Abstract
There are urgent demands for efficient treatment of heritable genetic diseases. The base editing technology has displayed its efficiency and precision in base substitution in human embryos, providing a potential early-stage treatment for genetic diseases. Taking advantage of this technology, we corrected a Marfan syndrome pathogenic mutation, FBN1T7498C. We first tested the feasibility in mutant cells, then successfully achieved genetic correction in heterozygous human embryos. The results showed that the BE3 mediated perfect correction at the efficiency of about 89%. Importantly, no off-target and indels were detected in any tested sites in samples by high-throughput deep sequencing combined with whole-genome sequencing analysis. Our study therefore suggests the efficiency and genetic safety of correcting a Marfan syndrome (MFS) pathogenic mutation in embryos by base editing.
Keywords: pathogenic mutation, base editing, human embryos
Huang and colleagues took advantage of recently developed base editing technology to precisely correct a Marfan syndrome pathogenic mutation, FBN1T7498C, providing a proof-of-principle for the technical feasibility of gene therapy for MFS and other genetic diseases.
Introduction
Nearly 10,000 genetic diseases have been identified, which affects millions of families around the world. However, less than 6% of genetic diseases have approved treatments.1 To reduce the burden of genetic diseases in affected families and individuals, breakthroughs in diagnosis and therapeutic methods are urgently needed. Although preimplantation genetic diagnosis (PGD) is useful to prevent the generational transmission of the mutant allele to embryos, the potential risk for diagnostic errors still exists. On the other hand, gene therapy potentially provides an active approach for correcting the genetic diseases.2
Genome editing technologies, especially those based on CRISRP/Cas9, have been successfully applied in genome manipulation,3 which has inspired a brilliant outlook that the pathogenic mutation can be precisely repaired to achieve therapeutic effects.4, 5 Moreover, recent successes in precise genome editing trials in early human embryos have suggested a potentially true cure for genetic diseases.6, 7 However, genome editing of human embryos caused huge concerns because of ethical issues and technical uncertainties regarding the efficiency and off-target effects.8, 9 It is well-known that genome editing through CRISPR/Cas9 generates double-strand breaks (DSBs) that evoke the error-prone non-homologous end joining (NHEJ) DNA repair pathway after, which causes off-target mutagenesis.10 The recently developed base editor (BE) system was constructed by fusing the deaminase to the dCas9 protein.11 BE efficiently edits the mutation without donor, which is required for precise genome editing through homologous recombination at low efficiency. BE edits specific sites by C-to-T or G-to-A conversion without DSB formation, providing a safer genome editing tool with low off-target effects.12, 13 BE has been applied in plants and animals, and has shown enormous advantages in precise base-level genome editing compared with CRISPR/Cas9.14 Based on the developments of the BE systems, several studies have been undertaken and have proven the efficiency and safety of BE in human embryos.15, 16, 17
These previous studies, by others and our own, have suggested the technical feasibility of correcting genetic diseases at the embryo stage.11 With this success, we set out to try to correct a pathogenic gene mutation in human embryos. To this end, we focused on the FBN1 mutation that is causative for Marfan syndrome (MFS), an autosomal dominant disorder with the frequency of 0.2‰ in the world.18 MFS was initially described over 100 years ago and can be diagnosed with the characteristic abnormalities in connective tissues.19 In this study, taking advantage of the BE3 base editing system, we achieved specific correction of a MFS pathogenic FBN1 mutation first in a human cell line and then in human zygotic embryos. Our study therefore provides a proof-of-principle for the technical feasibility of gene therapy for MFS.
Results
The T7498C Mutation of the FBN1 Was Created Using CRISPR/Cas9 Combined with ssODN in HEK293T Cells
An adult male patient diagnosed with MFS based on clinical characteristics, including funnel chest and flatfoot, was found to carry a reported causative heterozygous T7498C mutation at the FBN1 gene.18 We further confirmed the mutation of FBN1T7498C using the donated blood and sperm of the patient by sequencing (Figure S1). Meanwhile, we thoroughly analyzed the sequence around the mutation site and found it was suitable for correction using the BE system that introduces a C-to-T conversion guided by a targeting single guide RNA (sgRNA).
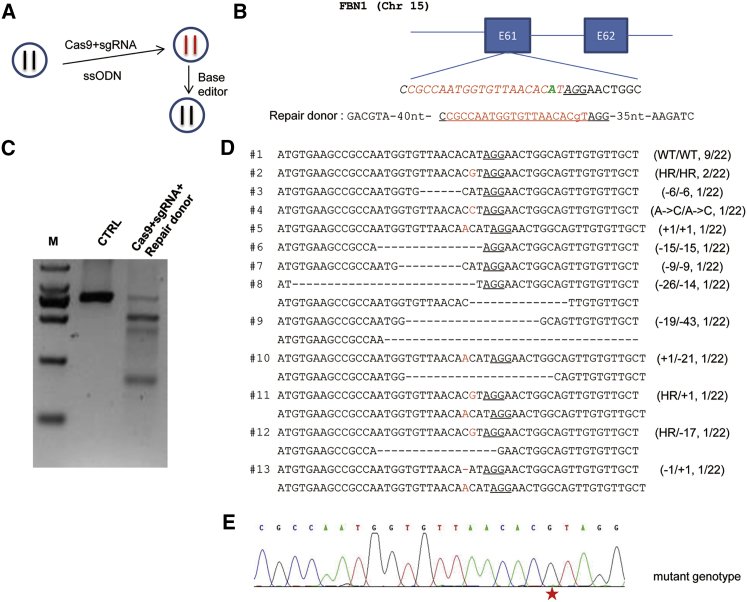
To demonstrate the feasibility, we created a cell model harboring a FBN1T7498C mutation using SpCas9 protein, sgRNA (hereafter mutational sgRNA), and the ssODN donor containing the sequence of the desired mutant allele (Figures 1A and 1B). The length of the ssODN was 110 nt to ensure efficient homologous recombination (HR).20 Three days after transfection of the ribonucleoprotein (RNP) complex containing SpCas9 protein/sgRNA and ssODN, the HEK293T cells were collected. Single cells were sorted into the wells of a 96-well plate for obtaining individual colonies. A portion of the unsorted cells was used for the DNA extraction and genotyping by T7EN1 assay. The results showed clear cleavage bands (Figure 1C), demonstrating that Cas9/sgRNA mediated efficient genome editing. With this, we further genotyped single-cell clones to confirm the mutations mediated by Cas9/sgRNA-assisted HR. A total of 22 colonies were analyzed by Sanger sequencing (Figure 1D). Among these colonies, 9 were genotyped with only the wild-type alleles, and the remaining 13 (about 60%) had 12 kinds of mutations detected, including 11 kinds of indels or desirable point mutation (Figure 1D). Two cell colonies harbored the intended mutation in homozygous fashion, as shown by the Sanger sequencing chromatogram (Figure 1E). One of the homozygous FBN1T7498C cell colonies was picked for further study.
Figure 1.
The Generation of Single-Cell Clones Harboring the FBN1T7498C Mutation by Cas9/sgRNA-Assisted Homologous Recombination
(A) Schematic illustration of the experimental procedure for the generation and correction of pathogenic mutation. The left flowchart shows the pathogenic FBN1T7498C mutation is first created by Cas9/sgRNA-assisted homologous recombination (HR) using an ssODN as a template in HEK293T cells; then the mutation is corrected by base editor. (B) Graphical representation of the Cas9/sgRNA-assisted HR. The T-to-C base substitution at exon 61 of the FBN1 gene located on chromosome 15 is achieved by Cas9/sgRNA-assisted HR using an ssODN as a template. The “A” highlighted in green at the sgRNA-targeted site was replaced by an “a” in lowercase in the template ssODN. The protospacer-adjacent motif (PAM) sequences are underlined; the targeting sequences are highlighted in red. (C) Detection of genome editing by T7EN1. The RNP was transfected into the HEK293T cells. Two days later, the cells were collected, and the DNA was extracted and used for analysis. (D) Genotype analysis of the single-cell clones with genome editing. The single cell was picked by fluorescence activated cell sorting (FACS) after genome editing. The cell clones from the single cell were collected. The genomic DNA was extracted and amplified by PCR. TA clones of the PCR products were analyzed by DNA sequencing. The PAM sequences are underlined; the modified bases are highlighted in red; deletions (−), insertions (+); the left N/N represents the genotypes of two alleles of the single clone, and the right represents the clone number harboring the genotype among the total 22 single clones sequenced. The left #N indicated the serial number of the genotype. (E) The representative Sanger sequencing chromatogram of genomic DNA from the single clone harboring the homozygous mutation. The red star indicates the substituted base.
Next, the off-target for the mutational sgRNA was examined to ensure the specificity. A total of seven off-target sites were predicted using published software. Using the genomic DNA from the homozygous FBN1T7498C cell colony, the sequences around the seven off-target sites were amplified and subjected for T7EN1 assay. No cleavage bands for any of these sites were observed (Figure S2A), suggesting that the off-target effects were likely to be minimal. Meanwhile, the PCR products were subjected to Sanger sequencing. As expected, no mutational peaks were observed (Figure S2B), further confirming the specificity.
Correction of the T7498C Mutation by BE3 in Mutant HEK293T Cells with High Specificity
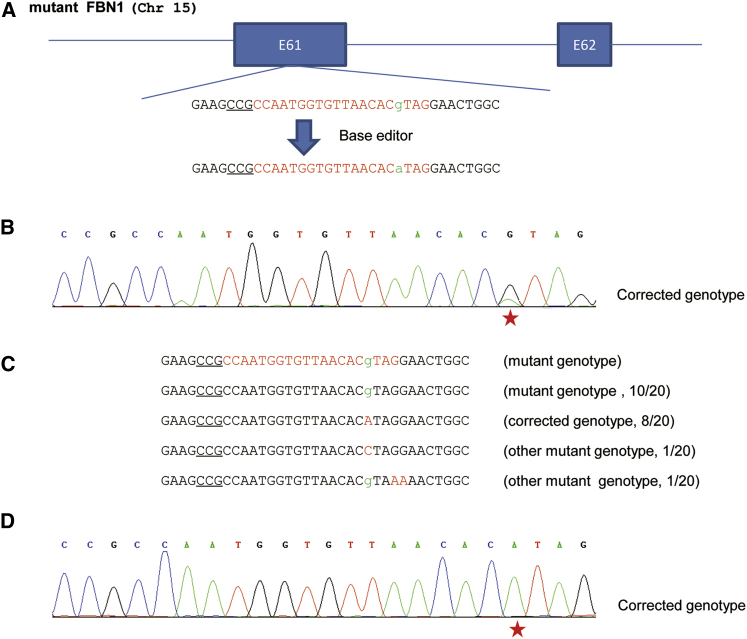
The high fidelity of base editing encouraged us to test the possibility of correcting the T7498C mutation in the FBN1T7498C cells using BE3. Therefore, we designed the sgRNA (hereafter correctional sgRNA) (Figure 2A). The FBN1T7498C cells described above were transfected with the correctional sgRNA and BE3-expressing plasmids. Three days later, the cells were collected, and the genomic DNA was extracted and used as the template to amplify the target sequence. The PCR products were subjected for Sanger sequencing. The results showed that, different from the single peaks detected in either the wild-type or FBN1T7498C cells, double peaks were observed in targeted cells (Figure 2B), indicative of occurrence of correction events. The PCR products were TA cloned and sequenced to confirm the correction of mutation. The results showed that 10 of 20 (50%) clones were edited. Among these edited clones, eight had desirable C-to-T correction at 7498 (Figure 2D), whereas the remaining two had unwanted C-to-G conversion at 7498 or C-to-T conversion at other positions (Figure 2C). Therefore, the overall efficiency of FBN1T7498C allele correction by the base editing system was high.
Figure 2.
Correction of the Pathogenic FBN1T7498C Mutation by Base Editing in Human Cells
(A) Schematic illustration of the correction of the pathogenic FBN1T7498C mutation. The “g” in lowercase highlighted in green at the targeted site was substituted by a “g” in lowercase in green. The PAM sequences are underlined; the targeting sequence is highlighted in red. (B) The representative chromatogram of the sequencing of PCR products. The cells harboring the FBN1T7498C mutation were transfected with sgRNA and BE3-expressing plasmids. The cells were collected, and the genomic DNA was extracted and amplified by PCR. The PCR products were analyzed by DNA sequencing. The red star indicates the substituted base. (C) Sequencing analysis of the base editing. TA clones of the PCR products from (B) were analyzed by DNA sequencing. The PAM sequences are underlined; the targeted bases in lowercase are highlighted in green; the modified bases in red; the N/N represents positive colonies out of the total sequenced. (D) The representative chromatogram of the sequencing of TA clones. TA clones from (C) were sequenced. The red star indicates the substituted base.
Correction of the T7498C Mutation by BE3 in Heterozygous Mutant Embryos with High Specificity
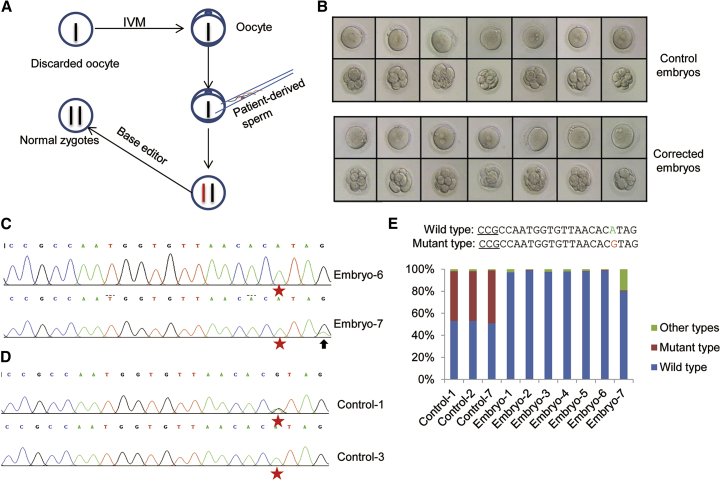
The efficient and specific correction of the MFS pathogenic FBN1T7498C mutation encouraged us to test this process in human embryos. The immature oocytes were collected for research with the informed consent from individual donors. The collected oocytes were first subjected to in vitro maturation. Then the matured oocytes were used for in vitro fertilization (IVF) by ICSI (intracytoplasmic sperm injection) of single sperm donated by the MFS patient with the heterozygous FBN1T7498C mutation (Figure 3A). 16–18 hr later, all of the zygotes were collected, and some of them were microinjected with the mRNA of BE3 and the correctional sgRNA at concentrations of 100 and 50 ng/μL, respectively. As the control, the other zygotes were microinjected similarly with the mRNA of BE3 and the scrambled sgRNA. The embryos were further cultured for 2 days and then collected for analysis. A total of seven testing and seven control embryos that showed apparently normal development were obtained (Figure 3B). All samples were used for whole-genome amplification and then genotyped by Sanger sequencing. As shown in the sequence chromatograms, all of the testing embryos showed A at the 7498 site (G4 site), whereas three of seven control embryos harbored G (about 50%, as expected), indicating that allele correction in testing embryos occurred at a rate of near 100% (Figures 3C and 3D; Embryo-1∼7 in Figures S3A and S3B). Nevertheless, an unwanted C-to-T conversion in addition to the desirable correction was detected in Embryo-7 (Figure 3C). To further characterize the BE3-mediated base editing, another 11 embryos were used to repeat the test by microinjection of BE3 and the correctional sgRNA as above. Sequencing the PCR products of the target site showed that 10 of the edited embryos yielded completed conversion at the 7498 site, besides Embryo-9, which harbored about 60% G-to-A at the 7498 site. These results showed BE3 mediated efficient correction of the pathogenic mutation in embryos.
Figure 3.
Correction of the Pathogenic FBN1T7498C Mutation by Base Editing in Heterozygous Human Embryos
(A) Schematic illustration of the experimental procedure for the correction of the pathogenic mutation by base editor in human embryos. (B) The development stage of the corrected embryos and the control embryos before and after treatment. (C) The representative chromatogram of the sequencing of PCR products from the corrected human embryos. The human embryos treated with base editor were collected, and the genomic DNA was extracted and amplified by PCR. The PCR products were analyzed by DNA sequencing. The red stars indicate the target base; the arrow indicates another base substitution in the target region. (D) The representative chromatogram of the sequencing of PCR products from the control human embryos. The control human embryos were collected, and the genomic DNA was extracted and amplified by PCR. The PCR products were analyzed by DNA sequencing. The red stars indicate the target base; the mixed peaks at the target site in Control-1 indicate the different kinds of nucleotides. (E) The genotyping analysis by deep sequencing. All of the samples, including three control and seven corrected human embryos, were collected, and the genomic DNA was extracted and amplified by PCR. The PCR products were analyzed by deep sequencing. The percentage of different genotypes of all samples was calculated.
To thoroughly characterize the editing effects, we comprehensively analyzed by deep sequencing all seven testing samples (Embryo-1∼7 in Figure S3A) and the three control samples from embryos with the heterozygous genotype. The results showed that all three control embryos harbored nearly half of the wild-type and half of the mutant alleles (Figure 3E). For the testing embryos, six embryos showed 100% wild-type alleles without other base conversion or indels, indicative of efficient and isogenic allele correction. As expected, about 20% of the genetic material from Embryo-7 showed two C-to-T conversions (C1 and C4). Taken together, these results demonstrated the high efficiency and precision of BE in correcting the MFS pathogenic FBN1T7498C mutation in human embryos.
The detection of unwanted conversion reminds us to test the YE1-BE3 and YEE-BE3 narrow editing window.21 The test in the FBN1T7498C cells by Sanger sequencing combined with deep sequencing PCR products showed, as expected, YE1-BE3 and YEE-BE3 did not mediate unwanted base editing at the G1 site as BE3, but introduced G-to-A conversion at the G4 site at lower efficiency than BE3 (40% and 30% for YE1-BE3 and YEE-BE3, respectively, versus 50% for BE3) (Figures S4A and S4B). We performed further analysis in embryos (10 for YE1-BE3 and 11 for YEE-BE3). Sequencing the PCR products showed no unwanted base conversion was detected over all 21 samples (Figure S5). Further analyses showed, besides wild-type genotypes, which indicate complete correction, three samples from YE1-BE3 (Embryo-19, -23, and -26) harbored pathogenic genotype and six samples from YEE-BE3 (Embryo-29, -32, -33, -36, -38, and -39) contained both wild genotype and pathogenic genotype. Taken together, BE3 introduces more efficient, whereas YE1-BE3 and YEE-BE3 perform more precise, in vivo base editing.
Off-Target Detection by Deep Sequencing and Whole-Genome Sequencing
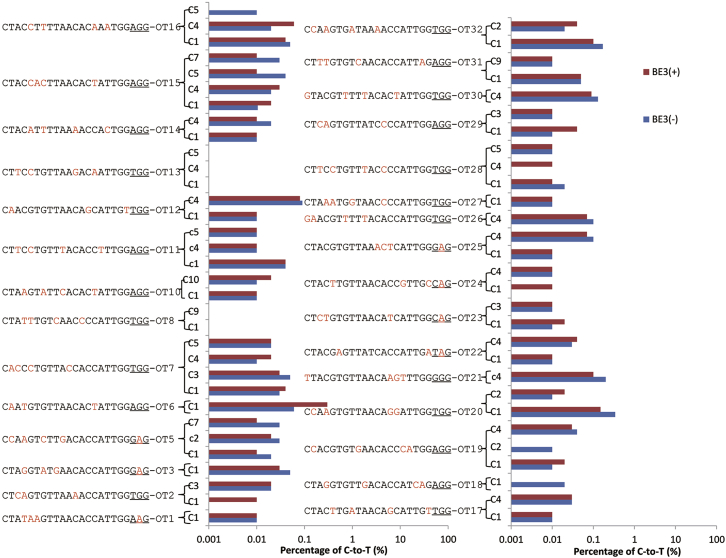
Given that off-target mutagenesis is the main concern of genome editing, 32 potential off-target sites with up to 4-nt mismatch for the correctional sgRNA were amplified with PCR from seven corrected samples (Embryo-1∼7) and three control samples (Control-1, Control-2, and Control-7) from heterozygous embryos. The PCR products for each site were equally mixed and subjected for deep sequencing. The data analysis of the deep sequencing showed that no off-target and indels were detected (Figure 4; Figure S6). We further performed whole-genome sequencing to identify the off-target mutation for Embryo-7, Control-1, and Control-2. All of the potential off-target sites (419) with up to five mismatches that differ from the correctional sgRNA were analyzed. After excluding the false-positive sites depending on the database of single nucleotide polymorphisms (dbSNPs) and the control embryos, no off-target was detected (Table 1). These results demonstrated the safety of correcting pathogenic mutation by base editing in human early embryos.
Figure 4.
Off-Target Analysis of the Corrected and Control Human Heterozygous Embryos
Genomic DNA from seven corrected and three control human embryos was collected, and the genomic DNA was extracted and amplified by PCR. The PCR products were analyzed by deep sequencing. The percentage of C-to-T within a 10-nt window near the PAM-distal end of the protospacer at 32 potential off-target sites was calculated. The PAM sequences are underlined; the mismatch sites are highlighted in red.
Table 1.
Genome-Wide Sequencing Analysis for the Off-Target
| Total SNPs | SNPs after Excluding dbSNPs | C→T SNPs | C→A SNPs | C→G SNPs | G→A SNPs | G→T SNPs | G→C SNPs | Possible Off-Target Sites (419 Sites) | |
|---|---|---|---|---|---|---|---|---|---|
| Control-5 | 2,217,143 | 589,148 | 89,395 | 27,787 | 41,748 | 90,286 | 27,415 | 41,279 | – |
| Control-6 | 2,235,681 | 614,848 | 101,895 | 27,701 | 41,909 | 102,163 | 27,578 | 41,325 | – |
| Embryo-7 | 2,048,936 | 592,219 | 98,086 | 25,945 | 39,552 | 97,888 | 25,877 | 39,209 | 0 |
| Uniquely assigned to Embryo-7 | 349,900 | 132,946 | 39,612 | 4,469 | 5,482 | 39,440 | 4,574 | 5,375 | 0 |
Discussion
With the development of DNA sequencing, accumulating diseases resulting from genetic mutations have been characterized. The emergence of CIRSPR/Cas9 genome editing technology has revolutionized the genomic researches. It has also reignited the enthusiasm to gene therapy, which aims at correcting the pathogenic mutation in affected patients.3 But the efficiency and safety of the genome editing in human embryos caused huge concerns.2
The recently developed base editing system featuring precise and specific base substitution capability is considered more suitable for genetic diseases that attributed to base mutation described in the ClinVar database.11 Taking advantage of BE, here we achieved correction of the MFS pathogenic FBN1 mutation by base editing in both human cells and heterozygous embryos. These results have instilled some hope that a genetic disease that has no current treatment is potentially correctable with a cutting-edge genome editing tool.
Besides efficiency, a successful gene therapy needs perfect precision and specificity. Conventional CRISPR/Cas9 triggers an error-prone, NHEJ DNA repair pathway after introducing a DSB, which causes uncontrollable indels.22 Base editing edits specific sites by base conversion without introduction of DSBs, providing a safer genome editing tool with low off-target effects.23, 24, 25 Consistent with this notion, by several types of assays, our results showed precise base substitution without detection of off-target and indels by BE in both the cell line and embryos, which may be caused by the specificity of the correctional sgRNA.14 Nevertheless, some unwanted base conversion events were detected, which shows that to further narrow down the editing window would be a focus of future technical improvement.21
In summary, we achieved correction of the MFS pathogenic mutation in the FBN1 gene in human embryos. The correction by BE-mediated base conversion was highly efficient and specific, demonstrating the feasibility of base editing in genetic correction and providing a possible treatment of MFS.
Materials and Methods
Ethical Statement
The study was approved by the Ethics Committee of the Third Affiliated Hospital of Guangzhou Medical University. Before collection of immature oocytes, all patients have signed the informed consent where they agreed to the use of their discarded oocytes for scientific research. The patient with MFS also signed the informed consent for donating his blood and semen samples for research. All related clinical and experimental procedures were conducted at the Center for Reproductive Medicine, the Third Affiliated Hospital of Guangzhou Medical University.
Plasmid Construction and In Vitro Transcription
The oligos for mutation and correctional sgRNA were synthesized, cloned into the pGL3-U6-sgRNA-PGK-puromycin (51133; Addgene) and pUC57-sgRNA expression vector (51132; Addgene), and in vitro transcribed as the reported protocol:17 mutation sgRNA, 5′-CGCCAATGGTGTTAACACATAGG-3′; correctional sgRNA, 5′-CCGCCAATGGTGTTAACACgTAG-3′. Besides, the correctional sgRNA was also cloned into the pGL3-U6-sgRNA-PGK-GFP plasmid for the comparison of BE3, YE1-BE3, and YEE-BE3. The plasmids of Cas9 (44758; Addgene), BE3 (73021; Addgene), YE1-BE3 (85174; Addgene), and YEE-BE3 (85177; Addgene) were used, and these plasmids were transcribed in vitro as the reported protocol.17 All of the transcribed RNA were stored at −80°C. The ssODN was synthesized at Sangon Biotech (http://www.sangon.com/) as the following sequence: 5′-GACGTATGGTGTTGGGTAAATCCGGGAGGACATTTGCATGTGAAGCCGCCAATGGTGTTAACACgTAGGAACTGGCAGTTGTGTTGCTTGGTTGCACACTCATCAAGATC-3′.
Cell Culture and Transfection
HEK293T cells were cultured in DMEM (Hyclone), supplemented with 10% fetal bovine serum (FBS) (v/v) (Gemini). The RNP complex was transfected using the Nuclear transfection with the Lonza protocol (https://www.lonza.com/) to make the mutant cells. In brief, the transfection buffer was prepared according to the protocol (V4XC-2024; Lonza). 1 M cells were picked and washed with PBS for one time; then the PBS was removed thoroughly. The quantity of the Cas9 protein, sgRNA, and ssODN was 3, 1.5, and 3 μg, respectively. The RNP-ssODN was mixed with the prepared buffer for electric shock with the program DS150 at the 4D instrument. The plasmids were transfected using Lipofectamine 2000 reagent (Life Technologies) according to manufacturer’s protocols, to correct the mutant cells.
Flow Cytometry and Identification of Positive Cell Colony
Three days after transfection of the RNP complex, a part of the cells was harvested and subjected to the flow cytometry for single-cell sorting. The single cell was cultured in a 96-well plate for about 10 days; then the cell colonies were divided into two parts. One part was used to identify the genotype, and another part was cultured in a 24-well plate. The cells used for genotyping were lysed in the lysis buffer (50 mM KCl, 1.5 mM MgCl2, 10 mM Tris [pH 8.0], 0.5% Nonidet P-40, 0.5% Tween 20, 100 μg/mL protease K) at 65°C for 30 min, then 98°C for 3 min. 1 μL lysate was used as the template to amplify the target sequences using the related primers (Table S1).
IVF and Microinjection
The immature oocytes were cultured in maturation medium (Cooper Surgical/SAGE, Trumbull, CT, USA) supplemented with 75 mIU/mL follicle-stimulating hormone (FSH) and luteinizing hormone (LH). The mature oocytes fertilized with the MFS patient’s sperm using ICSI. 16–18 hr later, the fertilization was confirmed by formation of two pronuclei. The concentrations of BE3 mRNA and sgRNA were adjusted to 100 and 50 ng/μL as previously reported.17 Microinjection was performed using an inverted microscope equipped with a microinjector and micromanipulators. The corrected embryos will be cultured for 2 days using standard procedures. Then these embryos were digested using acidic Tyrode’s solution (Solarbio, Beijing, China). The collected embryos were amplified using Discover-sc Single Cell Kit (N601-01; Vazyme) to obtain enough DNA for genotype identification.
Off-Target Analysis and Deep Sequencing
We used two softwares to analyze the off-target for mutation sgRNA and correctional sgRNA (http://crispr.mit.edu/ and https://crispr.cos.uni-heidelberg.de/), and 7 off-target sites for mutation sgRNA and 32 off-target sites for correctional sgRNA were obtained. The PCR products containing the on-target and the off-target were amplified using the related primers (Table S1). The purified PCR products were submitted to sequence using the Illumina Nextseq 500 (2 × 150) platform at CAS-MPG Partner Institute for Computational Biology Omics Core (Shanghai, China), and 1 M clean data was generated for each site. BWA and Samtools were used to process the data.
Whole-Genome Sequencing
Whole-genome sequencing of the genomic DNA from the embryos was performed at a sequencing depth of 30× to 40× using an Illumina HiSeq X Ten (2 × 150 PE) at the HuaGen Biotech Institute (Shanghai, China). The sequencing data were mapped using BWA v0.7.16 with a human reference genome (GRCh38/hg38). Sequence reads were marked for duplicates using Sambamba v0.6.7 and realigned using Genome Analysis Toolkit (GATK v3.7) IndelRealigner. Variants were identified by GATK HaplotypeCaller, and then the quality was evaluated using GATK VariantFiltration. To figure out the potential off-target sites, C-to-T and G-to-A were picked out. We also excluded the common SNPs between the corrected and the control embryos.
Author Contributions
X.H., J. Liu, and G.L. conceived and designed the project. G.L., J. Li, Y. Zeng, S.H., W.Y., Y. Zhang, and D.C. performed the experiments. X.H., J. Liu, D.C., and J.C. supervised the project. X.H. and G.L. analyzed the data and wrote the paper.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
We thank members of Huang and Liu labs for helpful discussions. We are grateful to Dr. Jianghuai Liu from Nanjing University for excellent language editing. This work is supported by the National Natural Science Foundation of China (grant 31471400), Innovation of Science and Technology Commission of Guangzhou, China (grant 201604020075), and China Postdoctoral Science Foundation (grant 2017M622659).
Footnotes
Supplemental Information includes six figures and one table and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.08.007.
Contributor Information
Jianqiao Liu, Email: liujqssz@gzhmu.edu.cn.
Xingxu Huang, Email: huangxx@shanghaitech.edu.cn.
Supplemental Information
References
- 1.Austin C.P., Dawkins H.J.S. Medical research: next decade’s goals for rare diseases. Nature. 2017;548:158. doi: 10.1038/548158c. [DOI] [PubMed] [Google Scholar]
- 2.Dunbar C.E., High K.A., Joung J.K., Kohn D.B., Ozawa K., Sadelain M. Gene therapy comes of age. Science. 2018;359:eaan4672. doi: 10.1126/science.aan4672. [DOI] [PubMed] [Google Scholar]
- 3.Komor A.C., Badran A.H., Liu D.R. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruan G.X., Barry E., Yu D., Lukason M., Cheng S.H., Scaria A. CRISPR/Cas9-mediated genome editing as a therapeutic approach for Leber congenital amaurosis 10. Mol. Ther. 2017;25:331–341. doi: 10.1016/j.ymthe.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y., Yang Y., Kang X., Lin B., Yu Q., Song B., Gao G., Chen Y., Sun X., Li X. One-step biallelic and scarless correction of a β-thalassemia mutation in patient-specific iPSCs without drug selection. Mol. Ther. Nucleic Acids. 2017;6:57–67. doi: 10.1016/j.omtn.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma H., Marti-Gutierrez N., Park S.W., Wu J., Lee Y., Suzuki K., Koski A., Ji D., Hayama T., Ahmed R. Correction of a pathogenic gene mutation in human embryos. Nature. 2017;548:413–419. doi: 10.1038/nature23305. [DOI] [PubMed] [Google Scholar]
- 7.Kang X., He W., Huang Y., Yu Q., Chen Y., Gao X., Sun X., Fan Y. Introducing precise genetic modifications into human 3PN embryos by CRISPR/Cas-mediated genome editing. J. Assist. Reprod. Genet. 2016;33:581–588. doi: 10.1007/s10815-016-0710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruzo A., Brivanlou A.H. At last: gene editing in human embryos to understand human development. Cell Stem Cell. 2017;21:564–565. doi: 10.1016/j.stem.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Kang X.J., Caparas C.I.N., Soh B.S., Fan Y. Addressing challenges in the clinical applications associated with CRISPR/Cas9 technology and ethical questions to prevent its misuse. Protein Cell. 2017;8:791–795. doi: 10.1007/s13238-017-0477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zong Y., Wang Y., Li C., Zhang R., Chen K., Ran Y., Qiu J.L., Wang D., Gao C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017;35:438–440. doi: 10.1038/nbt.3811. [DOI] [PubMed] [Google Scholar]
- 13.Kim K., Ryu S.M., Kim S.T., Baek G., Kim D., Lim K., Chung E., Kim S., Kim J.S. Highly efficient RNA-guided base editing in mouse embryos. Nat. Biotechnol. 2017;35:435–437. doi: 10.1038/nbt.3816. [DOI] [PubMed] [Google Scholar]
- 14.Kim D., Lim K., Kim S.T., Yoon S.H., Kim K., Ryu S.M., Kim J.S. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat. Biotechnol. 2017;35:475–480. doi: 10.1038/nbt.3852. [DOI] [PubMed] [Google Scholar]
- 15.Zhou C., Zhang M., Wei Y., Sun Y., Sun Y., Pan H., Yao N., Zhong W., Li Y., Li W. Highly efficient base editing in human tripronuclear zygotes. Protein Cell. 2017;8:772–775. doi: 10.1007/s13238-017-0459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang P., Ding C., Sun H., Xie X., Xu Y., Zhang X., Sun Y., Xiong Y., Ma W., Liu Y. Correction of β-thalassemia mutant by base editor in human embryos. Protein Cell. 2017;8:811–822. doi: 10.1007/s13238-017-0475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li G., Liu Y., Zeng Y., Li J., Wang L., Yang G., Chen D., Shang X., Chen J., Huang X., Liu J. Highly efficient and precise base editing in discarded human tripronuclear embryos. Protein Cell. 2017;8:776–779. doi: 10.1007/s13238-017-0458-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arbustini E., Grasso M., Ansaldi S., Malattia C., Pilotto A., Porcu E., Disabella E., Marziliano N., Pisani A., Lanzarini L. Identification of sixty-two novel and twelve known FBN1 mutations in eighty-one unrelated probands with Marfan syndrome and other fibrillinopathies. Hum. Mutat. 2005;26:494. doi: 10.1002/humu.9377. [DOI] [PubMed] [Google Scholar]
- 19.Pyeritz R.E. The Marfan syndrome. Annu. Rev. Med. 2000;51:481–510. doi: 10.1146/annurev.med.51.1.481. [DOI] [PubMed] [Google Scholar]
- 20.Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y.B., Komor A.C., Levy J.M., Packer M.S., Zhao K.T., Liu D.R. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang L., Zeng Y., Du H., Gong M., Peng J., Zhang B., Lei M., Zhao F., Wang W., Li X., Liu J. CRISPR/Cas9-mediated gene editing in human zygotes using Cas9 protein. Mol. Genet. Genomics. 2017;292:525–533. doi: 10.1007/s00438-017-1299-z. [DOI] [PubMed] [Google Scholar]
- 23.Hess G.T., Tycko J., Yao D., Bassik M.C. Methods and applications of CRISPR-mediated base editing in eukaryotic genomes. Mol. Cell. 2017;68:26–43. doi: 10.1016/j.molcel.2017.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komor A.C., Zhao K.T., Packer M.S., Gaudelli N.M., Waterbury A.L., Koblan L.W., Kim Y.B., Badran A.H., Liu D.R. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci. Adv. 2017;3:eaao4774. doi: 10.1126/sciadv.aao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of A⋅T to G⋅C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.