Abstract
We present a computational study of tissue transcriptomic data of 14 cancer types to address: what may drive cancer cell division? Our analyses point to that persistent disruption of the intracellular pH by Fenton reactions may be at the root of cancer development. Specifically, we have statistically demonstrated that Fenton reactions take place in cancer cytosol and mitochondria across all the 14 cancer types, based on cancer tissue gene-expression data integrated via the Michaelis–Menten equation. In addition, we have shown that (i) Fenton reactions in cytosol of the disease cells will continuously increase their pH, to which the cells respond by generating net protons to keep the pH stable through a combination of synthesizing glycolytic ATPs and consuming them by nucleotide syntheses, which may drive cell division to rid of the continuously synthesized nucleotides; and (ii) Fenton reactions in mitochondria give rise to novel ways for ATP synthesis with electrons ultimately coming from H2O2, largely originated from immune cells. A model is developed to link these to cancer development, where some mutations may be selected to facilitate cell division at rates dictated by Fenton reactions.
Keywords: cancer driver, Fenton reaction, gene-expression data, intracellular pH, Warburg effect
Introduction
pH is one of the most fundamental properties of living cells. It measures the proton concentration in a cell or a subcellular compartment where complex biochemistry takes place. Alterations in the cellular pH can have detrimental impacts on molecular structure, function, subcellular localization, and biochemical reactions, hence the overall cellular biology. For this reason, (human) cells have a designated system for maintaining the pH stability as well as for adjusting the pH needed for cell proliferation, which consists of a buffering system and a set of transporters for moving basic and/or acidic molecules in or out of the cells. It is known that normal human cells have a slightly alkaline extracellular pH and a mildly acidic intracellular pH. In comparison, cancer tissue cells have exactly the opposite, all having alkaline intracellular pH and acidic extracellular pH from the early onset of the disease (Damaghi et al., 2013).
A few proposals have been made regarding the possible cause of as well as the benefit from this fundamental change in cancer cells. Two benefits have been extensively studied: (i) a slightly alkaline intracellular pH enables more efficient cell proliferation as the key molecular machinery and the rate-limiting step, namely the ribosome, works most efficiently at pH = 7.4–7.6 (Johansson et al., 2011); and (ii) an acidic extracellular environment offers a few advantages to cancer cells such as weakening of the attacks by activated T-cells and induction of apoptosis in the neighboring normal cells (Fischer et al., 2007). However, it remains unknown how do cancer cells become intracellularly alkaline, even though there have been proposals regarding how cancer cells may achieve this change via pH-related transporters such as Na+/H+ exchanger (NHE), sodium bicarbonate cotransporter (NBC), and/or monocarboxylic acid transporters (MCT) (Spugnini et al., 2015). One issue with such proposals is that these transporters are all gradient driven, hence highly unlikely to accomplish the reversal of the pH levels at least by the transporters alone.
We report here a new discovery that offers a possible answer to the above question and beyond, which is rooted in the increased iron accumulation at a chronic inflammatory site. It has been widely observed that cancer tissues tend to have (local) iron accumulation across virtually all solid cancer types (Torti and Torti, 2013). An important consequence of this in an environment high in H2O2 released by innate immune cells, typical of chronic inflammatory sites, is the occurrence of Fenton reaction: (Imlay et al., 1988). The reaction produces the most reactive radical, i.e. hydroxyl radicals , and the damages they induce may attract more immune cells, leading to further increased H2O2 level and forming a vicious cycle. Numerous studies have established that cancer tissue cells have Fenton reactions (Stevens and Kalkwarf, 1990; Toyokuni, 2009; Akatsuka et al., 2012). Iron–sulfur clusters (Johnson et al., 2005), heme (Bian et al., 2003), and labile iron pool (LIP) (Kruszewski, 2003) have been identified as sources of Fe2+ for cellular Fenton reactions. A variety of molecules can reduce Fe3+ to Fe2+, such as NAD(P)H, superoxide , S2−, and ascorbic acid, which can lead to repeated Fenton reactions, also called the Haber–Weiss reaction (Fong et al., 1976; Kojima and Bates, 1979; Elmagirbi et al., 2012), giving rise to continuous production of and OH− if the cellular environment is rich in such molecules. For simplicity, we continue to call such repeated reactions as Fenton reactions.
Between the two products of Fenton reactions, several published studies have suggested the possible roles of hydroxyl radical in damaging lipids and DNA (Balasubramanian et al., 1998). To the best of our knowledge, none of the published studies has reported on the impact of OH− and its possible link to cancer. It is worth emphasizing that Fenton reaction, an inorganic reaction, is the result of increased concentrations of Fe2+ and H2O2 at a site enriched with macrophage, neutrophil and other innate immune cells, which does not require an enzyme to take place and hence is not regulated biologically, at least not actively. We present here a novel statistical approach to estimate the level of Fenton reactions using tissue-based transcriptomic data. By applying this approach to gene-expression data of tissues of 14 cancer types and 16 types of non-cancerous chronic inflammatory diseases, we address: (i) how do Fenton reaction-harboring cells respond to the persistent production of OH− and hence increased intracellular pH? (ii) how do the responses link to cancer development?
Our analyses strongly suggest that cancer tissue cells have Fenton reactions in their cytosol and mitochondria while the non-cancerous inflammatory diseases tend to have extracellular but no or low levels of intracellular Fenton reactions. We have predicted distinct roles of Fenton reactions in the two different subcellular locations of cancer cells: (i) cytosolic Fenton reactions can increase the intracellular pH that drives increased glycolytic ATP generation and nucleotide synthesis as a key response to the continuous production of OH−; and (ii) mitochondrial Fenton reactions give rise to two novel pathways for ATP production: an anaerobic respiration chain for ATP synthesis by using H2O2 rather than O2 as the final electron acceptor and another directly consuming protons within the mitochondrial inner membrane (or simply inside mitochondria), both leading to increased cross-membrane proton gradients and hence ATP synthesis.
Results
The basic framework
All cancers under study were found to have elevated levels of H2O2 production and iron accumulation compared with the control tissues, based on our analyses of gene-expression data (Supplementary Figures S1 and S2). Together they give rise to increased Fenton reactions in multiple subcellular locations in cancer tissue cells. To estimate the levels of Fenton reactions and their impact on the intracellular pH, we consider the following chemical reactions:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
Equations (2) and (3) are the two most common reactions for reducing intracellular Fe3+ in human cells (Fong et al., 1976; Kojima and Bates, 1979). Equation (4) is a key superoxide-producing reaction, which comes from two sources in cancer: Complexes I and III of the mitochondrial electron transport chain (ETC), and NADH oxidases in stromal and/or innate immune cells, both having been well established. Equation (5) represents a general class of reactions that reduce NADP+ to NADPH by a reducing agent (RA), represented as . It is noteworthy that some hydroxides ( produced by Fenton reactions will be neutralized by protons generated by some of the above reactions while others will consume protons in the pH buffer since they do not have matching protons generated by Fenton or related reactions, which will drive up the intracellular pH level unless they are neutralized by protons generated by other sources. Specifically, if a is reduced by or produced by Equation (4), the by Equation (1) will be neutralized by protons from Equation (3) or Equation (4). In comparison, if the is reduced by an exogenous or not reduced, the produced will consume a proton in the pH buffer, where an exogenous superoxide refers to those generated in mitochondria that diffuse into cytosol via the VDAC channels on the mitochondrial outer membrane (Han et al., 2003) or those generated by NADH oxidases on the surface of stromal and/or immune cells, which can get inside cancer cells via anion transporters (Meitzler et al., 2014). These explain why the amounts of net and produced by Fenton reactions could be different, which also complicates our analyses. Overall, persistent Fenton reactions can be rewritten as follows as Fe is not consumed and, therefore, treated as a catalyst:
| (6) |
for the former case,
| (7) |
for the latter case.
Hence, the rates of net and productions can be estimated by
| (8) |
| (9) |
where , , and represent the reaction rates of Equations (6), (7), and the accumulation of unreduced Fe3+, respectively. Using the Michaelis–Menten equation (Michaelis and Menten, 1913), , , and can be estimated as follows:
| (10) |
| (11) |
| (12) |
where are kinetic parameters that are collected from published studies; generally, the , , , , , , , and should be positive; [], , [H2O2], , [Fe2+], and [Fe3+] are concentrations of the relevant molecular species; are the gene-expression levels of ferritin, ferric reductases, transferrin, and other iron transporters, respectively, and are parameters to be estimated using gene-expression data of subcellular location specific proteins. By integrating Equations (8) and (10–12), we use the following Equation (13) to estimate the occurrence of Fenton reaction in a specific subcellular location:
| (13) |
where , , , , and denote the expression levels of the selected genes to estimate [Fe2+], [H2O2], , and [Fe3+] accumulation by the following models. We assume each of the to-be-estimated quantities can be linearly approximated by a linear function of the expression levels of the relevant genes:
where , , , , , and are regression parameters and is the regression error. When estimating the quantity of each parameter, we have used approximately the same set of genes across different cancer types to capture the most consistent relationships across the cancers, with the detailed gene lists given in Supplementary Table S1.
To check if a subcellular compartment may have Fenton reactions, we have assessed the correlation between the two sides of Equation (13) using gene expressions of proteins localized to that subcellular compartment. Three criteria are used to assess the statistical significance of the predicted occurrence of Fenton reactions: (i) the predicted level of ; (ii) the R2 values when fitting the two sides of Equation (13); and (iii) signs of the regression parameters (see Supplementary Methods B.9). The criteria are selected based on the following consideration: (i) can be generated only by Fenton reactions; (ii) the predicted levels of in non-cancerous disease tissues are considerably lower than in cancer; and (iii) for cancer, tends to strongly correlate with the R2 value between the two sides of Equation (13).
Fenton reactions in cytosol
The occurrence of Fenton reaction
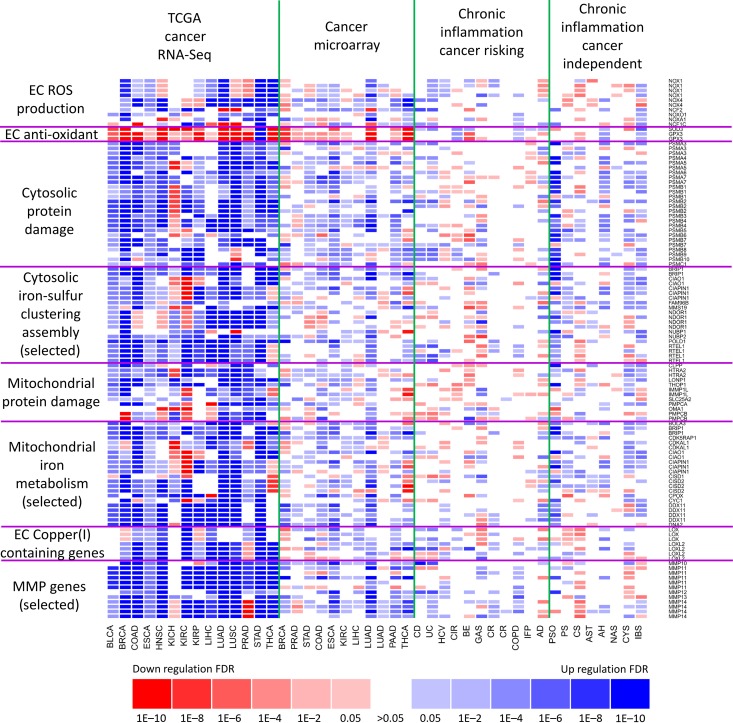
Table 1 shows the calculated values of all the three criteria along with their statistical significance. These data provide strong evidence for the occurrence of cytosolic Fenton reactions in all 14 cancer types under study; see Supplementary Table S1 for details. From the model fitting analyses, we noted: (i) extracellular superoxide makes substantial contributions to cytosolic Fenton reactions in breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), stomach adenocarcinoma (STAD), and thyroid carcinoma (THCA), while mitochondrial superoxide has considerable contributions in BRCA, ESCA, KIRC, KIRP, lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and STAD. For the other four cancer types, contributions from either source are moderate; these conclusions are drawn based on the levels of contributions to the observed R2 values by the relevant terms in the fitted model, as shown in Supplementary Figure S4; (ii) a substantial amount of unreduced Fe3+ from cytosolic Fenton reactions is accumulated in cancer cytosol as shown in Supplementary Figure S5, hence contributing to changing the intracellular proton concentration, where the predicted Fe3+ accumulation is consistent with published studies (Chen and Paw, 2012); and (iii) the two outcomes of Fe3+ given in Equations (10) and (11) that lead to net OH− generation correlates highly negatively with H+ exporter genes, as detailed in Supplementary Figure S6. Figure 2 (rows labeled with cytosolic) lists all the key genes and their expression levels, measured using both RNA-Seq and microarray, related to cytosolic Fenton reactions in cancer and in non-cancerous chronic inflammatory diseases, which are grouped into two groups: cancer prone and cancer independent.
Table 2.
Gene-expression data used in the current study, consisting of both RNA-Seq data of cancer vs. control tissues of 14 cancer types from the TCGA database and microarray data of both cancer and 16 types of non-cancerous inflammatory disease tissues from the GEO database.
| TCGA RNA-Seq data | ||
|---|---|---|
| Cancer type | Number of tumor samples | Number of control samples |
| BLCA | 408 | 19 |
| BRCA | 1095 | 113 |
| COAD | 285 | 41 |
| ESCA | 184 | 13 |
| HNSC | 520 | 44 |
| KICH | 66 | 25 |
| KIRC | 533 | 72 |
| KIRP | 290 | 32 |
| LIHC | 371 | 50 |
| LUAD | 515 | 59 |
| LUSC | 501 | 51 |
| PRAD | 497 | 52 |
| STAD | 238 | 33 |
| THCA | 505 | 59 |
| Chronic inflammation data | |||||
|---|---|---|---|---|---|
| GSE ID | Inflammation | Disease label | Number of samples | Number of controls | Relevance to cancer |
| GSE28619 | Alcohol hepatitis | AH | 15 | 7 | N |
| GSE4302 | Asthmatics | AST | 42 | 28 | N |
| GSE32924 | Atopic dermatitis | AD | 13 | 8 | L |
| GSE26886 | Barrett’s esophagus | BE | 20 | 19 | H |
| GSE36830 | Chronic rhinosinusitis | CR | 12 | 6 | L |
| GSE6764 | Cirrhosis | CIR | 13 | 10 | H |
| GSE16879 | Colitis (CD) | CD | 37 | 12 | H |
| GSE16879 | Colitis (UC) | UC | 40 | 12 | H |
| GSE32887 | Cutaneous sarcoidosis | CS | 15 | 5 | N |
| GSE5081 | Gastritis | GAS | 26 | 33 | H |
| GSE11190 | HCV | HCV | 16 | 2 | H |
| GSE36701 | IBS | IBS | 28 | 77 | N |
| GSE21369 | IFP | IFP | 23 | 6 | L |
| GSE11784 | Lung (COPD) | COPD | 35 | 63 | L |
| GSE63067 | Non-alcoholic steatohepatitis | NAS | 9 | 7 | N |
| GSE14905 | Psoriasis | PSO | 33 | 21 | N |
| The second column is for the name of a disease; the fourth column is the probability of a disease to become cancerous within the next 10 years, based on published data. H, highly relevant to cancer; L, moderately relevant to cancer; N, not relevant to cancer. | |||||
| Cancer microarray data | ||||
|---|---|---|---|---|
| GSE ID | Cancer type | Disease label | Number of samples | Number of controls |
| GSE31189 | Bladder carcinoma | BLCA | 52 | 40 |
| GSE42568 | Breast carcinoma | BRCA | 104 | 17 |
| GSE55945 | Prostate adenocarcinoma | PRAD | 12 | 7 |
| GSE13911 | Gastric adenocarcinoma | STAD | 38 | 31 |
| GSE22598 | Colorectal adenocarcinoma | COAD | 17 | 17 |
| GSE26886 | Esophageal carcinoma | ESCA | 21 | 20 |
| GSE11151 | Renal cell carcinoma | KIRC | 62 | 5 |
| GSE41804 | Hepatocellular carcinoma | LIHC | 10 | 10 |
| GSE19804 | Lung adenocarcinoma | LUAD | 60 | 60 |
| GSE16515 | Pancreatic adenocarcinoma | PAAD | 36 | 16 |
| GSE33630 | Thyroid carcinoma | THCA | 49 | 45 |
Figure 2.
Comparative analyses of expression levels of Fenton reaction-related genes in 14 cancer types (RNA-Seq data) and microarray data of 16 inflammatory diseases and 12 cancer types. Two types of gene-expression data, RNA-Seq and microarray, are used to show general consistencies between them. Inflammatory diseases are grouped into two classes: cancer prone and cancer independent. Genes covered here fall into eight functional categories with (i) two related to cytosolic Fenton reactions: cytosolic protein damages for reflecting the level of Fenton reactions and cytosolic iron–sulfur cluster assembly genes to reflect the level of iron available for Fenton reactions; (ii) two related to mitochondrial Fenton reactions; and (iii) four related to ECM Fenton reactions with MMP genes for assessing the level of ECM Fenton reactions, ECM copper-containing genes for the level of copper available for Fenton reactions; and ECM ROS and anti-oxidant genes to reflect the level of H2O2 available for Fenton reactions.
Impact of Fenton reactions to pH
To demonstrate that the amount of OH− produced by cytosolic Fenton reactions in cancer cells is large enough to overwhelm the pH buffer, we have conducted a computational analysis to show: (i) substantial fraction of the net protons originated from glycolysis in cancer stays inside the cell rather than secreted extracellularly; (ii) the intracellular accumulation rate of such protons can quickly reach a level that can change the intracellular pH; and (iii) the rate of the OH− produced by cytosolic Fenton reactions in cancer cells is higher than (ii).
While the detailed analysis and results are given in Supplementary Section A.2 and Figure S8, we highlight a few observations here, first focusing on the cytosolic pyruvate metabolism. Among the five main effluxes from glycolytic pyruvate, acetyl-CoA, oxaloacetate, amino acids (alanine, lysine or aspartate), lactate, and sialic acids, our analyses suggest that a substantial fraction of glycolytic pyruvate goes to non-lactate production, hence their associated protons will stay inside the cell. It will not take long for such protons to overwhelm the pH buffer and start to change the cytosolic pH. And from the literature, we know: (i) cancer cells have alkaline intracellular pH; (ii) the glucose catabolism makes the cells more acidic (and the cells have no other obvious metabolisms that persistently generate OH−); and (iii) protons are being imported rather than exported in cancer, which is the opposite in normal proliferating cells (unpublished data). All these indicate that the rate of OH− produced by cytosolic Fenton reactions is higher than the accumulation rate of protons originated from glycolysis, which completes our discussion that the amount of OH− produced by cytosolic Fenton reactions in cancer cells can overwhelm the pH buffer. See Supplementary Table S2 for details.
Cellular response to the continuous production of OH−
Now the question is how do cancer cells respond to the continuous production of OH−? Clearly, one response is the import of protons as mentioned above. But further analyses revealed more. Strong correlations were observed between the estimated rate of OH− production by Equation (9) and the rate of glycolytic ATP synthesis with high statistical significance across all 14 cancer types, as detailed in Supplementary Figures S8 and S9. While unexpected, this makes sense as it has been well established that glycolytic ATP synthesis: glucose + 2 ADP3− + 2 HPO42−→ 2 lactate− + 2 ATP4− is pH neutral while ATP synthesis by respiration: ADP3− + HPO42− → ATP4− + OH− each consumes one H+ (Swietach et al., 2014). Hence the consumption of each glycolytic ATP generates one net H+ but not the respiration-generated ATP since ATP hydrolysis: ATP4− + H2O = ADP3− + HPO42− + H+ releases one net H+. Based on this, we posit that the induction of glycolytic ATP synthesis in cancer is a cellular response for neutralizing the OH− produced by cytosolic Fenton reactions.
Our next question is do cancers tend to utilize specific biological processes to consume the glycolytic ATPs? A co-expression analysis revealed that cancer cells generally use the following to consume such ATPs: aminoacyl-tRNA synthesis, nucleotide synthesis, base-excision repair, and glyoxylate and dicarboxylate metabolism, as shown in Supplementary Table S3. As a comparison, we have also examined gene-expression data of 16 types of chronic inflammatory diseases and detected cytosolic Fenton reactions at moderate levels in 6 out of the 16 diseases. In these six diseases, the following strongly correlates with glycolysis: aminoacyl-tRNA synthesis, purine synthesis, base-excision repair, and the glyoxylate and dicarboxylate metabolism (Supplementary Figure S10). Hence, we conclude that nucleotide synthesis, DNA repair, and pH buffering represent a common set of processes used to consume glycolytic ATPs in both cancer and cytosolic Fenton reaction-harboring chronic inflammatory diseases.
Our explanation for ‘why nucleotide synthesis?’ is: it is most ATP-demanding, hence able to rapidly release protons through consuming glycolytic ATPs. In addition, the intracellular pool of nucleotides is very large, hence capable to keep very large quantities of synthesized nucleotides before they are channeled to somewhere. Furthermore, we speculate that import of protons may not be sustainable at a rate comparable to the rate of Fenton reactions since this needs to import a negatively charged molecule or export a positively charged molecule for each proton to keep intracellular electric neutrality.
Our next question is where do the continuously synthesized nucleotides go? Clearly, as the intracellular concentration of nucleotides increases, the reactions for nucleotide syntheses, hence generation of protons, will slow down and even stop. In addition, continuous export of such nucleotides will not be sustainable, knowing that nucleotides are negatively charged. Our hypothesis is: the cells channel the continuously synthesized nucleotides towards DNA synthesis and use cell division to remove the DNA (along with the positively charged histones) to the new daughter cells, which may represent a most sustainable way to rid of the nucleotides. This clearly requests substantial effort to validate or reject experimentally. Supplementary Section A.3 provides some discussion regarding the possible link between nucleotide synthesis/cellular accumulation and cell cycle progression. Very interestingly, a recent Nature publication (Bonora et al., 2015) provides a mechanic model that links increased glycolysis, specifically increased expression of glycolytic gene PFKFB4 to nucleotide synthesis and cell division via activation of the SRC gene by PFKFB4, hence providing an indirect evidence for the feasibility of our model here.
Fenton reactions in mitochondria
The occurrence of Fenton reactions
All 14 cancer types have strong correlations and ten types have very high statistical significance, namely BLCA, BRCA, COAD, KICH, KIRC, KIRP, LUAD, LUSC, prostate adenocarcinoma (PRAD), and stomach adenocarcinoma (STAD), as detailed in Table 1 and Supplementary Table S1. From the modeling and analyses, we have observed the following across all 14 cancer types: (i) the mitochondrial NADH and superoxide contribute substantially to the reduction of Fe3+ from mitochondrial Fenton reactions, as detailed in Supplementary Figure S11, hence driving the continuous Fenton reactions; (ii) the expression levels of ETC Complexes I and III both show strong correlations with our predicted levels of the mitochondrial Fenton reactions when Fe3+ being reduced by exogenous superoxide or unreduced (Supplementary Figure S13); and (iii) mitochondrial Fenton reactions contribute to ATP syntheses (Supplementary Figure S15). All these strongly suggest that mitochondrial Fenton reactions contribute to ATP synthesis. See Supplementary Section A.4 for details. Figure 2 (rows labeled with mitochondria) lists all the key genes and their expression levels, related to mitochondrial Fenton reactions in cancer and in non-cancerous chronic inflammatory diseases.
Impact of Fenton reactions to ATP synthesis
Our question is: do mitochondrial Fenton reactions directly contribute to the production of ATP? If yes, how? Compared with the normal aerobic respiration chain with electrons going from NADH to O2 through ETC Complexes I–IV, mutations in mitochondrial DNA and/or other relevant genes in cancer may cause increased electron leak from Complexes I–III (Shi et al., 2012) and decreased activity of Complex IV (Abdulhag et al., 2015), hence reducing the normal electron flow through the entire ETC.
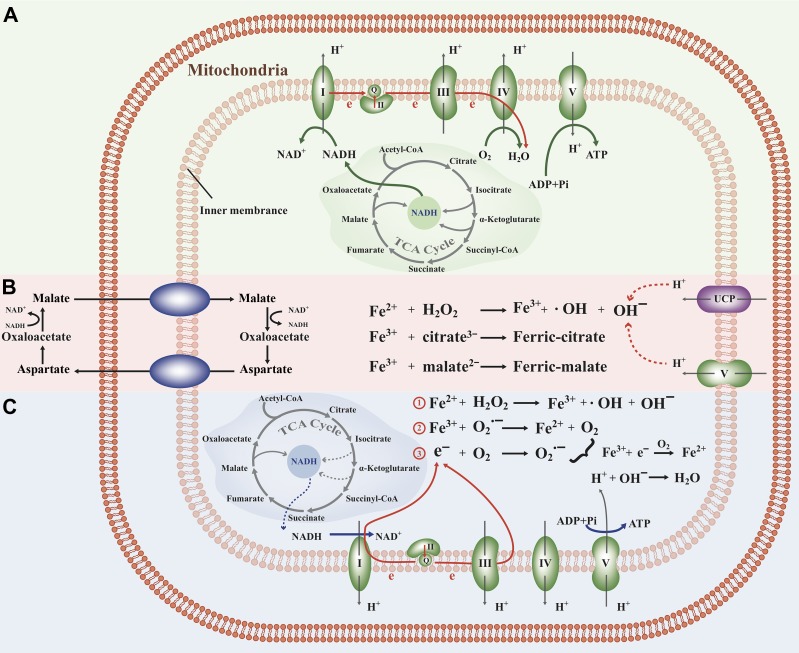
Our observations suggest that cancer cells use an alternative respiration chain with the following route for electron movement. It starts from NADH donating its electrons to Complex I and then to Complex III, which drives protons across the mitochondrial inner membrane to form a proton gradient for ATP synthesis by Complex V, as illustrated in Figure 1A. The main difference here vs. normal cells is that substantial fractions of electrons leak out of the ETC, specifically from CoQ in Complex I and cytochrome c in Complex III, to give electrons to nearby O2 and form superoxide as in Equation (4). Then the unpaired electrons in the superoxide flow to Fe3+ generated by mitochondrial Fenton reactions and reduce it to Fe2+, hence driving continuous Fenton reactions, as in Equation (2). Leaking of electrons from Complexes I and III has been widely observed (Wallace, 2005) but here our analysis suggests that they have become a main route of electrons to reach their terminal acceptor via Fenton reactions. The overall effect of the above is summarized by the following and shown in Figure 1C:
Figure 1.
(A) ETC under normal condition. (B) Fenton reactions directly drive ATP synthesis through the accumulation of Fe3+, generating proton gradients and hence ATP synthesis, which activates UCP genes as the response to the reduced concentration of protons. (C) Fenton reactions drive ATP synthesis by moving electrons from superoxide produced in Complexes I and III and using H2O2 as the final electron receiver.
One key difference between this altered and the normal ETC is that it uses H2O2 as the final electron acceptor instead of O2 as illustrated in Figure 1C. In these reactions, O2 is not consumed, instead, it serves as a catalyst along with Fe2+. It is Fenton reactions that involve the iron–sulfur clusters in these complexes that serve as a bridge to make this altered ETC route possible. It is noteworthy that Complexes I and III with electrons leaked to generate superoxide may still pump , that contribute to the cross-membrane proton gradient and ATP synthesis via Complex V as the final step in normal respiration chain, all suggested by co-expression data (Figure 1C).
One puzzling issue remains: it has been previously speculated that cancers seem to have unlimited ATPs (Gottesman et al., 2002; Zhou et al., 2012) although no reliable sources for electrons have been identified. Particularly, it has been widely observed that cancer cells generally have reduced TCA and respiratory activities (Cairns et al., 2011), with the impaired ETC complexes due to highly mutated mitochondrial DNA (Gaude and Frezza, 2014), hence reduced rates of mitochondrial NADH production to feed the ETC and ATP production, compared with normal proliferating cells. Our computational analysis has revealed: the glutaminolysis pathway remains unchanged while the malate-aspartate shuttle reaches saturation in multiple cancer types (Supplementary Sections A.5, B.7 and Table S3), hence strongly suggesting that cancer cells may utilize other electron sources to produce ATP in addition to NADH.
Recall that (mitochondrial) Fenton reaction with the Fe3+ unreduced will lead to OH− accumulation, directly consuming protons inside mitochondria, leading to increased proton gradient across the mitochondrial inner membrane and hence ATP production. The detailed information regarding unreduced Fe3+ in mitochondria and its link to ATP synthesis is given in Supplementary Section A.4 and Figure 1B. This represents a fundamentally different mechanism for ATP synthesis from the above since it does not use the respiration chain except for ATP synthase. The feasibility of this model has been established by Andre Jagendorf at 50 years ago: a proton gradient on the two sides of a membrane is sufficient to lead to ATP synthesis by the ATP synthase (Jagendorf and Uribe, 1966). Note that this model consumes protons inside mitochondria, compared with the respiration-based ATP production that only moves protons around.
A key supporting evidence for this model is that all cancer types have increased expression of UCP transporters, particularly UCP5, which move H+ inward across the mitochondrial inner membrane without producing ATPs. Activation of the UCPs strongly suggests that substantial amounts of protons inside mitochondria are consumed, hence the need for replenishment. We have identified the significant overexpression of UCP5 and strong positive correlations between UCP5 and ATP synthesis in all 14 cancer types (Supplementary Table S3). Additional supporting evidence can be found in Supplementary Section A.5.
Fenton reactions in extracellular matrix
Comparative analyses of Fenton reactions-related genes in non-cancerous inflammatory diseases and the 14 cancer types have revealed that the former have considerably lower levels of Fenton reactions in cytosol and mitochondria as shown in Figure 2. Further analyses discovered that the extracellular superoxide dismutase SOD3 is consistently overexpressed in inflammatory diseases but downregulated in cancer while hydrogen peroxide and superoxide productions in extracellular space are upregulated in both classes of diseases (Supplementary Figure S16). Our explanation is that cancer tissues may save the superoxide for reducing intracellular Fe3+ as the substantial accumulation of Fe3+ will cast a stress on the host cells. This is consistent with the previous studies showing that decreased SOD3 expression correlates with increased malignancy of cancer (O’Leary et al., 2015). Figure 2 (rows labeled with EC and MMPs) lists all the key genes and their expression levels, related to Fenton reactions in the extracellular matrix (ECM) of cancer and non-cancerous chronic inflammatory diseases.
A large amount of H2O2 and superoxide produced in extracellular space raises the possibility of Fenton reactions. To assess if there are indeed Fenton reactions in ECM, we have applied a similar approach to those in the previous sections to assess the possibility of Fenton reactions in ECM, knowing the following differences: (i) the level of superoxide does not correlate strongly with the predicted ECM Fenton reaction levels as detailed in Supplementary Table S1, suggesting that Fenton reactions in ECM may use other and yet-to-be-identified reducing elements; hence we did not include the relevant term in our modeling but assume that it is not rate limiting; (ii) it proves challenging to find marker genes for estimating the production rate of [OH−]; hence we consider the impact of only, knowing that the impact of OH− is probably limited in ECM where the pH is known to be low; (iii) a linear regression model without using the Michaelis–Menten equation is employed because of the relative simplicity of the problem; and (iv) no iron-containing proteins in ECM have been identified, which have strong correlations with but interestingly several copper-containing proteins are found to strongly correlate with the predicted —copper or any transient metal can have Fenton reactions like iron when H2O2 is high. Hence we used [Cu1+] to establish the occurrence of Fenton reactions, as summarized in Table 1 and Supplementary Section B.3.
Overall model
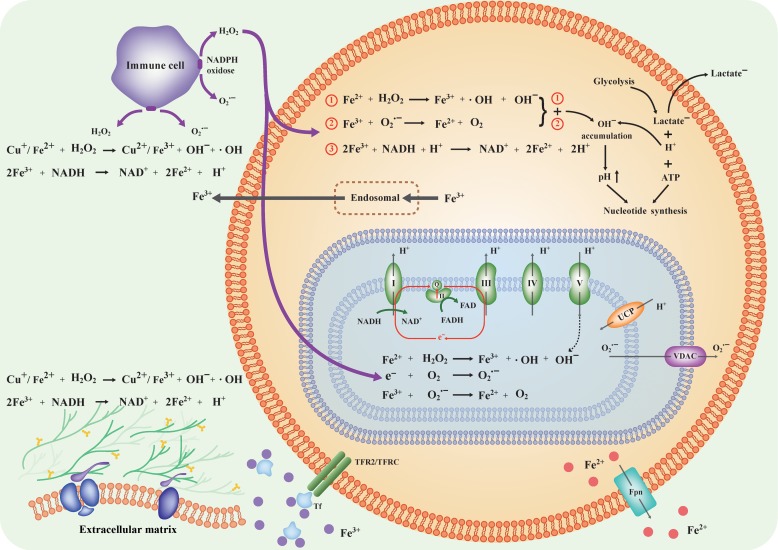
By integrating the information derived above, we have constructed a model for how Fenton reactions in the cytosol, mitochondria, ECM drive nucleotide synthesis, ATP production, and cell cycle progression as depicted in Figure 3 (with details in Table 1 and Supplementary Tables S1 and S2). Specifically, the cytosolic Fenton reactions with Fe3+ reduced by exogenous superoxide or unreduced will lead to the continuous production of net OH−, which will raise the pH and drive glycolytic ATP production and nucleotide synthesis as cells’ sustainable response. Mitochondrial Fenton reactions drive ATP syntheses through two novel pathways. Damages to extracellular components by hydroxyl radicals from Fenton reactions drive persistent inflammation and give rise to cell proliferation signals.
Figure 3.
Collective effects of Fenton reactions in three subcellular locations drive the altered intra-/extracellular pH levels and form a tumorigenesis environment. Specifically, cytosolic Fenton reactions drive glycolytic ATP production and nucleotide syntheses; mitochondrial Fenton reactions lead to new ways of mitochondrial ATP production; and Fenton reactions in ECM and space produce signals for cell cycle progression and inflammation.
As a validation, we have systematically compared the key upregulated genes used in our model with protein abundance data in the relevant cancer types (if available) in the Human Protein Atlas (Pontèn et al., 2008). We found that virtually all the major upregulated genes also have significant protein abundance in the relevant cancer types, hence providing one important piece of supporting evidence. See Supplementary Section B.11 and Figure S17 for details.
Discussion
What is cancer? Our analyses suggest that cancer cell proliferation is driven by cytosolic Fenton reactions and enabled by signals generated by cytosolic and ECM Fenton reactions as well as by plentiful ATPs enabled by mitochondrial Fenton reactions. Specifically, OH− produced on a continuous basis needs to be neutralized before they alter the cytosolic pH level. The affected cells find a sustainable way to neutralize them by synthesizing and consuming glycolytic ATPs through continuous syntheses of nucleotides. Then these nucleotides must have an exit since otherwise their increased intracellular concentration will slow down and even stop the production of the neutralizers, which will lead to cell death due to the continuous production of OH−; the affected cells have found cell division as a sustainable way for accomplishing this. Hence, cell proliferation is a way for cytosolic Fenton reaction-affected cells to survive.
Our model complements the view of cancer as a disease of genomic mutations, and explains the mutation-based model at a more fundamental level. Specifically, our model proposes that cancer is a way for the cells affected by Fenton reactions to survive through cell division. To do this, such cells must overcome numerous hurdles:
Cell proliferation at a rate comparable with the rate of cytosolic production. Early stage cancers may be able to achieve this when the level of cytosolic Fenton reactions is low, through utilizing tissue-repair signals and growth signals from damaged glycan and proteoglycan, plus utilization of general-purpose growth factors available in blood circulation such as insulin-like growth factors. As the disease evolves, the affected cells may continue to accumulate more irons and increase the H2O2 levels, leading to increased levels of Fenton reactions, which may require additional drivers for cell cycle progression to keep up with the increased needs for neutralizing. We posit that most of the oncogenic mutations provide such drivers. Previous studies have shown that cancers occur with oncogenic mutations, and our study here has shown: there are deeper reasons beneath the mutations for the cells to divide.
The proliferating cells need to protect themselves from destruction by apoptosis, the immune system, and other factors that may limit the survival of the diseased cells such as the limited number of cell division, contact-inhibition restraint or cell-cell competition in a growing tissue. Mutations in tumor suppressor genes might have been selected to help overcome these obstacles for the cells to remain viable.
While neutralizing the cytosolic represents a key challenge for the affected cells, they may face an increasingly wider range of other challenges created by these cells when they select genomic mutations and epigenomic alterations to overcome the above hurdles. For example, they may face secondary challenges such as acidosis due to glycolysis or hypoxia due to overgrowth as well as increased innate immune-responses. Additional mutations might be selected to help overcome these secondary hurdles.
Based on our analyses, the key reasons for the Warburg effect in cancer are: Fenton reaction-affected cells need to neutralize the continuous influx of OH−, and the cells respond by releasing net H+ at a rate comparable to that of the OH− generation through synthesizing glycolytic ATPs and consuming them via nucleotide synthesis as respiration-produced ATPs do not contribute to overcoming this challenge. In addition, ATPs can be produced by Fenton reaction-enabled anaerobic respiration and consumption of mitochondrial protons, and hence the cells may not have a need for aerobic respiration.
A number of other open questions in cancer research can also be naturally answered or addressed using our model, including (i) why cancer cells utilize (de novo) nucleotide synthesis rather than uptake of nucleotides from circulation for DNA synthesis; (ii) why cancer cells tend to have increased and truncated cell-surface glycan (Radhakrishnan et al., 2014); and (iii) why Warburg effect in cancer cells is fundamentally different from the Warburg effect in normal proliferating cells.
It has been reported that Fenton reactions can take place with chelated Fe2+ ions in the LIP (Kruszewski, 2004). However, we have not been able to study such reactions since we do not have a reliable way to estimate the level of such LIP-associated reactions using gene-expression data. We suspect that some of the four cancer types that do not show strong iron–sulfur cluster associated mitochondrial Fenton reactions, as well as those shown in Supplementary Figures S1 and S4, may involve these irons in their Fenton reactions.
What makes extracellular pH of cancer tissue cells to become acidic? Is lactic acid secretion solely responsible for the acidic extracellular space as widely believed? Our analysis suggests that the activation of the extracellular NADPH oxidases leads to proton accumulation, hence reducing the pH in extracellular space since some of their produced superoxides, as in Equation (4), will diffuse into cells and leave their matching protons behind. Similarly, the reduction of Fe3+ exported from the cytosol by Equation (3) will also generate extracellular protons. These two reactions may represent key mechanisms for decreasing the extracellular pH in addition to protons released from the cytosol by MCT transporters along with lactates.
It is noteworthy that a few recent articles provide strong evidence that cancer tissue cells may uptake substantial amount of lactate from the extracellular space (Kennedy et al., 2013; Xie et al., 2014; Hui et al., 2017). This makes sense, knowing that cancer tissues tend to have higher levels of lactate in the extracellular space than the intracellular space and the lactate transporters are all gradient driven.
Note that our model is developed based on correlations coupled with known chemistry associated with Fenton reactions, which are irreversible due to the rapid reactions involving hydroxyl radicals (typically within 1 ns), and the fact that Fenton reactions become increasingly stronger going from inflammatory diseases to cancer. The model is self-consistent and can explain a wide range of perplexing issues associated with cancer. It provides a fundamentally novel perspective of cancer formation and offers numerous testable predictions about the relationships between Fenton reactions and cancer.
Our study strongly suggests that neutralizing cytosolic Fenton reaction-produced OH− may represent the cancer-defining stress that the affected cells must overcome. Cancer tissues have all selected consumption of glycolytic ATPs via nucleotide synthesis as the main route to generate net protons. Cell division may represent the most sustainable way for the riding of the rapidly synthesized nucleotides. Two conditions are needed to make this happen: (i) signals for cell cycle progression and (ii) plentiful ATPs. Cancer tissues seem to have all selected cells having Fenton reactions in both mitochondria and ECM, to provide the needed ATPs and signals for cell cycle progression. Fenton reactions in these subcellular locations may represent necessary conditions for cancer to take place.
While Fenton reactions may play fundamental driving roles in cancer formation and development as we have computationally demonstrated here, we do not consider such reactions as the sole driver since still various cancer-related questions, such as why certain cancers are more drug-resistant than the others; why certain organs are less prone to cancer development compared to others; and what may drive cancer to metastasize; are yet to be understood. We expect that further development of our model, particularly in terms of more organic integration of the model with the existing knowledge such as that related to functional roles played by the adaptive immune system (vs. innate immune system discussed in this study) in cancer development, may prove to be a fruitful angle.
Materials and methods
Data
RNA-Seq data of tissue samples of 14 cancer types, microarray gene-expression data (Affymetrix UA133 plus 2.0) of 12 of the 14 cancer types and 16 types of chronic inflammatory diseases along with their normal controls are retrieved from the TCGA and GEO databases, respectively. The detailed information of these datasets is given in Table 2. The 14 cancer types are selected since they are all the cancer types each with a sufficiently large number of tissue samples with RNA-Seq data in TCGA. The same applies to the 16 non-cancerous inflammatory diseases. In addition, we have also included in our study all the cancer types in GEO that are also covered by the 14 cancer types, totaling 12 types. The reason we used both RNA-Seq- and microarray-based gene-expression data for some of the cancer types is: having both data types allowing us to make meaningful comparisons between gene-expression level changes in cancer and in inflammatory diseases as the latter has only microarray data available and RNA-Seq data are clearly more reliable and hence used whenever possible.
Table 1.
Agreement between model-based estimations of Fenton reactions and estimations directly based on gene expression, and associated statistical significance in cytosol, mitochondria, and extracellular matrix.
| Cancer | Cytosol | Mitochondria | Extracellular matrix | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| P-value of upregulated proteasome | Averaged R2 value | P-value of obtained R2 value (permuting x) | P-value of obtained R2 value (permuting y) | P-value of upregulated damage response | Averaged R2 value | P-value of obtained R2 value (permuting x) | P-value of obtained R2 value (permuting y) | P-value of upregulated damage response | 75% quantile of R2 values | P-value of obtained R2 value | |
| BLCA | 4.94E−07 | 0.557 | 0.000117 | 1.16E−06 | 0.91896 | 0.813 | 0.01402 | <1E−5 | 0.24 | 0.628 | 0.039 |
| BRCA | 1.12E−07 | 0.614 | 0.000505 | 1.06E−07 | 0.00024 | 0.841 | 5.63E−05 | <1E−5 | 0.0321 | 0.564 | 0.044 |
| COAD | 4.86E−07 | 0.681 | 3.75E−05 | 3.92E−09 | <1E−5 | 0.848 | 3.54E−11 | <1E−5 | 0.0053 | 0.6408 | 0.021 |
| ESCA | 1.02E−19 | 0.574 | 0.00144 | 0.345288 | 0.65235 | 0.781 | 0.3201 | <1E−5 | 2.00E−04 | 0.5755 | 0.027 |
| HNSC | 3.00E−04 | 0.62 | 0.00145 | 3.34E−07 | 0.00178 | 0.766 | 0.2179 | <1E−5 | <1E−5 | 0.5797 | 0.034 |
| KICH | 2.20E−07 | 0.777 | 0.00844 | 0.002735 | <1E−5 | 0.935 | 0.0056 | <1E−5 | 1 | 0.6324 | 0.071 |
| KIRC | 0.0794 | 0.779 | 7.50E−05 | 2.61E−05 | <1E−5 | 0.893 | 1.49E−06 | <1E−5 | 0.2021 | 0.4612 | 0.137 |
| KIRP | 3.28E−05 | 0.759 | 2.01E−06 | 2.56E−10 | <1E−5 | 0.881 | 0.0097 | <1E−5 | 0.5843 | 0.3639 | 0.6775 |
| LIHC | 0.0056 | 0.639 | 0.000702 | 7.70E−12 | 1 | 0.767 | 1 | <1E−5 | 0.306 | 0.5594 | 0.148 |
| LUAD | 8.50E−10 | 0.635 | 0.000431 | 2.41E−11 | 0.05117 | 0.841 | 4.30E−10 | <1E−5 | 0.001 | 0.4999 | 0.0305 |
| LUSC | 2.42E−09 | 0.559 | 0.0018 | 0.012747 | <1E−5 | 0.79 | 7.89E−07 | <1E−5 | 0.0041 | 0.5969 | 0.02 |
| PRAD | 6.02E−05 | 0.75 | 8.69E−06 | 2.05E−09 | 0.00621 | 0.887 | 8.05E−05 | <1E−5 | 1 | 0.5708 | 0.3145 |
| STAD | 2.28E−11 | 0.685 | 0.000877 | 6.83E−07 | 0.03263 | 0.861 | 0.0013 | <1E−5 | 0.0381 | 0.5466 | 0.027 |
| THCA | 8.41E−05 | 0.812 | 5.01E−05 | 2.77E−11 | <1E−5 | 0.884 | 0.1956 | <1E−5 | 0.0033 | 0.6562 | 0.021 |
It is noteworthy that our differential gene-expression data analyses throughout the study are always done between disease samples and control samples generated using the same platform. Hence batch effect is not an issue.
Genes selected for estimation of Fenton reactions in cytosol
We have examined differential expressions of Fenton reactions-related genes to estimate the levels of the five quantities involved in Fenton reaction: plus the level of accumulation of [Fe3+]. Consistencies between the differentially expressed Fenton reaction-related genes in the microarray and RNA-Seq data of the 12 cancer types are ensured. Detailed information of the analysis is given in Supplementary Table S4.
To estimate the production rate of in the cytosol, we have used gene expressions that reflect the amounts of protein-degradation by proteasome and mRNA degradation by the relevant enzymes in the cytosol, which are considerably upregulated in cancer vs. controls. The rationale is: (i) proteins and mRNA account for ~50%–60% of biomolecules in a cell; hence their level of damages gives a good estimate of the total level of cellular damages due to ; (ii) the total expressions of proteasome complexes and mRNA degradation enzymes would be proportional to the number of proteins and mRNA being degraded by them; and (iii) previous studies have reported gene-expression level alterations of protein-degradation genes in response to elevated oxidative stress (Friguet, 2006). In the end, is estimated using a non-linear model of the expressions of 65 proteasome and mRNA degradation genes, which form a highly co-expressed gene cluster in each cancer type. We have noted that normal tissue cells have substantially lower expressions of these genes than cancer cells.
All cytosolic iron–sulfur proteins are considered as the source of Fe2+ accessible to Fenton reactions; hence the expression levels for iron–sulfur assembly genes are used to estimate the level of Fe2+, . The rationale is that the total mRNA levels of these proteins should be proportional to the number of Fe2+ ions in the proteins, which should be the case, knowing that each such protein contains one such cluster and two Fe2+ ions. A linear model of expressions of cytosolic iron–sulfur cluster synthesis genes is used to estimate .
The total gene expressions of H2O2 scavenging enzymes are used to estimate cytosolic [H2O2]. The level of RA, that are involved in the reduction of ferric iron is estimated by gene expressions of cytosolic dehydrogenases. Cytosolic superoxide, has two possible sources: mitochondrial superoxide that diffuses into cytosol via the VDAC transporters and extracellular superoxide that diffuses into cytosol via some plasma membrane anion transporters; together the expression levels of the SOD and VDAC genes, as well as the NADPH oxidase and associated genes expressed in stromal and immune cells, are combined to estimate . The level of the cytosolic Fe3+ accumulation is estimated by transferrin, ferritin, cytosolic ferrous iron reductase, and iron ion exporter genes.
Similarly, genes selected to estimate mitochondrial and ECM Fenton reactions are detailed in Supplementary Section B.3, along with other analysis tools.
Supplementary Material
Acknowledgements
We thank Dr Fenglou Mao, Professor Dong Xu, Professor Dave Puett, Mr Liang Chen, and Ms Ying-nan Hou of the UGA and Professor Ruren Xu of Jilin University for helpful discussions regarding this study.
Funding
This work was supported by grants from Georgia Research Alliance, the National Natural Science Foundation of China (61572227), Projects of International Cooperation and Exchanges of the National Natural Science Foundation of China (81320108025), and Jilin University.
Conflict of interest
none declared.
Author contributions
H.S. and C.Z. did most of the development and analysis work that leads to the main discoveries of the paper. S.C. did statistical analyses on a number of issues presented in this study. N.D. and T.S. played key roles in conducting analyses of chemical reactions key to this study. Y.X. conceived the project and guided the research of the project.
References
- Abdulhag U.N., Soiferman D., Schueler-Furman O., et al. (2015). Mitochondrial complex IV deficiency, caused by mutated COX6B1, is associated with encephalomyopathy, hydrocephalus and cardiomyopathy. Eur. J. Hum. Genet. 23, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akatsuka S., Yamashita Y., Ohara H., et al. (2012). Fenton reaction induced cancer in wild type rats recapitulates genomic alterations observed in human cancer. PLoS One 7, e43403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian B., Pogozelski W.K., and Tullius T.D. (1998). DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proc. Natl Acad. Sci. USA 95, 9738–9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian K., Gao Z., Weisbrodt N., et al. (2003). The nature of heme/iron-induced protein tyrosine nitration. Proc. Natl Acad. Sci. USA 100, 5712–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonora M., Wieckowsk M.R., Chinopoulos C., et al. (2015). Molecular mechanisms of cell death: central implication of ATP synthase in mitochondrial permeability transition. Oncogene 34, 1608. [DOI] [PubMed] [Google Scholar]
- Cairns R.A., Harris I.S., and Mak T.W. (2011). Regulation of cancer cell metabolism. Nat. Rev. Cancer 11, 85–95. [DOI] [PubMed] [Google Scholar]
- Chen C., and Paw B.H. (2012). Cellular and mitochondrial iron homeostasis in vertebrates. Biochim. Biophys. Acta 1823, 1459–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaghi M., Wojtkowiak J.W., and Gillies R.J. (2013). pH sensing and regulation in cancer. Front. Physiol. 4, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmagirbi A., Sulistyarti H., and Atikah A. (2012). Study of ascorbic acid as iron(III) reducing agent for spectrophotometric iron speciation. J. Pure Appl. Chem. Res. 1, 11–17. [Google Scholar]
- Fischer K., Hoffmann P., Voelkl S., et al. (2007). Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood 109, 3812–3819. [DOI] [PubMed] [Google Scholar]
- Fong K.-L., McCay P.B., Poyer J.L., et al. (1976). Evidence for superoxide-dependent reduction of Fe3+ and its role in enzyme-generated hydroxyl radical formation. Chem. Biol. Interact. 15, 77–89. [DOI] [PubMed] [Google Scholar]
- Friguet B. (2006). Oxidized protein degradation and repair in ageing and oxidative stress. FEBS Lett. 580, 2910–2916. [DOI] [PubMed] [Google Scholar]
- Gaude E., and Frezza C. (2014). Defects in mitochondrial metabolism and cancer. Cancer Metab. 2, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman M.M., Fojo T., and Bates S.E. (2002). Multidrug resistance in cancer: role of ATP-dependent transporters. Nat. Rev. Cancer 2, 48–58. [DOI] [PubMed] [Google Scholar]
- Han D., Antunes F., Canali R., et al. (2003). Voltage-dependent anion channels control the release of the superoxide anion from mitochondria to cytosol. J. Biol. Chem. 278, 5557–5563. [DOI] [PubMed] [Google Scholar]
- Hui S., Ghergurovich J.M., Morscher R.J., et al. (2017). Glucose feeds the TCA cycle via circulating lactate. Nature 551, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay J.A., Chin S.M., and Linn S. (1988). Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240, 640–642. [DOI] [PubMed] [Google Scholar]
- Jagendorf A.T., and Uribe E. (1966). ATP formation caused by acid-base transition of spinach chloroplasts. Proc. Natl Acad. Sci. USA 55, 170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M., Ieong K.-W., Trobro S., et al. (2011). pH-sensitivity of the ribosomal peptidyl transfer reaction dependent on the identity of the A-site aminoacyl-tRNA. Proc. Natl Acad. Sci. USA 108, 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.C., Dean D.R., Smith A.D., et al. (2005). Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74, 247–281. [DOI] [PubMed] [Google Scholar]
- Kennedy K.M., Scarbrough P.M., Ribeiro A., et al. (2013). Catabolism of exogenous lactate reveals it as a legitimate metabolic substrate in breast cancer. PLoS One 8, e75154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima N., and Bates G. (1979). The reduction and release of iron from Fe3+•transferrin•CO32−. J. Biol. Chem. 254, 8847–8854. [PubMed] [Google Scholar]
- Kruszewski M. (2003). Labile iron pool: the main determinant of cellular response to oxidative stress. Mutat. Res. 531, 81–92. [DOI] [PubMed] [Google Scholar]
- Kruszewski M. (2004). The role of labile iron pool in cardiovascular diseases. Acta Biochim. Pol. 51, 471–480. [PubMed] [Google Scholar]
- Meitzler J.L., Antony S., Wu Y., et al. (2014). NADPH oxidases: a perspective on reactive oxygen species production in tumor biology. Antioxid. Redox Signal. 20, 2873–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis L., and Menten M.L. (1913). Die kinetik der invertinwirkung. Biochem. Z. 49, 352. [Google Scholar]
- O’Leary B.R., Fath M.A., Bellizzi A.M., et al. (2015). Loss of SOD3 (EcSOD) expression promotes an aggressive phenotype in human pancreatic ductal adenocarcinoma. Clin. Cancer Res. 21, 1741–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontèn F., Jirström K., and Uhlen M. (2008). The Human Protein Atlas—a tool for pathology. J. Pathol. 216, 387–393. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan P., Dabelsteen S., Madsen F.B., et al. (2014). Immature truncated O-glycophenotype of cancer directly induces oncogenic features. Proc. Natl Acad. Sci. USA 111, E4066–E4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Zhang Y., Zheng J., et al. (2012). Reactive oxygen species in cancer stem cells. Antioxid. Redox Signal. 16, 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spugnini E.P., Sonveaux P., Stock C., et al. (2015). Proton channels and exchangers in cancer. Biochim. Biophys. Acta 1848, 2715–2726. [DOI] [PubMed] [Google Scholar]
- Stevens R.G., and Kalkwarf D.R. (1990). Iron, radiation, and cancer. Environ. Health Perspect. 87, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swietach P., Vaughan-Jones R.D., Harris A.L., et al. (2014). The chemistry, physiology and pathology of pH in cancer. Phil. Trans. R. Soc. B Biol. Sci. 369, 20130099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torti S.V., and Torti F.M. (2013). Iron and cancer: more ore to be mined. Nat. Rev. Cancer 13, 342–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyokuni S. (2009). Role of iron in carcinogenesis: cancer as a ferrotoxic disease. Cancer Sci. 100, 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. (2005). Mitochondria and cancer: Warburg addressed. Cold Spring Harb. Symp. Quant. Biol. 70, 363–374. [DOI] [PubMed] [Google Scholar]
- Xie J., Wu H., Dai C., et al. (2014). Beyond Warburg effect—dual metabolic nature of cancer cells. Sci. Rep. 4, 4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Tozzi F., Chen J., et al. (2012). Intracellular ATP levels are a pivotal determinant of chemoresistance in colon cancer cells. Cancer Res. 72, 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.