Abstract
The life cycle of malaria parasites in both their mammalian host and mosquito vector consists of multiple developmental stages that ensure proper replication and progeny survival. The transition between these stages is fueled by nutrients scavenged from the host and fed into specialized metabolic pathways of the parasite. One such pathway is used by Plasmodium falciparum, which causes the most severe form of human malaria, to synthesize its major phospholipids, phosphatidylcholine, phosphatidylethanolamine, and phosphatidylserine. Much is known about the enzymes involved in the synthesis of these phospholipids, and recent advances in genetic engineering, single-cell RNA-Seq analyses, and drug screening have provided new perspectives on the importance of some of these enzymes in parasite development and sexual differentiation and have identified targets for the development of new antimalarial drugs. This Minireview focuses on two phospholipid biosynthesis enzymes of P. falciparum that catalyze phosphoethanolamine transmethylation (PfPMT) and phosphatidylserine decarboxylation (PfPSD) during the blood stages of the parasite. We also discuss our current understanding of the biochemical, structural, and biological functions of these enzymes and highlight efforts to use them as antimalarial drug targets.
Keywords: malaria, plasmodium, phospholipid metabolism, development, differentiation
Malaria and the challenging search for effective antimalarial chemotherapy
Malaria is a mosquito-borne parasitic disease caused by protozoan parasites of the genus Plasmodium and is one of the leading causes of death throughout human history. The disease is endemic in 91 countries with the World Health Organization African Region carrying the biggest burden of morbidity and mortality (1). Of the Plasmodium species that infect humans, Plasmodium falciparum and Plasmodium vivax account for the overall majority of malaria clinical cases, hospital stays, and death (1, 2). In 2016, these parasites were responsible for ∼216 million clinical cases and ∼445,000 deaths (1). Thanks to major international efforts aimed at implementing improved policies for control of mosquito populations, the wide use of bed nets, and the application of new therapeutic strategies, this mortality rate represents a drop of more than 50% from the ∼839,000 deaths recorded in 2000 (1, 3). However, despite this success, the death toll is still unacceptably high. Emerging drug resistance to first line therapies and the high cost of drugs continue to add more health and economic burden on the affected populations (1, 4, 5). The development of an effective vaccine continues to be both scientifically and technically challenging. Among multiple candidates currently in development, RTS,S/AS01 (or MosquirixTM), a recombinant protein-based vaccine that targets the major circumsporozoite protein of P. falciparum, has shown the most promise so far (6). In the absence of a vaccine with high effectiveness among the overall population, there continues to be a need for new and affordable therapies and novel therapeutic strategies to treat the disease.
Plasmodium parasites have a complex life cycle in the mosquito vector and humans, involving multiple developmental stages, different morphological, biochemical, and metabolic requirements, and well-controlled and highly coordinated gene expression and regulatory mechanisms (7, 8). Following injection of the parasite into the skin of the human host by an infected female Anopheles mosquito, the parasite undergoes rapid multiplication within liver hepatocytes to produce thousands of merozoites, which are packed into host cell membrane-derived vesicles (merosomes) and safely transported past the resident macrophages (Kupffer cells) into the liver sinusoids where their invasion of the erythrocytes begins (9–12).
Within the erythrocytes, each merozoite grows to several times its original size before dividing asexually via schizogony to produce 16–32 new blood merozoites (13, 14). The intraerythrocytic life cycle ends with the rupture of the host cell. The repeated cycles of invasion and destruction of host erythrocytes are directly linked to the pathology and symptoms of the disease, which include fever, chills, and fatigue (15, 16).
The orchestration of the intraerythrocytic schizogony requires a complete parasitic reorganization of the metabolically reduced and terminally differentiated host erythrocyte to ensure protection against immune attacks and to facilitate nutrient supply to fuel parasite development and replication (1, 17–20). While long recognized as critical structural components for parasite development and attractive therapeutic targets, phospholipids and their by-products have also emerged (21) as major signaling molecules that control development and differentiation processes during Plasmodium intraerythrocytic cycle (21–26). The rapid generation of abundant parasitic progeny requires the appropriate amount of suitable lipid species at the proper compartment and at the right time to establish an active membrane biogenesis, which leads to a dramatically elevated lipid metabolism during the intraerythrocytic schizogony (22, 27). To acquire the necessary lipid species for different compartments, the parasite either synthesizes them de novo from previously produced metabolites or uses exogenous sources such as the erythrocyte membrane or the human plasma. This results in a 6-fold increase in the relative levels of phospholipids in the infected erythrocyte (24, 27, 28). The dependence on phospholipids for rapid parasite multiplication and the uniqueness of some of the steps in the pathways of Plasmodium lipid metabolism create significant opportunities for the identification of antimalarial drug targets (22, 24, 27).
Structural, developmental, and signaling functions of phospholipid biosynthesis in P. falciparum
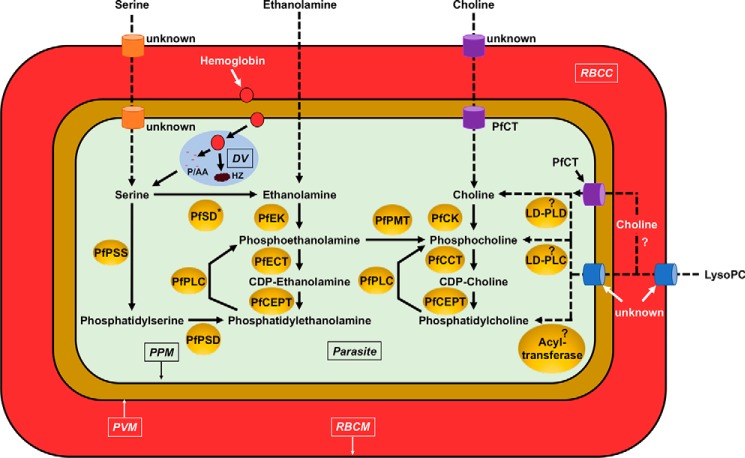
Asexual blood stages and gametocytes of Plasmodium parasites are able to scavenge or synthesize up to 300 different lipid species to facilitate growth, proliferation, transmission, and sexual reproduction (24). The phospholipid classes, phosphatidylcholine (PC),3 phosphatidylethanolamine (PE), and phosphatidylserine (PS), are the major lipid components that define Plasmodium membranes (21–23, 25, 29–32). In the uninfected erythrocytes, PC, PE, and PS constitute 30–40, 25–35, and 10–20% of the total phospholipids, respectively, whereas in P. falciparum-infected erythrocytes and in purified parasites, these major phospholipids constitute 20–55, 15–40, and 4–15% of the total phospholipids, respectively (Table 1) (25, 33). The reported higher levels of PE in P. falciparum membranes compared with the membranes of other eukaryotes (Table 1) have been proposed to be largely due to the inability of the parasite to directly convert PE to PC (22, 23). However, P. falciparum is able to generate phosphocholine, a precursor for the synthesis of PC, from phosphoethanolamine via the PMT pathway (see below). With the exception of a few pathways, which have been identified through metabolic and genetic analyses, most components of the PC, PE, and PS biosynthetic machineries have been identified by searching for homologs in the Plasmodium genome databases of well-characterized enzymes from yeast, plants, and other eukaryotes (22, 30). These pathways are outlined in Fig. 1.
Table 1.
Phospholipid composition of different eukaryotic cells
The comparison of lipid composition of different eukaryotic cells and uninfected erythrocytes is shown.
| Organism/cell | PC | PE | PS | Refs. |
|---|---|---|---|---|
| Uninfected erythrocytes | 30–40% | 25–35% | 10–20% | 33 |
| P. falciparum-infected erythrocytes and free parasites | 20–55% | 15–40% | 4–15% | 25, 33, 37 |
| Trypanosoma brucei | 45–60% | 10–20% | <4% | 36 |
| Toxoplasma gondii (free parasites) | 75% | 10% | 6% | 34 |
| Aspergillus niger | 51% | 28.5% | 5% | 35 |
| Candida albicans | 40% | 25.3% | 12% | 35 |
| Cryptococcus neoformans | 49% | 28% | 8% | 35 |
| Microsporum gypseum | 23.1% | 29.8% | 19.4% | 35 |
Figure 1.
Lipid synthetic pathways in the asexual stages of Plasmodium. The following abbreviations are used: RBCM, red blood cell membrane; RBCC, red blood cell cytosol; PPM, parasite plasma membrane; DV, digestive vacuole; HZ, hemozoin; P/AA, peptides and amino acids; PfPSS, P. falciparum PS synthase; PfSD, P. falciparum serine decarboxylase (asterisk indicates that enzyme activity has been reported but no corresponding gene has been identified); PfPLC, P. falciparum phospholipase C; PfPSD, P. falciparum PS decarboxylase; PfCT, P. falciparum choline transporter; PfCK, P. falciparum choline kinase; PfCCT, P. falciparum CTP:phosphocholine cytidylyltransferase; PfCEPT, P. falciparum choline/ethanolamine-phosphotransferase; PfPMT, P. falciparum phosphoethanolamine methyltransferase; LD-PLC, lyso-PC–dependent phospholipase C; PfNSM, P. falciparum neutral sphingomyelinase; LD-PLD, lyso-PC–dependent phospholipase D. Dashed arrows represent putative routes of transport and metabolism. The “?” indicates pathways and enzymes that might exist or catalyze certain reactions respectively but that have not been identified yet.
Phosphatidylcholine
In most eukaryotic membranes (see Table 1), PC accounts for more than 50% of phospholipids and spontaneously self-organizes to a planar bilayer (38, 39). In addition to its structural role, PC can also modulate cellular signaling functions because its hydrolysis by phospholipases leads to the formation of the second messenger diacylglycerol (DAG), which is critical for activation of specific classes of protein kinases (40, 41). Changes in cellular PC levels have been shown to alter cell proliferation, differentiation, as well as membrane movement (42). Studies in malaria parasites have shown that PC is the major phospholipid in Plasmodium membranes during both liver stage and intraerythrocytic schizogony (22, 30, 43, 44).
One of the two routes used to synthesize PC in Plasmodium parasites is the cytidine diphosphate- or CDP-choline branch of the Kennedy pathway (25, 28, 33, 38, 45). Metabolomic analyses showed that 89% of PC is generated by the parasite from choline via this particular pathway (33). To enter the Kennedy pathway, choline is first transported into the parasite by an as yet unidentified transporter and then phosphorylated by choline kinase to form phosphocholine, which is subsequently used as a precursor to form CDP-choline and finally PC, by the 1,2-diacylglycerol cholinephosphotransferase (45). PC can also be synthesized by the PMT pathway, which uses three sequential methylations of phosphoethanolamine (via S-adenosylmethionine donors) to form phosphocholine, catalyzed by the parasite phosphoethanolamine methyltransferase (PfPMT) (22, 46–48). Metabolomic analyses indicate that 11% of total PC in P. falciparum is generated from the PMT pathway (33). Analysis of available genome databases showed that among protozoan parasites, PfPMT homologs are found primarily among Plasmodium species that infect humans (P. vivax and P. knowlesi), primates (P. knowlesi and Plasmodium reichenowi), and birds (P. gallinaceum) (49–51). Orthologs of PfPMT have also been found in proteobacteria (Burkholderia pseudomallei and Burkholderia oklahomensis), many species of plants, two species of African clawed frogs (Xenopus laevis and Xenopus tropicalis), nematodes (Caenorhabditis elegans and Caenorhabditis briggsae), zebrafish (Danio rerio), the Florida lancelet (Brachiostoma floridae), and the protozoan fungus Phytophthora infestans (49). Interestingly, no PMT orthologs are found in mammals, making them attractive targets for selective therapies (49). The 3D structure of PfPMT and subsequently that of PvPMT (P. vivax phosphoethanolamine methyltransferase) and PkPMT (P. knowlesi phosphoethanolamine methyltransferase) showed high conservation, functional activity, and inhibition by the same drugs (29, 52, 53). Like other methyltransferases known to be inhibited by the 4-aminoquinoline, amodiaquine, PfPMT was also inhibited by this compound (Table 2) (54). The specificity of interaction of amodiaquine with the enzyme was further demonstrated using NMR spectroscopy (54). The compound was found to induce major conformational changes, whereas its analog chloroquine had no effect on the enzyme (54). Structural analyses of PvPMT with amodiaquine further showed that the drug is an allosteric inhibitor targeting a cleft located distal to the active site (43).
Table 2.
Overview of inhibitors and genetic knockouts of lipid synthetic enzymes
The following abbreviations are used: PfECT, P. falciparum CTP:phosphoethanolamine cytidylyltransferase; PfEK, P. falciparum ethanolamine kinase; PfCT, P. falciparum choline transporter; PfCCT, P. falciparum CTP:phosphocholine cytidylyltransferase; PfCEPT, P. falciparum choline/ethanolamine-phosphotransferase; PfPMT, P. falciparum phosphoethanolamine methyltransferase; LD-PLC, lyso-PC–dependent phospholipase C; PfNSM, lyso-PC–dependent phospholipase C (SM/LCPL-phospholipase C); PfPSS, P. falciparum PS synthase; PfPLC, P. falciparum phospholipase C; PfPSD, P. falciparum PS decarboxylase; PfCK, P. falciparum choline kinase; NA, not applicable.
| Enzyme | Knockout lethality | Pharmacological inhibition | Putative compounds | Refs. |
|---|---|---|---|---|
| PfEK | NA | + | 2-Amino-1-butanol, bis-thiazolium T3 | 78 |
| PfECT | NA | NA | NA | |
| PfCT | NA | + | Amiodarone, bepridil, possibly pentamine, P16, and HC-3 | 86 |
| PfCK | NA | + | Hemicholinium-3, bis-thiazolium T3 | 78–80 |
| PfCCT | + | + | PG12 | 81 |
| PfCEPT | NA | + | T4 | 81, 82 |
| PfPMT | + | + | Amodiaquine, NSC-158011 | 21, 23, 54 |
| PfPSS | NA | NA | NA | |
| PfPLC | NA | NA | NA | |
| PfPSD | + | + | 7CPQA analog (MMV007285) | 29, 85 |
| PfNSM (LD-PLC) | NA | + | Scyphostatin | 84 |
| PfSD | NA | NA | NA |
P. falciparum pfpmtΔ parasites lacking the PfPMT gene were found to be viable even in the absence of exogenous choline, although their growth was severely reduced (Table 2) (23). TLC analyses showed that these parasites produce PC in the absence of choline, suggesting that an alternative source of choline or a choline-containing precursor is also available to the parasite (23). Accordingly, a recent study by Brancucci et al. (55) showed that P. falciparum actively transports lysophosphatidylcholine (lyso-PC) from host plasma to generate PC (Fig. 1). Although the mechanism of uptake and mode of utilization of lyso-PC by the parasite remain to be determined, it is possible that lyso-PC is either hydrolyzed to form a precursor that then enters the CDP-choline pathway to form PC or is directly acylated to form PC (Fig. 1). In yeast, lyso-PC is transported by the phospholipid flippases, Dnf1p and Dnf2p, which are P4 ATPases (56, 57). A second protein, Lem3p, also plays a role in the transport of lyso-PC as a noncatalytic subunit of Dnf1p–Lem3p and Dnf2p–Lemp3p complex. Because the crystal structure of the P4 ATPase is not yet available, the structural basis of lyso-PC uptake remains unknown. However, several models have been proposed based on extensive mutagenesis and computer simulation analyses using crystal structures of cation-transporting P-type ATPases (58–61). Among them, the two-gate model has been proposed for phospholipid translocation by Dnf1p (58). First, the phospholipid headgroups slide through the entry gate formed by the transmembrane domains (TMs) on the extracellular side and then reach the exit gate formed through the other TMs toward the cytosol. Once inside the cell, acylation of lyso-PC is catalyzed by Ale1p (acyltransferase for lysophosphatidylethanolamine) (57, 62). A BLAST search for homologs of these proteins in the P. falciparum genome database (http://plasmodb.org/plasmo/)4 identified several homologs of Lem3p and Dnf1/2p, but no homologs of Ale1p could be found.
Phosphatidylethanolamine
PE can be found in the membranes of all eukaryotic and prokaryotic cells and is a key factor in several processes such as membrane fusion or cytokinesis and can further serve as a donor of the ethanolamine moiety that covalently modifies various proteins (63, 64). PE also increases the membrane curvature, which is crucial for membrane budding, as well as fusion and fission processes (29, 65). PE metabolism has been implicated in important cellular processes such as autophagy, signaling, and viral replication (66–70) and in human diseases such as Alzheimer's disease, Parkinson's disease, and nonalcoholic liver disease (71).The biosynthesis of PE in Plasmodium parasites can occur either from ethanolamine via the cytidine diphosphate- or CDP-ethanolamine (Kennedy) pathway or from PS via decarboxylation (Fig. 1) (29, 31, 33). The PE branch of the Kennedy pathway uses three sequential steps catalyzed by parasite enzymes to generate PE de novo from ethanolamine (72). The ethanolamine kinase of the Kennedy pathway uses ATP to phosphorylate ethanolamine, whereas the phosphoethanolamine cytidylyltransferase uses CTP to generate the high-energy intermediate CDP-ethanolamine (45). This high-energy intermediate is then used in conjunction with DAG, by the 1,2-diacylglycerol ethanolaminephosphotransferase, to generate PE (45). PE synthesis from PS is catalyzed by parasite-encoded PS decarboxylase (PSD). PSD genes of both P. falciparum (PfPSD) and P. knowlesi (PkPSD) have been shown to complement the ethanolamine auxotrophy of a yeast mutant lacking the PSD activity (29, 73). Biochemical characterization of PkPSD showed that the enzyme is less hydrophobic than other PSD enzymes and is recovered in both soluble and membrane-associated compartments when expressed in yeast (73). Deletion of the N-terminal membrane domain of the protein greatly improved its solubility while preserving catalytic activity (73). Recent studies have shown that the enzyme belongs to the DHS (Asp–His–Ser) protease family (74) and undergoes auto-endoproteolytic cleavage into a large β-subunit and a smaller α-subunit with a pyruvoyl prosthetic group (73, 75, 76).
Metabolic studies in P. knowlesi suggested that besides ethanolamine uptake, a parasite-encoded serine decarboxylase might exist which could produce ethanolamine from serine (31). This reaction has previously been shown to occur exclusively in plants (77). However, no homologs of plant serine decarboxylases could be found in the P. falciparum genome, and attempts to isolate this enzyme from parasite extracts or to identify the malarial gene by functional complementation in yeast have not been successful (73). Biochemical assays further excluded the PfPSD enzyme as a possible decarboxylase of serine (29). One possible explanation of the available data is that Plasmodium lacks the ability for direct decarboxylation of serine and that the formation of ethanolamine from serine as shown by metabolic analyses in P. knowlesi is a result of PE turnover. In this case, serine is first incorporated into PS, and the resulting PS is then converted into PE, which is subsequently hydrolyzed to produce ethanolamine.
Genetic studies in P. falciparum and rodent parasites suggest that the PSD function might be essential for parasite development in the absence of exogenous ethanolamine (Table 2) (29). This has so far been supported by pharmacological studies with inhibitors that target this enzyme (Table 2) (29). However, further studies using gene editing technologies, conditional knockouts, or more specific inhibitors are needed to evaluate the importance of this step in parasite development and differentiation.
Phosphatidylserine
PS is usually found in the inner leaflet of the lipid bilayer of the plasma membrane of healthy mammalian cells and plays a crucial role in apoptosis, erythrocyte senescence, blood coagulation, as well as targeting and function of various intracellular signaling proteins (64, 87–89). In malaria parasites, PS has been shown to play a role in P. falciparum-established erythrocyte cytoadhesion (32). Interestingly, PS is also particularly enriched in microvesicles that can be found in elevated levels within the blood of P. falciparum-infected patients (24). These microvesicles have been implicated in intercellular communication as well as gametocytogenesis (24). A recent study has further shown that the parasite actively hampers the presentation of PS on the outer membrane leaflet of the plasma membrane of liver cells during merosome formation to avoid the phagocytic clearance of the parasitic progeny (90).
The synthesis of PS by Plasmodium parasites requires serine that is either directly imported from the host plasma or obtained from hemoglobin degradation in the digestive vacuole by parasite proteases (Fig. 1) (22, 23, 31, 33). PfPSS (P. falciparum phosphatidylserine synthase) then uses the scavenged serine to generate PS. PS further serves as a precursor for the synthesis of PE.
Role of phospholipid synthesis in Plasmodium development and sexual differentiation
The first evidence that phospholipids play more than just a housekeeping function in P. falciparum came from studies by Witola et al. (23) on the characterization of knockout parasites lacking the PfPMT gene. These mutant parasites had severe growth defects, even in the presence of high concentrations of choline (23). Furthermore, mature schizonts isolated from these knockout parasites produced a smaller number of merozoites and displayed an altered pattern of nuclear division compared with WT parasites (23). Finally, these knockout parasites were severely altered in their commitment to sexual differentiation and could not be transmitted to mosquitoes (21). However, these defects could not be complemented by choline supplementation, implying that either pools of PC derived from the PMT pathway or intermediates of the methylation reactions may play a critical role in these processes.
Recent studies in P. falciparum have shown that lyso-PC is a major substrate of parasite phospholipid metabolism that further acts as an environmental sensor (55). By degradative processes, lyso-PC is also a major source of the Kennedy pathway components choline and fatty acids, which are needed to fuel the proper biosynthesis of PC (55). In case of lyso-PC limitation between 34 and 38 h after red blood cell invasion, the parasite responds with the production of less merozoites during the development of the blood stages (55, 91). Low lyso-PC conditions in the external medium further trigger the expression of the transcriptional regulator of irreversible sexual commitment and gametocytogenesis, Api2G, whereas the expression of other key players such as PfPMT (especially in already committed cells) and other transcriptional factors are up-regulated (55, 92, 93). These recent findings suggest that lyso-PC controls the fate of P. falciparum by facilitating the repression of sexual differentiation. Interestingly, depletion of lyso-PC in the rodent parasite Plasmodium berghei, which lacks PMT function and other factors that are regulated by lyso-PC levels, had no effect on the sexual differentiation of the parasite (50, 55).
Summary and future directions
Important milestones have been achieved over the past several years in understanding lipid metabolism and its role in P. falciparum development and differentiation. The recent advances in metabolomic and genetic analyses will undoubtedly unravel more secrets about these processes and identify ideal targets for the development of new antimalarials.
The unique enzyme PfPMT has already been identified as an important key player in the lipid metabolism of P. falciparum parasites by Witola et al. (23). The knockout of PMT had major effects on growth and survival of the parasites, suggesting that PfPMT could be a promising target for antimalarial chemotherapy (23). Two studies showed that the antimalarial 4-aminoquinoline, amodiaquine, and four additional compounds (NSC-641296, NSC-668394, NSC-323241, and NSC-158011) were able to inhibit PfPMT activity (Table 2) (21, 54). NSC-158011 appeared to be a particularly promising candidate because it acts as a competitive inhibitor in the transmethylation reaction and inhibits PfPMT activity as well as PC biosynthesis in vivo (21).
Recent studies by Brancucci et al. (55) highlighted the important role of lyso-PC in P. falciparum sexual differentiation. However, the mechanism by which lyso-PC is transported from the host serum into the parasite remains to be investigated. The lyso-PC uptake might depend on the developmental stage of the parasite or even the age of the infected erythrocyte (14, 94). The parasite might also encounter different lyso-PC concentrations within various tissues while traveling with the host erythrocyte during its journey through the body as a juvenile ring or during sequestration as a mature trophozoite or a schizont (18, 95). Mature P. falciparum blood stages secrete and present cytoadhesins on the surface of the infected erythrocytes, enabling these cells to become sequestered in the deep vascular bed of inner organs to avoid splenic clearance (18). Brancucci et al. (55) reported that parasite-induced sequestration in intra- and extravascular spaces might cause variation in lyso-PC exposure. They further observed dramatically reduced levels of lyso-PC in bone marrow fluids, where high densities of gametocytes can be found (96). However, gametocyte levels can also be high in the brain, gut, heart, and spleen, and it needs to be determined whether the localization of the sexual stages directly correlates with the lyso-PC levels within those organs (96).
Future studies are also needed to investigate why some of the asexual parasites appear to be unaffected when lyso-PC is removed from the medium, whereas others develop into gametocytes. There are certainly various additional factors involved in the sexual commitment of the parasites that are expressed at a certain time in the life cycle of the parasite or under certain conditions that are exclusively determined by the host. For example, the fact that the levels of some lyso-PC molecular species significantly decrease during the pyrogenic peak is one important host condition that needs to be examined in conjunction with parasite development and gene expression (97, 98).
The finding that lyso-PC plays an important role in parasite development, differentiation, and transmission makes the metabolic pathways and the involved enzymes very attractive targets for antimalarial drugs. A promising enzyme candidate might be the PfNSM, a lyso-PC–dependent phospholipase C (SM/LCPL-phospholipase C), which catalyzes the hydrolysis of sphingomyelin and lysophospholipids (Fig. 1) (27, 84). Hanada and co-workers (84) previously reported that the treatment with scyphostatin, a mycelial extract of the fungus Trichopeziza mollissima, inhibited enzyme activity (ID50 = 3–5 μm) as well as the growth of P. falciparum (∼7 μm) (99).
Targeting PE synthesis is another attractive avenue to fight malaria. An in vivo screen of inhibitors against PkPSD has been previously reported (29). The screen searched for compounds that inhibit growth of a yeast mutant that relies on malarial PSD activity for survival. 18 candidates were identified as PSD inhibitors, and 7-chloro-N-(4-ethoxyphenyl)-4-quinolinamine (7CPQA analog) (MMV007285) was chosen for further analysis because of the ready availability of precursors, ease of synthesis, and low toxicity toward mammalian cells. The drug has been shown to be effective against P. falciparum with IC50 values ranging between 0.5 and 1 μm (85), and it has a selectivity index of >8 in vitro. 7CPQA, an analog of MMV007285, has been shown to prevent Plasmodium yoelii growth in mice at 30 mg kg−1 following a daily oral administration for 3 consecutive days (29). 7CPQA further inhibits all stages of parasite intraerythrocytic development.
In vitro inhibitor screens amenable for high-throughput screening for decarboxylase enzymes have been challenging due to a lack of suitable enzyme assays. The conventional PSD enzyme assay is based on measuring captured radioactive CO2, which is released following decarboxylation of 14C-PS by a PSD enzyme (100). Recently, a fluorescence assay based on the interaction of PE with the water-soluble distyrylbenzene-bisaldehyde (DSB-3) has been developed to facilitate chemical screens for inhibitors of PSD enzymes. The assay is amenable to high-throughput screening and can measure PSD activity of recombinant enzymes as well as native activity from cell extracts and membrane preparations from bacteria and yeast (83). Efforts are underway to identify more potent inhibitors of the malarial PSD enzyme using this assay. Finally, targeting PS biosynthesis through inhibition of PfPSS is another opportunity to inhibit parasite growth. However, no promising antimalarial candidates have been reported so far.
In summary, the lipid metabolism of Plasmodium parasites is both complex and fascinating. Not all of the puzzle pieces have been identified, but the already emerging picture gives a very promising outlook of the future of antimalarial chemotherapy due to the uniqueness of some of the already identified factors.
Perspectives
Efforts needed to further advance knowledge about phospholipid metabolism in development, differentiation, and malaria therapy include the following.
Genetic analysis and target validation of phospholipid biosynthesis genes using CRISPR-Cas9 knockout and conditional knockout strategies.
Biochemical and genetic characterization of the transport and metabolism of lyso-PC and other phospholipid precursors.
Identification and development of new classes of inhibitors that target essential steps in membrane biogenesis.
The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- CDP
- cytidine diphosphate
- DAG
- diacylglycerol
- lyso-PC
- lysophosphatidylcholine
- PSD
- PS decarboxylase
- PfNSM
- lyso-PC–dependent phospholipase C (SM/LCPL-phospholipase C or PLC)
- PfPSD
- P. falciparum phosphatidylserine decarboxylase
- PfPSS
- P. falciparum phosphatidylserine synthase
- PfPMT
- P. falciparum phosphoethanolamine methyltransferase
- PS
- phosphatidylserine
- PvPMT
- P. vivax phosphoethanolamine methyltransferase
- PMT
- phosphoethanolamine methyltransferase
- TM
- transmembrane domain
- 7CPQA
- 7-chloro-N-(4-ethoxyphenyl)-4-quinolinamine.
References
- 1. World Health Organization (2017) World Malaria Report, World Health Organization, Geneva, Switzerland [Google Scholar]
- 2. Calderaro A., Piccolo G., Gorrini C., Rossi S., Montecchini S., Dell'Anna M. L., De Conto F., Medici M. C., Chezzi C., and Arcangeletti M. C. (2013) Accurate identification of the six human Plasmodium spp. causing imported malaria, including Plasmodium ovale wallikeri and Plasmodium knowlesi. Malar. J. 12, 321 10.1186/1475-2875-12-321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Muema J. M., Bargul J. L., Njeru S. N., Onyango J. O., and Imbahale S. S. (2017) Prospects for malaria control through manipulation of mosquito larval habitats and olfactory-mediated behavioural responses using plant-derived compounds. Parasit. Vectors 10, 184 10.1186/s13071-017-2122-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blasco B., Leroy D., and Fidock D. A. (2017) Antimalarial drug resistance: linking Plasmodium falciparum parasite biology to the clinic. Nat. Med. 23, 917–928 10.1038/nm.4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shretta R., Avanceña A. L., and Hatefi A. (2016) The economics of malaria control and elimination: a systematic review. Malar. J. 15, 593 10.1186/s12936-016-1635-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Draper S. J., Sack B. K., King C. R., Nielsen C. M., Rayner J. C., Higgins M. K., Long C. A., and Seder R. A. (2018) Malaria vaccines: recent advances and new horizons. Cell Host Microbe 24, 43–56 10.1016/j.chom.2018.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ben Mamoun C., Gluzman I. Y., Hott C., MacMillan S. K., Amarakone A. S., Anderson D. L., Carlton J. M., Dame J. B., Chakrabarti D., Martin R. K., Brownstein B. H., and Goldberg D. E. (2001) Co-ordinated programme of gene expression during asexual intraerythrocytic development of the human malaria parasite Plasmodium falciparum revealed by microarray analysis. Mol. Microbiol. 39, 26–36 10.1046/j.1365-2958.2001.02222.x [DOI] [PubMed] [Google Scholar]
- 8. Le Roch K. G., Zhou Y., Blair P. L., Grainger M., Moch J. K., Haynes J. D., De La Vega P., Holder A. A., Batalov S., Carucci D. J., and Winzeler E. A. (2003) Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301, 1503–1508 10.1126/science.1087025 [DOI] [PubMed] [Google Scholar]
- 9. Phillips M. A., Burrows J. N., Manyando C., van Huijsduijnen R. H., Van Voorhis W. C., and Wells T. N. (2017) Malaria. Nat. Rev. Dis. Primers 3, 17050 10.1038/nrdp.2017.50 [DOI] [PubMed] [Google Scholar]
- 10. Santos J. M., Egarter S., Zuzarte-Luís V., Kumar H., Moreau C. A., Kehrer J., Pinto A., Costa M. D., Franke-Fayard B., Janse C. J., Frischknecht F., and Mair G. R. (2017) Malaria parasite LIMP protein regulates sporozoite gliding motility and infectivity in mosquito and mammalian hosts. Elife 6, e24109 10.7554/eLife.24109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sinnis P., and Zavala F. (2012) The skin: where malaria infection and the host immune response begin. Semin. Immunopathol. 34, 787–792 10.1007/s00281-012-0345-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sturm A., Amino R., van de Sand C., Regen T., Retzlaff S., Rennenberg A., Krueger A., Pollok J. M., Menard R., and Heussler V. T. (2006) Manipulation of host hepatocytes by the malaria parasite for delivery into liver sinusoids. Science 313, 1287–1290 10.1126/science.1129720 [DOI] [PubMed] [Google Scholar]
- 13. Cowman A. F., and Crabb B. S. (2006) Invasion of red blood cells by malaria parasites. Cell 124, 755–766 10.1016/j.cell.2006.02.006 [DOI] [PubMed] [Google Scholar]
- 14. Grüring C., Heiber A., Kruse F., Ungefehr J., Gilberger T. W., and Spielmann T. (2011) Development and host cell modifications of Plasmodium falciparum blood stages in four dimensions. Nat. Commun. 2, 165 10.1038/ncomms1169 [DOI] [PubMed] [Google Scholar]
- 15. Srivastava A., Philip N., Hughes K. R., Georgiou K., MacRae J. I., Barrett M. P., Creek D. J., McConville M. J., and Waters A. P. (2016) Stage-specific changes in Plasmodium metabolism required for differentiation and adaptation to different host and vector environments. PLoS Pathog. 12, e1006094 10.1371/journal.ppat.1006094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bartoloni A., and Zammarchi L. (2012) Clinical aspects of uncomplicated and severe malaria. Mediterr. J. Hematol. Infect. Dis. 4, e2012026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tuteja R. (2007) Malaria-an overview. FEBS J. 274, 4670–4679 10.1111/j.1742-4658.2007.05997.x [DOI] [PubMed] [Google Scholar]
- 18. Cyrklaff M., Sanchez C. P., Kilian N., Bisseye C., Simpore J., Frischknecht F., and Lanzer M. (2011) Hemoglobins S and C interfere with actin remodeling in Plasmodium falciparum-infected erythrocytes. Science 334, 1283–1286 10.1126/science.1213775 [DOI] [PubMed] [Google Scholar]
- 19. Kilian N., Srismith S., Dittmer M., Ouermi D., Bisseye C., Simpore J., Cyrklaff M., Sanchez C. P., and Lanzer M. (2015) Hemoglobin S and C affect protein export in Plasmodium falciparum-infected erythrocytes. Biol. Open 4, 400–410 10.1242/bio.201410942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Spycher C., Rug M., Klonis N., Ferguson D. J., Cowman A. F., Beck H. P., and Tilley L. (2006) Genesis of and trafficking to the Maurer's clefts of Plasmodium falciparum-infected erythrocytes. Mol. Cell. Biol. 26, 4074–4085 10.1128/MCB.00095-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bobenchik A. M., Witola W. H., Augagneur Y., Nic Lochlainn L., Garg A., Pachikara N., Choi J. Y., Zhao Y. O., Usmani-Brown S., Lee A., Adjalley S. H., Samanta S., Fidock D. A., Voelker D. R., Fikrig E., and Ben Mamoun C. (2013) Plasmodium falciparum phosphoethanolamine methyltransferase is essential for malaria transmission. Proc. Natl. Acad. Sci. U.S.A. 110, 18262–18267 10.1073/pnas.1313965110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pessi G., Kociubinski G., and Mamoun C. B. (2004) A pathway for phosphatidylcholine biosynthesis in Plasmodium falciparum involving phosphoethanolamine methylation. Proc. Natl. Acad. Sci. U.S.A. 101, 6206–6211 10.1073/pnas.0307742101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Witola W. H., El Bissati K., Pessi G., Xie C., Roepe P. D., and Mamoun C. B. (2008) Disruption of the Plasmodium falciparum PfPMT gene results in a complete loss of phosphatidylcholine biosynthesis via the serine-decarboxylase-phosphoethanolamine-methyltransferase pathway and severe growth and survival defects. J. Biol. Chem. 283, 27636–27643 10.1074/jbc.M804360200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gulati S., Ekland E. H., Ruggles K. V., Chan R. B., Jayabalasingham B., Zhou B., Mantel P. Y., Lee M. C., Spottiswoode N., Coburn-Flynn O., Hjelmqvist D., Worgall T. S., Marti M., Di Paolo G., and Fidock D. A. (2015) Profiling the essential nature of lipid metabolism in asexual blood and gametocyte stages of Plasmodium falciparum. Cell Host Microbe 18, 371–381 10.1016/j.chom.2015.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Flammersfeld A., Lang C., Flieger A., and Pradel G. (2017) Phospholipases during membrane dynamics in malaria parasites. Int. J. Med. Microbiol. 2017, S1438-4221(17)30284-9 10.1016/j.ijmm.2017.09.015 [DOI] [PubMed] [Google Scholar]
- 26. Ramakrishnan S., Serricchio M., Striepen B., and Butikofer P. (2013) Lipid synthesis in protozoan parasites: a comparison between kinetoplastids and apicomplexans. Prog. Lipid Res. 52, 488–512 10.1016/j.plipres.2013.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mitamura T., and Palacpac N. M. (2003) Lipid metabolism in Plasmodium falciparum-infected erythrocytes: possible new targets for malaria chemotherapy. Microbes Infect. 5, 545–552 10.1016/S1286-4579(03)00070-4 [DOI] [PubMed] [Google Scholar]
- 28. Pessi G., and Mamoun C. B. (2006) Pathways for phosphatidylcholine biosynthesis: targets and strategies for antimalarial drugs. Future Lipidol. 1, 173–180 [Google Scholar]
- 29. Choi J. Y., Kumar V., Pachikara N., Garg A., Lawres L., Toh J. Y., Voelker D. R., and Ben Mamoun C. (2016) Characterization of Plasmodium phosphatidylserine decarboxylase expressed in yeast and application for inhibitor screening. Mol. Microbiol. 99, 999–1014 10.1111/mmi.13280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ben Mamoun C., Prigge S. T., and Vial H. (2010) Targeting the lipid metabolic pathways for the treatment of malaria. Drug Dev. Res. 71, 44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Elabbadi N., Ancelin M. L., and Vial H. J. (1997) Phospholipid metabolism of serine in Plasmodium-infected erythrocytes involves phosphatidylserine and direct serine decarboxylation. Biochem. J. 324, 435–445 10.1042/bj3240435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Eda S., and Sherman I. W. (2002) Cytoadherence of malaria-infected red blood cells involves exposure of phosphatidylserine. Cell. Physiol. Biochem. 12, 373–384 10.1159/000067908 [DOI] [PubMed] [Google Scholar]
- 33. Wein S., Ghezal S., Buré C., Maynadier M., Périgaud C., Vial H. J., Lefebvre-Tournier I., Wengelnik K., and Cerdan R. (2018) Contribution of the precursors and interplay of the pathways in the phospholipid metabolism of the malaria parasite. J. Lipid Res. 59, 1461–1471 10.1194/jlr.M085589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gupta N., Zahn M. M., Coppens I., Joiner K. A., and Voelker D. R. (2005) Selective disruption of phosphatidylcholine metabolism of the intracellular parasite Toxoplasma gondii arrests its growth. J. Biol. Chem. 280, 16345–16353 10.1074/jbc.M501523200 [DOI] [PubMed] [Google Scholar]
- 35. Gealt M. A., Abdollahi A., and Evans J. L. (1989) Lipids and lipoidal mycotoxins of fungi. Curr. Top. Med. Mycol. 3, 218–247 10.1007/978-1-4612-3624-5_9 [DOI] [PubMed] [Google Scholar]
- 36. Smith T. K., and Bütikofer P. (2010) Lipid metabolism in Trypanosoma brucei. Mol. Biochem. Parasitol. 172, 66–79 10.1016/j.molbiopara.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vial H. J., and Ancelin M. L. (1992) Malarial lipids. An overview. Subcell Biochem. 18, 259–306 10.1007/978-1-4899-1651-8_8 [DOI] [PubMed] [Google Scholar]
- 38. van Meer G., Voelker D. R., and Feigenson G. W. (2008) Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schuler M. H., Di Bartolomeo F., Böttinger L., Horvath S. E., Wenz L. S., Daum G., and Becker T. (2015) Phosphatidylcholine affects the role of the sorting and assembly machinery in the biogenesis of mitochondrial β-barrel proteins. J. Biol. Chem. 290, 26523–26532 10.1074/jbc.M115.687921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mohammadi A. S., Li X., and Ewing A. G. (2018) Mass spectrometry imaging suggests that cisplatin affects exocytotic release by alteration of cell membrane lipids. Anal. Chem. 90, 8509–8516 10.1021/acs.analchem.8b01395 [DOI] [PubMed] [Google Scholar]
- 41. Cooke M., Magimaidas A., Casado-Medrano V., and Kazanietz M. G. (2017) Protein kinase C in cancer: the top five unanswered questions. Mol. Carcinog. 56, 1531–1542 10.1002/mc.22617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Farine L., Niemann M., Schneider A., and Bütikofer P. (2015) Phosphatidylethanolamine and phosphatidylcholine biosynthesis by the Kennedy pathway occurs at different sites in Trypanosoma brucei. Sci. Rep. 5, 16787 10.1038/srep16787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Garg A., Lukk T., Kumar V., Choi J. Y., Augagneur Y., Voelker D. R., Nair S., and Ben Mamoun C. (2015) Structure, function, and inhibition of the phosphoethanolamine methyltransferases of the human malaria parasites Plasmodium vivax and Plasmodium knowlesi. Sci. Rep. 5, 9064 10.1038/srep09064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Itoe M. A., Sampaio J. L., Cabal G. G., Real E., Zuzarte-Luis V., March S., Bhatia S. N., Frischknecht F., Thiele C., Shevchenko A., and Mota M. M. (2014) Host cell phosphatidylcholine is a key mediator of malaria parasite survival during liver stage infection. Cell Host Microbe 16, 778–786 10.1016/j.chom.2014.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gibellini F., and Smith T. K. (2010) The Kennedy pathway–de novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 62, 414–428 [DOI] [PubMed] [Google Scholar]
- 46. Pessi G., Choi J. Y., Reynolds J. M., Voelker D. R., and Mamoun C. B. (2005) In vivo evidence for the specificity of Plasmodium falciparum phosphoethanolamine methyltransferase and its coupling to the Kennedy pathway. J. Biol. Chem. 280, 12461–12466 10.1074/jbc.M414626200 [DOI] [PubMed] [Google Scholar]
- 47. Reynolds J. M., Takebe S., Choi J. Y., El Bissati K., Witola W. H., Bobenchik A. M., Hoch J. C., Voelker D. R., and Mamoun C. B. (2008) Biochemical and genetic analysis of the phosphoethanolamine methyltransferase of the human malaria parasite Plasmodium falciparum. J. Biol. Chem. 283, 7894–7900 10.1074/jbc.M709869200 [DOI] [PubMed] [Google Scholar]
- 48. Witola W. H., Pessi G., El Bissati K., Reynolds J. M., and Mamoun C. B. (2006) Localization of the phosphoethanolamine methyltransferase of the human malaria parasite Plasmodium falciparum to the Golgi apparatus. J. Biol. Chem. 281, 21305–21311 10.1074/jbc.M603260200 [DOI] [PubMed] [Google Scholar]
- 49. Bobenchik A. M., Augagneur Y., Hao B., Hoch J. C., and Ben Mamoun C. (2011) Phosphoethanolamine methyltransferases in phosphocholine biosynthesis: functions and potential for antiparasite therapy. FEMS Microbiol. Rev. 35, 609–619 10.1111/j.1574-6976.2011.00267.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Déchamps S., Maynadier M., Wein S., Gannoun-Zaki L., Maréchal E., and Vial H. J. (2010) Rodent and nonrodent malaria parasites differ in their phospholipid metabolic pathways. J. Lipid Res. 51, 81–96 10.1194/jlr.M900166-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kantele A., and Jokiranta T. S. (2011) Review of cases with the emerging fifth human malaria parasite, Plasmodium knowlesi. Clin. Infect. Dis. 52, 1356–1362 10.1093/cid/cir180 [DOI] [PubMed] [Google Scholar]
- 52. Bezsonova I., Rujan I., Bobenchik A. M., Gorbatyuk V., Maciejewski M. W., Gorbatyuk O., Hao B., Arthanari H., Mamoun C. B., and Hoch J. C. (2013) (1)H, (13)C, and (15)N chemical shift assignments for PfPMT, a phosphoethanolamine methyltransferase from Plasmodium falciparum. Biomol. NMR Assign. 7, 17–20 10.1007/s12104-012-9372-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee S. G., Kim Y., Alpert T. D., Nagata A., and Jez J. M. (2012) Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite Plasmodium falciparum: an antiparasitic drug target. J. Biol. Chem. 287, 1426–1434 10.1074/jbc.M111.315267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bobenchik A. M., Choi J. Y., Mishra A., Rujan I. N., Hao B., Voelker D. R., Hoch J. C., and Mamoun C. B. (2010) Identification of inhibitors of Plasmodium falciparum phosphoethanolamine methyltransferase using an enzyme-coupled transmethylation assay. BMC Biochem. 11, 4 10.1186/1471-2091-11-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brancucci N. M. B., Gerdt J. P., Wang C., De Niz M., Philip N., Adapa S. R., Zhang M., Hitz E., Niederwieser I., Boltryk S. D., Laffitte M. C., Clark M. A., Grüring C., Ravel D., Blancke Soares A., et al. (2017) Lysophosphatidylcholine regulates sexual stage differentiation in the human malaria parasite Plasmodium falciparum. Cell 171, 1532–1544.e15 10.1016/j.cell.2017.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Riekhof W. R., and Voelker D. R. (2009) The yeast plasma membrane P4-ATPases are major transporters for lysophospholipids. Biochim. Biophys. Acta 1791, 620–627 10.1016/j.bbalip.2009.02.013 [DOI] [PubMed] [Google Scholar]
- 57. Riekhof W. R., Wu J., Gijón M. A., Zarini S., Murphy R. C., and Voelker D. R. (2007) Lysophosphatidylcholine metabolism in Saccharomyces cerevisiae: the role of P-type ATPases in transport and a broad specificity acyltransferase in acylation. J. Biol. Chem. 282, 36853–36861 10.1074/jbc.M706718200 [DOI] [PubMed] [Google Scholar]
- 58. Baldridge R. D., and Graham T. R. (2013) Two-gate mechanism for phospholipid selection and transport by type IV P-type ATPases. Proc. Natl. Acad. Sci. U.S.A. 110, E358–E367 10.1073/pnas.1216948110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Baldridge R. D., and Graham T. R. (2012) Identification of residues defining phospholipid flippase substrate specificity of type IV P-type ATPases. Proc. Natl. Acad. Sci. U.S.A. 109, E290–E298 10.1073/pnas.1115725109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vestergaard A. L., Coleman J. A., Lemmin T., Mikkelsen S. A., Molday L. L., Vilsen B., Molday R. S., Dal Peraro M., and Andersen J. P. (2014) Critical roles of isoleucine-364 and adjacent residues in a hydrophobic gate control of phospholipid transport by the mammalian P4-ATPase ATP8A2. Proc. Natl. Acad. Sci. U.S.A. 111, E1334–E1343 10.1073/pnas.1321165111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jensen M. S., Costa S. R., Duelli A. S., Andersen P. A., Poulsen L. R., Stanchev L. D., Gourdon P., Palmgren M., Günther Pomorski T., and López-Marqués R. L. (2017) Phospholipid flipping involves a central cavity in P4 ATPases. Sci. Rep. 7, 17621 10.1038/s41598-017-17742-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Riekhof W. R., Wu J., Jones J. L., and Voelker D. R. (2007) Identification and characterization of the major lysophosphatidylethanolamine acyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 282, 28344–28352 10.1074/jbc.M705256200 [DOI] [PubMed] [Google Scholar]
- 63. Emoto K., Kobayashi T., Yamaji A., Aizawa H., Yahara I., Inoue K., and Umeda M. (1996) Redistribution of phosphatidylethanolamine at the cleavage furrow of dividing cells during cytokinesis. Proc. Natl. Acad. Sci. U.S.A. 93, 12867–12872 10.1073/pnas.93.23.12867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vance J. E., and Tasseva G. (2013) Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim. Biophys. Acta 1831, 543–554 10.1016/j.bbalip.2012.08.016 [DOI] [PubMed] [Google Scholar]
- 65. McMahon H. T., and Boucrot E. (2015) Membrane curvature at a glance. J. Cell Sci. 128, 1065–1070 10.1242/jcs.114454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Emoto K., and Umeda M. (2000) An essential role for a membrane lipid in cytokinesis. Regulation of contractile ring disassembly by redistribution of phosphatidylethanolamine. J. Cell Biol. 149, 1215–1224 10.1083/jcb.149.6.1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Emoto K., Inadome H., Kanaho Y., Narumiya S., and Umeda M. (2005) Local change in phospholipid composition at the cleavage furrow is essential for completion of cytokinesis. J. Biol. Chem. 280, 37901–37907 10.1074/jbc.M504282200 [DOI] [PubMed] [Google Scholar]
- 68. Xie Z., and Klionsky D. J. (2007) Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9, 1102–1109 10.1038/ncb1007-1102 [DOI] [PubMed] [Google Scholar]
- 69. Paulick M. G., and Bertozzi C. R. (2008) The glycosylphosphatidylinositol anchor: a complex membrane-anchoring structure for proteins. Biochemistry 47, 6991–7000 10.1021/bi8006324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Xu K., and Nagy P. D. (2015) RNA virus replication depends on enrichment of phosphatidylethanolamine at replication sites in subcellular membranes. Proc. Natl. Acad. Sci. U.S.A. 112, E1782–E1791 10.1073/pnas.1418971112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Calzada E., Onguka O., and Claypool S. M. (2016) Phosphatidylethanolamine metabolism in health and disease. Int. Rev. Cell Mol. Biol. 321, 29–88 10.1016/bs.ircmb.2015.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Elabbadi N., Ancelin M. L., and Vial H. J. (1992) Use of radioactive ethanolamine incorporation into phospholipids to assess in vitro antimalarial activity by the semiautomated microdilution technique. Antimicrob. Agents Chemother. 36, 50–55 10.1128/AAC.36.1.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Choi J. Y., Augagneur Y., Ben Mamoun C., and Voelker D. R. (2012) Identification of gene encoding Plasmodium knowlesi phosphatidylserine decarboxylase by genetic complementation in yeast and characterization of in vitro maturation of encoded enzyme. J. Biol. Chem. 287, 222–232 10.1074/jbc.M111.313676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Choi J. Y., Duraisingh M. T., Marti M., Ben Mamoun C., and Voelker D. R. (2015) From protease to decarboxylase: the molecular metamorphosis of phosphatidylserine decarboxylase. J. Biol. Chem. 290, 10972–10980 10.1074/jbc.M115.642413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Satre M., and Kennedy E. P. (1978) Identification of bound pyruvate essential for the activity of phosphatidylserine decarboxylase of Escherichia coli. J. Biol. Chem. 253, 479–483 [PubMed] [Google Scholar]
- 76. Li Q. X., and Dowhan W. (1988) Structural characterization of Escherichia coli phosphatidylserine decarboxylase. J. Biol. Chem. 263, 11516–11522 [PubMed] [Google Scholar]
- 77. Kwon Y., Yu S. I., Lee H., Yim J. H., Zhu J. K., and Lee B. H. (2012) Arabidopsis serine decarboxylase mutants implicate the roles of ethanolamine in plant growth and development. Int. J. Mol. Sci. 13, 3176–3188 10.3390/ijms13033176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Alberge B., Gannoun-Zaki L., Bascunana C., Tran van Ba C., Vial H., and Cerdan R. (2009) Comparison of the cellular and biochemical properties of Plasmodium falciparum choline and ethanolamine kinases. Biochem. J. 425, 149–158 [DOI] [PubMed] [Google Scholar]
- 79. Serrán-Aguilera L., Denton H., Rubio-Ruiz B., López-Gutiérrez B., Entrena A., Izquierdo L., Smith T. K., Conejo-García A., and Hurtado-Guerrero R. (2016) Plasmodium falciparum choline kinase inhibition leads to a major decrease in phosphatidylethanolamine causing parasite death. Sci. Rep. 6, 33189 10.1038/srep33189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Choubey V., Maity P., Guha M., Kumar S., Srivastava K., Puri S. K., and Bandyopadhyay U. (2007) Inhibition of Plasmodium falciparum choline kinase by hexadecyltrimethylammonium bromide: a possible antimalarial mechanism. Antimicrob. Agents Chemother. 51, 696–706 10.1128/AAC.00919-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. González-Bulnes P., Bobenchik A. M., Augagneur Y., Cerdan R., Vial H. J., Llebaria A., and Ben Mamoun C. (2011) PG12, a phospholipid analog with potent antimalarial activity, inhibits Plasmodium falciparum CTP:phosphocholine cytidylyltransferase activity. J. Biol. Chem. 286, 28940–28947 10.1074/jbc.M111.268946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Le Roch K. G., Johnson J. R., Ahiboh H., Chung D. W., Prudhomme J., Plouffe D., Henson K., Zhou Y., Witola W., Yates J. R., Mamoun C. B., Winzeler E. A., and Vial H. (2008) A systematic approach to understand the mechanism of action of the bisthiazolium compound T4 on the human malaria parasite, Plasmodium falciparum. BMC Genomics 9, 513 10.1186/1471-2164-9-513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Choi J. Y., Surovtseva Y. V., Van Sickle S. M., Kumpf J., Bunz U. H. F., Ben Mamoun C., and Voelker D. R. (2018) A novel fluorescence assay for measuring phosphatidylserine decarboxylase catalysis. J. Biol. Chem. 293, 1493–1503 10.1074/jbc.RA117.000525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hanada K., Palacpac N. M., Magistrado P. A., Kurokawa K., Rai G., Sakata D., Hara T., Horii T., Nishijima M., and Mitamura T. (2002) Plasmodium falciparum phospholipase C hydrolyzing sphingomyelin and lysocholine phospholipids is a possible target for malaria chemotherapy. J. Exp. Med. 195, 23–34 10.1084/jem.20010724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Van Voorhis W. C., Adams J. H., Adelfio R., Ahyong V., Akabas M. H., Alano P., Alday A., Alemán Resto Y., Alsibaee A., Alzualde A., Andrews K. T., Avery S. V., Avery V. M., Ayong L., Baker M., et al. (2016) Open source drug discovery with the malaria box compound collection for neglected diseases and beyond. PLoS Pathog. 12, e1005763 10.1371/journal.ppat.1005763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Biagini G. A., Pasini E. M., Hughes R., De Koning H. P., Vial H. J., O'Neill P. M., Ward S. A., and Bray P. G. (2004) Characterization of the choline carrier of Plasmodium falciparum: a route for the selective delivery of novel antimalarial drugs. Blood 104, 3372–3377 10.1182/blood-2004-03-1084 [DOI] [PubMed] [Google Scholar]
- 87. Verhoven B., Schlegel R. A., and Williamson P. (1995) Mechanisms of phosphatidylserine exposure, a phagocyte recognition signal, on apoptotic T lymphocytes. J. Exp. Med. 182, 1597–1601 10.1084/jem.182.5.1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lentz B. R. (2003) Exposure of platelet membrane phosphatidylserine regulates blood coagulation. Prog. Lipid Res. 42, 423–438 10.1016/S0163-7827(03)00025-0 [DOI] [PubMed] [Google Scholar]
- 89. Freikman I., and Fibach E. (2011) Distribution and shedding of the membrane phosphatidylserine during maturation and aging of erythroid cells. Biochim. Biophys. Acta 1808, 2773–2780 10.1016/j.bbamem.2011.08.014 [DOI] [PubMed] [Google Scholar]
- 90. Burda P. C., Caldelari R., and Heussler V. T. (2017) Manipulation of the host cell membrane during Plasmodium liver stage egress. MBio 8, e00139-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Llinás M. (2017) Less lipid, more commitment. Cell 171, 1474–1476 10.1016/j.cell.2017.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bechtsi D. P., and Waters A. P. (2017) Genomics and epigenetics of sexual commitment in Plasmodium. Int. J. Parasitol. 47, 425–434 10.1016/j.ijpara.2017.03.002 [DOI] [PubMed] [Google Scholar]
- 93. Filarsky M., Fraschka S. A., Niederwieser I., Brancucci N. M. B., Carrington E., Carrió E., Moes S., Jenoe P., Bártfai R., and Voss T. S. (2018) GDV1 induces sexual commitment of malaria parasites by antagonizing HP1-dependent gene silencing. Science 359, 1259–1263 10.1126/science.aan6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lutz H. U., and Bogdanova A. (2013) Mechanisms tagging senescent red blood cells for clearance in healthy humans. Front. Physiol. 4, 387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Dembo E. G., Phiri H. T., Montgomery J., Molyneux M. E., and Rogerson S. J. (2006) Are Plasmodium falciparum parasites present in peripheral blood genetically the same as those sequestered in the tissues? Am. J. Trop. Med. Hyg. 74, 730–732 10.4269/ajtmh.2006.74.730 [DOI] [PubMed] [Google Scholar]
- 96. Gardiner D. L., and Trenholme K. R. (2015) Plasmodium falciparum gametocytes: playing hide and seek. Ann. Transl. Med. 3, 45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Orikiiriza J., Surowiec I., Lindquist E., Bonde M., Magambo J., Muhinda C., Bergström S., Trygg J., and Normark J. (2017) Lipid response patterns in acute phase paediatric Plasmodium falciparum malaria. Metabolomics 13, 41 10.1007/s11306-017-1174-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lakshmanan V., Rhee K. Y., Wang W., Yu Y., Khafizov K., Fiser A., Wu P., Ndir O., Mboup S., Ndiaye D., and Daily J. P. (2012) Metabolomic analysis of patient plasma yields evidence of plant-like α-linolenic acid metabolism in Plasmodium falciparum. J. Infect. Dis. 206, 238–248 10.1093/infdis/jis339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Nara F., Tanaka M., Hosoya T., Suzuki-Konagai K., and Ogita T. (1999) Scyphostatin, a neutral sphingomyelinase inhibitor from a discomycete, Trichopeziza mollissima: taxonomy of the producing organism, fermentation, isolation, and physico-chemical properties. J. Antibiot. 52, 525–530 10.7164/antibiotics.52.525 [DOI] [PubMed] [Google Scholar]
- 100. Trotter P. J., Pedretti J., and Voelker D. R. (1993) Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J. Biol. Chem. 268, 21416–21424 [PubMed] [Google Scholar]