Bacteria rotate propeller-like flagella to find and colonize environmental niches. The flagellum is a complex machine, and the understanding of its structure is still incomplete. Here, we characterize and biochemically define the assembly order of the subunits that make up the axle-like rod. The rod is a critical structure for the assembly of subsequent components and is central to our understanding of how the flagellum is anchored but still free spinning within the context of the cell envelope.
KEYWORDS: motility, flagella, rod, structure, FliE, swarming, swimming
ABSTRACT
Bacterial flagella contain an axle-like rod that transits the cell envelope and connects the transmembrane basal body to the extracellular hook and filament. Although the rod is a crucial component of the flagellum, its structure and assembly are poorly understood. Previous reports defining the order of rod assembly in Gram-negative bacteria suggest that the rod requires five proteins to successfully assemble, but assembly intermediates have not been well characterized due to metastability and periplasmic proteolysis. Bacillus subtilis is a Gram-positive, genetically tractable model bacterium that synthesizes flagella and lacks a true periplasm. Here, we genetically, biochemically, and cytologically determine the assembly order of the flagellar rod proteins from cell proximal to distal as FliE, FlgB, FlgC, FlhO, and FlhP. We further show that, under conditions in which rod structure cannot be completed, assembly intermediates are both metastable and subject to proteolysis. Finally, we support previous results that FliE serves as both a structural assembly platform for the rod and as an enhancer of flagellar type III secretion.
IMPORTANCE Bacteria rotate propeller-like flagella to find and colonize environmental niches. The flagellum is a complex machine, and the understanding of its structure is still incomplete. Here, we characterize and biochemically define the assembly order of the subunits that make up the axle-like rod. The rod is a critical structure for the assembly of subsequent components and is central to our understanding of how the flagellum is anchored but still free spinning within the context of the cell envelope.
INTRODUCTION
Bacterial motility is an important mechanism for nutrient acquisition and niche establishment in the environment. Various bacteria utilize different means of motility, including the assembly of helical flagella to swim through a liquid or swarm on a semisolid surface (1). Flagella are comprised of more than 20 distinct proteins that are assembled with precise stoichiometry and in a fixed order (2). Flagella are organized into three structural domains (Fig. 1A), as follows: (i) a basal body containing the type III secretion machinery and rotational apparatus and (ii) an extracellular hook that acts as a universal joint to transmit rotational energy to (iii) the helical filament that provides propulsion (3–5). The basal body is connected to the hook via an axle-like rod that transits the peptidoglycan and outer membrane (6–9). Whereas the flagellar hook and filament structures are comprised of a single repeating protein, the rod is assembled from as many as 4 to 5 different subunits (10, 11).
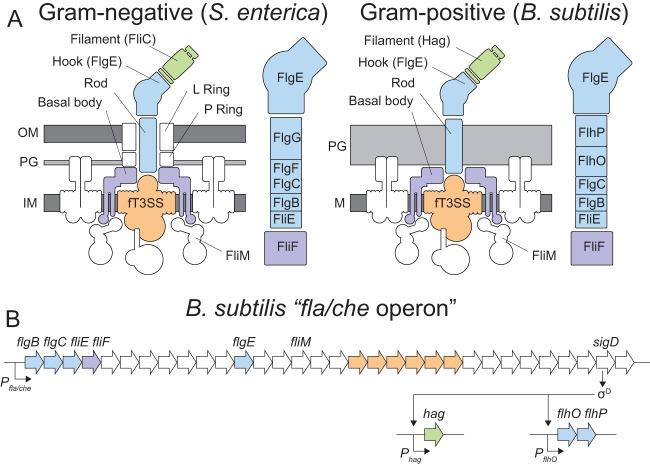
FIG 1.
Diagrams of B. subtilis flagellar structure and genetic architecture. (A) Cartoon model of the Gram-negative and Gram-positive flagellar architectures. To the right of each flagellum is a closeup depiction of the assembly order of the rod components (14). The position of FlgC in the Gram-negative rod has traditionally been considered cell proximal to FlgF, but a recent publication suggests that the orders may be reversed (25). The order of the rod components in B. subtilis is indicated based on the information in the present manuscript. The membrane is colored dark gray. Peptidoglycans are colored light gray. The flagellar basal body is colored purple, the flagellar type III secretion system (fT3SS) is colored orange, the rod-hook structure is colored blue, and the filament is colored green. (B) B. subtilis fla/che operon structure and flagellar genetic hierarchy. Genes mentioned in the manuscript are indicated in italics. Bent arrows indicate promoters, and open arrows indicate genes. Genes are color coded to match the relative locations of their gene products in panel A.
Rod assembly has mainly been characterized in the Gram-negative species Salmonella enterica serovar Typhimurium. Early reports showed that the Salmonella enterica rod structure required FlgB, FlgC, FlgF, and FlgG, and that the rod did not assemble unless all four proteins were present (11–14). Consistent with structural subunits of the rod, each protein was secreted by the flagellar type III secretion system, and each protein was found to polymerize in vitro (15–17). The rod spans the width of the cell envelope, passing through the peptidoglycan and the outer membrane by the “P” and “L” rings, which are thought to act as bushings and permit rod rotation in the context of the envelope (18–20). The assembly order of the Salmonella rod has been provisionally deduced. FlgB is thought to be the first rod protein polymerized, as it was found to interact with FliE, a protein shown to be in close association with the plasma membrane-bound basal body protein FliF (21–23). FlgG is thought to be the last rod protein polymerized, as particular mutants of FliF cause the rod to shear such that FlgG is released with the flagellar hook (10, 11, 24). Thus, the inferred rod order from cell proximal to cell distal is FlgB, FlgC/FlgF, and FlgG with recent evidence that FlgF may precede FlgC (25) (Fig. 1A, left). Prediction of the rod assembly order was necessarily indirect as mutation of any particular subunit prevented the assembly of intermediate structures.
The absence of intermediate rod structures in S. enterica mutants could potentially be explained in one of two ways. One explanation posits that the rod is intrinsically unstable, such that in the absence of the complete structure, intermediate states are metastable and prone to disassemble (11, 14). Another explanation posits that the rod is extrinsically unstable, such that in the absence of the complete structure, intermediate states are prone to proteolytic degradation in the periplasm (26). Recently, cryoelectron tomography structural analysis of intermediate rod structures was performed in mutants of the spirochete Borrelia burgdorferi, and these results supported the assembly order of the homologous proteins in S. enterica (27). That intermediate rod structures were observed and measured at high resolution might suggest that rod metastability and/or proteolysis is particular to S. enterica. Alternatively, Borrelia rod intermediates may also be unstable, but continuous resynthesis and instantaneous in vivo flash freezing may preserve the structure.
Rod assembly also requires the poorly understood protein FliE (22). Two possible roles of FliE have been reported. First, FliE may be structural and serve as a geometric adaptor protein that connects the intramembrane, ring-like polymer of the basal body protein FliF to the extracellular helical polymer formed by the secreted rod subunits. Consistent with a structural adaptor, FliE has been shown to interact with both FliF and FlgB (21, 22). Alternatively, FliE may be regulatory, as FliE has been shown to enhance flagellar type III secretion, perhaps by relieving steric inhibition of an inactive basal body conformation (16, 27, 28). Furthermore, FliE has been shown to be exported during flagellar assembly, suggesting that it is unlikely to be an integral part of the cytoplasmic and transmembrane secretion apparatus (29, 30). Given that the two functions are not mutually exclusive, it is unclear whether FliE is a structural subunit, a regulator of flagellum-specific secretion, or both.
The Gram-positive bacterium Bacillus subtilis encodes putative homologs of all four rod proteins, termed FlgB, FlgC, FlhO, and FlhP, as well as FliE (31, 32). Whereas FlgB, FlgC, and FliE are encoded in the fla/che operon, FlhO and FlhP are encoded as an unlinked dicistron (Fig. 1B) (32). Moreover, purification of intact flagellar basal bodies determined that some of the rod homologs were present in the complex (33), and mutation of FlhO or FlhP abolished motility and assembly of the hook (32). Here, we further characterize the putative rod proteins and show that FlgB, FlgC, and FliE are also required for motility and assembly of the hook. We take a biochemical approach and probe cell fractions in Western blot analysis to support the role of each protein as a structural component of the rod and set the order of assembly. Moreover, we show that intermediate rod structures can only be isolated from membrane fractions in the presence of a cross-linker and in the absence of secreted proteases. Thus, the rod is both metastable and susceptible to proteolysis. Finally, mutation of FliE abolished membrane retention and also reduced the secretion of all other rod structural elements, consistent with both structural and regulatory roles reported in S. enterica.
RESULTS
FlgB and FlgC are required for the assembly of both the hook and filament.
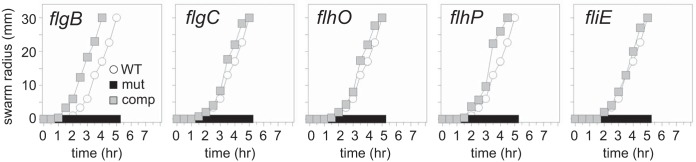
The protein composition of the rod in B. subtilis is unknown. Two candidate rod genes, flgB and flgC, encode protein sequences homologous to those of S. enterica rod proteins FlgB and FlgC (see Fig. S1 in the supplemental material) and are the first two genes located within the 32-gene-long fla/che operon that encodes components of the flagellar basal body (Fig. 1B). To determine whether flagellar motility requires FlgB and FlgC, we separately deleted flgB and flgC and tested the resulting mutants for swarming motility atop a soft agar surface. Mutation of either gene abolished swarming motility, and swarming motility was restored to the flgB and flgC mutants when a complementation construct, in which each gene was expressed from the Pfla/che promoter, was inserted at an ectopic site in the chromosome (amyE::Pfla/che-flgB or amyE::Pfla/che-flgC, respectively) (Fig. 2). Furthermore, no spontaneous suppressor mutations appeared upon prolonged incubation of the swarm plate of the uncomplemented mutants, consistent with mutations in structural components of the flagellum. We conclude that swarming motility requires both FlgB and FlgC.
FIG 2.
Rod homologs and FliE are required for swarming motility. Quantitative graphs of swarm expansion assays. Each graph contains wild-type strain 3610 (WT; data were generated at the same time as for the other strains and are in each panel for comparison), a strain containing a mutation in the gene indicated in the upper left corner (mut), and a strain containing both a mutation in the indicated gene and an ectopic complementation construct of that gene (comp). The swarm expansion assays were generated with the following strains: 3610 (WT), DS4680 (flgB), DS9555 (flgB [flgB+]), DS7307 (flgC), DK3720 (flgC [flgC+]), DS5161 (flhO), DS5944 (flhO [flhO+]), DS7351 (flhP), DK7360 (flhP [flhP+]), DS7308 (fliE), and DK3721 (fliE [fliE+]). Each data point is the average of three replicates. Each swarm was arbitrarily terminated at a radius of 30 mm to compensate for inoculation variation from the geometric center.
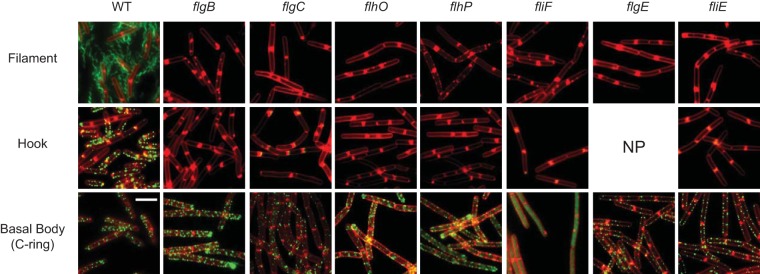
The flagellum assembles sequentially such that a failure to construct a cell-proximal component results in an inability to incorporate a more distal segment (12, 14, 27). To determine where in flagellar assembly FlgB and FlgC are required, we used fluorescence microscopy to assess the presence of various flagellar components in flgB and flgC mutant backgrounds. To determine if FlgB and FlgC affect filament assembly, each gene was mutated in a background containing a variant of the filament protein Hag that could be stained with a cysteine-reactive maleimide-conjugated fluorophore (HagT209C) (34, 35). Wild-type cells produced multiple flagellar filaments protruding from the cell surface, but cells mutated for flgB and flgC were defective in the assembly of the flagellar filament (Fig. 3). Next, we asked whether the structure preceding the filament, the hook, was assembled in the flgB or flgC mutants. To determine if FlgB and FlgC affect hook assembly, each gene was mutated in a background containing a variant of the hook protein FlgE that could be stained with a cysteine-reactive maleimide-conjugated fluorophore (FlgET123C) (32). Wild-type cells produced multiple fluorescent foci on the cell surface, but flgB and flgC mutant strains did not (Fig. 3). We conclude that FlgB and FlgC are required for flagellar hook and filament assembly.
FIG 3.
Rod homologs are required for hook and filament assembly. Fluorescent micrographs of strains representing the presence of filament, hook, and basal body. Membranes were stained with FM-4-64 dye (false colored red). Filaments and hooks were visualized by using a maleimide-conjugated dye to stain proteins modified to introduce a single surface-exposed cysteine residue. (HagT209C and FlgET123C, respectively; false colored green). Basal bodies were visualized using a strain background containing a fluorescently tagged FliM-GFP (false colored green). The filament panel was generated with the following strains: DS1916 (WT), DK5040 (ΔflgB), DK5041 (ΔflgC) DS5897 (ΔflhO), DS5042 (ΔflhP), DK5038 (ΔfliF), DS5896 (ΔflgE), and DK5039 (ΔfliE). The hook panel was generated with the following strains: DS7673 (WT), DK4895 (ΔflgB), DK4868 (ΔflgC) DS8839 (ΔflhO), DS8840 (ΔflhP), DK3135 (ΔfliF), and DK4896 (ΔfliE). The basal body panel was generated with the following strains: DS8521 (WT), DK5082 (ΔflgB), DK5268 (ΔflgC) DK5083 (ΔflhO), DK5084 (ΔflhP), DS8598 (ΔfliF), DK4892 (ΔflgE), and DK5081 (ΔfliE). NP, not possible, as the cysteine replacement is within the gene that was deleted.
Finally, we assessed the role of FlgB and FlgC in the assembly of the basal body, the membrane-bound portion of the flagellum. To determine if FlgB and FlgC affect basal body assembly, each gene was mutated in a background containing a variant of the basal body C-ring protein FliM fused to green fluorescent protein (FliM-GFP) (36). Fluorescent puncta were observed in wild-type cells, and a similar number of puncta were observed in cells mutated for flgB and flgC. As a control, fluorescent puncta were abolished in cells mutated for the basal body protein FliF (Fig. 3). We infer that the initiation of basal body construction does not require the presence of FlgB or FlgC. Taken together, these data indicate that flgB and flgC mutants complete flagellar assembly at least up to the integration of the FliM protein in the C-ring but fail to subsequently assemble detectable hook or filament structures. A structural defect between the basal body and flagellar hook is cytologically consistent with FlgB and FlgC being structural components of the rod.
The Bacillus subtilis rod consists of FlgB, FlgC, FlhO, and FlhP.
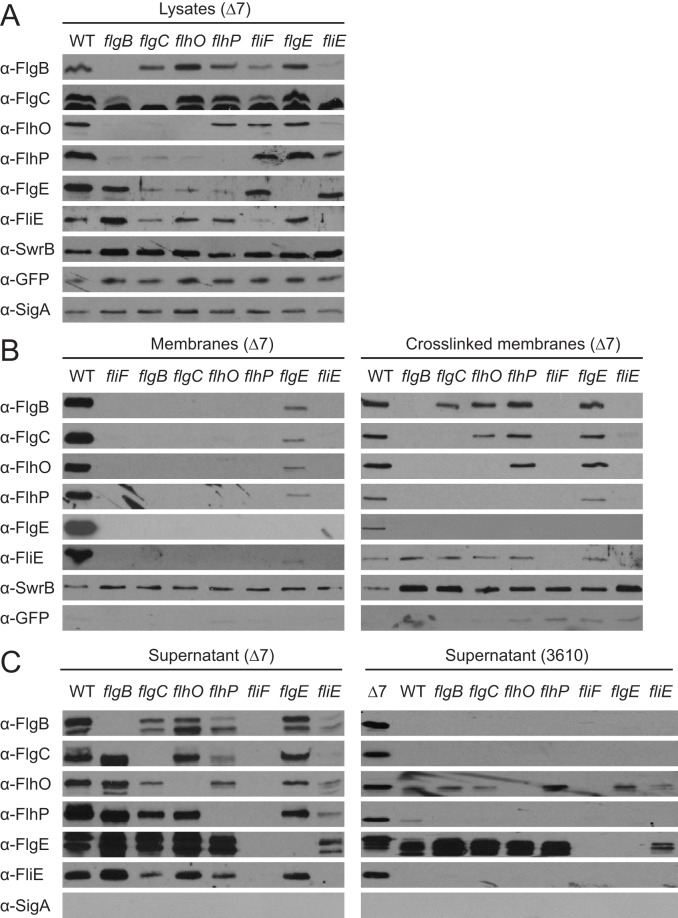
Two other rod homologs, FlhO and FlhP, were previously proposed to constitute structural components of the rod, and mutation of either protein results in swarming and flagellar assembly defects similar to those of mutants defective in flgB and flgC (Fig. 2, Fig. 3, and Fig. S1) (32, 33). To determine whether FlgB, FlgC, FlhO, and FlhP are structural components of the flagellum, each protein was purified and submitted for generation of polyclonal antibodies. Western blot analysis was performed on cell lysates of wild type and various mutants in a background also mutated for 7 extracellular proteases to improve protein detection (Δ7) (37). The cytoplasmic sigma factor sigma A (SigA) and the transmembrane protein SwrB were probed as loading controls and detected in each sample. Each putative rod protein was detected in whole-cell lysates of the wild-type strain and absent in their corresponding deletion mutant (Fig. 4A). However, some mutant cell lysates lacked more than just their cognate protein: FlhP was also reduced in cells mutated for flgB, flgC, or flhO; FlhO was also reduced in cells mutated for flgB and flgC; and FlgC was also reduced in cells mutated for flgB. We hypothesize that the stepwise absence of each protein observed in the mutants could indicate either the ordered assembly or ordered secretion of the four proteins in the flagellum.
FIG 4.
Rod homologs are secreted and sequentially assembled. Western blot analyses of putative rod proteins in whole-cell lysates (A), purified membrane fractions (B, left), formaldehyde-cross-linked purified membrane fractions (B, right), and trichloroacetic acid (TCA)-precipitated supernatants (C). Strains mutated for 7 extracellular proteases are denoted by Δ7. Each strain also encodes a cytoplasmic green fluorescent protein (GFP) induced with isopropyl-β-d-thiogalactopyranoside (IPTG) to serve as a loading and fraction control (Physpank-gfp). Genetic backgrounds are indicated in parentheses above each panel, and the bar indicates all strains below that share the indicated genetic background. Samples were generated as described in Materials and Methods using the following strains: DK4537 (Δ7), DK4540 (Δ7 ΔflgB), DK4541 (Δ7 ΔflgC), DK4542 (Δ7 ΔflhO), DK4543 (Δ7 ΔflhP), DK4544 (Δ7 ΔflgE), DK4538 (Δ7 ΔfliF), and DK4539 (Δ7 ΔfliE). Strain 3610-derived supernatant samples for the panel C (right) were generated via TCA precipitation with the following strains: DS6329 (Δ7), 3610 (WT), DS4680 (ΔflgB), DS7307 (ΔflgC), DS5161 (ΔflhO), DS7351 (ΔflhP), DS3968 (ΔfliF), DS4681 (ΔflgE), and DS7308 (ΔfliE).
To determine whether each putative rod protein assembles sequentially in the flagellum, membrane fractions from the wild type and putative rod mutants were isolated and resolved by Western blotting. Membrane fraction purity was supported by the absence of soluble, cytoplasmically expressed green fluorescent protein (GFP), as well as by the presence of SwrB as a loading control (Fig. 4B, left). Each putative rod protein was detected in the membranes of wild type isolations (Fig. 4B, left). However, none of the candidate rod proteins were detected in the membrane fractions of any strain from which a putative rod gene was deleted, seemingly inconsistent with their presence in the lysates. Previous reports suggest that rod structures are metastable in other organisms, such that the rod disassembles when any required component is absent (10). To aid in the detection of assembly intermediates, potential rod disassembly was impaired by treating cells with the protein cross-linking agent formaldehyde prior to membrane isolation (Fig. 4B, right). Cross-linked membranes recapitulated the stepwise inclusion of each putative rod component observed in whole-cell lysates after the cross-linking was reversed by heat treatment. We conclude that the structure formed by the putative rod proteins is metastable, that the metastable structure can be preserved by cross-linking, that incorporation of each subunit is sequential, and that the proximal to distal assembly order is FlgB, FlgC, FlhO, and FlhP.
If FlgB, FlgC, FlhO, and FlhP are structural components of the rod, we then expect that each protein would be secreted beyond the membrane via the flagellar secretion system. To detect protein secretion, trichloroacetic acid (TCA)-precipitated supernatants from wild-type and the candidate rod mutant strains were subjected to Western blot analysis. Each putative rod protein, but not the cytoplasmic control protein SigA, was detected in the supernatant of wild-type cells (Fig. 4C, left). Moreover, we found that each strain from which a candidate rod protein was deleted secreted all other candidate rod proteins. Thus, the nested absence of rod proteins seen in a particular mutant lysate was due to continuous export in the absence of retention by polymerization. For example, FlhP levels were low in flgB mutant lysates because FlhP was secreted and not retained (compare Fig. 4A with left panel of Fig. 4C). We conclude that the stepwise absence of putative rod proteins in mutant membranes was a reflection of sequential assembly and not of sequential secretion.
To determine the location of the structure formed by the putative rod proteins, cells were mutated for known structural components of the flagellum. One structural component, FliF, comprises the flagellar basal body base plate, houses and activates the flagellar type III secretion apparatus, and is the polymerization platform for the rod (38). No putative rod proteins were detected in either supernatants or cross-linked membranes of cells mutated for FliF (Fig. 4B and C). We conclude that the putative rod proteins are secreted in a flagellar type III-dependent manner. Flagellar type III secretion sustains two discrete classes of secreted cargo, constituting early and late flagellar proteins (10, 16). Completion of the flagellar hook structure is necessary for a substrate specificity switch to activate export of the late-class proteins (39–42). All putative rod proteins were detected in both the supernatants and membranes of cells mutated for the hook gene flgE but at reduced levels. We conclude that the putative rod proteins are early-class type III secretion substrates and are retained in the membrane fraction prior to hook completion. We hypothesize that the hook enhances, but is not essential for, stability of the proximal structure. Taking these data together, we conclude that FlgB, FlgC, FlhO, and FlhP are the proximal to distal structural components of the flagellar rod between FliF and FlgE in B. subtilis.
Rod structure can potentially be destabilized by extracellular proteolysis (26). To determine the role of extracellular proteolysis, cell supernatant fractions were harvested from a strain that was wild type for all 7 secreted proteases. The extracellular abundance of most rod proteins was substantially diminished in the wild type compared to those in the Δ7 protease mutant (Fig. 4C, right). Moreover, the abundance of most rod proteins was reduced to undetectable levels in wild-type strains mutated for any gene encoding a rod protein. We conclude that secreted but unpolymerized rod subunits are subject to extracellular proteolysis. Extracellular proteolysis, however, is separable from the metastability of rod assembly, as rod intermediates were not detected in the Δ7 mutant background in the absence of a fixing agent (Fig. 4B). Thus, difficulty in detecting rod assembly intermediates is confounded by both metastability and proteolysis, but the two processes are unrelated.
FliE.
A fifth protein, FliE, has been shown in other organisms to associate with the rod, but the role of FliE in flagellar assembly is poorly understood. One model suggests that FliE is structural, constituting either a fifth rod subunit or acting as an adaptor between the basal body protein FliF and the first rod structural subunit FlgB (21, 22). Another model suggests that FliE is either an essential component or activator of the flagellar type III secretion system (16, 21, 27). To explore FliE function in B. subtilis, we generated an in-frame markerless deletion of the fliE gene. The mutant was defective for swarming motility, and swarming was complemented when the fliE gene was cloned downstream of the Pfla/che promoter and inserted at an ectopic site in the chromosome (Fig. 2C). Moreover, the fliE mutant was defective in swimming motility, as observed in wet mounts, and was defective in the assembly of the filament and hook but not of the basal body (Fig. 3). We conclude that FliE is required for motility and that the fliE strain phenocopies a mutation in any of the four other rod proteins.
To determine the location of FliE relative to the other rod proteins, Western blot analysis was performed on various cellular fractions. FliE was detected in the TCA-precipitated supernatant of the wild type but not that of the fliF mutant, indicating that it was secreted in a type III-dependent manner (Fig. 4C). Moreover, FliE was retained in the membrane fraction like other rod subunits but only upon cross-linking, suggesting that it is part of the metastable structure (Fig. 4B). Retention of FliE in the cross-linked membrane, however, did not require any other rod protein, as it was present in all other rod mutant backgrounds. We conclude that FliE is part of the flagellar structure and may be the first extracellular flagellar component upon which the rod is subsequently assembled.
To determine whether FliE is required for subsequent rod assembly, the location of each rod protein was assessed in a fliE mutant background. When fliE was mutated, none of the four rod proteins were retained in the membrane fraction, regardless of whether a cross-linker was included (Fig. 4B). Instead, the rod proteins were secreted into the extracellular environment, suggesting a failure to polymerize and be retained in the membrane. In addition, the amounts of secreted rod proteins were reduced, and the reduction was not related to extracellular proteolysis, as the experiment was conducted in the Δ7 background. Consistent with a generalized defect in type III secretion, the extracellular pool of the flagellar hook protein FlgE was also substantially diminished when samples were normalized by loading based on total secreted protein (Fig. 4C; see also Fig. S2 in the supplemental material). We conclude that FliE is an early part of the bacterial flagellum that both serves as the structural base for subsequent rod assembly and enhances flagellar type III secretion.
DISCUSSION
The rod is an essential structural component for flagellar assembly and function. With respect to assembly, the rod is the part of the complex that transit the layers of the cell envelope, including the peptidoglycan, periplasm, and outer membrane. Moreover, the rod serves as a conduit for secretion and as a construction platform for distal domains, such as the hook and filament. With respect to function, the rod transmits rotational force generated by the rotor-basal body in the inner membrane to the exterior hook and filament. Despite its importance, the rod is the least-studied structural component of the flagellum, likely for technical reasons. Here, we show that FlgB, FlgC, FlhO, and FlhP are required for swarming motility and that they are structural proteins required for flagellar assembly in Bacillus subtilis. Moreover, our data indicate that these proteins assemble the rod in a stepwise and discrete order, consistent with predictions in Gram-negative bacteria and electron microscopy (EM) observations in Spirochaetes.
One technical difficulty in studying rod assembly is that the structure is metastable, such that in the absence of completion, the structure falls apart (11–14). Thus, intermediate rod structures could not be captured in purified basal bodies of rod mutants in Escherichia coli and S. enterica and instead, rod assembly order was indirectly inferred. Here, we biochemically demonstrate sequential assembly of the rod using advantages of Bacillus subtilis and the Gram-positive architecture. Like E. coli and S. enterica, intermediate rod structures were not isolated from B. subtilis membrane fractions in rod mutants. The absence of an outer membrane, however, allowed for the stabilization and recovery of such intermediates by using a cross-linking agent prior to purification. Another technical difficulty in studying rod assembly is that incomplete structures are proteolytically degraded in the Gram-negative periplasm. Gram-positive bacteria do not have a true periplasm, and while rod subunits were also degraded in wild-type B. subtilis, deleting seven extracellular proteases was sufficient to detect secreted rod proteins. We support the model generated in Gram-negative bacteria that the flagellar rod is both metastable and proteolytically sensitive prior to completion, suggesting that this structural property is likely conserved throughout phylogeny.
We took the opportunity to explore the function of a poorly understood rod-associated protein, FliE. FliE might constitute a fifth subunit of rod structure, despite a lack of sequence homology to other rod proteins (11, 43). Consistent with a structural role, FliE was secreted in a FliF-dependent manner and was retained in membrane fractions that had been treated with a chemical cross-linker. Moreover, B. subtilis FliE was the most cell-proximal secreted component and was required for the retention of the four other rod proteins. FliE has also been shown to enhance flagellar type III secretion, and cryoelectron tomography indicated that in the absence of FliE, the secretion channel appears to have an altered, likely defective, conformation (16, 27). Consistent with FliE being a regulator and not an essential part of the secretion system in B. subtilis, the secretion of rod and hook proteins was reduced but not abolished in the absence of FliE. Thus, FliE appears to be both a structural component of the rod and a regulatory component necessary for full secretion of flagellar proteins. We note that if FliE is regulatory, it must regulate its own secretion, as it too is secreted by the flagellar type III apparatus. Perhaps the two functions of FliE are spatially separable, whereby the structural role is extracellular and the regulatory role is cytoplasmic.
Although the rod of B. subtilis and other Gram-positive organisms appears structurally similar to that of Gram-negative organisms, the environment through which it assembles is quite distinct, and questions remain regarding these differences. Rod polymerization in Gram-negative bacteria is thought to halt when the structure transits the outer-membrane bushing, but Gram-positive bacteria lack both the outer membrane and bushing proteins (20, 44, 45). Thus, we infer that rod length in Gram-positive bacteria is restricted by a different mechanism. Furthermore, the rod is thought to penetrate the single layer of Gram-negative peptidoglycan by a rod-mounted hydrolase, but a homologous protein is absent in Gram-positive bacteria, and no hydrolase has yet been found to be required for flagellar assembly (46–49). Thus, how the flagellar rod transits the multilamellar peptidoglycan of the Gram-positive architecture, and how rod length matches peptidoglycan depth, is unknown. Finally, the flagellar hook and filament are each polymerized from a single protein subunit, and why the flagellar rod would require 4 or 5 different structural subunits is unclear unless the subunits are related to the different architectural environments of the cell envelope. Here, we begin to address these questions by determining the order of rod assembly in B. subtilis from cell proximal to distal, namely, FliE, FlgB, FlgC, FlhO, and finally FlhP.
MATERIALS AND METHODS
Strains and growth conditions.
B. subtilis strains were grown in lysogeny broth (LB) (10 g tryptone, 5 g yeast extract, and 5 g NaCl per liter) or on LB plates fortified with 1.5% Bacto agar at 37°C. When appropriate, antibiotics were included at the following concentrations: 10 μg/ml tetracycline, 100 μg/ml spectinomycin, 5 μg/ml chloramphenicol, 5 μg/ml kanamycin, and 1 μg/ml erythromycin plus 25 μg/ml lincomycin (macrolides-lincosamides-streptogramin B [MLS]). Isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma) was added to the medium at the indicated concentration when appropriate.
Strain construction.
Constructs were first introduced into either the domesticated strain PY79 by natural competence and then transferred to the 3610 background using SPP1-mediated generalized phage transduction or by transformation directly into the competent derivative of 3610, DK1042 (comIQ12L) (50, 59). All strains used in this study are listed in Table 1. All plasmids used in this study are listed in Table S1 in the supplemental material. All primers used in this study are listed in Table S2 in the supplemental material.
TABLE 1.
Strains
| Strain | Genotypea | Reference |
|---|---|---|
| 3610 | Wild type | |
| PY79 | sfp0 swrAfs | |
| DS1916 | amyE::Phag-hagT209C spec | 34 |
| DS3968 | ΔfliF | 58 |
| DS4680 | ΔflgB | 37 |
| DS4681 | ΔflgE | 32 |
| DS5161 | ΔflhO | 32 |
| DS5896 | ΔflgE amyE::Phag-hagT209C spec | 32 |
| DS5897 | ΔflhO amyE::Phag-hagT209C spec | 32 |
| DS5944 | ΔflhO thrC::PflhO-flhO erm | 32 |
| DS6329 | Δ7 (Δmpr ΔaprE ΔnprE Δbpr Δvpr Δepr ΔwprA) | 37 |
| DS7307 | ΔflgC | 37 |
| DS7308 | ΔfliE | 37 |
| DS7351 | ΔflhP | 32 |
| DS7360 | ΔflhP amyE::PflhO-flhP cat | 32 |
| DS7673 | ΔflgE amyE::Pfla/che-flgET123C cat | 32 |
| DS8521 | ΔfliM amyE::Pfla/che-fliM-gfp spec | 36 |
| DS8598 | ΔfliF ΔfliM amyE::Pfla/che-fliM-gfp spec | 36 |
| DS8839 | ΔflhO ΔflgE amyE::Pfla/che-flgET123C cat | 32 |
| DS8840 | ΔflhP ΔflgE amyE::Pfla/che-flgET123C cat | 32 |
| DS9555 | ΔflgB amyE::Pfla/che-flgB spec | |
| DK1042 | comIQ12L | 50 |
| DK3135 | ΔfliF ΔflgE amyE::Pfla/che-flgET123C cat | |
| DK3720 | ΔflgC amyE::Pfla/che-flgC cat | |
| DK3721 | ΔfliE amyE::Pfla/che-fliE cat | |
| DK4537 | Δ7 thrC::Physpank-gfp erm | |
| DK4538 | Δ7 ΔfliF thrC::Physpank-gfp erm | |
| DK4539 | Δ7 ΔfliE thrC::Physpank-gfp erm | |
| DK4540 | Δ7 ΔflgB thrC::Physpank-gfp erm | |
| gfp erm DK4541 | Δ7 ΔflgC thrC::Physpank-gfp erm | |
| DK4542 | Δ7 ΔflhO thrC::Physpank-gfp erm | |
| DK4543 | Δ7 ΔflhP thrC::Physpank-gfp erm | |
| DK4544 | Δ7 ΔflgE thrC::Physpank-gfp erm | |
| DK4868 | ΔflgC ΔflgE amyE::Pfla/che-flgET123C cat | |
| DK4892 | ΔflgE ΔfliM amyE::Pfla/che-fliM-gfp spec | |
| DK4895 | ΔflgB ΔflgE amyE::Pfla/che-flgET123C cat | |
| DK4896 | ΔfliE ΔflgE amyE::Pfla/che-flgET123C cat | |
| DK5038 | ΔfliF amyE::Phag-hagT209C spec | |
| DK5039 | ΔfliE amyE::Phag-hagT209C spec | |
| DK5040 | ΔflgB amyE::Phag-hagT209C spec | |
| DK5041 | ΔflgC amyE::Phag-hagT209C spec | |
| DK5042 | ΔflhP amyE::Phag-hagT209C spec | |
| DK5081 | ΔfliE ΔfliM amyE::Pfla/che-fliM-gfp spec | |
| DK5082 | ΔflgB ΔfliM amyE::Pfla/che-fliM-gfp spec | |
| DK5083 | ΔflhO ΔfliM amyE::Pfla/che-fliM-gfp spec | |
| DK5084 | ΔflhP ΔfliM amyE::Pfla/che-fliM-gfp spec | |
| DK5268 | ΔflgC ΔfliM amyE::Pfla/che-fliM-gfp spec |
spec, resistance gene for spectinomycin; cat, resistance gene for chloramphenicol; erm, resistance gene for macrolides-lincosamides-streptogramin B [MLS].
Complementation constructs.
The flgB complementation construct was constructed by amplifying the Pfla/che promoter and flgB gene from the B. subtilis 3610 chromosomal DNA with the primer pair 3041/3042. The resulting PCR product was digested with EcoRI and BamHI and ligated into the EcoRI and BamHI sites of the plasmid pAH25 containing a spectinomycin resistance cassette between two arms of the amyE gene to create pEV1 (51).
The flgC and fliE complementation strains were constructed by amplifying the Pfla/che promoter region from B. subtilis 3610 chromosomal DNA with primer pair 4797/4798. The resulting PCR product was digested with MfeI and BamHI and ligated into the EcoRI and BamHI sites of the plasmid pDG364 (52) containing a chloramphenicol acetyltransferase (cat) cassette between two arms of the amyE gene to generate pAMB1. The flgC and fliE genes were amplified from strain 3610 chromosomal DNA with the primer pairs 4773/4799 and 4775/4800, respectively. The resulting PCR products were ligated into the EcoRI and BamHI sites of pAMB1 downstream of the Pfla/che promoter to create plasmids pAMB3 and pAMB4, respectively.
Swarm expansion assay.
Cells were grown to mid-log phase at 37°C in LB and resuspended to an optical density at 600 nm (OD600) of 10 in pH 7.4 phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4) containing 0.5% India ink (Higgins). Freshly prepared LB containing 0.7% Bacto agar (25 ml/plate) was dried for 10 min in a laminar flow hood, centrally inoculated with 10 μl of the cell suspension, dried for another 10 min, and incubated in a humidity chamber at 37°C (53). The India ink demarks the origin of the colony, and the swarm radius was measured relative to the origin. For consistency, an axis was drawn on the back of the plate and swarm radius measurements were taken along this transect. For experiments including IPTG, cells were propagated in broth in the presence of 1 mM IPTG and 1 mM IPTG was included in the swarm agar plates.
Protein purification.
flgB, flgC, flhO, and fliE genes were amplified with the primer pairs 3150/3151, 3152/3153, 3154/3155, and 4198/4199, respectively, and ligated into pTB146 (54) using SapI and XhoI restriction enzymes to generate plasmids pEV2, pEV3, pEV4, and pKRH12, respectively. The flhP gene was amplified with the primer pair 3156/3157 and ligated into pTB146 using SalI and XhoI restriction enzymes to generate pEV5.
The Rosetta-gami strains containing each plasmid were cultured in 500 ml of Terrific Broth supplemented with 25 μg/ml chloramphenicol and 100 μg/ml ampicillin to an OD600 of ∼0.6 and induced with 1 mM IPTG for ∼16 h overnight. Cells were pelleted, resuspended in lysis buffer (50 mM Na2HPO4, 300 mM NaCl, and 10 mM imidazole), treated with lysozyme and protease inhibitor cocktail (Roche), and frozen overnight at −80°C. The frozen pellet was thawed at room temperature and lysed by sonication. Lysed cell debris was pelleted at 18,000 rpm in a JA-17 rotor (Beckman Coulter) for 30 min at 4°C, and clarified supernatant was incubated with Ni-nitrilotriacetic acid (Ni-NTA) resin (Novagen) overnight at 4°C. The bead lysate mixture was poured into a 1-cm separation column (Bio-Rad), and after packing the resin was washed three times with wash buffer (50 mM Na2HPO4, 300 mM NaCl, and 30 mM imidazole). Small ubiquitin-like modifier (SUMO)-tagged protein bound to the resin was eluted using wash buffer containing increasing concentrations of imidazole (50 mM to 500 mM). Elution fractions were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie brilliant blue to verify the presence of purified protein. Fractions with the highest concentrations of protein were dialyzed into dialysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, and 10% glycerol) overnight in the cold and stored at 4°C.
To purify SUMO-FliE protein from inclusion bodies, cells were sonicated and pelleted as described above. The pellet was resuspended in buffer A (50 mM Tris-HCl [pH 8.0], 500 mM NaCl, 5 mM imidazole, and 6 M guanidine-HCl) and rotated end over end at 4°C for 2 h to lyse inclusion bodies. Samples were centrifuged (14,000 rpm in a JA-17 rotor for 30 min at 4°C), and the supernatant was added to Ni-NTA resin and eluted as described above. Isolated protein was dialyzed in PBS with 10% glycerol overnight in the cold and stored at 4°C.
Antibodies.
Antibodies used in this study were generated from protein purified as described above. One milligram of each purified SUMO-tagged protein was sent to Cocalico Biologicals, Inc., and a host rabbit was immunized by serial injection. After multiple injections, crude serum was obtained and either used as is (anti-FlhO) or affinity purified on a column (anti-FlgB, anti-FlgC, anti-FliE, and anti-FlhP). Anti-FlgE, and anti-SigA antibodies are described elsewhere (32). Anti-GFP antibody was a generous gift from David Rudner (Harvard Medical School), and anti-SigA antibody was a generous gift from Masaya Fujita (University of Houston).
Western blotting.
B. subtilis and E. coli strains were grown in LB to an OD600 of ∼1.0; 1 ml was harvested by centrifugation and resuspended to an OD600 of 10 in lysis buffer (20 mM Tris [pH 7.0], 10 mM EDTA, 1 mg/ml lysozyme, 10 μg/ml DNase I, 100 μg/ml RNase I, and 1 mM phenylmethylsulfonyl fluoride [PMSF]) and incubated for 30 min at 37°C. Lysate (10 μl) was mixed with 2 μl of 6× SDS loading dye, and samples were boiled at 95°C for 10 min. Samples were separated by SDS-PAGE, using 12 to 15% polyacrylamide gels. The proteins were electroblotted onto nitrocellulose, developed with a specified dilution of primary antibody (1:1,000 for rabbit anti-FliE, 1:5,000 for rabbit anti-FlgC, 1:10,000 for rabbit anti-FlgB, anti-FlhO, anti-FlhP, and anti-GFP, 1:20,000 for rabbit anti-FlgE, and 1:120,000 for rabbit anti-SigA), washed with tris-buffered saline (PBS) with Tween 20, and counterprobed with a 1:10,000 dilution of secondary antibody (horseradish peroxidase-conjugated goat anti-rabbit IgG, catalog no. 31460; Thermo Scientific). Immunoblots were developed using Pierce enhanced chemiluminescence (ECL) Western blotting substrate (catalog no. 32106; Thermo Scientific).
Membrane fractionation.
Membrane fractions were purified using a modified version of a previously described protocol (33). B. subtilis strains were grown in 250 ml of LB broth to an OD600 of ∼1.0. Cultures were pelleted (6,000 × g in a JA-10 rotor, 45 min), and pellets were resuspended by gentle swirling in 10 ml lysis buffer (100 mM Tris [pH 8.0], 1 mg/ml lysozyme, and 0.5% Triton X-100, plus complete EDTA-free protease inhibitor cocktail tablet [catalog no. 4693132001; Sigma-Aldrich]) and incubated in a shaker at 37°C for 2 h. After 1 h, DNase I and MgCl2 were added to final concentrations of 5 μg/ml and 10 mM, respectively, to reduce the viscosity of the cell lysate. After lysis, cell debris were pelleted from lysates by centrifugation at 10,000 × g in a JA-17 rotor for 10 min, and clarified supernatant was further pelleted via ultracentrifugation at 100,000 × g in an SW-41 rotor (Beckman Coulter) for 90 min. The supernatant was discarded, membrane pellets were washed twice with saline citrate (100 mM NaCl [pH 7.3] and 10 mM sodium citrate), and then resuspended to an OD600 of 100 in saline citrate with a Potter-Elvehjem polytetrafluoroethylene (PTFE) pestle and glass tube (catalog no. P7734; Sigma-Aldrich). Twenty microliters of 6× SDS loading dye was added to 100 μl of membrane solution and boiled for 10 min at 95°C. For formaldehyde treatments, cells were pelleted and resuspended in 10 ml PBS containing 0.4% (wt/vol) formaldehyde for 60 min prior to lysis. All other procedures were carried out identically.
TCA precipitation of secreted proteins.
Culture (1 ml) was collected from B. subtilis strains grown in LB broth to an OD600 of ∼1.0. The culture was centrifuged at 14,500 × g for 10 min, and supernatant was collected, passed through a syringe with a 0.45-μm filter, and treated with 100 μl of freshly prepared 0.015% sodium deoxycholate for 10 min at room temperature. Proteins in the supernatant were precipitated by adding 50 μl chilled trichloroacetic acid (TCA) and incubating on ice for 2 h. The protein precipitate was isolated by pelleting at 14,500 × g in a JA-17 rotor for 10 min at 4°C, and washed with 1 ml ice-cold acetone. The pellet was resuspended in 0.1 M NaOH to an OD600 of 10 and mixed with 6× SDS loading dye to 1× concentration. Samples were boiled at 95°C for 10 min prior to subjecting to immunoblotting.
Microscopy.
Fluorescence microscopy was performed with a Nikon 80i microscope with a phase-contrast objective Nikon Plan Apo 100X and an Excite 120 metal halide lamp. FM4-64 was visualized with a C-FL HYQ Texas red filter cube (excitation filter, 532 to 587 nm; barrier filter, >590 nm), and Alexa Fluor 488 C5 maleimide and GFP fluorescent signals were visualized using a C-FL HYQ fluorescein isothiocyanate (FITC) filter cube (excitation filter, 460 to 500 nm; barrier filter, 515 to 550 nm). Images were captured with a Photometrics Coolsnap HQ2 camera in black and white, false colored, and superimposed using MetaMorph (Molecular Devices) image software.
To stain hook and filament structures, 0.5 ml of broth culture was harvested at an OD600 of 0.5 to 1.0, and washed once in 1.0 ml of PBS. The suspension was pelleted, resuspended in 50 μl of PBS containing 5 μg/ml Alexa Fluor 488 C5 maleimide dye (Molecular Probes), and incubated for 5 min at room temperature (34). Cells were pelleted and washed twice with 1 ml PBS. Membranes were stained by resuspension in 30 μl of PBS containing 5 μg/ml FM4-64 (Molecular Probes) and incubated for 5 min at room temperature. Cells were pelleted and washed twice more in 1 ml PBS and resuspended in 30 μl PBS. Suspension (4 μl) was placed on a microscope slide and immobilized with a poly-l-lysine-treated coverslip. The suspension was manually pressed into the slide to isolate single cells within the same viewing plane. Alternatively, samples were observed by spotting 4 μl of suspension on a slide containing an agarose pad and covered with a glass coverslip. Agarose pads were created by making a 1% solution of agarose electrophoresis grade; Fisher Scientific in S750 defined minimal growth medium (50 mM MOPS [morpholinepropanesulfonic acid, pH 7.0], 10 mM (NH4)2SO4, 5 mM potassium phosphate [Ph 7.0], 2 mM MgCl2, 0.7 mM CaCl2, 50 μM MnCl2, 1 μM ZnCl2, 1 μg/ml thiamine-HCl, 20 μM HCl, 5 μM FeCl3, and 1% [wt/vol] d-glucose, 0.1% [wt/vol] l-glutamic acid monohydrate, and 0.002% [wt/vol] Casamino Acids) (56, 57) and applying molten solution to a slide, covering with a glass microscope slide, and allowing to cool for 5 min prior to use.
Supplementary Material
ACKNOWLEDGMENTS
The work was funded by NIH grants GM093030 (to D.B.K.) and GM123635 (to A.M.B.).
We thank Masaya Fujita, Katherine Hummels, and David Rudner for material support.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00425-18.
REFERENCES
- 1.Jarrell KF, McBride MJ. 2008. The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol 6:466–476. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- 2.Macnab RM. 1992. Genetics and biogenesis of bacterial flagella. Annu Rev Genet 26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 3.Macnab RM. 2003. How bacteria assemble flagella. Annu Rev Microbiol 57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 4.Chevance FFV, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukherjee S, Kearns DB. 2014. The structure and regulation of flagella in Bacillus subtilis. Annu Rev Genet 48:319–340. doi: 10.1146/annurev-genet-120213-092406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DePamphilis ML, Alder J. 1971. Purification of intact flagella from Escherichia coli and Bacillus subtilis. J Bacteriol 105:376–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DePamphilis ML, Alder J. 1971. Fine structure and isolation of the hook-basal body complex of flagella from Escherichia coli and Bacillus subtilis. J Bacteriol 107:384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stallmeyer MJB, Aizawa S-I, Macnab RM, DeRosier DJ. 1989. Image reconstruction of the flagellar basal body of Salmonella typhimurium. J Mol Biol 205:519–528. doi: 10.1016/0022-2836(89)90223-4. [DOI] [PubMed] [Google Scholar]
- 9.Sosinsky GE, Francis NR, Stallmeyer MJB, DeRosier DJ. 1992. Substructure of the flagellar basal body of Salmonella typhimurium. J Mol Biol 223:171–184. doi: 10.1016/0022-2836(92)90724-X. [DOI] [PubMed] [Google Scholar]
- 10.Jones CJ, Macnab RM. 1990. Flagellar assembly in Salmonella typhimurium: analysis with temperature-sensitive mutants. J Bacteriol 172:1327–1339. doi: 10.1128/jb.172.3.1327-1339.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Homma M, Kutsukake K, Hasebe M, Iino T, Macnab RM. 1990. FlgB, FlgC, FlgF, and FlgG, a family of structurally related proteins in the flagellar basal body of Salmonella typhimurium. J Mol Biol 211:465–477. doi: 10.1016/0022-2836(90)90365-S. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki T, Iino T, Horiguchi T, Yamaguchi S. 1978. Incomplete flagellar structures in nonflagellate mutants of Salmonella typhimurium. J Bacteriol 133:904–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki T, Komeda Y. 1981. Incomplete flagellar structures in Escherichia coli mutants. J Bacteriol 145:1036–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa SI. 1992. Morphological pathway of flagellar assembly in Salmonella typhimurium. J Mol Biol 226:433–446. doi: 10.1016/0022-2836(92)90958-M. [DOI] [PubMed] [Google Scholar]
- 15.Jones CJ, Macnab RM, Okino H, Aizawa S-I. 1990. Stoichiometric analysis of the flagellar hook-basal body complex of Salmonella typhimurium. J Mol Biol 212:377–387. doi: 10.1016/0022-2836(90)90132-6. [DOI] [PubMed] [Google Scholar]
- 16.Minamino T, Macnab RM. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J Bacteriol 181:1388–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saijo-Hamano Y, Uchida N, Namba K, Oosawa K. 2004. In vitro characterization of FlgB, FlgC, FlgF, FlgG and FliE, flagellar basal body proteins of Salmonella. J Mol Biol 339:423–435. doi: 10.1016/j.jmb.2004.03.070. [DOI] [PubMed] [Google Scholar]
- 18.DePamphilis ML, Adler J. 1971. Attachment of flagellar basal bodies to the cell envelope: specific attachment to the outer, lipopolysaccharide membrane and the cytoplasmic membrane. J Bacteriol 105:396–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones CJ, Homma M, Macnab RM. 1989. L-, P-, and M-ring proteins of the flagellar basal body of Salmonella typhimurium: gene sequences and deduced proteins sequences. J Bacteriol 171:3890–3900. doi: 10.1128/jb.171.7.3890-3900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chevance FFV, Takahashi N, Karlinsey JE, Gnerer J, Hirano T, Samudrala R, Aizawa SI, Hughes KT. 2007. The mechanism of outer membrane penetration by the eubacterial flagellum and implications for spirochete evolution. Genes Dev 21:2326–2335. doi: 10.1101/gad.1571607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minamino T, Yamaguchi S, Macnab RM. 2000. Interaction between FliE and FlgB, a proximal rod component of the flagellar basal body of Salmonella. J Bacteriol 182:3029–3036. doi: 10.1128/JB.182.11.3029-3036.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller V, Jones CJ, Kawagishi I, Aizawa SI, Macnab RM. 1992. Characterization of the fliE geners of Escherichia coli and Salmonella typhimurium and identification of the FliE protein as a component of the flagellar hook-basal body complex. J Bacteriol 174:2298–2304. doi: 10.1128/jb.174.7.2298-2304.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ueno T, Oosawa K, Ueno T, Aizawa S-I. 1992. M ring, S ring, and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein FliF. J Mol Biol 227:672–677. doi: 10.1016/0022-2836(92)90216-7. [DOI] [PubMed] [Google Scholar]
- 24.Okino H, Isomura M, Yamaguchi S, Magariyama Y, Kudo S, Aizawa S-I. 1989. Release of flagellar filament-hook-rod complex by a Salmonella typhimurium mutant defective in the M ring of the basal body. J Bacteriol 171:2075–2082. doi: 10.1128/jb.171.4.2075-2082.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osorio-Valeriano M, de la Mora J, Camarena L, Dreyfus G. 2016. Biochemical characterization of the flagellar rod components of Rhodobacter sphaeroides: properties and interactions. J Bacteriol 198:544–552. doi: 10.1128/JB.00836-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hizukuri Y, Yakushi T, Kawagishi I, Homma M. 2006. Role of the intermolecular disulfide bond in FlgI, the flagellar P-ring component of Escherichia coli. J Bacteriol 188:4190–4197. doi: 10.1128/JB.01896-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao X, Zhang K, Boquoi T, Hu B, Motaleb MA, Miller KA, James ME, Charon NW, Manson MD, Norris SJ, Li C, Liu J. 2013. Cryoelectron tomography reveals the sequential assembly of bacterial flagella in Borrelia burgdorferi. Proc Natl Acad Sci U S A 110:14390–14395. doi: 10.1073/pnas.1308306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki H, Yonekura K, Murata K, Hirai T, Oosawa K, Namba K. 1998. A structural feature in the central channel of the bacterial flagellar FliF ring complex is implicated in type III protein export. J Struct Biol 124:104–114. doi: 10.1006/jsbi.1998.4048. [DOI] [PubMed] [Google Scholar]
- 29.Hirano T, Minamino T, Keiichi N, Macnab RM. 2003. Substrate specificity classes and the recognition signal for Salmonella type III flagellar export. J Bacteriol 185:2485–2492. doi: 10.1128/JB.185.8.2485-2492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singer HM, Erhardt M, Hughes KT. 2014. Comparative analysis of the secretion capability of early and late flagellar type III secretion substrates. Mol Microbiol 93:505–520. doi: 10.1111/mmi.12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuberi AR, Ying C, Bischoff DS, Ordal GW. 1991. Gene-protein relationships in the flagellar hook-basal body complex of Bacillus subtilis: sequences of the flgB, flgC, flgG, fliE and fliF genes. Gene 101:23–31. doi: 10.1016/0378-1119(91)90220-6. [DOI] [PubMed] [Google Scholar]
- 32.Courtney CR, Cozy LM, Kearns DB. 2012. Molecular characterization of the flagellar hook in Bacillus subtilis. J Bacteriol 194:4619–4629. doi: 10.1128/JB.00444-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kubori T, Okumura M, Kobayashi N, Nakamura D, Iwakura M, Aizawa SI. 1997. Purification and characterization of the flagellar hook-basal body complex of Bacillus subtilis. Mol Microbiol 24:399–410. doi: 10.1046/j.1365-2958.1997.3341714.x. [DOI] [PubMed] [Google Scholar]
- 34.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. 2008. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- 35.Wang F, Burrage AM, Postel S, Clark RE, Orlova A, Sundberg EJ, Kearns DB, Egelman EH. 2017. A structural model of flagellar filament switching across multiple bacterial species. Nat Commun 8:960. doi: 10.1038/s41467-017-01075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guttenplan SB, Kearns DB. 2013. The cell biology of peritrichous flagella in Bacillus subtilis. Mol Microbiol 87:211–229. doi: 10.1111/mmi.12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calvo RA, Kearns DB. 2015. FlgM is secreted by the flagellar export apparatus in Bacillus subtilis. J Bacteriol 197:81–91. doi: 10.1128/JB.02324-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Homma M, Aizawa SI, Dean GE, Macnab RM. 1987. Identification of the M-ring protein of the flagellar motor of Salmonella typhimurium. Proc Natl Acad Sci U S A 84:7483–7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 40.Kutsukake K, Minamino T, Yokoseki T. 1994. Isolation and characterization of FliK-independent flagellation mutants from Salmonella typhimurium. J Bacteriol 176:7625–7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferris HU, Furukawa Y, Minamino T, Kroetz MB, Kihara M, Namba K, Macnab RM. 2005. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J Biol Chem 280:41236–41242. doi: 10.1074/jbc.M509438200. [DOI] [PubMed] [Google Scholar]
- 42.Erhardt M, Singer HM, Wee DH, Keener JP, Hughes KT. 2011. An infrequent molecular ruler controls flagellar hook length in Salmonella enterica. EMBO J 30:2948–2961. doi: 10.1038/emboj.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujii T, Kato T, Hiraoka KD, Miyata T, Minamino T, Chevance FFV, Hughes KT, Namba K. 2017. Identical folds used for distinct mechanical functions of the bacterial flagellar rod and hook. Nat Commun 8:14276. doi: 10.1038/ncomms14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen EJ, Hughes KT. 2014. Rod-to-hook transition for extracellular flagellum assembly is catalyzed by the L-ring-dependent rod scaffold removal. J Bacteriol 196:2387–2395. doi: 10.1128/JB.01580-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cohen EJ, Ferreira JL, Ladinsky MS, Beeby M, Hughes KT. 2017. Nanoscale-length control of the flagellar driveshaft requires hitting the tethered outer membrane. Science 356:197–200. doi: 10.1126/science.aam6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nambu T, Minamino T, Macnab RM, Kutsukake K. 1999. Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium. J Bacteriol 181:1555–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nambu T, Inagaki Y, Kutsukake K. 2006. Plasticity of the domain structure in FlgJ, a bacterial protein involved in flagellar rod formation. Genes Genet Syst 81:381–389. doi: 10.1266/ggs.81.381. [DOI] [PubMed] [Google Scholar]
- 48.Chen R, Guttenplan SB, Blair KM, Kearns DB. 2009. Role of the sigmaD-dependent autolysins in Bacillus subtilis population heterogeneity. J Bacteriol 191:5775–5784. doi: 10.1128/JB.00521-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herlihey FA, Moynihan PJ, Clarke AJ. 2014. The essential protein for bacterial flagella formation FlgJ functions as a β-N-acetylglucosaminidase. J Biol Chem 45:31029–31042. doi: 10.1074/jbc.M114.603944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Konkol MA, Blair KM, Kearns DB. 2013. Plasmid-encoded ComI inhibits competence in the ancestral 3610 strain of Bacillus subtilis. J Bacteriol 195:4085–4093. doi: 10.1128/JB.00696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guérout-Fleury AM, Frandsen N, Stragier P. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57–61. doi: 10.1016/S0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- 52.Antoniewski C, Savelli B, Stragier P. 1990. The spoIIJ gene, which regulates early developmental steps in Bacillus subtilis, belongs to a class of environmentally responsive genes. J Bacteriol 172:86–93. doi: 10.1128/jb.172.1.86-93.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kearns DB, Losick R. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol 49:581–590. doi: 10.1046/j.1365-2958.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- 54.Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. 2009. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J 28:193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reference deleted.
- 56.Vasantha N, Freese E. 1980. Enzyme changes during Bacillus subtilis sporulation caused by deprivation of guanine nucleotides. J Bacteriol 144:1119–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grossman AD, Losick R. 1988. Extracellular control of spore formation in Bacillus subtilis. Proc Natl Acad Sci U S A 85:4369–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kearns DB, Losick R. 2005. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev 19:3083–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yasbin RE, Young FE. 1974. Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol 14:1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.