Abstract
In contrast to Z-DNA that was stabilized and well-studied for its structure by chemical approaches, the stabilization and structural study of Z-RNA remains a challenge. In this study, we developed a Z-form RNA stabilizer m8Gm, and demonstrated that incorporation of m8Gm into RNA can markedly stabilize the Z-RNA at low salt conditions. Using the m8Gm-contained Z-RNA, we determined the structure of Z-RNA and investigated the interaction of protein and Z-RNA.
Keywords: Z-RNA structure, synthesis of oligonucleotide, NMR, circular dichroism
1. Introduction
The RNA helixes mostly adopt a right-handed A-from geometry. Another RNA conformation is characteristic of left-handed RNA having alternating CG base pairs [1,2]. Zα, a Z-form-binding domain of RNA-editing enzyme ADAR1, has been found to specifically bind to Z-DNA and Z-RNA [3,4,5,6,7,8,9]. It has been reported that Z-RNA participates in the interferon-response pathway [10,11] or viral inhibition [10,11,12]. The physical, chemical and spectral data on Z-RNA have been collected, whereas the biological role of Z-RNA has long been in question due to the difficulty of obtaining stable Z-RNAs under physiological salt conditions. For example, an A- to Z-RNA transition requires a very high salt concentration, e.g., 6 M NaClO4 [1,2].
Several studies have been reported on incorporating modified nucleic acid residues for the stabilization of Z-DNA [13,14,15,16,17,18,19,20,21]. Notably modified guanine residues highly favor the formation of Z-DNA. We have previously reported that the methylation of the C8 position of guanine significantly stabilized the Z-DNA because the modification by methylation was favorable for the syn conformation of the nucleobases [17]. Based on the results of these past studies, we have now developed a Z-form RNA stabilizer that stabilizes Z-RNA under physiological salt conditions. We designed and synthesized a 2′-O-methyl-8-methyl guanosine (m8Gm) by insertion of a methyl group at the C8 position of 2′-O-methyl guanosine. We found that incorporation of the m8Gm in the RNA dramatically stabilized the Z-RNA, even under physiological salt concentrations, and facilitated the A- to Z-RNA transition, even for AU-containing sequences that do not favor the formation of Z-RNA. We then determined the solution structure of r(CGC[m8Gm]CG)2. This allowed us to see the effect of the incorporation of a m8Gm on RNA and allowed us to understand the Z-RNA structure. The Z-RNA stabilizer can also be used to investigate the interaction of the Zα domain and Z-RNA.
2. Results and Discussion
We synthesized the m8Gm-incorporating oligonucleotides by phosphoramidite chemistry, using 2′-O-methyl guanosine (Schemes S1, S2 and Figures S1–S7). The induction of a methyl group in the C8 position highly favors the syn conformation. These structural features can lead the m8Gm to greatly stabilize Z-RNA. Using nuclear Overhauser effect (NOE) between C1′H and 8CH3, we confirmed the syn conformation of m8Gm (Figure S8).
Circular dichroism (CD) spectroscopy has been used to study the Z-form conformation [17,22]. A-RNA, a negative Cotton effect appears around 295 nm, whereas in Z-RNA, a more positive band appears at 280 nm [23,24]. Thus, we performed CD spectroscopy experiments to study the conformation using various NaClO4 concentrations.
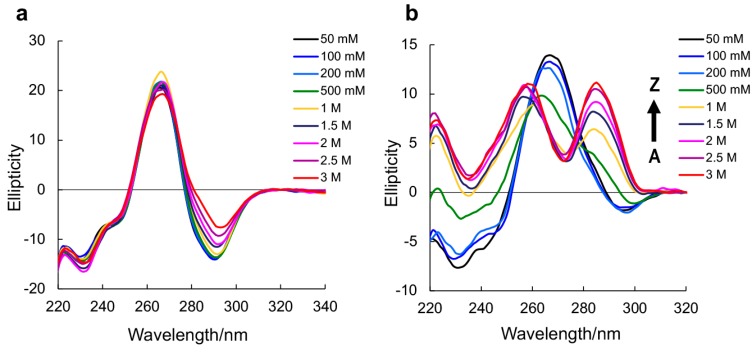
We used the CD spectroscopy to examine the A-Z transition under various salt concentrations. Native RNA r(CGCGCG)2 in A-form does not transfer to the Z-form, even in the presence of 3 M NaClO4 (Figure 1a). m8Gm-incorporated r(CGC[m8Gm]CG)2 showed a Z-form CD spectrum with increasing concentrations of NaClO4. (Figure 1b). The midpoint NaClO4 concentration for r(CGC[m8Gm]CG)2 was 880 mM, lower than that of the native RNA (4090 mM) (Table 1). r(C[m8Gm]C[m8Gm]CG)2 containing two m8Gms greatly stabilized the Z-form, showing a Z-form CD spectrum even in the presence of 50 mM NaClO4 (Figure S9) at a lower physiological salt concentration, and the midpoint was 100 mM (Table 1).
Figure 1.
CD spectra of native RNA r(CGCGCG)2 (a) and m8Gm-contained r(CGC[m8Gm]CG)2 (b) (0.15 mM base concentration) at 10 °C with various NaClO4 concentrations in 5 mM sodium phosphate buffer (pH 7.0).
Table 1.
Midpoint NaClO4 concentration in A-Z transition.
| Oligonucleotides | NaClO4 (mM) |
|---|---|
| r(CGCGCG)2 | 4090 |
| r(CGC[m8Gm]CG)2 | 880 |
| r(C[m8Gm]C[m8Gm]CG)2 | 100 |
| r(CGCGUGCG)/r(CGCACGCG) | 5430 |
| r(CGCGUGCG)/r(C[m8Gm]CAC[m8Gm]CG) | 2360 |
| r(C[m8Gm]CGU[m8Gm]CG)/r(C[m8Gm]CAC[m8Gm]CG) | 850 |
The incorporation of m8Gm can stabilize the Z-form containing an AU base pair. It showed the typical Z-form with 2360 mM midpoint compared to the native RNA (Figure S9 and Table 1). Octamer RNA containing four m8Gms in double strands also effectively stabilized the Z-form with 850 mM midpoint.
We designed a short duplex containing only one modified base, r(C[m8Gm]CGU[m8Gm]CG)/r(CGCACG), in which the underlined sequences can form a duplex containing only one m8Gm base. We compared its stability with native RNA r(CGCGUGCG)/r(CGCACG). Although the 6 base pairs duplex containing an AU base pair does not favor the formation of Z-RNA, it facilitated the A- to Z-RNA transition as compared to native 6 base pairs RNA (Figure S10). The midpoint for RNA containing one m8Gm was 4000 mM, lower than that of the native RNA (>6000 mM) (Figure S10), indicating that only one m8Gm can stabilize the Z-form, indicating that only one m8Gm can stabilize the Z-form.
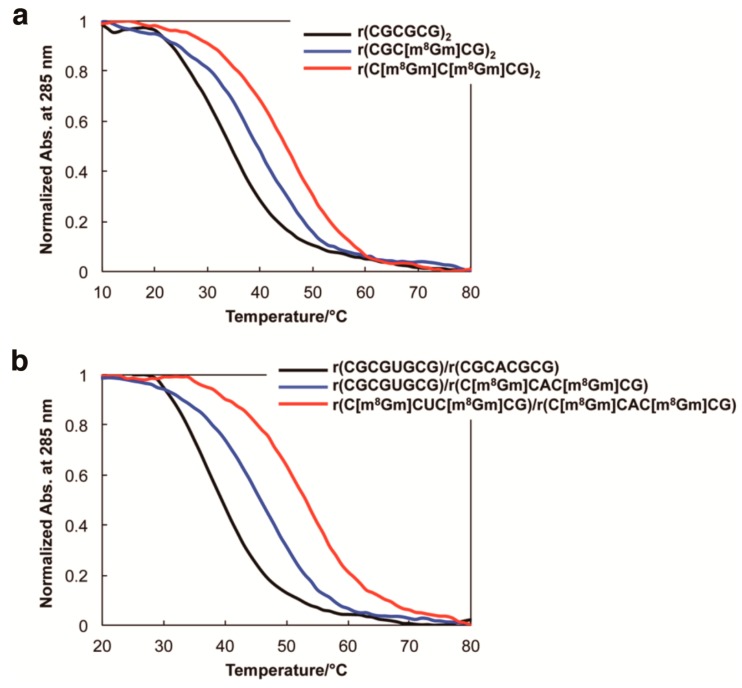
To further understand the m8Gm effect on the thermodynamic properties of Z-RNA, the melting temperature (Tm) and thermodynamic parameters were examined using CD melting experiments. (Tm) values of m8Gm-incorporated hexamer RNA increased by 6–15 °C (ΔTm), (Figure 2a and Table 2), and octamer RNA with four m8Gms in double strands containing an AU base pair increased even more, by 15 °C (Figure 2b and Table 2). Thermodynamic parameters revealed a very favorable free energy of formation for the m8Gm-incorporated Z-RNA compared to the native RNA (Table 2) suggesting that the syn conformation of m8Gm was thermodynamically favorable, and the preferred C3′ endo conformation of ribose induced by the 2′-O-methyl group also contributed to the stabilization of the Z-form, which is consistent with the previous study [1,25]. The advantages of 2′-O-methyl substituent include provision of an NMR signal from 2′-O-methyl group for NMR structure study, easy-to-prepare phosphoramidite reagent more than 2′-O-hydroxy group for the chemical introduction of m8Gm into RNA oligonucleotides, and efficient resistance to enzymatic cleavage of oligonucleotide [1,26,27].
Figure 2.
(a) CD melting curves for native RNA r(CGCGCG)2 (back line), r(CGC[m8Gm]CG)2 containing one m8Gm (blue line), and r(C[m8Gm]C[m8Gm]CG)2 containing two m8Gms (red line). (b) CD melting curves for RNA sequences containing an AU base pair, native RNA r(CGCGUGCG)/r(CGCACGCG) (back line), r(CGCGUGCG)/r(C[m8Gm]CAC[m8Gm]CG) having two m8Gms in one strand (blue line), r(C[m8Gm]CGU[m8Gm]CG)r(C[m8Gm]CAC[m8Gm]CG) having four m8Gms in two strands (red line). In 5 mM sodium phosphate buffer (pH 7.0), 7 M NaClO4 concentrations (0.15 mM base concentration).
Table 2.
Thermodynamic parameters for A-Z transition of various m8Gm-containing RNAs.
| Oligonucleotides | Tm1 (°C) | ΔTm (°C) | ∆G298K (kcal/mol) |
|---|---|---|---|
| r(CGCGCG)2 | 33.5 | − | −1.1 |
| r(CGC[m8Gm]CG)2 | 40.0 | +6.5 | −1.7 |
| r(C[m8Gm]C[m8Gm]CG)2 | 45.7 | +12.2 | −2.1 |
| r(CGCGUGCG)/r(CGCACGCG) | 37.7 | − | −1.7 |
| r(CGCGUGCG)/r(C[m8Gm]CAC[m8Gm]CG) | 45.7 | +8.0 | −2.5 |
| r(C[m8Gm]CGU[m8Gm]CG)/r(C[m8Gm]CAC[m8Gm]CG) | 52.7 | +15.0 | −3.3 |
1 Measurement condition: [RNA] = 0.15 mM base concentration, [sodium phosphate buffer (pH 7.0)] = 5 mM, [NaClO4] = 7 M.
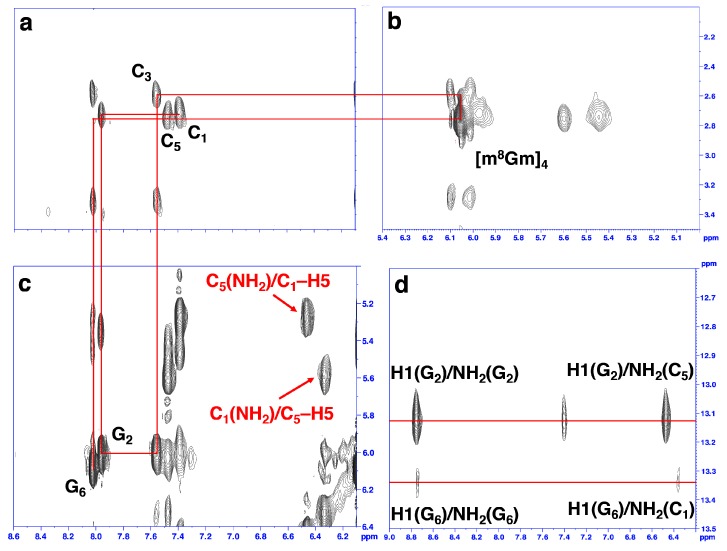
In order to obtain the structural information of Z-RNA, we performed an analysis of the 2D NOESY spectra of r(CGC[m8Gm]CG)2. A complete list of 1H chemical shifts was shown in Table S1. The NOE-restrained refinement provided an unequivocal demonstration that the structure of r(CGC[m8Gm]CG)2 is Z-RNA. We observed the strong cross-peaks in 2D-NOESY spectrum from intranucleotide H1′/H8 NOEs of G2 and G6 and a special NOE between C1′H and 8CH3 of m8Gm4. This indicated the syn conformation of all three guanosine residues (Figure 3b,c).
Figure 3.
(a–c) H6/H5′′ of C and H8/H1′ of G (8CH3/H1′ of m8Gm) proton region of NOESY spectra of r(CGC[m8Gm]CG)2 in NaClO4 solution. The NOE connectivity pathway is shown as red line. Intraresidue NOE cross-peaks are labeled with residue numbers. The Z-RNA structure specific cross-peaks were observed between C5 amino protons and C1H5 protons from the opposite strand (indicated as red words), as well as between C1 amino protons and C5H5 protons from the opposite strand (c). (d) The cross peaks of imino proton of G2 and amino proton of C5, as well as imino proton of G6 and amino proton of C1, indicated Watson–Crick-type base pairs of Z-RNA. Intraresidue NOE cross-peaks of imino and amino proton of G2 and G6 are shown.
The NOE connectivity path between H6/H5′′ of C and H8/H1′ of G (8CH3/H1′ of m8Gm4) allows for a cross-peak assignment of Z-RNA nonexchangeable resonances (Figure 3a–c). Sequential assignments of C1 to G6 for the Z-form can complete the pathway: C1(H6/H5′′)-G2(H8/H1′)-C3(H6/H5′′)-m8Gm4(8CH3/H1′)-C5(H6/H5′′)-G6(H8/H1′) (Figure 3a–c), indicating a sequence-specific connectivity for left-handed helices.
The H′2 resonance was assigned based on NOESY spectra for sequential connections between H6/H8 and H′2 of C and G (Figure S11). Moreover, the clear cross-peaks of the imino proton of G (around 13.0 ppm) and the amino proton of C suggested WC base pairs (Figure 3d). The 2D NOESY of the imino protons and other protons was used for assignment. For example, we note that the C5(NH2) amino proton has cross-peaks (C5(NH2)/C1–H5) to the C1–H5 proton, and the C1(NH2) amino proton has cross-peaks to the C5–H5 proton (C1(NH2)/C5–H5) (Figure 3c). Such cross-peaks can only happen between the two interstrand cytosines between C1 in one strand and C5 in another strand of the Z-RNA because of their base pair stacking pattern [28,29].
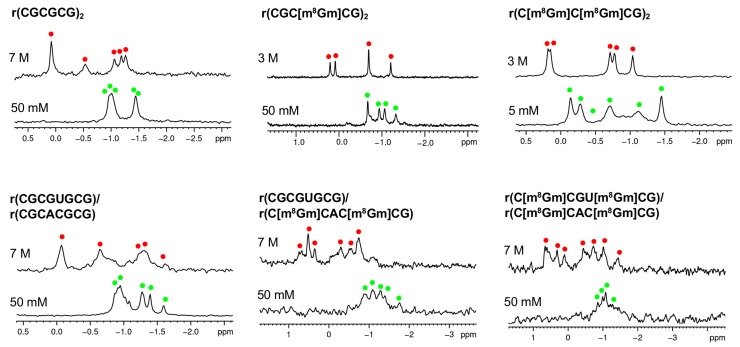
We further used 31P-NMR spectroscopy to confirm these observations. We found that these sequences as shown in Table 2 undergo a A-Z transition with increasing NaClO4 from 5 mM to 7 M (Figure 4). These results are consistent with the conformational data from the CD and proton NMR results.
Figure 4.
Monitoring of the A to Z-RNA transition by 31P NMR. 31P NMR spectra of the RNAs at 5 mM-7 M NaClO4. Green and red dots indicate the 31P peaks resulting from A-RNA and Z-RNA, respectively.
A model of r(CGC[m8Gm]CG)2 was constructed based on the reported Z-form structure and NOE-constrained refinement [2]. The molecular dynamics simulations were performed by the standard dynamics cascade in BIOVIA Discovery Studio 4.5 with some modifications. The conformation with the lowest energy was selected as shown in Figure 5. In the m8Gm-contanied Z-RNA, the C8-methyl groups were hydrophobic and located in the outside of the helix, which is consistent with the induction of a methyl group that strongly contributes to the increased stabilization of Z-RNA.
Figure 5.
The model for r(CGC[m8Gm]CG)2 Z-RNA structure. (a) m8Gm is expanded in green with the C8-methyl group, hydrogen and carbon of methyl group are coloured in yellow and purple. (b) Hydrogen and carbon of methyl group are coloured in green and white. C8-methyl groups were located outside of Z-RNA.
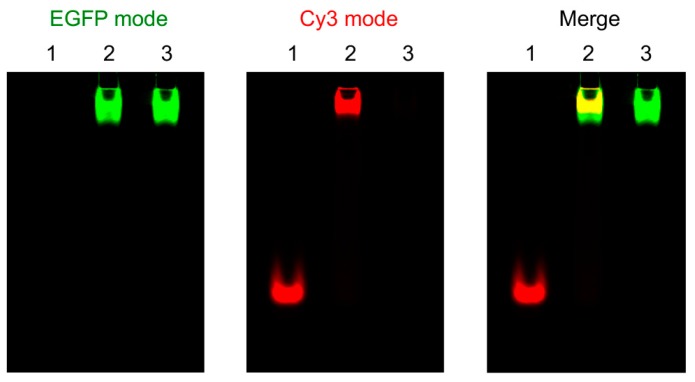
Encouraged by the ability to use m8Gm to stabilize Z-RNA, we study the interaction of the Zα domain and Z-RNA by using a m8Gm-containing a Z-RNA and Zα-EGFP fusion protein, in which the Zα domain is tagged with a green fluorescence protein [29]. Figure 6 indicates the visualized mode as EGFP-mode, Cy3-mode and Merge, respectively. Lane 1, lane 2 and lane 3 in Figure 5 show free Z-RNA labeling with fluorescent dye Cy3, a complex of Z-RNA and Zα-EGFP and free Zα-EGFP, respectively. Green and red fluorescence emission visualized under EGFP-mode and Cy3-mode distinguished Zα-EGFP and Z-RNA. Because of the large molecular weight of Zα-EGFP, we visualized the complex of the Zα-EGFP and Z-RNA in the upper. Lane 2 in merge mode of Figure 6 emitted yellow fluorescence and clearly demonstrated the complex of Zα with Z-RNA. These observations indicated that Zα-EGFP efficiently binds to the m8Gm-containing Z-RNA without severe steric clashes with the added methyl groups, suggesting the m8Gm stabilized Z-RNA can be used to investigate the interaction of proteins with Z-RNA.
Figure 6.
Visualization of Z-RNA and Zα-EGFP protein. Lane 1: Z-RNA only, Lane 2: RNA + Zα-EGFP, Lane 3: Zα-EGFP only. The different modes are indicated in upper. [RNA] = 1 μM, [Zα-EGFP] = 10 μM, [NaCl] = 100 mM, [Tris-HCl (pH7.0)] = 10 mM, [DTT] = 5 mM, 10% glycerol, 10 μg/mL BSA. Z-RNA: r(CGCGUGCG)-Cy3/r(C[m8Gm]CAC[m8Gm]CG).
Recent studies suggested that dsRNA pathways of RNA silencing and micro-RNA regulation were interfered with by ADAR1 [29]. Zα is also reported to enhance ADAR1 editing activity on RNAs with Z-forming sequences in vitro [29]. This suggested that Z-RNA plays important biological roles similar to Z-DNA.
In conclusion, the results described here reveal that the newly synthesized guanosine analogue 2′-O-methyl-8-methyl guanosine dramatically stabilizes Z-RNA, which arises from the syn conformation of the m8Gm base and the C3′ endo conformation of ribose. The oligonucleotides with AU base pairs can convert into Z-RNA. In addition, researchers can utilize the Z-RNA stabilizer to study the interaction of Z-RNA sequences with the Zα domain. Using the Z-RNA stabilizer, we can refine the structure of Z-RNA based on NMR and examine the molecular basis of Z-form-specific interactions and reactions.
3. Materials and Methods
3.1. Sample Preparation
We synthesized the RNA oligonucleotides by phosphoramidite chemistry with a DNA/RNA synthesizer. We purified by RP-HPLC and prepared Zα-EGFP fusion protein according to the method of Oyoshi et al. [15].
3.2. CD Experiments
We carried out CD experiments by a JASCO J-820 CD spectrophotometer (JASCO Corporation, Tokyo, Japan). The melting curves were obtained by monitoring 285 nm. 0.3 mL samples at 0.15 mM base concentration with 0.1–7 M NaClO4, 5 mM sodium phosphate buffer (pH 7.0).
3.3. NMR Experiments
We performed NMR experiments on a Bruker AV-400 M spectrometer (Bruker, Billerica, MA, USA). A special Micro Tube that is designed for use with reduced sample volumes was used (catalog no: NE-H5/3-Br, New Era NMR). Dissolve RNA samples (1.0 mM) in 150 μL of 90% H2O/10% D2O, 5 mM sodium phosphate buffer (pH 7.0), 50 mM or 3 M NaClO4. 2D NOE spectra were recorded with mixing times of 500 ms.
3.4. Molecular Modeling
The model of RNA structure was manually generated based on the reported structure using the BIOVIA Discovery Studio 4.5 (Accelrys, San Diego, CA, USA). Then molecular dynamics simulations were performed by the standard dynamics cascade in BIOVIA Discovery Studio 4.5 with some modifications. The structure was heating from 50 K to 300 K over 4 ps and equilibration at 300 K with 100 ps simulation time. The save results interval in the production step was 2 ps during 100 ps simulation time at 300 K. 10 best conformations generated by simulation were further energy minimized. The conformation with lowest energy was selected as shown in Figure 5.
3.5. Elctrophoretic Mobility Shift Assay (EMSA)
We performed the experiments using RNA containing m8Gm and Cy3-labeled RNA (1 μM each) with Zα-GFP fusion protein (10 μM) in a buffer, 10 μg/mL BSA and 10% (v/v) glycerol, 100 mM NaCl, 10 mM dithiothreitol, 10 mM Tris-HCl (pH 7.0). Nondenatured polyacrylamide gel (8%) is used and performed at 80 V and 4 °C in 1× TBE buffer with 20 mM NaCl.
Supplementary Materials
The following are available online. Schemes S1 and S2: Synthetic scheme of compound 5 and 6, Figures S1–S6: 1H-NMR spectra of synthesized compounds, Figure S7: HRMS spectra of synthesized compounds, Figure S8: NOESY spectrum of m8Gm, Figure S9: CD spectra of RNA shown in Table 1 at various NaClO4 concentrations, Figure S10: H6/H2′ of C and H8/H2′ of G (8CH3/H2′ of m8Gm) proton region of NOESY spectra of r(CGC[m8Gm]CG)2, Figure S11: Monitoring of the A to Z-RNA transition by 31P-NMR, Table S1: 1H-NMR chemical shifts δH (p.p.m. of r(CGC[m8Gm]CG)2 Z-RNA.
Author Contributions
Y.X. designed the research. T.B., T.I., C.-D.X., and H.-L.B. carried out the experiments. All authors analyzed the data and wrote the manuscript.
Funding
This work was supported by JSPS KAKENHI Grant Numbers 17H03091 (to Y.X.) and 16K17938 (to T.I.). T.B. is supported by the Otsuka Toshimi Scholarship Foundation. Support from the Takeda Science Foundation is also acknowledged.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds are not available from the authors.
References
- 1.Popenda M., Biala E., Milecki J., Adamiak R.W. Solution structure of RNA duplexes containing alternating CG base pairs: NMR study of r(CGCGCG)2 and 2′-O-Me(CGCGCG)2 under low salt conditions. Nucleic Acids Res. 1997;25:4589–4598. doi: 10.1093/nar/25.22.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Popenda M., Milecki J., Adamiak R.W. High salt solution structure of a left-handed RNA double helix. Nucleic Acids Res. 2004;32:4044–4054. doi: 10.1093/nar/gkh736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz T., Rould M.A., Lowenhaupt K., Herbert A., Rich A. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999;284:1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- 4.Ha S.C., Lowenhaupt K., Rich A., Kim Y.G., Kim K.K. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005;437:1183–1186. doi: 10.1038/nature04088. [DOI] [PubMed] [Google Scholar]
- 5.Kang Y.M., Bang J., Lee E.H., Ahn H.C., Seo Y.J., Kim K.K., Kim Y.G., Choi B.S., Lee J.H. NMR spectroscopic elucidation of the B-Z transition of a DNA double helix induced by the Z alpha domain of human ADAR1. J. Am. Chem. Soc. 2009;131:11485–11491. doi: 10.1021/ja902654u. [DOI] [PubMed] [Google Scholar]
- 6.Lee M., Kim S.H., Hong S.C. Minute negative superhelicity is sufficient to induce the B-Z transition in the presence of low tension. Proc. Natl. Acad. Sci. USA. 2010;7:4985–4990. doi: 10.1073/pnas.0911528107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bae S., Kim D., Kim K.K., Kim Y.G., Hohng S. Intrinsic Z-DNA is stabilized by the conformational selection mechanism of Z-DNA-binding proteins. J. Am. Chem. Soc. 2011;133:668–671. doi: 10.1021/ja107498y. [DOI] [PubMed] [Google Scholar]
- 8.Lee A.R., Park C.J., Cheong H.K., Ryu K.S., Park J.W., Kwon M.Y., Lee J., Kim K.K., Choi B.S., Lee J.H. Solution structure of the Z-DNA binding domain of PKR-like protein kinase from Carassius auratus and quantitative analyses of the intermediate complex during B-Z transition. Nucleic Acids Res. 2016;44:2936–2948. doi: 10.1093/nar/gkw025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S.H., Lim S.H., Lee A.R., Kwon D.H., Song H.K., Lee J.H., Cho M., Johner A., Lee N.K., Hong S.C. Unveiling the pathway to Z-DNA in the protein-induced B-Z transition. Nucleic Acids Res. 2018;46:4129–4137. doi: 10.1093/nar/gky200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz T., Behlke J., Lowenhaupt K., Heinemann U., Rich A. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Biol. 2001;8:761–765. doi: 10.1038/nsb0901-761. [DOI] [PubMed] [Google Scholar]
- 11.Rothenburg S., Deigendesch N., Dittmar K., Koch-Nolte F., Haag F., Lowenhaupt K., Rich A. A PKR-like eukaryotic initiation factor 2α kinase from zebrafish contains Z-DNA binding domains instead of dsRNA binding domains. Proc. Natl. Acad. Sci. USA. 2005;2:1602–1607. doi: 10.1073/pnas.0408714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kahmann J.D., Wecking D.A., Putter V., Lowenhaupt K., Kim Y.G., Schmieder P., Oschkinat H., Rich A., Schade M. The solution structure of the N-terminal domain of E3L shows a tyrosine conformation that may explain its reduced affinity to Z-DNA in vitro. Proc. Natl. Acad. Sci. USA. 2004;101:2712–2717. doi: 10.1073/pnas.0308612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugiyama H., Kawai K., Matsunaga A., Fujimoto K., Saito I., Robinson H., Wang A.H. Synthesis, structure and thermodynamic properties of 8-methylguanine-containing oligonucleotides: Z-DNA under physiological salt conditions. Nucleic Acids Res. 1996;24:1272–1278. doi: 10.1093/nar/24.7.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawai K., Saito I., Sugiyama H. Conformation-Dependent Photochemistry of 5-Halouracil-Containing DNA: Stereospecific 2′α-Hydroxylation of Deoxyribose in Z-form DNA. J. Am. Chem. Soc. 1999;121:1391–1392. doi: 10.1021/ja9827200. [DOI] [Google Scholar]
- 15.Oyoshi T., Kawai K., Sugiyama H. Efficient C2′alpha-hydroxylation of deoxyribose in protein-induced Z-form DNA. J. Am. Chem. Soc. 2003;125:1526–1531. doi: 10.1021/ja028388g. [DOI] [PubMed] [Google Scholar]
- 16.Tashiro R., Sugiyama H. A nanothermometer based on the different pi stackings of B- and Z-DNA. Angew. Chem. Int. Ed. 2003;42:6018–6020. doi: 10.1002/anie.200352752. [DOI] [PubMed] [Google Scholar]
- 17.Xu Y., Ikeda R., Sugiyama H. 8-Methylguanosine: A powerful Z-DNA stabilizer. J. Am. Chem. Soc. 2003;125:13519–13524. doi: 10.1021/ja036233i. [DOI] [PubMed] [Google Scholar]
- 18.Xu Y., Zhang Y.X., Sugiyama H., Umano T., Osuga H., Tanaka K. (P)-helicene displays chiral selection in binding to Z-DNA. J. Am. Chem. Soc. 2004;126:6566–6567. doi: 10.1021/ja0499748. [DOI] [PubMed] [Google Scholar]
- 19.Chen F.Y., Park S., Otomo H., Sakashita S., Sugiyama H. Investigation of B-Z transitions with DNA oligonucleotides containing 8-methylguanine. Artif. DNA PNA XNA. 2014;5:e28226. doi: 10.4161/adna.28226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park S., Otomo H., Zheng L., Sugiyama H. Highly emissive deoxyguanosine analogue capable of direct visualization of B-Z transition. Chem. Commun. 2014;50:1573–1575. doi: 10.1039/c3cc48297a. [DOI] [PubMed] [Google Scholar]
- 21.Vongsutilers V., Gannett P.M. C8-Guanine modifications: Effect on Z-DNA formation and its role in cancer. Org. Biomol. Chem. 2018;16:2198–2209. doi: 10.1039/C8OB00030A. [DOI] [PubMed] [Google Scholar]
- 22.Bothe J.R., Lowenhaupt K., Al-Hashimi H.M. Sequence-specific B-DNA flexibility modulates Z-DNA formation. J. Am. Chem. Soc. 2011;133:2016–2018. doi: 10.1021/ja1073068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown B.A., Lowenhaupt K., Wilbert C.M., Hanlon E.B., Rich A. The Zα domain of the editing enzyme dsRNA adenosine deaminase binds left-handed Z-RNA as well as Z-DNA. Proc. Natl. Acad. Sci. USA. 2000;97:13532–13536. doi: 10.1073/pnas.240464097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kypr J., Kejnovska I., Renciuk D., Vorlickova M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto S., Park S., Sugiyama H. Development of a visible nanothermometer with a highly emissive 2′-O-methylated guanosine analogue. RSC Adv. 2015;5:104601–104605. doi: 10.1039/C5RA24756J. [DOI] [Google Scholar]
- 26.Cummins L.L., Owens S.R., Risen L.M., Lesnik E.A., Freier S.M., McGee D., Guinosso C.J., Cook P.D. Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1995;23:2019–2024. doi: 10.1093/nar/23.11.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uesugi S., Ohkubo M., Urata H., Ikehara M., Kobayashi Y., Kyogoku Y. Ribooligonucleotides, r(C-G-C-G) analogs containing 8-substituted guanosine residues, form left-handed duplexes with Z-form-like structure. J. Am. Chem. Soc. 1984;6:3675–3676. doi: 10.1021/ja00324a047. [DOI] [Google Scholar]
- 28.Chen G., Znosko B.M., Kennedy S.D., Krugh T.R., Turner D.H. Solution structure of an RNA internal loop with three consecutive sheared GA pairs. Biochemistry. 2005;44:2845–2856. doi: 10.1021/bi048079y. [DOI] [PubMed] [Google Scholar]
- 29.Placido D., Brown B.A., 2nd, Lowenhaupt K., Rich A., Athanasiadis A. A left-handed RNA double helix bound by the Zα domain of the RNA-editing enzyme ADAR1. Structure. 2007;15:395–404. doi: 10.1016/j.str.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.