Abstract
Weedy rice (Oryza spp.) is a problematic weed of cultivated rice (O. sativa) around the world. Recent studies have established multiple independent evolutionary origins of weedy rice, raising questions about the traits and genes that are essential for the evolution of this weed. Among world regions, South Asia stands out due to the heterogeneity of its weedy rice populations, which can be traced to at least three origins: two through de-domestication from distinct cultivated rice varieties, and one from local wild rice (O. rufipogon/O. nivara). Here we examine five traits considered typical of or advantageous to weedy rice in weedy, cultivated and wild rice samples from South Asia. We establish that convergence among all three weed groups occurs for easy seed shattering, red pericarp color, and compact plant architecture, suggesting that these traits are essential for weed success in the South Asian agricultural environment. A high degree of convergence for black hull color is also seen among weeds with wild ancestors and weeds evolved from the aus cultivated rice group. We also examine polymorphism in five known domestication candidate genes, and find that Rc and Bh4 are associated with weed seed pericarp color and hull color, respectively, and weedy alleles segregate in the ancestral populations, as do alleles for the seed dormancy-linked gene Sdr4. The presence of a domestication related allele at the seed shattering locus, sh4, in weedy rice populations with cultivated ancestry supports a de-domestication origin for these weedy groups, and raises questions about the reacquisition of the shattering trait in these weedy populations. Our characterization of weedy rice phenotypes in South Asia and their associated candidate genes contribute to the emerging understanding of the mechanisms by which weedy rice evolves worldwide, suggesting that standing ancestral variation is often the source of weedy traits in independently evolved groups, and highlighting the reservoir of genetic variation that is present in cultivated varieties as well as in wild rice, and its potential for phenotypic evolution.
Keywords: red rice, weed evolution, candidate genes, seed shattering, seed dormancy, Genomic Prediction, GenPred, Shared Data Resources
Agricultural weeds, defined as unwanted plants growing in agricultural environments (Monaco et al. 2002), are responsible for ∼30% annual reduction in crop productivity worldwide (Oerke 2006). This negative impact of weeds can be attributed to various traits, which allow them to adapt and be competitive in the agricultural environment. Examples of such traits in diverse species can include rapid growth, efficient seed dispersal and seed dormancy (Baker 1965), and can be considered to be part of an agricultural weed syndrome (Vigueira et al. 2013b). Weed syndrome traits have been found to evolve repeatedly and independently in different weed lineages (Vigueira et al. 2013a; Kane and Rieseberg 2008; Cui et al. 2016; Huang et al. 2017), and are examples of what is known as parallel or convergent evolution (Arendt and Reznick 2008).
Weedy rice (Oryza spp.), an aggressive unwanted weed of cultivated rice (O. sativa), is a type of agricultural weed displaying parallel evolution of weediness traits (Ziska et al. 2015). Weedy rice occurs worldwide, and studies have established multiple independent evolutionary origins of this weed from non-weedy backgrounds (Reagon et al. 2010; Huang et al. 2017; Vigueira et al., 2017). Many typical traits of weedy rice resemble reversals to wild phenotypes. Despite diverse genetic backgrounds, red pericarp is a common feature of weedy rice, leading to the common name “red rice” (Gross et al. 2010; Cui et al. 2016). The fact that red pericarp is common likely reflects selection for dormancy in weedy rice seeds, since the gene which regulates proanthocyanidin pigment synthesis also pleiotropically regulates ABA regulated seed dormancy (Gu et al. 2011; Cui et al. 2016), and seed dormancy has been detected in several populations (Gianinetti and Cohn 2008; Ziska et al. 2015). Ease of seed shattering or dispersal has also been reported for several populations (Thurber et al. 2010; Zhu et al. 2012). The wealth of knowledge of candidate genes underlying many morphological traits in rice has recently made it possible to examine the origins of weedy traits in weedy rice populations (Gross et al. 2010; Vigueira et al. 2013b), as well as to examine the extent to which the same genes underlie convergent traits across weedy rice populations.
We recently characterized populations of weedy rice from South Asia (Huang et al. 2017), an area known for its great diversity of cultivated and wild Oryzas (Civáň et al. 2015; Garris et al. 2005; Huang et al. 2012; Zhu et al. 2007), that has also proven to contain a great diversity of weedy rice. Two weedy rice populations in this area trace their ancestry to cultivated rice varieties aus and indica through a process known as de-domestication (Huang et al. 2017). A third weedy population has evolved from local wild rice (the species complex known as O. rufipogon/nivara). Moreover, weeds presenting an admixture of all these backgrounds also occur (Huang et al. 2017). These South Asian weedy rice origins are also independent of the well-studied U.S. weedy rice populations de-domesticated from aus and indica varieties (Reagon et al. 2010; Huang et al. 2017). The diversity of weedy rice origins in a single world region raises questions about how each of these groups has acquired the traits that make them weedy.
To understand how weediness traits have evolved in each of the genetically distinct weedy populations of South Asia, we examine here the presence of iconic weedy rice traits in each of these populations. We additionally investigate the genetic mechanisms possibly underlying these traits by capitalizing on the extensive prior characterization of domestication trait candidate genes in cultivated rice, and deduce the evolutionary origins of alleles carried by weeds.
We first focus on pericarp pigmentation, as red pericarps have been proposed to be adaptive either by providing protection from abiotic or biotic stress (Ithal and Reddy 2004), and through association with seed dormancy (Gu et al. 2011). A red pericarp is also ubiquitous among wild rice, while during domestication most cultivated rice varieties were artificially selected to have white pericarps (Sweeney et al. 2006). The Rc gene, which codes for a basic helix-loop-helix protein in the proanthocyanin synthesis pathway, has been shown to underlie variation in pericarp color in Oryza (Sweeney et al. 2006).
The seed dormancy trait is also known to be affected by another gene in Oryza: Sdr4 has been shown to be involved in differences in the degree of seed dormancy between the two cultivated rice subspecies: japonica and indica (Sugimoto et al. 2010). Seed dormancy is a trait common in wild rice but selected against during domestication, and is often reported for weedy rice; the capacity for dormancy over multiple seasons aids in propagation and persistence of weedy rice in crop field.
Another seed-related trait shown to be common in some weedy rice populations is black colored hulls (e.g., Reagon et al. 2010). Wild rice tends to have dark hulls, while cultivated rice tends to have straw colored hulls. Dark hulls have been proposed as an adaptation aiding crypsis of the seed on the ground and avoidance of predators (Gu et al. 2011). The Bh4 gene, encoding an amino acid transporter, is known to affect hull color (Zhu et al. 2011).
Easy seed shattering is one of the traits that most consistently differentiates weedy rice from cultivated rice, and is also a trait strongly selected against during domestication. Seed shattering is beneficial to weeds as it increases seed dispersal. Variation in a transcription factor encoding gene, sh4, is strongly associated with incomplete development of an abscission layer in the seed-pedicel junction and subsequent decrease of shattering during rice domestication Li et al. (2006).
A last trait we examine is plant architecture, as cultivated rice differs from wild rice in having narrower tiller angle and fewer tillers (Jin et al. 2008). In weedy rice, compact architectures have been proposed as adaptive, as they make the plants harder to distinguish from the crop, allowing evasion of removal efforts. Also plants with compact architectures are able to compete on equal footing for light (Ziska et al. 2015). The PROG1 gene, encoding a zinc-finger nuclear transcription factor, has been implicated in this transition (Jin et al. 2008; Tan et al. 2008). The occurrence of PROG1 domestication alleles has recently been implicated in the de-domestication origins of US weedy rice (Li et al. 2017).
Our goals were to: 1) determine which of these typical weedy traits characterize each of the known weedy rice populations occurring in South Asia; 2) investigate whether known candidate genes may explain the trait values found in each weedy rice population; and 3) determine whether weedy rice populations in South Asia have acquired weediness traits and alleles from standing variation in their ancestors, de novo mutation, or introgression from other sources. We further discuss the evolution of weediness traits in South Asian in context of recent discoveries of weedy rice evolution in other world regions.
Material and Methods
Plant material and phenotypic measurements
A panel of 101 Oryza accessions of types occurring in South Asia (including the countries Bangladesh, Myanmar, India, Nepal, Pakistan, and Sri Lanka) was used in this study. These included 35 South Asian weedy rice samples, for which predominant ancestries were previously characterized (Huang et al. 2017), comprising 15 aus-like, five indica-like, five wild-like and ten admixed weeds; admixed weeds are most often a mix of indica and wild backgrounds or indica and aus backgrounds (Huang et al. 2017). The panel also included Oryza groups potentially contributing to the genetic make-up of weedy rice; these were 44 cultivar varieties (15 aus, 13 indica, 11 japonica and five admixed cultivars), 18 wild ancestors of cultivated Asian rice (O. rufipogon/O. nivara), and four outgroup species (two O. meridionalis and two O. barthii) (Table S1).
Plant DNA was extracted as described in Huang et al. (2017) and used for sequencing of candidate genes (see below). We recorded seed traits for all 101 accessions from the original seed storage, with pericarp color classified as red or white, hull color classified as black or straw and awn as present of absent (Table S1).
We planted a subset of 44 accessions, with three replicates per accession, in a randomized design in two Conviron PGW36 growth chambers, under 11 hr day length with 25° temperature until 30 days after flowering. Due to limited chamber availability, the growth experiment was carried out in two stages. The first batch was planted on Feb 15, 2013 and the second on June 28, 2013. Tiller number and tiller angle were recorded at flowering. We divided tiller angle into three categories, <30°, 30-60° and 60-90°, as determined by the maximum tiller angle of the accession among three replicates. Seed shattering was measured as breaking tensile strength, using the method described in Thurber et al. (2010), which measures the weight needed to detach seeds from panicles 30 days after flowering.
DNA sequencing and genotyping of candidate genes
Following the Agilent (Agilent, Santa Clara, CA) SureSelect Target Enrichment method (Gnirke et al. 2009) instructions, we designed baits of 120 bp length targeting the five candidate genes’ open reading frame (ORF). Additional baits were designed to target other known gene ORFs and gene fragments (∼1500 base pair (bp)) in upstream and downstream regions of each baited gene. A total of 128 loci were thus targeted and sequenced (Table S2). Baits were designed based on the rice reference genome (MSU 6.0 assembly; http://rice.plantbiology.msu.edu/). The SureSelect Target Enrichment method was applied to capture targeted genomic sequences in the Oryza panel, and Illumina Hi-seq 2500 sequencing was performed at the Whitehead Institute (Massachusetts Institute of Technology).
Raw sequencing reads were assessed for quality with FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The low quality reads were filtered using NGStoolkit (Patel and Jain 2012), keeping reads with depth >3 and mapping quality >30. Clean reads were mapped onto the MSU 6.0 reference genome using BWA (Li and Durbin 2009). SNP and indels were called with SAMtools (Li et al. 2009). A series of in-house Perl scripts were employed to convert the polymorphisms from variant call format (VCF) to FASTA format for sequence alignment. In the FASTA files, unmapped nucleotides were represented with N and low-quality nucleotides were represented with “?”.
Due to limitations of the SureSelect method to accurately detect moderate and large insertion/deletion (InDel) polymorphisms, we confirmed the presence of InDels with cleaved amplified polymorphisms (CAPS; Konieczny and Ausubel 1993) for two cases: the 14 bp InDel in exon 6 of Rc and the 22-bp InDel in exon 3 of Bh4. WebMap (http://pga.mgh.harvard.edu/web_apps/web_map_/start/) was used to create a map from the gene sequence files taken from the MSU database and Genbank. Restriction enzyme site complexity was set to greater than or equal to 5 residues in order to find an enzyme that would cut in the target region. Primer3 (http://frodo.wi.mit.edu/primer3/) was used to choose primers that would amplify the target region of the sequence.
The Rc gene was amplified using the primer pair Rc_4774_for (Sequence: tcaattcttgcatttcttttcca) and Rc_5299_rev (Sequence: acggctttatagaaatagagggagt). The Bh4 gene was amplified with the primer pair Bh4_1_for (Sequence: caatctggtgcataatcagaatgga) and Bh4_1_rev primers (Sequence: cccgaagatcctgacgtagcag). The PCR profiles were set for one cycle of 95° of 3 min (Rc) and 30 sec (Bh4), 40 cycles of melting, annealing, and extension (95° for 1 min, 55° (Rc) and 59° (Bh4) for 30 sec, 68° for 1 min) and one cycle of 72° for 10 min. PCR products were digested with 0.5 ul of 20 units/ul restriction enzyme Ban1 for Rc and ApaLI for Bh4 by incubating at 37° for 60 min. The products of the digestion were analyzed using gel electrophoresis to identify insertion and deletion alleles in Rc and Bh4.
Data Analysis
Sequence alignments among accessions were performed again for the 101 accessions using MUSCLE by EMBL-EBI (http://www.ebi.ac.uk/Tools/msa/muscle/). Aligned sequences were imported into BioEdit Ver.7.0.9 (Hall 1999) for manual refinement and localization of relevant polymorphisms. FASTA files of the alignments were exported and converted to PHYLIP format using PGDSpider software. We constructed haplotype trees for each candidate gene with the DNANJ tool implemented in the PHYLIP package (Felsenstein 1993) on CYVERSE (http://www.cyverse.org/), using Kimura 2-parameter distance model, with bootstrap values calculated via 1000 replicates, and two O. meridionalis samples (omd01 and omd02) designated as outgroups. MEGA and ITOL (http://itol.embl.de/) were used to visualize the trees.
The panel of accessions was separated into groups based on prior population structure analyses (Huang et al. 2017). For each candidate gene, information about un-translated regions, exons and introns were obtained from the Rice Genome Annotation Project database (http://rice.plantbiology.msu.edu/). Estimates of nucleotide diversity including Nei’s average pairwise nucleotide diversity (π) and Watterson’s estimator of theta (θw) were calculated with in-house Perl scripts on each of the 128 loci for synonymous, non-synonymous, silent and all sites within each group. Tajima’s D (Tajima 1989) was also calculated for each group with in-house Perl scripts. Pairwise genetic distances between each group, assessed by FST values (Hudson et al. 1992), were calculated for seven population pairs, including three between weed groups and their ancestors (aus-like vs. aus, indica-like vs. indica and wild-like vs. wild), one between cultivars (aus vs. indica) and three among weed groups (aus-like vs. indica-like, aus-like vs. wild-like and indica-like vs. wild-like). Negative FST values were converted into zeros.
For all candidate genes, allele types occurring in each of the weed groups were compared to those of their non-weedy ancestors as determined in Huang et al. (2017). When sequenced haplotypes in weed groups were identical or very closely related to haplotypes found in ancestral groups, weedy alleles were considered to have been inherited from standing ancestral variation. In contrast, weed alleles most closely related to alleles belonging to a group different than that weed’s genomic ancestry, were taken as a possible sign of introgression. All candidate genes were examined for possible novel mutations in weed that could have an effect on gene function.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Newly generated DNA sequences for candidate genes are available in GenBank (accessions MH771032 - MH771132, MH798903 - MH799306). DNA sequences for the 123 supplementary loci, as well as other supplementary material is available on figshare via the GSA portal. Supplementary tables contain genotype and phenotype of Oryza accessions included in the study, and genomic regions sequenced by target capture. Figure S1 contains the PROG1 phylogenetic tree based on SNPs. Scripts used to obtain population statistics are available at Github: https://github.com/Zhongyun-Huang/G3-2018-200605. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6853244.
Results
Phenotypic traits characterizing South Asian weedy rice groups
We had previously characterized several seed traits in a larger panel of South Asian weedy rice (Huang et al. 2017), and results for the subset of accessions characterized here confirm prior observations (Table 1, Table S1). Red pericarps are common in all groups of South Asian weedy rice (>60% of samples in weedy groups are red). This is in contrast to both progenitor cultivated rice groups, although they do show the presence of both pericarp types, but similar to wild rice (Table 1). Hull color varies among South Asian weedy rice, with aus-like, wild-like, and admixed weeds having primarily black hulls (>70%), while indica-like weeds have exclusively straw colored hulls (Table 1, Table S1). Cultivated groups tend to have straw colored hulls (>83% of our cultivated samples), and 83% of the wild rice samples we examined have black hulls.
Table 1. Categorical phenotypic traits in South Asian Oryza groups.
| Pericarp | Hull | Tiller angle | ||||||
|---|---|---|---|---|---|---|---|---|
| Group | red | white | black | straw | <30 | 30-60 | 60-90 | |
| Weedy rice | aus-like (15) | 15 (100%) | 0 (0%) | 12 (80%) | 3 (20%) | 4 | 4 | 0 |
| indica-like (5) | 3 (60%) | 2 (40%) | 0 (0%) | 5 (100%) | 2 | 2 | 0 | |
| wild-like (5) | 5 (100%) | 0 (0%) | 4 (80%) | 1 (20%) | 2 | 3 | 0 | |
| admixed (10) | 8 (80%) | 2 (20%) | 7 (70%) | 3 (30%) | 3 | 4 | 0 | |
| aus (14) | 5 (36%) | 9 (64%) | 2 (17%) | 12 (83%) | 3 | 3 | 0 | |
| indica (13) | 3 (23%) | 10 (77%) | 1 (8%) | 12 (92%) | 5 | 1 | 0 | |
| South Asian wild rice (18) | 17 (94%) | 1 (6%) | 15 (83%) | 3 (17%) | 1 | 1 | 4 | |
We additionally recorded the maximum tiller angle among replicates for each accession in our phenotyping study (Table S1). All South Asian weedy rice sub-groups have a relatively compact plant architecture, with angles <60 o (Table 1, Table S1). This is similar to the cultivated aus group, but not as compact as indica, which usually has tiller angles < 30°. As expected, wild rice has the most spread tillers, with the majority having angles between 60° and 90° (Table 1, Table S1). This result is consistent with previous observations on cultivars and wild rice (Jin et al. 2008).
We did not detect significant differences in tiller number among groups with the accession characterized for this study (P = 0.4), but trends confirmed prior significant observations from Huang et al. (2017), with higher tiller number occurring in wild rice and wild-like weeds (mean > 11), and lower numbers seen in cultivated aus and indica (mean < 7.71; Table 2). As previously reported (Huang et al. 2017), all weedy rice groups shatter easily, as does wild rice, and both cultivar-like weed groups shatter significantly more easily than their ancestors (P < 0.001; Table 2, Table S1).
Table 2. Quantitative phenotypic traits in Asian Oryza groups. Numbers in brackets beside means are standard deviations.
| Tiller number1 | Shattering2 | ||
|---|---|---|---|
| South Asian weed (25) | |||
| aus-like (8) | 8.44(3.35) | 2.14(5.79)d | |
| indica-like (5) | 11.11(6.68) | 17.46(16.70)bc | |
| wild-like (5) | 11.50(4.14) | 8.36(19.05)d | |
| admixed (7) | 7.57(3.95) | 14.70(16.58)c | |
| aus (7) | 7.71(1.50) | 25.61(17.92)ab | |
| indica (6) | 7.00(2.97) | 35.96(5.49)a | |
| South Asian wild rice (6) | 11.44(7.55) | 2.47(5.51)d | |
| Kuskal-Wallis | 0.4040086 | 9.42E-08 | |
Chamber effect detected, only data from Chamber 1 plants were used in the analysis.
No chamber effect detected.
Taken together, our results suggest convergence among South Asian weedy rice groups in ease of shattering, red pericarp color and compact plant architecture. Additionally, convergence occurs among aus-like, wild-like and admixed weeds for black hull color.
Patterns of polymorphism in Rc and Bh4
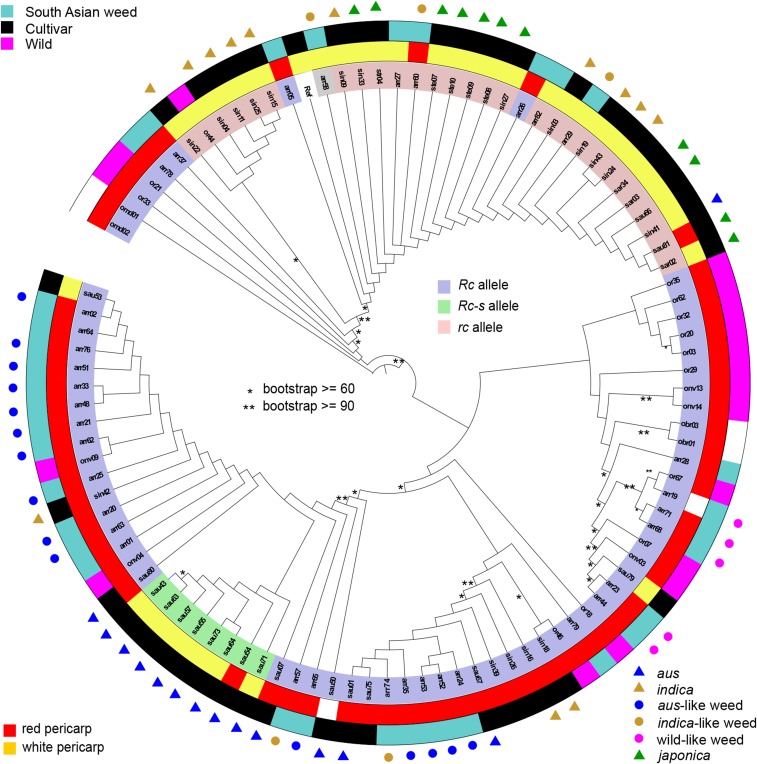
Rc is considered a rice improvement gene contributing to the prevalence of white pericarps in cultivated rice, with the recessive white pericarp phenotypes caused by at least two known loss-of-function alleles: a 14-bp deletion (rc) in exon 6 or a point mutation from C to A (Rc-s) (Sweeney et al. 2006). We first carried out a phylogenetic analysis of the Rc coding region based solely on SNP information, which verified a single origin for the white pericarp conferring Rc-s allele in aus cultivars (Figure 1). We mapped on to the tree the presence and absence of the 14-bp deletion, allowing us to distinguish Rc from rc alleles (Figure 1). Our data supports a single origin of the rc allele, which is prevalent in, but not exclusive to, cultivars. The two Rc haplotypes observed in the rc clade may have come about by recombination of alleles without the deletion into an rc SNP background. Our data also support the ancestral status of Rc alleles, and their prevalence in wild rice. Further examination of the association between Rc allelic variation and pericarp color confirms that discrepancies between genotype and pericarp color are rare (six cases out of 95 accessions): most accessions with red pericarps carry the Rc allele and most accession with white pericarps carry the rc or Rc-s allele across most Oryza groups (Table 3, Table S1). This supports prior observations that Rc is the major gene underlying pericarp color, although other genes may also have effects.
Figure 1.
Neighbor joining (NJ) tree of Rc haplotypes. The color range indicates the three major alleles of the Rc gene; the inner ring indicates the colors of the pericarp. The outer ring and the bullets indicate Oryza groups, as labeled in the key. Unmarked weeds are admixed according to results of Huang et al. (2017). Asterisks on branches indicate bootstrap values as labeled.
Table 3. Pericarp color phenotype and Rc gene variation in Oryza accessions.
| Pericarp color | ||||||
|---|---|---|---|---|---|---|
| Red | White | |||||
| Group | Rc | rc | Rc-s | Rc | rc | Rc-s |
| Cultivars (43) | ||||||
| aus (14) | 4 | 0 | 1 | 1 | 1 | 7 |
| indica (13) | 3 | 0 | 0 | 0 | 10 | 0 |
| japonica (11) | 0 | 1 | 0 | 1 | 9 | 0 |
| admixed cutivar (5) | 3 | 0 | 0 | 1 | 1 | 0 |
| South Asian weedy rice (34) | ||||||
| aus-like (15) | 15 | 0 | 0 | 0 | 0 | 0 |
| indica-like (5) | 2 | 1 | 0 | 0 | 2 | 0 |
| wild-like (4) | 4 | 0 | 0 | 0 | 0 | 0 |
| admixed (10) | 8 | 0 | 0 | 0 | 2 | 0 |
| South Asian wild rice (18) | 17 | 0 | 0 | 0 | 1 | 0 |
As expected due to the high frequency of red pericarps, South Asian weeds predominantly carry Rc alleles (Figure 1, Table 3). Furthermore, there is phylogenetic structure in Rc-type alleles, such that South Asian wild-like weeds most closely resemble wild rice Rc haplotypes, and aus-like weeds tend to possess alleles that are most closely related to haplotypes present in aus cultivars, be they Rc or Rc-s alleles. Indica-like and admixed weeds possess alleles falling in both the Rc and rc clades, but in all cases similar alleles from ancestral groups are also observed. Thus, for both weeds of wild and cultivated descent, the red pericarp trait in all South Asian weedy rice groups is likely inherited directly from ancestors where this trait is either common (wild rice) or rare but present (cultivated rice).
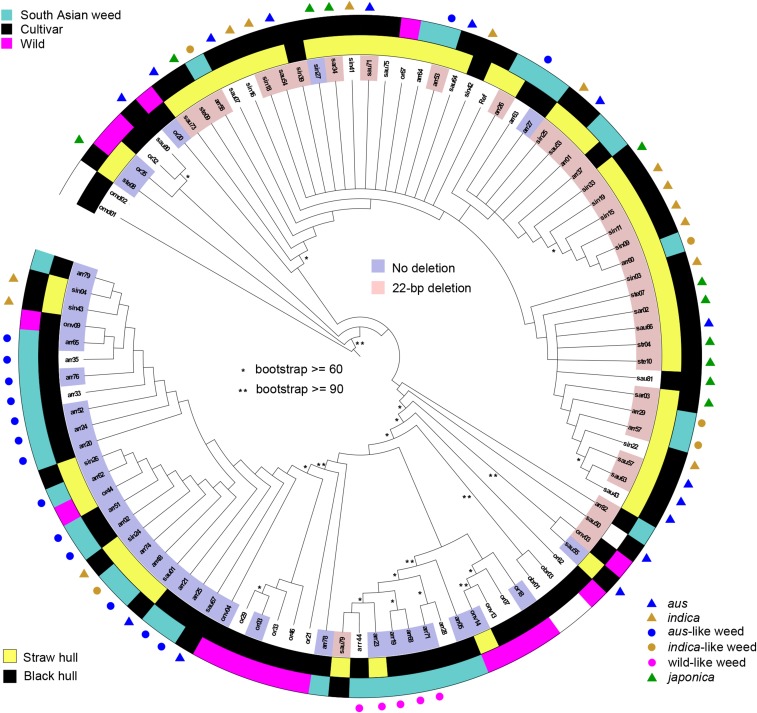
Like Rc, Bh4 is considered a rice improvement gene that contributes to the straw colored hull phenotype that is common, but not fixed, in cultivated rice (Zhu et al. 2011). A 22-bp frame-shift deletion in exon 3 has been shown to be the dominant functional polymorphism causing a straw hull phenotype (Zhu et al. 2011). Again we carried out a phylogenetic analysis based solely on SNP data in the ORF of the gene, and mapped the presence or absence of the deletion onto the tree (Figure 2). Phylogenetic SNP analysis broadly supports a single origin for Bh4 alleles carrying the 22 bp deletion, which are more common in crops, which tend to have straw-colored hulls, than in wild rice, which is predominantly black-hulled. A few deletion-carrying haplotypes occur outside the clade, as well as a few haplotypes without the deletion; recombination among alleles belonging to each clade could explain the rare lack of association between deletion status and Bh4 SNP background.
Figure 2.
Neighbor joining (NJ) tree of Bh4 haplotypes. The color range indicates the two major InDel alleles of Bh4; the inner ring indicates the hull color. The outer ring and the bullets indicate Oryza groups, as labeled in the key. Unmarked weeds are admixed according to results of Huang et al. (2017). Asterisks on branches indicate bootstrap values as labeled.
Across Oryza groups, there is a strong association between hull color and the presence or absence of the Bh4 deletion, with 75% of samples matching expected phenotype based on deletion status, but several exceptions were also observed (Table S3, Figure 2). For example three straw-hulled wild rice accessions and eight straw-hulled cultivars in our panel also have wild type alleles (Table S1, Table S3, Figure 2). This indicates that other loci can also affect hull color, as previously suggested (Kuriyama and Kudo 1967). In a previous survey of Bh4 sequences, two other loss-of-function mutations were detected leading to straw hulls: a 1-bp frame shift deletion in exon 1 and a SNP in exon 3 that causes an early stop codon (Zhu et al. 2011), but neither was observed in our panel.
South Asian weeds of different origins carry Bh4 haplotypes that primarily cluster with haplotypes of their ancestors (Figure 2). For example, wild-like weed alleles cluster together in the tree and seem to originate from a few closely related wild alleles. Aus-like and indica-like weeds with haplotypes carrying the 22 bp deletion cluster with various aus and indica haplotypes, and aus-like weeds lacking the 22 bp deletion also have Bh4 haplotypes similar to these rarer haplotypes occurring in aus cultivars.
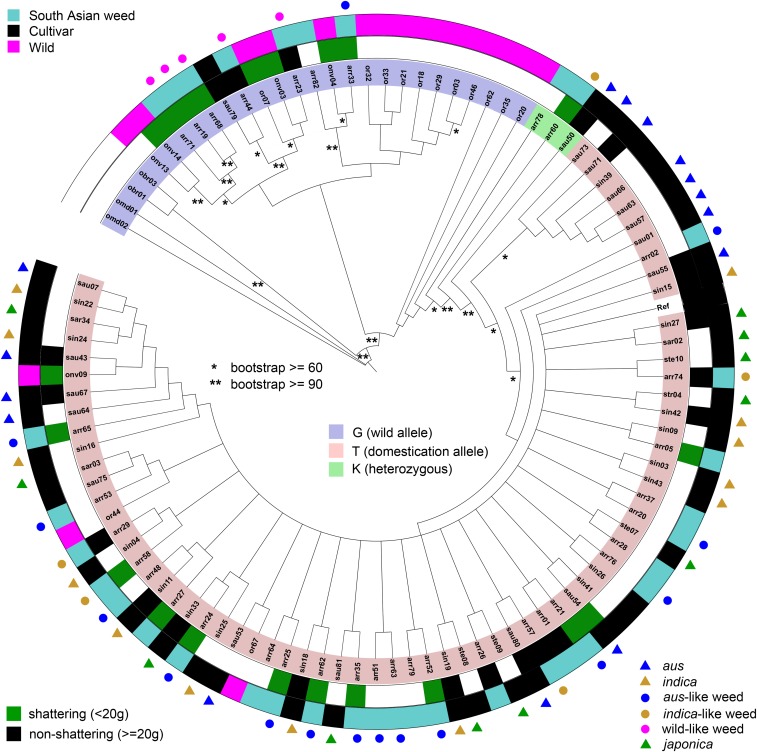
Patterns of polymorphism in sh4
A single nonsynonymous G to T substitution in the first exon of sh4 has been shown to lead to incomplete development of an abscission layer in the seed-pedicel junction in cultivated rice, and thus loss of seed shattering during domestication (Li et al. 2006). Our phylogenetic analysis of the sh4 gene sequences obtained confirms the ancestral status of the G wild-type allele, and the prevalence of the derived T allele that is associated with loss of shattering in cultivated rice (Figure 3). South Asian weeds varied in the type of sh4 alleles they possess. Wild-like weeds all bear the ancestral G allele, and aus-like and indica-like weeds (with only two exceptions) have the derived T allele, indicating that all weeds groups have inherited their sh4 alleles from their respective ancestors. Given the near fixation of the T sh4 allele during rice domestication (Li et al. 2006), these results support the origin of indica-like and aus-like weeds in South Asia through de-domestication (Huang et al. 2017).
Figure 3.
Neighbor joining (NJ) tree of Sh4 haplotypes. The color range indicates the allele of the functional SNP; the inner ring indicates the shattering phenotype. The outer ring and the bullets indicate Oryza groups, as labeled in the key. Unmarked weeds are admixed according to results of Huang et al. (2017). Asterisks on branches indicate bootstrap values as labeled.
Despite the fact that there is a strong correlation between seed shattering phenotype and sh4 genotype in wild and cultivated rice (i.e., all wild rice shatters to some degree and only one accession was found to carry the derived T allele; 93% of cultivars carry at least one T allele and 73% are non-shattering), this association breaks down when weedy rice groups are examined (Table S4, Table S1, Figure 3). The occurrence of easy shattering can easily be explained in wild-like weeds, as these all carry ancestral sh4 alleles very similar to those found in their wild rice ancestors. However, aus-like and indica-like weeds also have a prevalent shattering phenotype (93% are medium or strong shattering; Table S4), yet all but one examined individual carried the derived domestication allele of sh4 present in their cultivated ancestors. This suggests that these two weed-groups have evolved the shattering phenotype through an alternative genetic mechanism. This is similar to the case of U.S. weedy rice, which also arose through a process of de-domestication, and shatters despite fixation of the sh4 T domestication allele (Thurber et al. 2010).
Patterns of polymorphism in PROG1
The PROG1 gene has been implicated in changes in plant architecture during rice domestication, with a single amino acid haplotype said to be present in cultivated rice, but multiple haplotypes present in wild ancestors (Jin et al. 2008; Tan et al. 2008). The occurrence of PROG1 domestication alleles has also recently been implicated in the de-domestication origins of US weedy rice (Li et al. 2017). We used SNP variation in our PROG1 sequences to construct a phylogenetic tree, and examined the distribution of haplotypes among the various Oryza groups. The causative mutation leading to the prog1 allele associated with erect growth has not been isolated from the set of polymorphisms present in PROG1; however, wild rice has been shown to be polymorphic for PROG1 amino acid haplotypes, while most cultivated rice carries a single amino acid haplotype (Jin et al. 2008, Tan et al. 2008). We thus also examined the amino acid haplotypes present in our sequences (Figure S1). PROG1 does not display much nucleotide or amino acid haplotypic diversity in our set of accessions, and, as previously reported, most cultivated rice of all varieties shares the same or similar alleles. Only four out of 44 cultivated rice accessions had alleles that were basal in the tree. In contrast, wild rice contains more different types of haplotypes, some of which are similar to those of cultivated rice, and some that are more basal. The bulk of South Asian weedy rice from all groups contains PROG1 alleles that are similar or identical to the derived haplotypes predominating in cultivated rice. Weedy PROG1 alleles can thus be explained through inheritance from standing variation in ancestral groups.
Our data did not yield an obvious association between tiller angle and PROG1 alleles. All cultivated rice we measured has tiller angles that are less than 60°, and most accessions have similar PROG1 alleles. This is true for weedy rice too, yet one wild rice accession with cultivated PROG1 allele (or03) has broad tiller angle. Additionally, two wild-like weeds with very narrow tiller angels (<30°; arr23 and arr44) carry PROG1 alleles that are closely related to an allele carried by a wild rice with a very broad angle (Figure S1). Thus, though PROG1 allele identity in weeds can be explained by ancestral standing variation, our results suggest that tiller angle likely has other genetic determinants in our set of samples.
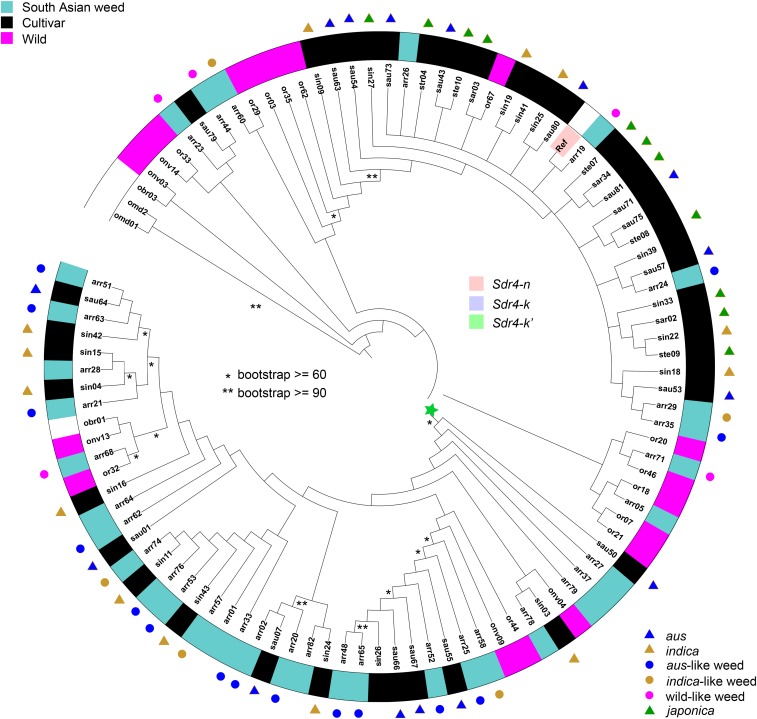
Patterns of polymorphism in Sdr4
The Sdr4 locus has been implicated in the seed dormancy trait, with Sdr4-k alleles being common in wild rice (O. rufipogon) and present in some indica cultivar varieties, and Sdr4-n alleles conferring reduced dormancy in almost all japonica and some indica cultivated rice (Sugimoto et al. 2010). We used SNP information to construct a phylogenetic tree of Sdr4, and overlay information of allele type, based on the ten SNPs and one 3 bp indels that differentiate the reported Sdr4-k and Sdr4-k haplotypes, as well as the two SNPs and one 3 bp indel that differentiate Sdr4-k from its closely related, but rarer, Sdr4-k’ allele (Sugimoto et al. 2010). We detected many variations of Sdr4, with multiple haplotype that could not be classified as either Sdr4-k or Sdr4-n (Figure 4, Table S1). For cultivars, consistent with previous reports (Sugimoto et al. 2010), japonica accessions only have the Sdr4-n allele, while Sdr4-n, Sdr4-k, Sdr4-k’ alleles, as well as various other haplotypes occur in aus and indica accessions.
Figure 4.
Neighbor joining (NJ) tree of Sdr4 haplotypes. The color range indicates the three Sdr4 haplotypes identified in Sugimoto et al. (2010). The ring and the bullets indicate Oryza groups, as labeled in the key. Unmarked weeds are admixed according to results of Huang et al. (2017). Asterisks on branches indicate bootstrap values as labeled.
We found that very few weeds have inherited the Sdr4-n allele despite its occurrence in all cultivated and wild groups. Instead, most weeds carry haplotypes that are more similar to Sdr4-k and k’. Unfortunately, for this candidate gene we had no phenotypic data. However, if the previous association between Sdr4 allele types and seed dormancy is taken at face value, our results suggest that most aus-like, indica-like, wild-like, and admixed South Asian weeds have some degree of seed dormancy conferred by the lack of Sdr4-n alleles. Moreover, although this trait may have been inherited from standing variation in cultivated and wild ancestors, it is significant that almost no weed derived from these groups has inherited the Sdr4-n variant that segregates in these groups, suggesting selection favoring Sdr4-k and similar alleles in South Asian weeds.
Genetic diversity analyses
If weediness candidate genes were targeted by selection during weed evolution, a decrease in nucleotide diversity should be evident at these loci. We compared nucleotide diversity and Tajima’s D values at silent sites in candidate genes with those obtained from all 128 targeted loci as a representation of genome-wide averages for groups of weeds and their putative ancestors. Mean genome-wide nucleotide diversity is highest in wild rice and in wild-like weeds (Table 4). The two cultivar groups and their derived weeds have similar levels of diversity, suggesting that these de-domesticated weed groups have not experienced strong bottlenecks upon evolving from cultivated ancestors.
Table 4. Summary statistics on silent sites for candidate genes and all loci obtained with target-capture.
| Oryza group | |||||||
|---|---|---|---|---|---|---|---|
| Locus | Summary statistics | aus | indica | aus-like weed | indica-like weed | wild-like weed | wild |
| Rc | π | 0.0025 | 0.0026 | 0.0021 | 0.0040 | 0.0029 | 0.0057 |
| θw | 0.0029 | 0.0033 | 0.0018 | 0.0039 | 0.0027 | 0.0089 | |
| Tajima’s D | −0.5243 | −0.9636 | 0.6020 | 0.2043 | 0.4154 | −1.5138 | |
| Bh4 | π | 0.0068 | 0.0092 | 0.0059 | 0.0096 | 0.0013 | 0.0141 |
| θw | 0.0074 | 0.0077 | 0.0074 | 0.0115 | 0.0015 | 0.0130 | |
| Tajima’s D | −0.2999 | 0.8084 | −0.7779 | −1.2185 | −0.9726 | 0.3387 | |
| sh4 | π | 0.00035 | 0 | 0.0009 | 0 | 0.0090 | 0.0058 |
| θw | 0.00023 | 0 | 0.0021 | 0 | 0.0083 | 0.0083 | |
| Tajima’s D | 1.1224 | NA | -2.124 | NA | 0.6457 | −1.2382 | |
| PROG1 | π | 0 | 0.0011 | 0.0010 | 0.0089 | 0.0089 | 0.0087 |
| θw | 0 | 0.0024 | 0.0023 | 0.0106 | 0.0071 | 0.0107 | |
| Tajima’s D | NA | −1.1491 | −1.1594 | −1.0485 | 1.4588 | −0.6036 | |
| Sdr4 | π | 0.0018 | 0.0022 | 0.0018 | 0.0036 | 0.0036 | 0.0037 |
| θw | 0.0011 | 0.0023 | 0.0033 | 0.0034 | 0.0035 | 0.0063 | |
| Tajima’s D | 1.3759 | −0.1269 | −1.3165 | 0.2431 | 0.2431 | −1.3434 | |
| All 128 genes | π | 0.0041 | 0.0040 | 0.0044 | 0.0048 | 0.0062 | 0.0090 |
| θw | 0.0040 | 0.0043 | 0.0044 | 0.0049 | 0.0060 | 0.0105 | |
| Tajima’s D | −0.0211 | −0.2047 | −0.0885 | −0.1591 | 0.2740 | −0.8180 | |
Nucleotide diversity (π or θw) of individual gene <0.0005.
Tajima’s D >2 or < -2.
Most candidate genes surveyed have levels of nucleotide diversity that are unremarkable compared to genomic averages, not exceeding two standard deviations from the mean. Sh4 stands out for its very low levels of diversity in aus, indica and indica-like groups, and this is consistent with selection for reduced shattering during the domestication process of cultivar groups (Li 2006; Konishi et al. 2006; Thurber et al. 2010). Low sh4 nucleotide diversity in indica-like weeds can be attributed to the lack of diversity at this gene in ancestral indica. Other candidate genes showing extremely low levels of nucleotide diversity are PROG1 and sh4 in the aus group, which suggests selection on some aspect of plant architecture in aus (Table 4).
Weedy groups derived from crops did not show bottleneck evidence on a genome-wide level (Table S5), but wild-like weeds on average possess about 60% of their ancestral diversity. Loss of diversity in wild-like weeds is particularly evident in Rc and Bh4 suggesting possible selection on these pericarp and hull coloration genes in wild-like weeds. In contrast, crop-derived weeds show remarkable increases in diversity for PROG1 compared to their ancestors. This could be due to strong selection on cultivars for domesticated plant architecture, coupled with release of selection during de-domestication of the weeds. However, it is notable that weeds resembled crops in their predominant tiller angles (Table 1). A similar situation is seen for sh4 in aus-like weeds, where high diversity could be due to release from selection.
Genome-wide FST values for the three weed-ancestor comparisons show that indica-like weeds have negligible amounts of differentiation from indica cultivars (FST = 0.014), while differentiation between aus-like weeds and aus (FST = 0.172) and wild-like weeds and wild rice (FST = 0.165) is moderate (Table 5). Among candidate genes, Bh4 stands out by exceeding genome-wide FST values by more than two standard deviations (SD) from the mean between aus-like weeds and aus. This is consistent with the higher frequency of black hulls observed in aus-like weeds compared to their cultivated ancestors and could indicate selection favoring black-hull Bh4 haplotypes in weeds (Table 1).
Table 5. Pairwise FST between weed groups and ancestors for each of the 15 candidate genes (Hudson et al. 1992).
| aus-like vs. aus | indica-like vs. indica | wild-like vs. wild | |
|---|---|---|---|
| Rc | 0.0123 | 0.0000 | 0.2917 |
| Bh4 | 0.6362 | 0.0000 | 0.3067 |
| Sh4 | 0.0884 | NA | 0.1047 |
| PROG1 | 0.1364 | 0.0124 | 0.2072 |
| Sdr4 | 0.2279 | 0.0000 | 0.1386 |
| All 128 genes | 0.1717 | 0.0142 | 0.1654 |
Discussion
We had previously investigated the evolutionary origins of weedy rice in South Asia, an area harboring great cultivated and wild Oryza diversity (Huang et al. 2017). Weedy rice in this region has a highly heterogeneous genetic background, more so than many other world regions (Reagon et al. 2010; Vigueira et al. 2017; Li et al. 2017), with contributions from two cultivated rice varieties (aus and indica) and from the wild ancestor of domesticated rice. Despite these very diverse origins, our current study finds that weedy rice groups in South Asia share the presence of several iconic weediness traits, supporting the concept of an agricultural weed syndrome (Vigueira et al. 2013b), and helping define the suite of traits that are most likely to be adaptive in weedy rice.
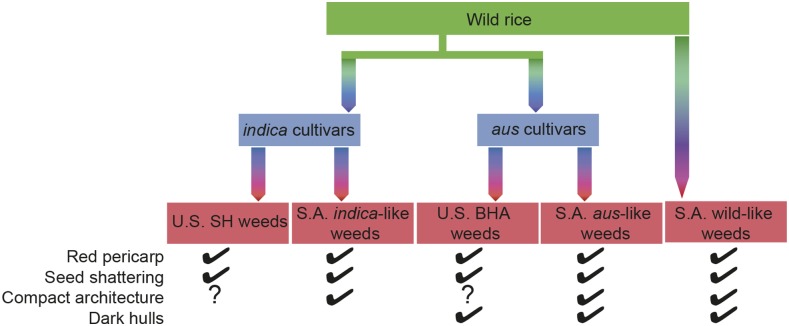
Of the traits examined, the greatest convergence among all three weed groups in South Asia occurred for easy seed shattering, red pericarp color, and compact plant architecture (Table 1; Table 2; Figure 1; Figure 3; Figure S1; Figure 5), suggesting that these traits are advantageous in the South Asian agricultural environment. Convergence among the wild and cultivar derived weedy rice groups is striking, given that for some of these traits weeds do not resemble their ancestors. For example, red pericarps and shattering phenotypes do not predominate in cultivars but are common in aus-like and indica-like weeds; likewise, wild rice tends to have a spreading architecture, yet wild-like weeds tend to be more compact. Convergence for seed shattering and red pericarp also occurs with weed groups in other parts of the world such as the U.S. (Thurber et al. 2010; Gross et al. 2010; Figure 5) and Malaysia (Song et al. 2014; Cui et al. 2016), supporting the idea that these are important traits in weedy rice evolution. Unfortunately, there is not enough information in the literature to gauge if compact plant architecture is also common in other weed groups. Our data suggest then, that among the traits surveyed, seed dormancy potentially related to red pericarps, and seed shattering are the most essential weediness traits in weedy rice worldwide.
Figure 5.
Summary figure of known convergent traits among the South Asian weedy rice groups in this study and previously studied U.S. weedy rice groups. Featured weedy groups have been shown in Huang et al. (2017) to have all evolved independently. Convergence in red pericarps may be indicative of convergence in seed dormancy, which was not directly evaluated.
Given separate origins, how have weedy groups in South Asia acquired convergent weedy traits? Our survey of PROG1 as a candidate gene for tiller angle is inconclusive, but our candidate genes for pericarp color and seed shattering suggest differing genetic mechanisms. Most weedy accessions with red pericarps carry wild-type Rc alleles that can explain the phenotype and were likely inherited from variation present in their ancestral groups (Figure 1). The similar nucleotide diversity in the Rc gene for all cultivar and weed groups, as well as the low pairwise FST between weed and ancestor, also serve as evidence of the inheritance of ancestral alleles (Table 4; Table 5). The very occasional discrepancies between pericarp color and Rc alleles (Figure 1, Table 3) indicate that other genes or mutations may affect pericarp color, but the Rc genotype in weeds is mostly sufficient to explain weed phenotype. Consistent with origins for ancestral standing variation, there are no strong signatures of selection observed for this gene in weed groups (Table 3).
A similar situation to pericarp color and Rc may be occurring with seed dormancy and its candidate gene Sdr4. We unfortunately do not have seed dormancy phenotypic data for our weedy populations, but seed dormancy has often been considered a characteristic weedy trait (Sugimoto et al. 2010). The lack of dormancy-associated Sdr4-n alleles in any weed population, while easily explained through inheritance from ancestral standing variation, suggests possible convergence in seed dormancy among all weed groups possibly mediated through the evolutionary advantage conferred by maintenance of the dormancy trait.
Convergence of seed shattering in South Asian weeds seems to have occurred in a different manner. In our South Asian panel, every wild-like weed has the same ancestral wild type sh4 allele as wild rice that has been shown to be associated with seed shattering (Li et al. 2006), thus sh4 inheritance from ancestors can explain the shattering phenotype in wild-like weeds. In contrast, almost all cultivar-like weeds have the derived sh4 allele common in crop rice (Table S4; Figure 3) that has been implicated in loss of shattering during domestication. Thus, although inheritance from cultivated ancestors explains the sh4 allele present in indica and aus-like weeds, this allele cannot explain the shattering phenotype. This is similar to what has been observed for US weedy rice (Thurber et al. 2010), where a novel genetic mechanism must account for seed shattering. Whether there is convergence in this novel mechanism between South Asian cultivar-derived weeds and any of the US groups is unknown, and we cannot yet draw conclusions about the roles of standing variation vs. new mutations in the evolution of shattering in these groups. The high levels of nucleotide diversity of aus-like weed in sh4 compared to aus cultivars, and the extremely negative Tajima’s D (Tables 4; Table S5), suggest that aus-like weed may be undergoing a release in selection for this gene.
While not common to all weedy groups in South Asia, a high degree of convergence for black hull color occurs among wild-like, admixed and aus-like weeds. This trait is also convergent with one group (BHA) of US weedy rice (Reagon et al. 2010). The fact that dark hulls and awns are present, but not prevalent in the aus ancestors of both BHA and aus-like South Asian weeds (Table S3), suggests there may be some adaptive value to these traits such as resistance to pests and predators. However, these cannot be considered essential weedy traits, as weedy rice groups that lack them nevertheless thrive around the world (Ziska et al. 2015; Song et al. 2014). Both wild-like and aus-like weeds likely inherited these phenotypes from variation present in ancestral groups. In the case of hull color, this is supported by Bh4 variation (Figure 2), with wild-like weeds carrying Bh4 alleles similar to wild rice, and aus-like weeds carrying alleles similar to the aus in our panel with no-deletion alleles. While the black hull phenotype is rare in the aus in our panel, a slightly higher frequency has been previously reported for a wider set of aus surveyed (Huang et al. 2017). Surprisingly, although aus-like weeds likely inherited Bh4 alleles from a small subset of aus cultivars with black hulls, both groups show similar nucleotide diversity at Bh4 (Table 4; Table S5), but very large differentiation as measured by FST (Table 5). This is compatible with a scenario whereby aus-like weeds were de-domesticated from aus cultivars early in the domestication process, before the majority of aus cultivars underwent selection for straw hulls with the 22-bp Bh4 deletion (Vigueira et al. 2013a).
Our characterization of weedy rice phenotypes in South Asia and the associated candidate genes contribute to the emerging understanding of the mechanisms by which weedy rice evolves worldwide. Whether evolving from crop ancestors, as inferred for BHA and SH weeds in the US and aus-like weeds and indica-like weeds in South Asia, or evolving from wild ancestors, such as the wild-like weeds of South Asia, standing variation present in the non-weedy ancestors often seems a sufficient explanation for the origin of weedy rice traits. Thus, for none of the genes surveyed, even those that underlie traits where weeds diverge from their ancestors, is introgression required to explain the alleles present in weedy rice (the only exception being the unknown genetic mechanism for shattering in cultivar-related weeds). The fact that cultivated rice varieties, which have been shown to have gone through domestication bottlenecks and loss of diversity (Caicedo et al. 2007), as well as wild rice, which is not a priori adapted to the agricultural environment, nevertheless harbor the necessary standing variation (i.e., allelic diversity) to give rise to troublesome weeds, may help explain the prevalence of the weedy rice problem worldwide. The possibility of continued repeated evolution of weedy rice presents an interesting conundrum to usual calls for maintenance of diversity in crop species.
Acknowledgments
The authors thank the University of Massachusetts greenhouse staff and S. Perera for plant care support. We also thank the larger working group of the Plant and Microbial Innovation theme in the Institute for Applied Life Sciences for discussions. The United States Department of Agriculture (USDA) is an equal opportunity provider and employer. This work was supported by a grant from the US National Science Foundation Plant Genome Research Program (IOS-1032023) to A.L.C, K.M.O and Y.J.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.6853244.
Communicating editor: J. Wendel
Literature Cited
- Arendt J., Reznick D., 2008. Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends Ecol. Evol. 23: 26–32. 10.1016/j.tree.2007.09.011 [DOI] [PubMed] [Google Scholar]
- Baker H. G., 1965. Characteristics and mode of origin of weeds. In H. G. Baker and G. L. Stebbins [eds.], The genetics of colonizing species, 147–172. Academic Press, New York, New York. [Google Scholar]
- Caicedo A. L., Williamson S. H., Hernandez R. D., Boyko A., Fledel-Alon A., et al. , 2007. Genome-Wide Patterns of Nucleotide Polymorphism in Domesticated Rice. PLoS Genet. 3: e163 10.1371/journal.pgen.0030163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civáň P., Craig H., Cox C. J., Brown T. A., 2015. Three geographically separate domestications of Asian rice. Nat. Plants 1: 15164 10.1038/nplants.2015.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Song B. K., Li L.-F., Li Y.-L., Huang Z., et al. (2016) Little White Lies: Pericarp Color Provides Insights into the Origins and Evolution of Southeast Asian Weedy Rice. G3 GenesGenomesGenetics (Bethesda) g3.116.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J., 1993. PHYLIP: phylogeny inference package (version 3.5c). University of Washington, Seattle. [Google Scholar]
- Garris A. J., Tai T. H., Coburn J., Kresovich S., McCouch S., 2005. Genetic Structure and Diversity in Oryza sativa L. Genetics 169: 1631–1638. 10.1534/genetics.104.035642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianinetti A., Cohn M. A., 2008. Seed dormancy in red rice. XIII: Interaction of dry-afterripening and hydration temperature. Seed Sci. Res. 18: 151–159. 10.1017/S0960258508037999 [DOI] [Google Scholar]
- Gnirke A., Melnikov A., Maguire J., Rogov P., LeProust E. M., et al. , 2009. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 27: 182–189. 10.1038/nbt.1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross B. L., Reagon M., Hsu S.-C., Caicedo A. L., Jia Y., et al. , 2010. Seeing red: the origin of grain pigmentation in US weedy rice. Mol. Ecol. 19: 3380–3393. 10.1111/j.1365-294X.2010.04707.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X.-Y., Foley M. E., Horvath D. P., Anderson J. V., Feng J., et al. , 2011. Association between seed dormancy and pericarp color is controlled by a pleiotropic gene that regulates abscisic acid and flavonoid synthesis in weedy red rice. Genetics 189: 1515–1524. 10.1534/genetics.111.131169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98. [Google Scholar]
- Huang X., Kurata N., Wei X., Wang Z.-X., Wang A., et al. , 2012. A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501. 10.1038/nature11532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Young N. D., Reagon M., Hyma K. E., Olsen K. M., et al. , 2017. All roads lead to weediness: Patterns of genomic divergence reveal extensive recurrent weedy rice origins from South Asian Oryza. Mol. Ecol. 26: 3151–3167. 10.1111/mec.14120 [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Slatkin M., Maddison W. P., 1992. Estimation of levels of gene flow from DNA sequence data. Genetics 132: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ithal N., Reddy A. R., 2004. Rice flavonoid pathway genes, OsDfr and OsAns, are induced by dehydration, high salt and ABA, and contain stress responsive promoter elements that interact with the transcription activator, OsC1-MYB. Plant Sci. 166: 1505–1513. 10.1016/j.plantsci.2004.02.002 [DOI] [Google Scholar]
- Jin J., Huang W., Gao J.-P., Yang J., Shi M., et al. , 2008. Genetic control of rice plant architecture under domestication. Nat. Genet. 40: 1365–1369. 10.1038/ng.247 [DOI] [PubMed] [Google Scholar]
- Kane N. C., Rieseberg L. H., 2008. Genetics and evolution of weedy Helianthus annuus populations: adaptation of an agricultural weed. Mol. Ecol. 17: 384–394. 10.1111/j.1365-294X.2007.03467.x [DOI] [PubMed] [Google Scholar]
- Konieczny A., Ausubel F. M., 1993. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. The Plant Journal 4: 403–410. 10.1046/j.1365-313X.1993.04020403.x [DOI] [PubMed] [Google Scholar]
- Konishi S., Izawa T., Lin S. Y., Ebana K., Fukuta Y., et al. , 2006. An SNP Caused Loss of Seed Shattering During Rice Domestication. Science 312: 1392–1396. 10.1126/science.1126410 [DOI] [PubMed] [Google Scholar]
- Kuriyama H., Kudo M., 1967. Complementary genes ph and bh controlling ripening-black coloration of rice hulls and their geographical distribution. Jpn. J. Breed. 17: 13–19. 10.1270/jsbbs1951.17.13 [DOI] [Google Scholar]
- Li C., Zhou A., Sang T., 2006. Rice Domestication by Reducing Shattering. Sci. 311: 1936–1939. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , and 1000 Genome Project Data Processing Subgroup, 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.-F., Li Y.-L., Jia Y., Caicedo A. L., Olsen K. M., 2017. Signatures of adaptation in the weedy rice genome. Nat. Genet. 49: 811–814. 10.1038/ng.3825 [DOI] [PubMed] [Google Scholar]
- Monaco T. J., Weller S. C., Ashton F. M., 2002. Weed Science: Principles and Practices, John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- Oerke E.-C., 2006. Crop losses to pests. J. Agric. Sci. 144: 31–43. 10.1017/S0021859605005708 [DOI] [Google Scholar]
- Patel R. K., Jain M., 2012. NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. PLoS One 7: e30619 10.1371/journal.pone.0030619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reagon M., Thurber C. S., Gross B. L., Olsen K. M., Jia Y., et al. , 2010. Genomic patterns of nucleotide diversity in divergent populations of U.S. weedy rice. BMC Evol. Biol. 10: 180 10.1186/1471-2148-10-180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B.-K., Chuah T.-S., Tam S. M., Olsen K. M., 2014. Malaysian weedy rice shows its true stripes: wild Oryza and elite rice cultivars shape agricultural weed evolution in Southeast Asia. Mol. Ecol. 23: 5003–5017. 10.1111/mec.12922 [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Takeuchi Y., Ebana K., Miyao A., Hirochika H., et al. , 2010. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl. Acad. Sci. USA 107: 5792–5797. 10.1073/pnas.0911965107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney M. T., Thomson M. J., Pfeil B. E., McCouch S., 2006. Caught Red-Handed: Rc Encodes a Basic Helix-Loop-Helix Protein Conditioning Red Pericarp in Rice. Plant Cell Online 18: 283–294. 10.1105/tpc.105.038430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Li X., Liu F., Sun X., Li C., et al. , 2008. Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40: 1360–1364. 10.1038/ng.197 [DOI] [PubMed] [Google Scholar]
- Thurber C. S., Reagon M., Gross B. L., Olsen K. M., Jia Y., et al. , 2010. Molecular evolution of shattering loci in U.S. weedy rice. Mol. Ecol. 19: 3271–3284. 10.1111/j.1365-294X.2010.04708.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigueira C. C., Li W., Olsen K. M., 2013a The role of Bh4 in parallel evolution of hull colour in domesticated and weedy rice. J. Evol. Biol. 26: 1738–1749. 10.1111/jeb.12171 [DOI] [PubMed] [Google Scholar]
- Vigueira C. C., Olsen K. M., Caicedo A. L., 2013b The red queen in the corn: agricultural weeds as models of rapid adaptive evolution. Heredity 110: 303–311. 10.1038/hdy.2012.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigueira C. C., Qi X., Song B.-K., Li L.-F., Caicedo A. L., et al. , 2017. Call of the wild rice: Oryza rufipogon shapes weedy rice evolution in Southeast Asia. Evol. Appl. 00: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B.-F., Si L., Wang Z., Zhou Y., Zhu J., et al. , 2011. Genetic control of a transition from black to straw-white seed hull in rice domestication. Plant Physiol. 155: 1301–1311. 10.1104/pp.110.168500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Zheng X., Luo J., Gaut B. S., Ge S., 2007. Multilocus Analysis of Nucleotide Variation of Oryza sativa and Its Wild Relatives: Severe Bottleneck during Domestication of Rice. Mol. Biol. Evol. 24: 875–888. 10.1093/molbev/msm005 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Ellstrand N. C., Lu B.-R., 2012. Sequence polymorphisms in wild, weedy, and cultivated rice suggest seed-shattering locus sh4 played a minor role in Asian rice domestication. Ecol. Evol. 2: 2106–2113. 10.1002/ece3.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziska L. H., Gealy D. R., Burgos N., Caicedo A. L., Gressel J., et al. , 2015. Chapter Three - Weedy (Red) Rice: An Emerging Constraint to Global Rice Production, pp. 181–228 in Advances in Agronomy, edited by Sparks D. L. Academic Press, Cambridge, MA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Newly generated DNA sequences for candidate genes are available in GenBank (accessions MH771032 - MH771132, MH798903 - MH799306). DNA sequences for the 123 supplementary loci, as well as other supplementary material is available on figshare via the GSA portal. Supplementary tables contain genotype and phenotype of Oryza accessions included in the study, and genomic regions sequenced by target capture. Figure S1 contains the PROG1 phylogenetic tree based on SNPs. Scripts used to obtain population statistics are available at Github: https://github.com/Zhongyun-Huang/G3-2018-200605. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6853244.