Summary
Systemic rheumatic diseases are characterized by abnormal B cell activation with autoantibody production and hypergammaglobulinaemia. Ro52/SSA, also denoted tripartite motif (TRIM)21, is a major autoantigen in Sjögren’s syndrome and systemic lupus erythematosus. Interestingly, TRIM21‐deficient mice develop systemic autoimmunity with B cell‐driven manifestations such as autoantibodies, hypergammaglobulinaemia and glomerulonephritis following tissue injury. The mechanisms by which TRIM21‐deficiency leads to enhanced B cell activation and antibody production are, however, not well understood, and to further elucidate the role of TRIM21 in systemic autoimmunity, we investigated the B cell phenotype and antibody responses of Trim21 –/– mice following immunization with thymus‐dependent (TD) and thymus‐independent (TI) antigens. We found that TRIM21‐deficient mice developed significantly higher specific antibody titres than their wild‐type counterparts upon B cell receptor (BCR) engagement by TD and TI type II antigens, and this was accompanied by an altered B cell phenotype. Furthermore, BCR cross‐linking, but not anti‐CD40 stimulation, in vitro resulted in a significantly higher proliferation of Trim21 –/– cells. We also observed that splenic follicular B cells were expanded not only in immunized mice but also already in young, unmanipulated Trim21 –/– mice, and transcriptomic analysis of these cells revealed an up‐regulation of genes associated with B cell differentiation, indicating a role for TRIM21 in their regulation. In conclusion, in this study we describe a link between the rheumatic autoantigen Ro52/TRIM21 and increased antibody production associated with follicular B cell expansion, implicating a potential role for Ro52/TRIM21 in the pathogenesis of systemic autoimmune diseases.
Keywords: B cell, immunoglobulin, Ro52, Sjögren’s syndrome, TRIM21
Introduction
Sjögren’s syndrome (SS) and systemic lupus erythematosus (SLE) are systemic autoimmune diseases, characterized immunologically and serologically by a pathological B cell phenotype, ectopic germinal centre formation and the presence of hypergammaglobulinaemia and autoantibodies to multiple autoantigens 1, 2, 3. The patients also have a substantially increased risk of developing non‐Hodgkin B cell lymphomas 4, 5, further highlighting the role of B cells in the pathogenesis of these diseases.
TRIM21, also known as Ro52 or SSA, is one of the major autoantigens in systemic rheumatic diseases 6, 7, 8. TRIM21 belongs to the tripartite motif (TRIM) protein family, thus denoted for the RING, B‐box and coiled‐coil motif common to its members 9, 10. The TRIM proteins function as E3 ligases in the ubiquitination cascade 11, 12 and have been implicated as regulators of immunity and cellular processes such as proliferation, differentiation and apoptosis 13, 14, 15. TRIM21 is predominantly expressed in haematopoietic cells and has been shown to ubiquitinate multiple interferon regulatory factors (IRFs), proteins of the nuclear factor kappa B (NF‐kB) pathway, as well as proteins regulating the cell cycle 12, 16, 17. We and others have previously observed that naive, unmanipulated Trim21‐deficient mice display no obvious phenotype. However, upon chronic injury they develop severe tissue inflammation that progresses into systemic autoimmunity with hypergammaglobulinaemia and autoantibody production, revealing the importance of TRIM21 as a negative regulator of inflammatory responses 18. More recently, Trim21‐deficient naive T cells have also been shown to have an augmented intrinsic capacity for T helper type 17 (Th17) differentiation 19. The B cell phenotype and the mechanisms by which TRIM21‐deficiency leads to enhanced B cell activation and antibody production are, however, not well characterized. To address this question, we utilized immunization models and found that significantly higher specific immunoglobulin (Ig)M and IgG titres developed in Trim21 –/– mice compared to Trim21 +/+ mice upon B cell receptor (BCR) activation. BCR‐specific stimulation in vitro resulted in significantly higher proliferation of Trim21 –/– B cells, and we observed a relative expansion of splenic follicular B cells in Trim21 –/– mice, both following immunization and in naive mice. In addition, these cells presented an up‐regulated expression of genes associated with B cell differentiation, suggesting a link between the rheumatic autoantigen Ro52/TRIM21 and increased antibody production driven by an expansion of follicular B cells.
Materials and methods
Animals and in‐vivo immunizations
Trim21 –/– mice were generated on a C57BL/6J background, as previously described 18. Littermate Trim21 –/– and Trim21 +/+ mice aged 8–16 weeks were used, except in the analysis of old mice, where mice were analysed at the age of 52–58 weeks. Mice were identified by ear punching at weaning, and metal ear clips were not used for any mice in the present study. For T cell‐independent stimulations, mice were injected intraperitoneally with 100 µl of 4‐hydroxy‐3‐nitrophenyl acetyl (NP) coupled with lipopolysaccharide (LPS) or Ficoll at 250 µg/ml in sterile phosphate‐buffered saline (PBS) (Biosearch Technologies, Novato, CA, USA). Serum was collected by tail vein puncture prior to immunization and at days 5, 7 and 14, and mice were killed at day 14 post‐immunization. The experiments were repeated (LPS) and four (Ficoll) times, respectively. T cell‐dependent stimulations were performed by immunization with NP‐ovalbumin (NP‐OVA) subcutaneously. Mice were immunized with 100 µg NP‐OVA dissolved in complete Freund’s adjuvant (CFA) at day 0 and boosted at day 14 with 25 µg NP‐OVA (Biosearch Technologies) in incomplete Freund’s adjuvant (IFA) (Sigma‐Aldrich, St Louis, MO, USA). Serum was collected for serological analysis prior to immunization and at days 5, 7, 14, 21 and 28. Mice were killed at day 28 post‐immunization and cellular composition of spleen and bone marrow was analysed by flow cytometry. The OVA immunizations were repeated three times. Individual experiments were performed on either female or male mice; however, similar results were generated in both sexes. The local Region North Ethics committee in Stockholm, Sweden, approved the animal studies. All experiments were carried out according to the 2010/63/EU directive for animal experiments and mice were housed in accordance with the Swedish National Board for Laboratory Animals.
Enzyme‐linked immunosorbent assay (ELISA)
Antibodies towards NP‐coupled antigens were measured by ELISA. High binding plates (Nunc, Roskilde, Denmark) were coated overnight with 1 μg NP‐bovine serum albumin (BSA) (Biosearch Technologies) per well diluted in carbonate buffer (pH 9.6). After blocking with 5% milk powder in Tween‐PBS, sera diluted in Tween‐PBS/1% milk powder were added and incubated for 2 h at room temperature. Sera were diluted 1 : 500 for IgG and 1 : 1000 for IgM analysis. Antibody binding was detected with an alkaline phosphatase‐conjugated rabbit anti‐human IgG or IgM antibody 1 : 1000 (Dako, Glostrup, Denmark), visualized using phosphatase substrate tablets (Sigma) and measured at 405 nm absorbance. Optical density values were compared to a standard curve generated from five pooled high‐titre samples. Data are presented as arbitrary units.
Enzyme‐linked immunospot (ELISPOT)
ELISPOT plates (Millipore, Billerica, MA, USA) were precoated with NP‐BSA in PBS (Biosearch Technologies). Freshly isolated spleen cells were plated in duplicates at 0·25 × 106 cells/well in 200 μl Dulbecco’s modified Eagle’s medium (DMEM) medium supplemented with 5% fetal calf serum (FCS), L‐glutamine, HEPES and penicillin–streptomycin. Cells were incubated overnight in 37°C before the plates were washed and stained. Developed plates were read in a CTL ELISPOT reader using the Immunocapture version 6.3 software and analysed with the ImmunoSpot version 5.0 Professional software (CTL, Shaker Heights, OH, USA).
Flow cytometry and cell sorting
Single‐cell suspensions from spleen were incubated with mouse Fc‐block (BD Biosciences, San Jose, CA, USA or Miltenyi Biotech, Bergisch Gladbach, Germany) for 15 min at 4°C and subsequently stained for 20 min at 4°C with fluorophore‐conjugated antibodies (all from Biolegend, San Diego, CA, USA). Samples were run on a Dako CyAn flow cytometer, apart from the cellular characterization of naive splenocytes, which was performed on a Gallios flow cytometer (both Beckman Coulter, Brea, CA, USA) and analysed with FlowJo version 10 software (TreeStar, Inc., Ashland, OH, USA. For microarray analysis, total B cells were sorted from single‐cell suspensions from spleen of Trim21 +/+ and Trim21 –/– mice using the Pan B cell isolation kit (Miltenyi Biotech). Cells were subsequently stained with CD19‐V450, CD23‐phycoerythrin (PE), CD21‐allophycocyanin (APC), CD11b‐peridinin chlorophyll cyanin 5.5 (PerCP Cy5.5) CD11b (BD Biosciences) and CD4‐PerCP Cy5.5 (Beckman Coulter), and CD19+CD21+CD23+CD4–CD11b– follicular B cells were isolated by fluorescence activated cell sorter (FACS) (MoFlo cell sorter; Beckman Coulter).
Proliferation and cell death assays
Lymphocyte proliferation was assessed in vitro by both [3H]‐thymidine incorporation and measurement of Ki67 expression. For the [3H]‐thymidine incorporation assay, total splenocytes or magnetic‐activated cell sorting (MACS)‐separated CD19+ cells (Miltenyi Biotech) were seeded at 0·5 × 106 cells/well in 96‐well round‐bottomed plates. Cells were stimulated with anti‐IgM (Jackson ImmunoResearch, Bar Harbor, ME, USA) or anti‐CD40 (BD Biosciences) at varying concentrations and cultured for three days. [3H]‐thymidine was added to the wells during the last 18 h of culture and plates were harvested and analysed using a scintillation beta‐counter. For the Ki67 assay, total splenocytes were cultured at 1 × 106 cells/ml in 24‐well plates and stimulated with anti‐IgM at 0·1 g/ml for 3 days. Cells were stained for CD3, CD11b and CD19 (BioLegend), fixed and permeabilized using the BD Fix/Perm kit (BD Biosciences), subsequently stained for intracellular expression of Ki67 (BioLegend) and analysed by flow cytometry. Activation‐induced cell death was assessed in splenocytes cultured at 1 × 106 cells/ml stimulated with 0·1 g/ml anti‐IgM for 3 days and then restimulated with anti‐IgM for an additional 24 h. Cells were then stained for annexin V and 7‐aminoactinomycin D (7‐AAD) expression (apoptosis detection kit; BioLegend), CD3 and CD19 (BioLegend), and expression was assessed by flow cytometry. Experiments were performed twice.
RNA isolation and microarray analysis
FACS‐sorted cells were pelleted and resuspended in TRIzol (Life Technologies, Carlsbad, CA, USA). Total RNA was isolated as previously described 20. Global gene expression was analysed with a Mouse Transcriptome Assay 1.0 microarray chip (Affymetrix, Santa Clara, CA, USA) following quality control at the BEA core facility, Karolinska Institutet (https://www.bea.ki.se). Genes with low (< 1) or unnaturally high (> 106) values were excluded. Genes differentially regulated between Trim21 –/– and Trim21 +/+ follicular B cells were identified using the CollapseDataset and ComparativeMarkerSelection (CMS) module in the Broad Institute GenePattern software on default setting, without permutations 21. Heat‐maps were generated using Morpheus software (software.broadinstitute.org/morpheus/). Pathway analysis was performed using GOrilla software for ranked list enrichment 22, utilizing predefined GeneOntology pathways.
Statistical analysis
The Mann–Whitney U‐test was used for comparison of groups. Prism GraphPad version 7 was used for all statistical analyses except for microarray data. P‐values < 0·05 were considered significant unless indicated otherwise.
Results
In‐vivo B cell activation results in augmented antibody production in Trim21–/– mice
Unmanipulated Trim21 –/– mice display no overt phenotype, and similar numbers of total CD19+ B cells in spleen and bone marrow are observed in Trim21 –/– and Trim21 +/+ mice 18. However, tissue damage induced by ear‐tagging results in systemic inflammation with hypergammaglobulinaemia and autoantibody production in Trim21 –/– mice. To study the mechanisms by which Trim21‐deficiency leads to enhanced B cell activation and antibody production in a more controlled setting, we first assessed the immune response of naive Trim21 –/– and Trim21 +/+ mice to immunization with NP‐labelled ovalbumin (NP‐OVA). We observed that Trim21 –/– mice developed significantly higher titres of both IgM‐ and IgG‐specific antibodies compared to their Trim21 +/+ littermates (Fig. 1a). Subclass analysis revealed that the higher NP‐OVA specific IgG response depended primarily on higher IgG1 and IgG2a subclass antibody titres (Fig. 1b). We further observed a significantly higher number of B cells producing NP‐OVA‐specific IgM, as detected by ELISPOT (Fig. 2a), and a higher proportion of NP‐specific IgM‐producing B cells, as assessed by flow cytometry (Fig. 2), in the spleen of Trim21 –/– mice compared to wild‐type mice. No differences in IgG‐producing NP‐specific B cells were noted in the spleen (Fig. 2). There was, however, a significantly higher proportion of plasma cells in the bone marrow of Trim21–/– compared to Trim21 +/+ mice (Fig. 2c). Trim21–/– mice also presented a relative increase of CD19+CD21+CD23+ follicular B cells and a reciprocal reduction of CD21–CD23– T1 B cells in the spleen compared to Trim21 +/+ mice, while total CD19+ cell counts were similar (Fig. 2). No differences in splenic T cell and CD11b+ cell numbers or in CD4/CD8 ratios and T cell activation as assessed by CD44 expression were observed between Trim21 –/– and Trim21 +/+ mice following immunization (Fig. 2f and data not shown).
Figure 1.
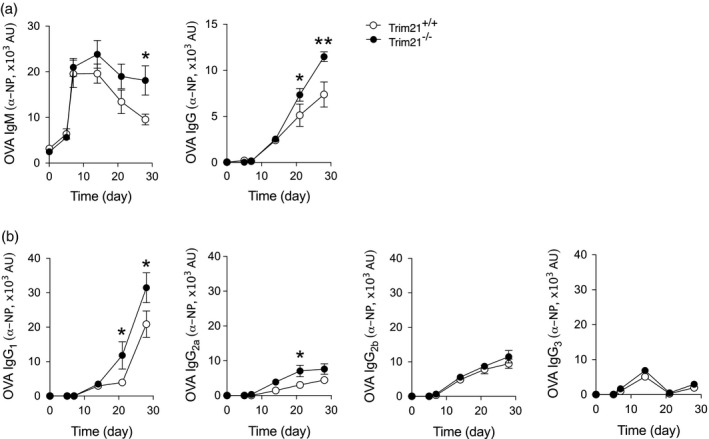
T cell‐dependent activation induces higher antibody titres in Trim21‐deficient mice. Trim21 +/+ and –/– mice were immunized subcutaneously with 4‐hydroxy‐3‐nitrophenyl ovalbumin (NP‐OVA) in complete Freund’s adjuvant on day 0 and boosted with incomplete Freund’s adjuvant on day 14. Blood was collected for serological analysis at days 0, 5, 7, 14, 21 and at the end of the experiment at day 28 (Trim21 +/+ n = 9, Trim21 –/– n = 10). NP‐specific antibodies were measured by enzyme‐linked immunosorbent assay (ELISA) in sera. (a) Anti‐NP‐OVA immunoglobulin (Ig)M and total IgG titres. (b) Anti‐NP‐OVA IgG1, IgG2a, IgG2b and IgG3 subclass analysis. Data are presented as mean ± standard error of the mean (s.e.m.). *p < 0·05, **P < 0·01 (Mann–Whitney U‐test)
Figure 2.
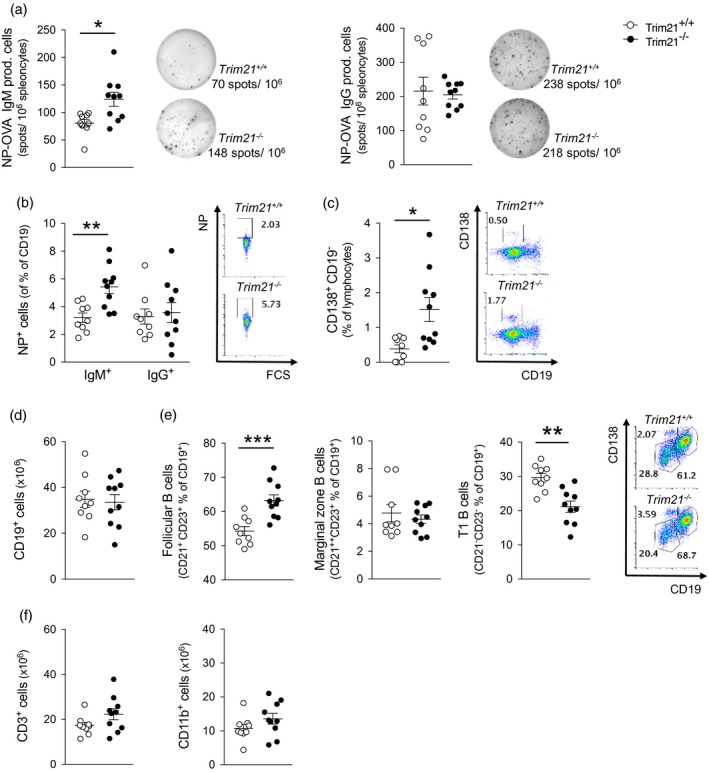
Increased antigen‐specific B cell response upon T cell‐dependent activation. Trim21 +/+ and –/– mice were immunized subcutaneously with 4‐hydroxy‐3‐nitrophenyl ovalbumin (NP‐OVA) in complete Freund’s adjuvant on day 0 and boosted with incomplete Freund’s adjuvant on day 14 (n = 9 and 10, respectively). Cellular composition of spleens and bone marrow was analysed at day 28. (a) NP‐specific immunoglobulin (Ig)M‐ and IgG‐producing B cells from spleen detected by enzyme‐linked immunospot (ELISPOT). Representative wells for each genotype are shown in the right panel. (b) Frequency of NP‐specific IgM‐ and IgG‐expressing B cells among total CD19+ cells in spleen. (c) Frequency of CD138+CD19– plasma cells in bone marrow. (d) Absolute numbers of CD19+ B cells and (e) frequency of B cell subsets in spleens. (f) Absolute numbers of CD3+ T cells and CD11b+ cells. Data are presented as mean ± standard error of the mean (s.e.m.). *P < 0·05, **P < 0·01, ***P < 0·001 (Mann–Whitney U‐test)
OVA represents a prototypical thymus dependent (TD) antigen, where B cell activation relies on interaction with major histocompatibility complex (MHC) class II‐restricted T cells. The higher specific antibody levels observed in the Trim21–/– mice may thus relate to either an increased response to activation via the BCR or to an increased response of the B cells upon T cell interaction. To test this directly, we next assessed the immune response to immunization with thymus‐independent (TI) antigens. TI antigens can be divided into type I and type II, with type I responses primarily mediated by mitogens such as Toll‐like receptor (TLR) agonists [e.g. lipopolysaccharide (LPS)], while type II responses relate to agents that convey their effect through cross‐linking the BCR (e.g. Ficoll) 23, 24. Notably, we observed that administration of NP‐LPS did not result in any difference in the specific Ig response between Trim21 –/– and Trim21 +/+ mice, whereas NP‐Ficoll, acting by cross‐linking the BCR, resulted in higher specific IgM and IgG responses in Trim21 –/– mice (Fig. 3). No differences in cell subsets, however, were observed by flow cytometry analysis between Trim21 –/– and Trim21 +/+ mice following NP‐Ficoll immunization (data not shown).
Figure 3.
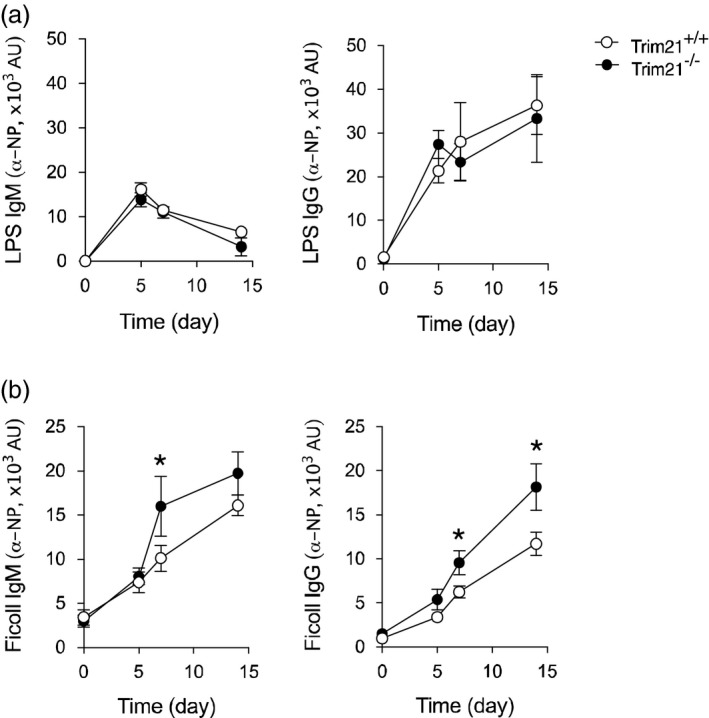
T cell‐independent type II activation results in a stronger response in Trim21 –/– mice than T cell independent type I stimulation. Mice were immunized intraperitoneally with 4‐hydroxy‐3‐nitrophenyl‐lipopolysaccharide (NP‐LPS) or NP‐Ficoll in phosphate‐buffered saline (PBS) at day 0 and serum was collected at days 0, 5, 7 and 14. NP‐specific antibody levels were assessed by enzyme‐linked immunosorbent assay (ELISA). (a) Anti‐NP‐LPS immunoglobulin (Ig)M and IgG titres (Trim21 +/+ n = 9, Trim21 –/– n = 10). (b) Anti‐NP‐Ficoll IgM and IgG titres (Trim21 +/+ n = 9, Trim21 –/– n = 10) (data are presented as mean ± standard error of the mean (s.e.m.). *P < 0·05 (Mann–Whitney U‐test)
In all, these data demonstrate that mice lacking Trim21 respond with higher titres of specific IgM and IgG antibodies to both TD and TI type II antigen stimulation, but not to TI type I antigen stimulation, indicating that this effect is driven at least in part by a differential response to BCR activation.
Trim21 deficiency results in increased B cell proliferation upon BCR, but not CD40, stimulation
Several previously published studies indicate that TRIM21 negatively regulates proliferation. Specifically, over‐expression of TRIM21 in the A20 B cell line results in reduced proliferation 12, TRIM21 mRNA expression correlates negatively to proliferation of peripheral blood mononuclear cells in vitro, and reduced expression of TRIM21 is associated with more aggressive disease in diffuse large B cell lymphomas (DLBCL) 25. We therefore hypothesized that the increased B cell responses observed after immunization with OVA and NP‐Ficoll could, at least in part, be driven by an inherent increased proliferative capacity of TRIM21‐deficient cells in response to BCR activation. To test this, we first stimulated splenocytes with B cell‐specific stimuli using anti‐IgM as a BCR‐activating factor or anti‐CD40, which induces proliferation via the CD40 pathway. We observed increased proliferation in cells derived from Trim21–/– mice compared to Trim21 +/+ mice when stimulating cells with anti‐IgM (Fig. 4a), but not when stimulating cells via the CD40 pathway with anti‐CD40 antibody up to a concentration of 5 g/ml 26 (Fig. 4b). Higher proliferation rates of Trim21 –/– cells were also observed when stimulating purified CD19+ cells (Fig. 4c), further pointing to an intrinsic increased capacity of Trim21 –/– cells to expand upon BCR‐specific activation by an increased expression of the proliferation marker Ki67 in CD19+ B cells upon anti‐IgM stimulation of Trim21 –/– splenocytes (Fig. 4d). We next investigated whether the increased cellular proliferation of B cells lacking TRIM21 could, in part, be explained by decreased activation‐induced cell death. Splenocytes were stimulated with anti‐IgM, restimulated on day 3, and the frequency of live, apoptotic and necrotic cells was assessed 24 h later. We could not, however, detect any altered susceptibility to activation‐induced cell death in CD19+ Trim21 –/– cells compared to Trim21 +/+ cells (Fig. 4e).
Figure 4.
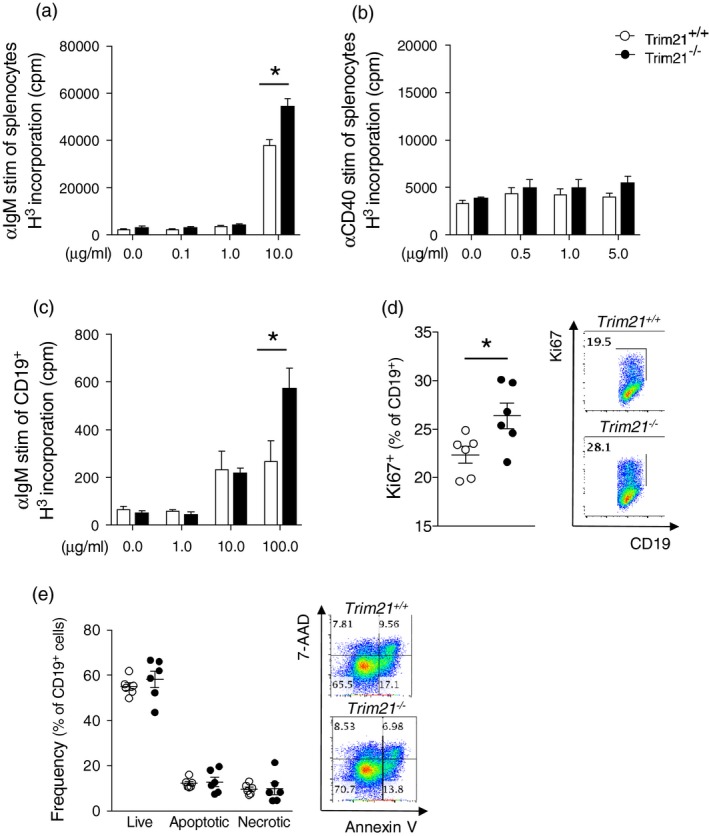
Trim21–/– B cells proliferate more readily upon anti‐immunoglobulin (Ig)M stimulation. Spleen cells were cultured with either anti‐IgM, anti‐CD40 or without stimuli. Incorporation of thymidine was measured after 3 days. (a) anti‐IgM and (b) anti‐CD40 stimulation of total splenocytes from Trim21 +/+ and –/– (five per group). (c) Anti‐IgM stimulation of B cells purified from spleens (three per group). (d) Total splenocytes were cultured with 0·1 g/ml anti‐IgM for 4 days and expression of the proliferation marker Ki67 in CD19+ B cells was assessed by flow cytometry (six per group). (e) Activation‐induced cell death was assessed in anti‐IgM stimulated cells at day 5, 2 days after restimulation. Frequencies of live, annexin V+ apoptotic and annexin V+ 7‐aminoactinomycin D (7‐AAD)+ necrotic cells among Trim21 +/+ and –/– cells (six per group). Data are presented as mean ± standard error of the mean (s.e.m.). *P < 0·05 (Mann–Whitney I‐test)
In summary, these data demonstrate directly that TRIM21 negatively regulates B cell proliferation downstream of BCR, but not CD40, stimulation, and suggest that the higher antibody responses observed in Trim21 –/– mice may relate to a higher proliferative capacity of B cells in the absence of TRIM21.
The follicular B cell population is expanded in naive Trim21–/– mice
Our observation that TRIM21 influences B cell proliferation after BCR activation raised the question of whether specific B cell populations would be expanded in Trim21 –/– mice, considering that no differences in frequency were observed when assessing the total splenic CD19+ B cell population in naive Trim21–/– versus Trim21 +/+ mice (18 and Fig. 5a). Interestingly, we found that the CD19+CD21+CD23+ follicular B cell population was indeed expanded in the spleen of Trim21 –/– mice compared to Trim21 +/+ mice, while the frequency of CD19+CD21++CD23+ marginal zone B cell was decreased (Fig. 5b). Furthermore, while similar proportions of total splenic CD19+ B cells were still observed in older Trim21–/– and Trim21 +/+ mice (aged approximately 1 year), the specific expansion of the CD19+CD21+CD23+ follicular B cell population persisted at that age (Fig. 5). In all, these data reveal an increase of the follicular B cell population in the spleen of naïve Trim21–/– mice, suggesting that TRIM21 contributes to the development and/or maintenance of normal B cell populations.
Figure 5.
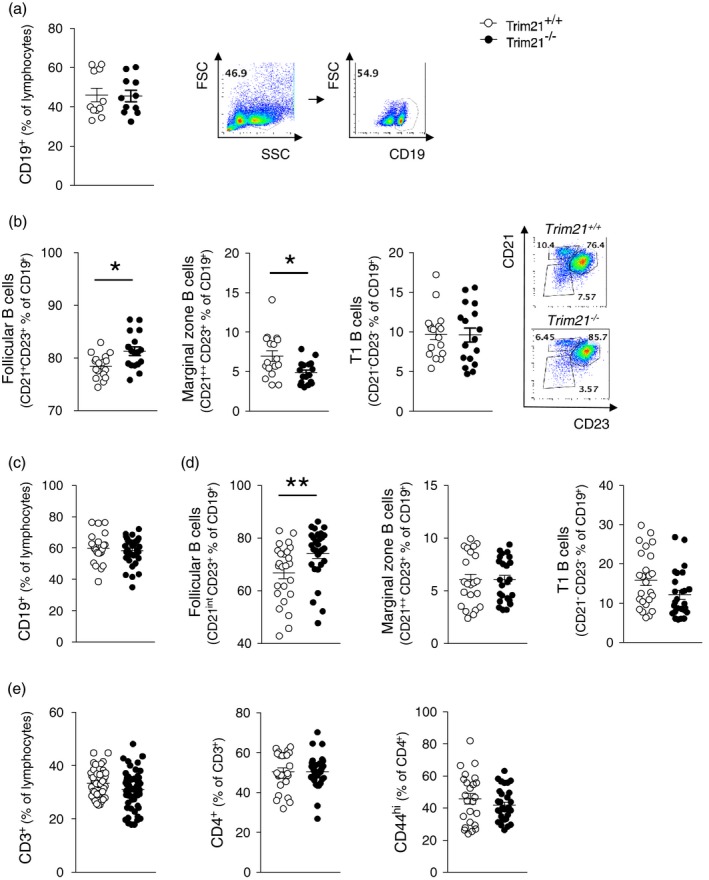
Follicular B cells are expanded in unmanipulated Trim21‐deficient mice. The cellular composition of spleens of naive, unmanipulated mice was assessed by flow cytometry. (a) Frequency of CD19+ cells and (b) CD19+CD21+CD23+ follicular B cells, CD19+CD21++CD23+ marginal zone B cells and CD19+CD21–CD23– T1 transitional B cells in mice aged 8–16 weeks (n = 17 per group, pooled data from four experiments). (c) Frequency of CD19+ cells and (d) CD19+CD21+CD23+ follicular B cells, CD19+CD21++CD23+ marginal zone B cells and CD19+CD21–CD23– T1 transitional B cells in mice aged 52–58 weeks (Trim21 +/+ n = 25, Trim21 –/– n = 29, pooled data). Data are presented as mean ± standard error of the mean (s.e.m.). *P < 0·05, **P < 0·01 (Mann–Whitney U‐test)
Gene expression analysis in Trim21‐deficient follicular B cells reveals highly significant differences in B cell differentiation pathways
To explore the role of TRIM21 in follicular B cells, we performed gene expression microarray analysis in FACS‐sorted CD19+CD21+CD23+ follicular B cell isolated from naive Trim21 –/– and Trim21 +/+ mice. Gene expression profiling revealed prominent differences between Trim21 –/– and Trim21 +/+ follicular B cells (Fig. 6a). Interestingly, pathway analysis based on GeneOntology terms showed that pathways altered in Trim21 –/– cells were related primarily to immunity, cellular metabolism and cellular homeostasis (Fig. 6b, Supporting information, Table S1). Specifically, the top processes were regulation of B cell differentiation, sterol biosynthetic process, cholesterol biosynthetic process, secondary alcohol biosynthetic process and cell cycle. The most differentially expressed genes (P < 0·01) in the top pathway are depicted in Fig. 6c and reveal a predominance of genes associated with B cell differentiation among up‐regulated genes in Trim21–/– cells. Interestingly, one of the top hits in this pathway was Ikzf1, a gene associated previously with systemic lupus erythematosus (SLE) 27, 28, and B cell malignancies 29. The three top metabolic pathways regulated differentially in Trim21–/– cells displayed a large overlap in associated genes, however, altogether revealing altered sterol metabolism. Finally, the cell cycle pathway was the last top differentially regulated pathway in Trim21–/– cells; this pathway includes a considerably higher number of genes than the other pathways, and accordingly also contained the majority of differentially expressed genes.
Figure 6.
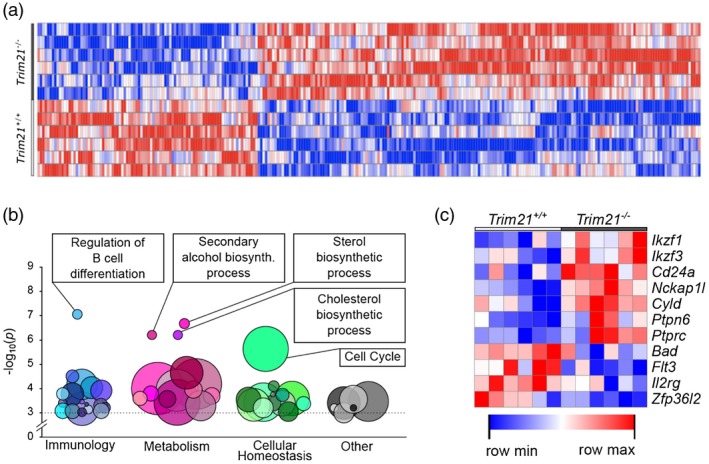
Distinct transcriptomic profiles in Trim21 –/– follicular B cells. Total RNA was extracted from CD19+CD21+CD23+ follicular B cells isolated from Trim21 +/+ and Trim21 –/– mice (six per group), and the whole‐genome transcriptome was analysed using the Mouse Transcriptome Assay 1.0 microarray chip (Affymetrix). (a) Gene expression profiling showing top differentially regulated genes (P‐value < 0·01). (b) Pathway analysis based on GO search terms. (c) Top differentially expressed genes (P < 0·05) belonging to the GO term GO:0045577 ‘GO: regulation of B cell differentiation’ pathway.
In conclusion, analysis of the transcriptome of naive Trim21 –/– and Trim21 +/+ follicular B cells revealed a significant association with pathways of B cell development, sterol metabolism and cell cycle, further supporting a role for TRIM21 in B cell proliferation and differentiation.
Discussion
Ro52/TRIM21 is a main autoantigen in Sjögren’s syndrome and SLE, and Trim21‐deficient mice develop systemic autoimmunity with hypergammaglobulinaemia and autoantibodies after tissue injury. In the present study, we therefore aimed to understand how TRIM21 influences B cell activation and antibody production.
To study B cell responses under standardized conditions, we first chose a T cell‐dependent NP‐OVA immunization system. We found that higher levels of specific IgM and IgG developed in Trim21 –/– mice and observed higher numbers of specific antibody‐producing cells in the spleen and a larger proportion of plasma cells in the bone marrow. The antibody subclasses IgG1 and to some extent IgG2a were the dominating IgG subclasses, similar to what had previously been observed in Trim21 –/– mice developing systemic autoimmunity following tissue injury 18. Interestingly, increased IgG levels and a shift to IgG1, the human equivalent to murine IgG2a, is seen in Sjögren’s syndrome and SLE patients 30.
Higher IgM antibody titres and higher numbers of IgM‐producing B cells were still detected in Trim21 –/– mice 28 days after the initial immunization with OVA. During infections, persisting IgM levels indicate incomplete clearance of the infectious agent, and in autoimmunity IgM antibodies specific for autoantigens can persist for long periods of time 31. Of note, elevated IgM levels are observed in both SLE and Sjögren’s syndrome patients. Also, the occurrence of IgM extra‐nuclear autoantibodies (ENA), of which anti‐Ro52/SSA is one, is seen commonly in systemic rheumatic diseases 32. While it is possible that the prolonged IgM response observed in immunized Trim21 –/– mice reflects a reduced antigen clearance capability, it may also relate to an expanded IgM‐producing B cell population.
The enhanced B cell activation and antibody production seen in Trim21 –/– mice after OVA immunization could be specifically the result of differential response to BCR activation. To test this directly, we challenged mice with LPS or Ficoll. Interestingly, only Ficoll, which acts by cross‐linking the BCR, generated higher specific antibody titres in Trim21 –/– mice compared to wild‐type controls. A potential differential regulation downstream of BCR activation was further investigated by stimulating cells with anti‐IgM or anti‐CD40, demonstrating that activation via the BCR, but not the CD40 pathway, at least at the concentrations tested, led to stronger proliferation of Trim21 –/– cells compared to Trim21 +/+ cells. BCR activation is essential for both B cell survival and differentiation 33, and we indeed observed not only increased proliferative activity, but also altered B cell subset ratios in Trim21–/– mice. Upon BCR activation, the proximate signalling complex, the BCR signalosome, is assembled, facilitating downstream activation 33. In a recent study by Satpathy et al. 34, the molecular components of the BCR signalosome were analysed using mass spectrometry, and BCR activation was observed to lead to immediate downstream ubiquitination and phosphorylation of multiple molecules. Interestingly, TRIM21, which is an E3 ligase mediating ubiquitination, was identified in this BCR signalosome. While it was not in the scope of the present study to investigate the potential targets of TRIM21 downstream of BCR activation, TRIM21 has previously been proposed to regulate the canonical NF‐κB pathway 35; this pathway is activated downstream of the BCR, and therefore represents a possible way in which TRIM21 might modulate BCR signalling.
TRIM21 has previously been associated with the regulation of both proliferation and apoptosis. Specifically, we have shown that ectopic expression of TRIM21 in the B cell line A20, but not TRIM21∆RING, where the functional domain has been deleted, results in retarded growth 12. Here, for the first time, by using Trim21–/– cells, we show that TRIM21 exerts a direct effect on cellular proliferation and that, in the context of B cells, this effect is BCR‐dependent. Interestingly, the TRIM21 gene is located in a tumour suppression region associated to multiple cancers 36, 37, 38. In addition to reducing the growth of A20 cells, ectopic expression of Trim21 was shown to enhance activation‐induced cell death when cells were stimulated with an agonistic anti‐CD40 antibody 12. Over‐expression of TRIM21 in HeLa cells has also been shown to have a pro‐apoptotic effect in a Bcl‐2‐dependent and p53‐independent manner 39. However, here, in primary B cells lacking Trim21, we could not detect any altered susceptibility to activation‐induced cell death. This may relate to specific properties of the tumour cell lines used in previous studies, as our experiments were performed in non‐transformed primary cells or to the choice and concentration of stimuli used.
We also noted a relative increase of splenic follicular B cells in Trim21 –/– mice that was present in all naive mice, upon immunization, and in aged mice. However, no difference in splenic CD3 subsets nor spleen histopathology, B or T cell distribution was noted in a previous study [ 18 ]. Considering the persistence of the increase of follicular B cells and the fact that the CD19+ pool of cells did not appear expanded, our findings nevertheless suggest that TRIM21 has a role in B cell differentiation and lineage commitment, and that differentiation towards a follicular B cell is favoured in the absence of TRIM21. The differentiation of naive circulating B2 cells into follicular B cells is primarily dependent on BCR signalling strength, as well as the activation of the canonical NF‐κB pathway 35. Interestingly, TRIM21 has been proposed to regulate the NF‐κB pathway by mono‐ubiquitination of nuclear factor kappa‐B kinase subunit beta (IKKβ), leading to increased levels of I‐κB 35, 40 which, in turn, inhibits the canonical NF‐κB pathway. Trim21‐deficiency could therefore affect the negative regulation of the canonical NF‐κB pathway and thereby favour follicular B cell differentiation. Furthermore, several IRFs, transcription factors shown previously to be directly regulated by TRIM21 18, 41, are implicated in B cell differentiation and homeostasis. IRF‐4 and IRF‐8 in particular have been associated repeatedly with B cell differentiation, from the early pre‐B cell to the plasma cell stages 42, 43, and IRF‐4 is involved in BCR expression during development, germinal centre formation and immunoglobulin class switch 42, 43, 44, 45. IRF‐5, a disease risk gene in SLE and progressive systemic sclerosis (pSS) 3, has also been implicated in the regulation of plasma cell differentiation 46. TRIM21 may therefore also modulate B cell differentiation via ubiquitination of IRFs. Collectively, these data suggest a role for TRIM21 in regulating multiple steps of B cell homeostasis, favouring follicular B cell differentiation and survival.
Whole genome expression analysis of unmanipulated CD21+CD23+ follicular B cells revealed prominent differences in B cell differentiation, sterol metabolism and cell cycle pathways between Trim21–/– and Trim21+/+ cells. Our observations of follicular B cell expansion and increased B cell proliferation in Trim21‐deficient mice is therefore supported by the altered expression of genes belonging to these pathways, which point to altered B cell differentiation and cell cycle activity in the absence of TRIM21. Interestingly, sterol metabolism has been observed to influence both immunoglobulin production and lymphocyte proliferation 47, 48, 49. In addition, we have recently described an enhanced susceptibility to high‐fat diet in Trim21–/– bone marrow‐transferred Ldlr–/– mice 19, with these mice developing significantly larger atherosclerotic plaques. This phenotype was linked to an increased differentiation and activation of the Th17 pathway in Trim21–/– mice, further emphasizing the link between altered sterol metabolism, the immune system and TRIM21.
In conclusion, our findings implicate the rheumatic autoantigen TRIM21/Ro52 in the regulation of B cell proliferation, follicular B cell differentiation and antibody responses, suggesting that this stems, at least in part, from a B cell intrinsic effect of TRIM21 downstream of BCR activation. In all, this offers a potential role for an autoantigen in the development of systemic inflammation.
Disclosures
The authors declare no financial or commercial conflicts of interest.
Author contributions
S. B. and M. W. H. conceived the study. S. B., M. I., G. E. T. and A. A. performed the experiments, analysed the data and generated the figures with input from M. W. H. S. B. and M. W. H. wrote the first version of the manuscript, and all authors participated in the editing until its final version.
Supporting information
Table S1. Genes differentially regulated between wild‐type and knockout mice (P < 0.05) belonging to the most significantly enriched GO terms.
Acknowledgements
We thank Marika Rönnholm and David Brodin at the BEA microarray facility for excellent technical assistance and computation. This study was supported by grants from the Swedish Research Council, the Heart–Lung Foundation, the Stockholm County Council, the Karolinska Institute, the Swedish Rheumatism Association, King Gustaf the Vth 80‐year Foundation, and the Torsten and Ragnar Söderberg Foundation.
References
- 1. Ambrosi A, Wahren‐Herlenius M. Update on the immunobiology of Sjogren’s syndrome. Curr Opin Rheumatol 2015;27:468–75. [DOI] [PubMed] [Google Scholar]
- 2. Ramirez Sepulveda JI, Kvarnstrom M, Eriksson P et al Long‐term follow‐up in primary Sjogren’s syndrome reveals differences in clinical presentation between female and male patients. Biol Sex Differ 2017;8:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wahren‐Herlenius M, Dorner T. Immunopathogenic mechanisms of systemic autoimmune disease. Lancet 2013;382:819–31. [DOI] [PubMed] [Google Scholar]
- 4. Bernatsky S, Ramsey‐Goldman R, Labrecque J et al Cancer risk in systemic lupus: an updated international multi‐centre cohort study. J Autoimmun 2013;42:130–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Theander E, Henriksson G, Ljungberg O, Mandl T, Manthorpe R, Jacobsson LT. Lymphoma and other malignancies in primary Sjogren’s syndrome: a cohort study on cancer incidence and lymphoma predictors. Ann Rheum Dis 2006;65:796–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Halse AK, Marthinussen MC, Wahren‐Herlenius M, Jonsson R. Isotype distribution of anti‐Ro/SS‐A and anti‐La/SS‐B antibodies in plasma and saliva of patients with Sjogren’s syndrome. Scand J Rheumatol 2000;29:13–9. [DOI] [PubMed] [Google Scholar]
- 7. Hennig J, Ottosson L, Andresen C et al Structural organization and Zn2+‐dependent subdomain interactions involving autoantigenic epitopes in the Ring‐B‐box‐coiled‐coil (RBCC) region of Ro52. J Biol Chem 2005;280:33250–61. [DOI] [PubMed] [Google Scholar]
- 8. Ottosson L, Hennig J, Espinosa A, Brauner S, Wahren‐Herlenius M, Sunnerhagen M. Structural, functional and immunologic characterization of folded subdomains in the Ro52 protein targeted in Sjogren’s syndrome. Mol Immunol 2006;43:588–98. [DOI] [PubMed] [Google Scholar]
- 9. Hennig J, Bresell A, Sandberg M et al The fellowship of the RING: the RING‐B‐box linker region interacts with the RING in TRIM21/Ro52, contains a native autoantigenic epitope in Sjogren syndrome, and is an integral and conserved region in TRIM proteins. J Mol Biol 2008;377:431–49. [DOI] [PubMed] [Google Scholar]
- 10. Reymond A, Meroni G, Fantozzi A et al The tripartite motif family identifies cell compartments. EMBO J 2001;20:2140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Espinosa A, Oke V, Elfving A, Nyberg F, Covacu R, Wahren‐Herlenius M. The autoantigen Ro52 is an E3 ligase resident in the cytoplasm but enters the nucleus upon cellular exposure to nitric oxide. Exp Cell Res 2008;314:3605–13. [DOI] [PubMed] [Google Scholar]
- 12. Espinosa A, Zhou W, Ek M et al The Sjogren’s syndrome‐associated autoantigen Ro52 is an E3 ligase that regulates proliferation and cell death. J Immunol 2006;176:6277–85. [DOI] [PubMed] [Google Scholar]
- 13. Crawford LJ, Johnston CK, Irvine AE. TRIM proteins in blood cancers. J Cell Commun Signal 2018;12:21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kimura T, Jain A, Choi SW, Mandell MA, Johansen T, Deretic V. TRIM‐directed selective autophagy regulates immune activation. Autophagy 2017;13:989–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ozato K, Shin DM, Chang TH, Morse HC 3rd. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol 2008;8:849–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Higgs R, Ni Gabhann J, Ben Larbi N, Breen EP, Fitzgerald KA, Jefferies CA. The E3 ubiquitin ligase Ro52 negatively regulates IFN‐beta production post‐pathogen recognition by polyubiquitin‐mediated degradation of IRF3. J Immunol 2008;181:1780–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kong HJ, Anderson DE, Lee CH et al Cutting edge: autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF‐8 and enhances cytokine expression in macrophages. J Immunol 2007;179:26–30. [DOI] [PubMed] [Google Scholar]
- 18. Espinosa A, Dardalhon V, Brauner S et al Loss of the lupus autoantigen Ro52/Trim21 induces tissue inflammation and systemic autoimmunity by disregulating the IL‐23‐Th17 pathway. J Exp Med 2009;206:1661–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brauner S, Jiang X, Thorlacius GE et al Augmented Th17 differentiation in Trim21 deficiency promotes a stable phenotype of atherosclerotic plaques with high collagen content. Cardiovasc Res 2018;114:158–67. [DOI] [PubMed] [Google Scholar]
- 20. Sjostrand M, Johansson A, Aqrawi L, Olsson T, Wahren‐Herlenius M, Espinosa A. The expression of BAFF is controlled by IRF transcription factors. J Immunol 2016;196:91–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reich M, Liefeld T, Gould J, Lerner J, Tamayo P. Mesirov JP. GenePattern 2.0. Nat Genet 2006;38:500–1. [DOI] [PubMed] [Google Scholar]
- 22. Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform 2009;10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mond JJ, Lees A, Snapper CM. T cell‐independent antigens type 2. Annu Rev Immunol 1995;13:655–92. [DOI] [PubMed] [Google Scholar]
- 24. Obukhanych TV, Nussenzweig MC. T‐independent type II immune responses generate memory B cells. J Exp Med 2006;203:305–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brauner S, Zhou W, Backlin C et al Reduced expression of TRIM21/Ro52 predicts poor prognosis in diffuse large B‐cell lymphoma patients with and without rheumatic disease. J Intern Med 2015;278:323–32. [DOI] [PubMed] [Google Scholar]
- 26. Haxhinasto SA, Bishop GA. Synergistic B cell activation by CD40 and the B cell antigen receptor: role of B lymphocyte antigen receptor‐mediated kinase activation and tumor necrosis factor receptor‐associated factor regulation. J Biol Chem 2004;279:2575–82. [DOI] [PubMed] [Google Scholar]
- 27. Han JW, Zheng HF, Cui Y et al Genome‐wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet 2009;41:1234–7. [DOI] [PubMed] [Google Scholar]
- 28. Wang C, Ahlford A, Jarvinen TM et al Genes identified in Asian SLE GWASs are also associated with SLE in Caucasian populations. Eur J Hum Genet 2013;21:994–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tefferi A, Vainchenker W. Myeloproliferative neoplasms: molecular pathophysiology, essential clinical understanding, and treatment strategies. J Clin Oncol 2011;29:573–82. [DOI] [PubMed] [Google Scholar]
- 30. Blanco F, Kalsi J, Ravirajan CT, Speight P, Bradwell AR, Isenberg DA. IgG subclasses in systemic lupus erythematosus and other autoimmune rheumatic diseases. Lupus 1992;1:391–9. [DOI] [PubMed] [Google Scholar]
- 31. Nyman U, Lundberg I, Hedfors E, Wahren M, Pettersson I. IgG and IgM anti‐snRNP reactivity in sequentially obtained serum samples from patients with connective tissue diseases. Ann Rheum Dis 1992;51:1307–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Duarte‐Rey C, Bogdanos DP, Leung PS, Anaya JM, Gershwin ME. IgM predominance in autoimmune disease: genetics and gender. Autoimmun Rev 2012;11:A404–12. [DOI] [PubMed] [Google Scholar]
- 33. Rickert RC. New insights into pre‐BCR and BCR signalling with relevance to B cell malignancies. Nat Rev Immunol 2013;13:578–91. [DOI] [PubMed] [Google Scholar]
- 34. Satpathy S, Wagner SA, Beli P et al Systems‐wide analysis of BCR signalosomes and downstream phosphorylation and ubiquitylation. Mol Syst Biol 2015;11:810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wada K, Niida M, Tanaka M, Kamitani T. Ro52‐mediated monoubiquitination of IKK{beta} down‐regulates NF‐{kappa}B signalling. J Biochem 2009;146:821–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schiebe M, Ohneseit P, Hoffmann W, Meyermann R, Rodemann HP, Bamberg M. Loss of heterozygosity at 11p15 and p53 alterations in malignant gliomas. J Cancer Res Clin Oncol 2001;127:325–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zenklusen JC, Weitzel JN, Ball HG, Conti CJ. Allelic loss at 7q31.1 in human primary ovarian carcinomas suggests the existence of a tumor suppressor gene. Oncogene 1995;11:359–63. [PubMed] [Google Scholar]
- 38. Zhao B, Bepler G. Transcript map and complete genomic sequence for the 310 kb region of minimal allele loss on chromosome segment 11p15.5 in non‐small‐cell lung cancer. Oncogene 2001;20:8154–64. [DOI] [PubMed] [Google Scholar]
- 39. Jauharoh SN, Saegusa J, Sugimoto T et al SS‐A/Ro52 promotes apoptosis by regulating Bcl‐2 production. Biochem Biophys Res Commun 2012;417:582–7. [DOI] [PubMed] [Google Scholar]
- 40. Sasaki Y, Derudder E, Hobeika E et al Canonical NF‐kappaB activity, dispensable for B cell development, replaces BAFF‐receptor signals and promotes B cell proliferation upon activation. Immunity 2006;24:729–39. [DOI] [PubMed] [Google Scholar]
- 41. Yoshimi R, Chang TH, Wang HS, Atsumi T, Morse HC, Ozato K. Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF‐kappa B‐dependent cytokine expression in fibroblasts. J Immunol 2009;182:7527–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ochiai K, Maienschein‐Cline M, Simonetti G et al Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity 2013;38:918–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Willis SN, Good‐Jacobson KL, Curtis J et al Transcription factor IRF4 regulates germinal center cell formation through a B cell‐intrinsic mechanism. J Immunol 2014;192:3200–6. [DOI] [PubMed] [Google Scholar]
- 44. Lu R. Interferon regulatory factor 4 and 8 in B‐cell development. Trends Immunol 2008;29:487–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Muto A, Ochiai K, Kimura Y et al Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. EMBO J 2010;29:4048–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lien C, Fang CM, Huso D, Livak F, Lu R, Pitha PM. Critical role of IRF‐5 in regulation of B‐cell differentiation. Proc Natl Acad Sci USA 2010;107:4664–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bauman DR, Bitmansour AD, McDonald JG, Thompson BM, Liang G, Russell DW. 25‐Hydroxycholesterol secreted by macrophages in response to Toll‐like receptor activation suppresses immunoglobulin A production. Proc Natl Acad Sci USA 2009;106:16764–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bensinger SJ, Bradley MN, Joseph SB et al LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell 2008;134:97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Spann NJ, Glass CK. Sterols and oxysterols in immune cell function. Nat Immunol 2013;14:893–900. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Genes differentially regulated between wild‐type and knockout mice (P < 0.05) belonging to the most significantly enriched GO terms.


