Abstract
Motivation
Prognostic models are widely used in clinical decision-making, such as risk stratification and tailoring treatment strategies, with the aim to improve patient outcomes while reducing overall healthcare costs. While prognostic models have been adopted into clinical use, benchmarking their performance has been difficult due to lack of open clinical datasets. The recent DREAM 9.5 Prostate Cancer Challenge carried out an extensive benchmarking of prognostic models for metastatic Castration-Resistant Prostate Cancer (mCRPC), based on multiple cohorts of open clinical trial data.
Results
We make available an open-source implementation of the top-performing model, ePCR, along with an extended toolbox for its further re-use and development, and demonstrate how to best apply the implemented model to real-world data cohorts of advanced prostate cancer patients.
Availability and implementation
The open-source R-package ePCR and its reference documentation are available at the Central R Archive Network (CRAN): https://CRAN.R-project.org/package=ePCR. R-vignette provides step-by-step examples for the ePCR usage.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
There is increasing interest in open sharing of clinical trial and patient registry data for improving clinical study designs as well as patient management and outcomes (Bertagnolli et al., 2017; Laajala et al., 2017). Application of prognostic models to integrated patient data with increased sample size has the potential to provide novel insights into disease pathophysiology and to identify clinical variables associated with patient outcomes that have been missed by earlier single-cohort investigations. These insights and factors have the potential to improve future clinical trial designs, for instance, by homogenizing risk groups, thus addressing trial questions of treatment efficacy or predictive markers more efficiently (Khozin et al., 2017).
In the recent DREAM 9.5 Prostate Cancer Challenge (PCC-DREAM), over 50 international teams participated to develop prognostic models for overall survival (OS) of mCRPC patients using data from four phase III clinical trials, containing >2000 advanced mCRPC patients treated with docetaxel (Guinney et al., 2017). Our top-ranked model was based on an ensemble of penalized Cox regressions (ePCR), and it significantly outperformed the other submitted models and a previous state-of-the-art model (Halabi et al., 2014). The ePCR model makes use of inter-variable interactions and advanced multi-variable machine learning to identify marker combinations with greatest predictive power for the patients’ treatment outcome.
In this Application Note, we make available an open-source CRAN-package of the ePCR methodology. The model was originally developed for the clinical trials in PCC-DREAM Challenge. Here, we provide novel extensions of the ePCR model and demonstrate how to best apply the model to more heterogeneous, real-world prostate cancer patient registry cohorts. In addition to testing the real-world performance of ePCR, we also investigate the modelling and data processing options that affect its prognostic accuracy in such real-world data. Our results show how the inherent differences between clinical trial data and real-world patient registry data should be taken into account when using prognostic models in clinical decision making.
2 Implementation of the ePCR package
The original ePCR model was developed using the clinical trial datasets from the PCC-DREAM Challenge, hosted by Project Data Sphere (PDS, https://www.projectdatasphere.org/), a broad-access research platform that collects and curates patient-level data from completed, phase III cancer clinical trials. The original model included 101 clinical variables, such as demographics, lab values and lesion measures. Compared to the LASSO-regularized reference model that modelled eight clinical variables only (Halabi et al., 2014), the ePCR model learning is based on a more comprehensive model optimization, which also selects correlated groups of predictors and their interactions. The ePCR methodology makes use of an ensemble structure, in which each patient cohort and their combinations were modelled as separate components to account for stratification factors due to intrinsic characteristics specific to each clinical dataset (Guinney et al., 2017). This ensemble approach is also useful when modelling multiple real-world patient cohorts, each including potentially different sets of clinical variables (e.g. completely missing variables that cannot be imputed).
The CRAN-package implements both the original ePCR model, estimated based on clinical trial datasets used in the PCC-DREAM Challenge, as well as two new models, based on the Finnish real-world prostate cancer patient cohorts presented in this report (see Section 3). The implementation enables the user to freely adjust various modeling parameters, including the type of missing value imputation method (median or k-NN imputation), L1/L2 regularization parameter optimization (α parameter to weight the L1 and L2 penalties), performance metrics [time-integrated area under curve (iAUC) or concordance-index], and the specific use of cross-validation and other modeling schema supporting the model estimation (see Appendix 2 of R-vignette for functionality comparison against the code provided in Guinney et al., 2017). Various diagnostic visualization options are also provided to investigate data clustering (PCA plots), model fitting (box plots), patient outcomes (Kaplan-Meier plots) and time-dependent prediction accuracy (iAUC plots). The package also contains benchmarking data simulated based on the real-world registry data, which have been made available to test the open-source implementation (see Supplementary Data for details).
3 Application to a real-world patient cohort
As an application use case, we investigated how to best use the ePCR model trained on the clinical trial data from the PCC-DREAM Challenge for prognostic modelling of survival of CRPC patients using independent, real-world hospital registry data (see Supplementary Data). This investigation was inspired by recent work by Seyednasrollah et al. (2017), where the authors made use of our original, PCC-DREAM Challenge winning model for prognostic prediction of mCRPC patients treated with docetaxel in the same hospital registry data. We used these published results as a baseline, and carried out further analyses to investigate which modelling options and patient selection criteria could enhance prediction accuracy for OS. These follow-up analyses revealed marked improvements in the ePCR model performance (Fig. 1A, left panel), which may partly be due to the differences in the number of patients selected based on the medication information only (Fig. 1D, n = 180 vs. n = 289).
Fig. 1.
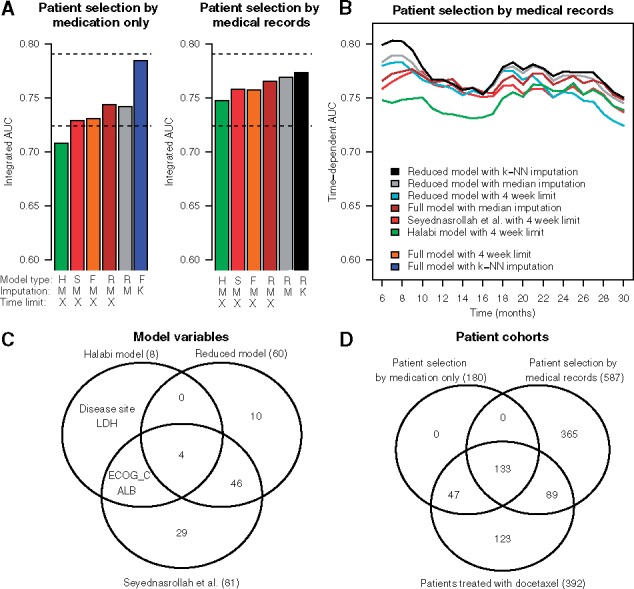
(A) Predictive accuracy evaluated in terms of integrated area under curve (iAUC) over 6–30 months follow-up period with various modelling options. The DREAM clinical trial estimated models were applied separately to two real-world CRPC patient cohorts, selected by medication information only (left barplot, n = 180) or based on the full medical records (right barplot, n = 587). Model type: H, Halabi et al. (2014); S, ePCR model with 81 variables used in Seyednasrollah et al. (2017); F, full ePCR model with all the 101 variables available in the real-world cohort; R, reduced ePCR model including those 60 variables that were available for at least 60% of patients in the cohort selected by medical records. Imputation: M, median imputation; K, k-nearest neighbor imputation (k = 10). Time limit: X, 4 week time limit for the baseline measurements before the docetaxel treatment (left) or castration resistance (right). The two horizontal dotted lines indicate the ePCR model accuracy reported in Seyednasrollah et al. (2017) (iAUC = 0.724), and the best accuracy obtained in the PCC-DREAM Challenge clinical trial data (Guinney et al., 2017) (iAUC = 0.791). The bar colors correspond to those of panel B. The rightmost bars use either the full model (blue) or the reduced model (black), with k-NN imputation and no time limit, which were selected in the same real-word patient data, so these results may be overly-optimistic. (B) Examples of the ePCR models’ accuracy for predicting OS at various follow-up time points when applied to the patient cohort selected by full medical records (n = 587). The colors correspond to those of panel A. (C) The number and overlap of variables in different models (see Supplementary Table S1 for the variable labels). (D) The number and overlap of patients in different patient cohorts (n)
Notably, a reduced model with 60 variables (out of the 101 in the original ePCR model) led to improved predictive accuracy in this real-world cohort, compared to the Seyednasrollah et al. (2017) version of ePCR model (81 variables) and the Halabi et al. model with 8 variables (Fig. 1C). We obtained further improvements by using all the available baseline clinical data, instead of limiting to 4 weeks prior to treatment (as was done in Seyednasrollah et al., 2017), and by using the k-nearest neighbor (k-NN) imputation, instead of median imputation (as was done in Seyednasrollah et al., 2017). Since the relatively small patient cohort selected using medication information (n = 180) may lead to over-optimistic results, we repeated the same analyses in a larger CRPC patient cohort (n = 587), which resulted in more consistent iAUC levels (Fig. 1A, right panel). Strikingly, the model that performed best in the smaller cohort was found to be sub-optimal in this larger patient cohort across various follow-up periods (Fig. 1B).
Although these optimized results in the real-world patient data are already close to those obtained in the PCC-DREAM Challenge external validation trials (ENTHUSE M1 placebo arm, iAUC = 0.768), there is still room for improvement, as compared to the best performance obtained in the Challenge scoring cohort (ENTHUSE 33 trial docetaxel arm, iAUC = 0.791; Fig. 1A, the top dotted line). For instance, the model-based imputation developed for the clinical trial datasets did not perform well in this real-world patient data, suggesting that better approaches to deal with larger blocks of missing values in registry data are required. For instance, aspartate aminotransferase (AST) was found as an important, novel factor in the PCC-DREAM clinical trial datasets (Guinney et al., 2017), but since it is rarely measured in laboratory tests in Finland, AST levels could not be accurately imputed or used in these real-world cohorts.
4 Conclusion
The ePCR methodology and its applications provide clinical researchers and practitioners with practical means and guidelines on how to best apply the prognostic model to real-world prostate cancer patient cohorts. We also encourage the community to test the ePCR model in other types of cancer cohorts to extend its real-world performance evaluation beyond the advanced PC. Although the ePCR model was originally developed for clinical trial data from docetaxel-treated mCRPC patients, the current results demonstrate its applicability also to more heterogeneous hospital registry cohorts of advanced prostate cancer patients (Fig. 1). Docetaxel has been the first line chemotherapy in Finland since 2004 for non-metastatic CRPC, and since 2015 for primary metastatic prostate cancer.
While the specific focus here was on Finnish prostate cancer patients, the implementation should prove useful more globally and in other cancers, and the results additionally provide guidance for other similar projects that aim to develop predictive models based on hospital registry data. We are happy to help researchers to guarantee the best use of the ePCR model. Even though the open-source implementation is available for anyone to apply or modify, it should be in our common interests to optimize modeling options to obtain the best possible results, especially as the findings may affect the management of patients with lethal cancers. It will also be important to further develop the model in larger representative cohorts, once available, to learn how to make the best use of real-world hospital registry data for improved prognostic modelling.
Supplementary Material
Acknowledgements
The authors thank the PCC-DREAM Challenge organizers for their help with the ePCR model post-challenge development and validation; James C. Costello (University of Colorado Denver) for providing comments on the manuscript; Anna Hammais (Turku University Hospital, Centre for Clinical Informatics) for extracting and harmonizing the clinical data used in the study; Tuomas Mirtti (Helsinki University Hospital and HUSLAB) for clinical expertise and consultation, as well as Fatemeh Seyednasrollah and Mehrad Mahmoudian (Turku Centre for Biotechnology) for their explanation on how they used our ePCR model in their work.
Funding
This work was supported by the Academy of Finland (grants 295504 and 310507), Cancer Society of Finland, Sigrid Juselius Foundation, Finnish Cultural Foundation and the National Cancer Institute (16X064).
Conflict of Interest: Teemu D. Laajala and Tero Aittokallio participated in the same DREAM 9.5 Prostate Cancer Challenge as the team of Seyednasrollah et al.
References
- Bertagnolli M.M., et al. (2017) Advantages of a truly open-access data-sharing model. N. Engl. J. Med., 376, 1178–1181. [DOI] [PubMed] [Google Scholar]
- Guinney J., et al. (2017) Prediction of overall survival for patients with metastatic castration-resistant prostate cancer: development of a prognostic model through a crowdsourced challenge with open clinical trial data. Lancet. Oncol., 18, 132–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halabi S., et al. (2014) Updated prognostic model for predicting overall survival in first-line chemotherapy for patients with metastatic castration-resistant prostate cancer. J. Clin. Oncol., 32, 671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khozin S., et al. (2017) From big data to smart data: FDA’S INFORMED initiative. Nat. Rev. Drug Discov., 16, 306. [DOI] [PubMed] [Google Scholar]
- Laajala T.D., et al. (2017) Community mining of open clinical trial data. Oncotarget, 8, 81721–81722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyednasrollah F., et al. (2017) How reliable are trial-based prognostic models in real-world patients with metastatic castration-resistant prostate cancer? Eur . Urol., 71, 838–840. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


