Abstract
Leishmania donovani is responsible for visceral leishmaniasis, a neglected and lethal parasitic disease with limited treatment options and no vaccine. The study of L. donovani has been hindered by the lack of a high-quality reference genome and this can impact experimental outcomes including the identification of virulence genes, drug targets and vaccine development. We therefore generated a complete genome assembly by deep sequencing using a combination of second generation (Illumina) and third generation (PacBio) sequencing technologies. Compared to the current L. donovani assembly, the genome assembly reported within resulted in the closure over 2,000 gaps, the extension of several chromosomes up to telomeric repeats and the re-annotation of close to 15% of protein coding genes and the annotation of hundreds of non-coding RNA genes. It was possible to correctly assemble the highly repetitive A2 and Amastin virulence gene clusters. A comparative sequence analysis using the improved reference genome confirmed 70 published and identified 15 novel genomic differences between closely related visceral and atypical cutaneous disease-causing L. donovani strains providing a more complete map of genes associated with virulence and visceral organ tropism. Bioinformatic tools including protein variation effect analyzer and basic local alignment search tool were used to prioritize a list of potential virulence genes based on mutation severity, gene conservation and function. This complete genome assembly and novel information on virulence factors will support the identification of new drug targets and the development of a vaccine for L. donovani.
Introduction
Visceral Leishmaniasis (VL) is the second most lethal parasitic disease following malaria and is prevalent throughout underdeveloped and tropical regions of the world. There are some 300,000 new cases each year1 and Leishmania donovani, transmitted by the infected sand fly, is the major causative agent of VL in the Indian and African continents. Although L. donovani is extensively studied, its genome remains poorly annotated because it is heavily fragmented and a complete assembly is crucial to understanding this parasite’s biology, metabolic pathways, tissue tropism and disease pathology.
The pathology of leishmaniasis is predominantly parasite species-specific, such as for example L. major that causes cutaneous leishmaniasis (CL) whereas L. donovani typically causes lethal visceral leishmaniasis (VL). Previous studies have compared genomes of L. major and L. donovani parasites to study virulence and disease tropism and have identified a number of species specific genes including A2 present in L. donovani that is a pseudogene in L. major2,3. More recently, as cases of atypical CL caused by L. donovani have emerged, studies have compared cutaneous and visceral disease-causing strains of L. donovani, as these strains provide a unique opportunity to study the genetic determinants of disease pathogenesis using more recently diverged strains4.
Second-generation sequencing technologies including Illumina, have made the sequencing of large genomes feasible through the mapping of short sequence reads of 50 to 250 nucleotides (nt) to a reference genome5. While human and many other well studied higher vertebrates have better assembled reference genomes6, the kinetoplastids suffer in this regard because most Leishmania species either lack sequencing information altogether or have incomplete reference genomes with sometimes thousands of sequence gaps7. The current L. donovani reference genome (ASM22713v2 from strain BPK282A8) was generated using second generation technologies and contains over 2,000 gaps and therefore there are many incomplete or inaccurate protein coding sequences. The first complete Leishmania genome generated is that of L. major by a consortium of laboratories employing large insert clone tiling paths to sequence each chromosome individually9,10. This genome was later improved by the reassembly of complex collapsed loci that were incorrect in the original reference genome11.
Since then however, advances in sequencing technologies have drastically reduced the cost of sequencing and eased genome assembly tasks by increasing the length of the generated sequences. Long read sequencing or “third-generation” sequencing refers to more recent technologies including Oxford NanoPore12 and Pacific Biosciences (PacBio)13 that can result in reads ranging up to 50 kb or 100 kb that are capable of generating more complete genomic assemblies, provided the read lengths traverse across repetitive elements. One such highly repetitive cluster is the A2 gene family from L. donovani considered to be an important virulence factor and is necessary for survival in visceral organs14,15 and protection against host response stress16,17. Due to its repetitive nature, the A2 gene cluster is misassembled in all Leishmania genomes generated using second generation sequencing, and only resolved in a recent resequencing effort targeted to L. infantum exploiting the long-read capabilities of PacBio sequencing which resulted in a complete genome assembly18. The current L. donovani genome however was obtained from second generation sequencing and consequently, no precise DNA or complete protein sequences are available for any A2 protein in L. donovani, hindering the comparison of A2 genes in visceral disease-causing strains or using mass spectrometry to identify A2 proteins which relies on accurate genome sequences for protein identification.
In this study, we have combined second and third generation sequencing to generate a complete assembly of the L. donovani genome from the strain responsible for cutaneous leishmaniasis (CL) in Sri Lanka4,19. This new assembly enabled the generation of an improved genome annotation and an unbiased analysis of chromosome synteny comparing L. donovani and L. major genes and strand switch transcription units. We have used this complete assembly to re-interrogate the genetic makeup of the visceral and cutaneous disease-causing L. donovani strains resulting in the identification of novel SNPs and indels generating a more complete and accurate chromosome map of the genetic differences between these phenotypically distinct L. donovani strains4,20. This study further enabled re-annotation of much of the genome highlighting the importance of a complete reference assembly to support future functional genomic and proteomic studies involving the L. donovani pathogen.
Results
A complete L. donovani genome assembly
The currently available assembly for L. donovani (ASM22713v2 from strain BPK282A8) contains over 2,000 gaps due to the presence of low complexity regions and the highly repetitive nature of the Leishmania genome21. This incomplete assembly makes it difficult to compare L. donovani genomes from strains with different phenotypic properties. DNA was therefore isolated from the attenuated cutaneous disease-causing strain of L. donovani from Sri Lanka4 and was subjected to deep sequencing using second and third generation sequencing. We reasoned that a complete assembly of the genome from this attenuated L. donovani strain will identify a more complete complement of genetic changes associated with loss of virulence of this strain. A total of 9 PacBio sequencing runs were performed generating 712,443 reads representing an estimated 107-fold coverage of the estimated 35 Mb genome. Importantly, there were 51,484 reads longer than 12 kb, representing a 20-fold coverage in very long reads. The long-read sequencing data was assembled using various assemblers as described in methods and merged using the longest chromosomes produced by each assembler followed by refinement using the high-quality short-read Illumina-generated data and iterative edge extension to close the remaining gaps.
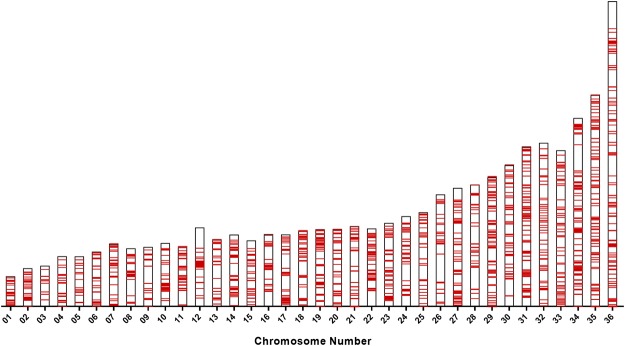
The previous L. donovani reference assembly (ASM22713v2 from strain BPK282A) had over 2000 gaps spread across the 36 chromosomes. Figure 1 depicts the location on each chromosome of the gaps that have been closed in the new assembly reported here. The new assembly now contains contiguous DNA sequences in all 36 chromosomes and a corresponding 22-fold increase in N50 indicating that a larger proportion of the data has been assembled into large contigs as 50% of the genome is contained in contigs >= N50, resulting in an N50 of over 1Mbp (Table 1). Further, using this completed assembly, we have generated annotations for more potential protein coding regions than previously annotated (8,633 compared to 7,969 proteins) and identified more transfer-RNA and ribosomal RNA genes as well as all 6 small nuclear RNA genes, all spliced leader RNA genes and close to a thousand small nucleolar RNA genes. An additional 13 genes were marked as pseudogenes due to multiple internal stop codons and/or frameshifts. (Supplementary Table S1). Alignment of the second-generation Illumina reads to the PacBio generated assembly was used to cross-validate and correct the assembly at the nucleotide level. Graphs of the coverage from the alignment of Illumina and PacBio data across the 36 chromosomes are available in Supplementary Fig. S1. Taken together, we consider this new assembly to be contiguous and complete.
Figure 1.
Location of the gaps along 36 chromosomes that have been closed in this new assembly. Chromosomal locations of gaps are indicated in red. No gaps remain in the current assembly.
Table 1.
Quality assessment metrics of the previous and current assemblies.
| Contigs | N50 (bp) | Protein coding | tRNA | rRNA | snRNA | SLRNA | snoRNA | Genes mapped | |
|---|---|---|---|---|---|---|---|---|---|
| Old Assembly | 2,154 | 45,436 | 7,969 | 64 | 11 | 4 | — | 31 | 8,081 |
| New Assembly | 36 | 1,067,468 | 8,633 | 90 | 51 | 6 | 68 | 910 | 9,758 |
Old assembly refers to ASM22713v2 from strain BPK282A, new assembly refers to the assembly presented in this work. Contigs denotes the number of genomic fragments uninterrupted by stretches of unknown bases (Ns) or chromosome ends. N50 is used as a measure of contiguity, 50% of the genome is contained in contigs of size N50 and above. Annotated genes were broken down into protein coding, transfer-RNA (tRNA), ribosomal RNA (rRNA), small nuclear RNA (snRNA), spliced leader RNA (SLRNA) and small nucleolar RNA (snoRNA) genes. The number of genes mapped indicates the number of annotated genes along the genome.
Assembly of the A2 virulence gene cluster and synteny comparison between L. major and L. donovani
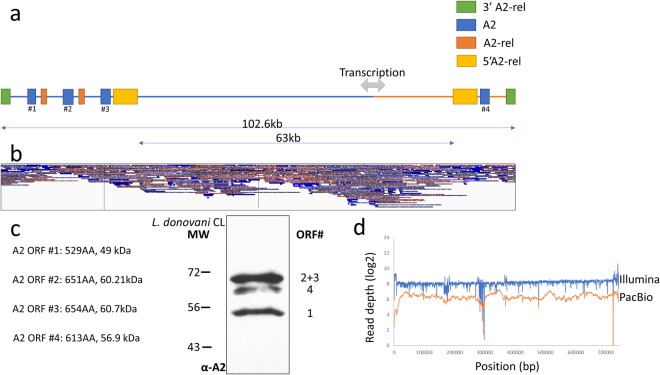
A2 is a major virulence factor required for L. donovani survival in visceral organs22. The A2 gene family cluster on chromosome 22 has recently been assembled for L. infantum18, however has not been for L. donovani. We therefore investigated whether the structure of this region could be determined with this revised assembly. It was advantageous that the attenuated cutaneous L. donovani strain used in this assembly has fewer copies of the A2 genes than other virulent strains of L. donovani4. As shown in Fig. 2a, the new assembly could read-through the entire cluster of highly repetitive A2 and flanking sequences and could position the A2 genes and interspersed flanking 3′ A2-rel and 5′ A2-rel genes. The A2 genes are contained in two opposite facing clusters on either side of a strand-switch locus consisting of one cluster of 3 copies and one cluster with a single A2 gene. The long sequence reads generated by the PacBio sequencing were crucial in generating the assembly of the A2 genes where reads of 11 kb and longer are shown spanning the repetitive cluster (Fig. 2b).
Figure 2.
Organization of the 4 copies of the A2 gene on chromosome 22 in the attenuated cutaneous L. donovani strain. (a) Locations of the 4 A2 genes are shown in blue and numbered 1–4. Interspaced A2-rel genes are labeled in orange, 3′ A2-rel genes are labeled in green and 5′ A2-rel genes are labeled in yellow. A2-rel genes have no homology with A2 genes15. Transcription direction is shown according to strandedness: blue represents reverse strand direction of transcription, red represents forward strand transcription. The genes located in the 63 kb region between opposing A2 clusters are not depicted for clarity. (b) Alignment of the longest (~11 kb+) PacBio reads to the A2 clusters. Reads in the 5′ to 3′ direction labeled in red; reads in the 3′ to 5′ direction labeled in blue. (c) Western blot analysis of A2 proteins in the attenuated cutaneous L. donovani strain. The sizes of the A2 proteins are consistent with the ORFs and number of A2 genes identified in this assembly. (d) Coverage graph of chromosome 22 using Illumina (blue) and PacBio (orange) reads.
To generate supporting evidence for this A2 gene assembly, Western blot analysis of the A2 proteins from this strain was performed to compare the number and sizes of the A2 proteins with the predicted molecular weights from this assembly (ORFs; Supplementary Fig. S2). As shown in Fig. 2c, the apparent molecular weights from Western blotting correspond to the sizes predicted from the sequenced ORFs. The 3 bands on the Western blot are consistent with the molecular weights of the 4 gene products as the A2 gene copies 2 and 3 encode proteins of a similar size (Supplementary Fig. S2). This represents the first complete structure and sequence for A2 genes in L. donovani, a prototype virulence factor. The difficulty in assembling this complex region is demonstrated in Fig. 2d, where a deviation from the average read coverage can be seen around the 300,000 bp position, in and around the A2 cluster, due to difficulties in the aligner assigning a unique position to similar reads across a repetitive region.
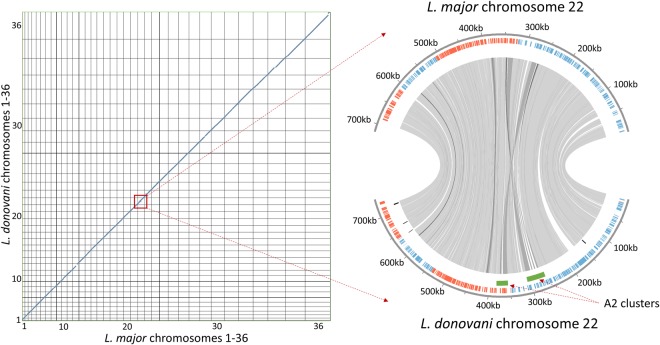
Directly comparing synteny at the chromosomal level was not possible with the previous L. donovani assembly due to the heavy fragmentation of the genome. With the new L. donovani assembly, it was possible to accurately compare chromosome synteny between L. donovani and L. major. As shown in Fig. 3, the genome of L. donovani, exhibited a very high level of synteny with the L. major. Chromosome 22 was highlighted here because this is the location of the A2 genes that have become pseudogenes in L. major and have therefore diverged between these old-world species15. The level of synteny demonstrated here for chromosome 22 was maintained on all other chromosomes (Supplementary Fig. S3). These results indicate that evolution between cutaneous and visceral pathologies by different Leishmania species resulted largely from SNPs, pseudogenes and copy number variation and not from large changes such as chromosome rearrangements or complete gene deletions/insertions.
Figure 3.
L. donovani maintains high levels of synteny with L. major including chromosome 22 where the A2 genes are located. Left: Dot plot of the coding DNA sequences of L. major compared to those of L. donovani generated from our assembly across the entire genome. Right: Synteny comparison of chromosome 22. The outer most circle represents the chromosomal location. The second circle is labelled with genes on the forward strand (blue) and genes on the reverse strand (red). The third circle represents genes that are only present in one of the two compared species. The inner association lines join syntenic genes between the two species.
Identification of new genes and improvements in annotations
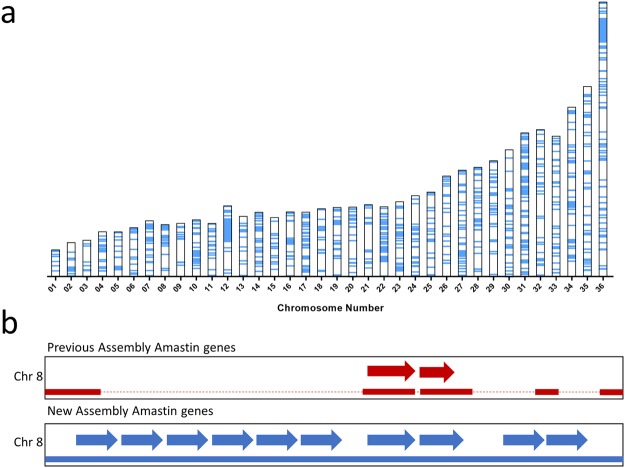
As this assembly was larger in terms of total number of bases covered and more contiguous due to the removal of sequence gaps, the impact this had on gene annotations was investigated. The genome from the new assembly was annotated using the Companion pipeline23 and the new and previous annotations (GenBank: GCF_000227135.1) were then aligned together and overlapping annotations were removed. Remarkably, close to 15% of the L. donovani protein coding genes had new or edited annotations as shown in Fig. 4a. Part of this increase in number of annotations resulted from the expansions of multi-copy gene families beyond the copy numbers in the previous annotation. An example is shown in Fig. 4b where there are 10 amastin genes identified in this new assembly compared to the previously identified 2. These results support the use of this assembly as the reference for bioinformatic analysis as it provides a more complete and accurate annotation of the L. donovani genome ORFs.
Figure 4.
The new L. donovani genome assembly results in a significant change in gene annotations. (a) New or improved gene annotations are highlighted in Blue along the 36 chromosomes. Compared to the previous L. donovani reference assembly (ASM22713v2 from strain BPK282A1), there were 1,087 protein coding genes unannotated or differently annotated in the current assembly. Unannotated or differently annotated genes were obtained by removing all annotations generated from our assembly that shared 95% or greater similarity to those previously available8. (b) Expansion of the amastin gene cluster on chromosome 8. Top track contains the previously two known coding sequences aligned to the previous L. donovani reference assembly (ASM22713v2 from strain BPK282A1). Gaps in the previous assembly depicted as dotted lines. Bottom track contains 10 amastin genes identified in the updated assembly. One previously identified Amastin gene has been aligned, 1 has been expanded and 8 have been annotated de novo.
Comparison of virulent and attenuated L. donovani parasites
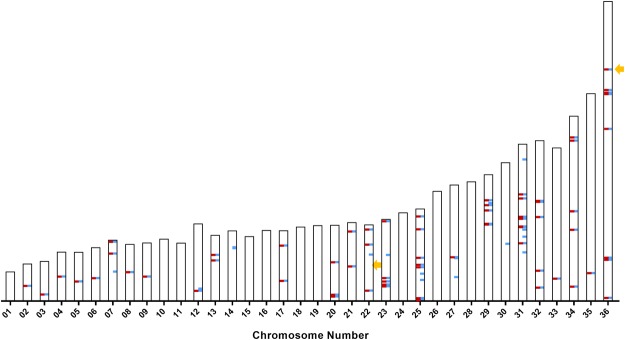
As indicated above, there are 2 distinct strains of L. donovani in Sri Lanka where one is responsible for visceral leishmaniasis (VL) and the other for cutaneous leishmaniasis (CL)4. Subsequently, the CL strain was experimentally passaged through the visceral organs of BALB/c mice to select for a gain-of-function strain with increased virulence (IV strain) for survival in visceral organs where it was revealed through proteomic analysis that the resulting IV strain had an increase in stress response and antioxidant proteins24. Illumina whole genome sequencing and comparative genomic analysis of the VL, CL and IV strains was performed to identify SNPs associated with a change in virulence for survival in the visceral organs. As shown in Fig. 5, all 70 of the previously identified homozygous SNP differences between the VL and CL strains4 were confirmed in this new assembly and an additional 15 novel SNPs within protein coding genes were found using this complete assembly. In addition, there were 12 mutations associated with the IV strain with gain-of-function for increased survival in visceral organs not labeled in Fig. 5; four were heterozygous but with frequencies changing towards the VL genotype (IV → VL), four were heterozygous but present only in the IV strain and four were homozygous deletions in the IV strain. The newly identified differences between the VL and CL strains and the ones contributed from the IV strain are summarized in Table 2.
Figure 5.
Verification of previously identified SNPs and location of new SNPs that differ between the virulent VL and attenuated CL strains of L. donovani. Chromosomal location of previously identified homozygous non-synonymous SNPs between the cutaneous and visceral disease derived L. donovani strains (Red)4 compared to the novel SNPs identified only in this study (Blue) (synonymous and heterozygous codon changes identified are not labeled). Note that all the previously identified SNPs were also identified, or confirmed, in this study. 70 SNPs were previously identified across 66 genes. The same 70 SNPs were identified in this study, with an additional 15 novel SNPs not previously seen specific to the cutaneous strain. Genomic locations of SNPs identified in the previous study were translated to new genomic coordinates based on the new assembly for consistency. Arrows in yellow highlight the position of the previously identified RagC SNP on chromosome 36 and the A2 copy number difference on chromosome 22.
Table 2.
Summary of novel mutations identified in this study.
| Chr | Gene | Mutation | PROVEAN | Protein Name |
|---|---|---|---|---|
| 7 | LdBPK_070700 LdCL_070011900 | Ala282Val | −0.743 | vacuolar-type Ca2 ± ATPase, putative |
| 12 | LdBPK_120275 LdCL_120008300 | Glu1157Asp | −0.258 | Myotubularin-related protein, putative |
| 14 | LdCL_140017600 | Ser2919fs | N/A | kinesin k39 |
| 14 | LdBPK_141190 LdCL_140017700 | Glu1034Asp | −1.06 | kinesin K39 |
| 22 | LdBPK_220840 LdCL_220015800 | Pro219F/S | N/A | hypothetical protein |
| 23 | LdCL_230017500 | INS:446Glua | −12.453 | sucrose hydrolase-like protein |
| 25 | LdBPK_250620 LdCL_250011400 | Ala969Glu | 0.736 | Raptor N-terminal CASPase like domain containing protein |
| 25 | LdBPK_250790 LdCL_250013200 | INS :110 Ala, Asn, Ser, Ala, Ala, Ala, Ala | N/A | hypothetical protein |
| 27 | LdBPK_270830 LdCL_270014900 | Ala1493Thr | −0.25 | ATP-binding cassette protein subfamily A |
| 29 | LdCL_290028400 | Thr208Ala | 0.4 | VIT family putative |
| 301 | LdBPK_301640 LdCL_300021700 | Gln334STOPa | N/A | hypothetical protein |
| 31 | LdBPK_311390 LdCL_310020800 | STOP1486Leu,Ser,His | 0 | hypothetical protein |
| 31 | LdBPK_311470 LdCL_310021600 | Thr498Alaa | −0.15 | hypothetical protein |
| 31 | LdBPK_311470 LdCL_310021600 | His497Arga | 0.942 | hypothetical protein |
| 31 | LdBPK_311470 LdCL_310021600 | Gly380Aspa | −0.383 | hypothetical protein |
| IV → VL mutations | ||||
| 23 | LdBPK_230830 LdCL_230014900 | Asp712Glu | −1.625 | hypothetical protein, unknown function |
| 31 | LdBPK_312870 LdCL_310037100 | Met189Thr | −4 | hypothetical protein, unknown function |
| 31 | LdBPK_313290 LdCL_310041200 | Val187Phe | −0.634 | Hypothetical protein |
| 34 | LdBPK_342210 LdCL_340029800 | Thr116DEL | −1.098 | hypothetical protein |
| IV-Only mutations | ||||
| 14 | LdBPK_140470 LdCL_140010000 | Gln89Lys | −0.044 | cystathionine beta-lyase-like protein |
| 31 | LdBPK_312810 LdCL_310036400 | Cys173Phe | −9.5 | regulator of chromosome condensation (RCC1) repeat, putative |
| 32 | LdBPK_312770 LdCL_310035800 | Gly667Ser | −1.292 | hypothetical protein |
| 32 | LdBPK_324000 LdCL_320046000 | Val250Ile | 0 | hypothetical protein, unknown function |
| 36 | LdBPK_361580 LdCL_360021300 | Gene deletion | N/A | Serine/Threonine Kinase, putative |
| 36 | LdBPK_361590 LdCL_360021400 | Gene deletion | N/A | Serine/Threonine Kinase, putative |
| 36 | LdBPK_361600 LdCL_360021500 | Gene deletion | N/A | Engulfment and cell motility domain 2, putative |
| 36 | LdBPK_361610 LdCL_360021600 | Gene deletion | N/A | Predicted tripartite motif protein |
All mutations are annotated using VL as the wild type amino acids and CL as the mutated amino acids. Genes with annotations in the previous assembly list the previous gene ID in italic, genes annotated only in this assembly list only one gene ID. The top segment lists fifteen attenuated cutaneous strain specific mutations identified in this study. Mutations marked witha appear at 50% but also co-occur with gene duplication event and are therefore possibly homozygous on one copy. ‘INS’ denotes amino acid insertions, ‘F/S’ denotes frameshifts, ‘DEL’ denotes amino acid deletions. The middle segment lists four mutations where the gain-of-function IV strain’s genotype changed towards that of the visceral genotype. The bottom segment lists eight mutations present only in the gain-of-function IV strain and likely represents adaptations specific to the murine host. Calculated PROVEAN scores are shown in the fourth column, scores below the −2.5 threshold for deleterious mutations are highlighted in bold25.
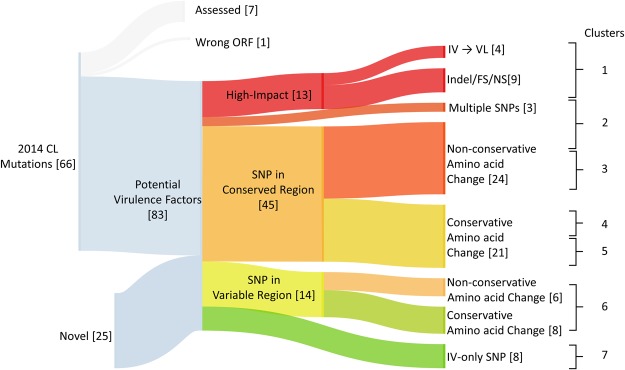
Combining data from the previous and current analysis, all the genes with genetic differences were organized into priority clusters based on the likelihood to affect protein function and phenotype (Fig. 6). A detailed list of the genes and cluster assignments is shown in Table 3. From the 66 genes containing 70 SNPs previously identified, 7 were previously experimentally assessed using gene replacement with a wildtype copy for virulence in visceral organs4 and one gene was identified as a misannotation and was therefore removed from the list. In decreasing order of priority, 13 genes in the highest impact cluster (red) were characterized as potentially having a major effect on protein function due to large amino acid changes or co-occurrence of mutations in both the VL and IV strains. SNPs in common between the IV and VL strains (4 IV → VL in the red cluster) indicate a selection associated with survival in visceral organs during the experimental passaging of the CL strain in mouse visceral organ24. Nine genes either with multiple co-occurring SNPs or non-conservative amino acid changes in conserved domains with a high score as assessed by PROVEAN software were placed in the second highest priority cluster. As detailed in methods, PROVEAN is a bioinformatic tool that classifies the significance of specific genetic mutations with respect to protein function25. Eighteen genes with non-conservative amino acid changes occurring in conserved domains but scored as unlikely to have a large effect on protein function by PROVEAN were placed in Cluster 3. Twenty-one genes with conservative amino acid changes in conserved domains were divided between Cluster 4 and Cluster 5 based on PROVEAN scores and 14 genes with mutations in domains with higher variability were placed in Cluster 6. Four mutations seen solely in the IV strain but not in the VL or CL strains are likely to be the result of random mutation or adaptation specific to the murine host were placed in Cluster 7.
Figure 6.
Summary of all genes with non-synonymous mutations between the cutaneous, visceral, and gain-of-function strains of L. donovani. All non-synonymous SNPs and Indels were classified as common to our previous study (2014 CL4) or identified in this study (Novel), as well as by their effect on amino acid changes from top to bottom, colored red to green in descending order of likelihood to affect the phenotype of the parasite. 66 genes were common to the previous data set. Of those genes, 7 were previously investigated4 and 1 was rejected due to an open reading frame misannotation. 25 genes were only listed in this study (Novel). Diagram created using SankeyMATIC (http://sankeymatic.com).
Table 3.
Summary of all genes containing mutations in the cutaneous isolates and classification into clusters.
| Cluster Number | Cluster Mutation Type | New annotation | Equivalents (when available) |
|---|---|---|---|
| Cluster 1 (13) | Nonsense, Frameshift, Insertions, Deletions, IV to VL | LdCL_300021700 | LdBPK_301640 |
| LdCL_310020800 | LdBPK_311390 | ||
| LdCL_250013200 | LdBPK_250790 | ||
| LdCL_310020100 | LdBPK_311320 | ||
| LdCL_310022200 | LdBPK_311510 | ||
| LdCL_080011700 | LdBPK_080670 | ||
| LdCL_340029800 | LdBPK_342210 | ||
| LdCL_230014900 | LdBPK_230830 | ||
| LdCL_310037100 | LdBPK_312870 | ||
| LdCL_220015800 | — | ||
| LdCL_140017600 | — | ||
| LdCL_230017500 | — | ||
| LdCL_310041200 | LdBPK_313290 | ||
| Cluster 2 (9) | Multiple SNPs in the same gene, Non-conservative amino acid change in conserved region with good PROVEAN score | LdCL_270015000 | LdBPK_270840 |
| LdCL_290026900 | LdBPK_292100 | ||
| LdCL_310021600 | LdBPK_311470 | ||
| LdCL_310028800 | LdBPK_312080 | ||
| LdCL_340046300 | LdBPK_343690 | ||
| LdCL_290022800 | LdBPK_291720 | ||
| LdCL_310024300 | LdBPK_311710 | ||
| LdCL_360006000 | LdBPK_360120 | ||
| LdCL_360062000 | LdBPK_365480 | ||
| Cluster 3 (18) | Non-conservative amino acid change in conserved region with poor PROVEAN score | LdCL_070018300 | LdBPK_071330 |
| LdCL_320013800 | LdBPK_320820 | ||
| LdCL_250016900 | LdBPK_251150 | ||
| LdCL_220022000 | LdBPK_221470 | ||
| LdCL_250015300 | LdBPK_251000 | ||
| LdCL_040011100 | LdBPK_040560 | ||
| LdCL_360016300 | LdBPK_361120 | ||
| LdCL_200014300 | LdBPK_200960 | ||
| LdCL_090011700 | LdBPK_090660 | ||
| LdCL_130016200 | LdBPK_131090 | ||
| LdCL_340009000 | LdBPK_340390 | ||
| LdCL_130017800 | LdBPK_131230 | ||
| LdCL_230009900 | LdBPK_230440 | ||
| LdCL_220018100 | LdBPK_221070 | ||
| LdCL_340044900 | LdBPK_343550 | ||
| LdCL_290028400 | — | ||
| LdCL_250011400 | LdBPK_250620 | ||
| LdCL_270014900 | LdBPK_270830 | ||
| Cluster 4 (8) | Conservative amino acid change in conserved region with good PROVEAN score | LdCL_350013100 | LdBPK_350830 |
| LdCL_360052700 | LdBPK_364550 | ||
| LdCL_230026600 | LdBPK_231940 | ||
| LdCL_230009400 | LdBPK_230400 | ||
| LdCL_320031100 | LdBPK_322560 | ||
| LdCL_020008200 | LdBPK_020280 | ||
| LdCL_310027700 | LdBPK_311990 | ||
| LdCL_320031200 | LdBPK_322570 | ||
| Cluster 5 (13) | Conservative amino acid change in conserved region with poor PROVEAN score | LdCL_330011900 | LdBPK_330640 |
| LdCL_170010200 | LdBPK_170470 | ||
| LdCL_070011900 | LdBPK_070700 | ||
| LdCL_210025000 | LdBPK_211930 | ||
| LdCL_290022900 | LdBPK_291730 | ||
| LdCL_200006300 | LdBPK_200140 | ||
| LdCL_290029000 | LdBPK_292290 | ||
| LdCL_030007500 | LdBPK_030250 | ||
| LdCL_250006200 | LdBPK_250110 | ||
| LdCL_360015800 | LdBPK_361070 | ||
| LdCL_310023400 | LdBPK_311630 | ||
| LdCL_360062700 | LdBPK_365540 | ||
| LdCL_140017700 | LdBPK_141190 | ||
| Cluster 6 (14) | Non-conservative amino acid change in less conserved region, Conservative amino acid change in less conserved region | LdCL_340022100 | LdBPK_341580 |
| LdCL_060011600 | LdBPK_060650 | ||
| LdCL_210015400 | LdBPK_211040 | ||
| LdCL_050010900 | LdBPK_050580 | ||
| LdCL_230011600 | LdBPK_230610 | ||
| LdCL_250024100 | LdBPK_251840 | ||
| LdCL_070015100 | LdBPK_071060 | ||
| LdCL_250005300 | LdBPK_250040 | ||
| LdCL_310022100 | LdBPK_311500 | ||
| LdCL_200006800 | LdBPK_200200 | ||
| LdCL_250014400 | LdBPK_250910 | ||
| LdCL_230010400 | LdBPK_230500 | ||
| LdCL_120008300 | LdBPK_120275 | ||
| LdCL_290028100 | LdBPK_292210 | ||
| Cluster 7 (4) | IV-only mutations | LdCL_140010000 | LdBPK_140470 |
| LdCL_310035800 | LdBPK_312770 | ||
| LdCL_310036400 | LdBPK_312810 | ||
| LdCL_320046000 | LdBPK_324000 | ||
| LdCL_360021300 | LdBPK_361580 | ||
| LdCL_360021400 | LdBPK_361590 | ||
| LdCL_360021500 | LdBPK_361600 | ||
| LdCL_360021600 | LdBPK_361610 |
Entries were not repeated in multiple lists.
Identified mutations were further classified into priority clusters for effect on protein function and future analysis for genes associated with survival in visceral organs. Mutations were prioritized by likelihood of contributing to visceral tissue tropism by severity of the coding change, accumulation of secondary mutations and conservation. Gene loci listed from the current assembly as well as previous ID numbers when available.
A 25 kb region on chromosome 36 containing 4 genes was found to be missing in the IV strain but present in the VL and CL strains. This deletion did not occur in a location previously identified on this chromosome where a fission can occur as seen in L. alderi26. Upon experimental verification, this deleted region was present in a subpopulation of the parental CL strain (Supplementary Fig. S4). The enrichment of this deletion in the IV strain could therefore be a consequence of selection in the mouse and likely to be unrelated to human visceral disease because this region is present in wild type or VL strains of L. donovani as well as L. major and therefore classified in cluster 7.
The classification of genetic differences in the CL, VL and IV genomes summarized in Fig. 6 and Table 3 represents a prioritization of genes to be empirically investigated for controlling the different phenotypes of these virulent and attenuated strains.
Discussion
It has been possible to generate a complete genome assembly for L. donovani through combining second and third generation sequencing technologies, similarly to a recent resequencing of the L. infantum genome resulting in a complete assembly, highlighting the usefulness of PacBio sequencing in regards to Leishmania genomes18. This resulted in a more accurate annotation of the genome increasing the number of potential protein coding genes and identifying novel mutations/polymorphisms associated with virulence. It was remarkable that the present assembly resulted in annotation changes in close to 15% of the genome representing 1087 protein coding genes. Although 13 degenerate pseudogenes are identified in Supplementary Table S1 more do exist since our annotations derived from functional genes in L. major and therefore genes functional in other species were not identified. Through this updated genome annotation, additional SNPs have been identified including in genes potentially involved in visceral disease and several non-coding genes have been annotated allowing future L. donovani research beyond protein coding genes. It has also been possible to assemble known virulence factor gene families in L. donovani including the A2 and Amastin gene families. This version of the L. donovani genome assembly will significantly improve genomic, functional genomic and proteomic research outcomes and support the identification of drug targets and the development of vaccines. This assembly further provides a larger repertoire of target DNA sequences to identify diagnostic and prognostic disease progression markers. Given the recent interest in generating genetically modified live attenuated parasites as vaccine candidates27, a complete genome assembly will permit the verification that genetic modifications target intended genes with no off target mutations.
Supported by a de novo assembly, this study provides the first direct evidence for synteny between chromosomes in L. donovani and L. major, two old world parasite species with different pathologies and reservoirs. Previously, due to the large number of gaps in the L. donovani genome, the segments were aligned to a reference assembly assumed to be syntenic and only gene synteny was possible. In contrast, the contiguous assembly presented within used an entirely reference-free and by extension, bias-free generation process. This assembly can be used in future sequencing efforts aimed at comparing genes and synteny of genomes of other Leishmania species with L. donovani. The strong gene level synteny further highlights the major phenotypic effects of SNPs and indel mutations when comparing genomes from L. donovani strains causing visceral and cutaneous pathologies. As no major chromosomal rearrangements or deletions are apparent between phenotypically different Leishmania species as previously reported2,4, and including this study, suggesting that virulence and tropism can be acquired or lost through relatively small coding changes at the amino acid level such as SNPs, indels and frameshifts without the need for chromosomal scale events.
This study reports the complete A2 gene continuous sequence and an assembly of an entire A2 cluster including the A2-rel flanking genes in L. donovani. While the organization of this A2 gene cluster was previously theorized based on available sequences and Southern blot analysis, no sequencing technology could accurately read-through an entire cluster4,28 prior to third-generation sequencers. Interestingly, a similar organization of A2 and A2-rel flanking genes was obtained during the resequencing and assembly of the L. infantum genome18, further supporting the genomic arrangement of this important virulence cluster. The present assembly contains entire A2 ORFs that are consistent with the corresponding protein sizes determined by Western blot analysis and provides novel insight into this elusive virulence factor through identifying 2 amino acid insertions between the 10 amino acid repeats at geometric intervals as well as a defined C-terminal sequence (Supplementary Fig. S2). The deviations from the 10-amino acid repeat sequence could contribute to the proper folding and function of the A2 protein.
In an attempt to identify additional genes associated with survival in visceral organs, the attenuated cutaneous L. donovani strain was experimentally passaged continuously through the visceral organs of BALB/c mice over an 8 month period to generate a gain-of-function strain with increased survival in the liver and spleen and was termed the IV strain24. Sequence analysis of the IV strain in this study did not identify homozygous SNP differences with the parental cutaneous strain but did identify four heterozygous SNPs with the same sequence in the virulent L. donovani strains, classified as high impact in Fig. 6. The corresponding SNP-containing genes with unknown function are of high priority for future studies. Nevertheless, although the gain of function IV strain had significantly increased survival in visceral organs24, it was surprising that this strain did not have more genetic differences associated with the increased virulence. It is possible that the selection process for survival in the visceral organs of mice is different from that in humans.
The Illumina sequence analysis of the cutaneous (CL) and visceral (VL) disease associated strains using the complete assembly identified 15 novel homozygous SNPs beyond the previously identified 70 SNPs4(Fig. 5). One of these new SNPs was in the Raptor gene that is part of the highly conserved Target Of Rapamycin (TOR) signaling pathway29. There are three TOR gene homologs in the Leishmania genome30 revealing this pathway is conserved in kinetoplastids. Interestingly, the RagC GTPase which is a binding partner of Raptor in the TOR pathway is also mutated in the attenuated cutaneous L. donovani strain and restoration of the wildtype RagC GTPase increased virulence in visceral organs4. Considering that there are two mutated genes (RagC and Raptor) in the TOR pathway in the attenuated cutaneous L. donovani strain strongly highlights this pathway as playing a role in determining disease tropism and virulence.
As both HIVE and VarScan were used to identify SNPs and indels, we are confident that the expanded list of 83 variable genes shown in Fig. 6 contains most if not all the genes associated with visceral disease, with the exception of UTR mutations that may influence protein expression levels. Since this number of genes is relatively small, we are currently investigating all genes in clusters 1–4 with respect to their involvement in visceral organ virulence using CRISPR-Cas9 gene editing recently developed for use in Leishmania31,32. It is noteworthy that the correct selection of gRNA sequences for CRISPR-Cas9 gene editing requires a complete genome and accurate annotations for precise gene editing with no off-target mutations that is now possible with the complete assembly reported here.
Methods
Whole genome sequencing
DNA extraction
Leishmania DNA for both Illumina and PacBio sequencing was derived from the attenuated cutaneous strain of L. donovani from Sri Lanka4 that was passaged through mice to increase survival in visceral organs24. DNA was extracted following the previously described phenol-chloroform methods for isolation of Trypanosomatid genomic material33.
Illumina
Sequencing library preparation (Kapa HTP) and 250 nt paired-end sequencing (Illumina MiSeq) was performed using manufacturers’ protocols.
PacBio sequencing
A total of 9 sequencing cells were prepared. 7 cells were prepared using the DNA Template kit v2.0 (3–10 kb) with DNA/Polymerase Binding Kit P4 and 2 using the DNA Template Prep Kit 3.0 with DNA/Polymerase Binding Kit P5. The cells were sequenced on a PacBio II RS instrument with BaseCaller v1 protocol.
Genome assembly
HGAP assembler
Raw reads from the 9 sequencing cells were loaded into the SMRT Analysis portal (Pacific Biosciences) in HD5 format. The Hierarchical Genome Assembly Process (HGAP) version 2 with Quiver polishing was chosen as version 3 is stated to improve speed at the detriment of assembly quality. Expected genome size was set to 36Mbp, minimum read length for pre-assembly was set to 500 bp and minimum read length for full assembly was set to 100 bp. Minimum Polymerase Read quality was set to 0.80, and the remainder of options remained at default settings.
Celera assembler
The PacBio corrected Reads (PBcR) module of the celera-assembler version 8.3 was used to assemble the long reads data34. The subreads were first extracted from the PacBio H5 files to FASTQ using bash5tools.py. The Bogart unitigger was used by specifying the “unitigger = bogart” option in the spec file. The consensus caller module was PBDAGCON. Due to the sequences originating from a non-clonal sample and the use of the DNA/Polymerase Binding kit P4 in some PacBio sequencing cells which produces lower quality data than P5 kits, error rate limits were relaxed for various variables, listed in the full spec file available in supplementary information (Supplementary Methods S1).
Canu assembler
The Canu v1.0 assembler is a modified version of the Celera Assembler designed to handle high noise data such as NanoPore and PacBio sequencing data. Canu has both the ability to assemble raw PacBio data by performing error correction using consensus sequence or assemble data in a hybrid mode where PacBio reads are pre-error corrected using short read Illumina data. In Raw mode, the trimmed PacBio reads were given to the assembler using default settings except for the expected genome size option which was set to 35Mbp using the option “genomeSize = 35 m”. In hybrid mode, the Illumina reads were first error corrected by internal consensus using Pollux35, the paired end reads were then merged together to form longer sequences with a high confidence core region using FLASh36, and used to correct the PacBio reads using Proovread37. The error-corrected PacBio data was then used by Canu to generate a draft assembly.
Pilon
The Pilon error correcting software was used to fix small errors present in the PacBio based assemblies using high depth and high accuracy Illumina data38. The entire MiSeq dataset in FASTQ format from the corresponding sample was aligned to the draft assembly using the Burrows-Wheeler Aligner (bwa) to generate SAM alignment files. Samtools was then used to convert and sort the files to a binary usable format as described in the Samtools section. This alignment was then passed to the Java Pilon executable for correction of small indels, SNPs, gap filling and assembly of unmapped reads using the command “java -Jar pilon.jar–genome [new-assembly.fasta] –frags [alignment.bam] –fix all,novel”.
GMCloser
GMcloser was used to merge the assemblies generated using the different assemblers39. Short read Illumina data was aligned to the contigs resulting from the different contigs from different assemblers with identical reads mapped to them were assumed to be part of the same chromosome. When a contig from one assembly encompassed a gap present in another assembly, the gap was filled with the missing information to generate a merged assembly with the least number of gaps. All the alignment and merging steps are handled internally to GMcloser using the command “gmcloser -t [assembly1.fasta] -q [assembly2.fasta] -r miseq_R1.fastq miseq_r2.fastq -et”.
IGV
The Broad Institute Integrative Genome Viewer40,41 was used to perform quality control on assemblies and manually inspect fragments in order to close gaps. The Pilon tools was used with the “-fix novel” option which assembled short contigs from unmapped data. The fragments were then placed on the appropriate likely chromosomes based on gene annotations and submitted to another round of gap filling using Pilon and GMCloser to find reads supporting this placement or were removed if no reads supported the join.
Annotations
Companion
The Companion webtool (https://companion.sanger.ac.uk/) was used to annotate genes on the assembly contigs and refine the assembly23. The closest available reference organism was chosen (L. major) with the following options: contiguate pseudochromosomes, align reference proteins to target sequence, perform pseudogene detection, use RATT Species transfer type, and the L. donovani taxon ID. Additional L. donovani and L. major genes not automatically transferred were manually verified and appended if necessary. An additional 3 genes were manually added from a search of all ribosomal protein transcripts in trypanosomes. The snRNAs U1 through U6, ribosomal RNAs and the spliced leader RNA were manually annotated as necessary from the sequences available for L. major on TriTrypDB42. Sequences for H/ACA and C/D box snoRNA were manually mapped using published L. major snoRNA research43.
Galaxy
The Galaxy webtool (https://usegalaxy.org/)44 was used to perform file conversions and data extraction such as moving a chromosome’s FASTA sequence from one assembly to another.
Identification of new genes
Genomic annotations from the Companion Pipeline were downloaded in General Feature Format (GFF) and gene annotations were extracted using the Galaxy tool “Extract features” set to look for the “CDS” keyword in column #3 of the GFF file. Known coding regions from the reference L. donovani strain BPK282A1, assembly ASM22713v2 were downloaded from GenBank and aligned to our improved assembly in BED format. Bedtools intersect intervals through Galaxy45,46 was used to identify annotations that were unique to our annotations or were not at least 95% covered previously using settings “-wa -f 0.95 -v -r”.
Synteny
The online SynMap2 software47 was used to generate the synteny dotplot across the entire genome using annotations from L. major and the annotations generated by Companion in this study. The chromosome to chromosome circular charts were generated by Companion as part of the annotation process.
Comparison of visceral (VL), cutaneous (CL) and increased virulence (IV) L. donovani strains
BWA
The Burrows-Wheeler Aligner (BWA) was used to process the FASTQ Illumina sequencing files obtained from Genome Quebec. The maximal exact match algorithm was used in paired-end mode using the command “bwa mem” and providing the matched pair read files and reference sequence as arguments in order to generate a SAM format alignment file of the reads on the reference48.
Samtools
The samtools package was used for file manipulations and conversions49. The commands “samtools view -b” was used to convert the BWA generated SAM file to the binary alignment BAM format. The file was then sorted by alignment location for compatibility with downstream analysis software using “samtools sort -@ 30 -o [output.file]”. The alignment files where then prepared for analysis using the mpileup modules which tabulates the base distribution at every position using the command “samtools mpileup -B -f [reference assembly] [strain specific position sorted BAM file] > [output.file]”.
VarScan
The VarScan v250 mutation caller was used to generate a list of mutations in Variant Call Format (VCF) using the mpileup file generated by samtools as described above using the command “java -jar VarScan.jar mpileup2snp –output-vcf 1 [mpileup.file] > [output.VCF]”. We also used VarScan to generate indel locations based on the same mpileup file using the command “java -jar VarScan.jar mpileup2indel –output-vcf 1 [mpileup.file] > [output.vcf]”.
SnpEff
To filter the VCF files generated by VarScan to a list of non-synonymous SNP, we used the SnpEff software51. The oriented and annotated assembly was downloaded from the Companion tool as described above along with the gene annotation file in GFF format containing the names, locations and amino acid sequences of identified genes. This GFF file was used to build a SnpEff database using the SnpEff.jar command “build” with argument “-gff” after installing the genome and GFF file in the appropriate locations according to the software instructions.
The SnpEff software was then used to annotate the 10th column of the VCF file with mutation effect codes. All the mutations were then examined manually for accuracy using the Integrative Genomics Viewer (IGV) with all raw Illumina data loaded.
Classification
Non-synonymous mutations were clustered according to the mutation effect in order to prioritize further gene function studies. Each cluster was further broken down based on the mutation’s PROVEAN score25. The PROVEAN software was designed to predict the magnitude of a mutation’s impact on protein function. To generate PROVEAN scores, we retrieved homologous sequences from other Leishmania species and kinetoplasts and generated a multiple sequence alignment (MSA). The MSA was then passed to the PROVEAN software which scored each SNP based on the alignment. We used PROVEAN scores below a threshold of −2.5 as an indication a SNP is likely to affect protein function. Cluster assignments were as follows:
Mutations likely to have the largest impact on protein function were included; non-sense, frameshift and amino acid insertion/deletions as well as all SNPs in the gain-of-function IV strain that were the same in the virulent visceral strain (VL) allele, indicating a selection pressure on those genes for visceral organ survival.
Genes in with multiple SNPs and genes where non-conservative mutations occurred in highly conserved Leishmania/Kinetoplastida regions.
Due to the high number of genes in cluster 2, split off poor PROVEAN scoring genes.
Cluster 4 comprised genes with conservative amino acid changes but occurring in Leishmania/Kinetoplastida conserved regions.
Due to the high number of genes in cluster 4, split off poor PROVEAN scoring genes.
Conservative and non-conservative amino acid changes in less conserved regions.
Changes present only in the gain of function IV strain. This cluster was considered low probability as it likely contains either random mutations or adaptations specific to survival in the murine host.
HIVE
HIVE52 was used to perform differential profiling of genomes from visceral (VL), gain of function increased virulence (IV), and (cutaneous) CL strains.
Reads from all the samples were aligned to the assembly of the genome using HIVE-hexagon53 parametrized for parasitic eukaryotic species and specifically adjusted to work with Leishmania analysis as demonstrated in previous studies27.
Coverage and variant calling analysis was performed using HIVE-heptagon54 to produce variant call frequencies and coverages for every genomic position.
HIVE differential profiler52 was used to analyze relative differences in SNP calls and variant coverages for multiple samples.
A2 Immunoblotting
A2 Immunoblotting was performed as previously described16. Briefly, 1 × 107 cutaneous CL strain promastigotes were collected at mid log-phase and resuspended in 1 mL fresh medium. The cells were then heat-shocked for 4 h at 40 °C to induce A2 protein expression, washed, lysed in SDS-PAGE loading buffer and loaded on a 10% (w/v) acrylamide gel. The proteins were transferred to nitrocellulose at 25 V overnight at 4 °C. The membrane was blocked for 1 h in 10% (w/v) skim milk powder dissolved in PBS with 0.1% (v/v) Tween-20. The membrane was then incubated for 1 h at RT with a 1:10,000 dilution of C9 Ascites fluid (anti-A2 Mab) in blocking solution followed by 6 × 5 min washes in PBS-T. Secondary HRP labeled anti-mouse IgG antibody (Thermo Fisher Scientific) was incubated at 1:10,000 in blocking buffer for 1 h at RT followed by 6 × 5 min washes in PBS-T. The membrane was incubated in ECL reagent (Zm Tech) for 1 min at RT before being exposed to x-ray film (Denville Scientific). Film images were captured using a Gel-Doc XR documentation system with Quantity One software (BioRad Laboratories).
Accession codes
GenBank BioProject PRJNA450813.
Electronic supplementary material
Acknowledgements
GM acknowledges the support of the Canadian Institutes of Health Research and the Global Health Innovative Technologies Fund. PL acknowledges receiving a doctoral training award from the Fonds de Recherche du Quebec Santé. LIM was funded by a graduate fellowship from the Canadian Institutes of Health Research (#MOP235928). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
P.L. wrote the main manuscript and performed the analysis. J.H., J.T.R. and A.M. performed analysis. W.Z., L.I.M., V.S. and K.D. provided materials and insight. G.M. planned the experiment and wrote the manuscript. All authors reviewed the manuscript.
Data Availability
All data used in this study have been deposited online with GenBank. Raw PacBio reads for the IV strain, Illumina MiSeq reads for the CL, VL and IV strains, the new genome assembly and the annotations generated in this study can be found under the PRJNA450813 BioProject accession number.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-34812-x.
References
- 1.Alvar J, et al. Leishmaniasis worldwide and global estimates of its incidence. PLoS One. 2012;7:e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peacock CS, et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat. Genet. 2007;39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang WW, Matlashewski G. Screening Leishmania donovani-specific genes required for visceral infection. Mol. Microbiol. 2010;77:505–517. doi: 10.1111/j.1365-2958.2010.07230.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhang WW, et al. Genetic Analysis of Leishmania donovani Tropism Using a Naturally Attenuated Cutaneous Strain. PLoS Pathog. 2014;10:e1004244. doi: 10.1371/journal.ppat.1004244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodwin S, McPherson JD, McCombie WR. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider VA, et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017;27:849–864. doi: 10.1101/gr.213611.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grisard EC, et al. Trypanosoma cruzi Clone Dm28c Draft Genome Sequence. Genome Announc. 2014;2:2–3. doi: 10.1128/genomeA.01114-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Downing T, et al. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res. 2011;21:2143–2156. doi: 10.1101/gr.123430.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivens AC, et al. The genome of the kinetoplastid parasite, Leishmania major. Science (80-.). 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogers MB, et al. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 2011;21:2129–2142. doi: 10.1101/gr.122945.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alonso, G., Rastrojo, A., López-Pérez, S., Requena, J. M. & Aguado, B. Resequencing and assembly of seven complex loci to improve the Leishmania major (Friedlin strain) reference genome. Parasites and Vectors9 (2016). [DOI] [PMC free article] [PubMed]
- 12.Mikheyev AS, Tin MMY. A first look at the Oxford Nanopore MinION sequencer. Mol. Ecol. Resour. 2014;14:1097–1102. doi: 10.1111/1755-0998.12324. [DOI] [PubMed] [Google Scholar]
- 13.Rhoads A, Au KF. PacBio Sequencing and Its Applications. Genomics, Proteomics Bioinforma. 2015;13:278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W-W, Matlashewski G. Loss of virulence in Leishmania donovani deficient in an amastigote-specific protein, A2. Proc. Natl. Acad. Sci. 1997;94:8807–8811. doi: 10.1073/pnas.94.16.8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang WW, et al. Comparison of the A2 gene locus in Leishmania donovani and Leishmania major and its control over cutaneous infection. J. Biol. Chem. 2003;278:35508–35515. doi: 10.1074/jbc.M305030200. [DOI] [PubMed] [Google Scholar]
- 16.McCall LI, Matlashewski G. Localization and induction of the A2 virulence factor in Leishmania: Evidence that A2 is a stress response protein. Mol. Microbiol. 2010;77:518–530. doi: 10.1111/j.1365-2958.2010.07229.x. [DOI] [PubMed] [Google Scholar]
- 17.McCall LI, Matlashewski G. Involvement of the Leishmania donovani virulence factor A2 in protection against heat and oxidative stress. Exp. Parasitol. 2012;132:109–115. doi: 10.1016/j.exppara.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 18.González-De La Fuente, S. et al. Resequencing of the Leishmania infantum (strain JPCM5) genome and de novo assembly into 36 contigs. Sci. Rep. 7, (2017). [DOI] [PMC free article] [PubMed]
- 19.Karunaweera ND, Pratlong F, Siriwardane HVYD, Ihalamulla RL, Dedet JP. Sri Lankan cutaneous leishmaniasis is caused by Leishmania donovani zymodeme MON-37. Trans. R. Soc. Trop. Med. Hyg. 2003;97:380–381. doi: 10.1016/S0035-9203(03)90061-7. [DOI] [PubMed] [Google Scholar]
- 20.Ranasinghe S, et al. Leishmania donovani zymodeme MON-37 isolated from an autochthonous visceral leishmaniasis patient in Sri Lanka. Pathog. Glob. Health. 2012;106:421–424. doi: 10.1179/2047773212Y.0000000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh N, Chikara S, Sundar S. SOLiDTM Sequencing of Genomes of Clinical Isolates of Leishmania donovani from India Confirm Leptomonas Co-Infection and Raise Some Key Questions. PLoS One. 2013;8:e55738. doi: 10.1371/journal.pone.0055738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCall Laura-Isobel, Zhang Wen-Wei, Matlashewski Greg. Determinants for the Development of Visceral Leishmaniasis Disease. PLoS Pathogens. 2013;9(1):e1003053. doi: 10.1371/journal.ppat.1003053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steinbiss S, et al. Companion: a web server for annotation and analysis of parasite genomes. Nucleic Acids Res. 2016;44:W29–W34. doi: 10.1093/nar/gkw292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCall LI, et al. Adaptation of leishmania donovani to cutaneous and visceral environments: In vivo selection and proteomic analysis. J. Proteome Res. 2015;14:1033–1059. doi: 10.1021/pr5010604. [DOI] [PubMed] [Google Scholar]
- 25.Choi Y, Chan AP. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coughlan, S. et al. The genome of Leishmania adleri from a mammalian host highlights chromosome fission in Sauroleishmania. Sci. Rep. 7 (2017). [DOI] [PMC free article] [PubMed]
- 27.Gannavaram, S. et al. Whole genome sequencing of live attenuated Leishmania donovani parasites reveals novel biomarkers of attenuation and enables product characterization. Sci. Rep. 7 (2017). [DOI] [PMC free article] [PubMed]
- 28.Zhang WW, Matlashewski G. Characterization of the A2-A2rel gene cluster in Leishmania donovani: Involvement of A2 in visceralization during infection. Mol. Microbiol. 2001;39:935–948. doi: 10.1046/j.1365-2958.2001.02286.x. [DOI] [PubMed] [Google Scholar]
- 29.Sancak Y, et al. The rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science (80-.). 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madeira da Silva L, Beverley SM. Expansion of the target of rapamycin (TOR) kinase family and function in Leishmania shows that TOR3 is required for acidocalcisome biogenesis and animal infectivity. Proc. Natl. Acad. Sci. 2010;107:11965–11970. doi: 10.1073/pnas.1004599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang WW, Matlashewski G. CRISPR-Cas9-mediated genome editing in Leishmania donovani. MBio. 2015;6:e00861–15. doi: 10.1128/mBio.00861-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang W-W, Lypaczewski P, Matlashewski G. Optimized CRISPR-Cas9 Genome Editing for Leishmania and Its Use To Target a Multigene Family, Induce Chromosomal Translocation, and Study DNA Break Repair Mechanisms. mSphere. 2017;2:1–15. doi: 10.1128/mSphere.00340-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medina-Acosta E, Cross GAM. Rapid isolation of DNA from trypanosomatid protozoa using a simple ‘mini-prep’ procedure. Mol. Biochem. Parasitol. 1993;59:327–329. doi: 10.1016/0166-6851(93)90231-L. [DOI] [PubMed] [Google Scholar]
- 34.Berlin K, et al. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 2015;33:623–630. doi: 10.1038/nbt.3238. [DOI] [PubMed] [Google Scholar]
- 35.Marinier, E., Brown, D. G. & McConkey, B. J. Pollux: Platform independent error correction of single and mixed genomes. BMC Bioinformatics16, (2015). [DOI] [PMC free article] [PubMed]
- 36.Magoč T, Salzberg SL. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hackl T, Hedrich R, Schultz J, Förster F. Proovread: Large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics. 2014;30:3004–3011. doi: 10.1093/bioinformatics/btu392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker Bruce J., Abeel Thomas, Shea Terrance, Priest Margaret, Abouelliel Amr, Sakthikumar Sharadha, Cuomo Christina A., Zeng Qiandong, Wortman Jennifer, Young Sarah K., Earl Ashlee M. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE. 2014;9(11):e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kosugi S, Hirakawa H, Tabata S. GMcloser: Closing gaps in assemblies accurately with a likelihood-based selection of contig or long-read alignments. Bioinformatics. 2015;31:3733–3741. doi: 10.1093/bioinformatics/btv465. [DOI] [PubMed] [Google Scholar]
- 40.Robinson JT, et al. Integrative genomics viewer. Nature Biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aslett M, Aurrecoechea C, Berriman M, Al. E. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38:D457–62. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eliaz D, et al. Genome-wide analysis of small nucleolar RNAs of leishmania major reveals a rich repertoire of RNAs involved in modification and processing of rRNA. RNA Biol. 2015;12:1222–1255. doi: 10.1080/15476286.2015.1038019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Afgan E, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gruening, B. A. Galaxy wrapper (2014).
- 47.Haug-Baltzell A, Stephens SA, Davey S, Scheidegger CE, Lyons E. SynMap2 and SynMap3D: Web-based whole-genome synteny browsers. Bioinformatics. 2017;33:2197–2198. doi: 10.1093/bioinformatics/btx144. [DOI] [PubMed] [Google Scholar]
- 48.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303 (2013).
- 49.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koboldt DC, et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin). 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simonyan V, Mazumder R. Genes (Basel). 2014. High-performance integrated virtual environment (hive) tools and applications for big data analysis; pp. 957–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Santana-Quintero Luis, Dingerdissen Hayley, Thierry-Mieg Jean, Mazumder Raja, Simonyan Vahan. HIVE-Hexagon: High-Performance, Parallelized Sequence Alignment for Next-Generation Sequencing Data Analysis. PLoS ONE. 2014;9(6):e99033. doi: 10.1371/journal.pone.0099033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simonyan V, et al. HIVE-heptagon: A sensible variant-calling algorithm with post-alignment quality controls. Genomics. 2017;109:131–140. doi: 10.1016/j.ygeno.2017.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study have been deposited online with GenBank. Raw PacBio reads for the IV strain, Illumina MiSeq reads for the CL, VL and IV strains, the new genome assembly and the annotations generated in this study can be found under the PRJNA450813 BioProject accession number.