Abstract
Deficiency of basement membrane heterotrimeric laminin 332 component, coded by LAMA3, LAMB3, and LAMC2 genes, causes junctional epidermolysis bullosa (JEB), a severe skin adhesion defect. Herein, we report the first application of CRISPR/Cas9-mediated homology direct repair (HDR) to in situ restore LAMB3 expression in JEB keratinocytes in vitro and in immunodeficient mice transplanted with genetically corrected skin equivalents. We packaged an adenovector carrying Cas9/guide RNA (gRNA) tailored to the intron 2 of LAMB3 gene and an integration defective lentiviral vector bearing a promoterless quasi-complete LAMB3 cDNA downstream a splice acceptor site and flanked by homology arms. Upon genuine HDR, we exploited the in vitro adhesion advantage of laminin 332 production to positively select LAMB3-expressing keratinocytes. HDR and restored laminin 332 expression were evaluated at single-cell level. Notably, monoallelic-targeted integration of LAMB3 cDNA was sufficient to in vitro recapitulate the adhesive property, the colony formation typical of normal keratinocytes, as well as their cell growth. Grafting of genetically corrected skin equivalents onto immunodeficient mice showed a completely restored dermal-epidermal junction. This study provides evidence for efficient CRISPR/Cas9-mediated in situ restoration of LAMB3 expression, paving the way for ex vivo clinical application of this strategy to laminin 332 deficiency.
Keywords: CRISPR/Cas9 gene editing, LAMB3 mutations, adhesion advantage
Benati et al. report the first application of CRISPR/Cas9-mediated LAMB3 editing to restore laminin 332 expression in keratinocytes from junctional epidermolysis bullosa patients (JEB) in vitro and in mice transplanted with genetically corrected human skin equivalents. This approach can be employed to tackle a wide range of JEB-causative LAMB3 mutations.
Introduction
Junctional epidermolysis bullosa (JEB) is a recessively inherited skin disorder characterized by fragility of the skin and mucous membranes and by tissue separation within the lamina lucida of the cutaneous basement membrane zone (BMZ). The majority of JEB cases are caused by defects in laminin 332, the major adhesion ligand of basal epithelial cells expressed in the BMZ of the skin and mucous epithelia. Laminin 332 is a multi-domain glycoprotein formed by 3 subunits, α3, β3, and γ2, coded by LAMA3, LAMB3, and LAMC2 genes, respectively. Restored expression of laminin 332 protein in defective skin keratinocytes by gene addition strategies results in a complete re-establishment of the adhesion property of keratinocytes in vitro1, 2 and in a permanent correction of a normal dermal-epidermal junction in vivo.3, 4
The gene addition strategy based on viral (e.g., adeno-associated virus [AAV] and lentiviral and retroviral vectors) delivery of an expression cassette for the correct gene has been the strategy of choice to treat severe genetic disorders, e.g., junctional3, 4 and dystrophic epidermolysis bullosa (DEB),5 hemophilia B,6 and X-linked severe combined immunodeficiency (SCID-X1).7 The gene addition strategy has shown excellent results over the years in many cell contexts and with multiple vectors and genes. However, permanent correction of disease-causing mutations or locus-specific insertion of a therapeutic cDNA remains preferable to allow long-term physiological and tissue-restricted expression of the introduced sequence.
In the last years, emerging genome-editing strategies, able to modify nucleic acids within disease-affected cells and tissues, have shown therapeutic potential for the treatment of monogenic, highly penetrant diseases, such as SCID-X18 and hemophilia B.9 Among the four major classes of nucleases, meganucleases,10 zinc finger nucleases (ZFNs),11 transcription activator-like effector nucleases (TALENs),12 and CRISPR-associated nuclease Cas9,13 CRISPR/Cas9 system from S. pyogenes (SpCas9) clearly boosted the applications of genome editing to gene therapy. It has turned out that Cas9-mediated homologous recombination (HR) of a donor DNA template offers a valuable option to correct the mutated genes associated to monogenic disorders.14, 15 This study aims for the first time to apply CRISPR/Cas9-triggered HR to in situ re-establish LAMB3 expression by integrating an almost complete LAMB3 cDNA. We employed, as therapeutic donor, a promoterless LAMB3 cDNA engineered to be transcriptionally silent unless a splicing event between genomic exon 2 and donor exon 3 occurs, upon HR event. This donor design overcomes most of the safety concerns of gene therapy and holds several advantages: (1) site-specific integration with transgene regulation and transcription controlled by the endogenous promoter; (2) quasi-complete LAMB3 cDNA (exon 3–23), thus tackling most of the mutations mapped in JEB patients; (3) lack of a promoter in the donor template vector, thus reducing the probability of activating neighboring oncogenes if undesired donor integration occurred; and (4) canonical splice acceptor site of LAMB3 gene conferring high expression of laminin 332, which is required to restore the in vitro adhesion property of corrected keratinocytes. Restoration of laminin 332 expression in the keratinocyte culture has been demonstrated to provide a biological system to positively select gene-corrected cells,16 avoiding cell sorting that could impair cell viability, or antibiotic selection, not feasible in many clinical applications. Thus, exploiting the in vitro adhesion advantage of laminin-332-corrected keratinocytes, we propose a novel and safe gene editing strategy to in situ restore LAMB3 expression upon canonical splicing event in the defective gene of JEB keratinocytes.
In this study, a LAMB3 editing strategy was applied to keratinocytes from two JEB patients: the first carrying a homozygous frameshift mutation (c.1945dupG) in the exon 14 of LAMB3 gene, leading to the formation of a premature stop codon within exon 15 (p.A649fsX38),17 and the second carrying a frequent (45%–63%) homozygous c.1903C > T transition in the exon 14 of LAMB3 gene, resulting in an Arg635-to-ter (R635X) substitution.18 To restore the expression of wild-type LAMB3 gene, we designed two molecular tools: a guide RNA (gRNA) for SpCas9 targeting LAMB3 intron 2 and a promoterless LAMB3 cDNA (exon 3–23) flanked by a splice acceptor and a poly-adenylation (pA) sites and embedded between two homology arms (HAs) flanking the target site. JEB keratinocytes were co-transduced with an adenoviral vector carrying expression cassettes for SpCas9 and gRNA (AdCas9gRNA) and non-integrating lentiviral vector bearing the therapeutic donor template (IDLV-L.B3). Transduced cells were positively selected in vitro exploiting physiological adhesion advantages of gene-corrected cells. Molecular analysis on single isolated clones demonstrated an in situ monoallelic and biallelic integration of the therapeutic donor template restoring the expression of laminin 332 heterotrimer. LAMB3-corrected clones showed morphology and adhesive properties comparable to wild-type (WT) keratinocytes and increased proliferation with respect to JEB cells. A functional reconstruction of dermal-epidermal junction was demonstrated in immunodeficient mice transplanted with skin equivalents from corrected clones. This study represents the first proof that in situ integration of wild-type LAMB3 cDNA spanning from exon 3 to 23 is able to correct the JEB phenotype in vitro and in a mouse model.
Results
Correction of Keratinocytes from a JEB Patient by CRISPR/Cas9-Mediated Homology Direct Repair of Promoterless LAMB3 cDNA
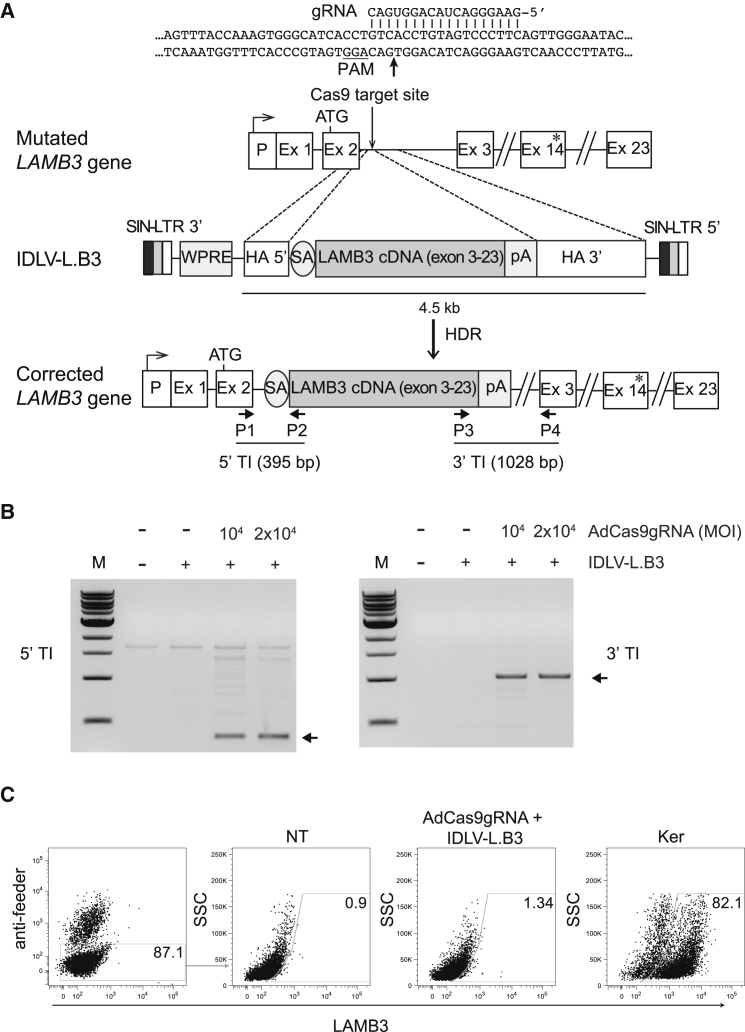
JEB disease exhibits heterogeneity in the location and number of mutations in LAMB3 gene. To tackle a wide range of JEB-causative mutations spread all along the transcription unit of LAMB3 gene, we employed CRISPR/SpCas9 system. We designed a gRNA tailored to the second intron of LAMB3 gene to trigger site-specific HR of a promoterless LAMB3 cDNA (Figure 1A). The therapeutic LAMB3 donor carried the splice acceptor site of exon 3 followed by a super exon consisting of correct cDNA from exon 3 to 23 and a bovine growth hormone (bGH) pA site. This promoterless therapeutic donor cassette, flanked by HAs, was cloned in reverse orientation into an integration defective lentiviral vector (IDLV-L.B3) in order to avoid cryptic splicing site in 5′ long terminal repeat (LTR),19 leading to expression of truncated proteins (Figure 1A). SpCas9 and gRNA were cloned into a first-generation adenoviral vector (AdCas9gRNA) able to efficiently transduce human keratinocytes.20, 21
Figure 1.
CRISPR/Cas9-Mediated HDR into Intron 2 of LAMB3 Gene in Keratinocytes Derived from a JEB Patient
(A) (Top) Endogenous mutated LAMB3 gene carrying 23 exons (Ex) is schematized (not in scale). The arrow above the promoter region (P) indicates the transcription start site; Cas9 target site and the ATG are depicted. Sequences of the selected gRNA and PAM on intron 2 are reported. The asterisk indicates LAMB3 mutations generating a stop codon. (Middle) Schematic representation (not in scale) of promoterless LAMB3 donor (4.5 kb in length) cloned in opposite orientation into IDLV backbone, generating IDLV-L.B3 vector, is shown. (Bottom) HDR event into the target site re-establishing the transcription of corrected LAMB3 cDNA is shown. HA, homology arm (90 bp HA5′ and 413 bp HA 3′); pA, polyadenylation site (278 bp); SA, splicing acceptor; SIN-LTR, self-inactivating long terminal repeat (LTR); WPRE, posttranscriptional regulatory element of woodchuck hepatitis virus. P1–4 represent primers used for targeted integration and allele discrimination PCRs. 5′ (395 bp) and 3′ (1,028 bp) TIs indicate the size of amplicons generated upon genuine HDR event. (B) PCR analyses on genomic DNA from JEB.A649fsX38 cells transduced with IDLV-L.B3 (4.32 × 102 vg/cell) in absence or in presence with AdCas9gRNA at indicated MOIs (vg/cell) or not transduced. Two couple of primers specific for 5′ and 3′ vector genome junctions (P1+P2 and P3+P4, respectively, as depicted in A) amplified 395- and 1,028-bp bands indicated by arrows, respectively, are shown. M, 1 kb molecular weight marker. (C) Flow cytometric analysis of laminin 332 expression in JEB.A649fsX38 cells not transduced (NT), co-transduced with IDLV-L.B3 (4.32 × 102 vg/cell) + AdCas9gRNA (MOI 104 vg/cell), and in WT keratinocytes (Ker), after feeder exclusion (left panel). The boxed gates represent the percentage of LAMB3-positive keratinocytes (0.9%, 1.34%, and 82.1%).
The nuclease-mediated in situ integration of the therapeutic LAMB3 donor was addressed in immortalized keratinocytes derived from a JEB patient carrying the c.1945dupG LAMB3 mutation (referred to as JEB.A649fsX38). This homozygous mutation maps on exon 14 and results in a frameshift starting at codon 649 and leading to the formation of a premature stop codon within exon 15 (p.A649fsX38).17 As a consequence, the mutant mRNA is degraded by nonsense-mediated mRNA decay mechanism and, in case of occurring protein synthesis, translated in a β3 chain truncated in the rod domain, which is required for laminin-332 assembly. Thus, the mutation leads to a complete loss of LAMB3 expression.
We first transduced JEB.A649fsX38 keratinocytes with increasing doses of AdCas9gRNA vector (MOI 104, 2 × 104, 5 × 104, and 105 vector genomes [vg]/cell) to evaluate gRNA editing efficiency. Surveyor assay of AdCas9gRNA-transduced cells clearly showed editing at all tested MOIs (Figure S1). To determine indels frequency, tracking of indels by decomposition (TIDE) analysis22 on PCR amplicons derived from transduced JEB.A649fsX38 cells was performed and showed 25%, 41%, 67%, and 76% (p < 0.05) of indels, respectively (Figure S2). Indels types upon AdCas9gRNA delivery (MOI 2 × 104 vg/cell) was more accurately evaluated by bioinformatics analysis (D.B., V. Marigo, and A.R., unpublished data) of single Sanger sequences (Figure S3) and resulted in 56.3% total indels, in particular 40.8% insertion, 14.1% deletion, and 1.4% mismatches. To note, higher doses resulted in toxic effect; thus, the MOI 104 and 2 × 104 vg/cell were selected for LAMB3-editing experiment. We co-transduced JEB.A649fsX38 keratinocytes with AdCas9gRNA (MOI 104 and 2 × 104 vg/cell) and IDLV-L.B3 (4.32 × 102 vg/cell) vectors. Targeted integration (TI) PCR confirmed that homology direct repair (HDR) events precisely occurred, as demonstrated by attended bands at both 5′ and 3′ vector-genome junctions (Figure 1B). However, flow cytometric analysis of laminin 332 expression in co-transduced JEB.A649fsX38 keratinocytes showed that, as previously observed,20 the HDR frequency was barely detectable (% LAMB3+ cells = 0.48 ± 0.19; Figures 1C and S4), remarking the requirement of positively selecting LAMB3-expressing cells.
To predict the most likely off-target sites of gRNA, we used public webservers (http://crispr.mit.edu; https://crispr.dbcls.jp)23 to assess and prioritize potential Cas9 activity at off-target loci based on predicted positional bias of a given mismatch in gRNA protospacer sequence and total number of mismatches to the intended target site. The CRISPR design tool scored a total of 186 potential off-target sites in the human genome (data not shown). The top 7 potential off-target sites (Table S1) were assessed by Surveyor assay and TIDE analysis on JEB.A649fsX38 cells transduced at MOI 2 × 104 vg/cell. None of the 7 predicted off-target loci showed detectable levels of off-target gene modification by Surveyor assay (Figure S5A). TIDE analyses scored a 0.9% frequency of indels in the OT7, although no significant (p < 0.05) level of indels was measured for the other off targets (OTs) (Figure S5B).
Selective Advantage of LAMB3-Corrected JEB Keratinocytes In Vitro
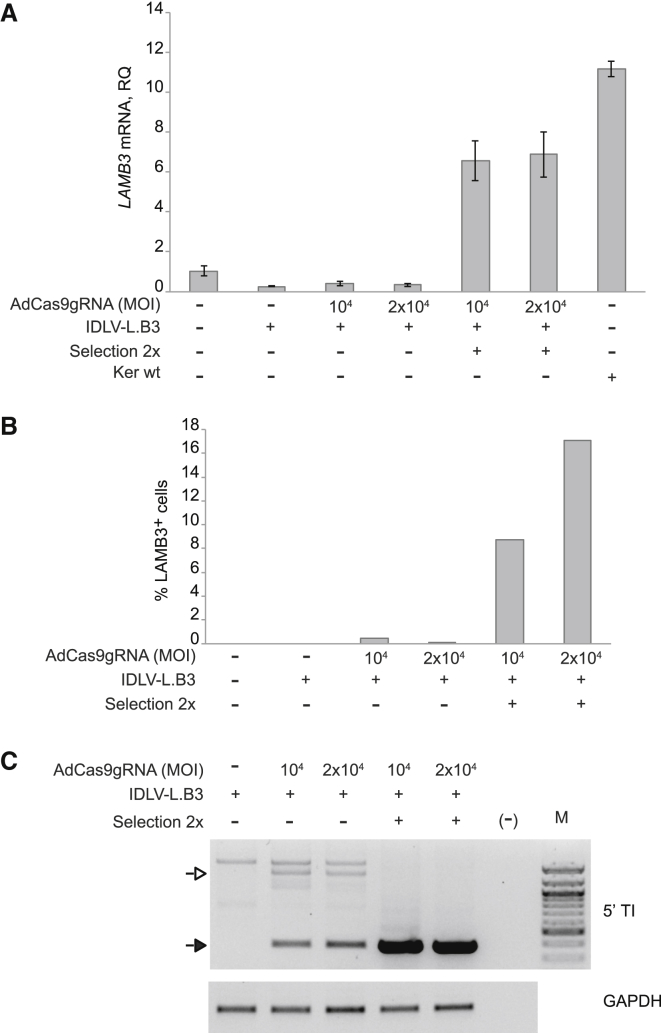
Taking advantage of in vitro adhesion properties of LAMB3-expressing cells,1 we selected bulk populations of LAMB3-corrected keratinocytes by 2 rounds of cell detachment from tissue culture plates using a trypsin solution. The restored expression of LAMB3 gene conferred to JEB.A649fsX38 keratinocytes an increased adhesion to a 3T3 feeder layer seeded onto the tissue culture plate with respect to untreated cells and to JEB.A649fsX38 treated with IDLV-L.B3 alone. Indeed, qRT-PCR analysis before selection showed a barely detectable increase of LAMB3 mRNA in co-transduced bulks compared to control JEB.A649fsX38. Conversely, after two rounds of selection, LAMB3 expression in co-transduced JEB.A649fsX38 cells was a 6- or 7-fold higher respect to untreated cells (Figure 2A). We further evaluated the enrichment of LAMB3-corrected keratinocytes by flow cytometry of laminin 332 expression. Up to 17% laminin-332-positive cells were detected in co-transduced JEB.A649fsX38 bulks with respect to untreated JEB.A649fsX38, indicating a robust enrichment of LAMB3-corrected cells after 2 rounds of selection (Figure 2B).
Figure 2.
Restored LAMB3 Expression in Gene-Corrected Keratinocytes Selected by In Vitro Adhesion Strategy
(A) qRT-PCR analysis on LAMB3 expression in JEB.A649fsX38 keratinocytes bulks transduced with IDLV-L.B3 (4.32 × 102 vg/cell) alone or in combination with AdCas9gRNA vector at increasing MOIs (vg/cell). LAMB3 expression was measured in JEB.A649fsX38 cells before and after 2 rounds of selections and in WT keratinocytes, as control. Data are representative of three independent PCR amplifications (mean ± SEM; n = 3). (B) Representative experiment of flow cytometric analysis of LAMB3+ cells before and after selection of JEB.A649fsX38 keratinocytes co-transduced with IDLV-L.B3 (4.32 × 102 vg/cell) and AdCas9gRNA (MOI 104 and 2 × 104 vg/cell). The frequency (%) of LAMB3-positive cells was determined after subtraction of background measured in not transduced cells. (C) Genomic analysis of 5′ TI in pre- and post-selected JEB.A649fsX38 keratinocytes co-transduced with IDLV-L.B3 (4.32 × 102 vg/cell) and AdCas9gRNA (MOI 104 and 2 × 104 vg/cell). The size of PCR bands corresponding to genuine HR (395 bp, black arrow) and HR-independent (approximately ∼1,400 bp, white arrow) integration of the donor template is indicated. The bottom gel shows PCR amplification targeting the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene. M, 100-bp molecular weight marker.
The positive selection was also confirmed at genomic level by 5′ TI PCR performed pre- and post-2 rounds of detachment (Figure 2C), pointing out that the selective adhesion procedure enriched cells undergone genuine HR event and splicing between the genomic splice donor site in exon 2 and the splice acceptor site of exon 3 cloned upstream LAMB3 cDNA. As shown in Figure 2C, the adhesion strategy negatively selected the HR-independent integration of the donor template represented by an ∼1,400-bp PCR band amplified in co-transduced JEB.A649fsX38 cells pre-selection. Sequencing of the PCR band indicated the in situ integration of the 3′ end of IDLV vector (Figure S6).
The selective advantage of LAMB3-corrected cells was also demonstrated in immortalized JEB keratinocytes carrying the R635X mutation (referred to as JEB.R635X),18 which is responsible for complete loss of laminin 332 expression. JEB.R635X keratinocytes were transduced with AdCas9gRNA (MOI 2 × 104 vg/cell) in presence or absence of IDLV-L.B3 (4.32 × 102 vg/cell). Three days post-transduction, we investigated the presence and frequency of indels in the target site by Surveyor assay and TIDE analysis. The results reported in Figures S7A and S7B showed a 56.7% of indels in JEB.R635X keratinocytes transduced with AdCas9gRNA vector. Co-transduction of JEB.R635X cells resulted in expected HDR event as demonstrated by 5′ and 3′ TI PCRs (Figure S7C). After 2 rounds of selection, we verified the enrichment of LAMB3-corrected cells by 5′ TI PCR, qRT-PCR, and flow cytometry as previously performed in JEB.A649fsX38 keratinocytes pre- and post-selection. The results (Figures S7D–S7F) demonstrated the positive selection of cells undergoing genuine HR events and the restored LAMB3 expression in co-transduced JEB.R635X keratinocytes respect to control cells.
Restored LAMB3 Expression in Selected Single-Cell Clones
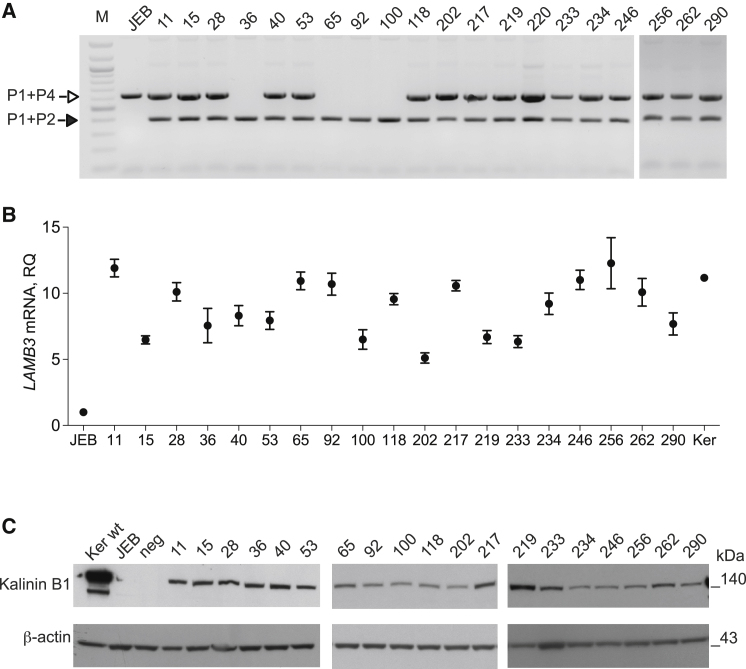
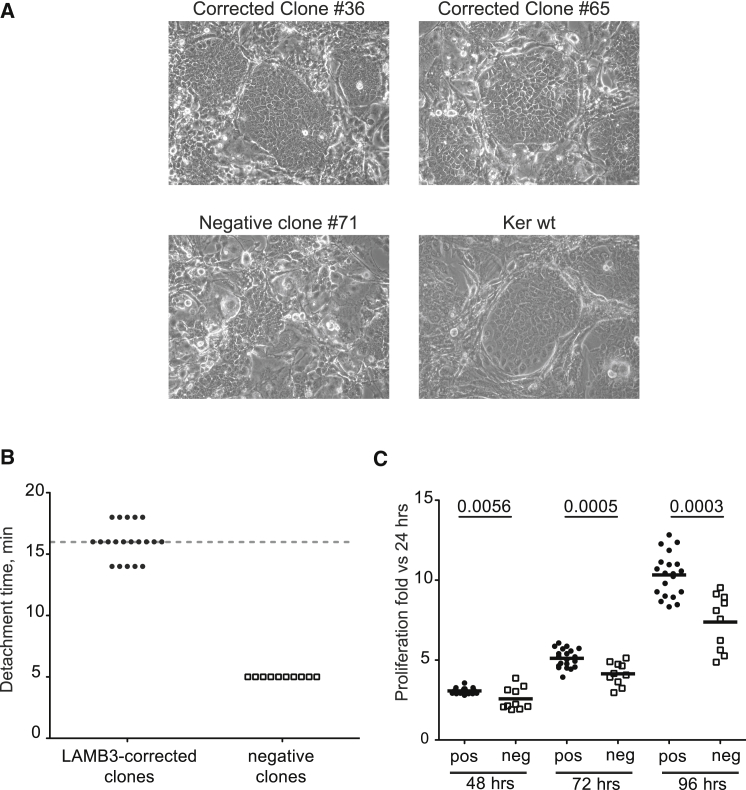
To molecularly characterize the in situ integration of LAMB3 cDNA, enriched JEB.A649fsX38 keratinocytes were cloned by limiting cell dilution. Twenty-four out of 54 clones derived from AdCas9gRNA (MOI 104 vg/cell) infection and 15 out of 34 clones from higher dose (MOI 2 × 104 vg/cell) resulted positive in 5′ and 3′ TI PCR analyses (Figures S8A and S8B), indicating that more than 2 rounds of selective adhesion would be required to fully enrich for LAMB3-corrected clones. Remarkably, the percentage of corrected clones was 44% from both bulk populations, indicating that AdCas9gRNA-tested doses did not influence the frequency of HR. Eight out of 39 clones, equally distributed in the two bulk keratinocytes, showed a 3′ TI PCR longer than expected (Figures S9A and S9B). Deeper investigation of this aberrant HR event revealed an in situ integration of the 5′ end of the IDLV vector (Figures S9C and S9D). Nevertheless, these clones correctly expressed LAMB3 mRNA (Figure S10A). For further analysis, we expanded 28 LAMB3-corrected clones and 10 negative clones for targeted integration PCR assay. First, we analyzed allelic distribution of targeted clones with a PCR-based allelic discrimination strategy employing 3 primers (P1, P2, and P4) schematized in Figure 1A. Twenty-four and four targeted clones displayed monoallelic and biallelic integration of the therapeutic donor, respectively (Figures 3A and S10B), meaning that, in the vast majority of clones, only one allele carrying the homozygous mutation hosted the therapeutic donor. Laminin 332 expression was rescued upon splicing between genomic and donor template splice sites in all 5′ and 3′ TI-positive clones as indicated by qRT-PCR (Figure 3B) and western blot analysis (Figure 3C) reported for representative 19 clones. Laminin 332 expression, both in terms of RNA and protein level, was variable but always stronger than in negative clones or JEB.A649fsX38 cells, ranging from 5- to 12-fold (Figure 3B). Microscopic appearance of representative corrected and control clones seeded on a feeder layer confirmed that LAMB3-corrected clones formed colonies comparable to normal keratinocytes (Figure 4A), as previously demonstrated.1 These clones seeded onto lethally irradiated feeder layer were characterized by the formation of large and round colonies surrounded by feeder cells, due to the expression of laminin 332 conferring adhesion to extracellular matrix proteins produced by feeder layer.24 In contrast, negative clones maintained the impairment to generate colonies, which is typical of JEB keratinocytes. Restoration of adhesive properties of LAMB3-expressing clones was also demonstrated by longer detachment time compared to negative clones. The detachment time for LAMB3-expressing clones was comparable to normal keratinocytes (16 min), and negative clones as well as JEB.A649fsX38 cells detached after 5 min of trypsin treatment (Figure 4B). LAMB3-expressing clones showed higher proliferation rate compared to negative clones, as assessed in vitro by 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) assay (Figure 4C). The kinetics of cell growth of laminin-332-expressing clones appeared faster at all time points considered (48, 72, and 96 hr) and was more pronounced at the latest time point.
Figure 3.
Molecular Characterization of LAMB3-Corrected Clones
(A) Allelic discrimination: a three-primer-based (P1, P2, and P4; see Figure 1A) PCR approach was employed to discriminate between parental allele (628 bp; P1+P4; white arrow) and targeted allele (395 bp; P1+P2; black arrow). Monoallelic clones hosting one copy of the therapeutic donor showed both PCR bands. Biallelic clones hosting two copies of the therapeutic donor displayed only 395-bp band. M, 100-bp molecular weight marker. (B) qRT-PCR analysis on LAMB3 expression in representative clones with respect to untreated JEB.A649fsX38 keratinocytes and WT keratinocytes (Ker). Data are representative of three independent PCR amplifications (mean ± SEM; n = 3). (C) Western blot analysis showing laminin 332 expression in lysates from representative clones using an antibody against Kalinin B1 (an alternative name of laminin subunit beta-3 protein). β-actin staining served as loading control. Wild-type keratinocytes (Ker wt) and JEB.A649fsX38 keratinocytes (JEB) are used as positive and negative controls, respectively. Neg. indicates a negative clone. Protein molecular weights are indicated (kDa).
Figure 4.
Phenotypic Analysis of LAMB3-Corrected Clones
(A) Microscopic appearance of representative LAMB3-corrected clones (no. 36 and no. 65) grown on feeder layer. As control, negative clone (no. 71) and WT keratinocytes are shown. (B) Detachment time graph indicates the minutes (min) required to 20 LAMB3-corrected clones and 10 negative clones to lose the adhesion properties upon trypsin solution treatment. WT keratinocytes’ detachment time is indicated by a dashed line. (C) In vitro proliferation of LAMB3-corrected clones (pos) versus negative clones (neg) during time (48, 72, and 96 hr). Results are expressed as fold change from untreated JEB.A649fsX38 keratinocytes at 24 hr, which is set at 1. The bars represent the mean of 3 independent experiments (mean ± SEM; n = 3). p values are indicated.
Laminin 332 Restoration in a Human-Murine Skin Graft Model
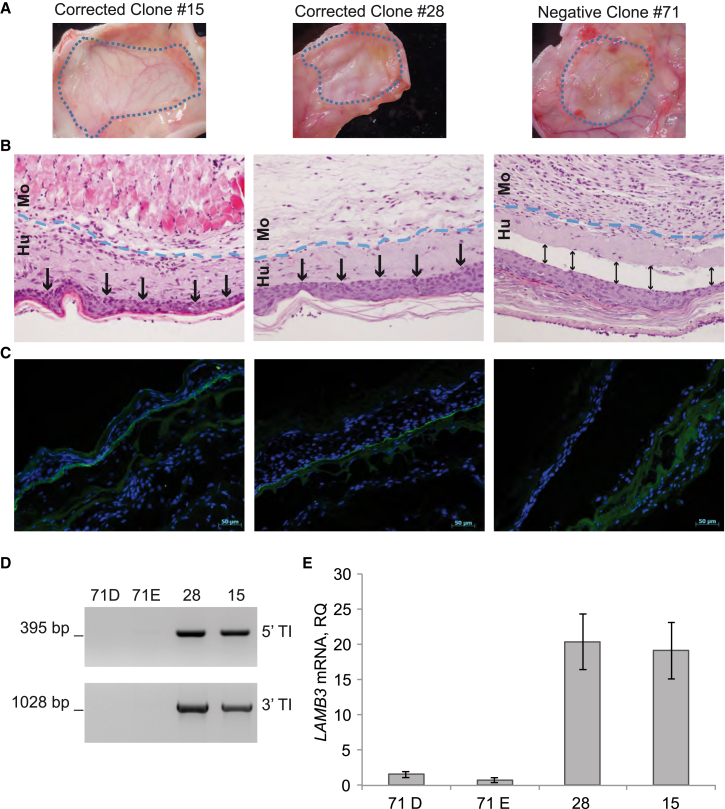
To determine whether LAMB3-corrected JEB.A649fsX38 keratinocytes sustain laminin 332 expression in vivo, skin equivalent (SE) generated from two clones displaying monoallelic integration of the therapeutic donor (no. 15 and no. 28), one un-corrected clone (no. 71), and untreated JEB.A649fsX38 keratinocytes (data on untreated cells are not shown) were grafted subcutaneously in the back of immunodeficient mice.
Four weeks post-transplantation, macroscopic analysis of engrafted areas clearly showed regenerated skin in mice grafted with LAMB3-corrected clones (no. 15 and no. 28) and heavily blistered skin in mice grafted with un-corrected clone (no. 71; Figure 5A). Skin biopsies were excised to perform histological, immunofluorescence, and molecular analysis. H&E staining of negative clone-derived SE showed JEB typical blistering phenotype, which was completely reversed in corrected clones. In SE derived from clone no. 71, epidermis (E) completely detached from dermis (D), and SEs derived from clones no. 28 and no. 15 showed continuous dermal-epidermal junction and visible stratification (Figure 5B). Staining using an antibody against human laminin 332 showed strong labeling along the basal membrane in the engrafted SEs derived from genetically corrected clones (no. 15 and no. 28) and undetectable labeling in the SE derived from the negative clone (no. 71; Figure 5C). Genomic DNA from E and D derived from clone no. 71 were included in the molecular assays run on biopsies from LAMB3-corrected SE. Targeted integration PCR analyses confirmed the genuine HR of the donor template into Cas9 target site only in biopsies derived from LAMB3-corrected SE (Figure 5D). qRT-PCR analysis showed more than 20-fold laminin 332 expression in LAMB3-corrected SE (Figure 5E) over SE from clone no. 71. These data indicate that monoallelic in situ integration of the therapeutic donor sustains sufficient laminin 332 expression to restore the dermal-epidermal junction in vivo, in a xenotransplantation model of human SEs on immunodeficient mice.
Figure 5.
In Vivo Skin Regeneration from LAMB3-Corrected Clones
(A) White light epi-illumination of grafts (cyan dotted lines) derived from 2 monoallelic corrected clones (no. 15 and no. 28) and 1 negative clone (no. 71) showed regeneration of human skin transplanted onto immunodeficient mice. (B) H&E staining of skin grafts 4 weeks after transplantation showed normal epidermal differentiation. Cyan dotted lines border the mouse (Mo) and the engrafted human skin (Hu). The arrows indicate BMZ. (C) Immunostaining of laminin 332 confirmed the complete restoration of epidermal-derma junction. The scale bar represents 50 μm. (D) 5′ and 3′ TI PCR on genomic DNA extracted from skin biopsies. The size of PCR bands is indicated. (E) LAMB3 expression in skin biopsies evaluated by qRT-PCR. D, derma; E, epidermis. Data are representative of three independent PCR amplifications (mean ± SEM; n = 3).
Discussion
Recently, gene editing approaches were proposed to treat severe diseases, viral infection,25 and monogenic disorders.26, 27, 28, 29, 30, 31, 32, 33, 34, 35 Any evidence on nuclease-mediated gene editing approach to restore laminin 332 protein, defective in JEB, has not been reported so far. The gene addition strategy based on viral vectors carrying an expression cassette for the correct gene has been the solely available strategy to correct genetic defects for many years. As regards to devastating skin disorders, successful gene therapy protocols based on autologous transplantation of skin keratinocytes transduced with a γ-retroviral vector expressing LAMB3 or COL7A1 cDNA were reported to treat JEB3, 4 and recessive DEB,5 respectively. However, gene editing approaches aimed to permanently correct disease-causing mutations or in situ integrate therapeutic cDNA remain an attractive and preferable alternative over the gene addition strategy.
Correction of a single mutation or replacement of quasi-complete gene employs the in situ HDR mechanism triggered by nuclease-mediated double-strand breaks (DSBs). A limiting drawback of HDR-mediated correction in clinically relevant cells, e.g., keratinocytes, is represented by the low efficiency of recombination, as reported also by other studies.20, 34 However, laminin 332 restoration in JEB keratinocytes confers adhesion and proliferation advantages to corrected cells,16 which could be therefore selected without the need of antibiotic selection or cell sorting. This observation prompted us to apply CRISPR-mediated HDR to safely in situ integrate a therapeutic LAMB3 cDNA in JEB keratinocytes.
To restore laminin 332 expression in JEB keratinocytes carrying A649fsX38 or R635X mutation in LAMB3 gene, we employed a first-generation adenoviral vector carrying SpCas9 coupled to a single gRNA tailored to intron 2 of LAMB3 gene and an IDLV vector bearing the promoterless LAMB3 cDNA (exon 3–23) flanked by HA. This strategy was designed to potentially address a variety of mutations mapped in LAMB3 gene,36, 37 to maintain the transcriptional regulatory elements of the endogenous gene, to optimize laminin 332 expression through a physiological splicing event, and to avoid the activation of neighboring oncogenes if undesired donor integration occurred. The choice of vectors to deliver Cas9 system and the donor template was based on our previous gene editing experience on human keratinocytes.20, 21 The top-ranked predicted off-targets associated to the designed gRNA did not show presence of indels by Surveyor assay, nevertheless 0.9% indel frequency was detected for OT7 by TIDE, suggesting that genome-wide analysis of off-targets performed by next generation sequencing (NGS)38, 39 or GUIDE sequencing (GUIDE-seq)40 will be necessary before clinical translation. Alternative Cas9 nucleases, e.g., SpCas9-HF141 or EvoCas9,42 should be considered too.
Genomic analysis of bulk keratinocytes revealed predominant genuine HDR events and rare undesired HDR-independent integration of IDLV-L.B3 donor, as previously observed using similar IDLV donors43, 44 (and unpublished data). After 2 rounds of cell detachment, we positively selected cells undergoing a predetermined splicing event, thus expressing the laminin 332 protein that confers adhesive and proliferative advantages. Cells carrying HDR-independent integration of IDLV-L.B3 were negatively selected as demonstrated by genomic analysis of treated JEB bulks and expanded JEB.A649fsX38 clones. However, these analyses do not rule out the presence of low-frequency (<1%) HDR-independent events in treated JEB keratinocytes, maybe undergoing aberrant splicing affecting LAMB3 mRNA expression and thereby poor in vitro adhesion. 5′ TI PCR on single isolated clones showed that 44% (39 out 88) of expanded clones correctly integrated the donor template by HDR, showing monoallelic correction in the vast majority of them (85%), independently from AdCas9gRNA doses. Based on our previous experience in wild-type keratinocytes20 and on flow cytometric analysis reported in this study, the HDR frequency without selection was <0.5%, thus remarking that selection of gene-corrected cells is needed. Eight clones out of 39 hosted the 5′ end of IDLV-L.B3 vector downstream bGH pA signal, further supporting the evidence that HDR-independent integration could be tolerated unless it does not affect LAMB3 mRNA expression, as demonstrated by RT-PCR analysis. However, the majority of clones screened by 5′ TI PCR resulted negative for genuine HDR events, indicating that an optimization of the adhesion-based selection protocol is required. Additional rounds of selection, different detachment time, or lower confluency of the transduced cultures could confer significant improvements to the procedure. Indeed, the experiments performed on JEB.R635X keratinocytes indicated that the adhesion selection protocol set up for JEB.A649fsX38 cells resulted in lower enrichment of LAMB3-expressing cells. Analysis at RNA and protein level on isolated JEB.A649fsX38 clones scored laminin 322 expression in all corrected clones. To note, laminin 332 expression level in monoallelic clones, which represent the vast majority of LAMB3-corrected clones, was sufficient to recapitulate the adhesive properties and the colony formation typical of normal keratinocytes, as well as their cell growth. Immunodeficient mice were transplanted with SE derived from 2 monoallelic LAMB3-corrected JEB.A649fsX38 clones, 1 un-corrected clone, and untreated JEB.A649fsX38 cells. Histological and immunofluorescence analysis showed a completely restored dermal-epidermal junction in biopsies of SE derived from LAMB3-corrected clones and a marked and complete detachment of human dermis to epidermis in un-corrected clone and in untreated JEB.A649fsX38 cells. Molecular analyses on DNA and RNA extracted from biopsies confirmed the genuine HR event and the physiological splicing event leading to 20-fold expression of LAMB3 mRNA over un-corrected or untreated JEB.A649fsX38 cells. Overall, these data provide evidence of the functional correction of laminin 332 defect in vivo.
This study represents the first promising application of CRISPR/Cas9-mediated gene editing to restore physiological LAMB3 expression in keratinocytes derived from JEB patients. The data demonstrated a safe in situ integration of the therapeutic LAMB3 cDNA in JEB keratinocytes, and although the adhesion selection protocol may require further improvement, this novel strategy moves gene editing for JEB closer to the clinic.
Materials and Methods
Vectors and Viral Production
For pCCL.LAMB3.HA.SA-cDNALAMB3pA.HA (IDLV-L.B3) construction, LAMB3 cDNA (3.8 kb in length), spanning from exon 3 to 23 and including the splice acceptor of exon 3, was amplified from MFG-LAMB3.3 Promoterless LAMB3 cDNA was cloned upstream of bGH pA signal, between 5′ and 3′ HA (90 and 413 bp in length, respectively), both from intron 2 of LAMB3 gene. The donor cassette was finally cloned in reverse orientation into pCCL.LV.polylinker vector,45 generating the transfer plasmid to be packaged into integration defective lentiviral vector (IDLV-L.B3).
For AdCas9gRNA construction, pShuttle.CRISPR.gRNA1LAMB3 was generated by cloning fragment U6.gRNA1LAMB3.CBH.hSpCas9.bGH.pA from plasmid pX330.gRNA1.LAMB3 in pShuttle from AdEasy Human Adenoviral Vector System (Stratagene). Briefly, pX330.gRNA1.LAMB3 was obtained by cloning gRNA1LAMB3 in pX330-U6-Chimeric_BB-CBh-hSpCas9 plasmid (Addgene no. 42230) by oligo annealing into BbsI sites (http://www.addgene.org). Then, pX330.gRNA1.LAMB3 was digested with PciI and NotI, and fragment U6.gRNA1Lamβ3.CBH.hSpCas9.bGH.pA was cloned in pShuttle from AdEasy (AdEasy Adenoviral Vector System; Stratagene), previously digested with NotI and filled in. pShuttle.CRISPR.gRNA1LAMB3 was linearized with PmeI, de-phosphorylated, and co-transformed with pAdEasy-1 from AdEasy system in BJ5183 bacteria to produce Ad5 genomic plasmid Ad.CRISPR.LAMB3gRNA1.
IDLV viral stocks pseudotyped with vesicular stomatitis virus G protein were prepared by transient co-transfection of HEK293T cells with transfer vector, pMD.Lg/pRRE.D64VInt packaging plasmid, pMD2.VSV-G envelope-encoding plasmid, and pRSV-Rev.20 Vector particles were concentrated by ultracentrifugation and titrated by Taqman RT-PCR with PCR primers and 6-carboxyfluorescein (FAM) probe for WPRE (Applied Biosystems). IDLV-L.B3 titer was 109 vg/mL by qPCR.
AdCas9gRNA vector was produced and titrated as previously described.46 Adenoviral titer was 1.9 × 1011 vg/mL by qPCR. The MOI reported for this virus indicates the amount of vg per cell.
Cell Culture
Primary keratinocytes from JEB patient carrying the homozygous LAMB3 mutation in exon 14 (c.1945dupG)17 were immortalized to facilitate clonal analysis. Early-passage primary JEB keratinocytes were infected with supernatants of pLXSN-based retroviral vector containing cDNA sequence of E6 and E7 proteins from papilloma virus type 1647 and referred to as JEB.A649fsX38 keratinocytes. The karyotype of JEB.A649fsX38 keratinocytes was analyzed (four metaphases) as described in Chamorro et al.47 and showed aneuploidy with redundant trisomy for chromosomes 9 and 20. Chromosome 1, which hosts LAMB3 gene, was diploid in 50% of analyzed metaphases, although in other metaphases, we detected an extra chromosome 1 (Figure S11). The second immortalized JEB keratinocytes line (JEB.R635X) used in this study was established as described in Hammersen et al.18 Feeder layer was required for JEB keratinocyte cell growth; thus, they were plated onto lethally irradiated 3T3-J2 cells. JEB.A649fsX38 keratinocytes were cultured in keratinocyte growth medium (cFAD medium and epidermal growth factor; KC medium), diluted 1:1 with Cnt-57 (progenitor cell-targeted [PCT] epidermal keratinocyte medium low bovine pituitary extract [BPE]; CellnTEC Advanced Cell Systems AG, Bern, Switzerland). JEB.R635X keratinocytes were cultured in keratinocyte growth medium (cFAD medium and epidermal growth factor; KC medium), diluted 1:1 with KGM-Gold (Lonza; Euroclone, Milan, Italy). Mouse 3T3-J2 cells48 were grown in DMEM supplemented with 10% donor bovine serum (Gibco; Thermo Fisher Scientific, Monza, Italy), 50 IU/mL penicillin-streptomycin, and 4 mmol/L glutamine.
Transduction of JEB Keratinocytes, Adhesion Selection, and Isolation of Single-Cell Clones
For editing analysis, JEB cells were plated 24 hr before transduction (without feeder layer) and then were transduced with AdCas9gRNA in serum-free medium containing viral particles at MOIs 104, 2 × 104, 5 × 104, or 105 vg/cell. After 1 hr at 37°C, the viral mixture was replaced with Cnt-57 + KC medium. For HR-mediated correction, JEB keratinocytes (2.5 × 105) were first infected with IDLV-L.B3 (4.32 × 102 vg/cell) in suspension, in the presence of polybrene (8 μg/mL), and seeded in 6-well plates onto lethally irradiated 3T3-J2 cells. After 6 hr, CnT-57 medium was added to cells. After 24 hr, AdCas9gRNA transduction was performed in serum-free medium containing viral particles at MOIs 104, 2 × 104, 5 × 104, or 105 vg/cell. After 1 hr at 37°C, the infection mixture was replaced with Cnt-57 + KC medium.
To enrich LAMB3-corrected cells, 4 days post-transduction JEB keratinocytes transduced with IDLV-L.B3 (4.32 × 102 vg/cell) and AdCas9gRNA (MOIs 104 and 2 × 104 vg/cell) were washed with PBS and incubated with trypsin solution at 37°C. Cells that detached after 4 min of incubation were removed and referred as un-corrected (negative) JEB cells. Adherent cells were further incubated with trypsin solution at 37°C for additional 4 min and named LAMB3-corrected JEB keratinocytes. After trypsin treatment, cells were plated onto lethally irradiated 3T3-J2 cells in KC medium diluted 1:1 with CnT-57. To isolate single-cell clones, JEB.A649fsX38 cells transduced with IDLV-L.B3 (4.32 × 102 vg/cell) and AdCas9gRNA (MOIs 104 and 2 × 104 vg/cell) were limiting diluted, plated onto lethally irradiated 3T3-J2 cells, and cultured in KC medium diluted 1:1 with CnT-57.
Editing Efficiency Analysis and DNA Sequence Analysis
To analyze editing efficiency and specificity, genomic DNA was extracted using DNeasy MiniKit (QIAGEN, Milano, Italy). To evaluate on-target editing, gRNA target region from JEB cells transduced at MOI 104, 2 × 104, 5 × 104, and 105 vg/cell or un-transduced (mock transduced) was amplified by PCR (Platinum Taq High Fidelity; Thermo Fisher Scientific, Monza, Italy), using primers Cel1LamB3for and Cel1LamB3rev (Table S1), and analyzed with TIDE software to evaluate indels frequency.
To amplify the off-targets predicted by two public webservers (http://crispr.mit.edu and CRISPR direct [https://crispr.dbcls.jp]), we designed several primers listed in Table S1. The amplification products were analyzed by Surveyor assay, according to SURVEYOR Mutation Detection Kit (Transgenomic, Omaha, NE) manufacturer’s protocol. Briefly, 72 hr after transduction, PCR amplifications were performed on genomic DNA and then PCR products were denatured, digested by Cel-I nuclease, and finally subjected to 2% agarose gel electrophoresis. 100 bp DNA ladder from New England Biolabs (Massachusetts, USA) was used as molecular weight marker. For DNA sequence analysis of on-target indels, PCR products were subcloned into a PCR2.1 TOPO vector (Thermo Fisher Scientific, Monza, Italy) and Sanger sequenced (Eurofin s.r.l Vimodrone, Italy). Sanger sequences were analyzed accordingly to a bioinformatics pipeline (D.B., V. Marigo, and A.R., unpublished data) to accurately evaluate the percentage and type of indels upon AdCas9gRNA treatment.
PCR and Semiquantitative and qRT-PCR Analyses
Genomic DNA was isolated from JEB cells or skin biopsies with a QIAmp DNA Mini kit (QIAGEN, S.p.A., Milan, Italy), according to manufacturer’s protocols. To analyze correct integration of the donor in LAMB3 locus, we performed target integration PCRs (PCR GoTaq, Promega, Milan Italy) on genomic DNA using primers TI5′LamB3for and TI5′LamB3rev to analyze the correct integration at 5′ region and primers TI3′LamB3for and TI3′LamB3rev to analyze the correct integration at 3′ region (see Table S2 for primers list). PCR cycles for TI5′ are 95°C 30 s, 57°C 30 s, and 72°C 30 s; PCR cycles for TI3′ are 95°C 30 s, 55°C 30 s, and 72°C 1 min. PCR products were loaded on 1% agarose gel, and 1 kb or 100 bp DNA ladders from New England Biolabs (Massachusetts, USA) were used as molecular weight markers.
To investigate the integration of the 5′ end of IDLV vector, we performed modified target integration 3′ PCR (TaKaRa LA Taq, USA) on genomic DNA using primers TI3′LamB3for and LamB3-Ex3-RC. PCR cycles are as follows: 98°C 30 s; 60°C 30 s; and 68°C 2 min. For allelic discrimination, a three-primer-based PCR (TaKaRa LA Taq, USA) was performed to discriminate between parental allele (TI5′LamB3for and TI3′LamB3rev) and donor-integrated allele (TI5′LamB3for and TI5′LamB3rev). PCR cycles are as follows: 98°C 20 s; 55°C 30 s; and 68°C 5 min.
Total RNA from JEB cells or skin biopsies was isolated with RNeasy Mini kit plus (QIAGEN, Milan, Italy), according to manufacturer’s protocols. cDNA from JEB cells was synthesized in a 20 μL reaction using 200 ng total RNA and ImProm-II Reverse Transcription System (Promega, Milan, Italy). cDNA from skin biopsies was synthesized in a 20-μL reaction using 400 ng and SuperScript III (Life Technologies, Monza, Italy).
To evaluate LAMB3 mRNA expression, semiquantitative RT-PCR was performed with primers LamB3Exon13_F and LamB3Exon15_R. PCR cycles are as follows: 95°C 30 s; 60°C 30 s; and 72°C 45 s. As housekeeping control, GAPDH mRNA was amplified with primers GAPDH-F and GAPDH-R. PCR cycles are as follows: 95°C 30 s; 60°C 30 s; and 72°C 45 s.
qRT-PCR was performed in 96-well plates on ABI Prism 7900 Sequence Detection System (Applied Biosystems, Monza, Italy) with TaqMan Universal PCR Master Mix and Taqman Gene Expression assay (Applied Biosystems), with a final reaction of 25 μL. PCR primers and 6-carboxyfluorescein (FAM) probes for GAPDH (Hs99999905_m1) or LAMB3 (Hs00989722_m1) were purchased from Applied Biosystems. Reactions were performed according to manufacturer’s instructions. The relative expression was normalized to the level of GAPDH in the same cDNA using 2-ΔΔCt quantification. The mean of replicated relative quantity (RQ) values for each sample was calculated.
Flow Cytometry
For flow cytometric analysis of LAMB3 expression, after washing with PBS + 5% fetal bovine serum (FBS) + 2.5 mM EDTA, JEB cells were stained with antigen-presenting cell (APC)-conjugated anti-feeder antibody (Miltenyi Biotec, Bologna, Italy) accordingly to manufacturer’s protocol. Cells were fixed and permeabilized using the Fix and Perm cell permeabilization reagents kit (Invitrogen, Monza, Italy), according to manufacturer’s protocol, before intracellular staining of LAMB3 with anti-β3 chain monoclonal antibody K140 (diluted 1:30; a kind gift from Prof. Castiglia, Istituto Dermopatico dell’Immacolata, IRCCS, Rome, Italy). As secondary antibody, Alexa 488 donkey anti-mouse antibody (Life Technologies, Monza, Italy; 1:1,000) was used. Fluorescence was acquired on a FACS CANTO cytofluorimeter (BD Biosciences, Milan, Italy). Intracellular LAMB3 expression was evaluated after feeder exclusion (APC− cells). The percentage of LAMB3-positive cells was determined after subtracting the percentage of LAMB3-positive events in untreated cells. All flow cytometry experiments were analyzed with the FlowJo software (Tree Star).
Western Blotting
Cell lysates were extracted with radioimmunoprecipitation assay (RIPA) buffer containing proteinase inhibitor cocktail (Complete Mini, EDTA-free; Roche Diagnostics, Monza, Italy), incubated for 30 min on ice and centrifuged at 14,000 rpm for 30 min. The amount of proteins was determined using Bradford assay (Bio-Rad, Milan, Italy). Sixty micrograms of JEB keratinocytes extracts and 10 μg of primary keratinocyte extracts were separated on 4%–12% gradient NuPAGE Bis-Tris Gel (Life Technologies, Monza, Italy) under reducing condition. Western blots were performed with a mouse monoclonal antibody against human Kalinin B1 (Kalinin B; 1:1,000; BD Transduction Laboratories, Milan, Italy). Kalinin B1 is an alternative name of laminin subunit beta-3 protein coded by LAMB3 gene according to Uniprot (https://www.uniprot.org/uniprot/Q13751). For protein loading normalization, a mouse monoclonal antibody anti-β-actin (sc-81178; 1:1,000; Santa Cruz Biotechnology, Heidelberg, Germany) was used. Horseradish-peroxidase-conjugated anti-mouse antibody (diluted 1:5,000) was used for chemiluminescent detection (Amersham; GE Healthcare Europe, Milan, Italy).
Keratinocyte Growth and Detachment Analysis
JEB clone proliferation was measured by MTT reduction colorimetric assay (Sigma-Aldrich, Milan, Italy). For the time course analysis, 4 × 104 cells/well were plated without feeder layer in KC medium diluted 1:1 with CnT-57, a well of a 24-well plate per time point, and incubated at 37°C for 24, 48, 72, or 96 hr. At each time point, cells were incubated with 500 ng/mL MTT solution at 37°C for 90 min, and then MTT solution was gently removed. The incorporated dye was solubilized in acidified isopropanol and determined by absorbance measurement (A570nm). Results are expressed as fold change from untreated control cells at 24 hr, which is set at 1 and as mean of 3 independent experiments.
Detachment time analysis was performed by measuring the time of incubation with trypsin solution necessary to all cells of a sub-confluent culture to lose adhesive properties.
Bioengineered Skin Preparation and Grafting on Immunodeficient Mice
The SE or bioengineered skin corresponds to a 3D culture resembling the basic architecture of skin. It comprises a dermal hydrogel matrix (fibrin) containing embedded live human fibroblasts and an epidermal component formed by human keratinocytes (genetically corrected or non-corrected) added to the dermal matrix and grown until confluence (keratinocyte layer). Bioengineered skin grafts were prepared from 4 samples: 2 LAMB3-corrected clones; 1 negative clone; and untreated JEB.A649fsX38 cells as previously described.49 Bioengineered SEs were grafted subcutaneously in the back of immunodeficient nu/nu mice (n = 3/sample) according to Barrandon et al.49 Four weeks after grafting, mice were sacrificed and grafts harvested for skin immunohistochemistry and molecular analysis. Animal studies were approved by our institutional animal care and use committee according to all legal regulations.
Immunofluorescence and Immunohistochemistry
Immunofluorescence analysis for laminin 332 expression on skin biopsies from mice was performed on 7-μm sections. After blocking with 10% fetal bovine serum + 3% BSA in PBS, sections were incubated with 50 μg/mL unconjugated Fab anti-immunoglobulin G (IgG) antibody. After wash, sections were incubated with mouse monoclonal GB3 antibody (diluted 1:5; a kind gift from Prof. Castiglia, Istituto Dermopatico dell’Immacolata, IRCCS, Rome, Italy). As secondary antibody, Alexa 488 donkey anti-mouse antibody (Life Technologies; 1:1,000) was used. Staining with 0.1 μg/mL DAPI was performed for nuclear staining. Slides were mounted with Dako Fluorescent Mounting Medium (Dako Italia SRL, Milan, Italy) and analyzed with a Zeiss Axioskop 40 FL fluorescence microscope equipped with a digital camera AxioCam and AxioVisionRel version 4.8 software for image processing (Carl Zeiss, Milan, Italy).
Histological samples of the whole mouse-human skin “sandwich” were formalin fixed and H&E stained as described in Chamorro et al.47
Statistical Analysis
Two-way ANOVA or Student’s t test was performed for statistical significance. All values in each group were expressed as mean ± SEM.
Author Contributions
D.B., F.M., and F.C. designed experiments; D.B. performed experiments in vitro and ex vivo on skin biopsies and analyzed data; F.M., F.C., C.P., and S.B. performed experiments in vitro; C.H. provided immortalized JEB carrying the R635X mutation; M.C., M.D.R., and F.L. were involved in SE production and grafting experiments; and S.C. and V.A. packaged the first generation AdCas9gRNA vector. D.B and A.R. wrote the manuscript.
Conflicts of Interest
The authors have no conflicts of interest to disclose.
Acknowledgments
This work was supported by grants from the European Commission (FP7, PERSIST) and the Italian Ministry of University and Research. M.D.R. and F.L. are supported by Spanish grants SAF2013-43475-R and SAF2017-86810-R from the Ministry of Economy and Competitiveness and PI14/00931 and PI17/01747 from Instituto de Salud Carlos III co-funded with European Regional Development Funds (ERDF). The authors thank Blanca Duarte (Epithelial Biomedicine Division, Cutaneous Disease Modelling Unit, CIEMAT, Madrid, Spain) for her technical assistance in the grafting experiments and Prof. Daniele Castiglia and Naomi De Luca (Istituto Dermopatico dell’Immacolata, IRCCS, Rome, Italy) for providing mouse monoclonal GB3 antibody used in the immunofluorescence analysis for laminin 332.
Footnotes
Supplemental Information includes eleven figures and two tables and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.07.024.
Supplemental Information
References
- 1.Dellambra E., Vailly J., Pellegrini G., Bondanza S., Golisano O., Macchia C., Zambruno G., Meneguzzi G., De Luca M. Corrective transduction of human epidermal stem cells in laminin-5-dependent junctional epidermolysis bullosa. Hum. Gene Ther. 1998;9:1359–1370. doi: 10.1089/hum.1998.9.9-1359. [DOI] [PubMed] [Google Scholar]
- 2.Melo S.P., Lisowski L., Bashkirova E., Zhen H.H., Chu K., Keene D.R., Marinkovich M.P., Kay M.A., Oro A.E. Somatic correction of junctional epidermolysis bullosa by a highly recombinogenic AAV variant. Mol. Ther. 2014;22:725–733. doi: 10.1038/mt.2013.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mavilio F., Pellegrini G., Ferrari S., Di Nunzio F., Di Iorio E., Recchia A., Maruggi G., Ferrari G., Provasi E., Bonini C. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nat. Med. 2006;12:1397–1402. doi: 10.1038/nm1504. [DOI] [PubMed] [Google Scholar]
- 4.Hirsch T., Rothoeft T., Teig N., Bauer J.W., Pellegrini G., De Rosa L., Scaglione D., Reichelt J., Klausegger A., Kneisz D. Regeneration of the entire human epidermis using transgenic stem cells. Nature. 2017;551:327–332. doi: 10.1038/nature24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siprashvili Z., Nguyen N.T., Gorell E.S., Loutit K., Khuu P., Furukawa L.K., Lorenz H.P., Leung T.H., Keene D.R., Rieger K.E. Safety and wound outcomes following genetically corrected autologous epidermal grafts in patients with recessive dystrophic epidermolysis bullosa. JAMA. 2016;316:1808–1817. doi: 10.1001/jama.2016.15588. [DOI] [PubMed] [Google Scholar]
- 6.Nathwani A.C., Reiss U.M., Tuddenham E.G., Rosales C., Chowdary P., McIntosh J., Della Peruta M., Lheriteau E., Patel N., Raj D. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 2014;371:1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hacein-Bey-Abina S., Pai S.Y., Gaspar H.B., Armant M., Berry C.C., Blanche S., Bleesing J., Blondeau J., de Boer H., Buckland K.F. A modified γ-retrovirus vector for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2014;371:1407–1417. doi: 10.1056/NEJMoa1404588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genovese P., Schiroli G., Escobar G., Tomaso T.D., Firrito C., Calabria A., Moi D., Mazzieri R., Bonini C., Holmes M.C. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510:235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan Y., Ma Y., Li Q., Sun Z., Ma L., Wu L., Wang L., Zeng L., Shao Y., Chen Y. CRISPR/Cas9-mediated somatic correction of a novel coagulator factor IX gene mutation ameliorates hemophilia in mouse. EMBO Mol. Med. 2016;8:477–488. doi: 10.15252/emmm.201506039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoddard B.L. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure. 2011;19:7–15. doi: 10.1016/j.str.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urnov F.D., Rebar E.J., Holmes M.C., Zhang H.S., Gregory P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 12.Bogdanove A.J., Voytas D.F. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 13.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwank G., Koo B.K., Sasselli V., Dekkers J.F., Heo I., Demircan T., Sasaki N., Boymans S., Cuppen E., van der Ent C.K. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Wild E.J., Tabrizi S.J. Therapies targeting DNA and RNA in Huntington’s disease. Lancet Neurol. 2017;16:837–847. doi: 10.1016/S1474-4422(17)30280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dellambra E., Pellegrini G., Guerra L., Ferrari G., Zambruno G., Mavilio F., De Luca M. Toward epidermal stem cell-mediated ex vivo gene therapy of junctional epidermolysis bullosa. Hum. Gene Ther. 2000;11:2283–2287. doi: 10.1089/104303400750035825. [DOI] [PubMed] [Google Scholar]
- 17.Castori M., Floriddia G., Pisaneschi E., Covaciu C., Paradisi M., Torrente I., Castiglia D. Complete maternal isodisomy causing reduction to homozygosity for a novel LAMB3 mutation in Herlitz junctional epidermolysis bullosa. J. Dermatol. Sci. 2008;51:58–61. doi: 10.1016/j.jdermsci.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Hammersen J., Has C., Naumann-Bartsch N., Stachel D., Kiritsi D., Söder S., Tardieu M., Metzler M., Bruckner-Tuderman L., Schneider H. Genotype, clinical course, and therapeutic decision making in 76 infants with severe generalized junctional epidermolysis bullosa. J. Invest. Dermatol. 2016;136:2150–2157. doi: 10.1016/j.jid.2016.06.609. [DOI] [PubMed] [Google Scholar]
- 19.Moiani A., Paleari Y., Sartori D., Mezzadra R., Miccio A., Cattoglio C., Cocchiarella F., Lidonnici M.R., Ferrari G., Mavilio F. Lentiviral vector integration in the human genome induces alternative splicing and generates aberrant transcripts. J. Clin. Invest. 2012;122:1653–1666. doi: 10.1172/JCI61852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coluccio A., Miselli F., Lombardo A., Marconi A., Malagoli Tagliazucchi G., Gonçalves M.A., Pincelli C., Maruggi G., Del Rio M., Naldini L. Targeted gene addition in human epithelial stem cells by zinc-finger nuclease-mediated homologous recombination. Mol. Ther. 2013;21:1695–1704. doi: 10.1038/mt.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latella M.C., Cocchiarella F., De Rosa L., Turchiano G., Gonçalves M.A.F.V., Larcher F., De Luca M., Recchia A. Correction of recessive dystrophic epidermolysis bullosa by transposon-mediated integration of COL7A1 in transplantable patient-derived primary keratinocytes. J. Invest. Dermatol. 2017;137:836–844. doi: 10.1016/j.jid.2016.11.038. [DOI] [PubMed] [Google Scholar]
- 22.Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naito Y., Hino K., Bono H., Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31:1120–1123. doi: 10.1093/bioinformatics/btu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Llames S., García-Pérez E., Meana Á., Larcher F., del Río M. Feeder layer cell actions and applications. Tissue Eng. Part B Rev. 2015;21:345–353. doi: 10.1089/ten.teb.2014.0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tebas P., Stein D., Tang W.W., Frank I., Wang S.Q., Lee G., Spratt S.K., Surosky R.T., Giedlin M.A., Nichol G. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M., Madhavan S., Pan X., Ran F.A., Yan W.X. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schiroli G., Ferrari S., Conway A., Jacob A., Capo V., Albano L., Plati T., Castiello M.C., Sanvito F., Gennery A.R. Preclinical modeling highlights the therapeutic potential of hematopoietic stem cell gene editing for correction of SCID-X1. Sci. Transl. Med. 2017;9:eaan0820. doi: 10.1126/scitranslmed.aan0820. [DOI] [PubMed] [Google Scholar]
- 28.Sharma R., Anguela X.M., Doyon Y., Wechsler T., DeKelver R.C., Sproul S., Paschon D.E., Miller J.C., Davidson R.J., Shivak D. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood. 2015;126:1777–1784. doi: 10.1182/blood-2014-12-615492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dever D.P., Bak R.O., Reinisch A., Camarena J., Washington G., Nicolas C.E., Pavel-Dinu M., Saxena N., Wilkens A.B., Mantri S. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diez B., Genovese P., Roman-Rodriguez F.J., Alvarez L., Schiroli G., Ugalde L., Rodriguez-Perales S., Sevilla J., Diaz de Heredia C., Holmes M.C. Therapeutic gene editing in CD34+ hematopoietic progenitors from Fanconi anemia patients. EMBO Mol. Med. 2017;9:1574–1588. doi: 10.15252/emmm.201707540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hainzl S., Peking P., Kocher T., Murauer E.M., Larcher F., Del Rio M., Duarte B., Steiner M., Klausegger A., Bauer J.W. COL7A1 editing via CRISPR/Cas9 in recessive dystrophic epidermolysis bullosa. Mol. Ther. 2017;25:2573–2584. doi: 10.1016/j.ymthe.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osborn M.J., Starker C.G., McElroy A.N., Webber B.R., Riddle M.J., Xia L., DeFeo A.P., Gabriel R., Schmidt M., von Kalle C. TALEN-based gene correction for epidermolysis bullosa. Mol. Ther. 2013;21:1151–1159. doi: 10.1038/mt.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chamorro C., Mencía A., Almarza D., Duarte B., Büning H., Sallach J., Hausser I., Del Río M., Larcher F., Murillas R. Gene editing for the efficient correction of a recurrent COL7A1 mutation in recessive dystrophic epidermolysis bullosa keratinocytes. Mol. Ther. Nucleic Acids. 2016;5:e307. doi: 10.1038/mtna.2016.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Izmiryan A., Danos O., Hovnanian A. Meganuclease-mediated COL7A1 gene correction for recessive dystrophic epidermolysis bullosa. J. Invest. Dermatol. 2016;136:872–875. doi: 10.1016/j.jid.2015.11.028. [DOI] [PubMed] [Google Scholar]
- 35.Kocher T., Peking P., Klausegger A., Murauer E.M., Hofbauer J.P., Wally V., Lettner T., Hainzl S., Ablinger M., Bauer J.W. Cut and paste: efficient homology-directed repair of a dominant negative KRT14 mutation via CRISPR/Cas9 nickases. Mol. Ther. 2017;25:2585–2598. doi: 10.1016/j.ymthe.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Posteraro P., De Luca N., Meneguzzi G., El Hachem M., Angelo C., Gobello T., Tadini G., Zambruno G., Castiglia D. Laminin-5 mutational analysis in an Italian cohort of patients with junctional epidermolysis bullosa. J. Invest. Dermatol. 2004;123:639–648. doi: 10.1111/j.0022-202X.2004.23302.x. [DOI] [PubMed] [Google Scholar]
- 37.Condorelli A.G., Fortugno P., Cianfarani F., Proto V., Di Zenzo G., Didona B., Zambruno G., Castiglia D. Lack of K140 immunoreactivity in junctional epidermolysis bullosa skin and keratinocytes associates with misfolded laminin epidermal growth factor-like motif 2 of the β3 short arm. Br. J. Dermatol. 2018;178:1416–1422. doi: 10.1111/bjd.15690. [DOI] [PubMed] [Google Scholar]
- 38.Tabebordbar M., Zhu K., Cheng J.K.W., Chew W.L., Widrick J.J., Yan W.X., Maesner C., Wu E.Y., Xiao R., Ran F.A. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casini A., Olivieri M., Petris G., Montagna C., Reginato G., Maule G., Lorenzin F., Prandi D., Romanel A., Demichelis F. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat. Biotechnol. 2018;36:265–271. doi: 10.1038/nbt.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holkers M., Maggio I., Henriques S.F., Janssen J.M., Cathomen T., Gonçalves M.A. Adenoviral vector DNA for accurate genome editing with engineered nucleases. Nat. Methods. 2014;11:1051–1057. doi: 10.1038/nmeth.3075. [DOI] [PubMed] [Google Scholar]
- 44.Lombardo A., Genovese P., Beausejour C.M., Colleoni S., Lee Y.L., Kim K.A., Ando D., Urnov F.D., Galli C., Gregory P.D. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 45.Cocchiarella F., Latella M.C., Basile V., Miselli F., Galla M., Imbriano C., Recchia A. Transcriptionally regulated and nontoxic delivery of the hyperactive sleeping beauty transposase. Mol. Ther. Methods Clin. Dev. 2016;3:16038. doi: 10.1038/mtm.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Catalucci D., Sporeno E., Cirillo A., Ciliberto G., Nicosia A., Colloca S. An adenovirus type 5 (Ad5) amplicon-based packaging cell line for production of high-capacity helper-independent deltaE1-E2-E3-E4 Ad5 vectors. J. Virol. 2005;79:6400–6409. doi: 10.1128/JVI.79.10.6400-6409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chamorro C., Almarza D., Duarte B., Llames S.G., Murillas R., García M., Cigudosa J.C., Espinosa-Hevia L., Escámez M.J., Mencía A. Keratinocyte cell lines derived from severe generalized recessive epidermolysis bullosa patients carrying a highly recurrent COL7A1 homozygous mutation: models to assess cell and gene therapies in vitro and in vivo. Exp. Dermatol. 2013;22:601–603. doi: 10.1111/exd.12203. [DOI] [PubMed] [Google Scholar]
- 48.Todaro G.J., Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barrandon Y., Li V., Green H. New techniques for the grafting of cultured human epidermal cells onto athymic animals. J. Invest. Dermatol. 1988;91:315–318. doi: 10.1111/1523-1747.ep12475646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.