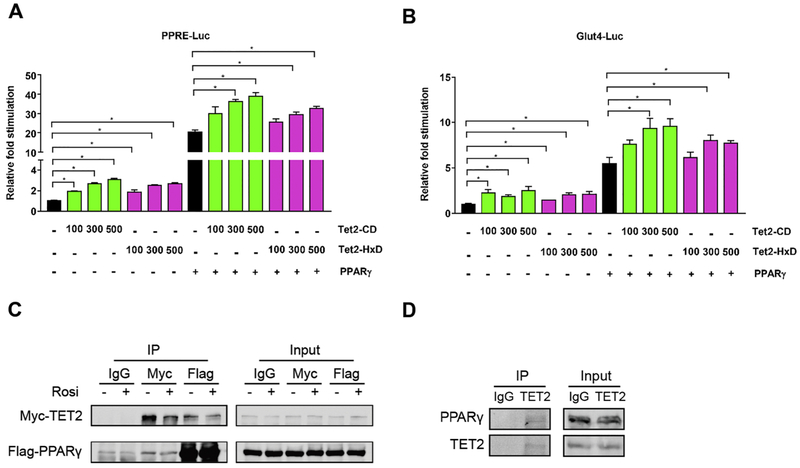
To directly assess whether TET2 regulates PPARγ-driven transcriptional activity, we used a reporter assay with PPARγ X3-TK-luc, which contains 3 copies of the PPARγ-binding DR1 site, and a plasmid with the Glut4 2.4-kb promoter (Fig. 5A, B). Overexpression of Tet2-CD or Tet2-HxD alone significantly increased the activity of both reporter plasmids, and the effect was additive when co-expressed with PPARγ (Fig. 5A, B).
Figure 5. TET2 facilitates the transcriptional activity of PPARγ via physical interaction.
(A, B) 293T cells were co-transfected with vectors expressing Tet2-CD, Tet2-HxD, PPARγ, and reporter plasmids containing 3 copies of PPREs (A) and 2.4 kb of the Glut4 promoter (B). We measured Luciferase and Renilla activity 24 hr after transfection. Shown is the relative fold stimulation of luciferase activity after assessing transfection efficiency with Renilla (n = 3, p < 0.05, Student’s t-test, mean ± s.e.m.). (C) 293T cells transfected with plasmids expressing Flag-PPARγ and Myc-TET2. At 24 hr after transfection, cells were treated with 1 μM Rosi or DMSO for 1 hour. Cell lysates were harvested and immunoprecipitated using anti-Flag, anti-Myc, or IgG and blotted with anti-Flag- and Myc-antibodies (n = 3, p < 0.05, Student’s t-test, mean ± s.e.m.). (D) Mature 3T3-L1 adipocytes were harvested, and endogenous TET2 protein was immunoprecipitated using anti-TET2 antibody and blotted with anti-PPARγ and TET2 antibodies (n = 3, p < 0.05, Student’s t-test, mean ± s.e.m.).
PPARγ colocalizes with 5hmC in adipocytes [20]; therefore, we reasoned that TET2 and PPARγ may physically interact, which could lead to TET2 regulating the transcriptional activity of PPARγ. Indeed, we found that the tagged version of TET2 and PPARγ co-immunoprecipitated in 293T cells in the presence and absence of Rosi (Fig. 5C). Furthermore, the physical interaction of the native proteins was confirmed in 3T3-L1 adipocytes (Fig. 5D). These results suggest that TET2 works in the same protein complex as PPARγ, modulating its transcriptional activity. Further, we investigated whether TET2 may affect post-translational modifications (PTM) of PPARγ [30-32], which are important for the regulation of transcriptional activity of PPARγ. In result, we did not see a major difference in the levels of Ser283 or acetylation of PPARγ in both TET2 gain- and loss-of-function settings (Supplemental Fig. 5F, G).
TET2 is required for PPARγ to maintain DNA binding at selective target loci in response to Rosi.
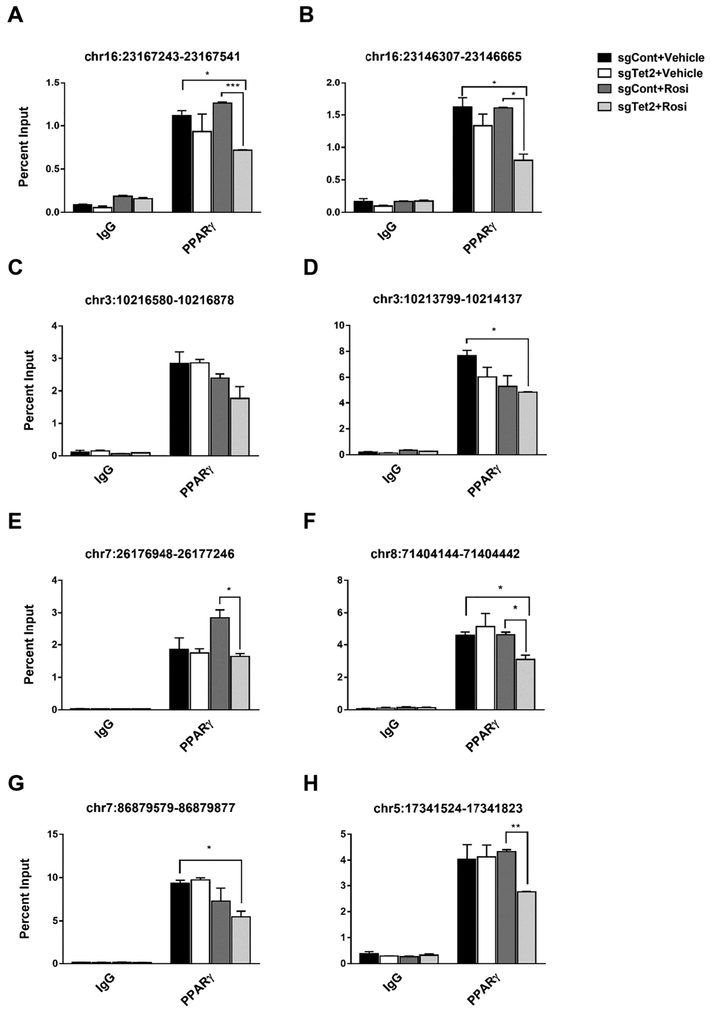
TET proteins and DNA demethylation at gene regulatory regions affects the DNA binding affinity of transcription factors. Thus, we examined PPARγ binding affinity at previously characterized PPARγ binding sites [23] after Rosi treatment using ChIP-PCR. PPARγ binding was mostly intact with TET2 gain-of-function (Supplemental Fig. 6), suggesting TET2 alone is not sufficient to increase PPARγ binding to target loci. Also, overall, the basal level of PPARγ binding was intact in Tet2-knockdown cells (Fig. 6). However, PPARγ binding affinity was greatly weakened by Rosi treatment in Tet2-knockdown cells compared to the control (Fig. 6), which suggests that TET2 is required to sustain PPARγ binding upon ligand activation. Together, these results suggest that TET2 plays an important role in facilitating the Rosi-stimulated transcriptional program by sustaining PPARγ binding.
Figure 6. TET2 is necessary to sustain PPARγ binding to target loci upon ligand activation with Rosi.
Mature 3T3-L1 adipocytes were lentivirally transduced with single-guide RNA specific for Tet2 (sgTet2) or with empty plasmid vector (sgCont). Two days after transduction, cells were treated with Rosi (1 μM) or DMSO for 1 hour. ChIP-qPCR analysis was performed using PPARγ antibody. Primers amplifying the individual PPREs are indicated (n = 3, p < 0.05, Student’s t-test, mean ± s.e.m.).
Discussion
We previously demonstrated a causal role for DNMT3a in the development of adipocyte insulin resistance both in vitro and in vivo [11]. Here, we revealed an opposing functional role for TET2 as a modulator of insulin sensitivity. In particular, we demonstrated that TET2 plays an important role in Rosi-mediated insulin sensitization and transcriptional regulation in adipocytes. Although the functional roles of Dnmt3a and TET2 are opposite in the regulation of insulin sensitivity, their underlying mechanisms, such as gene targets, do not seem to converge. We did not find obvious PPARγ target genes using RNA-Seq on DNMT3a gain- or loss-of-function models.
Emerging evidence indicates that epigenetic control directly contributes to metabolic regulation. For example, administering pan-inhibitors for histone deacetylase has beneficial metabolic effects in both mice and humans, such as increased energy expenditure and insulin sensitivity and secretion [33-36]. In line with pharmacological studies, mutant mouse models carrying loss-of-function histone modifiers (e.g., Mll2, Ehmt1, Jmhd2a, Lsd1) have profoundly different whole-body metabolism[37-41].
PPARγ is the master transcription factor of adipogenesis, as evidenced by its necessary and sufficient role [42,43]. The expression of PPARγ is highly induced during adipogenesis, and it is expressed primarily in adipose tissue [44,45], In addition to development, PPARγ plays a pivotal role in the regulation of numerous genes important for adipocyte functions such as lipid metabolism, adipokine secretion, and insulin sensitivity [46]. TZDs, which are synthetic ligands of PPARγ, exert a powerful insulin-sensitizing effect in vitro and in vivo [47], primarily working on the adipose tissue [48,49]. It has been proposed that the insulin-sensitizing effect of Rosi involves the regulation of key adipocyte target genes including adiponectin [50] and FGF-21 [51] while suppressing the expression of genes promoting insulin resistance including TNF-α [52], Resistin [53], and Rbp4 [54]. Thus, it is critical to fully understand how TZDs regulate the transcriptional program of PPARγ to achieve healthy adipose development and metabolism.
It is relatively well understood how TZDs activate PPARγ at the molecular level. For example, ligand activation of PPARγ recruits coactivators such as SRC-1, CBP, p300, and MED1 [55-58]. However, it is poorly understood whether ligand activation of PPARγ involves epigenetic mechanisms. Recent studies suggest that TET protein–mediated DNA demethylation is necessary for adipogenesis in vitro. In addition, 5hmC enrichment overlaps with enhancer peaks induced during adipogenesis [20], and it’s enriched in PPARγ-positive nuclei in association with other chromatin modifiers [21]. Consistent with this, PPARγ induces DNA demethylation at the Plin promoter region during adipogenesis [59]. These studies strongly suggest that TET-mediated DNA demethylation is important in adipocyte development. By contrast, our study focuses on the functional role of TET2 in the metabolic function of mature adipocytes.
During the revision of our manuscript, Wu et al published their work by revealing a novel axis between TET2 and AMPK in the regulation of glucose homeostasis [60]. In that study, authors demonstrated that hyperglycemia destabilizes the TET2 through the inhibition of AMPK-mediated TET2 phosphorylation at Ser99, which lead to downregulation of global 5hmC levels in diabetic patients. Further, hyperglycemia promoted tumor growth was suppressed by TET2 and that anti-tumor effect of Metformin requires the AMPK–TET2–5hmC axis. Together, their findings suggest that TET2 is an epigenetic sensor of glucose levels in tumor. It will be highly interesting to find out whether such regulatory loop operates in non-oncogenic state.
In summary, our study identified a novel role for TET2 as an epigenetic regulator of PPARγ ligand-mediated insulin sensitization and transcriptional regulation in adipocytes. Future studies will be required to fully elucidate the full range of TET2-dependent transcriptional effects on the genomic targets of PPARγ as well as the in vivo role of TET2 in adipose and whole-body metabolism.
Supplementary Material
Supplemental Figure 1. The relative expression of Tets in adipose vs. SVF fractions from fat depots. Tet expression was measured by q-PCR in adipocytes (AD) and SVF fractions from eWAT, iWAT, and BAT. Shown is the relative fold expression of Tet family members in AD (A) and SVF (B) when normalized to Tet1 expression (n = 3, p < 0.05, Student’s t-test, mean ± s.e.m.).
Supplemental Figure 2. TET2 knockdown and overexpression efficiency in TET2 gain- and loss-of-function models. Mature 3T3-L1 adipocytes were transduced with Tet2-knockdown (A) or -overexpressor lentiviruses (B). Two days after transduction, cell lysates were harvested and blotted using anti-TET2 antibody (A) and anti-Myc antibodies (B) (n = 3, p < 0.05, Student’s t-test, mean ± s.e.m.).
Supplemental Figure 3. TET2 gain- and loss-of-function leads to global changes in 5hmC level. (A) Genomic DNA was extracted from mature 3T3-L1 adipocytes transduced with Tet2-overexpressor or -knockdown lentiviruses. A dot blot was performed to assess the total amount of 5hmC using the indicated amount of genomic DNA. (B) Blots from (A) were processed with methylene blue staining. (C, D) Quantification of 5hmC dot blot by normalizing with methylene blue staining (n = 3, p < 0.05, Student’s t-test, mean ± s.e.m.).
Supplemental Figure 4. Insulin signal transduction is marginally changed in Tet2 gain- and loss-of-function models. Mature 3T3-L1 adipocytes were transduced with Tet2-knockdown (A-C) or -overexpressor lentiviruses (D-F). Three days after transduction, cells were treated with Rosi (1 μM), TNF (4 ng/ml), or both for two additional days. Serum-starved cells were stimulated with 20 nM insulin for 5 min, and cell lysates were subjected to immunoblotting with antibodies against total and phospho-AKT and -IRS-1. (B, C, E, F) Quantification of western blot in (A, C) (n = 3, p < 0.05, Student’s t-test, mean ± s.e.m.).
Supplemental Figure 5. Total- and post-translational levels of PPARγ is not grossly altered in TET2 gain- and loss-of-function models. Mature 3T3-L1 adipocytes were lentivirally transduced with Tet2-knockdown or overexpressor plasmids. Shown is protein (A-C) and mRNA expression (D, E) of PPARγ (n = 3, p < 0.05, Student’s t-test, mean ± s.e.m.). (F) Phosphorylation PPARγ at Ser283 in Tet2-knockdown and overexpressor. (G) PPARγ acetylation level was measured by immunoblotting of anti-acetyl-Lysine antibody from PPARγ IP from Tet2-knockdown and overexpressor.
Supplemental Figure 6. Overexpression of the TET2 catalytic domain does not increase the amount PPARγ binding to target loci. Mature 3T3-L1 adipocytes were lentivirally transduced with Tet2-CD or GFP. ChIP-qPCR assays was performed using PPARγ antibody. Primers amplifying the region as indicated in the figure were used for ChIP-qPCR analysis (n = 3, p<0.05, Student’s t-test, mean ± s.e.m.).
Highlights.
Tet2 expression is enriched in the adipose fraction, and adipocyte Tet2 expression is diminished in diet-induced obesity.
TET2 plays a necessary and sufficient role in a PPARγ agonist’s mediation of insulin sensitization in cultured adipocytes.
TET2 facilitates the transcriptional activity of PPARγ in both a catalytic-dependent and independent manner.
TET2-mediated gene regulation of PPARγ involves concordant changes in DNA demethylation at the promoter regions.
TET2 is required for PPARγ to maintain DNA binding at selective target loci in response to a PPARγ agonist.
Acknowledgement
The authors gratefully acknowledge members of the Kang lab for their helpful advice and discussion. We thank Dr. Yu-Hua Tseng (Joslin Diabetes Center, Boston) for sharing immortalized human preadipocytes and thank Dr. Alex Banks (Brigham and Women's Hospital, Boston) for sharing PPARγ S273 antibody. This work was supported by a China Scholarship Council Scholarship to [2017]3109 to FB and NIH R01 NIDDK DK116008-01 to SK.
Abbreviations:
- Rosi
Rosiglitazone
- Tet
Ten-eleven translocation
- Dnmts
DNA methyltransferases
- HFD
High fat diet
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Robertson KD. DNA methylation and human disease. Nature Reviews Genetics 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- [2].Xie M, Hong C, Zhang B, Lowdon RF, Xing X, Li D, et al. DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nature Genetics 2013;45:836–41. doi: 10.1038/ng.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ronn T, Poulsen P, Hansson O, Holmkvist J, Almgren P, Nilsson P, et al. Age influences DNA methylation and gene expression of COX7A1 in human skeletal muscle. Diabetologia 2008;51:1159–68. doi: 10.1007/s00125-008-1018-8. [DOI] [PubMed] [Google Scholar]
- [4].Barres R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T, et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab 2012;15:405–11. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- [5].Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proceedings of the National Academy of Sciences of the United States of America 2008; 105:17046–9. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ehrlich S, Weiss D, Burghardt R, Infante-Duarte C, Brockhaus S, Muschler MA, et al. Promoter specific DNA methylation and gene expression of POMC in acutely underweight and recovered patients with anorexia nervosa. J Psychiatr Res 2010;44:827–33. doi: 10.1016/j.jpsychires.2010.01.011. [DOI] [PubMed] [Google Scholar]
- [7].Feinberg AP, Irizarry RA, Fradin D, Aryee MJ, Murakami P, Aspelund T, et al. Personalized epigenomic signatures that are stable over time and covary with body mass index. Sci Transl Med 2010;2:49ra67–7. doi: 10.1126/scitranslmed.3001262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Multhaup ML, Seldin MM, Jaffe AE, Lei X, Kirchner H, Mondal P, et al. Mouse-human experimental epigenetic analysis unmasks dietary targets and genetic liability for diabetic phenotypes. Cell Metab 2015;21:138–49. doi: 10.1016/j.cmet.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Volkmar M, Dedeurwaerder S, Cunha DA, Ndlovu MN, Defrance M, Deplus R, et al. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. Embo J 2012;31:1405–26. doi: 10.1038/emboj.2011.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kim AY, Park YJ, Pan X, Shin KC, Kwak S-H, Bassas AF, et al. Obesity-induced DNA hypermethylation of the adiponectin gene mediates insulin resistance. Nature Communications 2015;6:7585. doi: 10.1038/ncomms8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].You D, Nilsson E, Tenen DE, Lyubetskaya A, Lo JC, Jiang R, et al. Dnmt3a is an epigenetic mediator of adipose insulin resistance. Elife 2017;6:e30766. doi: 10.7554/eLife.30766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Benner C, Isoda T, Murre C. New roles for DNA cytosine modification, eRNA, anchors, and superanchors in developing B cell progenitors. Proceedings of the National Academy of Sciences of the United States of America 2015;112:12776–81. doi: 10.1073/pnas.1512995112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 2011;480:490–5. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- [15].Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 2013;500:477–81. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nair VS, Song MH, Ko M, Oh KI. DNA Demethylation of the Foxp3 Enhancer Is Maintained through Modulation of Ten-Eleven-Translocation and DNA Methyltransferases. Mol Cells 2016;39:888–97. doi: 10.14348/molcells.2016.0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hon GC, Song C-X, Du T, Jin F, Selvaraj S, Lee AY, et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Molecular Cell 2014;56:286–97. doi: 10.1016/j.molcel.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shen L, Zhang Y. 5-Hydroxymethylcytosine: generation, fate, and genomic distribution. Curr Opin Cell Biol 2013;25:289–96. doi: 10.1016/j.ceb.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan J, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nature Genetics 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Serandour AA, Avner S, Oger F, Bizot M, Percevault F, Lucchetti-Miganeh C, et al. Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers. Nucleic Acids Research 2012;40:8255–65. doi: 10.1093/nar/gks595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yu P, Ji L, Lee KJ, Yu M, He C, Ambati S, et al. Subsets of Visceral Adipose Tissue Nuclei with Distinct Levels of 5-Hydroxymethylcytosine. PloS One 2016;11:e0154949. doi: 10.1371/journal.pone.0154949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kang S, Akerblad P, Kiviranta R, Gupta RK, Kajimura S, Griffin MJ, et al. Regulation of early adipose commitment by Zfp521. PLoS Biol 2012;10:e1001433. doi: 10.1371/journal.pbio.1001433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shamsi F, Tseng YH. Protocols for Generation of Immortalized Human Brown and White Preadipocyte Cell Lines. Methods Mol Biol. 2017; 1566:77–85.. doi: 10.1007/978-1-4939-6820-6_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- [25].Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 2010;468:839–43. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Montagner S, Leoni C, Emming S, Chiara Della G, Balestrieri C, Barozzi I, et al. TET2 Regulates Mast Cell Differentiation and Proliferation through Catalytic and Non-catalytic Activities. Cell Reports 2016;15:1566–79. doi: 10.1016/j.celrep.2016.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yoo Y, Park JH, Weigel C, Liesenfeld DB, Weichenhan D, Plass C, et al. TET-mediated hydroxymethylcytosine at the Pparγ locus is required for initiation of adipogenic differentiation. Int J Obes (Lond) 2017;41:652–9. doi: 10.1038/ijo.2017.8. [DOI] [PubMed] [Google Scholar]
- [28].Zhong X, Wang Q-Q, Li J-W, Zhang Y-M, An X-R, Hou J. Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro. Sci Rep 2017;7:43539. doi: 10.1038/srep43539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lio C-W, Zhang J, Gonzaláz-Avalos E, Hogan PG, Chang X, Rao A. Tet2 and Tet3 cooperate with B-lineage transcription factors to regulate DNA modification and chromatin accessibility. Elife 2016;5:417. doi: 10.7554/eLife.18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Banks AS, McAllister FE, Camporez JPG, Zushin P-JH, Jurczak MJ, Laznik-Bogoslavski D, et al. An ERK/Cdk5 axis controls the diabetogenic actions of PPARγ. Nature 2015;517:391–5. doi: 10.1038/nature13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Choi JH, Banks AS, Estall JL, Kajimura S, Boström P, Laznik D, et al. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARgamma by Cdk5. Nature 2010;466:451–6. doi: 10.1038/nature09291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, et al. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparγ. Cell 2012;150:620–32. doi: 10.1016/j.cell.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Christensen DP, Dahllöf M, Lundh M, Rasmussen DN, Nielsen MD, Billestrup N, et al. Histone deacetylase (HDAC) inhibition as a novel treatment for diabetes mellitus. Mol Med 2011; 17:378–90. doi: 10.2119/molmed.2011.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Daneshpajooh M, Bacos K, Bysani M, Bagge A, Ottosson Laakso E, Vikman P, et al. HDAC7 is overexpressed in human diabetic islets and impairs insulin secretion in rat islets and clonal beta cells. Diabetologia 2017;60:116–25. doi: 10.1007/s00125-016-4113-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sharma S, Taliyan R. Histone deacetylase inhibitors: Future therapeutics for insulin resistance and type 2 diabetes. Pharmacol Res 2016; 113:320–6. doi: 10.1016/j.phrs.2016.09.009. [DOI] [PubMed] [Google Scholar]
- [36].Ye J Improving insulin sensitivity with HDAC inhibitor. Diabetes 2013;62:685–7. doi: 10.2337/db12-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lee J, Saha PK, Yang QH, Lee S, Park JY, Suh Y, et al. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proceedings of the National Academy of Sciences of the United States of America 2008;105:19229–34. doi: 10.1073/pnas.0810100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature 2013;504:163–7. doi: 10.1038/nature12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sambeat A, Gulyaeva O, Dempersmier J, Tharp KM, Stahl A, Paul SM, et al. LSD1 Interacts with Zfp516 to Promote UCP1 Transcription and Brown Fat Program. Cell Reports 2016; 15:2536–49. doi: 10.1016/j.celrep.2016.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature 2009;458:757–61. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zeng X, Jedrychowski MP, Chen Y, Serag S, Lavery GG, Gygi SP, et al. Lysine-specific demethylase 1 promotes brown adipose tissue thermogenesis via repressing glucocorticoid activation. Genes Dev 2016;30:1822–36. doi: 10.1101/gad.285312.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, et al. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Molecular Cell 1999;4:611–7. [DOI] [PubMed] [Google Scholar]
- [43].Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 1994;79:1147–56. [DOI] [PubMed] [Google Scholar]
- [44].Chawla A, Schwarz EJ, Dimaculangan DD, Lazar MA. Peroxisome proliferator-activated receptor (PPAR) gamma: adipose-predominant expression and induction early in adipocyte differentiation. Endocrinology 1994;135:798–800. doi: 10.1210/endo.135.2.8033830. [DOI] [PubMed] [Google Scholar]
- [45].Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev 1994;8:1224–34. [DOI] [PubMed] [Google Scholar]
- [46].Rangwala SM, Lazar MA. Peroxisome proliferator-activated receptor gamma in diabetes and metabolism. Trends Pharmacol Sci 2004;25:331–6. doi: 10.1016/j.tips.2004.03.012. [DOI] [PubMed] [Google Scholar]
- [47].Nolan JJ, Ludvik B, Beerdsen P, Joyce M, Olefsky J. Improvement in glucose tolerance and insulin resistance in obese subjects treated with troglitazone. N Engl J Med 1994;331:1188–93. doi: 10.1056/NEJM199411033311803. [DOI] [PubMed] [Google Scholar]
- [48].Chao L, Marcus-Samuels B, Mason MM, Moitra J, Vinson C, Arioglu E, et al. Adipose tissue is required for the antidiabetic, but not for the hypolipidemic, effect of thiazolidinediones. The Journal of Clinical Investigation 2000;106:1221–8. doi: 10.1172/JCI11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].He W, Barak Y, Hevener A, Olson P, Liao D, Le J, et al. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proceedings of the National Academy of Sciences of the United States of America 2003;100:15712–7. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Maeda N, Takahashi M, Funahashi T, Kihara S, Nishizawa H, Kishida K, et al. PPARgamma ligands increase expression and plasma concentrations of adiponectin, an adipose-derived protein. Diabetes 2001;50:2094–9. [DOI] [PubMed] [Google Scholar]
- [51].Moyers JS, Shiyanova TL, Mehrbod F, Dunbar JD, Noblitt TW, Otto KA, et al. Molecular determinants of FGF-21 activity-synergy and cross-talk with PPARgamma signaling. J Cell Physiol 2007;210:1–6. doi: 10.1002/jcp.20847. [DOI] [PubMed] [Google Scholar]
- [52].Hofmann C, Lorenz K, Braithwaite SS, Colca JR, Palazuk BJ, Hotamisligil GS, et al. Altered gene expression for tumor necrosis factor-alpha and its receptors during drug and dietary modulation of insulin resistance. Endocrinology 1994;134:264–70. doi: 10.1210/endo.134.1.8275942. [DOI] [PubMed] [Google Scholar]
- [53].Steppan CM, Bailey ST, Bhat S, Brown EJ, Banerjee RR, Wright CM, et al. The hormone resistin links obesity to diabetes. Nature 2001;409:307–12. doi: 10.1038/35053000. [DOI] [PubMed] [Google Scholar]
- [54].Yang Q, Graham TE, Mody N, Preitner F, Peroni OD, Zabolotny JM, et al. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature 2005;436:356–62. doi: 10.1038/nature03711. [DOI] [PubMed] [Google Scholar]
- [55].Westin S, Kurokawa R, Nolte RT, Wisely GB, McInerney EM, Rose DW, et al. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- [56].Gelman L, Zhou G, Fajas L, Raspé E, Fruchart JC, Auwerx J. p300 interacts with the N-and C-terminal part of PPARgamma2 in a ligand-independent and -dependent manner, respectively. The Journal of Biological Chemistry 1999;274:7681–8. [DOI] [PubMed] [Google Scholar]
- [57].Ge K, Guermah M, Yuan C-X, Ito M, Wallberg AE, Spiegelman BM, et al. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature 2002;417:563–7. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- [58].Bugge A, Grøntved L, Aagaard MM, Borup R, Mandrup S. The PPARgamma2 A/B-domain plays a gene-specific role in transactivation and cofactor recruitment. Mol Endocrinol 2009;23:794–808. doi: 10.1210/me.2008-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Fujiki K, Shinoda A, Kano F, Sato R, Shirahige K, Murata M. PPARgamma-induced PARylation promotes local DNA demethylation by production of 5-hydroxymethylcytosine. Nature Communications 2013;4:2262. doi: 10.1038/ncomms3262. [DOI] [PubMed] [Google Scholar]
- [60].Wu D, Hu D, Chen H, Shi G, Fetahu IS, Wu F, et al. Glucose-regulated phosphorylation of TET2 by AMPK reveals a pathway linking diabetes to cancer. Nature 2018;559:637–41. doi: 10.1038/s41586-018-0350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. The relative expression of Tets in adipose vs. SVF fractions from fat depots. Tet expression was measured by q-PCR in adipocytes (AD) and SVF fractions from eWAT, iWAT, and BAT. Shown is the relative fold expression of Tet family members in AD (A) and SVF (B) when normalized to Tet1 expression (n = 3, p < 0.05, Student’s t-test, mean ± s.e.m.).
Supplemental Figure 2. TET2 knockdown and overexpression efficiency in TET2 gain- and loss-of-function models. Mature 3T3-L1 adipocytes were transduced with Tet2-knockdown (A) or -overexpressor lentiviruses (B). Two days after transduction, cell lysates were harvested and blotted using anti-TET2 antibody (A) and anti-Myc antibodies (B) (n = 3, p < 0.05, Student’s t-test, mean ± s.e.m.).
Supplemental Figure 3. TET2 gain- and loss-of-function leads to global changes in 5hmC level. (A) Genomic DNA was extracted from mature 3T3-L1 adipocytes transduced with Tet2-overexpressor or -knockdown lentiviruses. A dot blot was performed to assess the total amount of 5hmC using the indicated amount of genomic DNA. (B) Blots from (A) were processed with methylene blue staining. (C, D) Quantification of 5hmC dot blot by normalizing with methylene blue staining (n = 3, p < 0.05, Student’s t-test, mean ± s.e.m.).
Supplemental Figure 4. Insulin signal transduction is marginally changed in Tet2 gain- and loss-of-function models. Mature 3T3-L1 adipocytes were transduced with Tet2-knockdown (A-C) or -overexpressor lentiviruses (D-F). Three days after transduction, cells were treated with Rosi (1 μM), TNF (4 ng/ml), or both for two additional days. Serum-starved cells were stimulated with 20 nM insulin for 5 min, and cell lysates were subjected to immunoblotting with antibodies against total and phospho-AKT and -IRS-1. (B, C, E, F) Quantification of western blot in (A, C) (n = 3, p < 0.05, Student’s t-test, mean ± s.e.m.).
Supplemental Figure 5. Total- and post-translational levels of PPARγ is not grossly altered in TET2 gain- and loss-of-function models. Mature 3T3-L1 adipocytes were lentivirally transduced with Tet2-knockdown or overexpressor plasmids. Shown is protein (A-C) and mRNA expression (D, E) of PPARγ (n = 3, p < 0.05, Student’s t-test, mean ± s.e.m.). (F) Phosphorylation PPARγ at Ser283 in Tet2-knockdown and overexpressor. (G) PPARγ acetylation level was measured by immunoblotting of anti-acetyl-Lysine antibody from PPARγ IP from Tet2-knockdown and overexpressor.
Supplemental Figure 6. Overexpression of the TET2 catalytic domain does not increase the amount PPARγ binding to target loci. Mature 3T3-L1 adipocytes were lentivirally transduced with Tet2-CD or GFP. ChIP-qPCR assays was performed using PPARγ antibody. Primers amplifying the region as indicated in the figure were used for ChIP-qPCR analysis (n = 3, p<0.05, Student’s t-test, mean ± s.e.m.).