Abstract
Adhesive pili are external component of fibrous adhesive organelles and help bacteria attach to biotic or abiotic surfaces. The biogenesis of adhesive pili via the chaperone-usher pathway (CUP) is independent of external energy sources. In the classical CUP, chaperones transport assembly-competent pilins in a folded but expanded conformation. During donor-strand exchange, pilins subsequently collapse, producing a tightly packed hydrophobic core and releasing the necessary free energy to drive fiber formation. Here, we show that pilus biogenesis in non-classical, archaic, and alternative CUPs uses a different source of conformational energy. High-resolution structures of the archaic Csu-pili system from Acinetobacter baumannii revealed that non-classical chaperones employ a short donor strand motif that is insufficient to fully complement the pilin fold. This results in chaperone-bound pilins being trapped in a substantially unfolded intermediate. The exchange of this short motif with the longer donor strand from adjacent pilin provides the full steric information essential for folding, and thereby induces a large unfolded-to-folded conformational transition to drive assembly. Our findings may inform the development of anti-adhesion drugs (pilicides) to combat bacterial infections.
Keywords: chaperone, bacterial adhesion, bacterial pathogenesis, biofilm, protein folding, protein secretion, protein self-assembly, protein structure, X-ray crystallography, nuclear magnetic resonance (NMR), Acinetobacter baumannii, adhesive pili, chaperone-usher pathway, Csu pili, fimbriae
Introduction
Adhesive pili, the external component of fibrous adhesive organelles, mediate bacterial attachment to biotic or abiotic surfaces, playing a vital role in host and self-recognition. These adhesins mediate targeting to sites of infection, host cell invasion, and biofilm formation required to resist host defenses, environmental stress, and antibiotic treatment (1, 2). The major class of adhesive pili in Gram-negative bacteria are assembled via the classical, alternative, and archaic chaperone-usher pathways (CUPs)3 (3). The CUP pili comprise up to 7 distinct protein subunits (pilins) assembled into linear unbranched extracellular polymers (4, 5). Fibers can consist of adhesin subunits that act as a polyvalent adhesin, or display a specialized adhesin subunit at the tip to facilitate highly specific monovalent binding to host cell receptors (5–7). CUP pili assembly is mediated by a periplasmic chaperone and outer membrane assembly platform termed the usher (3, 4).
The biogenesis of CU pathways has been elucidated from the well-studied classical systems (Fig. S1). The periplasmic chaperone forms a binary chaperone-subunit complex in a process known as donor strand complementation (DSC) (8, 9). This traps a high-energy folding intermediate, and the collapse of which drives pilus polymerization and translocation (10–12). Fiber subunits are subsequently assembled by donor strand exchange (DSE), in which the N-terminal extension from an incoming subunit displaces the chaperone via a so-called “zip-in-zip-out” mechanism (11, 13) and provides the necessary β-strand to complete immunoglobulin-like fold of the subunit (11, 12). This process is facilitated by the usher assembly platform at the outer membrane and drives pilus assembly without the requirement of an external energy source (14, 15).
The increasing occurrence of antibiotic resistance has accelerated the search for novel therapeutic strategies and the CU pathways are currently being explored as novel drug targets. Non-classical (archaic and alternative) CUPs are of particular interest due to their wide distribution in multidrug-resistant pathogens and association with a broad spectrum of diseases (3, 16, 17). Archaic or σ CUPs, present in all Proteobacteria and Cyanobacteria and Deinococcus-Thermus phyla, are widespread compared with classical systems, which are restricted to pathogenic β- and γ-Proteobacteria (3). The alternative or α CU pathway is a highly divergent family with a wide phylogenetic distribution (3). This pathway includes CFA/I-like fimbriae, which are the primary adhesins of human enterotoxigenic Escherichia coli, a major cause of mortality in young children from developing countries (3, 18, 19). Although little sequence homology exists between proteins in classical, alternative and archaic CU pathways, a recent study revealed significant similarities in the structure of the assembly chaperones from the two non-classical pathways (20). However, our understanding of non-classical biogenesis is limited and studies are in their infancy compared with the classical CUPs. Acinetobacter baumannii, the most prevalent clinical Acinetobacter species, is a multidrug-resistant, opportunistic and highly persistent Gram-negative bacterium responsible for nosocomial infections (21). The formation of a dense biofilm is key to A. baumannii pathogenesis, as it confers survival on surfaces (16, 22–24). Biofilm formation is mediated by Csu pili assembled via the archaic chaperone-usher CsuC-CsuD pathway (16, 17). The Csu pilus comprises four distinct pili subunits, namely the major pilin subunit CsuA/B (16.1 kDa), adaptor subunits CsuA (17.3 kDa) and CsuB (16.9 kDa), and the CsuE tip adhesin (33.5 kDa). Three highly hydrophobic finger-like loops exposed at the N-terminal domain of CsuE are essential for the tight attachment of A. baumannii to various hydrophobic substrates, including the majority of plastics used in medical devices (25). Previously, we reported the high-resolution structure of the CsuC-CsuA/B chaperone-subunit pre-assembly complex (43 kDa) from archaic Csu pili (20, 26) and demonstrated that assembly is governed by the principles of DSC and DSE. We also highlighted that classical and non-classical chaperones exhibit significant structural differences (20, 27), employing distinct subunit-binding mechanisms (20). A shifted donor strand motif within non-classical chaperones contributes to an altered subunit-binding position (20). Most importantly, non-classical chaperones, unlike their classical counterparts, maintain the subunit in a state with significant conformational disorder (20). Consequently, the chaperone-subunit complex does not possess a preformed DSE initiation site, suggesting that the non-classical assembly pathway use an alternative mechanism of fiber assembly.
Here, we report the crystal structure of the CsuA/B self-complemented pilin, which completes our atomic resolution understanding of the final fiber-inserted conformation for archaic pilins. A comparison of the structure, dynamics, and thermodynamics of the pilin before and after DSE reveal that the subunit undergoes a dramatic folding transition akin to the conversion of a molten globule to a folded state. We found that non-classical chaperones use much shorter donor strand motifs than the sequence necessary for full complementation of the pilin. Hence, they provide only partial sequence information for the pilin to fold, which preserves substantial folding energy to drive pilus assembly. We also discovered that β sheet ABE of archaic pilins is interrupted in the middle with hairpin insertions. This unique structure undergoes a large conformational change upon DSE, which additionally drives the assembly.
Results and discussion
Non-classical chaperones maintain pilins in a highly disordered conformational state
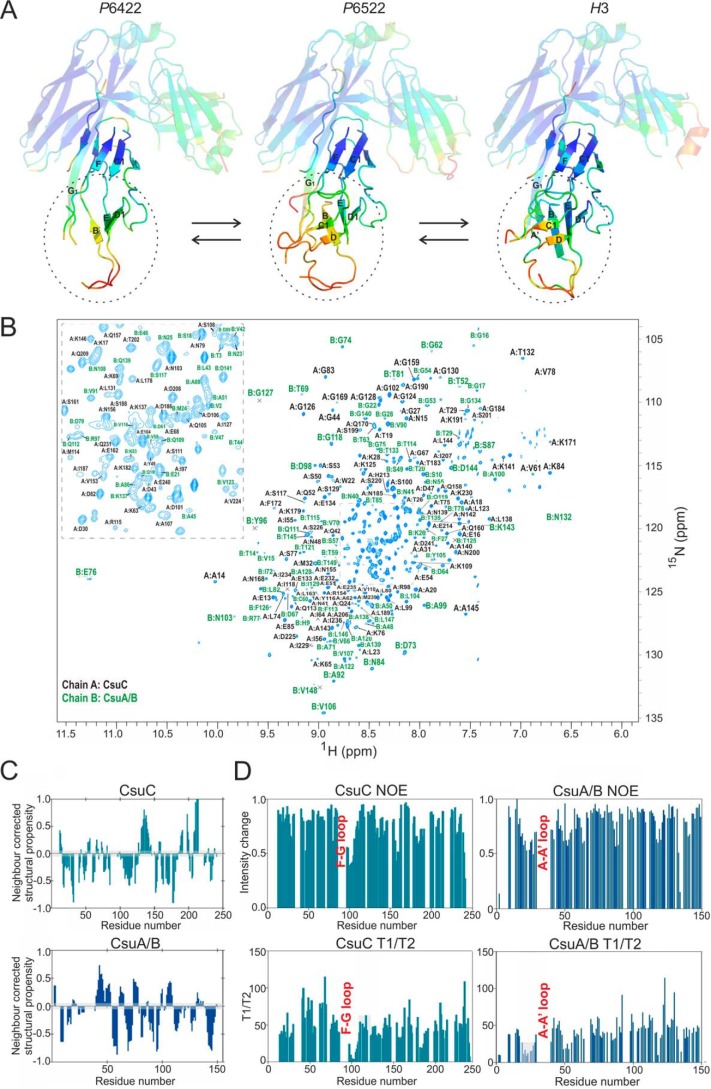
Our crystal structure of the CsuC-CsuA/B preassembly complex (PDB code 5D6H) suggested that CsuA/B exhibited significant disorder when in complex with CsuC (20). More than 40% of the CsuA/B sequence had no electron density in electron density maps, which is in sharp contrast to the well-ordered subunits bound to classical chaperones. We therefore decided to examine the importance of this phenomenon in the biogenesis of non-classical systems. We solved two additional crystal structures of the CsuC-CsuA/B preassembly complex in space groups P6522 and H3 (Table S1). The three CsuC molecules are highly similar and superimposable, with r.m.s. deviations on all Cα positions in the range of 0.5–0.6 Å (Fig. 1A). CsuA/B exhibits significantly higher structural heterogeneity (r.m.s. deviations of 1.2–1.3 Å), which is most pronounced in regions distant from CsuC (Fig. 1A, Fig. S2). Although some order is conferred to this segment in the P6522 and especially H3 crystal forms by stabilizing crystal contacts, the majority of this region is either completely disordered or has high thermal motion (B-factors) (Fig. S2). In addition, none of the structures exhibit electron density for the long stretch of sequence between β strands A and A′ in CsuA/B (Fig. 1A, Fig. S2).
Figure 1.
CsuC traps CsuA/B in a substantially disordered conformation. A, diagrams of crystal structures of CsuC-CsuA/B (43 kDa) in space groups P6422, P6522, and H3 colored by B-factor of Cα atoms with the color ranging from blue (lowest) to red (highest). CsuC is semi-transparent. β-Strands in CsuA/B and strand G1 in CsuC are labeled. B, 1H,15N-TROSY spectrum of CsuC-CsuA/B highlighting the 76% backbone resonance assignment. C, ncSCP chemical shift analysis of CsuC (a) and CsuA/B (b) in complex. Deviations from the expected disorder chemical shift range, within the gray boundary, predict helical (positive) or β (negative) structural propensity. D, relaxation analysis of CsuC and CsuA/B. Heteronuclear steady-state 1H,15N-NOE values (top) and T1/T2 values (bottom) are shown. Residues encompassing the loop upstream of the G1 strand in CsuC and loop between A and A′ strands in CsuA/B are indicated.
An NMR backbone resonance assignment of U-15N,13C,2H-labeled CsuC-CsuA/B was performed to characterize the dynamic nature of the archaic preassembly complex in solution (Fig. 1B). The 1H,15N-TROSY spectrum of CsuC-CsuA/B exhibited excellent peak dispersion but significant variations in peaks intensities were apparent, with fewer than expected resonances and some clustering of more intense signals at 8.5 ppm, indicative of native disorder. TROSY-based triple resonance spectra were recorded and facilitated ∼80% backbone assignment for the complex. This assignment covered 68% of CsuC chaperone sequence, in which 158 residues of the possible 233 could be assigned, and 89% of CsuA/B, where 126 residues of 141 were assigned. Many of unassigned amide peaks exhibited poor signal-to-noise in triple resonance experiments, which is likely due to the presence of conformational dynamics on an intermediate timescale. The NMR assignment covers several regions of CsuA/B that are absent from the crystallographic data, affording insight into the structure and dynamics of these regions.
NMR chemical shifts are sensitive local probes of structural and dynamics. The experimentally derived chemical shift assignment for the CsuC-CsuA/B complex were analyzed with TALOS-N (28) to probe the structural propensity of regions absent from the crystallographic data. Residues Gln-19 to Lys-26, spanning the region between the A and A′ β-strands, are considered dynamic along with Ala-48 and Ser-49, which lie upstream of the β-strand. CsuC residues Ile-97 to Asp-106 are also reported as dynamic and these are located within the loop after the G1 strand and are involved in CsuA/B binding. It should be noted that overall TALOS-N predictions of local secondary structure agree with the crystallographic data.
Further chemical shift analysis was performed using a disorder-specific neighbor-corrected structural propensity calculator, termed ncSPC (29, 30). Chemical shift data are compared with a chemical shift library of disordered proteins where deviation from the random coil state indicates structural propensity (29, 30). Scores of 1 and −1 reflect fully formed α-helical or β-sheet structure, respectively, whereas a score of less than 1 suggests that that a fraction of the conformers in the ensemble are structured at that position (i.e. 0.5 would 50% helical). Structural propensity reported by ncSPC is consistent with the crystallographic data and TALOS-N analysis (Fig. 1C). In agreement with TALOS-N analysis, the CsuA/B A–A′ region and the loop upstream of the G1 strand in CsuC fall within the range considered disordered. Both regions report slight strand propensity as expected. Taken together this suggests that residues between the A–A′ strands in CsuA/B and the loop upstream of the G1 strand in CsuC exhibit conformational plasticity in complex.
Further dynamic information was obtained by measuring backbone 15N-T1 and -T2 relaxation rates and the heteronuclear steady-state 15N-NOE (Fig. 1D). The CsuC chaperone exhibits NOE values consistently distributed around 0.82, the value expected for a rigid structured protein. Residues 99 to 108, in the loop upstream of the G1 strand, exhibit a significant decrease in NOE, indicating enhanced flexibility in this region consistent with the chemical shift analysis. This is observed to a lesser extent for residues 155 to 161, which relate to another loop missing from the original crystal structure. These findings are also in agreement with the 15N-T1 and 15N-T2 values, which are represented here as a T1/T2 plot (Fig. 1D). Deviation from the average value of 42.8 is most pronounced in the pre-G1 loop but is also observed for residues 155 to 161 and 182 to 187. Heteronuclear steady-state 15N-NOE values for CsuA/B exhibit a broader distribution with distinct regions of deviation from 0.8. The most pronounced deviation is observed in the region between the A and A′ strands that was absent in the crystal structure and indicated as disordered by chemical shift analysis. Deviation is observed in other loop regions but most significantly between residues 46 and 55, relating to a loop observed in the crystal structure. The 15N-T1 and 15N-T2 values, represented in a T1/T2 plot, agree with these observations. The average value is 40.4, with the largest deviation again observed in the A to A′ loop and to a lesser extent the loop from residues 46 to 55. Hence, consistent with the crystal structures, these observations highlight enhance flexibility of the chaperone-bound CsuA/B subunit in solution.
The authors were able to fully trace the polypeptide chain, but regions of CfaB outside of the subunit-chaperone interface, comprising 31% of the structure, exhibit a high degree of thermal motion (Figs. S2 and S3) and had very poor electron density. Many of these regions are characterized by very low values of real-space correlation coefficient between the model and the density (Fig. S2), suggesting that they contain serious errors resulting from the the poor experimental density. In general, these regions coincide with disordered/flexible regions in chaperone-bound CsuA/B, especially those between β-strands C and E, and E and F (Fig. 2). Both structures also have a poorly ordered region before β-strand B. However, CfaB has a well-ordered sequence that corresponds to the completely disordered region between β-strands A1 and A2 in CsuA/B. Closer inspection revealed a loosely packed structure lacking a traditional hydrophobic core (Fig. S3). Moreover, Bao et al. (14) had to substitute Thr-114 in the G1 donor strand with leucine to increase the stability of the complex, and likely inducing increased order in this unstructured region. Thus, a highly disordered structure of the chaperone-bound subunit appears to be a distinctive feature of both archaic and alternative CUPs.
Figure 2.
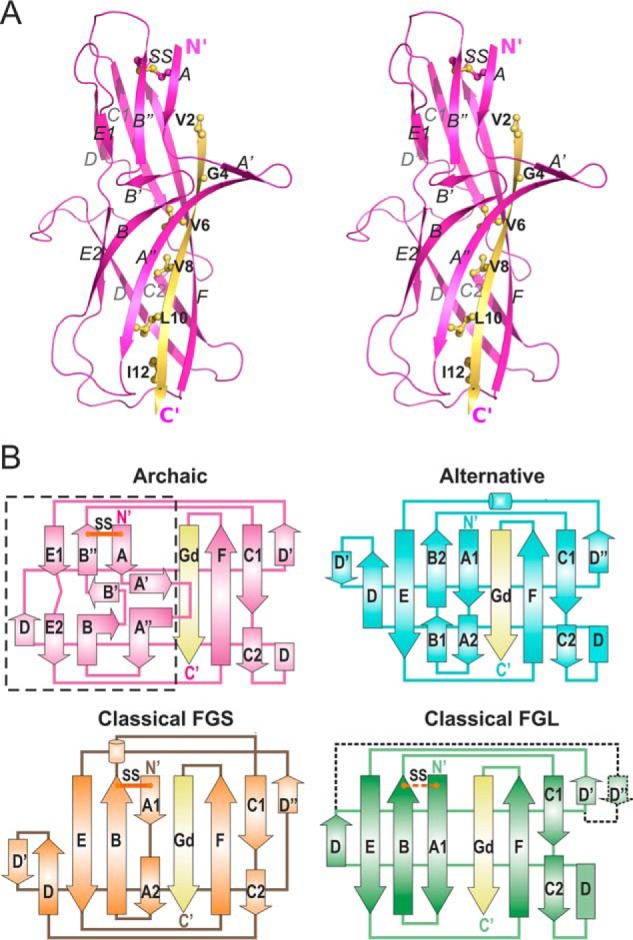
The first β-sheet of CsuA/B has a unique, non-Ig-like architecture. A, crystal structure of CsuA/Bsc (cartoon representation, stereo view). The Gd donor strand is colored yellow and donor residues are shown as balls-and-sticks and labeled. B, topology diagrams of pilins from archaic, classical FGS chaperone assembled, classical FGL chaperone assembled, and alternative CUP systems. Donor strands are shown in yellow. Positions of conserved disulfide bonds (SS) are shown with lines in red. In the classical FGL subfamily, the disulfide bond and additional D″ strand are present in some subunits and absent in others, and hence are shown using dashed lines. The unique for archaic systems β-sheet ABE is framed in a rectangle. Superpositions of pilins from different CUPs are shown in Fig. S4.
Structure of archaic pilins deviates from the standard Ig-fold
We have shown that CsuA/B is highly dynamic when in complex with CsuC. However, this structural snapshot has limited value when trying to decipher the overall pilus architecture. To evaluate the final pilus architecture of CsuA/B we produced and determined the crystal structure of a self-complemented CsuA/B (CsuA/Bsc). CsuA/Bsc was produced by moving the predicted N-terminal donor strand (Gd) sequence to the C terminus of CsuA/B, facilitating self-complementation and the formation of a monomeric subunit. The structure of CsuA/Bsc was solved to 1.5-Å resolution (Table S1).
The structure of CsuA/Bsc has a double β-sheet sandwich comprising strands A, B, E (β-sheet 1), and strands C, F, Gd (β-sheet 2), in which strand D switches between the sheets (Fig. 2A). Cys-16 and Cys-62 form a disulfide bond between strands A and B. The shifted Gd donor strand inserts six hydrophobic donor side chains into the acceptor cleft of the subunit. This structural organization is typical for all CU pilins and confirms that archaic CU assembly complies with the general principle of DSC and DSE. However, a closer inspection of the structure reveals that the ABE β-sheet of CsuA/Bsc adopts an unusual architecture featuring two prominent hairpins A′–A″ and B′–B″ protruding from the β-sheet. These hairpins break the β-sheet, splitting strands A and B into three shorter strands and strand E into two. Analysis of all available structures of polymerizing pilin subunits from the classical, both the FG short (FGS) chaperone assembled and highly divergent FG long (FGL) chaperone assembled or γ3 families (31, 32), and alternative CUPs reveal that this structure is unique to archaic subunits (Fig. 2B, Fig. S4). The A′–A″ hairpin is only found within the lectin domain of the FimH adhesin, but it runs parallel to the β-sheet and does not protrude from the β-barrel (Fig. S5).
The CsuA/B structure-based alignment of single-domain subunits (Fig. S6) suggests that other representatives of archaic fimbriae associated with animal pathogens also possess a similar arrangement of A′–A″ and B′–B″ hairpins. Indeed, glycine residues (Gly-20, Gly-25, Gly-31, and Gly-57) that introduce the turns responsible for disruption of β-strands are found in most sequences. Furthermore, the alignment contains conserved hydrophobic residues corresponding to Phe-30, Leu-33, and Phe-35 in strand A″ of CsuA/Bsc.
CsuA/Bsc combines some characteristic features of classical and alternative pili (Fig. 2B and Fig. S4). CsuA/B possess a disulfide bond between strands A and B similar to pilin subunits from classical FGS chaperone assembled and some representatives of FGS chaperone assembled subfamilies. The Gd strand of CsuA/Bsc interacts with both strands F and A, which is typical for subunits from classical FGS and alternative systems. The N-terminal part of strand D is hydrogen-bonded to strand C2, whereas its C-terminal part interacts with strand E, similarly to alternative and classical FGL CU systems. A Protein Data Bank (PDB) search with the Dali server (33) found a subunit from the alternative CUP as the closest structural homologue of CsuA/Bsc (Table S2). Surprisingly, this is not a self-polymerizing subunit, but the adhesin domain of the CfaE two-domain tip subunit from the alternative CFA/I fimbriae.
In conclusion, the structure of CsuA/Bsc demonstrated that pilin subunits from all CUPs, including the archaic pathway, have an incomplete Ig-like fold and assemble by the DSC-DSE mechanism. However, the structure of pilins from the archaic pathway deviate significantly from the standard Ig-fold as the ABE β-sheet is split and features a prominent twin-hairpin structure.
Nonclassical chaperones provide incomplete information for pilin folding
The stabilization of partially folded pilin subunits by non-classical chaperones is distinct and would likely influence downstream fimbrial assembly. We compared the self-complemented and chaperone-bound CsuA/B subunits to determine the structural consequences of this phenomenon (Fig. 3).
Figure 3.
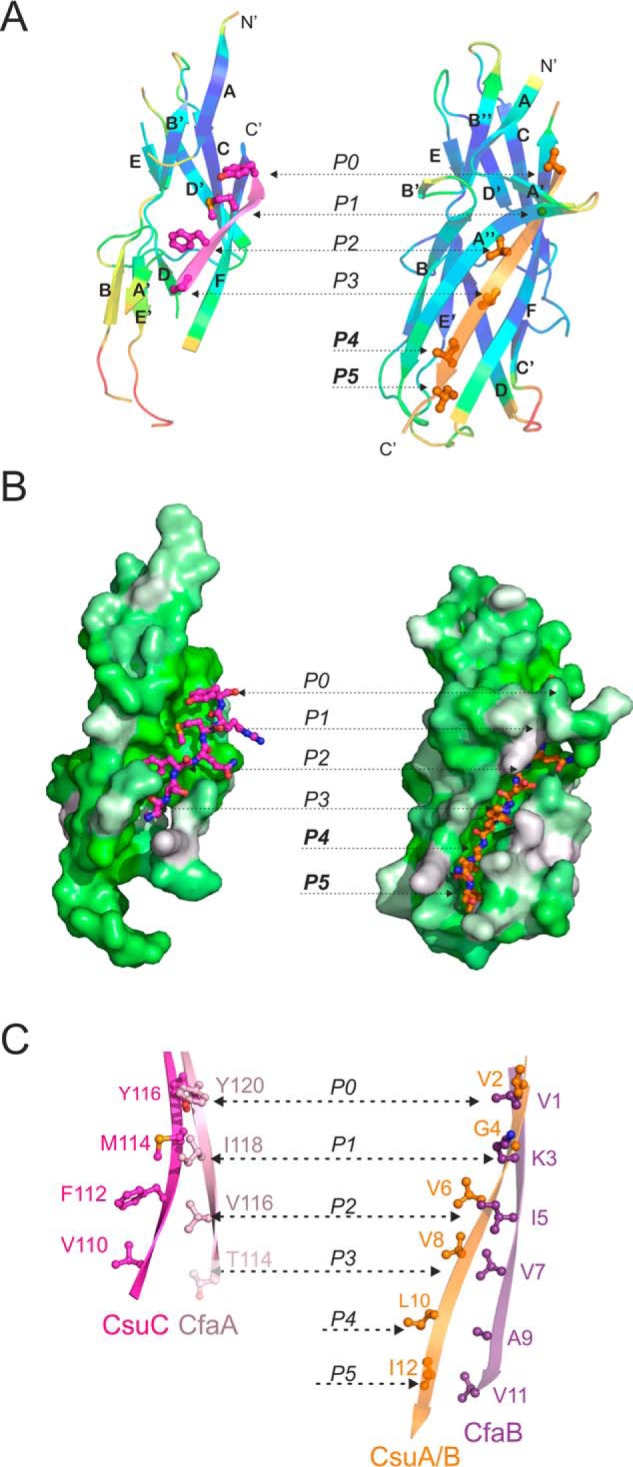
Comparison of the structure and donor strand complementation mechanism of the chaperone-bound and fiber inserted pilins from nonclassical systems. A and B, CsuA/B complemented with the G1 donor strand of CsuC (chaperone-bond conformation) is shown on the left and CsuA/Bsc (fiber-inserted conformation) is shown on the right. A, cartoon representation. CsuA/B is colored by B-factors. G1 and Gd donor strands are colored magenta and orange, respectively; donor residues are shown as ball-and-sticks. B, surface representation. CsuA/B is colored using the Eisenberg hydrophobicity scale. G1 and Gd donor strands are shown as ball-and-stick models. C, comparison of donor motifs in G1 strands of archaic CsuC and alternative CfaA chaperones (left diagram) and Gd strands of the respective pilins, CsuA/B and CfaB (right diagram). The images highlight fragments of structural superpositions of CsuC-CsuA/B and CfaA-CfaB (4Y2O) complexes, and self-complemented CsuA/B and CfaB (3F83) pilins. Donor residues are shown as ball-and-sticks and labeled. Pockets P0–P5 in the acceptor cleft of the pilin are indicated in A–C. Thr-114 in CfaA is shown in pale pink, because it only partially occupies acceptor pocket P3.
Structural comparison revealed that the CsuC chaperone utilizes a much shorter G1 donor strand than the complementing Gd strand in CsuA/B. The donor strand only occupies part of the full hydrophobic acceptor cleft in CsuA/B. Four large hydrophobic residues (Tyr-116, Met-114, Phe-112, and Val-110) are inserted into acceptor pockets P0–P3, respectively, whereas pockets P4 and P5 remain vacant and the proximal subunit is disordered. The CsuA/B Gd strand is four residues (or two donor residues) longer, facilitating full occupation of all acceptor pockets in the hydrophobic acceptor cleft and complete folding of the subunit (Fig. 3, A and B). A similar conformational change is observed in the alternative pathway. The donor strand motif in the alternative chaperone CfaA is also almost half the length of the CfaB Gd strand (Fig. 3C, Fig. S7). Moreover, the presence of a polar threonine at position 114 in the G1 strand of CfaA instead of a corresponding hydrophobic residue (as in CfaB) suggests that acceptor pocket P3 in CfaB is only partially occupied. Regions of chaperone-bound CfaB distant to the CfaA donor strand are disordered like chaperone-bound CsuA/B, despite the Thr-114 → Leu substitution. In agreement with the CsuA/Bsc structure, the fiber-inserted conformation of CfaB (CfaBsc, PDB code 3F83) is fully structured (Fig. S7).
Previously, we showed that non-classical systems utilize the register-shifted DSC, in which the positions of donor residues are shifted toward the C-terminal end of the G1 strand and the complementation of the acceptor cleft in the chaperone-bound subunit starts, not at pocket P1, but at pocket P0 (19). This distinct register shift explains why unstructured pilin subunits have not been observed in classical preassembly complexes. In the non-classical systems, the chaperone only partially occupies P3 and does not occupy pockets P4 and P5 in the subunit, providing incomplete information for the subunit folding.
DSE promote a striking unfolded-folded transition in non-classical pathways
The rigid arm-like protrusion in the in CsuA/Bsc structure, comprising two hairpins A′–A″ and B′–B″, is characterized by low B factors (Figs. 2A and 3A), whereas the corresponding region in chaperone-bound CsuA/B is highly disordered (Figs. 1, 3, and 4). This unique characteristic represents a secondary site that undergoes a major unfolded-folded transition during pilin assembly (Fig. 4).
Figure 4.
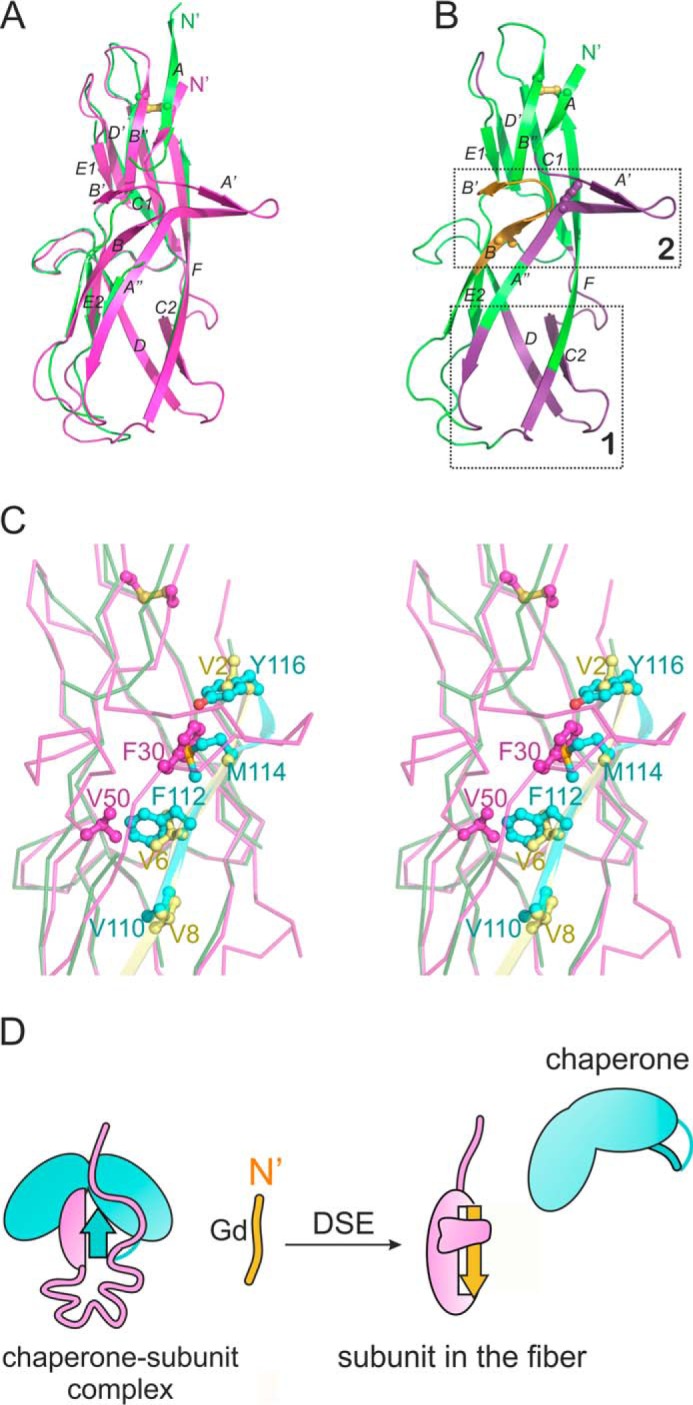
Archaic pilin assembly is accompanied by folding of two segments in the pilin. A, structural superposition of CsuC-bound CsuA/B (green) and CsuA/Bsc (magenta). B, cartoon diagram of CsuA/Bsc highlighting regions that do not significantly change (green), change dramatically (yellow), and undergo unfolded-folded transition (violet) during assembly. Two sites undergoing dramatic changes upon DSE are framed in rectangles. Phe-30 and Val-50 are shown as ball-and-sticks. C, steric clashes with CsuC prevent formation of A′–A″ and B–B′ hairpins in CsuC-bound CsuA/B subunit. Close up of the superposition shown in A (ribbon representation, stereo view). Donor strands G1 and Gd are shown as cartoon diagrams and colored blue and yellow, respectively. Phe-30 and Val-50 in CsuA/B and donor residues in G1 of CsuC are shown as ball-and-sticks and labeled. D, schematic of the mechanism of the archaic assembly.
The hairpins contribute several residues to the hydrophobic core of CsuA/Bsc, including conserved Phe-30 in hairpin A′–A″ and Val-50 in hairpin B′–B″ (Fig. 4B and 4C). Superposition of CsuA/Bsc and chaperone-bound CsuA/B showed that if these residues adopted the same position in the chaperone-bound subunit, they would collide with the bulky donor strand residues Phe-112 and Met-114 of CsuC (Fig. 4C, Fig. S8). Steric hindrance elicited by the donor strand residues inhibits proper folding of the two-hairpin structure in the chaperone-bound subunit. This mechanism is reminiscent of the intercalation of bulky donor residues of the chaperone between the subunit β-sheets of the classical systems (11). However, exchange of bulky donor strand residues in the classical chaperone to the smaller side chains of the Gd strand causes a global movement of β-sheets in a folded-to-folded transition within the subunit. The same process in the archaic system produces an unfolded-to-folded transition.
To examine if any folded-to-folded conformational changes occur in non-classical CUP, we compared the well-structured regions of chaperone-bound and fiber-inserted subunits. In this analysis, we use our new structures from the archaic Csu system and published structures from the alternative Cfa system (Fig. S9). The ordered regions of chaperone-bound CsuA/B superimposed with the corresponding regions of CsuA/Bsc with an r.m.s. deviation of less than 1 Å and revealed no unidirectional change in the structure (Fig. S9A). Superposition of the CfaBsc with chaperone-bound CfaB reveals a small rearrangement of the β-sheets of the subunit (Fig. S9B), but this is much less pronounced than for classical systems (Fig. S9C). Moreover, due to the highly disordered conformation of chaperone-bound CfaB, the number of residues involved in this rearrangement is significantly smaller than in classical fimbriae. Therefore, in contrast to classical fimbriae, the non-classical assembly involves either no rearrangement of the β-sheet or only a very modest conformational transition from folded to folded states.
These results suggest a distinct mechanism for non-classical pilus assembly (Fig. 4D). The short donor strand motif in non-classical chaperones provides incomplete structural information for the pilin to fold, resulting in an unstructured segment near acceptor pockets P4 and P5 (site 1 in Fig. 4B). The displacement of the short donor strand motif of the chaperone with a longer Gd strand of the attacking subunit provides the missing structural information for site 1 to fold. This process is common for all non-classical pili, but the DSE induced folding of the A′–A″ and B′–B″ hairpins is only found in archaic systems. In archaic systems, the bulky donor residues of the chaperone hinder the formation of the A′–A″ and B′–B″ hairpins and maintain disorder in strands A and B (site 2 in Fig. 4B).
Folding of unstructured regions in subunits drives assembly of non-classical pili
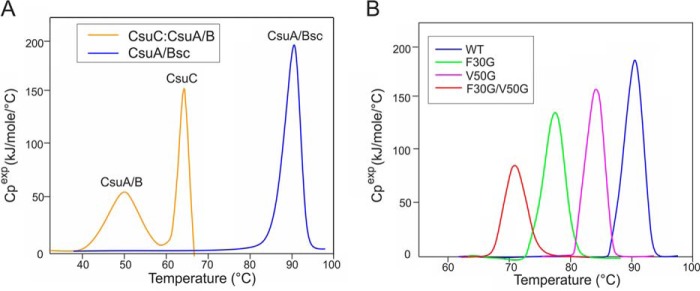
In classical CUPs, the collapse of the subunit β-sandwich releases energy that drives the fiber assembly (10, 11). The stability of CsuC-CsuA/B and CsuA/Bsc was analyzed with differential scanning calorimetry to examine whether archaic assembly is also driven by the energy of subunit folding. The excess heat capacity curve of CsuC-CsuA/B has two peaks with maxima at 48 and 62 °C, which correspond to the melting of CsuA/B within the complex followed by that for the dissociated CsuC, respectively (Fig. 5A, Table 1). CsuA/Bsc is extremely stable in comparison and melts as a single peak at 90 °C. The two proteins also exhibit a considerably different enthalpy of melting (ΔΔH of 423 kJ/mol). Both ΔTm and ΔΔH values are very similar to those obtained for the classical CUP Caf1 system from Yersinia pestis (20, 30), suggesting that classical and archaic pilins release an approximately similar amount of folding energy to drive fiber formation and gain similar stabilization upon DSE.
Figure 5.
Stability analysis of CsuC-CsuA/B, CsuA/Bsc, and CsuA/Bsc mutants with differential scanning calorimetry. A, transition excess heat capacity (Cpexp) of CsuC-CsuA/B and CsuA/Bsc. B, Cpexp of WT CsuA/Bsc and substitution variants F30G, V50G, and F30G/V50G.
Table 1.
Thermodynamic parameters of melting of CsuC-CsuA/B and CsuA/Bsc were determined from 2 to 3 thermograms
| Protein | Tm | ΔHcal |
|---|---|---|
| C | kJ/mol | |
| CsuC-CsuA/B | ||
| CsuA/B | 49.1 ± 0.6 | 363 ± 27 |
| CsuC | 62.8 ± 0.5 | |
| CsuA/Bsc, wildtype | 90.3 ± 0.4 | 786 ± 15 |
| CsuA/Bsc, V50G | 84 ± 0.4 | 665 ± 21 |
| CsuA/Bsc, F30G | 77.9 ± 0.3 | 538 ± 42 |
| CsuA/Bsc, F30G, V50G | 71.4 ± 0.2 | 454 ± 19 |
We hypothesized that the shorter, register-shifted donor strand motif in the G1 strand of non-classical chaperones accounts for the disordered conformation of pilins, preserving folding energy for the subsequent steps of the assembly. If this were the case, a longer donor strand motif would stabilize the preassembly complex and inhibit fiber formation. To test this hypothesis, we introduced 3-residue insertions INI, INL, or LNI at the N-terminal end of the G1 strand, extending the alternating hydrophobic-hydrophilic pattern of the donor motif by two hydrophobic positions. The WT chaperone and insertion variants were co-expressed with CsuA/B in the E. coli periplasm. Chaperone-assisted CsuA/B polymerization was analyzed by Western blotting of periplasmic extracts using anti-CsuA/B antibodies (Fig. 6). In the presence of WT CsuC, CsuA/B is efficiently polymerized, evidenced by an extensive long ladder in SDS-PAGE. Western blotting of samples obtained from cells co-expressing CsuA/B with insertion variants resulted in shorter ladders with no bands for longer CsuA/B polymers. The most dramatic effect was observed for the INL insertion mutant, whose appended donor residues are identical to those in the Gd strand of CsuA/B.
Figure 6.
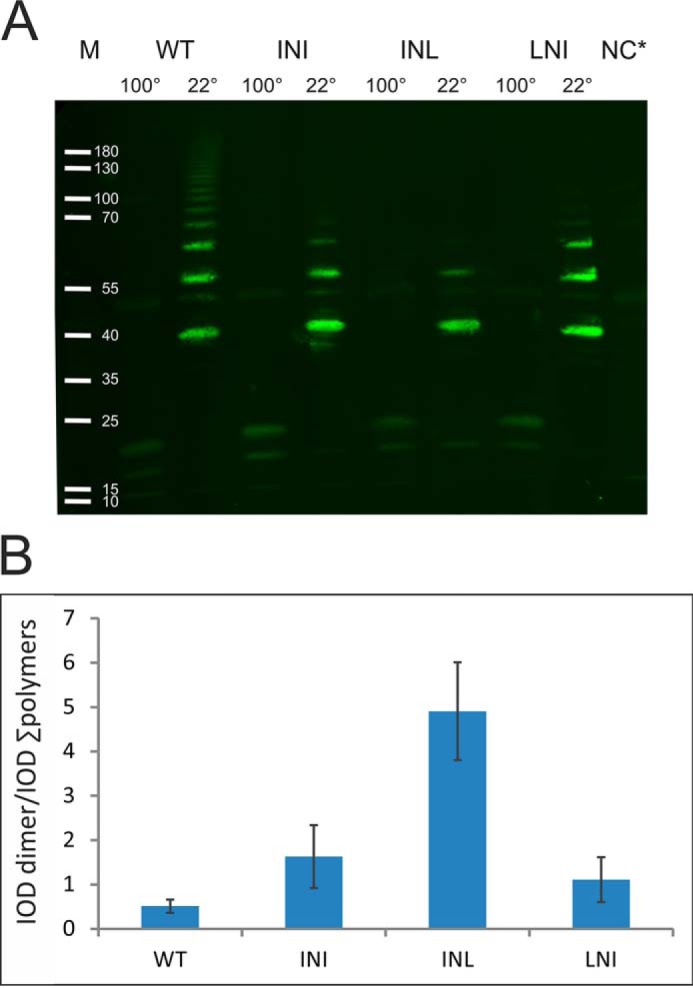
Extension of the CsuC G1 donor strand motif inhibits CsuA/B polymerization. A, Western blotting of periplasmic extracts of recombinant E. coli co-expressing CsuA/B and CsuC (wildtype (WT) and insertion mutants (INI, INL, LNI)). Complexes were incubated at 22 or 100 °C prior to electrophoresis. NC, negative control: periplasmic extract of noninduced cells; M, marker proteins, molecular weights are indicated. B, ratio of integrated optical density (IOD) of the first band (CsuA/B dimer) to the combined IOD of bands of larger CsuA/B polymers. Error bars indicate S.D. The results are representative of three independent experiments.
We compared binding of CsuA/B to WT and INL insertion mutant CsuC to confirm that complex stabilization was mediating the observed inhibition of assembly. A real-time surface plasmon resonance (SPR) method developed for Caf1M–Caf1 binding was implemented (30). Briefly, His6-CsuA/B complexed with CsuC was applied to a nickel-agarose-coated sensor chip. Prior to each binding cycle (testing a series of dilutions of the chaperone), the chip was fully regenerated to expose fresh CsuA/B. This method allowed us to accurately determine the forward and reverse rate constants and to estimate the equilibrium constant of binding using the classical 1:1 binding model (Table 2, Fig. S10). The forward and reverse rate constants obtained for the binding of WT CsuC to CsuA/B were 11 and 4.5 times lower than those found for the Caf1M–Caf1 binding, respectively, yielding only a 2-fold higher value for the equilibrium constant (∼160 nm). The archaic chaperone is considerably slower than its classical counterpart at capturing its subunit. The INL insertion increased the association rate constant 17-fold, exceeding the subunit binding capacity of Caf1M. The affinity of INL CsuC for CsuA/B increases 9-fold compared with WT, which confirm that increased subunit-chaperone complex stability mediates the potent inhibition of CsuA/B polymerization.
Table 2.
CsuC-CsuA/B association and dissociation kinetics were determined from two independent SPR experiments
| Protein | kon | koff | Kd |
|---|---|---|---|
| m−1 s−1 | ms−1 | nm | |
| CsuC-CsuA/B (WT) | 5,045 ± 329 | 0.8 ± 0.28 | 160 ± 67 |
| CsuC-CsuA/B (INL) | 84,358 ± 6,092 | 1.5 ± 0.4 | 18.0 ± 6.3 |
The high degree of disorder present in chaperone-bound CsuA/B suggests a destabilized preassembly complex, with the rigid twin-hairpin structure in CsuA/Bsc stabilizing the final fiber by locking the Gd strand in the acceptor cleft. To verify this, we substituted the stabilizing hairpin residues Phe-30 and Val-50 with glycine and analyzed the thermodynamic stability of the resultant mutants. As expected, the melting temperatures and enthalpy for all CsuA/B mutants were significantly lower than for the WT subunit (Fig. 5B, Table 1). The most pronounced difference was observed for the F30G/V50G double mutant, which melted at 71 °C releasing half as much heat as WT CsuA/B. The twin-hairpin structure provides a significant contribution to stabilization of the fiber-inserted conformation of CsuA/B.
Taken together these data suggest that subunit folding energy is the main driving force for fiber formation in non-classical systems. In classical systems, stored conformational energy is released through the collapse and efficient packing of two pilin β-sheets against each other. In non-classical systems, energy is released from the induced folding of the subunit around the extended donor strand Gd and not by the relative rearrangement of the β-sheets. Specifically, the folding of a unique twin-hairpin to form an arm-like structure drives the assembly of archaic systems.
Concluding remarks
The secretion and assembly of chaperone-usher pili are independent of external energy sources. In classical CUPs, chaperones transport folded pilins in an expanded, assembly-competent conformational state (11, 12). During DSE, pilins subsequently collapse into a conformation with a tightly packed hydrophobic core, releasing the necessary energy to drive fiber formation (10, 11, 34). Here, we showed that pilus biogenesis in non-classical CUPs utilizes a distinct form of conformational energy and a source. Non-classical chaperones bind to pilins using a truncated donor strand motif, in which it is 2 donor residues shorter for full complementation. Pilins remain in substantially unstructured conformations in the chaperone complex, having received only partial steric information required to fold. The exchange of this short donor strand motif with the longer Gd donor strand from adjacent pilin provides the full steric information essential for folding, and therefore induces a substantial unfolded-to-folded conformational transition to drive assembly.
We also showed that the first β sheet in archaic pilins is disrupted by an insertion of two unique hairpins. This two-hairpin structure, unusual for an Ig-fold, is only present in the final, fiber-inserted conformation and its folding during DSE provides additional energy to drive the assembly. In addition, the two-hairpin structure partially conceals the Gd donor strand, potentially playing a role in the stabilization of the DSC contact in the fiber. Future studies of assembled fibers will help to verify this exciting hypothesis.
Archaic and classical pre-assembly complexes exhibit comparable melting temperatures, enthalpy of melting values, and chaperone-subunit binding affinity. However, archaic chaperones are significantly slower at capturing their subunits. Non-classical subunits lack a pre-folded site for DSE initiation, which is critical for the triggering efficient assembly in the classical system (13, 35, 36). Consequently, the polymerization step is less efficient in non-classical systems. The absence of the DSE initiation site in archaic systems could facilitate a more flexible mechanism of polymerization, thereby allowing the insertion of a wider variety of donor strand sequences and incorporating a wider range of subunits sizes and shapes. Such a mechanism would have been beneficial at the early stages and expansion of fimbriae evolution, when polymers composed of various subunit sizes and shapes linked by different sequences of donor strands would have been sampled until the desired combinations were established. Therefore, the biogenesis of the more ancient non-classical systems is likely to be governed by a more fundamental assembly mechanism, and the evolution of classical fimbriae is an example of significant specialization.
CUPs are attractive targets for development of anti-adhesion drugs, known as pilicides (2, 37). Our result highlight fundamental differences in classical and non-classical CUPs assembly mechanisms, which must be taken in account in any future attempts to develop wide-spectrum pilicides. Furthermore, our work provides an essential resource to develop pilicides that specifically target the multidrug-resistant A. baumannii.
Experimental procedures
Design of expression constructs
Self-complemented CsuA/Bsc has been engineered by relocating the donor strand sequence (AVTGQVDVKLNIST) to the C′ terminus extended with a GG-linker. His6 tag has been added after the signal peptide cleavage site for purification purposes. The sequence coding 6HCsuA/Bsc has been ordered from GenScript and placed under the T7 promoter of the pET101D expression plasmid. Expression plasmid pET101-CsuC-6HCsuA/B has been constructed as detailed (38).
Mutagenesis
Mutagenesis of CsuC and CsuA/Bsc genes in plasmids pET101-CsuC6H-CsuA/B (20) and pET101-CsuC-6HCsuA/B, respectively, were performed by reverse PCR using primers listed in Table S3.
Protein expression and purification
The expression and purification of CsuC-6HCsuA/B is described in Ref. 38. The pET101–6HCsuA/Bsc expression plasmid was transformed into E. coli strain BL21-AI (Invitrogen). The transformants were cultivated in Luria-Bertani (LB) medium containing ampicillin (100 mg ml−1) at 310 K. Cells grown to an OD of 0.8–1 at 600 nm were induced with 1 mm isopropyl β-d-1-thiogalactopyranoside and 0.2% of arabinose for protein expression. The induced culture was further cultivated for 2.5 h. Expressed proteins were extracted by osmotic shock as described in Ref. 38.
Periplasmic extracts containing CsuA/Bsc were dialyzed against 20 mm sodium phosphate, 0.5 m sodium chloride, 5 mm imidazole buffer, pH 7.4, and then subjected to nickel-chelate chromatography at 277 K. A 5–500 mm imidazole gradient was used to elute the protein. Fractions containing the target protein were dialyzed against 50 mm HEPES buffer, pH 7.2. Further purification included cation-exchange chromatography with a 0–300 mm gradient of sodium chloride at 277 K and size-exclusion chromatography. Prior to size-exclusion chromatography, surface lysine methylation reaction of CsuA/Bsc was performed for crystallization purposes.
Surface lysine methylation reaction
The methylation reaction was performed overnight in 50 mm HEPES, pH 7.2, 250 mm sodium chloride containing 1 mg ml−1 of CsuA/Bsc, as described in Ref. 39. Briefly, 20 μl of freshly prepared dimethylamine-borane complex and 40 μl of 1 m formaldehyde were added to 1 ml of protein solution. After 2 h of incubation of the mixture at 277 K, the same amounts of dimethylamine-borane complex and 1 m formaldehyde were added. In 2 h, 10 μl of dimethylamine-borane complex was added per 1 ml of initial protein volume, and the reaction mixture was left overnight at 277 K. After the overnight incubation, the modified CsuA/Bsc is dialyzed against 50 mm HEPES buffer, pH 7.2, 150 mm NaCl prior to size-exclusion chromatography. The precipitated protein was removed by centrifugation.
SeMet incorporation
Incorporation of selenomethionine (SeMet) in CsuA/Bsc was performed as described in Ref. 40. Briefly, the plasmid pET101–6HCsuA/Bsc was transformed into E. coli BL21(DE3) cells (Invitrogen). Transformed cells were grown in LB medium supplemented with ampicillin (100 μg ml−1) overnight at 310 K and centrifuged (2000 rpm, 5 min) the following morning. Cell pellet was suspended in M9 minimal medium containing ampicillin (100 mg ml−1) at 310 K and cultivated at 110 rpm until an OD of 0.8–1 at 600 nm. Thirty minutes prior to induction, selenomethionine (50 mg liter−1), lysine (100 mg liter−1), phenylalanine (100 mg liter−1), threonine (100 mg liter−1), isoleucine (50 mg liter−1), leucine (50 mg liter−1), and valine (50 mg liter−1) were added to the growing cells. The expression of the target protein was induced with 1 mm isopropyl β-d-1-thiogalactopyranoside. After the induction, cells were further grown for 3 h at 310 K. Expressed CsuA/Bsc was extracted, purified, and subjected to the surface lysine methylation reaction as described above.
Crystallization and structure determination
Methylated CsuA/Bsc was concentrated to 34 mg ml−1 (SeMet incorporated CsuA/Bsc) and 24 mg ml−1 (native CsuA/Bsc) on a Vivaspin device (GE Healthcare) with molecular mass cut-off of 5 kDa. Crystals of both native and SeMet-incorporated CsuA/Bsc were obtained in 28–30% PEG 8000, 0.2 m ammonium sulfate.
Crystals were soaked for 30–60 s in cryoprotection solution (well solution complemented with 13% PEG 400) and then cooled by plunging them into liquid nitrogen. Diffraction data were collected under liquid-nitrogen cryoconditions at 100 K on beamlines ID23-1 and ID-29 at the European Synchrotron Radiation Facility (ESRF) (Grenoble, France) and processed using the Grenoble automatic data processing system (GrenADeS) at ESRF (41). Initial phases of SeMet CsuA/Bsc were determined by molecular replacement–single-wavelength anomalous diffraction (MR-SAD) phasing method using the coordinates of the subunit from the previously deposited structure of the CsuC-CsuA/B preassembly complex (PDB code 5D6H) as a search template. The model of CsuA/Bsc was constructed using the PHENIX software package (42). Manual corrections were done with molecular modeling program COOT (43). Diffraction data and refinement statistics are indicated in Table S1.
NMR spectroscopy
All NMR spectra were recorded with 500 μm [U-2H,13C,15N]CsuC-CsuA/B in 50 mm sodium phosphate, pH 6.0, 50 mm NaCl, 10% D2O. Experiments were performed at 298 K on Bruker Avance III HD 800 and Bruker Avance III HD 950 spectrometers, both equipped with a cryogenic triple resonance probe. Triple resonance experiments, namely 15N,1H-labeled TROSY-HNCACB, -HN(CO)CACB, -HNCO and -HN(CA)CO spectra (44, 45), were collected for the backbone assignment. Data were processed in NMRPipe (46) and analyzed with CCPN Analysis version 2.2 (47). The assignment was performed manually with this software and MARS (48) covering 68% of CsuC and 89% of CsuA/B excluding prolines and histidine tags. Secondary chemical shifts were analyzed with TALOS-N (28) to determine any structural propensity in portions of CsuA/B missing from the complexed structure. Dynamic analysis was performed on the complex using TROSY-based pulse sequences for T1, T2, and NOE measurements (49). Delay times were 5, 90, 220, 400, 630, 920, 1400, and 2100 ms for the T1 experiment and 0, 17, 34, 51, 68, 85, and 102 ms for the T2 experiment.
SPR assay
SPR analysis was performed on a Biacore X100 system (GE Healthcare) as described in Ref. 24. Briefly, the purified CsuC-CsuA/B preassembly complex carrying a His tag on CsuA/B (0.075 μm) was loaded on an nitrilotriacetic acid-sensor chip. CsuC was removed from the chip by continuous washing of the chip with loading buffer. To record the association and dissociation curves, samples of free CsuC at different concentrations (0.138–3 μm) were consecutively injected into the flow cell. The association phase was monitored for 180 s of the continuous infusion of CsuC, and the dissociation phase was recorded for 400 s of the constant buffer flow. After each cycle of the experiment the chip was regenerated. The software supplied with the device was used to determine the kon and koff rate constants assuming a 1:1 binding model. The model accounted for the balk effect observed in the data.
Stability analysis of CsuC-CsuA/B and CsuA/Bsc
Thermodynamic studies of CsuC-CsuA/B, WT CsuA/Bsc, and CsuA/Bsc mutants were performed by differential scanning calorimetry using a Micro-Cal VP-ITC microcalorimeter with a cell volume of 0.5 ml at a heating rate 1 °C/min. Protein samples (0.3 mg ml−1) in 20 mm HEPES, pH 7.2, and the reference buffer (20 mm HEPES, pH 7.2) were degased for 10 min and loaded into the sample and reference capillary, respectively.
Author contributions
N. P., H. M., S. Matthews, and A. V. Z. formal analysis; N. P., S. McKenna, M. T., S. P., H. M., Y. X., O. P., and A. V. Z. investigation; N. P. and A. V. Z. writing-original draft; S. McKenna, M. T., S. P., Y. X., S. Matthews, and A. V. Z. methodology; S. Matthews and A. V. Z. supervision; S. Matthews and A. V. Z. writing-review and editing; A. V. Z. conceptualization; A. V. Z. resources; A. V. Z. data curation; A. V. Z. funding acquisition; A. V. Z. validation; A. V. Z. visualization; A. V. Z. project administration.
Supplementary Material
Acknowledgments
We thank the staff at the ID29 and ID23-1 beamlines at the ESRF for their assistance during data collection. We also thank Dr. T. Nyholm for the help with differential scanning calorimetry.
This work was supported by Academy of Finland Grants 140959 and 273075 and Sigrid Juselius Foundation Grant 2014 (to A. V. Z.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S3 and Figs. S1–S10.
The atomic coordinates and structure factors (codes 6FM5, 6FQA, and 6FQ0) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The NMR chemical shifts of CsuC-CsuA/B were deposited to Biological Magnetic Resonance Data Bank (BMRB) with the BMRB accession number 27594.
- CUP
- chaperone-usher pathway
- DSC
- donor strand complementation
- DSE
- donor strand exchange
- TROSY
- transverse relaxation-optimized spectroscopy
- FGS
- FG short
- FGL
- FG long
- SPR
- surface plasmon resonance
- SeMet
- selenomethionine
- r.m.s.
- root mean square.
References
- 1. Kline K. A., Fälker S., Dahlberg S., Normark S., and Henriques-Normark B. (2009) Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5, 580–592 10.1016/j.chom.2009.05.011 [DOI] [PubMed] [Google Scholar]
- 2. Kostakioti M., Hadjifrangiskou M., and Hultgren S. J. (2013) Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 3, a010306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nuccio S. P., and Bäumler A. J. (2007) Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 71, 551–575 10.1128/MMBR.00014-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Busch A., and Waksman G. (2012) Chaperone-usher pathways: diversity and pilus assembly mechanism. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1112–1122 10.1098/rstb.2011.0206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zav'yalov V., Zavialov A., Zav'yalova G., and Korpela T. (2010) Adhesive organelles of Gram-negative pathogens assembled with the classical chaperone/usher machinery: structure and function from a clinical standpoint. FEMS Microbiol. Rev. 34, 317–378 10.1111/j.1574-6976.2009.00201.x [DOI] [PubMed] [Google Scholar]
- 6. De Greve H., Wyns L., and Bouckaert J. (2007) Combining sites of bacterial fimbriae. Curr. Opin. Struct. Biol. 17, 506–512 10.1016/j.sbi.2007.06.011 [DOI] [PubMed] [Google Scholar]
- 7. Moonens K., Bouckaert J., Coddens A., Tran T., Panjikar S., De Kerpel M., Cox E., Remaut H., and De Greve H. (2012) Structural insight in histo-blood group binding by the F18 fimbrial adhesin FedF. Mol. Microbiol. 86, 82–95 10.1111/j.1365-2958.2012.08174.x [DOI] [PubMed] [Google Scholar]
- 8. Choudhury D., Thompson A., Stojanoff V., Langermann S., Pinkner J., Hultgren S. L., and Knight S. D. (1999) X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285, 1061–1066 [DOI] [PubMed] [Google Scholar]
- 9. Sauer F. G., Fütterer K., Pinkner J. S., Dodson K. W., Hultgren S. J., and Waksman G. (1999) Structural basis of chaperone function and pilus biogenesis. Science 285, 1058–1061 10.1126/science.285.5430.1058 [DOI] [PubMed] [Google Scholar]
- 10. Zavialov A. V., Tischenko V. M., Fooks L. J., Brandsdal B. O., Aqvist J., Zav'yalov V. P., Macintyre S., and Knight S. D. (2005) Resolving the energy paradox of chaperone/usher-mediated fibre assembly. Biochem. J. 389, 685–694 10.1042/BJ20050426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zavialov A. V., Berglund J., Pudney A. F., Fooks L. J., Ibrahim T. M., MacIntyre S., and Knight S. D. (2003) Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell 113, 587–596 10.1016/S0092-8674(03)00351-9 [DOI] [PubMed] [Google Scholar]
- 12. Sauer F. G., Pinkner J. S., Waksman G., and Hultgren S. J. (2002) Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell 111, 543–551 10.1016/S0092-8674(02)01050-4 [DOI] [PubMed] [Google Scholar]
- 13. Remaut H., Rose R. J., Hannan T. J., Hultgren S. J., Radford S. E., Ashcroft A. E., and Waksman G. (2006) Donor-strand exchange in chaperone-assisted pilus assembly proceeds through a concerted beta strand displacement mechanism. Mol. Cell 22, 831–842 10.1016/j.molcel.2006.05.033 [DOI] [PubMed] [Google Scholar]
- 14. Phan G., Remaut H., Wang T., Allen W. J., Pirker K. F., Lebedev A., Henderson N. S., Geibel S., Volkan E., Yan J., Kunze M. B., Pinkner J. S., Ford B., Kay C. W., Li H., et al. (2011) Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate. Nature 474, 49–53 10.1038/nature10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nishiyama M., Ishikawa T., Rechsteiner H., and Glockshuber R. (2008) Reconstitution of pilus assembly reveals a bacterial outer membrane catalyst. Science 320, 376–379 10.1126/science.1154994 [DOI] [PubMed] [Google Scholar]
- 16. Tomaras A. P., Dorsey C. W., Edelmann R. E., and Actis L. A. (2003) Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149, 3473–3484 10.1099/mic.0.26541-0 [DOI] [PubMed] [Google Scholar]
- 17. Giraud C., Bernard C. S., Calderon V., Yang L., Filloux A., Molin S., Fichant G., Bordi C., and de Bentzmann S. (2011) The PprA-PprB two-component system activates CupE, the first non-archetypal Pseudomonas aeruginosa chaperone-usher pathway system assembling fimbriae. Environ. Microbiol. 13, 666–683 10.1111/j.1462-2920.2010.02372.x [DOI] [PubMed] [Google Scholar]
- 18. Isidean S. D., Riddle M. S., Savarino S. J., and Porter C. K. (2011) A systematic review of ETEC epidemiology focusing on colonization factor and toxin expression. Vaccine 29, 6167–6178 10.1016/j.vaccine.2011.06.084 [DOI] [PubMed] [Google Scholar]
- 19. Li Y. F., Poole S., Nishio K., Jang K., Rasulova F., McVeigh A., Savarino S. J., Xia D., and Bullitt E. (2009) Structure of CFA/I fimbriae from enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 106, 10793–10798 10.1073/pnas.0812843106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pakharukova N., Garnett J. A., Tuittila M., Paavilainen S., Diallo M., Xu Y., Matthews S. J., and Zavialov A. V. (2015) Structural insight into archaic and alternative chaperone-usher pathways reveals a novel mechanism of pilus biogenesis. PLoS Pathog. 11, e1005269 10.1371/journal.ppat.1005269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boucher H. W., Talbot G. H., Bradley J. S., Edwards J. E., Gilbert D., Rice L. B., Scheld M., Spellberg B., and Bartlett J. (2009) Bad bugs, no drugs: no ESKAPE! an update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48, 1–12 10.1086/595011 [DOI] [PubMed] [Google Scholar]
- 22. Cerqueira G. M., and Peleg A. Y. (2011) Insights into Acinetobacter baumannii pathogenicity. IUBMB Life 63, 1055–1060 10.1002/iub.533 [DOI] [PubMed] [Google Scholar]
- 23. Tomaras A. P., Flagler M. J., Dorsey C. W., Gaddy J. A., and Actis L. A. (2008) Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 154, 3398–3409 10.1099/mic.0.2008/019471-0 [DOI] [PubMed] [Google Scholar]
- 24. Giraud C., and de Bentzmann S. (2012) Inside the complex regulation of Pseudomonas aeruginosa chaperone usher systems. Environ. Microbiol. 14, 1805–1816 10.1111/j.1462-2920.2011.02673.x [DOI] [PubMed] [Google Scholar]
- 25. Pakharukova N., Tuittila M., Paavilainen S., Malmi H., Parilova O., Teneberg S., Knight S. D., and Zavialov A. V. (2018) Structural basis for Acinetobacter baumannii biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 115, 5558–5563 10.1073/pnas.1800961115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu M., Xu S., Zhu W., and Mao X. (2014) The archaic chaperone-usher pathways may depend on donor strand exchange for intersubunit interactions. Microbiology 160, 2200–2207 10.1099/mic.0.080457-0 [DOI] [PubMed] [Google Scholar]
- 27. Bao R., Fordyce A., Chen Y. X., McVeigh A., Savarino S. J., and Xia D. (2014) Structure of CfaA suggests a new family of chaperones essential for assembly of class 5 fimbriae. PLoS Pathog. 10, e1004316 10.1371/journal.ppat.1004316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shen Y., and Bax A. (2013) Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 56, 227–241 10.1007/s10858-013-9741-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tamiola K., Acar B., and Mulder F. A. (2010) Sequence-specific random coil chemical shifts of intrinsically disordered proteins. J. Am. Chem. Soc. 132, 18000–18003 10.1021/ja105656t [DOI] [PubMed] [Google Scholar]
- 30. Tamiola K., and Mulder F. A. (2012) Using NMR chemical shifts to calculate the propensity for structural order and disorder in proteins. Biochem. Soc. Trans. 40, 1014–1020 10.1042/BST20120171 [DOI] [PubMed] [Google Scholar]
- 31. Zavialov A., Zav'yalova G., Korpela T., and Zav'yalov V. (2007) FGL chaperone-assembled fimbrial polyadhesins: anti-immune armament of Gram-negative bacterial pathogens. FEMS Microbiol Rev. 31, 478–514 10.1111/j.1574-6976.2007.00075.x [DOI] [PubMed] [Google Scholar]
- 32. Hung D. L., Knight S. D., Woods R. M., Pinkner J. S., and Hultgren S. J. (1996) Molecular basis of two subfamilies of immunoglobulin-like chaperones. EMBO J. 15, 3792–3805 10.1002/j.1460-2075.1996.tb00753.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yu X. D., Fooks L. J., Moslehi-Mohebi E., Tischenko V. M., Askarieh G., Knight S. D., Macintyre S., and Zavialov A. V. (2012) Large is fast, small is tight: determinants of speed and affinity in subunit capture by a periplasmic chaperone. J. Mol. Biol. 417, 294–308 10.1016/j.jmb.2012.01.020 [DOI] [PubMed] [Google Scholar]
- 35. Verger D., Miller E., Remaut H., Waksman G., and Hultgren S. (2006) Molecular mechanism of P pilus termination in uropathogenic Escherichia coli. EMBO Rep. 7, 1228–1232 10.1038/sj.embor.7400833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Di Yu X., Dubnovitsky A., Pudney A. F., Macintyre S., Knight S. D., and Zavialov A. V. (2012) Allosteric mechanism controls traffic in the chaperone/usher pathway. Structure 20, 1861–1871 10.1016/j.str.2012.08.016 [DOI] [PubMed] [Google Scholar]
- 37. Pinkner J. S., Remaut H., Buelens F., Miller E., Aberg V., Pemberton N., Hedenström M., Larsson A., Seed P., Waksman G., Hultgren S. J., and Almqvist F. (2006) Rationally designed small compounds inhibit pilus biogenesis in uropathogenic bacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 17897–17902 10.1073/pnas.0606795103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pakharukova N., Tuittila M., Paavilainen S., and Zavialov A. (2015) Crystallization and preliminary X-ray diffraction analysis of the Csu pili CsuC-CsuA/B chaperone-major subunit pre-assembly complex from Acinetobacter baumannii. Acta Crystallogr. Sect. F Struct. Biol. Commun. 71, 770–774 10.1107/S2053230X15007955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Walter T. S., Meier C., Assenberg R., Au K. F., Ren J., Verma A., Nettleship J. E., Owens R. J., Stuart D. I., and Grimes J. M. (2006) Lysine methylation as a routine rescue strategy for protein crystallization. Structure 14, 1617–1622 10.1016/j.str.2006.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Doublié S. (2007) Production of selenomethionyl proteins in prokaryotic and eukaryotic expression systems. Methods Mol. Biol. 363, 91–108 10.1007/978-1-59745-209-0_5 [DOI] [PubMed] [Google Scholar]
- 41. Monaco S., Gordon E., Bowler M. W., Delagenière S., Guijarro M., Spruce D., Svensson O., McSweeney S. M., McCarthy A. A., Leonard G., and Nanao M. H. (2013) Automatic processing of macromolecular crystallography X-ray diffraction data at the ESRF. J. Appl. Crystallogr. 46, 804–810 10.1107/S0021889813006195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., and Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 10.1107/S0907444902016657 [DOI] [PubMed] [Google Scholar]
- 43. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Salzmann M., Pervushin K., Wider G., Senn H., and Wüthrich K. (1998) TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc. Natl. Acad. Sci. U.S.A. 95, 13585–13590 10.1073/pnas.95.23.13585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Salzmann M., Wider G., Pervushin K., Senn H., and Wüthrich K. (1999) TROSY-type triple-resonance experiments for sequential NMR assignments of large proteins. J. Am. Chem. Soc. 121, 844–848 10.1021/ja9834226 [DOI] [Google Scholar]
- 46. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 47. Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., and Laue E. D. (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696 10.1002/prot.20449 [DOI] [PubMed] [Google Scholar]
- 48. Jung Y. S., and Zweckstetter M. (2004) Mars: robust automatic backbone assignment of proteins. J. Biomol. NMR 30, 11–23 10.1023/B:JNMR.0000042954.99056.ad [DOI] [PubMed] [Google Scholar]
- 49. Zhu G., Xia Y., Nicholson L. K., and Sze K. H. (2000) Protein dynamics measurements by TROSY-based NMR experiments. J. Magn. Reson. 143, 423–426 10.1006/jmre.2000.2022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.