Summary
Periodontitis is a chronic inflammatory disease caused by the colonization of teeth by the bacterial plaque biofilm and the resultant host immune responses in adjacent periodontal tissues. Disease severity can vary dramatically between patients with periodontitis, with some subjects displaying inflammation without bony destruction (gingivitis), while others experience chronic progressive or rapidly aggressive gingival connective tissue damage and bone loss. To determine whether peripheral immune dysregulation is associated with periodontitis, we performed extensive analysis of immune cell subsets in peripheral blood from patients with chronic or aggressive periodontitis versus periodontally healthy control subjects. Peripheral blood mononuclear cells (PBMC) from patients with chronic periodontitis or aggressive periodontitis and from periodontally healthy controls were analysed by 8–10‐colour flow cytometry for the frequencies of various lymphocyte subsets, including interleukin (IL)‐17‐, interferon (IFN)‐γ‐, tumour necrosis factor (TNF)‐α‐ and IL‐10‐producing cells, and the frequencies and phenotype of monocytes. Cytokine levels in serum from the different groups were determined by Luminex assay. We found no significant differences in the frequencies of major immune cell populations [CD4+ T cells, CD8+ T cells, γδ T cells, CD4+CD45RO+CD25+CD127low regulatory T cells (Tregs), CD19+ B cells, CD14+ monocytes] or of cytokine‐producing T cells, or in the phenotype of CD14+ monocytes in peripheral blood from these patient cohorts. Additionally, no significant differences were observed in serum levels of prototypical inflammatory cytokines. These results suggest that the local gingival inflammatory response is not reflected by obvious changes in major blood immune cell subset frequencies.
Keywords: periodontal disease, peripheral blood mononuclear cells, T helper 17 cells, Tregs
Introduction
Periodontitis is a chronic inflammatory disease caused by the colonization of teeth by the bacterial plaque biofilm and the resultant host immune responses that occur in the surrounding periodontal tissues. The bacterial plaque biofilm induces an inflammatory infiltrate in periodontal tissue, in which the dominant immune cell populations may differ from person to person. There are wide variations in disease severity between patients with periodontal disease. Many subjects will have superficial gingival inflammation without any deeper tissue destruction (gingivitis), while other subjects will experience progressive destruction of the supporting tissues of the tooth, including connective tissue damage and loss of alveolar bone (periodontitis). In the majority of cases, progression of tissue damage occurs slowly over many years and is affected by risk factors such as oral hygiene and plaque levels, local predisposing factors, smoking, genetic factors, systemic risk factors such as diabetes and stress (chronic periodontitis). In a smaller number of cases tissue damage may occur rapidly, typically with an early age of onset (< 35 years), and may result in rapid tooth loss (aggressive periodontitis) 1.
Studies have shown increased levels of proinflammatory cytokines such as interleukin (IL)‐17 in serum from patients with periodontitis when compared to healthy controls 2, 3, 4. Furthermore, studies have reported a decreased percentage of IL‐17+CD4+ T cells in peripheral blood mononuclear cells (PBMC) as well as decreased levels of IL‐17 protein in serum from patients with periodontitis following non‐surgical periodontal treatment 5, 6, 7. These findings suggest that changes in cytokine levels in locally inflamed gingival tissue are mirrored by cytokine levels in the systemic circulation. Other studies have determined the frequencies of immune cell populations and cytokine‐producing T helper cells in peripheral blood from patients with different forms of periodontitis, i.e. chronic and aggressive periodontitis 8, 9, 10. However, the limited existing data regarding interferon (IFN)‐γ‐, IL‐4‐ or IL‐17‐producing cells in peripheral blood are not definitive, as both decreased and increased percentages of these cytokine‐expressing cells in PBMC from patients with periodontitis have been demonstrated relative to healthy controls 8, 9, 10. The conflicting conclusions from these studies may be because different classifications of periodontitis were utilized (e.g. differences in the number of teeth affected, or different age criteria for chronic or acute periodontitis), or because not all studies included patients with different forms of periodontitis.
Studies have suggested that monocytes during or after periodontal pathogen challenge could contribute to inflammation in periodontitis‐associated systemic disease, such as aortic inflammation 11, 12. One study has shown an increased percentage of CD14+CD16+ monocytes in the peripheral blood of patients with chronic periodontitis compared to healthy individuals 13 but, overall, data on peripheral blood monocyte populations in periodontitis are scarce.
Epidemiological data suggest an association between periodontal diseases and systemic conditions, such as atherosclerotic cardiovascular disease, an increased risk of preterm low birth‐weight babies and chronic inflammatory diseases such as rheumatoid arthritis, diabetes and respiratory diseases, although the mechanisms for the various relationships remain largely unknown (reviewed in 14, 15). A possible mechanism has been established by which locally produced proinflammatory cytokines, including IL‐6, IL‐1β and TNF‐α, in severe periodontitis can enter the systemic circulation and cause increases in the serum levels of these cytokines as well as elevated levels of C‐reactive protein (reviewed in 14).
To assess the effect of localized inflamed tissue on the systemic circulation in periodontitis, we performed a systematic analysis of immune cell subsets in peripheral blood from patients with chronic or aggressive periodontitis and periodontally healthy control subjects. Specifically, we investigated the frequencies of the main immune cell subsets [T cells, regulatory T cells (Tregs), B cells, monocytes, natural killer (NK) cells] and frequencies of IL‐17‐, IFN‐γ‐, TNF‐α‐ and IL‐10‐producing cells within CD4+ T cells, CD8+ T cells, γδ T cells and CD19+ B cells. We focused on these four cytokines as they are important in regulation of inflammation‐induced tissue destruction in periodontitis. Furthermore, we investigated the phenotype of peripheral blood CD14+ monocytes, and also determined the levels of a wide range of T cell‐derived or monocyte‐derived cytokines in the serum from patients with periodontitis and periodontally healthy controls. Overall, our data do not indicate significant changes in any of these parameters.
Materials and methods
Patient and healthy control samples
Healthy control samples were obtained from periodontally healthy volunteers. Newly diagnosed patients with periodontitis were recruited from the periodontal clinic at Guy’s Hospital (see Table 1 for clinical and demographic parameters). Fifty ml of peripheral venous blood was drawn from each participant from the antecubital fossa and collected in sodium heparin plasma tubes (Becton Dickinson, Franklin Lakes, NJ, USA). An additional 6 ml of blood was collected in anti‐coagulant‐free serum tubes (Becton Dickinson) and stored at 4°C (in a fridge) for 1 h before centrifugation at 1200 rpm for 10 min and harvesting of the serum. The harvested serum samples were aliquoted into 500‐μl amounts and stored at −80°C until further analysis. Oral and periodontal examination records, age, sex and smoking status were documented on the day of sample collection. Periodontal disease case definitions for chronic periodontitis were used according to those described by Page and Eke, and definitions for aggressive periodontitis according to those proposed by Demmer and Papapanou 16, 17, with the presence of two or more interproximal, non‐adjacent sites with attachment loss of ≥ 6 mm occurring at a minimum of two different teeth and accompanied by bleeding on probing. Age‐ and sex‐matched periodontally healthy control subjects were recruited and assessed with definitions such as no probing pocket depth (PPD) > 3 mm at any site, no proximal sites with attachment loss at any site and bleeding on probing (BOP) of < 20% of possible sites. All clinical investigations were conducted according to the Declaration of Helsinki principles. Ethics approval for this study was given by the NRES Committee London City Road and Hampstead Research Ethics Committee (12/LO/0376). Written informed consent was received from participants before their inclusion into the study.
Table 1.
Demographic and clinical characteristics of the study subjects
| HC (n = 13) | CP (n = 15) | AP (n = 15) | anova P‐valuea | ||
|---|---|---|---|---|---|
| Age, mean years (range) | 36·8 (20–63) | 42·1 (36–58)# | 30·5 (19–36)# | P < 0·001 | |
| Gender | Male, n (%) | 6 (46) | 7 (47) | 8 (53) | n.s. |
| Female, n (%) | 7 (54) | 8 (53) | 7 (47) | n.s. | |
| Number of teeth, mean | 28·7 | 28·1 | 28·7 | n.s. | |
| PPD, mean ± s.d. mm | 1·73 ± 0·24 | 3·53 ± 0·69*** | 3·19 ± 0·78*** | P < 0·0001 | |
| BOP, mean ± s.d. % | 10·9 ± 7·0 | 32·1 ± 14·9* | 34·2 ± 30·9** | P < 0·01 | |
| % sites of PPD > 5 mm | 0 ± 0 | 29·5 ± 12·8*** | 23·4 ± 16·6*** | P < 0·0001 | |
| PISA, median (IQR) mm2 | 91 (42–136) | 643 (337–906)*** | 255 (137–1336)*** | P < 0·0001 | |
AP = aggressive periodontitis patients; BOP = bleeding on probing; CP = chronic periodontitis patients; GI = gingivitis patients; HC = periodontally healthy controls; PISA = periodontal inflamed surface area; PPD = probing pocket depth. Data were tested for normality using D’Agostino and Pearson omnibus normality testing and where not normally distributed data were log10‐transformed prior to analysis. Data were analysed by one‐way analysis of variance (anova) and the overall P‐value shown as a. Individual groups were compared to healthy control (HC) values by Bonferroni’s multiple comparison test for parametric data, and significant differences indicated as *P < 0·05; **P < 0·01; ***P < 0·001. Significant differences between CP and AP groups are indicated as # P < 0·05; n.s. = not significant.
Cell isolation from peripheral blood
PBMC were isolated by density gradient centrifugation using lymphocyte separation medium (LSM 1077; PAA Laboratories, Pasching, Austria or Lymphoprep; Axis‐Shield, Oslo, Norway). PBMC were cryopreserved within 1 h of isolation and stored in liquid nitrogen in medium containing 90% fetal bovine serum (lot 030M3399; Sigma‐Aldrich, St Louis, MO, USA) and 10% dimethyl sulphoxide (Sigma‐Aldrich).
Ex‐vivo immune cell subset staining
The immune cell subsets and phenotypes that were determined are shown in Supporting information, Fig. S1. The identification of CD4+ T cells, CD8+ T cells, CD4+CD45RO+CD25+CD127low Tregs, γδ T cells, B cells, monocytes and NK cells was performed using an eight‐colour extracellular staining panel (Supporting information, Table S1). For the identification of cytokine‐expressing cells, PBMC were stimulated for 3 h with phorbol myristate acetate (PMA) (50 ng/ml) and ionomycin (750 ng/ml) (both from Sigma‐Aldrich) in the presence of GolgiStop, according to the manufacturer’s instructions (Becton Dickinson, Oxford, UK). The identification of IL‐17‐, IFN‐γ‐, TNF‐α‐ or IL‐10‐expressing cells within CD4+ T cells, CD8+ T cells, γδ T cells or CD19+ B cells was facilitated by staining with a 10‐colour intracellular cytokine staining panel (Supporting information, Table S2). Extracellular surface staining was performed using the following monoclonal antibodies: phycoerythrin‐cyanin 7 (PE‐Cy7)‐conjugated anti‐CD3 (clone UCHT1), peridinin chlorophyll (PerCP)‐Cy5.5‐conjugated anti‐CD4 (clone SK3), allophycocyanin (APC)‐Cy7‐conjugated anti‐CD14 (clone HCD14), Brilliant Violet 605‐conjugated anti‐CD19 (clone HIB19), APC‐conjugated anti‐CD56 (clone HCD56), PE‐conjugated anti‐CD25 (clone M‐A251), fluorescein isothiocyanate (FITC)‐conjugated anti‐CD127 (clone A019D5), APC‐Cy7‐conjugated anti‐CD45RA (clone HI100), Pacific Blue‐conjugated anti‐CD45RO (clone UCHL1), APC‐conjugated anti‐CD54 (clone HCD54), Pacific Blue‐conjugated anti‐CD86 (clone IT2.2) (all from BioLegend, London, UK), PE‐CF594‐conjugated anti‐CD8 (clone RPA‐T8), FITC‐conjugated anti‐γδ T‐cell receptor (TCR) (clone 11F2), PerCP‐Cy5.5‐conjugated anti‐human leucocyte antigen D‐related (HLA‐DR) (clone G46‐6) (all from BD Biosciences, Oxford, UK) and PE‐conjugated anti‐CD40 (clone LOB7/6; AbD Serotec, Kidlington, UK).
For intracellular staining, following appropriate cell surface staining, cells were fixed with 2% paraformaldehyde and permeabilized with 0·5% saponin (Sigma‐Aldrich) and stained intracellularly with Pacific Blue‐conjugated anti‐CD4 (clone SK3), Alexa Fluor 488‐conjugated anti‐IL‐10 (clone JES3‐9D7), PE‐conjugated anti‐IL‐17A (clone BL168), APC‐conjugated anti‐TNF‐α (clone MAb11) and PerCP‐Cy5.5‐conjugated anti‐IFN‐γ (clone 4S.B3) (all from BioLegend). For intranuclear staining, cells were extracellularly stained and fixed as described above followed by permeabilization with ×1 forkhead box protein 3 (FoxP3) Perm Buffer (BioLegend). Cells were then stained with Alexa Fluor 647‐conjugated FoxP3 (clone 259D; BioLegend). Cells were acquired by flow cytometry using a LSRFortessa (BD Biosciences) and analysed using FlowJo software (Tree Star, Ashland, OR, USA). Cytometer settings were tracked over time using BD Biosciences Cytometer Setup and Tracking software to monitor cytometer performance of the LSRFortessa at the NIHR Biomedical Research Centre Flow Cytometry Core Facility at Guy’s Hospital. Viable single cells were identified using a LIVE/DEAD® Fixable Blue Dead Cell stain kit (Life Technologies, Carlsbad, CA, USA).
Cytokine detection
Serum samples were collected and stored at −80°C until further use. Serum cytokine levels were determined by Luminex assay (Bio‐Plex ProTM Human Th17 Cytokine Assays, Bio‐Rad Laboratories, CA, USA).
Statistical analysis
Values are expressed as mean ± standard error of the mean (s.e.m.). Data were tested for normality using D’Agostino and Pearson omnibus normality testing, and where non‐normally distributed data were log10‐transformed prior to analysis (which provided normally distributed results). Comparisons between patients and healthy controls were made using one‐way analysis of variance followed by Bonferroni’s multiple comparison test for parametric data. Data were analysed using Prism version 5 (GraphPad Software Inc, La Jolla, CA, USA). For all tests, P‐values of less than 0·05 were considered significant. In addition, to allow for the possibility that variations in subject age might account for some of the differences seen, we carried out a bivariate linear regression analysis with diagnosis and age inputted as explanatory variables. This was analysed using spss version 23 (IBM, Armonk, NY, USA).
Results
Similar frequencies of major immune cell subsets in peripheral blood from patients with periodontal disease versus periodontally healthy subjects
To determine the frequencies of the major immune cell subsets, we applied the staining panel as described in Supporting information, Table S1 to cryopreserved cells from patients with chronic periodontitis, aggressive periodontitis and periodontally healthy donors. We found no statistically significant differences in the frequencies of CD4+, CD8+ and γδ T cell subsets, or in CD19+ B cells, CD14+ monocytes and CD56+ NK cells in peripheral blood between the three groups (Fig. 1). Frequencies of CD45RA+ or CD45RO+ cells within CD4+ T cell populations were also similar in the peripheral blood from the different groups (Fig. 2 and data not shown).
Figure 1.
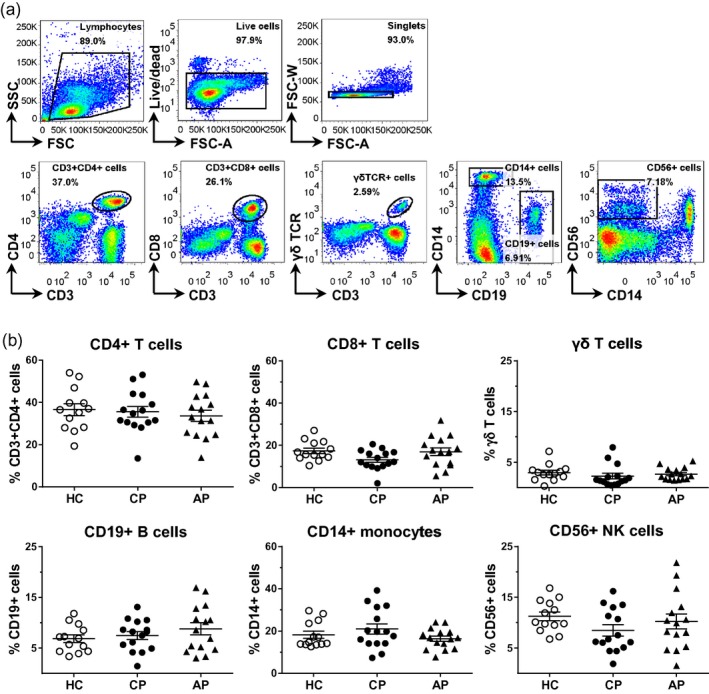
Frequencies of major immune cell populations in peripheral blood mononuclear cells (PBMC) from healthy controls and patients with periodontal disease. PBMC were isolated from peripheral blood from patients with chronic periodontitis (CP, n = 15) or aggressive periodontitis (AP, n = 15) as well as periodontally healthy controls (HC, n = 13). (a) Representative gating strategy and (b) cumulative data showing the frequencies of cell populations within live‐gated single cells. Each symbol represents an individual donor. Lines show the mean with standard error of the mean (s.e.m.). Log‐transformed data were analysed by one‐way analysis of variance followed by Bonferroni’s multiple comparison test.
Figure 2.
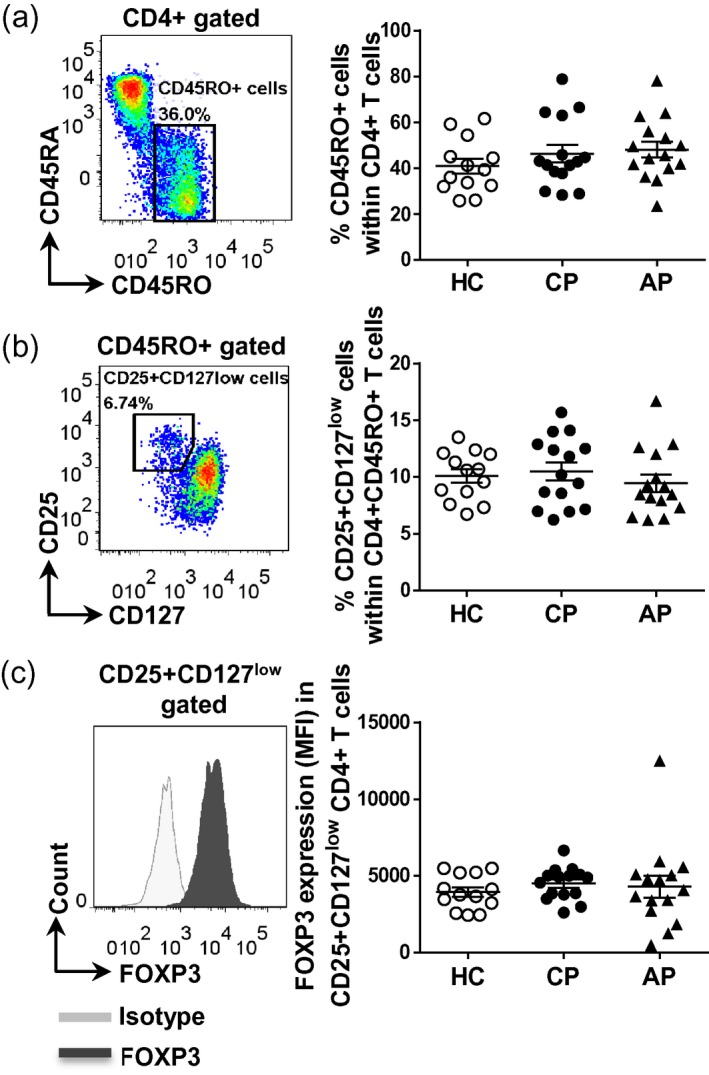
Frequencies of CD4+CD45RO+CD25+CD127low regulatory T cells in peripheral blood mononuclear cells (PBMC) from healthy controls and patients with periodontal disease. PBMC were isolated from peripheral blood from patients with chronic periodontitis (CP, n = 15) or aggressive periodontitis (AP, n = 15) as well as periodontally healthy controls (HC, n = 13). Cells were surface‐stained and then permeabilized and stained intranuclearly for forkhead box protein 3 (FoxP3). Representative dot‐plots and cumulative data showing the frequencies of CD4+CD45RO+ T cells (a) and CD25+CD127low cells within CD4+CD45RO+ T cells (b). (c) Representative histogram plots showing the expression levels of FoxP3 in CD4+CD25+CD127low T cells. Each symbol represents an individual donor. Lines show the mean with standard error of the mean (s.e.m.). Log‐transformed data were analysed by one‐way analysis of variance with Bonferroni’s multiple comparison test.
Regarding Tregs, as expected 18 there was a slightly higher percentage of CD25+CD127low T cells within the CD4+CD45RO+ T cell population (on average 10%) when compared to CD4+CD45RA+ or total CD4+ T cells (on average 5 and 7%, respectively), but the frequencies between the groups were not statistically significant different (Fig. 2 and data not shown). No significant difference was observed in FoxP3 expression in CD4+CD45RO+CD25+CD127low Tregs between the groups (Fig. 2).
CD14+ monocytes from peripheral blood of patients with periodontal disease and healthy control subjects are phenotypically similar
We investigated the phenotype of CD14+ monocytes from peripheral blood of patients with periodontal disease versus healthy control subjects. No significant differences in the expression of the markers CD14, CD16, CD40, CD54, CD86 or HLA‐DR by peripheral blood CD14+ monocytes were observed among the different groups (Fig. 3). We also investigated the frequency and phenotype (expression of the markers CD14, CD16, CD40, CD54, CD86 or HLA‐DR) of CD14+CD16+ monocytes in peripheral blood (often referred to as ‘inflammatory’ monocytes 19), but no significant differences were found between the groups (data not shown). The frequencies of CD14+CD16+ monocytes in the healthy control, chronic and aggressive periodontitis groups were also not significantly different [median 10.3%, interquartile range (IQR) = 3, 24; 4·2%, IQR = 3, 16; 4·9%, IQR = 2, 28; and 7·5%, IQR = 3, 25, respectively].
Figure 3.
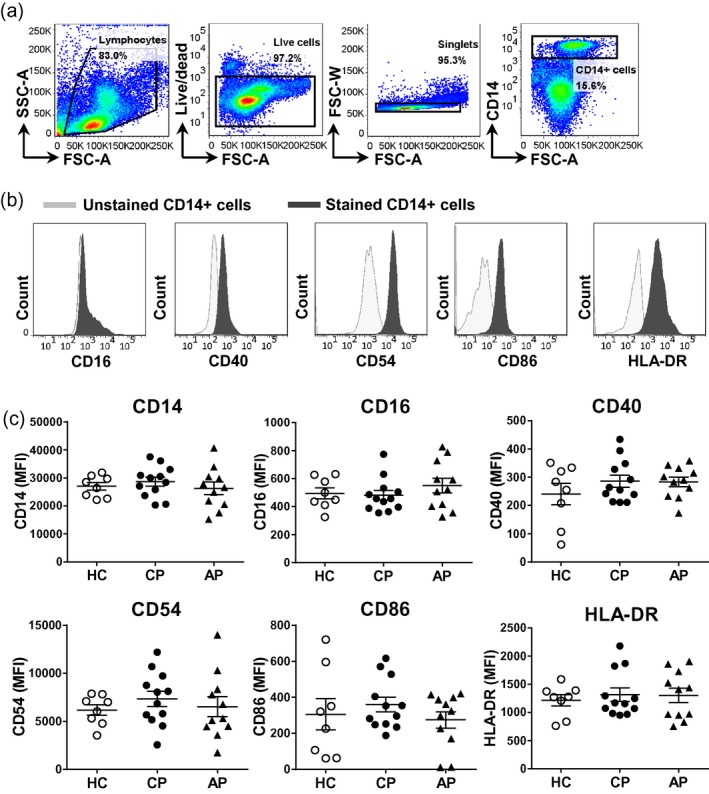
Expression of activation markers on CD14+ monocytes from healthy controls and patients with periodontal disease. Peripheral blood mononuclear cells (PBMC) were isolated from peripheral blood from patients with chronic periodontitis (CP, n = 12) or aggressive periodontitis (AP, n = 11), as well as periodontally healthy controls (HC, n = 8). (a) Representative gating strategy to identify CD14+ monocytes. (b) Representative histogram plots and (c) cumulative data showing the geometric mean fluorescence intensity (MFI) for the surface expression of indicated markers determined by flow cytometry on CD14+ monocytes. Each symbol represents an individual donor. Lines show the mean with standard error of the mean (s.e.m.). Log‐transformed data were analysed by one‐way analysis of variance with Bonferroni’s multiple comparison test.
Similar frequencies of cytokine‐producing cells within T cell populations from peripheral blood of patients with periodontal disease and healthy control subjects
Next, we investigated whether there were quantitative differences in cytokine producing CD4+ T cells in peripheral blood between the groups (Fig. 4a). In peripheral blood from patients with chronic periodontitis, on average 1 ± 0·1% of the CD4+ T cells expressed IL‐17, 12 ± 23% expressed IFN‐γ, a higher proportion expressed TNF‐α (on average 41 ± 3·8%) and a very low proportion expressed anti‐inflammatory IL‐10 (on average 0·3 ± 0·04%). However, these frequencies were not significantly different from patients with aggressive periodontitis or periodontally healthy control subjects (Fig. 4).
Figure 4.
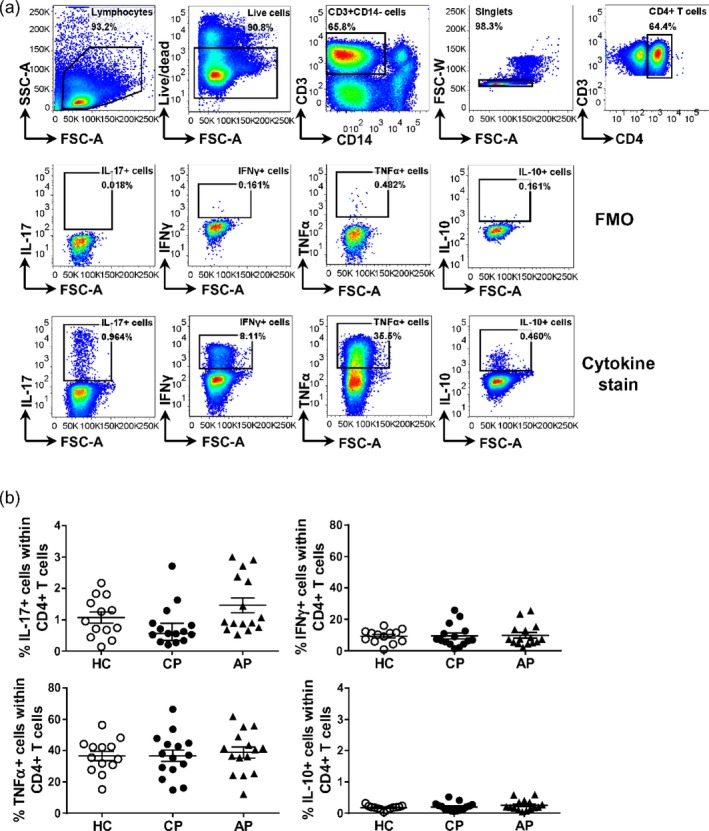
Frequencies of cytokine producing CD4+ T cells in PBMC from healthy controls and patients with periodontal disease. Peripheral blood mononuclear cells (PBMC) were isolated from peripheral blood from patients with chronic periodontitis (CP, n = 15) or aggressive periodontitis (AP, n = 15), as well as periodontally healthy controls (HC, n = 13). Cells were stimulated ex vivo with phorbol myristate acetate (PMA) and ionomycin for 3 h prior to intracellular cytokine staining. (a) Representative gating strategy and (b) cumulative data showing the frequencies for the interleukin (IL)‐17‐, interferon (IFN)‐γ‐, tumour necrosis factor (TNF)‐α‐ and IL‐10‐expressing cells within CD4+ T cells as determined based on fluorescence‐minus‐one (FMO) control. Each symbol represents an individual donor. Lines show the mean with standard error of the mean (s.e.m.). Log‐transformed data were analysed by one‐way analysis of variance with Bonferroni’s multiple comparison test.
In addition to the CD4+ T cell population, on average 0·4 ± 0·01% of the CD8+ T cells expressed IL‐17, 33 ± 4% expressed IFN‐γ, 36 ± 4% expressed TNF‐α, and a very low proportion expressed anti‐inflammatory cytokine IL‐10 (on average 0·1 ± 0·01%) in peripheral blood from patients with chronic periodontitis. In the γδ T cell population in peripheral blood from patients with chronic periodontitis, on average 0·7 ± 0·1% expressed IL‐17, 52 ± 6% expressed IFN‐γ and 52 ± 4% expressed TNF‐α (IL‐10 expression was not measured in γδ T cells). However, as for CD4+ T cells, there were no significant differences between the groups (Fig. 5 and data not shown). Very few CD19+ B cells expressed IL‐17, IFN‐γ or IL‐10, while a small proportion expressed TNF‐α (on average 15 ± 3% in chronic periodontitis), but this was not significantly different between groups.
Figure 5.
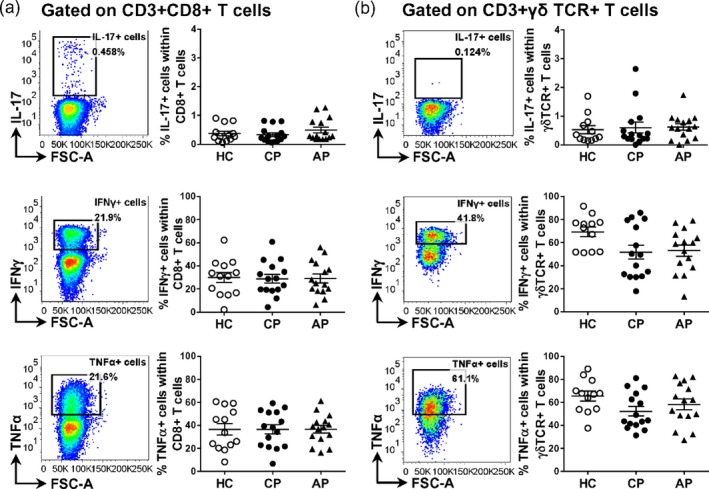
Frequencies of cytokine‐producing cells CD8+ and γδ T cells in peripheral blood mononuclear cells (PBMC) from healthy controls and patients with periodontal disease. PBMC were isolated from peripheral blood from patients with chronic periodontitis (CP, n = 15) or aggressive periodontitis (AP, n = 15) as well as periodontally healthy controls (HC, n = 13). (a,b) Representative dot‐plots and cumulative data for the frequencies of IL‐17+, IFN‐γ+ and TNF‐α+ cells within CD8+ T cells (a) or γδ T cells (b). Each symbol represents an individual donor. Lines show the mean with standard error of the mean (s.e.m.). Log‐transformed data were analysed by one‐way analysis of variance with Bonferroni’s multiple comparison test.
As chronic and aggressive periodontitis occur at different ages, and age itself may affect immune responses, we also carried out regression analysis to correct effects of disease diagnosis for age. The results are shown in Supporting information, Table S3 and show that age had a significant effect only on %γδ TCR+ cells and CD40 mean fluorescence intensity (MFI). There were no significant differences with periodontal diagnoses in all tests, with the exception of %γδ TCR+IFN‐γ+, which was significantly higher in healthy subjects compared to either aggressive or chronic periodontitis subjects.
Detection of T cell‐derived and monocyte‐derived mediators in serum
Finally, we determined the levels of CD4+ T helper cell‐derived (IFN‐γ, IL‐4, IL‐10, IL‐17A, IL‐17F, IL‐21, IL‐22, IL‐25, TNF‐α) and monocyte‐derived (IL‐1β, IL‐6, IL‐10, IL‐23, IL‐33 and TNF‐α) mediators in the sera of the aforementioned groups by Luminex assay. Protein levels of IFN‐γ, IL‐1β, IL‐17A, IL‐21, IL‐22, IL‐23 and IL‐25 (IL‐17E) were below the lower detection limit of the Luminex assay in the majority of samples. IL‐4, IL‐6, IL‐10, IL‐17F, IL‐33 and TNF‐α were detectable in most serum samples; however, no statistically significant differences between the groups were observed (Fig. 6 and data not shown).
Figure 6.
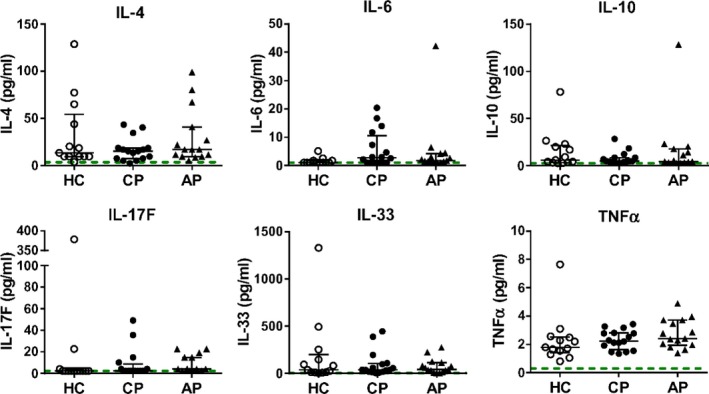
Cytokine levels in serum from patients with periodontitis or gingivitis, or from healthy controls. Serum from patients with chronic periodontitis (n = 15) or aggressive periodontitis (n = 15), as well as periodontally healthy controls (n = 13) was analysed. Cytokine levels in 25 µl of serum were determined by Luminex. Each symbol represents an individual subject; the median and interquartile range is shown, dotted lines indicate the minimum detectable concentration of that particular mediator. Log‐transformed data were analysed by one‐way analysis of variance with Bonferroni’s multiple comparison test. *P<0.05.
Discussion
In this study on newly diagnosed patients with chronic or aggressive periodontitis, we found no significant differences in the frequencies of major immune cell populations (CD4+ T cells, CD8+ T cells, Tregs, CD19+ B cells, CD14+ monocytes, CD56+ NK cells) and cytokine‐producing cells (IL‐17‐, IFN‐γ‐, TNF‐α‐ and IL‐10‐producing cells within CD4+ T cells, CD8+ T cells, γδ T cells and CD19+ B cells), or in the phenotype of CD14+ monocytes from the peripheral blood of chronic and aggressive periodontitis patient cohorts when compared to healthy controls.
Conflicting data have been reported regarding the frequencies of cytokine‐producing CD4+ T cells in periodontitis. A previous study reported a higher percentage of IL‐4 producing CD4+ T cells in PBMC from patients with chronic periodontitis versus healthy controls, a higher percentage of TNF‐α‐producing CD4+ T cells in PBMC from patients with aggressive periodontitis versus healthy controls, and no differences in the percentages of IFN‐γ‐ or IL‐10‐producing CD4+ T cells between groups 8. The authors suggested that CD4+ T cells from patients with chronic periodontitis are more committed to IL‐4 production, while CD4+ T cells from acute periodontitis are more committed to TNF‐α production; they did not measure the IL‐17‐producing CD4+ T cells in PBMC 8. Schmidt and colleagues reported slightly higher percentages of IFN‐γ‐ and IL‐4‐producing CD4+ T cells in PBMC from chronic versus aggressive periodontitis and healthy controls, and no differences in the percentages of IL‐17‐producing CD4+ T cells among the three groups 9. The authors also reported a slightly reduced T helper type 1 (Th1) response in patients with acute periodontitis defined by a lower Th1/Th2 ratio when compared with chronic periodontitis and healthy controls 9. Chen and colleagues recently reported significantly increased levels of IL‐17A+ and IFN‐γ+ CD4+ T cells in PBMC from chronic periodontitis versus healthy controls 10. Our data, however, show that CD4+ T cells from chronic or aggressive periodontitis patients did not contain significantly enhanced percentages of IL‐17+, IFN‐γ+ or TNF‐α+ cells compared to age‐ and sex‐matched healthy controls.
In terms of serum cytokine levels, previous studies have shown higher levels of IL‐17 and IL‐1β in serum from patients with periodontitis when compared to healthy controls 2, 3, 4, 20. In these studies, Gümüş et al. reported that the serum IL‐1β levels in chronic and aggressive periodontitis groups were higher than those in the healthy control group, while no significant differences in serum IL‐1β levels between the periodontitis groups were noted 20. A different study, however, reported no differences in the serum level of IL‐1β between chronic periodontitis and healthy control groups 21. In our data set, the serum levels of IL‐17A and IL‐1β were below the lower detection limit of the Luminex assay in the majority of samples. We detected IL‐4, IL‐6, IL‐10, IL‐17F, IL‐33 and TNF‐α; however, no statistically significant differences were found between the groups. Our data on IL‐4, IL‐6, IL‐10 and TNF‐α data are in line with several previously published papers 6, 21, 22, although significant differences in serum cytokines between healthy controls, aggressive and chronic periodontitis have been reported 6, 22, 23, 24.
Explanations for the differences in results between these studies are not immediately clear. Although there are some variations in study design, most particularly in differences in periodontal case definitions and in ages between groups in some of these studies, it seems unlikely that these account for the observed differences. Differences in experimental and technical approaches and standardization may explain some of these differences. In our study, to minimize interexperimental variation, cryopreserved samples were analysed in batches of five different samples that always consisted of a mix of healthy controls (n = 1 − 2), chronic periodontitis (n = 1−2) and aggressive periodontitis (n = 1−2); we also applied Cytometer Setup and Tracking (CS&T) settings to monitor and correct for flow cytometer performance over time. Furthermore, in addition to analysing cytokine‐expressing CD4+ T cells, we also analysed cytokine‐expressing CD8+ T cells, γδ T cells and B cells and FoxP3‐expressing Tregs. Recently, CD8+ T cells and γδ T cells have been reported to be able to produce IL‐17 25, 26 and B cells have been reported as a major source of IL‐17A and IL‐17F during Trypanosoma cruzi infection 27. However, we did not detect significant differences in the frequencies of any of these cell populations between groups. Furthermore, the data were not only not different between groups, but were also remarkably similar between groups. In a retrospective power calculation using the data we obtained in our results, we found that our study had 96% power to detect a mean increase of 50% in TNF+ cells with alpha as 0·05, and 74% power to detect a similar change in IFN‐γ+ cells. Thus, we can reasonably rule out the likelihood of a type II error in failing to detect a difference that actually existed.
Previous studies have suggested a role for monocytes from patients with periodontitis in aortic inflammation 11, 12. These data suggested that monocytes during or after periodontal pathogen challenge could contribute to inflammation in periodontitis‐associated systemic disease. In addition, we recently showed that in the presence of Porphyromonas gingivalis significantly higher IL‐17 production was observed in anti‐CD3 monoclonal antibody (mAb)‐stimulated monocyte/CD4+ T cell co‐cultures from patients with periodontitis compared to periodontally healthy controls 28. However, our current analysis did not reveal a significant difference in the frequencies of monocytes or expression of activation markers on monocytes among patients with different periodontal diseases and periodontally healthy controls.
Our data do not exclude the possibility that differences in immune cell subsets are present at the site of inflammation in chronic and/or acute periodontitis versus healthy gingival tissue. Indeed, studies have reported an increased percentage of IL‐17‐producing cells in gingival tissue from chronic periodontitis 28, 29, 30. Furthermore, in our study we only determined relative proportions of immune cell subsets in PBMC, and therefore we cannot rule out differences in absolute numbers of these immune cell types.
In conclusion, our data provide evidence that no significant alterations are present in the frequencies of major immune cell populations and cytokine‐producing cells in peripheral blood from patients with well‐defined periodontal disease.
Disclosures
The authors declare that they have no conflicts of interest in relation to this study.
Supporting information
Fig. S1. Preparation of peripheral blood mononuclear cells (PBMC) from patients with periodontitis and healthy volunteers and identification of immune cell subsets by antibody staining. PBMC were isolated from peripheral blood from patients with periodontitis and healthy volunteers. Isolated PBMC were cryopreserved for batch staining at a later date. Upon thawing, PBMC were stained with fluorochrome‐conjugated antibodies against the indicated markers for identification of immune cell subsets as indicated.
Fig. S2. Gating strategy to identify interleukin (IL)‐17‐, interferon (IFN)‐γ‐ or tumour necrosis factor (TNF)‐α‐producing CD8+ or γδ T cells in peripheral blood mononuclear cells (PBMC). PBMC were isolated from peripheral blood from patients with chronic periodontitis (CP, n = 15) or aggressive periodontitis (AP, n = 15) as well as periodontally healthy controls (HC, n = 13). Cells were stimulated ex vivo with phorbol myristate acetate (PMA) and ionomycin for 3 h prior to intracellular cytokine staining. A gate was set on cells that stained positive for CD3 and CD8 (a) or CD3 and γδ (b) and subsequently the percentages of IL‐17+, IFNγ+ or TNFα+ cells within these gates were determined. Representative gating strategy and typical dot‐plots are shown.
Table S1. Eight‐colour extracellular staining panels [CD3/CD4/CD8/regulatory T cells (Treg)/γδ T cell/B cell mono/natural killer (NK) cells].
Table S2. Ten‐colour intracellular cytokine staining panel interleukin (IL)‐17‐/IL‐10/interferon (IFN)‐γ‐/tumour necrosis factor (TNF)‐α‐expressing CD4+, CD8+ T cell, γδ T cell or B cells).
Table S3. Bivariate multiple regression with lymphocyte subsets as dependent variable and age and diagnosis as explanatory variables.
Acknowledgments
WCC was supported by a grant from Tri‐Service General Hospital, National Defense Medical Center. We are very grateful to the patients and volunteers who consented to participate in this study. We would like to thank Dr Emily Lu (Dental Institute, King’s College London) for help in patient recruitment and data collection, Dr Gina Walter and Dr Hayley Evans for flow panel advice, and Dr Susanne Heck, PJ Chana and Helen Graves for their help in flow cytometry at the Biomedical Research Centre Flow Cytometry Core Facility funded by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust and King's College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
References
- 1. Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol 1999; 4:1–6. [DOI] [PubMed] [Google Scholar]
- 2. Schenkein HA, Koertge TE, Brooks CN, Sabatini R, Purkall DE, Tew JG. IL‐17 in sera from patients with aggressive periodontitis. J Dent Res 2010; 89:943–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang H, Luo Z, Lei L et al Interaction between oral lichen planus and chronic periodontitis with Th17‐associated cytokines in serum. Inflammation 2013; 36:696–704. [DOI] [PubMed] [Google Scholar]
- 4. Qi Y, Feng W, Song A et al GAP > LAP > HC IL‐23/IL‐17 axis in the relationship between periodontitis and coronary heart disease. Int J Periodont Restor Dent 2013; 33:185–92. [DOI] [PubMed] [Google Scholar]
- 5. Zhao L, Zhou Y, Xu Y, Sun Y, Li L, Chen W. Effect of non‐surgical periodontal therapy on the levels of Th17/Th1/Th2 cytokines and their transcription factors in Chinese chronic periodontitis patients. J Clin Periodontol 2011; 38:509–16. [DOI] [PubMed] [Google Scholar]
- 6. Duarte PM, da Rocha M, Sampaio E et al Serum levels of cytokines in subjects with generalized chronic and aggressive periodontitis before and after non‐surgical periodontal therapy: a pilot study. J Periodontol 2010; 81:1056–63. [DOI] [PubMed] [Google Scholar]
- 7. Cifcibasi E, Koyuncuoglu C, Ciblak M et al Evaluation of local and systemic levels of interleukin‐17, interleukin‐23, and myeloperoxidase in response to periodontal therapy in patients with generalized aggressive periodontitis. Inflammation 2015; 38:1959–68. [DOI] [PubMed] [Google Scholar]
- 8. Lima PM, Souza PE, Costa JE, Gomez RS, Gollob KJ, Dutra WO. Aggressive and chronic periodontitis correlate with distinct cellular sources of key immunoregulatory cytokines. J Periodontol 2011; 82:86–95. [DOI] [PubMed] [Google Scholar]
- 9. Schmidt J, Jentsch H, Stingu CS, Sack U. General immune status and oral microbiology in patients with different forms of periodontitis and healthy control subjects. PLOS ONE 2014; 9:e109187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen XT, Chen LL, Tan JY, Shi DH, Ke T, Lei LH. Th17 and Th1 lymphocytes are correlated with chronic periodontitis. Immunol Invest 2016; 45:243–54. [DOI] [PubMed] [Google Scholar]
- 11. Miyajima S, Naruse K, Kobayashi Y et al Periodontitis‐activated monocytes/macrophages cause aortic inflammation. Sci Rep 2014; 4:5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buhlin K, Gustafsson A, Pockley AG, Frostegard J, Klinge B. Risk factors for cardiovascular disease in patients with periodontitis. Eur Heart J 2003; 24:2099–107. [DOI] [PubMed] [Google Scholar]
- 13. Jagannathan R, Lavu V, Rao SR. Comparison of the proportion of non‐classic (CD14+CD16+) monocytes/macrophages in peripheral blood and gingiva of healthy individuals and patients with chronic periodontitis. J Periodontol 2014; 85:852–8. [DOI] [PubMed] [Google Scholar]
- 14. Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol 2015; 15:30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Seymour GJ, Ford PJ, Cullinan MP, Leishman S, Yamazaki K. Relationship between periodontal infections and systemic disease. Clin Microbiol Infect 2007; 13(Suppl 4):3–10. [DOI] [PubMed] [Google Scholar]
- 16. Demmer RT, Papapanou PN. Epidemiologic patterns of chronic and aggressive periodontitis. Periodontol 2000 2010; 53:28–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Page RC, Eke PI. Case definitions for use in population‐based surveillance of periodontitis. J Periodontol 2007; 78:1387–99. [DOI] [PubMed] [Google Scholar]
- 18. Walter GJ, Fleskens V, Frederiksen KS et al Phenotypic, functional, and gene expression profiling of peripheral CD45RA+ and CD45RO+ CD4+CD25+CD127(low) Treg cells in patients with chronic rheumatoid arthritis. Arthritis Rheumatol 2016; 68:103–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rossol M, Kraus S, Pierer M, Baerwald C, Wagner U. The CD14(bright) CD16+ monocyte subset is expanded in rheumatoid arthritis and promotes expansion of the Th17 cell population. Arthritis Rheum 2012; 64:671–7. [DOI] [PubMed] [Google Scholar]
- 20. Gümüş P, Nizam N, Nalbantsoy A, Ozcaka O, Buduneli N. Saliva and serum levels of pentraxin‐3 and interleukin‐1beta in generalized aggressive or chronic periodontitis. J Periodontol 2014; 85:e40–6. [DOI] [PubMed] [Google Scholar]
- 21. Cetinkaya B, Guzeldemir E, Ogus E, Bulut S. Proinflammatory and anti‐inflammatory cytokines in gingival crevicular fluid and serum of patients with rheumatoid arthritis and patients with chronic periodontitis. J Periodontol 2013; 84:84–93. [DOI] [PubMed] [Google Scholar]
- 22. Miranda TS, Heluy SL, Cruz DF et al The ratios of pro‐inflammatory to anti‐inflammatory cytokines in the serum of chronic periodontitis patients with and without type 2 diabetes and/or smoking habit. Clin Oral Invest 2018. doi: 10.1007/s00784-018-2471-5. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 23. Keles ZP, Keles GC, Avci B, Cetinkaya BO, Emingil G. Analysis of YKL‐40 acute‐phase protein and interleukin‐6 levels in periodontal disease. J Periodontol 2014; 85:1240–6. [DOI] [PubMed] [Google Scholar]
- 24. Gümüş P, Nizam N, Lappin DF, Buduneli N. Saliva and serum levels of B‐cell activating factors and tumor necrosis factor‐alpha in patients with periodontitis. J Periodontol 2014; 85:270–80. [DOI] [PubMed] [Google Scholar]
- 25. Cua DJ, Tato CM. Innate IL‐17‐producing cells: the sentinels of the immune system. Nat Rev Immunol 2010; 10:479–89. [DOI] [PubMed] [Google Scholar]
- 26. Patel DD, Lee DM, Kolbinger F, Antoni C. Effect of IL‐17A blockade with secukinumab in autoimmune diseases. Ann Rheum Dis 2013; 72 (Suppl 2):ii116–23. [DOI] [PubMed] [Google Scholar]
- 27. Bermejo DA, Jackson SW, Gorosito‐Serran M et al Trypanosoma cruzi trans‐sialidase initiates a program independent of the transcription factors RORgammat and Ahr that leads to IL‐17 production by activated B cells. Nat Immunol 2013; 14:514–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheng WC, van Asten SD, Burns LA et al Periodontitis‐associated pathogens P. gingivalis and A. actinomycetemcomitans activate human CD14(+) monocytes leading to enhanced Th17/IL‐17 responses. Eur J Immunol 2016; 46:2211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cardoso CR, Garlet GP, Crippa GE et al Evidence of the presence of T helper type 17 cells in chronic lesions of human periodontal disease. Oral Microbiol Immunol 2009; 24:1–6. [DOI] [PubMed] [Google Scholar]
- 30. Adibrad M, Deyhimi P, Ganjalikhani Hakemi M, Behfarnia P, Shahabuei M, Rafiee L. Signs of the presence of Th17 cells in chronic periodontal disease. J Periodont Res 2012; 47:525–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Preparation of peripheral blood mononuclear cells (PBMC) from patients with periodontitis and healthy volunteers and identification of immune cell subsets by antibody staining. PBMC were isolated from peripheral blood from patients with periodontitis and healthy volunteers. Isolated PBMC were cryopreserved for batch staining at a later date. Upon thawing, PBMC were stained with fluorochrome‐conjugated antibodies against the indicated markers for identification of immune cell subsets as indicated.
Fig. S2. Gating strategy to identify interleukin (IL)‐17‐, interferon (IFN)‐γ‐ or tumour necrosis factor (TNF)‐α‐producing CD8+ or γδ T cells in peripheral blood mononuclear cells (PBMC). PBMC were isolated from peripheral blood from patients with chronic periodontitis (CP, n = 15) or aggressive periodontitis (AP, n = 15) as well as periodontally healthy controls (HC, n = 13). Cells were stimulated ex vivo with phorbol myristate acetate (PMA) and ionomycin for 3 h prior to intracellular cytokine staining. A gate was set on cells that stained positive for CD3 and CD8 (a) or CD3 and γδ (b) and subsequently the percentages of IL‐17+, IFNγ+ or TNFα+ cells within these gates were determined. Representative gating strategy and typical dot‐plots are shown.
Table S1. Eight‐colour extracellular staining panels [CD3/CD4/CD8/regulatory T cells (Treg)/γδ T cell/B cell mono/natural killer (NK) cells].
Table S2. Ten‐colour intracellular cytokine staining panel interleukin (IL)‐17‐/IL‐10/interferon (IFN)‐γ‐/tumour necrosis factor (TNF)‐α‐expressing CD4+, CD8+ T cell, γδ T cell or B cells).
Table S3. Bivariate multiple regression with lymphocyte subsets as dependent variable and age and diagnosis as explanatory variables.


