Abstract
The genus Sanguisorba, which contains about 30 species around the world and seven species in China, is the source of the medicinal plant Sanguisorba officinalis, which is commonly used as a hemostatic agent as well as to treat burns and scalds. Here we report the complete chloroplast (cp) genome sequences of four Sanguisorba species (S. officinalis, S. filiformis, S. stipulata, and S. tenuifolia var. alba). These four Sanguisorba cp genomes exhibit typical quadripartite and circular structures, and are 154,282 to 155,479 bp in length, consisting of large single-copy regions (LSC; 84,405–85,557 bp), small single-copy regions (SSC; 18,550–18,768 bp), and a pair of inverted repeats (IRs; 25,576–25,615 bp). The average GC content was ~37.24%. The four Sanguisorba cp genomes harbored 112 different genes arranged in the same order; these identical sections include 78 protein-coding genes, 30 tRNA genes, and four rRNA genes, if duplicated genes in IR regions are counted only once. A total of 39–53 long repeats and 79–91 simple sequence repeats (SSRs) were identified in the four Sanguisorba cp genomes, which provides opportunities for future studies of the population genetics of Sanguisorba medicinal plants. A phylogenetic analysis using the maximum parsimony (MP) method strongly supports a close relationship between S. officinalis and S. tenuifolia var. alba, followed by S. stipulata, and finally S. filiformis. The availability of these cp genomes provides valuable genetic information for future studies of Sanguisorba identification and provides insights into the evolution of the genus Sanguisorba.
Keywords: Sanguisorba, chloroplast genome, molecular structure, phylogenetic analysis
1. Introduction
The genus Sanguisorba belongs to the Rosaceae; there are about 30 species in the genus Sanguisorba in the world, mainly distributed in Asia, Europe, and North America (eFlora of China: http://www.eflora.cn/). There are seven species and six varieties of Sanguisorba in China [1], distributed in both northern and southern China, especially in the northeast provinces. Sanguisorba officinalis has been recorded as a medicinal plant that is commonly used to treat water and fire burns, hemorrhoidal bleeding, and hematochezia [2]. Diyu Shengbai Tablet, a Chinese patent medicine, is mainly composed of S. officinalis, and contains active chemical components including saponins, flavonoids and tannins [3]. It can protect the hematopoietic system, elevate the peripheral blood white blood cells, neutrophils, and platelets, improve bone marrow micro-circulation, and adjust and improve body immunity and other functions. It is also often clinically used as an adjuvant during chemotherapy [3].
The chloroplast genome is ~100–150 kb in length and contains a wealth of evolutionary information, which can be used to reveal phylogenetic relationships among closely related species and can also be valuable for species identification [4,5]. It has been widely used in species identification, phylogenetic evolution, and genetic engineering-related research [6,7]. With the rapid development of high-throughput sequencing technologies and bioinformatics tools, the cost of sequencing chloroplast genome has been significantly reduced, making the large-scale acquisition of chloroplast genomic sequences possible [8,9]. This has made possible the study of chloroplast genomes in terms of population genetic structure, phylogenetic evolution, and species identification.
However, molecular research on the genus Sanguisorba is still very scarce. Currently, there are no reports on the chloroplast genome sequence of the genus Sanguisorba, which seriously hampers molecular identification, phylogenetic, genetic, and breeding research involving the genus. In this study, we report the chloroplast genome assembly, annotation, and structural analysis of four Sanguisorba species (S. officinalis, S. filiformis, S. stipulata, and S. tenuifolia var. alba) as well as the complete chloroplast genome sequences of these species, which are the first four sequenced members of the genus Sanguisorba. In addition, we compared the chloroplast genomes of the four Sanguisorba species in detail (e.g., based on IR expansion/contraction and difference regions). From this we constructed a phylogenetic tree using the maximum parsimony (MP) method based on both the whole cp genome and on common protein-coding genes, respectively. Overall, our results provide useful genetic information on the chloroplast of Sanguisorba species, as well as their relative position in phylogenetic tree.
2. Results and Discussion
2.1. Chloroplast Genome Assembly and Features
Using an Illumina HiSeq X platform, four Sanguisorba species were sequenced to produce 11,554,422–18,828,898 paired-end raw reads. After screening these paired-end reads, 598,166 to 1,080,144 cp genome reads were successfully mapped with 569X to 1032X sequencing depth (Table 1). In this study, the sequencing depth was high enough to satisfy the technical requirements of an organelle genome assembly. In total, the complete cp genomes of the four Sanguisorba species were similar in length, ranging from 155,127 bp (S. stipulata) to 155,479 bp (S. officinalis) (Figure 1 and Figures S1–S3, and Table 1), with the typical quadripartite structure of angiosperms. All four cp genomes contained a large single-copy regions (LSC, 84,405–85,557 bp) and a small single-copy regions (SSC, 18,550–18,768bp), separated by a pair of inverted repeats regions (IRs, 25,576–25,615 bp).
Table 1.
Sequence information and Illumina next-generation sequencing (NGS) data of the four Sanguisorba chloroplast genomes.
| Species | Raw Reads No. | Mapped Reads No. | Sequencing Depth | Cp Genome Length (bp) | GC Content (%) | LSC a (bp) | SSC a (bp) | IRs a (bp) |
|---|---|---|---|---|---|---|---|---|
| S. officinalis | 11,554,422 | 609,666 | 581X | 155,479 | 37.19 | 85,547 | 18,768 | 25,582 |
| S. filiformis | 16,876,554 | 656,271 | 628X | 154,282 | 37.33 | 84,405 | 18,659 | 25,609 |
| S. stipulata | 18,828,898 | 1,080,144 | 1032X | 155,127 | 37.23 | 85,347 | 18,550 | 25,615 |
| S. tenuifolia var. alba | 18,366,336 | 598,166 | 569X | 155,457 | 37.20 | 85,557 | 18,748 | 25,576 |
a LSC (large single-copy regions), SSC (small single-copy regions), and IRs (inverted repeats regions).
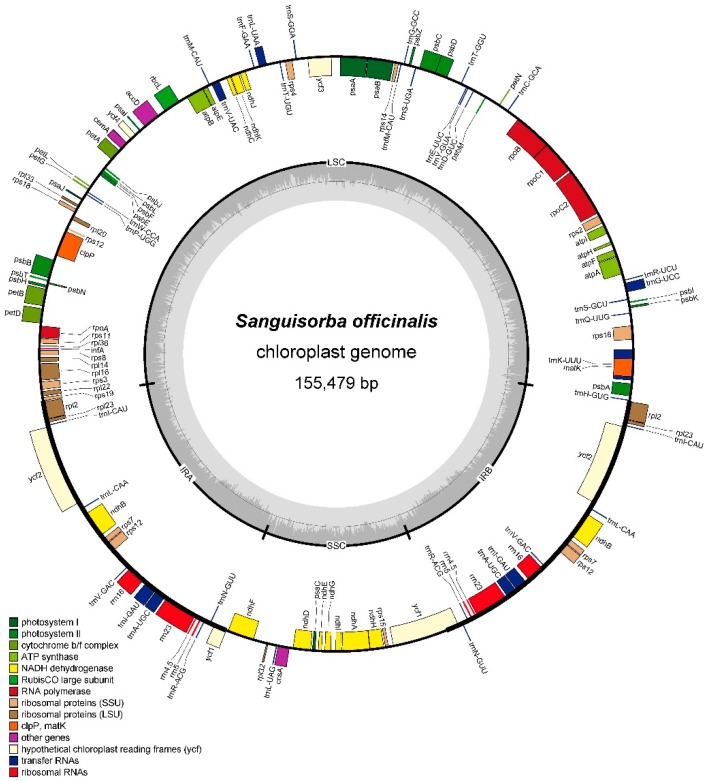
Figure 1.
Gene map of Sanguisorba officinalis chloroplast genome. Genes shown inside the circle are transcribed clockwise, and those outside are counterclockwise. Genes in different functional groups are color-coded.
The average GC content of the four Sanguisorba cp genomes was ~37.23%; in this respect they showed only minor differences from one another and resembled the cp genomes of other reported Rosaceae species [10,11,12]. Nevertheless, the GC content is unevenly distributed in the four Sanguisorba cp genomes. The GC content of the IR regions (~42.7%) is significantly higher than in the LSC region (~35.3%) or the SSC regions (~31.3%). We speculate that this may be a reason for the divergence of the conservation between the IR and SC regions [8,13].
All four Sanguisorba cp genomes possessed 112 unique genes including 78 protein-coding genes, 30 tRNA genes, and four rRNA genes (Table 2). Of these, six protein-coding genes, seven tRNA genes, and four rRNA genes are duplicated in the IR regions, making a total of 129 genes shared (Table 2). Our results showed that the four Sanguisorba cp genomes were highly conserved in gene type, order, and content. We classified the 112 genes into different categories according to their function, and the details are shown in Table 2. In addition, two pseudogenes (ycf1 and infA) were found in the four cp genomes. There were 18 genes located in the IR regions as follows: rrn16, rrn23, rrn5, rrn4.5, trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC, rps7, rps12, rpl2, rpl23, ndhB, ycf1, and ycf2 (Figure 1 and Figures S1–S3). rps12 is a trans-spliced gene, in which two 3’ end residues are located in the IR region and the 5’ end in the LSC region (Figure 1 and Figures S1–S3). This is a common phenomenon in the cp genomes of higher plants [14,15]. Significantly, the ycf15 gene is located in cp genome of most angiosperm while is absent from the Sanguisorba cp genomes. This phenomenon was also found to occur in Cedrela odorata [7], Schisandra chinensis [8], Cremastra appendiculata [16] and Aristolochia debilis [17].
Table 2.
List of genes encoded by the four Sanguisorba chloroplast genomes.
| Category | Group | Name |
|---|---|---|
| Self-replication | rRNA genes | rrn4.5a, rrn5a, rrn16a, rrn23a |
| tRNA genes | trnA-UGC *,a, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC *, trnH-GUG, trnI-CAU a | |
| trnI-GAU *,a, trnK-UUU *, trnL-CAA a, trnL-UAA *, trnL-UAG, trnM-CAU, trnN-GUU a, trnP-UGG, trnQ-UUG, trnR-ACG a, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC a, trnV-UAC *, trnW-CCA, trnY-GUA | ||
| Small subunit of ribosome | rps2, rps3, rps4, rps7a, rps8, rps11, rps12 **,a, rps14 rps15, rps16 *, rps18, rps19 | |
| Large subunit of ribosome | rpl2 *,a, rpl14, rpl16 *, rpl20, rpl22, rpl23 a, rpl32, rpl33, rpl36 | |
| DNA dependent RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 | |
| Genes for phytosynthesis | Subunits of NADH-dehydrogenase | ndhA *, ndhB *,a, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH ndhI, ndhJ, ndhK |
| Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunits of cytochrome b/f complex | petA, petB *, petD *, petG, petL, petN | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF, atpH, atpl | |
| Large subunit of RuBisCO | rbcL | |
| Other genes | Maturase | matK |
| Envelope membrane protein | cemA | |
| Subunit of Acetyl-CoA-carboxylase | accD | |
| C-type cytochrome synthesis gene | ccsA | |
| Protease | clpP ** | |
| Genes of unknown function | Open Reading Frames (ORF, ycf) | ycf1, ycf2a, ycf3 **, ycf4 |
| Pseudo genes | ycf1, infA |
* Gene with one intron, ** Gene with two introns, a Gene with two copies.
Introns play an important role in the regulation of alternative gene splicing [18,19]. We found that 17 genes contained introns in all four Sanguisorba cp genomes, of which 11 are protein-coding genes and six are tRNA genes. 14 of the 17 contain a single intron, whereas three (clpP, rps12, and ycf3) have two introns. The largest intron, located into the trnK-UUU gene, ranged 2508 bp to 2516 bp in the four species (Table 3 and Tables S1–S3). The matK gene is located in the intron of trnK-UUU gene.
Table 3.
The length of exons and introns in genes with introns in the Sanguisorba officinalis chloroplast genome.
| No. | Gene | Location | Exon I (bp) | Intron I (bp) | Exon II (bp) | Intron I (bp) | Exon III (bp) |
|---|---|---|---|---|---|---|---|
| 1 | clpP | LSC | 69 | 938 | 291 | 658 | 228 |
| 2 | ndhA | SSC | 563 | 1185 | 541 | ||
| 3 | ndhB | IR | 777 | 682 | 756 | ||
| 4 | petB | LSC | 6 | 761 | 657 | ||
| 5 | petD | LSC | 9 | 750 | 474 | ||
| 6 | rpl16 | LSC | 8 | 1011 | 403 | ||
| 7 | rpl2 | IR | 391 | 673 | 434 | ||
| 8 | rpoC1 | LSC | 435 | 749 | 1620 | ||
| 9 | rps12 * | LSC | 114 | - | 232 | 543 | 26 |
| 10 | rps16 | LSC | 39 | 899 | 228 | ||
| 11 | trnA-UGC | IR | 38 | 814 | 35 | ||
| 12 | trnG-UCC | LSC | 23 | 698 | 48 | ||
| 13 | trnI-GAU | IR | 42 | 949 | 35 | ||
| 14 | trnK-UUU | LSC | 37 | 2516 | 35 | ||
| 15 | trnL-UAA | LSC | 37 | 554 | 50 | ||
| 16 | trnV-UAC | LSC | 39 | 601 | 37 | ||
| 17 | ycf3 | LSC | 126 | 723 | 228 | 766 | 153 |
* rps12 is a trans-spliced gene, of which two 3′ end residues are located in the IR region and the 5′ end in the LSC region.
2.2. Codon Usage
The total length of the protein coding genes from the four Sanguisorba cp genomes is 78,582~78,612 bp, and these genes are encoded by 22,760~22,768 codons. Protein coding genes thus accounted for 50.6~50.9% of the whole genome sequence. The most frequent amino acid is leucine, with 2387~2400 (10.5%) of the codons, but cysteine is the least frequent in the four Sanguisorba cp genomes, with only 260~262 (1.1%) of all codons. Within the protein-coding sequences (CDS), the AT content of codons at the first to third positions is 54.5%, 61.9~62.0%, and 69.5~69.6%, respectively. The fact is that the AT content of the codons is the highest with the third position, and it’s common in land plants [7,13,20,21]. The same phenomenon was also found in the frequency of codon usage. All preferred synonymous codons (RSCU > 1) ended with A or U except the codons of trnL-CAA; however, most non-preferred synonymous codons (RSCU < 1) ended with G or C (Table 4 and Table S4–S6).
Table 4.
Codon usage in the Sanguisorba officinalis chloroplast genomes. RSCU: Relative Synonymous Codon Usage.
| Amino Acid | Codon | Count | RSCU | tRNA | Amino Acid | Codon | Count | RSCU | tRNA |
|---|---|---|---|---|---|---|---|---|---|
| Phe | UUU | 899 | 1.38 | Tyr | UAU | 682 | 1.61 | ||
| Phe | UUC | 401 | 0.62 | trnF-GAA | Tyr | UAC | 165 | 0.39 | trnY-GUA |
| Leu | UUA | 810 | 2.03 | trnL-UAA | Stop | UAA | 43 | 1.65 | |
| Leu | UUG | 466 | 1.17 | trnL-CAA | Stop | UAG | 20 | 0.77 | |
| Leu | CUU | 503 | 1.26 | His | CAU | 403 | 1.51 | ||
| Leu | CUC | 148 | 0.37 | His | CAC | 132 | 0.49 | trnH-GUG | |
| Leu | CUA | 306 | 0.77 | trnL-UAG | Gln | CAA | 616 | 1.53 | trnQ-UUG |
| Leu | CUG | 156 | 0.39 | Gln | CAG | 191 | 0.47 | ||
| Ile | AUU | 983 | 1.5 | Asn | AAU | 825 | 1.53 | ||
| Ile | AUC | 369 | 0.56 | trnI-GAU | Asn | AAC | 256 | 0.47 | trnN-GUU |
| Ile | AUA | 614 | 0.94 | Lys | AAA | 925 | 1.54 | trnK-UUU | |
| Met | AUG | 531 | 1 | trnfM-CAU, trnI-CAU, trnM-CAU | Lys | AAG | 280 | 0.46 | |
| Val | GUU | 471 | 1.48 | Asp | GAU | 712 | 1.62 | ||
| Val | GUC | 152 | 0.48 | trnV-GAC | Asp | GAC | 168 | 0.38 | trnD-GUC |
| Val | GUA | 474 | 1.48 | trnV-UAC | Glu | GAA | 904 | 1.52 | trnE-UUC |
| Val | GUG | 180 | 0.56 | Glu | GAG | 287 | 0.48 | ||
| Ser | UCU | 469 | 1.69 | Cys | UGU | 204 | 1.56 | ||
| Ser | UCC | 263 | 0.95 | trnS-GGA | Cys | UGC | 57 | 0.44 | trnC-GCA |
| Ser | UCA | 306 | 1.1 | trnS-UGA | Stop | UGA | 15 | 0.58 | |
| Ser | UCG | 171 | 0.61 | Trp | UGG | 396 | 1 | trnW-CCA | |
| Pro | CCU | 352 | 1.47 | Arg | CGU | 307 | 1.36 | trnR-ACG | |
| Pro | CCC | 198 | 0.83 | Arg | CGC | 95 | 0.42 | ||
| Pro | CCA | 257 | 1.08 | trnP-UGG | Arg | CGA | 312 | 1.38 | |
| Pro | CCG | 149 | 0.62 | Arg | CGG | 103 | 0.46 | ||
| Thr | ACU | 465 | 1.59 | Ser | AGU | 349 | 1.25 | ||
| Thr | ACC | 224 | 0.76 | trnT-GGU | Ser | AGC | 111 | 0.4 | trnS-GCU |
| Thr | ACA | 348 | 1.19 | trnT-UGU | Arg | AGA | 391 | 1.74 | trnR-UCU |
| Thr | ACG | 135 | 0.46 | Arg | AGG | 144 | 0.64 | ||
| Ala | GCU | 576 | 1.79 | Gly | GGU | 524 | 1.32 | ||
| Ala | GCC | 201 | 0.63 | Gly | GGC | 192 | 0.48 | trnG-GCC | |
| Ala | GCA | 348 | 1.08 | trnA-UGC | Gly | GGA | 568 | 1.43 | trnG-UCC |
| Ala | GCG | 161 | 0.5 | Gly | GGG | 305 | 0.77 | ||
| Average # codons = 22,768 | |||||||||
2.3. Long Repeats and SSR Analysis
For long repeats analysis, the four cp genomes enclose long repeats with a total number ranging from 39 to 53 with at least 30 bp per repeat unit. Taking S. officinalis as an example, a number of 49 repeats were detected. These included 24 palindromic repeats, 17 forward repeats, six reverse repeats, and two complement repeats. Most repeats showed lengths between 30 and 44 bp and are in intergenic regions or intron sequences.
SSRs, also called as microsatellites, are tandemly repeated sequences that consist of 1–6 nucleotide repeat units. SSRs are widely distributed in cp genomes in general and are important for studies of plant populations. Because of their high level of polymorphism, SSRs are widely used as molecular markers for species authentication, molecular breeding, and population genetics [22,23,24,25]. Here, we identified many SSRs in the cp genomes, ranging from 79 in S. tenuifolia var. alba to 91 in S. stipulata. Most of the SSRs are mononucleotide repeats, whose amount ranges from 55 (S. tenuifolia var. alba) to 69 (S. stipulata). The number of di-, tri-, tetra-, penta-, and hexanucleotide repeats found was 9~12, 3~4, 7~9, 0~1, and 1~2, respectively (Table 5). Most of the mononucleotide SSRs belonged to the A/T type in the four Sanguisorba species. The highest number of SSRs found was in S. stipulata, which showed 68 of 69 identified mononucleotide SSRs. The lowest number of SSRs found was 55 of the 59 found in S. officinalis. These results are consistent with those of previous studies that found that polyadenine (polyA) and polythymine (polyT) content were higher than polyguanine (polyG) and polycytosine (polyC) content in the cpSSRs of many plants [26]. We speculate that the abundance of A/T SSRs may be associated with the AT richness of these cp genomes [13,27].
Table 5.
Types and numbers of SSRs found in the four Sanguisorba chloroplast genomes.
| SSR Type | Repeat Unit | Number | |||
|---|---|---|---|---|---|
| S. officinalis | S. filiformis | S. stipulata | S. tenuifolia var. alba | ||
| Mono | A/T | 55 | 56 | 68 | 53 |
| C/G | 4 | 3 | 1 | 2 | |
| Di | AT/AT | 11 | 9 | 8 | 11 |
| AG/CT | 1 | 1 | 1 | 1 | |
| Tri | AAT/ATT | 3 | 4 | 3 | 4 |
| Tetra | AAAT/ATTT | 4 | 3 | 5 | 4 |
| AAAG/CTTT | 1 | 1 | 1 | 1 | |
| ACAT/ATGT | 1 | 1 | 1 | 1 | |
| AGAT/ATCT | 1 | 1 | 1 | 1 | |
| AATT/AATT | 0 | 1 | 1 | 0 | |
| Penta | AAATT/AATTT | 1 | 0 | 0 | 1 |
| Hexa | AAAGGG/CCCTTT | 0 | 2 | 0 | 0 |
| AAAATC/ATTTTG | 0 | 0 | 1 | 0 | |
| Total | 82 | 82 | 91 | 79 | |
2.4. IR Contraction and Expansion
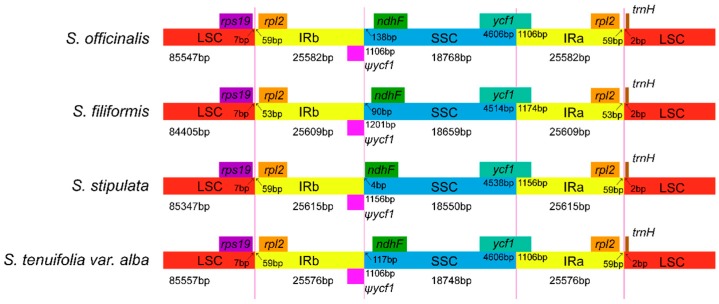
It is well known that IRs are the most conserved regions in chloroplast genomes, and the contraction and expansion at the borders of IR regions are common evolutionary events. It is also a main cause of length variation in the chloroplast genomes [28,29]. In this study, we compared the IR/SSC and IR/LSC boundaries of the four Sanguisorba cp genomes (Figure 2). In the four Sanguisorba species, the IRb/SSC boundary extends into functional ycf1 genes, yielding a pseudogene ycf1, which have a length of 1106~1201 bp in the four species. A previous study reported that the pseudogene ycf1 may be useful for researching variation among cp genomes in higher plants or algae [30]. In addition, we found no overlap between the ycf1 pseudogene and ndhF in the four species. The ndhF gene is found in the SSC region, and was 138 bp, 90 bp, four bp, and 117 bp away from the IRb/SSC boundary in S. officinalis, S. filiformis, S. stipulata, and S. tenuifolia var. alba, respectively. The trnH gene was found in the same position of the same LSC region in the four species, which is only two bp away from the IRb/SSC boundary. In the cp genome, variation in the IR/SSC and IR/LSC boundaries is governed by a dynamic and random process that is confined to conservative expansions and contractions [31,32]. There are many studies about the mechanisms responsible for IR expansion, and the leading view is that short IR expansions could be caused by gene conversion, but large IR expansions may be the result of double-strand DNA break repair (DSBR) [33,34]. In contrast, there are few reports on the mechanisms of IR contraction. However, Peery et al. proposed that DSBR theory was not only the main mechanism of IR region expansion, but also the main mechanism of IR region contraction [35].
Figure 2.
Comparison of the border regions of the LSC, SSC, and IR among four chloroplast genomes. Ψ: pseudogenes.
2.5. Comparative Chloroplast Genomic Analysis
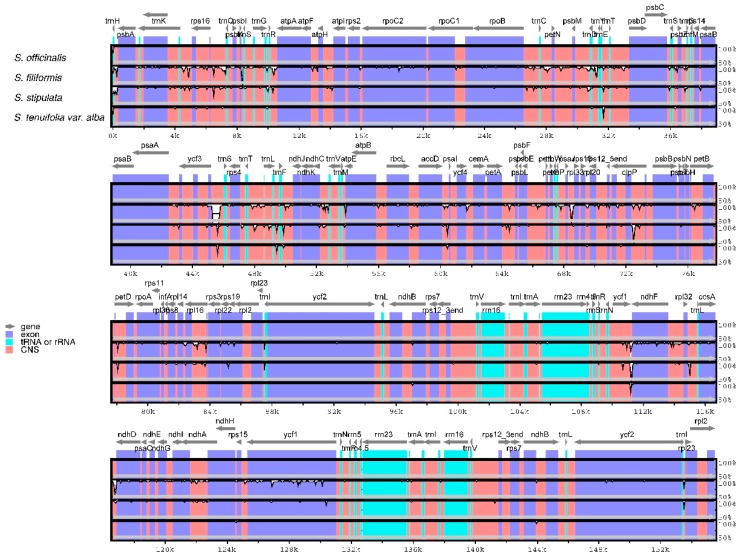
With the annotated S. officinalis cp genome as a reference, the whole cp genome of the four Sanguisorba species were compared and drawn by mVISTA to show sequence divergence (Figure 3), which is important for further phylogenetic analyses and species identification. Comparative genome analysis found that there is a high similarity between the cp genomes of all Sanguisorba species. The LSC and SSC regions are more divergent than the two IR regions, which is common in other higher plants and may be due to copy corrections between two IR regions by gene conversion [36]. Moreover, the coding regions have less variability proportions than the non-coding regions. The highest divergence among the four Sanguisorba cp genomes occurs in the intergenic spacers region, which contains trnE-trnT, trnS-psbZ, trnS-ycf3, trnF-ndhJ, accD-psal, and ycf1-ndhF. In this study, we found that the more conserved coding regions are the four rRNA located in IR region.
Figure 3.
Comparison of the four Sanguisorba chloroplast genomes using mVISTA. CNS indicates conserved noncoding sequences. The Y-scale represents the percent identity between 50% and 100%.
2.6. Phylogenetic Analysis
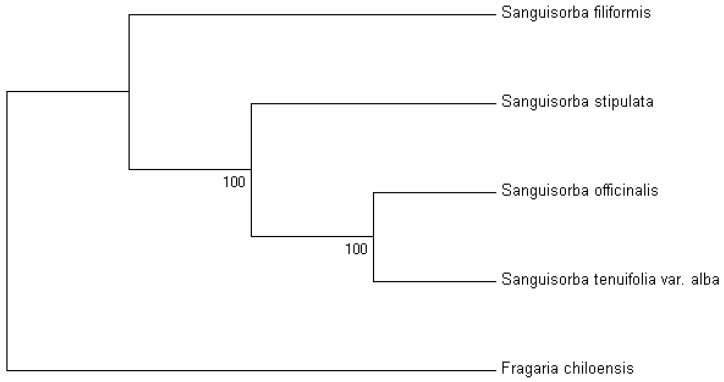
Chloroplast genomes contain abundant genetic information that is widely applied in plant identification and phylogenetic studies [6,37,38,39]. Sanguisorba belongs to the subfamily Rosoideae in the Rosaceae. Previous studies have reported phylogenetic relationships within the Rosaceae that were analyzed based on chloroplast regions [40,41]. Here, the availability of the completed cp genomes and protein coding genes of the four Sanguisorba species provide us with sequence and gene information for studying the molecular evolution and phylogeny of the genus Sanguisorba [9,42]. In this study, two datasets (i.e., the whole complete cp genome and the set of protein coding genes) from the cp genomes of the four Sanguisorba species and one outgroup (Fragaria chiloensis) were used to perform phylogenetic analysis. Phylogenetic trees were generated using the maximum parsimony (MP) method based on two datasets with the same topologies (Figure 4 and Figure S4). For the four Sanguisorba species, S. officinalis has the closest relationship with S. tenuifolia var. alba, followed by S. stipulata, and has the least close relationship with the S. filiformis. In addition, both S. stipulata and S. filiformis group into a monophyletic clade.
Figure 4.
Phylogenetic relationships between the four Sanguisorba species determined by whole cp genome sequences using the maximum parsimony (MP) method. Fragaria chiloensis was set as the outgroup.
3. Materials and Methods
3.1. Plant Materials and DNA Extraction
Fresh leaves of four Sanguisorba species were collected from Jilin and Yunan Provinces in China. Then we washed the leaves powder with HF buffer (100 mmol·L−1 Tris-HCl pH 8.0, 20 mmol·L−1 EDTA, 0.7 mol·L−1 NaCl, 2% PVP, and 0.2% 2-mercaptoethanol). HF buffer (600 μL) was added to leaves powder (~100 mg), the mixture vortexed vigorously for 3 min, centrifuged for 5 min at 12,000 rpm, and the supernatant discarded. Finally the total genomic DNA of each sample was isolated from the leaves powder by Plant Genomic DNA Kits (Tiangen Biotech Co., Beijing, China), according to the manufacturer’s instructions. The DNA quality and quantity of each sample was estimated by a NanoDrop 2000 Spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA) and a Qubit3.0 Fluorometer (Thermo Scientific, Waltham, MA, USA), as well as by agarose gel electrophoresis.
3.2. Chloroplast Genome Sequencing, Assembly and Annotation
After DNA was purified and prepared, ~2 μg was used to construct shotgun libraries. Genomic DNA was taken and sheared into 450 bp contigs with the Covaris M220 Focused-ultrasonicator (Covaris, Woburn, MA, USA). The library was constructed by TruSeqTM DNA Sample Prep Kit (Illumina Inc., San Diego, CA, USA), according to the manufacturer’s instructions. An Illumina HiSeq X platform was used for sequencing. Clean reads were obtained by using the Fastqc trim tool [43]. We then extracted cp-like reads from trimmed reads by performing BLASTs [44] using reference sequences (Rosa roxburghii, accession No.: NC_032038). Sequence assembly was performed by using SOAPdenovo [45], and the contigs were aligned using SSPACE [46]. The complete chloroplast genomes of the four Sanguisorba species were annotated using the CpGAVAS web service [47]. The tRNA genes were confirmed using tRNAscan-SE [48,49]. OGDRAW software (http://ogdraw.mpimp-golm.mpg.de/) [50] was used to draw circular cp genome maps for each species. The validated complete cp genome of the four Sanguisorba species were deposited in GenBank (https://www.ncbi.nlm.nih.gov/): S. officinalis, MF678801; S. filiformis, MF678800; S. stipulata, MF678798; and S. tenuifolia var. alba, MF678799.
3.3. Genome Comparison and Structural Analyses
The IR and SC boundary regions of the four Sanguisorba species were compared and examined. Comparison of the four cp genomes was performed using the Shuffle-LAGAN mode in mVISTA [51,52], with the annotation of S. officinalis used as the reference. In addition, we analyzed the codon usage, relative synonymous codon usage values (RSCU), and GC content using MEGA5 [53]. SSRs were identified by MISA (http://pgrc.ipk-gatersleben.de/misa/) [54] with minimum repeat numbers of 10, 5, 4, 3, 3, and 3 for mono-, di-, tri-, tetra-, penta-, and hexanucleotides, respectively. The forward and inverted repeats in the Sanguisorba cp genome were detected using REPuter [55] with a minimal repeat sequence of 30 bp and a sequence identity of 90%.
3.4. Phylogenetic Analyses
Phylogenetic analyses were performed for the four Sanguisorba species using Fragaria chiloensis (Rosaceae) as an outgroup. The complete cp genome sequences and protein coding genes shared in four Sanguisorba species and Fragaria chiloensis (accession No.: NC_019601) [56] were aligned by ClustalW2 [57]. Phylogenetic trees were constructed using the maximum parsimony (MP) method in PAUP*4.0b10 [58]. A heuristic search was performed using the MULPARS option, with the random stepwise addition of sequences in 1000 replications and tree bisection reconnection (TBR) branch swapping. The branch support of the phylogenetic tree was 1000 bootstrap replicates.
4. Conclusions
The complete cp genome sequences of four Sanguisorba species (S. officinalis, S. filiformis, S. stipulata, and S. tenuifolia var. alba), the first four sequenced members of the genus Sanguisorba, were assembled, annotated and analyzed in this study. The genome structure, gene content, and gene order were similar in the four species. Long repeats and SSRs reported here provide opportunities for the development of new molecular markers to study medicinal plants in the genus Sanguisorba. Phylogenetic analysis strongly supported that S. officinalis has the closest relationship with S. tenuifolia var. alba, followed by S. stipulata, and then S. filiformis. The available genome data presented in this paper provides a basis for further research on the evolution of the genus Sanguisorba, as well as for species identification.
Supplementary Materials
Supplementary materials are available online. Table S1. The length of exons and introns in genes with introns in the Sanguisorba filiformis chloroplast genome. Table S2. The length of exons and introns in genes with introns in the Sanguisorba stipulata chloroplast genome. Table S3. The length of exons and introns in genes with introns in the Sanguisorba tenuifolia var. alba chloroplast genome. Table S4. Codon usage in the Sanguisorba filiformis chloroplast genome. Table S5. Codon usage in the Sanguisorba stipulata chloroplast genomes. Table S6. Codon usage in the Sanguisorba tenuifolia var. alba chloroplast genomes. Figure S1. Gene map of Sanguisorba filiformis chloroplast genome. Genes shown inside the circle are transcribed clockwise, and those outside are counterclockwise. Genes in different functional groups are color-coded. Figure S2. Gene map of Sanguisorba stipulata chloroplast genome. Genes shown inside the circle are transcribed clockwise, and those outside are counterclockwise. Genes in different functional groups are color-coded. Figure S3. Gene map of Sanguisorba tenuifolia var. alba chloroplast genom. Genes shown inside the circle are transcribed clockwise, and those outside are counterclockwise. Genes in different functional groups are color-coded. Figure S4. Phylogenetic relationships of the four Sanguisorba species constructed by protein coding genes using the maximum parsimony (MP) method. Fragaria chiloensis was set as the outgroup.
Author Contributions
X.-X.M. and Y.-F.X. performed the experiments; X.-X.M. and M.-L.W. assembled sequences and analyzed the data; L.W. wrote the manuscript; W.S. and L.W. conceived the research framework. L.X., D.Z., Y.-H.S., G.-Q.D., S.-P.I. and Z.-X.L. made revisions to the final manuscript. All authors have read and approved the final manuscript.
Funding
This research was funded by the Major Scientific and Technological Special Project for “Major New Drug Creation” (No. 2014ZX09201021-008 and 2017ZX09101002-003-001), and the National Natural Science Foundation of China (No. 81503192).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Sequences data of four Sanguisorba species (S. officinalis, S. filiformis, S. stipulata, and S. tenuifolia var. alba) are available from the authors.
References
- 1.Yang B., Hu J., Zhang F., Li J., Liu Q., Pu G., Liu H., Zhang Y. Herbal Textural Study on Sanguisorba officinalis L. Shandong Univ. TCM. 2016;5:412–414. [Google Scholar]
- 2.State Pharmacopoeia Committee . Pharmacopoeia of the People’s Republic of China. Volume 1. Medical Science and Technology Press; Beijing, China: 2015. p. 126. [Google Scholar]
- 3.Jia L., Xi W., Jin G. Effect of Diyu Shengbai Tablets on Bone Marrow Depression Induced by Cyclophosphamide in Mice. Chin. J. Exp. Tradit. Med. Formul. 2012;18:251–254. [Google Scholar]
- 4.Freitas A., da Anunciação R., D’Oliveira-Matielo C., Stefenon V. Chloroplast DNA: A Promising Source of Information for Plant Phylogeny and Traceability. J. Mol. Biol. Methods. 2018;1:2. [Google Scholar]
- 5.Hong S.-Y., Cheon K.-S., Yoo K.-O., Lee H.-O., Cho K.-S., Suh J.-T., Kim S.-J., Nam J.-H., Sohn H.-B., Kim Y.-H. Complete chloroplast genome sequences and comparative analysis of Chenopodium quinoa and C. album. Front. Plant Sci. 2017;8:1696. doi: 10.3389/fpls.2017.01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leister D., Pesaresi P. The genomic era of chloroplast research. Annu. Plant Rev. 2018:1–29. doi: 10.1002/9781119312994.apr0121. [DOI] [Google Scholar]
- 7.Mader M., Pakull B., Blanc-Jolivet C., Paulini-Drewes M., Bouda Z.H.-N., Degen B., Small I., Kersten B. Complete Chloroplast Genome Sequences of Four Meliaceae Species and Comparative Analyses. Int. J. Mol. Sci. 2018;19:701. doi: 10.3390/ijms19030701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo H., Liu J., Luo L., Wei X., Zhang J., Qi Y., Zhang B., Liu H., Xiao P. Complete chloroplast genome sequences of Schisandra chinensis: Genome structure, comparative analysis, and phylogenetic relationship of basal angiosperms. Sci. China Life Sci. 2017;60:1286–1290. doi: 10.1007/s11427-017-9098-5. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y., Du L., Liu A., Chen J., Wu L., Hu W., Zhang W., Kim K., Lee S.-C., Yang T.-J. The complete chloroplast genome sequences of five Epimedium species: Lights into phylogenetic and taxonomic analyses. Front. Plant Sci. 2016;7:306. doi: 10.3389/fpls.2016.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng H., Li J., Zhang H., Cai B., Gao Z., Qiao Y., Mi L. The complete chloroplast genome sequence of strawberry (Fragaria × ananassa Duch.) and comparison with related species of Rosaceae. PeerJ. 2017;5:e3919. doi: 10.7717/peerj.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H.-W., Kim K.-J. The complete plastome sequence of Pentactina rupicola Nakai (Rosaceae), a genus endemic to Korea. Mitochondrial DNA Part B. 2016;1:698–700. doi: 10.1080/23802359.2016.1225523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao L., Li K., Teng Y., Zhang D. Characterization of the complete chloroplast genome of the wild Himalayan pear Pyrus pashia (Rosales: Rosaceae: Maloideae) Conserv. Genet. Resour. 2017;9:569–571. doi: 10.1007/s12686-017-0724-2. [DOI] [Google Scholar]
- 13.Yang Y., Yuanye D., Qing L., Jinjian L., Xiwen L., Yitao W. Complete chloroplast genome sequence of poisonous and medicinal plant datura stramonium: Organizations and implications for genetic engineering. PLoS ONE. 2014;9:e110656. doi: 10.1371/journal.pone.0110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howe C.J., Barbrook A.C., Koumandou V.L., Nisbet R.E.R., Symington H.A., Wightman T.F. Evolution of the chloroplast genome. Philos. Trans. R. Soc. B Biol. Sci. 2003;358:99–107. doi: 10.1098/rstb.2002.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M., Cui L., Feng K., Deng P., Du X., Wan F., Weining S., Nie X. Comparative analysis of Asteraceae chloroplast genomes: Structural organization, RNA editing and evolution. Plant Mol. Biol. Rep. 2015;33:1526–1538. doi: 10.1007/s11105-015-0853-2. [DOI] [Google Scholar]
- 16.Dong W.-L., Wang R.-N., Zhang N.-Y., Fan W.-B., Fang M.-F., Li Z.-H. Molecular Evolution of Chloroplast Genomes of Orchid Species: Insights into Phylogenetic Relationship and Adaptive Evolution. Int. J. Mol. Sci. 2018;19:716. doi: 10.3390/ijms19030716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J., Chen X., Cui Y., Sun W., Li Y., Wang Y., Song J., Yao H. Molecular Structure and Phylogenetic Analyses of Complete Chloroplast Genomes of Two Aristolochia Medicinal Species. Int. J. Mol. Sci. 2017;18:1839. doi: 10.3390/ijms18091839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith N.A., Singh S.P., Wang M.-B., Stoutjesdijk P.A., Green A.G., Waterhouse P.M. Gene expression: Total silencing by intron-spliced hairpin RNAs. Nature. 2000;407:319–320. doi: 10.1038/35030305. [DOI] [PubMed] [Google Scholar]
- 19.Graveley B.R. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/S0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 20.Clegg M.T., Gaut B.S., Learn G.H., Morton B.R. Rates and patterns of chloroplast DNA evolution. Proc. Natl. Acad. Sci. USA. 1994;91:6795–6801. doi: 10.1073/pnas.91.15.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi D.-K., Kim K.-J. Complete chloroplast genome sequences of important oilseed crop Sesamum indicum L. PLoS ONE. 2012;7:e35872. doi: 10.1371/journal.pone.0035872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dashnow H., Tan S., Das D., Easteal S., Oshlack A. Genotyping microsatellites in next-generation sequencing data. BMC Bioinform. 2015;16:A5. doi: 10.1186/1471-2105-16-S2-A5. [DOI] [Google Scholar]
- 23.Chmielewski M., Meyza K., Chybicki I.J., Dzialuk A., Litkowiec M., Burczyk J. Chloroplast microsatellites as a tool for phylogeographic studies: The case of white oaks in Poland. iForest. 2015;8:765. doi: 10.3832/ifor1597-008. [DOI] [Google Scholar]
- 24.Jiao Y., Jia H.-M., Li X.-W., Chai M.-L., Jia H.-J., Chen Z., Wang G.-Y., Chai C.-Y., van de Weg E., Gao Z.-S. Development of simple sequence repeat (SSR) markers from a genome survey of Chinese bayberry (Myrica rubra) BMC Genom. 2012;13:201. doi: 10.1186/1471-2164-13-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue J., Wang S., Zhou S.L. Polymorphic chloroplast microsatellite loci in Nelumbo (Nelumbonaceae) Am. J. Bot. 2012;99:240–244. doi: 10.3732/ajb.1100547. [DOI] [PubMed] [Google Scholar]
- 26.Kuang D.-Y., Wu H., Wang Y.-L., Gao L.-M., Zhang S.-Z., Lu L. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): Implication for DNA barcoding and population genetics. Genome. 2011;54:663–673. doi: 10.1139/g11-026. [DOI] [PubMed] [Google Scholar]
- 27.Raveendar S., Na Y.-W., Lee J.-R., Shim D., Ma K.-H., Lee S.-Y., Chung J.-W. The complete chloroplast genome of Capsicum annuum var. glabriusculum using Illumina sequencing. Molecules. 2015;20:13080–13088. doi: 10.3390/molecules200713080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim K.-J., Lee H.-L. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004;11:247–261. doi: 10.1093/dnares/11.4.247. [DOI] [PubMed] [Google Scholar]
- 29.Raubeson L.A., Peery R., Chumley T.W., Dziubek C., Fourcade H.M., Boore J.L., Jansen R.K. Comparative chloroplast genomics: Analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genom. 2007;8:174. doi: 10.1186/1471-2164-8-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Cambiaire J.-C., Otis C., Lemieux C., Turmel M. The complete chloroplast genome sequence of the chlorophycean green alga Scenedesmus obliquus reveals a compact gene organization and a biased distribution of genes on the two DNA strands. BMC Evol. Biol. 2006;6:37. doi: 10.1186/1471-2148-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma J., Yang B., Zhu W., Sun L., Tian J., Wang X. The complete chloroplast genome sequence of Mahonia bealei (Berberidaceae) reveals a significant expansion of the inverted repeat and phylogenetic relationship with other angiosperms. Gene. 2013;528:120–131. doi: 10.1016/j.gene.2013.07.037. [DOI] [PubMed] [Google Scholar]
- 32.Shen X., Wu M., Liao B., Liu Z., Bai R., Xiao S., Li X., Zhang B., Xu J., Chen S. Complete Chloroplast Genome Sequence and Phylogenetic Analysis of the Medicinal Plant Artemisia annua. Molecules. 2017;22:1330. doi: 10.3390/molecules22081330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goulding S.E., Wolfe K., Olmstead R., Morden C. Ebb and flow of the chloroplast inverted repeat. Mol. Gen. Genet. 1996;252:195–206. doi: 10.1007/BF02173220. [DOI] [PubMed] [Google Scholar]
- 34.Wang R.-J., Cheng C.-L., Chang C.-C., Wu C.-L., Su T.-M., Chaw S.-M. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol. 2008;8:36. doi: 10.1186/1471-2148-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peery R. Ph.D. Thesis. University of Illinois at Urbana-Champaign; Champaign, IL, USA: 2015. Understanding Angiosperm Genome Interactions and Evolution: Insights from Sacred Lotus (Nelumbo Nucifera) and the Carrot Family (Apiaceae) [Google Scholar]
- 36.Khakhlova O., Bock R. Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 2006;46:85–94. doi: 10.1111/j.1365-313X.2006.02673.x. [DOI] [PubMed] [Google Scholar]
- 37.Moore M.J., Bell C.D., Soltis P.S., Soltis D.E. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc. Natl. Acad. Sci. USA. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Y., Li X., Yang Z., Yang C., Yang J., Ji Y. Analysis of complete chloroplast genome sequences improves phylogenetic resolution in Paris (Melanthiaceae) Front. Plant Sci. 2016;7:1797. doi: 10.3389/fpls.2016.01797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang N., Erickson D.L., Ramachandran P., Ottesen A.R., Timme R.E., Funk V.A., Luo Y., Handy S.M. An analysis of Echinacea chloroplast genomes: Implications for future botanical identification. Sci. Rep. 2017;7:216. doi: 10.1038/s41598-017-00321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Potter D., Eriksson T., Evans R.C., Oh S., Smedmark J., Morgan D.R., Kerr M., Robertson K.R., Arsenault M., Dickinson T.A. Phylogeny and classification of Rosaceae. Plant Syst. Evol. 2007;266:5–43. doi: 10.1007/s00606-007-0539-9. [DOI] [Google Scholar]
- 41.Eriksson T., Hibbs M.S., Yoder A.D., Delwiche C.F., Donoghue M.J. The phylogeny of Rosoideae (Rosaceae) based on sequences of the internal transcribed spacers (ITS) of nuclear ribosomal DNA and the trnL/F region of chloroplast DNA. Int. J. Plant Sci. 2003;164:197–211. doi: 10.1086/346163. [DOI] [Google Scholar]
- 42.Jansen R.K., Cai Z., Raubeson L.A., Daniell H., Leebens-Mack J., Müller K.F., Guisinger-Bellian M., Haberle R.C., Hansen A.K., Chumley T.W. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andrews S.C. FastQC v0.11.3. Babraham Bioinformatics; Cambridge, MA, USA: 2015. [(accessed on 20 December 2017)]. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 44.Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boetzer M., Henkel C., Jansen H., Butler D., Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 47.Liu C., Shi L., Zhu Y., Chen H., Zhang J., Lin X., Guan X. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 2012;13:715. doi: 10.1186/1471-2164-13-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schattner P., Brooks A.N., Lowe T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lowe T.M., Eddy S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lohse M., Drechsel O., Bock R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 51.Mayor C., Brudno M., Schwartz J.R., Poliakov A., Rubin E.M., Frazer K.A., Pachter L.S., Dubchak I. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 52.Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang X.-M., Sun J.-T., Xue X.-F., Zhu W.-C., Hong X.-Y. Development and characterization of 18 novel EST-SSRs from the western flower thrips, Frankliniella occidentalis (Pergande) Int. J. Mol. Sci. 2012;13:2863–2876. doi: 10.3390/ijms13032863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurtz S., Choudhuri J.V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salamone I., Govindarajulu R., Falk S., Parks M., Liston A., Ashman T.L. Bioclimatic, ecological, and phenotypic intermediacy and high genetic admixture in a natural hybrid of octoploid strawberries. Am. J. Bot. 2013;100:939–950. doi: 10.3732/ajb.1200624. [DOI] [PubMed] [Google Scholar]
- 57.Larkin M.A., Blackshields G., Brown N., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 58.Swofford D.L. PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods) Sinauer Associates; Sunderland, MA, USA: 2002. Version 4.0b10. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.