Abstract
In humans, transport of food-derived cobalamin (vitamin B12) from the digestive system into the bloodstream involves three paralogous proteins: transcobalamin (TC), haptocorrin (HC), and intrinsic factor (IF). Each of these proteins contains two domains, an α-domain and a β-domain, which together form a cleft in which cobalamin binds. Zebrafish (Danio rerio) are thought to possess only a single cobalamin transport protein, referred to as Tcn2, which is a transcobalamin homolog. Here, we used CRISPR/Cas9 mutagenesis to create null alleles of tcn2 in zebrafish. Fish homozygous for tcn2-null alleles were viable and exhibited no obvious developmentally or behaviorally abnormal phenotypes. For this reason, we hypothesized that previously unidentified cobalamin-carrier proteins encoded in the zebrafish genome may provide an additional pathway for cobalamin transport. We identified genes predicted to code for two such proteins, Tcn-beta-a (Tcnba) and Tcn-beta-b (Tcnbb), which differ from all previously characterized cobalamin transport proteins as they lack the α-domain. These β-domain–only proteins are representative of an undescribed class of cobalamin-carrier proteins that are highly conserved throughout the ray-finned fishes. We observed that the genes encoding the three cobalamin transport homologs, tcn2, tcnba, and tcnbb, are expressed in unique spatial and temporal patterns in the developing zebrafish. Moreover, exogenously expressed recombinant Tcnba and Tcnbb bound cobalamin with high affinity, comparable with binding by full-length Tcn2. Taken together, our results suggest that this noncanonical protein structure has evolved to fully function as a cobalamin-carrier protein, thereby allowing for a compensatory cobalamin transport mechanism in the tcn2−/− zebrafish.
Keywords: zebrafish, one-carbon metabolism, transport, phylogenetics, protein evolution, affinity, cobalamin, transcobalamin, vitamin B12
Introduction
Vitamin B12, also referred to as cobalamin, is a micronutrient that acts as a cofactor for two enzymes: methionine synthase, which is involved in the one-carbon metabolism pathway, and methylmalonyl-CoA mutase, which is involved in the catabolism of amino acids and odd-chain fatty acids. As only bacteria and archaea are capable of cobalamin synthesis, animals must obtain vitamin B12 from their diet where it is generally found as a trace component or from their gastrointestinal microbiome. In humans, vitamin B12 deficiency can be caused by a variety of factors; these include increased demand (as is the case during pregnancy), decreased absorption and/or increased excretion (often observed in the elderly and those suffering from diseases involving malabsorption and renal dysfunction), low dietary intake, and inborn errors of the cobalamin transport and absorption pathways (1). Vitamin B12 deficiency has been shown to contribute to a variety of conditions, including, but not limited to, megaloblastic anemia (2), methylmalonic aciduria (3), autism and schizophrenia (4), age-related macular degeneration (5), and improper bone development (6). Maternal vitamin B12 deficiency has been demonstrated to increase risk for severe neural tube defects and omphalocele (7–9). Clinically, vitamin B12 deficiency has been found to manifest with symptoms including impaired cognition, depression, infertility, myelopathy, cardiomyopathy, and ataxia (1, 10).
In humans, the path of cobalamin transport through the digestive system, bloodstream, and into the cell requires a host of support proteins required for uptake, transport, and chemical modification. Three paralogous proteins act as carrier proteins for cobalamin: transcobalamin (TC)2, haptocorrin (HC), and intrinsic factor (IF) (11). The three-dimensional structures of all three carriers in humans have been resolved using X-ray crystallography. TC, HC, and IF all consist of two domains, a large α-domain and a smaller β-domain. Cobalamin binds in a cleft formed by these two domains, making contact with both (12–14). Although these proteins share similar domain structures, each has distinct roles in cobalamin transport. HC, produced in the saliva, is responsible for the transport of cobalamin through the upper digestive tract. HC is degraded in the duodenum, allowing IF to bind cobalamin. IF is responsible for carrying cobalamin through the remainder of the digestive tract to the terminal ileum, where receptor-mediated endocytosis moves the vitamin into epithelial cells before secreting it into the bloodstream. Both HC and TC bind cobalamin in the blood; however, only TC-bound cobalamin is available to most cells for uptake via the transcobalamin receptor CD320. HC is also present in the blood, where it carries ∼80% of the total cobalamin in plasma and is theorized to act as a scavenger for clearing cobalamin analogues from the bloodstream (15). HC and IF share greater protein sequence similarity with one another than either does with TC, and both are glycosylated, whereas TC is not (16, 17).
Previous work has carefully explored the evolution of vertebrate HC, TC, and IF in the context of whole-genome duplication events (18). Specifically, it has been proposed that a single cobalamin-carrier protein existed in early vertebrates prior to 2R, a whole genome duplication that occurred in the ancestral vertebrate after the divergence of cephalochordates but before the divergence of cartilaginous and bony vertebrates (18, 19). Furthermore, it has been theorized that one of the two paralogs resulting from 2R was then lost in the teleost lineage (infraclass of the ray-finned fish Actinopterygii), whereas a tandem duplication further expanded that gene into HC and IF in the ancestor of Sarcopterygii (the lobe-finned fish, which are ancestral to Tetrapoda and therefore all land-dwelling vertebrates) (18). This resulted in one and three predicted cobalamin-carrier proteins in these lineages, respectively (20, 21). HC is proposed to have been secondarily lost in amphibians, birds, and some mammalian species (18).
Although vitamin B12 deficiency is a serious worldwide health issue, nonclinical research has been primarily limited to mammalian models (22–28). Zebrafish (Danio rerio) are a potentially powerful model system with which to study vitamin B12 metabolism and biology. Zebrafish produce large numbers of offspring, are easily housed, and are amenable to genetic manipulation. Fish develop from fertilized eggs to free-swimming larva in less than a week. During the majority of this time, the embryos are transparent, allowing development to be monitored in real time. To date, only a single cobalamin-carrier protein has been described in zebrafish (20). Although it shares marginally higher amino acid similarity to tetrapod transcobalamin, it also displays properties that are characteristic of both HC and IF. For this reason, the protein was originally referred to as HIT (abbreviation of Haptocorrin, Intrinsic Factor, and Transcobalamin) and was thought to be more functionally similar to the ancestral carrier protein that duplicated and subfunctionalized into TC, HC, and IF (20). More recently, the gene that codes for this protein in zebrafish has been officially renamed tcn2 (Gene ID: 407646; NP_001116703) (18).
To develop a zebrafish model of vitamin B12 deficiency, we created two lines of zebrafish carrying homozygous tcn2 knockout mutations. These tcn2−/− fish demonstrate no obvious developmental or behavioral phenotypes, which is inconsistent with models of vitamin B12 deficiency in mammals (as reviewed in Ref. 22) (24). As cobalamin remains an essential enzymatic cofactor in fish (29), the benign effects of tcn2 knockout strongly suggests a compensatory, as yet undescribed, system of cobalamin transport in zebrafish. We have identified two putative proteins, Tcn-beta-a (Tcnba) (NP_001122207) and Tcn-beta-b (Tcnbb) (NP_001239578), that may be involved in such a compensatory system. Although homologous to Tcn2, these proteins are missing the α-domain associated with all other known cobalamin-carrier proteins; therefore, the bona fide nature of Tcnba and Tcnbb and their ability to bind cobalamin has been questioned without direct investigation by previous studies (18, 20).
In this study, we characterize the molecular and biochemical properties of zebrafish Tcnba and Tcnbb, demonstrating that they are able to bind cobalamin and are likely to provide a supporting role as cobalamin-carrier proteins. Despite the lack of an α-domain, these proteins bind cobalamin with an affinity approaching that of zebrafish Tcn2. Our findings shed light on the evolution of cobalamin-carrier proteins in nonmammalian species and demonstrate that the α-domain is not necessarily required for the ability to bind cobalamin.
Results
Development of CRISPR/Cas9 tcn2−/− mutant zebrafish lines
Single-guide RNAs (sgRNAs) were designed to target either the second or third exon of zebrafish tcn2 and injected into the yolk of fertilized eggs at the one-cell stage (Fig. S1A). Founder fish (F0) were outbred to WT fish and the resulting F1 progeny were inbred, 30 of which were screened for out-of-frame alleles. The founder fish chosen for the tcn2 exon 2 KO line possessed a 43-bp deletion/2-bp insertion overlapping the exon-intron boundary for exon 2. The resulting transcript is missing exon 2 (verified by RT-PCR and subsequent sequencing), which results in the presence of an early stop codon in the beginning of the fourth exon (Fig. S1B). The founder fish chosen for the tcn2 exon 3 KO line possessed a 51-bp deletion/4-bp insertion in exon 3, which results in the presence of an early stop codon at the end of the third exon (Fig. S1C). Both lines were bred to homozygosity and observed for developmental and behavioral phenotypes. For each of these characteristics, the homozygous null animals did not differ from each other and were indistinguishable from the WT fish.
Zebrafish Tcnba and Tcnbb share protein sequence characteristics with known cobalamin-binding proteins
Genes encoding transcobalamin-like proteins in zebrafish, tcnba (also known as tcn1; Gene ID: 566714; NM_001128735) and tcnbb (also known as si:ch211-117m20.5; Gene ID: 566658; NM_001252649), have been identified by previous studies but have not been extensively characterized (18, 20). Both of these genes are composed of four exons, and they are adjacent to one another on chromosome 5, separated by ∼2 kb of intergenic sequence. At the amino acid level, Tcnba and Tcnbb share 53% sequence identity and 68% sequence similarity, as measured by a Smith-Waterman local protein alignment.
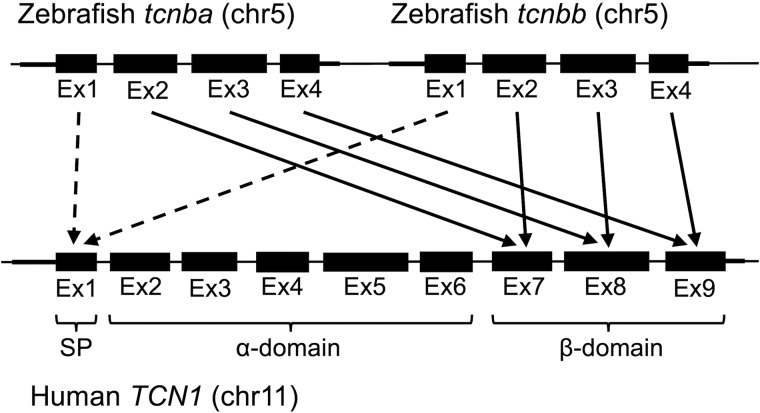
On an exon by exon basis, translated exons 2–4 in both zebrafish tcnba and tcnbb correspond to translated human TCN1 (gene encoding HC) exons 7, 8, and 9, respectively (Fig. 1), and the first exon from each of these genes encodes a predicted signal peptide for extracellular secretion. Exons 7–9 in human HC encode the β-domain (13), and the residues known to facilitate interaction with cobalamin (30) are widely conserved in both Tcnba and Tcnbb (Fig. S2); this suggests that zebrafish Tcnba and Tcnbb may form a structure similar to the β-domain of full-length cobalamin-carrier proteins and are potentially biochemically active.
Figure 1.
Alignment of zebrafish tcnba and zebrafish tcnbb to human TCN1. Exons of human TCN1, zebrafish tcnba, and zebrafish tcnbb were translated and compared using Smith-Waterman local protein alignments. Arrows indicate the highest alignment score for exons 2–4 in both tcnba and tcnbb compared with the exons of human TCN1. Dotted arrows demonstrate functional homology of the first exon in all three genes as a signal peptide. The exons encoding the signal peptide (SP), the α-domain, and the β-domain of human TCN1 are indicated. Exons, introns, and intergenic regions not to scale.
Tcn-beta class is specific to and well-conserved in ray-finned fishes
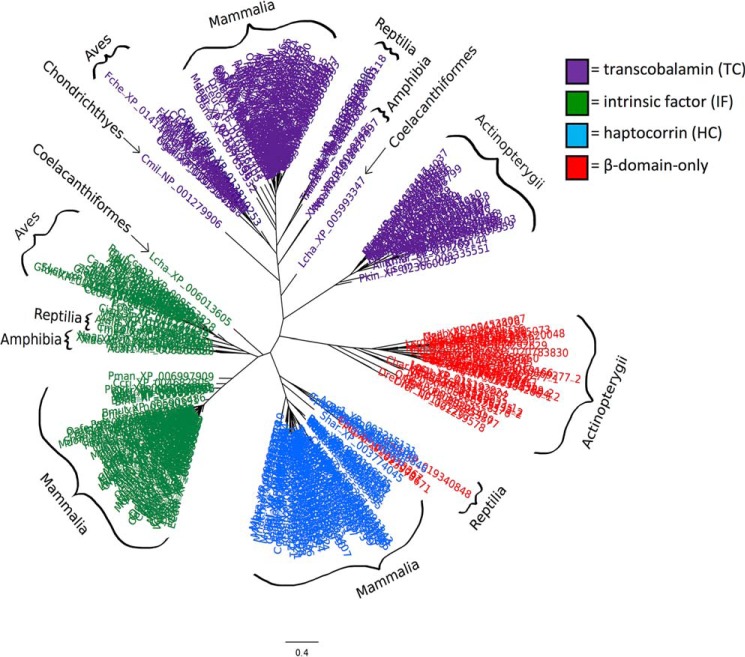
To investigate the prevalence of β-domain–only cobalamin-carrier homologs, we queried the RefSeq database using PSI-BLAST for the human HC α- and β-domains separately, restricting our results to vertebrate sequences. Any proteins which contained an α-domain were filtered from the results, leaving 48 β-domain–only homologs representing 29 species (Table S1). Only 4 of the β-domain–only homologs belonged to species not within Actinopterygii, confirming that the α-domain has been widely conserved outside these ray-finned fish. The β-domains of all 430 sequences identified in the initial PSI-BLAST search were used to calculate a multiple sequence alignment, from which a maximum-likelihood phylogeny was inferred (Fig. 2). All of the β-domain–only Actinopterygii sequences formed a monophyletic clade separate from vertebrate TC, HC, and IF, indicating that these proteins (including zebrafish Tcnba and Tcnbb) represent a novel class of putative cobalamin-binding proteins; for this reason, we have elected to update the original gene names (tcn1 and si:ch211-117m20.5) to ensure that an orthologous relationship is not inferred between zebrafish tcn1 and human TCN1. We will refer to the genes in this orthogroup collectively as tcn-beta, or tcnb. The proteins encoded by the genes in this orthogroup span 26 ray-finned fish species in both the Holostei (spotted gar) and Teleostei lineages. Whereas the majority of tcnb sequences were found to encode a protein consisting of a single β-domain, seven sequences were identified as encoding a protein that consists of two fused β-domains. Five of the seven were identified in Protacanthopterygii (comprising both Salmoniformes and Esociformes); the remaining two were identified in Clupea harengus (Atlantic herring). Syntenic analyses suggest that tandem duplication may have played a role in the evolutionary development of these unique proteins. Four additional β-domain–only homologs were identified in select reptilian species, all of which grouped with vertebrate HC. Further exploration of these genes was outside of the scope of this study, however, because their phylogenetic placement clearly indicates that they are not members of the tcnbb orthogroup.
Figure 2.
Tcnba and Tcnbb represent a novel class of cobalamin-carrier proteins specific to ray-finned fish. PSI-BLAST was used to identify cobalamin-carrier protein homologs restricted to the RefSeq database in vertebrate species. A multiple sequence alignment of 430 sequences was produced from which a maximum-likelihood phylogeny was inferred using PhyML software. Sequences marked in purple are categorized as TC homologs, sequences marked in green are categorized as IF homologs, and sequences marked in blue are categorized as HC homologs. Sequences marked in red are those identified from the search for homologs containing only the β-domain of full-length cobalamin-carrier proteins. All accession numbers, homolog classifications, and full species names can be found in Table S1.
Spatial and temporal expression of tcn2, tcnba, and tcnbb in vivo
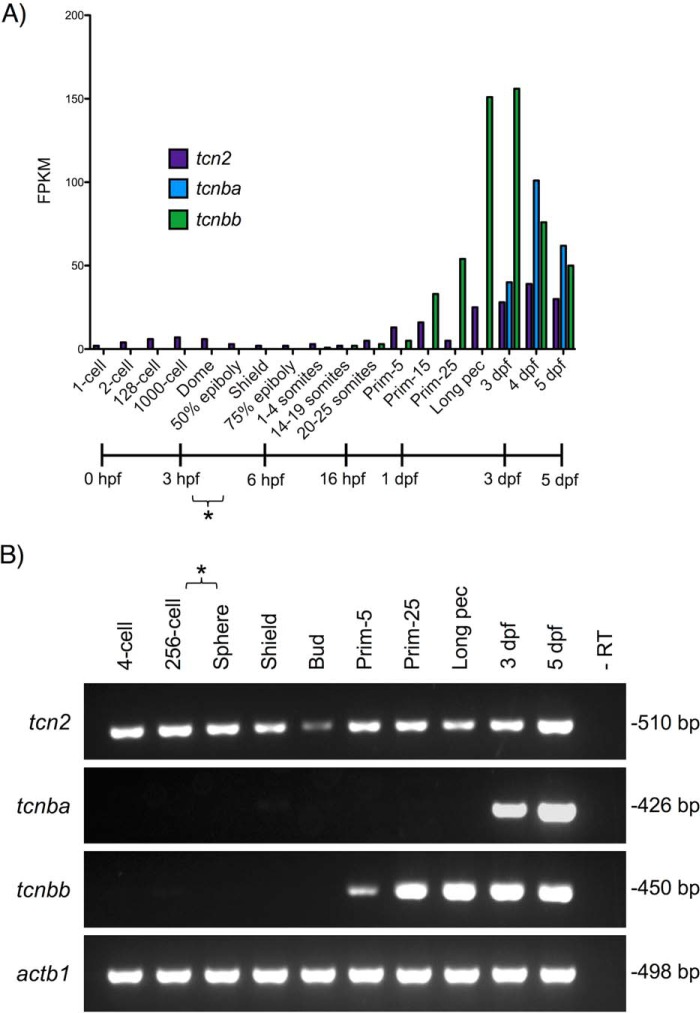
The developmental expression patterns of zebrafish tcn2, tcnba, and tcnbb were analyzed using the Zebrafish ESTs track in the UCSC Genome Browser (31). RNA-Seq libraries were found to contain abundant transcripts from these loci at multiple developmental stages. Time-course RNA-Seq data were obtained from the Expression Atlas server (32) and the levels of mRNA expression for zebrafish tcn2, tcnba, and tcnbb during various stages of development through 5 days post fertilization (dpf) were analyzed (33). The maternal to zygotic transition (MZT) occurs during the blastula period; therefore, any transcript present in the developing embryo prior to this stage is considered to be maternally loaded (34). These data demonstrated that tcn2 is a maternally loaded transcript, whereas tcnba and tcnbb are products of embryonic transcription. tcnbb is produced by the embryo at the onset of somitogenesis and tcnba is detectable by 3 dpf (Fig. 3A). RT-PCR was used to confirm the RNA-Seq results, using cDNA from WT TAB5 zebrafish embryos collected at various developmental stages ranging from four cells to 5 dpf (Fig. 3B).
Figure 3.
Temporal transcription analyses of zebrafish tcn2, tcnba, and tcnbb throughout early zebrafish development. A, RNA-Seq data from White et al. (33) were analyzed for expression of tcn2, tcnba, and tcnbb at various stages of early zebrafish development. y-Axis: fragments per kilobase million (FPKM); x-axis: developmental stage and corresponding timeline post fertilization (hours (hpf) or days (dpf)). Asterisk indicates estimated range of MZT transition. B, RT-PCR verification of RNA-Seq data. RNA was isolated from WT TAB5 zebrafish embryos at various stages of development and RT-PCR was performed to analyze expression of tcn2, tcnba, and tcnbb. Asterisk indicates estimated range of MZT transition. Invitrogen 1 Kb Plus DNA Ladder was used for evaluation of fragment size.
We then used whole-mount in situ hybridizations using embryos at 3 dpf and 5 dpf to determine spatial expression patterns of these transcripts (Fig. 4, A–I). At 3 dpf, tcn2 localized to the yolk syncytial layer, indicating a possible role in nutrient transport in the developing larva (Fig. 4A). At 5 dpf, expression of tcn2 was found to be concentrated in the intestinal tract and the liver, as well as residually in the yolk syncytial layer (Fig. 4B and 4C). We found tcnba to be specifically expressed in the primordial intestine at 3 dpf (Fig. 4D) and the mature intestinal bulb at 5 dpf (Fig. 4E). Finally, we found that tcnbb expression was largely restricted to the craniofacial region at 3 dpf (Fig. 4F and 4G) and 5 dpf (Fig. 4H and 4I) in a unique expression pattern that does not resemble any anatomical structures identified in the zebrafish to date.
Figure 4.
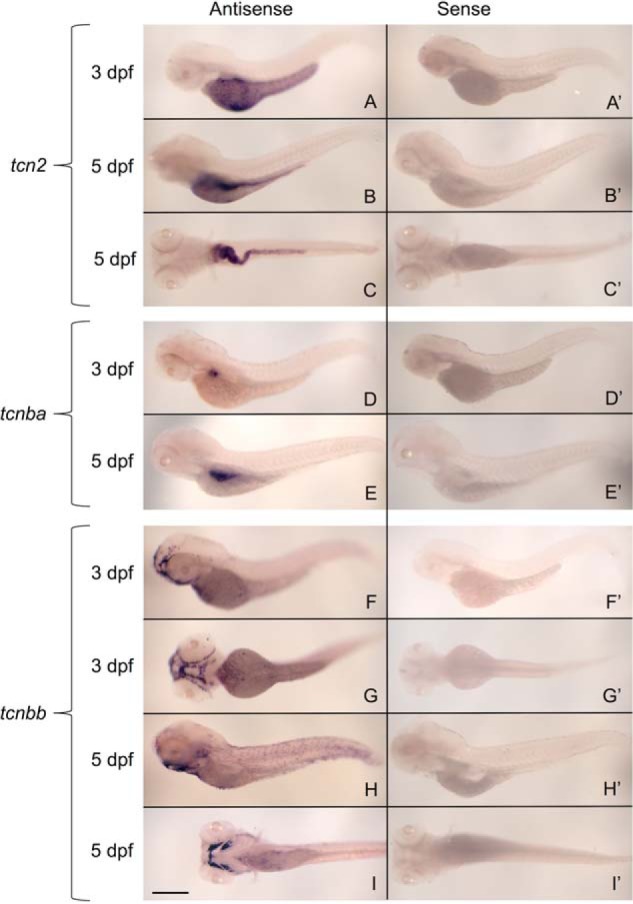
Whole-mount in situ hybridizations demonstrate specific and unique transcription expression patterns of zebrafish tcn2, tcnba, and tcnbb. Whole-mount in situ hybridizations were performed on zebrafish embryos at 3 dpf and 5 dpf with DIG-labeled RNA probes for tcn2, tcnba, and tcnbb to determine spatial expression patterns of these genes. A–I, sense probe controls (A′, B′, C′, etc.) for each antisense probe (A, B, C, etc.) are shown. Results are shown for tcn2 probe (A–C), tcnba probe (D and E), and tcnbb probe (F–I). A, lateral view of tcn2 expression at 3 dpf. B, lateral view of tcn2 expression at 5 dpf. C, ventral view of tcn2 expression at 5 dpf. D, lateral view of tcnba expression at 3 dpf. E, lateral view of tcnba expression at 5 dpf. F, lateral view of tcnbb expression at 3 dpf. G, ventral view of tcnbb expression at 3 dpf. H, lateral view of tcnbb expression at 3 dpf. I, ventral view of tcnbb expression at 5 dpf. Scale bar: 400 μm.
Embryos were sectioned to examine tcn2, tcnba, and tcnbb expression at higher resolution. We found tcn2 expression to be localized to the liver and the lumen of the intestinal bulb, the mid-intestine, and the posterior intestine (Fig. 5, A and B). The expression of tcnba localized to the intestinal bulb, with the strongest signal in the junction between the esophagus and the intestinal bulb. Additionally, select cells were shown to have strong signal in the outer lining of the intestinal bulb (Fig. 5, C and D). The expression pattern of tcnbb remained enigmatic after examination at the cellular level in histologic section. At this higher resolution, it was evident that expression is not in the craniofacial cartilage but is rather more superficial. Additionally, there appears to be an aggregation of expression in the interior of the pharynx which does not correspond to any known anatomical structure or gland (Fig. 5, E and F). Determination of spatial expression patterns was based on anatomical references on the ZFIN (35) and Bio-Atlas databases (http://bio-atlas.psu.edu/zf/)3 and by comparison to a liver-specific probe, fabp10a (Fig. S3).
Figure 5.
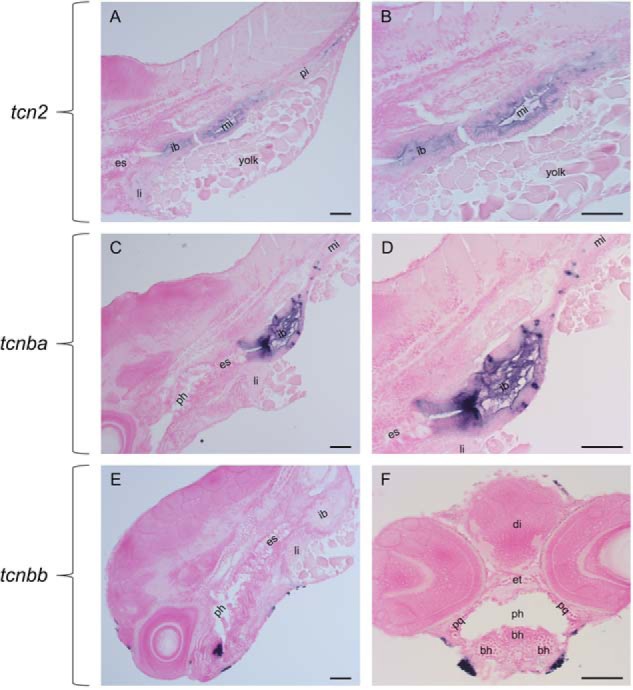
Sectioning and NFR staining of whole-mount in situ hybridizations reveal expression patterns at cellular resolution. RNA expression of tcn2, tcnba, and tcnbb was visualized by hybridization of DIG-labeled RNA probes and subsequent treatment with an anti-DIG antibody conjugated to alkaline phosphatase and BM-purple staining. Embryos were then sectioned and stained with Nuclear Fast Red to visualize at higher resolution. A, sagittal section of tcn2 expression at 5 dpf. B, sagittal section of tcn2 expression at 5 dpf. C, sagittal section of tcnba expression at 5 dpf. D, sagittal section of tcn2 expression at 5 dpf. E, sagittal section of tcnbb expression at 5 dpf. F, transverse section of tcnbb expression at 5 dpf. Abbreviations: es = esophagus, ib = intestinal bulb, mi = mid-intestine, pi = posterior intestine, li = liver, ph = pharynx, di = diencephalon, et = ethmoid plate, pq = palatoquadrate, bh = basihyal. Scale bar: 50 μm.
Expression of recombinant zebrafish tcn2, tcnba, and tcnbb in eukaryotic cells
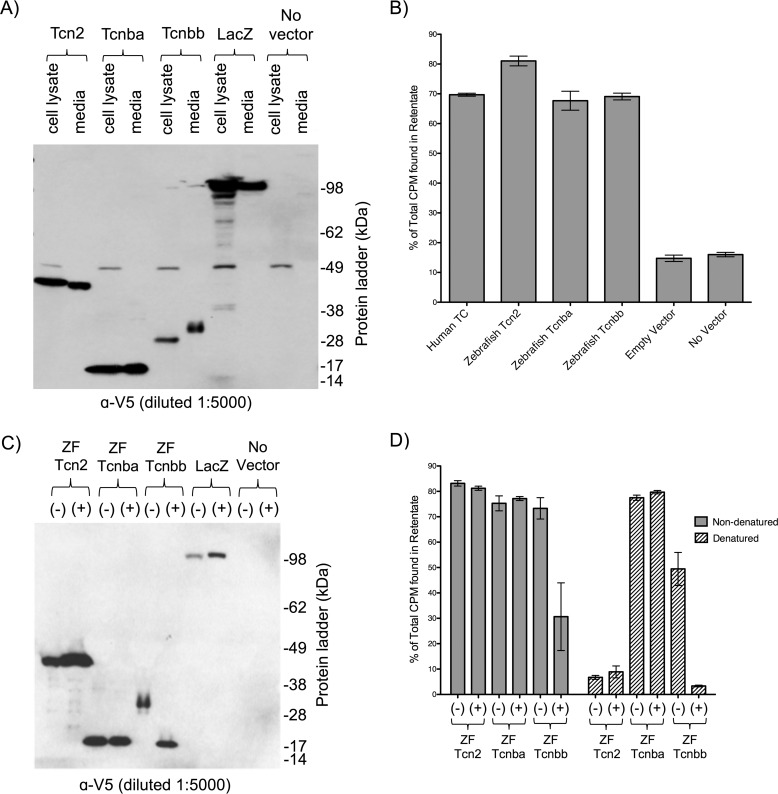
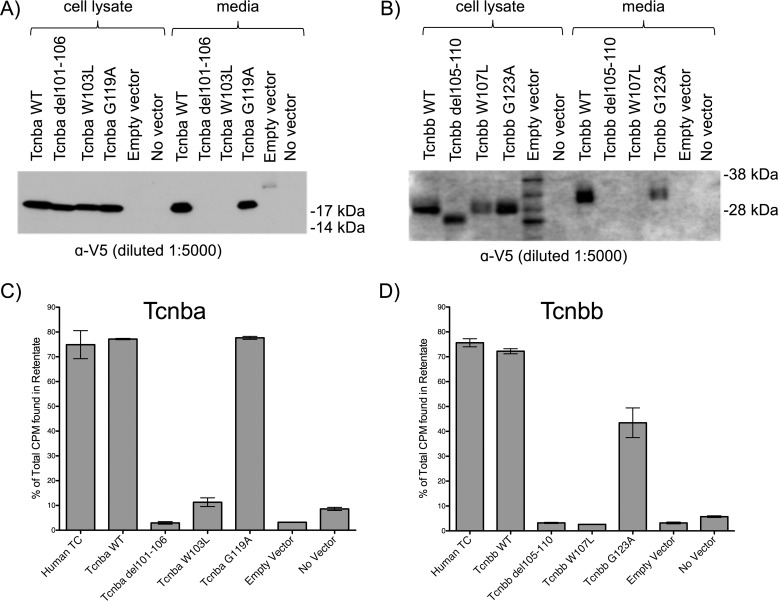
To test the ability of Tcnba and Tcnbb to bind cobalamin in vitro, expression constructs encoding zebrafish tcn2, tcnba, or tcnbb fused to C-terminal V5 and His6 epitope tags were transfected into Chinese Hamster Ovary K1 (CHO-K1) cells. An essential aspect of a functional cobalamin-carrier protein is its extracellular secretion; hence, extracellular media and cell lysate components from each individual transfection were harvested and compared. All three recombinant proteins were detectable in both the cell lysate and media. Recombinant Tcn2 and Tcnba were both observed at their predicted molecular weights (50 kDa and 18 kDa, respectively). However, recombinant Tcnbb was detectable at ∼30 kDa, which is larger than the 18 kDa predicted from the primary amino acid sequence (Fig. 6A). An unidentified band at ∼50 kDa was present in all cell lysate samples, which we believe to be an endogenous hamster protein cross-reacting with the anti-V5 antibody.
Figure 6.
Transfection of recombinant tcn2, tcnba, and tcnbb produces secreted proteins which are capable of binding cobalamin in vitro. A, SDS-PAGE Western blot analysis of recombinant zebrafish Tcn2, Tcnba, and Tcnbb expressed in CHO-K1 cells. For each sample, maximum volume (13 μl) was loaded. LacZ = empty vector. SeeBlue Plus2 pre-stained protein standard was used for molecular weight comparison. B, media samples were incubated with 57Co-containing cobalamin and spun through 10 kDa filters to separate protein-bound and unbound radioactive cobalamin. This assay was performed with media collected from cells transiently expressing recombinant proteins (zebrafish Tcn2, Tcnba, and Tcnbb). Positive control: Human TC media; negative controls: empty vector media, no vector media. Percent of total CPM in the retentate (protein-bound fraction) are shown, mean ± S.D. (n = 3). C, denaturing deglycosylation and SDS-PAGE Western blot analysis were performed on media samples to visualize the molecular weights of recombinant proteins either with (+) or without (−) deglycosylation treatment. SeeBlue Plus2 pre-stained protein standard was used for molecular weight comparison. D, untreated (−) and deglycosylated (+) media samples under nondenaturing or denaturing conditions were incubated with 57Co-containing cobalamin and spun through 10 kDa filters to separate protein-bound and unbound radioactive cobalamin. Percent of total CPM in the retentate (protein-bound fraction) are shown, mean ± S.D. (n = 3).
Cobalamin-binding ability of recombinant Tcn2, Tcnba, and Tcnbb
To directly assay cobalamin binding, radiolabeled vitamin B12 was mixed with samples of growth media harvested from tcn2-, tcnba-, and tcnbb-transfected CHO-K1 cells. The proportion of protein-bound cobalamin was measured by the percent of radioactivity present in the retentate separated via ultrafiltration. Protein-bound cobalamin in media containing zebrafish Tcnba and Tcnbb was comparable with that of media containing human TC and zebrafish Tcn2, indicating that both zebrafish Tcnba and Tcnbb are capable of binding cobalamin in vitro (Fig. 6B). Samples from both the empty vector and no vector control treatments demonstrated ∼3- to 4-fold lower levels of % CPM found in the retentate. To ensure that the His6 tag was not driving cobalamin interaction, we repeated this experiment with recombinant proteins containing only the V5 epitope tag and found that the results of the two binding experiments were concordant.
Glycosylation status of zebrafish Tcn2, Tcnba, and Tcnbb
Tcnbb had an apparent molecular weight that was nearly twice that predicted from its primary amino acid sequence. Given that human HC and IF are glycosylated in vivo, we hypothesized that the higher molecular weight was because of post-translationally added carbohydrates. To probe the nature of this anomaly, we asked if Tcnbb was glycosylated. Treatment of media with broad-spectrum deglycosylation reagents had no obvious effect on zebrafish Tcn2 or Tcnba, but it did reduce the secreted molecular weight of Tcnbb by ∼12 kDa (Fig. 6C). The after-treatment mass of ∼18 kDa is consistent with the expected molecular weight of Tcnbb, and clearly indicates that this protein is secreted as a glycoprotein. We expect that the mobility of Tcnbb differs between cell lysate and media because of as yet incomplete glycosylation of the protein in cell lysate.
To test whether glycosylation influences cobalamin-binding ability, each of our constructs were assayed with and without deglycosylation treatment under both native and denaturing conditions (Fig. 6D). As expected, the deglycosylation reagent had no effect on Tcn2 or Tcnba under native conditions, but it did reduce the fraction of protein-bound cobalamin in the Tcnbb samples by ∼2-fold. Under denaturing conditions, the effect of deglycosylation on Tcnbb was even more pronounced, although the ability to bind cobalamin was also markedly decreased by the denaturing environment alone. Denaturing conditions had no effect on Tcnba, while completely eliminating cobalamin binding by Tcn2.
Characterization of select motifs in Tcnba and Tcnbb
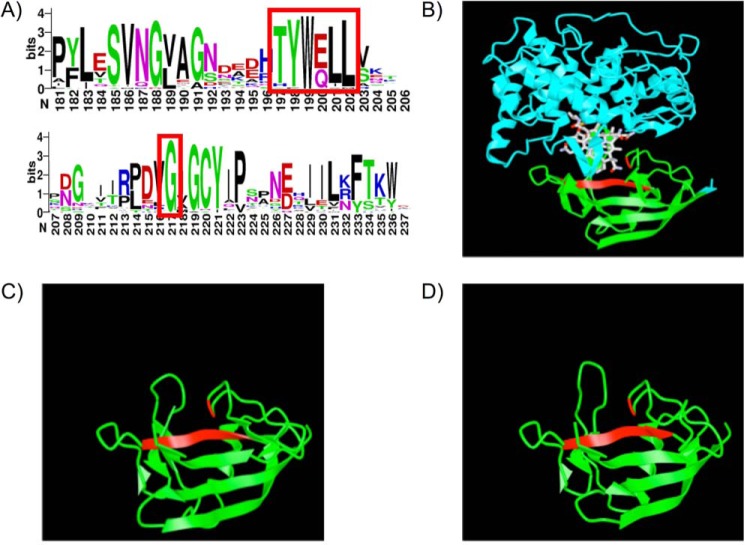
The proteins encoded by tcn-beta sequences were aligned and converted into a WebLogo (37) to visualize residue and motif conservation (Fig. 7A). For this analysis, only sequences consisting of a single β-domain were used to create the alignment and resulting WebLogo. Based on conservation identified here and analysis of the multiple sequence alignment of human and zebrafish cobalamin-carrier proteins (Fig. S2), we identified two areas of conservation to target with site-directed mutagenesis to characterize motifs essential for cobalamin binding in Tcnba and Tcnbb. The first was a six–amino acid sequence that was conserved between human HC and zebrafish Tcnba and Tcnbb (“TYWELL”) and the second was a conserved glycine residue downstream from this region. Both of these sites are located in the binding pocket of cobalamin in the crystal structure of human HC and contain residues that have been implicated in the interaction with cobalamin (Fig. 7B) (13, 30). In particular, the tryptophan residue of the TYWELL region is highly conserved in all vertebrate cobalamin-carrier proteins. SWISS-MODEL protein structure predictions of Tcnba (Fig. 7C) and Tcnbb (Fig. 7D) were produced using human HC as a reference structure, with the mutagenesis targets highlighted in red.
Figure 7.
Crystal structure of human HC and SWISS-MODEL predicted structures of zebrafish Tcnba and Tcnbb. A, the sequences identified as members of the Tcn-beta class of cobalamin-carrier proteins were used to create a WebLogo based on amino acid frequency at each position in the alignment to visualize highly conserved regions within this set of proteins. The two regions enclosed in a red rectangle were ultimately targeted for mutagenesis based on high conservation and predicted contact points with cobalamin based on data in Ref. 30. B, the crystal structure of human HC in complex with cyanocobalamin was obtained from the NCBI structure database (PDB ID: 4KKI) (13) and annotated using iCn3d. The α-domain of HC is shown in blue and the β-domain is shown in green. Mutagenesis targets for Tcnba and Tcnbb are conserved in human HC and are highlighted in red. C and D, SWISS-MODEL was used to create predicted protein structures for (C) Tcnba and (D) Tcnbb using human HC as a model (Tcnba: GMQE = 0.50, QMEAN = −2.82; Tcnbb: GMQE = 0.51, QMEAN = −2.87). Mutagenesis targets are highlighted in red (red ribbon = TYWELL, red line = glycine residue).
To test whether these motifs are essential for protein production, secretion, and cobalamin binding, site-directed mutagenesis was used to perform three types of mutagenesis in both Tcnba and Tcnbb: deleting TYWELL, mutagenizing the tryptophan in TYWELL to a leucine, and mutagenizing the downstream glycine to an alanine. SDS-PAGE analysis probed with anti-V5 antibody showed that the TYWELL deletion in Tcnba and Tcnbb (p.T101_L106del and p.T105_L110del, respectively) and the tryptophan to leucine mutagenesis in Tcnba and Tcnbb (p.W103L and p.W107L, respectively) resulted in the absence of the mutagenized protein secreted into the media. However, these mutagenized proteins could be detected in the cell lysate, indicating that protein secretion or stability may have been affected (Fig. 8, A and B). This was not the case for the mutagenesis of glycine to alanine in Tcnba and Tcnbb (p.G119A and p.G123A, respectively), which resulted in a recombinant protein that could be detected in both cell lysate and media samples (Fig. 8, A and B). We carried out a binding assay using cell lysate samples from these transfections to determine whether cobalamin-binding ability was maintained in the mutagenized proteins. The proteins mutagenized to delete TYWELL (Tcnba p.T101_L106del and Tcnbb p.T105_L110del) and the proteins with tryptophan mutagenized to leucine (Tcnba p.W103L and Tcnbb p.W107L) demonstrated a low percentage of protein-bound cobalamin, indicating an elimination in cobalamin-binding ability for those mutagenized proteins (Fig. 8, C and D). Conversely, Tcnba p.G119A maintained full cobalamin-binding ability compared with WT Tcnba, whereas Tcnbb p.G123A demonstrated only a slight reduction in cobalamin-binding ability compared with WT Tcnbb (Fig. 8, C and D).
Figure 8.
Site-directed mutagenesis of Tcnba and Tcnbb implicates the highly conserved TYWELL region in the ability to bind cobalamin in vitro. Site-directed mutagenesis was performed to delete the TYWELL region, mutagenize the TYWELL tryptophan to a leucine, and to mutagenize the highly conserved downstream glycine to an alanine in recombinant Tcnba and Tcnbb. A and B, SDS-PAGE Western blot analysis of cell lysate and media samples from CHO-K1 cells transfected with WT or mutagenized versions of Tcnba (A) or Tcnbb (B). SeeBlue Plus2 pre-stained protein standard was used for molecular weight comparison. C, cell lysate samples from WT and mutagenized Tcnba transfections were incubated with 57Co-containing cobalamin and spun through 10 kDa filters to separate protein-bound and unbound radioactive cobalamin. Percent of total CPM in the retentate (protein-bound fraction) are shown, mean ± S.D. (n = 3). D, this assay was repeated with the transfected WT and mutagenized Tcnbb cell lysate samples. Percent of total CPM in the retentate (protein-bound fraction) are shown, mean ± S.D. (n = 3).
Sample enrichment and binding-affinity analysis via microscale thermophoresis
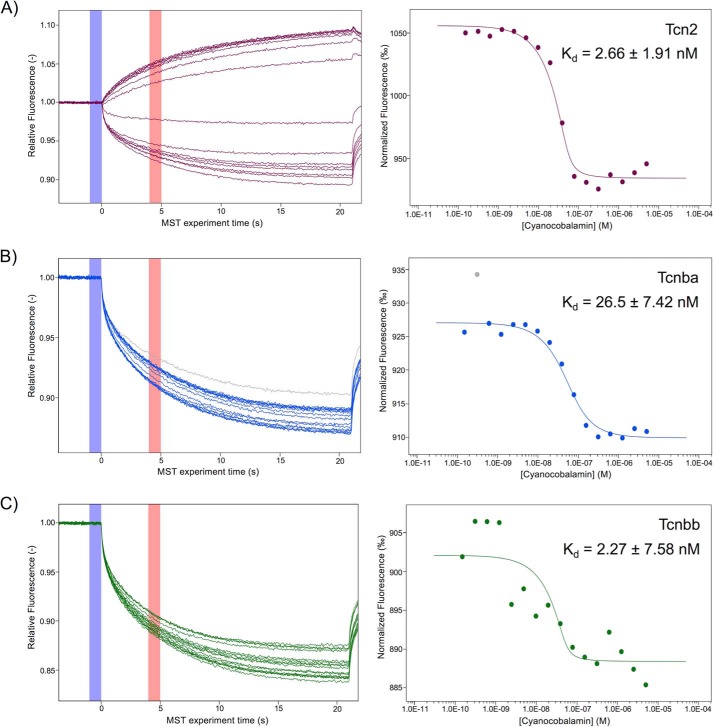
We next sought to measure the affinity of recombinant Tcn2, Tcnba, and Tcnbb for cyanocobalamin with microscale thermophoresis (MST) using enriched protein samples of Tcn2, Tcnba, and Tcnbb. All three proteins produced a shift in the MST trace when saturated with cyanocobalamin, indicating binding between the protein and its ligand. The dose response curve for zebrafish Tcn2 demonstrated a Kd of 2.66 ± 1.91 nm (Fig. 9A). The MST trace demonstrates that Tcn2 alone exhibits negative thermophoresis whereas the complex of Tcn2 and cyanocobalamin exhibits positive thermophoresis. The dose response curve for zebrafish Tcnba demonstrated a Kd of 26.5 ± 7.42 nm (Fig. 9B). Both Tcnba alone and the complex of Tcnba with cyanocobalamin exhibit positive thermophoresis. The dose response curve for zebrafish Tcnbb demonstrated a Kd of 2.27 ± 7.58 nm (Fig. 9C). Both Tcnbb alone and the complex of Tcnbb with cyanocobalamin exhibit positive thermophoresis. These measurements were performed at a target protein concentration of 50 nm. Ideally, the target protein concentration should be less than that of the measured Kd; however, lowering the target protein concentration to 25 nm for Tcn2, Tcnba, and Tcnbb produced MST traces demonstrating a low signal to noise ratio, indicating that the data were not reliable.
Figure 9.
Microscale thermophoresis of zebrafish Tcn2, Tcnba, and Tcnbb demonstrates all have comparable affinity for cyanocobalamin. Protein concentrations of enriched recombinant Tcn2, Tcnba, and Tcnbb were determined using a Bradford concentration assay to determine overall protein concentrations and a Coomassie gel to determine percent purity of the protein of interest. MST experiments were performed using a 50 nm concentration of each protein labeled with the fluorescent dye NT-647 and cyanocobalamin concentrations ranging from 5 μm to 150 pm. A, raw MST trace data and dose response curve for Tcn2. The fit yields a Kd of 2.66 ± 1.91 nm (S.E. of regression: 7.35, response amplitude: 121.50, signal to noise ratio: 17.76, target concentration: 50 nm). B, raw MST trace data and dose response curve for Tcnba. The fit yields a Kd of 26.5 ± 7.42 nm (S.E. of regression: 1.06; response amplitude: 17.13; signal to noise ratio: 17.51; target concentration: 50 nm). Gray dot indicates outlier removed from analysis because of irregularity in MST trace. C, raw MST trace data and dose response curve for Tcnbb. The fit yields a Kd of 2.27 ± 7.58 nm (S.E. of regression: 3.50, response amplitude: 13.67, signal to noise ratio: 4.19, target concentration: 50 nm).
Discussion
In mammals, severe cobalamin deficiency generally presents with a developmental and/or reproductive phenotype (22); therefore, the lack of obvious phenotypes in our homozygous tcn2−/− zebrafish led us to suspect the presence of other cobalamin-binding carrier proteins encoded in the zebrafish genome. We identified two zebrafish proteins that shared sequence similarity to tcn2. In previous analyses, these were not thought to be bona fide cobalamin-carrier proteins as they were considerably smaller than all known cobalamin-carrier proteins in vertebrates. Generally, cobalamin-carrier proteins have a length of roughly 400 amino acids, corresponding to about 40–50 kDa, and consist of two domains, a larger α-domain and a smaller β-domain (18, 38). However, tcnba and tcnbb were found to encode only the β-domain portion of other known cobalamin-carrier proteins, corresponding to a molecular weight of roughly 11–12 kDa after removal of the signal peptide and without consideration of the added C-terminal tags. Interestingly, we found that Tcnbb is the only zebrafish cobalamin-binding protein investigated here to exist as a glycoprotein, which may potentially speak to its functionality. Previous research has demonstrated that glycosylation of human IF is not essential for the binding of IF to cobalamin nor to its receptor; therefore, it has been theorized that the role of glycosylation is subtler via protection of the protein from proteolysis (39). From this, we theorize that zebrafish Tcnbb acts a carrier protein for cobalamin in an organ or tissue that has a high concentration of proteases, potentially the upper digestive tract.
After examining a large number of Actinopterygii species, it became evident that tcnba and tcnbb were well-conserved and found in many fish genomes. We hypothesized that they encode and represent a novel class of cobalamin-carrier protein, which we have termed Tcn-beta, or Tcnb. A paralogous pair can be found in the genomes of spotted gar (Lepisosteus oculatus, infraclass Holostei) and zebrafish (D. rerio, infraclass Teleostei), allowing us to conclude that the duplication event which led to the formation of the tcnb class of cobalamin-carrier proteins preceded the divergence of Holostei from Teleostei, which occurred roughly 350 million years ago and is one of the most ancient divisions within the class of ray-finned fish (40). Further investigation of the Tcnb sequences led to the observation that a subset of sequences was found to encode a protein consisting of two β-domains. Although a thorough investigation of these unique genes is beyond the scope of this study, we predict that these sequences may have arisen through a process of tandem gene duplication. The evolutionary timing of this duplication requires further investigation, as it is possible that this tandem duplication occurred in the common ancestor of Clupea and Protacanthopterygii, which diverged over 250 million years ago and was subsequently lost in the intermittent lineages; alternatively, it is possible that this tandem duplication occurred independently in both lineages (40).
We further demonstrated that tcn2, tcnba, and tcnbb are expressed in the developing zebrafish in unique spatial and temporal patterns. Notably, tcn2 is the only maternally expressed transcript of these three genes; it is therefore particularly surprising that tcn2−/− zebrafish do not display a developmental phenotype. Homozygous tcn2−/− zebrafish produced from parents with the same null genotype should not have access to a cobalamin-carrier protein until the expression of tcnbb begins at the onset of somitogenesis (∼16 h post fertilization), which may affect their viability. However, it is not clear if cobalamin-carrier proteins are required to reach this point in development, as the digestive tract and vasculature are immature until ∼5–6 dpf (41, 42). Therefore, it is possible that the yolk syncytial layer is capable of supplying embryos with cobalamin without the aid of a carrier protein up to this point in development. Additionally, we know very little about the bioavailability of cobalamin to zebrafish held under laboratory conditions. Further studies are needed to elucidate the mechanism of cobalamin absorption in early embryonic development.
The specific spatial expression patterns of tcn2, tcnba, and tcnbb identified in the developing zebrafish suggest possible functional divisions by comparing our observed expression patterns with the known tissue-specific expression of human tcn2 (encoding TC), tcn1 (encoding HC), and gif (encoding IF) (43). Expression of tcn2 in the liver and the entire digestive tract would allow this carrier protein to be distributed to the rest of the body via the circulation; although this expression pattern is more restricted than that of the near-ubiquitously expressed human TCN2 in adult tissues, this could be suggestive of an analogous function as supported by phylogeny. Notably, in contrast to mammals, zebrafish do not have a defined stomach; rather, the intestinal tract consists of an anterior intestinal bulb (which contains the majority of the digestive enzymes), a mid-intestine, and a posterior intestine (35). Expression of tcnba in the intestinal bulb, the portion of the intestine most similar to the human stomach, could suggest a role analogous to IF in carrying dietary cobalamin through the intestinal tract. Additionally, we observed strong expression in the most anterior portion of the intestinal bulb potentially corresponding to parietal cells, which are the site of IF production in humans. The expression pattern of tcnbb in the craniofacial region is more puzzling. The site of expression could correspond to the lining of the gills and could potentially allow for absorption of free cobalamin in the water through the gills. Alternatively, this site may be the equivalent of salivary glands, which have not been identified in zebrafish to date and are a major site of tcn1 (encoding HC) expression in humans. Synthesis at this location would allow tcnbb to be secreted into the pharynx and esophagus, thereby acting in a role analogous to HC. Additionally, tcnbb expression appears to be distributed throughout the body in a pattern that resembles the lymphatic or circulatory vasculature. In this case, it is possible that Tcnbb is secreted into the bodily fluids, potentially allowing for a HC-like function in the clearing of cobalamin analogues from the bloodstream and interstitium. In summary, our expression data suggest the potential for analogous functionality of the three cobalamin-carrier proteins in zebrafish and the three cobalamin-carrier proteins in humans despite having different evolutionary origins (Tcn2 = Tcn2; Tcnba = IF; Tcnbb = HC). However, it should be noted that because these are secreted proteins, they may be present in compartments and tissues other than those identified by mRNA expression. The lack of antibodies to the zebrafish cobalamin-carrier proteins limits our ability to study the proteins in vivo and requires us to infer expression and translation of Tcnba and Tcnbb based on mRNA expression data.
Despite the uncertainty discussed above, our data demonstrate that Tcnba and Tcnbb bind cobalamin with an affinity that is comparable with Tcn2. Our MST-based experiment to measure cobalamin binding was carried out using a target concentration that exceeded the potential Kd. Reduction of the concentration of our fluorescently labeled recombinant proteins on this platform resulted in unreliable data because of low signal intensity. As such, the true affinity values could be an order of magnitude smaller than the experimental values reported here. Given that the reported affinities of human cobalamin-carrier proteins for cyanocobalamin are reported in the subpicomolar range, future experiments using an alternative platform should be performed to determine more precisely how the affinities of the zebrafish cobalamin-binding proteins compare with those of the human cobalamin-carrier proteins (44). Our study recapitulates the findings in Ref. 30 in a zebrafish context, in which we implicate the hydrophobic interactions and hydrogen bonds made by residues in the TYWELL region (namely the tyrosine and tryptophan) as being essential for these proteins to bind cobalamin.
All vertebrate cobalamin-carrier proteins described to date consist of both a larger N-terminal α-domain and a smaller C-terminal β-domain. Previous studies have investigated the hypothesis that both domains are essential for cobalamin binding. Supporting this assumption, one study found that a highly conserved sequence located in the α-domain of human TC was required for cobalamin-binding ability in vitro (45). Our finding that proteins lacking the α-domain can readily bind cobalamin challenges the application of this rule to zebrafish. Although no β-domain–only proteins of this sort have been characterized thus far, our findings are not without precedent. A study characterizing the kinetics of the IF–cobalamin interaction found that the proteolytic fragment corresponding to the IF β-domain, IF20 (Kd = 0.2 ± 0.2 μm), was able to bind cobalamin independently, albeit several orders of magnitude less avidly than the full-length form, IF50 (Kd ≤1 pm) (46). In a more recent study, the crystal structure of the β-domain of human TC in complex with cyanocobalamin was solved, thereby demonstrating the ability of the β-domain to bind cobalamin in the absence of the α-domain (47).
Previous studies have also investigated the binding of these carrier proteins to their respective receptors. In the study above regarding the binding kinetics of IF, the researchers also found that the binding of IF to its receptor, cubilin, depended on contacts in both domains, which was later validated in the three-dimensional model of IF-cubilin (48). This interaction is not consistent with recognition of TC by its receptor in humans, CD320, which is driven by contacts only in the α-domain (49). It should be noted that homologs of cubilin, amnionless (which forms a complex with cubilin for holo-IF uptake) and asialoglycoprotein receptors 1 and 2 (responsible for the uptake of HC in the liver) have been identified in zebrafish. We have not been able to identify a homolog of the TC receptor, CD320, in zebrafish. The absence of CD320 and the presence of β-domain–only proteins in zebrafish suggest that zebrafish may use an alternative receptor or pathway to facilitate the uptake of cobalamin by cells.
In light of these results, and with the addition of our findings, we propose that the model of cobalamin binding to its carrier proteins should be re-evaluated to account for proteins that have evolved to consist solely of the β-domain. We propose that ∼400 million years of evolution in the Actinopterygii lineage (40) has allowed for the development of a novel class of cobalamin-carrier proteins consisting solely of the β-domain that are able to bind cobalamin with strong affinity. Our findings clearly demonstrate that tcnba and tcnbb are conserved, expressed, and produce functional proteins that are capable of binding cobalamin in vitro, providing strong evidence that these proteins play an active role in cobalamin transport in zebrafish. Further studies are required to determine whether tcnba or tcnbb are essential for zebrafish development and survival. Additionally, further biochemical analysis, including assessment of binding affinity for cobalamin analogues and stability at low pH, will give insight into the role of these proteins in cobalamin transport. Understanding the cobalamin transport pathway in zebrafish and its parallels to human cobalamin transport will aid in the creation of a zebrafish model for cobalamin deficiency and shed light on the evolutionary history of cobalamin-binding proteins in the vertebrate lineage.
Experimental procedures
Development of tcn2−/− mutant zebrafish lines
CRISPR/Cas9 mutagenesis was performed essentially as described in (50), using sgRNA molecules designed to target either the second or third exon of zebrafish tcn2. Briefly, primers were designed consisting of the T7 promoter sequence, 20 nucleotides of target sequence, and 20 nucleotides of sequence that overlapped with the generic sgRNA template (Integrated DNA Technologies, Coralville, IA). Each of the custom oligos was annealed with an oligo consisting of 80 nucleotides of sgRNA sequence, amplified using Phusion polymerase (New England Biolabs, Ipswich, MA), and transcribed into RNA. 50 picograms of sgRNA per target was injected into the yolk of fertilized embryos at the one-cell stage. All injections were performed in either the WT strain TAB-5 (ZIRC, Eugene, OR) or the transgenic HuC:eGFP strain demonstrating neuronal GFP expression (51). Injected founder fish (F0) were grown to adulthood and outbred to WT. F1 progeny were raised to adulthood and inbred. Thirty of the resulting progeny were then screened for deleterious mutations. Sequences of sgRNAs used are available upon request. Experimental protocols involving zebrafish were reviewed and approved by the National Human Genome Research Institute Animal Use and Care Committee under protocol G-08-1.
Bioinformatics in silico sequence analyses
Individual exons from human TCN1 and zebrafish tcnba and tcnbb were downloaded from the UCSC Genome Browser (31), translated to amino acid sequences using the ExPASy translate tool (52), and aligned (Smith-Waterman, PAM250 substitution matrix, 5.0 gap opening penalty, and 1.0 gap extension penalty) on the EMBOSS server (53). Human HC was chosen as a representative for this analysis as it demonstrated the highest local alignment score when Tcnba was aligned with each of the three human cobalamin-carrier proteins. Signal peptide predictions were performed using SignalP4.0 software with default parameters (54). Position-Specific Iterated Basic Local Alignment Search Tool (PSI-BLAST) was used to search the NCBI RefSeq database for other homologous cobalamin-carrier proteins using human HC α-domain and β-domain, separately, restricting our results to vertebrate sequences (taxonomy id: 7742). PSI-BLAST first performed a standard blastp search using the query sequence; a position-specific scoring matrix was then created from those results and iterative searches were performed using this matrix to expand the results. For this analysis, three iterations were performed and an e-value cut-off of 0.1 was used. Sequences were curated with DatabaseBuddy (55) to remove additional isoforms and low quality protein sequences. Multiple sequence alignments were predicted with Clustal Omega (56) and maximum likelihood phylogenies were inferred against the LG-gamma model using PhyML (57). Alignments and phylogenies were performed using AlignBuddy and PhyloBuddy, respectively (55). Time course RNA-Seq data published in Ref. 33 were retrieved from Expression Atlas on the EMBL-EBI server (32) and analyzed to search for zebrafish tcn2, tcnba, and tcnbb. Fragments per kilobase million (FPKM) values for each of the three genes at all 18 developmental stages were recorded and graphed.
RT-PCR of zebrafish embryos
Zebrafish embryos were isolated at various developmental stages (50 embryos/stage) and homogenized using a pellet pestle in TRI Reagent (Zymo Research, Irvine, CA). Total RNA was harvested and purified using the Direct-zol RNA Miniprep kit (Zymo Research). 1 μg of total RNA was reverse transcribed into cDNA for each developmental stage using the SuperScript III First-Strand Synthesis kit for RT-PCR according to manufacturer's instructions (Thermo Fisher Scientific). PCR was performed using HotStart Taq and standard PCR conditions for the primers used (available upon request).
Whole-mount in situ hybridization
Zebrafish embryos were collected at various developmental stages and fixed in 4% paraformaldehyde overnight at 4 °C. DIG-labeled probe synthesis and whole-mount in situ hybridizations were performed essentially as described in Ref. 36. An anti-DIG antibody conjugated to alkaline phosphatase was used to visualize staining with BM-purple. Embryos were fixed in 4% paraformaldehyde and mounted in 0.8% agarose for imaging.
Embryo sectioning and Nuclear Fast Red staining
Zebrafish embryos that had been previously treated for whole-mount in situ hybridization with DIG-labeled probes for our genes of interest were fixed in 4% paraformaldehyde, embedded in paraffin, and serial sectioned (5 μm per section) by Histoserv, Inc. (Germantown, MD). Sections were treated with Nuclear Fast Red stain and adhered on glass slides for imaging.
Gateway cloning system
Clones containing the cDNA of zebrafish tcn2 (Clone ID: 6797298), tcnba (Clone ID: 9037644), and tcnbb (Clone ID: 6901188) were obtained from GE Dharmacon (Lafayette, CO). The Gateway Cloning System from Invitrogen was utilized to clone these cDNAs into mammalian expression vectors, pcDNA-DEST40 or pcDNA3.2-DEST (Life Technologies, Thermo Fisher Scientific). Cloning was performed according to instructions in the Gateway Technology manual. Briefly, PCR primers were designed that contained an attB recombination site, a Kozak consensus sequence, and the first 20 bp of the gene of interest. These primers were used in a standard PCR reaction to create amplicons containing the gene of interest and flanking attB sites (primer sequences and PCR conditions available upon request). These amplicons were then used in a BP reaction with entry vector pDONR221 to produce an entry clone containing the gene of interest. The desired entry clones were isolated according to the protocol of the Qiagen Plasmid Miniprep kit (Qiagen, Hilden, Germany). Plasmids were Sanger-sequenced in both directions (T7 forward and reverse primers) by ACGT, Inc. (Germantown, MD). Entry clone sequencing demonstrated that all plasmid sequences were as expected. The entry clones were then used in an LR recombination reaction with the desired destination vector (pcDNA-DEST40 or pcDNA3.2-DEST). Potential destination clones were isolated according to the protocol of the Qiagen Plasmid Miniprep kit (Qiagen) for further use.
CHO-K1 cell maintenance and transfection
Chinese Hamster Ovarian K1 cells were obtained from liquid nitrogen stocks, thawed, and plated in liquid media consisting of DMEM supplemented with 10% FBS (Thermo Fisher, lot no.: 1901903), 1× Non-Essential Amino Acids (Thermo Fisher), and 2 mm l-glutamine (Thermo Fisher). The pcDNA-DEST40 and pcDNA3.2-DEST constructs were transfected according to the protocol in the Lipofectamine manual (Life Technologies). Briefly, Lipofectamine and plasmid were separately diluted in 150 μl DMEM according to a 3:1 ratio of Lipofectamine (μl): DNA (μg). The dilutions were mixed, allowed to incubate at room temperature for 5 min, and then added dropwise to CHO-K1 cells at 80–90% confluency in 6-well plates. The cell lysates and media from each individual transfection were harvested 48 h post transfection. Medium was applied to a 0.22 μm filter and stored at −80 °C. Cells were harvested using a Fisherbrand Cell Scraper (Fisher Scientific, Thermo Fisher Scientific) and resuspended in 1 ml cold 1× PBS. Cell lysates were then centrifuged at 4 °C, and the pellets were resuspended and incubated for 30 min in 500 μl RIPA–Nonidet P-40 buffer with 1× protease inhibitors. Cell lysates were then centrifuged at 4 °C to remove cell debris.
SDS-PAGE Western blot analysis
Media and cell lysate samples were tested for the presence of transfected protein using SDS-PAGE Western blot analysis. Negative controls included an empty vector cell lysate control (lacZ for pcDNA-DEST40 plasmids or GW-CAT for pcDNA3.2-DEST plasmids) and a no vector media control. For both cell lysate and media samples, maximum sample volume (13 μl) was loaded into each well. Samples were mixed with 5 μl NuPAGE LDS Sample Buffer (4×) (Thermo Fisher) and 2 μl NuPAGE Reducing Agent (10×) (Thermo Fisher) and incubated at 70 °C for 10 min. Transfected proteins were resolved using Novex NuPAGE 4–12% Bis-Tris gels and NuPAGE MOPS-SDS Running Buffer at 200 V for 50 min (Life Technologies). Protein transfer onto a PVDF membrane (0.2 μm pore size) was accomplished using iBlot (Life Technologies). The WesternBreeze anti-mouse chemiluminescent kit was used for immunoblotting according to the manufacturer's instructions (Life Technologies). Briefly, the membrane was incubated with blocking solution for 30 min at room temperature, then with primary antibody (anti-V5 diluted 1:5000; Sigma Aldrich) overnight at 4 °C, and finally with secondary antibody for 30 min at room temperature.
Protein deglycosylation
Deglycosylation was performed according to the protocol from Protein Deglycosylation Mix II, using either denaturing (Buffer 2) or nondenaturing (Buffer 1) conditions (New England Biolabs). Protein samples (40 μl) were aliquoted and 5 μl Deglycosylation Mix Buffer was added. For denaturing deglycosylation, samples were incubated at 75 °C for 10 min and allowed to cool before adding 5 μl Protein Deglycosylation Mix II. These samples were incubated at 25 °C for 30 min, then transferred to 37 °C and incubated for 1 h. For nondenaturing deglycosylation, 5 μl Protein Deglycosylation Mix II was added and the samples were incubated at 25 °C for 30 min and then transferred to 37 °C for 16 h. The samples were then loaded into a Novex NuPAGE 4–12% Bis-Tris gel (13 μl sample, 5 μl loading buffer, 2 μl reducing agent) with their respective untreated media samples in NuPAGE MOPS SDS Running Buffer (Life Technologies). Two identical protein gels were loaded and run; one gel was stained with Coomassie Blue for 1 h and then placed in destaining solution overnight at 4 °C and imaged under epi-white light to test for successful deglycosylation of the positive glycoprotein control fetuin. For the second, Western blot analysis with the anti-V5 antibody (diluted 1:5000, Sigma Aldrich) was performed as described above.
[57Co]Cobalamin-binding assays
Samples (normalized by input volume, 40 μl) were incubated with radioactive cobalamin tracer stock (MP Biomedicals, catalogue no. 06B-430000, specific activity 191 μCi/μg) at room temperature for 60 min and then transferred to 10 kDa filters (EMD Millipore) for separation of protein-bound and unbound cobalamin (free cobalamin has a molecular weight of ∼1.5 kDa). Filters were blocked with 5% milk for 30 min prior to loading the radioactive samples. Input concentration of radiolabeled cobalamin was 145 pm. Input counts ranged from 10,000 to 25,000 CPM for all assays. Radioactivity in retentate, flow through, and wash samples were then measured in an automatic gamma counter (Wallac Wizard 1470) and CPMs were transformed to percentages of total input CPM. Negative controls included empty vector media/cell lysate control (lacZ for pcDNA-DEST40 plasmids or GW-CAT for pcDNA3.2-DEST plasmids) and a no vector media/cell lysate control. The positive control for this experiment was media obtained from cells transfected with a construct encoding human TC. Binding specificity of the radiolabeled cobalamin was shown via a competition assay using unlabeled cobalamin competitor (Fig. S4). Unlabeled cobalamin concentrations included 10× (1.45 nm), 50× (7.25 nm), and 100× (14.5 nm).
Site-directed mutagenesis using QuikChange XL kit
The QuikChange XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) was used to mutagenize the tcnba and tcnbb cDNA clones according to manufacturer's instructions. Briefly, complementary primers containing the mutation of interest were designed and used in a PCR reaction to amplify the plasmid with the desired mutation. Unmutated, methylated template DNA was then digested with DpnI and the reaction was transformed into XL10-Gold ultracompetent cells. Individual clones were isolated, and the mutagenized plasmids were extracted using the Qiagen Miniprep kit (Qiagen). Mutagenized plasmids were then transfected into CHO-K1 cells according to the transfection protocol above and media/cell lysate samples were collected from each transfection. Radioligand-binding assays were performed on these samples as above, incubating 40 μl of cell lysate per replicate with radioactive cobalamin tracer stock and applying that solution to a 10 kDa spin filter.
Protein enrichment using anti-V5 agarose
Transfections were performed to obtain a sufficient quantity of media to enrich the desired protein for each construct (80 ml per construct). These samples were concentrated ∼50× using 10 kDa filters (EMD Millipore). 100 μl of anti-V5 agarose beads (Sigma Aldrich) was equilibrated using five 1 ml volumes of 1× PBS. Concentrated media samples were added to the agarose beads and the mixture was incubated on an end-over-end rotator at 4 °C for 16 h. The agarose beads were separated from the sample using Pierce Paper Filter Spin Cups (Thermo Scientific) and washed 10 times with 500 μl of 1× PBS. The proteins were eluted by incubating the agarose beads in 200 μl of 200 ng/μl V5 peptide (Sigma Aldrich) in 1× PBS on an end-over-end rotator at room temperature for 1 h. Affinity purification using anti-V5 agarose beads produced samples of ∼80% purity for Tcn2, 4% purity for Tcnba, and 50% purity for Tcnbb. Protein concentrations in samples enriched using anti-V5 agarose were determined by performing a Bio-Rad Protein Assay (Bio-Rad) and adjusted for approximate purity as determined by a Coomassie-stained SDS-PAGE gel. MST affinity analysis on the Monolith NT.115 platform using RED-tris-NTA dye allows for the use of impure samples as long as an estimate of the target protein concentration can be calculated; for that reason, an accurate purity estimation is more important than having a high purity sample.
Protein labeling with RED-tris-NTA dye
The protein was labeled with RED-tris-NTA dye, which labels His-tagged proteins, using the His-Tag Labeling Kit (Nanotemper, Munich, Germany) according to the manufacturer's instructions. In brief, the protein was diluted in PBS-T (1× PBS with 0.1% Tween 20) and mixed at a 1:1 molar ratio with the RED-tris-NTA dye. The protein-dye mixture was incubated for 30 min at room temperature and then centrifuged at 4 °C for 10 min at 15,000 × g to remove aggregates.
Microscale thermophoresis
MST experiments were performed on the Monolith NT.115 platform (Nanotemper) with 50% excitation power, 40% MST power, and temperature of 23 °C. Experiments were performed on the Monolith NT.115 platform (Nanotemper) following standard protocol as outlined in the Monolith NTTM His-Tag Labeling Kit RED-tris-NTA user manual. In brief, a serial dilution of cyanocobalamin (Cerilliant, Round Rock, TX) was prepared in PBS-T and fluorescently labeled protein was added for a final concentration of 50 nm protein. Dose response curves were drawn using relative fluorescence at 5 s after MST laser activation. A curve was fit to the data and the Kd was determined using the Kd setting on the MO Affinity Analysis software (Nanotemper). Outliers were manually excluded based on irregularity in the MST trace.
Author contributions
C. R. B., D. M. M., S. R. B., and L. C. B. conceptualization; C. R. B., A. E. S., and S. R. B. data curation; C. R. B. formal analysis; C. R. B., A. E. S., A. C. T., and L. C. B. validation; C. R. B., A. E. S., A. C. T., A. R. M., D. M. M., and S. R. B. investigation; C. R. B., A. E. S., A. C. T., A. R. M., D. M. M., and S. R. B. visualization; C. R. B., A. E. S., A. C. T., A. R. M., D. M. M., S. R. B., and L. C. B. methodology; C. R. B. writing-original draft; C. R. B., A. E. S., A. C. T., A. R. M., D. M. M., S. R. B., and L. C. B. writing-review and editing; S. R. B. and L. C. B. resources; S. R. B. software; S. R. B. and L. C. B. supervision; L. C. B. funding acquisition; L. C. B. project administration.
Supplementary Material
Acknowledgments
We thank the National Human Genome Research Institute (NHGRI) zebrafish core for experimental and technical guidance, specifically Kevin Bishop, Blake Carrington, and Raman Sood. We also thank Alberto Rissone for the donation of the fabp10a DIG-labeled RNA probe, and Shawn Burgess for consultation and advice. We thank Nadia Houerbi for microscopy imaging and analysis. We thank the Charles River staff for continued care and maintenance of zebrafish lines. We thank the National Heart Lung and Blood Institute (NHLBI) biophysics core for use of their Monolith NT.115 platform for MST experiments. We thank members of the NHLBI biophysics core, Dr. Grzegorz Piszczek and Dr. Di Wu, as well as Nanotemper supporting scientist Nicole Eckart for help in analysis of MST data. We extend gratitude to Faith Pangilinan and David Bernard for editorial contribution. Bio-Atlas is funded by National Institutes of Health Grant 5R24 RR01744, Jake Gittlen Cancer Research Foundation, and Pennsylvania Tobacco Settlement Fund.
This work was supported by the Division of Intramural Research of the National Human Genome Research Institute, National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4 and Table S1.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- TC
- transcobalamin
- HC
- haptocorrin
- IF
- intrinsic factor
- sgRNA
- single-guide RNA
- RefSeq
- Reference Sequence (database)
- dpf
- days post fertilization
- MZT
- maternal to zygotic transition
- CHO-K1
- Chinese Hamster Ovary K1 cells
- CPM
- counts per minute
- MST
- microscale thermophoresis.
References
- 1. Green R., Allen L. H., Bjørke-Monsen A. L., Brito A., Guéant J. L., Miller J. W., Molloy A. M., Nexo E., Stabler S., Toh B. H., Ueland P. M., and Yajnik C. (2017) Vitamin B12 deficiency. Nat. Rev. Dis. Primers 3, 17041 10.1038/nrdp.2017.40 [DOI] [PubMed] [Google Scholar]
- 2. Hakami N., Neiman P. E., Canellos G. P., and Lazerson J. (1971) Neonatal megaloblastic anemia due to inherited transcobalamin II deficiency in siblings. N. Engl. J. Med. 285, 1163–1170 10.1056/NEJM197111182852103 [DOI] [PubMed] [Google Scholar]
- 3. Higginbottom M. C., Sweetman L., and Nyhan W. (1978) A syndrome of methylmalonic aciduria, homocystinuria, megaloblastic anemica and neurological abnormalities in a vitamin B12-deficient breast-fed infant of a strict vegetarian. N. Engl. J. Med. 299, 317–323 10.1056/NEJM197808172990701 [DOI] [PubMed] [Google Scholar]
- 4. Zhang Y., Hodgson N. W., Trivedi M. S., Abdolmaleky H. M., Fournier M., Cuenod M., Do K. Q., and Deth R. C. (2016) Decreased brain levels of vitamin B12 in aging, autism and schizophrenia. PLoS One 11, e0146797 10.1371/journal.pone.0146797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gopinath B., Flood V. M., Rochtchina E., Wang J. J., and Mitchell P. (2013) Homocysteine, folate, vitamin B-12, and 10-y incidence of age-related macular degeneration. Am. J. Clin. Nutr. 98, 129–135 10.3945/ajcn.112.057091 [DOI] [PubMed] [Google Scholar]
- 6. Roman-Garcia P., Quiros-Gonzalez I., Mottram L., Lieben L., Sharan K., Wangwiwatsin A., Tubio J., Lewis K., Wilkinson D., Santhanam B., Sarper N., Clare S., Vassiliou G. S., Velagapudi V. R., Dougan G., and Yadav V. K. (2014) Vitamin B12-dependent taurine synthesis regulates growth and bone mass. J. Clin. Invest. 124, 2988–3002 10.1172/JCI72606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Molloy A. M., Kirke P. N., Troendle J. F., Burke H., Sutton M., Brody L. C., Scott J. M., and Mills J. L. (2009) Maternal vitamin B12 status and risk of neural tube defects in a population with high neural tube defect prevalence and no folic acid fortification. Pediatrics 123, 917–923 10.1542/peds.2008-1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mills J. L., Carter T. C., Kay D. M., Browne M. L., Brody L. C., Liu A., Romitti P. A., Caggana M., and Druschel C. M. (2012) Folate and vitamin B12-related genes and risk for omphalocele. Hum. Genet. 131, 739–746 10.1007/s00439-011-1117-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nasri K., Ben Fradj M. K., Touati A., Aloui M., Ben Jemaa N., Masmoudi A., Elmay M. V., Omar S., Feki M., Kaabechi N., Marrakchi R., and Gaigi S. S. (2015) Association of maternal homocysteine and vitamins status with the risk of neural tube defects in Tunisia: A case-control study. Birth Defects Res. 103, 1011–1020 10.1002/bdra.23418 [DOI] [PubMed] [Google Scholar]
- 10. Hunt A., Harrington D., and Robinson S. (2014) Vitamin B12 deficiency. BMJ 349, g5226 10.1136/bmj.g5226 [DOI] [PubMed] [Google Scholar]
- 11. Gherasim C., Lofgren M., and Banerjee R. (2013) Navigating the B12 road: Assimilation, delivery, and disorders of cobalamin. J. Biol. Chem. 288, 13186–13193 10.1074/jbc.R113.458810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wuerges J., Garau G., Geremia S., Fedosov S. N., Petersen T. E., and Randaccio L. (2006) Structural basis for mammalian vitamin B12 transport by transcobalamin. Proc. Natl. Acad. Sci. U.S.A. 103, 4386–4391 10.1073/pnas.0509099103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Furger E., Frei D. C., Schibli R., Fischer E., and Prota A. E. (2013) Structural basis for universal corrinoid recognition by the cobalamin transport protein haptocorrin. J. Biol. Chem. 288, 25466–25476 10.1074/jbc.M113.483271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mathews F. S., Gordon M. M., Chen Z., Rajashankar K. R., Ealick S. E., Alpers D. H., and Sukumar N. (2007) Crystal structure of human intrinsic factor: Cobalamin complex at 2.6-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 104, 17311–17316 10.1073/pnas.0703228104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nielsen M. J., Rasmussen M. R., Andersen C. B., Nexø E., and Moestrup S. K. (2012) Vitamin B12 transport from food to the body's cells—a sophisticated, multistep pathway. Nat. Rev. Gastroenterol. Hepatol. 9, 345–354 10.1038/nrgastro.2012.76 [DOI] [PubMed] [Google Scholar]
- 16. Li N., Seetharam S., and Seetharam B. (1995) Genomic structure of human transcobalamin II: Comparison to human intrinsic factor and transcobalamin I. Biochem. Biophys. Res. Commun. 208, 756–764 10.1006/bbrc.1995.1402 [DOI] [PubMed] [Google Scholar]
- 17. Alpers D., and Russell-Jones G. (1999) Intrinsic factor, haptocorrin, and their receptors. in Chemistry and Biochemistry of Vitamin B12 (Banerjee R., ed.), pp. 411–439, John Wiley & Sons, New York, NY [Google Scholar]
- 18. Lopes-Marques M., Ruivo R., Delgado I., Wilson J. M., Aluru N., and Castro L. F. C. (2014) Basal gnathostomes provide unique insights into the evolution of vitamin B12 binders. Genome Biol. Evol. 7, 457–464 10.1093/gbe/evu289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Putnam N. H., Butts T., Ferrier D. E. K., Furlong R. F., Hellsten U., Kawashima T., Robinson-Rechavi M., Shoguchi E., Terry A., Yu J. K., Benito-Gutiérrez E. L., Dubchak I., Garcia-Fernàndez J., Gibson-Brown J. J., Grigoriev I. V., et al. (2008) The amphioxus genome and the evolution of the chordate karyotype. Nature 453, 1064–1071 10.1038/nature06967 [DOI] [PubMed] [Google Scholar]
- 20. Greibe E., Fedosov S., and Nexo E. (2012) The cobalamin-binding protein in zebrafish is an intermediate between the three cobalamin-binding proteins in human. PLoS One 7, e35660 10.1371/journal.pone.0035660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Greibe E., Fedosov S., Sorensen B., Hojrup P., Poulsen S., and Nexo E. (2012) A single rainbow trout cobalamin-binding protein stands in for three human binders. J. Biol. Chem. 287, 33917–33925 10.1074/jbc.M112.398016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peng L., Dreumont N., Coelho D., Gueant J. L., and Arnold C. (2016) Genetic animal models to decipher the pathogenic effects of vitamin B12 and folate deficiency. Biochimie 126, 43–51 10.1016/j.biochi.2016.05.007 [DOI] [PubMed] [Google Scholar]
- 23. Van Tonder S. V., Metz J., and Green R. (1975) Vitamin B12 metabolism in the fruit bat (Rousettus aegyptiacus). The induction of vitamin B12 deficiency and its effect on folate levels. Br. J. Nutr. 34, 397–410 10.1017/S0007114575000463 [DOI] [PubMed] [Google Scholar]
- 24. Yamada K., Maeda N., Noguchi J., Yamada H., Morinaga E., Yatake H., Yamamoto Y., Tadokoro T., and Kawata T. (2013) Influences of maternal B12 and methionine intake during gestation and lactation on testicular development of offspring in rats. J. Nutr. Sci. Vitaminol. 59, 238–242 [DOI] [PubMed] [Google Scholar]
- 25. Khaire A., Rathod R., Kale A., and Joshi S. (2016) Vitamin b12 deficiency across three generations adversely influences long-chain polyunsaturated fatty acid status and cardiometabolic markers in rats. Arch. Med. Res. 47, 427–435 10.1016/j.arcmed.2016.10.006 [DOI] [PubMed] [Google Scholar]
- 26. Stangl G. I., Roth-Maier D. A., and Kirchgessner M. (2000) Vitamin B-12 deficiency and hyperhomocysteinemia are partly ameliorated by cobalt and nickel supplementation in pigs. J. Nutr. 130, 3038–3044 10.1093/jn/130.12.3038 [DOI] [PubMed] [Google Scholar]
- 27. Young P., Kennedy S., Molloy A., Scott J., Weir D., and Kennedy D. (1995) Effect of N2O treatment/vitamin B12 deficiency in pigs on tissue concentrations of odd-numbered, branched-chain fatty acids. Int. J. Vitam. Nutr. Res. 65, 255–260 [PubMed] [Google Scholar]
- 28. Cardinale G. J., Dreyfus P. M., Auld P., and Abeles R. H. (1969) Experimental vitamin B12 deficiency: Its effect on tissue vitamin B12-coenzyme levels and on the metabolism of methylmalonyl-CoA. Arch. Biochem. Biophys. 131, 92–99 10.1016/0003-9861(69)90108-8 [DOI] [PubMed] [Google Scholar]
- 29. Hansen A., Waagbo R., and Hemre G. (2015) New B vitamin recommendations in fish when fed plant-based diets. Aquaculture Nutr. 21, 507–527 10.1111/anu.12342 [DOI] [Google Scholar]
- 30. Wuerges J., Geremia S., and Randaccio L. (2007) Structural study on ligand specificity of human vitamin B12 transporters. Biochem. J. 403, 431–440 10.1042/BJ20061394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., and Haussler D. (2002) The human genome browser at UCSC. Genome Res. 12, 996–1006 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Petryszak R., Keays M., Tang Y. A., Fonseca N. A., Barrera E., Burdett T., Füllgrabe A., Fuentes A. M., Jupp S., Koskinen S., Mannion O., Huerta L., Megy K., Snow C., Williams E., et al. (2016) Expression Atlas update—an integrated database of gene and protein expression in humans, animals, and plants. Nucleic Acids Res. 44, D746–D752 10.1093/nar/gkv1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. White R. J., Collins J. E., Sealy I. M., Wali N., Dooley C. M., Digby Z., Stemple D. L., Murphy D. N., Billis K., Hourlier T., Fullgrabe A., Davis M. P., Enright A. J., and Busch-Nentwich E. M. (2017) A high-resolution mRNA expression time course of embryonic development in zebrafish. eLife 6, e30860 10.7554/eLife.30860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tadros W., and Lipshitz H. D. (2009) The maternal-to-zygotic transition: A play in two acts. Development 136, 3033–3042 10.1242/dev.033183 [DOI] [PubMed] [Google Scholar]
- 35. Howe D. G., Bradford Y. M., Conlin T., Eagle A. E., Fashena D., Frazer K., Knight J., Mani P., Martin R., Moxon S. A., Paddock H., Pich C., Ramachandran S., Ruef B. J., Ruzicka L., et al. (2013) ZFIN, the Zebrafish Model Organism Database: Increased support for mutants and transgenics. Nucleic Acids Res. 41, D854–D860 10.1093/nar/gks938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thisse C., and Thisse B. (2008) High-resolution in situ hybridizaiton to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 10.1038/nprot.2007.514 [DOI] [PubMed] [Google Scholar]
- 37. Crooks G. E., Hon G., Chandonia J. M., and Brenner S. E. (2004) WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fedosov S. N. (2012) Physiological and molecular aspects of cobalamin transport. in Water Soluble Vitamins: Clinical Research and Future Application (Stanger O. H., ed), pp. 347–367, Springer Netherlands, Berlin, Germany: 10.1007/978-94-007-2199-9 [DOI] [PubMed] [Google Scholar]
- 39. Gordon M., Hu C., Chokshi H., Hewitt J. E., and Alpers D. H. (1991) Glycosylation is not required for ligand or receptor binding by expressed rat intrinsic factor. Am. J. Physiol. 260, G736–G742 10.1152/ajpgi.1991.260.5.G736 [DOI] [PubMed] [Google Scholar]
- 40. Near T. J., Eytan R. I., Dornburg A., Kuhn K. L., Moore J. A., Davis M. P., Wainwright P. C., Friedman M., and Smith W. L. (2012) Resolution of ray-finned fish phylogeny and timing of diversification. Proc. Natl. Acad. Sci. U.S.A. 109, 13698–13703 10.1073/pnas.1206625109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wallace K. N., and Pack M. (2003) Unique and conserved aspects of gut development in zebrafish. Dev. Biol. 255, 12–29 10.1016/S0012-1606(02)00034-9 [DOI] [PubMed] [Google Scholar]
- 42. Jung H. M., Castranova D., Swift M. R., Pham V. N., Venero Galanternik M., Isogai S., Butler M. G., Mulligan T. S., and Weinstein B. M. (2017) Development of the larval lymphatic system in zebrafish. Development 144, 2070–2081 10.1242/dev.145755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Uhlen M., Fagerberg L., Hallstrom B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C. A., et al. (2015) Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- 44. Fedosov S. N., Grissom C. B., Fedosova N. U., Moestrup S. K., Nexø E., and Petersen T. E. (2006) Application of a fluorescent cobalamin analogue for analysis of the binding kinetics. A study employing recombinant human transcobalamin and intrinsic factor. FEBS J. 273, 4742–4753 10.1111/j.1742-4658.2006.05478.x [DOI] [PubMed] [Google Scholar]
- 45. Kalra S., Li N., Yammani R. R., Seetharam S., and Seetharam B. (2004) Cobalamin (vitamin B12) binding, phylogeny, and synteny of human transcobalamin. Arch. Biochem. Biophys. 431, 189–196 10.1016/j.abb.2004.08.011 [DOI] [PubMed] [Google Scholar]
- 46. Fedosov S. N., Fedosova N. U., Berglund L., Moestrup S. K., Nexø E., and Petersen T. E. (2005) Composite organization of the cobalamin binding and cubilin recognition sites of intrinsic factor. Biochemistry 44, 3604–3614 10.1021/bi047936v [DOI] [PubMed] [Google Scholar]
- 47. Bloch J. S., Ruetz M., Kräutler B., and Locher K. P. (2017) Structure of the human transcobalamin beta domain in four distinct states. PLoS One 12, e0184932 10.1371/journal.pone.0184932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Andersen C. B., Madsen M., Storm T., Moestrup S. K., and Andersen G. R. (2010) Structural basis for receptor recognition of vitamin-B(12)-intrinsic factor complexes. Nature 464, 445–448 10.1038/nature08874 [DOI] [PubMed] [Google Scholar]
- 49. Alam A., Woo J. S., Schmitz J., Prinz B., Root K., Chen F., Bloch J. S., Zenobi R., and Locher K. P. (2016) Structural basis of transcobalamin recognition by human CD320 receptor. Nat. Commun. 7, 12100 10.1038/ncomms12100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Varshney G. K., Pei W., LaFave M. C., Idol J., Xu L., Gallardo V., Carrington B., Bishop K., Jones M., Li M., Harper U., Huang S. C., Prakash A., Chen W., Sood R., Ledin J., and Burgess S. M. (2015) High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 25, 1030–1042 10.1101/gr.186379.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. St. John J. A., and Key B. (2012) HuC-eGFP mosaic labelling of neurons in zebrafish enables in vivo live cell imaging of growth cones. J. Mol. Histol. 43, 615–623 10.1007/s10735-012-9462-7 [DOI] [PubMed] [Google Scholar]
- 52. Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., and Bairoch A. (2003) ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 10.1093/nar/gkg563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rice P., Longden I., and Bleasby A. (2000) The European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 10.1016/S0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- 54. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Meth. 8, 785–786 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 55. Bond S. R., Keat K. E., Barreira S. N., and Baxevanis A. D. (2017) BuddySuite: Command-line toolkits for manipulating sequences, alignments, and phylogenetic trees. Mol. Biol. Evol. 34, 1543–1546 10.1093/molbev/msx089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sievers F., Wilm A., Dineen D. G., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J., Thompson J. D., and Higgins D. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., and Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.